Abstract
Multimodal image registration has many applications in diagnostic medical imaging and image-guided interventions, such as Transcatheter Arterial Chemoembolization (TACE) of liver cancer guided by intraprocedural CBCT and pre-operative MR. The ability to register peri-procedurally acquired diagnostic images into the intraprocedural environment can potentially improve the intra-procedural tumor targeting, which will significantly improve therapeutic outcomes. However, the intra-procedural CBCT often suffers from suboptimal image quality due to lack of signal calibration for Hounsfield unit, limited FOV, and motion/metal artifacts. These non-ideal conditions make standard intensity-based multimodal registration methods infeasible to generate correct transformation across modalities. While registration based on anatomic structures, such as segmentation or landmarks, provides an efficient alternative, such anatomic structure information is not always available. One can train a deep learning-based anatomy extractor, but it requires large-scale manual annotations on specific modalities, which are often extremely time-consuming to obtain and require expert radiological readers. To tackle these issues, we leverage annotated datasets already existing in a source modality and propose an anatomy-preserving domain adaptation to segmentation network (APA2Seg-Net) for learning segmentation without target modality ground truth. The segmenters are then integrated into our anatomy-guided multimodal registration based on the robust point matching machine. Our experimental results on in-house TACE patient data demonstrated that our APA2Seg-Net can generate robust CBCT and MR liver segmentation, and the anatomy-guided registration framework with these segmenters can provide high-quality multimodal registrations.
Keywords: Multimodal registration, Unsupervised segmentation, Image-guided intervention, Cone-beam Computed Tomography
Graphical Abstract
1. Introduction
Primary liver cancer is the fourth most common cancer and the third most common cause of cancer-related mortality worldwide with incidence rates rising across the globe and especially in the United States and Europe (Bray et al., 2018). Local image-guided therapies, such as Transcatheter Arterial Chemoembolization (TACE), are commonly used procedures that are performed in patients with intermediate to advanced stages as a palliative therapy option, capable of significantly prolonging patient survival (Pung et al., 2017). Most patients undergo multi-parametric multi-phasic contrast-enhanced MRI using gadolinium-enhanced T1 sequences both for diagnostic purposes as well as for the sake of therapy planning. This readily available multi-parametric information on tumor vascularity, size, location and even tissue properties is clinically underutilized for intra-procedural navigation primarily for technical reasons such as lack of practical image registration solutions. Intraprocedural navigation and targeting is instead achieved with serial, planar angiographic imaging as well as intra-procedural Cone-beam Computed Tomography (CBCT) imaging that provides a coarse cross-sectional dataset, which can then be used to map arterial supply of the tumor and allow for accurate catheter guidance and intra-procedural feedback. While CBCT utilizes an x-ray source and the high-resolution 2D flat panel detector enables fast 3D organ visualization during procedures, CBCT suffers from low contrast-to-noise ratio (CNR), narrow abdominal tissue dynamic range, limited field-of-view (FOV), and motion/metal-induced artifacts, making it challenging to directly visualize and localize targeted tumors (Tacher et al., 2015; Pung et al., 2017). Therefore, multimodal image registration, i.e. mapping preoperative MR imaging and associated liver segmentations to intraprocedural CBCT is essential for accurate liver/tumor localization, targeting and subsequent drug delivery. Current workflows do not apply any quantitative measurements on the acquired CBCT images and the predominant technique is mere gestalt assessment of the images. Automatic multimodal registration is therefore highly desirable in image-guided interventional procedures.
Previous multimodal/monomodal image registration algorithms can be categorized into two classes: conventional iterative based approaches (Wyawahare et al., 2009; Maes et al., 1997; Avants et al., 2008; Rohr et al., 2001; Heinrich et al., 2012) and deep learning based approaches (Hu et al., 2018; Qin et al., 2019; Lee et al., 2019; Zhou et al., 2020a; Wang and Zhang, 2020; Arar et al., 2020; Mok and Chung, 2020). Conventional approaches utilize iterative maximization of intensity similarity metrics, such as mutual information (Maes et al., 1997), cross correlation (Avants et al., 2008) and difference in MIND (Heinrich et al., 2012), to find the optimal registration transformation between images. If paired key points between the images are available, landmark-based thin-plate splines registration can be applied to estimate the transformation between images. Previously, Al-Saleh et al. (2015, 2017) demonstrated the feasibility of temporomandibular joints MRI-CBCT registration via the above mentioned intensity-based and landmark-based registration methods. In the application of head and neck CBCT-CT registration, Zhen et al. (2012); Park et al. (2017) proposed to integrate the intensity matching between CT and CBCT into conventional iterative based approaches for more accurate CT-CBCT registrations. However, the intensity matching approaches are suitable for either monomodal or multimodal with similar imaging physics and cannot be adapted to CBCT-MR registration. More recently, Solbiati et al. (2018) proposed a two-stage registration for CBCT-CT liver registration, where manually annotated key points in the first stage are used for coarse alignment and conventional iterative registration based on mutual information is subsequently performed to refine the alignment.
With the recent advances in data driven learning (Zhou et al., 2020b), deep learning based methods have achieved comparable registration performance with a significantly higher inference speed. For monomodal registration, Balakrishnan et al. (2019) proposed the first deep learning based registration method using a deep convolutional network to predict the registration transformation between monomodality images, called VoxelMorph. Mok and Chung (2020) further improved its registration performance by adding the symmetric diffeomorphic properties into the network design. Moreover, Wang and Zhang (2020) developed a learning-based registration framework, called DeepFLASH, that utilizes low dimensional band-limited space for efficient transformation field computing. For multimodal registration, Hu et al. (2018) proposed to use the organ segmentations for weakly supervised training the transformation estimation network, where intensity-neutral supervision makes the multimodal registration feasible. However, accurate manual organ segmentation is required for their approach and thus limits its applications. As an alternative to this approach, Qin et al. (2019) proposed to estimate the non-rigid transformation from disentangled representation of multimodal image contents. There are also recent studies of multimodal image registration for natural images (Arar et al., 2020), where source image appearance is first translated to fix image appearance, and then previously established monomodal registration methods (Balakrishnan et al., 2019) are applied. Although all the above methods achieve impressive results, they are limited to multimodal registration with no occluded FOV, sufficiently wide intensity range, or organ segmentations. Those conditions are hardly satisfied in many image-guided intervention procedures, such as TACE. More recently, Augenfeld et al. (2020) proposed to use manual CBCT liver annotation to train a CBCT segmenter and register based on the predicted CBCT segmentation and manually annotated MR. While demonstrating the feasibility of registration in TACE, segmenting liver on intraprocedural image is not clinical routine and training such segmenter from limited annotation data impede the segmentation and registration performance.
To tackle these issues, we present an anatomy-guided registration framework by learning segmentation without target modality ground truth. In previous works of learning segmentation without target modality, Zhang et al. (2018a) proposed a two-step strategy, where they first use CycleGAN (Zhu et al., 2017) to adapt the target domain image to the domain with a well-trained segmenter, and then predict the segmentation on the adapted image. However, the segmentation performance relies on the image adaptation performance, thus the two-step process may prone to error aggregation. To improve the CycleGAN performance in medical imaging, Zhang et al. (2018b) suggested adding two segmenters as additional discriminators for generating shape-consistent image adaption results. However, ground-truth segmentation is required for both source and target domains. Recently, Yang et al. (2020) proposed to add the Modality Independent Neighborhood Descriptor (MIND) loss (Heinrich et al., 2012) in the CycleGAN to constrain image structure during the adaptation. Similarly, Ge et al. (2019) proposed to incorporate correlation coefficient loss in the CyleGAN to constrain image structure. Both strategies demonstrated improvements in MR-to-CT translation. On the other hand, Huo et al. (2018) proposed Syn2Seg-Net that merges CycleGAN with a segmentation network on the target domain output, such that the segmentation network is trained on the target domain without target domain ground truth. However, the training image of the segmentation network relies on high-quality adapted images from the CycleGAN part of SynSeg-Net. Without anatomy-preserving constraint during the adaptation, the image could be adapted to a target domain image with incorrect anatomical contents, and negatively impact the subsequent segmentation network’s training. Inspired by Huo et al. (2018) and with large-scale manual liver segmentation on conventional CT available from public dataset, such as LiTS (Bilic et al., 2019) and CHAOS (Kavur et al., 2020), we propose an anatomy-preserving domain adaptation to segmentation network (APA2Seg-Net) for learning segmentation without CBCT/MR ground truth. Specifically, we aim to use only conventional CT segmentation to train robust CBCT/MR segmenters in an anatomy-preserving unpaired fashion. The extracted anatomic information of CBCT/MR, i.e liver segmentations, guides our Robust Point Matching (RPM) to estimate the multimodal registration transformation. Our experimental results on TACE patients demonstrate that our APA2Seg-Net based registration framework allows us to get robust target modality segmenters without ground truth, and enables accurate multimodal registration. Our code is available at https://github.com/bbbbbbzhou/APA2Seg-Net.
2. Methods
We propose a novel two-stage multimodal registration framework for mapping pre-operative MR to intraprocedural CBCT for liver image-guided interventions. In the first stage, our APA2Seg-Net is trained with 3 sources of images: paired conventional CT with liver segmentation and unpaired CBCT/MR, such that CBCT/MR segmenters can be extracted from our APA2Seg-Net for outputting the anatomic information. In the second stage, we extract the surface points of the outputted CBCT and MR segmentations, and input them into our RPM machine to predict MR to CBCT transformation. Finally, the transformation is applied to the pre-operative MR and the associated labels to register to the intraprocedural CBCT. The details are discussed in following sections.
2.1. Anatomy-preserving Adaptation to Segmentation Network
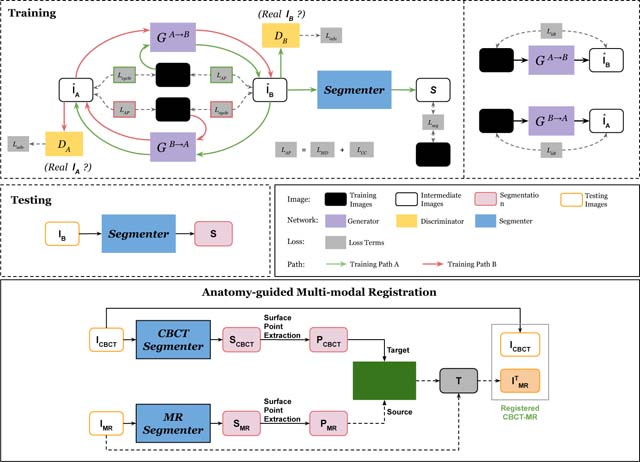
Our APA2Seg-Net consists of two parts - an anatomy-preserving domain adaptation network (APA-Net) and a segmentation network. The architecture and training/test stages are shown in Figure 1. The APA-Net is a cyclic adversarial network based on Zhu et al. (2017) with the addition of anatomy content consistency regularization. As illustrated in Figure 1, APA-Net adapts images between two domains: the conventional CT domain and the CBCT/MR domain . The anatomy consistency regularization ensures organ and tumor content information are not lost during the unpaired domain adaptation process, thus critical for training a robust segmenter in domain . Specifically, our APA2Seg-Net contains five networks, including two generators, two discriminators and one segmenter. The generator GA→B adapts images from the conventional CT domain to the CBCT/MR domain, the generator GB→A adapts the inverse way, the discriminator DB identifies real CBCT or the adapted ones from GA→B, the discriminator DA identifies real conventional CT or the adapted ones from GB→A, and the segmenter MB predicts the segmentation on adapted image from generator GA→B. There are two training paths in our APA2Seg-Net. Path A first adapts conventional CT images IA to in the CBCT/MR domain through GA→B. Then, is adapted back to the conventional CT domain as through GB→A. In parallel, is also feed into segmenter MB to generate segmentation prediction . Similarly, path B first adapts CBCT/MR images IB to in the conventional CT domain through GB→A. Then, is adapted back to the CBCT/MR domain as through GA→B.
Fig. 1.
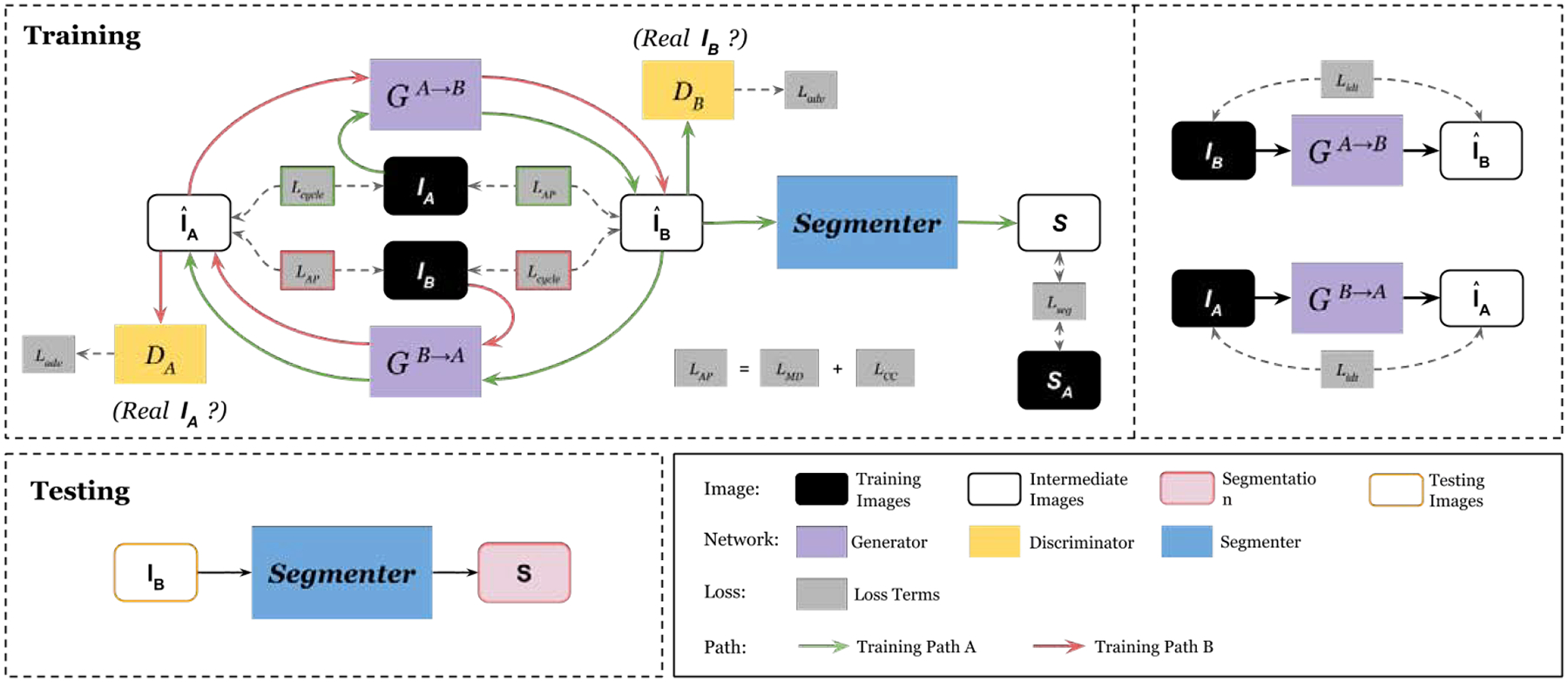
Illustration of our Anatomy-Preserving domain Adaptation to Segmentation Network (APA2Seg-Net). It consists of an anatomy-preserving domain adaptation network (left portion), and a segmenter for target domain. During test phase, the segmenter is extracted from APA2Seg-Net for predicting structural information, i.e., segmentation.
Training supervision comes from five sources:
(a) adversarial loss utilizes discriminators to classify if adapted image belong to specific domain. The adversarial objective aims to encourage G to generate adapted images that are indistinguishable to the discriminators. Two adversarial losses are introduced to train generators and discriminators:
| (1) |
| (2) |
(b) cycle-consistency loss constrains the image that returns to the original domain after passing through two generators to have minimal alternation to image content, such that a compound of two generators should be an identity mapping:
| (3) |
(c) segmentation loss on the segmentation prediction from image . The segmentation prediction should be consistent with the ground truth label from the conventional CT domain :
| (4) |
(d) identity loss regularizes the generators to be near an identity mapping when real samples of the target domain are provided. For example, if a given image looks like it is from the target domain, the generator should not map it into a different image. Therefore, the identity loss is formulated as:
| (5) |
(e) anatomy-preserving loss enforces the anatomical content is preserved before and after adaptation. Unlike conventional CycleGAN (Zhu et al., 2017) that does not use direct content constraint, we use both the MIND loss (Yang et al., 2020; Heinrich et al., 2012) and correlation coefficient loss (Ge et al., 2019) to preserve the anatomy in our unpaired domain adaptation process:
| (6) |
The first term is the correlation coefficient loss, and is formulated as:
| (7) |
where Cov is the variance operator and σ is the standard deviation operator. The second term is the MIND loss (Yang et al., 2020), and is formulated as:
| (8) |
where F is a modal-independent feature extractor defined by:
| (9) |
where Kx(I) is a distance vector of image patches around voxel x with all the neighborhood patches within a non-local region in image I. Vx(I) is the local variance at voxel x in image I. Here, dividing Kx(I) with Vx(I) aims to reduce the influence of image modality and intensity range, and Z is a normalization constant to ensure that the maximum element of Fx equals to 1. In our anatomy-preserving loss, the weight parameters are set to λcc = 1 and λmd = 1 to achieve balanced training. The anatomy-preserving loss ensures the adaptation only alter the appearance of image while maintaining the anatomical content, such that segmentation network MB can be trained correctly to recognize the anatomical content in the adapted image.
Finally, the overall objective is a weighted combination of all loss listed above:
| (10) |
where weight parameters are set to λ1 = 10 and λ2 = λ3 = λ4 = λ5 = 1 to achieve a balanced training and near-optimal performance according to our hyper-parameter search. The sub-networks’ details are shown in Figure 2. Specifically, we use a decoder-encoder network with 9 residual bottleneck for our generators, a 3-layer CNN for our discriminators. Our segmenter is a 5-level UNet with concurrent SE module Roy et al. (2018) concatenated to each level’s output.
Fig. 2.
Illustration of our segmenter, generator, and discriminator network structures in the APA2Seg-Net. 5-level U-Net with the concurrent squeeze and excitation module is used for our segmenter. An autoencoder with multiple residual bottleneck is used for our generator. The feature size shrinks in the encoder phase, stays constant in the transformer phase, and expands again in the decoder phase. The feature size of the layer outputs is listed below it, in terms of the input image size, H. On each layer is listed the number of filters, the size of those filters, and the stride.
2.2. Anatomy-guided Multimodal Registration
Using our APA2Seg-Net trained by conventional CT with liver segmentation, we can obtain CBCT and MR segmenters. Then, the segmenters are deployed in our anatomy-guided multimodal registration to guide the Robust Point Matching (RPM) machine to predict the transformation between MR and CBCT images. The registration pipeline is shown in Figure 3. The CBCT and MR segmenters from APA2Seg-Net predict the CBCT and MR segmentations. Then, we extract the surface points from the CBCT and MR segmentations and input them into the RPM.
Fig. 3.
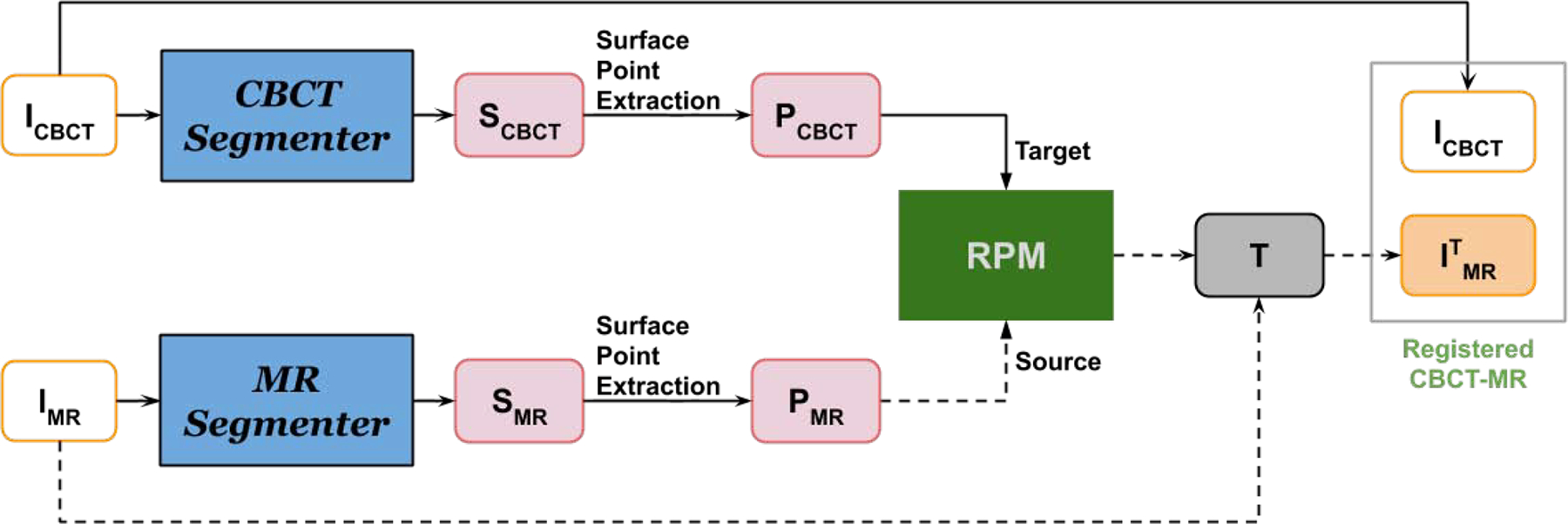
Our anatomy-guided multimodal registration pipeline. The segmenters are obtained from APA2Seg-Net in Figure 1.
RPM is a point-based registration framework based on deterministic annealing and soft assignment of correspondences between point sets (Gold et al., 1998), which is robust to point outliers. Specifically, given two point sets and , RPM aims to find the affine transformation T that best relates the two point sets. Reformulating the transform T into transformation matrix A and translation vector t form, we have T(m) = Am + t where A is composed of scale, rotation, horizontal shear, and vertical shear parameters, denoted as a, θ, b, and c, respectively. The registration cost function can be written as:
| (11) |
where μ is the point match matrix with uij = 1 if point mi corresponds to point sj and uij = 0 otherwise. The first term minimizes the distance between point sets, and the second term constrains the transformation to avoid large numbers or dramatic transformations. The third term biases the cost toward stronger point correlation by decreasing the cost function. Then, the cost function can be iteratively solved by the soft assignment algorithm. The soft assignment between point sets allows point registration with exclusion of the outlying points and avoids local minima, which fits the problem of CBCT-MR registration well, since the liver is often partially occluded in CBCT due to limited FOV. In addition, for the purpose of intraprocedural registration, RPM provides high-speed registration as it is based on points. The generated transformation T is then applied to original MR image to created registered CBCT-MR pair, which provides better visualization of tumor during image-guided intervention.
In our implementation, we extract the liver surface points from CBCT and MR segmentation for RPM. Other types of point features from segmentation can also be used in RPM, such as landmarks and skeletons.
3. Experimental Results
3.1. Data and Setup
In the conventional CT domain, we collected 131 and 20 CT volumes with liver segmentation from LiTS (Bilic et al., 2019) and CHAOS (Kavur et al., 2020), respectively. In the CBCT/MR domain, we collected 16 in-house TACE patients with both intraprocedural CBCT and pre-operative MR for our segmentation and registration evaluations. All the CBCT data were acquired using a Philips C-arm system with a reconstructed image size of 384 × 384 × 297 and voxel size of 0.65 × 0.65 × 0.65mm3. The MR data were acquired using different scanners with different spatial resolutions. Thus, we resampled all the CBCT, MR and conventional CT to an isotropic spatial resolution of 1 × 1 × 1mm3.
In the CBCT APA2Seg-Net setup, the conventional CT inputs were first randomly cropped in the axial view using a circular spherical mask on the liver region to simulate the limited FOV in CBCT, as demonstrated in Figure 4. The circular mask maintains the same cropping geometry observed in CBCT with a radius of 125mm. As a result, we obtained 13, 241 2D conventional CT images with liver segmentation, and 3, 792 2D CBCT images. In the MR APA2Seg-Net setup, both the conventional CT and MR input were zero-padded or cropped to keep an axial FOV of 410 × 410mm2. As a result, we obtained 13, 241 2D conventional CT images with liver segmentation, and 1, 128 2D MR images. All the 2D images were resized to 256 × 256 for APA2Seg-Net inputs. With 16 TACE patients in our dataset, we performed four-fold cross-validation with 12 TACE used as training and 4 patients used as testing in each validation.
Fig. 4.
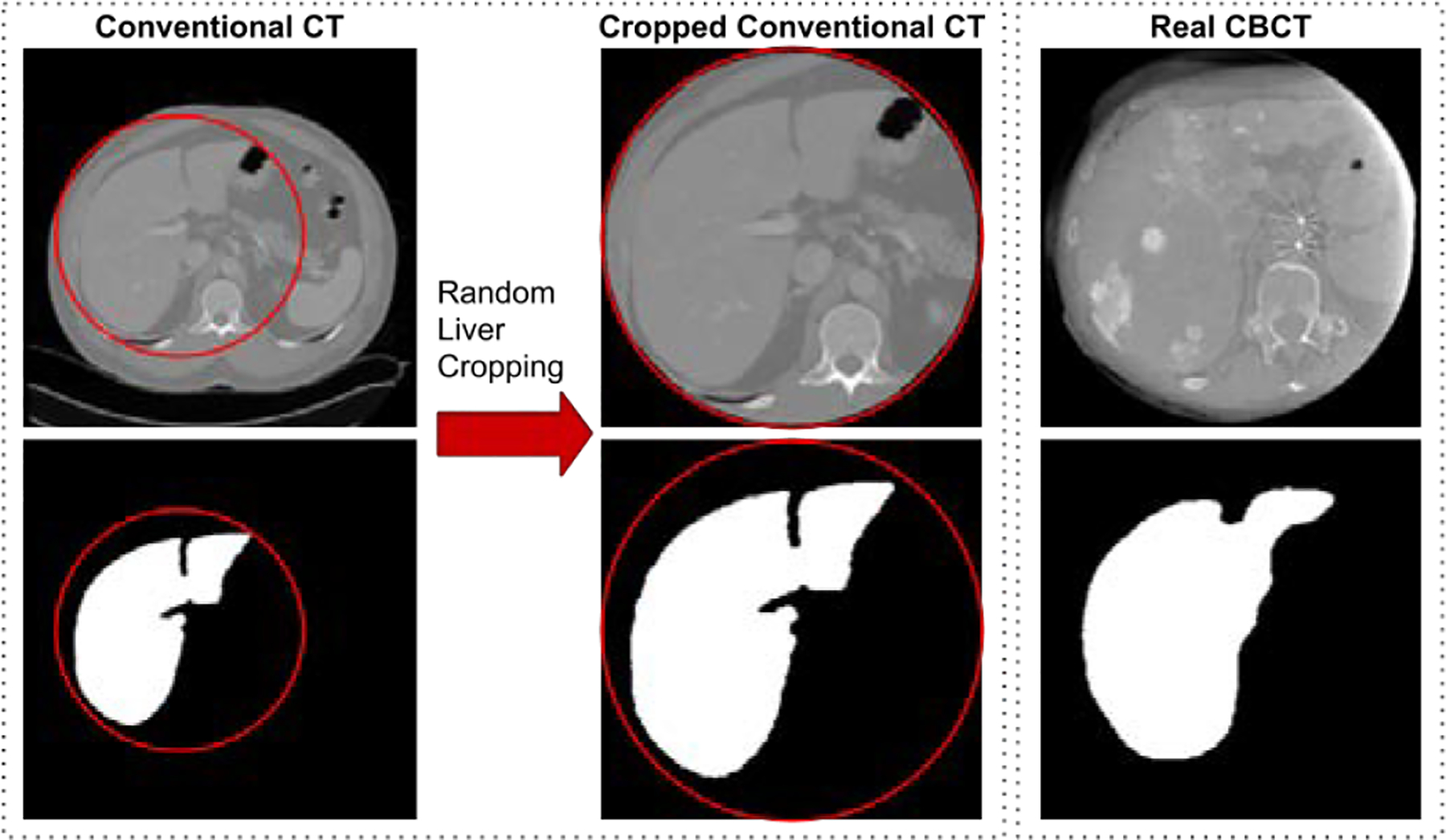
Generation of limited FOV CT from conventional CT as input for our APA2Seg-Net.
3.2. Segmentation Results
After training, we extracted the segmenters from the APA2Seg-Net for prediction of liver segmentations on both CBCT and MR. For qualitative study, we compared our segmentation performance with: i. CBCT/MR-to-CT CycleGAN concatenated with conventional CT segmenter (CycleGAN+SegCT), where the CT segmenter is trained on conventional CT images with liver annotations (SegCT); ii. APA2Seg-Net without MIND loss and CC loss for anatomy preserving constraint during the training (Ours-MD-CC); iii. APA2Seg-Net with MIND loss only for anatomy preserving constraint during the training (Ours+MD-CC); iv. APA2Seg-Net with CC loss only for anatomy preserving constraint during the training (Ours-MD+CC); v. APA2Seg-Net with both MIND loss and CC loss for anatomy preserving constraint during the training (Ours+MD+CC); and vi. the segmenter trained on target domain images with limited liver annotations (Supervised SegCBCT / SegMRI).
Qualitative comparison of CBCT segmentation results are shown in Figure 5. As we can see, CBCT in TACE suffers from limited FOV, metal artifacts, and low CNR. CycleGAN+SegCT is non-ideal because it requires adapting the input CBCT to conventional CT first, and the segmentation relies on the translated image quality. However, the unpaired and unconstrained adaption from CBCT to CT is difficult as it consists of metal artifact removal and liver boundary enhancement. The multi-stage inference in CycleGAN+SegCT aggregates the prediction error into the final segmentation. On the other hand, our APA2Seg-Net with anatomy-preserving constraint and one-stage inference mechanism achieved significantly better CBCT liver segmentation results. We found combining CC loss and MIND loss for our anatomy-preserving constraint in APA2Seg-Net yields the best results. We also found our identity loss that helps maintain the target domain feature during the adaptation process provides us better segmentation performance. Furthermore, compared to the segmenters trained on target domains using relatively limited annotation data (2844 2D images), our APA2Seg-Net trained from large-scale conventional CT data (13,241 2D images) can provide slightly better segmenters. Qualitative comparison of MR segmentation results are illustrated in Figure 6. Similar observations can be found in the MR segmentation results.
Fig. 5.
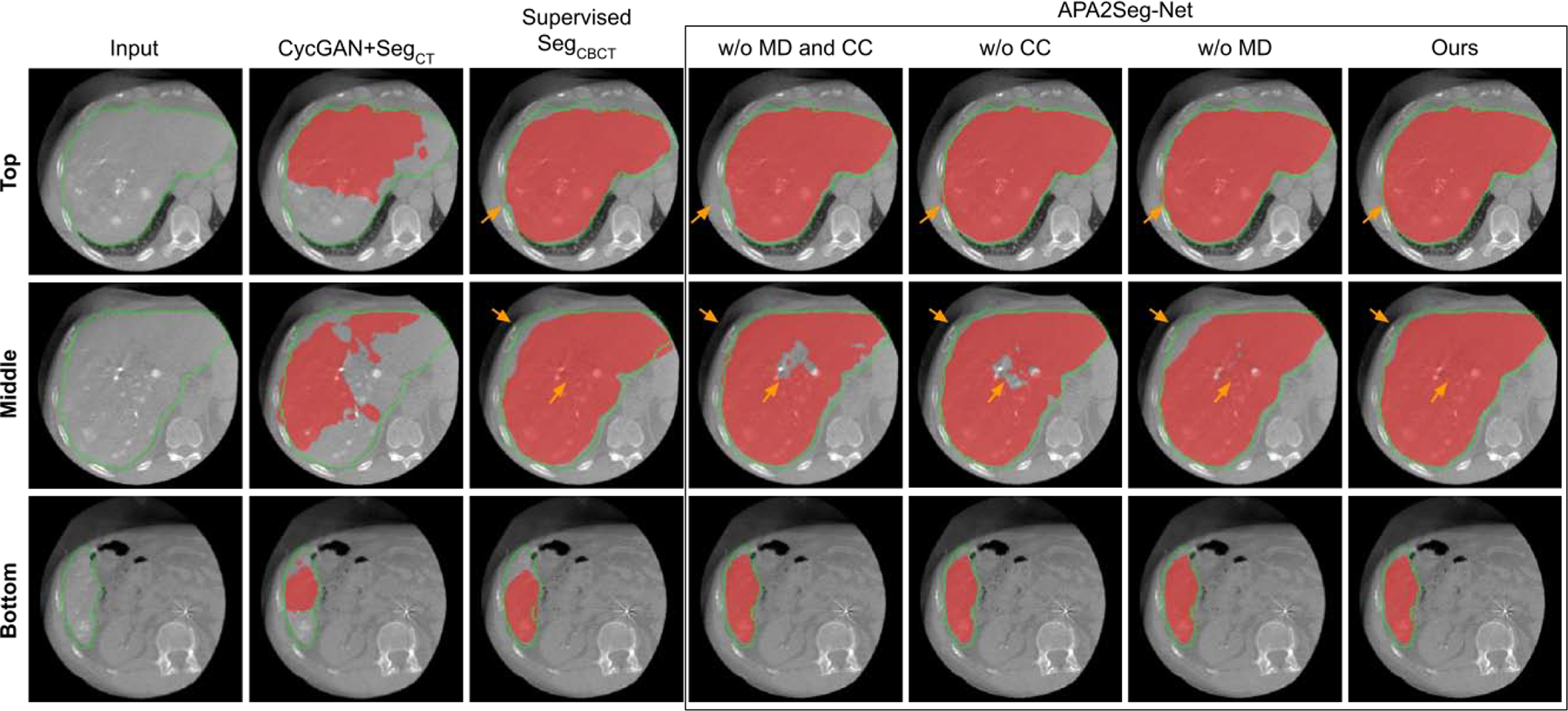
Comparison of CBCT segmentation at different liver latitudes. Red mask: liver segmentation prediction. Green contour: liver segmentation ground truth. Results on APA2Seg-Net with or without CC loss and MIND loss are shown in the box.
Fig. 6.
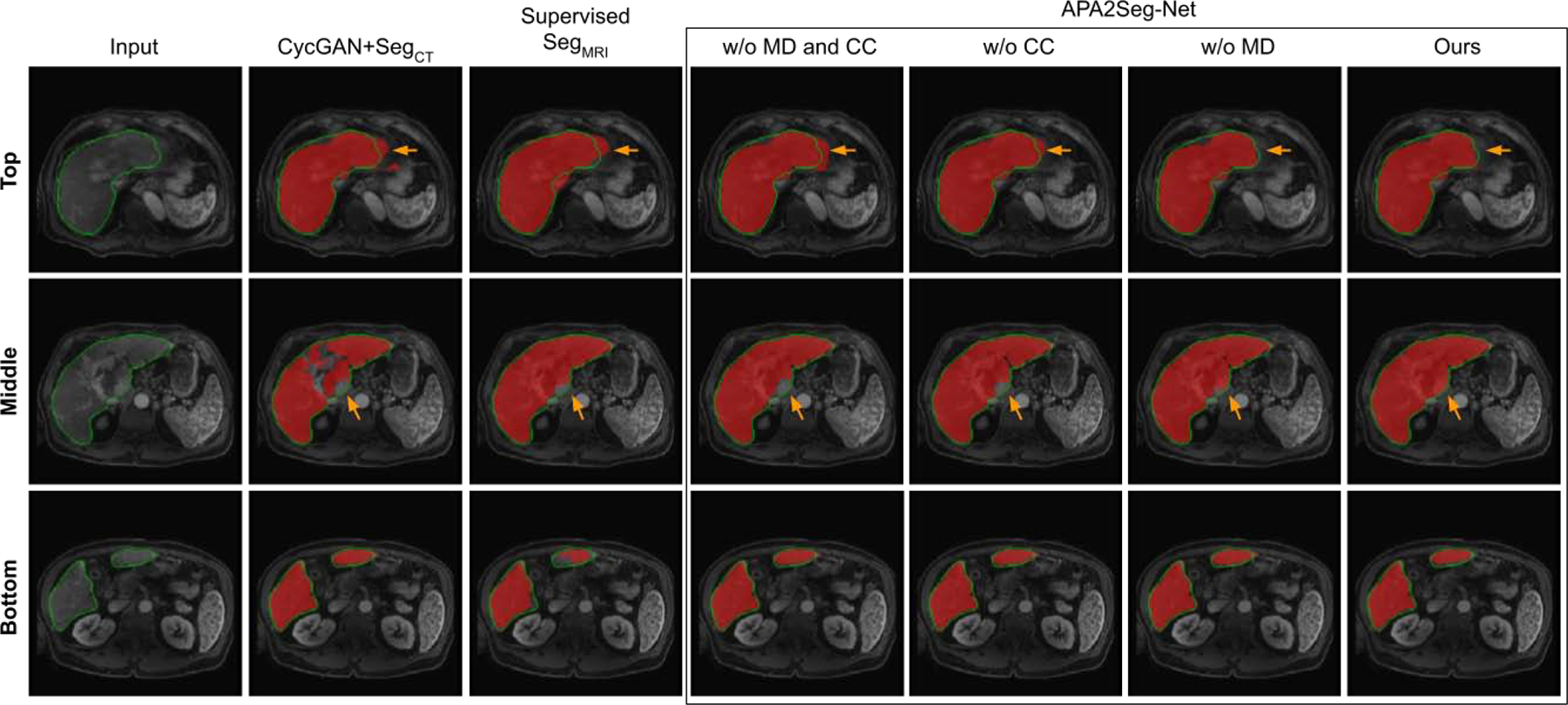
Comparison of MRI segmentation at different liver latitudes. Red mask: liver segmentation prediction. Green contour: liver segmentation ground truth. Results on APA2Seg-Net with or without CC loss and MIND loss are shown in the box.
Dice Similarity Coefficient (DSC) and Average Symmetric Surface Distance (ASD) were used to evaluate the quantitative segmentation performance. Table 1 summarizes the quantitative comparison of CBCT and MR segmentation results. As we can see, our APA2Seg-Net achieved the best CBCT and MR segmentation in terms of DSC and ASD, indicating the best overall liver segmentation.
Table 1.
Quantitative comparison of CBCT and MRI segmentation results using DSC and ASD(mm). Best results are marked in red. Underline means supervised training with ground truth segmentation on the target domain, i.e. CBCT or MRI segmentation. The negative sign ‘-’ means without the corresponding loss component in our APA2Seg-Net.
| CBCT | CycGAN+SegCT | SegCBCT | Ours-Idt | Ours-MD-CC | Ours-CC | Ours-MD | Ours |
|---|---|---|---|---|---|---|---|
| Median DSC | 0.685 | 0.882 | 0.877 | 0.870 | 0.878 | 0.887 | 0.903 |
| Mean±Std DSC | 0.695 ± 0.092 | 0.874 ± 0.035 | 0.873 ± 0.056 | 0.862 ± 0.051 | 0.871 ± 0.051 | 0.882 ± 0.038 | 0.893 ± 0.034 |
| Median ASD | 10.144 | 9.190 | 6.863 | 7.289 | 5.918 | 6.476 | 5.882 |
| Mean±Std ASD | 10.697 ± 2.079 | 10.742 ± 4.998 | 6.948 ± 2.138 | 7.459 ± 2.769 | 5.971 ± 1.823 | 6.086 ± 1.415 | 5.886 ± 1.517 |
| MRI | CycGAN+SegCT | SegMRI | Ours-Idt | Ours-MD-CC | Ours-CC | Ours-MD | Ours |
| Median DSC | 0.907 | 0.907 | 0.913 | 0.915 | 0.917 | 0.916 | 0.918 |
| Mean±Std DSC | 0.900 ± 0.044 | 0.859 ± 0.102 | 0.914 ± 0.028 | 0.912 ± 0.029 | 0.917 ± 0.026 | 0.916 ± 0.025 | 0.921 ± 0.022 |
| Median ASD | 1.632 | 2.838 | 1.619 | 1.681 | 1.498 | 1.532 | 1.491 |
| Mean±Std ASD | 2.328 ± 2.070 | 3.660 ± 2.522 | 1.932 ± 1.303 | 1.916 ± 1.142 | 1.976 ± 1.311 | 2.047 ± 1.382 | 1.860 ± 1.099 |
3.3. Registration Results
With the CBCT and MR segmenters extracted from APA2Seg-Nets, we integrate the segmenters into our anatomy-guided multimodal registration pipeline for registering MR to CBCT. The CBCT and MR liver segmentation from segmenters are inputted into RPM to generate the transformation parameters. For qualitative studies, we first compared our registration results with classical previous works of intensity-based affine registration and intensity-based B-spline registration (Wyawahare et al., 2009; Maes et al., 1997). We also compared our registration results with intensity-based affine/B-spline registration based on CT images translated from CBCT and MR using APA-Net - similar to the idea in Arar et al. (2020). Two examples are illustrated in Figure 7. The ground truth (GT) CBCT liver mask (green) and the transformed GT MR liver mask (blue) are overlaid on the CBCT image to qualitatively evaluate the registration performance. As we can observe, neither intensity-based registration methods can correctly estimate the MR transformation, while our anatomy-guided registration, as demonstrated in the last column of Figure 7, can more accurately map the MR to CBCT images. Compared to the RPM registration based on ground truth liver segmentations, our anatomy-guided registration based on APA2Seg-Net’s segmenter provides similar registration performance. Additional registration results using our method are shown in Figure 8.
Fig. 7.
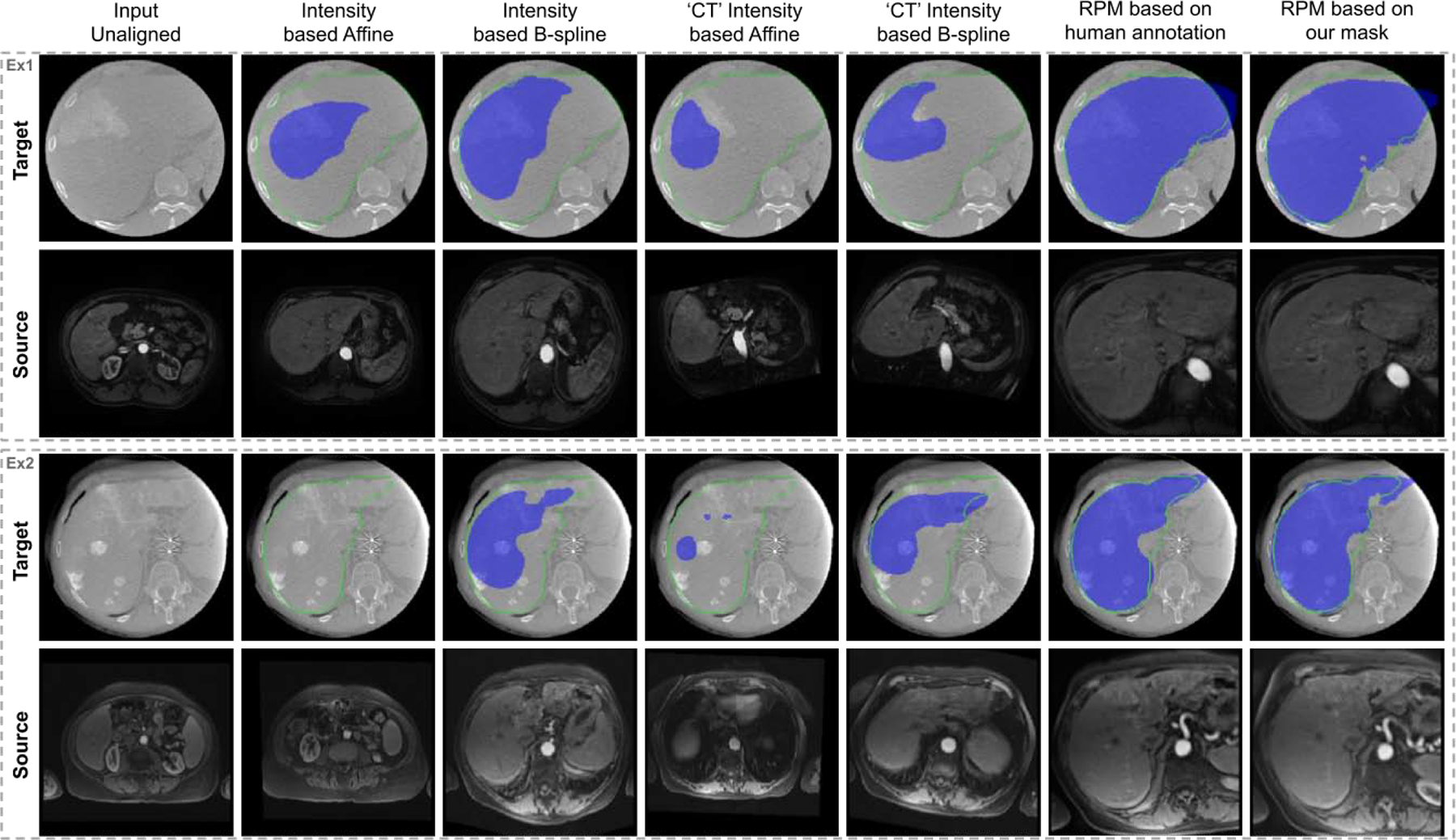
CBCT (target) and MRI (source) registration results. Deformation fields are applied on ground truth MRI liver mask and overlaid on CBCT images (blue). Ground truth CBCT liver mask (green contour) is overlaid on CBCT images as well. ‘CT’ Intensity based Affine means intensity-based affine registration based on CT images translated from CBCT and MR using APA-Net. ‘CT’ Intensity based BSpline means intensity-based BSpline registration based on CT images translated from CBCT and MR using APA-Net.
Fig. 8.
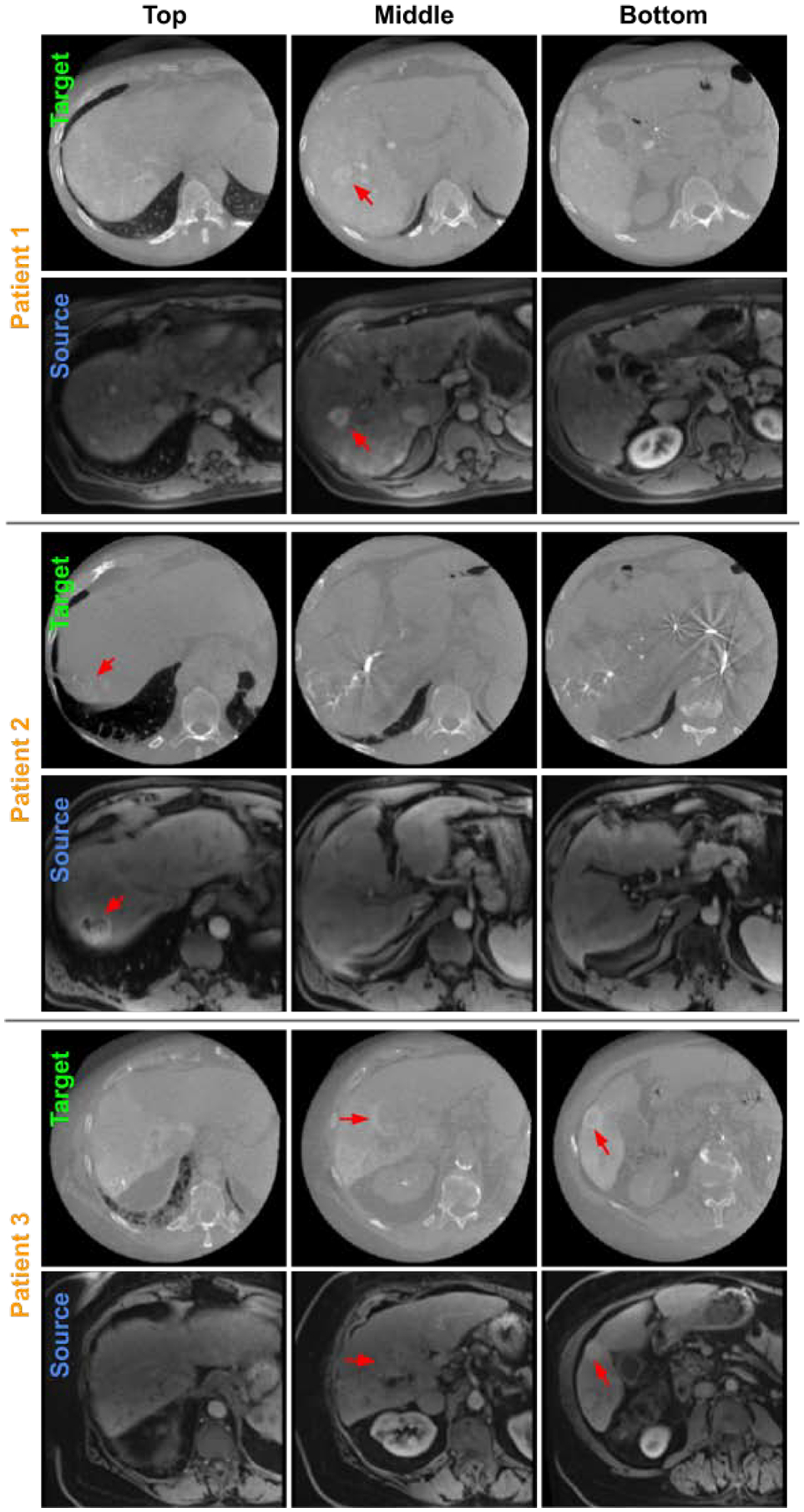
Three examples of CBCT (target) and MRI (source) registration results visualized at 3 liver latitudes. RPM registration is performed based on APA2Seg-Net segmentation. Liver tumors are located by red arrows in CBCT and registered MRI.
For quantitative registration evaluation, we first evaluated the averaged error of transformation parameters where the transformation from human annotation based RPM registration is used as ground truth. Based on 3D affine transformation equation:
| (12) |
twelve 3D affine transformation parameters were evaluated: sx, sy, sz are the scaling factors on the x,y,z directions, hxy, hxz, hyx, hyz, hzx, hzy are parameters that control the shear transformation, and Δx, Δy, and Δz are the translation on the x,y,z directions. The average errors of the parameters are reported in Table 2. As we can observe, our registration method achieves the least errors in estimating transformation parameters.
Table 2.
Quantitative comparison of average transformation parameter errors. Underline means supervised trained model using ground truth segmentation on the target domain.
| Names | Intst-Affine | ‘CT’-Intst-Affine | RPM(Ours) | RPM(Seg) |
|---|---|---|---|---|
| sx | 3.23 | 3.33 | 0.23 | 0.33 |
| sy | 0.51 | 0.62 | 0.07 | 0.15 |
| sz | 0.62 | 0.67 | 0.06 | 0.26 |
| hxy | 0.19 | 0.67 | 0.04 | 0.09 |
| hxz | 0.32 | 0.65 | 0.04 | 0.08 |
| hyx | 0.22 | 1.39 | 0.12 | 0.21 |
| hyz | 0.29 | 0.43 | 0.07 | 0.15 |
| hzx | 0.68 | 0.71 | 0.25 | 0.72 |
| hzy | 0.28 | 0.33 | 0.09 | 0.39 |
| Δx | 79.85 mm | 73.76 mm | 25.13 mm | 65.71 mm |
| Δy | 64.48 mm | 38.44 mm | 15.05 mm | 27.03 mm |
| Δz | 96.12 mm | 161.32 mm | 22.74 mm | 52.54 mm |
To further validate our registration, we computed the DSC and ASD metrics between the human annotated CBCT liver segmentation and the transformed human annotated MR liver segmentation using transformation generated from different methods. The results are summarized in Table 3. Please note that due to the limited FOV of CBCT, the liver mask in CBCT is often truncated while the liver mask in MR is intact. Therefore, the upper limit/gold standard of the metrics are not DSC=1 and ASD=0, but assumed to be the registration results based human annotated liver segmentation. As we can observe from the table, our segmenter-based RPM registration achieved a mean DSC of 0.847 that is comparable to the human annotation based RPM registration with a mean DSC of 0.853. Our anatomy-guided method is also significantly better than the intensity-based methods. In Figure 9, we visualize the case-by-case DSC/ASD differences between our method and the human annotation based registration. Our method can achieve similar registration performance across all 16 cases as compared to the human annotation based registration. A maximal DSC difference less than 0.07 and a maximal ASD difference less than 3mm can be observed. Furthermore, we compared our APA2Seg-Net segementer-based RPM registration to the target domain supervised segmenter-based RPM registration in Table 3. We found that our method also outperforms the supervised method that requires annotations on the target domains with the difference significant at p < 0.05 for both DSC and ASD.
Table 3.
Quantitative comparison of CBCT-MRI registration results using DSC and ASD(mm). Best and second best results are marked in red and blue, respectively. Underline means supervised trained model using ground truth segmentation on the target domain. IMG-BSpline and IMG-Affine mean intensity-based BSpline registration and affine registration, respectively. ‘CT’-IMG-BSpline means intensity-based BSpline registration based on CT images translated from CBCT and MR using APA-Net.
| IMG-BSpline | IMG-Affine | ‘CT’-IMG-BSpline | RPM(Human Seg) | RPM(Ours) | RPM(Seg) | |
|---|---|---|---|---|---|---|
| Median DSC | 0.366 | 0.156 | 0.332 | 0.852 | 0.848 | 0.769 |
| Mean±Std DSC | 0.401 ± 0.168 | 0.168 ± 0.049 | 0.304 ± 0.182 | 0.853 ± 0.054 | 0.844 ± 0.029 | 0.755 ± 0.101 |
| Median ASD | 25.225 | 40.365 | 28.424 | 4.921 | 5.853 | 9.347 |
| Mean±Std ASD | 25.016 ± 8.809 | 40.682 ± 7.187 | 33.645 ± 13.511 | 5.095 ± 1.934 | 5.629 ± 0.909 | 8.649 ± 2.818 |
Fig. 9.
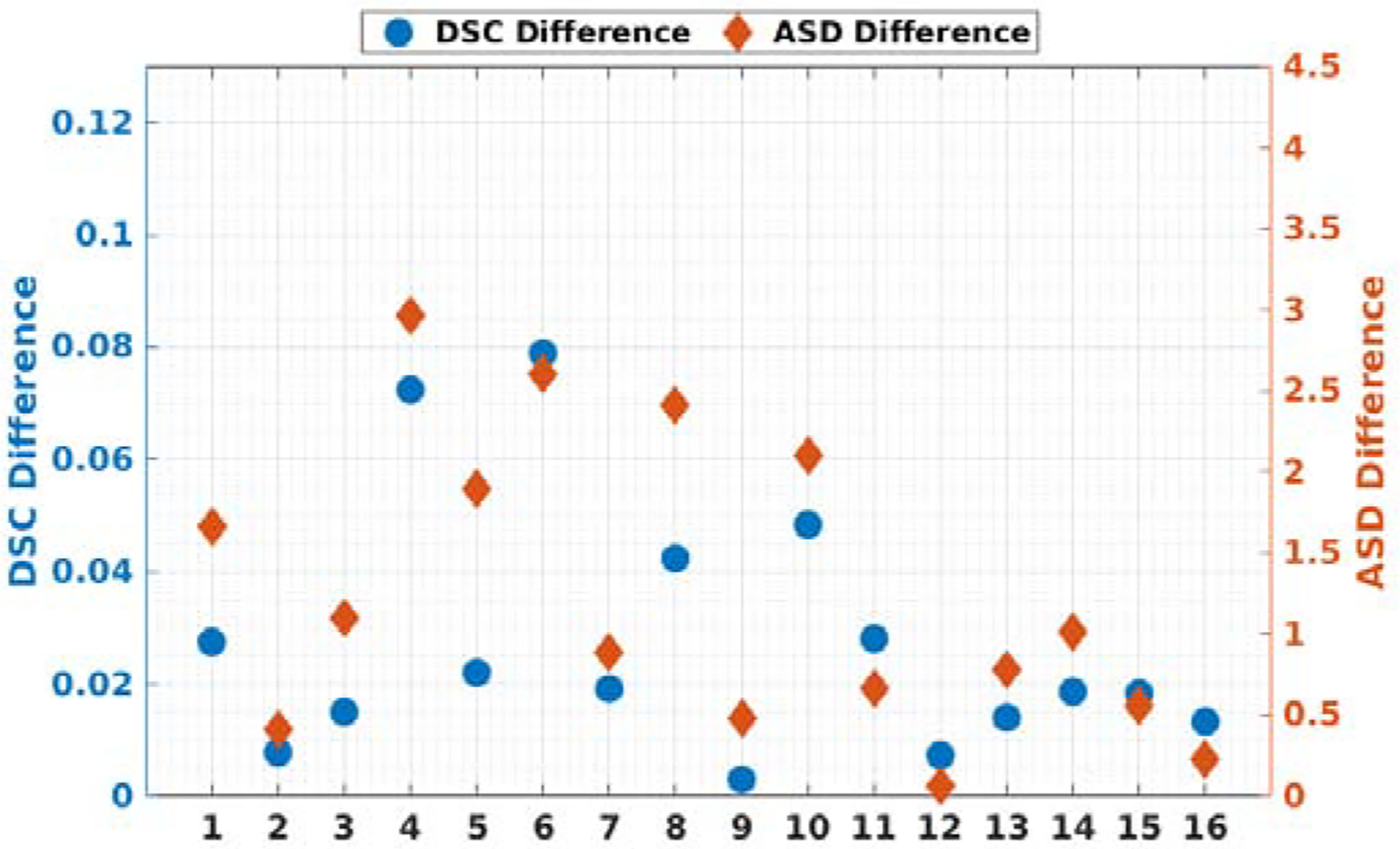
Plots of all 16 patients’ DSC and ASD differences between RPM registration based on human annotation and RPM registration based on our segmentation.
4. Conclusion and Discussion
In this work, we proposed an anatomy-guided registration framework by learning segmentation without target modality ground truth. Specifically, we developed an APA2Seg-Net to learn CBCT and MR segmenters without ground truth, which are then plugged into the anatomy-guided registration pipeline for mapping MR to CBCT. We overcame three major difficulties in multimodal image registration. First, we proposed an anatomy-based registration framework that utilizes point clouds of the segmented anatomy, instead of relying on multimodal image intensity which may have significant distribution differences. To obtain robust segmenters of target modality without ground truth, we proposed a segmentation network training scheme without using target modality ground truth, which mitigates the manual annotation requirement on the target modality. Then, we also proposed to use RPM-based point registration that is robust to partially occluded view (point outliers), a scenario commonly observed in TACE and other image-guided intervention procedures.
We demonstrated the successful application on TACE, in which pre-operative diagnostic MR are registered to intraprocedural CBCT for guiding TACE procedures. Firstly, our method achieved the superior segmentation performance even when compared to the fully supervised methods that requires annotations on the target domains. As annotating new domain data, i.e. intraprocedural CBCT, is not a clinical routine and is time-consuming, one may only obtain limited amount of labeled data for supervised training on the target domain. Thus, it cannot provide sufficient data variability for generating a robust model. On the other hand, our APA2Seg-Net utilizing large-scale conventional CT dataset offers much larger data variability, thus achieved superior segmentation performance even without using ground truth annotations from the target domain. Then, given the more robust segmenters from APA2Seg-Net, our registration pipeline based on these segmenters and RPM can also offer superior registration performance. In Table 3, our method is able to reduce the ASD between MR and CBCT liver segmentation from 4 cm based on previous intensity-based affine registration to 0.5cm, and reduce the translation difference from 9.6cm based on previous intensity-based affine registration to 2cm, as demonstrated in Table 2. With our method, the registration errors now fall within a more acceptable range. This allows MR to be more accurately registered to CBCT, reinforcing the utility of MR-derived information within the clinical TACE image guidance environment.
The presented work also has potential limitations. First of all, the CBCT segmentation performance is far from perfect with a mean DSC of 0.893. In our current APA2Seg-Net implementation, only 2D networks were considered in this study since the amount of training data is not large enough to train a robust 3D network. However, the proposed APA2Seg-Net can be extended to 3D with the expense of higher GPU memory consumption and longer computation time, which would potentially provide better segmentation results if a large amount of 3D training scans is available. As a matter of fact, Zhang et al. (2018b); Cai et al. (2019) had demonstrated the promising results from 3D synthesis and segmentation. On the other hand, our APA2Seg-Net is an open framework with flexibility in network components. While we used 2D Res-Net/Patch GAN/concurrent-SE-UNet as our generator/discriminator/segmenter, we do not claim optimality of the combination for segmentation. Other image segmentation networks, such as attention UNet (Oktay et al., 2018), multi-scale guided attention network (Sinha and Dolz, 2020), and adversarial image-to-image network (Yang et al., 2017), could be deployed in our framework and might yield better segmentation performance on different applications. Secondly, as the segmentation is imperfect from our APA2Seg-Net, it leads to difference between registration based on our segmentation and human segmentation. However, the impact of imperfect CBCT/MR segmentation is mitigated through our RPM based registration. As we can observe from Table 3 and Figure 7, the human segmentation based registration is very close to the APA2Seg-Net segmentation-based registration in terms of qualitative visualization and quantitative comparison (< 0.01 in terms of Dice). Thirdly, we considered affine transformation in our CBCT-MR liver registration, and incorporating non-rigid registration could potentially provide more accurate internal structure mappings. However, liver in CBCT is often truncated due to limited FOV. Therefore, using non-rigid registration would lead to incorrect matching on the truncated boundary. Future works on incorporating non-rigid registration while rejecting the point outliers outside of FOV is needed. Lastly, we evaluated the registration performance on the entire liver, while registration performance on other important landmarks, such as tumor location, is not included here. We will evaluate other landmark’s alignment in our future clinical feasibility studies.
The design of our anatomy-guided registration framework by learning segmentation without ground truth also suggests several interesting topics for future studies. First of all, our method could be adapted to other multimodal registration tasks that conventional registration techniques are not applicable, such as MR-Ultrasound (US) registration for neurosurgery (Rivaz et al., 2014) and prostate interventions (Hu et al., 2012), where occluded FOV and intensity inhomogeneity is often observed in US. There are several public datasets containing MR brain tumor and MR prostate with ground truth segmentation Simpson et al. (2019), which make it possible to adapt our method to these applications. More specifically, we could use APA2Seg-Net to obtain the US and MR segmenters, which are then embedded into our anatomy-guided registration pipeline for real-time MR-US alignments. Similar to our idea in registration, Sultana et al. (2019) recently proposed a prostate US-PET/CT registration algorithm based on segmentation for dose planning Dréan et al. (2016), in which our APA2Seg-Net could potentially provide the US and PET/CT prostate segmenter as well. Secondly, our method could also be adapted to landmark-based registration tasks. While anatomy-guided registration framework based on segmentation is demonstrated in this work, the segmenter in APA2Seg-Net could be replaced with a detector for learning keypoint detection without ground truth on target domain. Then, the keypoint detector could also be embedded into our anatomy-guided registration pipeline for keypoint based alignments.
In summary, we proposed an anatomy-guided registration framework by learning segmentation without target modality ground truth based on APA2Seg-Net. We demonstrated the successful application on intraprocedural CBCT-MR liver registration. In the future, we will assess the tumor-of-interest’s registration accuracy and evaluate the clinical impact of real-time intraprocedural MR-CBCT liver registration.
Highlights.
We proposed an anatomy-guided multimodal registration framework and demonstrated the successful application to CBCT-MR liver registration.
For obtaining the anatomic structure information from CBCT/MR, we proposed an anatomy-preserving domain adaptation to segmentation network (APA2Seg-Net) that allows us to learn segmentation without ground truth.
With the CBCT/MR segmenters obtained from APA2Seg-Net, we proposed a registration pipeline that allows MR and MR-derived information to be accurately registered to CBCT liver to facilitate the TACE procedure.
We believe our design can be extended to similar image-guided intervention procedures, when standard registration techniques are limited due to the non-ideal image quality conditions.
Acknowledgments
This work was supported by funding from the National Institutes of Health/ National Cancer Institute (NIH/NCI) under grant number R01CA206180. BZ was supported by the Biomedical Engineering Ph.D. fellowship from Yale University.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Al-Saleh M, Jaremko J, Alsufyani N, Jibri Z, Lai H, Major P, 2015. Assessing the reliability of mri-cbct image registration to visualize temporomandibular joints. Dentomaxillofacial Radiology 44, 20140244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Saleh MA, Alsufyani N, Lai H, Lagravere M, Jaremko JL, Major PW, 2017. Usefulness of mri-cbct image registration in the evaluation of temporomandibular joint internal derangement by novice examiners. Oral surgery, oral medicine, oral pathology and oral radiology 123, 249–256. [DOI] [PubMed] [Google Scholar]
- Arar M, Ginger Y, Danon D, Bermano AH, Cohen-Or D, 2020. Unsupervised multi-modal image registration via geometry preserving image-to-image translation, in: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 13410–13419. [Google Scholar]
- Augenfeld Z, Lin M, Chapiro J, Duncan J, 2020. Automatic multimodal registration via intraprocedural cone-beam ct segmentation using mri distance maps, in: 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI 2020), IEEE. pp. 1217–1220. [Google Scholar]
- Avants BB, Epstein CL, Grossman M, Gee JC, 2008. Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain. Medical image analysis 12, 26–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balakrishnan G, Zhao A, Sabuncu MR, Guttag J, Dalca AV, 2019. Voxelmorph: a learning framework for deformable medical image registration. IEEE transactions on medical imaging 38, 1788–1800. [DOI] [PubMed] [Google Scholar]
- Bilic P, Christ PF, Vorontsov E, Chlebus G, Chen H, Dou Q, Fu CW, Han X, Heng PA, Hesser J, et al. , 2019. The liver tumor segmentation benchmark (lits). arXiv preprint arXiv:1901.04056. [Google Scholar]
- Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A, 2018. Global cancer statistics 2018: Globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: a cancer journal for clinicians 68, 394–424. [DOI] [PubMed] [Google Scholar]
- Cai J, Zhang Z, Cui L, Zheng Y, Yang L, 2019. Towards cross-modal organ translation and segmentation: A cycle-and shape-consistent generative adversarial network. Medical image analysis 52, 174–184. [DOI] [PubMed] [Google Scholar]
- Dréan G, Acosta O, Lafond C, Simon A, de Crevoisier R, Haigron P, 2016. Interindividual registration and dose mapping for voxelwise population analysis of rectal toxicity in prostate cancer radiotherapy. Medical Physics 43, 2721–2730. [DOI] [PubMed] [Google Scholar]
- Ge Y, Xue Z, Cao T, Liao S, 2019. Unpaired whole-body mr to ct synthesis with correlation coefficient constrained adversarial learning, in: Medical Imaging 2019: Image Processing, International Society for Optics and Photonics. p. 1094905. [Google Scholar]
- Gold S, Rangarajan A, Lu CP, Pappu S, Mjolsness E, 1998. New algorithms for 2d and 3d point matching: Pose estimation and correspondence. Pattern recognition 31, 1019–1031. [Google Scholar]
- Heinrich MP, Jenkinson M, Bhushan M, Matin T, Gleeson FV, Brady M, Schnabel JA, 2012. Mind: Modality independent neighbourhood descriptor for multi-modal deformable registration. Medical image analysis 16, 1423–1435. [DOI] [PubMed] [Google Scholar]
- Hu Y, Ahmed HU, Taylor Z, Allen C, Emberton M, Hawkes D, Barratt D, 2012. Mr to ultrasound registration for image-guided prostate interventions. Medical image analysis 16, 687–703. [DOI] [PubMed] [Google Scholar]
- Hu Y, Modat M, Gibson E, Li W, Ghavami N, Bonmati E, Wang G, Bandula S, Moore CM, Emberton M, et al. , 2018. Weakly-supervised convolutional neural networks for multimodal image registration. Medical image analysis 49, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo Y, Xu Z, Bao S, Assad A, Abramson RG, Landman BA, 2018. Adversarial synthesis learning enables segmentation without target modality ground truth, in: 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), IEEE. pp. 1217–1220. [Google Scholar]
- Kavur AE, Gezer NS, Barış M, Conze PH, Groza V, Pham DD, Chat-terjee S, Ernst P, Ӧzkan S, Baydar B, et al. , 2020. Chaos challenge– combined (ct-mr) healthy abdominal organ segmentation. arXiv preprint arXiv:2001.06535. [DOI] [PubMed] [Google Scholar]
- Lee MC, Oktay O, Schuh A, Schaap M, Glocker B, 2019. Image-and-spatial transformer networks for structure-guided image registration, in: International Conference on Medical Image Computing and Computer-Assisted Intervention, Springer. pp. 337–345. [Google Scholar]
- Maes F, Collignon A, Vandermeulen D, Marchal G, Suetens P, 1997. Multimodality image registration by maximization of mutual information. IEEE transactions on Medical Imaging 16, 187–198. [DOI] [PubMed] [Google Scholar]
- Mok TC, Chung A, 2020. Fast symmetric diffeomorphic image registration with convolutional neural networks, in: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 4644–4653. [Google Scholar]
- Oktay O, Schlemper J, Folgoc LL, Lee M, Heinrich M, Misawa K, Mori K, McDonagh S, Hammerla NY, Kainz B, et al. , 2018. Attention u-net: Learning where to look for the pancreas. arXiv preprint arXiv:1804.03999. [Google Scholar]
- Park S, Plishker W, Quon H, Wong J, Shekhar R, Lee J, 2017. Deformable registration of ct and cone-beam ct with local intensity matching. Physics in Medicine & Biology 62, 927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pung L, Ahmad M, Mueller K, Rosenberg J, Stave C, Hwang GL, Shah R, Kothary N, 2017. The role of cone-beam ct in transcatheter arterial chemoembolization for hepatocellular carcinoma: a systematic review and meta-analysis. Journal of Vascular and Interventional Radiology 28, 334–341. [DOI] [PubMed] [Google Scholar]
- Qin C, Shi B, Liao R, Mansi T, Rueckert D, Kamen A, 2019. Unsupervised deformable registration for multi-modal images via disentangled representations, in: International Conference on Information Processing in Medical Imaging, Springer. pp. 249–261. [Google Scholar]
- Rivaz H, Chen SJS, Collins DL, 2014. Automatic deformable mr-ultrasound registration for image-guided neurosurgery. IEEE transactions on medical imaging 34, 366–380. [DOI] [PubMed] [Google Scholar]
- Rohr K, Stiehl HS, Sprengel R, Buzug TM, Weese J, Kuhn M, 2001. Landmark-based elastic registration using approximating thin-plate splines. IEEE Transactions on medical imaging 20, 526–534. [DOI] [PubMed] [Google Scholar]
- Roy AG, Navab N, Wachinger C, 2018. Concurrent spatial and channel squeeze & excitationin fully convolutional networks, in: International Conference on Medical Image Computing and Computer-Assisted Intervention, Springer. pp. 421–429. [Google Scholar]
- Simpson AL, Antonelli M, Bakas S, Bilello M, Farahani K, Van Ginneken B, Kopp-Schneider A, Landman BA, Litjens G, Menze B, et al. , 2019. A large annotated medical image dataset for the development and evaluation of segmentation algorithms. arXiv preprint arXiv:1902.09063. [Google Scholar]
- Sinha A, Dolz J, 2020. Multi-scale self-guided attention for medical image segmentation. IEEE Journal of Biomedical and Health Informatics. [DOI] [PubMed] [Google Scholar]
- Solbiati M, Passera KM, Goldberg SN, Rotilio A, Ierace T, Pedicini V, Poretti D, Solbiati L, 2018. A novel ct to cone-beam ct registration method enables immediate real-time intraprocedural three-dimensional assessment of ablative treatments of liver malignancies. Cardiovascular and interventional radiology 41, 1049–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultana S, Song DY, Lee J, 2019. Deformable registration of pet/ct and ultrasound for disease-targeted focal prostate brachytherapy. Journal of Medical Imaging 6, 035003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tacher V, Radaelli A, Lin M, Geschwind JF, 2015. How i do it: cone-beam ct during transarterial chemoembolization for liver cancer. Radiology 274, 320–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Zhang M, 2020. Deepflash: An efficient network for learning-based medical image registration, in: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 4444–4452. [Google Scholar]
- Wyawahare MV, Patil PM, Abhyankar HK, et al. , 2009. Image registration techniques: an overview. International Journal of Signal Processing, Image Processing and Pattern Recognition 2, 11–28. [Google Scholar]
- Yang D, Xu D, Zhou SK, Georgescu B, Chen M, Grbic S, Metaxas D, Comaniciu D, 2017. Automatic liver segmentation using an adversarial image-to-image network, in: International Conference on Medical Image Computing and Computer-Assisted Intervention, Springer. pp. 507–515. [Google Scholar]
- Yang H, Sun J, Carass A, Zhao C, Lee J, Prince JL, Xu Z, 2020. Unsupervised mr-to-ct synthesis using structure-constrained cyclegan. IEEE Transactions on Medical Imaging. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Miao S, Mansi T, Liao R, 2018a. Task driven generative modeling for unsupervised domain adaptation: Application to x-ray image segmentation, in: International Conference on Medical Image Computing and Computer-Assisted Intervention, Springer. pp. 599–607. [Google Scholar]
- Zhang Z, Yang L, Zheng Y, 2018b. Translating and segmenting multimodal medical volumes with cycle-and shape-consistency generative adversarial network, in: Proceedings of the IEEE conference on computer vision and pattern recognition, pp. 9242–9251. [Google Scholar]
- Zhen X, Gu X, Yan H, Zhou L, Jia X, Jiang SB, 2012. Ct to cone-beam ct deformable registration with simultaneous intensity correction. Physics in Medicine & Biology 57, 6807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B, Tsai YJ, Liu C, 2020a. Simultaneous denoising and motion estimation for low-dose gated pet using a siamese adversarial network with gate-to-gate consistency learning, in: International Conference on Medical Image Computing and Computer-Assisted Intervention, Springer. pp. 743–752. [Google Scholar]
- Zhou SK, Greenspan H, Davatzikos C, Duncan JS, van Ginneken B, Madabhushi A, Prince JL, Rueckert D, Summers RM, 2020b. A review of deep learning in medical imaging: Image traits, technology trends, case studies with progress highlights, and future promises. ArXiv preprint arXiv:2008.09104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JY, Park T, Isola P, Efros AA, 2017. Unpaired image-to-image translation using cycle-consistent adversarial networks, in: Proceedings of the IEEE international conference on computer vision, pp. 2223–2232. [Google Scholar]


