Abstract
Background
Inhibition of phosphoinositide 3-kinase δ (PI3Kδ) exerts corrective effects on the dysregulated migration characteristics of neutrophils isolated from patients with chronic obstructive pulmonary disease (COPD).
Objective
To develop novel, induced sputum endpoints to demonstrate changes in neutrophil phenotype in the lung by administering nemiralisib, a potent and selective inhaled PI3Kδ inhibitor, to patients with stable COPD or patients with acute exacerbation (AE) of COPD.
Methods
In two randomized, double-blind, placebo-controlled clinical trials patients with A) stable COPD (N=28, randomized 3:1) or B) AECOPD (N=44, randomized 1:1) received treatment with inhaled nemiralisib (1mg). Endpoints included induced sputum at various time points before and during treatment for the measurement of transcriptomics (primary endpoint), inflammatory mediators, functional respiratory imaging (FRI), and spirometry.
Results
In stable COPD patients, the use of nemiralisib was associated with alterations in sputum neutrophil transcriptomics suggestive of an improvement in migration phenotype; however, the same nemiralisib-evoked effects were not observed in AECOPD. Inhibition of sputum inflammatory mediators was also observed in stable but not AECOPD patients. In contrast, a placebo-corrected improvement in forced expiratory volume in 1 sec of 136 mL (95% Credible Intervals −46, 315mL) with a probability that the true treatment ratio was >0% (Pr(θ>0)) of 93% was observed in AECOPD. However, FRI endpoints remained unchanged.
Conclusion
We provide evidence for nemiralisib-evoked changes in neutrophil migration phenotype in stable COPD but not AECOPD, despite improving lung function in the latter group. We conclude that induced sputum can be used for measuring evidence of alteration of neutrophil phenotype in stable patients, and our study provides a data set of the sputum transcriptomic changes during recovery from AECOPD.
Keywords: nemiralisib, sputum, transcriptomics, COPD exacerbations, PI3Kdelta
Introduction
The mechanisms of action of novel immunomodulator drugs are far removed from established clinical efficacy endpoints, hence translational endpoints are helpful to build confidence in the pharmacology of the drug and enables better interpretation of study outcomes. We set out to explore the consequence of treatment with the inhaled phosphoinositide 3-kinase δ (PI3Kδ) inhibitor nemiralisib, which was administered to patients suffering from chronic obstructive pulmonary disease (COPD) and to see if in vitro observations translated into clinical immune modulation.
COPD is characterized by progressive airflow limitation in association with a chronic inflammatory response, and many patients have a persistent colonization with respiratory pathogens.1 COPD patients experience exacerbations resulting in increased risk of hospitalization and a reduction in life expectancy.2 Despite the existence of extensive clinical analyses of patients with COPD, little is known about the cellular mechanisms driving this disorder.3
Analysis of the longitudinal Acute Exacerbation and Respiratory Infections in COPD (AERIS) study,4,5 demonstrated the presence of bacteria and viruses in sputum taken from stable and exacerbating COPD patients. Neutrophils are required for the clearance of pathogens from the lung,6 and are increased in COPD patients.7 However, neutrophils from COPD patients display dysregulated migration characteristics which may interfere with their ability to adequately clear pathogens. This aberrant migration has been shown to be corrected in vitro by inhibitors of PI3Kδ,8 and increased expression and activation of the PI3Kδ pathway has been demonstrated in neutrophils from COPD patients, where it correlates with disease severity.9,10 As COPD exacerbations are often triggered by lung infections, PI3Kδ inhibitors may provide therapeutic benefit through their unique potential to improve neutrophil immune function by correcting the aberrant directionality of chemotaxis observed in COPD neutrophils,8 and by inhibiting inflammatory mediator release.11
Monitoring lung neutrophils in a clinical trial setting is currently largely limited to counting the abundance of neutrophils in sputum samples,12 or through imaging techniques.13 Neither can demonstrate if the phenotype of the neutrophil itself has been altered. Clinical trials have used changes in the messenger ribonucleic acid (mRNA) transcriptome to explore the effect of immune modulatory drugs,14–16 which could potentially be adopted to explore changes in neutrophils in the lung following treatment with nemiralisib.
Nemiralisib is a potent and highly selective inhaled PI3Kδ inhibitor,17 which has been shown to reduce the inflammatory mediators IL-6 and IL-8 in induced sputum from stable COPD patients.18 Not previously reported from that trial was genome scale unbiased transcriptomic analysis of induced sputum which was suggestive of an improvement in neutrophil migration phenotype.
The objective of the current study was to replicate induced sputum neutrophil transcriptomic observations in patients with an acute exacerbation (AE) of COPD (AECOPD), hence developing novel translational endpoints to confirm mechanism of action of a novel immunomodulator. The primary endpoint was a predefined subset of genes relating to neutrophil function. The effects of inhaled nemiralisib was additionally assessed using functional respiratory imaging (FRI), spirometry and other sputum and systemic biomarkers. To our knowledge, this study is the first comprehensive profiling of induced sputum and blood transcriptome following recovery from AECOPD.
Methods
Two double-blind, multicenter, randomized, placebo-controlled, parallel group studies (GSK protocols: PII115119 and 201928) were performed with nemiralisib, in stable COPD patients or those experiencing AECOPD. Written informed consent was obtained from each patient prior to any study-specific procedures. The studies were conducted in accordance with the principles of the Declaration of Helsinki and both were approved by the relevant ethics committees/institutional review boards and regulatory authorities (details given in Supplementary Materials, Appendix 1).
Study A (NCT02130635)
Study A dosed stable COPD patients with inhaled nemiralisib (1mg) daily for 14 days. Full details of the study inclusion/exclusion criteria and primary (safety) and secondary (pharmacokinetic (PK)) endpoints from this trial have been published previously.18 The exploratory endpoint of mRNA analysis at Day 1 and Day 14 is included in this report.
Study B (NCT02522299)
Study B evaluated treatment with inhaled nemiralisib (1mg or equivalent) in COPD patients experiencing AECOPD in Canada and Denmark between November 2015 and June 2018 (see Online Data Supplement for dosing device information, and patient visit schedule (Figure S1)). The primary endpoint was alterations (fold change >1.5 and p<0.05) in a panel of 258 predefined neutrophil-related genes determined by changes in mRNA transcriptomics in induced sputum. Secondary endpoints included safety, FRI with low dose high resolution computed tomography (LD-HRCT),19 lung function assessments and plasma PK.
To be eligible for recruitment, patients were male or female (of non-child bearing potential), aged 40–80 years with an established 6-month history of COPD,20 a previously documented post-bronchodilator forced expiratory volume in 1 sec (FEV1)/forced vital capacity (FVC) ratio of <0.70, an FEV1 ≤80% of predicted values,21 a smoking history of ≥10 pack years and a diagnosed AECOPD requiring escalation in therapy. A body weight ≥45 kg and body mass index (BMI) within the range 18–32 kg/m2 (inclusive) was also required. Subjects were excluded if they required mechanical ventilation, were suspected of having cardiac failure, had any abnormal clinical or laboratory observations, or ongoing liver or kidney disease. Subjects with active tuberculosis, lung cancer, clinically overt bronchiectasis, pulmonary fibrosis, asthma were also excluded.
An acute exacerbation was defined as described previously by Seemungal et al.22 Standard of care treatment was started after the diagnosis of an AECOPD and included both antibiotics and systemic corticosteroids. Sputum induction and LD-HRCT scan were conducted at the earliest opportunity and within 48 h of the diagnosis of the AECOPD. Patients were monitored for 14 days post final dose.
Induced Sputum Capture for mRNA, Total Cell Counts and Cytokine Analysis
Induced sputum samples were collected prior to randomization and again on Day 12, 28 and 84, and processed for cytokine measurement as described previously.18 RNA transcriptomics detection was conducted using a methodology as described previously.23 Additional methodology is provided in the Supplementary Materials-Methods.
Pharmacokinetic (PK) Assessments
Plasma samples were derived from sparse 2 mL blood samples, and analyzed using a previously described, validated, analytical method.17
In vitro Neutrophil Elastase and Reactive Oxygen Species Production
Blood neutrophils captured from exacerbating COPD patients and assayed for neutrophil elastase and reactive oxygen species were analyzed as previously described.24 Blood neutrophils from COPD patients experiencing an acute exacerbation were isolated to explore their responsiveness to treatment with a PI3Kδ inhibitor. Neutrophils were primed with LPS (25 µg/mL) and activated with fMLP (1 µM) in the presence of increasing concentrations of nemiralisib, and were normalized to vehicle control (0.1% DMSO).
Statistical Analysis
Detailed information on RNA transcriptomics analysis is provided in the Online Data Supplement. Sample size was based primarily on feasibility, but with a subject number similar to that used previously.14,15
The LD-HRCT images were processed centrally, by blinded radiologist as described previously.19,25 For a subset of the lung regions, the endpoints were analyzed in a multivariate model, under a Bayesian framework.
The change from baseline in the endpoints (FEV1 and FVC) were analyzed using a Bayesian repeated measures model. The change from baseline at each of the visits was represented via adjusted posterior medians for each of the treatment groups, as well as their associated 95% credible intervals.
Results
Study A
Treatment of Stable COPD Patients with Nemiralisib Alters Lung Neutrophil Phenotype
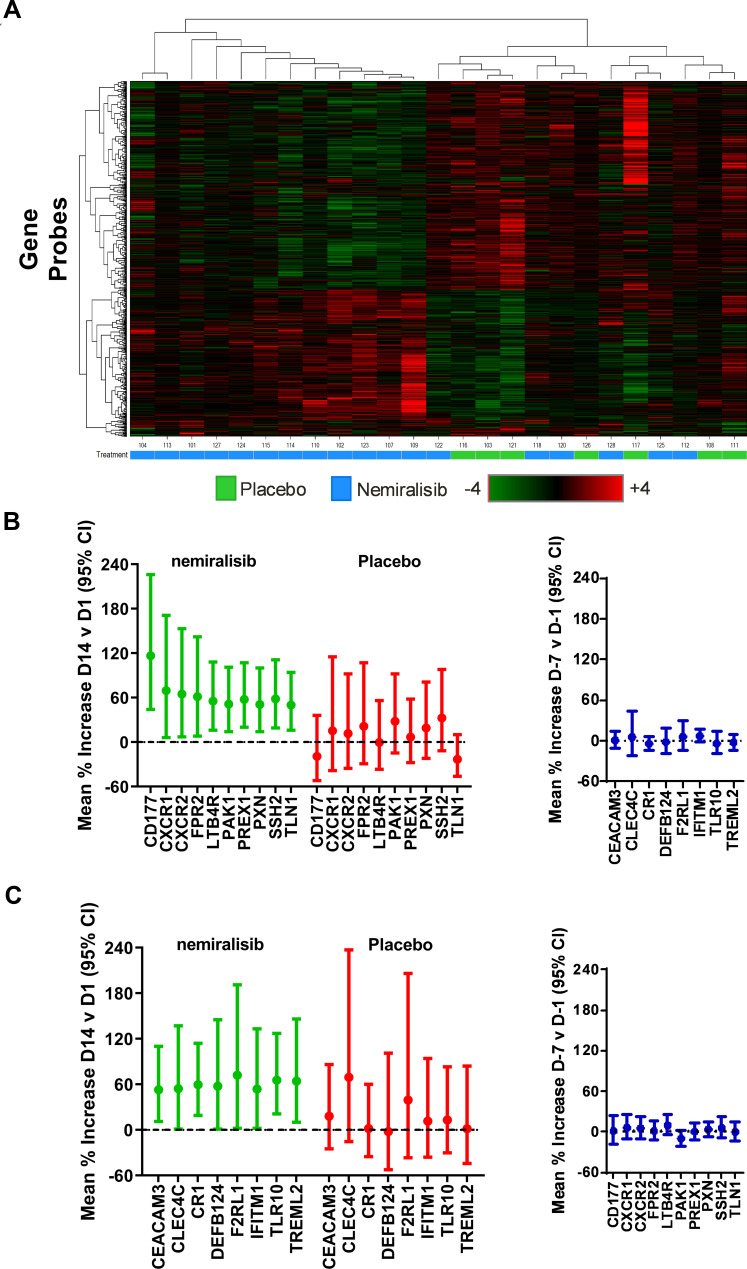
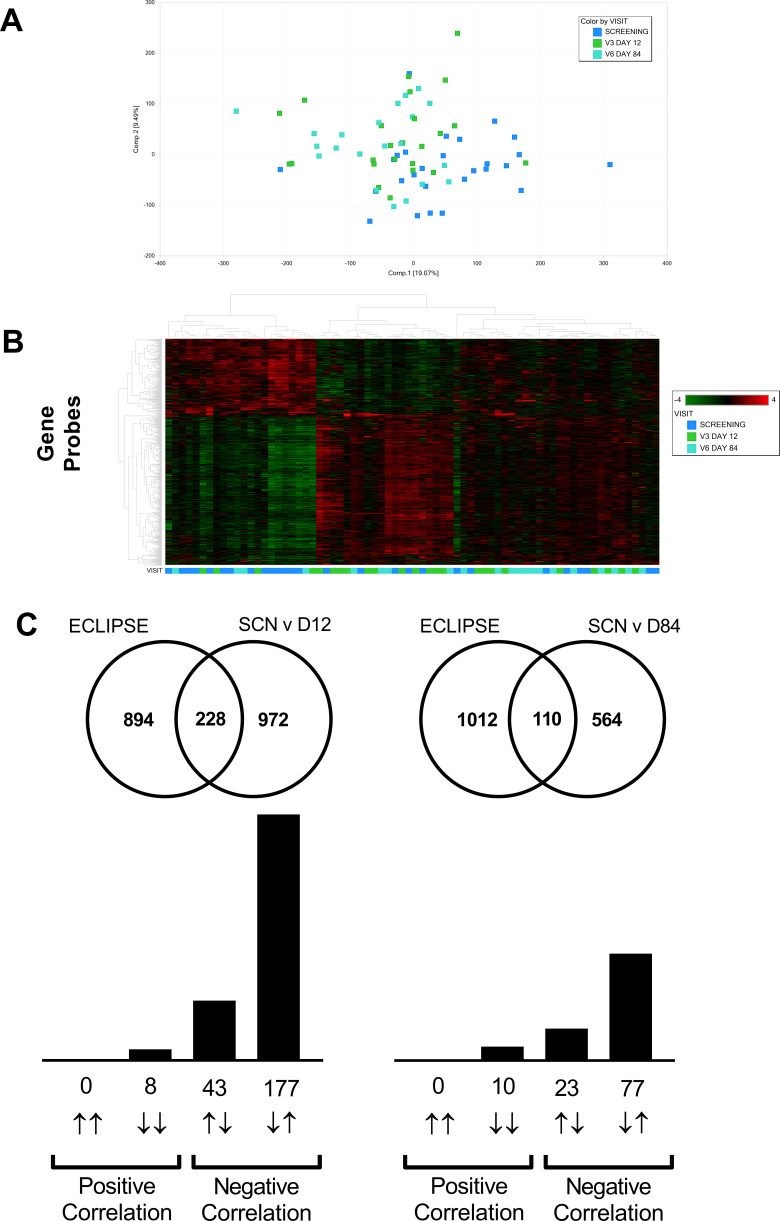
Study A dosed 28 stable COPD patients with nemiralisib (1mg) or placebo (3:1) for 14 days; baseline characteristics were similar between groups (Table S1, Online Data Supplement). A heat map of altered genes suggests a separation of subjects by treatment (Figure 1A). Nemiralisib altered 371 genes compared to 161 genes in the placebo-treated group, with an overlap of 15 genes suggesting unique profiles (fold change >1.5 and unadjusted p value <0.05), with no genes reaching cut offs when false discovery rate (FDR) adjusted. The top pathways following nemiralisib treatment using Metacore were annotated as neutrophil migration, neutrophil adhesion, and bacterial recognition pathways. The nemiralisib-evoked enriched genes relating to neutrophil migration (Figure 1B) and bacterial infection response (Figure 1C) were not observed in the placebo-treated group. These genes were not altered in the comparison over the 7 days before dosing commenced (Figure 1B and C, right panels). Only 24 probes changed when comparing samples taken over the 7 days before dosing commenced.
Figure 1.
mRNA transcriptomic alterations in induced sputum from stable COPD patients (Study A). Transcriptomic changes in induced sputum taken from stable COPD patients dosed for 14 days with nemiralisib show a partial clustering by treatment (Panel A). Unbiased pathway analysis highlighted nemiralisib-evoked alterations in neutrophil phenotype and bacterial infection response genes (Panels B and C, left), which were not observed in the samples taken 7 days apart prior to randomization (Panels B and C, right).
Nemiralisib Inhibited Reactive Oxygen Species and Elastase Production in vitro from Neutrophils Isolated from COPD Patients Experiencing an Acute Exacerbation
Neutrophil elastase, and reactive oxygen species were inhibited by nemiralisib in a concentration-dependent manner (Figure S3), suggesting that the effects of the inhibition of PI3Kδ in exacerbating patients was maintained.
Study B
Nemiralisib Treatment Did Not Alter Neutrophil Phenotype in COPD Patients Experiencing an Acute Exacerbation
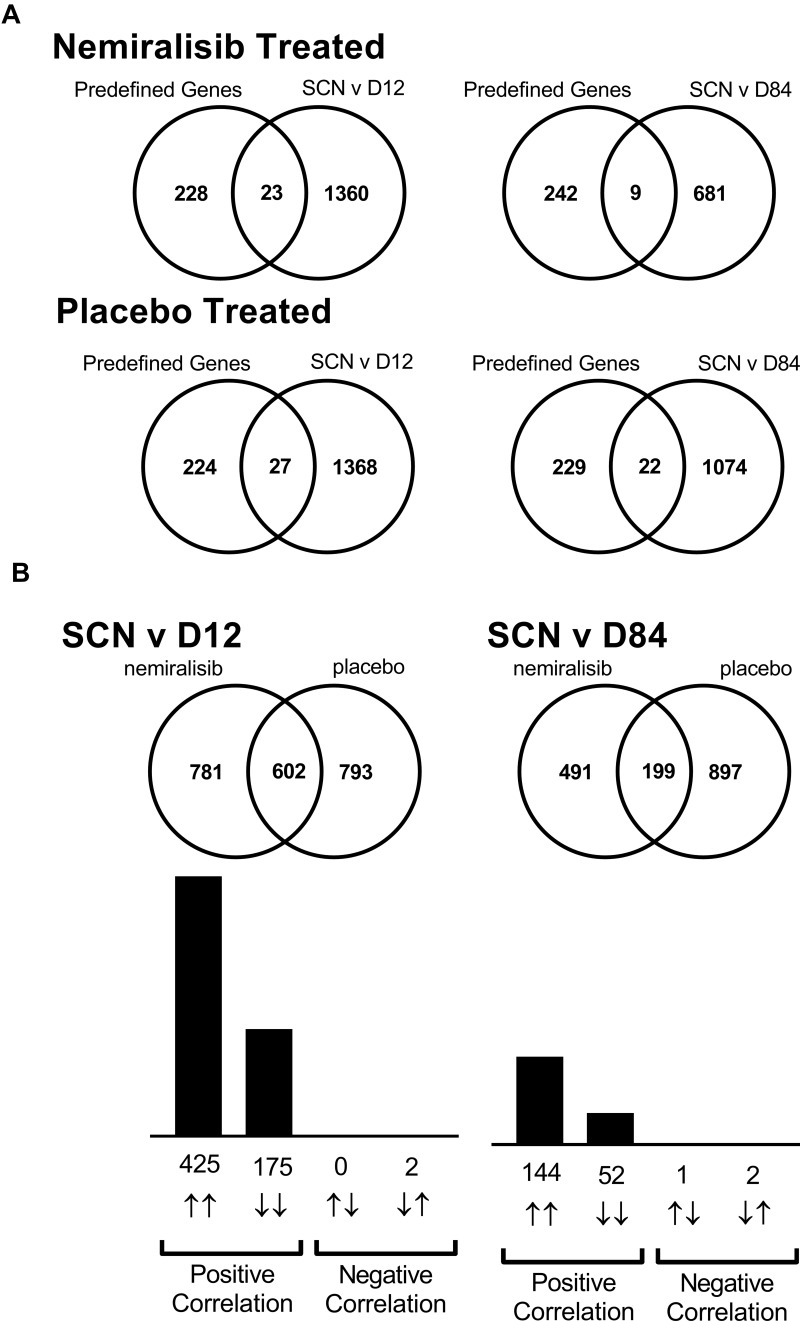
Study B dosed nemiralisib to COPD patients experiencing an AECOPD with daily treatment continued for 84 days. A total of 44 subjects were randomized (Figure S2), and baseline characteristics were similar between groups (Table S1, Online Data Supplement). Nemiralisib was generally well tolerated; see Table S2, Online Data Supplement. Due to the low number (20/44) of acceptable samples obtained at Day 28, further downstream analyses were not performed on this time point (see full list of number of samples obtained and those passing quality control in Table S3, Online Data Supplement). The list of 258 predefined neutrophil genes, and their origin is shown in Table S4, Online Data Supplement. Of these, 35 genes were altered in the placebo-treated group and 29 genes were altered in the nemiralisib-treated group for the Day 12 vs Screening comparison (Table 1), with no genes reaching cut offs when FDR adjusted. When comparing day 84 versus screening, 15 genes in the nemiralisib-treated group were altered and 10 genes in the placebo-treated group. In combination, these observations suggest a lack of unique nemiralisib-induced gene changes.
Table 1.
Summary of Number of Genes Reaching Cut Offs (Fold Change >1.5 and P<0.05) from the Predefined Gene List, and Entire Gene Array in Sputum and Blood
| Comparison | Sputum Gene Changes Predefined List | Sputum Gene Changes Total Set | Blood Gene Changes Total Set |
|---|---|---|---|
| Screening vs D12 nemiralisib | 29 | 1383 | 336 |
| Screening vs D12 placebo | 35 | 1395 | 90 |
| Screening vs D28 nemiralisib | 3 | 350 | 204 |
| Screening vs D28 placebo | 1 | 329 | 302 |
| Screening vs D84 nemiralisib | 15 | 690 | 529 |
| Screening vs D84 placebo | 29 | 1096 | 590 |
| D84 nemiralisib vs D84 placebo | 10 | 297 | 253 |
Abbreviations: D12, Day 12; D84, Day 84.
Nemiralisib Did Not Evoke Any Unique Changes in Sputum Transcriptomics Compared to Placebo-Treated Subjects
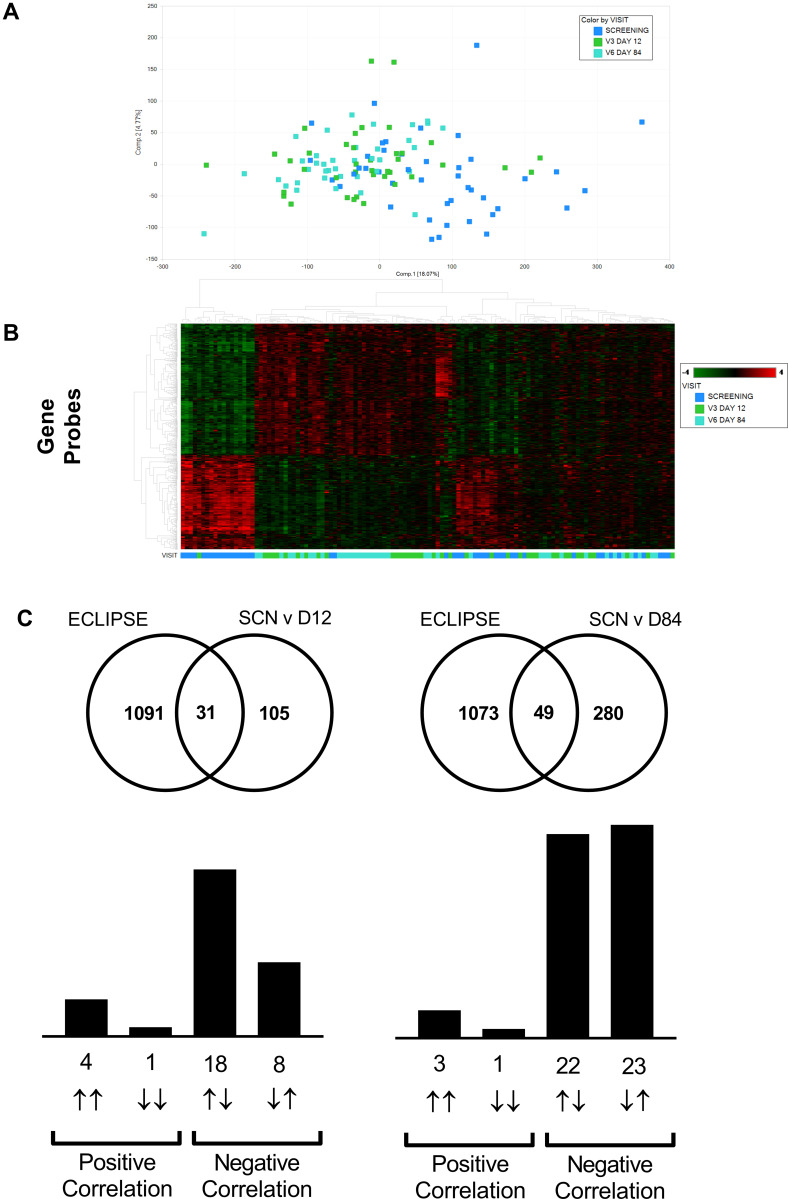
In addition to the predefined gene set, an unbiased analysis of all gene changes was also investigated (Table 1). When comparing the Screening visit and Day 12 visit there were 1383 gene changes for the nemiralisib group and 1395 genes changed in the placebo group (Figure 2A). There was a substantial overlap of 602 genes that were common between placebo- and nemiralisib-treated groups, with 600 genes changing in the same direction (Figure 2B). Furthermore, the apparent unique gene set for each treatment arm, almost without exception, changed in the same direction but did not reach predefined thresholds. Consistent with the pre-defined neutrophil-related transcripts observations, there were no detected nemiralisib-induced changes across the total gene panel. Table 1 also displays the genes changing in the blood data at each timepoint. Samples were compared between nemiralisib and placebo arms on Day 84, with 297 gene changes observed, however these did not map to coherent biological mechanisms.
Figure 2.
mRNA transcriptomic alterations in induced sputum from AECOPD patients (Study B). Transcriptomic changes in induced sputum taken from acutely exacerbating COPD patients. Of the predefined neutrophil-related genes a small number were altered in both the nemiralisib and placebo-treated groups (Panel A). The entire unbiased gene array output also showed a similar number of genes changing in the nemiralisib and placebo-treated groups when comparing both day 12 or day 84 to the screening time point (Panel A, SCN v D12). There was a significant overlap in the genes changing between screening and day 12 in the nemiralisib and placebo-treated groups at day 12 and day 84, with almost all genes changing in the same direction (Panel B).
Nemiralisib Treatment Did Not Impact Lung and Circulating Biomarkers
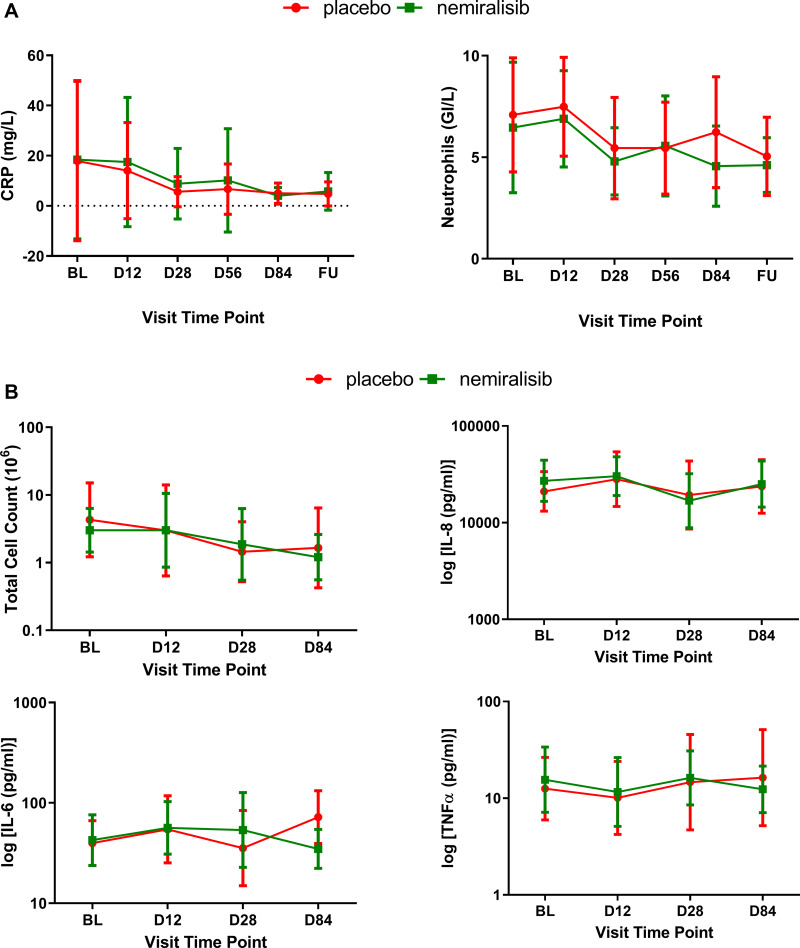
The mean circulating levels of C-reactive protein (CRP) were 17.9 mg/L in the placebo group and 18.4 mg/L in the nemiralisib group at screening, before decreasing to 5.04 mg/L in the placebo-treated group and 4.01 mg/L in the nemiralisib-treated group by day 84 (Figure 3A), consistent with resolution of the exacerbation.26 Similarly, mean circulating blood neutrophil count was 7.1 GI/L in the placebo group and 6.4 GI/L in the nemiralisib group at screening, before decreasing to 5.0 GI/L in the placebo-treated group and 4.6 GI/L in the nemiralisib-treated group by day 84 (Figure 3A), consistent with resolution of the exacerbation. Furthermore, the geometric mean (95% CI) total sputum cell counts (x106) at screening prior to randomization were similar between the placebo (4.297 [1.220, 15.132]) and nemiralisib (3.011 [1.435, 6.318]) groups, Figure 3B. During the study, the geometric mean (95% CI) total cell counts in the induced sputum in both placebo- and nemiralisib-treated groups were reduced to 1.656 (0.425, 6.455) and 1.207 (0.558, 2.612), respectively, representing approximately a 40% reduction in inflammatory cell counts in the sputum consistent with exacerbation recovery.
Figure 3.
Inflammatory biomarkers in induced sputum from AECOPD patients (Study B) Sputum and circulating biomarkers measured were also not impacted by treatment with nemiralisib. Circulating levels of CRP decreased (Panel A, left), as did neutrophil count (Panel A, right), presented as mean (95% CI) at baseline, Day 12, Day 28, Day 56, Day 84 and at follow-up. Total sputum cell count and the inflammatory cytokines IL-8, IL-6 and TNFα were also not inhibited following treatment with nemiralisib (Panel B), presented as geometric mean (95% CI) at baseline, Day 12, Day 28 and Day 84.
There was no meaningful difference between the reductions observed in both the placebo- and nemiralisib-treated groups.
Treatment with nemiralisib also had no effect on the levels of the sputum cytokines measured compared to placebo (Figure 3B). Cytokine levels also remained relatively unchanged in both placebo and nemiralisib-treated groups across the 84 days of treatment and despite treatment with oral corticosteroids.
Nemiralisib Treatment Had No Impact on HRCT Parameters
Functional HRCT scans were taken to study the time course of improvement in imaging parameters during recovery from an exacerbation. Imaging endpoints did not meaningfully change by treatment with nemiralisib compared to placebo, at any time points measured (Table 2).
Table 2.
Alterations in Functional Respiratory Imaging Endpoints Measured Using HRCT
| Endpoint | Parameter | Region | Placebo (Baseline vs D28, Treatment Ratio (95% Cr I)) | Nemiralisib (Baseline vs D28, Treatment Ratio (95% Cr I)) | Nemiralisib vs Placebo (Baseline vs D28, Treatment Ratio (95% Cr I)) | Probability (θ>1) or (θ<1)a |
|---|---|---|---|---|---|---|
| siVaw | Specific Imaging Airway Volume | Distal | 1.01 (0.83, 1.24) | 1.00 (0.83, 1.20) | 0.98 (0.75, 1.30) | 45.2% |
| siRaw | Specific imaging airway resistance | Distal | 0.97 (0.53, 1.77) | 1.13 (0.63, 2.02) | 1.17 (0.51, 2.72) | 35.6% |
| siVaww | Specific airway wall thickness | Distal | 0.96 (0.88, 1.06) | 0.92 (0.84, 1.00) | 0.95 (0.83, 1.09) | 77.1% |
| LAS | Low attenuation score | Total | 1.01 (0.84, 1.21) | 0.98 (0.83, 1.17) | 0.97 (0.75, 1.26) | 58.6% |
| IALD | Internal airflow lobar distribution | Upper | 1.03 (0.98, 1.09) | 0.99 (0.94, 1.04) | 0.96 (0.89, 1.03) | 59.7% |
| Lower | 0.98 (0.93, 1.02) | 0.99 (0.95, 1.04) | 1.02 (0.95, 1.09) | 97.9% | ||
| BVD | Blood vessel density | Total | 0.95 (0.88, 1.03) | 0.94 (0.88, 1.01) | 0.99 (0.89, 1.10) | 44.4% |
Note: aθ represents the true treatment ratio to placebo.
Abbreviations: BVD, blood vessel density; Cr l, credible interval; D28, day 28; HRCT, high resolution computed tomography; IALD, internal airflow lobar distribution; LAS, low attenuation score; siRaw, specific imaging airway resistance; siVaw, specific imaging airway volume; siVaww, specific imaging airway wall thickness.
Nemiralisib Treatment Evoked a Clinically Meaningful Improvement in FEV1
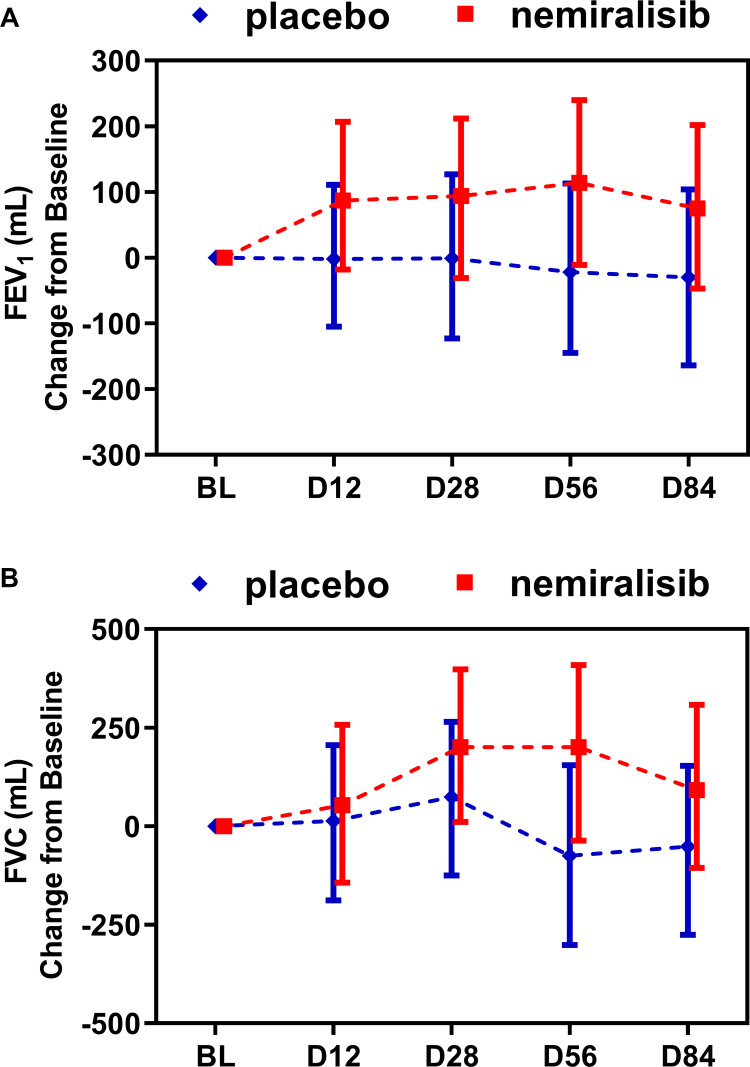
FEV1 remained relatively constant in the placebo-treated group throughout the study (Figure 4A). By contrast FEV1 increased in the nemiralisib-treated group at all time points, with the largest mean change from baseline of 114mL at day 56 (Figure 4A). This represents a placebo-corrected mean change of 136mL (95% credible interval (Cr. I.) −46, 315mL) with a probability that the true treatment ratio was >0% (Pr(θ>0)) of 93%. This was mirrored by improvements in FVC at all time points, with the largest placebo-corrected mean change from baseline occurring at Day 56 of 275mL (95% Cr. I. −55, 587mL) with a Pr(θ>0) of 95% (Figure 4B).
Figure 4.
Summary of statistical analysis of FEV1 and FVC in AECOPD patients (Study B). Treatment with nemiralisib showed an improvement in FEV1 and FVC at all time points. Panel A shows adjusted median change from baseline in FEV1 (95% Cr I), and Panel B shows adjusted median change from baseline in FVC (95% Cr I) measured at clinical visits at baseline, Day 12, Day 28, Day 56 and Day 84. Data presented as adjusted median ± 95% Credible Intervals.
Pharmacokinetics of Nemiralisib Were Similar to Previous Studies
A sparse PK sampling schedule was used in this study to confirm drug exposures which were similar to previously reported data.16,17 Importantly there were no meaningful differences in mean trough/pre-dose exposures achieved between each study (Table S5 Online Data Supplement).
Exploration of Lung and Blood Transcriptomics During Exacerbation Recovery Showed Overlap with Existing COPD Disease Severity Data Sets
Given the lack of unique nemiralisib-evoked changes in gene transcriptomic data, the sputum and blood data from both arms of the study were combined to provide a post-hoc analysis. The principle component analysis and heat maps of altered genes for each data set showed an element of separation between the screening time point and days 12 and 28 when adjusting for subject to subject variability (Figures 5 and 6A and B (sputum and blood, respectively)). Computational pathway analysis of the sputum data set showed an enrichment for genes associated with corticosteroid treatment, B-cell biology, cellular metabolism, peroxisome proliferator-activated receptor alpha (PPARα) agonists and mitochondrial dysfunction. In the combined blood analysis, there was an enrichment for corticosteroid regulated genes hand genes associated with treatment with granulocyte-CSF (Filgrastim).
Figure 5.
Lung transcriptomics during exacerbation recovery (Study B). Combined sputum transcriptomic data shows partial clustering by time point (Panels A and B). Unbiased overlay analysis showed a highly significant overlap with data from ECLIPSE .27,30 The degree of overlap and gene directionality for the matching genes are displayed in (Panel C).
Figure 6.
Blood transcriptomics during exacerbation recovery (Study B). Combined blood transcriptomic data shows partial clustering by time point (Panels A and B). Unbiased overlay analysis showed a highly significant overlap with data from ECLIPSE.27,30 The degree of overlap and gene directionality for the matching genes are displayed in (Panel C).
An unbiased comparison of the overlap of differentially expressed genes with publicly available data sets revealed significant overlap between the sputum data and the Evaluation of COPD Longitudinally to Identify Predictive Surrogate End-points (ECLIPSE) cohort.27–29 The current study (recovery from exacerbations) showed a strong inverse relationship to Gold 3 vs Gold 2 comparison in ECLIPSE (Figure 5C) suggesting recovery from an exacerbation is the inverse of an increase in disease severity. Downstream pathway enrichment focused on Phenethylamine metabolism, Liver X receptor (LXR), Retinoid X receptor (RXR), and lipopolysaccharide (LPS) signaling. Although not as strong, there was also a largely inverse relationship between ECLIPSE and the blood transcriptomic data sets (Figure 6C).
Discussion
In drug development, the mechanism of action of novel immunomodulators is removed from established clinical efficacy endpoints and outcomes. Translational endpoints, linking biomarkers to clinical endpoints, help enable the appropriate interpretation of clinical study outcomes, both positive and negative. Here we present the steps taken to translate the preclinical therapeutic rationale for the use of PI3Kδ inhibitors in COPD patients, initially in stable patients, then subsequently in patients experiencing an AECOPD. In this report, we demonstrate that treatment of stable COPD patients with the PI3Kδ inhibitor nemiralisib modulates neutrophil phenotype, inferred from changes in gene expression patterns. However, this modulation was not observed during recovery from an AECOPD despite a clinically significant improvement in lung function.
Neutrophils isolated from blood of COPD patients demonstrate a dysregulated migration pattern to chemoattraction which in vitro can be corrected using selective inhibitors of PI3Kδ.8 Given the major role of neutrophils in pathogen clearance in the lung, this observation provides a potential novel mechanism to target recurrent lung infections in COPD. Limited availability of clinical methodologies for demonstrating alterations in lung neutrophil characteristics prompted our application of gene transcriptomic pathway analysis in induced sputum to pursue changes in neutrophil biology that might result from nemiralisib treatment.
In stable COPD patients, treatment with nemiralisib evoked gene changes which focused on two key areas; neutrophil migration machinery and bacterial recognition/infection responses. Importantly, stability of transcription profile in sputum was demonstrated by the very few expression changes observed prior to randomization, the neutrophil changes were not observed in the placebo group, and absolute number of neutrophils did not change (see Figure 3 Cahn et al).18 This provides reassurance that exploring nemiralisib-evoked gene changes in sputum could provide a mechanistic clinical readout of improved neutrophil function observed in vitro, suitable for translation into a clinically meaningful endpoint.
Before moving to exacerbating patients, we confirmed that neutrophils isolated from COPD patients experiencing an AECOPD retained the ability to be inhibited by PI3Kδ. This was achieved by measuring inhibition of reactive oxygen species and neutrophil elastase production from isolated neutrophils with a similar concentration range to previous reports, consistent with another report of the ability of PI3Kδ to inhibit mediator production by neutrophils from AECOPD patients.30
There was no evidence for unique nemiralisib-evoked gene changes in the exacerbating patients, neither in the predefined neutrophil subset (primary endpoint), nor the unbiased whole-gene array (exploratory endpoint). In the predefined set, a subset of these genes did change following treatment with nemiralisib, but a similar number also changed in the placebo arm, with a modest number in common. Consistent with the predefined gene set there was also no apparent consequence of treatment with nemiralisib at the gene expression level when probing the entire array. This was concluded due to a similar number of genes changing in both treatment arms, a significant overlap between them, and a striking coherence in gene change directionality.
The breadth of genes changing proximal to the index exacerbation is likely reflective of the significant immune response in the lung at exacerbation which begins to resolve after 12 days.31 Furthermore, the data set is likely influenced by the oral steroids and antibiotics administered to every subject at entry to the study.32 Hence detecting subtle changes evoked by nemiralisib may not be achievable using an Affymetrix approach on a mixed cell population obtained from sputum. It could also be the case that due to the heterogeneity of exacerbations of COPD the population most likely to be PI3Kδ-dependent were not represented, or that overall, PI3Kδ-dependent neutrophil processes are not relevant during acute recovery from an exacerbation.
Several additional biomarker and lung function endpoints were measured in this study with the intent of exploring the relationship with the gene signature data. As expected, circulating CRP and total sputum cell counts were elevated at screening, suggestive of infection in the patients, and resolved during the duration of the study, however no meaningful effect of nemiralisib treatment was observed on either endpoint. At exacerbation, levels of IL-8, TNFα, and IL-6 in sputum were elevated, compared to our study A in stable COPD patients, but were not reduced following treatment with nemiralisib. In fact, levels of inflammation in the lung, as indicated by cytokines, remained elevated for the entire 3 months despite patients showing clinical improvement. This suggests that exacerbation may result in a prolonged inflammatory insult in the lung. HRCT has been used to measure subtle and regional changes in lung function in COPD patients experiencing an exacerbation. In the current study, no notable improvement in any of the HRCT parameters was detected following treatment with nemiralisib when compared to placebo. This is contrary to our previous study in exacerbating patients.25 Consistent however, with this previous study, a clinically meaningful improvement in FEV1 and FVC was observed following treatment with nemiralisib.
As there was no discernible difference between placebo and nemiralisib with respect to gene expression, gene data sets were combined to explore a larger recovery dataset as a post-hoc analysis. Pathway mapping of the sputum data suggested a strong overlap in genes associated with cellular metabolism and mitochondrial dysfunction, which are known to be dysregulated in COPD,33,34 and B-cell biology which in COPD is less well understood.35 There was also an overlap with PPARα agonist data sets which may be related to skeletal muscle dysregulation seen in COPD.36 Analysis of the blood data set suggested overlap with corticosteroid treatment and interestingly with treatment with granulocyte-CSF (Filgrastim) which is known to influence blood neutrophil numbers. This is likely to be a consequence of blood neutrophils being elevated at screening and reduced with resolution.
The sputum data in the current study strongly overlapped with data obtained from the ECLIPSE cohort,29,32 which provides support that the data from our small cohort study is reflective of a wider population of COPD patients. The current study (recovery from exacerbation) showed a strong inverse relationship to the Gold 3 vs Gold 2 comparison in the ECLIPSE cohort, suggesting recovery from exacerbation is possibly the transient inverse of an increase in disease severity. Highly enriched pathways were those related to LPS-activation which commonly appear in COPD data sets,28,37 and interestingly, those related to LXR and RXR pathways, which have been previously explored as potential mechanisms to treat COPD.38,39 From this data overlap it would be enticing to generate an overall physiological hypothesis as to why repeated acute exacerbation events lead to a decline in disease status and increased mortality.2
The investigation of local lung biology in exacerbating COPD patients presented several risks. Firstly, it was unknown if acutely unwell patients would be willing to participate in the clinical trial procedures, and if inducing sputum from this population would be safe and tolerated. Despite these challenges 44 patients were successfully recruited, producing a matched sample set of enough quality for downstream analysis. The study design also presented challenges due to a lack of stable baseline sampling before exacerbations, however our approach reflects real-world patient management,40 and enables assessment of drug mechanisms under appropriate clinical conditions. Exacerbations of COPD are heterogenous in clinical presentation, and the ability to accurately determine the drivers of an exacerbation at the point of presentation is currently not broadly feasible, and hence our study represents the current standard of patient management.
Conclusions
Our aim was to compare in vitro mechanistic observations of the inhibition of PI3Kδ with those seen in a clinical trial setting where inhaled nemiralisib was administered to COPD patients. This was achieved through the measuring of transcriptomic changes as a marker of drug action. In nemiralisib-treated stable COPD patients, a transcriptomic profile was obtained which was consistent with the modulation of neutrophil dysfunction observed in vitro. This neutrophil profile was not observed in the samples taken from exacerbating patients treated with nemiralisib despite an improvement in FEV1. Further research is required to explain, the absence of detectable transcriptomic changes during COPD exacerbations, and the lack of relationship between the reduction in FEV1 and the minimally meaningful HRCT endpoint changes in COPD exacerbating patients exposed to nemiralisib.
We have successfully translated preclinical in vitro observations in stable COPD patients and observed improvements in FEV1 in exacerbating COPD patients treated with nemiralisib. Furthermore, we have generated a comprehensive profile of the human lung transcriptome following recovery from an AECOPD which is a valuable resource that can be explored further.
Acknowledgments
DISKUS and ELLIPTA are owned by/licensed to the GSK group of companies. Editorial support was provided by Kate Hollingworth of Continuous Improvement Ltd, and funded by GlaxoSmithKline (GSK) R&D.
Funding Statement
This work was funded by GlaxoSmithKline (clinicaltrials.gov: NCT02130635 and NCT02522299). The sponsor/funder, as described under author contributions, was involved in the study design, data analysis, interpretation of data and writing of the manuscript.
Abbreviations
AECOPD, acute exacerbation of COPD; AERIS, Acute Exacerbation and Respiratory Infections in COPD; CI, confidence interval; COPD, chronic obstructive pulmonary disease; Cr I, credible interval; ECLIPSE, Evaluation of COPD Longitudinally to Identify Predictive Surrogate End-points; FDR, false discovery rate; FEV1, forced expiratory volume in 1 sec; FRI, functional respiratory imaging; FVC, forced vital capacity; LD-HRCT, low dose high resolution computed tomography; LPS, lipopolysaccharide; LXR, liver X receptor; mRNA, messenger ribonucleic acid; PI3Kδ, phosphoinositide 3-kinase δ; PK, pharmacokinetics; PPARα, peroxisome proliferator-activated receptor alpha; RXR, retinoid X receptor.
Data Sharing Statement
Information on GlaxoSmithKline R&D’s data sharing commitments and requesting access can be found at: https://www.clinicalstudydatarequest.com.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
MB, EJ, GB, SM, DM, ML, HW, AA, KS, RT, SA, TT and AC are employees and shareholders of GSK. JNH, RW & EMH are former employees and shareholders of GSK. JNH, EMH and AA are named on patents related to compound GSK2269557 (nemiralisib). JNH has patents WO 2010125082 and WO 2012055846 issued. EMH has a patent WO2015055691A1 issued. AA has patents US20160263109, US20160256466, EP3057588, EP3057587, WO2015055691, and WO2015055690 issued to GSK. CVH and JDB are employees of FLUIDDA, the company responsible for FRI analysis and interpretation. WV is a former employee of FLUIDDA. WV and JDB hold FLUIDDA shares. JMF has received research funding paid directly to the University of British Columbia from AstraZeneca, GSK, Novartis, Sanofi-Regeneron, Canadian Institutes of Health Research (CIHR), NIH and AllerGen; and has received honoraria from AstraZeneca, GSK, TEVA, Sanofi for participation in Advisory Boards and speaker bureaus. JB has received grants from CIHR, Canadian Respiratory Research Network, Foundation of the McGill University Health Centre (MUHC), Aerocrine, AstraZeneca, GSK, Boehringer Ingelheim, Novartis, Grifols and Trudell; and personal fees from Canadian Thoracic Society, CHEST, AstraZeneca, GSK, Boehringer Ingelheim, Novartis, Grifols, Pfizer, and Trudell for consultancy and lectures. CP has received personal fees from GSK, Boehringer Ingelheim, Novartis, AstraZeneca and Apnee Sante. FM has received grants paid to his institution from AstraZeneca, GSK, Boehringer Ingelheim, Sanofi, Novartis and Grifols; has received personal fees from GSK, Boehringer Ingelheim, Grifols, and Novartis for serving on speaker bureaus; and is financially involved with Oxynov, a company developing an oxygen delivery system. The current affiliation of JNH is Discovery, Charles River, Chesterford Research Park, Cambridge, UK. The current affiliation of DM is Benevolent AI Ltd, London, United Kingdom. The current affiliation of RW is DLRC, Letchworth Garden City, UK. The current affiliation of WV is OncoRadiomics, Liège, Belgium. The current affiliation of EMH is Eligo Bioscience, Paris, France. The authors report no other conflicts of interest in this work.
References
- 1.Dy R, Sethi S. The lung microbiome and exacerbations of COPD. Curr Opin Pulm Med. 2016;22:196–202. doi: 10.1097/MCP.0000000000000268 [DOI] [PubMed] [Google Scholar]
- 2.Ritchie AI, Wedzicha JA. Definition, causes, pathogenesis, and consequences of chronic obstructive pulmonary disease exacerbations. Clin Chest Med. 2020;41:421–438. doi: 10.1016/j.ccm.2020.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bafadhel M, McKenna S, Terry S, et al. Acute exacerbations of chronic obstructive pulmonary disease: identification of biologic clusters and their biomarkers. Am J Respir Crit Care Med. 2011;184:662–671. doi: 10.1164/rccm.201104-0597OC [DOI] [PubMed] [Google Scholar]
- 4.Wilkinson TMA, Aris E, Bourne SC, et al. Drivers of year-to-year variation in exacerbation frequency of COPD: analysis of the AERIS cohort. ERJ Open Res. 2019;5:00248–2018. doi: 10.1183/23120541.00248-2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.AERIS Study Group; Wilkinson TMA, Aris E, Bourne S, et al. A prospective, observational cohort study of the seasonal dynamics of airway pathogens in the aetiology of exacerbations in COPD. Thorax. 2017;72:919–927. doi: 10.1136/thoraxjnl-2016-209023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu J, Pang Z, Wang G, et al. Advanced role of neutrophils in common respiratory diseases. J Immunol Res. 2017;2017:6710278. doi: 10.1155/2017/6710278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sapey M, Rijkers GT, van Overveld FJ. Neutrophils and emerging targets for treatment in chronic obstructive pulmonary disease. Expert Rev Clin Immunol. 2013;9:1055–1068. doi: 10.1586/1744666X.2013.851347 [DOI] [PubMed] [Google Scholar]
- 8.Sapey E, Stockley JA, Greenwood H, et al. Behavioral and structural differences in migrating peripheral neutrophils from patients with chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2011;183:1176–1186. doi: 10.1164/rccm.201008-1285OC [DOI] [PubMed] [Google Scholar]
- 9.Milara J, Lluch J, Almudever P, Freire J, Xiaozhong Q, Cortijo J. Roflumilast N-oxide reverses corticosteroid resistance in neutrophils from patients with chronic obstructive pulmonary disease. J Allergy Clin Immunol. 2014;134:314–322. doi: 10.1016/j.jaci.2014.02.001 [DOI] [PubMed] [Google Scholar]
- 10.To Y, Ito K, Kizawa Y, et al. Targeting phosphoinositide-3-kinase-delta with theophylline reverses corticosteroid insensitivity in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2010;182:897–904. doi: 10.1164/rccm.200906-0937OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stark A-K, Sriskantharajah S, Hessel EM, Okkenhaug K. PI3K inhibitors in inflammation, autoimmunity and cancer. Curr Opin Pharmacol. 2015;2382–2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gupta V, Singh D. Critical assessment of the value of sputum neutrophils. COPD. 2013;10:107–114. doi: 10.3109/15412555.2013.766105 [DOI] [PubMed] [Google Scholar]
- 13.Tregay N, Begg M, Cahn A, et al. Use of autologous 99mTechnetium-labelled neutrophils to quantify lung neutrophil clearance in COPD. Thorax. 2019;74:659–666. doi: 10.1136/thoraxjnl-2018-212509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sims P, Coffman RL, Hessel EM. Biomarkers measuring the activity of Toll-like receptor ligands in clinical development programs. Methods Mol Biol. 2009;517:415–440. [DOI] [PubMed] [Google Scholar]
- 15.Gauvreau GM, Hessel EM, Boulet LP, Coffman RL, O’Byrne PM. Immunostimulatory sequences regulate interferon-inducible genes but not allergic airway responses. Am J Respir Crit Care Med. 2006;174:15–20. doi: 10.1164/rccm.200601-057OC [DOI] [PubMed] [Google Scholar]
- 16.Govoni M, Bassi M, Vezzoli S, et al. Sputum and blood transcriptomics characterisation of the inhaled PDE4 inhibitor CHF6001 on top of triple therapy in patients with chronic bronchitis. Respir Res. 2020;21:72. doi: 10.1186/s12931-020-1329-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Begg M, Wilson R, Hamblin JN, et al. Relationship between pharmacokinetics and pharmacodynamic responses in healthy smokers informs a once-daily dosing regimen for nemiralisib. J Pharmacol Exp Ther. 2019;369:337–344. doi: 10.1124/jpet.118.255109 [DOI] [PubMed] [Google Scholar]
- 18.Cahn A, Hamblin JN, Begg M, et al. Safety, pharmacokinetics and dose-response characteristics of GSK2269557, an inhaled PI3Kδ inhibitor under development for the treatment of COPD. Pulm Pharmacol Ther. 2017;46:69–77. doi: 10.1016/j.pupt.2017.08.008 [DOI] [PubMed] [Google Scholar]
- 19.van Geffen WH, Hajian B, Vos W, et al. Functional respiratory imaging: heterogeneity of acute exacerbations of COPD. Int J Chron Obstruct Pulmon Dis. 2018;13:1783–1792. doi: 10.2147/COPD.S152463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Global Initiative for Chronic Obstructive Lung Disease (GOLD). From the global strategy for the diagnosis, management and prevention of COPD; 2020. Available from: http://goldcopd.org. Accessed May6, 2021.
- 21.Quanjer PH, Tammeling GJ, Cotes JE, Pedersen OF, Peslin R, Yernault JC. Lung volumes and forced ventilatory flows. Report working party standardization of lung function tests, European community for steel and coal. Official statement of the European Respiratory Society. Eur Respir J. 1993;6(suppl 16):5–40. doi: 10.1183/09041950.005s1693 [DOI] [PubMed] [Google Scholar]
- 22.Seemungal TA, Donaldson GC, Bhowmik A, Jeffries DJ, Wedzicha JA. Time course and recovery of exacerbations in patients with chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2000;161:1608–1613. doi: 10.1164/ajrccm.161.5.9908022 [DOI] [PubMed] [Google Scholar]
- 23.Juss JK, House D, Amour A, et al. Acute respiratory distress syndrome neutrophils have a distinct phenotype and are resistant to phosphoinositide 3-kinase inhibition. Am J Respir Crit Care Med. 2016;194(8):961–973. doi: 10.1164/rccm.201509-1818OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vlahos R, Wark PA, Anderson GP, Bozinovski S. Glucocorticosteroids differentially regulate MMP-9 and neutrophil elastase in COPD. PLoS One. 2012;7:e33277. doi: 10.1371/journal.pone.0033277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cahn A, Hamblin JN, Robertson J, et al. An inhaled PI3δ inhibitor improves recovery in acutely exacerbating COPD patients, a randomized trial. Int J Chron Obstruct Pulmon Dis. In press 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Butler CC, Gillespie D, White P, et al. C-reactive protein testing to guide antibiotic prescribing for COPD exacerbations. N Engl J Med. 2019;381:111–120. doi: 10.1056/NEJMoa1803185 [DOI] [PubMed] [Google Scholar]
- 27.ECLIPSE investigators; Vestbo J, Anderson W, Coxson HO, et al. Evaluation of COPD longitudinally to identify predictive surrogate end-points (ECLIPSE). Eur Respir J. 2008;31:869–873. doi: 10.1183/09031936.00111707 [DOI] [PubMed] [Google Scholar]
- 28.Reinhold D, Morrow JD, Jacobson S, et al. Meta-analysis of peripheral blood gene expression modules for COPD phenotypes. PLoS One. 2017;12:e0185682. doi: 10.1371/journal.pone.0185682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.ECLIPSE Investigators; Singh D, Fox SM, Tal-Singer R, et al. Induced sputum genes associated with spirometric and radiological disease severity in COPD ex-smokers. Thorax. 2011;66(6):489–495. doi: 10.1136/thx.2010.153767. [DOI] [PubMed] [Google Scholar]
- 30.Gupta V, Khan A, Higham A, et al. The effect of phosphatidylinositol-3 kinase inhibition on matrix metalloproteinase-9 and reactive oxygen species release from chronic obstructive pulmonary disease neutrophils. Int Immunopharmacol. 2016;35:155–162. doi: 10.1016/j.intimp.2016.03.027 [DOI] [PubMed] [Google Scholar]
- 31.Pavord ID, Jones PW, Burgel PR, Rabe KF. Exacerbations of COPD. Int J Chron Obstruct Pulmon Dis. 2016;11(speciss):21–30. doi: 10.2147/COPD.S85978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Singh D, Fox SM, Tal-Singer R, Bates S, Riley JH, Celli B. Altered gene expression in blood and sputum in COPD frequent exacerbators in the ECLIPSE cohort. PLoS One. 2014;9(9):e107381. doi: 10.1371/journal.pone.0107381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.COPDMAP; Michaeloudes C, Kuo CH, Haji G, et al. Metabolic re-patterning in COPD airway smooth muscle cells. Eur Respir J. 2017;50:1700202. doi: 10.1183/13993003.00202-2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ryter SW, Rosas IO, Owen CA, et al. Mitochondrial dysfunction as a pathogenic mediator of chronic obstructive pulmonary disease and idiopathic pulmonary fibrosis. Ann Am Thorac Soc. 2018;15:S266–S272. doi: 10.1513/AnnalsATS.201808-585MG [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cosio MG, Saetta M, Agusti A. Immunologic aspects of chronic obstructive pulmonary disease. N Engl J Med. 2009;360:2445–2454. doi: 10.1056/NEJMra0804752 [DOI] [PubMed] [Google Scholar]
- 36.Remels AH, Gosker HR, Schrauwen P, Langen RC, Schols AM. Peroxisome proliferator-activated receptors: a therapeutic target in COPD? Eur Respir J. 2008;31:502–508. doi: 10.1183/09031936.00068207 [DOI] [PubMed] [Google Scholar]
- 37.Wan ES, Silverman EK. Genetics of COPD and emphysema. Chest. 2009;136:859–866. doi: 10.1378/chest.09-0555 [DOI] [PubMed] [Google Scholar]
- 38.Stolk J, Stockley RA, Stoel BC, et al. Randomised controlled trial for emphysema with a selective agonist of the γ-type retinoic acid receptor. Eur Respir J. 2012;40:306–312. doi: 10.1183/09031936.00161911 [DOI] [PubMed] [Google Scholar]
- 39.Higham A, Lea S, Plumb J, et al. The role of the liver X receptor in chronic obstructive pulmonary disease. Respir Res. 2013;14:106. doi: 10.1186/1465-9921-14-106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Criner RN, Han MK. COPD care in the 21st century: a public health priority. Respir Care. 2018;63:591–600. doi: 10.4187/respcare.06276 [DOI] [PMC free article] [PubMed] [Google Scholar]