Abstract
Background
This study aimed to establish machine learning models for preoperative prediction of the pathological types of acute appendicitis.
Methods
Based on histopathology, 136 patients with acute appendicitis were included and divided into three types: acute simple appendicitis (SA, n=8), acute purulent appendicitis (PA, n=104), and acute gangrenous or perforated appendicitis (GPA, n=24). Patients with SA/PA and PA/GPA were divided into training (70%) and testing (30%) sets. Statistically significant features (P<0.05) for pathology prediction were selected by univariate analysis. According to clinical and laboratory data, machine learning logistic regression (LR) models were built. Area under receiver operating characteristic curve (AUC) was used for model assessment.
Results
Nausea and vomiting, abdominal pain time, neutrophils (NE), CD4+ T cell, helper T cell, B lymphocyte, natural killer (NK) cell counts, and CD4+/CD8+ ratio were selected features for the SA/PA group (P<0.05). Nausea and vomiting, abdominal pain time, the highest temperature, CD8+ T cell, procalcitonin (PCT), and C-reactive protein (CRP) were selected features for the PA/GPA group (P<0.05). By using LR models, the blood markers can distinguish SA and PA (training AUC =0.904, testing AUC =0.910). To introduce additional clinical features, the AUC for the testing set increased to 0.926. In the PA/GPA prediction model, AUC with blood biomarkers was 0.834 for the training and 0.821 for the testing set. Combining with clinical features, the AUC for the testing set increased to 0.854.
Conclusions
Peripheral blood biomarkers can predict the pathological type of SA from PA and GPA. Introducing clinical symptoms could further improve the prediction performance.
Keywords: T cell subsets, preoperative predicting, pathological types of appendicitis, clinical features
Introduction
Acute appendicitis is the most common surgical acute abdominal disease, and its lifetime incidence is approximately 7% to 9% (1). A clinical diagnosis of acute appendicitis mainly depends on clinical symptoms, abdominal signs, laboratory data, and clinical imaging. Despite the high incidence of appendicitis, it is still difficult to accurately diagnose the preoperative pathological type of appendicitis (2). Histopathologic findings are the gold standards for the diagnosis of acute appendicitis. The histopathological type of appendicitis affects clinical treatment. Acute appendicitis is divided into acute simple appendicitis (SA), acute purulent appendicitis (PA), acute gangrenous or perforated appendicitis (GPA), and periappendiceal abscess according to histopathology (3). The clinical symptoms, abdominal signs, and peripheral blood biomarkers, including white blood cell (WBC), lymphocytes, and CRP, were used to predict the pathological types of acute appendicitis before the operation. This helped in making a preoperative clinical treatment plan (4-6).
Some studies have shown that a change in the immune system is one of the most important causes of appendicitis (7-9). Lymphocytes, especially T lymphocytes, play a key role in the immune system. Some studies focused on the aggregation of lymphocytes and subtypes in appendicitis (10,11), as well as the relationship between the decrease of peripheral blood lymphocytes and the aggregation of lymphocytes in appendicitis. However, there is no direct relationship between peripheral T cell subsets and pathological types of appendicitis. Reports show that T cell subsets of peripheral blood are rarely used to predict the pathological types of acute appendicitis before the operation alone. Peripheral blood T cell subsets are readily available. Obtaining peripheral blood T cell subsets and preoperative pathological types of acute appendicitis have important reference value for the clinical diagnosis and treatment decisions of appendicitis. This study is expected to reveal the relationship between peripheral blood T cell subsets and pathological types of acute appendicitis and to use a machine learning method to predict the pathological types of tissues based on previous experience in modeling with conventional statistical methods (4). We present the following article in accordance with the STARD reporting checklist (available at http://dx.doi.org/10.21037/atm-20-7883).
Methods
Patients
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by Ethics Committee of Beijing Rehabilitation Hospital of Capital Medical University. All patients signed preoperative informed consent and gave permission to use their data for research. The study initially included 146 patients with acute appendicitis who had operations from June 2016 to November 2018 and who had clinical, pathological, and laboratory data, and registered in the Chinese Clinical Trial Register website (www.chictr.org.cn, ChiCTR1900028241).
The inclusion criteria were as follows: (I) the patient agreed and signed informed consent; (II) histologically confirmed acute appendicitis, including acute SA, acute PA, and acute GPA; (III) the preoperative examination showed no surgical contraindications; (IV) the patient’s age was between 18–80.
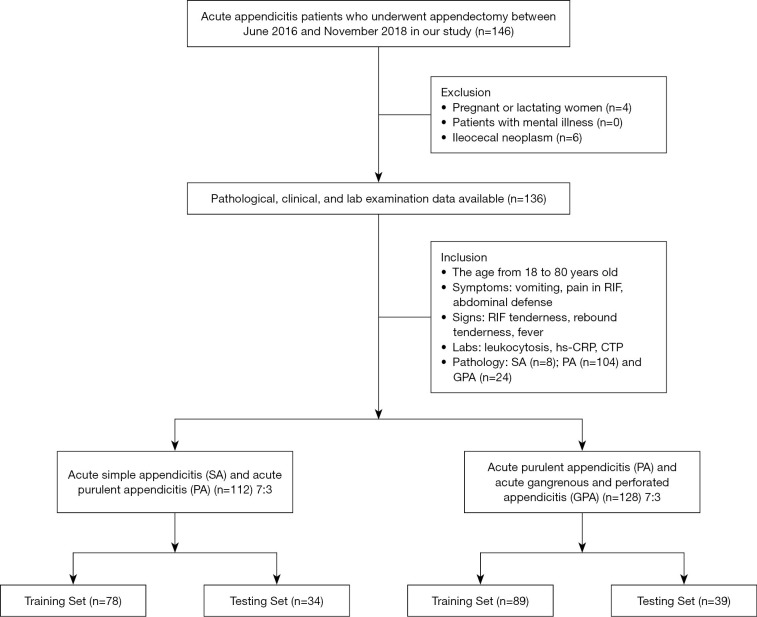
The exclusion criteria were as follows: (I) pregnant or lactating women; (II) patients with mental illness; (III) patients with any forms of cancer; (IV) patients with a history of hematopoietic stem cell, bone marrow, or solid organ transplantation. Based on these criteria, four pregnant or lactating women and six patients with mucinous adenocarcinoma were excluded, and 136 patients were enrolled in this study. Figure 1 illustrates the process of patient enrollment. These patients were randomly divided into training and testing groups at a ratio of 7:3 for further modeling.
Figure 1.
Flow chart of the patient selection and exclusion process.
We collected the basic information and preoperative clinical and laboratory data of all patients retrospectively from the electronic medical records system. This included: (I) age, gender, clinical signs and symptoms score, abdominal pain score, vomiting score, abdominal pain time, abdominal pain type, abdominal tenderness pain range, and the highest temperature. The clinical signs and symptoms of appendicitis were evaluated according to international standards (4); (II) laboratory records: blood routine, coagulation function, blood biochemistry, WBC, NE, CD3+ T, CD4+ T, CD8+ T, CD19+ T, CD16+56, NK, total T cell counts, helper T cell counts, inhibitors T, B cell counts, NK cell counts, CD4+/CD8+ ratio, CRP, PCT, and blood neutrophil to lymphocyte (NLR) ratio.
Biomarkers detection
Serum samples were collected from patients. CRP was measured by AU5400 (OLYMPUS) using an immunoturbidimetric assay (CRP VARIO). PCT levels were tested by Electrochemiluminescent Immunoassay (ELECYS BRAHMS PCT) performed on COBAS e411 analyzer (Roche Diagnostics).
Flow cytometry (FCM) analysis
FCM determined the frequencies of different cells in the patients’ peripheral blood. Immunofluorescent-labeled antibodies were all purchased from eBioscience (San Diego, CA): anti-human CD3 (11-0037-42), CD4 (69-0049-42), CD8 (MHCD0831), CD16 (15-0168-42), CD19 (13-0199-80), and CD56 (62-0566-41). Different fluorescence conjugated antibodies were added directly to the cell suspension for 20 minutes in the dark at 4 °C for cell surface antigens staining. After washing twice, cells were re-suspended in PBS containing 0.1% BSA and 1 mM EDTA. Data were acquired on a BD Fortessa X20 using BD FACSDiva8.0 (BD Bioscience), and data were analyzed using Flowjo software (Tree Star Inc., Asland, OR, USA).
Histopathology
Two pathologists examined all specimens. They reviewed all histopathological results individually and then together. The two readers discussed any discrepancies until they reached a final consensus. The pathological types of 136 cases of acute appendicitis were summarized as follows: acute SA (n=8), acute PA (n=104), acute GPA (GPA, n=24).
Data preprocessing
The study was designed to differentiate pathological types between SA and PA, and PA and GPA. Therefore, two datasets involving SA and PA, or PA and GPA data, were respectively organized, named SA/PA and PA/GPA group. For modeling, each dataset was randomly divided at a 7:3 ratio as the training set and testing set, respectively. All cases in the training set were used to train the predictive model, while the test set cases were used to evaluate the model’s type independently.
Before analyses, variables with zero variance were excluded. Then the missing values and outlier values were replaced by the median. Finally, the data were standardized.
Feature selection
Univariate logistic analysis was performed to select statistically significant clinical or laboratory features that were candidates for pathology differentiation between SA and PA or PA and GPA. P<0.05 indicated statistical significance.
Machine learning modeling
A logistic regression (LR) model was built from the selected feature subsets of the training dataset of the SA/PA group and the PA/GPA group. For each group, the LR model was established based on selected laboratory features. Then, selected clinical features were introduced to establish combined multivariate LR models. The models were validated in the testing sets.
Receiver operating characteristic (ROC) curve was calculated for the training and testing sets of the SA/PA and the PA/GPA groups to evaluate the performance of the machine learning model. The accuracy, sensitivity, specificity, and area under the curve (AUC) were derived to assess the prediction efficiency of the model.
Statistical analysis
The clinical and laboratory features were compared between the SA/PA or the PA/GPA group. A Chi-square test or Fisher’s exact test was used for the nominal variable, Kruskal-Wallis H-test for the ordinal variable, and Mann-Whitney U test for the continuous variable with abnormal distributions. Univariate logistic analysis was used for feature selection. In addition, ROC curve analyses were performed to determine the AUC, accuracy, sensitivity, and specificity for each predictive model. The statistical difference of AUC between any two of the machine learning models was analyzed by Delong’s test.
All statistical analyses for the present study were performed with R 3.5.1 (https:www.r-project.org) and Python 3.5.6. The reported statistical significance levels were all two-sided, and the statistical significance was set at 0.05. The multivariate LR analysis was performed with the “stats” package.
Results
Statistical analysis and feature selection of clinical and laboratory features
The statistical analysis of clinical and laboratory data of the 136 patients was shown in Tables 1,2. In addition, the clinical and laboratory features of SA/PA and PA/GPA group were selected as candidates for pathological subtyping, respectively. There were significant differences in nausea and vomiting, abdominal pain time, NE%, CD4+, T cells, B lymphocyte counts and CD4+/CD8+ ratio between acute SA and acute PA (P<0.05, as shown in Table 1). These are potential markers for SA and PA differentiation. Notably, the mean value of total T cell counts and helper T cell counts of acute SA were about two times higher than acute PA (P<0.05). For the PA/GPA dataset, nausea and vomiting, abdominal pain time, the highest temperature and PCT in peripheral blood were significantly different between acute PA and acute gangrenous and perforated appendicitis (P<0.05, as shown in Table 2). GPA, CD8+ T cells and CRP were much higher.
Table 1. Statistical difference analysis of clinical and laboratory features between acute simple appendicitis and acute purulent appendicitis.
| Variable | Sample | Acute simple appendicitis | Acute purulent appendicitis | Statisticsa | P value |
|---|---|---|---|---|---|
| Age (years, mean ± SD) | 112 | 39.12±20.00 | 42.02±17.31 | −0.451 | 0.653 |
| Gender | |||||
| M | 61 | 5 (62.50%) | 56 (53.85%) | 0.011 | 0.916 |
| F | 51 | 3 (37.50%) | 48 (46.15%) | ||
| Abdominal pain scoreb [1–10] | |||||
| 2 | 6 | 0 (0.00%) | 6 (5.77%) | 2.184 | 0.139 |
| 3 | 8 | 1 (12.50%) | 7 (6.73%) | ||
| 4 | 13 | 1 (12.50%) | 12 (11.54%) | ||
| 5 | 21 | 5 (62.50%) | 16 (15.38%) | ||
| 6 | 1 | 0 (0.00%) | 1 (0.96%) | ||
| 7 | 31 | 0 (0.00%) | 31 (29.81%) | ||
| 8 | 13 | 0 (0.00%) | 13 (12.50%) | ||
| 9 | 17 | 1 (12.50%) | 16 (15.38%) | ||
| 10 | 2 | 0 (0.00%) | 2 (1.92%) | ||
| Nausea and vomiting score [0–2] | |||||
| 0 | 31 | 7 (87.50%) | 24 (23.08%) | 8.833 | 0.003* |
| 1 | 60 | 0 (0.00%) | 60 (57.69%) | ||
| 2 | 21 | 1 (12.50%) | 20 (19.23%) | ||
| Abdominal pain type [1–3] | |||||
| Metastatic right lower abdominal pain | 85 | 4 (50.00%) | 81 (77.88%) | 2.902 | 0.088 |
| Lower right abdominal pain or lower abdominal pain | 25 | 4 (50.00%) | 21 (20.19%) | ||
| Upper abdominal pain | 2 | 0 (0.00%) | 2 (1.92%) | ||
| Abdominal tenderness range [1–10] | |||||
| 1 | 24 | 4 (50.00%) | 20 (19.23%) | 3.081 | 0.079 |
| 2 | 15 | 0 (0.00%) | 15 (14.42%) | ||
| 3 | 48 | 4 (50.00%) | 44 (42.31%) | ||
| 4 | 3 | 0 (0.00%) | 3 (2.88%) | ||
| 5 | 1 | 0 (0.00%) | 1 (0.96%) | ||
| 6 | 1 | 0 (0.00%) | 1 (0.96%) | ||
| 7 | 2 | 0 (0.00%) | 2 (1.92%) | ||
| 8 | 5 | 0 (0.00%) | 5 (4.81%) | ||
| 9 | 13 | 0 (0.00%) | 13 (12.50%) | ||
| Abdominal pain time (hours, mean ± SD) | 112 | 34.75±14.77 | 24.03±15.02 | 1.948 | 0.054 |
| Highest temperature (°C, mean ± SD) | 112 | 37.19±0.63 | 37.46±0.80 | −0.931 | 0.354 |
| WBC counts (×109) (mean ± SD) | 112 | 11.99±3.93 | 13.77±4.23 | −1.153 | 0.252 |
| NE% (mean ± SD) | 112 | 75.61±4.80 | 85.07±7.00 | −3.745 | <0.001* |
| CD3+ (%, mean ± SD) | 112 | 68.91±7.28 | 65.47±8.48 | 1.117 | 0.266 |
| CD4+ (%, mean ± SD) | 112 | 42.12±6.77 | 34.27±7.97 | 2.71 | 0.008* |
| CD8+ (%, mean ± SD) | 112 | 24.62±3.77 | 27.12±8.16 | −1.609 | 0.132 |
| CD19+ (%, mean ± SD) | 112 | 15.89±2.77 | 17.53±7.46 | −0.617 | 0.539 |
| CD16+56- (%, mean ± SD) | 112 | 14.11±6.25 | 15.34±8.50 | −0.52 | 0.615 |
| Total T cell counts (µL, mean ± SD) | 112 | 1417.30±342.76 | 877.45±493.14 | 3.034 | 0.003* |
| Helper T cell counts (µL, mean ± SD) | 112 | 865.75±252.75 | 468.22±279.91 | 3.894 | <0.001* |
| Inhibitor T (µL, mean ± SD) | 112 | 497.38±129.29 | 356.59±219.86 | 1.783 | 0.077 |
| B cell counts (µL, mean ± SD) | 112 | 317.62±42.96 | 224.16±130.27 | 4.709 | <0.001* |
| NK cell counts (µL, mean ± SD) | 112 | 294.38±154.49 | 199.51±149.45 | 1.726 | 0.087 |
| CD4+/CD8+ (mean ± SD) | 112 | 1.76±0.31 | 1.45±0.71 | 2.413 | 0.03* |
| CRP, mg/L (mean ± SD) | 112 | 79.07±49.32 | 52.74±51.87 | 1.388 | 0.168 |
| PCT, ng/L (mean ± SD) | 112 | 0.44±0.95 | 1.43±4.68 | −0.596 | 0.553 |
P value <0.05 indicated statistical significance. * indicated statistical significance. aFisher’s exact test was used for the nominal variable. Wilcoxon test was used for the ordinal variable, whose statistics is W. Student’s t-test was used for the continuous variable with abnormal distribution, whose statistics is t. bVisual analogue scale (VAS) is the most commonly used in pain assessment. The basic method is to use a swimming scale about 10 cm long, with 10 scales on one side. The two ends are “0” and “10” points respectively. 0 points means no pain, and 10 points means the most severe pain that is unbearable. Participants do not need to fill in complicated questionnaires, just look at a “pain ruler”, and then say a number between 0 and 10. In clinical use, the side with scale should be turned back to the patient, and the patient should mark the corresponding position on the ruler which can represent the pain degree of Baiji. The doctor should evaluate the score according to the position marked by the patient, and the clinical evaluation should be “0–2” as “excellent”, “3–5” as “good”, “6–8” as “OK”, and >“8” as “poor”. Before and after clinical treatment using the same method can be more objective to make a score, and the effect of pain treatment can be more objective evaluation. This method is simple, objective and sensitive. SD, standard deviation; WBC, white blood cell; NE, neutrophil; PCT, procalcitonin; hs-CRP, high-sensitivity C-reactive protein.
Table 2. Statistical difference analysis of clinical and laboratory features between acute purulent appendicitis and gangrenous or perforated appendicitis.
| Variable | Sample | Acute purulent appendicitis | Gangrenous or perforated appendicitis | Statisticsa | P value |
|---|---|---|---|---|---|
| Age (years, mean ± SD) | 127 | 42.16±17.34 | 40.54±15.31 | 0.419 | 0.676 |
| Gender | |||||
| M | 68 | 56 (53.40%) | 12 (54.17%) | 0.005 | 0.948 |
| F | 59 | 48 (46.15%) | 11 (45.83%) | ||
| Abdominal pain scoreb [1–10] | |||||
| 2 | 6 | 6 (5.77%) | 0 (0.00%) | 0.142 | 0.707 |
| 3 | 9 | 7 (6.73%) | 2 (8.33%) | ||
| 4 | 15 | 12 (11.54%) | 3 (12.50%) | ||
| 5 | 21 | 15 (14.56%) | 5 (20.83%) | ||
| 6 | 1 | 1 (0.96%) | 0 (0.00%) | ||
| 7 | 37 | 31 (29.81%) | 6 (25.00%) | ||
| 8 | 16 | 13 (12.50%) | 3 (12.50%) | ||
| 9 | 19 | 16 (15.38%) | 3 (12.50%) | ||
| 10 | 4 | 2 (1.94%) | 2 (8.33%) | ||
| Nausea and vomiting score [0–2] | |||||
| 0 | 31 | 24 (23.08%) | 7 (29.17%) | 1.671 | 0.196 |
| 1 | 65 | 59 (57.28%) | 6 (25.00%) | ||
| 2 | 31 | 20 (19.23%) | 11 (45.83%) | ||
| Abdominal pain type [1–3] | |||||
| 1 | 101 | 80 (77.67%) | 20 (83.33%) | 0.385 | 0.535 |
| 2 | 25 | 21 (20.19%) | 4 (16.67%) | ||
| 3 | 2 | 2 (1.94%) | 0 (0.00%) | ||
| Abdominal tenderness range [1–10] | |||||
| 1 | 25 | 20 (19.23%) | 5 (20.83%) | 1.738 | 0.187 |
| 2 | 18 | 15 (14.56%) | 3 (12.50%) | ||
| 3 | 49 | 43 (41.75%) | 5 (20.83%) | ||
| 4 | 5 | 3 (2.91%) | 2 (8.33%) | ||
| 5 | 1 | 1 (0.97%) | 0 (0.00%) | ||
| 6 | 1 | 1 (0.97%) | 0 (0.00%) | ||
| 7 | 2 | 2 (1.94%) | 0 (0.00%) | ||
| 8 | 5 | 5 (4.85%) | 0 (0.00%) | ||
| 9 | 22 | 13 (12.62%) | 9 (37.50%) | ||
| Abdominal pain time (hours, mean ± SD) | 127 | 23.80±14.90 | 36.00±14.26 | −3.641 | <0.001* |
| Highest temperature (°C, mean ± SD) | 127 | 37.46±0.80 | 38.04±0.94 | −3.094 | 0.002* |
| WBC counts (×109) (mean ± SD) | 127 | 13.73±4.23 | 15.26±3.33 | −1.656 | 0.1 |
| NE% (mean ± SD) | 127 | 85.03±7.02 | 87.09±4.63 | −1.76 | 0.084 |
| CD3+ (%, mean ± SD) | 127 | 65.47±8.52 | 64.37±12.17 | 0.525 | 0.6 |
| CD4+ (%, mean ± SD) | 127 | 34.21±7.98 | 31.70±7.23 | 1.409 | 0.161 |
| CD8+ (%, mean ± SD) | 127 | 27.16±8.20 | 34.71±10.67 | −3.83 | <0.001* |
| CD19+ (%, mean ± SD) | 127 | 17.53±7.50 | 17.98±7.37 | -0.263 | 0.793 |
| CD16+56- (%, mean ± SD) | 127 | 15.32±8.54 | 13.72±9.36 | 0.813 | 0.418 |
| Total T cell counts (µL, mean ± SD) | 127 | 880.42±494.62 | 835.49±418.38 | 0.412 | 0.681 |
| Helper T cell counts (µL, mean ± SD) | 127 | 469.24±281.09 | 396.50±213.86 | 1.189 | 0.237 |
| Inhibitor T (µL, mean ± SD) | 127 | 358.03±220.44 | 396.88±252.29 | −0.756 | 0.451 |
| B cell counts (µL, mean ± SD) | 127 | 224.89±130.69 | 203.17±90.04 | 0.772 | 0.442 |
| NK cell counts (L, mean ± SD) | 127 | 199.96±150.11 | 166.75±97.37 | 1.033 | 0.304 |
| CD4+/CD8+ (mean ± SD) | 127 | 1.45±0.71 | 1.22±0.65 | 1.423 | 0.157 |
| CRP, mg/L (mean ± SD) | 127 | 52.10±51.72 | 103.85±72.48 | −4.068 | <0.001* |
| PCT, ng/L (mean ± SD) | 127 | 1.44±4.70 | 4.46±9.21 | −1.559 | 0.131 |
P value <0.05 indicated statistical significance. * indicated statistical significance. aFisher’s exact test was used for the nominal variable. Wilcoxon test was used for the ordinal variable, whose statistics is W. Student’s t test was used for the continuous variable with abnormal distribution, whose statistics is t. bIt is same to Table 1. SD, standard deviation; WBC, white blood cell; NE, neutrophil; PCT, procalcitonin; hs-CRP, high-sensitivity C-reactive protein.
M-variate LR predicting model
The prediction model based on selected clinical and laboratory features and their combinations were developed. ROC analysis for the training and testing set were shown in Figure 2 and Figure 3. In addition, the peripheral blood biomarkers including NE%, CD4+ T cells, T cell counts, helper T cell counts, B lymphocyte counts and CD4+/CD8+ ratio can accurately distinguish SA and PA (training AUC =0.904, testing AUC =0.910). Introducing further selected clinical features, including nausea, vomiting, and pain time, the model showed enhanced prediction efficiency (training AUC =0.921, testing AUC =0.926). In the PA/GPA prediction model, AUC predicted by peripheral blood biomarkers, including CD8+ T cells, CRP, and PCT, was 0.834 for the training set and 0.821 for the testing set. When combined with nausea and vomiting, pain time, and the highest temperature, the AUC for training and testing sets increased to 0.867 and 0.854, respectively (Table 3).
Figure 2.
Performance of pathological type prediction model in acute appendicitis and purulent appendicitis. (A) ROC curves of acute appendicitis and purulent appendicitis prediction in the training and testing sets based on T cell subsets alone. (B) ROC curves of acute appendicitis and suppurative appendicitis prediction in the training and testing sets based on T cell subsets combined with clinical signs and symptoms.
Figure 3.
Performance of pathological type prediction model in acute purulent appendicitis and acute gangrenous or perforated appendicitis. (A) ROC curves of acute purulent appendicitis and acute gangrenous or perforated appendicitis prediction in the training and testing sets based on T cell subsets alone. (B) ROC curves of acute purulent appendicitis and acute gangrenous or perforated appendicitis prediction in the training and testing sets based on T cell subsets combined with clinical signs and symptoms.
Table 3. Comparison of the predictive performance of different machine learning method.
| Variable | T cell | Training/testing set | ACC | AUC | Sensitivity | Specification |
|---|---|---|---|---|---|---|
| Acute simple appendicitis vs. acute purulent appendicitis | T cell subsets | Training set | 0.875 | 0.904 | 0.750 | 1.0 |
| Testing set | 0.875 | 0.910 | 0.750 | 1.0 | ||
| T cell subsets with clinical | Training set | 0.910 | 0.921 | 0.819 | 1.0 | |
| Testing set | 0.906 | 0.926 | 0.812 | 1.0 | ||
| Acute purulent appendicitis vs. acute gangrenous or perforated appendicitis | T cell subsets | Training set | 0.826 | 0.834 | 0.819 | 0.833 |
| Testing set | 0.806 | 0.821 | 0.903 | 0.710 | ||
| T cell subsets with clinical | Training set | 0.806 | 0.867 | 0.736 | 0.875 | |
| Testing set | 0.774 | 0.854 | 0.903 | 0.645 |
ACC, accuracy; AUC, area under the curve.
Discussion
It is important to diagnose the pathological type accurately before surgery, not only to differentiate simple from perforated appendicitis but also to prevent negative appendectomy. Predicting the pathological type of acute appendicitis before the operation helps the treatment plan and prognosis of patients, including the surgical approach and the choice and use of antibiotics. Currently, there are no acknowledged serum or urine biomarkers for diagnosis of the pathological type of acute appendicitis. Circulating biomarkers in peripheral blood may carry informative changes that would reveal the pathological type of acute appendicitis.
In current practice, WBC and CRP are the most widely used peripheral blood biomarkers for suspected appendicitis. CRP is a non-specific acute phase reactant and mainly stimulates cell-mediated immunity and chemotaxis in inflammation. The increase of CRP levels in acute appendicitis is in proportion to the severity of infection, which was found to have higher sensitivity and diagnostic accuracy for acute appendicitis (12). To date, appendicitis protein biomarkers in the blood, such as bilirubin, CRP, and PCT, have been reported (13-15). Lymphocytes and confirmatory response factors are also involved in the pathogenesis and development of appendicitis. Several studies showed that the T lymphocyte subgroups are involved in the development of acute appendicitis, which is consistent with our findings (16,17). These results indicated that peripheral blood biomarkers have potential in acute appendicitis diagnosis or pathology subtyping. In the current study, we identified that the machine learning method could be used to predict the histopathological types of acute appendicitis by biomarkers in peripheral blood. The prediction model built on these peripheral blood biomarkers was able to differentiate SA and PA, or PA and GPA.
In the pathological process of acute appendicitis, T cell subsets in peripheral blood decreased. CD4+ T cells play a central role in the function of the immune system. CD4+ T cells not only help B lymphocytes to produce antibodies but also orchestrate CD8+ T cells and macrophages against a wide variety of pathogenic microorganisms (18). Immunohistochemical staining showed that CD8+ T lymphocytes are present in the appendix of all patients undergoing appendicectomy, whereas B lymphocytes, natural killer (NK) cells, and CD4+ T lymphocytes are found in a majority of perforated appendicitis specimens, in comparison to approximately 50% of non-perforated appendicitis samples (19). B lymphocytes, CD8+ T cells, and T helper cells participate in the inflammatory process of acute appendicitis (20,21). These results showed that T cell subsets might play an important role in the prediction of pathological types. Our results also showed that CD4+ total T cells, helper T cells, and B lymphocytes are significantly lower in acute PA than in acute SA. There is no significant difference for CRP between these pathological types. Compared to acute PA, the CRP and CD8+ T cells of acute GPA increased significantly.
These results indicated that peripheral blood biomarkers could have great potential to predict pathological types of acute appendicitis. We certificated that the machine learning model based on peripheral blood biomarkers can predict the pathological type of acute appendicitis before surgery, with prediction efficiency (AUC >0.80) in the training set and the AUC of the validation group exceeding 0.75. The prediction efficiency could be enhanced when further introducing clinical symptoms. Such a method is relatively simple but effective.
There were some limitations in our study as follows: (I) the limited number of clinical cases, which need to increase to improve the prediction efficiency; (II) data sources from a single center. Extending the generalization of the prediction model to multi-center research would improve the prediction model greatly; (III) due to the sample size and the possibility of false-positive results, this study can not fully prove that peripheral blood biomarkers and clinical features can predict the pathological type of appendicitis. In the future research, we hope to further expand the sample size and conduct multi center RCT study to further prove the scientific nature of the model.
Conclusions
Our current study established an easy-to-practice model which is convenient and can diagnose pathological types of acute appendicitis quickly using biomarkers in patients’ peripheral blood. This study provides a feasible method for predicting the acute appendicitis pathological before operation.
Supplementary
The article’s supplementary files as
Acknowledgments
We are particularly grateful to all the people who have given us help on our article.
Funding: This study was funded by the Scientific research project of Space Center Hospital (Approval Number: YN201429). The funding body had no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by Ethics Committee of Beijing Rehabilitation Hospital of Capital Medical University, and registered in the Chinese Clinical Trial Register website (www.chictr.org.cn, ChiCTR1900028241). All patients signed preoperative informed consent and gave permission to use their data for research.
Footnotes
Reporting Checklist: The authors have completed named STARD reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-7883
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-7883
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-7883). The authors have no conflicts of interest to declare.
References
- 1.Anderson JE, Bickler SW, Chang DC, et al. Examining a common disease with unknown etiology: trends in epidemiology and surgical management of appendicitis in California, 1995-2009. World J Surg 2012;36:2787-94. 10.1007/s00268-012-1749-z [DOI] [PubMed] [Google Scholar]
- 2.Resende F, Almeida AB, Maia JC, et al. Challenges in uncomplicated acute appendicitis. J Acute Dis 2016;5:109-13. 10.1016/j.joad.2015.11.002 [DOI] [Google Scholar]
- 3.Carr NJ. The pathology of acute appendicitis. Ann Diagn Pathol 2000;4:46-58. 10.1016/S1092-9134(00)90011-X [DOI] [PubMed] [Google Scholar]
- 4.Kang CB, Li WQ, Zheng JW, et al. Preoperative assessment of complicated appendicitis through stress reaction and clinical manifestations. Medicine (Baltimore) 2019;98:e15768. 10.1097/MD.0000000000015768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Karami MY, Niakan H, Zadebagheri N, et al. Which One is Better? Comparison of the Acute Inflammatory Response, Raja Isteri Pengiran Anak Saleha Appendicitis and Alvarado Scoring Systems. Ann Coloproctol 2017;33:227-31. 10.3393/ac.2017.33.6.227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ünal Y. A new and early marker in the diagnosis of acute complicated appendicitis: immature granulocytes. Ulus Travma Acil Cerrahi Derg 2018;24:434-39. [DOI] [PubMed] [Google Scholar]
- 7.Pedros C, Duguet F, Saoudi A, et al. Disrupted regulatory T cell homeostasis in inflammatory bowel diseases. World J Gastroenterol 2016;22:974-95. 10.3748/wjg.v22.i3.974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murphy CG, Glickman JN, Tomczak K, et al. Acute appendicitis is characterized by a uniform and highly selective pattern of inflammatory gene expression. Mucosal Immunol 2008;1:297-308. 10.1038/mi.2008.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheluvappa R, Thomas DG, Selvendran S. The Role of Specific Chemokines in the Amelioration of Colitis by Appendicitis and Appendectomy. Biomolecules 2018;8:59. 10.3390/biom8030059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Soo KS, Michie CA, Baker SR, et al. Selective recruitment of lymphocyte subsets to the inflamed appendix. Clin Exp Immunol 1995;100:133-8. 10.1111/j.1365-2249.1995.tb03614.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Watson Ng WS, Hampartzoumian T, Lloyd AR, et al. A murine model of appendicitis and the impact of inflammation on appendiceal lymphocyte constituents. Clin Exp Immunol 2007;150:169-78. 10.1111/j.1365-2249.2007.03463.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saucier A, Huang EY, Emeremni CA, et al. Prospective evaluation of a clinical pathway for suspected appendicitis. Pediatrics 2014;133:e88-e95. 10.1542/peds.2013-2208 [DOI] [PubMed] [Google Scholar]
- 13.D'Souza N, Karim D, Sunthareswaran R. Bilirubin; a diagnostic marker for appendicitis. Int J Surg 2013;11:1114-7. 10.1016/j.ijsu.2013.09.006 [DOI] [PubMed] [Google Scholar]
- 14.Kaya B, Sana B, Eris C, et al. The diagnostic value of D-dimer, procalcitonin and CRP in acute appendicitis. Int J Med Sci 2012;9:909-15. 10.7150/ijms.4733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huckins DS, Simon HK, Copeland K, et al. A novel biomarker panel to rule out acute appendicitis in pediatric patients with abdominal pain. Am J Emerg Med 2013;31:1368-75. 10.1016/j.ajem.2013.06.016 [DOI] [PubMed] [Google Scholar]
- 16.Mosayebi G, Alizadeh SA, Alasti A, et al. Is CD19 an immunological diagnostic marker for acute appendicitis? Iran J Immunol 2013;10:216-28. [PubMed] [Google Scholar]
- 17.Shao R, Li CS, Fang Y, et al. Low B and T lymphocyte attenuator expression on CD4+ T cells in the early stage of sepsis is associated with the severity and mortality of septic patients: a prospective cohort study. Crit Care 2015;19:308. 10.1186/s13054-015-1024-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu J, Yamane H, Paul WE. Differentiation of effector CD4 T cell populations (*). Annu Rev Immunol 2010;28:445-89. 10.1146/annurev-immunol-030409-101212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hanson NB, Lanning DK. Microbial induction of B and T cell areas in rabbit appendix. Dev Comp Immunol 2008;32:980-91. 10.1016/j.dci.2008.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ouyang W, Kolls JK, Zheng Y. The biological functions of T helper 17 cell effector cytokines in inflammation. Immunity 2008;28:454-67. 10.1016/j.immuni.2008.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Watson Ng WS, Hampartzoumian T, Lloyd AR, et al. A murine model of appendicitis and the impact of inflammation on appendiceal lymphocyte constituents. Clin Exp Immunol 2007;150:169-78. 10.1111/j.1365-2249.2007.03463.x [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as