Abstract
Background
Gallbladder cancer (GBC) is a highly aggressive biliary epithelial malignancy. The median survival time of GBC patients was less than 1 year. Tumor invasion and metastasis are the major cause of high mortality of GBC patients. However, the molecular mechanisms involved in GBC metastases are still unclear.
Methods
We performed 10X genomics single-cell RNA sequencing (scRNA-seq) on GBC liver metastasis tissue to evaluate the characteristics of the GBC liver metastasis microenvironment.
Results
In this study, 8 cell types, a total of 7,788 cells, including T cells, B cells, malignant cells, fibroblasts, endothelial cells, macrophages, dendritic cells (DCs), and mast cells were identified. Malignant cells displayed a high degree of intratumor heterogenicity, while neutrophils were found to promote GBC cell proliferation, migration, and invasion. Furthermore, cytotoxic cluster of differentiation (CD8+) T cells became exhausted and CD4+ regulatory T cells (Tregs) exhibited immunosuppressive characteristics. Macrophages played an important role in the tumor microenvironment (TME). We identified three distinct macrophage subsets and emergent M2 polarization. We also found that cancer-associated fibroblasts exhibited heterogeneity and may be associated with GBC metastasis.
Conclusions
Although preliminary in nature, our study provides a landscape view at the single-cell level. These results offer a unique perspective into understanding the liver metastasis of GBC.
Keywords: Gallbladder cancer (GBC), liver metastasis, single-cell transcriptome, tumor microenvironment (TME), heterogenicity
Introduction
Gallbladder cancer (GBC) is a common biliary tract tumor with extremely poor prognosis and high aggressiveness (1,2). The majority of GBC patients have already reached an advanced stage at the time of diagnosis, and only 10% of patients have access to curative surgery (3). Among those who undergo surgical resection, recurrence and metastasis rates remain high. Hepatic invasion and metastatic progression are common and represent a major cause of mortality in GBC patients (4,5). However, the mechanisms of GBC cell metastasis have not yet been elucidated.
The tumor microenvironment (TME) plays an important role in tumor metastasis (6). The GBC microenvironment consists of malignant cells, stromal cells, and extracellular components. Some factors such as tumor growth factor beta (TGFβ), interleukin 10 (IL-10), and vascular endothelial growth factor (VEGF) secreted by cellular elements in the TME contribute to desmoplasia, immunosuppression, and metastasis (7). The TME has become an attractive target for drug therapy strategies (8).
Single-cell RNA sequencing (scRNA-seq) provides a powerful tool to reveal cellular diversity and disclose microenvironment heterogeneity at a single-cell resolution (9,10). In recent years, scRNA-seq has been used to dissect the complicated TME of several cancers, such as hepatocellular carcinoma (11,12), intrahepatic cholangiocarcinoma (13), colorectal cancer (14,15), glioblastoma (16), and pancreatic ductal adenocarcinoma (17). However, microenvironment heterogeneity at the single-cell level in human GBC liver metastases is still poorly understood.
In this study, we used scRNA-seq to examine single cells from human GBC liver metastasis tissue and identified 8 cell types corresponding to 7,788 cells. Malignant cells were observed with intratumor heterogenicity while neutrophils were found to play an important role in GBC liver metastases. We proved that neutrophils facilitate GBC cell proliferation, migration, and invasion in vitro. Moreover, tumor-infiltrating regulatory T cells (Tregs) displayed immunosuppressive characteristics, while cytotoxic cluster of differentiation (CD)8+ T cells appeared to be exhausted. We also identified 3 macrophage subtypes. M2 polarization was common in the TME. Furthermore, RGS5+ cancer-associated fibroblasts showed a correlation with metastasis. Together, we hope this work can contribute to the better understanding of TME composition and promote GBC metastasis therapy.
We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/atm-21-2227).
Methods
Tumor sample collection
Human cancer tissue subjected to scRNA-seq was obtained from a female patient who was diagnosed as gallbladder adenosquamous carcinoma with liver metastasis. The H&E staining is shown in Figure S1. The tissue samples were collected by surgery with the approval of the institutional research ethics committee of the First Affiliated Hospital of Nanjing Medical University. All enrolled patients provided informed consent. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013).
Fresh tissue preparation and dissociation into single cells
The fresh tumor tissue was immediately washed with phosphate-buffered saline (PBS) and then conserved in MACS Tissue Storage Solution (Cat#130-100-008, Miltenyi Biotec) at 4 °C. Tissue digestion was performed according to the Tumor dissociation Kit user guide (Cat#130-093-237, Miltenyi Biotec). Cell suspensions were filtered using a 70-µm nylon mesh, and 10 µL of cell suspensions were counted by trypan blue to determine the concentration of live cells. The suspensions were then centrifuged at 300 ×g at 4 °C for 5 min, and the supernatant was discarded.
Library preparation and sequencing
We loaded viable cells that had undergone fluorescence-activated cell sorting (FACS) into a well of a microfluidic chip to generate a complementary DNA (cDNA) library using a Chromium Single-Cell Instrument (10X Genomics). Single-cell transcriptomic amplification and library preparation were performed by single-cell 3’ Reagent Kit v3 (10X Genomics) according to the manufacturer’s protocol. The library was loaded on an Illumina XTEN system.
Single cell sequencing data preprocessing, quality control, and subcluster detection
The CellRanger software (v.3.1.0, 10X Genomics) was used to process raw FASTQ files, align the sequencing reads to the GRCh38 reference, and generate a filtered feature-barcode matrix. The “Seurat” package in R (v3.1.5, R Foundation for Statistical Computing) (18) was used to analyze our data. Cells with over 10,000 genes and more than 25% of mitochondrial UMI counts were removed. The remaining high-quality cells were normalized and scaled. High variable genes were calculated using the FindVariableFeatures function in Seurat with default parameters. The selected features were used for principal components analysis (PCA) analysis. Clusters were found using the FindClusters function (resolution =0.8). t-distributed stochastic neighbor embedding (tSNE) analysis was used for dimension reduction and visualization of gene expression.
Cell type classification
We conducted tSNE dimensionality reduction with the first 50 principal components. Cell clusters were annotated to known biological cell types using canonical marker genes and the putative copy number variation (CNV) score.
CNV estimation
We used the “inferCNV” (v1.2.1) R package to estimate the CNV score for each single cell. All T cells served as the reference for inferCNV. The CNV score of each cell was calculated as a quadratic sum of the CNV region.
Estimation of the transcriptional diversity of each cell
CytoTRACE [Cellular (Cyto) Trajectory Reconstruction Analysis using gene Counts and Expression] is a computational framework used to predict the relative differentiation state of a single cell (19). We used R software (v.0.1.0) to predict the transcriptional diversity.
Pseudotime trajectory analysis
Trajectory analysis was performed using R package “Monocle 2” (v.2.14.0) (20). The differentialGeneTest function was used to identify significant genes (qval <0.01, num_cells_expressed >10). Cell ordering was then conducted on these genes. The trajectories were visualized using DDRTree method.
Cell-cell communication analysis
CellPhoneDB is a python-based tool used to analyze cell-cell communication at the molecular level (21). We used CellPhoneDB python package (v.2.1.1) to analyze the potential interaction networks of the 8 major cell types.
Gene regulatory network analysis
As described previously, we use “SCENIC” R package (v.1.1.2-2) to infer gene regulatory networks and cell types from single-cell RNA-seq data (22). Two gene-motif rankings including 10 kb around the transcription start site (TSS) and 500 bp upstream of the TSS were used to determine the search space around the TSS. A 20-thousand motif database was downloaded for RcisTarget and GENIE3 package.
Functional enrichment analysis
Differentially expressed genes (DEGs) were identified using the FindAllMarkers function of Seurat. The cutoff threshold values were used: adj.p.val <0.05, min.pct >0.25, and log fold change >0.25. R package clusterProfiler (23) was used to perform the Gene Ontology (GO) enrichment analysis. Pathways in which the adjusted P value <0.05 were considered significantly enriched. Gene set variation analysis (GSVA) was performed in the GSVA R package (v1.34.0) (24) to evaluate pathway activity of 50 hallmark gene sets.
Processing of GBC bulk RNA sequencing data
We downloaded the RNA sequencing data of GBC liver metastatic tumor, primary tumor, and adjacent tissues from GSE132223. STAR software was used to align the data to the hg38 reference genome. Fragments per kilobase million (FPKM) data were used for the further analysis.
Cell culture
The GBC cell lines GBC-SD (RRID: CVCL_6903) and SGC-996 (RRID: CVCL_M737) were purchased from the Procell Life Science & Technology (Wuhan, China). The cell lines were cultured in RPMI 1640 containing 2% fetal bovine serum (FBS).
Isolation and preparation of human neutrophils
Human neutrophils were isolated from the peripheral blood of healthy donors collected at the First Affiliated Hospital of Nanjing Medical University using a human neutrophils separation kit (Solarbio, Beijing, China). All donors signed he informed consent. Neutrophils were resuspended in RPMI 1640 containing 2% fetal FBS. Neutrophil-conditioned medium (NCM) was obtained as previously described (25).
Cell proliferation assay, and cell migration and invasion assays
The Cell Counting Kit-8 (CCK-8 Cell Counting Kit; Vazyme Biotech Co., Ltd) was used to assess the abilities of cell proliferation according to the manufacturer’s protocol. In brief, we planted cell lines with or without NCM into 96-well plates (2,000 cells/well) with the addition of CCK-8 solution (10 µL/well). After incubation for 2 hours in the incubator described above, the absorbance at 450 nm was measured to determine the number of variable cells in each well.
Cell migration was assessed through the scratch wound assay. We made a uniform linear scratch using a 10-µL pipet tip on the cell lines which had been cultured with or without NCM for 2 days. The distance between the wound edges was taken as a baseline. We then acquired the image of the same location after 24 hours. The change of the distance was measured to assess the cellular ability of migration.
Next, 24-well Transwell chambers (Millipore) were used to evaluate the invasive capabilities of the cells. In total, 1×105 cells were seeded in the upper chamber. The lower chamber was filled with 100 µL RPMI 1640 with 2% FBS or NCM. After incubation for 24 hours, the cells in the upper chamber were removed using cotton swabs. Cells on the lower surface of membrane were fixed and stained with crystal violet. The GBC cells were visualized under an inverted light microscope (Olympus), photographed, and counted in 5 microscopic fields. All experiments were performed 3 times.
Immunohistochemistry (IHC) staining
We performed IHC staining on the tissue sections of formalin-fixed, paraffin-embedded specimens. All sections were deparaffinized, rehydrated, and washed. Endogenous peroxidase was blocked using 3% hydrogen peroxide for 10 min. After water-bath heating was performed for antigen retrieval, slides were incubated with primary antibodies, which was followed by horseradish peroxidase (HRP)-linked secondary antibodies and diaminobenzidine staining. Counterstaining was done with hematoxylin. Slides were dehydrated with sequential ethanol washes (75%, 80%, and 100%) for 1 min each. Two pathologists blinded to clinical data independently assessed staining results for LAG3 (Cat # 16616-1-AP, RRID: AB_2133350; Proteintech), and CD163 (Cat #16646, RRID: AB_2756528; Proteintech).
Statistical analysis
All statistical analyses were performed using R platform (v.3.6.0). A P value <0.05 was considered statistically significant. Statistical differences between multiple groups were compared using Student’s t test or analysis of variance (ANOVA).
Results
Single cell transcriptomic analysis revealed cellular diversity in GBC liver metastasis
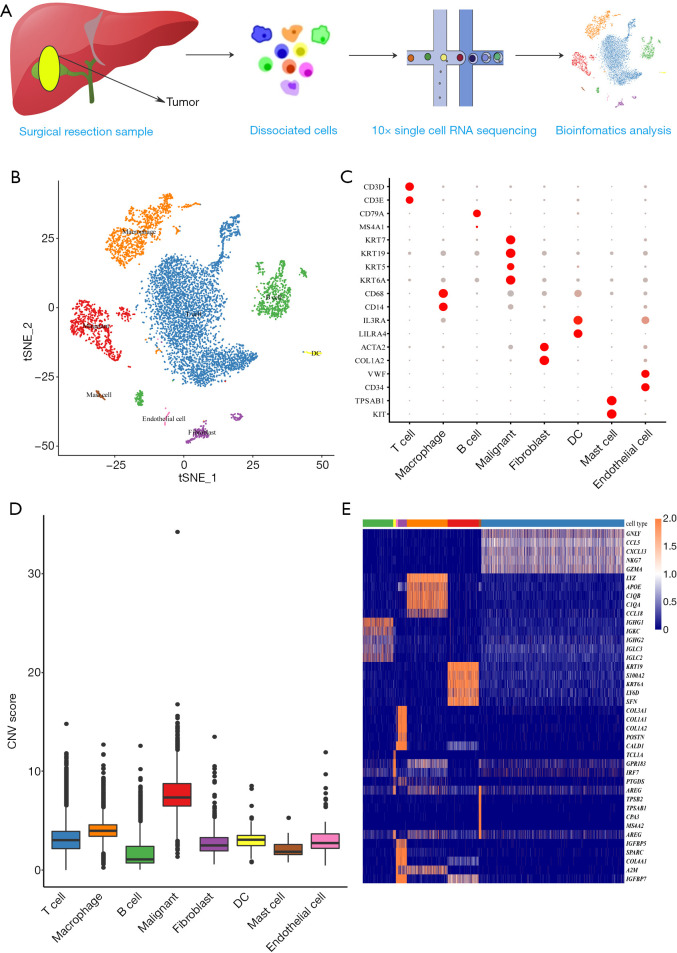
To explore the potential mechanisms and microenvironment of GBC liver metastasis, we generated a single-cell transcriptomic profile from GBC liver metastasis tissue using 10X Genomics (Figure 1A). A total of 7,788 cells passed quality control, and 8 distinct cell types were identified (Figure 1B and Figure S2) using the following conventional markers: malignant cells (925 cells, marked with KRT7 and KRT19 (markers of adenocarcinoma), KRT5 and KRT6A (markers of squamous cell carcinoma); T cells (4,258 cells, marked with CD2, CD3D, CD3E, and CD3G); B cells (907 cells, marked with CD79A and MS4A1); macrophages (1,214 cells; marked with CD68, CD163, and CD14); fibroblasts (264 cells, marked with ACTA2 and COL1A2); dendritic cells (DCs) (79 cells, marked with IL3RA and LILRA4); endothelial cells (64 cells, marked with VWF, ENG, and CD34) and mast cells (77 cells, marked with KIT and TPSAB1). The dot plots of marker genes are shown in Figure 1C. Further CNV analysis was conducted to distinguish malignant from nonmalignant cells (Figure 1D and Figure S3). The DEGs in 8 cell types, as shown in Figure 1E, confirmed the accuracy of cell recognition.
Figure 1.
The overview of cell type distribution in GBC liver metastasis. (A) The workflow of this scRNA-seq study. Metastatic liver tissue was dissociated into single cells and sequenced with 10X platform. (B) The tSNE plot of 8 cell types in this study and each color indicating an associated cell type. (C) Dot plots showing the expression of marker genes in 8 distinct cell types. (D) Box plots of CNV score for each cell type. (E) Heatmap showing the top 5 DEGs in each subtype. GBC, gallbladder cancer; CNV, copy number variation; DEGs, differentially expressed genes.
Transcriptomic intratumor heterogeneity of tumor cells in GBC liver metastasis
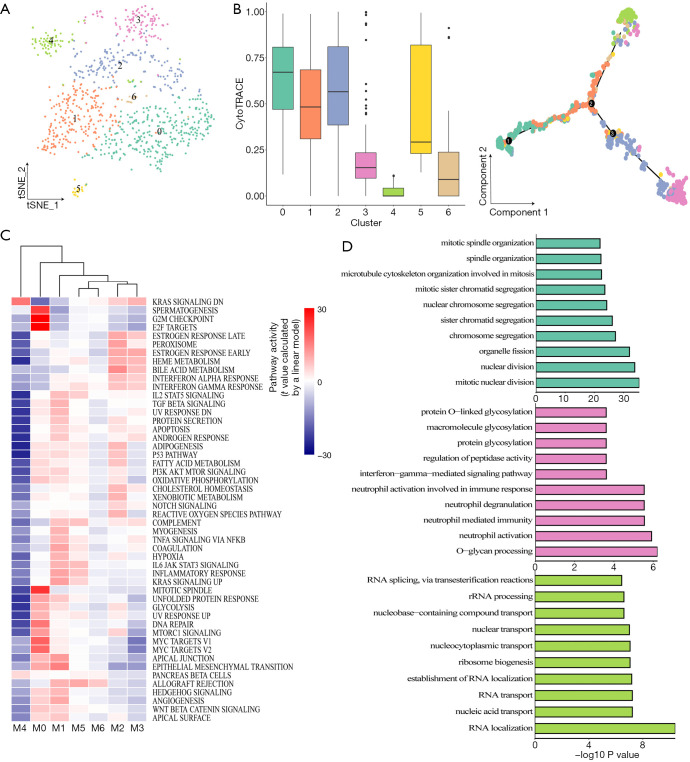
In this study, we identified 7 subclusters after the reclustering of malignant cells (Figure 2A). To investigate the heterogeneity of tumor cells, we used CytoTRACE to evaluate the transcriptional diversity of each cell, which is a hallmark of developmental potential. As shown in Figure 2B (left), the CytoTRACE score was diversely distributed, indicating that tumor cells were in different differentiation states. Monocle 2 was used to construct a trajectory. Consistent with the CytoTRACE result, we noticed that the trajectory’s root was populated by the majority of M0 cells (high CytoTRACE score). M3 and M4 cells (low CytoTRACE score) were present at the end of 2 differentiation branches [Figure 2B (right) and Figure S4A]. In addition, pathway analysis with GSVA revealed that hallmark pathways were partially activated in subpopulations M0, M1, or M2 with higher CytoTRACE scores, particularly, the M4 subcluster had the lowest GSVA score (Figure 2C). We conducted gene function enrichment analysis on DEGs of M0, M3, and M4 subclusters. As shown in Figure 2D and Figure S4B, G2M/S cell cycle-related genes were enriched in M0. In M3, neutrophil-associated immune response and interferon gamma (IFNγ) signaling were commonly observed. Furthermore, the M4 cluster was mainly correlated with nucleocytoplasmic transport pathways. In M3, M2, and M5 macrophages, neutrophil-related gene functions were enriched, suggesting that there was abundant crosstalk between neutrophils and cancer cells during the metastasis of GBC (Figure S4C,D,E,F). Different functional patterns of tumor cells may reflect diverse processes of the metastatic seeds adapted to the soil. Taken together, these findings indicated a high degree of intratumoral heterogeneity in malignant cells of GBC metastasis.
Figure 2.
Transcriptomic heterogeneity of malignant cells in metastatic liver tissue. (A) tSNE map showing 7 cancer cell clusters. (B) Boxplot showing transcriptional diversity of cancer cells estimated by CytoTRACE (left) and differentiation trajectory of cancer cells in GBC liver metastasis (right). (C) Differences in the activity of 50 hallmark pathways scored by GSVA software. t values were calculated by a linear model. (D) GO analysis of DEGs for cluster 0 (top), cluster 3 (middle), and cluster 4 (bottom). FDR <0.05 was considered as significantly enriched. GBC, gallbladder cancer.
Neutrophils promoted proliferation, migration, and invasion of GBC cells in vitro
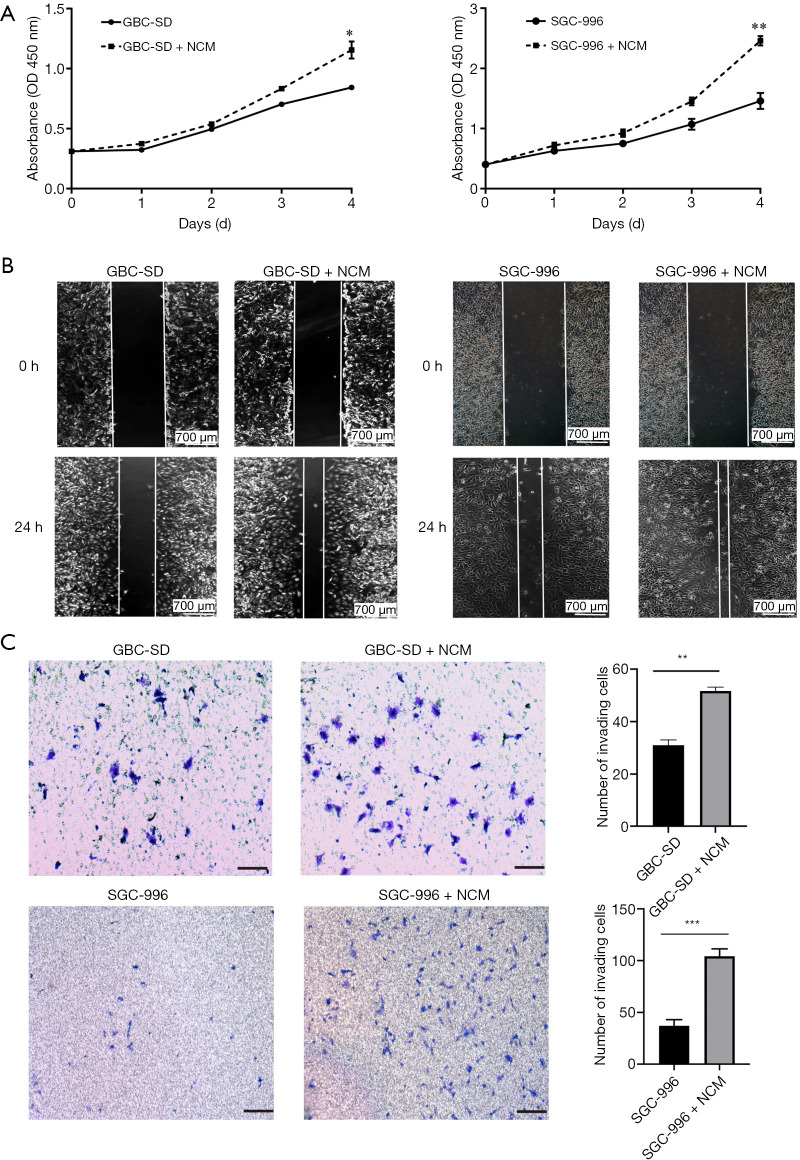
Significant neutrophil infiltration has previously been observed in human cancer metastatic sites. To examine the effect of neutrophils on GBC cells, we incubated the GBC cell lines (GBC-SD and SGC-996) with NCM. CCK-8 proliferation analysis showed that NCM promoted GBC cell growth in vitro (Figure 3A). Wound-healing migration assays and cell invasion assays also demonstrated that NCM significantly enhanced the migration and invasion abilities of GBC cells (Figure 3B,C).
Figure 3.
Neutrophils promoted proliferation, migration, and invasion of GBC cells in vitro. (A) Cell proliferation measured by CCK8 assay. NCM significantly promoted GBC proliferation. (B) Wound-healing migration assays showed that there was a significant increase in the wound closure rate of GBC cells cocultured with NCM compared with the controls. (C) In vitro invasive assays showed that the number of invasive GBC cells was higher when cells were cultured with NCM than with the control. Cells were fixed and stained with crystal violet. Scale bar =100 µm. **P<0.01, ***P<0.001. GBC, gallbladder cancer; NCM, neutrophil-conditioned medium.
Tumor-infiltrating Tregs and exhausted CD8+ T cells contributed to the immunosuppressive metastatic microenvironment
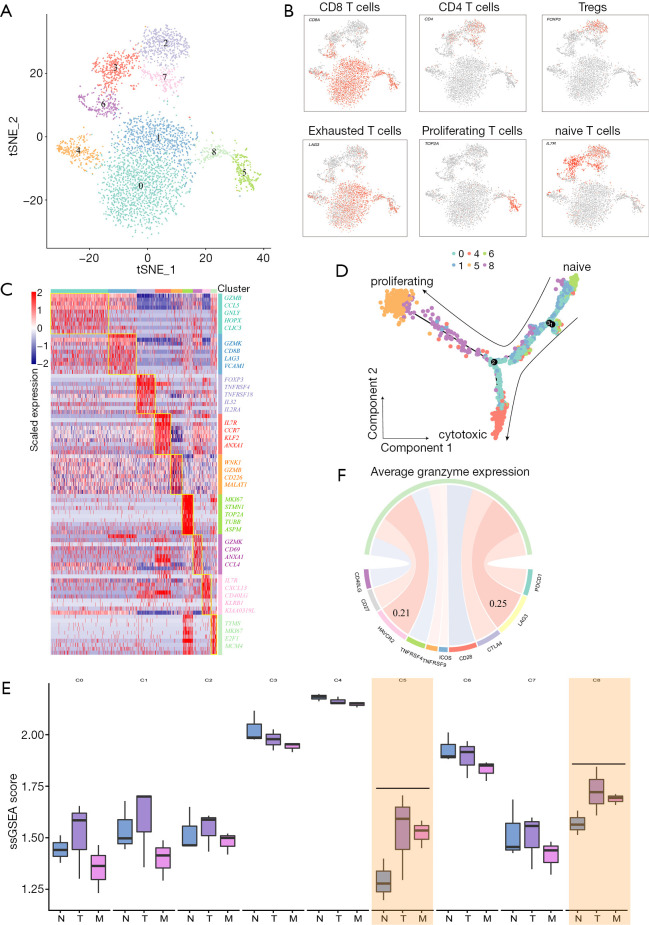
T cells were found to be the most prevalent cell type in the TME of liver metastasis tumor. It was found that the tumor-infiltrating immune cells played an important role in response to immunotherapy. As shown in Figure 4A, T cells were clustered into 9 distinct subgroups. According to the known markers of T cells, 9 subclusters were annotated as T0-CD8T-GZMB (CD8A+GZMB+, cluster 0 and 4), T1-CD8T-GZMK (CD8A+GZMK+, cluster 1), T2-Treg-FOXP3 (CD4+ FOXP3+, cluster 2), T3-CD4T-naïve (CD4+IL7R+, cluster 3, 7), T4-CD8T-proliferation (CD8A+MKI67+, cluster 5, 8), and T5-CD8T- naïve (CD8A+IL7R+, cluster 6; Figure 4B,C). In this study, we found that CD8+ T cells (cluster 0, 1, 4, 5, 6, 8) expressed different levels of cytotoxic markers, including GZMK, IFNG, and PRF1. In proliferating CD8+ T cell populations (cluster 5 and 8), a set of T cell exhaustion-related immune checkpoints including TIGIT, LAG3, HAVCR 2 (TIM3), and CTLA4 tended to be upregulated, indicating that the CD8+ T cells became dysfunctional (Figure S5A). We used Monocle 2 package to perform pseudotime analysis on CD8+ T cells. The trajectory was visualized as a DDRTree plot (Figure 4D). We noticed that naïve T cells tended to convert into proliferating CD8+ T cell subgroups that had become dysfunctional, suggesting that cytotoxic CD8+ T cells were being exhausted. In addition, Tregs (cluster 2) expressed a certain number of immunosuppression markers, including TIGIT, CTLA4, and TNFRSF18 (GITR). We extracted the gene signatures of T cell subclusters from scRNA-seq data. Through single-sample gene set enrichment analysis (ssGSEA) algorithm, we evaluated the infiltration of T cell subclusters in GSE132223. As shown in Figure 4E, the percentage of proliferating and exhausted CD8+ T cells was also significantly higher in primary and metastatic tumors than in adjacent tissues. Interestingly, the expression of LAG3 was found to be highly correlated with the activity of dysfunctional CD8+ T cells as was measured by the average expression of granzymes (GZMA, GZMB and GZMH; Spearman correlation R =0.25; Figure 4F). The IHC results also indicated higher LAG3 expression in tumor tissues (Figure S5B). Taken together, our analyses demonstrated that in the GBC metastatic TME, cytotoxic T cells tend to be exhausted and Tregs presented as a highly immunosuppressive phenotype. LAG3 may be a potential treatment target for patients with GBC liver metastasis.
Figure 4.
Distinct subpopulations of infiltrating T cells in the metastatic TME. (A) tSNE plot for 9 T cell subclusters. (B) t-SNE plots showing the expression of marker genes for each cell type. (C) Heatmap showing the expression of the top 10 DEGs in each cluster. (D) Differentiation trajectory of cytotoxic CD8+ T cells. (E) Box plots showing signature scores of T cell subclusters in GBC primary tumors, liver metastatic tumors, and adjacent tissues. (F) Spearman correlation between the activity of CD8+ T cells, as measured by the mean granzyme expression (GZMA, GZMB, and GZMH) and known immune checkpoint molecules. TME, tumor microenvironment; DEGs, differentially expressed genes; GBC, gallbladder cancer.
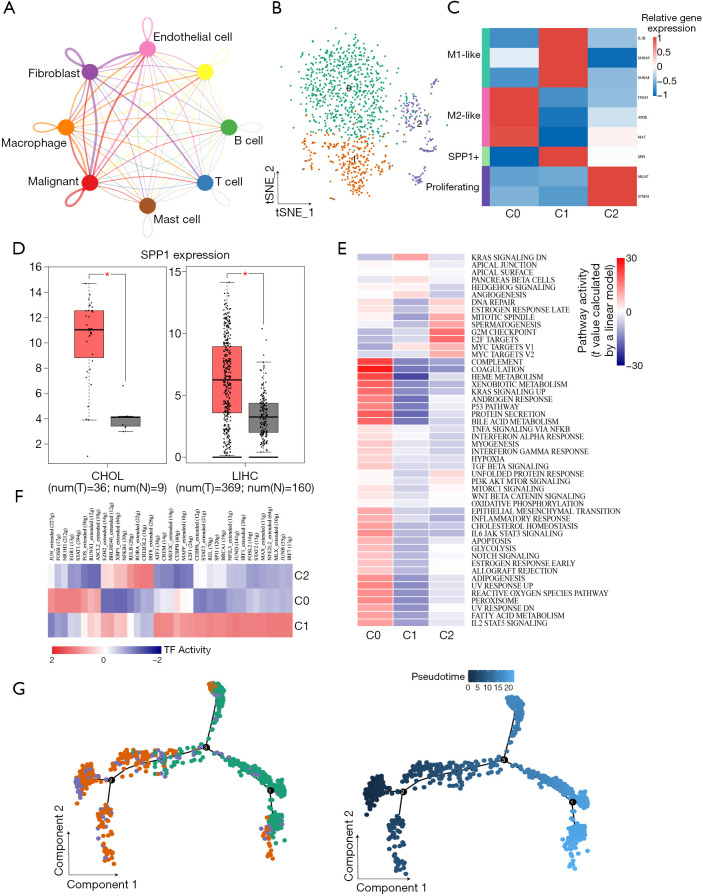
Macrophages exhibited greater crosstalk with malignant cells and tended to generate M2-type polarization
We used CellPhoneDB to infer intercellular communication in the TME. Macrophages were found to have more interactions with tumor cells, revealing an important role in the microenvironment (Figure 5A). To investigate the heterogeneity of macrophages, we clustered macrophages into 3 subclusters (Figure 5B). As shown in Figure 5C, subcluster 0 cells highly expressed M2-type macrophage markers including CD163, APOE, and MAF. Cells from cluster 1 exhibited the M1-like phenotype. SPP1, also known as secreted phosphoprotein 1, was also expressed in subcluster 1. For the lack of large-cohort GBC gene expression data, we checked the expression of SPP1 in liver hepatocellular carcinoma (LIHC) and cholangiocarcinoma (CHOL). SPP1 was highly expressed in tumor tissues (Figure 5D) and correlated with overall survival in LIHC (Figure S6A) and disease-free survival in CHOL (Figure S6B). In addition, MKI67 and STMN1, which correlated with the cell cycle process, were expressed by cells from subcluster 2. GSVA revealed that interferon (pathways were enriched in subcluster 0; however, the inflammatory response was also activated (Figure 5E). SCENIC analysis showed that genes regulated by FOS, JUN, STAT1, ERG1, and RUNX1 were upregulated in subcluster 0 (Figure 5F). Fos/Jun TFs has been reported to be able to enhance inflammatory responses in macrophages. Meanwhile, IRF7, which was activated in macrophage subcluster 1, participates in the regulation of the M1-to-M2 phenotype switch (26). Therefore, M2-like macrophages maintained some of their proinflammatory functions. Furthermore, trajectory analysis showed that M2-like macrophages were present at the end of the differentiation trajectory (Figure 5G). Further ligand-receptor analysis between malignant cells and macrophages showed that the TNFSF10-TNFRSF10D pair was enriched in the M2-like subcluster (Figure S6C), suggesting that blocking the TNFSF10-TNFRSF10D axis may affect the interaction of malignant cells with M2 macrophages. We adopted the same method as above to check the infiltration of each macrophage cluster. As shown in Figure S6D, the M2-like macrophage signature score was also higher in tumor sites. The same phenomenon was also observed in IHC results (Figure S6E), indicating that the phenomenon of M2-type polarization in GBC was common.
Figure 5.
M2 polarization of macrophages in the TME. (A) Interaction network of 8 cell types in the TME constructed by CellPhoneDB. At thicker line indicates greater interaction. (B) tSNE plot of 3 subclusters of macrophages. (C) The average expression of canonical marker genes for macrophage subpopulations. (D) SPP1 expression in the TCGA LIHC cohort and CHOL cohort. (E) Heatmap of differences in 50 hallmark pathways scored per cell by GSVA. (F) Heatmap showing the AUC scores of expression regulation by transcription factors estimated by SCENIC. (G) Differentiation trajectory of macrophages with each color coded for subclusters (left) and pseudotime (right). TME, tumor microenvironment; AUC, area under the curve.
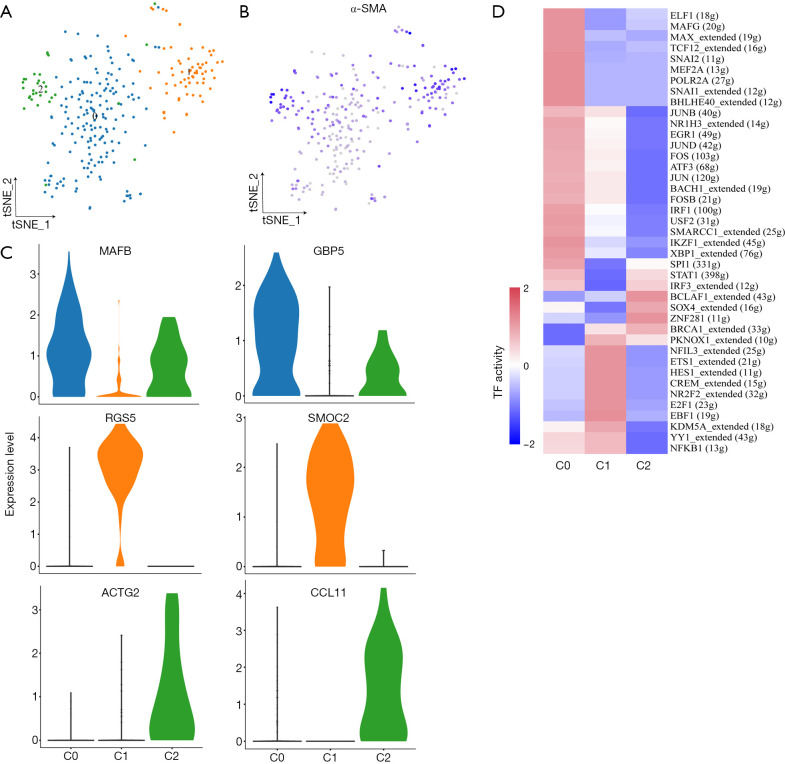
Fibroblasts exhibited heterogeneity and expressed markers correlated with cancer invasion or metastasis
As shown in Figure 6A,B, cancer-associated fibroblasts (CAFs) from three subgroups were positive for alpha smooth muscle actin (α-SMA; ACTA2) which is a classical marker of fibroblasts. These 3 subgroups expressed different markers (cluster 0: MAFB+ and GPB5+; cluster 1: RGS5+and SMOC2+; cluster 2: ACTG2+and CCL11+; Figure 6C). The Gene Ontology (GO) analysis results of DEGs in 3 distinct subgroups are shown in Figure S7. RGS5, a marker for vascular CAFs (vCAFs), has been reported to be related to invasion and metastasis of cancers such as liver cancer and lung cancer by inducing epithelial-mesenchymal transition (EMT) (27,28). The SCENIC analysis revealed that genes regulated by TF NR2F2 and NFIL3 were upregulated in cells from the RGS5+ fibroblasts subgroup (Figure 6D). Overall, our data suggest that RGS5+ fibroblasts are associated with the tumorigenesis, invasion, and metastasis of GBC.
Figure 6.
Three distinct fibroblast subpopulations detected in GBC liver metastasis. (A,B) tSNE plot of 3 fibroblast subpopulations (A) and the expression levels of α-SMA (ACTA2) (B). (C) Violin plots showing marker genes of 3 subclusters. (D) Differences of AUC scores in expression regulated by transcription factors. GBC, gallbladder cancer.
Discussion
GBC is a highly aggressive and lethal tumor (29). Currently, the therapy of GBC, especially metastatic GBC, is still a challenge (2,3,29). Only a few patients can benefit from curative resection, and even then, the prognosis is poor. The mechanisms underlying GBC metastasis are little known. Several studies have shown that TME participates in tumor progression and metastasis (30,31). Disrupting the protumorigenic TME is a promising therapeutic target for cancer patients. In this study, we employed scRNA-seq to delineate the transcriptomic heterogeneity of the TME in GBC liver metastasis at a single-cell resolution.
In this work, several key observations arose from examining the intratumoral heterogeneity of the TME in GBC liver metastasis. First, GBC cells exhibited high heterogeneity at the metastatic site. The existence of cell subpopulations in distinct differentiation states fully reflects the complexity and diversity of GBC in its evolution. In previous studies, neutrophil-to-lymphocyte ratio (NLR) showed prognostic significance in GBC patients (32) and increasing neutrophil levels have a significant influence on the TME by inducing various cytokines and chemokines, which could accelerate the proliferation and promote metastasis of tumor cells (33,34). Thus, targeting neutrophils might become a new direction for the treatment of GBC metastasis. By using pseudotime analysis and GO analysis, we found that neutrophil-associated gene ontology was enriched in the intermediate stage of the differentiation trajectory, indicating that tumor cells communicate extensively with immune cells at the intermediate stage of metastasis. We also confirmed the influence of neutrophils on GBC cells in vitro. The evidence from our study suggests that targeting tumor-associated neutrophils is a promising method to antitumor metastasis for GBC patients.
Second, the high expression of TIGIT, CTLA4, LAG3, and HAVCR2 in different T cells indicated a tumor-suppressive TME in GBC metastasis. Tregs are immunosuppressive T cells that participate in immune escape and block the antitumor immune response of cancer patients (35). A set number of Tregs in GBC liver metastasis was observed. The infiltrating level of Tregs is correlated with the disease progression of GBC patients (36,37). Additionally, LAG3, known as an exhausted marker, was found to be correlated with CD8+ T cell activity, suggesting that LAG3 is the most prominent immune checkpoint molecule in GBC metastasis. Akhilesh Pandey et al. found that a group of GBC patients had a high infiltration of CD8+ T cells and a high expression of LAG3 (38). LAG3 may serve as a better immunotherapeutic target in GBC patients with higher infiltration of CD8+ T cells.
Third, 3 types of macrophages were identified from the TME. M1-like macrophages exhibited the CD68+CD163+SPP1+ phenotype while M2-like macrophages exhibited the CD68+IL1B+S100A8+ phenotype. It has been reported that proinflammatory molecules, such as IFNγ and IL-1β could induce SPP1 expression, and these have been found to play an important role in colon cancer liver metastasis in previous studies (39). Tumor necrosis factor (TNF)-related apoptosis-inducing ligand (TRAIL, TNFSF10) and its receptors are key mediators of the innate immune response in tumor immune surveillance. One study showed that TRAIL-induced secretome could drive monocyte polarization to myeloid-derived suppressor cells (MDSCs) and M2-like macrophages (40). Targeting TRAIL is a promising method to break the immunosuppressive microenvironment in GBC metastatic sites. Using GSVA pathway analysis and SCENIC analysis, we revealed some underlying transcription factors (TFs) and potential mechanisms involved in the M2 polarization phenomenon. Furthermore, transcription factor IRF8 was also found activated in the proliferative macrophage population. IRF8 has been shown to suppress the macrophage-derived matricellular protein, osteopontin, thus making it a novel immunosuppressive checkpoint (41). Osteopontin also controls immunosuppression in the TME (42). Interestingly, M1-like macrophages highly expressed secreted phosphoprotein 1, indicating that SPP1+ M1-like macrophages are correlated with immunosuppression and may be associated with immunotherapy outcomes for patients with GBC metastasis.
Finally, several TFs related to metastasis were found to be activated in RGS5+ fibroblasts via SCENIC analysis. Previous studies have shown that NR2F2 (COUP-TFII) is a transcription factor closely associated with tumorigenesis, invasion, and metastasis in several digestive cancer such as pancreatic (43), colorectal (44), and gastric cancer (45). NFIL3, also known as E4-binding protein 4 (E4BP4), is a leucine zipper transcription factor in the immune system. Lin et al. found that cellular prion protein transcriptionally regulated by NFIL3 promotes lung cancer invasion and metastasis (46). A study indicated that RGS5+ CAFs (vCAFs) contribute to tumor progression in intrahepatic cholangiocarcinoma (13). The data supported the notion that RGS5+ CAFs is also a key factor associated with GBC metastasis.
Taken together, our findings provide a landscape view of the single-cell expression profiles in GBC liver metastasis and clarify the characteristics of certain cell subpopulations. However, some limitations to our study should be mentioned. Most importantly, the tissue sample sent for scRNA-seq was adenosquamous carcinoma, which is a rare subtype of GBC. Due to a lack of squamous cancer cell lines, we validated our results in gallbladder adenocarcinoma cell lines and clinical samples. Although we have proved the accuracy of our result in human cell lines and samples, these results still need to be further validated in scRNA-seq data of other types of GBC samples. We hope our study can help address microenvironment-mediated GBC progression and drug resistance and contribute to identifying novel therapeutic targets for GBC metastasis.
Supplementary
The article’s supplementary files as
Acknowledgments
Funding: This work was supported by funds from the National Natural Science Foundation of China (No. 81971495, 81571564, and 81522020), CAMS Innovation Fund for Medical Sciences (No. 2019-I2M-5-035), the National Science Foundation of Jiangsu Province (No. BRA2017533, BK20131024, and BE2016766), and the 863 Young Scientists Special Fund (No. SS2015AA0209322).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the institutional research ethics committee of the First Affiliated Hospital of Nanjing Medical University (No. 2019-SR-127) and informed consent was taken from all the patients.
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/atm-21-2227
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-21-2227
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-21-2227). LL serves as an unpaid associate editor-in-chief of Annals of Translational Medicine from June 2019 to May 2024. The other authors have no conflicts of interest to declare.
(English Language Editor: J. Gray)
References
- 1.Siegel RL, Miller KD, Fuchs HE, et al. Cancer Statistics, 2021. CA Cancer J Clin 2021;71:7-33. 10.3322/caac.21654 [DOI] [PubMed] [Google Scholar]
- 2.Kanthan R, Senger JL, Ahmed S, et al. Gallbladder Cancer in the 21st Century. J Oncol 2015;2015:967472. 10.1155/2015/967472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhu AX, Hong TS, Hezel AF, et al. Current management of gallbladder carcinoma. Oncologist 2010;15:168-81. 10.1634/theoncologist.2009-0302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wullstein C, Woeste G, Barkhausen S, et al. Do complications related to laparoscopic cholecystectomy influence the prognosis of gallbladder cancer? Surg Endosc 2002;16:828-32. 10.1007/s00464-001-9085-7 [DOI] [PubMed] [Google Scholar]
- 5.Bartlett DL, Fong Y, Fortner JG, et al. Long-term results after resection for gallbladder cancer. Implications for staging and management. Ann Surg 1996;224:639-46. 10.1097/00000658-199611000-00008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nat Med 2013;19:1423-37. 10.1038/nm.3394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ren B, Cui M, Yang G, et al. Tumor microenvironment participates in metastasis of pancreatic cancer. Mol Cancer 2018;17:108. 10.1186/s12943-018-0858-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roma-Rodrigues C, Mendes R, Baptista PV, et al. Targeting Tumor Microenvironment for Cancer Therapy. Int J Mol Sci 2019;20:840. 10.3390/ijms20040840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Levitin HM, Yuan J, Sims PA. Single-Cell Transcriptomic Analysis of Tumor Heterogeneity. Trends Cancer 2018;4:264-8. 10.1016/j.trecan.2018.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baslan T, Hicks J. Unravelling biology and shifting paradigms in cancer with single-cell sequencing. Nat Rev Cancer 2017;17:557-69. 10.1038/nrc.2017.58 [DOI] [PubMed] [Google Scholar]
- 11.Ma L, Hernandez MO, Zhao Y, et al. Tumor Cell Biodiversity Drives Microenvironmental Reprogramming in Liver Cancer. Cancer Cell 2019;36:418-30.e6. 10.1016/j.ccell.2019.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Q, He Y, Luo N, et al. Landscape and Dynamics of Single Immune Cells in Hepatocellular Carcinoma. Cell 2019;179:829-45.e20. 10.1016/j.cell.2019.10.003 [DOI] [PubMed] [Google Scholar]
- 13.Zhang M, Yang H, Wan L, et al. Single-cell transcriptomic architecture and intercellular crosstalk of human intrahepatic cholangiocarcinoma. J Hepatol 2020;73:1118-30. 10.1016/j.jhep.2020.05.039 [DOI] [PubMed] [Google Scholar]
- 14.Zhang Y, Song J, Zhao Z, et al. Single-cell transcriptome analysis reveals tumor immune microenvironment heterogenicity and granulocytes enrichment in colorectal cancer liver metastases. Cancer Lett 2020;470:84-94. 10.1016/j.canlet.2019.10.016 [DOI] [PubMed] [Google Scholar]
- 15.Li H, Courtois ET, Sengupta D, et al. Reference component analysis of single-cell transcriptomes elucidates cellular heterogeneity in human colorectal tumors. Nat Genet 2017;49:708-18. 10.1038/ng.3818 [DOI] [PubMed] [Google Scholar]
- 16.Patel AP, Tirosh I, Trombetta JJ, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 2014;344:1396-401. 10.1126/science.1254257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peng J, Sun BF, Chen CY, et al. Single-cell RNA-seq highlights intra-tumoral heterogeneity and malignant progression in pancreatic ductal adenocarcinoma. Cell Res 2019;29:725-38. 10.1038/s41422-019-0195-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stuart T, Butler A, Hoffman P, et al. Comprehensive Integration of Single-Cell Data. Cell 2019;177:1888-902.e21. 10.1016/j.cell.2019.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gulati GS, Sikandar SS, Wesche DJ, et al. Single-cell transcriptional diversity is a hallmark of developmental potential. Science 2020;367:405-11. 10.1126/science.aax0249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qiu X, Hill A, Packer J, et al. Single-cell mRNA quantification and differential analysis with Census. Nat Methods 2017;14:309-15. 10.1038/nmeth.4150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Efremova M, Vento-Tormo M, Teichmann SA, et al. CellPhoneDB: inferring cell-cell communication from combined expression of multi-subunit ligand-receptor complexes. Nat Protoc 2020;15:1484-506. 10.1038/s41596-020-0292-x [DOI] [PubMed] [Google Scholar]
- 22.Aibar S, Gonzalez-Blas CB, Moerman T, et al. SCENIC: single-cell regulatory network inference and clustering. Nat Methods 2017;14:1083-6. 10.1038/nmeth.4463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu G, Wang LG, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 2012;16:284-7. 10.1089/omi.2011.0118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics 2013;14:7. 10.1186/1471-2105-14-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou SL, Dai Z, Zhou ZJ, et al. CXCL5 contributes to tumor metastasis and recurrence of intrahepatic cholangiocarcinoma by recruiting infiltrative intratumoral neutrophils. Carcinogenesis 2014;35:597-605. 10.1093/carcin/bgt397 [DOI] [PubMed] [Google Scholar]
- 26.Cohen M, Matcovitch O, David E, et al. Chronic exposure to TGFbeta1 regulates myeloid cell inflammatory response in an IRF7-dependent manner. EMBO J 2014;33:2906-21. 10.15252/embj.201489293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hu M, Chen X, Zhang J, et al. Over-expression of regulator of G protein signaling 5 promotes tumor metastasis by inducing epithelial-mesenchymal transition in hepatocellular carcinoma cells. J Surg Oncol 2013;108:192-6. 10.1002/jso.23367 [DOI] [PubMed] [Google Scholar]
- 28.Xu Z, Zuo Y, Wang J, et al. Overexpression of the regulator of G-protein signaling 5 reduces the survival rate and enhances the radiation response of human lung cancer cells. Oncol Rep 2015;33:2899-907. 10.3892/or.2015.3917 [DOI] [PubMed] [Google Scholar]
- 29.Misra S, Chaturvedi A, Misra NC, et al. Carcinoma of the gallbladder. Lancet Oncol 2003;4:167-76. 10.1016/S1470-2045(03)01021-0 [DOI] [PubMed] [Google Scholar]
- 30.Brassart-Pasco S, Brezillon S, Brassart B, et al. Tumor Microenvironment: Extracellular Matrix Alterations Influence Tumor Progression. Front Oncol 2020;10:397. 10.3389/fonc.2020.00397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eble JA, Niland S. The extracellular matrix in tumor progression and metastasis. Clin Exp Metastasis 2019;36:171-98. 10.1007/s10585-019-09966-1 [DOI] [PubMed] [Google Scholar]
- 32.Zhang L, Wang R, Chen W, et al. Prognostic significance of neutrophil to lymphocyte ratio in patients with gallbladder carcinoma. HPB (Oxford) 2016;18:600-7. 10.1016/j.hpb.2016.03.608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gregory AD, Houghton AM. Tumor-associated neutrophils: new targets for cancer therapy. Cancer Res 2011;71:2411-6. 10.1158/0008-5472.CAN-10-2583 [DOI] [PubMed] [Google Scholar]
- 34.Liu K, Wang FS, Xu R. Neutrophils in liver diseases: pathogenesis and therapeutic targets. Cell Mol Immunol 2021;18:38-44. 10.1038/s41423-020-00560-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Togashi Y, Shitara K, Nishikawa H. Regulatory T cells in cancer immunosuppression - implications for anticancer therapy. Nat Rev Clin Oncol 2019;16:356-71. 10.1038/s41571-019-0175-7 [DOI] [PubMed] [Google Scholar]
- 36.Patil RS, Shah SU, Shrikhande SV, et al. IL17 producing gammadeltaT cells induce angiogenesis and are associated with poor survival in gallbladder cancer patients. Int J Cancer 2016;139:869-81. 10.1002/ijc.30134 [DOI] [PubMed] [Google Scholar]
- 37.Ozgur HH, Ercetin AP, Eliyatkin N, et al. Regulatory T cells and their prognostic value in hepatopancreatobiliary tumours. Hepatogastroenterology 2014;61:1847-51. [PubMed] [Google Scholar]
- 38.Pandey A, Stawiski EW, Durinck S, et al. Integrated genomic analysis reveals mutated ELF3 as a potential gallbladder cancer vaccine candidate. Nat Commun 2020;11:4225. 10.1038/s41467-020-17880-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee HO, Hong Y, Etlioglu HE, et al. Lineage-dependent gene expression programs influence the immune landscape of colorectal cancer. Nat Genet 2020;52:594-603. 10.1038/s41588-020-0636-z [DOI] [PubMed] [Google Scholar]
- 40.Hartwig T, Montinaro A, von Karstedt S, et al. The TRAIL-Induced Cancer Secretome Promotes a Tumor-Supportive Immune Microenvironment via CCR2. Mol Cell 2017;65:730-42.e5. 10.1016/j.molcel.2017.01.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Klement JD, Paschall AV, Redd PS, et al. Repression of osteopontin by IRF8 regulates a novel immunosuppressive checkpoint. J Immunol 2019;202:195.26.
- 42.Shurin MR. Osteopontin controls immunosuppression in the tumor microenvironment. J Clin Invest 2018;128:5209-12. 10.1172/JCI124918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Polvani S, Tarocchi M, Tempesti S, et al. COUP-TFII in pancreatic adenocarcinoma: clinical implication for patient survival and tumor progression. Int J Cancer 2014;134:1648-58. 10.1002/ijc.28502 [DOI] [PubMed] [Google Scholar]
- 44.Bao Y, Gu D, Feng W, et al. COUP-TFII regulates metastasis of colorectal adenocarcinoma cells by modulating Snail1. Br J Cancer 2014;111:933-43. 10.1038/bjc.2014.373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ding W, Zhang Y, Cai H, et al. Overexpression of COUPTFII suppresses proliferation and metastasis of human gastric cancer cells. Mol Med Rep 2018;17:2393-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lin SC, Lin CH, Shih NC, et al. Cellular prion protein transcriptionally regulated by NFIL3 enhances lung cancer cell lamellipodium formation and migration through JNK signaling. Oncogene 2020;39:385-98. 10.1038/s41388-019-0994-0 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as