Abstract
Head and neck squamous cell carcinomas (HNSCC) arising from the oral cavity, pharynx, and larynx constitute the 6th most common human cancer. Human papillomavirus (HPV)-positive tumours are distinct from HPV-negative counterparts, with HPV status affording clear clinical utility, prognostic benefit and better treatment outcomes. In contrast to their HPV-positive counterparts, HPV-negative tumours are characterized by high mutational load and chromosomal aberrations, with varying copy number alteration (CNA) profiles. HNSCC are distinct tumours at the chromosomal, gene and expression levels, with additional insight gained from immune profiling. Based on mutational analyses, HNSCC are categorized as HPV-positive, HPV-negative CNA-silent, and HPV-negative CNA-high tumours. Furthermore, gene expression profiling segregates these tumours into atypical, classical, basal, and mesenchymal, with clear differences observed between tumours of the oral cavity, oropharynx, hypopharynx and larynx. Additional immune profiling further classifies tumours as either immune-active or immune-exhausted. The clinical utility and impact of these tumour molecular subtypes however remains to be determined. HNSCC harbor high levels of somatic mutations. They display loss at 3p and 18q and gain at 3q and 8q, with mutations in CDKN2A, TP53, CCND1, EGFR, PIK3CA, PTEN, NOTCH1, NSD1, FAT1, AJUBA and KMT2D. Important pathways include the p53 and RB pathways which are involved in cell cycle control and are frequently lost in HPV-negative tumours, the WNT-β-catenin pathway related to the mesenchymal subtype and smoking etiology, and the PI3K pathway which includes the most common genetic alteration in HPV-positive HNSCC. Understanding the mutational, genomic and transcriptomic landscape of HNSCC has leveraged better therapeutic approaches to manage this group of diseases, and it is hoped that additional insight into the molecular subtypes of HNSCC and its specific subsites will further drive improved strategies to stratify and treat patients with this debilitating disease.
Keywords: Head and neck cancer, oral cancer, squamous cell carcinoma, molecular landscape, therapy
Introduction
Head and neck squamous cell carcinomas (HNSCC) arise from the squamous and lining epithelium of the oral cavity, pharynx and larynx. HNSCC is the 6th most common cancer globally with more than 700,000 new cases reported (1) accounting for between 0.5% (oropharynx) to 1.9% (lip and oral cavity) of deaths from all cancers combined. Age standardised risk (ASR) incidence for lip and oral cavity cancer is 5.8 and 2.3 compared to cancer of the oropharynx which is 0.21 and 0.4, for males and females respectively. ASR mortality for lip and oral cavity cancer is 2.8 and 1.2 for males and females respectively, compared to that of larynx which is 1.9 and 0.3 respectively (1).
HNSCC is a heterogeneous group of tumours involving distinct anatomical sites and subsites with varying etiological factors including smoking, alcohol consumption and infection with HPV (2,3). A growing number of low-risk younger patients have been reported to have a distinct clinical and histopathological pattern associated with poorer prognosis (4,5), although this has not been universally accepted. There are genetic and prognostic differences between HPV-positive and HPV-negative HNSCC (6). The heterogeneity of this cancer, suggests a critical role for genetic alterations contributing to its carcinogenesis (7).
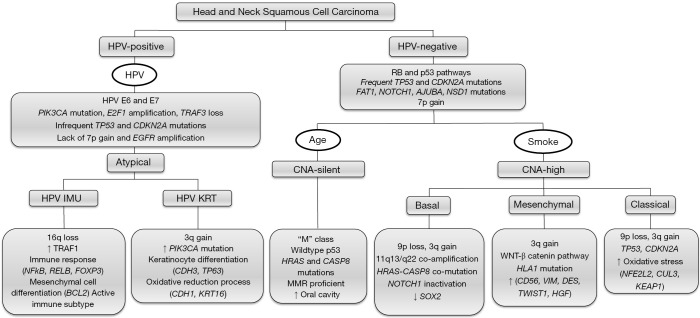
Clinical and histological differences are mirrored by diverse genomic and immunological subtypes. The varying presentations and subsite specific profiles has manifested in an ever evolving range of therapies including surgery, radiotherapy, chemotherapy and more recently immunotherapy in an effort to improve five-year survival rates which currently stand at approximately 50% across all subsites. Understanding the mutational, genomic and transcriptomic landscape of HNSCC has leveraged better therapeutic approaches to manage this group of diseases, and it is hoped that additional insight into the genomic and immunological profiles of HNSCC and its specific subsites will further drive improved strategies to stratify and treat patients with this debilitating disease. The most pertinent molecular features highlighted in this review are summarized in Figure 1.
Figure 1.
Composite schematic depicting the molecular landscape of head and neck squamous cell carcinoma. Tumours are grouped according to CNA alterations, gene mutations and gene expression profiles summarized from various studies described in this review. Only the most pertinent and discriminatory chromosomal, gene and immune profiles are highlighted. Based on mutational analyses, HNSCC are characterized as HPV-positive, HPV-negative CNA-silent, and HPV-negative CNA-high tumours. Furthermore, gene expression profiling segregates these tumours into atypical, basal, classical and mesenchymal, with clear differences observed between tumours of different anatomical sites and proposed etiology. Additional immune profiling further classifies tumours as either immune-active or immune-exhausted. The clinical utility and impact of these molecular tumour subtypes remains to be determined.
Gene characterization of head and neck squamous cell carcinoma
Prior to utilization of next generation sequencing (NGS) in HNSCC (8,9), only a few biomarkers of HNSCC had been revealed with studies on alterations in cellular signalling pathways (TP53 and CDK2NA) (10) and chromosomal abnormalities (amplification of 11q13, cyclin D1, and EGFR) (11). The first reports describing NGS findings in HNSCC were published simultaneously in 2011 (12,13). Stransky et al. performed whole exome sequencing on tumours and blood of 74 HNSCC patients (13) and found approximately 130 coding mutations per tumour. 321 mutations were queried further and 89.7% validated (13). Agrawal et al. sequenced 32 tumours and compared their results to findings from another 88 HNSCC and matched non-cancerous tissues (12). Both studies confirmed previously known HNSCC genomic alterations such as mutations in TP53, CDKN2A, PIK3CA, HRAS, and PTEN but also introduced NOTCH1, which was reported as the second most commonly involved gene in HNSCC (14). NOTCH1 had previously been shown to be important in cutaneous SCC (15), but it had not been identified by conventional Sanger sequencing due to its large size.
Mahjabeen et al. sequenced exons of XRCC1 in HNSCC and matched controls and found two missense mutations in 55% of cases and two silent mutations in 45% cases, accounting for a mutation frequency of 87% (16). Sequencing of RAD51C revealed five distinct heterozygous alterations in 5.8% of HNSCCs (17). Laborde et al. undertook whole transcriptome sequencing of matched tumour and cancer-free tissues from patients with oropharyngeal carcinoma, and showed elevated levels of expression of two gene targets in DNA damage repair (CHEK2 and ATR) of HPV-negative current smokers compared with past smokers and non-smokers (18).
The Cancer Genome Atlas (TCGA) characterized the molecular profile of 279 primary HNSCC in a cohort mostly composed of male heavy smokers. Tumours were mostly those from the oral cavity (62%), followed by the larynx (26%), and oropharynx (12%) with a greater majority being HPV-negative and only 13% classified as HPV-positive (19). HPV-positive tumours are clinically distinct from their HPV-negative counterparts, with HPV-negative HNSCCs largely distributed among different anatomic sites, occurring in the context of heavy alcohol and/or tobacco use relative to HPV-positive tumours which are focussed in the oropharynx.
The study validated previously identified frequently mutated genes, but also profiled copy number alterations (CNAs), gene and protein differential expression, and epigenetic changes (12,13,19). The comprehensive genetic profile of these tumours has been reviewed by Cho and colleagues (20). HNSCC display high instability, as noted by the presence of CNAs and chromosomal fusions and exhibit deletions of 3p and 8p as well as amplifications of 3q, 5q, and 8q (19). TP53, CDKN2A, CASP8, and NSD1 have been shown to be differentially mutated across all head and neck sites, but unlike other mutations, CASP8 are additionally concentrated within the oral cavity, and contain missense and other mutations in caspase peptidase and death effector domains (19).
Genes have been grouped into four categories including those significant for cell survival and proliferation (TP53, EGFR, PIK3CA, and HRAS), cell-cycle control (CDKN2A and CCND1), cellular differentiation (NOTCH1), and cellular adhesion and invasion signalling (FAT1) (12,13,19). The top 10 mutations being TP53, CDKN2A, FAT1, PIK3CA, NOTCH1, KMT2D (MLL2), NSD1, CASP8, AJUBA, and NFE2L2. The two most altered genes in the TCGA cohort were TP53 and CDKN2A, and these appear to play important roles in driving carcinogenesis. Inactivation of p53 protein, encoded by TP53 on 17p12, plays a vital role in the pathogenesis of HNSCC (21). Commonly detected TP53 mutations in HNSCC include those in exon 4 or intron 6 (19). DNA damage can cause translocation of p53 to the nucleus, inducing cell growth arrest or apoptosis. p16, encoded by CDKN2A on 9p21, blocks cell cycle progression from G1 to S phase by inhibiting Cyclin D1 (21). Deficiency of cell senescence results from disruption of p16 activity, ultimately contributing to development of dysplasia.
HPV-positive tumours lack mutations and alterations in TP53 and CDKN2A in contrast with their HPV-negative counterparts (19). In addition, there is a high proportion of mutations and CNAs in genes encoding constituents of the PI3 kinase (PI3K) pathway (19). HPV-positive HNSCC commonly present with PIK3CA mutations at higher levels than HPV-negative tumours, but both possess amplifications of 3q26/28, the region containing TP63 and SOX2 (13,19). HPV-positive tumours also display loss of TRAF3 and amplification of E2F1, additionally distinguishing them from their HPV-negative counterparts.
HPV-negative tumours demonstrate unique structural abnormalities and somatic mutations including amplifications in receptor tyrosine kinases such as EGFR, ERBB2, FGFR1, and deletions in NSD1 and tumour suppressor genes including CDKN2A, NOTCH1, SMAD4, and FAT1 (19). They also display alterations in oxidative stress regulators (NFE2L2, KEAP1, CUL3). NOTCH1 inactivating mutations are present in up to 20% of HNSCCs, with increased mortality noted in patients with OSCC associated with Notch activation and FGF1 transcriptional upregulation (22). NOTCH1 has a proto-oncogenic role in other cancers but is thought to act as a tumour suppressor in HNSCC. Other novel findings include mutations in genes involved in chromatin remodelling (KMT2D/MLL2) and immune evasion (HLA-A), with novel co-amplifications of 11q13 (CCND1, CTTN, FADD) and 11q22 (YAP1, BIRC2) identified (19). Somatic mutations such as non-synonymous inactivating mutations were notable in TP53, CDKN2A, FAT1and AJUBA.
Based on these mutational data, HNSCCs have been divided into HPV-positive, HPV-negative CNA-high, and HPV-negative CNA-silent tumours (Figure 1) (23). These are further divided based on gene expression profiling as detailed below. The p53 and RB pathways are frequently abrogated in HPV-negative HNSCC, but appear to remain active in CNA-silent tumours, with mutations in HRAS and CASP8 (23). The etiology of this subgroup of tumours remains unclear, but ageing is thought to be a risk factor. Smoking is a key etiological risk factor for HPV-negative CNA-high tumours, with many cancer genes (FAT1, NOTCH1) and pathways (WNT-β-catenin) being involved in their progression (Figure 1). The role of microRNA (24) and lncRNA (25,26) in HNSCC have been reviewed elsewhere.
Gene expression subtypes of head and neck squamous cell carcinoma
As additional gene expression data becomes available, HNSCC have been categorized not only based on their clinical, histopathological and genetic features, but also based on gene expression subtypes with purported clinical and biological implications (4,27). Currently four subtypes have been proposed namely: atypical, basal, classical, and mesenchymal (4,27-30).
The atypical subtype involves a majority of HPV-positive tumours with activating mutations of PIK3CA and a lack in 7p amplifications (encoding EGFR). The latter observation is consistent with findings correlating HPV-positivity and low EGFR expression (27,31). Unlike the atypical subtype, the other subtypes exhibit gain-of-function of 7p. The classical and basal subtypes are categorized by loss at 9p (CDKN2A) encoding p16.
The basal subtype demonstrates NOTCH1 inactivation, HRAS-CASP8 co-mutation, co-amplification of 11q13/q22, and fewer alterations of 3q (SOX2).
The classical subtype is distinguished for TP53 mutation, loss of CDKN2A, 3q amplification, changes in oxidative stress genes (KEAP1, CUL3, and NFE2L2), and is correlated with a significant smoking history, and a high proportion of laryngeal SCCs (19).
The mesenchymal subtype demonstrates a distinct molecular profile which involves modifications to immune-related genes including higher expression of CD56, and low frequency mutations in HLAI (19).
Classical and basal nomenclature has been chosen based on gene expression patterns in HNSCC subtypes which show strong similarities to classical and basal subtypes of lung SCC (27). The classical subtype exhibits well recognised genomic alterations associated with SCC specifically 3p and 9p deletion, 3q amplification, and focal amplification of CCND1 and EGFR. The mesenchymal subtype is so named because of the prominence of the epithelial to mesenchymal transition (EMT) pathway, while the atypical subtype was selected because of the distinct absence of 9p deletion or EGFR amplification (27).
In OSCC, the basal (42.7%) and mesenchymal (34.8%) are the two main subtypes, compared to atypical (50.7%) and classical (22%) in non-OSCC tumours including those of the larynx, oropharynx and hypopharynx (32).
Additional subtypes based on immune profiling have also being reported (33,34), with further analysis of the TCGA and other datasets undertaken to characterize the immune landscape of HNSCC (33-35). Saloura et al. (35) classified tumours into low or high CD8+ T cell inflamed phenotype (TCIP-L vs. TCIP-H) based on their chemokine signature. They showed that TCIP-H tumours were enriched for mutations in CASP8, HRAS, EP300, and EPHA2 and demonstrated more frequent amplifications of CD274, JAK2, PDCD1LG2, and KDM4C. TCIP-L tumours were more likely to display higher rates of mutation in NSD1, deletion of CDKN2A, and amplification of EGFR and YAP1.
Zhang et al. identified two distinct subtypes of HPV-positive SCC which they differentiated based on RNA sequencing data into those with upregulated genes in mesenchymal and immune response (named HPV-IMU) and those enriched for keratinocyte differentiation and oxidative reduction process (named HPV-KRT) (36). More recently, Chen and colleagues (34) found that nearly 40% of HNSCCs showed enriched inflammatory response, greater cytolytic activity, and active IFN-γ signalling. This new molecular class of tumours was termed “Immune” class. This contained two distinct microenvironment-based immune response subtypes, identified by “Active” (characterized by an enriched pro-inflammatory M1 macrophage signature, heightened cytolytic activity, abundant tumour-infiltrating lymphocytes, high HPV infection, and better prognosis) and “Exhausted” (characterized by enrichment of stroma and anti-inflammatory M2 macrophage profiles, WNT/TGF-β pathway signalling and poorer survival). The “Active” immune class was more common in the less aggressive atypical subtype, and showed potential response to PD-L1 blockade. Approximately 50–60% of HNSCC tumour cells overexpress PD-L1, which increases to nearly 85% when immune cells are included. PD-L1 expression has been associated with younger age, higher tumour grade, and HPV-positivity (37). There is no association between PD-L1 status and overall survival in HNSCC patients overall or in the oropharyngeal SCC sub-population.
Genomic signature of oral squamous cell carcinoma
The molecular basis of OSCC has recently been summarized (38). There is significant genetic diversity seen in OSCC, and as such it remains a significant challenge to identify molecular drivers of this cancer subtype. The TCGA cohort included a prominent subgroup of HPV-negative tumours primarily focussed in the oral cavity demonstrating mutations but lacking CNAs enriched for wild-type TP53, mutant HRAS and CASP8. This subset (referred to as “M” class for Mutation) suggests that some OSCC occur in a p53-independent tumourigenesis pathway (19). This subtype is DNA mismatch repair proficient and is thought to be more prominent in older females without a history of smoking or alcohol consumption (23,39). Other subtypes show abrogation of the p53 and RB pathways with mutations in TP53 and CDKN2A, and are typically associated with a history of smoking. Through field cancerization, OSCCs developing along this pathway are thought to commonly pass through a precancerous stage such as oral leukoplakia and harbour mutations in FAT1 and NOTCH1 (40) and display deficiency in DNA damage repair pathway genes such as BRCA1, BRCA2, FANCA and other double strand break (DSB) repair Fanconi anaemia (FA)/BRCA pathway genes (41,42). A more comprehensive analysis of molecular biomarkers of oral leukoplakia and their utility in malignant transformation are detailed elsewhere (40,43,44).
The most common molecular subtypes of OSCC are basal and mesenchymal, followed by classical. The classical subtype is characterized by high expression of genes in oxidative stress response pathways enriched for genes such as NFE2L2 (nuclear factor erythroid 2-related factor 2; also known as NRF1). Tumours predominantly located on the oral tongue may be appropriate targets for drugs against p53 cell-cycle pathway genes TP53 and CCND1. Recently, inhibitors of the PI3K/AKT/mTOR pathway have demonstrated efficacy in preclinical studies of OSCC (45-47), with rapamycin; the canonical inhibitor of mTOR, shown to inhibit OSCC growth (48).
Whole-exome sequencing of OSCCs from Taiwanese males confirmed mutations in genes involved in cell cycle regulation (TP53, CDKN2A, and CCND1), but also identified other mutational signatures including new driver genes, such as ELAVL1 and CHUK, which are involved in altering the functions of various tumour suppressor genes and oncogenes (49). Other genes that were less frequently mutated included ASXL1 which codes for a transcription factor, and RPTN which is associated with epithelial differentiation.
Several driver genes that confer phenotypic malignancy have been mapped to 11q13, and are implicated in OSCC (50,51). Specifically, within 11q13.2-q13.4, an amplicon core has been identified including genes CCND1, CTTN, FADD, and ORAOV1. CCND1 (encoding cyclin D1 that promotes G1-S phase transition) and CTTN (encoding cortactin, a F-actin binding protein) are two frequently associated drivers of locus 11q13. Amplification of CTTN confers resistance to gefitinib, while amplification of CCND1 is associated with resistance to cisplatin (52). These oncogenes influence clinicopathological characteristics of OSCC including poor tumour differentiation, lymph node involvement, and low survival (53). Expression levels of CCND1 have been used to select patients who may benefit from induction chemotherapy. Genomic amplification of 11q13.3 also impacts FADD (Fas-associated death domain-containing protein) regulation and expression, which is correlated with gender, and may account for the two-fold increased frequency of OSCC in males compared to females (54). FADD acts as an adaptor to convey apoptotic signals initiated by death receptors such as Fas, and is highly expressed in tongue OSCC compared to adjacent tissue, in addition to being associated with metastatic carcinoma (54). Another oncogene located at this locus is ORAOV1 (oral cancer overexpressed 1). Gain of function in ORAOV1 has been linked to apoptotic inhibition, cell cycle progression, and angiogenesis (55,56).
Gingivobuccal squamous cell carcinoma (GB-SCC) is a subtype of OSCC, prevalent in the Indian sub-continent and parts of Asia, where tobacco and betel chewing is prevalent (51). The most common anatomical site is the buccal mucosa, with synergistic contributions of major driver genes such as NOTCH1, PIK3CA, CASP8, HRAS, FAT1, MAP4K2, EPHA2, and RASA1 contributing to tumour formation (51). Mutations in several genes including TRPM3, KMT2D, ARID2, USP9X, and UNC13C appear to be specific to GB-SCC and seem be functionally involved in tumour suppression (49,57). Some genes such as TP53, NOTCH1, CASP8, HRAS, and FAT1, are common to both GB-SCC and HNSCC. Alterations in some novel genes such as YAP1, DROSHA, and DDX3X have also been reported in GB-SCC (51).
More recently, the genomic and transcriptomic profile of areca nut-related OSCC has been explored in a Chinese cohort which identified a set of 11 mutated genes including four novel genes (ATG2A, WEE1, DST, TSC2), of which ATG2A and WEE1 were more significantly mutated more commonly in areca nut-related SCC. Areca nut-related OSCCs are typified by genomic deficiency of mismatch repair (MMR) genes, which could also predict prognosis (58), affirming once again the role of genetic instability in oral carcinogenesis (59,60).
In addition to genetic mutations observed in oral cancer, epigenetic modifications to the genome (61) are commonly seen in OSCC (62-64) and severe epithelial dysplasia (65), with p16 frequently inactivated due to promoter hypermethylation in OSCC, and in tobacco users with premalignant oral lesions where up to 58% of these demonstrate genome-wide DNA methylation, with levels rising with progression to OSCC (65).
Implications for therapy
Although many predictive biomarkers have been discovered and reported for various HNSCCs through genomic research, only a few have been translated into clinical oncology practice. Among the frequently mutated genes in HNSCC, EGFR remains a positive target for cancer therapy. Over expression and mutation of EGFR is associated with cancer of the breast, lung, ovary, prostate and colorectum (66-68). EGFR is a transmembrane receptor belonging to the human epidermal receptor (HER) family of growth factor receptors (HER2, HER3 and HER4). Formation of EGFR homodimers or heterodimers triggers intracellular pathways that result in cancer cell proliferation, blockade of apoptosis, activation of invasion and metastasis, and stimulation of tumour-induced neo-angiogenesis (69,70). Anti-EGFR monoclonal antibodies (cetuximab and panitumumab) recognise EGFR exclusively and bind to its extracellular domain, compete for receptor binding and block ligand-induced EGFR tyrosine kinase activation. Small-molecule EGFR tyrosine kinase inhibitors (erlotinib and gefitinib) compete reversibly with ATP to bind to the intracellular catalytic domain of EGFR tyrosine kinase, and inhibit EGFR autophosphorylation and downstream signalling. Small-molecule EGFR tyrosine kinase inhibitors can also block different growth factor receptor tyrosine kinases, including vascular endothelial growth factor receptor (VEGFR).
High expression of EGFR is associated with tumour aggressiveness and poor survival in OSCC patients (71,72), specifically oral tongue SCC (73,74). The elevated expression of EGFR in OSCC makes it an attractive molecular target for treatment. Cetuximab, is an approved targeted therapy for HNSCC, including advanced OSCC. The combination of cetuximab with platinum-based chemotherapy is approved as first-line treatment for recurrent or metastatic HNSCC, including OSCC, and cetuximab alone is approved as second-line treatment for platinum-resistant HNSCC (75,76). Adding cetuximab to platinum-based chemotherapy significantly prolongs median overall survival of HNSCC from 7.4 to 10.1 months, as well as median progression-free survival (5.6 vs. 3.3 months) (76).
Immune checkpoint inhibitors such as pembrolizumab and nivolumab have demonstrated efficacy as first-line therapy for patients with advanced HNSCC, either alone or in combination with chemotherapy, particularly in HPV-positive disease. For pembrolizumab therapy alone, there has been significant improvement in median overall survival compared to the EXTREME regime in PD-L1 positive patients but no significant benefit in progression-free survival (77). Patients treated with pembrolizumab and chemotherapy demonstrated a significant increase in overall survival, and a modest improvement in progression-free survival compared to controls. This has led to pembrolizumab being approved as first-line monotherapy for patients with PD-L1 positive recurrent or metastatic HNSCC, and for combination pembrolizumab and chemotherapy approved for recurrent or metastatic HNSCC regardless of PD-L1 expression. Patients who fail platinum chemotherapy may also benefit from nivolumab or pembrolizumab as second-line therapy regardless of PD-L1 expression. A recent systematic review also points to the effective use of immune checkpoint inhibitor therapy in management of OSCC (78). Despite advances in therapeutic options for patients with HNSCC based on immune profiling, those with low or negative expression of PD-L1 are more likely to still benefit from cetuximab and chemotherapy to avoid rapid progression and local disease recurrence.
So far, other molecular targets have not been successfully translated into clinical practice, and much work remains to be done before a true personalised approach to clinical head and neck oncology can be achieved. Understanding the genotype-phenotype relationships that underpin the heterogeneous presentations of HNSCCs is required, as is better resolution of what will determine efficacy of targeted therapies as the latter become available. A glimpse at possible targets is provided in this paper, but their advancement is dependent on large scale laboratory, clinical and population studies.
Conclusion
HNSCC is a heterogeneous group of tumours involving distinct anatomical sites and with varying etiological factors including smoking, alcohol consumption and infection with HPV. The clinical and histological differences are mirrored by diverse genomic and immunological subtypes. The tumour microenvironment is characterized by alterations in immune cell populations in addition to local factors favouring immunosuppression, driving tumour evasion or loss of immune surveillance. As the mutational, genomic and transcriptomic landscape of HNSCC is better resolved, our understanding of therapeutic approaches for effect control of the disease has increased. These advances have enabled effective therapeutic options for advanced tumours with cetuximab, and more recently with immune checkpoint inhibitors. Improved understanding of the genomic and immunological profiles of HNSCC and its specific subsites is poised to drive improved strategies to stratify and treat patients with HNSCC.
Supplementary
The article’s supplementary files as
Acknowledgments
Funding: None.
Ethical Statement: The author is accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work is appropriately investigated and resolved.
Footnotes
Provenance and Peer Review: This article was commissioned by the Guest Editor (Dr. Mukund Seshadri) for the series “Head and Neck Cancers – Disease Biology, Diagnostics, Prevention and Management” published in Annals of Translational Medicine. The article has undergone external peer review.
Conflicts of Interest: The author has completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-6264). The series “Head and Neck Cancers – Disease Biology, Diagnostics, Prevention and Management” was commissioned by the editorial office without any funding or sponsorship. The author has no conflicts of interest to declare.
References
- 1.Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 2.Hashibe M, Boffetta P, Zaridze D, et al. Contribution of tobacco and alcohol to the high rates of squamous cell carcinoma of the supraglottis and glottis in central Europe. Am J Epidemiol 2007;165:814-20. 10.1093/aje/kwk066 [DOI] [PubMed] [Google Scholar]
- 3.Hashibe M, Brennan P, Benhamou S, et al. Alcohol drinking in never users of tobacco, cigarette smoking in never drinkers, and the risk of head and neck cancer: Pooled analysis in the international head and neck cancer epidemiology consortium. J Natl Cancer Inst 2007; 99:777-89. 10.1093/jnci/djk179 [DOI] [PubMed] [Google Scholar]
- 4.Chung CH, Parker JS, Karaca G, et al. Molecular classification of head and neck squamous cell carcinomas using patterns of gene expression. Cancer Cell 2004; 5:489-500. 10.1016/S1535-6108(04)00112-6 [DOI] [PubMed] [Google Scholar]
- 5.Dahlstrom KR, Little JA, Zafereo ME, et al. Squamous cell carcinoma of the head and neck in never smoker-never drinkers: A descriptive epidemiologic study. Head Neck 2008;30:75-84. 10.1002/hed.20664 [DOI] [PubMed] [Google Scholar]
- 6.Chung CH, Gillison ML. Human Papillomavirus in Head and Neck Cancer: Its Role in Pathogenesis and Clinical Implications. Clin Cancer Res 2009;15:6758-62. 10.1158/1078-0432.CCR-09-0784 [DOI] [PubMed] [Google Scholar]
- 7.Liang XH, Lewis J, Foote R, et al. Prevalence and significance of human papillomavirus in oral tongue cancer: The Mayo Clinic experience. J Oral Maxillofac Surg 2008;66:1875-80. 10.1016/j.joms.2008.04.009 [DOI] [PubMed] [Google Scholar]
- 8.Jessri M, Farah CS. Harnessing Massively Parallel Sequencing in Personalized Head and Neck Oncology. J Dent Res 2014;93:437-44. 10.1177/0022034514524783 [DOI] [PubMed] [Google Scholar]
- 9.Jessri M, Farah CS. Next generation sequencing and its application in deciphering head and neck cancer. Oral Oncol 2014;50:247-53. 10.1016/j.oraloncology.2013.12.017 [DOI] [PubMed] [Google Scholar]
- 10.Leemans CR, Braakhuis BJM, Brakenhoff RH. The molecular biology of head and neck cancer. Nat Rev Cancer 2011;11:9-22. 10.1038/nrc2982 [DOI] [PubMed] [Google Scholar]
- 11.Berenson JR, Yang J, Mickel RA. Frequent Amplification of the Bcl-1 Locus in Head and Neck Squamous-Cell Carcinomas. Oncogene 1989;4:1111-6. [PubMed] [Google Scholar]
- 12.Agrawal N, Frederick MJ, Pickering CR, et al. Exome Sequencing of Head and Neck Squamous Cell Carcinoma Reveals Inactivating Mutations in NOTCH1. Science 2011;333:1154-7. 10.1126/science.1206923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stransky N, Egloff AM, Tward AD, et al. The mutational landscape of head and neck squamous cell carcinoma. Science 2011;333:1157-60. 10.1126/science.1208130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Loyo M, Li RJ, Bettegowda C, et al. Lessons learned from next-generation sequencing in head and neck cancer. Head Neck 2013;35:454-63. 10.1002/hed.23100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dotto GP. Notch tumor suppressor function. Oncogene 2008;27:5115-23. 10.1038/onc.2008.225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mahjabeen I, Masood N, Baig RM, et al. Novel mutations of OGG1 base excision repair pathway gene in laryngeal cancer patients. Fam Cancer 2012;11:587-93. 10.1007/s10689-012-9554-2 [DOI] [PubMed] [Google Scholar]
- 17.Scheckenbach K, Baldus SE, Balz V, et al. RAD51C--a new human cancer susceptibility gene for sporadic squamous cell carcinoma of the head and neck (HNSCC). Oral Oncol 2014;50:196-9. 10.1016/j.oraloncology.2013.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laborde RR, Wang VW, Smith TM, et al. Transcriptional Profiling by Sequencing of Oropharyngeal Cancer. Mayo Clin Proc 2012;87:226-32. 10.1016/j.mayocp.2011.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Network CGA. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature 2015;517:576-82. 10.1038/nature14129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cho J, Johnson DE, Grandis JR. Therapeutic Implications of the Genetic Landscape of Head and Neck Cancer. Semin Radiat Oncol 2018;28:2-11. 10.1016/j.semradonc.2017.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hsu PJ, Yan K, Shi H, et al. Molecular biology of oral cavity squamous cell carcinoma. Oral Oncol 2020;102:104552. 10.1016/j.oraloncology.2019.104552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weaver AN, Burch MB, Cooper TS, et al. Notch Signaling Activation Is Associated with Patient Mortality and Increased FGF1-Mediated Invasion in Squamous Cell Carcinoma of the Oral Cavity. Mol Cancer Res 2016;14:883-91. 10.1158/1541-7786.MCR-16-0114 [DOI] [PubMed] [Google Scholar]
- 23.Leemans CR, Snijders PJF, Brakenhoff RH. The molecular landscape of head and neck cancer. Nat Rev Cancer 2018;18:269-82. 10.1038/nrc.2018.11 [DOI] [PubMed] [Google Scholar]
- 24.John K, Wu J, Lee BW, et al. MicroRNAs in Head and Neck Cancer. Int J Dent 2013;2013:650218. 10.1155/2013/650218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pentenero M, Bowers LM, Jayasinghe R, et al. World Workshop on Oral Medicine VII: Clinical evidence of differential expression of lncRNAs in oral squamous cell carcinoma: A scoping review. Oral Dis 2019;25 Suppl 1:88-101. 10.1111/odi.13076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pentenero M, Bowers L, Jayasinghe R, et al. World Workshop on Oral Medicine VII: Functional pathways involving differentially expressed lncRNAs in oral squamous cell carcinoma. Oral Dis 2019;25 Suppl 1:79-87. 10.1111/odi.13051 [DOI] [PubMed] [Google Scholar]
- 27.Walter V, Yin X, Wilkerson MD, et al. Molecular subtypes in head and neck cancer exhibit distinct patterns of chromosomal gain and loss of canonical cancer genes. PLoS One 2013;8:e56823. 10.1371/journal.pone.0056823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.De Cecco L, Nicolau M, Giannoccaro M, et al. Head and neck cancer subtypes with biological and clinical relevance: Meta-analysis of gene-expression data. Oncotarget 2015;6:9627-42. 10.18632/oncotarget.3301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rickman DS, Millon R, De Reynies A, et al. Prediction of future metastasis and molecular characterization of head and neck squamous-cell carcinoma based on transcriptome and genome analysis by microarrays. Oncogene 2008;27:6607-22. 10.1038/onc.2008.251 [DOI] [PubMed] [Google Scholar]
- 30.Keck MK, Zuo Z, Khattri A, et al. Integrative analysis of head and neck cancer identifies two biologically distinct HPV and three non-HPV subtypes. Clin Cancer Res 2015;21:870-81. 10.1158/1078-0432.CCR-14-2481 [DOI] [PubMed] [Google Scholar]
- 31.Kang H, Kiess A, Chung CH. Emerging biomarkers in head and neck cancer in the era of genomics. Nat Rev Clin Oncol 2015;12:11-26. 10.1038/nrclinonc.2014.192 [DOI] [PubMed] [Google Scholar]
- 32.Foy JP, Saintigny P, Goudot P, et al. The promising impact of molecular profiling on treatment strategies in oral cancers. J Stomatol Oral Maxillofac Surg 2017;118:242-7. 10.1016/j.jormas.2017.05.004 [DOI] [PubMed] [Google Scholar]
- 33.Wondergem NE, Nauta IH, Muijlwijk T, et al. The Immune Microenvironment in Head and Neck Squamous Cell Carcinoma: on Subsets and Subsites. Curr Oncol Rep 2020;22:81. 10.1007/s11912-020-00938-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen YP, Wang YQ, Lv JW, et al. Identification and validation of novel microenvironment-based immune molecular subgroups of head and neck squamous cell carcinoma: implications for immunotherapy. Ann Oncol 2019;30:68-75. 10.1093/annonc/mdy470 [DOI] [PubMed] [Google Scholar]
- 35.Saloura V, Izumchenko E, Zuo Z, et al. Immune profiles in primary squamous cell carcinoma of the head and neck. Oral Oncol 2019;96:77-88. 10.1016/j.oraloncology.2019.06.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Y, Koneva LA, Virani S, et al. Subtypes of HPV-Positive Head and Neck Cancers Are Associated with HPV Characteristics, Copy Number Alterations, PIK3CA Mutation, and Pathway Signatures. Clin Cancer Res 2016;22:4735-45. 10.1158/1078-0432.CCR-16-0323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tang H, Zhou X, Ye Y, et al. The different role of PD-L1 in head and neck squamous cell carcinomas: A meta-analysis. Pathol Res Pract 2020;216:152768. 10.1016/j.prp.2019.152768 [DOI] [PubMed] [Google Scholar]
- 38.Farah CS, Kujan O, Prime S, et al. Oral Mucosal Malignancies. In: Farah CS, Balasubramaniam R, McCullough MJ, editors. Contemporary Oral Medicine: A Comprehensive Approach to Clinical Practice. Cham, Switzerland: Springer Nature; 2019. p. 1570-786. [Google Scholar]
- 39.Smeets SJ, Braakhuis BJM, Ylstra B, et al. TP53 mutations are associated with a particular pattern of genomic imbalances in head and neck squamous cell carcinoma. Cell Oncol 2007;29:160. [Google Scholar]
- 40.Farah CS. Molecular, genomic and mutational landscape of oral leukoplakia. Oral Dis 2021;27:803-12. 10.1111/odi.13608 [DOI] [PubMed] [Google Scholar]
- 41.Farah CS, Jessri M, Bennett NC, et al. Exome sequencing of oral leukoplakia and oral squamous cell carcinoma implicates DNA damage repair gene defects in malignant transformation. Oral Oncol 2019;96:42-50. 10.1016/j.oraloncology.2019.07.005 [DOI] [PubMed] [Google Scholar]
- 42.Jessri M, Dalley AJ, Farah CS. Deficient double-strand break repair in oral squamous cell carcinoma cell lines. J Oral Pathol Med 2017;46:695-702. 10.1111/jop.12576 [DOI] [PubMed] [Google Scholar]
- 43.Celentano A, Glurich I, Borgnakke WS, et al. World Workshop on Oral Medicine VII: Prognostic biomarkers in oral leukoplakia and proliferative verrucous leukoplakia-A systematic review of retrospective studies. Oral Dis 2021;27:848-80. 10.1111/odi.13363 [DOI] [PubMed] [Google Scholar]
- 44.Villa A, Celentano A, Glurich I, et al. World Workshop on Oral Medicine VII: Prognostic biomarkers in oral leukoplakia: A systematic review of longitudinal studies. Oral Dis 2019;25 Suppl 1:64-78. 10.1111/odi.13087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smolensky D, Rathore K, Bourn J, et al. Inhibition of the PI3K/AKT Pathway Sensitizes Oral Squamous Cell Carcinoma Cells to Anthracycline-Based Chemotherapy In Vitro. J Cell Biochem 2017;118:2615-24. 10.1002/jcb.25747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hsu CM, Lin PM, Lin HC, et al. NVP-BEZ235 Attenuated Cell Proliferation and Migration in the Squamous Cell Carcinoma of Oral Cavities and p70S6K Inhibition Mimics its Effect. Int J Mol Sci 2018;19:3546. 10.3390/ijms19113546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lam TG, Jeong YS, Kim S-A, et al. New metformin derivative HL156A prevents oral cancer progression by inhibiting the insulin-like growth factor/AKT/mammalian target of rapamycin pathways. Cancer Sci 2018;109:699-709. 10.1111/cas.13482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Day TA, Shirai K, O'Brien PE, et al. Inhibition of mTOR Signaling and Clinical Activity of Rapamycin in Head and Neck Cancer in a Window of Opportunity Trial. Clin Cancer Res 2019;25:1156-64. 10.1158/1078-0432.CCR-18-2024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Su SC, Lin CW, Liu YF, et al. Exome Sequencing of Oral Squamous Cell Carcinoma Reveals Molecular Subgroups and Novel Therapeutic Opportunities. Theranostics 2017;7:1088-99. 10.7150/thno.18551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ramos-Garcia P, Ruiz-Avila I, Gil-Montoya JA, et al. Relevance of chromosomal band 11q13 in oral carcinogenesis: An update of current knowledge. Oral Oncol 2017;72:7-16. 10.1016/j.oraloncology.2017.04.016 [DOI] [PubMed] [Google Scholar]
- 51.Chakraborty P, Karmakar T, Arora N, et al. Immune and genomic signatures in oral (head and neck) cancer. Heliyon 2018;4:e00880. 10.1016/j.heliyon.2018.e00880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang P, Zhang Z, Zhou X, et al. Identification of genes associated with cisplatin resistance in human oral squamous cell carcinoma cell line. BMC Cancer 2006;6:224. 10.1186/1471-2407-6-224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wilkerson PM, Reis-Filho JS. The 11q13-q14 amplicon: clinicopathological correlations and potential drivers. Genes Chromosomes Cancer 2013;52:333-55. 10.1002/gcc.22037 [DOI] [PubMed] [Google Scholar]
- 54.Prapinjumrune C, Morita K, Kuribayashi Y, et al. DNA amplification and expression of FADD in oral squamous cell carcinoma. J Oral Pathol Med 2010;39:525-32. [DOI] [PubMed] [Google Scholar]
- 55.Jiang L, Zeng X, Yang H, et al. Oral cancer overexpressed 1 (ORAOV1): a regulator for the cell growth and tumor angiogenesis in oral squamous cell carcinoma. Int J Cancer 2008;123:1779-86. 10.1002/ijc.23734 [DOI] [PubMed] [Google Scholar]
- 56.Xia J, Chen Q, Li B, et al. Amplifications of TAOS1 and EMS1 genes in oral carcinogenesis: association with clinicopathological features. Oral Oncol 2007;43:508-14. 10.1016/j.oraloncology.2006.05.008 [DOI] [PubMed] [Google Scholar]
- 57.India Project Team of the International Cancer Genome C. Mutational landscape of gingivo-buccal oral squamous cell carcinoma reveals new recurrently-mutated genes and molecular subgroups. Nat Commun 2013;4:2873. 10.1038/ncomms3873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang WF, Qin N, Song X, et al. Genomic Signature of Mismatch Repair Deficiency in Areca Nut-Related Oral Cancer. J Dent Res 2020;99:1252-61. 10.1177/0022034520930641 [DOI] [PubMed] [Google Scholar]
- 59.Jessri M, Dalley AJ, Farah CS. MutSalpha and MutLalpha immunoexpression analysis in diagnostic grading of oral epithelial dysplasia and squamous cell carcinoma. Oral Surg Oral Med Oral Pathol Oral Radiol 2015;119:74-82. 10.1016/j.oooo.2014.06.017 [DOI] [PubMed] [Google Scholar]
- 60.Jessri M, Dalley AJ, Farah CS. hMSH6: a potential diagnostic marker for oral carcinoma in situ. J Clin Pathol 2015;68:86-90. 10.1136/jclinpath-2014-202411 [DOI] [PubMed] [Google Scholar]
- 61.Fox SA, Tiwari L, Farah CS. Epigenetics and oral disease. In: Sonis ST, Villa A, editors. Translational Systems Medicine and Oral Disease. Academic Press, Elsevier; 2020. p. 163-206. [Google Scholar]
- 62.Viswanathan M, Tsuchida N, Shanmugam G. Promoter hypermethylation profile of tumor-associated genes p16, p15, hMLH1, MGMT and E-cadherin in oral squamous cell carcinoma. Int J Cancer 2003;105:41-6. 10.1002/ijc.11028 [DOI] [PubMed] [Google Scholar]
- 63.Shaw RJ, Liloglou T, Rogers SN, et al. Promoter methylation of P16, RARbeta, E-cadherin, cyclin A1 and cytoglobin in oral cancer: quantitative evaluation using pyrosequencing. Br J Cancer 2006;94:561-8. 10.1038/sj.bjc.6602972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ogi K, Toyota M, Ohe-Toyota M, et al. Aberrant methylation of multiple genes and clinicopathological features in oral squamous cell carcinoma. Clin Cancer Res 2002;8:3164-71. [PubMed] [Google Scholar]
- 65.Kresty LA, Mallery SR, Knobloch TJ, et al. Alterations of p16(INK4a) and p14(ARF) in patients with severe oral epithelial dysplasia. Cancer Res 2002;62:5295-300. [PubMed] [Google Scholar]
- 66.Mendelsohn J, Baselga J. The EGF receptor family as targets for cancer therapy. Oncogene 2000;19:6550-65. 10.1038/sj.onc.1204082 [DOI] [PubMed] [Google Scholar]
- 67.Mellinghoff IK, Wang MY, Vivanco I, et al. Molecular determinants of the response of glioblastomas to EGFR kinase inhibitors. N Engl J Med 2005;353:2012-24. 10.1056/NEJMoa051918 [DOI] [PubMed] [Google Scholar]
- 68.Yarden Y. The EGFR family and its ligands in human cancer: signalling mechanisms and therapeutic opportunities. Eur J Cancer 2001;37:S3-S8. 10.1016/S0959-8049(01)00230-1 [DOI] [PubMed] [Google Scholar]
- 69.Citri A, Yarden Y. EGF-ERBB signalling: towards the systems level. Nat Rev Mol Cell Biol 2006;7:505-16. 10.1038/nrm1962 [DOI] [PubMed] [Google Scholar]
- 70.Hynes NE, Lane HA. ERBB receptors and cancer: The complexity of targeted inhibitors. Nat Rev Cancer 2005;5:341-54. 10.1038/nrc1609 [DOI] [PubMed] [Google Scholar]
- 71.Huang SF, Cheng SD, Chien HT, et al. Relationship between epidermal growth factor receptor gene copy number and protein expression in oral cavity squamous cell carcinoma. Oral Oncol 2012;48:67-72. 10.1016/j.oraloncology.2011.06.511 [DOI] [PubMed] [Google Scholar]
- 72.Xia W, Lau YK, Zhang HZ, et al. Combination of EGFR, HER-2/neu, and HER-3 is a stronger predictor for the outcome of oral squamous cell carcinoma than any individual family members. Clin Cancer Res 1999; 5:4164-74. [PubMed] [Google Scholar]
- 73.Ono Y, Nakanishi Y, Ino Y, et al. Clinocopathologic significance of laminin-5 gamma2 chain expression in squamous cell carcinoma of the tongue: immunohistochemical analysis of 67 lesions. Cancer 1999;85:2315-21. [DOI] [PubMed] [Google Scholar]
- 74.Ulanovski D, Stern Y, Roizman P, et al. Expression of EGFR and Cerb-B2 as prognostic factors in cancer of the tongue. Oral Oncol 2004;40:532-7. 10.1016/j.oraloncology.2003.11.004 [DOI] [PubMed] [Google Scholar]
- 75.Vermorken JB, Herbst RS, Leon X, et al. Overview of the efficacy of cetuximab in recurrent and/or metastatic squamous cell carcinoma of the head and neck in patients who previously failed platinum-based therapies. Cancer 2008;112:2710-9. 10.1002/cncr.23442 [DOI] [PubMed] [Google Scholar]
- 76.Vermorken JB, Mesia R, Rivera F, et al. Platinum-based chemotherapy plus cetuximab in head and neck cancer. N Engl J Med 2008;359:1116-27. 10.1056/NEJMoa0802656 [DOI] [PubMed] [Google Scholar]
- 77.Burtness B, Harrington KJ, Greil R, et al. Pembrolizumab alone or with chemotherapy versus cetuximab with chemotherapy for recurrent or metastatic squamous cell carcinoma of the head and neck (KEYNOTE-048): a randomised, open-label, phase 3 study. Lancet 2019;394:1915-28. 10.1016/S0140-6736(19)32591-7 [DOI] [PubMed] [Google Scholar]
- 78.Kujan O, van Schaijik B, Farah CS. Immune Checkpoint Inhibitors in Oral Cavity Squamous Cell Carcinoma and Oral Potentially Malignant Disorders: A Systematic Review. Cancers (Basel) 2020;12:1937. 10.3390/cancers12071937 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as