Abstract
Regulation of transcription is a tightly choreographed process. The establishment of RNA polymerase II promoter proximal pausing soon after transcription initiation and the release of Pol II into productive elongation are key regulatory processes that occur in early elongation. We describe the techniques and tools that have become available for the study of promoter proximal pausing and their utility for future experiments. We then provide an overview of the factors and interactions that govern a multipartite pausing process and address emerging questions surrounding the mechanism of RNA polymerase II’s subsequent advancement into the gene body. Finally, we address remaining controversies and future areas of study.
Keywords: Pol II, DSIF, NELF, TFIID, P-TEFb
Introduction
Eukaryotic transcription is a highly regulated process that requires precise temporal and spatial coordination of a multitude of factors at the initiation, elongation, and termination stages. Promoter proximal pausing of RNA polymerase II (Pol II) has emerged as a significant step in the canonical transcription cycle. In metazoans, transcription initiation is followed by an accumulation of Pol II ~30–60 nt downstream of the transcription start site[1]. This step is not present in Saccharomyces cerevisiae and Caenorhabditis elegans, likely owing to the absence of the key negative elongation factor (NELF). The first evidence of metazoan Pol II pausing in vivo emerged when the Chambon lab, using a nuclear run-on assay, observed a concentration of Pol II at the 5′end of the beta-globin gene in nuclei from mature hen erythrocytes that were anticipated to be transcriptionally silent[2]. Similar phenomena were later observed on mammalian c-myc[3–5], HIV-1 [6] and on non-induced Drosophila heat shock genes[7, 8]. The study of the Drosophila hsp70 heat shock gene by Gilmour and Lis was particularly groundbreaking. Using protein-DNA crosslinking with UV light, a predecessor to the widely used chromatin immunoprecipitation techniques of today, they determined that a single Pol II molecule associates with the region between −12 and +65 on the non-induced hsp70 gene[7]. Subsequent work by Rougvie and Lis determined that the Pol II at the 5′end of genes in Drosophila is transcriptionally engaged[8] and subsequent permanganate footprinting analyses in living cells revealed the transcription bubble associated with these Pol II[9]. The question remained, however, as to whether the paused Pol II was an idiosyncrasy of a small number of genes or a more general phenomenon. The advent of genomic methods provided resounding support for the latter, leading to the notion that promoter proximal pausing is a ubiquitous step in the transcription cycle for most if not all protein encoding genes in mammals and Drosophila[10–13]. Pausing has been linked to a number of regulatory functions, including developmental control[12, 14–16] and the maintenance of transcriptionally permissive, nucleosome-free regions around promoters[17, 18]. Indeed, it is this latter observation that can explain the seemingly contradictory finding that depletion of the pausing factor NELF results in a decrease rather than an increase in expression of some genes[18].
Promoter proximal pausing requires at least two factors which function cooperatively: DRB sensitivity-inducing factor (DSIF) and NELF[19–22]. The cyclin dependent kinase positive transcription elongation factor b (P-TEFb) appears to facilitate pause release by phosphorylating Pol II, DSIF, and NELF, resulting in the dissociation of NELF from the elongation complex and the transition of DSIF from a negative elongation factor to a positive elongation factor[21, 23, 24]. Extensive genome-wide and biochemical studies have provided a general framework for how pausing occurs, yet many of the underlying mechanisms remain an area of inquiry. First, the exact mechanism through which DSIF and NELF promote pausing, particularly with regards to the role of DSIF, has yet to be fully elucidated. Several factors in addition to NELF and DSIF have been implicated in pausing, most notable among them transcription factor II D (TFIID), GAGA factor, and RNA polymerase II-associated factor 1 complex (PAF1C), yet the exact role of each of these factors and the nature of their interactions with the paused complex requires further study. Finally, additional considerations, such as the role of DNA/RNA sequence, the duration and stability of the pause, and the relationship between Pol II pausing and chromatin architecture remain areas of continuing or emerging research and even debate.
Promoter proximal pausing of Pol II is governed by several intrinsic and extrinsic elements. Increasing evidence points to DNA and RNA sequence playing a role in regulating promoter proximal pausing. Enrichment of promoter elements and pause motifs, as well as GC content, likely influence the stability and efficiency of the pause[25–27]. Experiments in Drosophila show that disrupting downstream promoter elements can shift the location of the pause on the hsp70 gene, suggesting that pause position is heavily reliant on DNA sequence, but it is unclear whether this result can be widely extrapolated for other promoters[28]. Indeed, contrasting evidence has shown that the location of the pause depends on the kinetics of the elongating Pol II; lowering nucleotide concentrations or replacing the wild type Pol II with a slowly elongating mutant shifts the location of the pause upstream[29]. Thus, the role of intrinsic Pol II-nucleic acid interactions in pausing remains a topic of ongoing controversy.
Extrinsic interactions, such as those between Pol II and DSIF, NELF, and P-TEFb, as well as between Pol II and accessory factors such as TFIID and others play critical roles in establishing the pause and governing the dynamics of pause stability and pause release. Interactions between the nucleic acid scaffold and DSIF and NELF are also likely significant contributors to the pausing mechanism. Moreover, since the discovery of Pol II pausing, the role of chromatin architecture has been a subject of controversy. Of particular interest has been the +1 nucleosome, which may play a significant role in enhancing Pol II pausing in humans[30], but likely affects Drosophila Pol II promoter proximal pausing in a promoter-specific manner[31].
Taken together, the current body of knowledge about Pol II promoter proximal pausing suggests a multipartite mechanism that relies on the interplay between intrinsic and extrinsic interactions, as well as on the promoter and chromatin context of the pause. This review distinguishes itself from other recent reviews on Pol II elongation and promoter proximal pausing in several ways[32–36]. First, we provide an overview of the broad range of the tools available for studying promoter proximal pausing, including in vivo, cell free, and genomic techniques. Furthermore, we describe the factors involved in promoter proximal pausing with a particular emphasis on the structural and molecular mechanisms of DSIF and NELF function. We also discuss the emerging evidence that suggests TFIID plays a significant and context-dependent role as a regulator of promoter proximal pausing. Additionally, we discuss and attempt to resolve some of the controversies regarding the role of chromatin architecture in promoter proximal pausing, with an emphasis on the differences between human and Drosophila systems. Finally, we provide a holistic model for Pol II pausing that illustrates the various protein factors implicated in pausing and their interactions and reconciles some of the apparent contradictions in the current body of literature.
Tools to study pausing
In the almost three decades since the discovery of promoter proximal pausing, numerous tools have emerged to facilitate its study. Genome-wide techniques like ChIP-seq[37] and the higher resolution ChIP-exo[38, 39] and ChIP-nexus[40, 41] have emerged as powerful tools that can be utilized to determine the location of Pol II and its binding partners. These techniques involve crosslinking proteins to DNA in vivo followed by immunoprecipitation of protein-DNA adducts. ChIP-exo and ChIP-nexus include a digestion step with lambda exonuclease, allowing near-nucleotide resolution of the borders of the DNA-bound protein[38–41]. Permanganate footprinting as well as genome-wide permanganate ChIP-seq[9, 29, 31, 42] detect transcription bubbles associated with transcriptionally engaged Pol II. Staining of Drosophila polytene chromosomes has proved a useful technique for observing the effects of perturbations on Pol II, NELF and DSIF association with chromosomes[21, 43].
A shortcoming of these procedures is that they do not reveal which strand Pol II is transcribing. Strand-specific information is provided by several methods that monitor the 3′ ends of nascent transcripts. These approaches fall into two classes. One class involves isolating nuclei and allowing transcriptionally engaged Pol II molecules to resume elongation; the second class involves isolating nascent transcripts. Global run-on sequencing (GRO-seq)[13] and precision run-on sequencing (PRO-seq)[28] facilitate precise mapping of Pol II that is transcriptionally engaged, allowing the generation of moderate- and high-resolution pausing profiles. These two methods involve prompting paused Pol II to resume elongation following treatment of isolated nuclei with sarkosyl, which dissociates NELF and DSIF from the paused Pol II. Both methods map the location at which Pol II stops transcribing during the run-on reaction[13, 28]. PRO-seq provides notably better assessment of the location of Pol II on DNA in isolated nuclei than GRO-seq because because the run-on reaction is performed in the presence of biotinylated nucleotides, which dramatically slow further elongation once incorporated at the 3’ end of the nascent transcript[28]. In contrast, GRO-seq involves incorporation of bromodeoxyuridine and the distance the Pol II transcribes upon addition of sarkosyl is limited by the duration of the run-on reaction[13]. Incorporation of either nucleotide analog into the RNA provides an affinity tag for purification of nascent transcripts and subsequent sequencing from the 3′ end. Recently, a simplified version of PRO-seq called ChRO-seq was introduced. This technique bypasses the nuclear isolation step and instead involves lysing cells in the presence of urea and detergent to facilitate the isolation of chromatin. Poll II elongation complexes remain intact and can be prompted to resume elongation in the presence of biotinylated nucleotides as is done for PRO-seq[44].
Recent results indicate that the accuracy of Pol II mapping by run-on reactions in cells may depend on the method of nuclear isolation. Pol II has the potential to creep when nuclei are isolated in the presence of magnesium due to the non-biotinylated nucleotides present in cell lysates[45]. As a potential resolution to this, the Price lab has developed a “nuclear walk-on assay” in which nuclei are rapidly isolated from cells in the presence of EDTA, halting incorporation of NTPs and facilitating more accurate Pol II mapping[45, 46]. Nuclei isolated using this rapid method can also be incubated with α-32P-CTP to radiolabel nascent transcripts, allowing a quantitative assessment of promoter proximal pausing[46, 47].
The second class of techniques that provide strand specific information relies on the isolation of nascent transcripts. These techniques rely on immunoprecipitation of Pol II and its associated nascent transcript (NET-seq) [48, 49] or isolation of nascent transcripts stably bound to chromatin [50–53]. Unlike the run-on assays, these nascent transcript mapping techniques do not require Pol II to resume elongation; this allows the detection of Pol II molecules that are not transcriptionally competent, such as those that have backtracked into an arrested state[50]. Furthermore, techniques that rely on Pol II immunoprecipitation, like NET-seq, can take advantage of the multiple phosphorylation-specific antibodies available against Pol II to select for certain post-translational modifications[49].
The methods for monitoring Pol II behavior based on the associated transcript allow for high resolution mapping and strand specificity. They also tend to offer a wider dynamic range for quantifying levels of Pol II in specific regions than ChIP-based methods. However, these techniques do have an important shortcoming. Transcript mapping methods are unable to monitor Pol II in preinitiation complexes due to the absence of an associated nascent transcript. They are also unable to monitor transcriptionally engaged Pol II located within the first ~20 nucleotides of the transcription start site because these sequences are too short to be uniquely mapped to the genome. Thus, a combination of these approaches is worth considering when investigating the effects of inhibitors or depletion of specific cellular factors. For example, ChIP-nexus allows detection of the Pol II that becomes trapped in a preinitiation complex when cells are treated with the transcription initiation inhibitor triptolide[40].
Pol II mapping techniques have facilitated the quantification of promoter proximal pausing. To do this, researchers calculate the ratio of the level of Pol II in the promoter region to the level of Pol II in the gene body (pausing index)[12, 13] or its reciprocal (traveling ratio)[54]. These ratios provide values that can be used to sort genes and identify correlations with other genomic features. This type of quantification also allows evaluation of the effects of experimental perturbations such as RNAi-mediated depletion of protein factors on promoter proximal pausing. However, the changes that might occur must be judged with caution because the pausing index depends on both the numerator and the denominator. A perturbation causing a decrease in the Pol II level in the promoter region (numerator) accompanied by an increase in the body of the gene (denominator) could be attributed to a decrease in the efficiency of promoter proximal pausing. Alternatively, the perturbation could be slowing the rate of elongation in the body of the gene, which would in turn result in a buildup of Pol II in the gene body. This scenario at least in part appears to be the case when PAF1C is depleted from mouse myoblasts[55]. Hence, the assessment of the changes in traveling ratio must be accompanied by assessments of changes in mRNA levels, which provide a measure of productive elongation and elongation rates.
The pausing index is also highly sensitive to perturbations affecting initiation and premature termination rates. Altering initiation frequency influences the level of Pol II in the promoter region (numerator), because the zone defined as the promoter encompasses initiating Pol II, paused Pol II, and elongating Pol II. The hsp70 gene of Drosophila illustrates this complication. Early analyses indicated that there is an average of one molecule of paused Pol II per hsp70 gene prior to heat shock [7, 8]. Upon heat shock induction of hsp70, there is marked increase in the level of Pol II detected in the promoter proximal region[56], yet much of this is upstream and downstream from the pause[57]. Increased levels of premature termination downstream from the pause can result in decreased levels of Pol II in the promoter region (numerator) without the accompanying increase of Pol II in the gene body (denominator), resulting in an artificially deflated pausing index and giving the misleading impression that Pol II is being efficiently released into productive elongation. Finally, since the density of Pol II in the body of the gene is often significantly lower than in the promoter region, nonspecific background inherent to the technique being employed can dampen changes that might otherwise be occurring in the body of the gene, particularly in the case of lowly transcribed genes.
A recent technique developed by the Zeitlinger lab, reporter-ChIP-nexus, uses high resolution ChIP-nexus to capture endogenous Pol II pausing on transfected plasmids in Drosophila cells. This tool is particularly powerful because it facilitates the analysis of the effects of sequence changes on Pol II pausing without the need to edit endogenous sequences, which can be both costly and deleterious to the system[27].
In addition to techniques that capture Pol II pausing in vivo, a number of cell-free systems have been developed. Early experiments by the Price lab used a pulse/chase transcription system with preinitiation complexes reconstituted on immobilized DNA templates to show both pausing during early elongation and a regulated transition from pausing to productive elongation. The Price lab was also able to use a similar pulse/chase method in nuclear extract to show the occurrence of pausing on several promoters. These experiments led to speculation that pausing could be regulated by so-called “negative transcription elongation factors[58].” The Price and Gilmour labs have since both reconstituted Pol II pausing using HeLa and Drosophila nuclear extract respectively, allowing for the study of individual pausing factors such as NELF and DSIF in a defined context[23, 29, 43]. In vitro reconstitution of a complex containing Pol II, DSIF, and NELF by the Gilmour lab has enabled the examination of individual domains of NELF and DSIF and their effects on paused complex formation[22, 43]. Finally, the Taatjes lab recently reconstituted promoter proximal pausing in vitro without nuclear extract using an HSP70 promoter, purified Pol II, DSIF, NELF, heat shock factor 1 (HSF1), and preinitiation complex (PIC) components[59].
As a complement to the abundance of in vivo and in vitro techniques, several small molecules have been used to gain insight into Pol II pausing mechanisms. Among these are inhibitors of the pause release factor P-TEFb such as 5,6-Dichloro-1-beta-D-ribofuranosylbenzimidazole (DRB)[60] and flavopiridol[61], both of which have proved useful in the measurement of elongation rates [55, 62, 63]. However, experiments using these kinase inhibitors should be interpreted with caution because both are known to have off-target effects[64, 65]. This limitation has prompted the development of increasingly specific P-TEFb inhibitors, including the adenine analog 1-NA-PP1 coupled with an analog-sensitive mutation in the CDK9 subunit of P-TEFb[26], and the drugs Atuveciclib[66] and NVP-2[67]. Triptolide, an inhibitor of the ATPase activity of the XPB subunit of transcription factor II H (TFIIH), has been widely used to inhibit new Pol II initiation in the measurement of paused Pol II half-life[40, 68–70]. A recent small molecule screen by the Dikstein lab also identified several Pol II-DSIF inhibitors, some of which had a marked effect on pausing in cells[71]. Expanding the use of Pol II-DSIF inhibitors into the various cell-free systems available could provide significant insight into the mechanism of pausing.
Finally, major breakthroughs like the cryo-electron microscopy (cryo-EM) structures of the human paused (Pol II-DSIF-NELF) and active (Pol II-DSIF-Spt6-PAF1C) elongation complexes published by the Cramer lab[72–74] provide excellent templates for designing targeted mutagenesis experiments to be carried out using the cell-free systems described above.
DSIF
DSIF is a two-subunit transcription factor composed of Spt4 and Spt5 and is widely conserved across eukaryotes (reviewed by Decker 2020[75]). In metazoans, DSIF acts to stimulate or repress transcription elongation, depending on its phosphorylation state. The pausing function of DSIF was first discovered by the Handa lab as an activity that rendered Pol II transcription sensitive to inhibition by DRB, a nucleoside analog[19]. DSIF associates with the Pol II elongation complex after the transcription of at least eighteen nucleotides and was later found to be the linchpin of promoter proximal pausing due to its critical role in recruiting NELF to the elongation complex[22, 72], but the mechanism through which this occurs has yet to be fully elucidated.
A recent structure of the human paused elongation complex containing Pol II, DSIF and NELF from the Cramer group provides insight into the mechanism of NELF recruitment by DSIF. While the structure reveals that NELF and DSIF bind to opposite faces of Pol II, the unstructured C-terminal “tentacles” of the NELF-A and NELF-E subunits may drive the NELF-DSIF association. The tentacles wrap around either side of the elongation complex, contacting Pol II as well as the Spt5 and Spt4 subunits of DSIF. These tentacles are not visible in the structure derived from cryo-EM but their locations have been inferred from crosslinks identified by mass spectrometry (Fig. 1)[72]. The Spt5 subunit of DSIF is composed of several domains, including an unstructured N-terminal domain, a NusG N-terminal (NGN) domain, multiple Kyprides, Ouzounis, Woese (KOW) domains, as well as a disordered C-terminal region (CTR) (Figs. 1 and 4). The NELF-A tentacle crosslinks to the KOW1 and KOW4 domains of Spt5 in addition to contacting Spt4. Contacts made by the NELF-E tentacle are more extensive: several residues in the KOW1 and KOW2-3 domains as well as a significant portion of the KOW4 domain of Spt5 were found to crosslink to the tentacle[72]. The lysine-specific crosslinker used to identify these contacts, bis(sulfosuccinimidyl)suberate (BS3), has a long spacer arm of 11.4 Å[76]. Given the flexible nature of the NELF-A and NELF-E tentacles, it is unclear whether some of the crosslinks identified are fortuitous or whether they represent bona fide NELF-DSIF interactions. In vitro biochemical studies are needed to shed further light on the mechanism of NELF recruitment to the elongation complex. The available structural information provides an excellent template for targeted mutagenesis of both NELF and DSIF.
Figure 1: Surface representation of paused Pol II-DSIF-NELF complex structure.
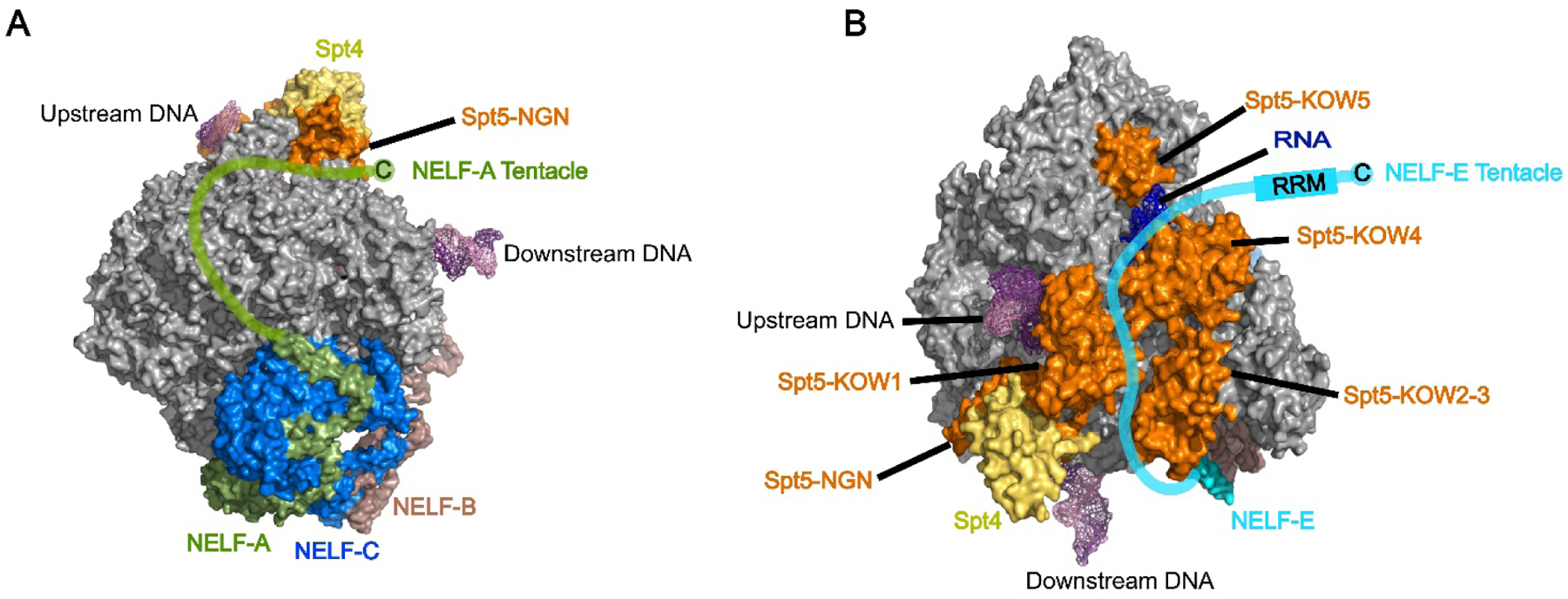
A) Side view of complex illustrating modeled path of the unstructured NELF-A C-terminal tentacle (green)[72]. B) Top view of complex illustrating modeled path of the unstructured NELF-E C-terminal tentacle (blue)[72]. PDB ID: 6GML.
Figure 4: Conserved Spt5 and NELF P-TEFb phosphorylation sites.
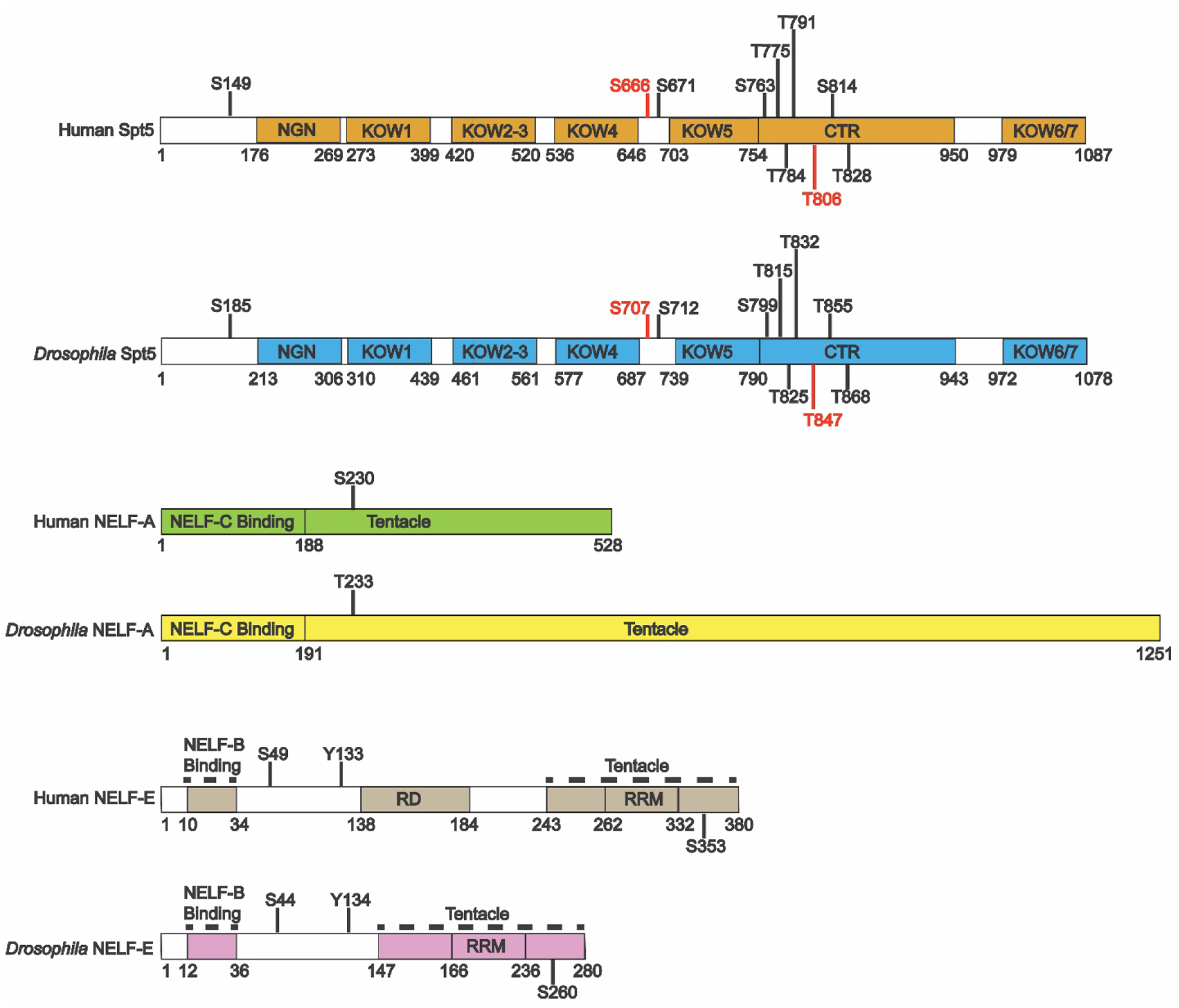
Ten of the residues identified as putative P-TEFb phosphorylation sites on human Spt5 are conserved or semi-conserved (Ser or Thr) in Drosophila Spt5. In contrast, only one putative NELF-A phosphorylation site is semi-conserved from human to Drosophila and three putative NELF-E phosphorylation sites are conserved from human to Drosophila. The S666 and T806 in Spt5 (red) have been identified as critical participants in the phosphatase-kinase switch that governs pausing and pause release dynamics[78]; both residues are conserved in Drosophila.
Whether DSIF contributes to promoter proximal pausing outside its role in NELF binding remains an open question. Comparing structures of paused and active elongation complexes hint at additional possible roles of DSIF in pausing (Fig. 2). The Spt5 subunit of DSIF forms two nucleic acid clamps; the NGN and KOW1 domains frame the upstream DNA duplex while the KOW4 and KOW5 domains bracket the nascent RNA. Both nucleic acid clamps undergo a conformational change during the transition from the paused to active state. The KOW1 domain rotates into an “open” conformation, accompanied by a tilt in the upstream DNA. The RNA clamp undergoes a similar opening as a result of repositioning of the KOW4 domain (Fig. 2A)[72, 73]. For Pol II to elongate, the downstream DNA must be allowed to unwind and move into the active site while the nascent RNA and upstream DNA move out of their respective exit channels. The Spt5 DNA and RNA clamps may promote pausing by restricting the movement of the nucleic acids out of the exit channels, thus impairing the forward motion of the elongation complex (Fig. 2). Moreover, in the paused state, the RNA-interacting face of the KOW4 domain also interacts with the NELF-E tentacle, which could serve to stabilize the RNA clamp’s closed conformation[72]. Three phosphorylation sites have been identified in the flexible linker region between the KOW4 and KOW5 domains of Spt5[73, 77]. Phosphorylation in this region could play a role in pause release by structuring the linker region and facilitating the RNA clamp opening[78]. While further biochemical work is required to fully understand the function of these nucleic acid clamps, the structural data suggests that the Spt5-nucleic acid interactions play roles in establishing or stabilizing the pause.
Figure 2: Spt5 DNA and RNA clamps.
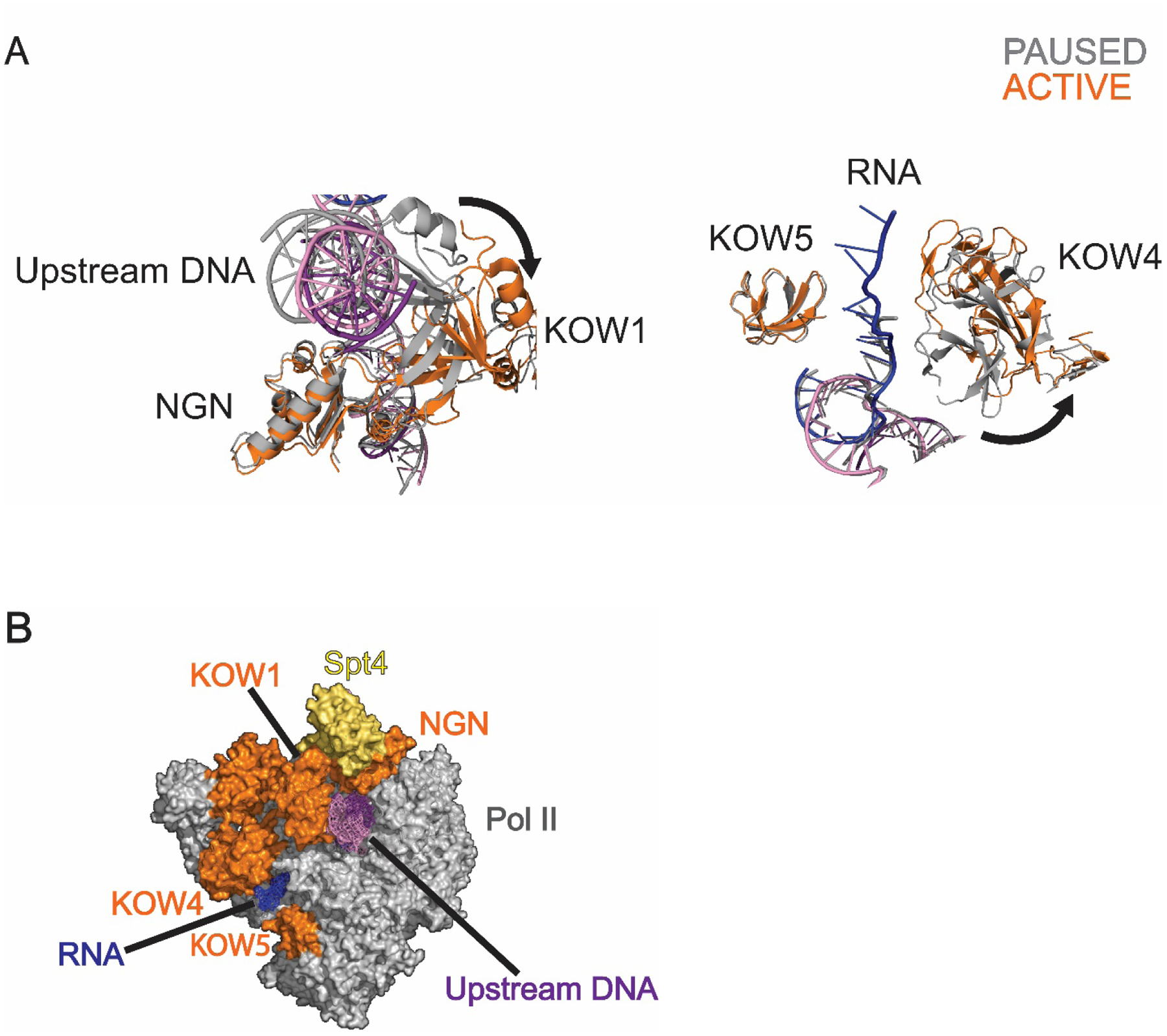
A) Superimposed structures of the Spt5 nucleic acid clamps in their paused (grey, PDB ID: 6GML) and active (orange, PDB ID: 6GMH). The KOW1 and KOW4 domains change conformation upon pause release, resulting in clamp opening. B) Surface model of Pol II-DSIF. The Spt5 NGN and KOW1 domains (shown in orange) form a clamp around the upstream DNA (purple) and the Spt5 KOW4 and KOW5 domains (orange) form a clamp around the nascent RNA (blue). PDB ID: 6GML.
NELF
NELF was first identified as a four-subunit complex (NELF-A, -B, -C/D isoforms, -E) that works cooperatively with DSIF to repress Pol II elongation[20, 79]. NELF is recruited to the elongation complex by DSIF[22] and plays a multifaceted role in establishing and maintaining promoter proximal pausing.
Structural model
The structure of the human Pol II-DSIF-NELF complex provides several insights into the mechanism of promoter proximal pausing. Most notably, binding of NELF to the elongation complex stabilizes the formation of a half-translocated or “tilted” DNA-RNA duplex in the Pol II active site (Fig. 3). This partially-translocated state prevents the incorporation of new nucleotides, forcing the complex into a paused state[72]. Such half-translocated DNA-RNA hybrids are also a feature of hairpin-stabilized paused bacterial RNA polymerase (RNAP) complexes (Fig. 3)[80, 81]. While the structure of the human paused complex was obtained using a template containing a hairpin sequence[72], the observed half-translocated hybrid is likely a key feature of NELF-containing elongation complexes rather than an artifact of the nucleic acid scaffold; NELF preferentially binds Pol II-DSIF complexes with the RNA-DNA duplex in the tilted conformation (Seychelle Vos, personal communication). Therefore, stabilization of the tilted nucleic acid duplex in the paused Pol II active site is likely a direct effect of NELF binding to the elongation complex and may reflect a conserved facet of a multipartite pausing mechanism.
Figure 3: Half-translocated RNA-DNA duplex in the paused RNA polymerase active site.
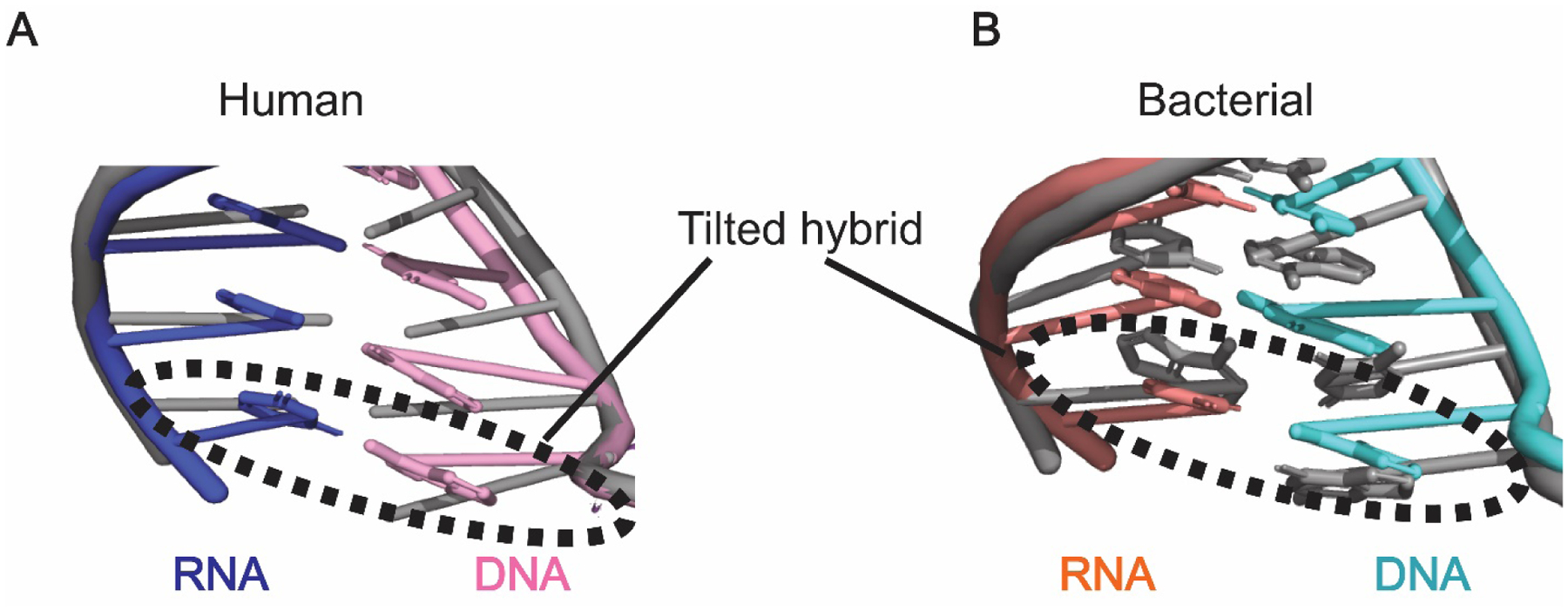
A) The RNA-DNA duplex observed in the Pol II-DSIF-NELF structure (colored, PDB ID: 6GML) is in a “tilted” conformation relative to the RNA-DNA duplex observed in the Pol II-DSIF structure (grey, PDB ID: 5OIK). B) The RNA-DNA hybrid in the active site of the paused bacterial RNA polymerase (colored, PDB ID: 6ASX) adopts a similarly tilted conformation compared to the hybrid in the active site of a post-translocated RNA polymerase (grey, PDB ID: 6ALF).
While bacterial RNAP pausing differs significantly from pausing in metazoans—bacteria, for instance, lack NELF and NELF-dependent promoter-proximal pausing—the two systems share common features that could point to a generalized mechanism. Pausing of bacterial RNAP is generally hairpin-stabilized and multiple cryo-EM structures have shown that this pause is accompanied by a tilted RNA-DNA duplex akin to the one observed in the NELF-bound human paused complex (Fig. 3)[80–82]. Similarly, in human cells, “transcription units” with longer pause durations tend to adopt more RNA secondary structure in vivo and in silico[26]. This is suggestive of conserved roles for RNA secondary structures in bacteria and metazoans. It is important to note, however, that the degree of influence of RNA secondary structure on NELF-dependent pausing is likely gene-specific and influenced by the distance between the secondary structure sequence and the Pol II pause site. For instance, the classic example of hairpin-mediated pausing in metazoans is the human immunodeficiency virus type 1 (HIV-1) provirus, which contains a regulatory motif called the TAR element. The fully transcribed TAR element forms a hairpin secondary structure that has been proposed to facilitate NELF recruitment[83, 84]. However, permanganate footprinting shows that NELF-dependent pausing occurs well before the TAR element hairpin is fully transcribed[85, 86]. Thus, while RNA secondary structure may be a contributing factor to promoter proximal pausing in metazoans, it is not likely to be a primary driver as it is in bacteria. Moreover, there is no evidence of substantial RNA secondary structure in the promoter proximal region of numerous genes in Drosophila[42], suggesting that a pausing mechanism driven by RNA secondary structure is likely not a widespread and conserved phenomenon in metazoans. Thus, it will be interesting to see whether the tilted hybrid can be observed in paused elongation complexes assembled in the absence of the RNA hairpin.
Another shared aspect of metazoan promoter proximal pausing and bacterial pausing is the inhibition of trigger loop folding. The trigger loop of RNA Polymerases generally adopts an “open” conformation, but folds over the incoming NTP to close off the active site and facilitate catalytic chain extension[87, 88]. In the human paused Pol II structure, NELF-C contacts the open trigger loop, potentially preventing the closure of the Pol II active site[72]. In bacteria, trigger loop inhibition is achieved by the hairpin-stabilized rotation of an RNAP swivel module, which results in allosteric inhibition of trigger loop folding[80]. Thus, NELF appears to carry out functions in metazoans that bacterial systems achieve without NELF homologs.
Despite the clear differences between bacterial and metazoan pausing, understanding the commonalities between the two systems could provide significant insight into the mechanism of NELF-dependent pausing. While NELF plays a major role in keeping Pol II in the paused state, increasing evidence suggests that pausing is a multipartite mechanism. Looking to other systems will facilitate the creation of a more complete mechanistic picture.
RRM and NELF-RNA contacts
Much of the work on NELF has focused on its interactions with RNA. Early experiments by the Handa group showed that human NELF-E binds RNA, including HIV-1 TAR, through an RNA recognition motif (RRM) at the subunit’s C-terminal end. Deleting this RRM domain impaired the transcription inhibition activity of NELF in nuclear extract and point mutations in the RRM diminished NELF’s inhibitory activity [79]. The Kim lab later observed that enhancer RNAs (eRNAs) can facilitate the transition of Pol II from the paused to the active state by binding and redirecting NELF in an RRM-dependent manner in mouse neuronal cells. Furthermore, the Kim group showed that deleting the RRM results in decreased NELF occupancy at some promoters[89]. Consistent with these studies, SELEX experiments from the Lis group using Drosophila NELF-E and a dNELF-E RRM domain identified a NELF-E Binding Element (NBE) that is both necessary and sufficient for dNELF-E binding as long as the motif is accessible as single-stranded RNA. The Lis group also found that both human and Drosophila NELF-E bind specifically to the NBE found in HIV-1 TAR RNA and that the NBE is enriched in Drosophila promoter regions[90]. The Cramer group later found that NELF contains additional RNA binding faces on NELF-B and the NELF-A/C duplex[91]. However, work performed by the Gilmour lab using paused elongation complexes reconstituted with purified Pol II, NELF, and DSIF failed to detect NELF-RNA contacts in elongation complexes with transcripts of 22–31 nucleotides, lengths which correspond to the pausing region in Drosophila[22]. More recently, the Cramer group showed that deletion of the C-terminal domain of NELF-E, which includes the unstructured DSIF-interacting tentacle as well as the RRM domain, does not affect pausing in an in vitro system[72]. The NELF-E RRM may assist in NELF binding to the elongation complex in an in vivo context, perhaps by guiding NELF to regions where RNA resides; its contribution would therefore depend heavily on cellular concentrations and Pol II elongation kinetics, parameters that in vitro systems would have difficulty recapitulating.
Additional considerations
Recent work by the Shilatifard group found that acute depletion of NELF using an auxin-mediated degradation system in human cells does not result in release of Pol II into the gene body. Instead, upon NELF depletion, Pol II shifts downstream to a “second pause” site. This transition is independent of P-TEFb kinase activity and is consistent with a strongly positioned +1 nucleosome blocking Pol II elongation[92]. Single-nucleotide resolution mapping of elongating and arrested Pol II in Drosophila cells performed by the Henikoff lab yielded a similar observation: the +1 nucleosome acts as a significant barrier for transcription, even at genes that pause upstream from the nucleosome[53]. Although two pauses are evident in Drosophila, it is unclear whether the second pause, observed after NELF depletion by Shilatifard and colleagues, is normally present in mammalian cells. The NELF-dependent pause may act as a critical checkpoint to prime the elongation complex to transverse the +1 nucleosome.
NELF-E has also been shown to interact with the cap-binding complex (CBC)[93, 94]. ChIP-seq data indicates that the CBC subunit CBP80 co-occupies promoters with NELF. Acute depletion of NELF results in a reduction of CBP80 located at pause regions[92]. This suggests that the NELF complex possesses additional transcriptional regulatory roles that complement its pausing function. Indeed, promoter proximal pausing may act as a kinetic facilitator for the recruitment of additional regulatory factors to the elongation complex.
Phosphorylation-dependent regulation of the pause
P-TEFb, predominantly composed of Cdk9 [95] and Cyclin T in Drosophila [96] and Cdk9[95] and Cyclin T1 or Cyclin T2 in human cells[97] is broadly considered to be critical for the release of Pol II from the pause and its transition into productive elongation. P-TEFb is frequently found as a part of larger complexes. In its inhibited form, P-TEFb is associated with the 7SK noncoding RNA and the MeCPE, LARP7, and HEXIM factors[98]. In its most active form, P-TEFb is part of the multi-subunit super-elongation complex (SEC), which facilitates the transition of Pol II from the paused state to productive elongation[99–103]. The SEC has also been found to interact directly with TFIID, suggesting a potential mechanism for its recruitment to the elongation complex (discussed in the next section)[104].
P-TEFb was discovered by the Price lab as the activity required for the transition of Pol II into productive elongation[95]. P-TEFb phosphorylation of the C-terminal region (CTR) of Spt5 is required for processive elongation[24]. Moreover, adding pre-phosphorylated Spt5 to a transcription reaction prevents NELF-mediated suppression of Pol II elongation and abolishes the effect of P-TEFb inhibition on elongation[23]. This suggests that the phosphorylation of the Spt5 CTR by P-TEFb plays a critical role in the transition of DSIF from a pausing factor to an elongation factor. Indeed, phosphorylation of the Spt5 CTR may be critical for the recruitment of elongation factors such as the PAF1C[74]. Additional phosphorylation sites on Spt5 could also contribute to the transition to productive elongation; P-TEFb phosphorylation sites have been identified in the N-terminal region of human Spt5 and in the unstructured linker between the human Spt5 KOW4 and KOW5 domains that bracket the nascent RNA (Table 2, Fig. 4)[73, 77].
Table 2:
Phosphorylation of Spt5, NELF-A, and NELF-E
| Human | Drosophila |
|---|---|
| Spt5 | |
| S148 | E184 |
| S149 | S185** |
| T153 | R189 |
| S666 | S707** |
| S671 | S712** |
| S686 | A722 |
| S763 | S799** |
| T775 | T815** |
| T784 | T825** |
| T791 | T832** |
| T799 | R840 |
| T806 | T847** |
| S814 | T855* |
| T828 | T868** |
| NELF-A | |
| S3 | N3 |
| T157 | P160 |
| S225 | G228 |
| T227 | R230 |
| S230 | T233* |
| S233 | G236 |
| T239 | R241 |
| S244 | R246 |
| T246 | P248 |
| T277 | Q279 |
| T297 | -- |
| S363 | K363 |
| NELF-E | |
| S49 | S44** |
| S131 | N132 |
| Y133 | Y134** |
| S140 | R141 |
| S179 | -- |
| S181 | -- |
| S185 | -- |
| S187 | -- |
| S191 | -- |
| S353 | S260** |
Conserved residue
Semi-conserved Residue
Recent work by the Fisher lab provides further support for the hypothesis that DSIF phosphorylation by P-TEFb is a critical component of the pause release mechanism. The group defined a kinase-phosphatase switch that governs pausing and pause release dynamics. The phosphates installed by P-TEFb at Ser666 between the KOW4 and KOW5 domains and atThr806 in the CTR (Fig. 4) are removed by two protein phosphatase complexes called PP1 and PP4. PP1 only acts on the Spt5 CTR and functions primarily in the cleavage and polyadenlylation region of genes. In contrast, PP4 acts on both Spt5 Ser666 and the CTR and affects Spt5 activity at the 5՛ ends of genes. Reducing cellular levels of PP4 increases Spt5 phosphorylation on Ser666 and the CTR and results in a shift in Pol II distribution from the pause region to the gene body at several genes. In addition to phosphorylating Spt5, P-TEFb also phosphorylates PP1 and PP4, resulting in an inhibition of their phosphatase activities. Thus, in the absence of P-TEFb, PP4 enforces the paused state by dephosphorylating Spt5. P-TEFb recruitment results in the simultaneous inhibition of PP4 and activation of Spt5. Metagene analysis of over 20,000 genes reveals an increase in the pSer666/Spt5 ratio, but not the pThr806/Spt5 ratio, in the gene body compared to the region near the transcription start site[78].
Both phosphorylation sites are conserved in Drosophila (Fig. 4), but the phosphorylation of the linker between the KOW4 and KOW5 regions may serve a more critical role in pause release. Recent work by the Adelman lab shows that pSer666 and its orthologous residue in Drosophila, pSer707, are substrates for another phosphatase called protein phosphatase 2A (PP2A) that also associates with the Integrator complex. Adelman and colleagues propose that dephosphorylation of Ser666 by Integrator-PP2A prevents pause release and primes the elongation complex for premature termination[105]. Phosphorylation of human Ser666 (Drosophila Ser707) therefore appears to be a critical switch that facilitates transition into productive elongation and may promote pause release by stimulating the opening of the Spt5 RNA clamp (Fig. 2A).
P-TEFb has also been shown to phosphorylate several sites on both the human NELF-A and NELF-E subunits (Table 2)[73, 83, 106, 107], but only four of the twenty-two sites are also conserved in Drosophila (Table 2, Fig. 4). Notably, the sites that were implicated in elongation control of HIV through NELF release are absent in Drosophila[83]. Thus, a generalized mechanism in which pause release also depends on P-TEFb phosphorylation of NELF likely involves only a fraction of the sites previously identified. Further biochemical studies should focus on the functions of these conserved sites.
Experiments by the Price lab using Pol II lacking the carboxy-terminal domain (CTD) of the largest subunit showed that the CTD is required for productive elongation. The Price group also found that P-TEFb can phosphorylate the CTD of a Pol II in an early elongation complex[108]. Further studies ultimately showed that P-TEFb phosphorylates the Pol II CTD on the Ser2 position[109, 110]. However, recent work by the Roeder lab, in which an inducible Pol II degradation system allowed generation of transcriptionally-engaged CTD-less Pol II in vivo, found that the Pol II CTD is not essential for pausing or for the transition into productive elongation[111]. Thus, the role of the Pol II CTD in pausing, and by extension the role of P-TEFb phosphorylation of the CTD in pause release, remains a topic in need of further investigation.
TFIID
TFIID is a general transcription factor that provides the foundation for assembling a preinitiation complex (PIC). The relationship between TFIID and Pol II promoter proximal pausing remains a topic of some controversy. In 2019, the Biswas lab used co-immunoprecipitation experiments to show that the TAF subunit of TFIID interacts with components of the SEC both in mammalian cells and in vitro. AF9 and EAF1, two SEC subunits, both have serine-rich domains that are required for this interaction. This data yields a model in which TFIID plays a role in pause release by recruiting the SEC through the serine-rich domain of AF9[104].
In contrast, the Taatjes lab recently showed that TFIID enables Pol II pausing in a cell free system as well as in human and Drosophila cells. The human PIC (composed of purified TFIIA, TFIIB, TFIID, TFIIF, TFIIH, Mediator) was found to be sufficient to establish pausing in vitro on an HSP70 template in the presence of HSF1. The observed pause occurs primarily between +60 and +80 and, to a lesser extent, between +24 and +44. DSIF and NELF enhance the pausing between +24 and +44. While most of the PIC components are dispensable for pausing Pol II, TFIID was found to be essential for reconstituting in vitro pausing. Knockdown of the TAF 1 and 2 subunits in human and Drosophila cells results in increased pause release[59].
Thus, the work regarding the role of TFIID in pausing points to dual yet conflicting roles. A promoter-dependent function of TFIID could explain the apparent contradiction. Increased pause release upon knockdown of TAFs was not observed at all genes, suggesting differential functions[59]. TFIID exhibits structural plasticity that could facilitate multiple functions around and beyond the promoter regions of genes[112–114]. Protein-DNA crosslinking data indicates that TAF2 interacts directly with DNA downstream of the transcription start site[115, 116]. ChIP-nexus and ChIP-exo data further demonstrate that TFIID binding extends beyond the paused region, at least in Drosophila cells[40, 117], a likely result of conformational flexibility and possibly indicative of dual pause-promoting and pause release functions.
A role for TFIID in pausing and pause release can also be reconciled with the enrichment of key promoter elements at some genes. For example, in Drosophila, NELF-associated genes with paused Pol II are also enriched for TFIID binding sites[42]. TFIID can contact promoter elements such as Initiator (Inr), motif ten element (MTE), downstream promoter element (DPE), and the TATA box. Downstream promoter sequences, the presence of a TATA box, and variations in the Inr sequence (discussed later in this review) strongly influence Pol II pause stability in Drosophila[27]. The structural flexibility of TFIID is consistent with promoter-specific roles that are at least partially determined by the combination of promoter elements at each gene.
GAGA factor
GAGA factor (GAF) is a sequence-specific DNA binding factor that has been shown to promote promoter proximal pausing in Drosophila. Early work from the Lis lab indicated that point mutations in the GAGA element significantly decrease pausing on the Drosophila hsp70 promoter[118, 119]. However, it was unclear whether the loss of paused Pol II was due to disruption of pausing or to diminished transcription initiation, a prerequisite for pausing. The Gilmour lab later established that the GAGA element can promote Pol II pausing[120]. A later genome-wide analysis of promoter elements in the Drosophila embryo by the Levine lab observed a positive correlation between Pol II pausing and the presence of a GAGA element at promoters[25].
Cell-free transcription experiments in nuclear extract performed by the Gilmour lab established a direct link between GAF and pausing. Using GAF antibodies to disrupt GAF function at various stages of the transcription cycle, they showed that GAF contributes to initiation and the establishment of the pause after initiation, but not to the stability of the pause state once it has been established. Immunodepletion of GAF from nuclear extract decreases Pol II pausing, but this pausing can be re-established by adding back purified GAF to the extract[29]. In accordance with these biochemical results, GAF knockdown experiments by the Lis group showed a similar effect genome-wide[121] and the Huang lab later showed that overexpression of GAF increases paused Pol II globally[122]. Additional results show that reducing GAF levels in cells shifts the location of the pause downstream and that GAF-associated genes exhibit pausing that is closer to the TSS than on non-GAF genes[29]. All together, these results suggest that GAF facilitates a kinetically favorable association between NELF and Pol II by recruiting NELF to the promoter region [29].
Though GAF has been extensively studied in Drosophila, GAF homologs have not been extensively studied in mammalian systems. Some vertebrate GAF homologs have been identified[123], but further investigation is required to determine whether the pausing function is conserved across species. As exemplified by Drosophila GAF, these analyses must be designed to distinguish between functions in initiation and pausing since initiation is a prerequisite for promoter proximal pausing.
PAF1C
The multi-subunit Paf1 complex (PAF1C) has been implicated in promoter proximal pausing but its role is controversial. Initial studies by the Shilatifard group showed that PAF1C loss results in release of paused Pol II into gene bodies in both Drosophila and human HCT116 and MCF7 cell lines, with a greater effect observed at highly paused genes. PAF1 knockdown also results in increased SEC recruitment and Pol II CTD phosphorylation at the Ser2 position[124]. Hence, they proposed that PAF1C helps enforce the pause by inhibiting SEC interaction with the gene. In contrast, results from the Roeder group indicate PAF1C promotes pause release in human THP1 cells. They noted that high PAF1C occupancy correlates with high gene expression and depletion of PAF1C results in an increase in Pol II occupancy near promoters. The Roeder group also demonstrated a direct interaction between P-TEFb and PAF1C using pulldown assays, suggesting that PAF1C stimulates pause release by recruiting P-TEFb to the paused elongation complex [125]. The contradicting observations could initially be reconciled by a cell-specific model of PAF1C function given that the Shilatifard and Roeder groups used different cell lines for their experiments. The Roeder group noted that depletion of PAF1C from a different cell line, CCRF-CEM, had the opposite effect from what they observed in THP1 cells[125]. However, follow-up work by the Shilatifard lab showed that PAF1C maintains the Pol II paused state even in THP1 cells, in direct contrast to the results obtained by the Roeder group[126]. Thus, the differences in the outcomes of these studies remain unresolved.
Dynlacht and colleagues have also investigated the function of PAF1C in murine myoblasts. By measuring rates of elongation before and after depletion of PAF1, they provided strong evidence that PAF1C functions as a positive elongation factor promoting both pause-release and elongation in the body of genes. Depletion of PAF1 increases Pol II occupancy in both the gene body and in the promoter region. The latter observation comports with the results seen by the Shilatifard group. However, the Dynlacht lab ascribe the changes in Pol II levels to reductions in elongation rate, which would be expected to result in a buildup of Pol II in the gene body[55].
The Shilatifard group also provided evidence that PAF1C can enforce promoter proximal pausing indirectly by repressing the action of a gene’s enhancer. Depletion of PAF1 increases promoter-enhancer interactions and decreases promoter proximal pausing, resulting in a release of Pol II into gene bodies. Strikingly, knockout of PAF1C-regulated enhancers reduces pause release in PAF1 knockdown conditions—reversing the effect of PAF1 knockdown on paused Pol II. This suggests that in some cases, rather than acting directly as a pausing factor at a gene’s promoter, PAF1C prevents pause release by inhibiting the activation of certain enhancers. Indeed, enhancers have a higher ratio of PAF1 to Pol II occupancy when compared to promoters[126], suggesting that PAF1C may have distinct functions at promoters and enhancers.
Further work is needed to sort out the role of PAF1C in promoter proximal pausing. Structures of an activated elongation complex containing Pol II, DSIF, Spt6, and PAF1C clearly demonstrate that NELF and PAF1C are mutually exclusive because they bind to the same surface of Pol II (Fig. 5)[73, 74].Therefore, the contradicting observations about its role can be reconciled by PAF1C having two different states: enhancer-bound and promoter-bound. Enhancer-bound PAF1C (with greater occupancy and possibly without binding directly to Pol II) helps maintain pausing at certain gene promoters by inhibiting enhancer activation. Promoter-bound PAF1C, in contrast, binds Pol II and acts as an elongation factor.
Figure 5: Summary of the factors involved in promoter proximal pausing.
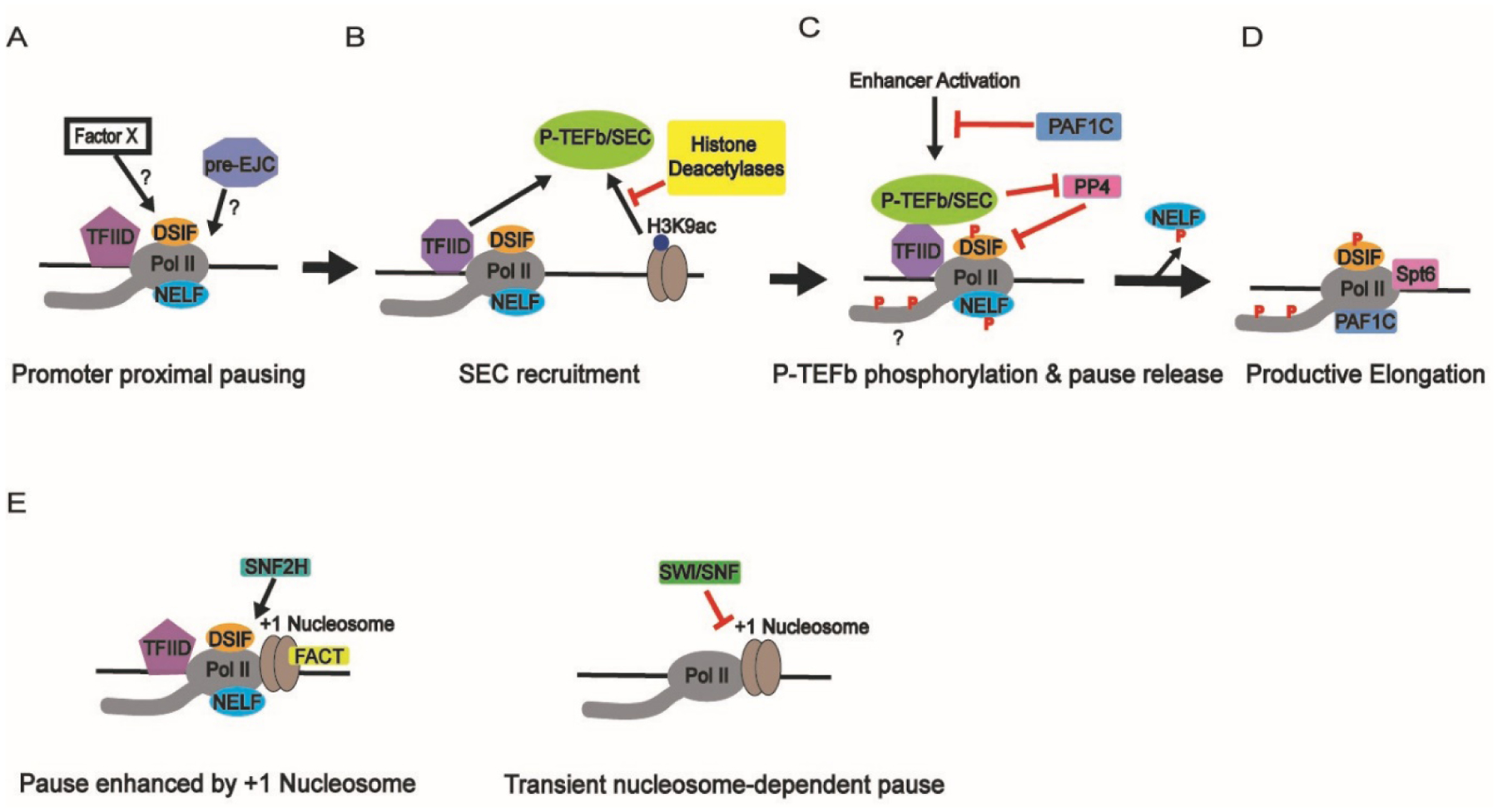
A) Pol II pausing occurs ~30–60 nt downstream of the transcription start site and requires DSIF and NELF, which function cooperatively; TFIID helps promote pausing[59]. The pre-EJC helps maintain pausing at genes with long introns[62]. Additional “Factor X” proteins may contribute to pause stabilization. B) TFIID, perhaps in a different conformation than in (A), helps recruit P-TEFb and the SEC to the elongation complex[104]. H3K9ac can also facilitate recruitment of the SEC[140], a process that can be inhibited by histone deacetylases[139, 141]. C) The phosphatase PP4 promotes pausing by maintaining the dephosphorylated state of DSIF[78]. P-TEFb inactivates PP4 through phosphorylation and phosphorylates DSIF, NELF, and potentially phosphorylates the Pol II CTD. This results in the ejection of NELF from the elongation complex. For some genes, pause release can be triggered by enhancer activation, which is in turn inhibited by PAF1C[126]. D) Pause release is accompanied by association of elongation factors like Spt6 and PAF1C with the elongation complex, allowing the transition into productive elongation[73, 74]. TFIIS binds and rescues backtracked Pol II (not shown)[128, 129]. E) At human and some Drosophila promoters, the +1 nucleosome acts to stabilize paused Pol II[30, 31]. In Drosophila, FACT promotes pausing, possibly by stabilizing the +1 nucleosome[127]. In humans, SNF2H promotes nucleosome-stabilized pausing[30]. In the absence of pausing factors, nucleosomes act as barriers to productive transcription. These barriers can be relieved by chromatin remodelers SWI/SNF[135].
Additional proteins
Since the discovery of DSIF and NELF, many more proteins have been implicated in promoter proximal pausing of Pol II and it is likely that more will be discovered in the future. We discuss some of them in the following section but note that the complete picture remains to be fully resolved.
Recently, the pre-exon junction complex (pre-EJC), which regulates expression of long genes in Drosophila has also been identified as an intron length-dependent facilitator of Pol II pausing. Knockdown of the Mago subunit of the pre-EJC results in global decrease in pausing, with the strongest effect observed at highly paused genes. Depletion of pre-EJC components also results in a redistribution of Ser2-phosphorylated (Ser2P) Pol II. As with DSIF, pre-EJC association with the Pol II elongation complex requires nascent RNA. This RNA association appears to contribute significantly to pausing; tethering the Mago subunit to the 5′ UTR of nascent RNA results in increased Pol II occupancy at several genes. However, the interplay between the pre-EJC and NELF remains unclear. Of the genes found to have pre-EJC occupancy, 45% do not overlap with NELF binding, suggesting independent mechanisms of action. Depletion of the Mago subunit results in increased Cdk9 occupancy near promoters, so its role may primarily be to maintain rather than establish pausing at long genes[62].
Histone-associated factors also work to promote pausing. For example, in Drosophila, the histone chaperone Facilitates Chromatin Transcription (FACT) is enriched at 5′ ends of genes with high Pol II occupancy. Knockdown of FACT results in a decrease in Pol II pausing and decreases Pol II pause half-life. FACT may promote pausing by stabilizing the +1 nucleosome[127]. This stabilization is likely promoter-specific since nucleosome organization in Drosophila differs based on promoter class (discussed further in a later section)[31].
Transcription elongation factor IIS (TFIIS) has previously been viewed as a prominent player in the regulation of promoter proximal pausing. Early in vitro studies described TFIIS as a factor that facilitates rescue of backtracked polymerases by stimulating Pol II’s intrinsic RNA endonuclease activity[128, 129]. Later work by the Lis lab led to the observation that TFIIS is critical to maintaining the activity of paused Pol II on the hsp70 gene in Drosophila. In the absence of TFIIS, Pol II was found to have impaired elongation through pause regions, resulting in weaker activation of hsp70 upon heat shock[130]. More recent genome-wide studies in Drosophila and in human cells have emphasized the role of TFIIS in re-setting the elongation competence of stalled Pol II and have indicated that stalled Pol II has a propensity to be backtracked[50, 131]. However, the mechanistic and temporal relationship between backtracked Pol II and NELF-dependent pausing remains unclear. Structural information indicates that NELF prevents the binding of TFIIS[72], in line with previous biochemical studies by Price and Landick showing that NELF inhibits the anti-arrest activity of TFIIS[132]. A problem with cellular studies that involve knocking down TFIIS is that levels of backtracked Pol II can accumulate because overcoming the backtracked state becomes a rate limiting step. Thus, it is unclear to what extent backtracking normally contributes to the promoter proximal pause and what role TFIIS normally plays in pause release.
Role of sequence in pausing
Promoter elements such as GAGA, Inverted GAGA, Inr, and DPE are enriched at paused genes in Drosophila[25, 42]. Some of these, such as the Inr and DPE elements, are TFIID binding sites and are likely key players in regulating differential TFIID function, as discussed above. In Drosophila, sequence can act as a particularly strong contributor to Pol II pause stability. Reporter-ChIP-nexus experiments, which detect pausing on plasmids transfected into cells, have shown that an Inr variant with a G at the +2 position results in a significantly more stable pause. Promoters with the Inr-G variant lack TATA boxes and have longer paused Pol II half-lives[27]. An additional element called the Pause Button (PB), which is present at approximately 25% of paused genes, also has strongly positioned CpG motifs. Indeed, Drosophila paused promoters have significantly higher GC content just downstream of the TSS when compared to promoters with low levels of pausing [25].
While mounting evidence points to DNA sequence as a key contributor to pausing, the degree of influence remains a matter of controversy. Disrupting or altering downstream elements of Drosophila promoters affects the position and stability of the pause[27, 28]. However, the Gilmour lab showed that slowing transcription by lowering nucleotide concentrations in vitro shifts the position of the pause closer to the transcription start site[29]. Even more compellingly, permanganate footprinting analysis of Drosophila salivary glands expressing a slow mutant of Pol II showed an upstream shift in the location of the pause[29]. Since slowing the rate of transcription can alter the position of the paused Pol II, sequence likely plays an accessory role in establishing the pause, either by recruiting factors like TFIID or by slowing Pol II movement to allow pausing factors such as DSIF and NELF to bind.
In human cells, evidence connecting sequence motifs and pausing is more limited. However, recent PRO-seq analysis of several thousand genes in multiple human cell types reveals that Pol II tends to pause at the same nucleotide positions across individuals and across cell types[133]. Pause sites have high GC content, as previously observed in Drosophila, and are enriched in a high GC content 9-mer motif; a high percentage of the pause sites are at cytosines. Mutagenesis of sequences at or near the identified pause sites alters gene expression[133]. Recent analysis of promoter sequences in HeLa cells by Luse and Price also found that promoter proximal pause sites are GC rich. More notably, they found that a preferred sequence signature encompassing the TFIID binding site is present at over 177,000 genes[45]. Thus, TFIID-DNA interactions may play a role in regulating promoter proximal pausing in Drosophila as well as humans, but further work is required to elucidate the mechanistic role of these human sequence features in Pol II pausing.
Promoter Proximal Pausing and chromatin architecture
Drosophila vs. human and the role of the +1 nucleosome
The role of nucleosome organization, in particular the positioning of the +1 nucleosome, in establishing and maintaining Pol II promoter proximal pausing has been the subject of several contradictory studies. Early experiments by the Luse and Kingston labs established that partially purified Pol II elongation complexes from HeLa cells pause on templates containing assembled nucleosomes[134, 135]. Mapping of micrococcal nuclease cuts in HeLa nuclei also determined that human hsp70 is nucleosomal in the transcribed region[135]. These studies led to the conclusion that nucleosomes must be necessary to establish promoter proximal pausing. However, experiments by the Price lab during the same period established that pausing could occur on soluble, non-nucleosomal templates in nuclear extract from Drosophila Kc cells. The Price group also showed that preinitiation complexes assembled from nuclear extract on immobilized DNA templates are able to pause in the absence of nucleosomes[58]. Soon after, Benjamin and Gilmour were also able to reconstitute promoter proximal pausing on the Drosophila hsp70 gene in the absence nucleosomes[136]. At least some of this discrepancy can be attributed to a major difference in the experimental models used. The Price and Gilmour groups both used nuclear extracts in their transcription assays, so the pausing they observed was likely due to Pol II-associated pausing factors like NELF and DSIF[58, 136]. In contrast, the Kingston group used partially purified elongation complexes from HeLa cells that were washed with 1% Sarkosyl prior to nucleosome assembly[134, 135]. Sarkosyl likely stripped the purified elongation complexes of pausing factors such as DSIF and NELF, making the pause sites observed in the human system nucleosome-dependent rather than NELF-dependent[29].
Later studies in human cells and Drosophila indicate that the role for the +1 nucleosome in promoter proximal pausing is context specific. ChIP experiments using human cell lines expressing versions of the mouse c-Myc where various nucleosome positioning sequences were inserted into the pause region established a link between a strongly positioned +1 nucleosome and increased NELF-dependent pausing[30]. Genome-wide studies by the Henikoff lab detected two closely spaced pauses in the 5′ region of genes in Drosophila with the proximal one located where NELF-dependent pausing occurs and the distal one mapping to the edge of the +1 nucleosome[53]. The Shilatifard lab observed that acute NELF depletion in mammalian cells results in a similar nucleosome-mediated “second pause” by Pol II in human cells [92]. Recently, Luse and Price used digestion with DNA Fragmentation Factor (DFF) coupled with pulse/chase transcription “run-off” to map the location of the +1 nucleosome in HeLa cells. Unlike micrococcal nuclease, DFF does not produce sub-nucleosomal fragments and does not digest RNA. They observed that some, but not all, of the paused Pol II abuts the +1 nucleosome; the remaining paused Pol II were separated from the +1 nucleosome. Importantly, unlike other studies, Luse and Price were able to show a direct interaction between Pol II and the +1 nucleosome, rather than a mere correlation between their positions. It remains unclear whether these two classes of paused Pol II are found on the same or on distinct types of promoters[45].
Genome-wide data generated using Drosophila embryos also indicates an overlap between the locations of the paused Pol II and the +1 nucleosome[137]. However, the role of the +1 nucleosomes in Drosophila depends on the chromatin structure established at different promoter types. Well over a thousand genes in Drosophila associate with a transcription factor called M1BP. M1BP-bound genes have well-positioned nucleosomes downstream from the transcription start site and the location of the paused Pol II correlates with the location of the +1 nucleosome. GAF also associates with well over a thousand genes, but few genes are co-occupied by M1BP and GAF. GAF genes, in contrast to M1BP genes, lack well-positioned nucleosomes[31]. GAF-associated nucleosomes have also been shown to be highly dynamic in composition [138]. GAF genes have a greater overall pausing index as well as a greater range of pausing indices compared to M1BP genes. Thus, GAF and M1BP genes exemplify two distinct pausing mechanisms. At M1BP genes, pausing is less efficient and facilitated in large part by the strongly positioned downstream nucleosome. Notably, M1BP genes are housekeeping genes and include most of the ribosomal protein genes, so a short pause duration is expected because they are actively transcribed. GAF genes, on the other hand, have more efficient pausing that is likely mediated by additional factors[31]. Since GAF associates with NELF, GAF-associated genes may have a high local concentration of NELF that allows Pol II to be captured prior to Pol II encountering the first nucleosome, resulting in more efficient pausing.
Chromatin remodelers and histone modifiers
Chromatin remodelers modulate nucleosome-dependent pausing in the human system. In vitro transcription experiments by the Kingston lab showed that addition of fractions with hSWI/SNF activity reduced nucleosome-dependent pausing and resulted in read-through of Pol II in the presence of HSF activator[135]. Additionally, in vivo experiments by Reyes lab showed that SNF2H facilitates promoter proximal pausing if the sequence downstream of the pause site has an affinity for nucleosomes[30].
Studies in both mammalian and Drosophila systems have shown that histone deacetylases promote promoter proximal pausing. Knockdown of the histone deacetylase SIRT6, which deacetylates H3K9ac and H3K56ac in embryonic stem cells decreases Pol II pausing and NELF-E coverage in promoter proximal paused regions[139]. In HeLa cells, H3K9ac has been shown to recruit the SEC[140], raising the possibility that SIRT6 promotes pausing by removing a chromatin mark recognized by SEC. In Drosophila cells, inhibition of histone deacetylases by Trichostatin A (TSA) also results in Pol II release from the promoter proximal pause[141], suggesting a generalized role for histone deacetylases in maintaining the Pol II pause.
Duration of the pause
The duration of the pause and whether Pol II, upon release, transitions to productive elongation or prematurely terminates transcription impacts the level of gene expression. These dynamics were initially studied by blocking transcription initiation with the chemical triptolide, which covalently inhibits the ATPase activity of the TFIIH subunit XPB[142].This ATPase activity plays a critical role in overcoming a block to initiation imposed by XPB[143, 144]. The stability of the paused Pol II is then measured by monitoring the rate of its disappearance following the application of the drug[40, 63, 68–70, 145]. This analysis has been applied specifically to the hsp70 heat shock gene of Drosophila[69] and genome-wide in Drosophila and mammalian cells[40, 63, 68, 70, 145]. These investigations reveal that there is considerable variation among genes in the half-lives of the paused Pol II. Values for paused Pol II half-life range from less than 5 minutes at some genes to as long as 60 minutes at others [40, 63, 68, 69, 145]. It is unclear what allows for the stability observed at genes harboring a paused Pol II with a half-life beyond 30 minutes [40, 63, 68, 145]. It has been argued that triptolide treatment regimens may be overestimating the duration of the pause due to slow and concentration-dependent inhibition of initiation[47, 146]. For instance, an analysis using photobleaching of endogenous GFP-tagged Pol II estimated the Pol II pause half-life to be on the order of mere seconds, contradicting earlier triptolide-based studies[147]. Indeed, no study to date has been able to demonstrate rapid inhibition of initiation by triptolide in cells. These issues have been discussed in previous reviews[34, 146]. Nevertheless, even for the case of the hsp70 heat shock gene of Drosophila, results vary between studies and fall within a range from 1 to 5 minutes[69, 70]. The basis for these differences is unclear but may be due to the different techniques used. The Lis lab tracked the decay of photoreactive GFP-Pol II at the hsp70 gene after triptolide treatment [69] while the Schübeler lab used single-molecule footprinting to measure the level of paused Pol II on hsp70 after triptolide treatment[70].
The more significant and perhaps more controversial issue is what happens to the Pol II after it departs from the promoter proximal pause. Does Pol II proceed to produce a mature transcript or does it prematurely terminate transcription? Since it appears that promoter proximal pausing is a ubiquitous step in the transcription cycle, some portion of the Pol II must transition into productive elongation; yet what portion of the released Pol II gets diverted into an unproductive course is debated. Based on the level of Pol II detected in the body of the genes and the turnover rate of the paused Pol II, some studies have concluded that premature termination is infrequent[63, 68]. An exception appears to be those genes with a short-lived pause whose premature termination is mediated by the Integrator complex[105, 148]. In contrast, other studies have concluded that premature termination is frequent [70, 147]. This remains an important issue to resolve as it impacts the understanding of the mechanism of activation. On the one hand, if the bulk of Pol II departing from the pause enters into productive elongation, then the regulatory mechanism is directed at pause release. On the other hand, if the bulk of the Pol II departing from the pause is prone to premature termination, then pause release must be accompanied by mechanisms that prevent premature termination.
Open questions and future directions
Promoter proximal pausing of Pol II requires NELF and DSIF and may also involve accessory factors such as TFIID, PAF1, GAF (in Drosophila), the pre-EJC, and FACT, as well as chromatin remodelers and histone modifiers (Fig. 5). However, for many of these factors, how they promote pausing in the context of the Pol II-DSIF-NELF complex remains a mystery. Do they associate directly with the elongation complex or do some of them, such as FACT and other chromatin-associated factors, mediate pausing by organizing the neighboring chromatin architecture (Fig. 5)? Which of these factors function at which genes? The differences in sequence, paused Pol II half-life, and chromatin architecture make it unlikely that promoter proximal pausing occurs in a uniform manner at each gene. It is clear that many of these additional factors, like the pre-EJC, are enriched only at specific promoters, and do not always co-exist with NELF and DSIF. Also unclear is the role of DSIF and NELF phosphorylation in pause release. If the Pol II CTD is not required for pausing or pause release, it follows that DSIF and NELF phosphorylation must play a significant role. To what degree does Spt5 phosphorylation occur in vivo? Which Spt5 phosphorylation sites are most critical for regulation of pausing and pause release and are these functions conserved across species? Does phosphorylation of the NELF-A and NELF-E subunits play a significant, generalized role in pause release, given that most of these sites are not evolutionarily conserved between human and Drosophila (Table 2, Fig. 4). These questions merit significant consideration and require further study to arrive at a complete understanding of the variables that govern promoter proximal pausing. Finally, future analyses should take into account recent evidence that endogenous nucleotides present in cell lysates could allow Pol II to elongate during some nuclear isolation procedures[45]. This is particularly important when knowing the precise location of Pol II is desired.
Table 1:
Overview of Tools and Techniques for the Study of Promoter Proximal Pausing
| Type | Tool/Technique | Utility |
|---|---|---|
| Genome-wide Pol II and transcription factor mapping | ChIP, ChIP-seq, ChIP-exo, ChIP-nexus | Determination of the location of Pol II and its binding partners |
| Genome-wide Pol II mapping | Permanganate ChIP-seq, GRO-seq, PRO-seq, ChRO-seq, NET-seq, chromatin-associated nascent transcript sequencing | Mapping of transcriptionally engaged Pol II |
| Pol II mapping | Nuclear walk-on assay | Quantitative assessment of pausing |
| Pol II mapping | Reporter ChIP-nexus | Evaluation of Pol II pausing on transfected plasmids |
| Genome-wide Pol II and transcription factor mapping | Drosophila polytene chromosome staining | Observing association of Pol II and other factors such as NELF and DSIF with chromosomes |
| Cell free | Nuclear extract pausing assays | Study of individual factors in semi in vivo conditions |
| Cell free | Electrophoretic mobility shift assay | Examination of binding effectiveness between Pol II and pausing factors; useful for targeted mutagenesis studies of NELF and DSIF domains |
| Cell free | Reconstituted pausing assay using purified Pol II, DSIF, NELF. HSF1, and PIC components | Potentially transformative for in vitro mutagenesis-function studies of pausing factors |
| Drug | Triptolide | Inhibition of Pol II initiation; useful for estimates of Pol II pause half life |
| Drug | Flavopiridol, DRB, other P-TEFb inhibitors | Inhibition of Pol II pause release |
| Drug | Pol II-DSIF inhibitors | May provide significant insight into the role of DSIF in pausing |
| Structural data | Structures of Pol II-DSIF-NELF and Pol II-DSIF-Spt6-PAF | Critical roadmaps for designing biochemical studies |
Highlights:
Promoter proximal pausing of RNA Polymerase II is a critical early elongation step
Pausing is governed by a variety of intrinsic and extrinsic interactions
DSIF and NELF are critical for establishing and maintaining pausing
TFIID regulates both promoter proximal pausing and pause release
P-TEFb regulates release of promoter proximal pausing by phosphorylation of DSIF
Acknowledgements:
Funding for this work was provided by NIH grant GM047477 to David S. Gilmour. We thank Seychelle M. Vos for thoughtful reading of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession numbers:
The following structures were used to generate the figures in this review: PDB ID: 6GML, structure of human Pol II-NELF-DSIF complex; PDB ID: 6GMH, structure of human Pol II-DSIF-SPT6-PAF1C; PDB ID: 5OIK, structure of Pol II-DSIF; PDB ID: 6ASX, structure of paused bacterial RNA Polymerase; PDB ID: 6ALF, structure of post-translocated bacterial RNA Polymerase.
Declarations of interest: none
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References:
- [1].Jonkers I, Lis JT. (2015) Getting up to speed with transcription elongation by RNA polymerase II. Nat Rev Mol Cell Biol. 16:167–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Gariglio P, Bellard M, Chambon P. (1981) Clustering of RNA polymerase B molecules in the 5’ moiety of the adult beta-globin gene of hen erythrocytes. Nucleic Acids Res. 9:2589–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Bentley DL, Groudine M. (1986) A block to elongation is largely responsible for decreased transcription of c-myc in differentiated HL60 cells. Nature. 321:702–6. [DOI] [PubMed] [Google Scholar]
- [4].Eick D, Bornkamm GW. (1986) Transcriptional arrest within the first exon is a fast control mechanism in c-myc gene expression. Nucleic Acids Res. 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Nepveu A, Marcu KB. (1986) Intragenic pausing and anti-sense transcription within the murine c-myc locus. EMBO J. 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Kao S, Calman AF, Luciw PA, Peterlin BM. (1987) Anti-termination of transcription within the long terminal repeat of HIV-1 by tat gene product. Nature. 330:489–93. [DOI] [PubMed] [Google Scholar]
- [7].Gilmour DS, Lis JT . (1986) RNA Polymerase II Interacts with the Promoter Region of the Noninduced hsp7O Gene in Drosophila melanogaster Cells. Molecular and Cellular Biology. 6:3984–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Rougvie AE, Lis JT . (1988) The RNA Polymerase II Molecule at the 5’ End of the Uninduced hsp70 Gene of D. melanogaster Is Transcriptionally Engaged. Cell. 54:795–804. [DOI] [PubMed] [Google Scholar]
- [9].Giardina C, Perez-Riba M, Lis JT. (1992) Promoter melting and TFIID complexes on Drosophila genes in vivo. Genes Dev. 6:2190–200. [DOI] [PubMed] [Google Scholar]
- [10].Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. (2007) A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 130:77–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Muse GW, Gilchrist DA, Nechaev S, Shah R, Parker JS, Grissom SF, et al. (2007) RNA polymerase is poised for activation across the genome. Nat Genet. 39:1507–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Zeitlinger J, Stark A, Kellis M, Hong JW, Nechaev S, Adelman K, et al. (2007) RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat Genet. 39:1512–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Core LJ, Waterfall JJ, Lis JT. (2008) Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 322:1845–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Liu X, Gogate AA, Tastemel M, Malladi VS, Yao H, Nguyen K, et al. (2017) Dynamic Change of Transcription Pausing through Modulating NELF Protein Stability Regulates Granulocytic Differentiation. Blood Adv. 1:1358–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Wang X, Lee C, Gilmour DS, Gergen JP. (2007) Transcription elongation controls cell fate specification in the Drosophila embryo. Genes Dev. 21:1031–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Boettiger AN, Levine M. (2009) Synchronous and stochastic patterns of gene activation in the Drosophila embryo. Science. 325:471–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Gilchrist DA, Nechaev S, Lee C, Ghosh SK, Collins JB, Li L, et al. (2008) NELF-mediated stalling of Pol II can enhance gene expression by blocking promoter-proximal nucleosome assembly. Genes Dev. 22:1921–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Gilchrist DA, Dos Santos G, Fargo DC, Xie B, Gao Y, Li L, et al. (2010) Pausing of RNA polymerase II disrupts DNA-specified nucleosome organization to enable precise gene regulation. Cell. 143:540–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Wada T, Takagi T, Yamaguchi Y, Ferdous A, Imai T, Hirose S, et al. (1998) DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev. 12:343–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Yamaguchi Y, Takagi T, Wada T, Yano K, Furuya A, Sugimoto S, et al. (1999) NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell. 97:41–51. [DOI] [PubMed] [Google Scholar]
- [21].Wu CH, Yamaguchi Y, Benjamin LR, Horvat-Gordon M, Washinsky J, Enerly E, et al. (2003) NELF and DSIF cause promoter proximal pausing on the hsp70 promoter in Drosophila. Genes Dev. 17:1402–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Missra A, Gilmour DS. (2010) Interactions between DSIF (DRB sensitivity inducing factor), NELF (negative elongation factor), and the Drosophila RNA polymerase II transcription elongation complex. Proc Natl Acad Sci U S A. 107:11301–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Cheng B, Price DH. (2007) Properties of RNA polymerase II elongation complexes before and after the P-TEFb-mediated transition into productive elongation. J Biol Chem. 282:21901–12. [DOI] [PubMed] [Google Scholar]
- [24].Yamada T, Yamaguchi Y, Inukai N, Okamoto S, Mura T, Handa H. (2006) P-TEFb-mediated phosphorylation of hSpt5 C-terminal repeats is critical for processive transcription elongation. Mol Cell. 21:227–37. [DOI] [PubMed] [Google Scholar]
- [25].Hendrix DA, Hong JW, Zeitlinger J, Rokhsar D, Levine MS. (2008) Promoter elements associated with RNA Pol II stalling in the Drosophila embryo. PNAS. 105:7762–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Gressel S, Schwalb B, Decker TM, Qin W, Leonhardt H, Eick D, et al. (2017) CDK9-dependent RNA polymerase II pausing controls transcription initiation. Elife. 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Shao W, Alcantara SG, Zeitlinger J. (2019) Reporter-ChIP-nexus reveals strong contribution of the Drosophila initiator sequence to RNA polymerase pausing. Elife. 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Kwak H, Fuda NJ, Core LJ, Lis JT. (2013) Precise maps of RNA Polymerase reveal how promoters direct initiation and pausing. Science. 339:950–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Li J, Liu Y, Rhee HS, Ghosh SK, Bai L, Pugh BF, et al. (2013) Kinetic competition between elongation rate and binding of NELF controls promoter-proximal pausing. Mol Cell. 50:711–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Jimeno-Gonzalez S, Ceballos-Chavez M, Reyes JC. (2015) A positioned +1 nucleosome enhances promoter-proximal pausing. Nucleic Acids Res. 43:3068–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Li J, Gilmour DS. (2013) Distinct mechanisms of transcriptional pausing orchestrated by GAGA factor and M1BP, a novel transcription factor. EMBO J. 32:1829–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Mayer A, Landry HM, Churchman LS. (2017) Pause & go: from the discovery of RNA polymerase pausing to its functional implications. Curr Opin Cell Biol. 46:72–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Chen FX, Smith ER, Shilatifard A. (2018) Born to run: control of transcription elongation by RNA polymerase II. Nat Rev Mol Cell Biol. 19:464–78. [DOI] [PubMed] [Google Scholar]
- [34].Core L, Adelman K. (2019) Promoter-proximal pausing of RNA polymerase II: a nexus of gene regulation. Genes Dev. 33:960–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Wissink EM, Vihervaara A, Tippens ND, Lis JT. (2019) Nascent RNA analyses: tracking transcription and its regulation. Nat Rev Genet. 20:705–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Noe Gonzalez M, Blears D, Svejstrup JQ. (2021) Causes and consequences of RNA polymerase II stalling during transcript elongation. Nat Rev Mol Cell Biol. 22:3–21. [DOI] [PubMed] [Google Scholar]
- [37].Johnson DS, Mortazavi A, Myers RM, Wold B. (2007) Genome-wide mapping of in vivo protein-DNA interactions. Science. 316. [DOI] [PubMed] [Google Scholar]
- [38].Rhee HS, Pugh BF. (2011) Comprehensive genome-wide protein-DNA interactions detected at single-nucleotide resolution. Cell. 147:1408–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Rossi MJ, Lai WKM, Pugh BF. (2018) Simplified ChIP-exo assays. Nat Commun. 9:2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Shao W, Zeitlinger J. (2017) Paused RNA polymerase II inhibits new transcriptional initiation. Nat Genet. 49:1045–51. [DOI] [PubMed] [Google Scholar]
- [41].He Q, Johnston J, Zeitlinger J. (2015) ChIP-nexus enables improved detection of in vivo transcription factor binding footprints. Nat Biotechnol. 33:395–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Lee C, Li X, Hechmer A, Eisen M, Biggin MD, Venters BJ, et al. (2008) NELF and GAGA factor are linked to promoter-proximal pausing at many genes in Drosophila. Mol Cell Biol. 28:3290–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Qiu Y, Gilmour DS. (2017) Identification of Regions in the Spt5 Subunit of DRB Sensitivity-inducing Factor (DSIF) That Are Involved in Promoter-proximal Pausing. J Biol Chem. 292:5555–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Chu T, Rice EJ, Booth GT, Salamanca HH, Wang Z, Core LJ, et al. (2018) Chromatin run-on and sequencing maps the transcriptional regulatory landscape of glioblastoma multiforme. Nat Genet. 50:1553–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Luse DS, Parida M, Spector BM, Nilson KA, Price DH. (2020) A unified view of the sequence and functional organization of the human RNA polymerase II promoter. Nucleic Acids Res. 48:7767–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Ball CB, Nilson KA, Price DH. (2019) Use of the nuclear walk-on methodology to determine sites of RNA polymerase II initiation and pausing and quantify nascent RNAs in cells. Methods. 159–160:165–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Nilson KA, Lawson CK, Mullen NJ, Ball CB, Spector BM, Meier JL, et al. (2017) Oxidative stress rapidly stabilizes promoter-proximal paused Pol II across the human genome. Nucleic Acids Res. 45:11088–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Churchman LS, Weissman JS. (2011) Nascent transcript sequencing visualizes transcription at nucleotide resolution. Nature. 469:368–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Nojima T, Gomes T, Grosso ARF, Kimura H, Dye MJ, Dhir S, et al. (2015) Mammalian NET-Seq Reveals Genome-wide Nascent Transcription Coupled to RNA Processing. Cell. 161:526–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Nechaev S, Fargo DC, Santos G, Liu L, Gao Y, Adelman K. (2010) Global analysis of short RNAs reveals widespread promoter-proximal stalling and arrest of Pol II in Drosophila. Science. 327:335–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Henriques T, Scruggs BS, Inouye MO, Muse GW, Williams LH, Burkholder AB, et al. (2018) Widespread transcriptional pausing and elongation control at enhancers. Genes Dev. 32:26–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Mayer A, di Iulio J, Maleri S, Eser U, Vierstra J, Reynolds A, et al. (2015) Native elongating transcript sequencing reveals human transcriptional activity at nucleotide resolution. Cell. 161:541–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Weber CM, Ramachandran S, Henikoff S. (2014) Nucleosomes are context-specific, H2A.Z-modulated barriers to RNA polymerase. Mol Cell. 53:819–30. [DOI] [PubMed] [Google Scholar]
- [54].Reppas NB, Wade JT, Church GM, Struhl K. (2006) The transition between transcriptional initiation and elongation in E. coli is highly variable and often rate limiting. Mol Cell. 24:747–57. [DOI] [PubMed] [Google Scholar]
- [55].Hou L, Wang Y, Liu Y, Zhang N, Shamovsky I, Nudler E, et al. (2019) Paf1C regulates RNA polymerase II progression by modulating elongation rate. Proc Natl Acad Sci U S A. 116:14583–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Boehm AK, Saunders A, Werner J, Lis JT. (2003) Transcription factor and polymerase recruitment, modification, and movement on dhsp70 in vivo in the minutes following heat shock. Mol Cell Biol. 23:7628–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Weber JA, Taxman DJ, Lu Q, Gilmour DS. (1997) Molecular architecture of the hsp70 promoter after deletion of the TATA box or the upstream regulation region. Mol Cell Biol. 17:3799–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Marshall NF, Price DH. (1992) Control of formation of two distinct classes of RNA polymerase II elongation complexes. Mol Cell Biol. 12:2078–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Fant CB, Levandowski CB, Gupta K, Maas ZL, Moir J, Rubin JD, et al. (2020) TFIID Enables RNA Polymerase II Promoter-Proximal Pausing. Mol Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Egyhazi E, Ossoinak A, Pigon A, Holmgren C, Lee JM, Greenleaf AL (1996) Phosphorylation dependence of the initiation of productive transcription of Balbiani ring 2 genes in living cells. Chromosoma. 104:422–33. [DOI] [PubMed] [Google Scholar]
- [61].Chao SH, Price DH. (2001) Flavopiridol inactivates P-TEFb and blocks most RNA polymerase II transcription in vivo. J Biol Chem. 276:31793–9. [DOI] [PubMed] [Google Scholar]
- [62].Akhtar J, Kreim N, Marini F, Mohana G, Brune D, Binder H, et al. (2019) Promoter-proximal pausing mediated by the exon junction complex regulates splicing. Nat Commun. 10:521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Jonkers I, Kwak H, Lis JT. (2014) Genome-wide dynamics of Pol II elongation and its interplay with promoter proximal pausing, chromatin, and exons. Elife. 3:e02407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Mancebo HS, Lee G, Flygare J, Tomassini J, Luu P, Zhu Y, et al. (1997) P-TEFb kinase is required for HIV Tat transcriptional activation in vivo and in vitro. Genes Dev. 11:2633–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Morales F, Giordano A. (2016) Overview of CDK9 as a target in cancer research. Cell Cycle. 15:519–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Lucking U, Scholz A, Lienau P, Siemeister G, Kosemund D, Bohlmann R, et al. (2017) Identification of Atuveciclib (BAY 1143572), the First Highly Selective, Clinical PTEFb/CDK9 Inhibitor for the Treatment of Cancer. ChemMedChem. 12:1776–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Olson CM, Jiang B, Erb MA, Liang Y, Doctor ZM, Zhang Z, et al. (2018) Pharmacological perturbation of CDK9 using selective CDK9 inhibition or degradation. Nat Chem Biol. 14:163–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Henriques T, Gilchrist DA, Nechaev S, Bern M, Muse GW, Burkholder A, et al. (2013) Stable pausing by RNA polymerase II provides an opportunity to target and integrate regulatory signals. Mol Cell. 52:517–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Buckley MS, Kwak H, Zipfel WR, Lis JT. (2014) Kinetics of promoter Pol II on Hsp70 reveal stable pausing and key insights into its regulation. Genes Dev. 28:14–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Krebs AR, Imanci D, Hoerner L, Gaidatzis D, Burger L, Schubeler D. (2017) Genome-wide Single-Molecule Footprinting Reveals High RNA Polymerase II Turnover at Paused Promoters. Mol Cell. 67:411–22 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Bahat A, Lahav O, Plotnikov A, Leshkowitz D, Dikstein R. (2019) Targeting Spt5-Pol II by Small-Molecule Inhibitors Uncouples Distinct Activities and Reveals Additional Regulatory Roles. Mol Cell. 76:617–31 e4. [DOI] [PubMed] [Google Scholar]
- [72].Vos SM, Farnung L, Urlaub H, Cramer P. (2018) Structure of paused transcription complex Pol II-DSIF-NELF. Nature. 560:601–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Vos SM, Farnung L, Boehning M, Wigge C, Linden A, Urlaub H, et al. (2018) Structure of activated transcription complex Pol II-DSIF-PAF-SPT6. Nature. 560:607–12. [DOI] [PubMed] [Google Scholar]
- [74].Vos SM, Farnung L, Linden A, Urlaub H, Cramer P. (2020) Structure of complete Pol II-DSIF-PAF-SPT6 transcription complex reveals RTF1 allosteric activation. Nat Struct Mol Biol. 27:668–77. [DOI] [PubMed] [Google Scholar]
- [75].Decker TM. (2020) Mechanisms of Transcription Elongation Factor DSIF (Spt4-Spt5). J Mol Biol. [DOI] [PubMed] [Google Scholar]
- [76].Merkley ED, Rysavy S, Kahraman A, Hafen RP, Daggett V, Adkins JN. (2014) Distance restraints from crosslinking mass spectrometry: mining a molecular dynamics simulation database to evaluate lysine-lysine distances. Protein Sci. 23:747–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Sansó M, Levin RS, Lipp JJ, Wang VY, Greifenberg AK, Quezada EM, et al. (2015) P-TEFb regulation of transcription termination factor Xrn2 revealed by a chemical genetic screen for Cdk9 substrates. Genes Dev. 30:117–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Parua PK, Kalan S, Benjamin B, Sanso M, Fisher RP. (2020) Distinct Cdk9-phosphatase switches act at the beginning and end of elongation by RNA polymerase II. Nat Commun. 11:4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Yamaguchi Y, Inukai N, Narita T, Wada T, Handa H. (2002) Evidence that negative elongation factor represses transcription elongation through binding to a DRB sensitivity-inducing factor/RNA polymerase II complex and RNA. Mol Cell Biol. 22:2918–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Kang JY, Mishanina TV, Bellecourt MJ, Mooney RA, Darst SA, Landick R. (2018) RNA Polymerase Accommodates a Pause RNA Hairpin by Global Conformational Rearrangements that Prolong Pausing. Mol Cell. 69:802–15 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Guo X, Myasnikov AG, Chen J, Crucifix C, Papai G, Takacs M, et al. (2018) Structural Basis for NusA Stabilized Transcriptional Pausing. Mol Cell. 69:816–27 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Kang JY, Olinares PD, Chen J, Campbell EA, Mustaev A, Chait BT, et al. (2017) Structural basis of transcription arrest by coliphage HK022 Nun in an Escherichia coli RNA polymerase elongation complex. Elife. 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Fujinaga K, Irwin D, Huang Y, Taube R, Kurosu T, Peterlin BM. (2004) Dynamics of human immunodeficiency virus transcription: P-TEFb phosphorylates RD and dissociates negative effectors from the transactivation response element. Mol Cell Biol. 24:787–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Rao JN, Neumann L, Wenzel S, Schweimer K, Rosch P, Wohrl BM. (2006) Structural studies on the RNA-recognition motif of NELF E, a cellular negative transcription elongation factor involved in the regulation of HIV transcription. Biochem J. 400:449–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Zhang Z, Klatt A, Gilmour DS, Henderson AJ. (2007) Negative elongation factor NELF represses human immunodeficiency virus transcription by pausing the RNA polymerase II complex. J Biol Chem. 282:16981–8. [DOI] [PubMed] [Google Scholar]
- [86].Kulinski T, Olejniczak M, Huthoff H, Bielecki L, Pachulska-Wieczorek K, Das AT, et al. (2003) The apical loop of the HIV-1 TAR RNA hairpin is stabilized by a cross-loop base pair. J Biol Chem. 278:38892–901. [DOI] [PubMed] [Google Scholar]
- [87].Wang D, Bushnell DA, Westover KD, Kaplan CD, Kornberg RD. (2006) Structural basis of transcription: role of the trigger loop in substrate specificity and catalysis. Cell. 127:941–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Vassylyev DG, Vassylyeva MN, Perederina A, Tahirov TH, Artsimovitch I. (2007) Structural basis for transcription elongation by bacterial RNA polymerase. Nature. 448:157–62. [DOI] [PubMed] [Google Scholar]
- [89].Schaukowitch K, Joo JY, Liu X, Watts JK, Martinez C, Kim TK. (2014) Enhancer RNA facilitates NELF release from immediate early genes. Mol Cell. 56:29–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Pagano JM, Kwak H, Waters CT, Sprouse RO, White BS, Ozer A, et al. (2014) Defining NELF-E RNA binding in HIV-1 and promoter-proximal pause regions. PLoS Genet. 10:e1004090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Vos SM, Pollmann D, Caizzi L, Hofmann KB, Rombaut P, Zimniak T, et al. (2016) Architecture and RNA binding of the human negative elongation factor. Elife. 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Aoi Y, Smith ER, Shah AP, Rendleman EJ, Marshall SA, Woodfin AR, et al. (2020) NELF Regulates a Promoter-Proximal Step Distinct from RNA Pol II Pause-Release. Mol Cell. 78:261–74 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Narita T, Yung TM, Yamamoto J, Tsuboi Y, Tanabe H, Tanaka K, et al. (2007) NELF interacts with CBC and participates in 3’ end processing of replication-dependent histone mRNAs. Mol Cell. 26:349–65. [DOI] [PubMed] [Google Scholar]
- [94].Schulze WM, Cusack S. (2017) Structural basis for mutually exclusive co-transcriptional nuclear cap-binding complexes with either NELF-E or ARS2. Nat Commun. 8:1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Marshall NF, Price DH. (1995) Purification of P-TEFb, a transcription factor required for the transition into producitve elongation. J Biol Chem. 270:12335–8. [DOI] [PubMed] [Google Scholar]
- [96].Peng J, Marshall NF, Price DH. (1998) Identification of a cyclin subunit required for the function of Drosophila P-TEFb. J Biol Chem. 273:13855–60. [DOI] [PubMed] [Google Scholar]
- [97].Peng J, Zhu Y, Milton JT, Price DH. (1998) Identification of multiple cyclin subunits of human P-TEFb. Genes Dev. 12:755–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Zhou Q, Li T, Price DH. (2012) RNA polymerase II elongation control. Annu Rev Biochem. 81:119–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Shilatifard A, Lane WS, Jackson KW, Conaway CR, Conaway JW. (1996) An RNA Polymerase II Elongation Factor Encoded by the Human ELL Gene. Science. 271:1873–6. [DOI] [PubMed] [Google Scholar]
- [100].He N, Liu M, Hsu J, Xue Y, Chou S, Burlingame A, et al. (2010) HIV-1 Tat and host AFF4 recruit two transcription elongation factors into a bifunctional complex for coordinated activation of HIV-1 transcription. Mol Cell. 38:428–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Lin C, Smith ER, Takahashi H, Lai KC, Martin-Brown S, Florens L, et al. (2010) AFF4, a component of the ELL/P-TEFb elongation complex and a shared subunit of MLL chimeras, can link transcription elongation to leukemia. Mol Cell. 37:429–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Sobhian B, Laguette N, Yatim A, Nakamura M, Levy Y, Kiernan R, et al. (2010) HIV-1 Tat assembles a multifunctional transcription elongation complex and stably associates with the 7SK snRNP. Mol Cell. 38:439–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Luo Z, Lin C, Shilatifard A. (2012) The super elongation complex (SEC) family in transcriptional control. Nat Rev Mol Cell Biol. 13:543–7. [DOI] [PubMed] [Google Scholar]
- [104].Yadav D, Ghosh K, Basu S, Roeder RG, Biswas D. (2019) Multivalent Role of Human TFIID in Recruiting Elongation Components at the Promoter-Proximal Region for Transcriptional Control. Cell Rep. 26:1303–17 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Huang KL, Jee D, Stein CB, Elrod ND, Henriques T, Mascibroda LG, et al. (2020) Integrator Recruits Protein Phosphatase 2A to Prevent Pause Release and Facilitate Transcription Termination. Mol Cell. 80:345–58 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Lu X, Zhu X, Li Y, Liu M, Yu B, Wang Y, et al. (2016) Multiple P-TEFbs cooperatively regulate the release of promoter-proximally paused RNA polymerase II. Nucleic Acids Res. 44:6853–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Rawat P, Boehning M, Hummel B, Aprile-Garcia F, Pandit AS, Eisenhardt N, et al. (2021) Stress-induced nuclear condensation of NELF drives transcriptional downregulation. Molecular Cell. 81:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Marshall NF, Peng J, Xie Z, Price DH. (1996) Control of RNA Polymerase II elongation potential by a novel carboxyl-terminal domain kinase. J Biol Chem. 271:27176–83. [DOI] [PubMed] [Google Scholar]
- [109].Komarnitsky P, Cho EJ, Buratowski S. (2000) Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 14:2452–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Zhou M, Halanski MA, Radonovich MF, Kashanchi F, Peng J, Price DH, et al. (2000) Tat modifies the activity of CDK9 to phosphorylate Serine 5 of the RNA Polymerase II carboxyl-terminal domain during Human Immunodeficiency Virus Type 1 transcription. Molecular and Cellular Biology. 20:5077–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Gerber A, Roeder RG. (2020) The CTD Is Not Essential for the Post-Initiation Control of RNA Polymerase II Activity. J Mol Biol. 432:5489–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Guermah M, Tao Y, Roeder RG. (2001) Positive and negative TAF(II) functions that suggest a dynamic TFIID structure and elicit synergy with traps in activator-induced transcription. Mol Cell Biol. 21:6882–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Grob P, Cruse MJ, Inouye C, Peris M, Penczek PA, Tjian R, et al. (2006) Cryo-electron microscopy studies of human TFIID: conformational breathing in the integration of gene regulatory cues. Structure. 14:511–20. [DOI] [PubMed] [Google Scholar]
- [114].Nogales E, Fang J, Louder RK. (2017) Structural dynamics and DNA interaction of human TFIID. Transcription. 8:55–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Sypes MA, Gilmour DS. (1994) Protein/DNA crosslinking of a TFIID complex reveals novel interactions downstream of the transcription start. Nucleic Acids Res. 22:807–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Wu CH, Madabusi L, Nishioka H, Emanuel P, Sypes M, Arkhipova I, et al. (2001) Analysis of core promoter sequences located downstream from the TATA element in the hsp70 promoter from Drosophila melanogaster. Mol Cell Biol. 21:1593–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Baumann DG, Gilmour DS. (2017) A sequence-specific core promoter-binding transcription factor recruits TRF2 to coordinately transcribe ribosomal protein genes. Nucleic Acids Res. 45:10481–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Lee H, Kraus KW, Wolfner MF, Lis JT. (1992) DNA sequence requirements for generating paused polymerase at the start of hsp70. Genes Dev. 6:284–95. [DOI] [PubMed] [Google Scholar]
- [119].Shopland LS, Hirayoshi K, Fernandes M, Lis JT. (1995) HSF access to heat shock elements in vivo depends critically on promoter architecture defined by GAGA factor, TFIID, and RNA Polymerase II binding sites. Genes Dev. 9:2756–69. [DOI] [PubMed] [Google Scholar]
- [120].Wang YV, Tang H, Gilmour DS. (2005) Identification in vivo of different rate-limiting steps associated with transcriptional activators in the presence and absence of a GAGA element. Mol Cell Biol. 25:3543–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Fuda NJ, Guertin MJ, Sharma S, Danko CG, Martins AL, Siepel A, et al. (2015) GAGA factor maintains nucleosome-free regions and has a role in RNA polymerase II recruitment to promoters. PLoS Genet. 11:e1005108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Tsai SY, Chang YL, Swamy KB, Chiang RL, Huang DH. (2016) GAGA factor, a positive regulator of global gene expression, modulates transcriptional pausing and organization of upstream nucleosomes. Epigenetics Chromatin. 9:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Matharu NK, Hussain T, Sankaranarayanan R, Mishra RK. (2010) Vertebrate homologue of Drosophila GAGA factor. J Mol Biol. 400:434–47. [DOI] [PubMed] [Google Scholar]
- [124].Chen FX, Woodfin AR, Gardini A, Rickels RA, Marshall SA, Smith ER, et al. (2015) PAF1, a Molecular Regulator of Promoter-Proximal Pausing by RNA Polymerase II. Cell. 162:1003–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Yu M, Yang W, Ni T, Tang Z, Nakadai T, Zhu J, et al. (2015) RNA polymerase II-associated factor 1 regulates the release and phosphorylation of paused RNA polymerase II. Science. 350:1383–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Chen FX, Xie P, Collings CK, Cao K, Aoi Y, Marshall SA, et al. (2017) PAF1 regulation of promoter-proximal pause release via enhancer activation. Science. 357:1294–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Tettey TT, Gao X, Shao W, Li H, Story BA, Chitsazan AD, et al. (2019) A Role for FACT in RNA Polymerase II Promoter-Proximal Pausing. Cell Rep. 27:3770–9 e7. [DOI] [PubMed] [Google Scholar]
- [128].Luse DS, Izban MG. (1992) The RNA polymerase II ternary complex cleaves the nascent transcript in a 3’----5’ direction in the presence of elongation factor SII. Genes Dev. 6:1342–56. [DOI] [PubMed] [Google Scholar]
- [129].Reines D (1992) Elongation Factor-dependent Transcript Shortening by Template-engaged RNA Polymerase II. J Biol Chem. 267:3795–800. [PMC free article] [PubMed] [Google Scholar]
- [130].Adelman K, Marr MT, Werner J, Saunders A, Ni Z, Andrulis ED, et al. (2005) Efficient release from promoter-proximal stall sites requires transcript cleavage factor TFIIS. Mol Cell. 17:103–12. [DOI] [PubMed] [Google Scholar]
- [131].Sheridan RM, Fong N, D’Alessandro A, Bentley DL. (2019) Widespread Backtracking by RNA Pol II Is a Major Effector of Gene Activation, 5’ Pause Release, Termination, and Transcription Elongation Rate. Mol Cell. 73:107–18 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Palangat M, Renner DB, Price DH, Landick R. (2005) A negative elongation factor for human RNA polymerase II inhibits the anti-arrest transcript-cleavage factor TFIIS. Proc Natl Acad Sci U S A. 102:15036–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Watts JA, Burdick J, Daigneault J, Zhu Z, Grunseich C, Bruzel A, et al. (2019) cis Elements that Mediate RNA Polymerase II Pausing Regulate Human Gene Expression. Am J Hum Genet. 105:677–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Izban MG, Luse DS. (1991) Transcription on nucleosomal templates by RNA polymerase II in vitro: inhibition of elongation with enhancement of sequence specific pausing. Genes Dev. 5:683–96. [DOI] [PubMed] [Google Scholar]
- [135].Brown SA, Imbalzano AN, Kingston RE. (1996) Activator-dependent regulation of transcriptional pausing on nucleosomal templates. Genes Dev. 10:1479–90. [DOI] [PubMed] [Google Scholar]
- [136].Benjamin LR, Gilmour DS. (1998) Nucleosomes are not necessary for promoter-proximal pausing in vitro on the Drosophila hsp70 promoter. Nucleic Acids Res. 26:1051–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Mavrich TN, Jiang C, Ioshikhes IP, Li X, Venters BJ, Zanton SJ, et al. (2008) Nucleosome organization in the Drosophila genome. Nature. 453:358–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Deal RB, Henikoff JG, Henikoff S. (2010) Genome-wide kinetics of nucleosome turnover determined by metabolic labeling of histones. Science. 328:1161–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Etchegaray JP, Zhong L, Li C, Henriques T, Ablondi E, Nakadai T, et al. (2019) The Histone Deacetylase SIRT6 Restrains Transcription Elongation via Promoter-Proximal Pausing. Mol Cell. 75:683–99 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Gates LA, Shi J, Rohira AD, Feng Q, Zhu B, Bedford MT, et al. (2017) Acetylation on histone H3 lysine 9 mediates a switch from transcription initiation to elongation. J Biol Chem. 292:14456–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Vaid R, Wen J, Mannervik M. (2020) Release of promoter-proximal paused Pol II in response to histone deacetylase inhibition. Nucleic Acids Res. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Titov DV, Gilman B, He QL, Bhat S, Low WK, Dang Y, et al. (2011) XPB, a subunit of TFIIH, is a target of the natural product triptolide. Nat Chem Biol. 7:182–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Alekseev S, Nagy Z, Sandoz J, Weiss A, Egly JM, Le May N, et al. (2017) Transcription without XPB Establishes a Unified Helicase-Independent Mechanism of Promoter Opening in Eukaryotic Gene Expression. Mol Cell. 65:504–14 e4. [DOI] [PubMed] [Google Scholar]
- [144].Verma D, Church TM, Swaminathan S. (2020) Epstein-Barr virus co-opts TFIIH component XPB to specifically activate essential viral lytic promoters. Proc Natl Acad Sci U S A. 117:13044–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Chen F, Gao X, Shilatifard A. (2015) Stably paused genes revealed through inhibition of transcription initiation by the TFIIH inhibitor triptolide. Genes Dev. 29:39–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Price DH. (2018) Transient pausing by RNA polymerase II. Proc Natl Acad Sci U S A. 115:4810–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Steurer B, Janssens RC, Geverts B, Geijer ME, Wienholz F, Theil AF, et al. (2018) Live-cell analysis of endogenous GFP-RPB1 uncovers rapid turnover of initiating and promoter-paused RNA Polymerase II. Proc Natl Acad Sci U S A. 115:E4368–E76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Elrod ND, Henriques T, Huang KL, Tatomer DC, Wilusz JE, Wagner EJ, et al. (2019) The Integrator Complex Attenuates Promoter-Proximal Transcription at Protein-Coding Genes. Mol Cell. 76:738–52 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]


