Abstract
Immunomodulatory medications are a mainstay of treatment for autoimmune diseases and malignancies. In addition to their direct effects on immune cells, these medications also impact the gut microbiota. Drug-induced shifts in commensal microbes can lead to indirect but important changes in the immune response. We performed a comprehensive literature search focusing on immunotherapy/microbe interactions. Immunotherapies were categorized into five subtypes based on their mechanisms of action: cell trafficking inhibitors, immune checkpoint inhibitors, immunomodulators, anti-proliferative drugs, and inflammatory cytokine inhibitors. Although no consistent relationships were observed between types of immunotherapy and microbiota, most immunotherapies were associated with shifts in specific colonizing bacterial taxa. The relationships between colonizing microbes and drug efficacy were not well-studied for autoimmune diseases. In contrast, the efficacy of immune checkpoint inhibitors for cancer was tied to the baseline composition of the gut microbiota. There was a paucity of high-quality data; existing data were generated using heterogeneous sampling and analytic techniques, and most studies involved small numbers of participants. Further work is needed to elucidate the extent and clinical significance of immunotherapy effects on the human microbiome.
Keywords: autoimmune, microbiome, immunomodulatory drug
Introduction
Once a “forgotten organ”, the human microbiome is increasingly recognized as integral to health and disease [1]. Trillions of microbiota colonize all mucosal and barrier surfaces, including the gut, oral cavity, nasopharyngeal, respiratory tract, urogenital tract and skin [2]. Commensal microbiota directly and indirectly shape immune cell development and phenotype, with a well-established impact on the pathophysiology of immune-mediated diseases [3, 4]. The microbiome also contributes to medication efficacy, for example by metabolizing drugs [5-7]. Medications, in turn, can impact the microbiota and cause shifts in downstream immune phenotypes which may then impact the microbial composition in a bi-directional manner.
In this review, we explore how immunomodulatory drugs impact the gut microbiota, as other microbial niches, such as the oral cavity, lung, and skin, are poorly studied in this context. We first summarize relevant aspects of the immune system, highlighting components that are relevant to the host-commensal relationship. Next, we discuss the characterization of the healthy gut microbiome and describe important bacterial taxa that modulate host health and disease. Lastly, we examine the complex multidirectional relationships between the host, immunomodulatory drugs, and the gut microbiota (Figure 1).
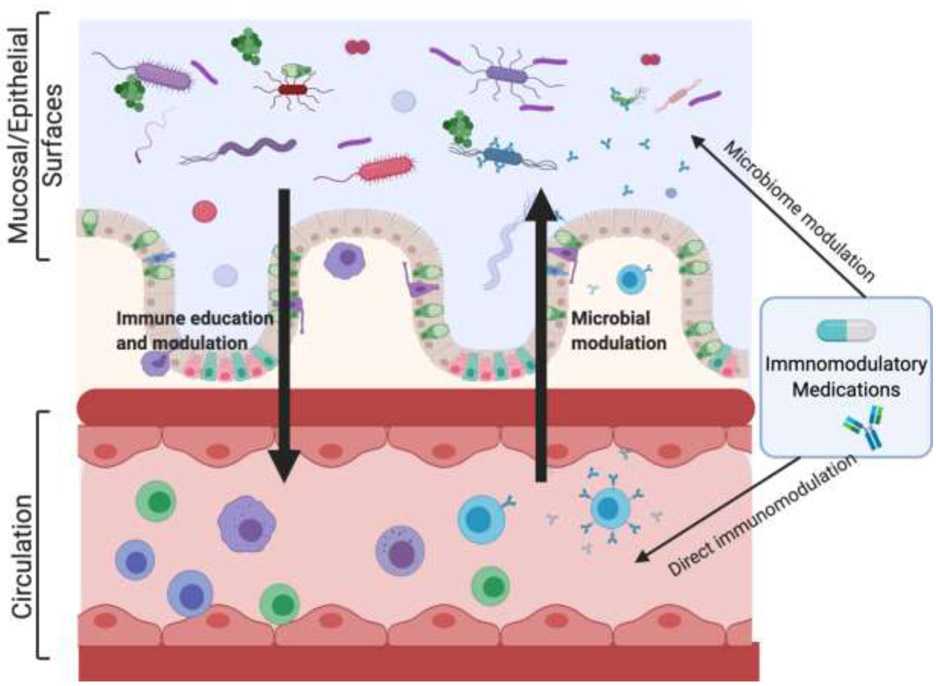
Figure 1. Host-Drug-Microbiota Interactions.
The interface between microorganisms and the immune is complex and multidirectional, and it is further influenced by immunomodulatory medications. Commensal organisms at mucosal/epithelial surfaces influence immune education and modulation both locally and systemically. These microbes also influence drug metabolism and efficacy. Inversely, the immune system shapes the composition of the microbiome. Immunomodulatory medications impact circulating immune cells directly, via a variety of mechanisms. Emerging data suggest that these medications also function indirectly, by shifting the composition of the microbiome. These multi-directional relationships are complex and remain poorly understood. Figure created with BioRender.com
The Innate Immune System
The innate immune system is characterized by fast, non-specific responses against pathogens. Key players include neutrophils and myeloid-derived cells (e.g. monocyte/macrophages, dendritic cells) which express pattern recognition receptors (PRRs) that bind to microbe-associated molecular patterns (MAMPs) such as Toll-like receptors (TLRs) and Nod-like receptors [8-10]. Natural killer cells are another circulating innate immune cell; these express major histocompatibility complex (MHC) Class I and kill infected cells by releasing cytotoxic granules [11].
Most organ systems include resident innate immune cells, which assist in screening for infections and mounting a rapid immune response [12]. These specialized cells include microglia (CNS) [13], Kupffer cells (liver) [14], alveolar macrophages (lungs) [15], innate lymphoid cells (ILC; mucosal surfaces)[16], and Langerhans cells (skin) [17]. The gut has a particularly robust resident immune system, including innate as well as adaptive cells. The gut-associated lymphoid tissue (GALT), discussed below, contains ILCs, Paneth cells and intestinal epithelial cells which express TLRs and release antimicrobial peptides (α-defensins, β-defensins, C-type lectins) to protect against extracellular pathogens. Commensal gut microbes (e.g. Lactobacillus) interact with PRRs on these cells to induce expression of antimicrobial peptides [18, 19].
The Adaptive Immune System
Unlike the general recognition of the innate immune response, the adaptive response is antigen specific. T cells express a unique T cell receptor, and cluster of differentiation (CD) 3. They are divided into two main classes: CD4 helper T (Th) cells and CD8 cytotoxic T cells. Helper T cells are generally categorized into Th1, Th2, Th17, or regulatory (Treg) subsets which express the transcription factors T-bet, GATA-3, RORγT, and Foxp3, respectively [20-25]. Th1 cells provide immunity against intracellular microbes (such as bacteria or viruses) characterized through the release of IFNγ [26, 27]. The cytokine interleukin-4 (IL)-4 skews T-cells towards a Th2 response. These cells protect against parasites/allergens and promote tissue repair through production of IL-4, IL-5, and IL-13 [28-30]. Th17 cells promote immunity against extracellular pathogens, such as fungi, and are pathogenic in a number of autoimmune diseases [31, 32]. Finally, Tregs are important for establishing self-tolerance, suppressing overactive immune responses, and maintaining homeostasis through the production of anti-inflammatory molecules such as IL-10 and TGF-beta [33, 34]. Similar to Th1 cells, CD8 cytotoxic T cells combat intracellular microbes and tumor cells. They do so by secreting IFN-γ/TNF-α, producing cytotoxic granules, and activating the Fas-mediated apoptosis pathway [35]. After their target antigen has been eliminated from the host, effector T cells either die via apoptosis or differentiate into antigen-specific memory T cells [36, 37].
Regardless of induction, T cells communicate with a variety of immune and non-immune cells to drive a robust, protective immune response, which is tightly regulated to limit damage off-target damage. Protective T cell responses can become pathogenic, especially during states of extremely acute or unresolved inflammation, and these phenotypes are influenced by the microbiome [38, 39]. Helper T cells exhibit profound plasticity in response to the cytokine milieu; for example, healthy human Tregs become dysfunctional and express IFNy, a Th1 cytokine, in the presence of IL-12 [40]. Functional and phenotypic imbalances in these T-cell subsets, especially Th17 and Tregs, have been implicated in autoimmune diseases [33, 40, 41]. Conversely, chronically stimulated antigen-specific CD4 and CD8 T cells may become “exhausted”, a state in which the cells are physically present, but no longer functionally active [36, 42]. As a result, antigen clearance and anti-tumor immunity is impaired. These T cell responses can be altered through selective interaction with the microbiome.
B cells comprise the humoral arm of the adaptive immune system. This lineage is best known for antibody production, which can occur in a T cell dependent- or independent manner. B cells also act as professional antigen presenting cells and produce cytokines that further support the adaptive and innate immune responses [43]. The gut microbiome educates the humoral immune response and conversely, mucosal B-cells secrete IgA, which coats potentially pathologic members of the gut microbiome, keeping them in check [44, 45].
The Mucosal Immune System
T cell responses are commonly studied in the context of the blood and secondary lymphoid organs (spleen, lymph nodes), however, the mucosal immune system represents the body’s largest lymphoid organ and directly interfaces with the gut microbiota [46] (Figure 1). Mucosal-associated lymphoid tissue comprises 80% of all immunocytes and is subdivided into gut-associated lymphoid tissue (GALT), bronchus-associated-lymphoid-tissue, and nasal-associated lymphoid tissue [46-48].
The GALT forms a protective immune-barrier surface and is organized within the connective tissue of the lamina propria and dome-shaped Peyer’s patches, which are enriched with antigen-experienced T cells and naïve B cells, respectively [49]. Specialized epithelial cells (M cells) in Peyer’s patches constantly sample gut bacterial epitopes and initiate lymphocyte activation [50]. Plasma cells in Peyer’s patches and mesenteric lymph nodes secrete dimeric secretory immunoglobulin A (IgA), which coats pathogenic/immunogenic bacteria, prevents their adhesion to the intestinal barrier [51] and promotes host-commensal homeostasis [52]. After activation in Peyer’s patches, experienced IgA-secreting B cells travel through the lymphatic system and into the systemic circulation, then return to the lamina propria via the mesenteric lymph nodes [53].
The host immune response has evolved to tolerate commensals. Tolerogenic mechanisms, such as Treg induction and IL-10 secretion, allow beneficial microbes to chronically colonize the host, but the host retains the ability to mount effective immune responses against microbes that breach these barrier surfaces [54, 55].
Commensal gut microbes are necessary for developing normal host mucosal immunity. Germ free mice have severe mucosal, immune, and anatomic abnormalities [56], including shorter ileal villi [57], reduced Peyer’s patch size [58], underdeveloped mesenteric lymph nodes [59], fewer IgA-producing plasma cells [60], diminished Th17 and Treg subpopulations [61, 62], and a skewed Th2 to Th1 ratio [63]. These immunological defects impact immune-mediated diseases and highlight the role of commensals in local and systemic immune development and responses [64].
The Human Gut Microbiome
Commensal gut bacteria fall into six main phyla: Bacteroidetes, Firmicutes, Actinobacteria, Proteobacteria, Fusobacteria, and Verrucomicrobia [65, 66]. Bacteroidetes and Firmicutes are the dominant organisms, making up the majority of the gut microbiota with Firmicutes predominantly composed of the Clostridium genera [65, 66].
The human gut offers a limited number of ecologic niches, which may be filled by different organisms in different individuals. This variation culminates in a high degree of inter-individual heterogeneity in commensal organisms [65, 66]. Moreover, the composition and stability of the gut microbiome is dynamic across a lifetime. In childhood, Firmicutes are enriched, while with increasing age Bacteroidetes become the dominant phylum [67]. Nevertheless, during adulthood the composition of the gut microbiome remains stable unless it is externally perturbed by common medications (e.g. antibiotics, proton-pump inhibitors, metformin [68, 69]) or other factors including drastic dietary changes [70] and geographic relocation [71, 72].
The individual-level heterogeneity in colonizing microbiota necessitates specialized computational techniques for studying the microbiome [73]. Species richness quantifies the number of different species represented within an ecologic niche. Diversity incorporates both the number of bacterial species and the abundance of each. More specifically, alpha diversity quantifies the richness and evenness of bacterial communities within a sample, while beta diversity measures diversity between samples (e.g. between different anatomic locations or different individuals).
Alterations in the composition of the gut microbiota, or gut dysbiosis, have been implicated in many diseases including obesity, diabetes, asthma, allergies, inflammatory bowel disease, cancer, autoimmunity, and even neurodegenerative diseases [4]. In this section, we highlight commensals with immune-modulatory capabilities, some of which have been implicated in disease, and discuss how they are known to modulate the host immune system.
Bacteroidetes
Bacteroidetes are gram-negative anaerobes that comprise a substantial portion of the adult gut microbiota [66]. This phyla contains many immunomodulatory genera. One of the best-studied is Bacteroides. Bacteroides thetaiotamicron, a genetically tractable organism, is frequently used to study host-commensal interactions [74, 75]. Another well-studied species is B. fragilis, which interacts indirectly with the pattern recognition receptor TLR2 via production of polysaccharide A to induce Treg differentiation and tolerance. Recolonization with B. fragilis alone ameliorates the Th1/Th2 imbalance observed in germ free mice [76]. Conversely, some strains of B. fragilis produce a pathogenic enterotoxin; these have been implicated in inflammatory bowel disease and colorectal cancer [77]. This phenomenon illustrates the importance of strain-level taxonomic resolution when studying the microbiome.
Prevotella are considered commensals due their contributions to glucose metabolism, although a few strains are opportunistic pathogens. Members of this genus can help prevent inflammation and autoimmune diseases; P. histicola strains have been shown to suppress experimental autoimmune encephalitis [78] and collagen-induced arthritis in mice [79] and have recently been discussed as a novel therapeutic option for patients with multiple sclerosis [80]. Prevotella have an important role in polysaccharide degradation and energy extraction, and members of this genus are expanded in African children with fiber-rich diets as compared to Italian children [81]. Nevertheless, emerging data have linked mucosal Prevotella to low-grade inflammation and a variety of diseases, including periodontitis and rheumatoid arthritis [82, 83]. The strain-specific nature of bacterial immunogenic potential remains to be fully elucidated. Firmicutes
Firmicutes represent the other dominant phylum in the human gut. Clostridia are gram-positive endospore-forming bacteria that comprise a large proportion of the Firmicutes. Butyrate-producers in Clostridium clusters IV and XIVa, such as Ruminococcus, Lachnospira, and Roseburia, promote Tregs [84]. They accomplish this directly, by stimulating Treg proliferation, and indirectly, by fermenting dietary fiber to produce butyrate [84]. Butyrate is a histone deacetylase inhibitor that has been shown to boost the generation and function of Tregs and secretion of IL-10 [85-88]. Indeed, gnotobiotic mice reconstituted with Clostridium strains have enriched populations of Foxp3+ Treg cells in the colon [62]. However, Clostridial species can also promote disease. For example, Clostridium difficile is a gram-positive spore-forming microbe that causes colitis when antibiotics or chemotherapeutic drugs kill other members of the gut flora allowing Clostridial overgrowth. C. difficile toxins A and B damage colonic epithelial cells and lead to abdominal pain and non-remitting diarrhea. Fecal microbiota transplantation from healthy individuals is an effective treatment for C. difficile colitis [89]. Other lactic acid-producing Firmicutes, such as Lactobacillus and Enterococcus, are also important immune modulators which induce Treg activity and suppress Th1 and Th2 cells [90, 91].
Proteobacteria
Proteobacteria are gram-negative organisms that populate the normal gut in small quantities. Its members are often pathogenic (e.g. gastric ulcer-inducing Helicobacter pylori [92]), recognized as immunogenic by the host immune system and are coated by secretory IgA. Proteobacteria dysregulation is associated with pathology, and the Enterobacteriaceae family has been associated with obesity, metabolic diseases and colitis [93]. However, members of this phylum also provide important immune education. For example, Alicaligenes spp, are tolerogenic Proteobacteria that inhabit Peyer’s patches, where they stimulate secretory IgA and help establish mutualism [94, 95].
Sex Differences in the Human Gut Microbiome
There are sex differences in relative microbial abundance at the phyla level [96]. In humans, Bacteroidetes are decreased in females compared to males [97]. Differences in microbial diversity and composition have also been correlated with serum levels of sex hormones, with high estradiol/testosterone producing individuals hosting more diverse microbial communities [98].
Animal models have provided insight into microbial sex differences in the context of autoimmune disease [96, 99]. Male nonobese diabetic (NOD) mice had expansion of the following bacterial families: Porphyromonadaceae, Veillonellaceae, Kineosporiaceae, Peptococcaceae, Enterobacteriaceae, Lactobacillaceae, Cytophagaceae, Peptostreptococcaceae, and Bacteroidaceae [99], Commensal colonization increased serum testosterone and protected males against type I diabetes. Fecal microbial transfer from adult male NOD mice to immature females also protected against the development of type I diabetes by an androgen-dependent mechanism [96].
In humans, androgen deprivation depleted testosterone-metabolizing Corynebacterium species and enriched Akkermansia muciniphila [100]. These microbial shifts appear to underlie the efficacy of the androgen inhibitor abiraterone acetate in androgen-independent prostate cancer. The full impact of sex hormones on the microbiota and vice versa remains to be well-characterized.
Drug-Microbial Relationships
Gut microbiota directly metabolize many oral drugs via reduction and hydrolysis [101-103]. This may occur either prior to (first-pass metabolism) or directly following absorption (enterohepatic circulation) in the small/large-intestine [101]. Well-studied examples include activation of sulfasalazine by azo-reductase containing bacteria [104], inactivation of digoxin by Eggerthella lenta strains [105], and toxification of mycophenolate motefil (MMF) as a result of bacterial β-glucorinodiase (GUS) activity [106]. Leveraging microbial metabolism has major clinical implications for improving therapeutic responses in patients with cancer and autoimmune diseases. For example, inhibition of GUS-containing bacteria via antibiotics ameliorates GI-related side-effects and improves antitumor effect in mice treated with MMF [106], or irinotecan [107], respectively.
In addition to metabolizing medications, commensal gut bacteria can be directly impacted by oral and parenteral medications. Antibiotics substantially reshape the gut microbiome [108], but many other drugs also have downstream effects on the microbiota. Well documented examples include metformin, which leads to expansion of Akkermansia muciniphila and other short chain fatty acid producing microbes [109], and proton pump inhibitors, which lead to expansion of Lactobacillus species [110]. Immunomodulatory medications are beginning to be recognized as another class of medications that impacts the gut microbiota with downstream effects on the underlying disease. We will review the existing literature on interactions between immunomodulatory medications and commensal microbiota.
Anti-Proliferative Immunotherapies
Cyclophosphamide
Cyclophosphamide is a cytotoxic alkylating agent that impairs transcription and translation in rapidly proliferating cells [111]. It is used as a chemotherapeutic agent for a variety of malignancies and as an immunosuppressant for numerous autoimmune conditions. Cyclophosphamide profoundly depletes circulating B and T cells, with a predilection for CD4+ T cells. Rodent research demonstrated that cyclophosphamide caused translocation of gram-positive commensals (mainly Lactobacillus and Enterococcus species) to secondary lymphoid organs, where they promoted Th1 and Th17 differentiation. These immune cells were necessary for the antitumor effect of cyclophosphamide [112]. To date, there have been no human studies investigating its impact on composition or function of the microbiome.
Mycophenolate Mofetil
Mycophenolate mofetil (MMF) inhibits de novo guanine nucleotide synthesis, eliciting a cytostatic effect on B- and T-lymphocytes [113]. It is an important immunosuppressant used for transplant recipients and a variety of autoimmune conditions [114]. MMF-treated mice had decreased gut alpha diversity. This was driven by increased pathogenic Escherichia/Shigella and a decrease in three gut-protective genera: Clostridium, Akkermansia, and Parabacteroides. Concurrently, mice had rapid weight loss and increased colonic inflammation that was dependent on the colonizing microbiota [115]. MMF has been associated with erosive enterocolitis in human transplant recipients, but no specific drug effects have yet been shown on the human microbiome [116]. Of note, MMF is directly metabolized by B-glucuronidase (GUS)-expressing gut microbiota during enterohepatic circulation [117]. Inhibiting GUS-expressing bacteria can improve MMF-associated GI toxicity [106].
Methotrexate
Methotrexate (MTX) is a first-line drug in treating RA and other autoimmune diseases and an anti-neoplastic agent. It is a folate antimetabolite that competitively inhibits dihydrofolate reductase, interfering with purine and pyrimidine nucleotide synthesis and suppressing rapidly dividing cells. At lower doses, MTX inhibits 5-aminoimidazole-4-carboxamide ribonucleotide, which leads to adenosine accumulation [118]. Adenosine can suppress neutrophil and macrophage recruitment and reduce proinflammatory cytokines such as TNFα and IFNγ [119, 120]. Microbiota containing carboxypeptidase glutamate 2 enzymes (CPG2) catabolize MTX and help decrease nephrotoxicity [121, 122].
MTX may modulate the composition of the gut microbiota in both mice and humans, but the patterns of change have been inconsistent [123-126]. In mice, Bacteroides fragilis decreased after MTX treatment in a time-dependent manner [123]. In another study, MTX-associated changes to the microbiome were dose-dependent. At low doses, the relative abundance of Firmicutes compared to Bacteroidetes increased, but the opposite trend was observed at high doses [121]. In humans, a small cohort of MTX-treated RA patients had significantly decreased Enterobacteriales [125] compared to treatment-naive patients (Table 1); these findings need to be validated in a larger cohort. In a metagenomic sequencing study, researchers compared the effects of MTX on the oral, salivary, and gut microbiomes [126]. In the dental plaque microbiome, MTX responders had increases in healthy control-enriched microbial linkage groups (MLGs) such as Prevotella maculosa. In the salivary microbiome, MTX responders had reductions in Veillonella MLGs, which were elevated in RA patients prior to treatment. In the fecal microbiome, Holdemania filiformis MLGs were increased after treatment with MTX compared to baseline.
Table 1.
Bacterial effects in the first comparison group are reported. The changes in bacterial composition in the human gut microbiome are reported as changes in terms of phyla and their specific class and family names are in parentheses. Numbers of patients in each comparison group is written in parentheses. R = Responder; NR= Nonresponder; Tx=Treated; BL = Baseline; UTx = Untreated; HC = Healthy Control; SE = side effects; NSE = no side effects; NS = not significant; NR = not reported; Δ = Changed; ↑ = Increased; ↓ = Decreased.
| Drug Name | Disease | Immunomodulatory Type |
Comparison (n) |
Bacterial Phyla (Family, Genus) | Diversity |
|---|---|---|---|---|---|
| Methotrexate | RA | Anti-Proliferative | Tx (11) vs. UTx (11) [85] | ↓ Proteobacteria (Enterobacteriales) | NS |
| Tx (9) vs. BL (9) [86] | ↑ Firmicutes (Erysipelotrichaceae, Holdemania) microbial linkage groups | NS | |||
| Tx (24) vs. HC (32)[84] † | NR | ↑ Alpha | |||
| Anti-PD1 | Cancer | Immunoenhancing | Tx (42) vs. BL (36) [102] | ↑ Firmicutes (Clostridium; Streptococcus; Eubacterium) ↑ Bacteroidetes (Alistipes) ↓ Firmicutes (Roseburia; Oscillibacter; Lachnoclostridium) ↓ Bacteroidetes (Alistipes; Coprobacter) |
↑ Richness |
| R (6) vs. NR (11) [103] | ↑ Firmicutes (Lactobacillaceae, Lactobacillus; Clostridiaceae, Clostridium; Lachnospiraceae, Syntrophococcus) ↓ Bacteroidetes (Parabacteroides) ↓ Proteobacteria (Bilophila; Sutterella) |
NS | |||
| Anti-CTLA-4 | Cancer | Immunoenhancing | Tx (26) vs BL (26)[104] | Shifts in bacterial proportions were not linked to treatment | NS |
| Colitis (7) vs. BL (7) [104] | ↓ Firmicutes (Ruminococcus; Lachnospiraceae, Blautia; Clostridium IV; Eubacterium; Pseudoflavonifractor) | ↓Alpha | |||
| Tx (25) vs. BL (25)[101] | Bidirectional shifts in Bacteroides | NR | |||
| Glatiramer Acetate | MS | Cellular | Tx (60) vs. UTx (75) [111] | ↓ Firmicutes (Lachnospiraceae, Roseburia; Veillonellaceae) ↓ Proteobacteria (Sutterella, Aggregatibacter, Haemophilus) ↑ Firmicutes (Enterococcus, Acidaminococcus) ↑ Proteobacteria (Enterobacter, Pseudomonas, Sphingoblum, Burkholderiales) |
NS |
| Dimethyl Fumarate | MS | Cellular | Tx (33) vs. UTx (75) [111] | ↓ Firmicutes (Anaerococcus, Finegoldia, Peptoniphilius; Lachnospiraceae, Blautia; Veillonellaceae, Megasphaera) ↑ Bacteroidetes ↓ Fusobacterilia (Fusobacterium) ↓ Proteobacteria (Campylobacter) ↓ Actinobacteria (Varibaculum, Corynebacterium, Rothia) |
NS |
| Tx (23) vs. BL (25) [116] | ↑ Firmicutes (Faecalibacterium) ↓ Bacteroidetes ↓ Actinobacteria (Bifidobacterium) * |
NS | |||
| Interferon-β | MS | Cellular | Tx (15) vs. UTx (15) [129] | ↑ Bacteroidetes (Prevotellaceae, Prevotella) | NR |
| Tx (24) vs. UTx (20-24) [128] | Δ Bacteroidetes enterotype distribution | ↓ Richness | |||
| Combined INF-β/GA | Tx (32) vs. UTx (28) [112] | ↑ Bacteroidetes (Prevotellaceae, Prevotella) ↑ Proteobacteria (Sutterellaceae, Sutterella) Firmicutes (Clostridiaceae, Sarcina) |
NS | ||
| TNF inhibitors | IBD | Anti-Cytokine | Tx (20) vs. BL (20) R (13) vs. NR (7) [133] † |
↓ Proteobacteria | NR |
| R (9) vs. NR (7) [134] | ↑ Firmicutes (Lachnospiraceae, Anaerostipes; Veillonellaceae, Veillonella; Acidaminococcaceae, Acidominococcus) | ↑ Alpha | |||
| Tx (12) vs BL (12) R (9) vs. NR (3) [136] † |
↑ Firmicutes (Lachnospiraceae, Coprococcus, Roseburia) NS |
↑ Alpha, beta NS |
|||
| RA | Tx (17) vs BL (17) R (11) vs. NR (6) [136] † |
↑ Firmicutes (Erysiopelotriahaceae; Lachnospiraceae, Dorea) NS |
↓ Beta NS |
||
| Tx (10) vs. UTx (11) [85] | ↓ Proteobacteria (Class: Deltaproteobacteria) ↓ Firmicutes (Clostridiaceae) ↑ Cyanobacteria (Class: Nostocophycideae, Order: Nostocales) |
NS |
A subset of patients was on other immunomodulators including azathioprine, leflunomide.
Changes were transient and reversed with longer times on drug
Some MTX-associated bacterial shifts decrease the gut microbiota’s capacity for drug detoxification, leading to gastrointestinal toxicity [121]. The complex interplay between drug and bacteria may explain the inter-individual heterogeneity of patient responses and outcomes, but the multi-directional relationships make this difficult to study. Overall, MTX treatment appeared to partially restore the microbial composition of RA patients to resemble that of healthy controls. Immunomodulatory therapies thus have widespread effects on the microbiota, impacting many environmental niches.
Targeted Immunoablative Therapies
Rituximab/Ocrelizumab
Rituximab and ocrelizumab are monoclonal antibodies that target CD20, a surface antigen expressed on most of the B cell lineage [127]. They are effective against hematologic malignancies and a variety of autoimmune conditions including multiple sclerosis, vasculitis, myasthenia gravis and some types of autoimmune encephalitis. B cells are a major contributor to the GALT and are necessary for ongoing IgA production/secretion; the tight relationship between the mucosal immune system and the gut microbiota might suggest that these medications would shift the composition of the microbiome. However, parenteral anti-CD20 monoclonals may incompletely deplete tissue-resident lymphoid cells [128, 129].
No studies have yet examined whether anti-CD20 medications directly impact the gut microbiota in humans. Germ-free mice mono-colonized with B. fragilis and treated with anti-CD20 had decreased IgA coating of intestinal bacteria and could be readily invaded by wild-type bacteria, losing their single-strain stability [130]. This study reveals that IgA coating is not only important for pathogen clearance, but also is required to maintain stable colonization by commensal bacteria such as B. fragilis. Additional research is needed to identify how these medications impact other members of the gut microbiome.
Alemtuzumab
Alemtuzumab (ALZ) targets the surface molecule CD52, which is expressed on B cells, T cells, and a variety of innate immune cells. It is used for hematologic malignancies, organ transplantation and multiple sclerosis [131]. Antibody binding to CD52 induces lysis of circulating cells; the affected cellular populations then recover gradually over time. Monocytes recover first, after about one month. B cells begin to repopulate around three months, while CD8/CD4 T cells subsets can take a year or longer to repopulate [132].
A mouse study has suggested that ALZ increases intestinal permeability by decreasing intestinal intraepithelial lymphocytes [133]. Similarly, a study using Cynomolgus monkeys observed transient changes in gut microbial composition. These shifts appeared at 1 day post ALZ-treatment, normalized by 9 days and were maintained until day 56 [134]. In the ileal mucosa, there was an enrichment in Enterobacteriales (E. coli, S. flexneri) and Prevotella (P. copri, P. dentalis) directly following treatment and on day 6, respectively. The colonic mucosa showed similar increases in Enterobacteriales. In the fecal microbiota, members of the Clostridiales order increased between days 1-9, except for Faecalibacterium prausnitzii which tended to decrease after lymphocyte depletion. ALZ may affect intestinal permeability via depleting intraepithelial lymphocytes, precipitating concurrent effects on microbial composition.
Immunoenhancing Therapies
Immune Checkpoint Inhibitors: Anti-PD1 and Anti-CTLA-4
Immune checkpoint proteins are cell-surface markers that prevent immune overactivation. Checkpoints such as cytotoxic T-lymphocyte-associated protein 4 (CTLA-4) and programmed cell death protein 1 (PD-1) are activated by interaction with specific ligands. Following activation, these checkpoints inhibit T cell proliferation and effector function [135]. Tumors that express the corresponding ligands (e.g. PD-L1, PD-L2) are able to evade immune surveillance by activating these checkpoints and avoiding anti-tumor immunity.
Immune checkpoint inhibitors (ICIs) such as anti-PD1 and anti-CTLA-4 agents are monoclonal antibodies that block these checkpoints and restore anti-tumor immunity. Unlike the immunoablative therapies, ICIs upregulate the adaptive immune system [135, 136]. By unleashing the full power of the adaptive immune response against tumor cells, ICIs have revolutionized cancer therapy, dramatically improving outcomes in melanoma and other malignancies [135, 136].
While recent studies have been investigating the impact of the baseline gut microbiome on immunotherapy efficacy [137-140], there is a paucity of research exploring how the microbial composition is impacted by treatment. Anti-CTLA4 antibodies led to an increase in Clostridiales, and a rapid decline in Bacteroidales and Burkholderiales. The therapeutic effect of the anti-CTLA4 antibody was dependent upon bacterial colonization of the gut mucosa [141]. When multiple melanoma patients were clustered into distinct enterotypes (A, B, C), anti-CTLA4 therapy tended to increase the proportion of patients in cluster C and decrease those in cluster B (clusters were characterized by distinct Bacteroides species). In a human study, ICI treatment for renal cell carcinoma resulted in an increase in stool richness after 2 months [142].
Perhaps more importantly, gut microbial composition appears to play an important role in ICI efficacy [142-144]. In one retrospective study in patients with advanced non-small lung cancer, ICI responders (those with longer times to treatment failure) were significantly enriched with Lactobacillus, Clostridium, and Syntrophococcus, although the time at which stool samples were obtained post-ICI treatment was not specified [143]. In another study, ICI responders were characterized by increased baseline abundance of Firmicutes and Bacteroidetes (Alistipes) [142]. In both cancer patient cohorts, responders had overrepresentation of Akkermansia muciniphila. In order to better examine a cause-effect relationship between anti-tumor effects of ICIs and specific microbiota, researchers transplanted fecal microbiota from human anti-PD-1 responders and nonresponders to germ-free/antibiotic-treated mice. Only mice colonized from anti-PD-1 responders were subsequently sensitive to PD-1 blockade. Oral gavage with A. muciniphila was also able to rescue the efficacy of the cancer immunotherapy in mice [142].
Gut microbial composition also influences ICI-related side effects. Despite being associated with therapeutic efficacy, baseline abundance of Firmicutes has been associated with ICI-induced colitis and other GI-related side effects [140, 144]. In contrast, higher levels of Bacteroidetes have been observed in individuals who experience no side effects [140, 144]. The ratio of Bacteroidetes/Firmicutes may predict not only the clinical response to immunotherapy, but also whether a patient experiences side effects.
Lymphocyte Trafficking Inhibitors
Natalizumab
Natalizumab (NTZ), a monoclonal antibody against alpha4beta1 integrin, blocks lymphocyte adhesion and extravasation [145, 146]. It is a highly effective therapy for MS and Crohn’s disease. The single published study of NTZ-associated microbial effects demonstrated that NTZ reduced lipopolysaccharide binding protein and lipopolysaccharide (LPS) levels in the blood, brain and spinal cord of rats with experimental autoimmune encephalitis, an animal model of MS [147]. High levels of LPS lead to mucosal barrier dysfunction and a proinflammatory state [148]. Since LPS is a major enterotoxin secreted by gram negative bacteria, especially those belonging to Proteobacteria phyla, this suggests that NTZ may alter Proteobacteria composition. Further work specifically examining bacterial taxonomy and metabolic function in response to NTZ, is needed.
Fingolimod
Fingolimod is a modulator of sphingosine-1-phosphate receptors 1, 3, 4 and 5; it sequesters lymphocytes in secondary lymphoid tissue, thus preventing CNS infiltration [149]. It is used for the treatment of multiple sclerosis. Very little is known about fingolimod’s effect on gut microbiota. A small Russian study observed that fingolimod-treated patients tended to have an increased Bacteroidetes/Firmicutes ratio [150], which is consistent with findings seen for other MS immunomodulators [151, 152]. They also suggested shifts in E. coli strains in response to fingolimod (Table 1); however these results remain to be replicated.
Immunomodulatory Therapies
Dimethyl Fumarate
Dimethyl fumarate (DMF) and its active metabolite monomethyl fumarate are small molecules used for the treatment of relapsing MS. DMF activates the Nrf2 antioxidant stress pathways, reducing axonal degradation and tissue damage [153]. It also shifts the immune system from a Th1/Th17 state to a Th2 state [153, 154]. This is accompanied by selective depletion of lymphocytes, predominantly CD8+ and CD4+ T cells [153, 155].
DMF affects both gut microbial composition and intestinal barrier integrity [147, 151, 156-158]. The specific bacterial species affected by DMF vary across studies (Table 1). One study found that Bifidobacterium decreased and Faecalibacterium increased after two and twelve weeks of DMF treatment, respectively [156]. In contrast, Sand and colleagues reported that DMF reduced Firmicutes (e.g. Lachnospiraceae, Veillonellaceae, Clostridiales) and Fusobacteria with a concurrent increase in Bacteroidetes [151]. Metabolic changes, including changes in retinols, amino acids, methane metabolism and ethylbenzene degradation, were also associated with DMF. In mice, DMF shifted the relative abundance of both Bacteroidetes and Firmicutes phyla depending on the region of the intestines studied [158]. DMF treatment also increased villi height in the jejunum and the ileum, suggesting improved absorption efficiency. These studies suggest that there are multidirectional interactions between DMF and the microbiota, but the influence of microbial composition on drug efficacy is unknown. While there are global microbial shifts occurring between treated and untreated individuals, there may be more subtle shifts that could be tied to responders or non-responders within a treatment group. Existing studies were small and lacked the power to address these patient outcomes. Future longitudinal studies may help dissect the relationship between microbial composition and treatment response.
Glatiramer Acetate
Glatiramer acetate (GA) is a synthetic myelin analogue used for the treatment of MS. Like DMF, GA promotes differentiation of anti-inflammatory Th2 cells and reduces the activity of autoreactive T cells [159, 160]. It also induces differentiation and proliferation of Tregs [161, 162] and CD8 T cells [163, 164]. Several human studies have investigated the influence of GA on the gut microbiota [150-152, 165] (Table 1). Katz-Sand and colleagues observed that GA treatment was associated with enrichment in seven Firmicutes/Proteobacteria genera and concurrent reductions in seven other Bacteriodetes/Proteobacteria/Firmicutes genera [151]. Others corroborated shifts in Firmicutes (Clostridia, Faecalibacterium, Clostridium, Ruminococcus; Lactobacillaceae) and Bacteriodetes (Bacteriodaceae) among GA-treated patients [165]. A small Russian study noted that patients treated with GA were more likely to report constipation, which they attributed to higher levels of atypical forms of Proteobacteria (e.g. Proteus species) [150]. However, all these studies were small and lacked the power to identify significant effects of GA on the microbiome.
Interferon Beta
Interferon beta (IFNβ), a type 1 interferon, treats MS through multiple mechanisms of action including reducing T cell activation, promoting Treg differentiation, inducing cytokine shifts, altering matrix metalloprotease expression, strengthening the blood-brain-barrier, and regulating B cell activity [166, 167]. Treatment was associated with increased Bacteroidetes among RRMS patients [168, 169] (Table 1). A small Spanish study found a trend for reduced Prevotella (P. copri) in MS patients that reversed with IFNβ treatment [169]. In order to further discern the effects of IFN, Reynders and colleagues sub-classified multiple sclerosis patients based on their disease phenotype. Microbial richness appeared to be lower in IFNβ-treated relapsing-remitting MS patients compared to benign and primary progressive MS patients [168]. Specific microbial composition changes also differed based on the phenotype subclassification with the Bacteriodetes 2 enterotype more prevalent in IFN β-treated relapsing-remitting MS patients compared to other clinical subgroups. Butyricicoccus, a genus in the Clostridia order, was observed to be inversely correlated with patient-reported symptoms. This study highlights the importance of studying microbial changes on a disease-subtype level in order to discern effects that IFN may be having on the gut microbiome and its impact in patient outcomes.
Anti-Cytokine Immunotherapies
TNF inhibitors
TNF inhibitors (TNFi) revolutionized the treatment of RA and other systemic autoimmune disorders including ankylosing spondylitis, Crohn’s disease, ulcerative colitis, and psoriasis [170]. This class of molecules antagonizes tumor necrosis factor alpha (TNFα), a proinflammatory cytokine secreted by activated macrophages and T cells that contributes to the pathophysiology of multiple systemic autoimmune diseases. TNFi include neutralizing monoclonal antibodies, fusion proteins, and pegylated fragments. In general, TNFi bind and inhibit both soluble and transmembrane TNFα [171]. These have divergent functions. Soluble TNFα is implicated in inflammatory diseases and expressed by activated cytotoxic T cells. In contrast, transmembrane TNFα is expressed on many adaptive immune cells. The lack of receptor specificity may contribute to the lack of TNFi efficacy in certain diseases. Indeed, TNF inhibition worsens central demyelinating diseases like MS [172]. One study reported that Cyanobacteria increased, while Deltaproteobacteria and Clostridiaceae decreased after TNFi treatment in RA patients [125]. Decreased Proteobacteria and increased Clostridiales were associated with successful TNFi treatment in inflammatory bowel disease [173, 174] (Table 1).
Relatively little, however, is known about TNFi and microbiota interactions with respect to drug efficacy. Blocking TNF alpha systemically has been linked to increase in fungal infections [175] and may possibly increase susceptibility to other types of pathobionts. A study found that TNFi may shift the diversity of the fecal microbiome in patients with inflammatory bowel disease (IBD) to resemble that of healthy controls [176]. The researchers also showed that the levels of butyrate and substrates involved in butyrate synthesis were diminished among IBD patients who were non-responders to TNFis but were enriched in IBD patients in clinical remission. This shows the advantage of using functional assays like metabolomics in addition to taxonomic characterization of the microbiome to parse out differences between responders and non-responders.
Discussion
Host-Drug-Microbiota Interactions
Immunotherapies directly affect the host immune system, but indirect drug effects on the gut microbiota may also contribute to their therapeutic efficacy. Moreover, the composition of the gut microbiome may impact whether individuals respond to immunotherapy. Most immunomodulators were not associated with measurable changes in alpha or beta diversity. However, shifts in the relative abundance of bacterial taxa were observed in response to most drugs studied.
The overall shifts in commensal microbiota differed depending on the immunomodulator. Medications used to treat autoimmune diseases have been associated with overall reductions of Firmicutes (e.g. Clostridium) [125, 151, 152] and increases in Bacteroidetes (e.g. Bacteroides) [151, 152, 169], leading some to hypothesize that the Firmicutes:Bacteroides ratio was fundamentally altered in autoimmune diseases. However, with time, it has become clear that both Firmicutes and Bacteroides play divergent roles and generalizing at the phylum level will likely not be scientifically accurate. Multiple studies reported that immunotherapy led to “normalization” of the gut microbiome; in other words, treated patients’ microbiota assumed a taxonomic distribution more similar to that of healthy controls than to untreated, diseased individuals [124-126, 151, 152, 156, 165, 169, 173, 176]. In several studies, Prevotella species increased among patients taking interferon beta [152, 169]. Counterintuitively, these organisms have sometimes been associated with inflammation and autoimmune pathology [82]. Changes in Prevotella were not observed with the other immunomodulatory medications studied to date (Table 1).
Perhaps unsurprisingly, ICIs and immune suppressing medications had distinct effects on gut microbiota. Although existing data have not demonstrated consistent relationships between ICI administration and downstream changes in gut microbial ecology, the baseline composition of the microbiome substantially impacted the therapeutic efficacy and side effect profile of ICIs. High baseline levels of Firmicutes were associated with greater therapeutic efficacy and increased progression-free survival [142, 143], while the relationship between Bacteroidetes and anti-PD-1 treatment response was more heterogeneous [142-144]. For instance, high levels of baseline Alistipes and Akkermansia were observed in responders to PD-1 targeted therapies, while Parabacteroides were abundant in non-responders [142]. Further elucidation of the mechanisms underlying the relationship between baseline gut ecology and treatment efficacy could ultimately lead to treatments designed to optimize the gut microbiota in advance of immunotherapy. This may further improve treatment efficacy.
Prophylactic antibiotics are common among cancer patients, which further complicates interpretation of microbiota-ICI interactions. Antibiotic administration before, during, or after the initiation of immunotherapy has generally been associated with negative outcomes in cancer patients [177-180]. A recent meta-analysis concluded that ICI-treated non-small cell lung cancer patients on antibiotics had shorter progression-free survival and overall survival compared to those with no antibiotics [178]. A negative association was also observed between antibiotics and survival in urothelial carcinoma patients treated with atezolizumab, an anti PDL-1 immunotherapy, but not for patients taking traditional chemotherapy [179]. This suggests that gut microbes are necessary for maximal ICI efficacy. However, these observations could be confounded by patient characteristics. Those needing antibiotics are often weaker, more immunodeficient and have previous infections/comorbidities. The heterogeneity of the cancer patient cohorts, the variable time-window when antibiotics were taken, and the retrospective nature of these studies also confound interpretation of these results.
Comprehending the functional significance of immunomodulatory drug-induced changes in microbial populations will require better understanding of host-commensal interactions, specifically the immune interactions. There are a plethora of immunologic mechanisms that may explain the positive or detrimental functional relevance of drug-induced microbiome shifts. For instance, specific bacteria and metabolites have been demonstrated to alter barrier permeability both in the gut (alternatively referred to as ‘leaky gut) [181] and non-gut barriers, such as the blood-brain barrier [182]. In mice, the gut and the blood-brain barrier have been altered in the presence of microbiome-derived metabolites, notably, tryptophan-derived aryl hydrocarbon receptor reactive metabolites [183]. Short chain fatty acids are bacterial metabolites that promote intestinal epithelial barrier integrity [184-186], B cell IgA production [187], regulatory T cell differentiation [188], and anti-inflammatory IL-10 production [189]. Many of the bacteria apparently impacted by immunotherapies are short-chain fatty acid producers, including multiple Clostridial species [190] Lactobacillus and Bifidobacterium. In addition to generating immunoactive metabolites, commensals themselves can translocate across mucosal barrier surfaces and directly promote inflammation and T cell polarization [191-194]. Another mechanism by which commensals could mediate differential anti-tumor and autoimmune responses is cross-reactivity, or molecular mimicry. Commensal antigens, by chance or homology, have linear and conformational epitopes with enough similarity to host antigens that commensal-specific T and B cell responses are able to recognize and react to host tissue, thereby promoting autoimmunity and anti-tumor immunity [195-202]. Taken together it is plausible that microbial alterations due to immunomodulators may have unintended side-effects on host-immune physiology.
The rapid evolution of sequencing technologies and analysis methods used for microbiome research has led to methodological challenges in data interpretation (Figure 2). Most researchers utilize 16S rRNA sequencing, but the hypervariable regions used (e.g. V1-3, V3-V4, V3-V5, V4) and the selected primers vary between studies with different amplicons preferentially identifying different bacterial genera [203, 204]. Moreover, alternate sequencing methodologies, such as shotgun metagenomics and long-amplicon 16S sequencing, are emerging. This methodological variance makes it extremely challenging to compare results between studies. Another pitfall of human microbiome research is that subjects are often incompletely characterized; variables like diet, medical comorbidities and medication usage substantially impact the microbiome yet are rarely captured by the study design [70, 73, 205]. Cross-sectional studies therefore become problematic, as it is difficult to assure comparable controls. Statistical tools to calculate power for microbiome experiments and analyze the data are also still evolving. The human studies reviewed were all relatively small. Given the heterogeneity of the substrate, it is likely that substantially more patients will be needed to detect differences between groups. Indeed, a similar phenomenon was observed during the advent of genetic research for immune diseases. For years, studies found only minimal genetic substrate for diseases such as MS and RA [206, 207]. It was only when multicenter, multinational consortia standardized data collection techniques and pooled thousands of cases that a clear picture emerged [208, 209], and now well over 230 genes have been linked to MS risk [210]. As with all biomarkers, best practice would mandate that microbiota of interest should be first identified in a discovery cohort and subsequently verified using a separate validation cohort. However, to date human microbiome research has not achieved this level of rigor. Consensus regarding best practices for microbiome experimental design and collaboration across centers will be needed to fully elaborate the role of the microbiome in human disease. Such large-scale studies will additionally allow meta-analyses to define reproducible microbiota-host interactions [211].
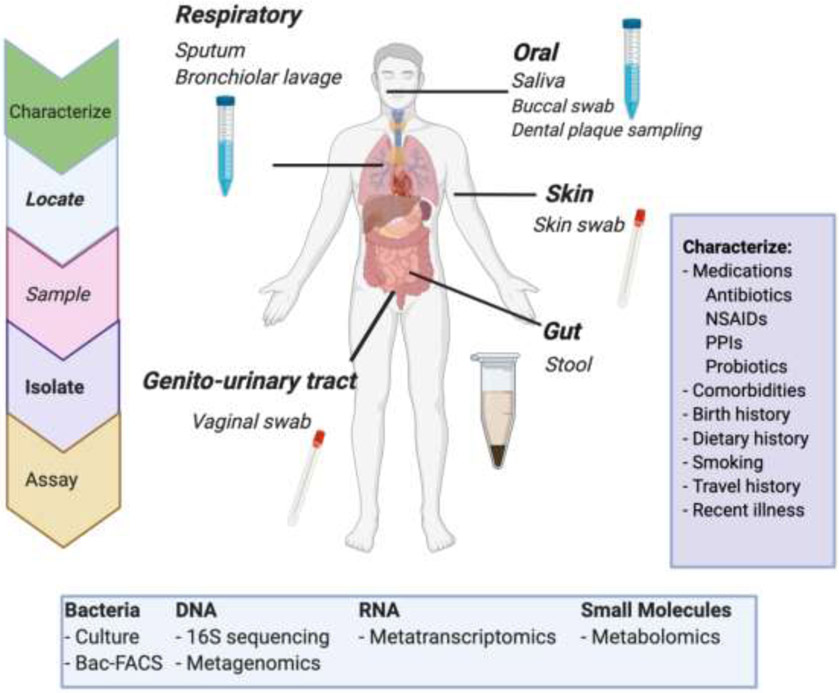
Figure 2. Microbiome Study Design.
Human microbiome studies require identifying the anatomic region of interest, selecting a sampling methodology, and isolating the specimen. Analytic assays will be targeted to the research question and may include microbial characterization at the cellular, DNA, RNA, or metabolic level. Human microbiome studies require subject-level characterization to adequately control for environmental variables known to impact the microbiome. NSAIDs: non-steroidal anti-inflammatory drugs. PPIs: proton pump inhibitors. Figure created with BioRender.com
We hypothesized that immunotherapies with shared mechanisms of action might elicit similar changes in the gut microbiota. This was not substantiated by the existing data. However, the literature in this field is striking for its heterogeneity. Many immunomodulators have never been studied in the context of the gut microbiome, or they have been examined in only one or two small studies. Given the expected level of microbial heterogeneity between individuals, the variability imposed by nonstandard methodology, and disease-specific microbial shifts , we expect that as the field matures, more clarity will emerge about the bidirectional relationships between immunotherapies and the gut microbiota. We anticipate that pharmacomicrobiomics is an important component of immunomodulator efficacy. Although the field is in its infancy, further elucidating drug/microbial interactions and how these impact the host immune response will afford opportunities to personalize treatment and achieve better treatment efficacy for autoimmune diseases and malignancies.
Acknowledgements:
This study was supported by NIH K23NS107624 and Race to Erase MS.
Abbreviations:
- CD
cluster of differentiation
- Th
helper T cells
- Treg
regulatory T-cell subsets
- GALT
gut-associated lymphoid tissue
- TLR2
Toll-like receptor-2
- MMF
Mycophenolate mofetil
- MTX
Methotrexate
- RA
Rheumatoid Arthritis
- MS
Multiple Sclerosis
- MLGs
microbial linkage groups
- ALZ
Alemtuzumab
- CTLA-4
cytotoxic T-lymphocyte-associated protein 4
- PD-1
programmed cell death protein 1
- PD-L1
programmed cell death protein 1 ligand 1
- PD-L2
programmed cell death protein 1 ligand 2
- ICIs
Immune checkpoint inhibitors
- NTZ
Natalizumab
- LPS
lipopolysaccharide
- DMF
Dimethyl fumarate
- GA
Glatiramer acetate
- IFNβ
Interferon beta
- TNFi’s
TNF inhibitors
- TNFα
tumor necrosis factor alpha
- IgA
immunoglobulin A
- IL
interleukin
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures:
EEL received honoraria from Genentech, Genzyme, Alexion, Biogen, Celegene/Bristol Myers Squibb & EMD Serono.
WER and IC have no disclosures to report
All authors have read the journal’s authorship agreement, and the manuscript has been reviewed by and approved by all authors. All authors have disclosed their conflicts of interest, above, and have read the journal’s policy on conflicts of interest.
References
- [1].Cho I, Blaser MJ. The human microbiome: at the interface of health and disease. Nat Rev Genet. 2012;13:260–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Davis CP. Normal flora. Medical Microbiology 4th edition: University of Texas Medical Branch at Galveston; 1996. [PubMed] [Google Scholar]
- [3].Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation. Cell. 2014;157:121–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Ruff WE, Greiling TM, Kriegel MA. Host–microbiota interactions in immune-mediated diseases. Nature Reviews Microbiology. 2020. [DOI] [PubMed] [Google Scholar]
- [5].Alexander JL, Wilson ID, Teare J, Marchesi JR, Nicholson JK, Kinross JM. Gut microbiota modulation of chemotherapy efficacy and toxicity. Nat Rev Gastroenterol Hepatol. 2017;14:356–65. [DOI] [PubMed] [Google Scholar]
- [6].Zimmermann M, Zimmermann-Kogadeeva M, Wegmann R, Goodman AL. Mapping human microbiome drug metabolism by gut bacteria and their genes. Nature. 2019;570:462–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Zimmermann M, Zimmermann-Kogadeeva M, Wegmann R, Goodman AL. Separating host and microbiome contributions to drug pharmacokinetics and toxicity. Science. 2019;363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Charles A, Janeway J, Medzhitov R. Innate Immune Recognition. Annual Review of Immunology. 2002;20:197–216. [DOI] [PubMed] [Google Scholar]
- [9].Akira S, Takeda K. Toll-like receptor signalling. Nature reviews immunology. 2004;4:499–511. [DOI] [PubMed] [Google Scholar]
- [10].Proell M, Riedl SJ, Fritz JH, Rojas AM, Schwarzenbacher R. The Nod-like receptor (NLR) family: a tale of similarities and differences. PloS one. 2008;3:e2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Vivier E, Tomasello E, Baratin M, Walzer T, Ugolini S. Functions of natural killer cells. Nature immunology. 2008;9:503–10. [DOI] [PubMed] [Google Scholar]
- [12].Davies LC, Jenkins SJ, Allen JE, Taylor PR. Tissue-resident macrophages. Nat Immunol. 2013;14:986–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Kettenmann H, Hanisch U-K, Noda M, Verkhratsky A. Physiology of microglia. Physiological reviews. 2011;91:461–553. [DOI] [PubMed] [Google Scholar]
- [14].Bilzer M, Roggel F, Gerbes AL. Role of Kupffer cells in host defense and liver disease. Liver International. 2006;26:1175–86. [DOI] [PubMed] [Google Scholar]
- [15].Joshi N, Walter JM, Misharin AV. Alveolar macrophages. Cellular immunology. 2018;330:86–90. [DOI] [PubMed] [Google Scholar]
- [16].Spits H, Artis D, Colonna M, Diefenbach A, Di Santo JP, Eberl G, et al. Innate lymphoid cells—a proposal for uniform nomenclature. Nature reviews immunology. 2013;13:145–9. [DOI] [PubMed] [Google Scholar]
- [17].Merad M, Ginhoux F, Collin M. Origin, homeostasis and function of Langerhans cells and other langerin-expressing dendritic cells. Nature Reviews Immunology. 2008;8:935–47. [DOI] [PubMed] [Google Scholar]
- [18].Cheng HY, Ning MX, Chen DK, Ma WT. Interactions Between the Gut Microbiota and the Host Innate Immune Response Against Pathogens. Front Immunol. 2019;10:607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Santaolalla R, Fukata M, Abreu MT. Innate immunity in the small intestine. Curr Opin Gastroenterol. 2011;27:125–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Zhu J T Helper Cell Differentiation, Heterogeneity, and Plasticity. Cold Spring Harb Perspect Biol. 2018;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Murphy KM, Reiner SL. The lineage decisions of helper T cells. Nature Reviews Immunology. 2002;2:933–44. [DOI] [PubMed] [Google Scholar]
- [22].Szabo SJ, Kim ST, Costa GL, Zhang X, Fathman CG, Glimcher LH. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell. 2000;100:655–69. [DOI] [PubMed] [Google Scholar]
- [23].Zheng W, Flavell RA. The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell. 1997;89:587–96. [DOI] [PubMed] [Google Scholar]
- [24].Ivanov II, McKenzie BS, Zhou L, Tadokoro CE, Lepelley A, Lafaille JJ, et al. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 2006;126:1121–33. [DOI] [PubMed] [Google Scholar]
- [25].Fontenot JD, Gavin MA, Rudensky AY. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nature Immunology. 2003;4:330–6. [DOI] [PubMed] [Google Scholar]
- [26].Windhagen A, Anderson DE, Carrizosa A, Williams RE, Hafler DA. IL-12 induces human T cells secreting IL-10 with IFN-gamma. J Immunol. 1996;157:1127–31. [PubMed] [Google Scholar]
- [27].Hsieh CS, Macatonia SE, Tripp CS, Wolf SF, O'Garra A, Murphy KM. Development of TH1 CD4+ T cells through IL-12 produced by Listeria-induced macrophages. Science. 1993;260:547–9. [DOI] [PubMed] [Google Scholar]
- [28].Amsen D, Blander JM, Lee GR, Tanigaki K, Honjo T, Flavell RA. Instruction of distinct CD4 T helper cell fates by different notch ligands on antigen-presenting cells. Cell. 2004;117:515–26. [DOI] [PubMed] [Google Scholar]
- [29].Chen F, Liu Z, Wu W, Rozo C, Bowdridge S, Millman A, et al. An essential role for TH2-type responses in limiting acute tissue damage during experimental helminth infection. Nat Med. 2012;18:260–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Gause WC, Wynn TA, Allen JE. Type 2 immunity and wound healing: evolutionary refinement of adaptive immunity by helminths. Nat Rev Immunol. 2013;13:607–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Bettelli E, Korn T, Oukka M, Kuchroo VK. Induction and effector functions of T(H)17 cells. Nature. 2008;453:1051–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Lee Y, Awasthi A, Yosef N, Quintana FJ, Xiao S, Peters A, et al. Induction and molecular signature of pathogenic TH17 cells. Nat Immunol. 2012;13:991–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Dominguez-Villar M, Hafler DA. Regulatory T cells in autoimmune disease. Nat Immunol. 2018;19:665–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Lucca LE, Dominguez-Villar M. Modulation of regulatory T cell function and stability by co-inhibitory receptors. Nat Rev Immunol. 2020. [DOI] [PubMed] [Google Scholar]
- [35].Wong P, Pamer EG. CD8 T Cell Responses to Infectious Pathogens. Annual Review of Immunology. 2003;21:29–70. [DOI] [PubMed] [Google Scholar]
- [36].Saeidi A, Zandi K, Cheok YY, Saeidi H, Wong WF, Lee CYQ, et al. T-Cell Exhaustion in Chronic Infections: Reversing the State of Exhaustion and Reinvigorating Optimal Protective Immune Responses. Frontiers in Immunology. 2018;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Mueller SN, Gebhardt T, Carbone FR, Heath WR. Memory T cell subsets, migration patterns, and tissue residence. Annu Rev Immunol. 2013;31:137–61. [DOI] [PubMed] [Google Scholar]
- [38].Honda K, Littman DR. The microbiota in adaptive immune homeostasis and disease. Nature. 2016;535:75–84. [DOI] [PubMed] [Google Scholar]
- [39].Brown EM, Kenny DJ, Xavier RJ. Gut Microbiota Regulation of T Cells During Inflammation and Autoimmunity. Annu Rev Immunol. 2019;37:599–624. [DOI] [PubMed] [Google Scholar]
- [40].Dominguez-Villar M, Baecher-Allan CM, Hafler DA. Identification of T helper type 1-like, Foxp3+ regulatory T cells in human autoimmune disease. Nat Med. 2011;17:673–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Noack M, Miossec P. Th17 and regulatory T cell balance in autoimmune and inflammatory diseases. Autoimmun Rev. 2014;13:668–77. [DOI] [PubMed] [Google Scholar]
- [42].McKinney EF, Lee JC, Jayne DR, Lyons PA, Smith KG. T-cell exhaustion, co-stimulation and clinical outcome in autoimmunity and infection. Nature. 2015;523:612–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].LeBien TW, Tedder TF. B lymphocytes: how they develop and function. Blood. 2008;112:1570–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Palm NW, de Zoete MR, Cullen TW, Barry NA, Stefanowski J, Hao L, et al. Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell. 2014;158:1000–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Cebra JJ, Periwal SB, Lee G, Lee F, Shroff KE. Development and maintenance of the gut-associated lymphoid tissue (GALT): the roles of enteric bacteria and viruses. Dev Immunol. 1998;6:13–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Holmgren J, Czerkinsky C. Mucosal immunity and vaccines. Nature Medicine. 2005;11:S45–S53. [DOI] [PubMed] [Google Scholar]
- [47].Cesta MF. Normal structure, function, and histology of mucosa-associated lymphoid tissue. Toxicol Pathol. 2006;34:599–608. [DOI] [PubMed] [Google Scholar]
- [48].Randall TD, Mebius RE. The development and function of mucosal lymphoid tissues: a balancing act with micro-organisms. Mucosal Immunol. 2014;7:455–66. [DOI] [PubMed] [Google Scholar]
- [49].Ahluwalia B, Magnusson MK, Öhman L. Mucosal immune system of the gastrointestinal tract: maintaining balance between the good and the bad. Scandinavian Journal of Gastroenterology. 2017;52:1185–93. [DOI] [PubMed] [Google Scholar]
- [50].Neish AS. Mucosal immunity and the microbiome. Ann Am Thorac Soc. 2014;11 Suppl 1:S28–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Woof JM, Kerr MA. The function of immunoglobulin A in immunity. The Journal of Pathology. 2006;208:270–82. [DOI] [PubMed] [Google Scholar]
- [52].Peterson DA, McNulty NP, Guruge JL, Gordon JI. IgA response to symbiotic bacteria as a mediator of gut homeostasis. Cell Host Microbe. 2007;2:328–39. [DOI] [PubMed] [Google Scholar]
- [53].Lycke NY, Bemark M. The regulation of gut mucosal IgA B-cell responses: recent developments. Mucosal Immunology. 2017;10:1361–74. [DOI] [PubMed] [Google Scholar]
- [54].Maynard CL, Weaver CT. Diversity in the contribution of interleukin-10 to T-cell-mediated immune regulation. Immunol Rev. 2008;226:219–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Sansonetti PJ, Medzhitov R. Learning tolerance while fighting ignorance. Cell. 2009;138:416–20. [DOI] [PubMed] [Google Scholar]
- [56].Smith K, McCoy KD, Macpherson AJ. Use of axenic animals in studying the adaptation of mammals to their commensal intestinal microbiota. Seminars in Immunology. 2007;19:59–69. [DOI] [PubMed] [Google Scholar]
- [57].Glaister JR. Factors Affecting the Lymphoid Cells in the Small Intestinal Epithelium of the Mouse. International Archives of Allergy and Immunology. 1973;45:719–30. [DOI] [PubMed] [Google Scholar]
- [58].Pollard M, Sharon N. Responses of the Peyer's Patches in Germ-Free Mice to Antigenic Stimulation. Infection and Immunity. 1970;2:96–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Bauer H, Horowitz RE, Levenson SM, Popper H. The response of the lymphatic tissue to the microbial flora. Studies on germfree mice. Am J Pathol. 1963;42:471–83. [PMC free article] [PubMed] [Google Scholar]
- [60].Hamada H, Hiroi T, Nishiyama Y, Takahashi H, Masunaga Y, Hachimura S, et al. Identification of Multiple Isolated Lymphoid Follicles on the Antimesenteric Wall of the Mouse Small Intestine. The Journal of Immunology. 2002;168:57–64. [DOI] [PubMed] [Google Scholar]
- [61].Ivanov II, de Llanos Frutos R, Manel N, Yoshinaga K, Rifkin DB, Sartor RB, et al. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell host & microbe. 2008;4:337–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, et al. Induction of Colonic Regulatory T Cells by Indigenous <em>Clostridium</em> Species. Science. 2011;331:337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122:107–18. [DOI] [PubMed] [Google Scholar]
- [64].Tlaskalová-Hogenová H, Štěpánková R, Kozáková H, Hudcovic T, Vannucci L, Tučková L, et al. The role of gut microbiota (commensal bacteria) and the mucosal barrier in the pathogenesis of inflammatory and autoimmune diseases and cancer: contribution of germ-free and gnotobiotic animal models of human diseases. Cellular & Molecular Immunology. 2011;8:110–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Rinninella E, Raoul P, Cintoni M, Franceschi F, Miggiano GAD, Gasbarrini A, et al. What is the Healthy Gut Microbiota Composition? A Changing Ecosystem across Age, Environment, Diet, and Diseases. Microorganisms. 2019;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Human Microbiome Project C Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Mariat D, Firmesse O, Levenez F, Guimarăes VD, Sokol H, Doré J, et al. The Firmicutes/Bacteroidetes ratio of the human microbiota changes with age. BMC Microbiology. 2009;9:123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Dethlefsen L, Relman DA. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proceedings of the National Academy of Sciences. 2011;108:4554–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Vich Vila A, Collij V, Sanna S, Sinha T, Imhann F, Bourgonje AR, et al. Impact of commonly used drugs on the composition and metabolic function of the gut microbiota. Nat Commun. 2020;11:362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Johnson AJ, Vangay P, Al-Ghalith GA, Hillmann BM, Ward TL, Shields-Cutler RR, et al. Daily Sampling Reveals Personalized Diet-Microbiome Associations in Humans. Cell Host Microbe. 2019;25:789–802 e5. [DOI] [PubMed] [Google Scholar]
- [71].Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Vangay P, Johnson AJ, Ward TL, Al-Ghalith GA, Shields-Cutler RR, Hillmann BM, et al. US Immigration Westernizes the Human Gut Microbiome. Cell. 2018;175:962–72.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Knight R, Vrbanac A, Taylor BC, Aksenov A, Callewaert C, Debelius J, et al. Best practices for analysing microbiomes. Nat Rev Microbiol. 2018;16:410–22. [DOI] [PubMed] [Google Scholar]
- [74].Peterson DA, McNulty NP, Guruge JL, Gordon JI. IgA Response to Symbiotic Bacteria as a Mediator of Gut Homeostasis. Cell Host & Microbe. 2007;2:328–39. [DOI] [PubMed] [Google Scholar]
- [75].Xu J, Bjursell MK, Himrod J, Deng S, Carmichael LK, Chiang HC, et al. A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science. 2003;299:2074–6. [DOI] [PubMed] [Google Scholar]
- [76].Troy EB, Kasper DL. Beneficial effects of Bacteroides fragilis polysaccharides on the immune system. Front Biosci (Landmark Ed). 2010;15:25–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Zakharzhevskaya NB, Tsvetkov VB, Vanyushkina AA, Varizhuk AM, Rakitina DV, Podgorsky VV, et al. Interaction of Bacteroides fragilis Toxin with Outer Membrane Vesicles Reveals New Mechanism of Its Secretion and Delivery. Frontiers in Cellular and Infection Microbiology. 2017;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Shahi SK, Freedman SN, Murra AC, Zarei K, Sompallae R, Gibson-Corley KN, et al. Prevotella histicola, A Human Gut Commensal, Is as Potent as COPAXONE(R) in an Animal Model of Multiple Sclerosis. Front Immunol. 2019;10:462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Marietta EV, Murray JA, Luckey DH, Jeraldo PR, Lamba A, Patel R, et al. Suppression of Inflammatory Arthritis by Human Gut-Derived Prevotella histicola in Humanized Mice. Arthritis Rheumatol. 2016;68:2878–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Mangalam AK, Murray J. Microbial monotherapy with Prevotella histicola for patients with multiple sclerosis. Expert Rev Neurother. 2019;19:45–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S, et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci U S A. 2010;107:14691–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Larsen JM. The immune response to Prevotella bacteria in chronic inflammatory disease. Immunology. 2017;151:363–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Scher JU, Sczesnak A, Longman RS, Segata N, Ubeda C, Bielski C, et al. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. Elife. 2013;2:e01202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Lopetuso LR, Scaldaferri F, Petito V, Gasbarrini A. Commensal Clostridia: leading players in the maintenance of gut homeostasis. Gut Pathog. 2013;5:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Ratajczak W, Ryl A, Mizerski A, Walczakiewicz K, Sipak O, Laszczynska M. Immunomodulatory potential of gut microbiome-derived short-chain fatty acids (SCFAs). Acta Biochim Pol. 2019;66:1–12. [DOI] [PubMed] [Google Scholar]
- [86].Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, deRoos P, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504:451–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504:446–50. [DOI] [PubMed] [Google Scholar]
- [88].Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly YM, et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341:569–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Bakken JS, Borody T, Brandt LJ, Brill JV, Demarco DC, Franzos MA, et al. Treating Clostridium difficile infection with fecal microbiota transplantation. Clin Gastroenterol Hepatol. 2011;9:1044–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Ding YH, Qian LY, Pang J, Lin JY, Xu Q, Wang LH, et al. The regulation of immune cells by Lactobacilli: a potential therapeutic target for anti-atherosclerosis therapy. Oncotarget. 2017;8:59915–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Hanchi H, Mottawea W, Sebei K, Hammami R. The Genus Enterococcus: Between Probiotic Potential and Safety Concerns-An Update. Front Microbiol. 2018;9:1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Martinez E, Marcos A. Helicobacter pylori and peptic ulcer disease. N Engl J Med. 1991;325:737–8. [DOI] [PubMed] [Google Scholar]
- [93].Rizzatti G, Lopetuso LR, Gibiino G, Binda C, Gasbarrini A. Proteobacteria: A Common Factor in Human Diseases. Biomed Res Int. 2017;2017:9351507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Obata T, Goto Y, Kunisawa J, Sato S, Sakamoto M, Setoyama H, et al. Indigenous opportunistic bacteria inhabit mammalian gut-associated lymphoid tissues and share a mucosal antibody-mediated symbiosis. Proc Natl Acad Sci U S A. 2010;107:7419–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Fung TC, Bessman NJ, Hepworth MR, Kumar N, Shibata N, Kobuley D, et al. Lymphoid-Tissue-Resident Commensal Bacteria Promote Members of the IL-10 Cytokine Family to Establish Mutualism. Immunity. 2016;44:634–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Markle JG, Frank DN, Mortin-Toth S, Robertson CE, Feazel LM, Rolle-Kampczyk U, et al. Sex differences in the gut microbiome drive hormone-dependent regulation of autoimmunity. Science. 2013;339:1084–8. [DOI] [PubMed] [Google Scholar]
- [97].Dominianni C, Sinha R, Goedert JJ, Pei Z, Yang L, Hayes RB, et al. Sex, body mass index, and dietary fiber intake influence the human gut microbiome. PLoS One. 2015;10:e0124599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Shin JH, Park YH, Sim M, Kim SA, Joung H, Shin DM. Serum level of sex steroid hormone is associated with diversity and profiles of human gut microbiome. Res Microbiol. 2019;170:192–201. [DOI] [PubMed] [Google Scholar]
- [99].Yurkovetskiy L, Burrows M, Khan AA, Graham L, Volchkov P, Becker L, et al. Gender bias in autoimmunity is influenced by microbiota. Immunity. 2013;39:400–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Daisley BA, Chanyi RM, Abdur-Rashid K, Al KF, Gibbons S, Chmiel JA, et al. Abiraterone acetate preferentially enriches for the gut commensal Akkermansia muciniphila in castrate-resistant prostate cancer patients. Nat Commun. 2020;11:4822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Weersma RK, Zhernakova A, Fu J. Interaction between drugs and the gut microbiome. Gut. 2020;69:1510–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Spanogiannopoulos P, Bess EN, Carmody RN, Turnbaugh PJ. The microbial pharmacists within us: a metagenomic view of xenobiotic metabolism. Nat Rev Microbiol. 2016;14:273–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Wilson ID, Nicholson JK. Gut microbiome interactions with drug metabolism, efficacy, and toxicity. Transl Res. 2017;179:204–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Sousa T, Yadav V, Zann V, Borde A, Abrahamsson B, Basit AW. On the colonic bacterial metabolism of azo-bonded prodrugsof 5-aminosalicylic acid. J Pharm Sci. 2014;103:3171–5. [DOI] [PubMed] [Google Scholar]
- [105].Haiser HJ, Gootenberg DB, Chatman K, Sirasani G, Balskus EP, Turnbaugh PJ. Predicting and manipulating cardiac drug inactivation by the human gut bacterium Eggerthella lenta. Science. 2013;341:295–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Taylor MR, Flannigan KL, Rahim H, Mohamud A, Lewis IA, Hirota SA, et al. Vancomycin relieves mycophenolate mofetil-induced gastrointestinal toxicity by eliminating gut bacterial beta-glucuronidase activity. Sci Adv. 2019;5:eaax2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Bhatt AP, Pellock SJ, Biernat KA, Walton WG, Wallace BD, Creekmore BC, et al. Targeted inhibition of gut bacterial beta-glucuronidase activity enhances anticancer drug efficacy. Proc Natl Acad Sci U S A. 2020;117:7374–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Dethlefsen L, Relman DA. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci U S A. 2011;108 Suppl 1:4554–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Vallianou NG, Stratigou T, Tsagarakis S. Metformin and gut microbiota: their interactions and their impact on diabetes. Hormones (Athens). 2019;18:141–4. [DOI] [PubMed] [Google Scholar]
- [110].Hojo M, Asahara T, Nagahara A, Takeda T, Matsumoto K, Ueyama H, et al. Gut Microbiota Composition Before and After Use of Proton Pump Inhibitors. Dig Dis Sci. 2018;63:2940–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Stankiewicz JM, Kolb H, Karni A, Weiner HL. Role of immunosuppressive therapy for the treatment of multiple sclerosis. Neurotherapeutics. 2013;10:77–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Viaud S, Saccheri F, Mignot G, Yamazaki T, Daillere R, Hannani D, et al. The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science. 2013;342:971–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Allison AC, Eugui EM. Mycophenolate mofetil and its mechanisms of action. Immunopharmacology. 2000;47:85–118. [DOI] [PubMed] [Google Scholar]
- [114].Ransom JT. Mechanism of action of mycophenolate mofetil. Ther Drug Monit. 1995;17:681–4. [DOI] [PubMed] [Google Scholar]
- [115].Flannigan KL, Taylor MR, Pereira SK, Rodriguez-Arguello J, Moffat AW, Alston L, et al. An intact microbiota is required for the gastrointestinal toxicity of the immunosuppressant mycophenolate mofetil. The Journal of Heart and Lung Transplantation. 2018;37:1047–59. [DOI] [PubMed] [Google Scholar]
- [116].Maes BD, Dalle I, Geboes K, Oellerich M, Armstrong VW, Evenepoel P, et al. Erosive enterocolitis in mycophenolate mofetil-treated renal-transplant recipients with persistent afebrile diarrhea. Transplantation. 2003;75:665–72. [DOI] [PubMed] [Google Scholar]
- [117].Pellock SJ, Redinbo MR. Glucuronides in the gut: Sugar-driven symbioses between microbe and host. J Biol Chem. 2017;292:8569–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Friedman B, Cronstein B. Methotrexate mechanism in treatment of rheumatoid arthritis. Joint Bone Spine. 2019;86:301–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Hasko G, Cronstein B. Regulation of inflammation by adenosine. Front Immunol. 2013;4:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Gerards AH, de Lathouder S, de Groot ER, Dijkmans BA, Aarden LA. Inhibition of cytokine production by methotrexate. Studies in healthy volunteers and patients with rheumatoid arthritis. Rheumatology (Oxford). 2003;42:1189–96. [DOI] [PubMed] [Google Scholar]
- [121].Letertre MPM, Munjoma N, Wolfer K, Pechlivanis A, McDonald JAK, Hardwick RN, et al. A Two-Way Interaction between Methotrexate and the Gut Microbiota of Male Sprague-Dawley Rats. J Proteome Res. 2020;19:3326–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Valerino DM, Johns DG, Zaharko DS, Oliverio VT. Studies of the metabolism of methotrexate by intestinal flora. I. Identification and study of biological properties of the metabolite 4-amino-4-deoxy-N 10 -methylpteroic acid. Biochem Pharmacol. 1972;21:821–31. [DOI] [PubMed] [Google Scholar]
- [123].Zhou B, Xia X, Wang P, Chen S, Yu C, Huang R, et al. Induction and Amelioration of Methotrexate-Induced Gastrointestinal Toxicity are Related to Immune Response and Gut Microbiota. EBioMedicine. 2018;33:122–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Chen J, Wright K, Davis JM, Jeraldo P, Marietta EV, Murray J, et al. An expansion of rare lineage intestinal microbes characterizes rheumatoid arthritis. Genome Med. 2016;8:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Picchianti-Diamanti A, Panebianco C, Salemi S, Sorgi ML, Di Rosa R, Tropea A, et al. Analysis of Gut Microbiota in Rheumatoid Arthritis Patients: Disease-Related Dysbiosis and Modifications Induced by Etanercept. Int J Mol Sci. 2018;19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Zhang X, Zhang D, Jia H, Feng Q, Wang D, Liang D, et al. The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nature Medicine. 2015;21:895–905. [DOI] [PubMed] [Google Scholar]
- [127].Gelfand JM, Cree BAC, Hauser SL. Ocrelizumab and Other CD20(+) B-Cell-Depleting Therapies in Multiple Sclerosis. Neurotherapeutics. 2017;14:835–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Ramwadhdoebe TH, van Baarsen LGM, Boumans MJH, Bruijnen STG, Safy M, Berger FH, et al. Effect of rituximab treatment on T and B cell subsets in lymph node biopsies of patients with rheumatoid arthritis. Rheumatology (Oxford, England). 2019;58:1075–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Ellwardt E, Ellwardt L, Bittner S, Zipp F. Monitoring B-cell repopulation after depletion therapy in neurologic patients. Neurology - Neuroimmunology Neuroinflammation. 2018;5:e463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Donaldson GP, Ladinsky MS, Yu KB, Sanders JG, Yoo BB, Chou WC, et al. Gut microbiota utilize immunoglobulin A for mucosal colonization. Science (New York, NY). 2018;360:795–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Evan JR, Bozkurt SB, Thomas NC, Bagnato F. Alemtuzumab for the treatment of multiple sclerosis. Expert Opin Biol Ther. 2018;18:323–34. [DOI] [PubMed] [Google Scholar]
- [132].Finkelsztejn A Multiple sclerosis: overview of disease-modifying agents. Perspect Medicin Chem. 2014;6:65–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Qu L, Li Q, Jiang H, Gu L, Zhang Q, Wang C, et al. Effect of anti-mouse CD52 monoclonal antibody on mouse intestinal intraepithelial lymphocytes. Transplantation. 2009;88:766–72. [DOI] [PubMed] [Google Scholar]
- [134].Li QR, Wang CY, Tang C, He Q, Li N, Li JS. Reciprocal Interaction Between Intestinal Microbiota and Mucosal Lymphocyte in Cynomolgus Monkeys After Alemtuzumab Treatment. American Journal of Transplantation. 2013;13:899–910. [DOI] [PubMed] [Google Scholar]
- [135].Seidel JA, Otsuka A, Kabashima K. Anti-PD-1 and Anti-CTLA-4 Therapies in Cancer: Mechanisms of Action, Efficacy, and Limitations. Front Oncol. 2018;8:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Couzin-Frankel J Breakthrough of the year 2013. Cancer immunotherapy. Science. 2013;342:1432–3. [DOI] [PubMed] [Google Scholar]
- [137].Frankel AE, Coughlin LA, Kim J, Froehlich TW, Xie Y, Frenkel EP, et al. Metagenomic Shotgun Sequencing and Unbiased Metabolomic Profiling Identify Specific Human Gut Microbiota and Metabolites Associated with Immune Checkpoint Therapy Efficacy in Melanoma Patients. Neoplasia. 2017;19:848–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Gopalakrishnan V, Spencer CN, Nezi L, Reuben A, Andrews MC, Karpinets TV, et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science. 2018;359:97–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Matson V, Fessler J, Bao R, Chongsuwat T, Zha Y, Alegre ML, et al. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science. 2018;359:104–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Liu T, Xiong Q, Li L, Hu Y. Intestinal microbiota predicts lung cancer patients at risk of immune-related diarrhea. Immunotherapy. 2019;11:385–96. [DOI] [PubMed] [Google Scholar]
- [141].Vetizou M, Pitt JM, Daillere R, Lepage P, Waldschmitt N, Flament C, et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science. 2015;350:1079–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Routy B, Le Chatelier E, Derosa L, Duong CPM, Alou MT, Daillère R, et al. Gut microbiome influences efficacy of PD-1–based immunotherapy against epithelial tumors. Science. 2018;359:91–7. [DOI] [PubMed] [Google Scholar]
- [143].Katayama Y, Yamada T, Shimamoto T, Iwasaku M, Kaneko Y, Uchino J, et al. The role of the gut microbiome on the efficacy of immune checkpoint inhibitors in Japanese responder patients with advanced non-small cell lung cancer. Transl Lung Cancer Res. 2019;8:847–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Chaput N, Lepage P, Coutzac C, Soularue E, Le Roux K, Monot C, et al. Baseline gut microbiota predicts clinical response and colitis in metastatic melanoma patients treated with ipilimumab. Annals of Oncology. 2017;28:1368–79. [DOI] [PubMed] [Google Scholar]
- [145].Hutchinson M Natalizumab: A new treatment for relapsing remitting multiple sclerosis. Ther Clin Risk Manag. 2007;3:259–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Ransohoff RM. Natalizumab for Multiple Sclerosis. New England Journal of Medicine. 2007;356:2622–9. [DOI] [PubMed] [Google Scholar]
- [147].Escribano BM, Medina-Fernandez FJ, Aguilar-Luque M, Aguera E, Feijoo M, Garcia-Maceira FI, et al. Lipopolysaccharide Binding Protein and Oxidative Stress in a Multiple Sclerosis Model. Neurotherapeutics. 2017;14:199–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Riccio P The molecular basis of nutritional intervention in multiple sclerosis: A narrative review. Complementary Therapies in Medicine. 2011;19:228–37. [DOI] [PubMed] [Google Scholar]
- [149].Chun J, Hartung HP. Mechanism of action of oral fingolimod (FTY720) in multiple sclerosis. Clin Neuropharmacol. 2010;33:91–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Abdurasulova IN, Tarasova EA, Nikiforova IG, Il'ves AG, Ivashkova EV, Matsulevich AV, et al. [The intestinal microbiota composition in patients with multiple sclerosis receiving different disease-modifying therapies DMT]. Zh Nevrol Psikhiatr Im S S Korsakova. 2018;118:62–9. [DOI] [PubMed] [Google Scholar]
- [151].Katz Sand I, Zhu Y, Ntranos A, Clemente JC, Cekanaviciute E, Brandstadter R, et al. Disease-modifying therapies alter gut microbial composition in MS. Neurol Neuroimmunol Neuroinflamm. 2019;6:e517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Jangi S, Gandhi R, Cox LM, Li N, von Glehn F, Yan R, et al. Alterations of the human gut microbiome in multiple sclerosis. Nat Commun. 2016;7:12015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Mills EA, Ogrodnik MA, Plave A, Mao-Draayer Y. Emerging Understanding of the Mechanism of Action for Dimethyl Fumarate in the Treatment of Multiple Sclerosis. Front Neurol. 2018;9:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Linker RA, Gold R. Dimethyl Fumarate for Treatment of Multiple Sclerosis: Mechanism of Action, Effectiveness, and Side Effects. Current Neurology and Neuroscience Reports. 2013;13:394. [DOI] [PubMed] [Google Scholar]
- [155].Longbrake EE, Cantoni C, Chahin S, Cignarella F, Cross AH, Piccio L. Dimethyl fumarate induces changes in B- and T-lymphocyte function independent of the effects on absolute lymphocyte count. Mult Scler. 2018;24:728–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Storm-Larsen C, Myhr KM, Farbu E, Midgard R, Nyquist K, Broch L, et al. Gut microbiota composition during a 12-week intervention with delayed-release dimethyl fumarate in multiple sclerosis - a pilot trial. Mult Scler J Exp Transl Clin. 2019;5:2055217319888767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Rumah KR, Vartanian TK, Fischetti VA. Oral Multiple Sclerosis Drugs Inhibit the In vitro Growth of Epsilon Toxin Producing Gut Bacterium, Clostridium perfringens. Front Cell Infect Microbiol. 2017;7:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Ma N, Wu Y, Xie F, Du K, Wang Y, Shi L, et al. Dimethyl fumarate reduces the risk of mycotoxins via improving intestinal barrier and microbiota. Oncotarget. 2017;8:44625–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Duda PW, Schmied MC, Cook SL, Krieger JI, Hafler DA. Glatiramer acetate (Copaxone®) induces degenerate, Th2-polarized immune responses in patients with multiple sclerosis. The Journal of clinical investigation. 2000;105:967–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Neuhaus O, Farina C, Yassouridis A, Wiendl H, Bergh FT, Dose T, et al. Multiple sclerosis: comparison of copolymer-1-reactive T cell lines from treated and untreated subjects reveals cytokine shift from T helper 1 to T helper 2 cells. Proceedings of the National Academy of Sciences. 2000;97:7452–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Jee Y, Piao WH, Liu R, Bai XF, Rhodes S, Rodebaugh R, et al. CD4(+)CD25(+) regulatory T cells contribute to the therapeutic effects of glatiramer acetate in experimental autoimmune encephalomyelitis. Clin Immunol. 2007;125:34–42. [DOI] [PubMed] [Google Scholar]
- [162].Hong J, Li N, Zhang X, Zheng B, Zhang JZ. Induction of CD4+CD25+ regulatory T cells by copolymer-I through activation of transcription factor Foxp3. Proc Natl Acad Sci U S A. 2005;102:6449–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Karandikar NJ, Crawford MP, Yan X, Ratts RB, Brenchley JM, Ambrozak DR, et al. Glatiramer acetate (Copaxone) therapy induces CD8+ T cell responses in patients with multiple sclerosis. The Journal of clinical investigation. 2002;109:641–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Tennakoon DK, Mehta RS, Ortega SB, Bhoj V, Racke MK, Karandikar NJ. Therapeutic induction of regulatory, cytotoxic CD8+ T cells in multiple sclerosis. The Journal of Immunology. 2006;176:7119–29. [DOI] [PubMed] [Google Scholar]
- [165].Cantarel BL, Waubant E, Chehoud C, Kuczynski J, DeSantis TZ, Warrington J, et al. Gut microbiota in multiple sclerosis: possible influence of immunomodulators. J Investig Med. 2015;63:729–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Dhib-Jalbut S, Marks S. Interferon-beta mechanisms of action in multiple sclerosis. Neurology. 2010;74 Suppl 1:S17–24. [DOI] [PubMed] [Google Scholar]
- [167].Markowitz CE. Interferon-beta: mechanism of action and dosing issues. Neurology. 2007;68:S8–11. [DOI] [PubMed] [Google Scholar]
- [168].Reynders T, Devolder L, Valles-Colomer M, Van Remoortel A, Joossens M, De Keyser J, et al. Gut microbiome variation is associated to Multiple Sclerosis phenotypic subtypes. Annals of Clinical and Translational Neurology.n/a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Castillo-Alvarez F, Perez-Matute P, Oteo JA, Marzo-Sola ME. The influence of interferon beta-1b on gut microbiota composition in patients with multiple sclerosis. Neurologia. 2018. [DOI] [PubMed] [Google Scholar]
- [170].Lis K, Kuzawinska O, Balkowiec-Iskra E. Tumor necrosis factor inhibitors - state of knowledge. Arch Med Sci. 2014;10:1175–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Lim H, Lee SH, Lee HT, Lee JU, Son JY, Shin W, et al. Structural Biology of the TNFalpha Antagonists Used in the Treatment of Rheumatoid Arthritis. Int J Mol Sci. 2018;19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Kemanetzoglou E, Andreadou E. CNS Demyelination with TNF-alpha Blockers. Curr Neurol Neurosci Rep. 2017;17:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [173].Ribaldone DG, Caviglia GP, Abdulle A, Pellicano R, Ditto MC, Morino M, et al. Adalimumab Therapy Improves Intestinal Dysbiosis in Crohn's Disease. J Clin Med. 2019;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [174].Zhou Y, Xu ZZ, He Y, Yang Y, Liu L, Lin Q, et al. Gut Microbiota Offers Universal Biomarkers across Ethnicity in Inflammatory Bowel Disease Diagnosis and Infliximab Response Prediction. mSystems. 2018;3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [175].Ali T, Kaitha S, Mahmood S, Ftesi A, Stone J, Bronze MS. Clinical use of anti-TNF therapy and increased risk of infections. Drug Healthc Patient Saf. 2013;5:79–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].Aden K, Rehman A, Waschina S, Pan WH, Walker A, Lucio M, et al. Metabolic Functions of Gut Microbes Associate With Efficacy of Tumor Necrosis Factor Antagonists in Patients With Inflammatory Bowel Diseases. Gastroenterology. 2019;157:1279–92 e11. [DOI] [PubMed] [Google Scholar]
- [177].Gori S, Inno A, Belluomini L, Bocus P, Bisoffi Z, Russo A, et al. Gut microbiota and cancer: How gut microbiota modulates activity, efficacy and toxicity of antitumoral therapy. Crit Rev Oncol Hematol. 2019;143:139–47. [DOI] [PubMed] [Google Scholar]
- [178].Lurienne L, Cervesi J, Duhalde L, de Gunzburg J, Andremont A, Zalcman G, et al. NSCLC Immunotherapy Efficacy and Antibiotic Use: A Systematic Review and Meta-Analysis. J Thorac Oncol. 2020;15:1147–59. [DOI] [PubMed] [Google Scholar]
- [179].Hopkins AM, Kichenadasse G, Karapetis CS, Rowland A, Sorich MJ. Concomitant Antibiotic Use and Survival in Urothelial Carcinoma Treated with Atezolizumab. Eur Urol. 2020;78:540–3. [DOI] [PubMed] [Google Scholar]
- [180].Elkrief A, Derosa L, Kroemer G, Zitvogel L, Routy B. The negative impact of antibiotics on outcomes in cancer patients treated with immunotherapy: a new independent prognostic factor? Ann Oncol. 2019;30:1572–9. [DOI] [PubMed] [Google Scholar]
- [181].Mu Q, Kirby J, Reilly CM, Luo XM. Leaky Gut As a Danger Signal for Autoimmune Diseases. Front Immunol. 2017;8:598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [182].Braniste V, Al-Asmakh M, Kowal C, Anuar F, Abbaspour A, Toth M, et al. The gut microbiota influences blood-brain barrier permeability in mice. Sci Transl Med. 2014;6:263ra158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [183].Agus A, Planchais J, Sokol H. Gut Microbiota Regulation of Tryptophan Metabolism in Health and Disease. Cell Host Microbe. 2018;23:716–24. [DOI] [PubMed] [Google Scholar]
- [184].Macia L, Tan J, Vieira AT, Leach K, Stanley D, Luong S, et al. Metabolite-sensing receptors GPR43 and GPR109A facilitate dietary fibre-induced gut homeostasis through regulation of the inflammasome. Nature communications. 2015;6:6734. [DOI] [PubMed] [Google Scholar]
- [185].Zhao Y, Chen F, Wu W, Sun M, Bilotta AJ, Yao S, et al. GPR43 mediates microbiota metabolite SCFA regulation of antimicrobial peptide expression in intestinal epithelial cells via activation of mTOR and STAT3. Mucosal immunology. 2018;11:752–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [186].Mathewson ND, Jenq R, Mathew AV, Koenigsknecht M, Hanash A, Toubai T, et al. Gut microbiome-derived metabolites modulate intestinal epithelial cell damage and mitigate graft-versus-host disease. Nat Immunol. 2016;17:505–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [187].Wu W, Sun M, Chen F, Cao AT, Liu H, Zhao Y, et al. Microbiota metabolite short-chain fatty acid acetate promotes intestinal IgA response to microbiota which is mediated by GPR43. Mucosal immunology. 2017;10:946–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [188].Park J, Kim M, Kang SG, Jannasch AH, Cooper B, Patterson J, et al. Short-chain fatty acids induce both effector and regulatory T cells by suppression of histone deacetylases and regulation of the mTOR-S6K pathway. Mucosal immunology. 2015;8:80–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [189].Sun M, Wu W, Chen L, Yang W, Huang X, Ma C, et al. Microbiota-derived short-chain fatty acids promote Th1 cell IL-10 production to maintain intestinal homeostasis. Nature communications. 2018;9:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [190].Stilling RM, van de Wouw M, Clarke G, Stanton C, Dinan TG, Cryan JF. The neuropharmacology of butyrate: The bread and butter of the microbiota-gut-brain axis? Neurochemistry International. 2016;99:110–32. [DOI] [PubMed] [Google Scholar]
- [191].Nakamoto N, Sasaki N, Aoki R, Miyamoto K, Suda W, Teratani T, et al. Gut pathobionts underlie intestinal barrier dysfunction and liver T helper 17 cell immune response in primary sclerosing cholangitis. Nat Microbiol. 2019;4:492–503. [DOI] [PubMed] [Google Scholar]
- [192].Schwab L, Goroncy L, Palaniyandi S, Gautam S, Triantafyllopoulou A, Mocsai A, et al. Neutrophil granulocytes recruited upon translocation of intestinal bacteria enhance graft-versus-host disease via tissue damage. Nat Med. 2014;20:648–54. [DOI] [PubMed] [Google Scholar]
- [193].Manfredo Vieira S, Hiltensperger M, Kumar V, Zegarra-Ruiz D, Dehner C, Khan N, et al. Translocation of a gut pathobiont drives autoimmunity in mice and humans. Science. 2018;359:1156–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [194].Costa FR, Francozo MC, de Oliveira GG, Ignacio A, Castoldi A, Zamboni DS, et al. Gut microbiota translocation to the pancreatic lymph nodes triggers NOD2 activation and contributes to T1D onset. J Exp Med. 2016;213:1223–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [195].Hebbandi Nanjundappa R, Ronchi F, Wang J, Clemente-Casares X, Yamanouchi J, Sokke Umeshappa C, et al. A Gut Microbial Mimic that Hijacks Diabetogenic Autoreactivity to Suppress Colitis. Cell. 2017;171:655–67 e17. [DOI] [PubMed] [Google Scholar]
- [196].Tanoue T, Morita S, Plichta DR, Skelly AN, Suda W, Sugiura Y, et al. A defined commensal consortium elicits CD8 T cells and anti-cancer immunity. Nature. 2019;565:600–5. [DOI] [PubMed] [Google Scholar]
- [197].Ruff WE, Dehner C, Kim WJ, Pagovich O, Aguiar CL, Yu AT, et al. Pathogenic Autoreactive T and B Cells Cross-React with Mimotopes Expressed by a Common Human Gut Commensal to Trigger Autoimmunity. Cell Host Microbe. 2019;26:100–13 e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [198].Greiling TM, Dehner C, Chen X, Hughes K, Iniguez AJ, Boccitto M, et al. Commensal orthologs of the human autoantigen Ro60 as triggers of autoimmunity in lupus. Sci Transl Med. 2018;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [199].Horai R, Zarate-Blades CR, Dillenburg-Pilla P, Chen J, Kielczewski JL, Silver PB, et al. Microbiota-Dependent Activation of an Autoreactive T Cell Receptor Provokes Autoimmunity in an Immunologically Privileged Site. Immunity. 2015;43:343–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [200].Tai N, Peng J, Liu F, Gulden E, Hu Y, Zhang X, et al. Microbial antigen mimics activate diabetogenic CD8 T cells in NOD mice. J Exp Med. 2016;213:2129–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [201].Planas R, Santos R, Tomas-Ojer P, Cruciani C, Lutterotti A, Faigle W, et al. GDP-l-fucose synthase is a CD4(+) T cell-specific autoantigen in DRB3*02:02 patients with multiple sclerosis. Sci Transl Med. 2018;10. [DOI] [PubMed] [Google Scholar]
- [202].Fluckiger A, Daillere R, Sassi M, Sixt BS, Liu P, Loos F, et al. Cross-reactivity between tumor MHC class I-restricted antigens and an enterococcal bacteriophage. Science. 2020;369:936–42. [DOI] [PubMed] [Google Scholar]
- [203].Tremblay J, Singh K, Fern A, Kirton ES, He S, Woyke T, et al. Primer and platform effects on 16S rRNA tag sequencing. Front Microbiol. 2015;6:771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [204].Bukin YS, Galachyants YP, Morozov IV, Bukin SV, Zakharenko AS, Zemskaya TI. The effect of 16S rRNA region choice on bacterial community metabarcoding results. Sci Data. 2019;6:190007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [205].McDonald D, Hornig M, Lozupone C, Debelius J, Gilbert JA, Knight R. Towards large-cohort comparative studies to define the factors influencing the gut microbial community structure of ASD patients. Microbial Ecology in Health and Disease. 2015;26:26555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [206].Ebers GC, Kukay K, Bulman DE, Sadovnick AD, Rice G, Anderson C, et al. A full genome search in multiple sclerosis. Nat Genet. 1996;13:472–6. [DOI] [PubMed] [Google Scholar]
- [207].Seldin MF, Amos CI, Ward R, Gregersen PK. The genetics revolution and the assault on rheumatoid arthritis. Arthritis & Rheumatism. 1999;42:1071–9. [DOI] [PubMed] [Google Scholar]
- [208].Gourraud P-A, Harbo HF, Hauser SL, Baranzini SE. The genetics of multiple sclerosis: an up-to-date review. Immunological Reviews. 2012;248:87–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [209].Kochi Y, Suzuki A, Yamamoto K. Genetic basis of rheumatoid arthritis: A current review. Biochemical and Biophysical Research Communications. 2014;452:254–62. [DOI] [PubMed] [Google Scholar]
- [210].Cotsapas C, Mitrovic M. Genome-wide association studies of multiple sclerosis. Clin Transl Immunology. 2018;7:e1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [211].Bisanz JE, Upadhyay V, Turnbaugh JA, Ly K, Turnbaugh PJ. Meta-Analysis Reveals Reproducible Gut Microbiome Alterations in Response to a High-Fat Diet. Cell Host Microbe. 2019;26:265–72.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]