Abstract
Artificial intelligence (AI) has recently become a very popular buzzword, as a consequence of disruptive technical advances and impressive experimental results, notably in the field of image analysis and processing. In medicine, specialties where images are central, like radiology, pathology or oncology, have seized the opportunity and considerable efforts in research and development have been deployed to transfer the potential of AI to clinical applications. With AI becoming a more mainstream tool for typical medical imaging analysis tasks, such as diagnosis, segmentation, or classification, the key for a safe and efficient use of clinical AI applications relies, in part, on informed practitioners. The aim of this review is to present the basic technological pillars of AI, together with the state-of-the-art machine learning methods and their application to medical imaging. In addition, we discuss the new trends and future research directions. This will help the reader to understand how AI methods are now becoming an ubiquitous tool in any medical image analysis workflow and pave the way for the clinical implementation of AI-based solutions.
1. Introduction
For the last decade, the locution Artificial Intelligence (AI) has progressively flooded many scientific journals, including those of image processing and medical physics. Paradoxically, though, AI is an old concept, starting to be formalized in the 1940s, while the term of artificial intelligence itself was coined in 1956 by John McCarthy. In short, AI refers to computer algorithms that can mimic features that are characteristic of human intelligence, such as problem solving or learning. The latest success of AI has been made possible thanks to tremendous growths of both computational power and data availability. In particular, AI applications based on machine learning (ML) algorithms have experienced unprecedented breakthroughs during the last decade in the field of computer vision. The medical community has taken advantage of these extraordinary developments in order to build AI applications that get the most of medical images, automating different steps of the clinical practice or providing support for clinical decisions. Papers relying on AI and ML report promising results in a wide range of medical applications [1–7]. Disease diagnosis, image segmentation or outcome prediction are some of the tasks that are experiencing a disruptive transformation thanks to the latest progress of AI.
More recently, ML tools have become mature enough to fulfill clinical requirements and, thus, research and clinical teams, as well as companies are working together to develop clinical AI solutions. Today, we are closer than ever to the clinical implementation of AI and, therefore, getting to know the basics of this technology becomes a “must” for every professional in the medical field. Helping the medical physics community to acquire such a solid background knowledge about AI and learning methods, including their evolution and current state of the art, will certainly result in higher quality research, facilitate the first steps of new researchers in this field, and inspire novel research directions.
The goal of this review article is to briefly walk the reader through some basic AI concepts with focus on medical imaging processing (Section 2); followed by a presentation of the state-of-the-art methods and current trends in the domain (Section 3). To finish, we discuss the future research directions that will make possible the next generation of AI-based solutions for medical image applications (Section 4).
2. Building blocks of AI methods for medical imaging
The field of AI evolves rapidly, with new methods published at a high pace. However, there are several central concepts that have settled for good. This section presents a brief overview of these building blocks for AI methods, with a focus on medical imaging. For more detailed descriptions we refer to relevant books [8–11] and publications [12,13].
2.1. Artificial intelligence, machine learning, and deep learning
As mentioned previously, AI broadly refers to any method or algorithm that mimics human intelligence. Historically, AI has been approached from two directions: computationalism and connectionism. The former attempts to mimic formal reasoning and logic directly, regardless of its biological implementation. Mostly based on hardcoded axioms and rules that are combined to deduce new conclusions, computationalism is conceptually similar to computers, storing and processing symbols. Connectionism, on the other hand, rather follows a bottom-up approach, starting from models of biological neurons that are interconnected in large numbers and from which intelligence is intended to emerge by learning from experience. Expert systems [14–16], which started to be very popular in the 1980, are a classical example of computationalism. Some famous applications of expert systems to the medical field are MYCIN (diagnosis of bacterial infection in the blood) [17], PUFF (interpretation of pulmonary function data) [18], or INTERNIST-1 (diagnosis for internal medicine) [19]. However, the bottleneck of expert systems is the complexity of acquiring the required knowledge in the form of production rules and, thus, interest in computationalist algorithms started to fade since the 1990’s in favor of connectionism approaches [20,21]. The appeal of connectionism and learning-based AI holds in that it delegates the responsibility for accuracy and exhaustiveness to data instead of human experts, who might be poorly available, prone to error, bias, or subjectivity. The ever growing abundance of data, including medical images, then typically tilts the scales in favor of learning techniques, and the community has focused successively on two nested subfamilies (Figure 1): machine learning and deep learning.
Figure 1.
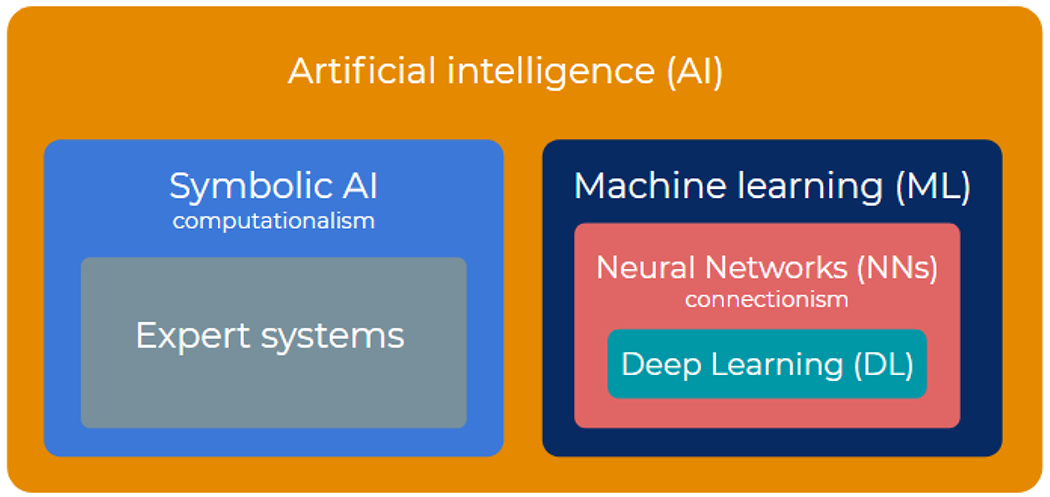
Artificial intelligence, machine learning, and deep learning can be seen as matryoshkas nested in each other. Artificial intelligence gathers both symbolic (top down) and connectionist (bottom up) approaches. Machine learning is the dominant branch of connectionism, combining biological (neural networks) and statistical (data-driven learning theory) influences. Deep learning focuses mainly on large-size neural networks, with functional specificities to process images, sounds, videos, etc.
The specificity of Machine learning (ML) is to be driven by data, which gives machines (computers) “the ability to learn without being explicitly programmed”, as formulated by Arthur Samuel in 1959, a pioneer in the ML field. ML typically works in two phases, training and inference. Training allows patterns to be found in previously collected data, whereas inference compares these patterns to new unseen data to then carry out a certain task like prediction or decision making. Since the 1990’s, ML algorithms have continuously evolved and improved, becoming more sophisticated and including hierarchical structures, which gave rise to the popular Deep Learning. The term Deep Learning (DL) was first coined by Aizenberg et al.[22] in the 2000s, and refers to a subset of ML algorithms, with the particularity of being organized hierarchically, on multiple levels, hence the term “deep”, to automatically extract meaningful features from data.
Although ML encompasses DL, DL is often opposed to classical “shallow” ML, the latter relying on algorithms that have a flatter architecture and depend on previous feature engineering to extract data representations. This distinction also reflects the evolution from ML to DL, namely, from specific feature engineering to generic feature learning. While ML generally relies on domain knowledge and expertise to define relevant features, DL involves generic, trainable features. In other words, despite the modeling power of ML, global performance remains limited by the adequacy of manually picked features. Alternatively, DL replaces these fixed specialized features with generic, trainable, low-level features that are involved in the learning procedure, thereby offering better performance guarantees. Sophistication is here achieved by stacking layers of simple features, leading to a hierarchical model structure. As training in DL concerns both the low-level features and the higher-level model, DL is often referred to as an end-to-end approach. For image data, this approach typically allows DL to learn optimal filters.
Today, ML models have reached important milestones, in some cases being able to accomplish tasks with an accuracy that is similar to or even better than human experts. For instance, the diagnostic performance of DL models has demonstrated to be equivalent to that of health-care professionals for certain applications [23], such as skin-cancer detection [24] or breast cancer detection [25]. In particular, the latter reported a DL model that not only reached an excellent performance in mammogram classification, but also outperformed five out of five full-time breast-imaging specialists with an average increase in sensitivity of 14% [25]. Image segmentation is another task that has experienced a transformation with the advent of ML algorithms. For instance, a recent study has described a DL model that can perform organ segmentation in the head and neck region from CT images with performance comparable to experienced radiographers [26]. For more detailed examples of the performance of state-of-the-art ML and DL methods for medical applications we refer to Section 3.
2.2. Learning frameworks and strategies
Machine learning can be broadly split into two complementary categories, supervised and unsupervised, which are inspired from human learning (Table 1). Supervised learning is the simplest and provides the tightest framework with strongest guarantees. It formalizes learning with a parent or teacher, providing the inputs and controlling the outputs. In supervised learning, the training data consists thus of labelled or annotated (input,output) pairs, and the model is trained to yield the right desired output when presented with some input. When data is not annotated, unsupervised learning, also known as self-organization, aims at discovering patterns in data (Figure 2).
Table 1.
Different learning frameworks and strategies, together with some of the most popular algorithms or techniques that are used for each of them, as well as a few examples of common applications in the field of medical imaging. The table is divided in three parts: the basic learning frameworks (supervised, unsupervised and reinforcement learning), the hybrid learning frameworks blending supervised and unsupervised, and finally common learning strategies that solve consecutive learning problems or combine several models together.
| Learning style | Common algorithms / methods | Examples |
|---|---|---|
| BASIC LEARNING FRAMEWORKS | ||
| Supervised learning | • Linear or logistic regression • Decision trees and random forests • Support vector machines • Convolutional neural networks • Recurrent neural networks |
• Cancer diagnosis [78–81] • Organ segmentation [26,82–86] • Radiotherapy dose denoising [33] • Radiotherapy dose prediction [87,88] • Conversion between image modalities [89,90] |
| Unsupervised learning | • (Variational) Auto encoders • Dimensionality reduction (e.g., Principal component analysis) • Clustering (e.g., K-means) |
• Domain adaptation tasks[35–37,91,92] • Classification of patient groups [93] • Image reconstruction [94] |
| Reinforcement learning | • Q-learning • Markov Decision Processes |
• Tumor segmentation [54,55] • Image reconstruction [95] • Treatment planning [50–53,96] |
| HYBRID LEARNING FRAMEWORKS | ||
| Semi-supervised learning | • Generative Adversarial Networks | • Tumor classification [45,46] • Organ segmentation [46] • Synthetic image generation [97,98] |
| Self-supervised learning | • Pretext task: distortion (e.g. rotation), color- or intensity-based, patch extraction | • Image classification or segmentation [76] |
| LEARNING STRATEGIES | ||
| Transfer learning | • Inductive • Transductive • Unsupervised |
• Radiotherapy toxicity prediction [58] • Adaptation to different clinical practices [62] Improving model generalization[99] |
| Ensemble learning | • Bagging - Bootstrap AGGregatING - (e.g. random forests) • Boosting (e.g. AdaBoost, gradient boosting) |
• Radiotherapy dose prediction [100,101] • Estimation of uncertainty [102] • Stratification of patients[103] |
Figure 2.
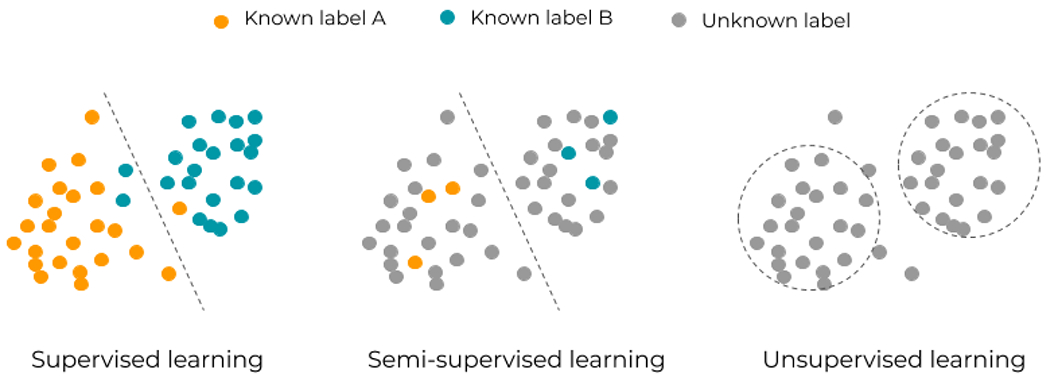
Three classical learning frameworks in artificial intelligence: supervised, semi-supervised, and unsupervised learning. Supervised learning relies on known input-output pairs. If some output labels are difficult or expensive to get, semi-supervised learning can apply. If no labels are available, unsupervised learning allows for a more exploratory approach of data.
Typical supervised tasks involve function approximation, like regression and classification. Classification can be binary, like in determining whether a pathology is present or not in an image [25,27], involve multiple classes, as in determining a particular pathology among several labels [28–30], or concern not the whole image but each pixel, as done for image segmentation [31,32]. On the regression side, also in a pixel-wise way, image enhancement (e.g. improving a low-quality image, the input, by mapping it to its higher quality counterpart, the output label or annotation) [33] or image-to-image mapping (e.g. mapping a CT image, the input, to the corresponding dose distribution, the output) [34]. More examples of clinical applications of supervised learning and the common ML methods used within this learning framework are presented in Table 1.
In contrast, most unsupervised tasks relate to probability density estimation, like clustering (finding separated groups of similar data items), outlier or anomaly detection (isolated items), or even manifold learning and dimensionality reduction (subspaces on which data concentrate). The use of unsupervised learning has been, so far, much more limited than its supervised counterpart, although useful applications for medical imaging exist, such as domain adaptation (e.g., adapting a segmentation model trained on an image modality to work on a different image modality) [35–37], data generation (e.g., generate artificial realistic images) [38–40] or even image segmentation [41]. Table 1 presents some of the main ML methods that work in an unsupervised framework.
Semi-supervised learning is a hybrid framework halfway between supervised and unsupervised and thus it involves data for which desired outputs are only partly known. Groups identified as clusters by unsupervised learning can be used as possible class labels [42] (Figure 2). Some examples of clinical applications for semi-supervised learning include the generation or translation of images from a specific class to another in a semi-supervised setting (e.g., generation of synthetic CTs from MR images) [43,44], and segmentation or classification of images with partially labelled data [45,46].
So far, supervised learning has been the most used learning framework for medical imaging applications, as it is totally univocal and models are very easy to train. However, it is well-known that data labelling in the medical domain is an extremely time-consuming task, subject to costly inspection by human experts. Therefore, more and more researchers are now exploring semi-supervised learning techniques because they are an excellent alternative to complement small sets of carefully labelled data with large amounts of cheap unlabelled data collected automatically [42,47]. In fact, many of the current limitations of ML/DL algorithms come from the use of labelled data (e.g., errors in labels [48], limited size labelled databases, etc) and thus, although the use of fully unsupervised learning in the medical field is still very limited, we believe that future research will focus on unsupervised techniques in order to unlock the full potential of ML. Very recently, unsupervised models are achieving improved performances over supervised models for computer vision tasks [49], and the same is likely to happen for medical imaging applications.
Yet another type of learning is by interacting with an environment where an agent gets feedback from its actions over the course of time, which is known as reinforcement learning. After each action towards a new state, the environment can either reward or punish the agent who has then to best predict the longer-term consequences of future actions in a trial and error fashion. The use of reinforcement learning for medical imaging is still not very extended, but has increased in the last couple of years, with promising applications that allow mimicking physician behaviour for typical tasks such as the design of a treatment [50–55], among others (Table 1).
On top of these three basic learning frameworks (supervised, unsupervised and reinforcement learning), there are other strategies that enable us to reuse previously trained models (transfer learning) or combine models (ensemble learning). Transfer learning [56,57] reuses blocks and layers from a model that was pre-trained with some data and for a certain task (source domain and task) and fine-tune it to be applied to different data and/or task (target domain and task). For example, a classification model pre-trained on ImageNet (a big collection of natural images) can be partly reused and fine-tuned for medical imaging applications, such as organ segmentation or treatment outcome prediction [58–60]. Transfer learning allows us to exploit knowledge from different but related domains, mitigating the necessity of a big dataset for the target task, and improving the model performance [60–62]. Ensemble learning methods are also a way to improve the overall performance and the stability of the model, by combining the output of multiple models or algorithms to perform a task [63]. Some examples of medical applications include the mapping of patient anatomy to dose distribution for radiotherapy treatments [64], image segmentation [65], or classification [66].
Last but not least, self-supervised learning is a recent hybrid framework that has become state-of-the-art in natural language processing [67–69]. It is gaining attention for computer vision tasks [70–73] and it could play an important role in future research directions for medical imaging applications. Self-supervised learning can be seen as a variant of unsupervised learning, in the sense that it works with unlabelled data. However, the trick here is to exploit labels that come for “free” with the data, namely, those that can be extracted from the structure of data itself. Self-supervised algorithms work in two steps. First, the model is pre-trained to solve a “pretext task” where the aim is to obtain those supervisory signals from the data. Second, the acquired knowledge is transferred and the model is fine tuned to solve the main or “downstream task”. The literature on self-supervision for medical imaging applications is still scarce [74–77], but for instance, a recent work used context restoration as a pretext task [76]. Especifically, small patches in the image were randomly selected and swapped to obtain a new image with altered spatial information, and the pretext task consisted in predicting or restoring the original version of the image. They later used this knowledge to tune the model for image classification of 2D fetal ultrasound images; organ localization on abdominal CT images, and segmentation on brain MR images.
The existence of several hybrid learning frameworks shows that the boundaries between supervised and unsupervised learning has been progressively blurred to accommodate hybrid framework and combined strategies (Table 1), which can address real-world problems and data sets pragmatically (see Figure 3).
Figure 3.
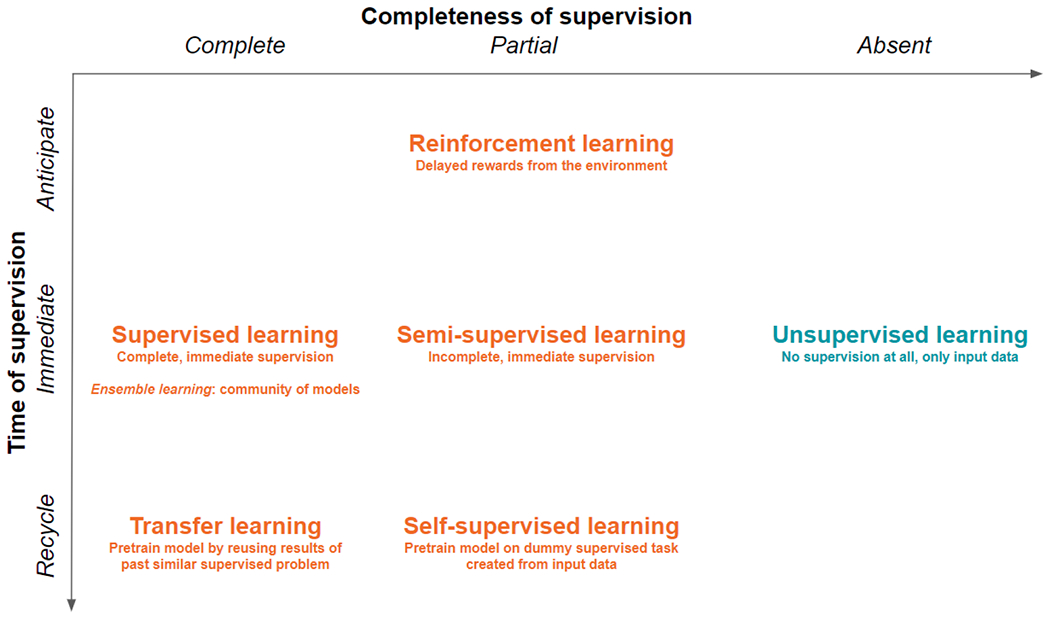
The tight framework of supervised learning can be hybridized with unsupervised learning to make room for practical cases and problems, as well as to accommodate temporality. Delaying supervision in future times leads towards reinforcement learning. Incompletely labelled data fosters semi-supervised learning, whereas small data sets encourage reusing (parts of) models trained previously on similar but bigger data sets, like in transfer learning. In self-supervision, pretraining relies on solving dummy supervised problems, where fake labels are created based on the inherent structure of image or sound data.
2.3. Typical AI-based medical imaging analysis workflow
Reviewing past works in the AI and ML literature shows that common blocks are used in most workflows for medical imaging processing (Figure 4). As ML is driven by data, preliminary steps are to extract and select relevant features from data, that is, quantitative characteristics that summarize information conveyed by data into vectors or arrays. Then, this information is fed to generic predictive models, like classifiers or regressors, which learn to perform a certain task. An example of this strategy is the field of radiomics [104,105], where “-omics-like” features are extracted from radiological images in order to predict some indicator of interest like a disease grade or a patient’s survival.
Figure 4.
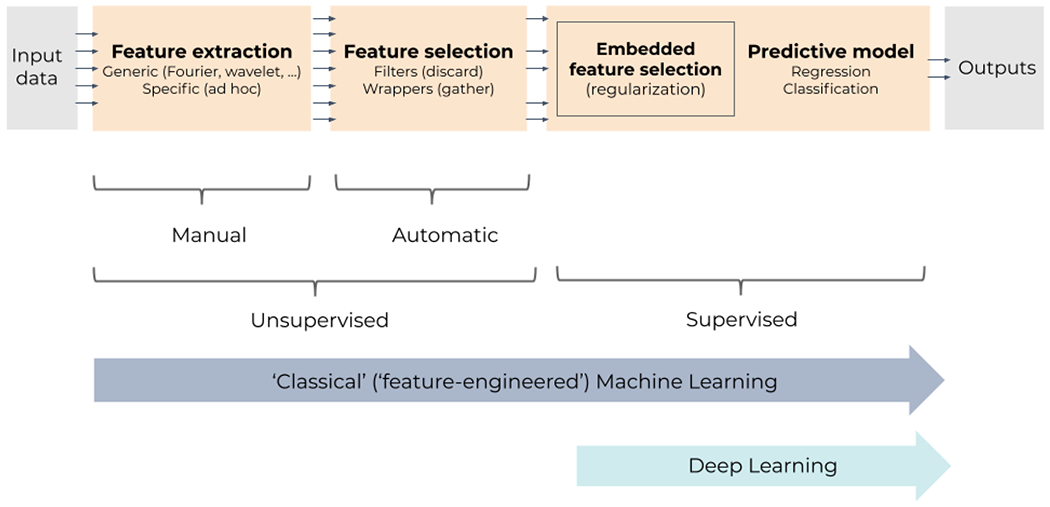
General ML pipeline for supervised learning: supervised predictive models are fed with features that are extracted and/or selected beforehand in an unsupervised way. Feature selection can, however, be embedded in some models, using regularization, for instance; selection then becomes supervised and therefore often improved. Classical (shallow) models tend to critically depend on unsupervised feature extraction and selection to preprocess data. In contrast, deep learning drops unsupervised feature extraction and selection; instead, it embeds multiple trainable layers of feature extractors and selectors, allowing the full pipeline to be supervised, end to end.
Feature engineering, extraction, and selection
Feature engineering, extraction, and selection are key steps to channel data to an AI method [8]. Feature engineering refers to crafting features by hand, either in ad hoc fashion or by relying on generic features from the literature. For images, the former could be gray level, color statistics, or shape descriptors (volume, diameter, curvature, sphericity, …). Image features are often classified in local or low-level features (specific to a small group of pixels in the image) and global or high-level features (characterizing the full image). For the latter, generic features would for instance result from applying Gabor or Laplace filters, edge detectors like Sobel operators, texture descriptors, Zernike moments, or popular transforms like Fourier’s or wavelet bases. In radiomics, all the above mentioned features can be used together, like ad hoc tumor shape and intensity descriptors, as well as textural descriptors (typically, Haralick’s gray level co-occurrence matrix [106]).
As an alternative or in a second time, higher-level features can be extracted in a more data-driven way, using dimensionality reduction. Methods like Principal Component Analysis [107]. Linear Discriminant Analysis [108], auto-associative [109] networks can reduce the number of input variables according to some unsupervised or supervised criterion. For images more specifically, the convolutional filters involved in CNNs bear similarity with the filters above: they extract local features, but their parameters are learnt from data and stacking them allows the global higher-level features to emerge. When features are not extracted in a supervised, data-driven way, it might happen that some of them are redundant or not relevant. To address this issue, a feature selection step can discard those to focus on a reduced set of features. Feature selection can follow several strategies, by either selecting or discarding. Wrappers [110] use a supervised predictive model to score subsets of features. To avoid the burden of a full fledged predictive model, feature filters [111], not to be confused with image filters above, use an unsupervised surrogate to score feature subsets, like their correlation or mutual information. Embedded methods [112] are directly integrated into the predictive model. For instance, feature weight regularization can favor sparse configurations, where irrelevant features get null weights. Examples of features selection in radiomics can be found in [113–119], for instance. Deep neural networks typically rely on this last approach with regularization. In the example of radiomics, embedded feature selection can be implemented with deep neural networks [120] and regularization, hence allowing for end-to-end learning instead of combining manually engineered features with shallow predictive models.
Predictive models
Common tasks in AI are regression and classification. The AI/ML model then attempts to predict either continuous values (e.g., a dose or a survival time) or class probabilities (e.g., benign vs malignant) starting from input features. In the following, we describe the main methodological aspects of the basic predictive ML models, state-of-the-art ML/DL methods and examples of their clinical applications for medical imaging are presented in the next section.
Regression is the most generic task in supervised learning. Linear regression is well known but other mathematical models can involve exponential or polynomial functions. ML generalizes this concept to universal approximators that can fit data sampled from almost any smooth function, with also possibly many input and output variables. Artificial neural networks (NNs) are the most iconic universal approximators (Figure 5). They consist of interconnected formal models of neurons, a mathematical ‘cell’ combining several ‘dendritic’ inputs into a weighted sum that triggers an ‘axonal’ output through a nonlinear activation function, like a step, a sigmoid, or a hinge (Rectified Linear Unit, ReLU).
Figure 5.
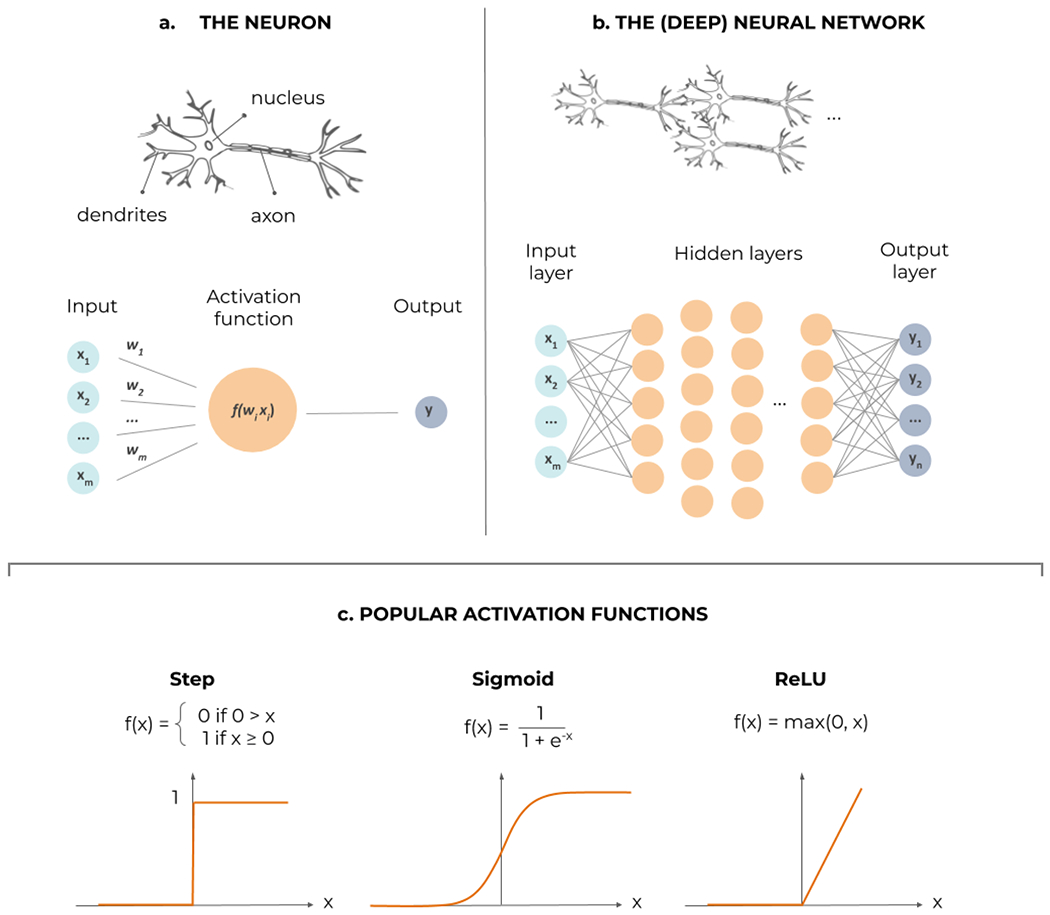
Artificial neural networks in a nutshell. (a) The formal neuron, processing several dendritic inputs through a nonlinear activation function f, to produce its actional output. (b) The neurons can be interconnected in a feed-forward way, into successive layers; as soon as a nonlinear ‘hidden’ layer is inserted in between the inputs and outputs, the network can potentially approximate any function; specific activation functions can be fitted in the output layer to achieve either regression or classification. (c) Examples of nonlinear activation functions in the hidden layers: the step function, from biological inspiration, the sigmoid, its continuous and differentiable surrogate, and the rectified linear unit (ReLU), that improves training of deep layers.
As soon as a hidden layer of neurons with nonlinear activation functions is inserted between the input and the output layers, a NN becomes a universal approximator [121]. However, a notion of capacity is associated with the NN architecture: the more neurons the hidden layer counts, the more complex functions can be approximated. The capacity is roughly proportional to the number of synaptic weights (parameters) in the NN and it is analogous to the polynomial order in polynomial regression (the number of weights in the terms). Deep NNs are obviously also universal approximators [121,122]. Their interest lies in trading width of a single hidden layer for depth, as stacks of hidden layers allow functional difference (e.g., convolutive neurons for image data) and thus hierarchical processing, explaining the later success of deep networks compared to shallow ones. Most NNs are feed-forward, meaning that data flows unidirectionally from inputs to outputs. Recurrent NNs (RNNs) add feedback loops to feedforward connections, allowing them to process sequences of data (text, videos) and somehow to keep memories of past inputs, which then gives context to new inputs.
Training of NNs relies on minimizing a loss function between the desired output and the one provided by the NN in its current parameter configuration. The partial derivatives, or gradient, of the loss function with respect to these parameters indicates the direction in which tuning the parameters is likely to decrease the loss. In a feedforward NN, this derivative information flows back from layer to layer and is therefore called gradient backpropagation.
For regression, typical loss functions can be the mean square error or mean absolute error. With a suitable change of the output layer (softmax or normalized exponential) and loss function (the cross entropy), the NN can approximate class probabilities.
Classification is the other prominent task in ML. Classifiers are simply algorithms that can sort data into groups or categories, and there exists a large variety of them [123]. Some of the most popular ones are very intuitive and easy to interpret, such as decision trees [124], where input data is classified by going through a hierarchical, tree-like process including different branching tests of the data features (Figure 6.a). Growing several complementary decision trees together, in an ensemble learning strategy, leads to random forests (Figure 6.b see also Section 3). Other simple algorithms for classification include the linear classifier, the Bayesian classifier, or the Perceptron (Figure 5.a). More sophisticated algorithms can actually be used for both regression and classification tasks. Some examples are NNs (Figure 5.b), which can yield class probabilities with suitable output layers; or support vector machines [125], which can be seen as an improved linear classifier that works in a higher-dimensional space and try to fit the separation (hyper)plane with the thickest margin in between points of two classes (Figure 7).
Figure 6.
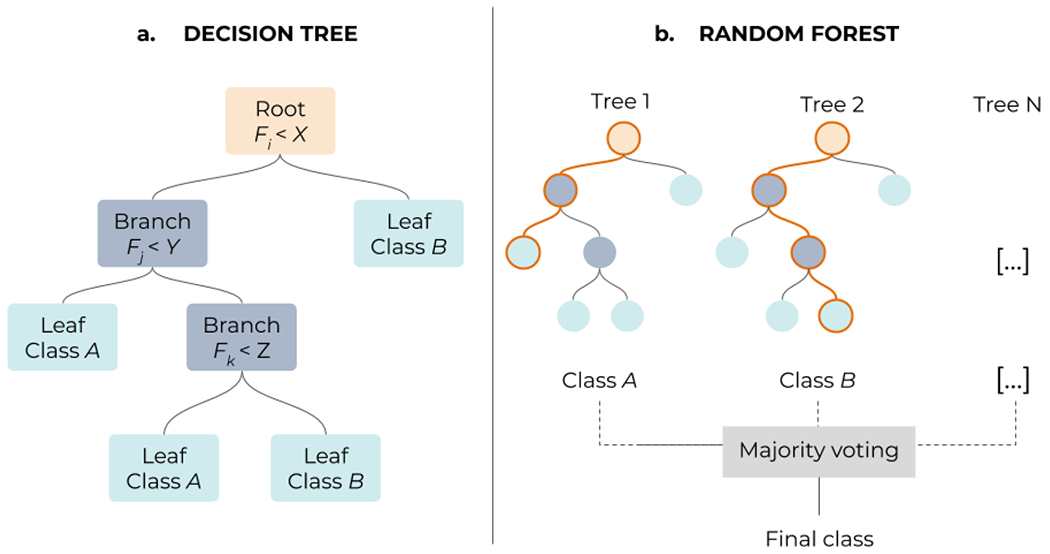
(a) Decision trees assign labels (leafs) to a given sample by going through a multi-level structure where different features (root nodes) and solutions (branches) are tested. (b) In a Random Forest algorithm, decision trees are combined, following an ensemble learning approach, which enables to get more accurate predictions than a single tree. Each individual tree in the forest spits out a class prediction and the class with the most votes becomes the final model’s prediction.
Figure 7.
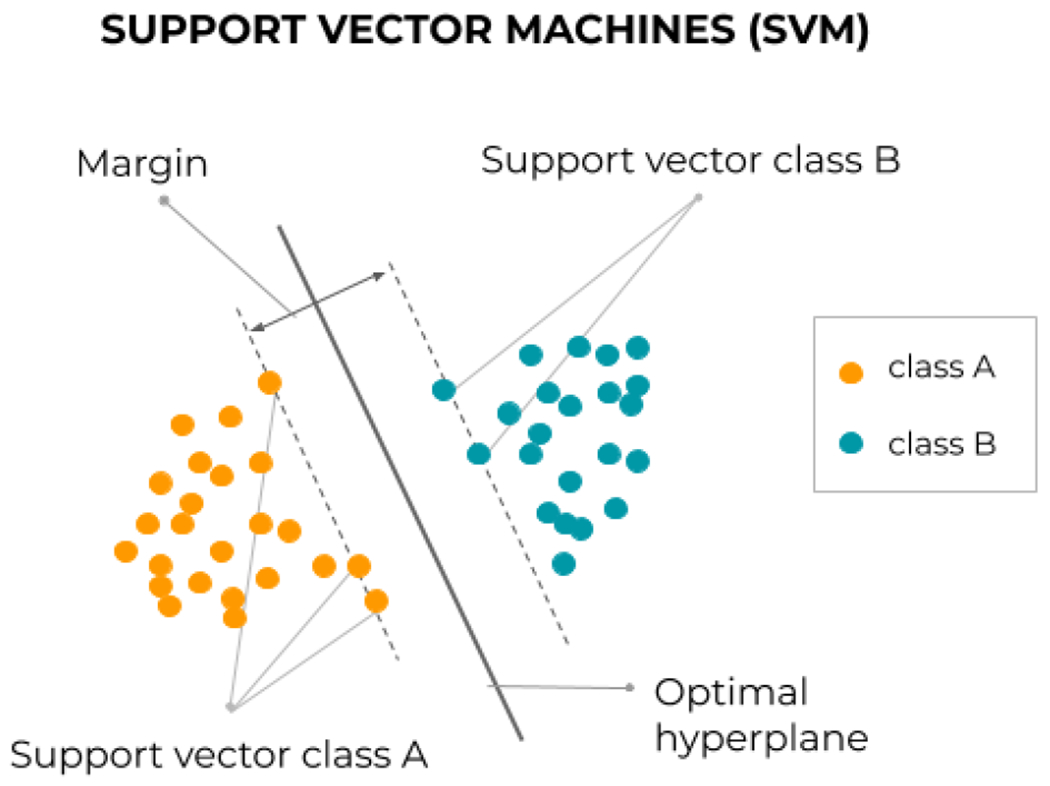
Principle of the linear support vector machine, which lifts the indeterminacy of separable classification by fitting the thickest margin, stuck in between a few ‘support vectors’. The principle can be extended to nonlinear class separation by using Mercer kernels [125].
3. State-of-the-art AI methods for medical image analysis
In the last decade, intensive research in AI methods for medical applications, and specifically in ML/DL (Figure 8, left), yielded thousands of publications reporting the performance of new algorithms and/or original variants of the existing ones. The number of publications using some of the most popular ML/DL methods is presented in Figure 8. In particular, in recent years, attention has moved from ML methods such as SVMs and Random Forests to Convolutional Neural Networks (Figure 8, right). In addition, since 2018, the use of other DL methods such as Generative Adversarial Networks or reinforcement learning algorithms is rapidly increasing. Notice that this section is not intended to be an exhaustive review of the application of AI methods to the medical field, but rather an illustration of the potential of these methods. Thus, in the following, we describe the basic methodological aspects of two of the most widely used algorithms (Random Forests and CNNs), as well as the increasingly popular GANs, and we provide some examples of recent applications of these methods to the field of medical image processing.
Figure 8.
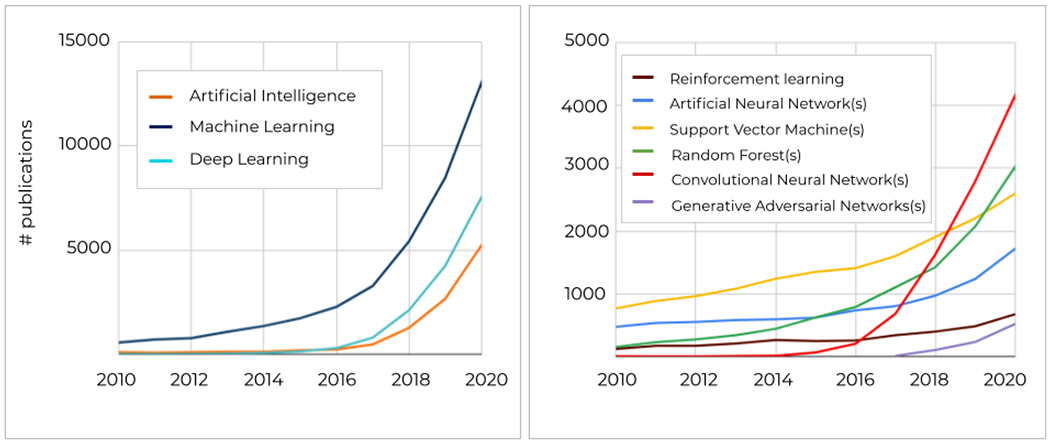
Number of publications since 2010 till 2020 in the PubMed repository, containing keywords related to AI/ML/DL methods in the title and/or abstract.
Random Forests (RFs)
Random forests (RFs) [126,127] use an ensemble of uncorrelated binary decision trees (multiple learning models) to find the best predictive model (Figure 6). Each decision tree can be seen as a base model (binary classifier) with its respective decision, where a combination of such decisions leads to the final output. This is achieved in RFs by using two distinctive mechanisms, i.e., internal feature selection and voting [128]. The RFs algorithm extracts a multitude of low-level (simple) data representations and uses the feature selection mechanism on all collected features to find the most informative ones. After feature selection, a majority vote on selected classifiers yields the final decision. For a full detailed description of the RF algorithm we refer to [128].
The earliest applications of RFs date from a decade ago for organ localization [129] and delineation [130]. Since then, RFs have been applied to numerous tasks, including detection and localization, segmentation, and image-based prediction [128]. For some specific applications, RFs have demonstrated an improved performance over other classical ML methods. For instance, Deist et al. [131,132] compared six different classification algorithms (decision tree, RFs, NNs, SVM, elastic net logistic regression, LogitBoost) on 12 datasets with a total of 3496 patients, for outcome and toxicity prediction in (chemo)radiotherapy. They concluded that RFs was the algorithm achieving higher discriminative performance (on 6 over 12 datasets). This goes in line with the findings from more fundamental ML research studies, which have reported RFs as one of the best classical learning algorithms [133]. However, many other works in the medical field have also compared the accuracy of RFs against more complex or simpler ML classifiers, and it is well known that their performance may vary for different applications [103,113,132,134–139] and even for different datasets within the same application [131,132]. This makes it hard to conclude on the absolute superiority of RFs algorithm over other ML classifiers. Nevertheless, the work of Deist et al. included one of the largest datasets investigated so far for radiotherapy outcome prediction, which is a strong argument in favor of considering RFs as one of the first options to investigate for this kind of application. In addition, RFs keep achieving very promising results in recent applications related to outcome prediction [135,139–143], but also for other domains like image classification [113,144] or automatic treatment planning [100,145–147]. Regarding other tasks where RFs were among the state-of-the-art methods a few years ago, like image synthesis [148–150] or segmentation [151,152], the community has now fully switched the attention to CNNs [5,153,154]. Nevertheless, in favor of RFs one could argue that they are easy to implement and less computationally expensive than CNNs (i.e., they can work in regular CPU). Therefore, they still deserve an important place in the ML toolbox for medical imaging.
Convolutional Neural Networks (CNNs)
Convolutional neural networks (CNNs) are inspired by the human visual system and exploit the spatial arrangement of data within images. Their remarkable capacity to detect hierarchical data representations has made CNNs the most popular architecture for current medical image processing applications.
Traditionally, CNNs stack successive layers of convolutions and down-sampling, and fully connected layers towards the output (Figure 9). Sequential applications of multiple convolutions enable the network to extract first simple features, like edges, in the deepest layers, which are next combined and refined into richer, more complex, hierarchical features, like full organs. Within each convolutional layer, feature saliency is determined by scanning a fixed-size convolution kernel (typically 3x3) all over the image to yield a feature map. This allows for an economy of parameters (weight sharing) and hence easier training. Downsampling layers are inserted between convolutional layers to reduce the size of feature maps, typically by applying a max-pooling operation, which keeps the maximum pixel value out of all non-overlapping 2-by-2 blocks in the feature map. To some extent, successive max-pooling allows for some shift invariance with respect to image content, as the salient maximum might stem from anywhere in the block. Downsampling trades resolution for number, as more convolution filters can be applied to smaller features maps within the same memory footprint. Eventually, fully connected layers generate the outputs, where all neurons are interconnected.
Figure 9.
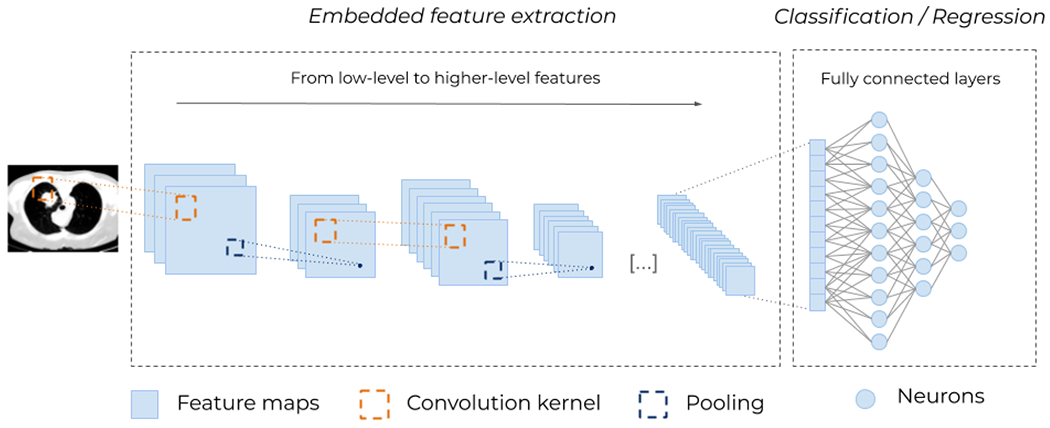
Typical architecture for a (deep) Convolutional Neural Network (CNN). Different convolutional kernels scan the input images leading to several feature maps. Then, down-sampling operations, such as max-pooling (i.e., taking the maximum value of a block of pixels), are applied to reduce the size of the feature maps. These two operations, convolution and pooling, are applied multiple times to extract higher-level features. At the end, the feature maps are flattened and passed through fully connected layers of neurons (see Figure 5), to obtain a final prediction. The embedded (automatic and unsupervised) feature extraction (Figure 4) is what enables CNNs to remove all handcrafted operations and makes them so powerful.
Fully convolutional networks (FCNs) [155] were proposed to efficiently perform image-to-image tasks like segmentation. In CNNs, repeated convolution and max-pooling layers lead to low resolution abstract outputs. In order to return to full-resolution images, fully connected layers in CNNs are replaced in FCNs with operations that revert convolution and max-pooling. Following the same line, U-net [156] was presented for biomedical image segmentation, and is now widely used in medical imaging. It is an encoder-decoder styled network, where the encoder can be seen as a feature extraction block, and the decoder as output generation block. Within medical imaging, FCNs are used in both supervised, and unsupervised settings depending on the respective architecture. In supervised training, FCNs are mostly used for discriminative tasks, such as detection, localization, classification, segmentation, and denoising. Note that CNNs and FCNs are often used interchangeably.
For certain applications, such as image segmentation [154,157,158] or synthesis [5], CNNs are now considered the state-of-the-art methods [4]. Although the comparison of different algorithms on the same dataset is not so common, an excellent way to track the evolution of the state-of-the-art algorithms is to look at the challenges and competitions organised around specific topics. In certain cases, CNNs have clearly surpassed the performance of more classical methods. A good example is the Challenge on Liver Ultrasound Tracking (CLUST): the winning team in the first edition (2014) achieved a tracking error of 1.51 ± 1.88 mm using an approach based on image registration algorithms [159]; whereas the current best performing algorithm, based on CNNs, achieves under 1 mm accuracy (0.69 ± 0.67 mm), demonstrating a more robust model [160]. Another example is the database from the MICCAI Head and Neck Auto-segmentation Challenge 2015 [161], for which the most recent methods based on CNNs [26,162–164] has improved the Dice coefficients obtained at that time with model- and atlas-based algorithms by more than 3% on average. In particular, the work of Nikolov et al [26] has recently reported a U-Net architecture with an accuracy equivalent to experienced radiographers. These are just two of the many competitions organised around medical imaging tasks [165], but year after year CNNs are becoming the backbone of the best performing algorithms.
Some of the latest methodological improvements in the architecture of CNNs that have contributed to more robust and accurate models include coarse-to-fine cascade of two CNNs [166] to address class-imbalance issues; the addition of squeeze-and-excitation (SE)-blocks to allow the network to model the channel and spatial information separately [167], increasing the model capacity; or the implementation of attention mechanisms, which enables the network to focus only on most relevant features [168–170].
Besides image segmentation, other recent successful applications of CNNs include classification [171,172], outcome prediction [120,173,174], automatic treatment planning [62,87,175], motion tracking [176,177] or image enhancement [33]. In numerous applications, CNNs have either demonstrated an accuracy similar to human experts [26,80,178,179], decreased the interobserver variability [180,181] or reduced the physician’s workload for a specific task [157]
Generative adversarial networks (GANs)
Generative adversarial networks (GANs) Generative adversarial networks (GANs) [182] are popular archi-tectures used for generative modeling. GANs consists of two networks: generator ℊ and discriminator (Fig. 10). The intuition is that ℊ iteratively tries to map a random input distribution to a given data distribution to generate new data, which D evaluates. Depending on the feedback from D, ℊ tends to minimize the loss between the two distributions, thus generating similar samples as input data. The goal is to trick D into classifying generated data as real. Both networks are trained simultaneously to get better at their respective tasks: while ℊ is learning to fool D, D is concurrently learning to better distinguish generated data from real input data. Note that both D and ℊ are generally CNNs trained in an adversarial setup.
Figure 10.
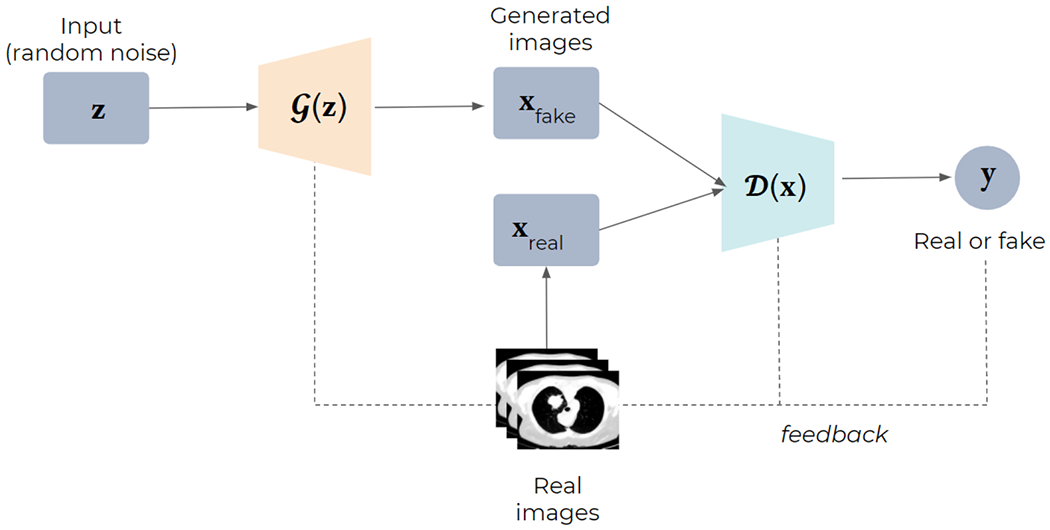
Structure of Generative Adversarial Networks (GANs). Starting from random noise, the generator (G) uses the feedback from the discriminator (D) and learns to create images that are similar to the provided ground truth.
Unlike CNNs, which have relatively old foundations dating back to 1980 (Section 2), adversarial learning is a rather new concept. However, it has rapidly rooted in the medical imaging field, leading to numerous publications in the last few years [183,184]. The initially proposed architecture for GANs [182] suffered from several drawbacks, such as a very unstable training, but the intensive research in the field of computer vision has lead substantial improvements by either changing the architecture of D and ℊ, or investigating new loss functions [183,185]. A way to better control the data generation process in GANs is to provide extra information about the desired properties of the output (e.g. ex amples of the desired real images or labels). This is known as conditional GANs (cGANs) [186], and it can be categorized as a form of supervised learning since it requires aligned training pairs. However, we believe that the real strength of GANs relies on their ability to learn in a semi-supervised or fully unsupervised manner. Specifically, in the medical imaging field, where aligned and properly annotated image pairs are seldom available, GANs are starting to play a very important role. In this context, cycleGANs [187] is probably one of the most famous architectures allowing bidirectional mapping between two domains using unpaired input data.
So far, in the medical imaging field, GANs have been mostly applied to synthetic image generation for data augmentation [188–190] and multi-modality image translation (e.g. MR to CT [90,191–193], CBCT to CT [97,194], among others [5,183]). Regarding data augmentation applications, we believe that GAN-based models have the potential to better sample the whole data distribution and generate more realistic images than traditional approaches (e.g. rotation, flipping, etc), which may contribute to a higher generalizability of the model [188] and more efficient training [195]. For multi-modality image translation, although cGANs have achieved good results [90,191,193,196], cycleGANs usually outperforms in terms of accuracy, in addition to overcome the issues related to paired image training (i.e. inaccurate aligning or labelling) [39,97,194,197]. Besides image translation, GANs have also been applied to other tasks, such as segmentation [198–203] or radiotherapy dose prediction [204–207], or artifact reduction [208], among others [183].
All above-mentioned applications have explored the generative capacity of GANs, but we believe that their discriminate capacity may also have some potential, since it can be used as regularized or detector when provided with abnormal images [209], which might be an excellent application for quality assurance tasks in radiation oncology, for instance.
4. Discussion and concluding remarks: where do we go next?
This article provided an overview of AI with a focus on medical imaging analysis, paying attention to key methodological concepts and highlighting the potential of the state-of-the-art ML and DL methods to automate and improve different steps of the clinical practice. Incorporating such knowledge into the clinical practice and making it accessible to the medical community will definitely help to demystify this technology, inspire new and high quality research directions, and facilitate the adoption of AI methods in the clinical environment.
Looking at the evolution of AI methods, one can certainly conclude that shifting from computationalism to connectionism, together with the transition from shallow to deep architectures, has brought a disruptive transformation to the medical field. However, an important part of the research so far has focused on simply translating the latest ML/DL advances in the field of computer vision to medical applications, in order to demonstrate the potential of these methods and the feasibility to use them to improve the clinical practice. It is the case of some of the papers cited in this manuscript, such as the first proof-of-concepts of the use of CNNs for organ segmentation [32] and for dose prediction for radiotherapy treatments [34], or the use of GANs for conversion between image modalities [97]. Although the technological transfer from computer science to the medical field will certainly continue to bring important progress, the next generation of AI methods for medical applications will only emerge if the medical community steps up to embrace AI technology and integrate all the domain-specific knowledge into the state-of-the-art AI methods [210,211]. This can be done in several ways, such as adding extra information in the input channels of the models or using dedicated loss functions during the model training. Some groups have already started to explore these research directions. For instance, instead of using generic loss functions from computer vision tasks, like the mean squared error, one could use loss functions that better target the specificities of our medical problem, such as including mutual information for the conversion of different image modalities [90,193] or dose-volume histograms for radiotherapy dose predictions [212]. Regarding the injection of domain-specific knowledge as input to the models, some examples include the addition of electronic health records and clinical data, like text and laboratory results, to the image data [213–215], or having first-order prior or approximations of the expected output [175,216–219]
Integrating domain-specific knowledge cannot only serve to improve the performances of state-of-the-art AI models, but also to increase the interpretability of the results, which is one of the well-acknowledged limitations of the current ML/DL methods [220–223]. This is the idea behind the so-called Expert Augmented Machine Learning (EAML), whose goal is to develop algorithms capable of extracting human knowledge from a panel of experts and use it to establish constraints for the model’s prediction [224]. This “human-in-the-loop” approach is also useful to train our AI models more efficiently. Indeed, some preliminary studies have reported that blindly increasing the training databases will not bring much improvement to our AI model’s performance [225]. In contrast, active learning [226] is a type of iterative supervised learning that follows this human-in-the-loop concept, where the algorithm itself query the user to obtain new data points where they are most needed, in order to build up an optimally balanced training dataset [227,228]. Nevertheless, although a human-centered approach for AI models is certainly the way to go in the close future, parallel research should focus on implementing strategies that leverage the problem of data labelling with semi-supervised, unsupervised or yet the increasingly popular self-supervised learning.
The quality of the data itself is certainly another important aspect that is worth discussion. Data collection and curation are indeed of paramount importance, since errors, biases, or variability in the training databases are often directly reflected in the model behavior and can have dramatic consequences in the model performances and its clinical outcome. Some examples of these issues include gender imbalance [229], racial bias [230], or data heterogeneity due to changes in treatment protocols overtime [225]. Despite progress in AI methods, data collection remains poorly automatized and the time dedicated to data collection and curation is often overly long. In fact, most state-of-the-art AI algorithms can be trained in a few hours, whereas building a large-scale well curated database can take months. Therefore, the same way physicians are familiar with planning protocols or delineation guidelines, the clinical teams should start being familiar with guiding principles for data management and curation in the era of AI. The FAIR (Findability, Accessibility, Interoperability, and Reusability) Data Principles [231] are the most popular and general ones, but the medical community should focus efforts on adapting those principles to the specificities of the medical domain [232–234]. Only in this way, we will manage to have a safe and efficient clinical implementation of AI methods. In addition, federated learning approaches [235–237] can be used to train AI models across institutions while ensuring data privacy protection, sharing the clinical knowledge and getting the advantages of collaborative AI solutions.
Investing time and collaborative effort in high-quality databases is certainly the way to move forward. So far, two aspects have played an important role in the recent development of AI and ML, namely, data repositories [238] and contests [239,240], the former feeding the latter. Competitions generate emulation among actors of the domain and allow state-of-the-art models to be benchmarked. A few examples have been cited in this manuscript, but there are multiple competitions every year that lead to public data repositories [241–243]. A very recent example is the breakthrough of AI in the CASP competition [244]. However, the results and rankings from competitions must be interpreted carefully when transferring the acquired knowledge into clinical applications [245,246]. Due to the high stakes in the medical domain, the community should devote even stronger efforts and international organizations should emit recommendations for data collection and curation, as well as for the design of contests and competitions. In the long term, this would lead to a much more structured and uniform clinical practice, with reduced differences between centres. Bigger, more homogeneous data could then potentially allow for another level of AI, by extracting much finer information, at the level of large populations. If data for AI is still in its infancy, so are also the methods. In spite of amazing progress and wowing results, current AI remains cast within tight frameworks. AI for images has been dealt here. Other application domains focus on natural language and speech processing with related but quite different approaches. So far, computer vision and audition follow different specialized approaches, although some works attempt to bridge the gap, like automatic image captioning. Nevertheless, these blocks remain mostly separated and current AI blocks lack integration and are still considered as weak AI. For instance, typical CNNs can boil down to just big filter banks, without any notion of time, and thus no memory and no experience. Strong AI is going to emerge when AI for images and speech, as well as active learning [227,228], will be combined into a sort of Frankensteinian brain, in which specialized lobes for the different senses get interconnected. This will allow for richer interaction, explainability through speech, reference to past experience, and continuous improvement. Such a strategy has been paved for autonomous driving, with different levels of automation. Confidence into ever more complex AI will grow only if AI can get more anthropomorphic, at least from a functional point of view.
In conclusion, artificial intelligence methods, and in particular, machine and deep learning methods, have reached important milestones in the last few years, demonstrating their potential to improve and automate the medical practice. However, a safe and full integration of these methods into the clinical workflow still requires a multidisciplinary effort (computer science, IT, medical experts, …) to enable the next generation of strong AI methods, ensuring robust and interpretable AI-based solutions.
Highlights.
Artificial intelligence (AI) has transformed the field of medical image analysis
Gathering key knowledge about AI becomes a must for the medical community
This review presents the basic technological pillars of AI for medical image analysis
We also discuss how the state-of-the-art AI methods and the new trends in the field
Acknowledgements
Ana Barragán is funded by the Walloon region in Belgium (PROTHERWAL/CHARP, grant 7289). Gilmer Valdés was supported by the National Institute of Biomedical Imaging and Bioengineering of the National Institutes of Health under Award Number K08EB026500. Dan Nguyen is supported by the National Institutes of Health (NIH) R01CA237269 and the Cancer Prevention & Research Institute of Texas (CPRIT) IIRA RP150485. Liesbeth Vandewinckele is supported by a Ph.D. fellowship of the research foundation - Flanders (FWO), mandate 1SA6121N. Kevin Souris is funded by the Walloon region (MECATECH / BIOWIN, grant 8090). John A. Lee is a Senior Research Associate with the F.R.S.-FNRS.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Singh R, Wu W, Wang G, Kalra MK. Artificial intelligence in image reconstruction: The change is here. Phys Med 2020;79:113–25. [DOI] [PubMed] [Google Scholar]
- [2].Wang M, Zhang Q, Lam S, Cai J, Yang R. A Review on Application of Deep Learning Algorithms in External Beam Radiotherapy Automated Treatment Planning. Front Oncol 2020;10:580919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Wang C, Zhu X, Hong JC, Zheng D. Artificial Intelligence in Radiotherapy Treatment Planning: Present and Future. Technol Cancer Res Treat 2019; 18:1533033819873922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Litjens G, Kooi T, Bejnordi BE, Setio AAA, Ciompi F, Ghafoorian M, et al. A survey on deep learning in medical image analysis. Medical Image Analysis 2017;42:60–88. 10.1016/j.media.2017.07.005. [DOI] [PubMed] [Google Scholar]
- [5].Wang T, Lei Y, Fu Y, Wynne JF, Curran WJ, Liu T, et al. A review on medical imaging synthesis using deep learning and its clinical applications. J Appl Clin Med Phys 2021;22:11–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Thompson RF, Valdes G, Fuller CD, Carpenter CM, Morin O, Aneja S, et al. Artificial intelligence in radiation oncology: A specialty-wide disruptive transformation? Radiotherapy and Oncology 2018;129:421–6. 10.1016/j.radonc.2018.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Hosny A, Parmar C, Quackenbush J, Schwartz LH, Aerts HJWL. Artificial intelligence in radiology. Nat Rev Cancer 2018;18:500–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Morra L, Delsanto S, Correale L. Artificial Intelligence in Medical Imaging 2019. 10.1201/9780367229184. [DOI] [Google Scholar]
- [9].Ranschaert ER, Morozov S, Algra PR, editors. Artificial Intelligence in Medical Imaging: Opportunities, Applications and Risks. Springer, Cham; 2019. [Google Scholar]
- [10].Friedman J, Hastie T, Tibshirani R. The elements of statistical learning, vol. 1. Springer series in statistics New York; 2001. [Google Scholar]
- [11].Kuhn M, Johnson K. Applied Predictive Modeling. Springer, New York, NY; 2013. [Google Scholar]
- [12].Shen C, Nguyen D, Zhou Z, Jiang SB, Dong B, Jia X. An introduction to deep learning in medical physics: advantages, potential, and challenges. Phys Med Biol 2020;65:05TR01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Cui S, Tseng H, Pakela J, Ten Haken RK, El Naqa I. Introduction to machine and deep learning for medical physicists. Medical Physics 2020;47. 10.1002/mp.14140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Holman JG, Cookson MJ. Expert systems for medical applications. J Med Eng Technol 1987;11:151–9. [DOI] [PubMed] [Google Scholar]
- [15].Haug PJ. Uses of diagnostic expert systems in clinical care. Proc Annu Symp Comput Appl Med Care 1993:379–83. [PMC free article] [PubMed] [Google Scholar]
- [16].Miller RA. Medical diagnostic decision support systems--past, present, and future: a threaded bibliography and brief commentary. J Am Med Inform Assoc 1994;1:8–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Buchanan BB, . Buchanan BG, Buchanan BG, Shortliffe EH, Heuristic S. Rule-based Expert Systems: The MYCIN Experiments of the Stanford Heuristic Programming Project. Addison Wesley Publishing Company; 1984. [Google Scholar]
- [18].Aikins JS, Kunz JC, Shortliffe EH, Fallat RJ. PUFF: an expert system for interpretation of pulmonary function data. Comput Biomed Res 1983;16:199–208. [DOI] [PubMed] [Google Scholar]
- [19].Miller RA, Pople HE Jr, Myers JD. Internist-1, an experimental computer-based diagnostic consultant for general internal medicine. N Engl J Med 1982;307:468–76. [DOI] [PubMed] [Google Scholar]
- [20].Buchanan BG. Can Machine Learning Offer Anything to Expert Systems? In: Marcus S, editor. Knowledge Acquisition: Selected Research and Commentary: A Special Issue of Machine Learning on Knowledge Acquisition, Boston, MA: Springer US; 1990, p. 5–8. [Google Scholar]
- [21].Su MC. Use of neural networks as medical diagnosis expert systems. Comput Biol Med 1994;24:419–29. [DOI] [PubMed] [Google Scholar]
- [22].Aizenberg IN, Aizenberg NN, Vandewalle J. Multi-Valued and Universal Binary Neurons 2000. 10.1007/978-1-4757-3115-6. [DOI] [Google Scholar]
- [23].Liu X, Faes L, Kale AU, Wagner SK, Fu DJ, Bruynseels A, et al. A comparison of deep learning performance against health-care professionals in detecting diseases from medical imaging: a systematic review and meta-analysis. Lancet Digit Health 2019;1:e271–97. [DOI] [PubMed] [Google Scholar]
- [24].Esteva A, Kuprel B, Novoa RA, Ko J, Swetter SM, Blau HM, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017;542:115–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Lotter W, Diab AR, Haslam B, Kim JG, Grisot G, Wu E, et al. Robust breast cancer detection in mammography and digital breast tomosynthesis using an annotation-efficient deep learning approach. Nat Med 2021;27:244–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Nikolov S, Blackwell S, Zverovitch A, Mendes R, Livne M, De Fauw J, et al. Deep learning to achieve clinically applicable segmentation of head and neck anatomy for radiotherapy. arXiv [csCV] 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Shen L, Margolies LR, Rothstein JH, Fluder E, McBride R, Sieh W. Deep Learning to Improve Breast Cancer Detection on Screening Mammography. Sci Rep 2019;9:12495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Komura D, Ishikawa S. Machine Learning Methods for Histopathological Image Analysis. Computational and Structural Biotechnology Journal 2018;16:34–42. 10.1016/j.csbj.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Yadav SS, Jadhav SM. Deep convolutional neural network based medical image classification for disease diagnosis. Journal of Big Data 2019;6:113. [Google Scholar]
- [30].Zhang X, Zhao H, Zhang S, Li R. A Novel Deep Neural Network Model for Multi-Label Chronic Disease Prediction. Front Genet 2019; 10:351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Hesamian MH, Jia W, He X, Kennedy P. Deep Learning Techniques for Medical Image Segmentation: Achievements and Challenges. J Digit Imaging 2019;32:582–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Ibragimov B, Xing L. Segmentation of organs-at-risks in head and neck CT images using convolutional neural networks. Med Phys 2017;44:547–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Javaid U, Souris K, Dasnoy D, Huang S, Lee JA. Mitigating inherent noise in Monte Carlo dose distributions using dilated U-Net. Med Phys 2019;46:5790–8. [DOI] [PubMed] [Google Scholar]
- [34].Nguyen D, Long T, Jia X, Lu W, Gu X, Iqbal Z, et al. A feasibility study for predicting optimal radiation therapy dose distributions of prostate cancer patients from patient anatomy using deep learning. Sci Rep 2019;9:1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Chen C, Dou Q, Chen H, Qin J, Heng PA. Unsupervised Bidirectional Cross-Modality Adaptation via Deeply Synergistic Image and Feature Alignment for Medical Image Segmentation. IEEE Trans Med Imaging 2020;39:2494–505. [DOI] [PubMed] [Google Scholar]
- [36].Perone CS, Ballester P, Barros RC, Cohen-Adad J. Unsupervised domain adaptation for medical imaging segmentation with self-ensembling. NeuroImage 2019;194:1–11. 10.1016/j.neuroimage.2019.03.026. [DOI] [PubMed] [Google Scholar]
- [37].Dou Q, Ouyang C, Chen C, Chen H, Glocker B, Zhuang X, et al. PnP-AdaNet: Plug-and-Play Adversarial Domain Adaptation Network at Unpaired Cross-Modality Cardiac Segmentation. IEEE Access 2019;7:99065–76. 10.1109/access.2019.2929258. [DOI] [Google Scholar]
- [38].Liu Y, Lei Y, Wang Y, Wang T, Ren L, Lin L, et al. MRI-based treatment planning for proton radiotherapy: dosimetric validation of a deep learning-based liver synthetic CT generation method. Phys Med Biol 2019;64:145015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Liu Y, Lei Y, Wang T, Fu Y, Tang X, Curran WJ, et al. CBCT-based synthetic CT generation using deep-attention cycleGAN for pancreatic adaptive radiotherapy. Med Phys 2020;47:2472–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Lei Y, Harms J, Wang T, Liu Y, Shu H, Jani AB, et al. MRI-only based synthetic CT generation using dense cycle consistent generative adversarial networks. Medical Physics 2019;46:3565–81. 10.1002/mp.13617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Aganj I, Harisinghani MG, Weissleder R, Fischl B. Unsupervised Medical Image Segmentation Based on the Local Center of Mass. Sci Rep 2018;8:13012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Peikari M, Salama S, Nofech-Mozes S, Martel AL. A Cluster-then-label Semi-supervised Learning Approach for Pathology Image Classification. Scientific Reports 2018;8. 10.1038/S41598-018-24876-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Jin C-B, Kim H, Liu M, Han IH, Lee JI, Lee JH, et al. DC2Anet: Generating Lumbar Spine MR Images from CT Scan Data Based on Semi-Supervised Learning. Applied Sciences 2019;9:2521. 10.3390/app9122521. [DOI] [Google Scholar]
- [44].Wang Z, Lin Y, Cheng K-TT, Yang X. Semi-supervised mp-MRI data synthesis with StitchLayer and auxiliary distance maximization. Med Image Anal 2020;59:101565. [DOI] [PubMed] [Google Scholar]
- [45].Ge C, Gu IY-H, Jakola AS, Yang J. Deep semi-supervised learning for brain tumor classification. BMC Med Imaging 2020;20:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Burton W 2nd, Myers C, Rullkoetter P. Semi-supervised learning for automatic segmentation of the knee from MRI with convolutional neural networks. Comput Methods Programs Biomed 2020; 189:105328. [DOI] [PubMed] [Google Scholar]
- [47].Cheplygina V, de Bruijne M, Pluim JPW. Not-so-supervised: A survey of semi-supervised, multi-instance, and transfer learning in medical image analysis. Med Image Anal 2019;54:280–96. [DOI] [PubMed] [Google Scholar]
- [48].Frénay B, Verleysen M. Classification in the presence of label noise: a survey. IEEE Trans Neural Netw Learn Syst 2014;25:845–69. [DOI] [PubMed] [Google Scholar]
- [49].Chen T, Kornblith S, Norouzi M, Hinton G. A Simple Framework for Contrastive Learning of Visual Representations. In: lii HD, Singh A, editors. Proceedings of the 37th International Conference on Machine Learning, vol. 119, PMLR; 2020, p. 1597–607. [Google Scholar]
- [50].Zhang J, Wang C, Sheng Y, Palta M, Czito B, Willett C, et al. An interpretable planning bot for pancreas stereotactic body radiation therapy. Int J Radiat Oncol Biol Phys 2020. 10.1016/j.ijrobp.2020.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Shen C, Gonzalez Y, Klages P, Qin N, Jung H, Chen L, et al. Intelligent inverse treatment planning via deep reinforcement learning, a proof-of-principle study in high dose-rate brachytherapy for cervical cancer. Phys Med Biol 2019;64:115013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Shen C, Nguyen D, Chen L, Gonzalez Y, McBeth R, Qin N, et al. Operating a treatment planning system using a deep-reinforcement learning-based virtual treatment planner for prostate cancer intensity-modulated radiation therapy treatment planning. Med Phys 2020;47:2329–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Watts J, Khojandi A, Vasudevan R, Ramdhani R. Optimizing Individualized Treatment Planning for Parkinson’s Disease Using Deep Reinforcement Learning. Conf Proc IEEE Eng Med Biol Soc 2020;2020:5406–9. [DOI] [PubMed] [Google Scholar]
- [54].Winkel DJ, Weikert TJ, Breit H-C, Chabin G, Gibson E, Heye TJ, et al. Validation of a fully automated liver segmentation algorithm using multi-scale deep reinforcement learning and comparison versus manual segmentation. Eur J Radiol 2020; 126:108918. [DOI] [PubMed] [Google Scholar]
- [55].Li Z, Xia Y. Deep Reinforcement Learning for Weakly-Supervised Lymph Node Segmentation in CT Images. IEEE J Biomed Health Inform 2020;PP. 10.1109/JBHI.2020.3008759. [DOI] [PubMed] [Google Scholar]
- [56].Thrun S, Pratt L. Learning to Learn: Introduction and Overview. In: Thrun S, Pratt L, editors. Learning to Learn, Boston, MA: Springer US; 1998, p. 3–17. [Google Scholar]
- [57].Pan SJ, Yang Q. A Survey on Transfer Learning. IEEE Trans Knowl Data Eng 2010;22:1345–59. [Google Scholar]
- [58].Zhen X, Chen J, Zhong Z, Hrycushko B, Zhou L, Jiang S, et al. Deep convolutional neural network with transfer learning for rectum toxicity prediction in cervical cancer radiotherapy: a feasibility study. Phys Med Biol 2017;62:8246–63. [DOI] [PubMed] [Google Scholar]
- [59].Morid MA, Borjali A, Del Fiol G. A scoping review of transfer learning research on medical image analysis using ImageNet. Comput Biol Med 2021;128:104115. [DOI] [PubMed] [Google Scholar]
- [60].Shin H-C, Roth HR, Gao M, Lu L, Xu Z, Nogues I, et al. Deep Convolutional Neural Networks for Computer-Aided Detection: CNN Architectures, Dataset Characteristics and Transfer Learning. IEEE Trans Med Imaging 2016;35:1285–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].van Opbroek A, Ikram MA, Vernooij MW, de Bruijne M. Transfer learning improves supervised image segmentation across imaging protocols. IEEE Trans Med Imaging 2015;34:1018–30. [DOI] [PubMed] [Google Scholar]
- [62].Kandalan RN, Nguyen D, Rezaeian NH, Barragán-Montero AM, Breedveld S, Namuduri K, et al. Dose Prediction with Deep Learning for Prostate Cancer Radiation Therapy: Model adaptation to Different Treatment Planning Practices. Radiother Oncol 2020. 10.1016/j.radonc.2020.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Schapire RE. The strength of weak learnability. 30th Annual Symposium on Foundations of Computer Science 1989. 10.1109/sfcs.1989.63451. [DOI] [Google Scholar]
- [64].Zhang J, Wu QJ, Xie T, Sheng Y, Yin F-F, Ge Y. An Ensemble Approach to Knowledge-Based Intensity-Modulated Radiation Therapy Planning. Front Oncol 2018;8:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Jia H, Xia Y, Song Y, Cai W, Fulham M, Feng DD. Atlas registration and ensemble deep convolutional neural network-based prostate segmentation using magnetic resonance imaging. Neurocomputing 2018;275:1358–69. 10.1016/j.neucom.2017.09.084. [DOI] [Google Scholar]
- [66].An N, Ding H, Yang J, Au R, Ang TFA. Deep ensemble learning for Alzheimer’s disease classification. J Biomed Inform 2020; 105:103411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Lan Z, Chen M, Goodman S, Gimpel K, Sharma P, Soricut R. ALBERT: A Lite BERT for Self-supervised Learning of Language Representations. arXiv [csCL] 2019. [Google Scholar]
- [68].Devlin J, Chang M-W, Lee K, Toutanova K. BERT: Pre-training of Deep Bidirectional Transformers for Language Understanding. arXiv [csCL] 2018. [Google Scholar]
- [69].Conneau A, Khandelwal K, Goyal N, Chaudhary V, Wenzek G, Guzman F, et al. Unsupervised Cross-lingual Representation Learning at Scale. Proceedings of the 58th Annual Meeting of the Association for Computational Linguistics 2020. 10.18653/v1/2020.acl-main.747. [DOI] [Google Scholar]
- [70].Jing L, Tian Y. Self-supervised Visual Feature Learning with Deep Neural Networks: A Survey. IEEE Trans Pattern Anal Mach Intell 2020;PP. 10.1109/TPAMI.2020.2992393. [DOI] [PubMed] [Google Scholar]
- [71].Kolesnikov A, Zhai X, Beyer L. Revisiting self-supervised visual representation learning. Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, 2019, p. 1920–9. [Google Scholar]
- [72].He K, Fan H, Wu Y, Xie S, Girshick R. Momentum contrast for unsupervised visual representation learning. Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, 2020, p. 9729–38. [Google Scholar]
- [73].Goyal P, Caron M, Lefaudeux B, Xu M, Wang P, Pai V, et al. Self-supervised Pretraining of Visual Features in the Wild. arXiv [csCV] 2021. [Google Scholar]
- [74].Taleb A, Loetzsch W, Danz N, Severin J, Gaertner T, Bergner B, et al. 3D Self-Supervised Methods for Medical Imaging. arXiv [csCV] 2020. [Google Scholar]
- [75].Hatamizadeh A, Yang D, Roth H, Xu D. UNETR: Transformers for 3D Medical Image Segmentation. arXiv [eessIV] 2021. [Google Scholar]
- [76].Chen L, Bentley P, Mori K, Misawa K, Fujiwara M, Rueckert D. Self-supervised learning for medical image analysis using image context restoration. Med Image Anal 2019;58:101539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Nguyen X-B, Lee GS, Kim SH, Yang HJ. Self-Supervised Learning Based on Spatial Awareness for Medical Image Analysis. IEEE Access 2020;8:162973–81. 10.1109/access.2020.3021469. [DOI] [Google Scholar]
- [78].Han Z, Wei B, Zheng Y, Yin Y, Li K, Li S. Breast Cancer Multi-classification from Histopathological Images with Structured Deep Learning Model. Sci Rep 2017;7:4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Wang H, Zhou Z, Li Y, Chen Z, Lu P, Wang W, et al. Comparison of machine learning methods for classifying mediastinal lymph node metastasis of non-small cell lung cancer from F-FDG PET/CT images. EJNMMI Res 2017;7:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Becker AS, Mueller M, Stoffel E, Maroon M, Ghafoor S, Boss A. Classification of breast cancer in ultrasound imaging using a generic deep learning analysis software: a pilot study. Br J Radiol 2018;91:20170576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Cheng J-Z, Ni D, Chou Y-H, Qin J, Tiu C-M, Chang Y-C, et al. Computer-Aided Diagnosis with Deep Learning Architecture: Applications to Breast Lesions in US Images and Pulmonary Nodules in CT Scans. Sci Rep 2016;6:24454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Guo Z, Li X, Huang H, Guo N, Li Q. Deep Learning-Based Image Segmentation on Multimodal Medical Imaging. IEEE Transactions on Radiation and Plasma Medical Sciences 2019;3:162–9. 10.1109/trpms.2018.2890359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Balagopal A, Kazemifar S, Nguyen D, Lin M-H, Hannan R, Owrangi A, et al. Fully automated organ segmentation in male pelvic CT images. Phys Med Biol 2018;63:245015. [DOI] [PubMed] [Google Scholar]
- [84].Javaid U, Dasnoy D, Lee JA. Multi-organ Segmentation of Chest CT Images in Radiation Oncology: Comparison of Standard and Dilated UNet. Advanced Concepts for Intelligent Vision Systems 2018:188–99. 10.1007/978-3-030-01449-0_16. [DOI] [Google Scholar]
- [85].Moradi S, Oghli MG, Alizadehasl A, Shiri I, Oveisi N, Oveisi M, et al. MFP-Unet: A novel deep learning based approach for left ventricle segmentation in echocardiography. Phys Med 2019;67:58–69. [DOI] [PubMed] [Google Scholar]
- [86].Nemoto T, Futakami N, Yagi M, Kunieda E, Akiba T, Takeda A, et al. Simple low-cost approaches to semantic segmentation in radiation therapy planning for prostate cancer using deep learning with non-contrast planning CT images. Phys Med 2020;78:93–100. [DOI] [PubMed] [Google Scholar]
- [87].Nguyen D, Jia X, Sher D, Lin M-H, Iqbal Z, Liu H, et al. 3D radiotherapy dose prediction on head and neck cancer patients with a hierarchically densely connected U-net deep learning architecture. Phys Med Biol 2019;64:065020. [DOI] [PubMed] [Google Scholar]
- [88].Fan J, Wang J, Chen Z, Hu C, Zhang Z, Hu W. Automatic treatment planning based on three-dimensional dose distribution predicted from deep learning technique. Med Phys 2019;46:370–81. [DOI] [PubMed] [Google Scholar]
- [89].Han X MR-based synthetic CT generation using a deep convolutional neural network method. Med Phys 2017;44:1408–19. [DOI] [PubMed] [Google Scholar]
- [90].Kazemifar S, McGuire S, Timmerman R, Wardak Z, Nguyen D, Park Y, et al. MRI-only brain radiotherapy: Assessing the dosimetric accuracy of synthetic CT images generated using a deep learning approach. Radiother Oncol 2019;136:56–63. [DOI] [PubMed] [Google Scholar]
- [91].Madani A, Moradi M, Karargyris A, Syeda-Mahmood T. Semi-supervised learning with generative adversarial networks for chest X-ray classification with ability of data domain adaptation. 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018) 2018. 10.1109/isbi.2018.8363749. [DOI] [Google Scholar]
- [92].Jiang J, Hu Y-C, Tyagi N, Rimner A, Lee N, Deasy JO, et al. PSIGAN: Joint Probabilistic Segmentation and Image Distribution Matching for Unpaired Cross-Modality Adaptation-Based MRI Segmentation. IEEE Trans Med Imaging 2020;39:4071–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Lynch CM, van Berkel VH, Frieboes HB. Application of unsupervised analysis techniques to lung cancer patient data. PLoS One 2017;12:e0184370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Mehta J, Majumdar A. RODEO: Robust DE-aliasing autoencOder for real-time medical image reconstruction. Pattern Recognit 2017;63:499–510. [Google Scholar]
- [95].Shen C, Gonzalez Y, Chen L, Jiang SB, Jia X. Intelligent Parameter Tuning in Optimization-Based Iterative CT Reconstruction via Deep Reinforcement Learning. IEEE Trans Med Imaging 2018;37:1430–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Barkousaraie AS, Bohara G, Jiang SB, Nguyen D. A reinforcement learning application of a guided Monte Carlo Tree Search algorithm for beam orientation selection in radiation therapy. Mach Learn: Sci Technol 2021. 10.1088/2632-2153/abe528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Liang X, Chen L, Nguyen D, Zhou Z, Gu X, Yang M, et al. Generating synthesized computed tomography (CT) from cone-beam computed tomography (CBCT) using CycleGAN for adaptive radiation therapy. Phys Med Biol 2019;64:125002. [DOI] [PubMed] [Google Scholar]
- [98].Yang X, Lin Y, Wang Z, Li X, Cheng K-T. Bi-Modality Medical Image Synthesis Using Semi-Supervised Sequential Generative Adversarial Networks. IEEE J Biomed Health Inform 2020;24:855–65. [DOI] [PubMed] [Google Scholar]
- [99].Liang X, Nguyen D, Jiang SB. Generalizability issues with deep learning models in medicine and their potential solutions: illustrated with cone-beam computed tomography (CBCT) to computed tomography (CT) image conversion. Machine Learning: Science and Technology 2020;2:015007. 10.1088/2632-2153/abb214. [DOI] [Google Scholar]
- [100].McIntosh C, Welch M, McNiven A, Jaffray DA, Purdie TG. Fully automated treatment planning for head and neck radiotherapy using a voxel-based dose prediction and dose mimicking method. Phys Med Biol 2017;62:5926–44. [DOI] [PubMed] [Google Scholar]
- [101].McIntosh C, Purdie TG. Voxel-based dose prediction with multi-patient atlas selection for automated radiotherapy treatment planning. Phys Med Biol 2017;62:415–31. [DOI] [PubMed] [Google Scholar]
- [102].Nguyen D, Sadeghnejad Barkousaraie A, Bohara G, Balagopal A, McBeth R, Lin M-H, et al. A comparison of Monte Carlo dropout and bootstrap aggregation on the performance and uncertainty estimation in radiation therapy dose prediction with deep learning neural networks. Phys Med Biol 2021;66:054002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Valdes G, Luna JM, Eaton E, Simone CB 2nd, Ungar LH, Solberg TD. MediBoost: a Patient Stratification Tool for Interpretable Decision Making in the Era of Precision Medicine. Sci Rep 2016;6:37854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Lambin P, Leijenaar RTH, Deist TM, Peerlings J, de Jong EEC, van Timmeren J, et al. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol 2017;14:749–62. [DOI] [PubMed] [Google Scholar]
- [105].Lambin P, Rios-Velazquez E, Leijenaar R, Carvalho S, van Stiphout RGPM, Granton P, et al. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer 2012;48:441–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Textural Features for Image Classification n.d. https://doi.org/10.1109%2FTSMC.1973.4309314 (accessed March 29, 2021).
- [107].Jolliffe IT. Principal Component Analysis. Springer Science & Business Media; 2013. [Google Scholar]
- [108].Cristianini N Fisher Discriminant Analysis (Linear Discriminant Analysis). Dictionary of Bioinformatics and Computational Biology 2004. 10.1002/9780471650126.dob0238.pub2. [DOI] [Google Scholar]
- [109].Hinton GE, Salakhutdinov RR. Reducing the dimensionality of data with neural networks. Science 2006;313:504–7. [DOI] [PubMed] [Google Scholar]
- [110].Kohavi R, John GH. The Wrapper Approach. Feature Extraction, Construction and Selection 1998:33–50. 10.1007/978-1-4615-5725-8_3. [DOI] [Google Scholar]
- [111].Guyon I, Gunn S, Nikravesh M, Zadeh LA. Feature Extraction: Foundations and Applications. Springer; 2008. [Google Scholar]
- [112].Lai TN, Chapelle O, Weston J, Elisseeff A. Embedded Methods. Feature Extraction n.d.: 137–65. 10.1007/978-3-540-35488-8_6. [DOI] [Google Scholar]
- [113].Yang F, Chen W, Wei H, Zhang X, Yuan S, Qiao X, et al. Machine Learning for Histologic Subtype Classification of Non-Small Cell Lung Cancer: A Retrospective Multicenter Radiomics Study. Front Oncol 2020; 10:608598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Liu T, Wu G, Yu J, Guo Y, Wang Y, Shi Z, et al. A mRMRMSRC feature selection method for radiomics approach. 2017 39th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC) 2017. 10.1109/embc.2017.8036900. [DOI] [PubMed] [Google Scholar]
- [115].Yuan R, Tian L, Chen J. An RF-BFE algorithm for feature selection in radiomics analysis. Medical Imaging 2019: Imaging Informatics for Healthcare, Research, and Applications 2019. 10.1117/12.2512045. [DOI] [Google Scholar]
- [116].Oubel E, Beaumont H, lannessi A. Mutual information-based feature selection for radiomics. Medical Imaging 2016: PACS and Imaging Informatics: Next Generation and Innovations 2016. 10.1117/12.2216746. [DOI] [Google Scholar]
- [117].Wei B-Y, Bing-Yan WEI, Song J-L, Li-Xu GU. The Research of Reproducibility and Non-redundancy Feature Selection Methods in Radiomics. DEStech Transactions on Computer Science and Engineering 2017. 10.12783/dtcse/aice-ncs2016/5661. [DOI] [Google Scholar]
- [118].Sun P, Wang D, Mok VC, Shi L. Comparison of Feature Selection Methods and Machine Learning Classifiers for Radiomics Analysis in Glioma Grading. IEEE Access 2019;7:102010–20. 10.1109/access.2019.2928975. [DOI] [Google Scholar]
- [119].van Timmeren JE, Leijenaar RTH, van Elmpt W, Reymen B, Lambin P. Feature selection methodology for longitudinal cone-beam CT radiomics. Acta Oncologica 2017;56:1537–43. 10.1080/0284186x.2017.1350285. [DOI] [PubMed] [Google Scholar]
- [120].Xu Y, Hosny A, Zeleznik R, Parmar C, Coroller T, Franco I, et al. Deep Learning Predicts Lung Cancer Treatment Response from Serial Medical Imaging. Clin Cancer Res 2019;25:3266–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Cybenko G Approximations by Superpositions of a Sigmoidal Function. 1989. [Google Scholar]
- [122].Hanin B Universal Function Approximation by Deep Neural Nets with Bounded Width and ReLU Activations. Mathematics 2019;7:992. 10.3390/math7100992. [DOI] [Google Scholar]
- [123].Erickson BJ, Korfiatis P, Akkus Z, Kline TL. Machine Learning for Medical Imaging. Radiographics 2017;37:505–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Hartmann C, Varshney P, Mehrotra K, Gerberich C. Application of information theory to the construction of efficient decision trees. IEEE Transactions on Information Theory 1982;28:565–77. 10.1109/tit.1982.1056522. [DOI] [Google Scholar]
- [125].VAPNIK V Pattern recognition using generalized portrait method. Autom Remote Control 1963;24:774–80. [Google Scholar]
- [126].Amit Y, Geman D. Shape Quantization and Recognition with Randomized Trees. Neural Computation 1997;9:1545–88. 10.1162/neco.1997.9.7.1545. [DOI] [Google Scholar]
- [127].Breiman L Random Forests. Mach Learn 2001;45:5–32. [Google Scholar]
- [128].Konukoglu E, Glocker B. Random forests in medical image computing. Handbook of Medical Image Computing and Computer Assisted Intervention, Elsevier; 2020, p. 457–80. [Google Scholar]
- [129].Criminisi A, Shotton J, Bucciarelli S. Decision forests with long-range spatial context for organ localization in CT volumes. Med Image Comput Comput Assist Interv 2009. [Google Scholar]
- [130].Lempitsky V, Verhoek M, Noble JA, Blake A. Random Forest Classification for Automatic Delineation of Myocardium in Real-Time 3D Echocardiography. Functional Imaging and Modeling of the Heart, Springer Berlin Heidelberg; 2009, p. 447–56. [Google Scholar]
- [131].Deist TM, Dankers FJWM, Valdes G, Wijsman R, Hsu I-C, Oberije C, et al. Machine learning algorithms for outcome prediction in (chemo)radiotherapy: An empirical comparison of classifiers. Med Phys 2018;45:3449–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Differentiation of glioblastoma from solitary brain metastases using radiomic machine-learning classifiers. Cancer Lett 2019;451:128–35. [DOI] [PubMed] [Google Scholar]
- [133].Caruana R, Niculescu-Mizil A. An empirical comparison of supervised learning algorithms. Proceedings of the 23rd international conference on Machine learning, New York, NY, USA: Association for Computing Machinery; 2006, p. 161–8. [Google Scholar]
- [134].Ma C, Wang R, Zhou S, Wang M, Yue H, Zhang Y, et al. The structural similarity index for IMRT quality assurance: radiomics-based error classification. Med Phys 2020. 10.1002/mp.14559. [DOI] [PubMed] [Google Scholar]
- [135].Akcay M, Etiz D, Celik O. Prediction of Survival and Recurrence Patterns by Machine Learning in Gastric Cancer Cases Undergoing Radiation Therapy and Chemotherapy. Adv Radiat Oncol 2020;5:1179–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Dean JA, Wong KH, Welsh LC, Jones A-B, Schick U, Newbold KL, et al. Normal tissue complication probability (NTCP) modelling using spatial dose metrics and machine learning methods for severe acute oral mucositis resulting from head and neck radiotherapy. Radiother Oncol 2016;120:21–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Qiu X, Gao J, Yang J, Hu J, Hu W, Kong L, et al. A Comparison Study of Machine Learning (Random Survival Forest) and Classic Statistic (Cox Proportional Hazards) for Predicting Progression in High-Grade Glioma after Proton and Carbon Ion Radiotherapy. Front Oncol 2020;10:551420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Dean J, Wong K, Gay H, Welsh L, Jones A-B, Schick U, et al. Incorporating spatial dose metrics in machine learning-based normal tissue complication probability (NTCP) models of severe acute dysphagia resulting from head and neck radiotherapy. Clin Transl Radiat Oncol 2018;8:27–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Luo Y, Chen S, Valdes G. Machine learning for radiation outcome modeling and prediction. Med Phys 2020;47:e178–84. [DOI] [PubMed] [Google Scholar]
- [140].Luna JM, Chao H-H, Diffenderfer ES, Valdes G, Chinniah C, Ma G, et al. Predicting radiation pneumonitis in locally advanced stage II–III non-small cell lung cancer using machine learning. Radiotherapy and Oncology 2019;133:106–12. 10.1016/j.radonc.2019.01.003. [DOI] [PubMed] [Google Scholar]
- [141].Valdes G, Chang AJ, Interian Y, Owen K, Jensen ST, Ungar LH, et al. Salvage HDR Brachytherapy: Multiple Hypothesis Testing Versus Machine Learning Analysis. Int J Radiat Oncol Biol Phys 2018;101:694–703. [DOI] [PubMed] [Google Scholar]
- [142].Meti N, Saednia K, Lagree A, Tabbarah S, Mohebpour M, Kiss A, et al. Machine Learning Frameworks to Predict Neoadjuvant Chemotherapy Response in Breast Cancer Using Clinical and Pathological Features. JCO Clin Cancer Inform 2021;5:66–80. [DOI] [PubMed] [Google Scholar]
- [143].Zhou G-Q, Wu C-F, Deng B, Gao T-S, Lv J-W, Lin L, et al. An optimal posttreatment surveillance strategy for cancer survivors based on an individualized risk-based approach. Nature Communications 2020;11. 10.1038/s41467-020-17672-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Novak J, Zarinabad N, Rose H, Arvanitis T, MacPherson L, Pinkey B, et al. Classification of paediatric brain tumours by diffusion weighted imaging and machine learning. Sci Rep 2021. ;11:2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Sheng Y, Li T, Yoo S, Yin F-F, Blitzblau R, Horton JK, et al. Automatic Planning of Whole Breast Radiation Therapy Using Machine Learning Models. Front Oncol 2019;9:750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].McIntosh C, Purdie TG. Contextual Atlas Regression Forests: Multiple-Atlas-Based Automated Dose Prediction in Radiation Therapy. IEEE Trans Med Imaging 2016;35:1000–12. [DOI] [PubMed] [Google Scholar]
- [147].Babier A, Mahmood R, McNiven AL, Diamant A, Chan TCY. The importance of evaluating the complete automated knowledge-based planning pipeline. Phys Med 2020;72:73–9. [DOI] [PubMed] [Google Scholar]
- [148].Jog A, Carass A, Roy S, Pham DL, Prince JL. Random forest regression for magnetic resonance image synthesis. Med Image Anal 2017;35:475–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Lei Y, Harms J, Wang T, Tian S, Zhou J, Shu H-K, et al. MRI-based synthetic CT generation using semantic random forest with iterative refinement. Phys Med Biol 2019;64:085001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Yang X, Lei Y, Shu H-K, Rossi P, Mao H, Shim H, et al. Pseudo CT Estimation from MRI Using Patch-based Random Forest. Proc SPIE Int Soc Opt Eng 2017;10133. 10.1117/12.2253936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Polan DF, Brady SL, Kaufman RA. Tissue segmentation of computed tomography images using a Random Forest algorithm: a feasibility study. Phys Med Biol 2016;61:6553–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Gao Y, Shao Y, Lian J, Wang AZ, Chen RC, Shen D. Accurate Segmentation of CT Male Pelvic Organs via Regression-Based Deformable Models and Multi-Task Random Forests. IEEE Trans Med Imaging 2016;35:1532–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Wang T, Lei Y, Fu Y, Curran WJ, Liu T, Nye JA, et al. Machine learning in quantitative PET: A review of attenuation correction and low-count image reconstruction methods. Phys Med 2020;76:294–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Seo H, Badiei Khuzani M, Vasudevan V, Huang C, Ren H, Xiao R, et al. Machine learning techniques for biomedical image segmentation: An overview of technical aspects and introduction to state-of-art applications. Med Phys 2020;47:e148–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Long J, Shelhamer E, Darrell T. Fully convolutional networks for semantic segmentation. 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) 2015. 10.1109/cvpr.2015.7298965. [DOI] [PubMed] [Google Scholar]
- [156].Ronneberger O, Fischer P, Brox T. U-Net: Convolutional Networks for Biomedical Image Segmentation. Medical Image Computing and Computer-Assisted Intervention – MICCAI 2015, Springer International Publishing; 2015, p. 234–41. [Google Scholar]
- [157].Lustberg T, van Soest J, Gooding M, Peressutti D, Aljabar P, van der Stoep J, et al. Clinical evaluation of atlas and deep learning based automatic contouring for lung cancer. Radiother Oncol 2018;126:312–7. [DOI] [PubMed] [Google Scholar]
- [158].Vrtovec T, Močnik D, Strojan P, Pernuš F, Ibragimov B. Auto-segmentation of organs at risk for head and neck radiotherapy planning: From atlas-based to deep learning methods. Med Phys 2020;47:e929–50. [DOI] [PubMed] [Google Scholar]
- [159].König L, Kipshagen T, Rühaak J. A non-linear image registration scheme for real-time liver ultrasound tracking using normalized gradient fields. Proc MICCAI CLUST14, Boston, USA: 2014:29–36. [Google Scholar]
- [160].Liu F, Liu D, Tian J, Xie X, Yang X, Wang K. Cascaded one-shot deformable convolutional neural networks: Developing a deep learning model for respiratory motion estimation in ultrasound sequences. Med Image Anal 2020;65:101793. [DOI] [PubMed] [Google Scholar]
- [161].Raudaschl PF, Zaffino P, Sharp GC, Spadea MF, Chen A, Dawant BM, et al. Evaluation of segmentation methods on head and neck CT: Auto-segmentation challenge 2015. Medical Physics 2017;44:2020–36. 10.1002/mp.12197. [DOI] [PubMed] [Google Scholar]
- [162].Zhu W, Huang Y, Zeng L, Chen X, Liu Y, Qian Z, et al. Anatomy Net: Deep learning for fast and fully automated whole-volume segmentation of head and neck anatomy. Med Phys 2019;46:576–89. [DOI] [PubMed] [Google Scholar]
- [163].Gou S, Tong N, Qi S, Yang S, Chin R, Sheng K. Self-channel-and-spatial-attention neural network for automated multi-organ segmentation on head and neck CT images. Phys Med Biol 2020;65:245034. [DOI] [PubMed] [Google Scholar]
- [164].Liang S, Thung K-H, Nie D, Zhang Y, Shen D. Multi-View Spatial Aggregation Framework for Joint Localization and Segmentation of Organs at Risk in Head and Neck CT Images. IEEE Trans Med Imaging 2020;39:2794–805. [DOI] [PubMed] [Google Scholar]
- [165].Grand Challenge n.d. https://grand-challenge.org/(accessed February 17, 2021).
- [166].Gerard SE, Patton TJ, Christensen GE, Bayouth JE, Reinhardt JM. FissureNet: A Deep Learning Approach For Pulmonary Fissure Detection in CT Images. IEEE Transactions on Medical Imaging 2019;38:156–66. 10.1109/tmi.2018.2858202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].Liu P, Dou Q, Wang Q, Heng P-A. An Encoder-Decoder Neural Network With 3D Squeeze-and-Excitation and Deep Supervision for Brain Tumor Segmentation. IEEE Access 2020;8:34029–37. 10.1109/access.2020.2973707. [DOI] [Google Scholar]
- [168].Hu H, Li Q, Zhao Y, Zhang Y. Parallel Deep Learning Algorithms with Hybrid Attention Mechanism for Image Segmentation of Lung Tumors. IEEE Transactions on Industrial Informatics 2020:1–1. 10.1109/tii.2020.3022912. [DOI] [Google Scholar]
- [169].Zhou T, Ruan S, Guo Y, Canu S. A Multi-Modality Fusion Network Based on Attention Mechanism for Brain Tumor Segmentation. 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI) 2020. 10.1109/isbi45749.2020.9098392. [DOI] [Google Scholar]
- [170].Schlemper J, Oktay O, Schaap M, Heinrich M, Kainz B, Glocker B, et al. Attention gated networks: Learning to leverage salient regions in medical images. Med Image Anal 2019;53:197–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Jiang H, Gao F, Xu X, Huang F, Zhu S. Attentive and ensemble 3D dual path networks for pulmonary nodules classification. Neurocomputing 2020;398:422–30. 10.1016/j.neucom.2019.03.103. [DOI] [Google Scholar]
- [172].McKinney SM, Sieniek M, Godbole V, Godwin J, Antropova N, Ashrafian H, et al. International evaluation of an AI system for breast cancer screening. Nature 2020;577:89–94. [DOI] [PubMed] [Google Scholar]
- [173].Ibragimov B, Toesca D, Chang D, Yuan Y, Koong A, Xing L. Development of deep neural network for individualized hepatobiliary toxicity prediction after liver SBRT. Medical Physics 2018;45:4763–74. 10.1002/mp.13122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [174].Sekaran K, Chandana P, Murali Krishna N, Kadry S. Deep learning convolutional neural network (CNN) With Gaussian mixture model for predicting pancreatic cancer. Multimedia Tools and Applications 2020;79:10233–47. 10.1007/s11042-019-7419-5. [DOI] [Google Scholar]
- [175].Barragán- Montero AM, Nguyen D, Lu W, Lin M-H, Norouzi- Kandalan R, Geets X, et al. Three- dimensional dose prediction for lung IMRT patients with deep neural networks: robust learning from heterogeneous beam configurations. Medical Physics 2019;46:3679–91. 10.1002/mp.13597. [DOI] [PubMed] [Google Scholar]
- [176].Roggen T, Bobic M, Givehchi N, Scheib SG. Deep Learning model for markerless tracking in spinal SBRT. Phys Med 2020;74:66–73. [DOI] [PubMed] [Google Scholar]
- [177].Mori S, Hirai R, Sakata Y. Simulated four-dimensional CT for markerless tumor tracking using a deep learning network with multi-task learning. Physica Medica 2020;80:151–8. 10.1016/j.ejmp.2020.10.023. [DOI] [PubMed] [Google Scholar]
- [178].Schreier J, Genghi A, Laaksonen H, Morgas T, Haas B. Clinical evaluation of a full-image deep segmentation algorithm for the male pelvis on cone-beam CT and CT. Radiother Oncol 2020;145:1–6. [DOI] [PubMed] [Google Scholar]
- [179].Wong J, Fong A, McVicar N, Smith S, Giambattista J, Wells D, et al. Comparing deep learning-based auto-segmentation of organs at risk and clinical target volumes to expert inter-observer variability in radiotherapy planning. Radiother Oncol 2020;144:152–8. [DOI] [PubMed] [Google Scholar]
- [180].Gatos I, Tsantis S, Spiliopoulos S, Karnabatidis D, Theotokas I, Zoumpoulis P, et al. Temporal stability assessment in shear wave elasticity images validated by deep learning neural network for chronic liver disease fibrosis stage assessment. Med Phys 2019;46:2298–309. [DOI] [PubMed] [Google Scholar]
- [181].Kagadis GC, Drazinos P, Gatos I, Tsantis S, Papadimitroulas P, Spiliopoulos S, et al. Deep learning networks on chronic liver disease assessment with fine-tuning of shear wave elastography image sequences. Phys Med Biol 2020;65:215027. [DOI] [PubMed] [Google Scholar]
- [182].Goodfellow I, Pouget-Abadie J, Mirza M, Xu B, Warde-Farley D, Ozair S, et al. Generative adversarial networks. Communications of the ACM 2020;63:139–44. 10.1145/3422622. [DOI] [Google Scholar]
- [183].Yi X, Walia E, Babyn P. Generative adversarial network in medical imaging: A review. Med Image Anal 2019;58:101552. [DOI] [PubMed] [Google Scholar]
- [184].Lan L, You L, Zhang Z, Fan Z, Zhao W, Zeng N, et al. Generative Adversarial Networks and Its Applications in Biomedical Informatics. Front Public Health 2020;8:164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [185].Gonog L, Zhou Y. A Review: Generative Adversarial Networks. 2019 14th IEEE Conference on Industrial Electronics and Applications (ICIEA), 2019, p. 505–10. [Google Scholar]
- [186].Isola P, Zhu J-Y, Zhou T, Efros AA. Image-to-image translation with conditional adversarial networks. Proceedings of the IEEE conference on computer vision and pattern recognition, 2017, p. 1125–34. [Google Scholar]
- [187].Zhu J-Y, Park T, Isola P, Efros AA. Unpaired Image-to-Image Translation Using Cycle-Consistent Adversarial Networks. 2017 IEEE International Conference on Computer Vision (ICCV) 2017. 10.1109/iccv.2017.244. [DOI] [Google Scholar]
- [188].Sandfort V, Yan K, Pickhardt PJ, Summers RM. Data augmentation using generative adversarial networks (CycleGAN) to improve generalizability in CT segmentation tasks. Sci Rep 2019;9:16884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [189].Shao S, Wang P, Yan R. Generative adversarial networks for data augmentation in machine fault diagnosis. Computers in Industry 2019;106:85–93. 10.1016/j.compind.2019.01.001. [DOI] [Google Scholar]
- [190].Shorten C, Khoshgoftaar TM. A survey on Image Data Augmentation for Deep Learning. Journal of Big Data 2019;6. 10.1186/s40537-019-0197-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [191].Maspero M, Savenije MHF, Dinkla AM, Seevinck PR, Intven MPW, Jurgenliemk-Schulz IM, et al. Dose evaluation of fast synthetic-CT generation using a generative adversarial network for general pelvis MR-only radiotherapy. Phys Med Biol 2018;63:185001. [DOI] [PubMed] [Google Scholar]
- [192].Wolterink JM, Dinkla AM, Savenije MHF, Seevinck PR, van den Berg CAT, Išgum I. Deep MR to CT Synthesis Using Unpaired Data. Simulation and Synthesis in Medical Imaging 2017:14–23. 10.1007/978-3-319-68127-6_2. [DOI] [Google Scholar]
- [193].Kazemifar S, Barragán Montero AM, Souris K, Rivas ST, Timmerman R, Park YK, et al. Dosimetric evaluation of synthetic CT generated with GANs for MRI-only proton therapy treatment planning of brain tumors. J Appl Clin Med Phys 2020;21:76–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [194].Kurz C, Maspero M, Savenije MHF, Landry G, Kamp F, Pinto M, et al. CBCT correction using a cycle-consistent generative adversarial network and unpaired training to enable photon and proton dose calculation. Phys Med Biol 2019;64:225004. [DOI] [PubMed] [Google Scholar]
- [195].Bowles C, Chen L, Guerrero R, Bentley P, Gunn R, Hammers A, et al. GAN Augmentation: Augmenting Training Data using Generative Adversarial Networks 2018. [Google Scholar]
- [196].Qi M, Li Y, Wu A, Jia Q, Li B, Sun W, et al. Multi-sequence MR image-based synthetic CT generation using a generative adversarial network for head and neck MRI-only radiotherapy. Med Phys 2020;47:1880–94. [DOI] [PubMed] [Google Scholar]
- [197].Liu Y, Lei Y, Wang Y, Shafai-Erfani G, Wang T, Tian S, et al. Evaluation of a deep learning-based pelvic synthetic CT generation technique for MRI-based prostate proton treatment planning. Phys Med Biol 2019;64:205022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [198].Wang Y, Wang S, Chen J, Wu C. Whole mammographic mass segmentation using attention mechanism and multiscale pooling adversarial network. J Med Imaging (Bellingham) 2020;7:054503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [199].Rezaei M, Yang H, Meinel C. Recurrent generative adversarial network for learning imbalanced medical image semantic segmentation. Multimedia Tools and Applications 2020;79:15329–48. 10.1007/s11042-019-7305-1. [DOI] [Google Scholar]
- [200].Khosravan N, Mortazi A, Wallace M, Bagci U. PAN: Projective Adversarial Network for Medical Image Segmentation. Lecture Notes in Computer Science 2019:68–76. 10.1007/978-3-030-32226-7_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [201].Jia H, Xia Y, Song Y, Zhang D, Huang H, Zhang Y, et al. 3D APA-Net: 3D Adversarial Pyramid Anisotropic Convolutional Network for Prostate Segmentation in MR Images. IEEE Trans Med Imaging 2020;39:447–57. [DOI] [PubMed] [Google Scholar]
- [202].Huo Y, Xu Z, Bao S, Bermudez C, Plassard AJ, Liu J, et al. Splenomegaly Segmentation using Global Convolutional Kernels and Conditional Generative Adversarial Networks. Proc SPIE Int Soc Opt Eng 2018;10574. 10.1117/12.2293406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [203].Dong X, Lei Y, Wang T, Thomas M, Tang L, Curran WJ, et al. Automatic multiorgan segmentation in thorax CT images using U-net-GAN. Med Phys 2019;46:2157–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [204].Kearney V, Chan JW, Wang T, Perry A, Descovich M, Morin O, et al. DoseGAN: a generative adversarial network for synthetic dose prediction using attention-gated discrimination and generation. Sci Rep 2020; 10:11073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [205].Babier A, Mahmood R, McNiven AL, Diamant A, Chan TCY. Knowledge-based automated planning with three-dimensional generative adversarial networks. Med Phys 2020;47:297–306. [DOI] [PubMed] [Google Scholar]
- [206].Murakami Y, Magome T, Matsumoto K, Sato T, Yoshioka Y, Oguchi M. Fully automated dose prediction using generative adversarial networks in prostate cancer patients. PLoS One 2020;15:e0232697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [207].Mahmood R, Babier A, McNiven A, Diamant A, Chan TCY. Automated Treatment Planning in Radiation Therapy using Generative Adversarial Networks. In: Doshi-Velez F, Fackler J, Jung K, Kale D, Ranganath R, Wallace B, et al. , editors. Proceedings of the 3rd Machine Learning for Healthcare Conference, vol. 85, Palo Alto, California: PMLR; 2018, p. 484–99. [Google Scholar]
- [208].Koike Y, Anetai Y, Takegawa H, Ohira S, Nakamura S, Tanigawa N. Deep learning-based metal artifact reduction using cycle-consistent adversarial network for intensity-modulated head and neck radiation therapy treatment planning. Phys Med 2020;78:8–14. [DOI] [PubMed] [Google Scholar]
- [209].Schlegl T, Seeböck P, Waldstein SM, Schmidt-Erfurth U, Langs G. Unsupervised Anomaly Detection with Generative Adversarial Networks to Guide Marker Discovery. Information Processing in Medical Imaging, Springer International Publishing; 2017, p. 146–57. [Google Scholar]
- [210].Xie X, Niu J, Liu X, Chen Z, Tang S, Yu S. A survey on incorporating domain knowledge into deep learning for medical image analysis. Med Image Anal 2021. ;69:101985. [DOI] [PubMed] [Google Scholar]
- [211].Deng C, Ji X, Rainey C, Zhang J, Lu W. Integrating Machine Learning with Human Knowledge. iScience 2020;23:101656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [212].Nguyen D, McBeth R, Sadeghnejad Barkousaraie A, Bohara G, Shen C, Jia X, et al. Incorporating human and learned domain knowledge into training deep neural networks: A differentiable dose-volume histogram and adversarial inspired framework for generating Pareto optimal dose distributions in radiation therapy. Med Phys 2020;47:837–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [213].Zhen S-H, Cheng M, Tao Y-B, Wang Y-F, Juengpanich S, Jiang Z-Y, et al. Deep Learning for Accurate Diagnosis of Liver Tumor Based on Magnetic Resonance Imaging and Clinical Data. Front Oncol 2020; 10:680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [214].Shehata M, Shalaby A, Switala AE, El-Baz M, Ghazal M, Fraiwan L, et al. A multimodal computer-aided diagnostic system for precise identification of renal allograft rejection: Preliminary results. Med Phys 2020;47:2427–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [215].Huang S-C, Pareek A, Seyyedi S, Banerjee I, Lungren MP. Fusion of medical imaging and electronic health records using deep learning: a systematic review and implementation guidelines. NPJ Digit Med 2020;3:136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [216].Hu J, Song Y, Wang Q, Bai S, Yi Z. Incorporating historical sub-optimal deep neural networks for dose prediction in radiotherapy. Med Image Anal 2021;67:101886. [DOI] [PubMed] [Google Scholar]
- [217].Xing Y, Zhang Y, Nguyen D, Lin M-H, Lu W, Jiang S. Boosting radiotherapy dose calculation accuracy with deep learning. J Appl Clin Med Phys 2020;21:149–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [218].Kontaxis C, Bol GH, Lagendijk JJW, Raaymakers BW. DeepDose: Towards a fast dose calculation engine for radiation therapy using deep learning. Phys Med Biol 2020;65:075013. [DOI] [PubMed] [Google Scholar]
- [219].Muralidhar N, Islam MR, Marwah M, Karpatne A, Ramakrishnan N. Incorporating Prior Domain Knowledge into Deep Neural Networks. 2018 IEEE International Conference on Big Data (Big Data), 2018, p. 36–45. [Google Scholar]
- [220].Luo Y, Tseng H-H, Cui S, Wei L, Ten Haken RK, El Naqa I. Balancing accuracy and interpretability of machine learning approaches for radiation treatment outcomes modeling. BJR Open 2019; 1:20190021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [221].Reyes M, Meier R, Pereira S, Silva CA, Dahlweid F-M, von Tengg-Kobligk H, et al. On the Interpretability of Artificial Intelligence in Radiology: Challenges and Opportunities. Radiol Artif Intell 2020;2:e190043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [222].Murdoch WJ, James Murdoch W, Singh C, Kumbier K, Abbasi-Asl R, Yu B. Definitions, methods, and applications in interpretable machine learning. Proceedings of the National Academy of Sciences 2019;116:22071–80. 10.1073/pnas.1900654116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [223].Barredo Arrieta A, Díaz-Rodríguez N, Del Ser J, Bennetot A, Tabik S, Barbado A, et al. Explainable Artificial Intelligence (XAI): Concepts, taxonomies, opportunities and challenges toward responsible AI. Inf Fusion 2020;58:82–115. [Google Scholar]
- [224].Gennatas ED, Friedman JH, Ungar LH, Pirracchio R, Eaton E, Reichmann LG, et al. Expert-augmented machine learning. Proc Natl Acad Sci U S A 2020; 117:4571–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [225].Barragán-Montero AM, Thomas M, Defraene G, Michiels S, Haustermans K, Lee JA, et al. Deep learning dose prediction for IMRT of esophageal cancer: The effect of data quality and quantity on model performance. Phys Med 2021;83:52–63. [DOI] [PubMed] [Google Scholar]
- [226].Blanch MG. Active Deep Learning for Medical Imaging Segmentation. 2017. [Google Scholar]
- [227].Smailagic A, Costa P, Noh HY, Walawalkar D, Khandelwal K, Galdran A, et al. MedAL: Accurate and Robust Deep Active Learning for Medical Image Analysis. 2018 17th IEEE International Conference on Machine Learning and Applications (ICMLA) 2018. 10.1109/icmla.2018.00078. [DOI] [Google Scholar]
- [228].Kim T, Lee K, Ham S, Park B, Lee S, Hong D, et al. Active learning for accuracy enhancement of semantic segmentation with CNN-corrected label curations: Evaluation on kidney segmentation in abdominal CT. Sci Rep 2020; 10:366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [229].Larrazabal AJ, Nieto N, Peterson V, Milone DH, Ferrante E. Gender imbalance in medical imaging datasets produces biased classifiers for computer-aided diagnosis. Proc Natl Acad Sci U S A 2020; 117:12592–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [230].Z O, Obermeyer Z, Powers B, C V et al. Dissecting racial bias in an algorithm used to manage the health of populations. Yearbook of Paediatric Endocrinology 2020. 10.1530/ey.17.12.7. [DOI] [PubMed] [Google Scholar]
- [231].Wilkinson MD, Dumontier M, Aalbersberg IJJ, Appleton G, Axton M, Baak A, et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci Data 2016;3:160018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [232].Kohli MD, Summers RM, Raymond Geis J. Medical Image Data and Datasets in the Era of Machine Learning—Whitepaper from the 2016 C-MIMI Meeting Dataset Session. Journal of Digital Imaging 2017;30:392–9. 10.1007/s10278-017-9976-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [233].Harvey H, Glocker B. A Standardised Approach for Preparing Imaging Data for Machine Learning Tasks in Radiology. Artificial Intelligence in Medical Imaging 2019:61–72. 10.1007/978-3-319-94878-2_6. [DOI] [Google Scholar]
- [234].Kortesniemi M, Tsapaki V, Trianni A, Russo P, Maas A, Källman H-E, et al. The European Federation of Organisations for Medical Physics (EFOMP) White Paper: Big data and deep learning in medical imaging and in relation to medical physics profession. Phys Med 2018;56:90–3. [DOI] [PubMed] [Google Scholar]
- [235].Sheller MJ, Anthony Reina G, Edwards B, Martin J, Bakas S. Multi-institutional Deep Learning Modeling Without Sharing Patient Data: A Feasibility Study on Brain Tumor Segmentation. Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries 2019:92–104. 10.1007/978-3-030-11723-8_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [236].Sheller MJ, Edwards B, Anthony Reina G, Martin J, Pati S, Kotrotsou A, et al. Federated learning in medicine: facilitating multi-institutional collaborations without sharing patient data. Scientific Reports 2020;10. 10.1038/s41598-020-69250-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [237].Chang K, Balachandar N, Lam C, Yi D, Brown J, Beers A, et al. Distributed deep learning networks among institutions for medical imaging. J Am Med Inform Assoc 2018;25:945–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [238].Prior F, Almeida J, Kathiravelu P, Kurc T, Smith K, Fitzgerald TJ, et al. Open access image repositories: high-quality data to enable machine learning research. Clin Radiol 2020;75:7–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [239].Challenges n.d. https://grand-challenge.org/challenges/ (accessed December 3, 2020).
- [240].Prevedello LM, Halabi SS, Shih G, Wu CC, Kohli MD, Chokshi FH, et al. Challenges Related to Artificial Intelligence Research in Medical Imaging and the Importance of Image Analysis Competitions. Radiology: Artificial Intelligence 2019;1:e180031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [241].Diaz O, Kushibar K, Osuala R, Linardos A, Garrucho L, Igual L, et al. Data preparation for artificial intelligence in medical imaging: A comprehensive guide to open-access platforms and tools. Phys Med 2021;83:25–37. [DOI] [PubMed] [Google Scholar]
- [242].Menze BH, Jakab A, Bauer S, Kalpathy-Cramer J, Farahani K, Kirby J, et al. The Multimodal Brain Tumor Image Segmentation Benchmark (BRATS). IEEE Trans Med Imaging 2015;34:1993–2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [243].Babier A, Zhang B, Mahmood R, Moore KL, Purdie TG, McNiven AL, et al. OpenKBP: The open-access knowledge-based planning grand challenge. arXiv [physics.med-Ph] 2020. [DOI] [PubMed] [Google Scholar]
- [244].Callaway E “It will change everything”: DeepMind’s AI makes gigantic leap in solving protein structures. Nature 2020. 10.1038/d41586-020-03348-4. [DOI] [PubMed] [Google Scholar]
- [245].Maier-Hein L, Eisenmann M, Reinke A, Onogur S, Stankovic M, Scholz P, et al. Why rankings of biomedical image analysis competitions should be interpreted with care. Nat Commun 2018;9:5217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [246].Reinke A, Eisenmann M, Onogur S, Stankovic M, Scholz P, Full PM, et al. How to Exploit Weaknesses in Biomedical Challenge Design and Organization. Medical Image Computing and Computer Assisted Intervention – MICCAI 2018. 2018:388–95. 10.1007/978-3-030-00937-3_45. [DOI] [Google Scholar]


