Summary
Here we report a molecular docking-based approach to identify small molecules that can target the β-catenin (β-cat)-TCF4 protein-protein interaction (PPI), a key effector complex for nuclear Wnt signaling activity. Specifically, we developed and optimized a computational model of β-cat using publicly available β-cat protein crystal structures, and existing β-cat-TCF4 interaction inhibitors as the training set. Using our computational model to an in silico screen predicted 27 compounds as good binders to β-cat, of which 3 were identified to be effective against a Wnt-responsive luciferase reporter. In vitro functional validation experiments revealed GB1874 as an inhibitor of the Wnt pathway that targets the β-cat-TCF4 PPI. GB1874 also affected the proliferation and stemness of Wnt-addicted colorectal cancer (CRC) cells in vitro. Encouragingly, GB1874 inhibited the growth of CRC tumor xenografts in vivo, thus demonstrating its potential for further development into therapeutics against Wnt-associated cancer indications.
Subject areas: Pharmacology, Biochemistry, Structural biology, Cancer, Omics
Graphical abstract

Highlights
-
•
A computational docking model for β-catenin was developed and optimized.
-
•
The model was trained using protein crystal structures and known inhibitor compounds.
-
•
Compound GB1874 was identified by in silico screening and functionally validated.
-
•
GB1874 inhibited colon cancer growth in vitro and in vivo by blocking Wnt activity.
Pharmacology; Biochemistry; Structural biology; Cancer; Omics
Introduction
The canonical Wnt/β-catenin (β-cat) signaling pathway is an important signal transduction pathway required for proper embryonic development and adult tissue self-renewal (Clevers, 2006). Activation of the pathway occurs through the binding of Wnt ligand proteins to Frizzled and low-density lipoprotein receptor-related protein 6. Upon activation, the destruction complex (DC), comprising Axin, casein kinase 1α, glycogen synthase kinase 3β, and adenomatous polyposis coli (APC), is recruited to the membrane, resulting in its inactivation. Inhibition of the DC leads to cytoplasmic accumulation of non-phosphorylated β-cat and its subsequent translocation to the nucleus, where it binds to the lymphoid enhancer factor/T-cell factor (LEF/TCF) family of transcription factors, along with a host of other co-factors including Bcl-9 and Pygopus, to activate expression of downstream target genes (Nusse and Clevers, 2017).
Numerous studies have established that aberrant activation of the Wnt pathway, especially in stem cells often serves as a driver of disease, such as cancer (Anastas and Moon, 2013). Particularly, in the vast majority of colorectal cancers (CRCs), loss of function of the APC tumor suppressor gene is an early driver of tumor initiation, resulting in the formation of adenomas (Bienz and Clevers, 2000). Consequently, various Wnt pathway targeting agents including small molecule inhibitors, and antibody-based therapeutics have been reported over the years (Barker and Clevers, 2006; Chen et al., 2009; Grandy et al., 2009; Huang et al., 2009; Madan et al., 2016; Pavlovic et al., 2018; Takebe et al., 2015). Despite the large number of promising preclinical drug entities targeting Wnt signaling, most have failed to proceed to human clinical trials, and none have been approved for clinical use. One reason is that some of the proteins with driver mutations are not directly actionable (e.g. APC and Axin), and targeting components of the Wnt pathway that are epistatically upstream of these proteins would render those molecules ineffective (Hankey et al., 2018; Tanaka et al., 2017).
Consequently, we and several other groups envision that effective and specific downregulation of the Wnt pathway can be achieved by inhibiting the β-cat-TCF4 interaction (Gonsalves et al., 2011; Hahne and Grossmann, 2013), which is absolutely essential for the activation of Wnt-associated downstream target genes. The advantage of this strategy is two-fold. First, the β-cat-TCF4 interaction is the most downstream component of the Wnt signaling cascade. Hence effective inhibition of Wnt signaling can be achieved regardless of the presence of driver mutations in the upstream components. Second, this strategy could provide an opportunity to specifically target the transcriptional function of β-cat without affecting its interaction with E-cadherin (ECAD) at the cell-cell adherens junctions. This is important as loss of β-cat-ECAD interaction leads to downregulation of ECAD and is associated with epithelial-mesenchymal transition and increased metastasis of cancer cells (Valenta et al., 2012). We propose that specificity could be achieved by selectively targeting the β-cat-TCF4 binding pocket that is distinct from the β-cat-ECAD binding pocket.
Previously we reported a cell-based RNA interference chemical genetics screen to identify compounds that inhibit β-cat-TCF4 interaction (Gonsalves et al., 2011). The screen identified 3 compounds; iCRT3, iCRT5, and iCRT14 that disrupted the β-cat-TCF4 interaction without affecting β-cat’s interaction with ECAD. In silico docking of iCRT3 onto β-cat predicted that iCRT3 binds to the site on β-cat that binds to the extended region of TCF4 (residues 13–25) (Gonsalves et al., 2011). This binding site on β-cat contains a K435 residue that forms a salt bridge with D16 of TCF4. Single amino acid substitution of D16 on TCF4 or K435 on β-cat has been shown to be sufficient to disrupt their interaction and markedly reduce downstream transcriptional activity (Fasolini et al., 2003; Liu et al., 2006; Poy et al., 2001), thus highlighting the importance of this binding site.
Taking an alternative approach toward hit identification, in this study, we adopted the strategy of in silico docking to identify inhibitors of β-cat-TCF4 interaction, with a particular focus on the critical TCF4 binding site on β-cat. Over the years, several other groups have also adopted similar strategies to target this binding pocket to inhibit β-cat-TCF4 interaction (Huang et al., 2014; Tian et al., 2012; Trosset et al., 2006; Yu et al., 2013). However, unlike previous studies, our approach made use of a combination of existing protein x-ray crystal structures, as well as structures of known β-cat-TCF4 interaction inhibitors to develop and further optimize the computational model of the β-cat protein. To demonstrate the predictability of our computational model system, we carried out in silico screening of a pharmacological diversity set of compounds from Enamine and prioritized 27 compounds as potential binders of β-cat. Experimental validation of the hit compounds identified 3 potent inhibitors of the β-cat-TCF-dependent Wnt reporter activity, of which compound GB1874 was found to be the most promising in eliciting robust Wnt tumor inhibitory phenotypes, both in vitro and in vivo.
Results
Generation of β-cat structural models
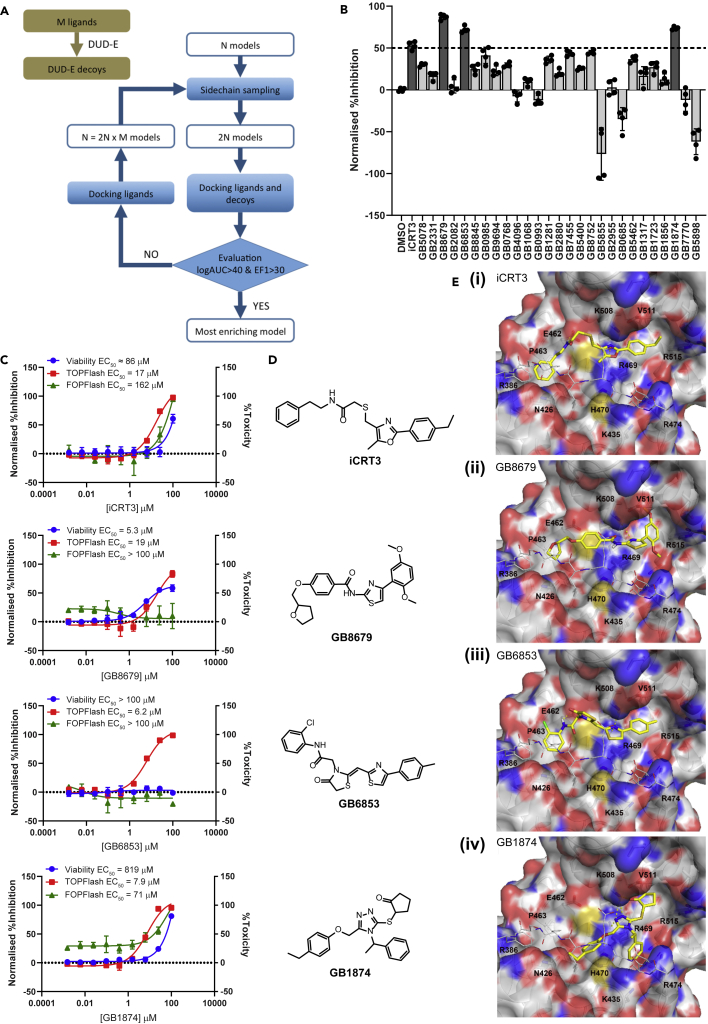
The final structure model of β-cat used in prospective ligand predictions was derived from three β-cat crystal structures including β-cat-TCF3, β-cat-TCF4, and β-cat-BCL9-TCF4 complexes [PDB codes: 1G3J (Graham et al., 2000), 1JPW (Poy et al., 2001), and 2GL7 (Sampietro et al., 2006), respectively] through two stages: (1) optimization (training), and (2) blind testing (validation) on the basis of ligand enrichment measured by EF1 and logAUC. The optimization was accomplished by an in-house automated modeling and docking platform (Figure 1A). This platform is composed of three key components: binding-site sidechain sampling in the presence of known ligands, docking of a training library containing both ligands and decoys, and ligand enrichment evaluation. In this study, a putative ligand binding site was first predicted by MetaPocket (Zhang et al., 2011). This binding site contained the charged groove created by β-cat armadillo repeats 4–9, which also overlapped with the β-cat-TCF4 interaction interface. Second, the three crystal structures of β-cat were subjected to sidechain sampling by SCWRL (Krivov et al., 2009), introducing three alternative models. This set of 6 β-cat structures (3 crystal structures + 3 sidechain-perturbed models) served as the starting point of optimization. Third, to allow a broader exploration of β-cat conformational space, we docked a training library of 153 compounds including 3 confirmed ligands (iCRT3, iCRT5, and iCRT14) (Gonsalves et al., 2011) and 150 property-matching decoys extracted from ZINC (Irwin and Shoichet, 2005) using the DUD-E approach (Mysinger et al., 2012) to each of the 6 β-cat structures and evaluated each β-cat structure’s docking performance in terms of the ligand enrichment. Fourth, for each of the 6 β-cat structures, the binding site sidechains were optimized using Protein Local Optimization Program (Sherman et al., 2006) in the presence of each docked ligand (iCRT3, iCRT5 and iCRT14), yielding in total 18 (6 x 3) new β-cat structures. The third and fourth steps were iterated till there are optimized structures showing ligand enrichment performance better than the following, empirical thresholds: logAUC ≥ 40 and EF1 ≥ 30.
Figure 1.
In silico docking studies on β-cat identified small molecule inhibitors of Wnt signaling
(A) The modeling and docking platform used in the generation of β-cat structures for ligand discovery. Three crystal structures were used as the starting models (N = 3). The training library of small molecules contains 3 known inhibitors of β-cat (M = 3) and 150 decoys. Within one iteration, every target structure would be further optimized in the presence of each of the 3 known ligands docked.
(B) TOPFlash reporter inhibitory activities of predicted compounds in HEK293T STF cells. Cells were treated with compounds at 10 μM and simultaneously stimulated with Wnt3A conditioned medium for 24 hr prior to measuring luciferase activity. TOPFlash reporter activities of compound-treated cells were normalized to cell viability and presented as fold change over DMSO treated cells. Error bars represent mean ± SD of 4 replicates.
(C) Secondary dose response TOPFlash reporter inhibition and toxicity curves. STF cells were treated with different concentrations of compounds and simultaneously stimulated with Wnt3A conditioned medium for 24 hr prior to measuring luciferase activity or cell viability. Luciferase activities of cells treated with compounds at the various concentrations were first normalized to cell viability at the same concentration and presented as percentage inhibition compared to DMSO treated cells. EC50 values were calculated using a three-parameter nonlinear regression. Error bars represent mean ± SD of 3 replicates.
(D) Chemical structures of iCRT3 and the top 3 hit compounds.
(E) Predicted binding modes of iCRT3 (i), and 3 novel hits found in this study: GB8679 (ii), GB6853 (iii), and GB1874 (iv) in the optimized, most enriching β-cat structure (partial transparent surface). The docked ligands (yellow solid sticks), and the critical β-cat binding site residues (partial transparent sticks) are displayed. The TCF4 peptide fragment (solid white lines) from β-cat-TCF4 crystal complex structure (PDBID 1JPW) is also shown here to indicate the PPI interface.
Prediction of small molecule inhibitors of β-cat-TCF4 interaction
In the training, the best β-cat structure generated by the above-described modeling and docking platform, showed the best overall ligand enrichment (logAUC = 44.2), and the best early ligand enrichment (EF1 = 33.3). Of the known inhibitors, iCRT3, was ranked the best amongst the 153 compounds in the training library. This most enriching β-cat structure was subjected to a blind testing with the aid of a validation library containing 63 iCRT3 analogs (Table S1). Nineteen ligand candidates were selected from this virtual screen, taking into consideration both docking ranks by DOCK energy scores, and docking poses by human inspection (Table S1). In the meantime, all the 63 compounds were tested in a Super-TOPFlash Wnt reporter assay (STF cells) (Xu et al., 2004) at a single concentration. Twelve of 63 compounds showed activity (data not shown). These 12 compounds were further tested for dose response inhibition against the STF cells, and 5 compounds showed comparable or lower EC50 values than the known inhibitor iCRT3 (Table S1). Importantly, all 5 inhibitors were predicted as top ligand candidates, and 4 of these 5 compounds (A03, A06, A45, A48) received better docking ranks than iCRT3. This suggested that the most enriched structure generated by training can predict for novel ligands.
Next, we computationally screened a library of 10240 small molecules (purchased from Enamine) against this most enriched β-cat structure. Five hundred (4.0% of the screened library) top scoring hits were analyzed manually. Twenty-seven compounds were selected for experimental testing based on three criteria: (1) the degree of overlap with TCF4 at the binding site on β-cat, (2) the formation of favorable interactions with β-cat residues, e.g. hydrogen bonds, and (3) the chemical novelty of their scaffolds.
Functional validation of predicted inhibitors using TOPFlash/Wnt-reporter assay
The 27 compounds identified from the docking studies were first screened at 10 μM for their ability to inhibit Wnt signaling in the STF reporter cells (Figure 1B). From this screen, we identified 3 compounds (i.e. GB8679, GB6853, and GB1874) which on average, inhibited Wnt signaling by more than 50% compared to DMSO control (Figure 1B) while having less than 25% toxicity on the cells (Figure S1A). Dose response studies of the 3 hit compounds on the STF TOPFlash reporter revealed that they could inhibit the Wnt signaling pathway with low micromolar EC50 values (Figure 1C). Amongst the 3 hit compounds, only GB8679 affected the viability of the reporter cells to the same extent as its effect on the reporter activity. Encouragingly, when we compared the selectivity of the compounds to TOPFlash reporter vs FOPFlash reporter (containing mutated TCF/LEF binding sites) (Veeman et al., 2003), the compounds were at least 8-fold more selective for the TOPFlash reporter (Figure 1C). GB1874 (EC50 = 7.9 μM) and GB6853 (EC50 = 6.2 μM) were also more potent against the TOPFlash reporter compared to iCRT3 (EC50 = 17 μM, Figure 1C).
To investigate the selectivity of the hit compounds on Wnt pathway inhibition, we tested the compounds in 3 other reporter cell lines. Amongst these 3 reporter cell lines were the STF3A reporter cells, which are STF cells with stable and constitutive expression of Wnt3A (Coombs et al., 2010). Other reporter cell lines include Notch CSL (CBF1, Suppressor of Hairless, Lag-1), and Hippo TEAD. The hit compounds and iCRT3 were tested against these reporter cell lines at a concentration of 20 μM. The results showed that while GB6853 was a potent inhibitor of Wnt STF reporter, its potency in Wnt STF3A reporter cells was greatly diminished (Figure S1B). This suggested that GB6853 would be less effective in targeting cells displaying constitutive Wnt pathway activation. Although GB8679 and GB1874 were both less effective inhibitors of Wnt reporter activity in the STF3A cells compared to the STF cells, they were still more potent compared to iCRT3. Apart from differences in Wnt pathway inhibition, GB8679 and GB1874 also varied in terms of pathway specificity. GB8679 inhibited both Notch and Hippo pathway while GB1874 affected the Notch pathway but not the Hippo pathway. These are also in contrast to the selectivity of iCRT3 whereby it has no effect on Notch pathway but is an activator of the Hippo pathway (Figure S1B). The effect of the inhibitors on the Notch pathway is not entirely unexpected since the cross talk between Wnt and Notch signaling have been documented by several previous studies (Collu et al., 2014; Kay et al., 2017). We hypothesize that while the compounds have a direct effect on the Wnt pathway, they may exert indirect effects on cross-regulatory pathways, such as Notch.
In silico modeling predicts distinct modes of β-cat binding of the top 3 candidate small molecule binders
Closer examination of the top 3 hit compounds revealed structural differences amongst the compounds (Figure 1D). However, the predicted β-cat binding modes of GB8679 and GB6853 were highly similar to that of iCRT3. The bulky, hydrophobic groups of all 3 compounds were predicted to occupy two pockets on β-cat (Figure 1E, i to iii). One of the pockets, lined by R386, N426, and P463, has been shown to interact with I19 and F21 of TCF4. In our docking model, the phenyl group of iCRT3, the tetrahydrofuran group of GB8679, and the chlorophenyl group of GB6853 were all predicted to occupy this pocket (Figure 1E, i to iii). The other pocket, lined by K508, V511, R515, was filled by the ethylphenyl group of iCRT3, the 2,5-dimethoxybenzyl group of GB8679, and the p-tolyl group of GB6853 (Figure 1E, i to iii). Meanwhile the cleft between K508 and R469, which interacts with E17 of TCF4, formed hydrogen bonds and salt bridges with the oxazole group of iCRT3 and with the thiazole groups of GB8679 and GB6853, respectively (Figure 1E, i to iii). Lastly, hydrogen bonding interactions between E462 of β-cat and the amide groups of iCRT3 and GB6853 were predicted to further stabilize the small molecule-protein interaction (Figure 1E, i and ii).
The third hit, GB1874, presented a different binding mode from iCRT3 and the other two hits. It still packed into the cleft between K508 and R469 with its triazole group, and filled the two pockets lined by R386, N426, P463, and K508, V511, R515 with its hydrophobic groups (Figure 1E, iv). In addition, the ethylphenyl group of GB1874 packed into the pocket lined by H470, R474, and K435 (Figure 1E, iv). This is also the pocket on β-cat that is occupied by TCF4 D16 residue, forming a salt bridge with K435 in β-cat, and representing a key β-cat-TCF4 interaction (Amit et al., 2002). Given that the predicted binding modes of iCRT3, GB8679, and GB6853 did not access this pocket, we hypothesized that this specific binding mode of GB1874 would allow it to more potently disrupt β-cat-TCF4 interactions, as compared to iCRT3 and the other two hits.
Notably, GB8679, GB6853, and GB1874 were all identified through docking studies with our most enriched β-cat structural model. To investigate whether these compounds could still be identified through standard docking studies, we docked the library of 10240 small molecules against the individual crystal structures of β-cat (i.e. PDB codes: 1G3J, 1JPW, and 2GL7) and ranked the compounds according to their docking scores. Interestingly the 3 hit compounds ranked much more poorly when docked against the individual crystal structures of β-cat as shown in Table S2. This comparative study therefore demonstrates the advantage of using our computational strategy of using the best-fit structure (from training stage) over standard docking studies in screening for novel small molecule inhibitors of protein-protein interactions (PPIs).
Hit compounds downregulate β-cat- and TCF-dependent Wnt signaling pathway
To investigate if the inhibitors of Wnt reporter activity could inhibit Wnt signaling in a more biologically relevant model, we tested the compounds in HCT116 CRC cells. HCT116 cells were chosen as they are wild-type for APC mutation but possess a heterozygous deletion of a Ser45 residue in β-cat (Ilyas et al., 1997). Ser45 is the site of phosphorylation by casein kinase 1, and this phosphorylation activates β-cat for subsequent phosphorylation and degradation (Amit et al., 2002). Consequently, Wnt signaling in HCT116 cells is constitutively active due to the presence of the mutant β-cat, and the inhibition of Wnt signaling in HCT116 cells would have to occur downstream of activated β-cat. We hypothesized that if our hit compounds inhibit the β-cat-TCF4 interaction, we should observe downregulation of Wnt target genes upon treatment with the compounds.
We first carried out gene expression profiling of HCT116 cells treated separately with the 3 hit compounds. Based on our previous experience with iCRT3 (Gonsalves et al., 2011), we decided to treat the cells with compounds at 50 μM. For comparison, we also profiled the gene expression changes upon transient depletion of β-cat by siRNA (i.e. siCTNNB1). RNA isolated from these cells was analyzed through RNA sequencing and read counts obtained were compared with the respective control-treated samples (DMSO for compounds and negative siRNA for siCTNNB1). Statistically significant and differentially expressed genes (Table S3) were analyzed via Gene Set Enrichment Analysis (GSEA) against Wnt signaling related gene sets (Figure 2A and Table S4). Included in the gene sets analyzed were gene sets reported in a recent study in which the authors used β-cat knockout and TCF/LEF knockout HEK293T cells to identify β-cat-dependent target genes that can be TCF/LEF-dependent or TCF/LEF-independent (Doumpas et al., 2019). Our analyses showed that differentially expressed genes upon β-cat knockdown and GB1874 treatment were significantly enriched for β-cat-dependent target genes identified in the abovementioned study. Notably, the genes were enriched for the TCF/LEF-dependent subset of β-cat-dependent target genes and not the TCF/LEF-independent subset (Figure 2A and Table S4). Gene expression changes upon GB1874 treatment in the β-cat- and TCF/LEF-dependent set of genes were also comparable to that of β-cat knockdown (Figure 2B). All these suggest that GB1874 targets the β-cat-TCF/LEF complex in HCT116 cells.
Figure 2.
Hit compounds downregulate β-cat- and TCF-dependent Wnt signaling pathway
(A) GSEA of differentially expressed genes in HCT116 cells upon β-cat knockdown (siCTNNB1) or treatment with hit compounds (50 μM). Gene sets for which p value <0.05 are shown.
(B) Heatmap showing gene expression changes of β-cat and TCF4 dependent genes upon β-cat knockdown (siCTNNB1) or treatment with hit compounds (50 μM).
(C) qPCR gene expression validation of Wnt target genes in HCT116 cells (top panel), DLD-1 cells (middle panel), and SW480 cells (bottom panel) upon compound treatment. Cells were treated for 18 hr with either DMSO or compounds at 50 μM after which RNA was harvested and analyzed for gene expression changes. Relative expression was normalized to DMSO control. Error bars represent mean ± SD of 3 replicates.
(D and E) Immunoblot analyses for expression of Wnt target proteins (D) or Wnt pathway related proteins (E) in HCT116 cells upon treatment with hit compounds. HCT116 cells were treated for 18 hr with either DMSO or compounds at 50 μM after which protein extracts were analyzed by Western blot.
Quantitative polymerase chain reaction (qPCR) validation of RNA sequencing results showed that canonical Wnt target genes such as AXIN2, CCND1, and NKD1 were all downregulated to different extents upon compound treatment in HCT116 cells (Figure 2C, top panel). Amongst the hit compounds, GB1874 was the most potent inhibitor of Wnt target gene expression. This downregulation of Wnt target genes was also evident, to a certain extent, in CRC cell lines DLD-1 and SW480 (Figure 2C, middle and bottom panels). Additionally, downregulation of CRC stem cell markers RNF43 and LGR5 were also observed upon GB1874 treatment in all the CRC cell lines (Figure 2C). HCT116 cells had very low expression of LGR5 hence it was not investigated in this cell line. Validation of protein expression changes via Western blot in HCT116 cells also showed that iCRT3 and GB1874 were the most effective at downregulating Wnt target protein expression (Figure 2D). Notably, while GB1874 strongly reduced the expression of Wnt target proteins, it had minimal effect on the expression of β-cat itself, or the expression of its partner proteins, such as ECAD and TCF4 (Figure 2E). These results suggested that GB1874 downregulates Wnt signaling via the inhibition of β-cat-TCF-dependent gene expression activity, and not by regulating the expression of these effector proteins themselves.
GB1874 inhibits Wnt signaling via disruption of β-cat-TCF4 PPIs
As GB1874 and the other hit compounds were identified through in silico docking studies on the β-cat protein, we investigated whether they were able to disrupt the β-cat-TCF4 PPIs. Co-immunoprecipitation (co-IP) of β-cat and its interacting partners was carried out in HCT116 cells. Western blot analyses of the proteins co-immunoprecipitated with β-cat upon compound treatment revealed that only GB1874 could significantly reduce β-cat-TCF4 interaction to the same extent as iCRT3 in HCT116 cells (Figures 3A and 3B). Encouragingly, we observed that while GB1874 affected the β-cat-TCF4 interaction, it has minimal effect on the β-cat-ECAD interaction (Figures 3A and 3B).
Figure 3.
GB1874 inhibits Wnt signaling by disrupting β-cat-TCF4 interaction
(A) Co-IP of β-cat and its endogenous binding partners. HCT116 cells were treated for 18 hr with the hit compounds at the indicated concentrations. β-cat was immunoprecipitated from protein lysates of the treated cells and the amount of TCF4 and ECAD bound to β-cat was analyzed via Western blot.
(B) Quantification of TCF4 (left panel) or ECAD (right panel) protein co-immunoprecipitated with β-cat under different treatment conditions. Normalized TCF4 and ECAD pull-down for each compound treatment is presented as fold-change with respect to DMSO control. Error bars represent mean ± SEM of n = 4 independent experiments. Two-tailed paired Student's t test was carried out between DMSO control and each compound treatment. ∗p < 0.05, ns p > 0.05.
(C) Dissociation constants KD of iCRT3 and GB1874 against β-cat as determined from SPR studies. The KD values were calculated using a steady state affinity model.
(D) Dose response inhibition of β-cat-TCF4 binding by compound GB1874 (left panel, n = 2) or a TCF4-fluorescein-5-isothiocyanate peptide (right panel, n = 1) based on SPR studies. The IC50 value was calculated using a four-parameter nonlinear regression. Error bars represent mean ± SEM.
See also Figures S2 and S3.
Next, we investigated whether GB1874 could interact with purified β-cat in vitro and inhibit its interaction with TCF4. We employed surface plasmon resonance (SPR) assays using purified proteins encoding for the armadillo repeat domain of β-cat (amino acids 134–668), and the β-cat-interacting TCF4 N-terminal domain (amino acids 1–55). We first determined the binding affinity of GB1874 to β-cat by flowing different concentrations of analyte GB1874 over the β-cat ligand. From the sensorgrams (Figure S2A), we observed dose dependent binding of GB1874 to β-cat. A plot of steady state response values against the different concentrations of GB1874 indicated that the compound binds to β-cat with a KD of 0.58 ± 0.07 mM (n = 2, Figure 3C). In comparison, iCRT3 binds to β-cat with a KD of 3.32 ± 2.22 mM (n = 2, Figure 3C). Apart from SPR, we also investigated the interaction between GB1874 and β-cat through microscale thermophoresis (MST) and found that the KD of this interaction was similar (0.42 mM–0.57 mM) to that measured through SPR (Figure S3). iCRT3 showed strong autofluorescence (data not shown) in the MST assay, hence no KD value for iCRT3 could be determined from this assay. The low binding affinity of GB1874 to β-cat also suggests that while the ethylphenyl group of GB1874 could potentially access the pocket lined by K435, the hydrophobic nature of the ethylphenyl group is unable to form strong interactions with K435. Replacement of the ethylphenyl group with a pyridinyl or phenol group could potentially improve the binding affinity of GB1874 to β-cat.
We also sought to ascertain if GB1874 can directly inhibit the interaction between β-cat and TCF4 in cell free assays in vitro. Toward this aim, we designed a competition SPR experiment in which the TCF4 N-terminal domain replaced β-cat as the ligand in this experiment. 50 nM of the β-cat analyte, pre-incubated with different concentrations of GB1874, was then injected over the TCF4 N-terminal domain ligand. As evident from the sensorgrams (Figure S3B), GB1874 was found to inhibit the interaction between β-cat and TCF4 with an IC50 value of 24 μM (n = 2) (Figure 3D). On the other hand, a fluorescein-5-isothiocyanate labeled TCF4 peptide (amino acids 7–51) in the same assay resulted in an IC50 value of 29 nM (n = 1) (Figure 3D).
GB1874 affected proliferation and stemness of “Wnt-addicted” cancer cells
Having established that GB1874 could disrupt the β-cat-TCF4 interaction and reduce expression of Wnt target genes, we wanted to determine whether GB1874 elicit phenotypes that are reminiscent of reduced Wnt activity in biologically relevant cancer cell lines. Firstly, we treated HCT116 cells daily with 10 μM of GB1874 and monitored cell proliferation over 4 days. For comparison, we included compounds such as IWP-2, XAV 939, and iCRT3 (Kahn, 2014), all of which target different components along the Wnt signaling transduction cascade. As shown in Figure 4A, treatment with GB1874 effectively inhibited the growth of HCT116 cells. Notably GB1874 was more effective than iCRT3 in the growth inhibition assay. Compounds IWP-2 and XAV939 (inhibitors of Porcupine and Tankyrase, respectively), which inhibit Wnt signaling upstream of the β-cat-TCF4 interaction, were less effective at affecting the growth of HCT116 cells. Dose response viability studies with CRC cells lines including multiple patient-derived primary CRC cell lines also indicated that GB1874 was more potent against these cell lines compared to other Wnt inhibitors, such as XAV939 (Figure 4B).
Figure 4.
GB1874 inhibited growth of Wnt-driven cells in vitro and in vivo
(A) Growth curves of HCT116 cells treated daily with compounds at 10 μM. One-way repeated measures ANOVA with Dunnett's multiple comparison test was carried out to determine the significance between compound treatment and DMSO control. ∗p < 0.05, ns p > 0.05. Error bars represent mean ± SD of 3 replicates.
(B) Dose response EC50 values of the different compounds against CRC cell lines, HCT116, DLD-1, SW480, and patient-derived primary CRC cell lines. Cells were treated with different concentrations of the various compounds for 72 hr after which cell viability was determined. EC50 values were calculated using a four-parameter nonlinear regression. n.d.: not determined.
(C) Effects of GB1874 on colony formation of HCT116 cells. HCT116 cells were grown in 6-well plates and treated with either DMSO or hit compounds at 30 μM for 7 days. Cells were then fixed and stained with crystal violet. The number of colonies obtained were counted. Two-tailed Student's t test was carried out between each compound treatment and DMSO control. ∗∗∗p < 0.001. Error bars represent mean ± SD of 3 replicates.
(D) Effects of GB1874 on spheroid formation of HCT116 cells. HCT116 cells were grown in ultra-low attachment 96-well plates and treated with either DMSO or the compounds at 30 μM for 14 days. The number of spheroids ≥200 μm in size was then determined. Two-tailed Student's t test was carried out between each compound treatment and DMSO control. ∗∗∗p < 0.001, ∗∗p < 0.01. Scale bar represents 1 mm. (A), (C), and (D), error bars represent mean ± SD of 6 replicates.
(E) NSG mice xenografted with HCT116 cells were treated with vehicle control (n = 6) or 50 mg/kg GB1874 (n = 6) via i.p. every other day for 2 weeks. Tumor volumes (left panel) were monitored over time. Two-way ANOVA was carried out between vehicle control and treatment tumor volumes. ∗p < 0.05. Error bars represent mean ± SEM. Images of tumors at the end of treatment are shown in the right panel.
(F) Representative IHC staining of tumors for Ki67 and cyclin D1 expression (left panel). Scale bar represents 100 μm. Quantification of the number of cyclin D1 and Ki67 expressing cells per unit tumor area (right panel). One-tailed Student's t test was carried out between vehicle control and treatment for each marker. ∗∗p < 0.01, ∗p < 0.05, ns p > 0.05. Error bars represent mean ± SD of 3 tumors.
See also Figure S4.
We next investigated the ability of GB1874 to affect the survival of cancer stem cells. Through HCT116 colony formation (Figure 4C) and spheroid formation (Figure 4D) assays, we demonstrated the potent inhibitory activity of GB1874 on cancer stem-like cells. This effect was not limited to HCT116 cells as similar phenotypes were observed upon treatment of DLD-1 (Figure S4A) and SW480 (Figure S4B) CRC cells with GB1874, both of which display enhanced Wnt activity due to APC loss of function, and thus increased levels of non-phosphorylated, active form of β-cat.
GB1874 inhibits growth of mouse tumor xenografts
Our results thus far suggest that GB1874 is a potent compound against Wnt driven CRC cells. To investigate its efficacy in vivo, we generated HCT116 xenografts in immunocompromised NSG mice. Tumor-bearing mice were then treated with either vehicle control or GB1874 at 50 mg/kg every other day (q.a.d.) via i.p injection. We found that GB1874 at this dosage effectively inhibited the growth of HCT116 xenografts in vivo (Figure 4E) while causing minimal to no systemic toxicity in the mice (Figures S4C and S4D). Tumors harvested at the end of the treatment cycle were also fixed and stained for expression of Wnt target protein cyclin D1 and the proliferation marker Ki67. Based on our immunohistochmemistry (IHC) staining results, we observed a significant reduction in the number of cyclin D1 expressing and Ki67 expressing cells per unit area of tumor (Figure 4F). Notably, GB1874-treated tumors displayed markedly larger necrotic regions compared to control tumors (Figure S4E). Altogether, these results corroborated the effect of GB1874 in stemming the growth and proliferation of activated β-cat-associated tumors in vivo by downregulating expression of Wnt target genes.
Analogs of GB1874 are also potent inhibitors of Wnt signaling
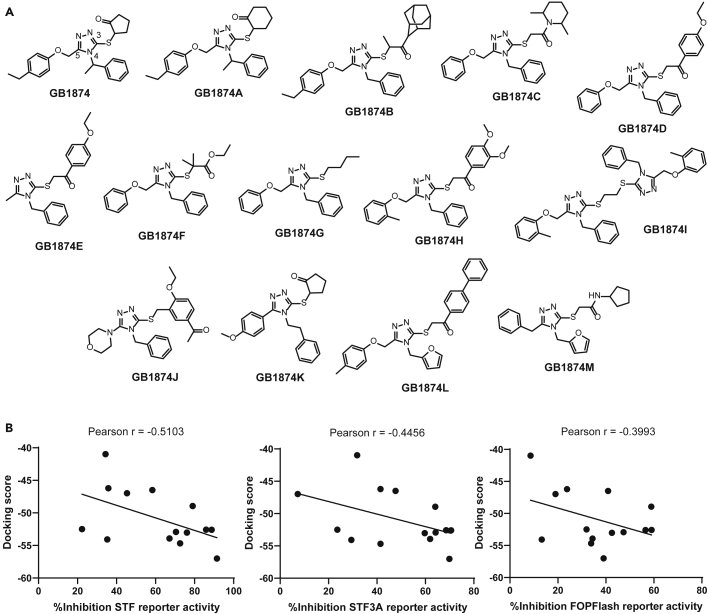
Having identified GB1874 as our most promising inhibitor of Wnt signaling through disruption of the β-cat-TCF4 interaction, we sought to use our computational model to screen for and identify structural analogs of GB1874 that can inhibit Wnt signaling. We obtained 215 GB1874 analogs by running the “Fingerprint Tanimoto-based 2D similarity search” provided by PubChem with a cutoff of Tanimoto Threshold 90% (the default by PubChem). Of these 215 analogs, we identified 13 analogs that are “in-stock” and with better, comparable, and worse docking scores with respect to that of GB1874 against β-cat (Figure 5A and Table S5). A screen at 10 μM against STF and STF3A Wnt reporter cell lines, as well as FOPFlash reporter identified varying inhibitory potencies and selectivity for the Wnt reporters (Table S5).
Figure 5.
Structural analogs of GB1874 were also potent inhibitors of Wnt signaling
(A) Structures of GB1874 and 13 structural analogs of GB1874 identified from in silico docking studies.
(B) Correlation plots and Pearson correlation scores between STF reporter (left panel), STF3A reporter (middle panel), and FOPFlash reporter (right panel) percentage inhibition scores of GB1874 analogs, tested at 10 μM, and their corresponding docking scores.
Based on the results obtained, some preliminary structure-activity relationships (SARs) could be deduced from the results. For example, GB1874A differs from GB1874 by replacing the cyclopentanone with a cyclohexanone functional group at the C3 position of the 1,2,4-triazole scaffold. Based on the docking scores as well as the reporter inhibition scores, this modification was tolerated (GB1874 inhibition of STF = 71%, STF3A = 64% vs GB1874A inhibition of STF = 86%, STF3A = 69%). In agreement with this, the docked pose of GB1874A was also highly similar to that of GB1874 (Figure S5). Bulky groups at the C3 position of the 1,2,4-triazole scaffold were also mostly tolerated when we compared GB1874 with analogs such as GB1874B (inhibition of STF = 89%, STF3A = 71%), GB1874H (inhibition of STF = 79%, STF3A = 64%), GB1874I (inhibition of STF = 91%, STF3A = 70%), and GB1874L (inhibition of STF = 76%, STF3A = 60%). The docked poses of these compounds (except GB1874B) were also similar in that the less bulky substituent on either C3 or C5 of the 1,2,4-triazole scaffold preferentially binds to the pocket lined by R386, N426, and P463 residues (Figure S5). Interestingly, for GB1874B, the bulkier adamantyl substituent on the C3 carbon was predicted to bind near to the pocket line by K508, V511, and R515 residues instead. GB1874D and GB1874M were also predicted to bind with conformations similar to GB1874H, GB1874I, and GB1874L (Figure S5), but they were significantly less effective against the TOPFlash reporter (GB1874D inhibition of STF = 35%, STF3A = 29%; GB1874M inhibition of STF = 22%, STF3A = 24%). Further investigation would need to be carried out to probe for the cause of this discrepancy. We also observed that replacement of either the C3 or C5 substituent on the 1,2,4-triazole scaffold with less bulky alkyl groups such as in GB1874E and GB1874G reduced the potency of the compounds, as predicted by docking (GB1874E inhibition of STF = 36%, STF3A = 41%; GB1874G inhibition of STF = 58%, STF3A = 48%). Lastly, having a bulky substituent directly bonded to the 1,2,4-triazole scaffold (i.e. GB1874J), or increasing the size of the substituent at the N4 nitrogen of the 1,2,4-triazole scaffold (i.e. GB1874K) were both unfavorable to the inhibitory activities of the compounds against the TOPFlash reporter (GB1874J inhibition of STF = 45%, STF3A = 7%; GB1874K inhibition of STF = 34%, STF3A = 32%).
We also correlated the docking scores of the 13 analogs against their STF, STF3A, and FOPFlash percentage inhibition scores (Figure 5B). Given that a lower docking score represents a more potent predicted binding affinity and hence a higher inhibition score, negative correlations between the docking scores and the percentage inhibition scores against STF and STF3A reporters were expected. Encouragingly the correlations with STF and STF3A percentage inhibition scores (Pearson correlation r = -0.5103 and -0.4456, respectively) were more negative compared to the correlation with FOPFlash percentage inhibition scores (Pearson correlation r = -0.3993) (Figure 5B).
Discussion
Effective targeting of the Wnt signaling pathway has been the goal of many drug discovery programs. However, till date none of the Wnt pathway inhibitors have reached clinical approval. Interestingly several recent studies have implicated a role for β-cat-driven immune exclusion as a mechanism for immune escape. These findings have revealed that cancers with elevated expression β-cat are strongly correlated with impaired immune cell recruitment, leading to poor prognosis (Luke et al., 2019). There is thus a renewed interest in identifying novel anti-β-cat agents and the possible use of such inhibitors as potential adjuvants to immune-checkpoint inhibitors (anti-PD1, anti-CTLA4, or anti-PD-L1). In this study, we chose to target the β-cat transcription complex and in particular, the β-cat-TCF4 PPIs. We envisioned that efficacious inhibitors that function most downstream in the linear signaling cascade would be broadly applicable for abrogating Wnt/β-cat activity in a variety of contexts where activating mutations are present either in β-cat itself or in the components upstream of the transcription complex.
Using the computational model of β-cat that we developed, we managed to screen for and identify novel scaffolds that are potent inhibitors of Wnt reporter activity. Out of the 27 compounds identified, GB8679, GB6853, and GB1874 showed promising Wnt inhibitory activities and were selected for downstream functional studies. Subsequent functional studies identified GB1874 as our most potent hit compound with similar biological activity as iCRT3. Phenotypically, GB1874 robustly affected the proliferation and stemness of Wnt-dependent CRC cells. Through co-IP and western blotting, we showed that the phenotypic effects were due to disruption of the β-cat-TCF4 interaction which subsequently affected the expression of Wnt target genes. Furthermore, we demonstrated the ability of GB1874 to bind to β-cat in vitro and disrupt the β-cat-TCF4 interaction through SPR studies. However, we also observed that the SPR response values (i.e. RU values) measured upon compound binding to β-cat were higher than expected for a small molecule-protein interaction. Based on the equation:
and with RUβ-cat = 7800, MWGB1874 = 422, MWβ-cat = 60 kDa, the theoretical RUmax for a 1:1 binding would be about 55. Instead, we observed RUmax values of up to 120 (Figure S2A), suggesting that the binding ratio of GB1874 to β-cat in vitro was greater than 1:1. While it may be possible that there are other sites on β-cat for GB1874 binding, the super stoichiometric binding observed in our SPR experiments could also be due to aggregation of GB1874 in aqueous solution, even in the presence of detergents such as Tween-20. For the SPR studies, the maximum solubility of GB1874 in the running buffer was around 100 μM. This low aqueous solubility of GB1874 also precludes the use of higher concentrations of the compound to obtain more accurate binding curves. Further studies with analogs of GB1874 that have improved aqueous solubility could potentially help to address this issue.
While the in vitro β-cat-TCF4 interaction efficacy of GB1874 can be further optimized, GB1874 was able to strongly inhibit the growth of HCT116 tumor xenografts at 50 mg/kg in vivo. Immunohistochemistry staining of HCT116 tumors also showed reduction in cell proliferation (Ki67), as well as Wnt target protein expression (cyclin D1) in GB1874-treated tumors compared to vehicle control-treated tumors. This suggested that the in vivo effect of GB1874 was, at least in part, via inhibition of Wnt signaling. We believe that with further optimization, we would be able to improve upon the potency and selectivity of GB1874 as a β-cat-TCF4 interaction inhibitor. Future studies would also need to be carried out to determine how well GB1874 fares in comparison with other β-cat-TCF4 interaction inhibitors.
PPI targets are often difficult to modulate with small molecules as the PPI interfaces tend to be large with shallow pockets (Arkin et al., 2014). Despite the interest, only a few potent and selective small molecule inhibitors have been discovered for the β-cat-TCF4 interface (Kahn, 2014). In this study, we attempted to find new inhibitor chemotypes for the β-cat-TCF4 interface with the aid of a structure-based computational approach that enables us to investigate the underlying molecular interactions involved in protein-ligand binding and thus interpret experimental results in atomic-level detail. We leveraged on existing protein structures determined by X-ray crystallography and biological activities of known β-cat-TCF4 interaction inhibitors to develop a more predictive computational model. The differentiating factor of our approach relies on the fact that it explicitly considers conformational changes in β-catenin surface due to the difference between a native partner protein and a small molecule inhibitor (e.g. to reveal transient pockets at PPI for the latter).
The advantage of our computational model over standard docking screens against single protein crystal structures was demonstrated by docking the same library of 10240 compounds against the various structures. As mentioned earlier, our top 3 hit compounds, GB8679, GB6853, and GB1874 fared poorly when screened against the individual protein crystal structures of β-cat (Table S2). We also observed better correlation of GB1874 analogs’ docking scores with Wnt pathway inhibitory activities in STF and STF3A cells compared with non-Wnt pathway related FOPFlash inhibitory activities (Figure 5B). Notably, our computational model can also be further optimized and improved through successive rounds of iteration using more known inhibitors of β-cat-TCF4 interaction. This would enable us to develop a more predictive model and increase the success rate of identifying different scaffolds as potent inhibitors of β-cat-TCF4 interaction. This approach, focusing on a particular target binding site with structure information, can potentially discover specific ligands with greater hit rates than from high-throughput screening (Bajorath, 2002; Shoichet, 2004). Altogether our data suggests that our predictive computational platform can indeed successfully screen for, and identify effective and functional small molecule inhibitors of PPI surfaces.
Limitations of the study
There are some limitations to this study, one of which is that GB1874 is only about 8-fold more selective for the TOPFlash reporter compared to the FOPFlash reporter. This might be due to off-target effects of GB1874 and requires further investigation such as conducting rescue experiments to determine the specificity of the effects. Another drawback of this study is that GB1874 demonstrated low affinity for β-cat in vitro (i.e. KD = 0.58 ± 0.08 mM). Based on our docking studies, the C4 ethylphenyl group of GB1874 was predicted to pack into the pocket lined by H470, R474, and K435 of β-cat. However, the nonpolar ethylphenyl group is unable to form hydrogen bonding interactions with K435 of β-cat. We speculate that this is one of the factors contributing to the moderate affinity of GB1874 for β-cat. While we were able to identify 13 structural analogs of GB1874 for preliminary SAR studies (Figure 5), none of the compounds had the appropriate substituent at the C4 position of the 1,2,4-trizole scaffold to test our hypothesis. Another potential factor contributing to the poor affinity of GB1874 to β-cat in vitro could be the low aqueous solubility of the compound. This low aqueous solubility of GB1874 limits the range of compound concentrations available for in vitro studies, both for SPR and MST assays, and could also contribute to the super stoichiometric binding observed in the SPR studies. Therefore, future studies involving synthesis of structural analogs carrying the desired functional groups, and also downstream functional validation would be required to develop more potent and selective inhibitors of β-cat-TCF4 interaction.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| IRDye 800CW goat anti-rabbit IgG antibody | LI-COR Biosciences | Cat# 926-32211; RRID: AB_621843 |
| IRDye 680RD goat anti-mouse IgG antibody | LI-COR Biosciences | Cat# 926-68070; RRID: AB_10956588 |
| Met (D1C2) XP rabbit mAb antibody | Cell Signaling Technology | Cat# 8198; RRID: AB_10858224 |
| Cyclin D1 (92G2) rabbit mAb antibody | Cell Signaling Technology | Cat# 2978; RRID: AB_2259616 |
| E-cadherin (24E10) rabbit mAb antibody | Cell Signaling Technology | Cat# 3195; RRID: AB_2291471 |
| E-cadherin antibody | Abcam | Cat# ab15148; RRID: AB_301693 |
| TCF4 (C48H11) rabbit mAb antibody | Cell Signaling Technology | Cat# 2569; RRID: AB_2199816 |
| β-Catenin (D10A8) XP® rabbit mAb antibody | Cell Signaling Technology | Cat# 8480; RRID: AB_11127855 |
| Mouse anti-beta-catenin monoclonal antibody, unconjugated, clone 15B8 | Sigma-Aldrich | Cat# C7207; RRID: AB_476865 |
| Survivin (FL-142) antibody | Santa Cruz Biotechnology | Cat# sc-10811; RRID: AB_2227956 |
| Mouse anti-actin, beta monoclonal antibody, unconjugated, clone mAbcam 8226 | Abcam | Cat# ab8226; RRID: AB_306371 |
| Ki-67 recombinant rabbit monoclonal antibody (SP6) | Thermo Fisher Scientific | Cat# MA5-14520; RRID: AB_10979488 |
| Chemicals, peptides, and recombinant proteins | ||
| iCRT3 | ChemDiv | Cat# C523-1410; PubChem CID: 6622273 |
| XAV939 | Selleckchem | Cat# S1180; PubChem CID: 135418940 |
| IWP-2 | Sigma Aldrich | Cat# I0536; PubChem CID: 2155128 |
| GB8679 | Enamine | Cat# Z29898679; PubChem CID: 5122310 |
| GB6853 | Enamine | Cat# Z55536853; PubChem CID: 3995787 |
| GB1874 | Enamine | Cat# Z24601874; PubChem CID: 24242759 |
| GB1874A | MolPort | Cat# MolPort-004-185-459; PubChem CID: 16327389 |
| GB1874B | MolPort | Cat# MolPort-004-538-480; PubChem CID: 16390759 |
| GB1874C | Mcule | Cat# MCULE-6610918918; PubChem CID: 3170257 |
| GB1874D | Mcule | Cat# MCULE-5311573371; PubChem CID: 22585269 |
| GB1874E | Mcule | Cat# MCULE-6862400688; PubChem CID: 4003259 |
| GB1874F | Mcule | Cat# MCULE-4508524154; PubChem CID: 1163424 |
| GB1874G | Mcule | Cat# MCULE-5066540532; PubChem CID: 3170244 |
| GB1874H | Mcule | Cat# MCULE-4103276183; PubChem CID: 5302987 |
| GB1874I | Ambinter | Cat# Amb9057739; PubChem CID: 3184567 |
| GB1874J | Mcule | Cat# MCULE-8753858694; PubChem CID: 33962720 |
| GB1874K | Mcule | Cat# MCULE-9767805728; PubChem CID: 24242757 |
| GB1874L | Mcule | Cat# MCULE-4930244106; PubChem CID: 2045984 |
| GB1874M | Ambinter | Cat# Amb9052521; PubChem CID: 646976 |
| C-terminal FITC-labeled TCF4 peptide (GGGDDLGANDELI SFKDEGEQEEKSSENSSAERD LADVKSSLVNE-FITC) |
1st BASE | N/A |
| Critical commercial assays | ||
| Steady-Glo luciferase assay system | Promega | Cat# E2550 |
| CellTiter-Glo luminescent cell viability assay | Promega | Cat# G7573 |
| PrestoBlue cell viability reagent | Invitrogen | Cat# A13262 |
| Cell counting Kit-8 | Dojindo | Cat# CK04 |
| RNeasy plus mini kit | Qiagen | Cat# 74,136 |
| Illumina stranded mRNA prep | Illumina | Cat# 20040532 |
| Series S sensor chip CM5 | Cytiva | Cat# 29104988 |
| Amine coupling kit | Cytiva | Cat# BR-1000-50 |
| Deposited data | ||
| Raw and analyzed RNA sequencing data | This paper | GEO: GSE152958 |
| Experimental models: cell lines | ||
| Wnt STF reporter | Laboratory of Dr David Virshup | N/A |
| Wnt STF3A reporter | Laboratory of Dr David Virshup | N/A |
| Hippo pathway TEAD reporter | BPS Bioscience | Cat# 60618 |
| Notch CSL reporter | BPS Bioscience | Cat# 60652 |
| HCT116 | ATCC | CCL-247 |
| DLD-1 | ATCC | CCL-221 |
| SW480 | ATCC | CCL-228 |
| CRC948 | Laboratory of Ramanuj DasGupta | N/A |
| CRC1177 | Laboratory of Ramanuj DasGupta | N/A |
| CRC1258 | Laboratory of Ramanuj DasGupta | N/A |
| CRC1414 | Laboratory of Ramanuj DasGupta | N/A |
| CRC1463 | Laboratory of Ramanuj DasGupta | N/A |
| CRC1489 | Laboratory of Ramanuj DasGupta | N/A |
| CRC1671 | Laboratory of Ramanuj DasGupta | N/A |
| CRC1707 | Laboratory of Ramanuj DasGupta | N/A |
| CRC1775 | Laboratory of Ramanuj DasGupta | N/A |
| CRC1837 | Laboratory of Ramanuj DasGupta | N/A |
| CRC1846 | Laboratory of Ramanuj DasGupta | N/A |
| CRC1850 | Laboratory of Ramanuj DasGupta | N/A |
| CRC2001 | Laboratory of Ramanuj DasGupta | N/A |
| CRC2255 | Laboratory of Ramanuj DasGupta | N/A |
| CRC2367 | Laboratory of Ramanuj DasGupta | N/A |
| CRC2413 | Laboratory of Ramanuj DasGupta | N/A |
| CRC2423 | Laboratory of Ramanuj DasGupta | N/A |
| CRC2440 | Laboratory of Ramanuj DasGupta | N/A |
| Experimental models: organisms/strains | ||
| Mouse: NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ | The Jackson Laboratory | JAX: 005557; RRID: IMSR_JAX:005557 |
| Oligonucleotides | ||
| Silencer™ select negative control No. 1 siRNA | Invitrogen | Cat# 4390843 |
| Silencer™ select siRNAs targeting CTNNB1 | Invitrogen | s436, s437, s438 |
|
CTNNB1_F: TTCGAAATCTT GCCCTTTGTCCCG |
Fang et al., 2016 | N/A |
|
CTNNB1_R: AATTCGGTTGTGA ACATCCCGAGC |
Fang et al., 2016 | N/A |
|
AXIN2_F: TCAAGTGCAAAC TTTCGCCAACCG |
Fang et al., 2016 | N/A |
|
AXIN2_R: TGGTGCAAAGAC ATAGCCAGAACC |
Fang et al., 2016 | N/A |
| CCND1_F: CAATGACCCCGCACGATTTC | PrimerBank | 77628152c3 |
| CCND1_R: CATGGAGGGCGGATTGGAA | PrimerBank | 77628152c3 |
|
NKD1_F: GAAGATGGAGA GAGTGAGCGAAC |
Origene | Cat# HP216324 |
| NKD1_R: GTCATACAGGGTGAAGGTCCAC | Origene | Cat# HP216324 |
| RNF43_F: GGTTACATCAGCATCGGACTTGC | Origene | Cat# HP212575 |
| RNF43_R: ATGCTGGCGAATGAGGTGGAGT | Origene | Cat# HP212575 |
| LGR5_F: TGCTTACCAGTGCTGTGCATTTGG | Fang et al., 2016 | N/A |
|
LGR5_R: TGCACTGAATGA AGGGCTTTCAGG |
Fang et al., 2016 | N/A |
| GAPDH_F: ACCCAGAAGACTGTGGATGG | Mani et al., 2008 | N/A |
| GAPDH_R: TCTAGACGGCAGGTCAGGTC | Mani et al., 2008 | N/A |
| Recombinant DNA | ||
| M51 super 8x FOPFlash (TOPFlash mutant) | Veeman et al., 2003 | Addgene Plasmid #12457 |
| GST-tagged TCF4 N-terminal domain (aa 1-55) | Gonsalves et al., 2011 | N/A |
| His-tagged β-cat (ARM domain, aa 134-668) | Gonsalves et al., 2011 | N/A |
| Software and algorithms | ||
| Prism | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| QuPath image analysis software | Bankhead et al., 2017 | https://qupath.github.io/ |
| nfcore/rnaseq v1.0 | Ewels et al., 2020 | https://github.com/nf-core/rnaseq |
| Trim Galore! v0.4.4_dev | Babraham Bioinformatics | https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/ |
| STAR v2.5.3a | Dobin et al., 2013 | https://github.com/alexdobin/STAR |
| featureCounts v1.6.2 | Liao et al., 2014 | http://subread.sourceforge.net/ |
| R v3.6.1 | R Core Team, 2017 | https://www.r-project.org/ |
| DOCK 3.6 | Mysinger and Shoichet, 2010 | http://dock.compbio.ucsf.edu/DOCK3.6/ |
| SCWRL | Krivov et al., 2009 | http://dunbrack.fccc.edu/SCWRL3.php/ |
| PLOP | Sherman et al., 2006 | http://www.jacobsonlab.org/plop_manual/plop_overview.htm |
| MetaPocket | Zhang et al., 2011 | https://projects.biotec.tu-dresden.de/metapocket/index.php |
| OpenBabel | O’Boyle et al., 2011 | https://github.com/openbabel/openbabel |
| Other | ||
| Monolith™ NT.LabelFree zero background standard treated capillaries | Nano Temper | Cat# MO-Z022 |
| Monolith NT.LabelFree system | Nano Temper | https://nanotempertech.com/monolith/ |
| Operetta CLS high-content analysis system | Perkin Elmer | Cat# HH16000000 |
| Vectra Polaris | Perkin Elmer | Cat# CLS143455 |
| HiSeq 4000 | Illumina | https://www.illumina.com/systems/sequencing-platforms/hiseq-3000-4000.html |
| Biacore T200 system | Cytiva | Cat# 28975001 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Ramanuj DasGupta (dasguptar@gis.a-star.edu.sg).
Materials availability
This study did not generate new unique reagents.
Data and code availability
The RNA sequencing data generated during this study are available in GEO via accession number GEO: GSE152958.
Experimental model and subject details
Cell lines
Wnt STF reporter cells (HEK 293 cells stably transfected with TOPFlash reporter) and STF3A reporter cells (STF cells with constitutive Wnt3A secretion) were kind gifts from David Virshup (Duke-NUS Graduate Medical School, Singapore). HCT116, DLD-1, and SW480 cell lines were obtained from ATCC. Reporter cell lines Notch CSL, and Hippo TEAD were obtained from BPS Bioscience. Primary patient-derived CRC cell lines were derived in the laboratory of Ramanuj DasGupta (manuscript under preparation). Cell lines were regularly tested for Mycoplasma using MycoAlert mycoplasma detection kit (Lonza).
Culture media
Reporter cells
STF, STF3A cells were cultured in DMEM (Gibco) supplemented with 10% FBS (Hyclone), and 1 mM sodium pyruvate (Gibco). Hippo TEAD reporter, and Notch CSL reporter cells were cultured in minimum essential medium (MEM) with Earle's balanced salts (EBSS) (Hyclone) supplemented with 10% FBS (Hyclone), 1% MEM non-essential amino acids (NEAA, Gibco), 1 mM sodium pyruvate (Gibco), and 400 μg/mL G-418 sulfate. The medium for Hippo TEAD reporter cells was additionally supplemented with 10 μg/mL Insulin (Sigma Aldrich). The medium for Notch CSL reporter cells was additionally supplemented with 100 μg/mL Hygromycin B (Invitrogen).
CRC cells
DLD-1 cells were cultured in DMEM (Gibco) supplemented with 10% FBS (Hyclone), and 1 mM sodium pyruvate (Gibco). HCT116 and SW480 cells were cultured in McCoy's 5A (Gibco) supplemented with 10% FBS, and 1 mM sodium pyruvate. Primary CRC cell lines were grown in 6-well tissue-culture treated plates (Falcon) precoated with Coating Matrix Kit (Gibco), and were maintained in DMEM/F12 medium (Gibco) supplemented with B27 (without vitamin A, Gibco), 20 ng/mL EGF (Gibco), and 10 ng/mL bFGF (Gibco).
The medium for spheroid formation was DMEM/F-12 (Gibco) supplemented with B27 (without Vitamin A, Gibco), 20 ng/mL EGF (Gibco), 20 ng/mL bFGF (Gibco), and 3% matrigel (Corning cat. # 354234). All media were supplemented with 100 U/mL penicillin, 100 μg/mL streptomycin (Pen-Strep, Gibco).
Animal models
5–7 weeks old female NSG mice (NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ, Jackson Laboratory) were purchased from InVivos and housed under controlled conditions at the Biological Resource Center (BRC), A∗STAR.
For in vivo xenograft studies, HCT116 cells (1 × 106 cells per site) and were implanted subcutaneously into the right flanks of healthy NSG mice. When tumors reached a size of 60–100 mm3 in volume, the animals were randomized to either the treatment or control group. Mice in the treatment group were administered with compound GB1874 at 50 mg/kg every other day via intraperitoneal injection. Mice in the control group were administered with the diluent in the absence of compound. Compound GB1874 was prepared by dissolving it in DMSO to a concentration of 60 mg/mL and diluting it in saline buffer containing 5% PEG 300 (Sigma Aldrich), 5% Tween-80 (Sigma Aldrich) to a final concentration of 3 mg/mL, 5% DMSO. Tumor sizes were measured by caliper once every 2 days and tumor volumes were estimated using the following formula: Tumor volume = 1/2(length × width2). The body weights and health of the mice were also monitored once every 2 days. Mice were euthanized when tumors in the control group reaches 2000 mm3. All in vivo studies were carried out at BRC with approval from A∗STAR Institutional Animal Care and Use Committee (IACUC protocol number 151065).
Method details
Generation of virtual chemical libraries
The training library is composed of three known inhibitors of β-cat-TCF4 complex including iCRT3, iCRT5 and iCRT14, and 150 property-matching computational decoys selected from ZINC database (Irwin and Shoichet, 2005) by the DUD-E approach (Mysinger et al., 2012) as negative controls. The validation library is composed of 63 iCRT3 analogs synthesized in the DasGupta laboratory. The virtual screening library is composed of 10,240 compounds that were purchased from Enamine (https://enamine.net/).
Molecular docking screens
Spheres and grids were generated prior to docking. 45 matching spheres serving to orient database compounds in the site were generated by augmenting the ligand-derived spheres with the receptor-derived spheres. The ligand-derived spheres were represented by the positions of non-hydrogen atoms of the crystal structures of TCF4 peptide fragments at the beginning and replaced by docked poses of iCRT ligands in the following rounds. The receptor-derived spheres were calculated using the program SPHGEN (Kuntz et al., 1982) from the molecular surface of the binding site. Docking screens was performed with DOCK version 3.6 (Mysinger and Shoichet, 2010). The docked compounds were ranked by the docking energy that is the sum of van der Waals, Poisson-Boltzmann electrostatic, and ligand desolvation penalty terms.
Docking performance evaluation
The accuracy of the structural models in ligand prediction was evaluated by the enrichment for the known ligands among the top scoring compounds, measured by the early enrichment factor EF1 and the overall enrichment logAUC which is analogous to the area under the curve (AUC) of the receiver operating characteristic (ROC) but gives early enrichment more weight (Fan et al., 2009; Mysinger and Shoichet, 2010).
Chemical similarity search
The analogs of GB1874 were searched using PubChem "Fingerprint Tanimoto-based 2D similarity search" with the default cutoff of Tanimoto Threshold 90%. The pairwise Tanimoto Coefficient between each tested analog (Table S5) and GB1874 was calculated using the MACCS method available in the OpenBabel program (Xue and Bajorath, 2000; O'Boyle et al., 2011; Sawada et al., 2014) that is considered similar to the PubChem method as they both use substructure key-based fingerprint.
Compounds
The list of compounds used in this study can be found in the Key Resources Table.
TOPFlash (Wnt signaling) reporter screen
STF cells were seeded at 20,000 cells in 100 μL culture medium per well in 96-well plates (Corning). The next day, compounds in DMSO were added to the cells at a final concentration of 10 μM, 1% DMSO. The cells were also stimulated with Wnt3A conditioned medium. After 24 hr, the viability of the cells was determined with PrestoBlue cell viability reagent (Invitrogen) while the TOPFlash reporter activity was determined with Steady-Glo luciferase system (Promega) according to the manufacturers’ protocols. Percentage inhibition of reporter activity and percentage toxicity scores were calculated according to equations below:
TOPFlash reporter dose response
STF cells were seeded as for the reporter screen. The next day, 5-fold dilutions of the compounds in DMSO were added to the cells at a final concentration of 1% DMSO. The cells were also stimulated with Wnt3A conditioned medium. After 24 hr, the viability of the cells was determined with PrestoBlue cell viability reagent according to the manufacturer’s protocol while the TOPFlash reporter activity was determined with Steady-Glo luciferase system. Percentage inhibition of reporter activity and percentage toxicity scores at each concentration was carried out as per the TOPFlash reporter screen. The percentage inhibition and percentage toxicity scores obtained were then plotted against the concentrations tested using GraphPad Prism, and the EC50 values were determined through three-parameter nonlinear regression.
FOPFlash reporter inhibition and dose response
M51 Super 8x FOPFlash (TOPFlash mutant) was a gift from Randall Moon (Addgene plasmid # 12457) (Veeman et al., 2003). HEK293T cells (30,000 cells per well) were reverse transfected with 150 ng of FOPFlash plasmid using Lipofectamine 3000 (0.3 μL per well). For dose response studies, 5-fold dilutions of the compounds in DMSO were added to the cells at a final concentration of 1% DMSO the next day. For single concentration inhibition studies, compounds in DMSO were added to the cells at a final concentration of 10 μM, 1% DMSO. After 24 hr, the viability of the cells was determined with PrestoBlue cell viability reagent according to the manufacturer’s protocol while the TOPFlash reporter activity was determined with Steady-Glo luciferase system. Percentage inhibition of reporter activity and percentage toxicity scores at each concentration was carried out as per the TOPFlash reporter screen. The percentage inhibition and percentage toxicity scores obtained were then plotted against the concentrations tested using GraphPad Prism, and the EC50 values were determined through three-parameter nonlinear regression.
Inhibition of Wnt STF3A reporter
STF3A cells were seeded at 20,000 cells in 100 μL culture medium per well in 96-well plates. The next day, compounds in DMSO were added to the cells at a final concentration of 20 μM, 1% DMSO. After 24 hr, the viability of the cells was determined with Cell Counting Kit-8 (CCK-8, Dojindo) cell viability reagent while the TOPFlash reporter activity was determined with Steady-Glo luciferase system. Percentage inhibition of reporter activity and percentage toxicity analyses were carried out as per the TOPFlash reporter screen.
Inhibition of hippo TEAD reporter
Cells were seeded at 100,000 cells in 100 μL assay medium per well in 96-well plates. The assay medium consists of the growth medium without G-418. The next day, compounds in DMSO were added to the cells at a final concentration of 20 μM, 1% DMSO. After 24 hr incubation, the viability of the cells was determined with CCK-8 cell viability reagent while the luciferase reporter activity was determined with Steady-Glo luciferase system. Percentage toxicity analysis was carried out as per the TOPFlash reporter screen. For percentage inhibition of reporter activity analysis, as activators of the Hippo pathway reduce luciferase expression, the following equation was used:
Inhibition of notch signaling reporter
Cells were seeded at 25,000 cells in 100 μL assay medium per well in 96-well plates. The assay medium consists of the growth medium without G-418 and Hygromycin B. The next day, compounds in DMSO were added to the cells at a final concentration of 20 μM, 1% DMSO. After 24 hr incubation, the viability of the cells was determined with CCK-8 cell viability reagent while the luciferase reporter activity was determined with Steady-Glo luciferase system. Percentage inhibition of reporter activity and percentage toxicity analyses were carried out as per the TOPFlash reporter screen.
RNA sequencing and analysis
HCT116 cells were either treated with compounds for 18 hr at a final concentration of 50 μM, 1% DMSO, or were transfected with 20 nM pooled siRNAs (Silencer Select) against CTNNB1 (s436, s437, and s438) or 20 nM Silencer Select negative control No. 1 siRNA for 72 hr. RNA from treated cells were extracted using RNeasy plus mini kit (Qiagen) according to the manufacturer's protocol. RNA quality was checked using Agilent RNA 6000 nano kit, and only RNA with RIN >9.5 were selected for library preparation using the standard Illumina stranded mRNA library prep kit with 150 bp paired-end sequencing. Data was generated on the Illumina HiSeq 4000 system by the GIS Integrated Genomics Platform. 34–46.8 million reads were obtained per sample (38.5 million average). The data was processed with the RNA-seq pipeline version 1.0 from nf-core (Ewels et al., 2020). Briefly, raw reads were trimmed (Trim Galore!) before alignment to GRCh38 reference genome using STAR v2.5.3a (Dobin et al., 2013). Read counts were then generated using featureCounts1.6.2 (Liao et al., 2014).
Differentially expressed genes (DEGs) were identified via limma package (Ritchie et al., 2015). Samples from compound treated group was compared against the DMSO control while siCTNNB1 group was compared against negative control siRNA. DEGs obtained were p value ranked for gene set enrichment analysis (GSEA) using the fgsea package (Sergushichev, 2016) using relevant gene sets collated from multiple sources to ensure robustness of results. This consisted of MSigDB (Liberzon et al., 2015): Hallmark, Canonical pathways (REACTOME, BIOCARTA, PID), non-canonical pathways and GO biological process as well as KEGG (Kanehisa and Goto, 2000) (https://www.kegg.jp/dbget-bin/www_bget?hsa04310) and Wnt homepage (http://wnt.stanford.edu/).
Additionally, gene sets for BCAT_DEP, BCAT_DEP_TCF_DEP, and BCAT_DEP_TCF_INDEP, were generated from data downloaded from ArrayExpress database (accession number E-MTAB-7029) and performing the comparisons described in the paper (Doumpas et al., 2019). Specifically, we obtained a list of differentially expressed genes upon CHIRON99021 (CHIR) stimulation in WT and dBcat cells. BCAT_DEP genes were then identified as genes that are differentially expressed in WT but not dBcat cells. Next, we obtained a list of differentially expressed gene upon CHIR stimulation in d4TCF cells. BCAT_DEP_TCF_DEP genes were identified as a subset of BCAT_DEP genes that are not differentially express in d4TCF cells upon CHIR stimulation. Similarly, BCAT_DEP_TCF_INDEP genes were identified as a subset of BCAT_DEP genes that are differentially expressed in d4TCF cells upon CHIR stimulation. The summary of GSEA analysis was plotted using the tmod package (Weiner and Domaszewska, 2016) and heatmap visualized by ComplexHeatmap package (Gu et al., 2016). All analysis was performed using R version 3.6.1 (https://cran.r-project.org/).
Quantitative polymerase chain reaction
500 ng of total RNA was reverse transcribed using SuperScript IV VILO master mix (Invitrogen). 10 ng of cDNA was used per reaction for quantitative polymerase chain reaction (qPCR) analysis using KAPA SYBR FAST qPCR kit (Sigma). The sequences of the qPCR primers (Fang et al., 2016; Mani et al., 2008) can be found in the Key Resources Table.
Western blotting
HCT116 cells were treated with compounds for 18 hr at a final concentration of 50 μM, 1% DMSO. Cells were lysed on ice in RIPA buffer (Thermo Scientific) containing cOmplete protease inhibitor cocktail (Roche) and PhosSTOP phosphatase inhibitor cocktail (Roche). 30 μg per sample of total cellular protein was resolved by SDS-PAGE and blotted onto PVDF membrane (Millipore). The blots were incubated overnight at 4°C with the respective primary antibodies followed by detection with either IRDye 800CW goat anti-rabbit or IRDye 680RD goat anti-mouse secondary antibodies (LI-COR) at 1:5000 dilution. Signals were visualized with the LI-COR Odyssey CLx imaging system. The primary antibodies used were Met (Cell Signaling Technology cat. # 8198, 1:1000), cyclin D1 (Cell Signaling Technology cat. # 2978, 1:1000), survivin (Santa Cruz cat. # sc-10811, 1:500), ECAD (Cell Signaling Technology cat. # 3195, 1:1000), TCF4/TCF7L2 (Cell Signaling Technology cat. # 2569, 1:1000), β-cat (Cell Signaling Technology cat. # 8480, 1:1000), β-actin (Abcam cat. # ab8226, 1:3000).
Co-immunoprecipitation
HCT116 cells were treated for 18 hr with compounds at a final concentration of 50 μM, 1% DMSO. Cells were lysed on ice in lysis buffer containing 20 mM HEPES, pH 7.5, 137 mM NaCl, 1.5 mM MgCl2, 1 mM EGTA, 1 mM dithiothreitol (DTT), 1% Triton X-100, 10% glycerol, cOmplete protease inhibitor cocktail (Roche), and 1 mM Na3VO4. Each sample of cleared protein lysate was incubated overnight at 4°C with 4 μg anti-β-cat antibody (Sigma cat. #C7207) followed by 1 hr incubation with a 50 μL slurry of Dynabeads Protein G (Invitrogen) at 4°C. The beads were then washed three times with lysis buffer at 4°C, and the bound protein was eluted by heating the beads with 30 μL (per sample) of SDS sample buffer at 95°C for 7 min. The pull-down proteins were resolved by SDS-PAGE and blotted onto PVDF membrane (Millipore). The blots were incubated overnight at 4°C with the respective primary antibodies followed by detection with either IRDye 800CW goat anti-rabbit or IRDye 680RD goat anti-mouse secondary antibodies (LI-COR) at 1:5000 dilution. Signals were visualized with the LI-COR Odyssey CLx imaging system and bands were quantified using ImageJ. The primary antibodies used were E-cadherin (Abcam cat. # ab15148, 1:1000), TCF4/TCF7L2 (Cell Signaling Technology cat. # 2569, 1:1000), β-cat (Cell Signaling Technology cat. # 8480, 1:1000).
Surface plasmon resonance studies
Protein expression and purification
His-tagged β-cat (ARM domain, aa 134–668) and GST-tagged TCF4 N-terminal domain (aa 1–55) proteins were expressed and purified according to the protocol described previously (Gonsalves et al., 2011). Specifically, BL21 Escherichia coli cells containing either the His-tagged β-cat vector or the GST-tagged TCF4 N-terminal domain vector were expanded and induced for protein expression with 0.5 mM IPTG (1st Base). After induction the proteins were extracted by lysing the cells in buffer containing 20 mM Tris, pH 8.8, 250 mM NaCl, 0.1% Triton-X, 5% glycerol, 2 mM DTT, and complete EDTA-free protease inhibitor (Roche). His-tagged β-cat was purified from the cell lysate using Ni-NTA agarose beads (Thermo) and eluted in buffer containing 20 mM Tris, pH 8.8, 250 mM NaCl, 5% glycerol, 0.5 mM TCEP, and 250 mM imidazole (Sigma). Similarly, GST-tagged TCF4 N-terminal domain was purified from the cell lysate using glutathione agarose beads (Pierce) and eluted in buffer containing 20 mM Tris, pH 8.8, 250 mM NaCl, 5% glycerol, 0.5 mM TCEP, and 10 mM reduced glutathione (Thermo). The purified proteins were then dialyzed against a buffer containing 20 mM HEPES, pH 7, 200 mM NaCl, 10% glycerol and 5 mM DTT before using them for surface plasmon resonance (SPR) studies.
Binding to β-cat
SPR analysis was performed on a Biacore T200 system (Cytiva). HEPES-buffered saline containing 0.05% Tween-20 and 5% DMSO was used as running buffer. By using a series S CM5 sensor chip and the Amine-coupling Kit (Cytiva) as per manufacturer's instructions, purified beta-catenin was amine-coupled to flow cells (FC) #2 and #4 at 7800 and 10779 RU, respectively. Analyte samples, iCRT3 and GB1874, diluted in running buffer were then injected over all four FCs so that FC(2-1) and FC(4-3) referenced data were collected at 25 degrees Celcius. Concentration-dependent response was observed although the sensorgrams suggest a degree of sample aggregation in the running buffer. Biacore T200 Evaluation Software 3.0 was used for steady-state affinity analysis with the assumption of one-to-one binding model. For each set of sensorgrams, a time point at which steady state signal is attained is chosen, the response levels plotted against concentration, and the KD values derived.
Inhibition of β-cat-TCF4 interaction
GST-tagged TCF4 N-terminal domain (31 kDa, >90% pure based on SDS–PAGE) was immobilized on a CM5 series S sensor chip using amine-coupling chemistry. The ligand at a concentration of 0.5 μM in 10 mm citrate buffer, pH 4.0, was immobilized at a density of 400 RU on flow cell 4 and flow cell 3 was left blank to serve as a reference surface. For inhibition studies, 50 nM His-tagged β-cat was pre-incubated with different concentrations of compound GB1874 or different concentrations of a C-terminal FITC-labeled TCF4 peptide (aa 7 – 51) in PBS containing 5% DMSO for 15 min at room temperature. The mixture was injected over the two flow cells at a flow rate of 30 μL/min, 25°C. The complex was allowed to associate and dissociate for 60 s each and the surfaces were regenerated with a 30 s injection of 50 mM NaOH. The RU responses were collected and plotted against the concentration of GB1874 tested using GraphPad Prism. The inhibitory IC50 value of compound GB1874 was obtained through a four-parameter nonlinear regression. The C-terminal FITC-labeled TCF4 peptide (GGGDDLGANDELISFKDEGEQEEKSSENSSAERDLADVKSSLVNE-FITC) was obtained through custom peptide synthesis by 1st Base.
Microscale thermophoresis studies
Purified His-tagged β-cat (ARM domain) was diluted to a concentration of 500 nM in a buffer containing 20 mM Tris (pH 8.0), 200 mM NaCl, 10% glycerol, 5 mM DTT and 0.01% Triton X-100. Serial dilutions of GB1874 were then mixed with the diluted β-cat solution to obtain final concentrations of 0.46 μM – 200 μM, 2% DMSO. The mixtures were then loaded into Monolith™ NT.LabelFree zero background standard treated capillaries (NanoTemper). Microscale thermophoresis (MST) analyses were carried out on the samples using the Monolith NT.LabelFree system (NanoTemper) using excitation power at 20% and MST power at 20% and 40%. Dose response Kd affinity fit was carried out using the software MO.Affinity Analysis v2.3 (NanoTemper).
Dose response studies with CRC cell lines
HCT116, DLD-1, SW480, and primary CRC cells were seeded at 5,000 – 7,000 cells per well in 100 μL growth medium in 96-well plates (Corning). The next day, 5-fold dilutions of the compounds in DMSO were added to the cells at a final concentration of 1% DMSO. After 72 hr incubation, the viability of the cells was determined with CellTiter-Glo luminescent cell viability assay (Promega) according to the manufacturer’s protocol. The signals from the treatment wells were normalized against the DMSO treatment wells. The normalized values were plotted against the concentrations tested using GraphPad Prism, and the EC50 values were determined through four-parameter nonlinear regression.
Proliferation assay
HCT116 cells were seeded at 3,000 cells in 100 μL growth medium per well in 96-well plates (Corning). The next day, cells were treated with compounds at a final concentration of 10 μM, 0.1% DMSO, for 4 days with daily medium and compound change. Each day’s cell viability was determined using CellTiter-Glo cell viability assay.
Spheroid formation assay
HCT116, DLD-1, and SW480 cells were seeded at 150 cells in 200 μL spheroid medium per well in 96-well ultra-low attachment plates (Corning). The compounds were added on the day of cell seeding at a final concentration of 30 μM, 0.5% DMSO. The cells were grown at 37°C, 5% CO2 and images of the spheroids formed were taken on day 14 after cell seeding using the Operetta CLS system (PerkinElmer).
Immunohistochemistry
Tumors were harvested at the end of treatment, 1 day after the last dosage, and fixed in 4% paraformaldehyde for paraffin block sectioning. Immunohistochemistry staining was carried out at the Advanced Molecular Pathology Laboratory (AMPL) using antibodies against Ki67 (Thermo Fisher, cat # MA5-14520) or cyclin D1 (Cell Signaling Technology, cat # 2978). Stained tumor sections were imaged using Leica SCN400 slide scanner and image analysis was carried out using QuPath image analysis software (Bankhead et al., 2017).
Mice organs were harvested at the end of treatment and fixed in 10% neutral buffered formalin (Sigma) for paraffin block embedding and sectioning. Hematoxylin and eosin (H&E) staining was carried out at the Advanced Molecular Pathology Laboratory (AMPL). Stained tissue sections were imaged using Vectra Polaris from PerkinElmer.
Quantification and statistical analysis
Whereby statistical tests are presented as asterisks in the figures, details of the statistical test are described in the figure legends. Details of statistical analysis of RNA sequencing data are also described in the RNA sequencing and analysis subsection in the method details. Except for RNA sequencing analysis, all statistical analyses and non-linear regression analyses were carried out using GaphPad Prism software.
Acknowledgments
We would like to thank Prof. David Virshup for providing us with the STF and STF3A Wnt reporter cell lines. We would also like to thank Dr Bharath Sakshibeedu Rajegowda and Prof. Song Haiwei for their kind assistance with the microscale thermophoresis studies. Many thanks to Michelle Lim and Joon Cheng Seng for their assistance in optimizing the conditions for dose response in the reporter cell lines. We would also like to thank the Center for High Throughput Phenomics - GIS (CHIP-GIS) for the compound library used in this study. All animal work was carried out at the Biological Resource Center (BRC), A∗STAR under IACUC protocol number 151065. R.D.G. was supported in part by NIH/NCI #1R01CA155125-01, and by the Genome Institute of Singapore (GIS) core funds (BMRC/A∗STAR). H.F. was supported by the Bioinformatics Institute (BII) core funds (BMRC/A∗STAR). J.-L.L. was supported by the National Research Foundation (NRF), Singapore Competitive Research Program (CRP) grant (NRF-CRP-2017-02).
Author contributions
Conceptualization, R.D., H.F. and J.L.L.; Methodology, H.F. and W.D.; Investigation, J.L.L., W.D., T.G., X.Z., M.W.C., S.M., and D.G.R.Y.; Formal analysis, A.R. and G.O.; Resources, B.H.I.T.; Writing – Original Draft, J.L.L; Writing – Review & Editing, R.D. and H.F.; Funding Acquisition, R.D. and H.F.; Supervision, R.D. and H.F.
Declaration of interests
The authors declare that they have no conflict of interest.
Inclusion and diversity
We worked to ensure diversity in experimental samples through the selection of the cell lines. The author list of this paper includes contributors from the location where the research was conducted who participated in the data collection, design, analysis, and/or interpretation of the work.
Published: June 25, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.102544.
Contributor Information
Hao Fan, Email: fanh@bii.a-star.edu.sg.
Ramanuj DasGupta, Email: dasguptar@gis.a-star.edu.sg.
Supplemental information
References
- Amit S., Hatzubai A., Birman Y., Andersen J.S., Ben-Shushan E., Mann M., Ben-Neriah Y., Alkalay I. Axin-mediated CKI phosphorylation of beta-catenin at Ser 45: a molecular switch for the Wnt pathway. Genes Dev. 2002;16:1066–1076. doi: 10.1101/gad.230302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anastas J.N., Moon R.T. WNT signalling pathways as therapeutic targets in cancer. Nat. Rev. Cancer. 2013;13:11–26. doi: 10.1038/nrc3419. [DOI] [PubMed] [Google Scholar]
- Arkin M.R., Tang Y., Wells J.A. Small-molecule inhibitors of protein-protein interactions: progressing toward the reality. Chem. Biol. 2014;21:1102–1114. doi: 10.1016/j.chembiol.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajorath J. Integration of virtual and high-throughput screening. Nat. Rev. Drug Discov. 2002;1:882–894. doi: 10.1038/nrd941. [DOI] [PubMed] [Google Scholar]
- Bankhead P., Loughrey M.B., Fernández J.A., Dombrowski Y., McArt D.G., Dunne P.D., McQuaid S., Gray R.T., Murray L.J., Coleman H.G. QuPath: open source software for digital pathology image analysis. Sci. Rep. 2017;7:16878. doi: 10.1038/s41598-017-17204-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker N., Clevers H. Mining the Wnt pathway for cancer therapeutics. Nat. Rev. Drug Discov. 2006;5:997–1014. doi: 10.1038/nrd2154. [DOI] [PubMed] [Google Scholar]
- Bienz M., Clevers H. Linking colorectal cancer to Wnt signaling. Cell. 2000;103:311–320. doi: 10.1016/s0092-8674(00)00122-7. [DOI] [PubMed] [Google Scholar]
- Chen B., Dodge M.E., Tang W., Lu J., Ma Z., Fan C.-W., Wei S., Hao W., Kilgore J., Williams N.S. Small molecule–mediated disruption of Wnt-dependent signaling in tissue regeneration and cancer. Nat. Chem. Biol. 2009;5:100–107. doi: 10.1038/nchembio.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H. Wnt/β-Catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- Collu G.M., Hidalgo-Sastre A., Brennan K. Wnt–Notch signalling crosstalk in development and disease. Cell. Mol. Life Sci. 2014;71:3553–3567. doi: 10.1007/s00018-014-1644-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coombs G.S., Yu J., Canning C.A., Veltri C.A., Covey T.M., Cheong J.K., Utomo V., Banerjee N., Zhang Z.H., Jadulco R.C. WLS-dependent secretion of WNT3A requires Ser209 acylation and vacuolar acidification. J. Cell Sci. 2010;123:3357–3367. doi: 10.1242/jcs.072132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doumpas N., Lampart F., Robinson M.D., Lentini A., Nestor C.E., Cantù C., Basler K. TCF/LEF dependent and independent transcriptional regulation of Wnt/β-catenin target genes. EMBO J. 2019;38:e98873. doi: 10.15252/embj.201798873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewels P.A., Peltzer A., Fillinger S., Patel H., Alneberg J., Wilm A., Garcia M.U., di Tommaso P., Nahnsen S. The nf-core framework for community-curated bioinformatics pipelines. Nat. Biotechnol. 2020;38:276–278. doi: 10.1038/s41587-020-0439-x. [DOI] [PubMed] [Google Scholar]
- Fan H., Irwin J.J., Webb B.M., Klebe G., Shoichet B.K., Sali A. Molecular docking screens using comparative models of proteins. J. Chem. Inf. Model. 2009;49:2512–2527. doi: 10.1021/ci9003706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang L., Zhu Q., Neuenschwander M., Specker E., Wulf-Goldenberg A., Weis W.I., von Kries J.P., Birchmeier W. A small-molecule antagonist of the β-catenin/TCF4 interaction blocks the self-renewal of cancer stem cells and suppresses tumorigenesis. Cancer Res. 2016;76:891–901. doi: 10.1158/0008-5472.CAN-15-1519. [DOI] [PubMed] [Google Scholar]
- Fasolini M., Wu X., Flocco M., Trosset J.Y., Oppermann U., Knapp S. Hot spots in Tcf4 for the interaction with beta-catenin. J. Biol. Chem. 2003;278:21092–21098. doi: 10.1074/jbc.M301781200. [DOI] [PubMed] [Google Scholar]
- Gonsalves F.C., Klein K., Carson B.B., Katz S., Ekas L.A., Evans S., Nagourney R., Cardozo T., Brown A.M.C., DasGupta R. An RNAi-based chemical genetic screen identifies three small-molecule inhibitors of the Wnt/wingless signaling pathway. Proc. Natl. Acad. Sci. U S A. 2011;108:5954–5963. doi: 10.1073/pnas.1017496108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham T.A., Weaver C., Mao F., Kimelman D., Xu W. Crystal structure of a beta-catenin/tcf complex. Cell. 2000;103:885–896. doi: 10.1016/s0092-8674(00)00192-6. [DOI] [PubMed] [Google Scholar]
- Grandy D., Shan J., Zhang X., Rao S., Akunuru S., Li H., Zhang Y., Alpatov I., Zhang X.A., Lang R.A. Discovery and characterization of a small molecule inhibitor of the PDZ domain of dishevelled. J. Biol. Chem. 2009;284:16256–16263. doi: 10.1074/jbc.M109.009647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z., Eils R., Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32:2847–2849. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- Hahne G., Grossmann T.N. Direct targeting of β-catenin: inhibition of protein-protein interactions for the inactivation of Wnt signaling. Bioorg. Med. Chem. 2013;21:4020–4026. doi: 10.1016/j.bmc.2013.02.050. [DOI] [PubMed] [Google Scholar]
- Hankey W., Frankel W.L., Groden J. Functions of the APC tumor suppressor protein dependent and independent of canonical WNT signaling: implications for therapeutic targeting. Cancer Metastasis Rev. 2018;37:159–172. doi: 10.1007/s10555-017-9725-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S.-M.A., Mishina Y.M., Liu S., Cheung A., Stegmeier F., Michaud G.A., Charlat O., Wiellette E., Zhang Y., Wiessner S. Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature. 2009;461:614–620. doi: 10.1038/nature08356. [DOI] [PubMed] [Google Scholar]
- Huang Z., Zhang M., Burton S.D., Katsakhyan L.N., Ji H. Targeting the Tcf4 G13ANDE17 binding site to selectively disrupt β-Catenin/T-Cell factor protein-protein interactions. ACS Chem. Biol. 2014;9:193–201. doi: 10.1021/cb400795x. [DOI] [PubMed] [Google Scholar]
- Ilyas M., Tomlinson I.P.M., Rowan A., Pignatelli M., Bodmer W.F. β-Catenin mutations in cell lines established from human colorectal cancers. Proc. Natl. Acad. Sci. U S A. 1997;94:10330–10334. doi: 10.1073/pnas.94.19.10330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin J.J., Shoichet B.K. ZINC − A free database of commercially available compounds for virtual screening. J. Chem. Inf. Model. 2005;45:177–182. doi: 10.1021/ci049714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn M. Can we safely target the WNT pathway? Nat. Rev. Drug Discov. 2014;13:513–532. doi: 10.1038/nrd4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay S.K., Harrington H.A., Shepherd S., Brennan K., Dale T., Osborne J.M., Gavaghan D.J., Byrne H.M. The role of the Hes1 crosstalk hub in Notch-Wnt interactions of the intestinal crypt. PLoS Comput. Biol. 2017;13:e1005400. doi: 10.1371/journal.pcbi.1005400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krivov G.G., Shapovalov M.V., Dunbrack R.L. Improved prediction of protein side-chain conformations with SCWRL4. Proteins. 2009;77:778–795. doi: 10.1002/prot.22488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuntz I.D., Blaney J.M., Oatley S.J., Langridge R., Ferrin T.E. A geometric approach to macromolecule-ligand interactions. J. Mol. Biol. 1982;161:269–288. doi: 10.1016/0022-2836(82)90153-x. [DOI] [PubMed] [Google Scholar]
- Liao Y., Smyth G.K., Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- Liberzon A., Birger C., Thorvaldsdóttir H., Ghandi M., Mesirov J.P., Tamayo P. The molecular signatures database Hallmark gene set collection. Cell Syst. 2015;1:417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Wang H., Zuo Y., Farmer S.R. Functional interaction between peroxisome proliferator-activated receptor and β-catenin. Mol. Cell. Biol. 2006;26:5827–5837. doi: 10.1128/MCB.00441-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luke J.J., Bao R., Sweis R.F., Spranger S., Gajewski T.F. WNT/β-catenin pathway activation correlates with immune exclusion across human cancers. Clin. Cancer Res. 2019;25:3074–3083. doi: 10.1158/1078-0432.CCR-18-1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madan B., Ke Z., Harmston N., Ho S.Y., Frois A.O., Alam J., Jeyaraj D.A., Pendharkar V., Ghosh K., Virshup I.H. Wnt addiction of genetically defined cancers reversed by PORCN inhibition. Oncogene. 2016;35:2197–2207. doi: 10.1038/onc.2015.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani S.A., Guo W., Liao M.J., Eaton E.N., Ayyanan A., Zhou A.Y., Brooks M., Reinhard F., Zhang C.C., Shipitsin M. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mysinger M.M., Carchia M., Irwin John.J., Shoichet B.K. Directory of useful decoys, enhanced (DUD-E): better ligands and decoys for better benchmarking. J. Med. Chem. 2012;55:6582–6594. doi: 10.1021/jm300687e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mysinger M.M., Shoichet B.K. Rapid context-dependent ligand desolvation in molecular docking. J. Chem. Inf. Model. 2010;50:1561–1573. doi: 10.1021/ci100214a. [DOI] [PubMed] [Google Scholar]
- Nusse R., Clevers H. Wnt/β-Catenin signaling, disease, and emerging therapeutic modalities. Cell. 2017;169:985–999. doi: 10.1016/j.cell.2017.05.016. [DOI] [PubMed] [Google Scholar]
- O’Boyle N.M., Banck M., James C.A., Morley C., Vandermeersch T., Hutchison G.R. Open Babel: an open chemical toolbox. J. Cheminf. 2011;3:33. doi: 10.1186/1758-2946-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlovic Z., Adams J.J., Blazer L.L., Gakhal A.K., Jarvik N., Steinhart Z., Robitaille M., Mascall K., Pan J., Angers S. A synthetic anti-Frizzled antibody engineered for broadened specificity exhibits enhanced anti-tumor properties. MAbs. 2018;10:1157–1167. doi: 10.1080/19420862.2018.1515565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poy F., Lepourcelet M., Shivdasani R.A., Eck M.J. Structure of a human Tcf4-β-catenin complex. Nat. Struct. Biol. 2001;8:1053–1057. doi: 10.1038/nsb720. [DOI] [PubMed] [Google Scholar]
- Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampietro J., Dahlberg C.L., Cho U.S., Hinds T.R., Kimelman D., Xu W. Crystal structure of a β-catenin/BCL9/tcf4 complex. Mol. Cell. 2006;24:293–300. doi: 10.1016/j.molcel.2006.09.001. [DOI] [PubMed] [Google Scholar]
- Sawada R., Kotera M., Yamanishi Y. Benchmarking a wide range of chemical descriptors for drug-target interaction prediction using a chemogenomic approach. Mol. Inf. 2014;33:719–731. doi: 10.1002/minf.201400066. [DOI] [PubMed] [Google Scholar]
- Sergushichev A.A. An algorithm for fast preranked gene set enrichment analysis using cumulative statistic calculation. bioRxiv. 2016 doi: 10.1101/060012. [DOI] [Google Scholar]
- Sherman W., Day T., Jacobson M.P., Friesner R.A., Farid R. Novel procedure for modeling ligand/receptor induced fit effects. J. Med. Chem. 2006;49:534–553. doi: 10.1021/jm050540c. [DOI] [PubMed] [Google Scholar]
- Shoichet B.K. Virtual screening of chemical libraries. Nature. 2004;432:862–865. doi: 10.1038/nature03197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takebe N., Miele L., Harris P.J., Jeong W., Bando H., Kahn M., Yang S.X., Ivy S.P. Targeting Notch, Hedgehog, and Wnt pathways in cancer stem cells: clinical update. Nat. Rev. Clin. Oncol. 2015;12:445–464. doi: 10.1038/nrclinonc.2015.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka N., Mashima T., Mizutani A., Sato A., Aoyama A., Gong B., Yoshida H., Muramatsu Y., Nakata K., Matsuura M. APC mutations as a potential biomarker for sensitivity to Tankyrase inhibitors in colorectal cancer. Mol. Cancer Ther. 2017;16:752–762. doi: 10.1158/1535-7163.MCT-16-0578. [DOI] [PubMed] [Google Scholar]
- Tian W., Han X., Yan M., Xu Y., Duggineni S., Lin N., Luo G., Li Y.M., Han Xiaobing, Huang Z. Structure-based discovery of a novel inhibitor targeting the β-catenin/Tcf4 interaction. Biochemistry. 2012;51:724–731. doi: 10.1021/bi201428h. [DOI] [PubMed] [Google Scholar]
- Trosset J.-Y., Dalvit C., Knapp S., Fasolini M., Veronesi M., Mantegani S., Gianellini L.M., Catana C., Sundström M., Stouten Inhibition of protein–protein interactions: the discovery of druglike β-catenin inhibitors by combining virtual and biophysical screening. Proteins. 2006;64:60–67. doi: 10.1002/prot.20955. [DOI] [PubMed] [Google Scholar]
- Valenta T., Hausmann G., Basler K. The many faces and functions of β-catenin. EMBO J. 2012;31:2714–2736. doi: 10.1038/emboj.2012.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veeman M.T., Slusarski D.C., Kaykas A., Louie S.H., Moon R.T. Zebrafish prickle, a modulator of noncanonical wnt/fz signaling, regulates gastrulation movements. Curr. Biol. 2003;13:680–685. doi: 10.1016/s0960-9822(03)00240-9. [DOI] [PubMed] [Google Scholar]
- Weiner J., 3rd, Domaszewska T. TMOD: an R package for general and multivariate enrichment analysis. PeerJ Preprints. 2016;4:e2420v1. [Google Scholar]
- Xu Q., Wang Y., Dabdoub A., Smallwood P.M., Williams J., Woods C., Kelley M.W., Jiang L., Tasman W., Zhang K. Vascular development in the retina and inner ear: control by Norrin and Frizzled-4, a high-affinity ligand-receptor pair. Cell. 2004;116:883–895. doi: 10.1016/s0092-8674(04)00216-8. [DOI] [PubMed] [Google Scholar]
- Xue L., Bajorath J. Molecular descriptors in chemoinformatics, computational combinatorial chemistry, and virtual screening. Comb. Chem. High Throughput Screen. 2000;3:363–372. doi: 10.2174/1386207003331454. [DOI] [PubMed] [Google Scholar]
- Yu B., Huang Z., Zhang M., Dillard D.R., Ji H. Rational design of small-molecule inhibitors for beta-catenin/T-cell factor protein-protein interactions by bioisostere replacement. ACS Chem. Biol. 2013;8:524–529. doi: 10.1021/cb300564v. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Li Y., Lin B., Schroeder M., Huang B. Identification of cavities on protein surface using multiple computational approaches for drug binding site prediction. Bioinformatics. 2011;27:2083–2088. doi: 10.1093/bioinformatics/btr331. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA sequencing data generated during this study are available in GEO via accession number GEO: GSE152958.