Abstract
The gonadotropin-releasing hormone (GnRH) signaling pathway controls reproductive functions and cancer growth and progression. However, few studies investigated roles of genetic variants of GnRH pathway genes in survival of patients with non-small cell lung cancer (NSCLC). Therefore, we first evaluated associations between 22,528 single-nucleotide polymorphisms (SNPs) in 101 GnRH pathway genes and survival of 1,185 NSCLC patients using a dataset from Prostate, Lung, Colorectal and Ovarian (PLCO) Cancer Screening Trial. We found 572 SNPs to be significantly associated with overall survival (OS) of NSCLC (P ≤ 0.05, Bayesian false discovery probability ≤ 0.80). We then validated these SNPs in another dataset from 984 NSCLC patients in Harvard Lung Cancer Susceptibility (HLCS) Study. Finally, two independent SNPs (HBEGF rs4150236G>A and ITPR3 rs116454384C>T) remained significantly associated with NSCLC OS with a combined hazards ratio of 0.84 (95% confidence interval=0.76–0.92, P=0.0003) and 0.85 (0.78–0.94, 0.0012), respectively, and their genetic score (the number of protective genotypes) was associated with better OS and disease-specific survival (DSS) (Ptrend=0.0002 and 0.0001, respectively). Further expression quantitative trail loci analysis showed significant correlations between ITPR3 rs116454384T genotypes, and higher mRNA expression levels in both whole blood and normal lung, and high ITPR3 mRNA expression levels in tumors was associated with better survival of NSCLC patients. Because ITPR3 mutations were rare in tumors, ITPR3 rs116454384C>T likely had an effect on cancer progression by regulating gene expression. Therefore, genetic variants of HBEGF rs4150236G>A and ITPR3 rs116454384C>T may be predictors for NSCLC survival, but HBEGF rs4150236G>A functional relevance remains to be determined.
Keywords: Non-small cell lung cancer (NSCLC), genome-wide association study (GWAS), single-nucleotide polymorphism (SNP), Gonadotropin releasing hormone (GnRH), overall survival (OS), disease specific survival (DSS)
Introduction
Lung cancer is the leading cause of cancer-related mortality in both men and women, with more than one million deaths each year worldwide1. In the USA in 2020, it is estimated that there were approximately 228,820 new cases diagnosed with and 135,720 deaths from lung cancer2. Non-small cell lung cancer (NSCLC) is the primary histological type, accounting for approximately 85% of all lung cancer patients. Although there have been advances in the treatment of NSCLC patients, including surgery, chemo-radiotherapy, molecular targeted therapy and immunotherapy, the 5-year overall survival (OS) rate of NSCLC remains only 18.1% in the United States3. Therefore, the discovery of suitable biomarkers would help improve early diagnosis and predict clinical outcomes as well as personalizing therapy of patients with NSCLC. It is well known that clinical characteristics, including age, sex, smoking status, histology, stage, and treatment options, are the main factors to influence lung cancer survival4. Moreover, increasing evidence suggests that genetic variations in critical genes play an important role in the tumorigenesis and progression of NSCLC4–6. Indeed, recently, by using the pathway-based analytic approaches, several novel and biologically functional variants of the cancer-related pathway genes have been identified to be associated with lung cancer survival4,7,8.
Accumulating evidence has demonstrated that the gonadotropin-releasing hormone (GnRH) signaling pathway not only is a key regulator of the reproductive system9, but also plays an important role in the control of tumorigenesis and progression in human cancers, including both reproductive and non-reproductive cancers9–11. It is well known that GnRH triggers the synthesis and release of luteinizing hormone (LH) and follicular stimulating hormone (FSH) by pituitary gonadotropes and plays a central role in the regulation of gonadial development12. GnRH specifically binds to its receptor (GnRH-R), which is an incipient key node in the GnRH signaling pathway and activates various intracellular mechanisms, having effects on cellular function by the GTP-binding protein-coupled receptor (GPCR) signaling13. It was reported that GnRH and GnRH-R were expressed in several types of cancer tissues, including NSCLC, indicating that the expression of GnRH may be associated with tumor progression14. Although the effect of GnRH on tumor progress is controversial, some studies revealed that GnRH has strong anti-proliferation and anti-metastasis properties in human cancers and is considered as a promising candidate for novel molecular-targeted strategies for the treatment of cancers10.
In addition, GnRH also activates and regulates multiple signaling pathways, such as the pathways of JNK/AP-1, calcineurin/NFAT, cAMP/PKA/CREB and mitogen-activated protein kinase (MAPK), all of which may play important roles in tumorigenesis and tumor progression in humans10,15–17. For instance, GnRH is coupled to Gαq/11 G proteins to activate phospholipase C, which catalyzes the hydrolysis of phosphatidylinositol 4,5-bisphosphate to inositol 1,4,5-trisphosphate (IP3) and diacylglycerol (DAG)18; IP3 can stimulate the release of intracellular calcium and activate conventional protein kinase C (PKC) isoforms, and the inositol 1,4,5-triphosphate receptor type 3 (IP3R3 or ITPR3), a modulator for diverse cellular functions, is responsive to the binding of IP3, whereas the generation of DAG can lead to the activation of novel PKC isoforms19,20. Furthermore, activation of PKC could result in transactivation of the EGF and MAPK signaling pathway9,18. It is also demonstrated that GnRH causes the activation of phosphor tyrosine phosphatase (PTP), which leads to the de-phosphorylation of activated EGF-R and inhibition of EGF-R signal transduction as well as inhibition of the heparin binding-epidermal growth factor (HBEGF) signaling in ERα-negative breast cancer cells10. Taken together, these data suggest that aberrant activation of the GnRH pathway has a significant impact on tumorigenesis or tumor progression.
Therefore, it is likely that genetic variation, including SNPs, in some key genes in the GnRH signaling pathway may be involved in the disorder or over-activation of the entire GnRH signaling pathway, modulating tumor growth and progression, but such genetic effects and their biological functions remain largely unknown. Therefore, in the present study, we investigated associations between potentially functional genetic variants in the GnRH signaling pathway genes and survival of NSCLC patients in a two-stage analysis of genotyping datasets extracted from two previously published genome-wide association studies (GWASs).
Materials and methods
Study populations
In the discovery stage, we obtained a genotyping dataset from the Prostate, Lung, Colorectal and Ovarian (PLCO) Cancer Screening Trial, which enrolled nearly 155,000 participants aged 55–74 from ten centers across the United States between 1993 and 200121. Among all the participants, 1,185 Caucasian NSCLC patients with complete personal information including age, sex, smoking status, histology, clinical stage, treatment options, follow-up information and genotyping data were available for survival analysis. We used OS as the primary endpoint and also examined disease-specific survival (DSS). The follow-up time was defined from the diagnosis of NSCLC to the last follow-up or the time of death. In the PLCO trial, we extracted genomic DNA samples from the blood and genotyped with Illumina HumanHap240Sv1.0, HumanHap300v1.1 and HumanHap550v3.0 (dbGaP accession: phs000093.v2.p2 and phs000336.v1.p1)22,23. To expand the genotyping data, we performed imputation with IMPUTE2 according to the CEU data from the 1000 Genomes Project (phase 1 release V3).
In the validation stage, we used another genotyping dataset that includes 984 histologically confirmed Caucasian NSCLC patients from the GWAS dataset of the Harvard Lung Cancer Susceptibility (HLCS) study24, in which genomic DNA samples extracted from the patients’ blood were genotyped with Illumina Humanhap610-Quad arrays, and the genotyping data were also imputed by using MaCH1.0 based on the 1000 Genomes Project. Details of the patients from the HLCS study have also been described elsewhere24.
The use of these two GWAS datasets was approved by both the Internal Review Board of Duke University School of Medicine (#Pro00054575) and the dbGAP database administration (#6404). The comparison of the characteristics between the PLCO trial (n=1,185) and the HLCS study (n=984) is presented in Supplementary Table 1.
Gene and SNP selection
We searched the GnRH pathway genes by using the Molecular Signatures Database with the keyword “GnRH” (http://software.broadinstitute.org/gsea/msigdb/index.jsp) and included 101 genes as the candidates for further analysis (Supplementary Table 2). SNPs within these genes and their ± 500-kb flanking regions were selected by the following quality control criteria: (1) a genotyping rate ≥ 95%, (2) a minor allelic frequency (MAF) ≥ 0.05, (3) Hardy-Weinberg equilibrium (HWE) P value ≥ 1×10−5, and (4) an imputation info score ³ 0.8. As a result, 2,378 genotyped SNPs were selected from the PLCO GWAS dataset and additional 20,150 SNPs were imputed (Supplementary Figure 1).
Statistical analysis
In the discovery stage, we performed multivariate Cox proportional hazards regression analysis in an additive genetic model with adjustments for age, sex, smoking status, histology, tumor stage, chemotherapy, radiotherapy, and surgery as well as the first four principal components identified from the GWAS dataset. We estimated associations between SNPs in the GnRH pathway genes and NSCLC survival by calculating hazards ratio (HR) and its 95% confidence interval (CI) with the GenABEL package of R software25. For multiple testing correction, the false discovery rate (FDR) with a cut-off value of 0.200 was first used to assess the probability of false positives26. Since the majority of SNPs were imputed with a high level of linkage disequilibrium (LD), we also used Bayesian false discovery probability (BFDP) with a cut-off value of 0.80 for multiple testing correction to reduce the probability of false positive findings as recommended27. In the LD analysis, we selected representative SNPs in the identified important genes in high LD (r2>0.8) and functional SNPs according to functional annotation based on RegulomeDB (http://www.regulomedb.org/) and HaploReg v4.1 (https://pubs.broadinstitute.org/mammals/haploreg/haploreg.php). We assigned a prior probability of 0.10 to detect an HR of 3.0 for an association with variant genotypes or minor alleles of the SNPs with P ≤ 0.05. In the validation stage, we used Cox regression analysis with adjustment for age, sex, smoking status, histology, tumor stage, chemotherapy, radiotherapy, surgery and the first three principal components to validate the findings from the discovery stage.
Next, we performed a meta-analysis to combine the results of both discovery and validation datasets by using PLINK 1.07, for which Cochran’s Q-test and the heterogeneity statistic (I2) were performed to assess the inter-study heterogeneity. If no heterogeneity was found between the two studies (Q-test P-value > 0.10 and I2 < 50.0%), a fixed-effects model was implemented; otherwise, a random-effects model was applied. Pairwise LD was also estimated by using the data from 373 European individuals in the 1000 Genomes Project. To further identify novel and independent SNPs among the validated SNPs, we constructed a multivariate stepwise Cox model that included the first four principal components of the PLCO dataset and 23 SNPs previously published, in addition to the adjustment for available demographic and clinical variables. We also used the combined genotypes to evaluate the cumulative effects of the identified significant SNPs and the Kaplan-Meier curves to depict survival probability associated with their genotypes.
In the stratified analysis, we performed the heterogeneity test of associations among subgroups of each clinical characteristic by using the Chi-square-based Q-test, with P < 0.05 considered statistically significant for differences among the subgroups of each clinical characteristic. All statistical analyses were performed with SAS software (version 9.4; SAS Institute, Cary, NC, USA), if not specified otherwise. To illustrate the prediction accuracy of the model integrating clinical and genetic variables on NSCLC survival28, we performed the receiver operating characteristic (ROC) curve and time-dependent area under the curve (AUC) with ROC-time package of R software (version 3.5.0). We also generated the LD map and haplotype blocks by Haploview software19, the Manhattan plot with the −log10 (Padj) for all SNPs that passed QC, and the regional association plots by using Locus Zoom (http://http://locuszoom.sph.umich.edu).
Finally, we performed expression quantitative trait loci (eQTL) analysis29 to evaluate correlations between genotypes of SNPs and mRNA expression levels of their corresponding genes by using RNA-sequencing data from lymphoblastoid cells derived from the same 373 individuals of European descent in the 1000 Genomes Project, and 369 whole blood samples and 383 normal lung tissue included in the Genotype-Tissue Expression (GTEx Analysis V7, dbGaP Accession phs000424.v7.p2) project30,31. We examined the differences in mRNA expression levels in 111 pairs of lung cancer tissues and adjacent normal tissues from The Cancer Genome Atlas (TCGA) dataset by using a paired Student’s t-test. We also assessed the differences in mRNA expression levels in a larger, but not paired, dataset from TCGA (http://ualcan.path.uab.edu), and performed Kaplan-Meier survival analysis to assess the associations between mRNA expression levels of the important genes and survival probability (http://kmplot.com) of lung cancer patients. To assess the mutation rates of those identified important genes in lung tumor tissues, we also used the publicly available database of the cBioPortal for cancer Genomics (http://www.cbioportal.org). All statistical analyses were performed with the SAS software (version 9.4; SAS Institute, Cary, NC, USA) unless specified otherwise.
Results
Associations of SNPs in the GnRH signaling pathway genes with survival of NSCLC
Basic characteristics of the discovery dataset with 1,185 NSCLC patients from the PLCO trial and the validation dataset of 984 NSCLC patients from the HLCS study have been described elsewhere4. As shown in the working flowchart (Figure 1), we first used a single-locus multivariate Cox regression analysis in the discovery dataset to evaluate associations between 22,528 SNPs of GnRH signaling pathway genes and NSCLC OS with adjustment for age, sex, smoking status, histology, tumor stage, chemotherapy, radiotherapy, surgery and the first four principal components (Supplementary Table 3). After multiple testing correction by FDR first and then by BFDP, we identified 572 SNPs to be significantly associated with NSCLC OS (P < 0.05, BFDP ≤ 0.8). All these significant SNPs were further validated by the HLCS GWAS dataset, and finally, seven SNPs remained significantly associated with NSCLC OS. Further combined analysis of the two datasets for these seven SNPs showed their associations with a better NSCLC OS without heterogeneity. The details of associations between these seven SNPs and NSCLC OS are described in Table 1.
Figure 1. The flowchart of the present study.
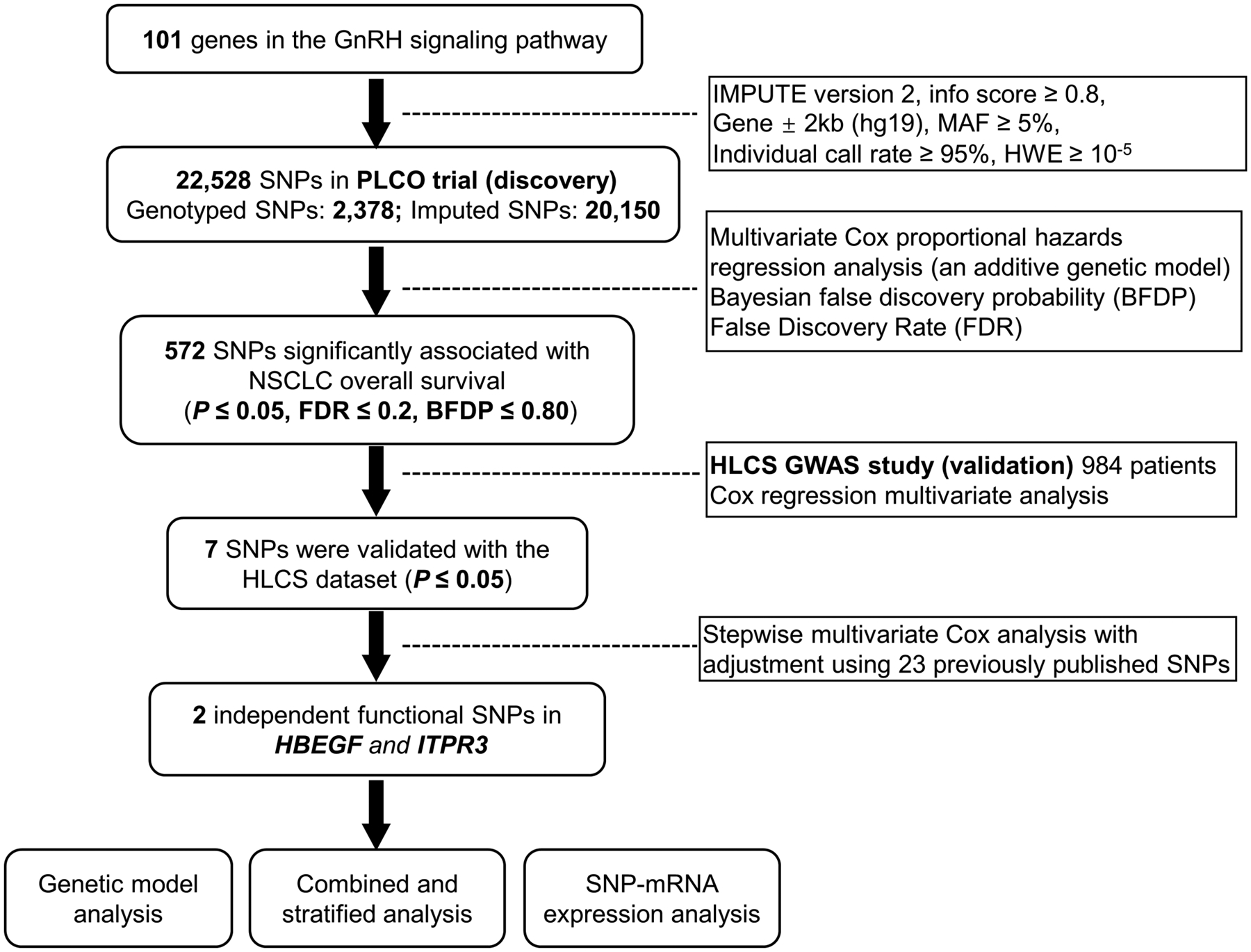
Abbreviations: SNP, single- nucleotide polymorphism; PLCO, Prostate, Lung, Colorectal and Ovarian cancer screening trial; NSCLC, non- small cell lung cancer; HBEGF, heparin binding-epidermal growth factor; ITPR3, inositol 1,4,5-triphosphate receptor type 3.
Table 1.
Associations of seven validated significant SNPs with overall survival in both discovery and validation datasets from two previously published NSCLC GWAS datasets
| SNP | Allelea | Gene | PLCO (n=1185) | HLCS (n=984) | Combined-analysis | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FDRa | BFDPa | MAF | HR (95% CI)b | Pb | MAF | HR (95% CI)c | Pc | Phetd | I2 | HR (95% CI)e | Pe | |||
| rs2237078f | G>C | HBEGF | 0.555 | 0.735 | 0.22 | 0.85 (0.74–0.96) | 0.010 | 0.21 | 0.86 (0.75–0.99) | 0.033 | 0.904 | 0 | 0.85 (0.78–0.94) | 0.0010 |
| rs4150230f | G>A | HBEGF | 0.547 | 0.747 | 0.22 | 0.84 (0.74–0.96) | 0.009 | 0.21 | 0.84 (0.73–0.96) | 0.013 | 1.000 | 0 | 0.84 (0.76–0.92) | 0.0003 |
| rs4150232f | G>A | HBEGF | 0.547 | 0.747 | 0.22 | 0.84 (0.74–0.96) | 0.009 | 0.21 | 0.83 (0.72–0.96) | 0.012 | 0.904 | 0 | 0.84 (0.76–0.92) | 0.0002 |
| rs4150234f | G>A | HBEGF | 0.547 | 0.747 | 0.22 | 0.84 (0.74–0.96) | 0.008 | 0.21 | 0.84 (0.73–0.96) | 0.011 | 1.000 | 0 | 0.84 (0.76–0.92) | 0.0003 |
| rs4150236f | G>A | HBEGF | 0.546 | 0.747 | 0.22 | 0.84 (0.74–0.96) | 0.008 | 0.21 | 0.84 (0.73–0.96) | 0.012 | 1.000 | 0 | 0.84 (0.76–0.92) | 0.0003 |
| rs13385f | G>A | HBEGF | 0.556 | 0.735 | 0.22 | 0.85 (0.75–0.96) | 0.011 | 0.21 | 0.86 (0.75–0.99) | 0.030 | 0.902 | 0 | 0.85 (0.78–0.94) | 0.0007 |
| rs116454384 | C>T | ITPR3 | 0.556 | 0.747 | 0.17 | 0.84 (0.74–0.96) | 0.012 | 0.19 | 0.87 (0.75–1.00) | 0.049 | 0.723 | 0 | 0.85 (0.78–0.94) | 0.0012 |
Abbreviations: SNP, single nucleotide polymorphism; NSCLC, non-small cell lung cancer; GWAS, genome-wide association study; PLCO, Prostate, Lung, Colorectal and Ovarian cancer screening trial; HLCS: Harvard Lung Cancer Susceptibility; MAF, minor allele frequency; HR, hazards ratio; CI, confidence interval; FDR: false discovery rate; BFDP: Bayesian false discovery probability
FDR and BFDP were available in the PLCO dataset because the HLCS study provided only the summary data
Obtained from an additive genetic model with adjustment for age, sex, stage, histology, smoking status, chemotherapy, radiotherapy, surgery, PC1, PC2, PC3, and PC4;
Obtained from an additive genetic model with adjustment for age, sex, stage, histology, smoking status, chemotherapy, radiotherapy, surgery, PC1, PC2, and PC3;
Phet: P value for heterogeneity by Cochrane’s Q test;
Meta-analysis in the fixed-effects model.
SNPs rs2237078, rs4150230, rs4150232, rs4150234, rs13385 are high LD with rs4150236.
In further LD analysis of these seven replicated SNPs, except for rs116454384 in ITPR3, other six SNPs in HBEGF were in high LD with each other (all r2 > 0.8) (Supplementary Figure 3a) by in silico SNP functional prediction (SNPinfo, RegulomeDB and HaploReg 4.1). In particular, we observed that the SNPs of rs4150236 in HBEGF and rs116454384 in ITPR3 are located in the enhancer-like H3K4me1 and H3K27ac in lung tissue or A549 EtOH 0.02pct lung carcinoma cell line, which is predicted to have putative regulatory sites, such as the enhancer histone modification (Supplemental Table 4). Thus, we selected these two representative SNPs (i.e. rs4150236 in HBEGF and rs116454384 in ITPR3) for further analyses and summarized the results in the Manhattan plot (Supplementary Figure 2) and the regional association plot (Supplementary Figure 3b and 3c). To further identify independent SNPs associated with NSCLC survival, we performed analysis with the multivariate stepwise Cox regression model, including the first four principal components of the PLCO dataset. When the two validated SNPs (rs4150236 and rs116454384) were added to the model with adjustment for the 23 previously published significant SNPs in the same PLCO GWAS dataset, both SNPs remained significantly and independently associated with NSCLC OS (Table 2).
Table 2.
Two independent SNPs in multivariate Cox hazards regression analysis with adjustment for other covariates and previous published SNPs in the PLCO Trial GWAS dataset
| Variables | Category | Frequency | HR (95% CI)a | Pa | HR (95% CI)b | Pb |
|---|---|---|---|---|---|---|
| Age | Continuous | 1185 | 1.03 (1.02–1.05) | <0.0001 | 1.04 (1.02–1.05) | <0.0001 |
| Sex | Male | 698 | 1.00 | 1.00 | ||
| Female | 487 | 0.81 (0.69–0.94) | 0.005 | 0.79 (0.68–0.93) | 0.004 | |
| Smoking status | Never | 115 | 1.00 | 1.00 | ||
| Current | 647 | 1.64 (1.25–2.16) | 0.0004 | 1.90 (1.43–2.54) | <0.0001 | |
| Former | 423 | 1.71 (1.27–2.29) | 0.0003 | 1.96 (1.45–2.66) | <0.0001 | |
| Histology | AD | 577 | 1.00 | 1.00 | ||
| SC | 285 | 1.20 (0.99–1.44) | 0.061 | 1.25 (1.03–1.51) | 0.026 | |
| Others | 323 | 1.29 (1.08–1.53) | 0.004 | 1.33 (1.11–1.59) | 0.002 | |
| Stage | I-IIIA | 655 | 1.00 | 1.00 | ||
| IIIB-IV | 528 | 2.82 (2.32–3.42) | <0.0001 | 3.00 (2.46–3.66) | <0.0001 | |
| Chemotherapy | No | 639 | 1.00 | 1.00 | ||
| Yes | 538 | 0.58 (0.49–0.69) | <0.0001 | 0.58 (0.48–0.70) | <0.0001 | |
| Radiotherapy | No | 762 | 1.00 | 1.00 | ||
| Yes | 415 | 0.92 (0.78–1.09) | 0.335 | 0.94 (0.79–1.11) | 0.448 | |
| Surgery | No | 637 | 1.00 | 1.00 | ||
| Yes | 540 | 0.21 (0.16–0.27) | <0.0001 | 0.19 (0.15–0.25) | <0.0001 | |
| HBEGF rs4150236 | GG/GA/AA | 719/409/56 | 0.84 (0.74–0.95) | 0.007 | 0.85 (0.74–0.97) | 0.014 |
| ITPR3 rs116454384 | CC/CT/TT | 807/334/37 | 0.85 (0.75–0.98) | 0.021 | 0.85 (0.74–0.97) | 0.020 |
Abbreviations: HR: hazards ratio; CI: confidence interval; SNP: single-nucleotide polymorphisms.
Stepwise analysis included age, sex, smoking status, tumor stage, histology, chemotherapy, radiotherapy, surgery, PC1, PC2, PC3, PC4 and SNPs.
23 published SNPs were used for post-stepwise adjustment: rs779901, rs3806116, rs199731120, rs10794069, rs1732793, rs225390, rs3788142, rs73049469, rs35970494, rs225388, rs7553295, rs1279590, rs73534533, rs677844, rs4978754, rs1555195, rs11660748, rs73440898, rs13040574, rs469783, rs36071574, rs7242481, rs1049493.
As shown in Table 3 for the 1,185 NSCLC patients in the PLCO dataset with complete adjustment, patients with the HBEGF rs4150236 A allele or ITPR3 rs116454384 T allele had a reduced risk of death (rs4150236: Ptrend = 0.008 for OS and Ptrend = 0.004 for DSS; rs116454384: Ptrend = 0.012 for OS and Ptrend = 0.011 for DSS). More specifically, compared with their wild genotypes in a dominant genetic model, patients with HBEGF rs4150236 GA/AA or ITPR3 rs116454384 CT/TT genotypes had a significantly reduced risk of death (HBEGF rs4150236: HR = 0.80, 95% CI = 0.69–0.92 and P = 0.002 for OS; 0.79, 0.67–0.92 and 0.003 for DSS; ITPR3 rs116454384: 0.84, 0.72–0.98 and 0.025 for OS; 0.82, 0.70–0.97 and 0.018 for DSS) (Table 3 and Figure 2).
Table 3.
Associations between two significantly independent SNPs and survival of NSCLC patients in the PLCO Trial
| Genotype | No. of patients | OS | DSS | ||||
|---|---|---|---|---|---|---|---|
| Death (%) | Multivariate analysisa | Death (%) | Multivariate analysisa | ||||
| HR (95% CI) | P | HR (95% CI) | P | ||||
| HBEGF | |||||||
| rs4150236 G>Ab | 1175 | ||||||
| GG | 713 | 500 (70.1) | 1.00 | 453 (63.5) | 1.00 | ||
| GA | 406 | 255 (62.8) | 0.79 (0.67–0.92) | 0.002 | 229 (56.4) | 0.79 (0.67–0.93) | 0.004 |
| AA | 56 | 34 (60.7) | 0.88 (0.62–1.26) | 0.492 | 27 (48.2) | 0.78 (0.53–1.16) | 0.217 |
| Ptrend test | 0.008 | 0.004 | |||||
| GA/AA | 462 | 289 (62.6) | 0.80 (0.69–0.92) | 0.002 | 256 (55.4) | 0.79 (0.67–0.92) | 0.003 |
| ITPR3 | |||||||
| rs116454384 C>Tb | 1175 | ||||||
| CC | 804 | 550 (68.4) | 1.00 | 499 (62.1) | 1.00 | ||
| CT | 333 | 220 (66.1) | 0.86 (0.74–1.01) | 0.072 | 192 (57.7) | 0.84 (0.71–1.00) | 0.048 |
| TT | 38 | 19 (50.0) | 0.64 (0.40–1.01) | 0.055 | 18 (47.4) | 0.66 (0.41–1.05) | 0.082 |
| Ptrend test | 0.012 | 0.011 | |||||
| CT/TT | 371 | 239 (64.4) | 0.84 (0.72–0.98) | 0.025 | 210 (56.6) | 0.82 (0.70–0.97) | 0.018 |
| Number of protective genotypes (NPGs)b,c | |||||||
| 0 | 489 | 354 (72.4) | 1.00 | 324 (66.3) | 1.00 | ||
| 1 | 539 | 342 (63.5) | 0.76 (0.65–0.88) | 0.0003 | 304 (56.4) | 0.75 (0.64–0.88) | 0.0005 |
| 2 | 147 | 93 (63.3) | 0.71 (0.56–0.89) | 0.004 | 81 (55.1) | 0.68 (0.53–0.87) | 0.003 |
| Ptrend test | 0.0002 | 0.0001 | |||||
| 0 | 489 | 354 (72.4) | 1.00 | 324 (66.3) | 1.00 | ||
| 1–2 | 686 | 435 (63.4) | 0.75 (0.65–0.86) | <0.0001 | 385 (56.1) | 0.74 (0.63–0.86) | <0.0001 |
Abbreviations: SNP, single nucleotide polymorphism; NSCLC, non-small cell lung cancer; PLCO, Prostate, Lung, Colorectal and Ovarian cancer screening trial; OS, overall survival; DSS, disease-specific survival; HR, hazards ratio; CI, confidence interval.
Adjusted for age, sex, smoking status, histology, tumor stage, chemotherapy, surgery, and principal components.
10 missing date were excluded.
Protective genotypes were HBEGF rs4150236 GA/AA and ITPR3 rs116454384 CT/TT.
Figure 2.
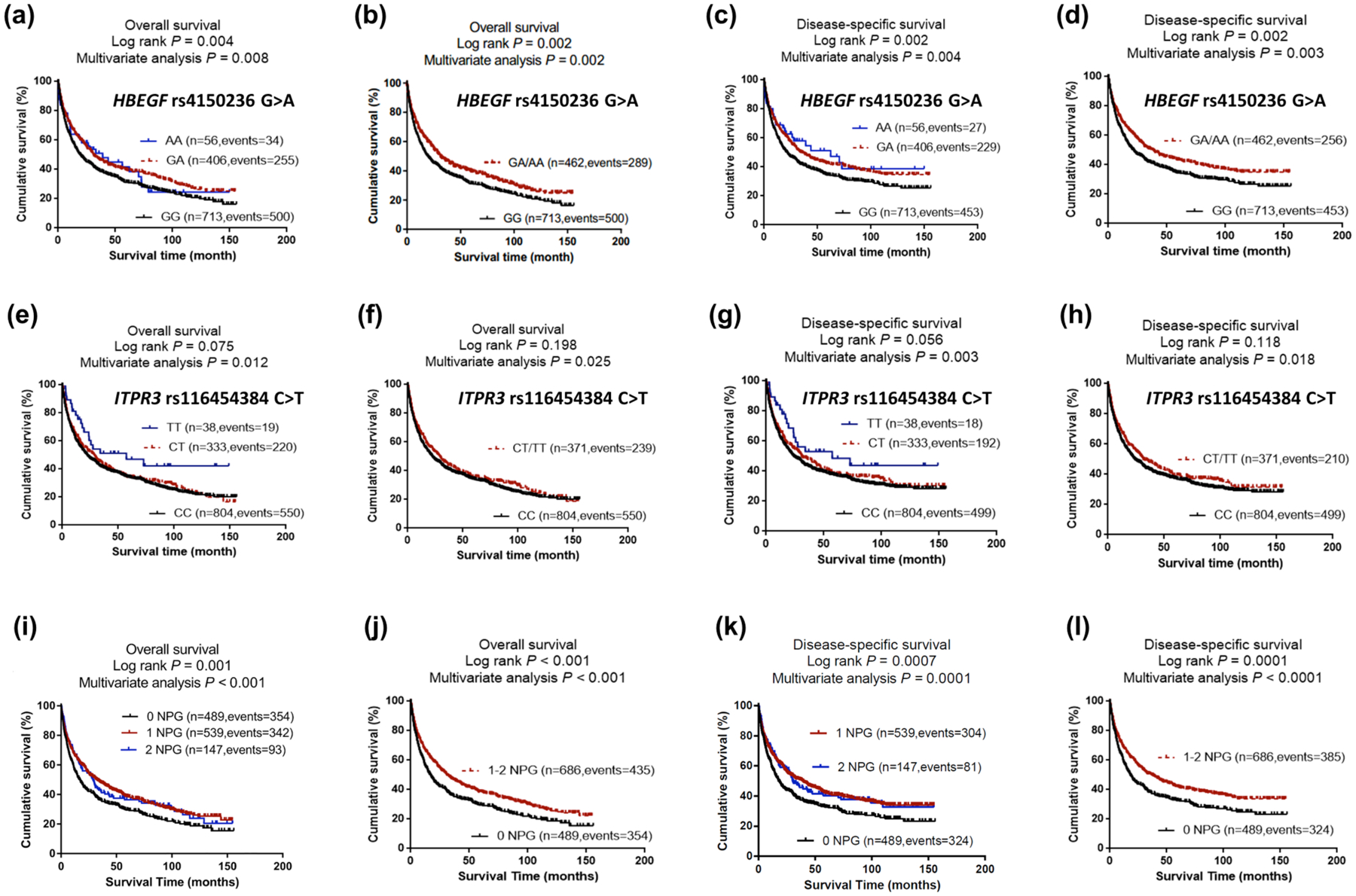
Prediction of survival with genotypes of HBEGF rs4150236, ITPR3 rs116454384 and combined protective genotypes.
(a) Kaplan–Meier survival curves with additive model for the overall survival of HBEGF rs4150236 genotypes and (b) dominate model; (c) Kaplan–Meier survival curves with additive model for the disease- specific survival of HBEGF rs4150236 genotypes and (d) dominate model; (e) Kaplan–Meier survival curves with additive model for the overall survival of ITPR3 rs116454384 genotypes and (f) dominate model; (g) Kaplan–Meier survival curves with additive model for the disease- specific survival of ITPR3 rs116454384 genotypes and (h) dominate model; (i) Kaplan–Meier survival curves for the overall survival of the combined protective genotypes and (j) dichotomized groups of the NPG in the PLCO dataset; (k) Kaplan–Meier survival curves for the disease- specific survival of the combined protective genotypes and (l) dichotomized groups of the NPG in the PLCO dataset.
Abbreviations: NPG, number of protective genotypes; PLCO, The Prostate, Lung, Colorectal and Ovarian Cancer Screening Trial.
Combined effects of the two independent SNPs in the PLCO dataset
To provide a better estimation of the hazards of survival, we combined the protective genotypes (i.e., rs4150236 GA/AA and rs116454384 CT/TT) into a genetic score as the number of protective genotypes (NPGs), which divided all NSCLC patients into three groups: zero, one, and two NPGs. As shown in Table 3, an increased NPG was associated with better survival after adjustment for other covariates (Ptrend = 0.0002 and 0.0001 for OS and DSS, respectively). To dichotomize for better survival analysis, we re-grouped all the patients into a low-protective-genotypes group (0 NPGs) and a high-protective-genotypes group (1–2 NPGs). Compared with the 0 NPGs, 1–2 group NPGs were associated with significantly better survival (adjHR = 0.75, 95% CI = 0.65–0.86, P < 0.0001 for both OS and DSS), which were further depicted in Kaplan-Meier survival curves (Figure 2).
Stratified analysis for associations between NPGs and NSCLC survival
We further performed the stratified analysis to evaluate the possible modification effect of protective genotypes on survival of NSCLC by age, sex, smoking status, histology, tumor stage, chemotherapy, radiotherapy and surgery in the PLCO dataset. As a result, compared with patients with 0 NPGs, patients with 1–2 NPGs exhibited a significantly better survival for OS and DSS in subgroups of males (P = 0.0006 and 0.0005, respectively); age < 71 or ≥ 71 (0.012 or 0.020 for OS, and 0.023 or 0.008 for DSS); former or current smokers (0.010 or 0.034 for OS and 0.012 or 0.024 for DSS); adenocarcinoma or squamous cell carcinoma (0.013 or 0.004 for OS and 0.033 or 0.0004 for DSS); I-IIIA or IIIB-IV tumor stage (0.028 or 0.011 for OS and 0.008 or 0.023 for DSS); no or received chemotherapy (0.019 or 0.003 for OS and 0.014 or 0.004 for DSS); no or received radiotherapy (0.004 or 0.005 for OS and 0.007 or 0.005 for DSS); and without or with surgery (0.014 or 0.011 for OS and 0.016 or 0.006 for DSS). However, no significant interaction was found between genotypes/genetic score and other covariates on NSCLC OS and DSS (Pinter > 0.05 for all, Supplementary Table 5).
The ROC curves and time-dependent AUC
We further assessed predictive values of the two SNPs with time-dependent AUC and ROC curves at the 12th, 24th, and 60th month (5-year) in the PLCO dataset (because we only had the summary genotyping data from the HLCS study). Compared with the model for all covariates including age, sex, smoking status, histology, tumor stage, chemotherapy, radiotherapy, surgery and first four principal components, the time-dependent AUC plot with addition of the independent SNPs did not improve prediction performance of the model at the 12th (1st year) and 60th month (5th year). That is, the AUCs for OS changed from 85.73% to 85.84% (P = 0.600) for the 1st year and from 88.59% to 88.75% (P = 0.512) for the 5th year (Supplementary Figure 4b and 4d, respectively); the AUCs for DSS changed from 86.07% to 86.26% (P = 0.425) for the 1st year and from 88.54% to 88.76% (P = 0.451) for the 5th years) (Supplementary Figure 4f and 4h, respectively). However, the AUC and ROC curves at the 24th month (or the 2nd year) suggested that the prediction performance of the model was improved significantly: the AUCs changed from 86.65% to 87.12% (P = 0.016) for OS and from 86.61% to 87.25% (P = 0.033) for DSS (Supplementary Figure 4c and 4g, respectively).
The eQTL analysis
To further explore potential functions of the two independent SNPs, we performed the eQTL analysis to identify the correlations between genotypes of the SNPs (HBEGF rs4150236 and ITPR3 rs116454384) and mRNA expression levels of their corresponding genes by using genomic data from 373 lymphoblastoid cell lines derived from individuals of European descent in the 1000 Genomes Project and 369 whole blood and 283 lung normal tissues in the GTEx project. We found that the rs4150236 variant AA genotype was borderline significantly correlated with an increased expression level of HBEGF mRNA (P = 0.052, Supplementary Figure 5a), compared with the GG/GA genotypes in the recessive model in the 1000 Genomes Project, while this was not the case in the additive and dominant models, nor in whole blood data (Figure 3a) and lung normal tissues (Figure 3b) of the GTEx project. For the ITPR3 rs116454384 variant T allele, there was also no significant difference in the 1000 Genomes Project (Supplementary Figure 6a). However, in the whole blood data and lung normal tissues of the GTEx project, the variant rs116454384 T allele was associated with higher expression levels of ITPR3 mRNA (in additive model, P = 0.9×10−5 for 369 whole blood individuals and P = 0.010 for 383 lung normal tissues) (Figure 3d and 3e, respectively).
Figure 3. Correlations between genotypes of the significant SNPs and their corresponding mRNA expression levels.
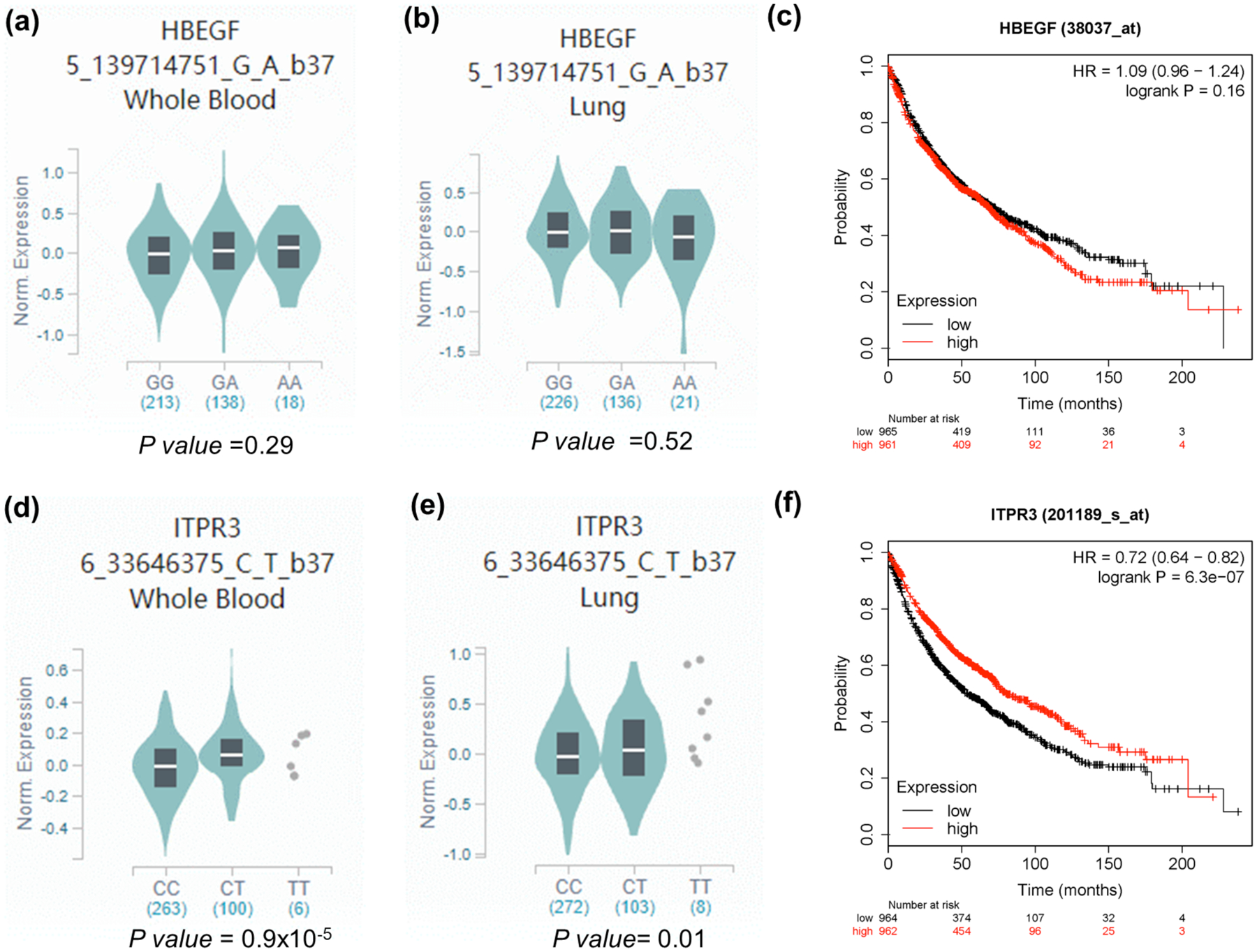
For GTEx project, The HBEGF rs4150236 A allele was no associated with mRNA expression levels of HBEGF in (a) normal lung tissue and (b) whole blood; (c) higher expression levels of HBEGF were associated with a better survival in patients with lung cancer (http://kmplot.com/analysis/index.php?p=service&cancer=lung); (d) The ITPR3 rs116454384 T allele was associated with higher mRNA expression levels in normal lung tissue and (e) whole blood from the GTEx project; (f) higher expression levels of ITPR3 were associated with a better survival in patients with lung cancer (http://kmplot.com/analysis/index.php?p=service&cancer=lung).
Differential mRNA expression analysis and survival of NSCLC
To find molecular mechanisms of the HBEGF and ITPR3 genes in the progression and survival of NSCLC, we first assessed mRNA expression levels of the two genes in 111 pairs of lung cancer tissues including 60 lung adenocarcinoma (LUAD) and 51 lung squamous cell carcinoma (LUSC) and adjacent normal tissue samples in NSCLC obtained from the TCGA database. As shown in Supplementary Figure 5b, compared with adjacent normal tissues, tumor tissues had a lower mRNA expression level of HBEGF in 60 LUAD and 51 LUSC (all P = 0.001). Meanwhile, the mRNA expression levels of ITPR3 were not significantly different in the pairs of LUAD and LUSC NSCLC tissues (P = 0.862) (Supplementary Figure 6b).
We then compared mRNA expression levels of these two genes in 59 adjacent normal lung tissues, 515 LUAD tissue samples, and 503 LUSC obtained from The Cancer Genome Atlas (TCGA) database (ualcan.path.uab.edu/home). As shown in Supplementary Figure 5c, the mRNA expression levels of HBEGF were all significantly lower in LUSC and LUAD than that in normal lung tissues (P = 1.69×10−12 for LUAD and P = 1.64×10−9 for LUSC), while the mRNA expression levels of ITPR3 were significantly higher in LUAD (P = 1.11×10−10) and non-significantly lower in LUSC than that in normal lung tissues (P = 0.302) (Supplementary Figure 6c).
Additionally, we also evaluated the correlation between mRNA expression levels of these two genes and OS of NSCLC patients from the TCGA dataset (www.kmplot.com) and found that the patients with high mRNA expression levels of ITPR3 had a better NSCLC OS (HR = 0.72; 95% CI = 0.64–0.82; Log-rank P = 6.3×10−7, Figure 3f). However, the impact of mRNA expression levels of HBEGF on OS was not statistically significant in the TCGA database (HR = 1.09; 95% CI = 0.96–1.24, Log-rank P = 0.16, Figure 3.c). Taken all the data together, we do not have sufficient evidence to judge whether these genes are oncogenes.
Mutation analysis
Because frequently mutated genes in tumor tissues would have a much greater impact on patients’ survival than SNPs in the same genes, we further investigated the mutation status of HBEGF and ITPR3 in lung tumor tissues by using the public database of the cBioPortal for Cancer Genomics. As shown in Supplemental Figure 5d, HBEGF had a much low somatic mutation rate in different NSCLC datasets (0.56% in TCGA pub, 0.21% in PanCan and 0.09% in TCGA 2016), while ITPR3 also displayed a low mutation rate in different NSCLC datasets (6.25% in MSKCC, 4.11% in TCGA 2016 and 1.33% in MSKCC 2018, Supplemental Figure 6d). These low mutation frequencies in both HBEGF and ITPR3 unlikely had a significant effect on the expression levels of these two genes in NSCLC tumors. Therefore, the roles of SNPs in HBEGF and ITPR3 in regulating gene expression and NSCLC survival need further research.
Discussion
In the present study, we investigated associations between 22,528 genetic variants of 101 genes in the GnRH signaling pathway and NSCLC survival using genotyping data 1,185 NSCLC patients from the PLCO trial and another 984 NSCLC patients from the HLCS study. We found that HBEGF rs4150236A and ITPR3 rs116454384T variant genetypes were significantly associated with a better NSCLC survival in US Caucasian populations. We also provided biological evidence that the protective ITPR3 rs116454384 variant T allele, but not HBEGF rs4150236 variant A allele, was associated with high mRNA expression levels of ITPR3 in both whole blood cells and normal lung tissues from individuals of European descent. Furthermore, patients with high mRNA expression levels of HBEGF and ITPR3 had a better NSCLC OS. These data imply that the HBEGF rs4150236 variant A or ITPR3 rs116454384 variant T alleles may play a role in survival of NSCLC patients, possibly by modulating the mRNA expression levels of their related genes, particularly for ITPR3 rs116454384 variant T allele. These provide further support for biological plausibility for the observed SNP-survival associations.
HBEGF is located on chromosome 5q31.3, contains six exons and encodes HBEGF, a 19–23 kDa protein of 208 amino acids32. As a member of the EGF family, HBEGF was initially found in human macrophages and identified as a protective cytokine on different target cells in the intestine33–35. Subsequently, the expression of HBEGF was detected in several human cancers. For example, one study found that HBEGF mRNA was expressed in seven human glioma cell lines with expression levels of two- to five-fold higher than that of normal brain tissues in eight of 11 glioblastoma patients36, while another study reported that HBEGF enhanced the growth of human pancreatic cancer cells in an allelic dose-dependent manner37. In a study of 108 Japanese patients, investigators found that HBEGF expression to be significantly increased in advanced ovarian cancer, compared with that in normal ovaries, and that the higher expression levels of HBEGF were significantly associated with poor clinical outcomes38,39.
For lung cancer, one study reported that HBEGF was highly expressed in a subset of lung cancer patients, in which proliferation was dependent on the HBEGF signaling and that silencing of HBEGF with RNA interference suppressed cell growth, leading to G1/S cell cycle arrest in HBEGF-positive lung cancer cells, which supports an oncogenic role of HBEGF in lung cancer cells40. The same study also found that the expression of HBEGF correlated with EGFR expression in primary lung tumors was associated with a poor survival of 287 Taiwanese lung cancer patients, suggesting the involvement of the EGFR/HBEGF signaling in lung tumor progression40. Although it was not clear whether the HBEGF rs4150236G>A had an effect of its gene expression, we observed that mRNA expression levels of HBEGF were higher in normal lung tissues than in tumor tissues and that the higher HBEGF mRNA expression was associated with a better survival in over 1,000 NSCLC patients in the TCGA database. In particular, such observations are also consistent with our findings, in that NSCLC patients with the HBEGF rs4150236 variant A allele had a better survival. However, we did not have the evidence for an association between HBEGF rs4150236 variant A allele and mRNA expression levels of the gene, this SNP was associated with considerable levels of monomethylation of lysine 4 (H3K4Me1) and histone modification of H3K27 acetylation (H327Ac) enrichment, which may have some effects on gene activation and expression41. Taken together, the exact molecular mechanisms of HBEGF in lung cancer need to be further explored.
ITPR3 encodes a receptor for the inositol 1,4,5-trisphosphate, a second messenger that mediates the release of intracellular calcium, that plays a key role in exocrine secretion underlying the energy metabolism and cell growth, and ITPR3 also plays a critical role in regulating cellular proliferation, activation and apoptosis of cancer cells20,42. However, the functions of ITPR3 in tumor progression, for lung cancer in particular, remain unclear43. One study reported that FBXL2 knockdown caused accumulation of ITPR3, which is a major player in Ca2+−dependent apoptosis, causing an increase in Ca2+mediated apoptosis in lung cancer A549 cell lines and limiting tumor growth44. In contrast, one recently published study observed that overexpression of ITPR3 could result in reduced apoptosis in breast and colon cancer cells20,42, while knockdown or silencing of ITPR3 enhanced apoptosis in breast, colon and renal cancer cells20,42,43.
In the present study, we found that a better survival of NSCLC was associated with the ITPR3 rs116454384T allele that was also associated with significantly higher mRNA expression levels of ITPR3 in whole blood and normal lung tissues, which in turn were associated with better survival in NSCLC. Furthermore, ITPR3 rs116454384 is located in the intron region of ITPR3 with considerable levels of H3K4Me1 and H3K27ac enrichment, a possible mechanism by which this SNP regulates gene expression41. Therefore, the roles of ITPR3 and related SNPs in tmor progression and the mechanisms involved in survival of NSCLC patients remain to be determined.
To our knowledge, the present study is the first that focuses on associations of potentially functional genetic variants in the GnRH pathway genes with survival of NSCLC patients by a two-stage analysis of two previously published GWAS datasets. However, there were some limitations. First, because both available GWAS datasets came from populations of European descendants, it is uncertain whether these results are generalizable to other ethnic populations. Second, detailed genotype information and clinical outcomes data of the HLCS study were not accessible for us to do additional combined modeling and stratified analysis. Third, because of no direct evidence of biological experiments, the biological mechanisms by which the identified SNPs may influence tumor progression and thus survival of NSCLC needs to be further explored. Finally, our results need to be validated in larger patient populations.
Supplementary Material
Acknowledgments
The authors thank all the participants of the PLCO Cancer Screening Trial. The authors also thank the National Cancer Institute for providing access to the data collected by the PLCO trial. The statements contained herein are solely those of the authors and do not represent or imply concurrence or endorsement by the National Cancer Institute. The authors would also like to acknowledge the dbGaP repository for providing cancer genotyping datasets. The accession numbers for the datasets for lung cancer are phs000336.v1.p1 and phs000093.v2.p2. A list of contributing investigators and funding agencies for those studies can be found in the Supplemental Data. Qingyi Wei was supported by the V Foundation for Cancer Research (D2017-19) and also partly supported by the Duke Cancer Institute as part of the P30 Cancer Center Support Grant (Grant ID: NIH/NCI CA014236). Sheng Luo was supported by NIH grants R01NS091307, R56AG062302). The Harvard Lung Cancer Susceptibility Study was supported by NIH grants U01CA209414, CA092824, CA074386 and CA090578 to David C. Christiani.
Abbreviations:
- GnRH
Gonadotropin releasing hormone
- NSCLC
non-small cell lung cancer
- OS
overall survival
- DSS
disease specific survival
- GWAS
genome-wide association studies
- SNP
single-nucleotide polymorphisms
- PLCO
Prostate, Lung, Colorectal and Ovarian Cancer Screening Trial
- HLCS
Harvard Lung Cancer Susceptibility
- NCI
National Cancer Institute
- MsigDB
Molecular Signatures Database
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- HR
hazards ratio
- CI
confidence interval
- MAF
minor allelic frequency
- HWE
Hardy-Weinberg equilibrium
- FDR
false discovery rate
- BFDP
Bayesian false discovery probability
- LD
linkage disequilibrium
- eQTL
expression quantitative trait loci
- TCGA
the Cancer Genome Atlas
- AUC
area under the receiver operating characteristic curve
- ROC
receiver operating characteristic curve
- HBEGF
heparin binding-epidermal growth factor
- ITPR3
inositol 1,4,5-triphosphate receptor type 3
Footnotes
Conflict of interest statement
None declared.
References
- 1.Global Burden of Disease Cancer C, Fitzmaurice C, Akinyemiju TF, Al Lami FH, Alam T, Alizadeh-Navaei R, Allen C, Alsharif U, Alvis-Guzman N, Amini E, Anderson BO, Aremu O, et al. Global, Regional, and National Cancer Incidence, Mortality, Years of Life Lost, Years Lived With Disability, and Disability-Adjusted Life-Years for 29 Cancer Groups, 1990 to 2016: A Systematic Analysis for the Global Burden of Disease Study. JAMA oncology 2018;4: 1553–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA: a cancer journal for clinicians 2019;69: 7–34. [DOI] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA: a cancer journal for clinicians 2017;67: 7–30. [DOI] [PubMed] [Google Scholar]
- 4.Wang Y, Liu H, Ready NE, Su L, Wei Y, Christiani DC, Wei Q. Genetic variants in ABCG1 are associated with survival of nonsmall-cell lung cancer patients. International journal of cancer 2016;138: 2592–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guan X, Yin M, Wei Q, Zhao H, Liu Z, Wang LE, Yuan X, O’Reilly MS, Komaki R, Liao Z. Genotypes and haplotypes of the VEGF gene and survival in locally advanced non-small cell lung cancer patients treated with chemoradiotherapy. BMC cancer 2010;10: 431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu H, Liu Z, Wang Y, Stinchcombe TE, Owzar K, Han Y, Hung RJ, Brhane Y, McLaughlin J, Brennan P, Bickeboller H, Rosenberger A, et al. Functional variants in DCAF4 associated with lung cancer risk in European populations. Carcinogenesis 2017;38: 541–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu Y, Liu H, Liu S, Wang Y, Xie J, Stinchcombe TE, Su L, Zhang R, Christiani DC, Li W, Wei Q. Genetic variant of IRAK2 in the toll-like receptor signaling pathway and survival of non-small cell lung cancer. International journal of cancer 2018;143: 2400–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen K, Liu H, Liu Z, Luo S, Patz EF Jr., Moorman PG, Su L, Shen S, Christiani DC, Wei Q. Genetic variants in RUNX3, AMD1 and MSRA in the methionine metabolic pathway and survival in nonsmall cell lung cancer patients. International journal of cancer 2019;145: 621–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harrison GS, Wierman ME, Nett TM, Glode LM. Gonadotropin-releasing hormone and its receptor in normal and malignant cells. Endocrine-related cancer 2004;11: 725–48. [DOI] [PubMed] [Google Scholar]
- 10.Grundker C, Emons G. The Role of Gonadotropin-Releasing Hormone in Cancer Cell Proliferation and Metastasis. Frontiers in endocrinology 2017;8: 187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jayaram S, Gupta MK, Raju R, Gautam P, Sirdeshmukh R. Multi-Omics Data Integration and Mapping of Altered Kinases to Pathways Reveal Gonadotropin Hormone Signaling in Glioblastoma. Omics : a journal of integrative biology 2016;20: 736–46. [DOI] [PubMed] [Google Scholar]
- 12.Stamatiades GA, Carroll RS, Kaiser UB. GnRH-A Key Regulator of FSH. Endocrinology 2019;160: 57–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Teng LH, Ahmad M, Ng WT, Sabaratnam S, Rasan MI, Parhar I, Khoo AS. Gonadotropinreleasing hormone inhibits the proliferation and motility of nasopharyngeal carcinoma cells. Molecular medicine reports 2015;12: 4909–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Limonta P, Montagnani Marelli M, Mai S, Motta M, Martini L, Moretti RM. GnRH receptors in cancer: from cell biology to novel targeted therapeutic strategies. Endocrine reviews 2012;33: 784–811. [DOI] [PubMed] [Google Scholar]
- 15.Wang Y, Sun T, Sun H, Yang S, Li D, Zhou D. SCF/C-Kit/JNK/AP-1 Signaling Pathway Promotes Claudin-3 Expression in Colonic Epithelium and Colorectal Carcinoma. International journal of molecular sciences 2017;18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Quang CT, Leboucher S, Passaro D, Fuhrmann L, Nourieh M, Vincent-Salomon A, Ghysdael J. The calcineurin/NFAT pathway is activated in diagnostic breast cancer cases and is essential to survival and metastasis of mammary cancer cells. Cell death & disease 2015;6: e1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Najafi M, Ahmadi A, Mortezaee K. Extracellular-signal-regulated kinase (ERK)/mitogen-activated protein kinase (MAPK) signaling as a target for cancer therapy: An updated review. Cell biology international 2019. [DOI] [PubMed] [Google Scholar]
- 18.Ruf F, Fink MY, Sealfon SC. Structure of the GnRH receptor-stimulated signaling network: insights from genomics. Frontiers in neuroendocrinology 2003;24: 181–99. [DOI] [PubMed] [Google Scholar]
- 19.Yang YC, Chang TY, Chen TC, Lin WS, Chang SC, Lee YJ. ITPR3 gene haplotype is associated with cervical squamous cell carcinoma risk in Taiwanese women. Oncotarget 2017;8: 10085–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Szatkowski C, Parys JB, Ouadid-Ahidouch H, Matifat F. Inositol 1,4,5-trisphosphate-induced Ca2+ signalling is involved in estradiol-induced breast cancer epithelial cell growth. Molecular cancer 2010;9: 156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hocking WG, Hu P, Oken MM, Winslow SD, Kvale PA, Prorok PC, Ragard LR, Commins J, Lynch DA, Andriole GL, Buys SS, Fouad MN, et al. Lung cancer screening in the randomized Prostate, Lung, Colorectal, and Ovarian (PLCO) Cancer Screening Trial. Journal of the National Cancer Institute 2010;102: 722–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tryka KA, Hao L, Sturcke A, Jin Y, Wang ZY, Ziyabari L, Lee M, Popova N, Sharopova N, Kimura M, Feolo M. NCBI’s Database of Genotypes and Phenotypes: dbGaP. Nucleic acids research 2014;42: D975–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mailman MD, Feolo M, Jin Y, Kimura M, Tryka K, Bagoutdinov R, Hao L, Kiang A, Paschall J, Phan L, Popova N, Pretel S, et al. The NCBI dbGaP database of genotypes and phenotypes. Nature genetics 2007;39: 1181–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhai R, Yu X, Wei Y, Su L, Christiani DC. Smoking and smoking cessation in relation to the development of co-existing non-small cell lung cancer with chronic obstructive pulmonary disease. International journal of cancer 2014;134: 961–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aulchenko YS, Ripke S, Isaacs A, van Duijn CM. GenABEL: an R library for genome-wide association analysis. Bioinformatics 2007;23: 1294–6. [DOI] [PubMed] [Google Scholar]
- 26.Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the false discovery rate in behavior genetics research. Behavioural brain research 2001;125: 279–84. [DOI] [PubMed] [Google Scholar]
- 27.Wakefield J A Bayesian measure of the probability of false discovery in genetic epidemiology studies. American journal of human genetics 2007;81: 208–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chambless LE, Diao G. Estimation of time-dependent area under the ROC curve for long-term risk prediction. Statistics in medicine 2006;25: 3474–86. [DOI] [PubMed] [Google Scholar]
- 29.Nica AC, Dermitzakis ET. Expression quantitative trait loci: present and future. Philosophical transactions of the Royal Society of London Series B, Biological sciences 2013;368: 20120362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lappalainen T, Sammeth M, Friedlander MR, t Hoen PA, Monlong J, Rivas MA, Gonzalez-Porta M, Kurbatova N, Griebel T, Ferreira PG, Barann M, Wieland T, et al. Transcriptome and genome sequencing uncovers functional variation in humans. Nature 2013;501: 506–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Consortium GT. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 2015;348: 648–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fen Z, Dhadly MS, Yoshizumi M, Hilkert RJ, Quertermous T, Eddy RL, Shows TB, Lee ME. Structural organization and chromosomal assignment of the gene encoding the human heparin-binding epidermal growth factor-like growth factor/diphtheria toxin receptor. Biochemistry 1993;32: 7932–8. [DOI] [PubMed] [Google Scholar]
- 33.Higashiyama S, Abraham JA, Miller J, Fiddes JC, Klagsbrun M. A heparin-binding growth factor secreted by macrophage-like cells that is related to EGF. Science 1991;251: 936–9. [DOI] [PubMed] [Google Scholar]
- 34.Besner G, Higashiyama S, Klagsbrun M. Isolation and characterization of a macrophage-derived heparin-binding growth factor. Cell regulation 1990;1: 811–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang J, Su Y, Zhou Y, Besner GE. Heparin-binding EGF-like growth factor (HB-EGF) therapy for intestinal injury: Application and future prospects. Pathophysiology : the official journal of the International Society for Pathophysiology 2014;21: 95–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mishima K, Higashiyama S, Asai A, Yamaoka K, Nagashima Y, Taniguchi N, Kitanaka C, Kirino T, Kuchino Y. Heparin-binding epidermal growth factor-like growth factor stimulates mitogenic signaling and is highly expressed in human malignant gliomas. Acta neuropathologica 1998;96: 322–8. [DOI] [PubMed] [Google Scholar]
- 37.Kobrin MS, Funatomi H, Friess H, Buchler MW, Stathis P, Korc M. Induction and expression of heparin-binding EGF-like growth factor in human pancreatic cancer. Biochemical and biophysical research communications 1994;202: 1705–9. [DOI] [PubMed] [Google Scholar]
- 38.Tanaka Y, Miyamoto S, Suzuki SO, Oki E, Yagi H, Sonoda K, Yamazaki A, Mizushima H, Maehara Y, Mekada E, Nakano H. Clinical significance of heparin-binding epidermal growth factor-like growth factor and a disintegrin and metalloprotease 17 expression in human ovarian cancer. Clinical cancer research : an official journal of the American Association for Cancer Research 2005;11: 4783–92. [DOI] [PubMed] [Google Scholar]
- 39.Miyata K, Yotsumoto F, Fukagawa S, Kiyoshima C, Ouk NS, Urushiyama D, Ito T, Katsuda T, Kurakazu M, Araki R, Sanui A, Miyahara D, et al. Serum Heparin-binding Epidermal Growth Factor-like Growth Factor (HB-EGF) as a Biomarker for Primary Ovarian Cancer. Anticancer research 2017;37: 3955–60. [DOI] [PubMed] [Google Scholar]
- 40.Hsieh CH, Chou YT, Kuo MH, Tsai HP, Chang JL, Wu CW. A targetable HB-EGF-CITED4 axis controls oncogenesis in lung cancer. Oncogene 2017;36: 2946–56. [DOI] [PubMed] [Google Scholar]
- 41.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell 2007;129: 823–37. [DOI] [PubMed] [Google Scholar]
- 42.Shibao K, Fiedler MJ, Nagata J, Minagawa N, Hirata K, Nakayama Y, Iwakiri Y, Nathanson MH, Yamaguchi K. The type III inositol 1,4,5-trisphosphate receptor is associated with aggressiveness of colorectal carcinoma. Cell calcium 2010;48: 315–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rezuchova I, Hudecova S, Soltysova A, Matuskova M, Durinikova E, Chovancova B, Zuzcak M, Cihova M, Burikova M, Penesova A, Lencesova L, Breza J, et al. Type 3 inositol 1,4,5-trisphosphate receptor has antiapoptotic and proliferative role in cancer cells. Cell death & disease 2019;10: 186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kuchay S, Giorgi C, Simoneschi D, Pagan J, Missiroli S, Saraf A, Florens L, Washburn MP, Collazo-Lorduy A, Castillo-Martin M, Cordon-Cardo C, Sebti SM, et al. PTEN counteracts FBXL2 to promote IP3R3- and Ca(2+)-mediated apoptosis limiting tumour growth. Nature 2017;546: 554–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


