Abstract
Background
To investigate the expression, function, and related mechanisms of circHIPK3 in oral squamous cell carcinoma (OSCC).
Methods
CircHIPK3 expression was determined by quantitative reverse transcription polymerized chain reaction (QRT-PCR) in OSCC and adjacent tissues, and the correlation between the circHIPK3 level and clinicopathological indexes of OSCC was analyzed. CircHIPK3 expressions in different OSCC cell lines were detected, cell counting kit-8 (CCK-8) and 5-bromodeoxyuridine (BrdU) assays were utilized to monitor cell proliferation and activity. Flow cytometry was adopted to detect apoptosis and transwell assay was used to detect cell invasion. The expressions of nuclear protein 1 (NUPR1), phosphatidylinositol 3 kinase (PI3K)/protein kinase B (AKT) (PI3K/AKT) pathway proteins, and E-cadherin, Vimentin, and N-cadherin markers of epithelial-mesenchymal transformation (EMT) were detected by Western blot or Quantitative Real-time PCR (QRT-PCR).
Results
Upregulated circHIPK3 was noted in OSCC tissues (compared with adjacent tissues), and its overexpression was related to OSCC size and histopathological grade. Functionally, overexpressed circHIPK3 can significantly promote EMT, proliferation, and invasion of OSCC cells and can inhibit cell apoptosis in vivo and in vitro. In addition, CircHIPK3 upregulated the activation of NUPR1 and PI3K/AKT. Bioinformatics analyses showed that miR-637 was the common target of circHIPK3 and NUPR1, while a dual luciferase reporting assay and RIP assay further demonstrated that circHIPK3 targeted miR-637 and bound to 3' UTR of NUPR1.
Conclusions
CircHIPK3 demonstrates potential as a prognostic marker of OSCC and mediates OSCC progression via the miR-637-mediated NUPR1/PI3K/AKT axis.
Keywords: Oral squamous cell carcinoma (OSCC), circHIPK3, miR-637, NUPR1, ceRNA
Introduction
Oral squamous cell carcinoma (OSCC) ranks eleventh in worldwide cancer incidence (1). The characteristics of OSCC include local invasiveness, high recurrence, and ease of metastasis. Its 5-year survival rate is worse than that of multiple solid tumors, at only 50–60% (2). However, the molecular mechanism of OSCC remains unclear, and there is no highly sensitive and specific biomarker that can be used as a monitoring indicator for early diagnosis, treatment, and prognosis.
CircRNA is a novel member of the lncRNA family (3). Unlike linear RNA, circRNA does not have the characteristics of the 5'methylguanosine cap and 3'polyadenylate tail but has a covalent closed-loop structure. Therefore, circRNA is not sensitive to ribonuclease (Rnase), which makes it more stable than linear RNA (4). CircRNA is widely involved in the pathogenesis of various diseases (5). Previous research indicates that circRNAs exert an impact on tumors via regulation of downstream-targeted miRNAs (6). For example, in lung cancer, circ-001569 can sponge miR-145 and upregulate the expression of downstream genes such as E2F5, BAG4, and FMNl2, resulting in an increase in G2/M phase cells and a decrease in tumor cell apoptosis (7). The abnormal regulation of circITCH can enhance the expression of its parent tumor suppressor gene ITCH through a sponge effect on miR-7 and miR-214, thus regulating the proliferation of cancer cells (8). Circ0000140 plays an anti-tumor role in OSCC through the miR-31/LATS2 axis of the HIPPO signaling pathway (9). Recent studies have found that CircHIPK3 is related to tumor progression, indicating that CircHIPK3 can regulate tumor progression. An increasing number of studies have demonstrated the involvement of circHIPK3 in the progression of cervical cancer (CC), non-small-cell lung cancer (NSCLC), and OSCC as an oncogene (10-12). However, the mechanism of circHIPK3 in OSCC remains unknown.
MiRNAs and circRNAs belong to the same kind of non-coding RNA, and the length of miRNAs is only 18–25 nucleotides. Multiple miRNAs can modulate the pathological processes of various diseases (13). For example, miR-504, a tumor inhibitor, weakens OSCC growth and metastasis via CDK6 (14). In addition, miR-495 impedes the proliferation of OSCC via Notch1 (15). MiR-637 is a crucial molecule in many tumors. For instance, in hepatocellular carcinoma, miR-637 negatively mediates USP21 expression by directly targeting 3'-UTR in USP21, thus exerting a tumor inhibitory effect (16). In addition, miR-637 hinders tumor cells in OSCC by targeting nuclear protein 1 (NUPR1) (17). This indicates that by targeting NUPR1, miR-637 can block the progress of OSCC. However, the mechanism of miR-637 in OSCC remains unclear.
Nuclear protein 1 (NUPR1/p8/Com1) was first described by Mallo in the acute stage of pancreatitis more than 20 years ago (18). NUPR1 was a small nuclear protein associated with various stress stimuli (19,20). Many previous studies had shown that many factors or molecules could lead to overexpression of NUPR1, such as endothelin and angiotensin. Attention has been paid to the research of NUPR1 in cancer. Yu et al. (21) have shown that nupr1 is up-regulated in ovarian cancer, which can promote the progress of cancer, our aim in this study was to investigate the functions of a new circHIPK3/miR-637/NUPR1/PI3K/AKT axis in OSCC, in order to identify a molecular marker for OSCC diagnosis and treatment in the clinic.
We present the following article in accordance with the ARRIVE reporting checklist (available at http://dx.doi.org/10.21037/atm-21-1908).
Methods
Clinical tissue samples
Tumor and adjacent tissues were taken from 40 OSCC patients admitted to School and Hospital of Stomatology, Kunming Medical University from February 2016 to February 2019. The National Comprehensive Cancer Network (NCCN) Oncology Clinical Practice Guidelines (V.1. 2012) and the Tumor Lymph Node Metastasis (TNM) staging system were used to make a definite diagnosis. No patients received adjuvant therapy pre-operatively. The normal specimens (cancer-free cells) were taken at least 3 cm away from the surgical margin in each patient. OSCC was diagnosed pathologically based on the World Health Organization (WHO) standards. Specimens were immediately stored in liquid nitrogen at −196 °C for RNA extraction. The Ethics Committee of the Affiliated Hospital of Binzhou Medical College approved the study and eligible patients signed informed consent. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013).
Cell lines, cell culture, and cell transfection
Normal oral keratinocyte lines (NOK) and OSCC lines (OSCC-15, Tca8113, SCC-9, SCC-25, and HSC-2) were procured from the American Type Culture Collection (ATCC) cell bank (Rockville, MD, USA), and cultured in Dulbecco’s Modified Eagle Medium (DMEM) (Gibco; Thermo Fisher Science, Inc.) with 10% (v/v) fetal bovine serum (FBS) (Invitgen; Thermo Fisher Science, Inc.), 100 IU/mL penicillin, and 100 mg/mL streptomycin (Baoman Biotechnology Co., Ltd. shanghai, China). Cell lines were stored in a humid atmosphere of 37 °C containing 5% CO2.
CircHIPK3 overexpression plasmid (pcDNA3.1-circHIPK3) and its sh-circHIPK3, miR-637 mimics, as well as NUPR1 overexpression plasmid (pcDNA3.1-NUPR1) were all derived from GenePharma (Shanghai, China). The sequence of sh-circHIPK3 was designed and synthesized by Ruibo Biology Co., Ltd. (Guangzhou, China). The Tca8113 and SCC-9 cells were inoculated in a 24-well plate (3×105/well) at 37 °C and 5% CO2 for 24 hours, and then transfected using Lipofectamine® 3000 (Invitgen; Thermo Fisher Science, Inc.). The transfection efficiency was then determined by quantitative reverse transcription polymerized chain reaction (QRT-PCR).
QRT-PCR
Total RNA was extracted with TRIzol reagent (Invitgen, Waltham, MA, USA). The concentration and purity of RNA were determined by a nanodroplet spectrophotometer. Subsequently, a PrimeScript-RT Kit (Madison, WI, USA) was adopted to reverse transcribe 1µg of the total RNA to synthesize its complementary gene. Afterwards, QRT-PCR was performed using SYBR® PreMix-Ex-Taq™ (Takara, TX, USA) and ABI7300 systems. The total volume of the PCR system was 30 µL, and each sample contained 300 µg of cDNA. Amplification was initially performed at 95 °C for 10 minutes for 45 cycles (95 °C for 10 seconds, 60 °C for 30 seconds, and 85 °C for 20 seconds). We converted all fluorescence data into relative quantization. GAPDH was the internal reference for circHIPK3 and NUPR1. U6 was the internal reference for miR-637. The primer was designed and synthesized by Guangzhou Ruibo Biotechnology Co., Ltd. CircRNA HIPK3: upstream primer 5'-TATGTTGGTGGATCCTGTTCGGCA-3', downstream primer 5'-TGGTGGTAGACCAAGACTTGTGA-3'; NUPR1: upstream primer 5'-GCACGAGAGGAAACTGGTGA-3', downstream primer 5'-GTCCCGTCTCTATTGCTGGGGG-3'; MiR-637: upstream primer 5'-ACUGGGGGCUUUCGGGGGCUCUGCGU-3', downstream primer 5'-ACGCAGAGCCCGAAAGCCCCCAGU-3'; GAPDH: upstream primer 5'-GGGGAGCCAAAAGGGTCAT-3', downstream primer 5'-GAGTCCTCCCGATACCAA-3'; U6: upstream primer 5'-CTCGCTTCGGCAGCACA-3', downstream primer 5'-AACGCTTCACGAATTTGCGT-3'
RNase R treatment
The stability of circHIPK3 was determined by RNase R. In short, 2 mg of total RNA was incubated at 37 °C for 30 minutes with or without 5 U/µg RNaseR (Epicentre; Illumina Inc), and then purified with the RNase MinElute cleaning kit (Jianlun Biotechnology Co., Ltd. Guangzhou, China) and analyzed with QRT-PCR.
Actinomycin D assay
Tca8113 and SCC-9 cells in logarithmic growth phase were added actinomycin D to the final concentration of 2 µg/mL (Sigma-Aldrich, Germany) and incubated at 37 °C (4, 8, 12, and 24 hours). Total RNA was extracted at the different time points, and the expressions of circHIPK3 and linear HIPK3 mRNA were detected by QRT-PCR.
CCK 8 assay
Tca8113 and SCC-9 cells were inoculated in 96-well plates (2×103 cells/well) with a 200 µL culture medium at 37 °C and 5% CO2. On days 1, 2, 3, and 4, 10 µL CCK8 solution (Dojindo Molecular Technologies, Inc.) was added to each well and the incubation was continued at 37 °C for 2 hours. The absorbance was measured at 450 nm using a miniature flat panel reader (Bio-Rad Laboratory, Inc.).
BrdU assay
When the cells grew to exponential phase, BrdU was added to the culture medium to make the final concentration of 10 µg/mL. After DNA denaturation, the cells were incubated with a BrdU primary antibody (Abcam, AB6326, 1:1,000, California, USA) at room temperature for 2 hours, and then 10 fluorescent secondary antibodies were added to incubate at room temperature for 2 hours. Subsequently, the nucleus was labeled with 10 mol/L of Hoechst 33342. Finally, an inverted fluorescence microscope was used for the imaging and statistical analyses.
Flow cytometry
The AnnexinV-FITC double staining method was used to detect apoptosis. After 24 hours of transfection, the cells were treated by pancreatin, collected, and then inoculated into 6-well plates, and the cell density was adjusted to 2×106 cells/well. Cells were continuously cultured for 24 hours. Afterwards, the supernatant was discarded, and the cells were resuspended with 1× binding buffer after being washed twice with pre-cooled phosphate-buffered saline (PBS). Next, 5 µL human AnnexinV-FITC and 5 µL PI (BD Biosciences, New Jersey, USA) were added to the cell suspension, and the cells were fully mixed and incubated at room temperature for 15 minutes. The apoptosis rate was detected by flow cytometry within 1 hour.
Transwell assay
A Transwell chamber (Corting, NY, USA) was covered with 200 mg/mL Matrigel (BD, SanJose, USA) and incubated overnight. Saos-2 and U2OS cells were then added to the upper chamber of the serum-free medium. DMEM (500 µL) containing 10% FBS was placed in the lower cavity as a chemotactic agent. After 24 hours of incubation, all non-invasive cells were removed. The Matrigel membrane was fixed with paraformaldehyde and stained with a crystal violet solution. The number of invasive cells was counted by a phase contrast microscope (Olympus, Tokyo, Japan).
RNA pull-down assay
The OSCC cells were co-transfected with pcDNA3-FlagMS2bp and pcDNA3-HIPK3-MS2bs, and the cells were collected 2 days later. Approximately 1×107 cells were dissolved in a soft lysis buffer containing 80 U/mL RNasin (Promega Madison, WI, USA). An amount of 50 µg/mL of anti-FLAG M-280 magnetic beads (Invitgen, USA) was added to each bonded reaction tube, incubated for 4 hours, and then washed six times in lysis buffer. The recovered supernatant was detected by QRT-PCR.
RIP assay
Magna RIP™ detection was performed using a Magna RNA binding protein immunoprecipitation kit (Merck Millipore, USA). Approximately 2×107 OSCC cells transfected with miR-637 or its control were collected and added to 200 µL of RIP lysis buffer. Afterwards, they were cracked on ice for 4 minutes and centrifuged (1,500 rpm) for 15 minutes. The extract was then incubated overnight with anti-Ago2 or anti-IgG (Sigma, Germany). Subsequently, the magnetic beads were rinsed four times and the supernatant was discarded, then protease K lysate was added to the magnetic beads and lyzed at 55 °C for 30 minutes. Finally, the supernatant was put into a new centrifuge tube, and the total RNA was extracted with phenol-chloroform-isoamyl alcohol and purified by isopropanol centrifugation. The expressions of circHIPK3 and NUPR1 were detected by QRT-PCR.
Dual-luciferase reporter assay
HIPK3 or NUPR1 sequences containing the predicted miR-637 binding sites were constructed into a pGL3 vector (Promega, USA). Subsequently, the OSCC cells were transfected with pGL3 plasmid and miR-637 mimic for the luciferase reporter assay. Luciferase activity was detected 48 hours later.
Western blot
Total cell proteins were extracted with a RIPA lysis and extraction buffer (Thermo Science™) and quantified by biochemical methods (Beyotime, China). Protein lysate (30 g/lane) was separated by SDS-PAGE and transferred to a polyvinylidene fluoride (Millipore, MA, USA). The primary antibody was added and incubated overnight at 4 °C. After washing the PVDF, an HRP-labeled secondary antibody (Jackson, USA) was added and incubated at room temperature for 2 hours. Western blotting data were quantified with the Alpha Innotech (Jackson, USA) imaging software. The primary antibodies, including NUPR1, p-PI3K/PI3K, p-AKT/AKT, E-cadherin, Vimentin, N-cadherin, and GAPDH were purchased from Abcam Trading (Shanghai) Co., Ltd.
Tumor formation assay
BALB/c-nu mice (4–6 weeks) were selected to establish the tumor models. Tca8113 and SCC-9 cells (in logarithmic growth phase) were selected, and the concentration was adjusted to 2×108/mL. Subsequently, 0.1 mL cell suspension was injected into the armpit of each nude mouse’s left forelimb. There were five mice in each group. The survival of mice was monitored, and the tumor size and weight of the newly dead mice were measured within 25 days of injection.
Tumorigenesis in nude mice
Animals Experiments were performed under a project license (No.: 201403024) granted by Stomatological Hospital of Xi’an Jiaotong University, in compliance with Stomatological Hospital of Xi’an Jiaotong University guidelines for the care and use of animals. According to previous studies (22), C57BL/6 nude mice were taken from Hunan relaxing Jingda experimental animals (Changsha, China) and inoculated with oral cancer cells with high or low expression of CircRNA-HIPK3 (20 nude mice were randomly divided into 4 groups with 5 mice in each group). The growth of transplanted oral cancer was monitored every 5 days after inoculation until 30 days after inoculation. The mice were killed and their xenotransplantation was photographed and measured.
Statistical analyses
SPSS17.0 statistical software (SPSS Inc., Chicago, IL, USA) was used to analyze the measurement data. The measurement data was expressed as mean ± standard deviation (). T test was used for comparison between the two groups, and ANOVA was used for comparison between multiple groups. P<0.05 was considered as the difference with statistical significance.
Results
Highly expressed circHIPK3 was noted in both OSCC tissues and OSCC cells
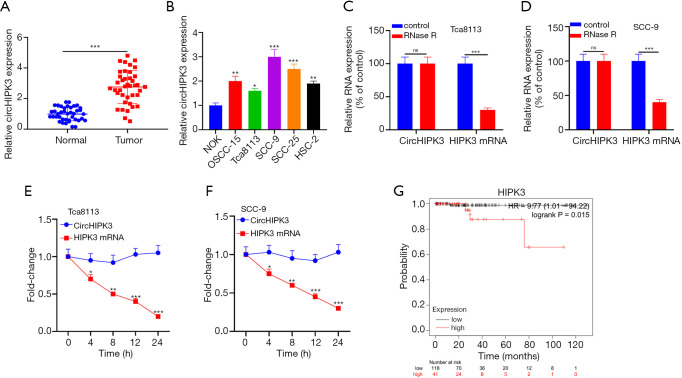
QRT-PCR showed that HIPK3 was upregulated in OSCC tissues (P<0.05, Figure 1A). Additionally, HIPK3 in OSCC cell lines (OSCC-15, Tca8113, SCC-9, SCC-25, and HSC-2) was notably upregulated compared with NOK (P<0.05, Figure 1B). CircRNA was a stable nucleic acid. As shown in Figure 1C and D, RNase R resulted in a notable decrease in HIPK3 mRNA expression compared with the control group, while CircHIPK3 expression did not change notably. Additionally, the actinomycin D assay revealed that circHIPK3 was more stable than linear HIPK3 mRNA (P<0.05, Figure 1E,F). Distant metastasis and higher tumor staging was associated with OSCC patients with highly expressed circHIPK3, who exhibited a shorter survival than patients with low-expressed circHIPK3 (Figure 1G and Table 1). Overall, circHIPK3 was related to the poor prognosis of OSCC patients and exerted a cancer-promoting effect.
Figure 1.
The expression of CircHIPK3 in normal and cancerous tissues of OSCC. (A) QRT-PCR is used to detect circHIPK3 expression in tumor and adjacent tissues. ***P<0.001; (B) QRT-PCR is used to detect HIPK3 expression in NOK and OSCC-15, Tca8113, SCC-9, SCC-25 and HSC-2. Compared with NOK group, *P<0.05, **P<0.01, ***P<0.001; (C,D) the stability of circHIPK3 in Tca8113 and SCC-9 cells is determined by the RNase R method. ***P<0.001; (E,F) the stability of circHIPK3 in Tca8113 and SCC-9 cells is detected by the actinomycin D method. *P<0.05, **P<0.01, ***P<0.001; (G) the survival curve of circHIPK3 in OSCC is obtained from the Kaplan-Meier Plotter library. circHIPK3, circular RNA HIPK3; OSCC, oral squamous cell carcinoma, QRT-PCR, quantitative real-time PCR.
Table 1. Relationship between circHIPK3 level and clinical characteristics in OSCC patients.
| Characteristics | n | CircHIPK3 high expression (n=40) | CircHIPK3 low expression (n=40) | P |
|---|---|---|---|---|
| Age | 0.648 | |||
| <60 | 32 | 15 | 17 | |
| ≥60 | 48 | 25 | 23 | |
| Gender | 0.496 | |||
| Male | 47 | 25 | 22 | |
| Female | 33 | 15 | 18 | |
| Smoke | 0.469 | |||
| Yes | 35 | 14 | 11 | |
| No | 45 | 26 | 29 | |
| Differentiation | 0.654 | |||
| Well | 42 | 20 | 22 | |
| Moderate/poorly | 38 | 20 | 18 | |
| Lymph node metastasis | 0.007 | |||
| Yes | 34 | 23 | 11 | |
| No | 46 | 17 | 29 | |
| TNM stage | 0.000 | |||
| I–II | 46 | 15 | 31 | |
| III–IV | 34 | 25 | 9 |
Circular RNA, circHIPK3; OSCC, oral squamous cell carcinoma;
Effects of circHIPK3 on the proliferation, apoptosis, invasion, and epithelial-mesenchymal transformation (EMT) of OSCC cells
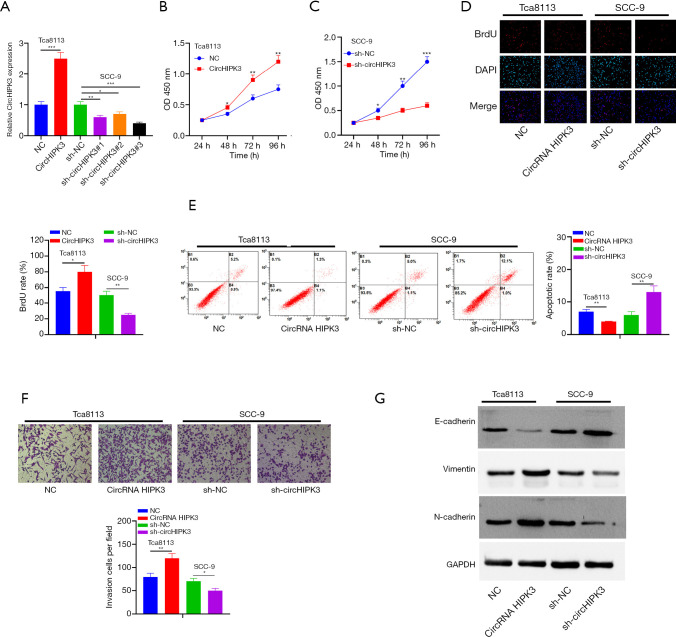
Models of overexpressed and underexpressed circHIPK3 were constructed in Tca8113 and SCC-9 cells to investigate the effects of circHIPK3 on OSCC (P<0.05, Figure 2A). Sh-circHIPK3#3, with the highest knockdown efficiency of circHIPK3 in SCC-9 cells, was selected for the following functional experiments: CCK8 and BrdU confirmed a notable enhancement of cell proliferation and viability after HIPK3 overexpression, while the HIPK3 gene knockdown produced an opposite trend (P<0.05, Figure 2B,C,D). Moreover, flow cytometry showed that the apoptosis rate decreased after overexpression of circHIPK3, while it increased after knockdown of the circHIPK3 gene (P<0.05, Figure 2E). The Transwell assay showed that overexpressed circHIPK3 accelerated cell invasion, while downregulated circHIPK3 exhibited an opposite effect (P<0.05, Figure 2F). As shown by Western blot, overexpressed circHIPK3 significantly reduced E-cadherin expression and increased the expressions of Vimentin and N-cadherin. On the contrary, after the HIPK3 gene was knocked down, E-cadherin was upregulated and the expressions of Vimentin and N-cadherin were downregulated (P<0.05, Figure 2G). In summary, circHIPK3 was involved in OSCC progression by accelerating the growth, invasion, and EMT of tumor cells and by blocking cell apoptosis.
Figure 2.
The effect of circHIPK3 on the proliferation, invasion, apoptosis and EMT of OSCC cells. (A) CircHIPK3 overexpression and gene knockdown models are established in OSCC cell lines Tca8113 and SCC-9, respectively. *P<0.05, **P<0.01, ***P<0.001; (B,C,D) cell proliferation is detected by the CCK-8 method and BrdU colorimetry. *P<0.05, **P<0.01, ***P<0.001. The magnification of (D) is 100×; (E) the apoptosis of Tca8113 and SCC-9 cells is detected by flow cytometry. **P<0.01; (F) Transwell assay is used to detect the invasion ability of Tca8113 and SCC-9 cells. *P<0.05, **P<0.01. Dyeing with crystal violet, the magnification is 100×; (G) EMT markers of E-cadherin, Vimentin and N-cadherin are detected by Western blot. circHIPK3, circular RNA HIPK3; OSCC, oral squamous cell carcinoma; EMT, epithelial-mesenchymal transition.
CircHIPK3 can facilitate the growth of OSCC and EMT in vivo
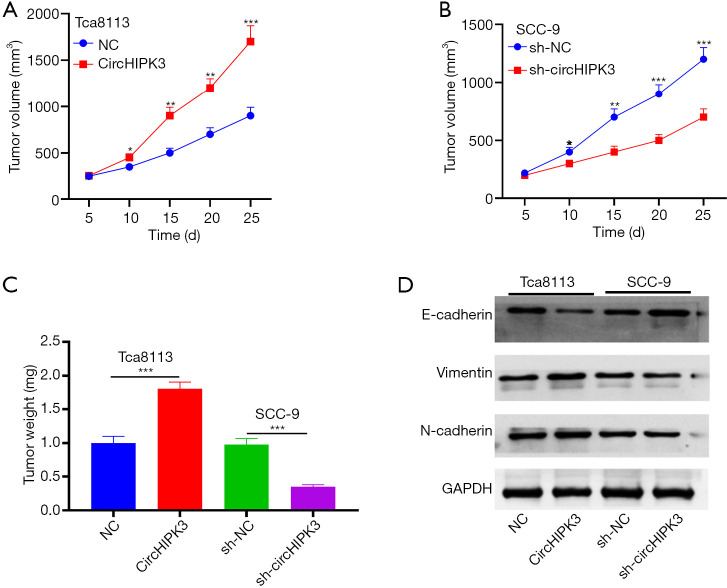
We established cell lines with overexpressed and knocked-down circHIPK3 in Tca8113 and SCC-9 cells, respectively, and verified the effect of circHIPK3 on the growth of OSCC in vivo through nude mouse tumorigenesis experiments. Overexpressed circHIPK3 increased tumor size and weight, while downregulated circHIPK3 had the opposite effect (P<0.05, Figure 3A,B,C). Western blot showed that E-cadherin was downregulated, and expressions of Vimentin and N-cadherin were upregulated, after HIPK3 overexpression. However, knocked-down circHIPK3 exerted an opposite effect (Figure 3D). Therefore, circHIPK3 enhanced the growth and EMT of OSCC cells.
Figure 3.
CircHIPK3 promotes OSCC growth and EMT transformation in vivo. (A,B,C) The volume and weight of tumor nodules are calculated. *P<0.05, **P<0.01, ***P<0.001; (D) Western blot is used to detect expressions of E-cadherin, Vimentin, and N-cadherin. OSCC, oral squamous cell carcinoma. circHIPK3, circular RNA HIPK3; OSCC, oral squamous cell carcinoma; EMT, epithelial-mesenchymal transition.
MiR-637 contains conserved binding sites for circRNA HIPK3 and NUPR1
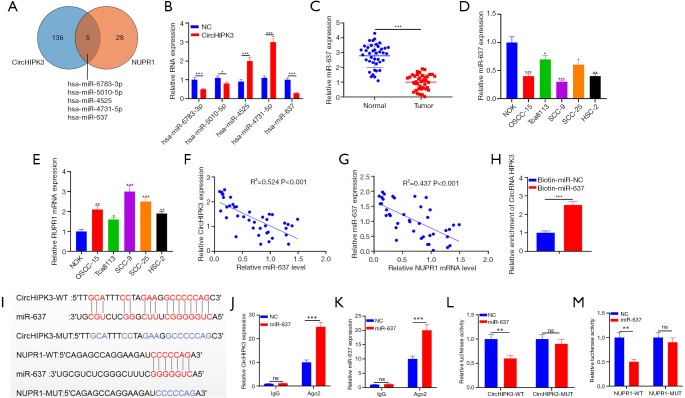
From Starbase, five miRNAs had conserved binding sites to circHIPK3 and NUPR1 (Figure 4A). MiR-637 expression was significantly downregulated (Figure 4B). QRT-PCR indicated that miR-637 was notably lower in OSCC tissues than adjacent tissues (P<0.05, Figure 4C). Additionally, miR-637 in OSCC cell lines (OSCC-15, Tca8113, SCC-9, SCC-25, and HSC-2) was notably downregulated compared with NOK, while NUPR1 upregulated (P<0.05, Figure 4D,E). Pearson analysis showed that in OSCC tissues, circHIPK3 showed a negative correlation with miR-637 (R2=0.524, P<0.001, Figure 4F), while NUPR1 was negatively correlated with miR-637 (R2=0.437, P<0.001, Figure 4G). To clarify the targeting relationship between these three molecules, we carried out the following experiments: First, we captured circHIPK3 with biotin-labeled miR-637 mimics and NC, and miR-637 mimics captured more circHIPK3 (P<0.05, Figure 4H). Figure 4I shows the binding sites between circRNA HIPK3 and miR-637, miR-637, and NUPR1. Next, RIP and dual luciferase assay revealed that after miR-637 transfection, the precipitation amount of circHIPK3 and NUPR1 was notably higher in the Ago2 antibody group than that in the IgG group, suggesting that circRNA HIPK3 and NUPR1 bind to Ago2 via miR-637 (P<0.05, Figure 4J,K). Moreover, miR-637 significantly impeded the luciferase activity of circHIPK3-WT and NUPR1-WT, but no notable effect was evident on that of circHIPK3-mut and NUPR1-mut (P<0.05, Figure 4L,M). In summary, miR-637 is the target of circRNA HIPK3, and NUPR1 is the target of miR-637 in OSCC.
Figure 4.
Targeting the relationships among circHIPK3, NUPR1, and miR-637. (A) The miRNA targets of circHIPK3 and NUPR1 are analyzed by Starbase, and five miRNAs are found to contain conserved binding sites to circHIPK3 and NUPR1; (B) QRT-PCR is used to detect expressions of five miRNAs in overexpressed cells; (C) QRT-PCR is used to detect expressions of miR-637 in OSCC tissues and normal tissues adjacent to cancer; (D,E) QRT-PCR is used to detect miR-637 and NUPR1 expression in NOK and OSCC-15, Tca8113, SCC-9, SCC-25 and HSC-2. (F,G) Pearson correlation analysis is used to detect the correlation between miR-637 and circHIPK3, miR-637, and NUPR1 in OSCC tissues; (H) the relative level of circHIPK3 in biotin-labeled miR-637 or NC-captured cell lysates is detected by an RNA pull-down assay; (I) the binding relationship between miR-637, circHIPK3, and NUPR1; (J,K) the binding of circHIPK3 and NUPR1 to Ago2 antibody in SCC-9 cells is detected by the RIP method; (L,M) the binding relationship between miR-637 and circHIPK3, and miR-637 and NUPR1 in SCC-9 cells is confirmed by dual luciferase reporter gene analysis. nsP>0.05, *P<0.05, **P<0.01, ***P<0.001. circHIPK3, circular RNA HIPK3; miR-637, microRNA-637; NUPR1, Nucleoprotein 1; OSCC, oral squamous cell carcinoma; QRT-PCR, quantitative real-time PCR.
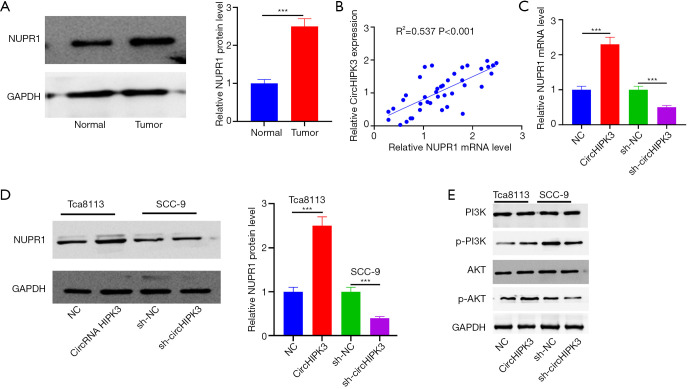
CircHIPK3 enhanced NUPR1 expression and activation of the PI3K/AKT signaling pathway
Western blot showed upregulated NUPR1 in OSCC tissues (P<0.05, Figure 5A). Interestingly, HIPK3 was positively correlated with NUPR1 in OSCC tissues (R2=0.537, P<0.001, Figure 5B). In the model, upregulated circHIPK3 increased NUPR1 expression and its mRNA, whereas downregulated circHIPK3 showed the opposite effect (Figure 5C,D). Western blot revealed that overexpressed circHIPK3 augmented expressions of p-PI3K and p-AKT, while low-expressed circHIPK3 produced an opposite effect (P<0.05, Figure 5E). HIPK3 exerted a positive regulatory effect on the NUPR1 and PI3K/AKT pathways.
Figure 5.
CircHIPK3 promotes the expression of the NUPR1 and PI3K/AKT signaling pathways. (A) Western blot is used to detect expressions of NUPR1 in OSCC and normal tissues adjacent to cancer. ***P<0.001; (B) Pearson correlation analysis shows a correlation between CircHIPK3 and NUPR1 in OSCC tissue; (C,D) the expression of NUPR1 mRNA and protein is detected by QRT-PCR and Western blot. ***P<0.001; (E) the activation of PI3K/AKT is detected by Western blot. OSCC, oral squamous cell carcinoma. circHIPK3, circular RNA HIPK3; NUPR1, Nucleoprotein 1; OSCC, oral squamous cell carcinoma; QRT-PCR, quantitative real-time PCR.
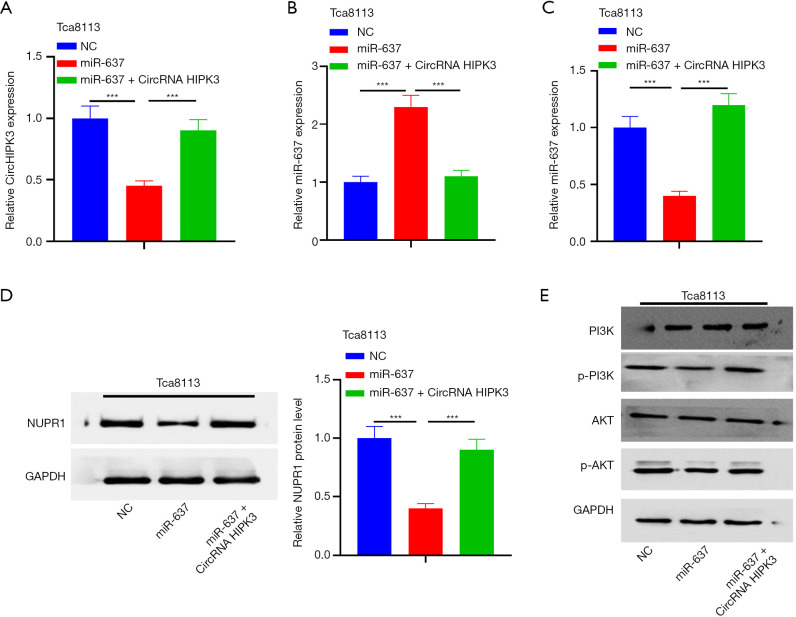
CircRNA HIPK3 regulates the expression of the NUPR1/PI3K/Akt gene by sponging miR-637
To confirm the regulation of circHIPK3/miR-637/NUPR1/PI3K/Akt in OSCC, a rescue experiment was carried out. The results showed that in the miR-637 mimic group, circHIPK3 was downregulated and miR-637 was upregulated. On the other hand, overexpressed circRNA HIPK3 resulted in upregulated circHIPK3 and downregulated miR-637 (P<0.05, Figure 6A,B). Next, QRT-PCR and Western blot showed that overexpressed miR-637 downregulated expressions of NUPR1 mRNA and protein, while supplementation of circHIPK3 increased the level of NUPR1 protein and its mRNA (P<0.05, Figure 6C,D). In addition, Western blot showed that upregulated miR-637 could inhibit the expressions of p-PI3K and p-AKT compared with NC, while supplementation of circHIPK3 in the miR-637 group could increase expressions of p-PI3K and p-AKT (P<0.05, Figure 6E). In summary, circHIPK3 regulated NUPR1 by sponging miR-637, thus activating the PI3K/AKT pathway.
Figure 6.
CircHIPK3 regulates the expression of NUPR1/PI3K/AKT by sponging miR-637. (A,B,C) QRT-PCR is used to detect expressions of circHIPK3, miR-637, and NUPR1 mRNA in Tca8113 cells transfected with circHIPK3 overexpression plasmid or miR-637 mimic. ***P<0.001; (D,E) Western blot is used to detect expressions of NUPR1 and PI3K/AKT. ***P<0.001. circHIPK3, circular RNA HIPK3; miR-637, microRNA-637; NUPR1, Nucleoprotein 1; QRT-PCR, quantitative real-time PCR.
Discussion
In the present study, the role of a new circRNA HIPK3 gene was explored in the progression of OSCC. CircRNA HIPK3 was shown to be upregulated in OSCC, which predicted a poor prognosis in OSCC patients. Moreover, CircHIPK3 plays a cancer-promoting role in OSCC through the NUPR1-PI3K/AKT axis.
CircRNA plays a significant role in the progression of various tumors, contain OSCC (23). For instance, circSEPT9 promoted OSCC via PKN2 and miR-1225 (24). Guo et al. pointed out that circ0000140 impeded the multiplication of OSCC cells by upregulating miR-182-5p and inhibiting CDC73 expression (25). Several recent studies have shown that circRNA HIPK3 exerts a strong regulatory effect in various cancers. For example, circHIPK3 was upregulated in CC and accelerated multiplication of CC cells via the miR-485-3p/FGF2 axis (10). Meanwhile, circHIPK3 enhanced the progression of NSCLC via the miR-107/BDNF signaling pathway (11). CircRNA HIPK3 upregulated CCND2 expression in OSCC via miR-124, thus promoting cell proliferation and invasion (12). Based on these findings, we speculated that circHIPK3 may exert a crucial regulatory role in OSCC. Interestingly, circHIPK3 was overexpressed in OSCC. Our experiments showed that circHIPK3 promoted proliferation, invasion, and EMT of OSCC cells, while inhibiting OSCC cell apoptosis, suggesting that circHIPK3 may play a role as a prognostic factor and therapeutic target in OSCC. The role of circRNA HIPK3 as an oncogene was further verified.
A growing number of studies have reported that miR-637 plays a role as a tumor suppressor gene in cancer. For instance, miR-637, which was negatively regulated by c5orf66-AS1, was downregulated in CC, thus exerting an anti-tumor role by downregulating RING1 (26). Similarly, miR-637 impeded the proliferation and metastasis of melanoma cells by directly targeting P-REX2a and blocking the PTEN/AKT signaling pathway (27). In this study, miR-637 was downregulated in OSCC cells and negatively correlated with circHIPK3, consistent with the above reports. Several studies confirmed that circRNAs were ceRNAs of miRNA. For instance, circBICD2 enhanced IGF2BP1 as ceRNA via miR-149-5p, thus accelerating the proliferation and migration of OSCC cells (28). In addition, overexpressed circPHIP downregulated miR-142-5p and upregulated PHIP and ACTN4, thus aggravating OSCC (29). Here, binding sites between circHIPK3 and miR-637 were determined through Starbase. Thus, we speculated that miR-637 may be a downstream molecule of circHIPK3 in OSCC. Additionally, overexpressed circHIPK3 weakened the inhibition of miR-637. Therefore, circHIPK3 exerted its biological function by sponge miR-637.
NUPR1 is a kind of nucleoprotein, which is synthesized in cytoplasm and then transported to the nucleus to play a role. NUPR1 has transcriptional activity and can play different functions by participating in different intracellular signaling pathways (30-32). NUPR1 is abnormally expressed in various tumor cells such as NSCLC, pancreatic cancer, breast cancer, thyroid cancer, and prostate cancer, and participates in the tumorigenesis and progression of tumors (33,34). It is reported that NUPR1 is involved in regulating cell growth and differentiation and has carcinogenicity. For instance, downregulating NUPR1 can inhibit the growth, migration, and invasion of ovarian cancer and promote apoptosis (21). Zhou et al. pointed out that overexpressed NUPR1 can weaken the inhibitory effects of miR-4443 on osteosarcoma cell metastasis (35). NUPR1 activates PI3K/AKT, thus affecting tumor progression. For instance, NUPR1 silencing induced autophagy-induced apoptosis of multiple myeloma cells via the PI3K/AKT/mTOR pathway (36). In the present study, we confirmed through in vitro experiments that NUPR1 was upregulated in OSCC, which is consistent with previous studies (17). The relationship between miR-637 and NUPR1 was analyzed by bioinformatics database. We further explored whether circRNA HIPK3 can indirectly up regulate nupr1 through miR-637 and play a role in promoting cancer. The information database showed that miR-637 negatively regulated the expression of NUPR1. In addition, circRNA HIPK3 regulates the NUPR1 and PI3K/Akt pathways. Based on these findings, circRNA HIPK3 promotes OSCC progression through miR-637-NUPR1-PI3K/AKT axis.
In conclusion, circHIPK3 accelerates the proliferation, metastasis, and EMT of OSCC cells via the miR-637-NUPR1-PI3K/AKT axis and impedes cell apoptosis. However, our sample size in this study was small. Additionally, the role of the circRNA HIPK3-miR-637-NUPR1-PI3K/AKT axis in OSCC progression needs to be further studied.
Supplementary
The article’s supplementary files as
Acknowledgments
Funding: This study was supported by The Shenzhen Fundamental Research Fund of Science and Technology Foundation of Shenzhen City (Grant No. JCYJ20190808155005649); the Natural Science Foundation of SZC (Grant No. 00000330, 000002110148); the SZU Medical Young Scientists Program (Grant No. 71201-000001); the SZU Top Ranking Project (Grant No. 86000000210); and the Medical and Health Science and Technology Development Project of Shandong Province (Grant No. 2011HW004).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Our study was approved by the Ethics Review Board of The Affiliated Hospital of Binzhou Medical College and eligible patients signed informed consent. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). Animals Experiments were performed under a project license (No.: 201403024) granted by Stomatological Hospital of Xi’an Jiaotong University, in compliance with Stomatological Hospital of Xi'an Jiaotong University guidelines for the care and use of animals.
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at http://dx.doi.org/10.21037/atm-21-1908
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-21-1908
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-21-1908). The authors have no conflicts of interest to declare.
(English Language Editor: D. Fitzgerald)
References
- 1.Global Burden of Disease Cancer Collaboration , Fitzmaurice C, Dicker D, et al. The Global Burden of Cancer 2013. JAMA Oncol 2015;1:505-27. Erratum in: JAMA Oncol 2015;1:690. 10.1001/jamaoncol.2015.0735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brocklehurst PR, Baker SR, Speight PM. Oral cancer screening: what have we learnt and what is there still to achieve. Future Oncol 2010;6:299-304. 10.2217/fon.09.163 [DOI] [PubMed] [Google Scholar]
- 3.Wu H, Yang L, Chen LL. The diversity of long noncoding RNAs and their generation. Trends Genet 2017;33:540-52. 10.1016/j.tig.2017.05.004 [DOI] [PubMed] [Google Scholar]
- 4.Li J, Yang J, Zhou P, et al. Circular RNAs in cancer: novel insights into origins, properties, functions and implications. Am J Cancer Res 2015;5:472-80. [PMC free article] [PubMed] [Google Scholar]
- 5.Liu J, Liu T, Wang X, et al. Circles reshaping the RNA world: from waste to treasure. Mol Cancer 2017;16:58. 10.1186/s12943-017-0630-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hansen TB, Kjems J, Damgaard CK. Circular RNA and miR-7 in cancer. Cancer Res 2013;73:5609-12. 10.1158/0008-5472.CAN-13-1568 [DOI] [PubMed] [Google Scholar]
- 7.Xie H, Ren X, Xin S, et al. Emerging roles of circRNA_001569 targeting miR-145 in the proliferation and invasion of colorectal cancer. Oncotarget 2016;7:26680-91. 10.18632/oncotarget.8589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pei W, Tao L, Zhang LW, Zhang S, et al. Circular RNA profiles in mouse lung tissue induced by radon. Environ Health Prev Med 2017;22:36. 10.1186/s12199-017-0627-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peng QS, Cheng YN, Zhang WB, et al. circRNA_0000140 suppresses oral squamous cell carcinoma growth and metastasis by targeting miR-31 to inhibit Hippo signaling pathway. Cell Death Dis 2020;11:112. 10.1038/s41419-020-2273-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu S, Liu S, Song H, et al. Circular RNA HIPK3 plays a carcinogenic role in cervical cancer progression via regulating miR-485-3p/FGF2 axis. J Investig Med 2021;69:768-74. 10.1136/jim-2020-001537 [DOI] [PubMed] [Google Scholar]
- 11.Hong W, Zhang Y, Ding J, et al. circHIPK3 Acts as Competing Endogenous RNA and Promotes Non-Small-Cell Lung Cancer Progression through the miR-107/BDNF Signaling Pathway. Biomed Res Int 2020;2020:6075902. 10.1155/2020/6075902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu Z, Guo S, Sun H, et al. Circular RNA CircHIPK3 Elevates CCND2 Expression and Promotes Cell Proliferation and Invasion Through miR-124 in Glioma. Front Genet 2020;11:1013. 10.3389/fgene.2020.01013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem 2010;79:351-79. 10.1146/annurev-biochem-060308-103103 [DOI] [PubMed] [Google Scholar]
- 14.Wang X, Chang K, Gao J, et al. MicroRNA-504 functions as a tumor suppressor in oral squamous cell carcinoma through inhibiting cell proliferation, migration and invasion by targeting CDK6. Int J Biochem Cell Biol 2020;119:105663. 10.1016/j.biocel.2019.105663 [DOI] [PubMed] [Google Scholar]
- 15.Lv L, Wang Q, Yang Y, et al. MicroRNA-495 targets Notch1 to prohibit cell proliferation and invasion in oral squamous cell carcinoma. Mol Med Rep 2019;19:693-702. [DOI] [PubMed] [Google Scholar]
- 16.Yang TB, Yi F, Liu WF, et al. Identification of hsa_circ_0039053 as an up-regulated and oncogenic circRNA in hepatocellular carcinoma via the miR-637-mediated USP21 activation. J Cancer 2020;11:6950-9. 10.7150/jca.48998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang W, Cao J, Peng X. LINC01234 facilitates growth and invasiveness of oral squamous cell carcinoma through regulating the miR-637/NUPR1 axis. Biomed Pharmacother 2019;120:109507. 10.1016/j.biopha.2019.109507 [DOI] [PubMed] [Google Scholar]
- 18.Mallo GV, Fiedler F, Calvo EL, et al. Cloning and expression of the rat p8 cDNA, a new gene activated in pancreas during the acute phase of pancreatitis, pancreatic development, and regeneration, and which promotes cellular growth. J Biol Chem 1997;272:32360-9. 10.1074/jbc.272.51.32360 [DOI] [PubMed] [Google Scholar]
- 19.Chowdhury UR, Samant RS, Fodstad O, et al. Emerging role of nuclear protein 1 (NUPR1) in cancer biology. Cancer Metastasis Rev 2009;28:225-32. 10.1007/s10555-009-9183-x [DOI] [PubMed] [Google Scholar]
- 20.Cano CE, Hamidi T, Sandi MJ, et al. Nupr1: the Swiss-knife of cancer. J Cell Physiol 2011;226:1439-43. 10.1002/jcp.22324 [DOI] [PubMed] [Google Scholar]
- 21.Yu J, Zhu H, Li R, et al. Oncogenic Role of NUPR1 in Ovarian Cancer. Onco Targets Ther 2020;13:12289-300. 10.2147/OTT.S262224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li X, Ma N, Zhang Y, et al. Circular RNA circNRIP1 promotes migration and invasion in cervical cancer by sponging miR-629-3p and regulating the PTP4A1/ERK1/2 pathway. Cell Death Dis 2020;11:399. 10.1038/s41419-020-2607-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang Y, Ci HS, Mao YL, et al. CircRNA_002178 promotes the proliferation and migration of oral squamous cell carcinoma cells by activating the Akt/mTOR pathway. Eur Rev Med Pharmacol Sci 2020;24:6122-30. [DOI] [PubMed] [Google Scholar]
- 24.Ai Y, Tang Z, Zou C, et al. circ_SEPT9, a newly identified circular RNA, promotes oral squamous cell carcinoma progression through miR-1225/PKN2 axis. J Cell Mol Med 2020;24:13266-77. 10.1111/jcmm.15943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo J, Su Y, Zhang M. Circ_0000140 restrains the proliferation, metastasis and glycolysis metabolism of oral squamous cell carcinoma through upregulating CDC73 via sponging miR-182-5p. Cancer Cell Int 2020;20:407. 10.1186/s12935-020-01501-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rui X, Xu Y, Jiang X, et al. Long non-coding RNA C5orf66-AS1 promotes cell proliferation in cervical cancer by targeting miR-637/RING1 axis. Cell Death Dis 2018;9:1175. 10.1038/s41419-018-1228-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang J, Liu WL, Zhang L, et al. MiR-637 suppresses melanoma progression through directly targeting P-REX2a and inhibiting PTEN/AKT signaling pathway. Cell Mol Biol (Noisy-le-grand) 2018;64:50-7. 10.14715/cmb/2018.64.11.10 [DOI] [PubMed] [Google Scholar]
- 28.Qiu L, Zheng L, Gan C, et al. circBICD2 targets miR-149-5p/IGF2BP1 axis to regulate oral squamous cell carcinoma progression. J Oral Pathol Med 2020. [Epub ahead of print]. doi:. 10.1111/jop.13156 [DOI] [PubMed] [Google Scholar]
- 29.Su W, Shen Y, Wang Y, et al. circPHIP promotes oral squamous cell carcinoma progression by sponging miR-142-5p and regulating PHIP and ACTN4 expression. Mol Ther Nucleic Acids 2020;23:185-99. 10.1016/j.omtn.2020.10.038 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 30.Emma MR, Iovanna JL, Bachvarov D, et al. NUPR1, a new target in liver cancer: implication in controlling cell growth, migration, invasion and sorafenib resistance. Cell Death Dis 2016;7:e2269. 10.1038/cddis.2016.175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grasso D, Garcia MN, Hamidi T, et al. Genetic inactivation of the pancreatitis-inducible gene Nupr1 impairs PanIN formation by modulating Kras (G12D)-induced senescence. Cell Death Differ 2014;21:1633-41. 10.1038/cdd.2014.74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen D, Kluz T, Fang L, et al. Hexavalent Chromium (Cr(VI)) Down-Regulates Acetylation of Histone H4 at Lysine 16 through Induction of Stressor Protein Nupr1. PLoS One 2016;11:e0157317. 10.1371/journal.pone.0157317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lopez MB, Garcia MN, Grasso D, et al. Functional characterization of Nupr1L, a novel p53-regulated isoform of the high-mobility group (HMG)-related protumoral protein Nupr1. J Cell Physiol 2015;230:2936-50. 10.1002/jcp.25022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang R, Yao F, Cheng C, et al. Expression and roles of As-NUPR1 protein from Artemia sinica during embryo development and in response to salinity stress. Mol Biol Rep 2014;41:3465-73. 10.1007/s11033-014-3208-4 [DOI] [PubMed] [Google Scholar]
- 35.Zhou C, Xu J, Lin J, et al. Long Noncoding RNA FEZF1-AS1 Promotes Osteosarcoma Progression by Regulating the miR-4443/NUPR1 Axis. Oncol Res 2018;26:1335-43. 10.3727/096504018X15188367859402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li A, Li X, Chen X, et al. NUPR1 Silencing Induces Autophagy-Mediated Apoptosis in Multiple Myeloma Cells Through the PI3K/AKT/mTOR Pathway. DNA Cell Biol 2020;39:368-78. 10.1089/dna.2019.5196 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as