Abstract
Background
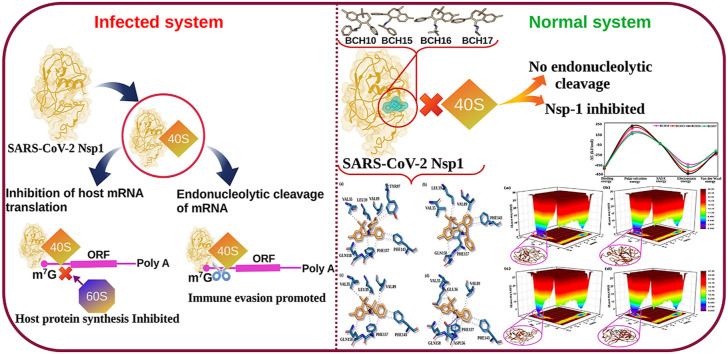
Non-structural protein 1 (Nsp1), a virulence agent of SARS-CoV-2, has emerged as an important target for drug discovery. Nsp1 shuts down the host gene function by associating with the 40S ribosomal subunit.
Methods
Molecular interactions, drug-likeness, physiochemical property predictions, and robust molecular dynamics (MD) simulations were employed to discover novel Nsp1 inhibitors. In this study, we evaluated a series of molecules based on the plant (Cedrus deodara) derived α,β,γ-Himachalenes scaffolds.
Results
The results obtained from estimated affinity and ligand efficiency suggested that BCH10, BCH15, BCH16, and BCH17 could act as potential inhibitors of Nsp1. Moreover, MD simulations comprising various MD driven time-dependent analyses and thermodynamic free energy calculations also suggested stable protein-ligand complexes and strong interactions with the binding site. Furthermore, the selected molecules passed drug likeliness parameters and the physiochemical property analysis showed acceptable bioactivity scores.
Conclusion
The structural parameters of dynamic simulations revealed that the reported molecules could act as lead compounds against SARS-CoV-2 Nsp1 protein.
Keywords: Nsp1, Non-structural protein 1, FEL, Free energy landscape, MM-PBSA, COVID-19, SARS-CoV-2
Graphical abstract
1. Introduction
It is the third time that the 21st century is experiencing a coronavirus (CoV) outbreak. However, the current pandemic is most endangering in terms of the global economy and health [[1], [2], [3]]. CoV, a diversified group of viruses (enveloped and pleomorphic configuration) with a 100 nm diameter, comprises of single 30 kb of RNA 5′-capped and 3′-polyadenylated genome. This genome represents two broad overlapping open reading frames (ORF) in gene 1 (1a and 1b), as well as several structural and non-structural proteins at the 3′ end. After infection, the proteins of ORF1a and b are translated and then proteolytically cleaved into functional proteins. Most of these proteins have a role in viral replication [4]. The non-structural protein 1 (Nsp1) is expressed only in α (~9 kDa) and β (~20 kDa) CoVs.
Nsp1 has crucial and conserved functions such as host mRNA degradation, suppressing the host antiviral signaling pathways and interferon expression. Due to these functions, Nsp1 is admitted as one of the prominent virulence factors. SARS-CoV-2 Nsp1 shares high structural similarity and sequence identity with SARS-CoV Nsp1; thus, the data of SARS-CoV Nsp1 functions could be highly informative to know the biological processes of SARS-CoV-2 Nsp1 [[5], [6], [7], [8]]. An extension of β4 strand and the appearance of an extra β-strand in NSp1 of SARS-CoV-2 are among the remarkable differences between Nsp1 structures of SARS-CoV and SARS-CoV-2 [9,10]. The change in amino acid (K to V) and (V to M) at positions 84 and 85 between SARS-CoV and SARS-CoV-2 Nsp1 resulted in β4 extension on the N-terminus by three residues [9,10]. This extension resulted in an increased number of hydrogen bonds between β4 and β7 strands in SARS-CoV-2 Nsp1 in comparison to the bonding pattern between β4 and β6 strands of SARS-CoV Nsp1. This extension also promoted the creation of the β5-strand, which was absent in Nsp1 of SARS-CoV. The β5 and β4 strands are part of a small β-sheet stabilizing the loop region between β5 and β6 equivalent to β4 and β5 in SARS-CoV Nsp1 [9].
Nsp1 promotes the complete cessation of host protein translation through two strategies. First, it binds with the small ribosomal subunit and delays mRNA translation at several stages of initiation [6,11]. Secondly, the binding of Nsp1 to the ribosome directs the endonucleolytic division and host mRNA degradation [6]. Nsp1 of SARS-CoV-2 disrupts the mRNA export machinery to inhibit host gene expression [12]. In addition, Nsp1 could be able to reduce cellular translation considerably; however, remaining ribosomes would still be able to translate viral mRNAs with high efficiency [5]. Besides, a new strain of SARS-CoV-2 was reported to lack specific amino acids from Nsp1. The lack of these specific amino acids indicated profound genomic changes in SARS-CoV-2 [13]. The complex of Nsp1 bound to the human 40S ribosomal subunit showed that Nsp1 blocked the messenger RNA entry tunnel [6]. The cessation of the essential innate immune response may promote immune evasion and an increased rate of viral replication [14]. The prominent role of Nsp1 of SARS-CoV-2 in reducing the antiviral immune response makes it a potential therapeutic target [5,6]. There are three vaccines approved at the moment and an emergency drug remdesivir has been authorized for emergency use against COVID-19 by FDA [15]. Although, no specific FDA-approved drug is present for the treatment of COVID-19 patients. The requisite demand for developing efficient drugs against SARS-CoV-2 necessitates the use of computational tools for the fast screening of potential drug-like molecules.
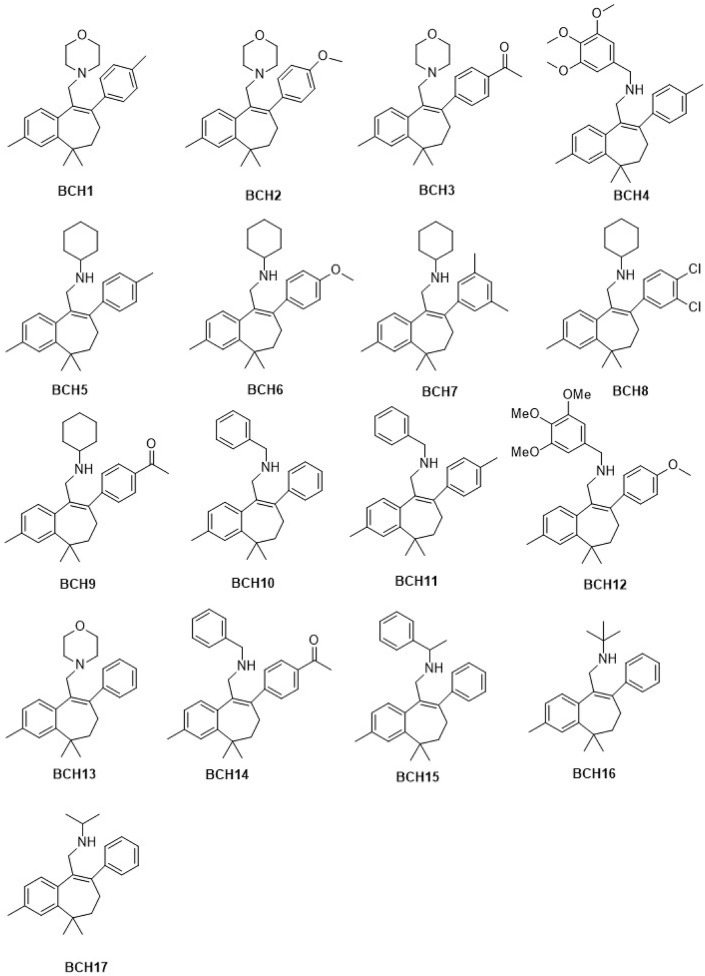
The primary goal of the present study is to identify potential Nsp1 inhibitors by applying structure-based drug design strategies, MD simulations, and thermodynamic binding free energy evaluations. Medicines from the natural origin are deemed to exert fewer side effects than synthetic drugs. Natural and novel molecules as inhibitors of SARS-CoV-2 are also being examined by practicing in-silico approaches [[16], [17], [18], [19], [20], [21], [22], [23]]. In earlier studies, Himachalenes scaffolds were recognized for their anti-inflammatory and antiviral activities [[24], [25], [26]]. A polymer-stabilized Pd(0) nanoparticle-catalyzed Suzuki-Miyaura crosscoupling reaction of amino vinyl bromide substituted benzosuberenes were used for aminoarylbenzosuberenes (AABs) synthesis [27]. The semi‐synthetically derived AAB from Himachalenes (isomeric mixture of sesquiterpenes found in Cedrus deodara oil) [27] were used in this study (Fig. 1 ) for elucidating their therapeutic potential against COVID-19. The aminoaryl functionality of these molecules is rare and provides a great opportunity to outspread binding capacity with the target receptors.
Fig. 1.
2d structures of 17 selected aminoarylbenzosuberene molecules.
2. Material and methods
2.1. Datasets
Iterative Threading ASSEmbly Refinement server (I-TASSER) used to model the SARS-CoV-2 Nsp1 protein of 180 amino acid residues. 17 aminoarylbenzosuberene (AAB) molecules were employed in computational analysis from in-house database, as depicted in Fig. 1 [27]. The structure of the protein was prepared through the Discovery studio software by making the use of its “prepare protein” methodology [28]. The optimization of Ligand geometry of all molecule was performed by the energy minimization protocols of Gaussian16 (with DFT) [29]. The “Find unoccupied pocket” function of SeeSAR was adopted to create the binding sites on the Nsp1 protein. Pocket containing C-terminal residues was selected for docking with AABs molecules.
2.2. Modeling and molecular docking
Since, the only available x-ray diffraction crystal structure of Nsp1 from SARS-CoV-2 (PDB ID:7K3N) [9] was incomplete due to missing residues 117–180, so our foremost step was to build a complete, robust, and validated modeled structure. The C-terminal region is important for inhibition of Nsp1. Hence, we used the SWISS-MODEL that uses homology modeling to render protein model by taking recently submitted Nsp1 structure (PDB ID: 7K3N) as a template. However, the structure generated by SWISS-MODEL was not a complete structure. Thus, modeling was performed through I-TASSER which uses a hierarchical path to predict the protein structure [30]. For this step the NMR structure of Nsp1 (PDB ID: 2GDT) was used as a template. The FASTA sequence of SARS-CoV-2 Nsp1 with NCBI sequence reference: YP_009725297.1 submitted in I-TASSER to built the protein model. Multiple threading method LOMETS is used to identify the structural templates from the protein data bank. Iterative template-based fragment modeling simulations assembled the optimized atomic model of the Nsp1 sequence. Protein function database (BIOLIP) was used to thread the 3D models and determine the targets function insights. The best model was selected on the basis of C- score.
Subsequently, the software SeeSAR v9.2 was utilized to dock the modeled SARS-CoV-2 Nsp1 protein with the selected molecules. The parameter settings were kept default in SeeSAR software. The scoring feature (equation (1)) of hydrogen dehydration (HYDE) was used in SeeSAR to test the estimated affinity of docking [31]. The estimated affinity in SeeSAR has been reflected in mM < pM order. The top molecules were picked based on their estimated affinity, Ligand Efficiency (LE), and Torsion participation toward protein-ligand binding.
| (1) |
In equation (1), ΔGdehydration represents the change in dehydration energy, whereas ΔGH-bonds represents the change in hydrogen bond (H-bond) energy for each atom i in the protein–ligand complex.
HYDE's scoring function is determined in terms of desolvation and hydration that have been conventionally linked with Octanol-water Partition Coefficients of molecules. The ligand molecule is completely dehydrated during the binding process. Even the H-bonds linking protein/ligand and water molecules are broken during the process, prompting a disfavored enthalpic augmentation, although the mass release of water atoms [32]. To cover the energy loss, new H-bonds are produced between protein-ligand. Moreover, the protein-ligand hydrophobic interactions in the water molecules region begin to crack the water H-bond formation and present adverse energy. The removal of these water molecules from the hydrophobic surfaces and their discharge into the bulk water causes an expansion in energy heading to a hydrophobic impact phenomenon. To ascertain the estimated affinity of a ligand in agreement with the amount of non-hydrogen atoms was presented by Kuntz et al. (1999) [33]. Extension of their fundamental approach of LE numerically expressed as the quotient of ΔG and the amount of non-hydrogen atoms of the molecule. The interaction analysis was assuredly done by the Protein-Ligand Interaction profiler (PLIP) server [34].
2.3. MD simulations, MM-PBSA, and Gibbs free energy
The best technique for gaining protein-ligand equilibrium is the Molecular Dynamics (MD) Simulation. It is established upon the law of molecular motions by Newton (equation (2)).
| (2) |
Fi = force exerted on particle i, mi = mass of particle i, and ai = acceleration of particle i. V = potential energy of the system. r = position. δ = change in velocity. Fi shows the force working on i given by the partial spatial derivative of V, which depends upon the positions rN (r1, r2, …., rN) of all N particles in the system [35]. We have produced four systems of the Nsp1 complex with molecules BCH10, BCH15, BCH16, and BCH17 for 250 ns MD simulation in Gromacs 4.6.7 [[36], [37], [38]]. The GROMOS 43a1 forcefield was adopted for protein topology creation, while the PRODRG 2.0 server was used to create ligand topology [39]. The systems neutralization was done by adding 98888 water and 5Cl- ions. The cubic box of 24.33 nm3 volume was used in systems solvation. Next, to eliminate the steric clashes during the energy minimization conjugant gradient and the steepest descent approach was adopted. After that, NPT and NVT simulation of 1 ns was performed to fix the systems temperature and pressure. The LINCS algorithm exercised to constrain the bond's length and the PME method to estimate absolute electrostatics [40,41]. The thermostat (V-rescale) method was applied to maintain the 300 K temperature [42]. The MD trajectories were kept for several structural interpretations like RMSD, RMSF, and H-bonds analysis.
The g_mmpbsa tool was utilized to determine binding free energy applying molecular mechanics Poisson-Boltzmann surface area (MM-PBSA) approach. The binding free energy gives the three dynamic terms. First is the standard molecular mechanics energy. Second is the entropic supplement to solvation and free energy in a vacuum [43]. The low-energy state of selected complexes captured via free energy landscape analysis (FEL). The g_sham module was used for FEL calculations.
2.4. Physiochemical properties prediction
Molinspiration, an automatic server, was utilized for the molecular property prediction that divines both pharmacological and physiochemical attributes such as LogP, molecular size, rotatable bonds, and hydrogen bonding features (https://www.molinspiration.com). The molecules were converted into SMILE format using OpenBabel server to analyze the molecular properties [44].
3. Results
3.1. Molecular modeling
Recently, a crystallographic structure of Nsp1 was submitted to the PDB database. However, the structure was missing the residues of the C-terminal domain. Hence, we adopted the molecular modeling approach to built the full length structure of Nsp1. Molecular modeling includes tactics (conceptual/computational) to reproduce a structure comparable to the original three-dimensional (3D) crystal structure. We first employed the SWISS-MODEL to build a full length protein model by providing Nsp1 structure (PDB ID: 7K3N) as a template. However, SWISS-MODEL was not able to generate a full length model of Nsp1 protein. Further, we employed the I-TASSER server that uses a hierarchical path to predict the protein structure. The full length 3D model of SARS-CoV-2 Nsp1 was developed, as displayed in Fig. S1a. The predicted structure was submitted to Github (https://github.com/Rahulbioinfo/Structural-Bionformatics-Lab.git). The predicted model showed a −2.53 C-score with a 0.42 ± 0.14 estimated TM-align score. The structure validation was done on the basis of Ramachandran plot analysis. The Ramachandran plot of the modeled protein confirmed that 92.5% and 96.4% of all residues were in the favored and allowed areas respectively (Fig. S1b). These results recommended that the predicted protein model was of high-quality and appropriate for further in-silico studies.
3.2. Binding mode and intermolecular interactions
The predicted full length model of Nsp1 was subjected to molecular docking. The “Find unoccupied pocket” of SeeSAR was used to define the binding site of Nsp1. A total of five binding pockets were produced using “Find unoccupied pocket” function of SeeSAR. The second binding pocket containing 21 residues of the C-terminal region was considered for docking, while other pockets were found away from the C-terminal area.
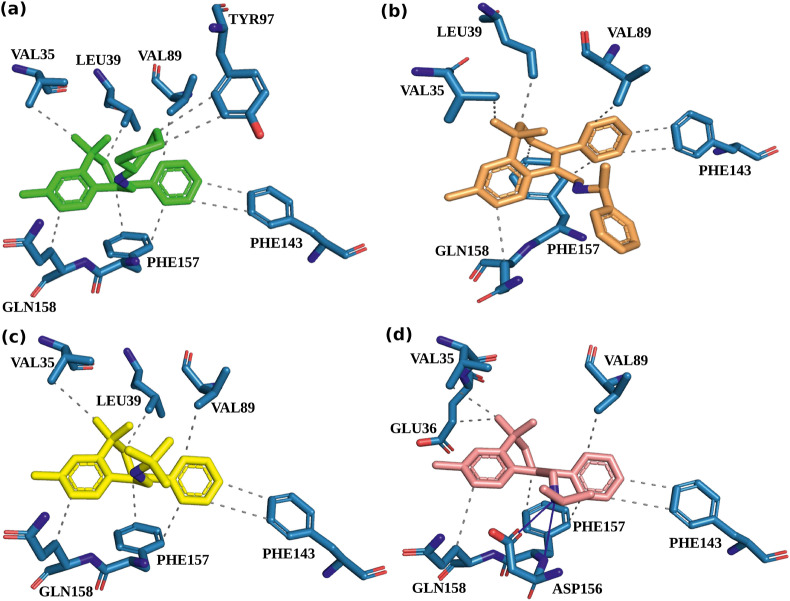
Molecular docking is a method that predicts the stable interaction profiles between a ligand and a protein. The AABs analogues were docked with SARS-CoV-2 Nsp1 to estimate the characteristics of protein-ligand binding, including the binding affinity, LE, and Torsion (Table S1). Based on these characteristics, BCH10 (N-((-6,7-dihydro-3,5,5-trimethyl-8phenyl5Hbenzo [7] annulen-9-yl)methyl)(phenyl)methan amine), BCH15 (1-Phenyl-N-((3,5,5-trimethyl-8-phenyl-6,7-dihydro-5H-benzo [7]annulen-9-yl)methyl)ethan amine), BCH16 (2-Methyl-N-((3,5,5-trimethyl-8-phenyl-6,7-dihydro-5H-benzo [7]annulen-9-yl)methyl)propan-2-amine), and BCH17 (N-((3,5,5-trimethyl-8-phenyl-6,7-dihydro-5H-benzo [7]annulen-9-yl)methyl)propan-2-amine) molecules were selected from seventeen AAB molecules. Molecular docking results were presented in Table 1 . The docking poses were produced from the PLIP server. In complex with Nsp1 molecules, BCH10, BCH15, BCH16, and BCH17 showed hydrophobic interactions with key residues Phe143, Phe157, and Gln158 of C-terminal domain. Other residues involved in hydrophobic interactions were Val35, Glu36, Leu39, Val89, and Tyr97. H-bonds bonds were observed only in Nsp1-BCH17 complex. The residues involved in H-bonds were Asp156 and Phe157 (Fig. 2 ).
Table 1.
Complexes selected based on Estimated Affinity, LE, and Torsion.
| Complexes | Estimated Affinity (Ki) | LE | Torsion |
|---|---|---|---|
| Nsp1-BCH10 | 8.81 mM-88.67 μM | 0.26 |  |
| Nsp1-BCH15 | 0.22mM-2.24 μM | 0.30 |  |
| Nsp1-BCH16 | 0.95mM-9.62 μM | 0.29 |  |
| Nsp1-BCH17 | 0.90mM-9.07 μM | 0.30 |  |
# Below 0.20 LE is low.
Fig. 2.
The 3D interaction poses of the Nsp1 binding site. (a) BCH10, (b) BCH15, (c)BCH16, and (d) BCH17.
3.3. Structural dynamics of protein-ligand complexes
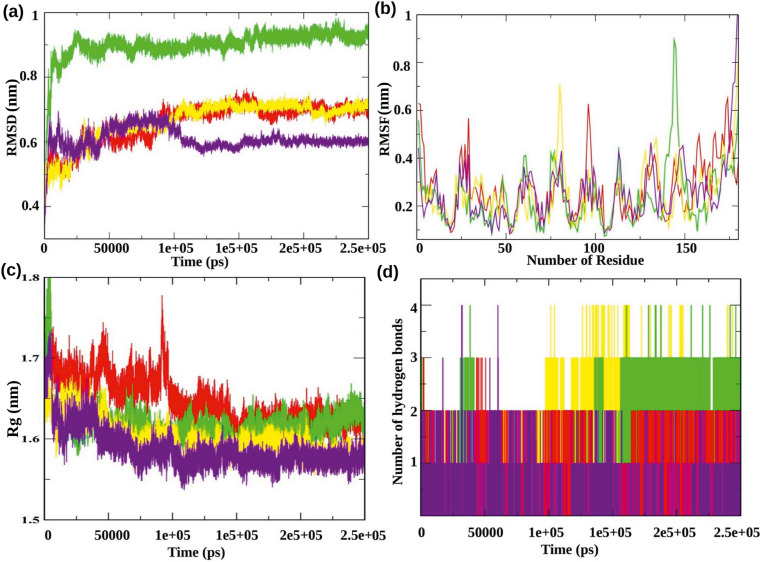
The protein-ligand interactions are static poses and their binding pattern tends to alter with physiological conditions and time. To get an insight into the correct dynamic status and estimation of different bond formations between protein and ligand, the selected protein-ligand complexes were subjected to long term MD simulations. To analyze the structural stability of the four selected protein-ligand complexes, we measured the root mean square deviation (RMSD) of backbone C-α-atoms. RMSD analysis of the Nsp1 in complex with BCH10, BCH15, BCH16, and BCH17 revealed that all the complexes achieved stability around ~100 ns, and afterwards, the RMSD trajectories were stable till the end of simulations. During the entire simulation, selected complexes had an average RMSD value between 0.5 and 0.69 nm, except for BCH15. BCH15 showed an average RMSD value between 0.8 and 0.9 nm (Fig. 3 a). Further, the fluctuations at a local level were analyzed through Root Mean Square Fluctuation (RMSF) calculations. The average fluctuations of all four complexes were low and comparable to each other (Fig. 3b). Moreover, we also measured other structural parameters, including the Radius of Gyration (Rg), Cluster analysis, and calculated the number of Inter-molecular H-bonds based on the fundamental dynamics strategy. The average Rg value was estimated to lie between 1.6 and 1.68 nm for all the four complexes (Fig. 3c). The rate of fluctuations and minute variations in the intermediate Rg and RMSD values steered to conversion of trajectories. Additionally, our MD simulations interpretation recorded an average of two H-bonds for all the selected complexes (Fig. 3d). These results demonstrated that all the complexes were fully equilibrated and structurally stable.
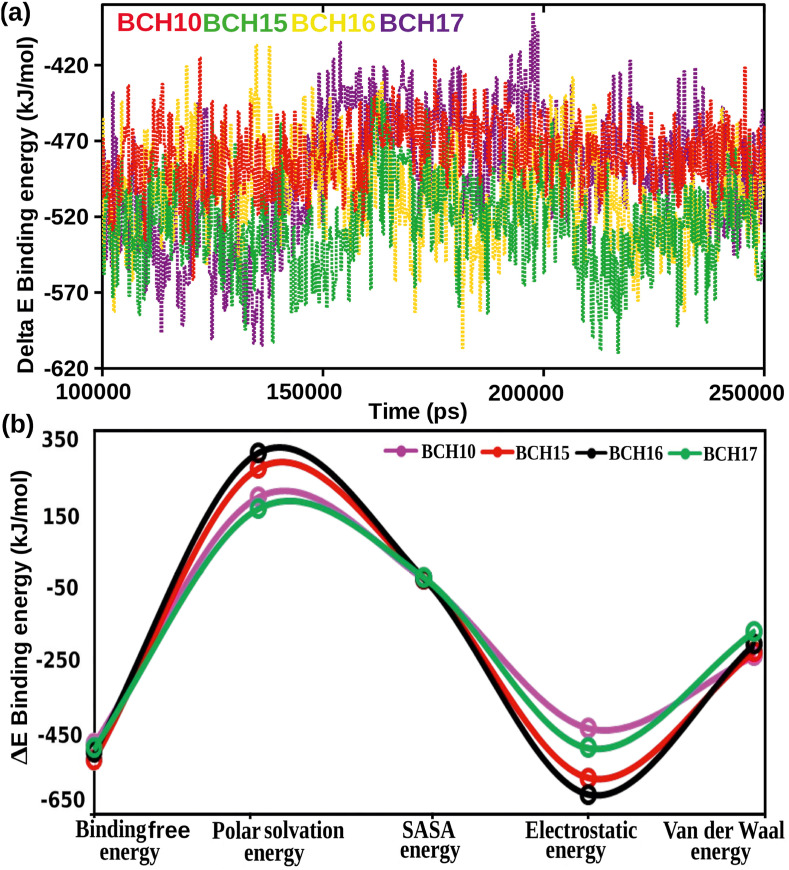
Fig. 3.
(a) The RMSDs of backbone C-α-atoms are shown as a function of time of the Nsp1 complexes. (b) Graphical representation of RMSF for the backbone C-α-atoms of Nsp1 complexes. (c) Change in Rg of the Nsp1 protein complexes. (d) The analysis of Hydrogen bonds formed between protein-ligand during the simulation. The symbol coding scheme is as follows: BCH10 (red color), BCH15 (green color), BCH16 (yellow color), and BCH17 (violet color).
The MD simulations can be used to observe protein-ligand interactions at different time intervals throughout the simulation run. We extracted the scripts of the top four complexes (BCH10, 15, 16, and 17) at different time intervals (50 ns, 100 ns, 150 ns, 200 ns, and 250 ns) to perceive the stability of the interactions between the ligand and protein. The results revealed that the interactions with C-terminal residues (mainly Phe143, Phe157, and Gln158) and other residues were present throughout the simulation run. The residues Phe143, Phe157, and Gln158 showed hydrophobic interactions throughout the simulation in each complexes, while Asp156 and Asn162 involved in both hydrogen and hydrophobic interactions (Fig. S2).
3.4. Thermodynamic binding energy and free energy landscape analysis
Besides, we estimated the end-state binding free energies for Nsp1 protein complexes to assess their binding affinities in a dynamic environment. The binding free energy results were presented in Fig. 4 . MD scripts from 100 to 250 ns time frame of the selected complexes were extracted to measure the binding free energies. The summary of binding free energy results showed that ΔE binding (kJ/mol) was −477.981 for BCH10, -524.819 for BCH15, -504.165 for BCH16, and -490.889 for BCH17. Molecules BCH16 and BCH15 proffered higher binding free energy among four selected molecules. The electrostatic energy contribution was the pre-eminent factor in total binding free energy, the VdW energy was another significant donor. However, the SASA energy participated with a small contribution to binding free energy. In contrast, the polar solvation energy was not contributing favorably to the binding free energy. Moreover, the two-time repetition of MM-PBSA calculations was carried out for the top two Nsp1 complexes at the time interval of 0–100 ns (Table S2, Fig. S3). BCH15 complex showed −537.468 in 1st run and −549.833 in 2nd run. BCH16 complex showed −510.643 in 1st run and −456.137 in 2nd run. The results were consistent with our first run of MM-PBSA analysis where BCH15 exhibited −524.819, while BCH16 showed −504.165, thereby validating our adapted protocol to calculate binding free energies.
Fig. 4.
(a) Graphical representation of binding free energy in kJ/mol, (b) MM-PBSA calculations of all constituents of binding free energy for all the selected Nsp1 complexes.
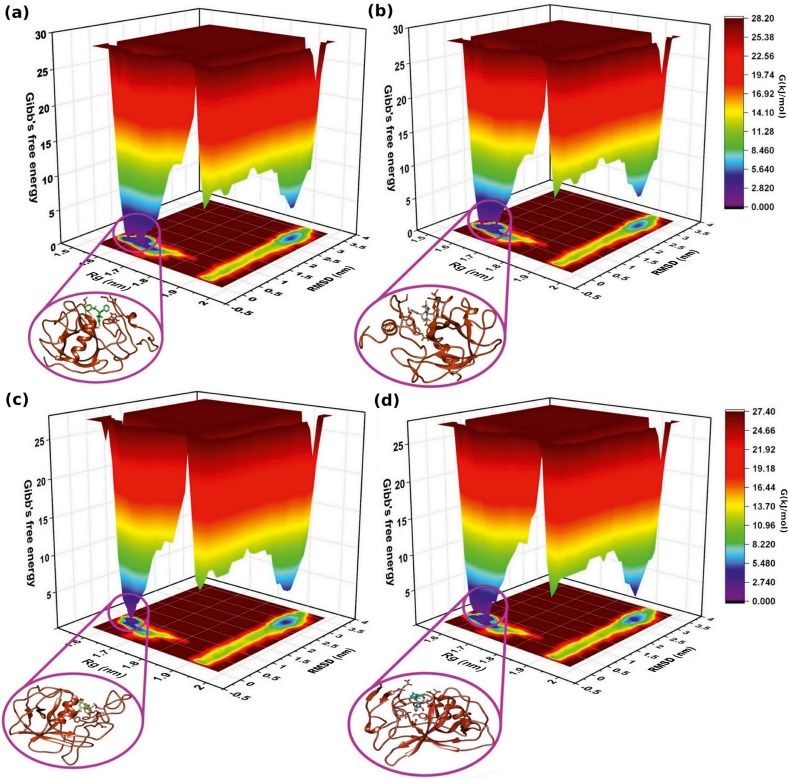
We also conducted the FEL interpretation for all the complexes to examine the minima basins (low-energy) obtained during the simulations. FEL analysis showed that the complexes attained a minimum energy structure during the simulation period with 0.5 ± 0.07 RMSD and 1.6 ± 0.05 nm Rg values, as shown in Fig. 5 . BCH10 showed three different broad and deep valleys, while BCH15 showed one consistent energy minima. BCH16 revealed a cluster and one energy basins adjacent to each other. BCH17 exhibited two energy basins with broad valleys. Every protein-ligand complex had a distinct pattern for the FEL. Additionally, the protein-ligand complexes were extracted from the minima states for geometrical shape analysis of the binding cavity. The initial structure (before docking) showed 421.105 cavity volume while the structures at minima state extracted structures showed 323.846, 370.219, 272.554, 401.161 for BCH10, BCH17, BCH16, and BCH15 respectively. These geometrical shape analysis outcomes acknowledged the decreased cavity space volume in minima structures compared to the initial structure.
Fig. 5.
The 2D and 3D free energy landscapes from MD trajectories for four Nsp1 complexes (a) BCH10, (b) BCH15, (c) BCH16, and (d) BCH17. The violet color region in circle shows the minimum energy conformation.
3.5. Physiochemical properties
Measuring the molecular properties is meaningful in discovering excellent drug candidates, and it is an essential hallmark in drug design and development. Usually, molecules contain functional groups that have specific features which are comparable to known drugs. In this context, the pharmaceutical and physiochemical attributes such as water/octanol value (LogP), H-bond acceptors/donor, molecular weight, and the number of rotatable bonds for seventeen molecules were examined. The physiochemical property analysis of selected molecules showed ample number of rotatable bonds and bioactivity scores of 0.27, 0.15, 0.19, and 0.24 for BCH10, BCH15, BCH16, and BCH17 respectively (Table S3).
4. Discussion
The Nsp1 of SARS-CoV-2 has emerged as an attractive target for drug development due to its involvement in viral factors that determine the virulence and pathogenesis of CoVs [5,6]. Nsp1 is primarily involved in the down-regulation of host innate immune responses to CoV infections [5,6]. A library of in-house synthesized AABs analogues was screened for its potential to interact with the binding site of Nsp1 protein. We followed a robust computational approach to select and rank these analogues based on their capacity to effectively bind with Nsp1 of SARS-CoV-2.
A fully resolved model of Nsp1 is not yet available, so we predicted a full length model of Nsp1 by I-TASSER server that showed 92.5% and 96.4% of all residues in the favored and allowed regions, respectively. The models with more than 88% of amino acids in the allowed region of Ramachandran plot are considered for computational studies. Additionally, we also validated our predicted model by superimposing it with the newly deposited Nsp1 structure of SARS-CoV-2 and template structure (Nsp1 of SARS-Co-V) as depicted in Fig. S4. The superimposition outcome showed 0.3 Å RMSD between all the compared structures. The low RMSD values reaffirmed that the predicted structure was robust and not producing any discrepancy.
Already published studies in well reputed journals proclaimed that the C-terminal region of SARS-CoV-2 Nsp1 containing 143–180 residues perform an essential role in the inhibition of host protein synthesis [5,6,45]. Hence, we adopted a similar binding site (C-terminal region) for docking predicted by the “Find unoccupied pocket” of SeeSAR. The “Find unoccupied pocket” is made on a heuristic prototype that utilizes the Gaussian variations on a 3-D grid to search the probability of administering with a pocket pattern. Furthermore, other notable parameters, such as the global H-bond features, lipophilic nature, and the hypothetical pockets solvent accessible surface area, were also acknowledged. Moreover, the prediction was additionally improved with local actions such as ranges between sets of functional group atoms. Thus, the binding site similarity between existing literature and our predicted site strengthened our proposed approach.
From molecular docking, best four docked complexes were selected based on LE, Estimated affinity, and torsion (Table 1). LE is described as evaluation of per atom participation in binding affinity of a protein-ligand complex [46,47] Interaction analysis of top four selected molecules (BCH10, 15, 16 and 17) exhibited interactions with C-terminal residues of Nsp1 of SARS-CoV-2. Most of the interactions observed were hydrophobic interactions. These interactions are essential in protein folding, stability and biological activity. Our docking results suggested that the selected molecules could be developed to target Nsp1 of SARS-CoV-2. A molecular docking study reported five phytochemical molecules (Austrocortilutein A, Austrocortirubin, Ganomycin A, Glabridin, and Neogrifolin) against Nsp1 of SARS-CoV-2 [48]. Apart from it, another study reported molecules Glycyrrhizic acid, Garcinolic acid, Lobaric acid, and Tirilazad as potential Nsp1 inhibitors. These molecules were also of natural origin screened by preliminary computational studies [49].
The selected protein-ligand complexes from docking results were subjected to MD simulations and binding free energy calculations. RMSD computations can probe the conformational changes and the stability of a protein at a global level. RMSD furnishes a quantitative degree of similarity of concerned systems by defining the deviations in backbone C-ɑ-atoms of the proteins [50]. Our results showed that the RMSD trajectories of all the selected complexes were stable and well-equilibrated. These results reported that all the simulated complexes were equivalent to experimental structures and were fit for further computational explorations. The protein-ligand complexes containing potential molecules suggested by a recent computational study [51] showed higher (>2 Å) RMSD values as compared to low value (<1 Å) shown by our complexes.
During the simulation, all the atoms were allowed to communicate with their surroundings, which allowed to measure their trajectories in the protein-ligand complexes. The RMSF values of every backbone atom in the Nsp1-BCH complexes were examined to analyze the flexibility of the protein backbone structure. The high RMSF value conferred flexibility, whereas the lower RMSF value indicated confined drifts. All the complexes achieved a low level of fluctuation in the C-terminal region (binding site). RMSF results for the binding site region of all the complexes revealed that the amino acids involved in binding of the molecules were stable throughout the simulation. In previously published article the protein-ligand complexes bestowed higher fluctuations (1.0–~4.0 Å), while our protein-ligand complexes showed low fluctuations between 0 and ~0.9 Å [51]. We also calculated the Rg values, which give insights into the protein overall shape and dimension [50]. The low average Rg values intimated that the protein was stable upon binding of the molecules.
Moreover, it was imperative to explore the protein-ligand interactions at different intervals during the simulations. The protein-ligand interaction poses at different time intervals revealed that the interactions generated in docked poses were maintained throughout the simulation run. The MD simulations signified that the C-terminal residues showed stable interactions and the selected molecules remained in the binding pocket till the end of the simulation.
The intermolecular H-bond interactions played a vital role in the stabilization of the protein-ligand complexes. The H-bond interactions stability between protein-ligand complexes were calculated throughout the 250 ns simulation period. The time-averaged results suggested that all the selected molecules also showed H-bonds with Nsp1. The residues Asp156 and Asn162 showed H-bonds at different time intervals during the simulation, hence, we explored their H-bond profiles. The results of H-bond analysis (Fig. S5) for residues Asp156 and Asn162 supported our time interval analysis.
Based on the MD simulations, binding free energies were computed using the MM-PBSA approach to estimate the strength of interaction between selected ligands and Nsp1. The more precise binding affinity prediction can be acquired through binding free energy computations, which is reliant on the thermodynamically significant parameter that are involved in the protein-ligand interactions [52,53]. MM-PBSA results pointed out that electrostatic and VdW interactions were the favorable contributors in the binding free energy. To validate these results, MD scripts of the top two Nsp1 complexes (BCH15 and BCH16) at the time interval of 0–100 ns were extracted to monitor the robustness of our approach to estimate binding free energies. The binding free energies obtained from the replication studies demonstrated no artifact in the values of binding free energy as depicted in Table S2 and Fig. S3.
Further, to acquire the Gibb's free energy, 2D and 3D FEL plots were generated, as shown in Fig. 5. The energy range for BCH10 and BCH17 was between 0 and 28.20 kJ/mol, while for the BCH15 and BCH16 the value ranges from 0 to 27.40 kJ/mol. Over higher free energy space with deep basins, the violet color localities symbolized the local energy minima and actively favored conformations (Fig. 5). The FEL approach revealed that all the complexes were highly stable. The protein-ligand poses were extracted from their respective minima states and subjected to geometrical shape analysis of the binding cavity. The decrease in cavity space showed stable and deep attachment of molecules in the cavity. The geometrical shape analysis results further complemented our binding free energy analysis.
The MD driven results suggested that the selected molecules could be developed to target the binding site of Nsp1 protein of SARS-CoV-2. Hence, we calculated the pharmaceutical and physiochemical properties of these molecules. These attributes were estimated on the basis of Lipinski's Rule of Five that predicts the drug-likeness and recognizes the pharmacological action that would support the making of an excellent orally effective drug [54,55]. All the selected molecules passed the criteria of rule of five. Moreover, we also accessed the bioavailability of selected molecules through molinspiration server. The molinspiration server compares the molecules with an average number of 100000 drug-like molecules, and the bioactivity score permits adequate separation of inactive and active molecules. The physiochemical results showed that all the selected molecules were biologically active. To conclude, the selected top four molecules demonstrated feasible drug-like properties and considerable bioavailability to be developed as therapeutic agents.
5. Conclusion
The Nsp1 of SARS-CoV-2 is an attractive target for development of inhibitors. The Nsp1 has a functional pocket that is linked with substrate binding and enzymatic action. We screened 17 AAB molecules for their potency to inhibit Nsp1 of SARS-CoV-2 and selected top four molecules based on LE and Estimated Affinity. Employing robust long-term simulations, we suggested four molecules BCH10, BCH15, BCH16, and BCH17 to possess the potential to inhibit Nsp1 of SARS-CoV-2. All the selected molecules passed the Lipinski's Rule of Five. Moreover, the drug-likeness properties revealed a positive bioactivity score for the selected molecules. The top-ranked molecules warrant further experimental and clinical testing against Nsp1 of SARS-CoV-2.
Consent for publication
All the authors have read and approved the manuscript in all respects for publication.
Declaration of competing interest
There are no competing interests declared by the authors.
Acknowledgment
We gratefully acknowledge to the Director, CSIR-Institute of Himalayan Bioresource Technology, Palampur for providing the facilities to carry out this work. This manuscript represents CSIR-IHBT communication no. 4832.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.compbiomed.2021.104555.
Appendix A. Supplementary data
The following is the supplementary data to this article:
References
- 1.Shang J., Ye G., Shi K., et al. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Walls A.C., Park Y.J., Tortorici M.A., et al. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292. doi: 10.1016/j.cell.2020.02.058. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yuan M., Wu N.C., Zhu X., et al. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science. 2020;368:630–633. doi: 10.1126/science.abb7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Masters P.S. The molecular biology of coronaviruses. Adv. Virus Res. 2006;65:193–292. doi: 10.1016/S0065-3527(06)66005-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schubert K., Karousis E.D., Jomaa A., et al. SARS-CoV-2 Nsp1 binds the ribosomal mRNA channel to inhibit translation. Nat. Struct. Mol. Biol. 2020;27:959–966. doi: 10.1038/s41594-020-0511-8. [DOI] [PubMed] [Google Scholar]
- 6.Thoms M., Buschauer R., Ameismeier M., et al. Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2. Science. 2020;369:1249–1256. doi: 10.1126/SCIENCE.ABC8665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Min Y.-Q., Mo Q., Wang J., et al. SARS-CoV-2 nsp1: bioinformatics, potential structural and functional features, and implications for drug/vaccine designs. Front. Microbiol. 2020;11:29. doi: 10.3389/fmicb.2020.587317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu C., Liu Y., Yang Y., et al. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm. Sin. B. 2020;10:766–788. doi: 10.1016/j.apsb.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Semper C., Watanabe N., Savchenko A. Structural characterization of nonstructural protein 1 from SARS-CoV-2. iScience. 2021;24:101903. doi: 10.1016/j.isci.2020.101903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clark L.K., Green T.J., Petit C.M. Structure of nonstructural protein 1 from SARS-CoV-2. J. Virol. 2020;95 doi: 10.1128/jvi.02019-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lokugamage K.G., Narayanan K., Huang C., Makino S. Severe acute respiratory syndrome coronavirus protein nsp1 is a novel eukaryotic translation inhibitor that represses multiple steps of translation initiation. J. Virol. 2012;86:13598–13608. doi: 10.1128/jvi.01958-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang K., Miorin L., Makio T., et al. Nsp1 protein of SARS-CoV-2 disrupts the mRNA export machinery to inhibit host gene expression. Sci. Adv. 2021;7 doi: 10.1126/sciadv.abe7386. eabe7386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Benedetti F., Snyder G.A., Giovanetti M., et al. Emerging of a SARS-CoV-2 viral strain with a deletion in nsp1. J. Transl. Med. 2020;18:329. doi: 10.1186/s12967-020-02507-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Narayanan K., Huang C., Lokugamage K., et al. Severe acute respiratory syndrome coronavirus nsp1 suppresses host gene expression, including that of type I interferon, in infected cells. J. Virol. 2008;82:4471–4479. doi: 10.1128/jvi.02472-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li G., De Clercq E. Therapeutic options for the 2019 novel coronavirus (2019-nCoV) Nat. Rev. Drug Discov. 2020;19:149–150. doi: 10.1038/d41573-020-00016-0. [DOI] [PubMed] [Google Scholar]
- 16.Bhardwaj V.K., Singh R., Sharma J., et al. Identification of bioactive molecules from tea plant as SARS-CoV-2 main protease inhibitors. J. Biomol. Struct. Dyn. 2020:1–10. doi: 10.1080/07391102.2020.1766572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sharma J., Kumar Bhardwaj V., Singh R., et al. An in-silico evaluation of different bioactive molecules of Tea for their inhibition potency against non structural protein-15 of SARS-CoV-2. Food Chem. 2020;128933 doi: 10.1016/j.foodchem.2020.128933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bhardwaj V.K., Singh R., Das P., Purohit R. Evaluation of acridinedione analogs as potential SARS-CoV-2 main protease inhibitors and their comparison with repurposed anti-viral drugs. Comput. Biol. Med. 2021;128:104117. doi: 10.1016/j.compbiomed.2020.104117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ghosh R., Chakraborty A., Biswas A., Chowdhuri S. Depicting the inhibitory potential of polyphenols from Isatis indigotica root against the main protease of SARS CoV-2 using computational approaches. J. Biomol. Struct. Dyn. 2020:1–12. doi: 10.1080/07391102.2020.1858164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ghosh R., Chakraborty A., Biswas A., Chowdhuri S. Identification of alkaloids from Justicia adhatoda as potent SARS CoV-2 main protease inhibitors: an in silico perspective. J. Mol. Struct. 2020;1229:129489. doi: 10.1016/j.molstruc.2020.129489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parashar A., Shukla A., Sharma A., et al. Reckoning γ-Glutamyl-S-allylcysteine as a potential Main protease (M pro ) inhibitor of novel SARS-CoV-2 virus identified using docking and molecular dynamics simulation. Drug Dev. Ind. Pharm. 2021:1–32. doi: 10.1080/03639045.2021.1934857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bhardwaj V.K., Singh R., Sharma J., et al. Bioactive molecules of Tea as potential inhibitors for RNA-dependent RNA polymerase of SARS-CoV-2. Front. Med. 2021;8:645. doi: 10.3389/FMED.2021.684020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Singh R., Bhardwaj V.K., Sharma J., et al. In-silico evaluation of bioactive compounds from tea as potential SARS-CoV-2 nonstructural protein 16 inhibitors. J. Tradit. Compl. Med. 2021 doi: 10.1016/j.jtcme.2021.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loizzo M.R., Saab A., Tundis R., et al. Phytochemical analysis and in vitro evaluation of the biological activity against herpes simplex virus type 1 (HSV-1) of Cedrus libani A. Rich. Phytomed. 2008;15:79–83. doi: 10.1016/j.phymed.2007.03.013. [DOI] [PubMed] [Google Scholar]
- 25.Saab A.M., Gambari R., Sacchetti G., et al. Phytochemical and pharmacological properties of essential oils from Cedrus species. Nat. Prod. Res. 2018;32:1415–1427. doi: 10.1080/14786419.2017.1346648. [DOI] [PubMed] [Google Scholar]
- 26.Liu A.L., Du G.H. Dietary Phytochemicals and Microbes. Springer; Netherlands: 2012. Antiviral properties of phytochemicals; pp. 93–126. [Google Scholar]
- 27.Bharti R., Bal Reddy C., Kumar S., Das P. Supported palladium nanoparticle-catalysed Suzuki–Miyaura cross-coupling approach for synthesis of aminoarylbenzosuberene analogues from natural precursor. Appl. Organomet. Chem. 2017;31 doi: 10.1002/aoc.3749. [DOI] [Google Scholar]
- 28.Studio D. Dassault systemes BIOVIA, discovery studio modelling environment. Release 4.5. Accelrys Softw. Inc. 2015:98–104. [Google Scholar]
- 29.Zheng J., Frisch M.J. Efficient geometry minimization and transition structure optimization using interpolated potential energy surfaces and iteratively updated hessians. J. Chem. Theor. Comput. 2017;13:6424–6432. doi: 10.1021/acs.jctc.7b00719. [DOI] [PubMed] [Google Scholar]
- 30.Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinf. 2008;9 doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schneider N., Lange G., Hindle S., et al. A consistent description of HYdrogen bond and DEhydration energies in protein-ligand complexes: methods behind the HYDE scoring function. J. Comput. Aided Mol. Des. 2013;27:15–29. doi: 10.1007/s10822-012-9626-2. [DOI] [PubMed] [Google Scholar]
- 32.Reulecke I., Lange G., Albrecht J., et al. Towards an integrated description of hydrogen bonding and dehydration: decreasing false positives in virtual screening with the HYDE scoring function. ChemMedChem. 2008;3:885–897. doi: 10.1002/cmdc.200700319. [DOI] [PubMed] [Google Scholar]
- 33.Kuntz I.D., Chen K., Sharp K.A., Kollman P.A. The maximal affinity of ligands. Proc. Natl. Acad. Sci. U. S. A. 1999;96:9997–10002. doi: 10.1073/pnas.96.18.9997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Salentin S., Schreiber S., Haupt V.J., et al. PLIP: fully automated protein-ligand interaction profiler. Nucleic Acids Res. 2015;43:W443–W447. doi: 10.1093/nar/gkv315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shukla R., Singh T.R. Identification of small molecules against cyclin dependent kinase-5 using chemoinformatics approach for Alzheimer's disease and other tauopathies. J. Biomol. Struct. Dyn. 2020:1–13. doi: 10.1080/07391102.2020.1844050. [DOI] [PubMed] [Google Scholar]
- 36.Abraham M.J., Murtola T., Schulz R., et al. Gromacs: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1(2):19–25. doi: 10.1016/j.softx.2015.06.001. [DOI] [Google Scholar]
- 37.Hess B., Kutzner C., Van Der Spoel D., Lindahl E. Grgmacs 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theor. Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 38.Van Der Spoel D., Lindahl E., Hess B., et al. GROMACS: fast, flexible, and free. J. Comput. Chem. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 39.Schüttelkopf A.W., Van Aalten D.M.F. PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr. Sect. D Biol. Crystallogr. 2004;60:1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- 40.Hess B., Bekker H., Berendsen H.J.C., Fraaije J. LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 1997;18:1463–1472. doi: 10.1002/(SICI)1096-987X(199709)18:12<1463::AID-JCC4>3.0.CO;2-H. [DOI] [Google Scholar]
- 41.Darden T., York D., Pedersen L. Particle mesh Ewald: an N·log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. doi: 10.1063/1.464397. [DOI] [Google Scholar]
- 42.Berendsen H.J.C., Postma J.P.M., Van Gunsteren W.F., et al. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984;81:3684–3690. doi: 10.1063/1.448118. [DOI] [Google Scholar]
- 43.Kumari R., Kumar R., Lynn A. G-mmpbsa -A GROMACS tool for high-throughput MM-PBSA calculations. J. Chem. Inf. Model. 2014;54:1951–1962. doi: 10.1021/ci500020m. [DOI] [PubMed] [Google Scholar]
- 44.O'Boyle N.M., Banck M., James C.A., et al. Open babel: an open chemical toolbox. J. Cheminf. 2011;3:33. doi: 10.1186/1758-2946-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.de Lima Menezes G., da Silva R.A. Identification of potential drugs against SARS-CoV-2 non-structural protein 1 (nsp1) J. Biomol. Struct. Dyn. 2020:1–11. doi: 10.1080/07391102.2020.1792992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hopkins A.L., Groom C.R., Alex A. Ligand efficiency: a useful metric for lead selection. Drug Discov. Today. 2004;9:430–431. doi: 10.1016/S1359-6446(04)03069-7. [DOI] [PubMed] [Google Scholar]
- 47.Schärfer C., Schulz-Gasch T., Ehrlich H.C., et al. Torsion angle preferences in druglike chemical space: a comprehensive guide. J. Med. Chem. 2013;56:2016–2028. doi: 10.1021/jm3016816. [DOI] [PubMed] [Google Scholar]
- 48.Hossain A. A computational approach for identifying potential phytochemicals against non-structural protein 1 (Nsp1) of SARS-CoV-2. Comb. Chem. High Throughput Screen. 2020;23 doi: 10.2174/1386207323999201103211106. [DOI] [PubMed] [Google Scholar]
- 49.Vankadari N., Jeyasankar N.N., Lopes W.J. Structure of the SARS-CoV-2 nsp1/5′-untranslated region complex and implications for potential therapeutic targets, a vaccine, and virulence. J. Phys. Chem. Lett. 2020;11:9659–9668. doi: 10.1021/acs.jpclett.0c02818. [DOI] [PubMed] [Google Scholar]
- 50.Bhardwaj V.K., Purohit R. Structural changes induced by substitution of amino acid 129 in the coat protein of Cucumber mosaic virus. Genomics. 2020;112:3729–3738. doi: 10.1016/j.ygeno.2020.04.023. [DOI] [PubMed] [Google Scholar]
- 51.Sharma A., Tiwari V., Sowdhamini R. Computational search for potential COVID-19 drugs from FDA-approved drugs and small molecules of natural origin identifies several anti-virals and plant products. J. Biosci. 2020;45:1–18. doi: 10.1007/s12038-020-00069-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bhardwaj V.K., Singh R., Sharma J., et al. Structural based study to identify new potential inhibitors for dual specificity tyrosine-phosphorylation- regulated kinase. Comput. Methods Progr. Biomed. 2020;194:105494. doi: 10.1016/j.cmpb.2020.105494. [DOI] [PubMed] [Google Scholar]
- 53.Singh R., Bhardwaj V.K., Sharma J., et al. Identification of selective cyclin-dependent kinase 2 inhibitor from the library of pyrrolone-fused benzosuberene compounds: an in silico exploration. J. Biomol. Struct. Dyn. 2021 doi: 10.1080/07391102.2021.1900918. [DOI] [PubMed] [Google Scholar]
- 54.Lombardo F., Desai P.V., Arimoto R., et al. In silico absorption, distribution, metabolism, excretion, and pharmacokinetics (ADME-PK): utility and best practices. An industry perspective from the international consortium for innovation through quality in pharmaceutical development. J. Med. Chem. 2017;60:9097–9113. doi: 10.1021/acs.jmedchem.7b00487. [DOI] [PubMed] [Google Scholar]
- 55.Ertl P., Rohde B., Selzer P. Fast calculation of molecular polar surface area as a sum of fragment-based contributions and its application to the prediction of drug transport properties. J. Med. Chem. 2000;43:3714–3717. doi: 10.1021/jm000942e. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.