Abstract
Objectives:
To investigate if the nicotine metabolite ratio (NMR, the ratio of nicotine metabolites 3’-hydroxycotinine/cotinine) is a reliable phenotypic biomarker for nicotine clearance across races, and as a function of differences in the rate of nicotine, cotinine and 3’-hydroxycotinine glucuronidation and UGT genotypes.
Methods:
Participants (Caucasians (Whites), African Americans (Blacks) and Asian-Americans (Asians)) received an oral solution of deuterium-labeled nicotine and its metabolite cotinine. Plasma and saliva concentrations of nicotine and cotinine were used to determine oral clearances. Rates of glucuronidation were assessed from urine glucuronide/parent ratios, and UGT2B10 and UGT2B17 genotypes from DNA.
Results:
Among the 227 participants, 96 (42%) were White, 67 (30%) Asian, and 64 (28%) Black. Compared to the other two races, Whites had higher nicotine and cotinine total oral clearance, Blacks had lower nicotine and cotinine glucuronidation rates, and Asians had lower 3’-hydroxycotinine glucuronidation rates. A strong positive correlation (correlations coefficients 0.77–0.84, p<0.001) between NMR and nicotine oral clearance was found for all three races, and NMR remained a strong predictor for the nicotine oral clearance while adjusting for race, sex, and age. Neither the metabolite glucuronidation ratios nor the UGT genotypes had significant effects on the ability of NMR to predict nicotine oral clearance.
Conclusions:
NMR appears to be a reliable phenotypic biomarker for nicotine clearance across races, glucuronidation phenotypes and genotypes. Racial differences in the relationships between NMR, smoking behaviors and addiction are unlikely to be related to an inadequate estimation of nicotine clearance based on NMR.
Keywords: nicotine, nicotine clearance, nicotine metabolite ratio, NMR, cotinine, racial differences, glucuronidation
Introduction
Nicotine is metabolized primarily by the hepatic cytochrome P450 enzyme CYP2A6, with approximately 80% of nicotine converted to its inactive metabolite cotinine (COT), which is in turn further metabolized exclusively by CYP2A6 to 3’-hydroxycotinine (3HC) [1]. The CYP2A6 gene is genetically polymorphic, with a number of variants associated with slower metabolism [2]. The ratio of 3HC/COT (usually based on unconjugated levels), also called the nicotine metabolite ratio (NMR), is a phenotypic biomarker of CYP2A6 activity that can be measured in plasma, urine and saliva of users of nicotine products and has been shown to be correlated with the rate of nicotine clearance [3]. The NMR accounts for both genetic and non-genetic influences on CYP2A6 activity, is reproducible within subjects, and generally independent of the time since last cigarette [4–6].
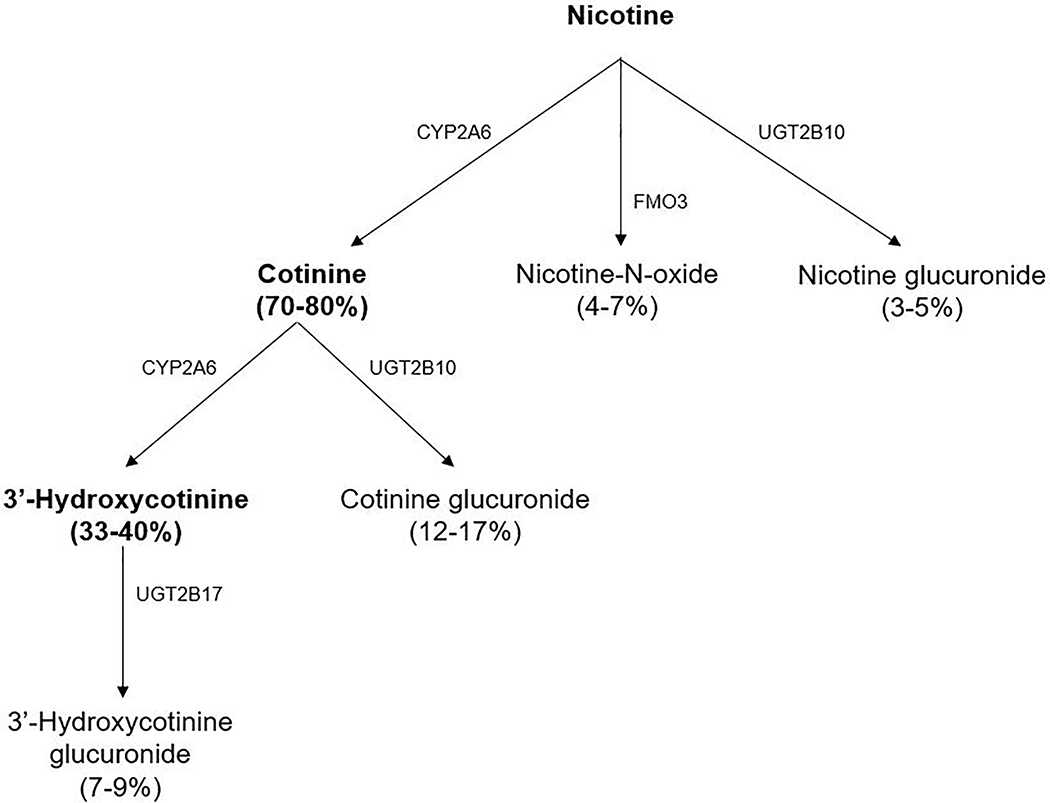
In addition to CYP2A6, nicotine is also metabolized by the flavin monooxygenase 3 (FMO3) to nicotine-N-oxide (NNO), with FMO3 variations not shown to substantially influence nicotine metabolism in previous studies [7], and by the uridine diphosphate-glucuronosyltransferase UGT2B10 to nicotine-glucuronide [2, 8–10] (Figure 1).
Figure 1.
Nicotine metabolism pathways (adapted from [2, 10], data from [2, 8, 10]); CYP: hepatic cytochrome P450 enzyme, FMO3: flavin monooxygenase 3, UGT: uridine diphosphate-glucuronosyltransferase
Glucuronidation of nicotine is usually a minor metabolic pathway (<10%) [9], but can be a larger determinant of nicotine clearance in people with reduced CYP2A6 activity [1, 11]. UGT2B10 also catalyzes the glucuronidation of COT (as well as nicotine), while UGT2B17 catalyzes formation of 3HC-glucuronide [1, 2] (Figure 1). It is therefore possible that differences in rate of glucuronidation might affect COT and 3HC levels and thus also the NMR. Furthermore, due to the larger impact on COT than on nicotine (Figure 1), UGT2B10 variants might affect the NMR more than nicotine clearance, while, due to the effect on 3HC, UGT2B17 variants might affect the NMR with no impact on nicotine clearance. Thus, the reliability of the NMR as a phenotypic marker of total nicotine clearance might be impacted by variation in glucuronidation of COT and/or 3HC and variation in the genes for these UGT enzymes, which differs among races [12–14].
Individual differences in the rate of nicotine metabolism (proxied by the NMR) have been shown to influence cigarette smoking behavior, and biomarkers of toxicant exposure and inflammation in smokers [15]. Rapid metabolism of nicotine is associated with smoking more cigarettes per day, greater nicotine dependence and lower rates of quitting smoking in the absence of pharmacotherapy and with nicotine transdermal patch compared to slower metabolizers [16–21]. However, it is possible that racial differences, such as the slower glucuronidation of nicotine and cotinine in African Americans (referred to here as Blacks) due to higher frequencies of slow metabolism variants in UGT2B10 [12–14], might affect the NMR and its correlation with the nicotine clearance. Previously racial differences in the relationship between NMR and nicotine exposure have been observed, with a greater influence of NMR on nicotine intake in Caucasians (referred to here as Whites) compared to Blacks, suggesting the potential for NMR to predict nicotine clearance less accurately across racial groups [22]. On the other hand, although previous studies in Black smokers found no significant impact of UGT2B17 reduced function alleles on NMR [23,24], a potential association between UGT2B17 and NMR was found in a Genome-wide association meta-analysis in European smokers, indicating a possible inter-ethnic variation regarding this relationship [25].
The main aim of the present study was to investigate the reliability of NMR as a phenotypic biomarker to predict nicotine clearance among different races and as a function of differences in the glucuronidation rates of nicotine, COT and 3HC and variation in UGT genotypes by comparing Whites, Blacks, and Asian-Americans (referred to here as Asians), using oral doses of nicotine and COT as metabolic probes. Due to the larger impact on COT than on nicotine, UGT2B10 variants might affect the NMR more than nicotine clearance. Slower COT glucuronidation could be associated with higher COT levels, but without an effect on NMR, since more COT should generate proportionately more 3HC. However, others have hypothesized that slower COT glucuronidation would be associated with higher COT levels and thus lower NMR [9], so we tested that hypothesis. Although no impact of 3HC glucuronidation on NMR was found in previous studies with Black participants [23], we retested the hypothesis that slower 3HC glucuronidation, which may result in higher 3HC levels, could alter NMR, while also including Asians, a racial group with higher prevalence of UGT2B17 deletion alleles than Whites or Blacks [23, 26].
Methods
Participants:
The study included male and female Whites, Blacks and Asians (four grandparents of the same race), ages 18–70. Participants were selected as healthy by medical history and taking no regular medication other than vitamins. Women of reproductive capacity had to have a negative pregnancy test. Participants with current alcoholism or illicit drug use were excluded. Potential participants were recruited by advertisements in San Francisco newspapers and campuses in the Bay Area, and postings on Craigslist, the Research-Online and our own website. The study was approved by the Committees on Human Research at the University of California, San Francisco and at the University of Toronto.
Screening and Consenting:
Potential participants were initially screened in a telephone interview. If suitable for the study, they were invited to the General Clinic Research Center or the Tobacco Research Clinic of the Zuckerberg San Francisco General Hospital for a screening visit. At this visit, they were asked to sign the consent form, and then fill out questionnaires inquiring about demographic, smoking, alcohol, caffeine, drug and medication, and general medical histories. A simple physical examination was performed (height and weight, vital signs and electrocardiogram), a saliva sample was collected for determination of COT level (to confirm smoking status), and a urine specimen was requested from females of childbearing potential for pregnancy testing.
Study procedures:
The study was conducted at the Clinical Study Center at Zuckerberg San Francisco General Hospital. Participants were asked not to eat or use tobacco starting at 10 pm on the previous night and to refrain from grapefruit or grapefruit juice for 48 hours prior to and for the duration of the study. At 8 am on study day, participants were given in solution a 2 mg oral dose of deuterium-labeled nicotine (nicotine-d2) and a 5 or 10 mg dose of either deuterium-labeled COT (cotinine-d4) for smokers or unlabeled COT (COT-d0) for non-smokers. These labeled compounds were synthetized as described previously [27, 28]. The 2 mg dose of nicotine was selected as a dose that is well tolerated by non-smokers but results in plasma nicotine concentrations that are easily measurable. The dose of COT was selected as a dose that would result in adequate saliva concentrations of COT over a 60-hour period, to allow us to determine the terminal elimination half-life of COT. The first 116 participants received 10 mg COT; afterwards the dose was reduced to 5 mg since pharmacokinetic analyses showed that this would not affect the ability to calculate the desired data. Deuterium-labeled COT was given to smokers because smokers already have unlabeled COT in their bodies which makes it impossible to do a kinetic study without a label. Blood samples were collected at 0 (i.e. before), 0.5, 1, 1.5, 2, 3, 4, 6, and 8 hours after dosing. One blood sample was also collected for genotyping. Samples of saliva (3–5 ml) were collected at 0, 6, 12, 24, 36, 48, and 60 hours following dosing. Participants could choose between staying overnight and have only three saliva samples taken at home, or get discharged after eight hours, in which case they were requested to provide five additional saliva samples taken at home. Urine samples were collected for eight hours after dosing and assayed for concentrations of nicotine, COT and 3HC and their glucuronides.
Analytical Chemistry:
Measurement of nicotine and COT in blood and saliva for the pharmacokinetic analysis was performed by gas chromatography–mass spectrometry, as described previously [29], modified for tandem mass spectrometry (MS/MS) for improved sensitivity. The limit of quantitation (LOQ) for nicotine was 0.5 ng/mL and for COT 1 ng/mL. For the calculation of plasma NMR, 3HC and COT were determined by liquid chromatography– tandem mass spectrometry (LC-MS/MS), as described previously [30]. The LOQ for 3HC and COT was 1 ng/mL. Urine concentrations were measured by LC-MS/MS. The LOQ for the urine analytes was 10 ng/mL. Nicotine, COT and 3HC glucuronides were generated by subtraction of the free form (measured without adding the beta-glucuronidase enzyme) from total form (measured after adding beta-glucuronidase enzyme type IXA from Escherichia coli and incubated overnight to deconjugate the molecule).
Genotyping was performed at the University of Toronto. Four single nucleotide polymorphisms (SNPs) within the UGT2B10 gene, i.e. rs2331559, rs11726322, rs835309, and the splice site variant rs2942857 (merged with rs116294140), that have been previously shown to be associated with nicotine and/or COT glucuronidation in at least one of the racial groups examined here [9, 31], were genotyped. The UGT2B17 copy number variant assay for the UGT2B17*2 deletion allele, that has been associated with impaired 3HC glucuronidation in smokers [12, 23, 32], was also genotyped. Individuals with a UGT2B17 duplication of intron 1 (i.e. UGT2B17 *1/*1×2, n=3), detected by the copy number variant assay, were merged with the UGT2B17 *1/*1 genotype, since there is currently no data indicating that this results in a gain of function. The CYP2A6 genotyping data are provided as supplementary material (Supplementary Document S1).
Data Analysis:
The main measure of the rate of nicotine metabolism was the oral plasma clearance of nicotine-d2, determined as the dose divided by the area under the plasma nicotine concentration–time curve extrapolated to infinity. The oral saliva clearance of COT (high correlation of plasma and saliva clearance of COT shown in previous studies [33]) was computed in a similar manner by use of the area under the saliva COT concentration–time curve. Both nicotine and cotinine clearances were normalized by subject body weights (kg). Elimination half-lives were determined by nonlinear least squares fitting of the log concentration versus time using Phoenix WinNonlin (Pharsight Corporation, Mountain View, CA). The plasma NMR, based on the ratio of free (unconjugated) COT and 3HC, was determined from the six hours post-dose plasma sample of dosed labeled (for smokers) or unlabeled (for non-smokers) COT. A high correlation between the plasma NMR ratio derived at this time point and oral nicotine clearance has been shown in previous studies (r=0.9, p<0.01 for the unlabeled (3HC-d0/COT-d0) and r=0.79, p<0.01 for the deuterium-labeled (3HC- d4/COT-d4) ratios) [3]. Urine data were used to estimate the glucuronidation activity as the ratio of glucuronide (ng/ml)/free (ng/ml) analyte and glucuronide/total (i.e. free and glucuronide) analyte. These were computed for nicotine and COT, phenotypic markers of UGT2B10 activity, and for 3HC, a phenotypic marker of UGT2B17 activity.
Numerical data are presented as arithmetic mean and standard deviation if normally distributed or median and range if not normally distributed, and nominal data as a proportion (%). Missing data were not imputed and not available glucuronidation ratios due to values below LOQ were not included in the analysis. Spearman’s correlations were used to describe variable associations between NMR, clearances and glucuronidation ratios. Between-group differences were tested using the chi-square or Fisher’s exact test for categorical variables, one-way analysis of variance (ANOVA) for continuous normally distributed variables, and the Kruskal-Wallis test for non-normally distributed variables. Additional analyses were performed using general linear model (GLM) where the log-transformed nicotine or cotinine clearance were predicted as a function of plasma NMR, sex [34, 35], age [2, 20], race, body mass index (BMI) [20], and smoking status [36]. Since glucuronidation might affect the nicotine clearance and NMR, but has a different role in this relationship than the CYP2A6 genotype, which is an established contributor to NMR and nicotine clearance (e.g. [3, 37]), the CYP2A6 genotype was not included in the analyses regarding the relationship between clearance and NMR. Analyses were conducted using SPSS statistical software (IBM SPSS Statistics 25.0) or R (version 3.5). A p<0.05 was considered statistically significant.
Results
A total of 227 participants were included in the study. The NMR and pharmacokinetic data for nicotine were available for all participants (N=227) while COT pharmacokinetic data were missing (e.g. due to missing saliva samples) in eight cases. Due to a missing sample in one case and values below LOQ in the rest of the cases, the glucuronidation ratios were not available in 11, 1, and 1 cases for nicotine, COT, and 3HC, respectively. The glucuronidation ratios were zero (i.e. no glucuronide present) in 17, 17, and 29 cases for COT, nicotine, and 3HC, respectively. The UGT2B10 genotyping was available for all participants while UGT2B17 genotype was missing in one case.
Among the 227 participants, 96 (42%) were White, 67 (30%) Asian, and 64 (28%) Black. The participants’ demographics and pharmacokinetic data are shown in Table 1; compared to the other two races, Whites had higher nicotine and cotinine total oral clearance, Blacks had lower nicotine and cotinine glucuronidation rates, and Asians had lower 3’-hydroxycotinine glucuronidation rates.
Table 1.
Participants’ demographics and pharmacokinetic parameters (shown as median (range) unless otherwise indicated)
| Measure | All (n=227) | Whites (n=96) | Blacks (n=64) | Asians (n=67) |
|---|---|---|---|---|
| Demographics | ||||
| Age, mean (SD) a,b,c | 33.2 (11.2) | 33.4 (11.2) | 38.1 (11.7) | 28.3 (8.3) |
| Male, n (%) | 128 (56.4) | 54 (56.2) | 38 (59.4) | 36 (53.7) |
| BMI, kg/m2, mean (SD) c | 25.1 (3.7) | 25.2 (3.8) | 26.1 (4.0) | 24.0 (3.0) |
| Non-Smoker, n (%) | 133 (58.6) | 50 (52.1) | 36 (56.2) | 47 (70.1) |
| Nicotine pharmacokinetics | ||||
| Nicotine Clearance, mL/min/kg a,b | 39.7 (7.0 – 230.3) | 48.9 (11.8 – 230.3) | 32.3 (7.0 – 174.3) | 27.4 (7.0 – 159.6) |
| Nicotine Half-Life, min b | 122.2 (31.9 – 623.4) | 113.2 (31.8 – 569.1) | 133.2 (38.2 – 585.6) | 144.3 (61.7 – 623.4) |
| Cotinine pharmacokinetics | ||||
| Cotinine Clearance, mL/h/kg a, c | 54.9 (6.8 – 341.4) | 58.9 (21.4 – 341.4) | 36.4 (6.8 – 146.8) | 54.6 (19.7 – 245.9) |
| Cotinine Half-Life, ha,b | 14.2 (6.9 – 59.9) | 13.5 (8.0 – 33.9) | 16.0 (8.2 – 59.9) | 14.8 (6.9 – 30.4) |
| Urinary Glucuronidation Ratios | ||||
| Nicotine Glucuronide / Total Nicotine a, c | 0.25 (0.00 – 0.77) | 0.29 (0.00 – 0.77) | 0.14 (0.00 – 0.69) | 0.29 (0.00 – 0.68) |
| Nicotine Glucuronide / Free Nicotine a, c | 0.33 (0.00 – 3.37) | 0.40 (0.00 – 3.37) | 0.16 (0.00 – 2.11) | 0.40 (0.00 – 2.14) |
| Cotinine Glucuronide / Total Cotinine a,c | 0.19 (0.00 – 0.76) | 0.23 (0.00 – 0.76) | 0.13 (0.00 – 0.60) | 0.20 (0.00 – 0.57) |
| Cotinine Glucuronide / Free Cotinine a,c | 0.24 (0.00 – 3.20) | 0.29 (0.00 – 3.20) | 0.15 (0.00 – 1.52) | 0.25 (0.00 – 1.30) |
| 3HC Glucuronide / Total 3HC b,c | 0.13 (0.00 – 0.68) | 0.14 (0.00 – 0.30) | 0.14 (0.00 – 0.68) | 0.10 (0.00 – 0.46) |
| 3HC Glucuronide / Free 3HC b,c | 0.15 (0.00 – 2.11) | 0.16 (0.00 – 0.44) | 0.16 (0.00 – 2.11) | 0.11 (0.00 – 0.86) |
| Phenotypic biomarker of nicotine clearance | ||||
| Plasma NMR (6h post-Cotinine Dosage)a,b | 0.18 (0.02 – 0.65) | 0.23 (0.05 – 0.63) | 0.17 (0.02 – 0.65) | 0.15 (0.02 – 0.50) |
Significant differences detected between Blacks and Whites
Significant differences detected between Asians and Whites
Significant differences detected between Blacks and Asians.
BMI: Body Mass Index; NMR: nicotine metabolite ratio; 3HC: 3’-hydroxycotinine
Plasma NMR was strongly correlated with nicotine clearance within each of the three race categories (White: correlation coefficient ρ= 0.77, Black: ρ=0.83, Asian: ρ= 0.84; all p <0.001) (Figure 2); the correlation remained significant among all three race categories after adjusting for age and sex.
Figure 2.
Correlation of plasma nicotine metabolite ratio (NMR) to oral plasma nicotine clearance for all three races (Whites, n=96; Blacks, n=64; Asians, n=67; clearance values on square root transformed scales; CI: Confidence Interval, COT: cotinine, 3HC: 3’-hydroxycotinine)
In the multivariate GLM, the NMR’s correlation with nicotine clearance (F(1,220)= 380.1, p <0.001) continued after adjustment for age, sex, race and smoking status (Table 2). In the unadjusted model, NMR, sex and race were predictors of nicotine clearance, while in the adjusted model, only NMR and race remained significant predictors (Table 2). A stronger relationship to nicotine clearance was found for NMR compared to race in both models.
Table 2.
Investigation of predictor effects on oral plasma nicotine clearance (ml/min/kg)
| Unadjusted Model | Multivariate Modela | |
|---|---|---|
| Variable | β (SE) | β (SE) |
| NMR | 4.48 (0.20)*** | 4.27 (0.23)*** |
| Age | 0.07 (0.05) | 0.01 (0.03) |
| Sex (Male) | −0.24 (0.01)* | −0.04 (0.06) |
| Race – Black£ | −0.41 (0.11)*** | −0.17 (0.07)** |
| Race – Asian£ | −0.52 (0.11)*** | −0.15 (0.07)* |
| Smoking Status (Positive for “Smoker”) | −0.13 (0.09) | −0.06 (0.05) |
F (6,220) = 83.4, p <0.001 Adj. R2 = 0.69
p< 0.05
p<0.01
p<0.001
White (largest population) used as reference
β: beta coefficient; NMR: Nicotine Metabolite Ratio; SE: Standard Error
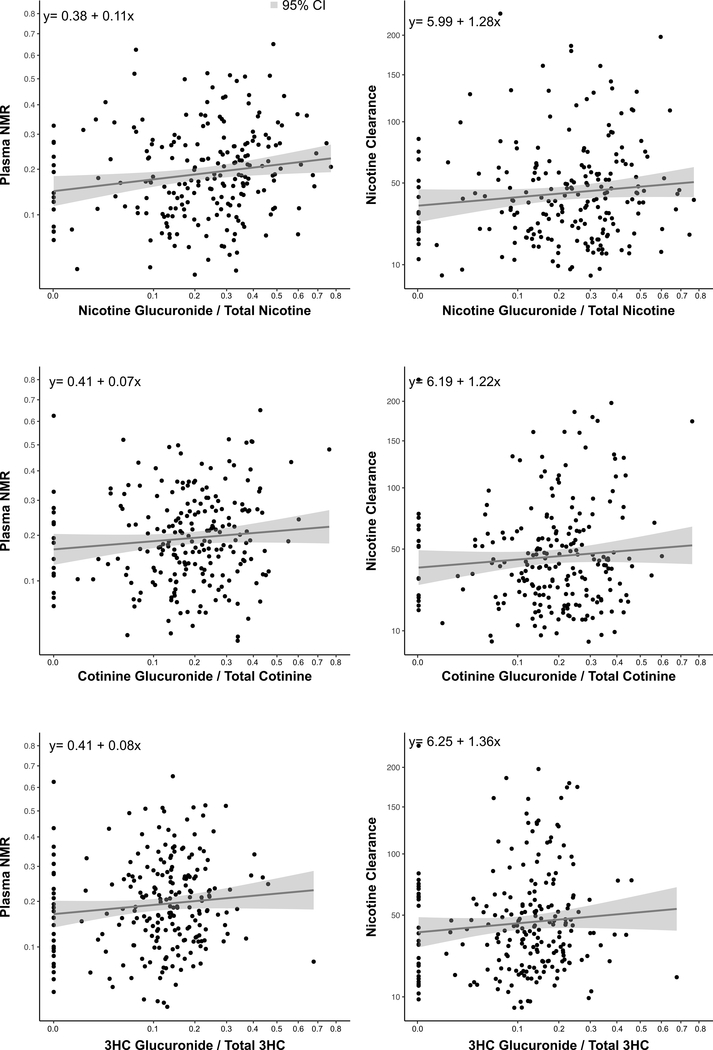
Investigation of correlations of the nicotine, COT and 3HC glucuronidation activity showed little evidence of a relationship with nicotine clearance (rho-value=0.12, p=0.09 for nicotine; rho-value=0.06, p=0.38 for COT; rho-value=0.11, p=0.09 for 3HC) or NMR (rho-value=0.19, p<0.001 for nicotine; rho-value=0.11, p=0.10 for COT; rho-value=0.12, p=0.07 for 3HC) (Figure 3, Table S1).
Figure 3.
Correlations between nicotine (n=216), cotinine (n=226) and 3’-hydroxycotinine (3HC; n=226) glucuronidation activity (assessed from urine glucuronide/parent ratios) and plasma nicotine metabolite ratio (NMR) or oral plasma nicotine clearance (values on square root transformed scales)
The correlations remained very weak or weak (all rho-values <0.4) also when investigating each race separately (Supplementary Table S1).
When examining the relationships in Figure 3, some individuals demonstrated very low rates of glucuronidation, most likely due to reduced activity or null UGTs. A categorical analysis based on the % of the measured nicotine, COT and 3HC glucuronidation activity, to also address the cases with very low or no glucuronidation activity, showed little evidence of a relationship with either nicotine clearance or NMR (Figure 4); the results remained insignificant also when analysed by race (analysis not shown).
Figure 4.
Relationship between nicotine (n=216), cotinine (n=226) and 3’-hydroxycotinine (3HC; n=226) glucuronidation activity (assessed from urine glucuronide/parent ratios), and oral plasma nicotine clearance and plasma nicotine metabolite ratio (NMR), based on the percentage of the measured glucuronidation activity
Table 3 shows the number of individuals with no glucuronidation activity for the total study population and also by race.
Table 3.
Number of participants with no glucuronidation activity by race
| All (n=227) | Whites (n=96) | Blacks (n=64) | Asians (n=67) | |
|---|---|---|---|---|
| Nicotine (n, %) | 16 (7.0%) | 4 (4.2%) | 10 (15.6%) | 2 (3.0%) |
| Cotinine (n, %) | 17 (7.5%) | 5 (5.2%) | 9 (14.1%) | 3 (4.5%) |
| 3’-hydroxycotinine (3HC; n, %) | 29 (12.8%) | 8 (8.3%) | 10 (15.6%) | 11 (16.4%) |
As a secondary approach to examining the impact of variation in glucuronidation on NMR’s ability to predict nicotine clearance, the impact of UGT2B10 and UGT2B17 genotype groups was examined. As the UGT genotypes variant frequencies vary by race, these are provided in Table 4; the allele frequencies were in Hardy-Weinberg equilibrium and similar to those found in 1000 Genomes and/or previously published [23, 24, 31].
Table 4.
Prevalence of UGT variants by race (shown as n (%) within race; p-values: comparisons by race; bold: values with adjusted residuals of <−2 or >2 for comparisons by race; Whites, n=96; Blacks, n=64; Asians, n=67)
| Race | ||||||
|---|---|---|---|---|---|---|
| N | White (n=96) | Black (n=64) | Asian (n=67) | P | ||
| UGT2B10 rs2331559 | G/^ | 118 | 73 (61.9) | 10 (8.5) | 35 (29.7) | <0.001 |
| C/G | 78 | 21 (26.9) | 30 (38.5) | 27 (34.6) | ||
| C/C | 31 | 2 (6.5) | 24 (77.4) | 5 (16.1) | ||
| UGT2B10 rs835309 | G/^ | 146 | 74 (50.7) | 15 (10.3) | 57 (39.0) | <0.001 |
| T/G | 60 | 21 (35.0) | 29 (48.3) | 10 (16.7) | ||
| T/T | 21 | 1 (4.8) | 20 (95.2) | 0 (0.0) | ||
| UGT2B10 rs11726322 | G/^ | 158 | 77 (48.7) | 33 (20.9) | 48 (30.4) | <0.001 |
| G/C | 57 | 18 (31.6) | 22 (38.6) | 17 (29.8) | ||
| C/C | 12 | 1 (8.3) | 9 (75.0) | 2 (16.7) | ||
| UGT2B10 rs2942857 (splice variant, previously rs116294140) | A/^^ | 175 | 89 (50.9) | 30 (17.1) | 56 (32.0) | <0.001 |
| A/C | 47 | 7 (14.9) | 29 (61.7) | 11 (23.4) | ||
| C/C | 5 | - | 5 (100) | - | ||
| UGT2B17 | *1/*1^^ | 77 | 43 (55.8) | 30 (39.0) | 4 (5.2) | <0.001 |
| *1/*2 | 89 | 45 (50.6) | 27 (30.3) | 17 (19.1) | ||
| *2/*2 | 60 | 7 (11.7) | 7 (11.7) | 46 (76.7) | ||
| Missing | 1 | 1 (100) | - | - | ||
The UGT variants had different SNP frequencies, linkage disequilibrium structures, and impact on nicotine and cotinine glucuronidation ratios between races (Supplementary Table S2), as expected and published previously (e.g. [9, 23]).
The UGT variants, as well as the glucuronidation ratios, were examined for their impact on NMR’s ability to predict nicotine clearance using regression analyses while adjusting for age, sex, and race (Table 5). None of the glucuronidation activities, nor any UGT genotype, affected the ability of NMR to predict nicotine clearance. The portion of variation in nicotine clearance predicted by NMR vary less than 2% among the models.
Table 5:
Regression analyses of plasma nicotine metabolite ratio (NMR)’s ability to predict oral plasma nicotine clearance, adjusted for an individual glucuronidation rate (assessed from urine glucuronide/parent ratios), an individual UGT variant, as well as age, sex, and race (Whites, n=95; Blacks, n=64; Asians, n=67)
| GLMs (Models a-g) Predicting Nicotine Clearance from Plasma NMR | β | 95 % CI | Scaled β | ANOVA Model Parameters (also includes adjustment for age and sex) |
|---|---|---|---|---|
| a: R2 = 0.69, p < 0.001 | ||||
| Plasma NMR | 4.32 | 3.88 – 4.75 | 0.80 | F(1,219) = 385.2 , p < 0.001 |
| Cot-Gluc/Total Ratio | F(1,219) = 0.69, p = 0.41 | |||
| Race | F(2.219) = 4.15, p = 0.02 | |||
| b: R2 = 0.69, p < 0.001 | ||||
| Plasma NMR | 4.29 | 3.86 – 4.72 | 0.79 | F(1,219) = 383.4, p < 0.001 |
| 3HC-Gluc/Total Ratio | F(1,219) = 0.08, p = 0.77 | |||
| Race | F(2.219) = 3.88, p = 0.02 | |||
| c: R2 = 0.69, p < 0.001 | ||||
| Plasma NMR | 4.31 | 3.87 – 4.74 | 0.79 | F (1,219) = 383.8, p < 0.001 |
| UGT2B10 rs2331559 Genotype | F(2,219) = 0.75, p = 0.48 | |||
| Race | F(2.219) = 2.43, p = 0.09 | |||
| d: R2 = 0.69, p < 0.001 | ||||
| Plasma NMR | 4.31 | 3.88 – 4.73 | 0.79 | F(1,219) = 397.2, p < 0.001 |
| UGT2B10 rs835309 Genotype | F(2,219) = 2.67, p = 0.07 | |||
| Race | F(2.219) = 2.27, p = 0.11 | |||
| e: R2 = 0.68, p < 0.001 | ||||
| Plasma NMR | 4.28 | 3.84 – 4.71 | 0.79 | F(1,219) = 380.3, p < 0.001 |
| UGT2B10 rs11726322 Genotype | F(2,219) =0.35, p = 0.70 | |||
| Race | F(2.219) = 3.91, p = 0.02 | |||
| f: R2 = 0.69, p < 0.001 | ||||
| Plasma NMR | 4.34 | 3.91 – 4.77 | 0.80 | F(1,219) = 399.6, p < 0.001 |
| UGT2B10 rs2942857 Genotype | F(2,219) = 2.53, p = 0.08 | |||
| Race | F(2.219) = 2.23, p = 0.11 | |||
| g: R2 = 0.69, p < 0.001 | ||||
| Plasma NMR | 4.31 | 3.89 – 4.74 | 0.80 | F(1,219) = 392.5, p < 0.001 |
| UGT2B17 Genotype | F(2,218) = 1.94, p = 0.14 | |||
| Race | F(2.218) = 3.25, p = 0.04 | |||
Nicotine Clearance (ml/min/kg); Not displayed are parameters for age and sex, which were also included in all models. ANOVA: Analysis of Variance; CI: Confidence Interval; GLM: General Linear Model; NMR: Nicotine Metabolite Ratio
Discussion
We present novel data showing a significant and strong correlation between NMR and nicotine oral clearance in three major racial groups, thus confirming that NMR is a reliable phenotypic biomarker for nicotine clearance across races. Further, none of the metabolite glucuronidation ratios had a significant effect on NMR and its ability to predict nicotine oral clearance. The UGT genotype frequencies and linkage disequilibrium structure varied between races as expected, but none of the UGT variants altered the ability of NMR to predict nicotine clearance (Table 5). Based on our findings, reports showing racial differences in the relationship between NMR, smoking behavior, levels of addiction and response to smoking cessation treatment are not likely to be related to inadequate estimation of nicotine clearance based on NMR.
As also reported by others, we found significant differences by race in pharmacokinetic measures, with higher nicotine and COT oral clearance in Whites compared to Blacks or Asians [2, 11], and lower nicotine and COT glucuronidation rates in Blacks, the latter in line with the higher frequency of slow UGT2B10 variants in this population [12, 38]. In a recent study using COT glucuronidation as a phenotypic marker of UGT2B10 activity approximately 15% of the Black participants excreted essentially no glucuronide [38], in line with the higher frequency of the UGT2B10 splice variant rs2942857 (formerly rs116294140) seen in this population [9], which was included in our study. The lower 3HC glucuronidation rates in Asians is likely related to the higher frequency of low activity UGT2B17 variants [23, 39].
In final regression models we found no effect of the glucuronidation ratios or of the UGT2B10 and UGT2B17 genotypes on the ability of NMR to predict nicotine clearance (Table 5). Similarly to our findings, a previous study investigating the impact of UGT2B10 and UGT2B17 variation on nicotine pharmacokinetics in Blacks also found no significant effect on NMR [24]. Lower nicotine and cotinine glucuronidation ratios in Blacks compared to Whites have also been reported previously [31]. However, these studies included either only Blacks [24], or only Whites and Blacks [31], whereas our study addressed these questions while including three different races, thus expanding the previously reported findings and also investigated the impact of race and glucuronidation on the ability of NMR to predict nicotine clearance across races.
In a recent study, smokers with essentially no UGT2B10 activity, as assessed by no quantifiable COT glucuronide, had lower NMR values compared to smokers with phenotypic higher UGT2B10 activity, but the difference did not remain significant after adjusting for cigarettes smoked per day, urinary total nicotine equivalents and UGT2B10 activity [38]. A large genotyping study including five racial groups (Blacks, Whites, Native Hawaiians, Latino and Japanese Americans) investigated the effect of UGT2B10 variants on nicotine metabolic pathways and found that, as expected, UGT2B10 variants associated with reduced activity in Blacks had a significant effect on the extent of nicotine glucuronidation [9]. The authors suggested that NMR might be less useful in Blacks, because the slower glucuronidation related to the high frequency of low activity UGT2B10 variants might impact the NMR and lead to misclassification of the CYP2A6 genotype. However, even in case of slower COT glucuronidation and higher COT levels, no effect on NMR is expected, since more COT is expected to generate proportionately more 3HC, thus leaving the 3HC/COT ratio (i.e. NMR) unaffected. In line with this theoretical prediction, we found no significant effect of the COT glucuronidation rate on NMR.
In contrast to the extent of COT glucuronidation, it would be expected that the 3HC glucuronidation might affect the NMR, since slower 3HC glucuronidation could result in higher free 3HC levels thus affecting the ratio. However, neither the 3HC glucuronidation ratios nor the UGT2B17 genotypes were significant predictors for NMR in our study, or in a previous study [23]. Zhu et al. [23] only included Blacks, while a higher prevalence of UGT2B17 deletion occurs in Asians [39]. We cannot exclude the possibility that a significant effect of 3HC glucuronidation on NMR would have been observed if we had a larger sample size of Asians in our study.
Limitations of our study include the smaller number of Black and Asian participants compared to Whites, the fact that these designations can represent heterogenous populations with variation in enzyme activity levels, as well as some missing values. However, although previous studies have demonstrated effects of race on NMR and/or nicotine clearance, this study is the first to investigate the relationship between NMR and nicotine oral clearance across races, thus providing a basis for the reliability and use of this phenotypic biomarker in clinical practice in different populations. In addition to the inclusion of three races, another strength was the detailed analyses on the effect of individual differences in rates of glucuronidation, assessed both phenotypically and by UGT2B17 and UGT2B10 genotypes, on the relationship between nicotine clearance and NMR.
In summary, our findings show that neither race nor differences in the glucuronidation rates of COT and 3HC appear to have a significant effect on NMR or the NMR’s ability to predict nicotine clearance, thus confirming that NMR is a reliable phenotypic biomarker for nicotine clearance and supporting its future use in smoking cessation studies and clinical practice.
Supplementary Material
Acknowledgements:
We would like to thank Kevin Delucchi for statistical support and advice, Sandy Tinetti for performing clinical studies, Gina Lowry and Rebecca Lenox for participant recruitment, Lisa Yu, Minjiang Duan, and Sylvia Wu for performing analytical chemistry, and the nursing staff of the Clinical Research Center at San Francisco General Hospital for their excellent care of the research participants. EL’s research fellowship was supported by the Bangerter-Rhyner Foundation. We acknowledge the support of a Canada Research Chair in Pharmacogenomics to RFT. We would like to thank Ewa Hoffmann for her work genotyping the samples. Laboratory resources were supported by the National Institute on Drug Abuse, P30 DA012393 and CIHR grant FDN-154294, and PJY-159710.
Footnotes
Potential conflicts of interest: N.L.B. serves as a paid consultant to pharmaceutical companies that are developing or that market smoking cessation medications. He also has been a paid expert witness in litigation against tobacco companies, including on issues related to light cigarettes. R.F.T. has served as a consultant to Quinn Emmanuel and as a paid consultant to pharmaceutical companies on unrelated topics. For the remaining authors there are no conflicts of interest.
References
- 1.Benowitz NL. Pharmacology of nicotine: addiction, smoking-induced disease, and therapeutics. Annu Rev Pharmacol Toxicol 2009;49:57–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tanner JA, Tyndale RF. Variation in CYP2A6 Activity and Personalized Medicine. J Pers Med 2017;7(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dempsey D, Tutka P, Jacob P 3rd, Allen F, Schoedel K, Tyndale RF, et al. Nicotine metabolite ratio as an index of cytochrome P450 2A6 metabolic activity. Clin Pharmacol Ther 2004;76(1):64–72. [DOI] [PubMed] [Google Scholar]
- 4.Lea RA, Dickson S, Benowitz NL. Within-subject variation of the salivary 3HC/COT ratio in regular daily smokers: prospects for estimating CYP2A6 enzyme activity in large-scale surveys of nicotine metabolic rate. J Anal Toxicol 2006;30(6):386–9. [DOI] [PubMed] [Google Scholar]
- 5.Mooney ME, Li ZZ, Murphy SE, Pentel PR, Le C, Hatsukami DK. Stability of the nicotine metabolite ratio in ad libitum and reducing smokers. Cancer Epidemiol Biomarkers Prev 2008;17(6):1396–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.St Helen G, Novalen M, Heitjan DF, Dempsey D, Jacob P 3rd, Aziziyeh A, et al. Reproducibility of the nicotine metabolite ratio in cigarette smokers. Cancer Epidemiol Biomarkers Prev 2012;21(7):1105–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chenoweth MJ, Zhu AZ, Sanderson Cox L, Ahluwalia JS, Benowitz NL, Tyndale RF, Variation in P450 oxidoreductase (POR) A503V and flavin-containing monooxygenase (FMO)-3 E158K is associated with minor alterations in nicotine metabolism, but does not alter cigarette consumption. Pharmacogenet Genomics 2014;24(3):172–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benowitz NL, Hukkanen J, Jacob P 3rd. Nicotine chemistry, metabolism, kinetics and biomarkers. Handb Exp Pharmacol 2009(192):29–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murphy SE, Park SS, Thompson EF, Wilkens LR, Patel Y, Stram DO, et al. Nicotine N-glucuronidation relative to N-oxidation and C-oxidation and UGT2B10 genotype in five ethnic/racial groups. Carcinogenesis 2014;35(11):2526–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hukkanen J, Jacob P 3rd, Benowitz NL. Metabolism and disposition kinetics of nicotine. Pharmacol Rev 2005;57(1):79–115. [DOI] [PubMed] [Google Scholar]
- 11.Benowitz NL, St Helen G, Dempsey DA, Jacob P 3rd, Tyndale RF, Disposition kinetics and metabolism of nicotine and cotinine in African American smokers: impact of CYP2A6 genetic variation and enzymatic activity. Pharmacogenet Genomics 2016;26(7):340–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berg JZ, Mason J, Boettcher AJ, Hatsukami DK, Murphy SE. Nicotine metabolism in African Americans and European Americans: variation in glucuronidation by ethnicity and UGT2B10 haplotype. J Pharmacol Exp Ther 2010;332(1):202–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaivosaari S, Toivonen P, Hesse LM, Koskinen M, Court MH, Finel M. Nicotine glucuronidation and the human UDP-glucuronosyltransferase UGT2B10. Mol Pharmacol 2007;72(3):761–8. [DOI] [PubMed] [Google Scholar]
- 14.Benowitz NL, Perez-Stable EJ, Fong I, Modin G, Herrera B, Jacob P 3rd. Ethnic differences in N-glucuronidation of nicotine and cotinine. J Pharmacol Exp Ther 1999;291(3):1196–203. [PubMed] [Google Scholar]
- 15.Carroll DM, Murphy SE, Benowitz NL, Strasser AA, Kotlyar M, Hecht SS, et al. Relationships between the Nicotine Metabolite Ratio and a Panel of Exposure and Effect Biomarkers: Findings from Two Studies of U.S. Commercial Cigarette Smokers. Cancer Epidemiol Biomarkers Prev 2020;29(4):871–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rubinstein ML, Benowitz NL, Auerback GM, Moscicki AB. Rate of nicotine metabolism and withdrawal symptoms in adolescent light smokers. Pediatrics 2008;122(3):e643–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lerman C, Tyndale R, Patterson F, Wileyto EP, Shields PG, Pinto A, et al. Nicotine metabolite ratio predicts efficacy of transdermal nicotine for smoking cessation. Clin Pharmacol Ther 2006;79(6):600–8. [DOI] [PubMed] [Google Scholar]
- 18.Patterson F, Schnoll RA, Wileyto EP, Pinto A, Epstein LH, Shields PG, et al. Toward personalized therapy for smoking cessation: a randomized placebo-controlled trial of bupropion. Clin Pharmacol Ther 2008;84(3):320–5. [DOI] [PubMed] [Google Scholar]
- 19.Schnoll RA, Patterson F, Wileyto EP, Tyndale RF, Benowitz N, Lerman C. Nicotine metabolic rate predicts successful smoking cessation with transdermal nicotine: a validation study. Pharmacol Biochem Behav 2009;92(1):6–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ho MK, Mwenifumbo JC, Al Koudsi N, Okuyemi KS, Ahluwalia JS, Benowitz NL, et al. Association of nicotine metabolite ratio and CYP2A6 genotype with smoking cessation treatment in African-American light smokers. Clin Pharmacol Ther 2009;85(6):635–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lerman C, Schnoll RA, Hawk LW Jr, Cinciripini P, George TP, Wileyto EP, et al. Use of the nicotine metabolite ratio as a genetically informed biomarker of response to nicotine patch or varenicline for smoking cessation: a randomised, double-blind placebo-controlled trial. Lancet Respir Med 2015;3(2):131–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ross KC, Gubner NR, Tyndale RF, Hawk LW Jr, Lerman C, George TP, et al. Racial differences in the relationship between rate of nicotine metabolism and nicotine intake from cigarette smoking. Pharmacol Biochem Behav 2016;148:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu AZ, Zhou Q, Cox LS, Ahluwalia JS, Benowitz NL, Tyndale RF. Variation in trans-3’-hydroxycotinine glucuronidation does not alter the nicotine metabolite ratio or nicotine intake. PLoS One 2013;8(8):e70938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Taghavi T, St Helen G, Benowitz NL, Tyndale RF. Effect of UGT2B10, UGT2B17, FMO3, and OCT2 genetic variation on nicotine and cotinine pharmacokinetics and smoking in African Americans. Pharmacogenet Genomics 2017;27(4):143–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buchwald J, Chenoweth MJ, Palviainen T, Zhu G, Benner C, Gordon S, et al. Genome-wide association meta-analysis of nicotine metabolism and cigarette consumption measures in smokers of European descent. Mol Psychiatry 2020. March 10: 10.1038/s41380-020-0702-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xue Y, Sun D, Daly A, Yang F, Zhou X, Zhao M, et al. Adaptive evolution of UGT2B17 copy-number variation. Am J Hum Genet 2008. September;83(3):337–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Benowitz NL, Jacob P 3rd. Metabolism of nicotine to cotinine studied by a dual stable isotope method. Clin Pharmacol Ther 1994;56(5):483–93. [DOI] [PubMed] [Google Scholar]
- 28.Jacob P, Benowitz N, Shulgin AT. Synthesis of optically pure deuterium‐labelled nicotine, nornicotine and cotinine. J Labelled Comp Radiopharmaceut 1988;25:1117–28. [Google Scholar]
- 29.Jacob P 3rd, Wu S, Yu L, Benowitz NL. Simultaneous determination of mecamylamine, nicotine, and cotinine in plasma by gas chromatography-mass spectrometry. J Pharm Biomed Anal 2000;23(4):653–61. [DOI] [PubMed] [Google Scholar]
- 30.Jacob P 3rd, Yu L, Duan M, Ramos L, Yturralde O, Benowitz NL. Determination of the nicotine metabolites cotinine and trans-3’-hydroxycotinine in biologic fluids of smokers and non-smokers using liquid chromatography-tandem mass spectrometry: biomarkers for tobacco smoke exposure and for phenotyping cytochrome P450 2A6 activity. J Chromatogr B Analyt Technol Biomed Life Sci 2011;879:267–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wassenaar CA, Conti DV, Das S, Chen P, Cook EH, Ratain MJ, et al. UGT1A and UGT2B genetic variation alters nicotine and nitrosamine glucuronidation in european and african american smokers. Cancer Epidemiol Biomarkers Prev 2015;24(1):94–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen G, Giambrone NE Jr, Dluzen DF, Muscat JE, Berg A, Gallagher CJ, et al. Glucuronidation genotypes and nicotine metabolic phenotypes: importance of functional UGT2B10 and UGT2B17 polymorphisms. Cancer Res 2010;70(19):7543–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zevin S, Jacob P, Geppetti P, Benowitz NL. Clinical pharmacology of oral cotinine. Drug Alcohol Depend 2000;60(1):13–8. [DOI] [PubMed] [Google Scholar]
- 34.Benowitz NL, Lessov-Schlaggar CN, Swan GE, Jacob P 3rd. Female sex and oral contraceptive use accelerate nicotine metabolism. Clin Pharmacol Ther 2006;79(5):480–8. [DOI] [PubMed] [Google Scholar]
- 35.Higashi E, Fukami T, Itoh M, Kyo S, Inoue M, Yokoi T, et al. Human CYP2A6 is induced by estrogen via estrogen receptor. Drug Metab Dispos 2007;35(10):1935–41. [DOI] [PubMed] [Google Scholar]
- 36.Benowitz NL, Jacob P 3rd. Nicotine and cotinine elimination pharmacokinetics in smokers and nonsmokers. Clin Pharmacol Ther 1993;53(3):316–23. [DOI] [PubMed] [Google Scholar]
- 37.El-Boraie A, Taghavi T, Chenoweth MJ, Fukunaga K, Mushiroda T, Kubo M, et al. Evaluation of a weighted genetic risk score for the prediction of biomarkers of CYP2A6 activity. Addict Biol 2020. January;25(1):e12741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murphy SE, Sipe CJ, Choi K, Raddatz LM, Koopmeiners JS, Donny EC, et al. Low Cotinine Glucuronidation Results in Higher Serum and Saliva Cotinine in African American Compared to White Smokers. Cancer Epidemiol Biomarkers Prev 2017;26(7):1093–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xue Y, Sun D, Daly A, Yang F, Zhou X, Zhao M, et al. Adaptive evolution of UGT2B17 copy-number variation. Am J Hum Genet 2008;83(3):337–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.