Abstract
The highly conserved C-terminal domain (CTD) of the largest subunit of RNA polymerase II comprises a consensus heptad (Y1S2P3T4S5P6S7) repeated multiple times. Despite the simplicity of its sequence, the essential CTD domain orchestrates eukaryotic transcription and co-transcriptional processes, including transcription initiation, elongation, and termination, and mRNA processing. These distinct facets of the transcription cycle rely on specific post-translational modifications (PTM) of the CTD, in which five out of the seven residues in the heptad repeat are subject to phosphorylation. A hypothesis termed the “CTD code” has been proposed in which these PTMs and their combinations generate a sophisticated landscape for spatiotemporal recruitment of transcription regulators to Pol II. In this review, we summarize the recent experimental evidence understanding the biological role of the CTD, implicating a context-dependent theme that significantly enhances the ability of accurate transcription by RNA polymerase II. Furthermore, feedback communication between the CTD and histone modifications coordinates chromatin states with RNA polymerase II-mediated transcription, ensuring the effective and accurate conversion of information into cellular responses.
Keywords: RNA polymerase II, phosphorylation, transcription, proline isomerization, crosstalk, histone
Introduction
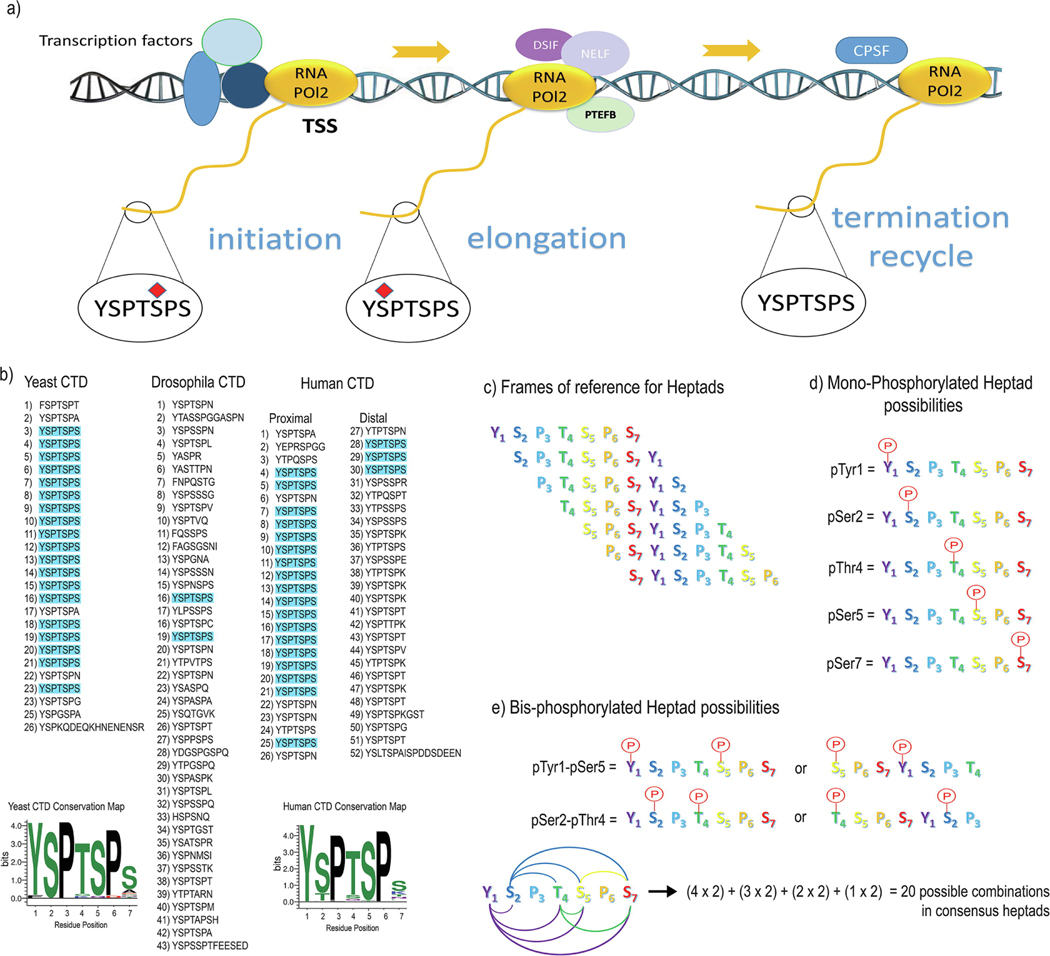
RNA polymerase II (Pol II) is a multi-subunit enzyme involved in transcribing all protein-coding genes in eukaryotes [1,2]. The largest subunit of Pol II (RPB1) contains a series of seven residue (Heptad) repeats as its Carboxyl-Terminal domain (CTD). The CTD interacts with various general and specific transcription factors, regulators, and other proteins to aid and regulate transcription [3–5]. Across eukaryotes, the number of repeats and the level of conservation differ among species. For example, Saccharomyces cerevisiae contains 26 repeats, which mostly conform to the consensus sequence of Tyrosine (Tyr1), Serine (Ser2), Proline (Pro3), Threonine (Thr4), Serine (Ser5), Proline (Pro6), and Serine (Ser7) [3] (Figure 1a and 1b). The positioning of heptads can be viewed differently with alternative frames of references though (Figure 1c). Through the evolution tree, complicated eukaryotes contain Pol II with the CTD heptads deviating from the consensus, mostly in the 7th position [6] (Figure 1b). The most extreme example is Drosophila, which has a CTD consisting of 42 repeats of highly divergent sequence with only two heptads as consensus [7]. Even though the CTD doesn’t directly contribute to Pol II’s catalytic activity, its function is required for the effective transcription of genetic information ― cells with the CTD deleted cannot survive [8,9].
Figure 1:
RNA Pol II exhibits different post-translational modifications at various stages of transcription. (a) The CTD is highly phosphorylated at Ser5 at the beginning of transcription. Ser2 gets phosphorylated during the pausing release. At the end of transcription, it is believed that both phosphorylation marks are removed. (b) Sequences of the yeast, Drosophila and human CTD are shown in a heptad-wise manner. Heptads with light blue background are consensus heptads (Sequence - YSPTSPS). Conservation maps for yeast and human CTD are shown using LogOdds Sequence Logo.
https://www.ncbi.nlm.nih.gov/CBBresearch/Yu/logoddslogo/
(c) Frames of references for heptads are shown with potential starting residue for each heptad. (d) All mono-phosphorylated heptad possibilities are shown. (e) All bis-phosphorylated heptad possibilities with the total number of combinations are shown.
The CTD heptad repeats undergo extensive post-translational modifications (PTMs) such as phosphorylation [10], acetylation [11], methylation [12], and glycosylation [13]. Phosphorylation is the most prominent modification in which kinases phosphorylate Ser2 and Ser5 of the heptad transiently. They are returned to their dephosphorylated form by the end of the cycle [14] (Figure 1a). When RNA Pol II binds to the promoter, it is free of phosphate modifications. Ser5 is among the first residues to be phosphorylated after transcriptional initiation [15]. After initiation, the DRB-sensitivity inducing factor (DSIF) and the negative elongation factor (NELF) block Pol II at ~60bp downstream of the transcription start site, called promoter-proximal pausing [16,17]. Pausing is released by the positive transcription elongation factor (P-TEFb) when negative elongation factors (DSIF and NELF), as well as Ser2 of the CTD, are phosphorylated [18]. These phosphorylation marks are removed at the end of the transcriptional cycle, which prepares Pol II to initiate another transcription cycle.
Spatiotemporal phosphorylation of the CTD is critical for Pol II’s biological function since mutations on the residues of heptads or the enzymes modifying the phosphorylation states compromise transcriptional functions [19,20]. The high abundance of PTM sites on the flexible CTD exhibits resemblance to the flexible histone tails that are also heavily modified by methylation, acetylation, ubiquitination, and phosphorylation [21–23]. In 2003, a CTD code hypothesis was proposed, highlighting its potential to encode a large amount of information through its PTMs and their combinations to recruit binding partners [24]. In a similar fashion to proteins interacting with histones, the proteins interacting with the CTD are identified as writers (such as kinases that place phosphate groups), modifiers (such as proline isomerases that change proline isomeric states), readers (such as proteins physically recruited to Pol II by interacting with the CTD), and erasers (such as phosphatases that remove phosphates) (Table 1). The interplay between this extensive collection of writers/modifiers/readers/erasers can generate all combinations of modification species, leading to various functions during transcription (Figure 1d and 1e). It has been twenty years since the CTD code was proposed, yet the exact mechanism of CTD-mediated transcription still evades our understanding.
Table 1:
Characterization of proteins bound to CTD.
| Proteins | Binding affinity (μM) | Peptide | Sequence | Method | Ref. |
|---|---|---|---|---|---|
| Set2 | |||||
| Sc Set2 SRI | ~6 | S2,5-PCTD peptide | YSPTSPSYSPTSPSYSPTSPSC | SPR | [106] |
| Hs Set2 SRI | 5.4±0.5 | S212-P/S512-P CTD peptide | TSPSYSPTSPSYSPTSPSYSPT | SPR | [108] |
| Hs Set2 SRI | 17.8±0.9 | S212-P/S51-P CTD peptide | TSPSYSPTSPSYSPTSPS | SPR | [108] |
| Hs Set2 SRI | 12.5±1.5 | S22-P/S512-P CTD peptide | SYSPTSPSYSPTSPSYSPT | SPR | [108] |
| Hs Set2 SRI | 5.5±0.5 | S2123-P/S5123-P CTD peptide | YSPTSPSYSPTSPSYSPTSPS | SPR | [108] |
| Pcf11 | |||||
| Sc Pcf11 CID | 54±6 | S2-p CTD peptide | SYSPTSPSYSPTSPS | FA | [114] |
| Sc Pcf11 CID | 180 | S2-p CTD peptide | PTSPSYSPTSPS | ITC | [55] |
| Sc Pcf11 CID | 1500±300; >1000 | Unphosp. CTD peptide | YSPTSPSYSPTSPS | NMR; FA | [115] |
| Sc Pcf11 CID | 1400±650; NBD | S21-P CTD peptide | YSPTSPSYSPTSPS | NMR; FA | [115] |
| Sc Pcf11 CID | 240±60; NBD | S22-P CTD peptide | YSPTSPSYSPTSPS | NMR; FA | [115] |
| Sc Pcf11 CID | 160±50; 130±35; | S212-P CTD peptide | YSPTSPSYSPTSPS | NMR; FA | [115] |
| Sc Pcf11 CID | 630±170; 370±160/110 | S212-P/ S512-P CTD peptide | YSPTSPSYSPTSPS | NMR; FA | [115] |
| Sc Pcf11 CID | 1200±500; >1000 | S512-P CTD peptide | YSPTSPSYSPTSPS | NMR; FA | [115] |
| Sc Pcf11 CID | 127±15 | S212-P CTD peptide | YSPTSPSYSPTSPS | FA | [116] |
| Sc Pcf11 CID | 269±57 | T412-P CTD peptide | YSPTSPSYSPTSPS | FA | [116] |
| Sp Pcf11 CID | 48.1±27 | S212-P CTD peptide | YSPTSPSYSPTSPS | FA | [116] |
| Sp Pcf11 CID | 51.1±5.2 | T412-P CTD peptide | YSPTSPSYSPTSPS | FA | [116] |
| Sc Rtt103 | |||||
| Sc Rtt103 CID | 12±2 | S2-p CTD peptide | SYSPTSPSYSPTSPS | FA | [114] |
| Sc Rtt103 CID | 1200±300; >1000 | Unphosp. CTD peptide | YSPTSPSYSPTSPS | NMR; FA | [115] |
| Sc Rtt103 CID | 420±70; NBD | S21-P CTD peptide | YSPTSPSYSPTSPS | NMR; FA | [115] |
| Sc Rtt103 CID | 76±16; NBD | S22-P CTD peptide | YSPTSPSYSPTSPS | NMR; FA | [115] |
| Sc Rtt103 CID | 15±10; 2.1±0.1 | S212-P CTD peptide | YSPTSPSYSPTSPS | NMR; FA | [115] |
| Sc Rtt103 CID | 41±19; 53±10 | S212-P/ S512-P CTD peptide | YSPTSPSYSPTSPS | NMR; FA | [115] |
| Sc Rtt103 CID | 600±300; 122±2/3 | S512-P CTD peptide | YSPTSPSYSPTSPS | NMR; FA | [115] |
| Sc Rtt103 CID | 32.3±6.8 | S2-P CTD peptide | YSPTSPSYSPTSPS | FA | [116] |
| Sc Rtt103 CID | 34.8±3.2 | T4-p CTD peptide | YSPTSPSYSPTSPS | FA | [116] |
| Sc Rtt103 CID | >1000 | S5-p CTD peptide | YSPTSPSYSPTSPS | FA | [116] |
| Sc Rtt103 CID | 6.0±0.2 | S2-P CTD peptide | PSYSPTSPSYSPTSPS | FA | [117] |
| Sc Rtt103 CID | 43±2 | S2,T4-P CTD peptide | PSYSPTSPSYSPTSPS | FA | [117] |
| Sc Rtt103 CID | 64±2 | Unphosp. CTD peptide | PSYSPTSPSYSPTSPS | FA | [117] |
| Sc Rtt103 CID | NBD | Y1-P CTD peptide | PSYSPTSPSYSPTSPS | FA | [117] |
| Sc Rtt103 CID | 15±1 | T4-P CTD peptide | PSYSPTSPSYSPTSPS | FA | [117] |
| Sc Rtt103 CID | 6.0±0.2 | T412-P CTD peptide | PSYSPTSPSYSPTSPS | FA | [117] |
| Sc Rtt103 CID | 1200±300 | Unphosp. CTD peptide | YSPTSPSYSPTSPS | NMR | [118] |
| Sc Rtt103 CID | 50±10 | T412-P CTD peptide | YSPTSPSYSPTSPS | NMR | [118] |
| Sc Rtt103 CID | 9±6 | S212-P CTD peptide | YSPTSPSYSPTSPS | NMR | [118] |
| Sc Spt6 | |||||
| Sc Spt6 SH2 | 3.6±0.15 | Y1-P CTD peptide | PSYSPTSPSYSPTSPS | FA | [114] |
| Sc Spt6 SH2 | 1.9±0.04 | Y1-P-S2-P CTD peptide | PSYSPTSPSYSPTSPS | FA | [114] |
| Sc Spt6 SH2 | 1.3±0.06 | Y1-P-S5-P CTD peptide | PSYSPTSPSYSPTSPS | FA | [114] |
| Sc Spt6 SH2 | 5.2±0.09 | S5-P CTD peptide | PSYSPTSPSYSPTSPS | FA | [114] |
| Sc Spt6 SH2 | 8.4±0.19 | S2-P CTD peptide | SYSPTSPSYSPTSPS | FA | [114] |
| Sc Spt6 tSH2 | 375±69 | Unphosp. CTD peptide | PSYSPTSPSYSPTSPS | FA | [119] |
| Sc Spt6 tSH2 | 38.7±2.4 | (Y1-P CTD) peptide | PSYSPTSPS | FA | [119] |
| Sc Spt6 tSH2 | 45.4±1.7 | Y11-P CTD peptide | PSYSPTSPSYSPTSPS | FA | [119] |
| Sc Spt6 tSH2 | 36.8±2.6 | Y12-P CTD peptide | PSYSPTSPSYSPTSPS | FA | [119] |
| Sc Spt6 tSH2 | 5.1±0.4 | Y112-P CTD peptide | PSYSPTSPSYSPTSPS | FA | [119] |
| Sc Spt6 tSH2 | 5.7±0.4 | S2,T4-P CTD peptide | PSYSPTSPSYSPTSPS | FA | [119] |
| Sc Spt6 tSH2 | 7.2±0.3 | T412-P CTD peptide | PSYSPTSPSYSPTSPS | FA | [119] |
| Sc Spt6 tSH2 | 0.3±0.1 | Y112,S512-P CTD peptide | PSYSPTSPSYSPTSPS | FA | [119] |
| Sc Spt6 tSH2 | 0.3±0.1 | Y112,S212-P CTD peptide | PSYSPTSPSYSPTSPS | FA | [119] |
| Sc Spt6 tSH2 | 1.0±0.1 | S212,S712-P CTD peptide | PSYSPTSPSYSPTSPS | FA | [119] |
| Sc Spt6 tSH2 | 0.0152±0.0009 | (Y1-P CTD) × 13 | PS(YSPTSPS) × 13 | MST | [119] |
| Nrd1 | |||||
| Sc Nrd1 CID | 85±25 | S5-p CTD peptide | PSYSPTSPSYSPTSPS | FA | [114] |
| Sc Nrd1 CID | 126±4 | S2-P CTD peptide | YSPTSPSYSPTSPS | FA | [116] |
| Sc Nrd1 CID | 148±7 | T4-p CTD peptide | YSPTSPSYSPTSPS | FA | [116] |
| Sc Nrd1 CID | 65.8±3 | S5-p CTD peptide | YSPTSPSYSPTSPS | FA | [116] |
| Sc Nrd1 CID | 40±3 | S5-P CTD peptide | YSPTSPSYSPTSPS | FA | [120] |
| Sc Nrd1 CID | 390±30 | S2-P CTD peptide | YSPTSPSYSPTSPS | FA | [120] |
| Sc Nrd1 CID | 16±1 | S2,5-P CTD peptide | YSPTSPSYSPTSPS | FA | [120] |
| Sc Nrd1 CID | 39±3 | (S5-P CTD) × 3 peptide | YSPTSPSYSPTSPSYSPTSPS | FA | [120] |
| Sc Nrd1 CID | 700±100 | Unphosp. CTD peptide | YSPTSPSYSPTSPS | FA | [121] |
| Sc Nrd1 CID | 113±5 | S512-P CTD peptide | YSPTSPSYSPTSPS | FA | [121] |
| Sc Nrd1 CID | 140±20 | S51/72-P CTD peptide | YSPTSPSYSPTSPS | FA | [121] |
| Sc Nrd1 CID | >1000 | S712-P CTD peptide | YSPTSPSYSPTSPS | FA | [121] |
| Sp Seb1 CID | 72.3±6.4 | S2-P CTD peptide | YSPTSPSYSPTSPS | FA | [116] |
| Sp Seb1 CID | 153.2±12.1 | T4-P CTD peptide | YSPTSPSYSPTSPS | FA | [116] |
| Sp Seb1 CID | 280.9±23.1 | S5-P CTD peptide | YSPTSPSYSPTSPS | FA | [116] |
| FluA and FluB polymerase | |||||
| Bat FluA Pol | 0.9± 0.1 | (S5-P)4 CTD peptide | (YSPTSPS) × 4 | FA | [122] |
| Bat FluA Pol | 6.1± 0.7 | (S5-P)2 CTD peptide | (YSPTSPS) × 2 | FA | [122] |
| Hs FluB Pol | 2.9± 0.3 | (S5-P)4 CTD peptide | (YSPTSPS) × 4 | FA | [122] |
| Hs FluB Pol | 4.2± 0.8 | (S5-P)2 CTD peptide | (YSPTSPS) × 2 | FA | [122] |
| Prolyl-isomerase | |||||
| Sc ESS1 WW | >3000 | Unphosp. CTD peptide | ASYSPTSPSYS | FA | [123] |
| Sc ESS1 WW | 241±23 | S2-P CTD peptide | ASYSPTSPSYS | FA | [123] |
| Sc ESS1 WW | 61±4.9 | S5-P CTD peptide | ASYSPTSPSYS | FA | [123] |
| Sc ESS1 WW | >300 | Unphosp. CTD peptide | GGSGGSYSPTSPSYS | BLI | [124] |
| Sc ESS1 WW | 2.6±0.7 | S5-P CTD peptide | GGSGGSYSPTSPSYS | BLI | [124] |
| Sc ESS1 WW | NBD | Unphosp. CTDpeptide21 | YSPTSPSYSPTSPSYSPTSPS | CD | [124] |
| Sc ESS1 WW | NBD | Unphosp. CTD peptide11 | SPTSPSYSPTS | CD | [125] |
| Sc ESS1 WW | 76±4 | S2-P CTD9 peptide | TSPSYSPTS | CD | [125] |
| Sc ESS1 WW | 76±4 | S2-P CTD11 peptide | SPTSPSYSPTS | CD | [125] |
| Sc ESS1 WW | 98±18 | S5-P CTD9 peptide | SPTSPSYSP | CD | [125] |
| Sc ESS1 WW | 79±13 | S5-P CTD11 peptide | SPTSPSYSPTS | CD | [125] |
| Sc ESS1 WW | 21 ± 3 | S5,2-P CTD11 peptide | SPTSPSYSPTS | CD | [125] |
| Sc ESS1 WW | 16 ±2 | S2,5-P CTD14 peptide | TSPSYSPTSPSYSPS | CD | [125] |
| Sc ESS1 WW | 17±2 | S5,5,5-P CTD21 peptide | YSPTSPSYSPTSPSYSPTSPS | CD | [125] |
| Hs Pin1 WW | NBD | Unphosp. CTD peptide | YSPTSPS | FA | [126] |
| Hs Pin1 WW | 61 ±6.3 | S2-P CTD peptide | YSPTSPS | FA | [126] |
| Hs Pin1 WW | 30±0.39 | S5-P CTD peptide | YSPTSPS | FA | [126] |
| Hs Pin1 WW | 10±0.8 | S2-P-S5-P CTD peptide | YSPTSPS | FA | [126] |
| Hs Pin1 WW | NBD | Unphosp. CTD peptide | YSPTSPS | FA | [126] |
| Hs Pin1 WW | 110± 23 | S2-P CTD peptide | YSPTSPS | FA | [126] |
| Hs Pin1 WW | 34 ± 5.9 | S5-P CTD peptide | YSPTSPS | FA | [126] |
| Hs Pin1 WW | 34 ± 6.2 | S2-P-S5-P CTD peptide | YSPTSPS | FA | [126] |
| Hs Pin1 PPIase | NBD | Unphosp. CTD peptide | YSPTSPS | FA | [126] |
| Hs Pin1 PPIase | NBD | S2-P CTD peptide | YSPTSPS | FA | [126] |
| Hs Pin1 PPIase | > 500 | S5-P CTD peptide | YSPTSPS | FA | [126] |
| Hs Pin1 PPIase | 390 ±6.82 | S2-P-S5-P CTD peptide | YSPTSPS | FA | [126] |
| Capping enzymes | |||||
| Sp Pce1 WT | 0.21 ± 0.03 | (S5-P)4 CTD peptide | (YSPTSPS)×4 | FA | [127] |
| Hs Mce 1 WT | NBD | Unphosp. CTD peptide | SPTSPSYSPTS | FA | [71] |
| Hs Mce1 WT | 139 ± 8.5 (pH=7.0) | S2-P-S5-P CTD peptide | SPTSPSYSPTS | FA | [71] |
| Hs Mce1 WT | 221 ± 19.5 (pH=8.0) | S2-P-S5-P CTD peptide | SPTSPSYSPTS | FA | [71] |
| Sc Ceg-1 WT | 460.7 | S5-P CTD peptide | YSPTSPSYSPTSPSYSPTSPS | FA | [128] |
| SCAF8 | |||||
| Hs SCAF8 CID | >1000 | Unphosp. CTD peptide | YSPTSPSYSPTSPS | FA | [129] |
| Hs SCAF8 CID | 68 ± 8/6 | S2-P CTD peptide | YSPTSPSYSPTSPS | FA | [129] |
| Hs SCAF8 CID | 330± 50/30 | S5-P CTD peptide | YSPTSPSYSPTSPS | FA | [129] |
| Hs SCAF8 CID | 19 ± 2 | S2/5-P CTD peptide | YSPTSPSYSPTSPS | FA | [129] |
| Hs SCAF8 CID | 90±4/3 | S2/S7-P CTD peptide | YSPTSPSYSPTSPS | FA | [129] |
| Hs TDRD3 Tudor domain | 770±30 | R1810-Methylated CTD peptide | YSPSSPR(Me2a)YTPQSP | FA | [130] |
| PHF3 | |||||
| Hs PHF3 SPOC | 1.6± 0.3 | S2-P CTD peptide | PSYSPTSPSYSPTSPS | FA | [131] |
| Hs PHF3 SPOC | 0.8± 0.1 | S2,7-P CTD peptide | PSYSPTSPSYSPTSPS | FA | [131] |
| Hs PHF3 SPOC | 4.8± 0.3 | S2,5-P CTD peptide | PSYSPTSPSYSPTSPS | FA | [131] |
| Hs PHF3 SPOC | 20.0± 4.0 | S5-P CTD peptide | PSYSPTSPSYSPTSPS | FA | [131] |
| Hs PHF3 SPOC | 26.0± 2.9 | S7-P CTD peptide | PSYSPTSPSYSPTSPS | FA | [131] |
| Hs PHF3 SPOC | 5.7± 0.4 | S5,7-P CTD peptide | PSYSPTSPSYSPTSPS | FA | [131] |
| RPRD family | |||||
| Hs RPRD1A CID | 339±56 | Unphosp. CTD peptide | SPSYSPTSPSYSPTSPSYS | ITC | [132] |
| Hs RPRD1A CID | 8.4±0.7 | S2-P CTD peptide | SPSYSPTSPSYSPTSPSYS | ITC | [132] |
| Hs RPRD1A CID | >1000 | S5-P CTD peptide | SPSYSPTSPSYSPTSPSYS | ITC | [132] |
| Hs RPRD1A CID | 49.8±13.7 | S7-P CTD peptide | SPSYSPTSPSYSPTSPSYS | ITC | [132] |
| Hs RPRD1A CID | 5.2±0.5 | S2,7-P CTD peptide | SPSYSPTSPSYSPTSPSYS | ITC | [132] |
| Hs RPRD1A CID | >1000 | S2,5-P CTD peptide | SPSYSPTSPSYSPTSPSYS | ITC | [132] |
| Hs RPRD1A CID | >1000 | S5,7-P CTD peptide | SPSYSPTSPSYSPTSPSYS | ITC | [132] |
| Hs RPRD1A CID | 48.3±5.5 | S7,5-P CTD peptide | SPSYSPTSPSYSPTSPSYS | ITC | [132] |
| Hs RPRD1A CID | 1.0±0.1 | S2-P, K7-P CTD peptide | SPKYSPTSPKYSPTSPKYS | ITC | [132] |
| Hs RPRD1A CID | 7.42±5.37 | S2-P CTD peptide | SPSYSPTSPSYSPTSPSYS | MST | [132] |
| Hs RPRD1B CID | 114±2 | Unphosp. CTD peptide | SPSYSPTSPSYSPTSPSYS | ITC | [132] |
| Hs RPRD1B CID | 6.8±0.2 | S2-P CTD peptide | SPSYSPTSPSYSPTSPSYS | ITC | [132] |
| Hs RPRD1B CID | >1000 | S5-P CTD peptide | SPSYSPTSPSYSPTSPSYS | ITC | [132] |
| Hs RPRD1B CID | 23.6±3.2 | S7-P CTD peptide | SPSYSPTSPSYSPTSPSYS | ITC | [132] |
| Hs RPRD1B CID | 2.6±0.2 | S2,7-P CTD peptide | SPSYSPTSPSYSPTSPSYS | ITC | [132] |
| Hs RPRD1B CID | >1000 | S2,5-P CTD peptide | SPSYSPTSPSYSPTSPSYS | ITC | [132] |
| Hs RPRD1B CID | >1000 | S5,7-P CTD peptide | SPSYSPTSPSYSPTSPSYS | ITC | [132] |
| RPRD1B CID | 30.0±4.9 | S7,5-P CTD peptide | SPSYSPTSPSYSPTSPSYS | ITC | [132] |
| RPRD1B CID | 8.3±0.5 | S2-P, K7-P CTD peptide | SPKYSPTSPKYSPTSPKYS | ITC | [132] |
| HsRPRD2 CID | 355±30 | Unphosp. CTD peptide | SPSYSPTSPSYSPTSPSYS | ITC | [132] |
| Hs RPRD2 CID | 8.3±0.5 | S2-P CTD peptide | SPSYSPTSPSYSPTSPSYS | ITC | [132] |
| Hs RPRD2 CID | >1000 | S5-P CTD peptide | SPSYSPTSPSYSPTSPSYS | ITC | [132] |
| Hs RPRD2 CID | 82.8±28.7 | S7-P CTD peptide | SPSYSPTSPSYSPTSPSYS | ITC | [132] |
| Hs RPRD2 CID | 7.2±0.1 | S2,7-P CTD peptide | SPSYSPTSPSYSPTSPSYS | ITC | [132] |
| Hs RPRD2 CID | >1000 | S2,5-P CTD peptide | SPSYSPTSPSYSPTSPSYS | ITC | [132] |
| Hs RPRD2 CID | >1000 | S5,7-P CTD peptide | SPSYSPTSPSYSPTSPSYS | ITC | [132] |
| Hs RPRD2 CID | 112±13 | S7,5-P CTD peptide | SPSYSPTSPSYSPTSPSYS | ITC | [132] |
| Hs RPRD2 CID | 5.2±0.1 | S2-P, K7-P CTD peptide | SPKYSPTSPKYSPTSPKYS | ITC | [132] |
| Hs CREPT RPR | 79.2±59.3 | Unphosp. CTD peptide | YSPTSPSYSPTSPS | FA | [133] |
| Hs CREPT RPR | 12.3±0.9 | S212-CTD peptide | YSPTSPSYSPTSPS | FA | [133] |
| Hs CREPT RPR | NBD | S512-CTD peptide | YSPTSPSYSPTSPS | FA | [133] |
| Hs CREPT RPR | 44.9±6.3 | S712-CTD peptide | YSPTSPSYSPTSPS | FA | [133] |
| Hs p15RS RPR | 40.5±8.8 | Unphosp. CTD peptide | YSPTSPSYSPTSPS | FA | [133] |
| Hs p15RS RPR | 13.7±0.8 | S212-CTD peptide | YSPTSPSYSPTSPS | FA | [133] |
| Hs p15RS RPR | >1000 | S512-CTD peptide | YSPTSPSYSPTSPS | FA | [133] |
| Hs p15RS RPR | 62.2±3.3 | S712-CTD peptide | YSPTSPSYSPTSPS | FA | [133] |
| FF domain | |||||
| Hs TCERG1 FF4–6 | 102±33 | S2,5-P CTD peptide | YSPTSPSYSPTSPSYSPTSPS | NMR | [134] |
| Hs TCERG1 FF4–6 | 13±5 | S2,5,7-P CTD peptide | SPSYSPTSPSYSPTSPSYSPT | NMR | [134] |
| Hs JMJD5 NTD | 1.22±0.63 | Unphosp. CTD peptide | SPSYSPTSPSYSPTSPSYS | MST | [135] |
| Hs JMJD5 NTD | 0.48±0.14 | S212-P CTD peptide | SPSYSPTSPSYSPTSPSYS | MST | [135] |
| Hs JMJD5 NTD | 1.41±0.84 | S512-P CTD peptide | SPSYSPTSPSYSPTSPSYS | MST | [135] |
| Hs JMJD5 NTD | 0.12±0.03 | S21,51-S22-P CTD peptide | SPSYSPTSPSYSPTSPSYS | MST | [135] |
| Hs JMJD5 NTD | 1.98±0.85 | S2-P CTD peptide | SPTSPSYSPTSPSYSPYSK | MST | [135] |
| Hs JMJD5 NTD | 9.92±2.63 | S21, -S22,52-P CTD peptide | SPSYSPTSPSYSPTSPSYS | MST | [135] |
| Hs SMN Tudor domain | NBD | Unmethylated CTD peptide | YSPSSPRYTPQSP | FA | [136] |
| Hs SMN Tudor domain | 717±66 | R1810me2a Methylated CTD | YSPSSPR(me2a)YTPQSP | FA | [136] |
| Hs SMN Tudor domain | 127±15 | R1810me2s Methylated CTD | YSPSSPR(me2s)YTPQSP | FA | [136] |
| Hs SMN Tudor domain | 175±14 | R1810me2s-S Methylated CTD | SPSYSPSSPR(me2s)YTPQs | FA | [136] |
Footnote: Abbreviation used in the table.
BLI: Bio-Layer Interferometry
CD: Circular Dichroism
FA: Fluorescence Anisotropy
ITC: Isothermal Titration Calorimetry
MST: Microscale Thermophoresis
NMR: Nuclear Magnetic Resonance
SPR: Surface Plasmon Resonance
Hs: Homo sapiens
Sc: Saccharomyces cerevisiae
Sp: Schizosaccharomyces pombe
NBD: No Binding Detected
NTD: N-terminal domain
Detection of CTD phosphorylation
The combinatorial coding system is highly attractive, with great potential for information encryption. However, experimental evidence which supports the combinatorial PTMs of CTD has been elusive due to the lack of adequate tools for detecting and identifying these chemical modifications. Historically, the detection of phosphorylation species can be achieved using electrophoretic mobility assays [25,26]. While fast and robust, these assays are unable to identify the specific phosphorylated locations. Identification was substantially improved when high specificity PTM-specific antibodies were introduced [27]. However, PTM-specific antibodies cannot specify the heptad position in the CTD where the modification has occurred. Furthermore, these antibodies are not precise for quantifying the phosphorylation level of the CTD for several reasons. First, antibodies are generated against the consensus sequence; thus, deviation from these repeats might compromise antibody recognition and affect binding strength. This issue frequently arises in metazoans. For example, the regions in human CTD containing the last 24 of the 52 overall heptad repeats deviate from the consensus sequence. Another complication in quantification is the chemical modification on flanking residues which affect antibody recognition. In human CTD, where doubly phosphorylated heptads have been detected [14], the levels of these bis-phosphorylated heptads cannot be accurately quantified but estimated to be at least 25% of the mono-phosphorylated heptad levels [28].
Direct detection methods such as mass spectrometry show significant advantages in deciphering the modification states of Pol II. The gold standard for site-specific characterization of PTMs is tandem mass spectrometry (MS/MS), which provides single residue resolution without context interference. However, several properties of the CTD heptad present hurdles for interpretation through conventional bottom-up MS/MS analyses: (1) the scarcity of Arg and Lys residues that serve both as protonation sites and proteolytic sites, (2) the potentially large number of labile phosphate groups, and (3) the repetitive nature of the CTD sequence [3,29]. The need for high-resolution detection of PTMs in the CTD motivated the recent MS/MS analyses of the CTD in yeast [30] and human cells [28] using conventional collision-induced dissociation (CID). The difficulty in proteolyzing the CTD to generate small peptides for bottom-up MS/MS analysis was circumvented by introducing mutations to the CTD sequence to facilitate proteolytic digestion and modification site localization [28,30]. Most mutations are generated at the heptad’s 7th position since it is the least conserved residue in the entire heptad. Such analyses show that the phosphorylation of Ser5 and Ser2 dominates the mono-phosphorylated species [30]. Furthermore, in human cells, both mono-phosphorylated and bis-phosphorylated heptads were mapped, and a substantial percentage of bis-phosphorylated heptads were detected [28]. The identification of phosphorylation marks on the CTD provides insights into the existence of different phosphorylation species within cells.
Mass spec strategies have pushed forward to allow a direct interpretation of the endogenous CTD phosphorylation during transcription without introducing mutations [31,32]. Certain regions of the human and Drosophila CTD have endogenous Lys and Arg residues that allow lysis of long polypeptides into fragments suitable for tandem MS analysis [31–33]. For the CTD consensus sequence, novel proteases such as chymotrypsin and proteinase K cut at the peptide bond following tyrosine residues. These strategies alleviate the concern of introducing mutation bias, presenting a similar phosphorylation pattern as the phosphorylation mapping done by mutating the heptad’s 7th residue [31,32].
The application of ultraviolet photodissociation (UVPD) has overcome the issue of low ionization/activation issue [32–34]. Unlike collision and electron-based activation methods, UVPD presents obvious advantages for identifying phosphorylation of Pol II [35–42]. As a one-step, high energy activation method, the use of UVPD retains labile modifications [42]. Next, UVPD provides high coverage of the sequence, allowing it to generate rich fragmentation patterns for positively and negatively charged peptides. Therefore, UVPD is highly applicable to acidic phosphoryl peptides [37]. Lastly, UVPD provides a significant advantage over electron-based MS/MS methods by characterizing even singly charged peptides. When considering the many benefits of UVPD, it becomes a natural choice for CTD analysis. Indeed, UVPD-mass spec was able to map the Drosophila CTD, which has proven difficult for analysis via PTM-specific antibodies due to its divergence from the consequence sequence from which those antibodies were derived [31,32].
Crosstalk of different residues on the CTD heptad
The histone modification hypothesis describes the generation of sophisticated PTM patterns for precise transcriptional control, achieved by the crosstalk between different histone PTMs. For example, Histone-2-B Lysine-120 ubiquitination (H2BK120ub) regulates the methylation of H3K79 and H3K4, which activates gene expression [43,44]. In turn, the tri-methylation of H3K4me3 and H3K36me3 both inhibit the Protein Regulator of Cytokinesis (PRC) 2 activity, ensuring gene expression [45]. In contrast, ubiquitination marks placed by PRC1 on H2AK119 activates PRC2 to methylate H3K27 and silences gene expression [46]. The combinatorial diversity of modification patterns allows recruitments of precise transcription regulators and factors that toggle specific gene expression for cell differentiation, development, or disease progression.
Histone crosstalks inspire the investigation of a similar network arising from PTMs of various residues in CTD heptads. Indeed, the development and use of mass spec methods to map CTD phosphorylation reveal the abundance of bis-phosphorylated heptads in the human CTD [28]. Bis-phosphorylated heptad levels are even higher when considering a span of two heptads since double heptad repeats are more likely to be the functional unit for protein recruitment [47]. Because five of the seven residues are capable of being phosphorylated, this leads to a potential of 20 different combinations of double phosphorylation marks, which would act as an excellent way of signaling during various transcriptional steps (Figure 1e).
Crosstalk between proline isomerization and serine phosphorylation
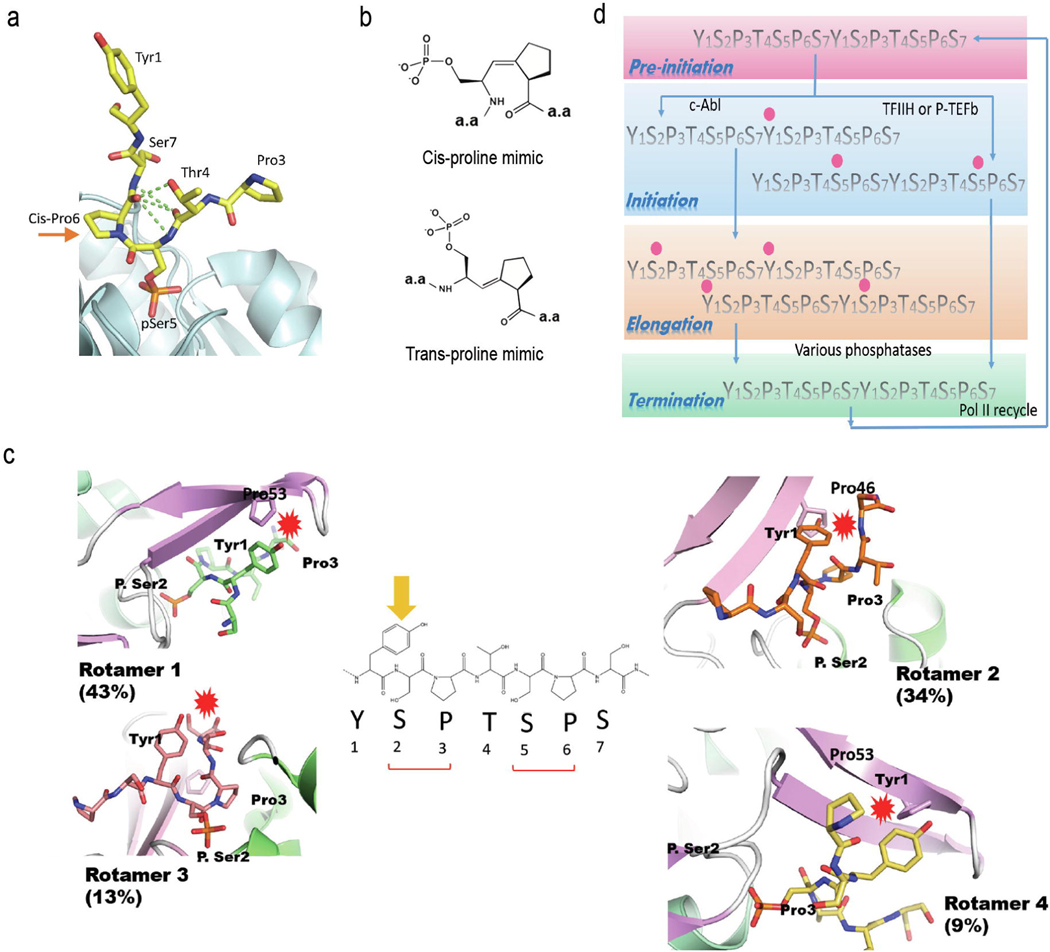
Only two residues (Pro3 and Pro6) out of the heptad sequence cannot undergo phosphorylation. Instead, proline’s unique chemical structure makes it the only natural amino acid exhibiting a high percentage of cis conformation in proteins (~10% to 25%). This natural chemical diversity exists in biological systems as a novel mechanism to trigger different signaling transduction pathways. In this case, cis- and trans- proline behave differently to activate distinct downstream signals [48]. The intrinsic transition between cis- and trans- proline occurs in solution at a slow rate. Thus, it is the rate-limiting step for some signaling pathways, especially when the phosphorylation of a flanking residue poses an additional hindrance to natural isomerization [49]. Under such a situation, Pin1, a prolyl-peptidyl isomerase, isomerizes the phosphoryl Ser/Thr-Pro motif by re-balancing cis- and trans- proline levels and guaranteeing the availability of cis-proline for downstream effectors [50]. Pin1 is a CTD binding protein that recognizes the hyperphosphorylated Ser-Pro motif on the CTD [49]. Its impact on the phosphorylation state of CTD became apparent when the structures of Ssu72, a CTD phosphatase, showed that only cis-proline fits into the CTD binding site of Ssu72 [51–53] (Figure 2a). Ssu72 is the only cis-specific phosphatase ever reported. Its selectivity is well established when tested with two peptidomimetic chemical compounds with proline that cannot undergo isomerization to mimic cis- or trans- proline [54] (Figure 2b). Ssu72 only binds and dephosphorylates the cis isostere-containing peptide but showed no activity against the trans version. Since cis-proline only accounts for a fraction of the natural population in proteins [55] and the isomerization conversion is slow, Pin1 can replenish the cis-proline pool when depleted by Ssu72, thus promoting its phosphatase activity. In contrast, Scp1, a trans-specific phosphatase, is not subject to Pin1 regulation because of the ample availability of trans-proline as a substrate [54], thereby circumventing the rate-limiting isomerization of proline. Thus, Pin1 activity selectively accelerates the dephosphorylation by Ssu72.
Figure 2:
Ssu72 dephosphorylates Ser5 of the CTD with the requirement of Pro6 is in the cis configuration. (a) The complex structures of Ssu72 bound to its substrates reveal that the proline residue is always in the cis configuration next to the dephosphorylation sites. (b) Chemical structures of the Isostere homologs mimic proline residues locked in cis or trans configurations. (c) Ssu72 has little activity towards Ser2 of the CTD because the flanking residues would cause steric clashes. (d) Crosstalk of Tyr1 and Ser2 phosphorylation leads to differentiated outcomes in transcription. Tyr1 phosphorylation primes the P-TEFb mediated Ser2 phosphorylation to promote elongation.
Ssu72’s requirement for cis-proline binding also explains its high specificity towards the dephosphorylation of Ser5 [56,57]. During Pol II’s transition from promoter pause-release to productive elongation, the level of phosphor-Ser5 plummets while the Ser2 phosphate level gradually accumulates [29]. Ssu72 needs to discriminate between the subtle differences of Ser2 and Ser5 to ensure elongation. High specificity is encoded in the unique active site of Ssu72, in which the flanking proline residue needs to be in cis-configuration, as seen in all Ssu72 structures [51]. Such a requirement precludes the binding of Ser2, whose preceding residue (Tyr1) is too bulky to fit into the active site (Figure 2c). Thus, Tyr1 prevents the inappropriate dephosphorylation of Ser2 during elongation. Consequently, Ser2 phosphorylation keeps accumulating and recruits splicing factors, polyadenylation, and termination protein complexes until its removal by Fcp1 at the end of the transcription cycle. Overall, the crosstalk between proline isomerization and serine phosphorylation ensures accurate phosphate removal during the transcription process.
Crosstalk between Tyr1 and Ser2 phosphorylation
Due to the possible combinations of bis-phosphorylated heptads, it is only natural to consider if different phosphorylation sites affect each other. The first evidence pointing to crosstalk among different CTD phosphorylation sites arose from kinetic studies of P-TEFb [58]. The physiological role of P-TEFb has been established in overriding the promoter-proximal pausing by phosphorylating Ser2 along with DSIF and NELF [59]. Surprisingly, purified P-TEFb only shows activity towards Ser5 of the CTD in kinetic assays, raising doubts about its identity as a Ser2 kinase [58]. CTD peptides with Tyr1, Ser5, and Ser7 phosphorylated were used as substrates for P-TEFb. None of these alter the in vitro specificity of P-TEFb, but Ser7 phosphorylation seems to increase its kinase activity [58]. The alternative candidates for Ser2 kinases, CDK12/13, exhibit Ser2 phosphorylation activity, but they are not responsible for the Ser2 phosphorylation occurring at the promoter-proximal pausing release [60,61].
The more careful experimental design allowed for a re-evaluation of the role of phosphorylated Tyr1 in P-TEFb kinase activity. Tyr1 phosphorylation is one of the understudied marks partly because of its low abundance, constituting <0.01% of the total phosphorylation in the yeast CTD [62,63]. Tyr1 phosphorylation is more prevalent in human cells and occurs at the beginning of transcription, with a pattern preceding increased Ser2 phosphate levels [64,65]. In vitro reconstruction of the P-TEFb-directed CTD phosphorylation was carefully analyzed to mimic physiological scenarios [34]. Tyr1 phosphorylation is achieved biochemically by treating the CTD with c-ABL, a tyrosine kinase in humans [34]. Significantly, only half of the CTD is phosphorylated, and, unlike synthetic CTD heptads with every Tyr1 phosphorylated, this in vitro phosphorylated CTD is susceptible to further phosphorylation by CDKs [34]. Three different biochemical methods were used to determine the location of these phosphates [34]. The most direct detection method is mass spectrometry using UVPD’s positive and negative activation/ionization modes. The results show that P-TEFb generates Ser5 phosphorylation when the substrate CTD is not modified. However, when CTD is first phosphorylated at Tyr1 by c-Abl, Tyr1 and Ser2 bis-phosphorylated heptads become the major products (Figure 2d). This result is further corroborated by immunoblotting and biochemical assay. Phosphorylation on other CTD residues showed no significant change to P-TEFb’s specificity. Thus, Tyr1 phosphorylation crosstalks with Ser2 and promotes its phosphorylation by P-TEFb (Figure 2d).
Crosstalk between Ser5 and Ser7 phosphorylation
Phosphorylation of Ser5 is among the first modification marks placed on Pol II during active transcription. The kinase responsible for the phosphorylation of Ser5, CDK7 kinase module, is part of Transcription Factor II-H (TFIIH), a general transcription factor. The kinase module contains CDK7 kinase, cyclin H, and MAT1, which doubles as a transcriptional regulator and a cell cycle regulator [66–68]. Ser5 phosphorylation is associated with Pol II’s release from the promoter by breaking up its interactions with the Mediator [69,70]. Ser5 phosphorylation is required for capping enzyme recruitment [71]. Alternatively, engineering the capping enzyme’s interaction with Pol II can bypass the need for Ser5 phosphorylation in S. pombe [72]. Although the identification of Ser7 phosphorylation lags behind that of Ser5 phosphorylation by two decades, the timing of Ser7 phosphorylation during the transcription cycle is very similar. Interestingly, in vitro phosphorylation of Ser7 requires the existence of a Mediator [73,74]. In vivo studies pinpoint a crucial role of CDK7 and identify it as the kinase for Ser7 when TFIIH phosphorylates both Ser5 and Ser7 at the beginning of transcription [74–76]. Ser7 phosphorylation is functionally essential for transcription of snRNA and other non-coding RNA [27,77]. Thus, TFIIH itself is an effector of the interplay between Ser5 phosphorylation and Ser7 phosphorylation.
Ser5 and Ser7 use the same kinase to place the phosphate group, and they also get dephosphorylated by the same phosphatase. In vitro and in vivo, Ssu72 dephosphorylates Ser5 and Ser7 with a strong preference for Ser5 dephosphorylation [56]. Phosphorylated Ser7 heptads also recruit RNA Pol II Associated protein 2 (RPAP2), another Ser5 phosphatase, to Pol II and help it to dephosphorylate Ser5 in non-coding RNAs [78]. This suggests that Ser7 phosphorylation can regulate the existence of Ser5 phosphorylation [79,80]. Further down the transcriptional process, the integrator complex responsible for the termination of U-rich small nuclear RNAs (UsnRNA) demands the phosphorylation of both Ser2 and Ser7 for proper recruitment and termination [81]. In such genes, the crosstalk between Ser5 and Ser7 allows the removal of Ser5 phosphorylation and the phosphorylation of Ser2, eventually leading to Ser2 – Ser7 double phosphoryl marks for integrator’s recognition.
Crosstalk of Ser5 and the residues at the 7th position
Although simpler eukaryotic organisms have CTD sequences faithfully conforming with consensus sequence, metazoans show some divergence in mostly in the 7th position of the heptad repeats. For human Pol II, the first half of the CTD (called the proximal region) consists primarily of consensus sequences. However, the second half of the CTD (distal region) shows a lot of deviation from consensus, with the most frequent replacements for Ser7 being polar and charged residues [lysine (in 8 repeats), threonine (6 repeats), glutamate (1 repeat), arginine (1 repeat) and asparagine (1 repeat)]. The role of divergent CTD heptads in transcription is not fully understood since the consensus sequence is sufficient to perform transcriptional functions in cells [7,82]. This was demonstrated in an engineered Drosophila CTD with ~42 heptad repeats heavily diverging from the consensus sequence, which was strategically altered to identify its functional components [82,83]. Heptads containing consensus sequence at half the Drosophila CTD length were sufficient to rescue lethality and support normal development, but replacing the entire Drosophila CTD with the consensus sequence led to deleterious development [83]. These studies suggest a role of divergent sequence in balancing Pol II distribution in various membrane-less clusters.
The distal CTD in human has eight lysine residues located at the 7th position (Figure 1b). Since lysine is heavily modified post-translationally in histones, its chemical states in the CTD were of great interest. Indeed, methylation and acetylation of lysine in the CTD were detected in human cells [11,12,84]. The acetylation results in an enhancement of binding between RPRD proteins and phosphorylated CTD by about fivefold, as demonstrated by quantitative measurement using Isothermal titration calorimetry [11]. RPRD family members, RPRD1A and 1B, form heterodimers to interact with Ser5 phosphatase RPAP2. Their association to acetylated lysine directs the dephosphorylation of Ser5 mediated by RPAP2. This physiological effect was corroborated when the inhibition of lysine acetylation was shown to cause the accumulation of Pol II species with phosphorylated Ser5 [11]. Thus, the divergent residues and their modification states have the potential to affect Ser5 phosphorylation states.
On the other hand, Lys7 methylation seems to control expression levels in specific gene classes. Competition between Lysine methylation and acetylation is another layer of gene regulation built into the CTD that requires further experimental investigation [12,84].
Crosstalk between Tyr1 phosphorylation and the residues at the 7th heptad position
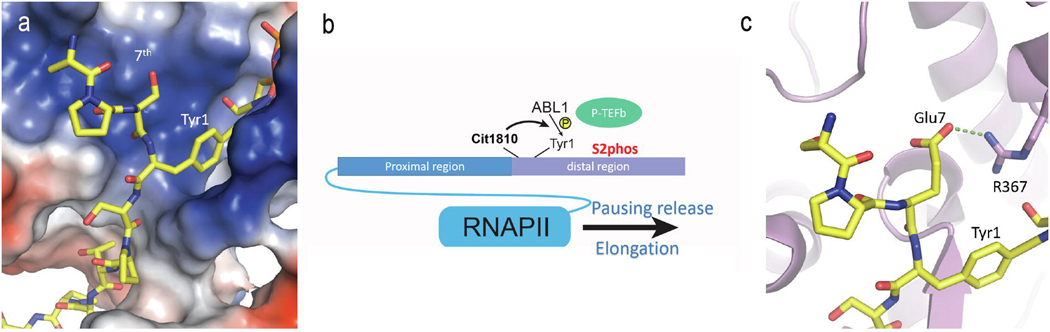
Although the 7th residue of the heptad is frequently changed, the residue following it on the polypeptide, Tyr1 of the next heptad repeat, is rarely altered in all species [6,29] (Figure 1b). Since Tyr1 gets phosphorylated during active transcription, physical proximity raises the possibility that the neighboring 7th residue in the previous repeat can influence the phosphorylation of the following Tyr1. Tyr1 is phosphorylated in vitro and in cells by Abl kinase, c-Abl, and its homolog ABL2 [85,86]. In the human distal CTD, eight lysines and one arginine are located at the 7th position of different heptad repeats (Figure 1b). When mapped by mass spectrometry, the tyrosines next to these positively-charged residues are rarely phosphorylated by c-Abl [33]. The structural analysis provides a reasonable explanation for this observation — the positively charged residue pocket close to the c-Abl active site expels positive residues at the 7th position and disfavors the following tyrosine as the substrate [33] (Figure 3a). Based on this analysis, neutralization of the positively charged residue would eliminate bias. Indeed, a PTM called citrullination occurs on Arg1830, which removes the positive charge on the arginine side chain and promotes promoter-proximal pausing release [87]. Mass spec analysis of this region shows that the tyrosine residue (Tyr1831) next to the citrullinated Arg (Arg1830) is resistant to phosphorylation [33]. However, upon arginine citrullination, a new peak of phosphorylated species appears and is mapped to the phosphorylation on the neighboring tyrosine [33]. Tyrosine phosphorylation consequently promotes the P-TEFb phosphorylation on Ser2, which releases pausing. Thus, the crosstalk of arginine at the 7th position with phosphorylated Tyr1 modulates transcriptional pausing [87].
Figure 3:
Cross-talks of the identities and PTMs of the 7th residues with Tyr1. (a) The conserved active sites of c-Abl and ABL2 show a highly positively charged pocket close to the reaction center. Modeling reveals close proximity of the pocket with the binding site of the 7th residue from the previous repeat. (b) A schematic model of how citrullination of Arg1810 promotes transcription elongation. (c) Favorable electrostatic interactions between the CTD and c-Abl when the 7th residue has a negatively-charged side chain such as glutamate.
Mass spec analysis for the preference of the residue proceeding Tyr1 also implies the possible existence of Ser7-Tyr1 bis-phosphorylated heptad (Figure 3c). Structural analysis reveals that tyrosine kinase’s active site favors a negatively charged residue in front of tyrosine due to a potential salt bridge formed with a conserved Arg close to the active site (Figure 3c). This preference suggests that phosphorylation of Ser7 in the previous repeat can promote phosphorylation of Tyr1 in the next heptad. Testing this prediction in vivo is not yet feasible since the kinase responsible for Ser7 phosphorylation, TFIIH, has coupled activity to Ser5. Using glutamate to mimic phosphorylated Ser7 has shown robust, preferential phosphorylation of neighboring Tyr1 [33]. However, the direct detection of endogenous Ser7-Tyr1 bis-phosphorylated heptads has yet to be carried out since previous research was done by mutating the heptad’s 7th residues to facilitate proteolysis for mass spec analysis [28,30].
Crosstalk between RNA Polymerase II and histone
Histones control the compactness and accessibility of DNA, which is key to Pol II’s function in transcription. Crosstalks between histones and Pol II ensures that transcription can progress efficiently without delay. Communication with histones is likely to be one of the CTD’s essential functions since in vitro reconstruction shows that a transcript can be produced without the CTD modifications [88–90]. Yet, the loss of the CTD in cells is fatal [19]. One reasonable explanation is that the CTD provides a medium for communication between the chromatin state and transcription progress, coordinating gene accessibility for transcription. Like Pol II, histone subunits have long tails extending from the nucleosome, subject to sophisticated PTMs. Communication between histone tails and the CTD is mediated by proteins recognizing both histone PTMs and CTD phosphorylation.
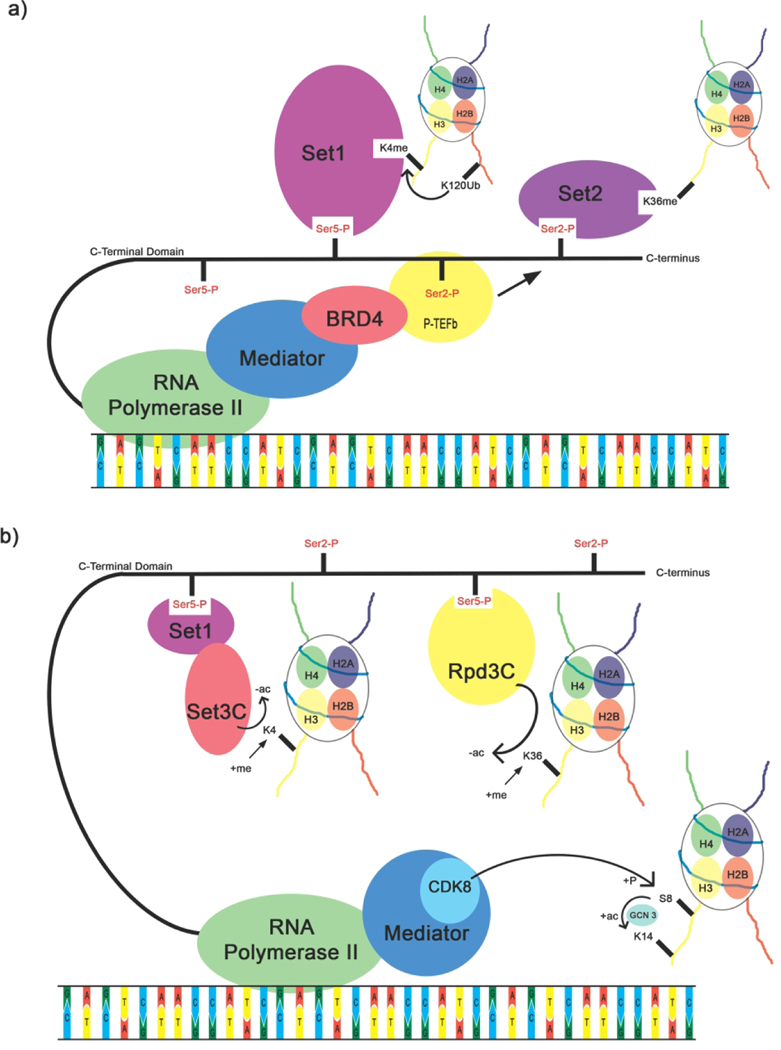
Communication between histones and Pol II specifies the methylation states for histone marks on histone H3. As the essential CTD kinase responsible for Ser2 phosphorylation, P-TEFb, functions as a central node in this communication (Figure 4a). P-TEFb activity is carefully modulated with cellular P-TEFb kept inactive by its association with 7SK/HEXIM1 until it is recruited by the epigenetic regulator, Bromodomain-containing protein 4 (BRD4), to Mediator and actively transcribing Pol II [91]. Disruption of the Mediator-BRD4-P-TEFb relay results in the release and eviction of Mediator from specific enhancers and promoters [92]. Bromodomain and Extra-Terminal motif (BET) inhibitors, which interrupt the interaction of Bromodomain-containing protein with protein complexes, have also been designed as therapeutics for cancers [92–94]. For example, the application of a small-molecule BET inhibitor successfully elicited cytotoxic effects in leukemia cells [92]. In turn, P-TEFb recruits mono-ubiquitination enzyme H2Bub1 [95], which ubiquitinates human H2BK120 (H2BK123 in yeast), a histone PTM required for SET1 methylation on H3K4 [96] and H3K79 [97,98]. Interestingly, the regulatory modes for H2BK120ub are different for these two lysines on H3. The binding of ubiquitin to Disruptor of telomeric silencing 1-like H3K79 methyltransferase (DOT1L) induces a conformational change that allows the rotation of substrate H3K79 from an inaccessible conformation to fit into the active site [99]. In contrast, ubiquitin interaction with the SET1/COMPASS (COMplex Proteins ASsociated with Set1) complex is more extensive for H3K4 methylation [100]. The binding of ubiquitin to the SET1 catalytic domain causes local denaturation of a helix that would have clashed with H2A. Instead, the helix loses its secondary structure and becomes a coil forming a favorable salt bridge to promote SET1 binding [100].
Figure 4:
Crosstalks between the histone code and the CTD code. (a) The phosphorylation states of the CTD regulates histone methylation. (b) crosstalks of the CTD and histone acetylation controls elongation speed and prevents cryptic transcription.
Phosphorylation states of the CTD also directly regulate histone methylation without mono-ubiquitin modification marks (Figure 4a). CTD phosphorylation sets the pattern of H3 methylation for active genes co-transcriptionally, for example, in the methylation pattern in H3K4 [101–103]. TFIIH-phosphorylated Ser5 recruits SET1/COMPASS through its N-terminal CTD-Interacting domain (CTD-ID) and activates its methyltransferase activity towards H3K4 [104]. Interruption of communication by mutating residues of the COMPASS complex reduces H3K4 methylation even though the enzymatic activity is not affected [105]. A transplant experiment establishes the causative effect of CTD phosphorylation on the histone methylation pattern of H3K4 [105]. When a different CTD-ID replaces the Set1 CTD-ID, histone methylation at H3K4 is restored right away [105]. The authors hypothesize that the interaction between SET1 CTD-ID and CTD of Pol II opens up the active site for methylation [105]. This experiment emphasizes the central role of CTD in histone methylation.
H3K36 methylation is another vital mark for an actively transcribed gene. Phosphorylated Ser2 of the CTD binds to the N-terminal CTD-ID of SET2 (Figure 4a), which in turn places methylation on H3K36 [106]. The interruption of this connection between histone and Pol II affects the H3K36 methylation level [107]. Mutations on Pol II, which slow down transcription elongation, cause both phosphorylated Ser2 of the CTD and H3K36me marks to shift closer to the 5’ end of the gene. This shows the coupling effect between these two PTMs during late transcriptional events [104]. SET2 CTD-ID specifically binds to Ser2 phosphorylation but not Ser5 phosphorylation [108,109]. Thus, SET2 protein mediates Pol II and histone communication in both directions of H3K36 methylation and Ser2 phosphorylation to modulate elongation speed.
Phosphorylation of the CTD also affects the acetylation states of histones that regulate nucleosome eviction in yeast (Figure 4b), but evidence for a general mechanism in metazoans is still lacking. A balance of acetylation and deacetylation controls co-transcriptional nucleosome eviction. This balance is monitored by histone acetylation complex (HAT) and histone deacetylase complex (HDAC), which are both recruited to the promoters by phosphorylated CTD. SAGA, a HAT complex, shows enhanced recruitment upon Ser5 phosphorylation by CDK7/Kin28, which activates gene expression and promotes nucleosome eviction [110]. In a separate pathway, the CDK8 kinase module within the Mediator phosphorylates H3S10 [111]. This phosphorylation crosstalks with H3K14, allowing the placement of acetylation marks by Gcn5 acetyltransferase to increase nucleosome eviction and stimulate elongation. Phosphorylation of Ser5 by CDK7/Kin28 can also recruit histone deacetylase complexes (HDACs) SET3C [112] and RPD3C(S) [113]. Recruitment was effective with SET1, but direct physical interaction between the CTD and the HDACs was also detected [113]. The balance between histone acetylation and deacetylation carefully reduces nucleosome density at promoters for gene expression while suppressing cryptic expression.
Perspective
The C-terminal domain of the largest subunit of Pol II is enriched with sites for post-translational modification. In combination with the existence of divergent heptads, there is a massive potential for variation. Crosstalk between modifications on both consensus and divergent sequence leads to a staggering capacity of transmitting information, which may ultimately alter transcription outcomes. By developing mass spectrometry techniques, we can more accurately identify the positions of post-translational marks and enable the investigation of crosstalks between highly chemically modified proteins. Future investigations from a proteomic standpoint can help us recognize transcription regulators that are recruited to on-going transcription machinery by these specific CTD modifications. Then, biological studies with these different combinations may ultimately lead us to a better understanding of the fundamental mechanisms governing eukaryotic transcription.
Research Highlights:
The C-terminal domain of the largest subunit of RNA polymerase II is heavily post-translational modified during transcription to coordinate transcription.
These modifications now can be accurately mapped by mass spectrometry.
The modifications on specific residues on this domain can influence the subsequent modification steps, resulting in differentiated transcriptional outcomes.
The modifications on Pol II and histones communicate to ensure a smooth progression in transcription.
Acknowledgment
We are grateful for Dr. Blasé LeBlanc’s critiques on our manuscript. We thank the National Institutes of Health (R01GM104896 and R01GM125882 to Y.J.Z) and Welch Foundation (F-1778 to Y.Z.) for supporting our research.
Footnotes
Declaration of interests
☒ The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- [1].Dignam JD, Lebovitz RM, Roeder RG, Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei, Nucleic Acids Res. 11 (1983) 1475–1489. 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Roeder RG, Rutter WJ, Multiple Forms of DNA-dependent RNA Polymerase in Eukaryotic Organisms, Nature. 224 (1969) 234–237. 10.1038/224234a0. [DOI] [PubMed] [Google Scholar]
- [3].Corden JL, RNA Polymerase II C-Terminal Domain: Tethering Transcription to Transcript and Template, Chem. Rev. 113 (2013) 8423–8455. 10.1021/cr400158h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Harlen KM, Trotta KL, Smith EE, Mosaheb MM, Fuchs SM, Churchman LS, Comprehensive RNA Polymerase II Interactomes Reveal Distinct and Varied Roles for Each Phospho-CTD Residue, Cell Rep. 15 (2016) 2147–2158. 10.1016/j.celrep.2016.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Phatnani HP, Greenleaf AL, Phosphorylation and functions of the RNA polymerase II CTD, Genes Dev. 20 (2006) 2922–2936. 10.1101/gad.1477006. [DOI] [PubMed] [Google Scholar]
- [6].Chapman RD, Heidemann M, Hintermair C, Eick D, Molecular evolution of the RNA polymerase II CTD, Trends Genet. 24 (2008) 289–296. 10.1016/j.tig.2008.03.010. [DOI] [PubMed] [Google Scholar]
- [7].Lu F, Gilmour DS, Genetic analysis of the RNA polymerase II CTD in Drosophila, Methods. 159–160 (2019) 129–137. 10.1016/j.ymeth.2019.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].West ML, Corden JL, Construction and analysis of yeast RNA polymerase II CTD deletion and substitution mutations., Genetics. 140 (1995) 1223–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Bartolomei MS, Halden NF, Cullen CR, Corden JL, Genetic analysis of the repetitive carboxyl-terminal domain of the largest subunit of mouse RNA polymerase II., Mol. Cell. Biol. 8 (1988) 330–339. 10.1128/MCB.8.1.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Zhang J, Corden JL, Identification of phosphorylation sites in the repetitive carboxyl-terminal domain of the mouse RNA polymerase II largest subunit., J. Biol. Chem. 266 (1991) 2290–2296. [PubMed] [Google Scholar]
- [11].Schröder S, Herker E, Itzen F, He D, Thomas S, Gilchrist DA, Kaehlcke K, Cho S, Pollard KS, Capra JA, Schnölzer M, Cole PA, Geyer M, Bruneau BG, Adelman K, Ott M, Acetylation of RNA Polymerase II Regulates Growth-Factor-Induced Gene Transcription in Mammalian Cells, Mol. Cell. 52 (2013) 314–324. 10.1016/j.molcel.2013.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Dias JD, Rito T, Torlai Triglia E, Kukalev A, Ferrai C, Chotalia M, Brookes E, Kimura H, Pombo A, Methylation of RNA polymerase II non-consensus Lysine residues marks early transcription in mammalian cells, ELife. 4 (2015) e11215. 10.7554/eLife.11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Kelly WG, Dahmus ME, Hart GW, RNA polymerase II is a glycoprotein. Modification of the COOH-terminal domain by O-GlcNAc., J. Biol. Chem. 268 (1993) 10416–10424. [PubMed] [Google Scholar]
- [14].Buratowski S, Progression through the RNA Polymerase II CTD Cycle, Mol. Cell. 36 (2009) 541–546. 10.1016/j.molcel.2009.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Ho CK, Shuman S, Distinct Roles for CTD Ser-2 and Ser-5 Phosphorylation in the Recruitment and Allosteric Activation of Mammalian mRNA Capping Enzyme, Mol. Cell. 3 (1999) 405–411. 10.1016/S1097-2765(00)80468-2. [DOI] [PubMed] [Google Scholar]
- [16].Ping Y-H, Rana TM, DSIF and NELF Interact with RNA Polymerase II Elongation Complex and HIV-1 Tat Stimulates P-TEFb-mediated Phosphorylation of RNA Polymerase II and DSIF during Transcription Elongation, J. Biol. Chem. 276 (2001) 12951–12958. 10.1074/jbc.M006130200. [DOI] [PubMed] [Google Scholar]
- [17].Adelman K, Lis JT, Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans, Nat. Rev. Genet. 13 (2012) 720–731. 10.1038/nrg3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Zhou Q, Li T, Price DH, RNA Polymerase II Elongation Control, Annu. Rev. Biochem. 81 (2012) 119–143. 10.1146/annurev-biochem-052610-095910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Schauer T, Tombácz I, Ciurciu A, Komonyi O, Boros IM, Misregulated RNA Pol II C-terminal domain phosphorylation results in apoptosis, Cell. Mol. Life Sci. 66 (2009) 909–918. 10.1007/s00018-009-8670-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Friedl EM, Lane WS, Erdjument-Bromage H, Tempst P, Reinberg D, The C-terminal domain phosphatase and transcription elongation activities of FCP1 are regulated by phosphorylation, Proc. Natl. Acad. Sci. 100 (2003) 2328–2333. 10.1073/pnas.2628049100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Strahl BD, Allis CD, The language of covalent histone modifications, nature. 403 (2000) 41–45. 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- [22].Prakash K, Fournier D, Histone code and higher-order chromatin folding: A hypothesis, (n.d.) 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Rando OJ, Combinatorial complexity in chromatin structure and function: revisiting the histone code, Curr. Opin. Genet. Dev. 22 (2012) 148–155. 10.1016/j.gde.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Buratowski S, The CTD code, Nat. Struct. Biol. N. Y. 10 (2003) 679–680. 10.1038/nsb0903-679. [DOI] [PubMed] [Google Scholar]
- [25].Cheng B, Price DH, Analysis of factor interactions with RNA polymerase II elongation complexes using a new electrophoretic mobility shift assay, Nucleic Acids Res. 36 (2008) e135–e135. 10.1093/nar/gkn630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Corden JL, Cadena DL, Ahearn JM, Dahmus ME, A unique structure at the carboxyl terminus of the largest subunit of eukaryotic RNA polymerase II., Proc. Natl. Acad. Sci. 82 (1985) 7934–7938. 10.1073/pnas.82.23.7934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Chapman RD, Heidemann M, Albert TK, Mailhammer R, Flatley A, Meisterernst M, Kremmer E, Eick D, Transcribing RNA Polymerase II Is Phosphorylated at CTD Residue Serine-7, Science. 318 (2007) 1780–1782. 10.1126/science.1145977. [DOI] [PubMed] [Google Scholar]
- [28].Schüller R, Forné I, Straub T, Schreieck A, Texier Y, Shah N, Decker T-M, Cramer P, Imhof A, Eick D, Heptad-Specific Phosphorylation of RNA Polymerase II CTD, Mol. Cell. 61 (2016) 305–314. 10.1016/j.molcel.2015.12.003. [DOI] [PubMed] [Google Scholar]
- [29].Eick D, Geyer M, The RNA Polymerase II Carboxy-Terminal Domain (CTD) Code, Chem. Rev. 113 (2013) 8456–8490. 10.1021/cr400071f. [DOI] [PubMed] [Google Scholar]
- [30].Suh H, Ficarro SB, Kang U-B, Chun Y, Marto JA, Buratowski S, Direct Analysis of Phosphorylation Sites on the Rpb1 C-Terminal Domain of RNA Polymerase II, Mol. Cell. 61 (2016) 297–304. 10.1016/j.molcel.2015.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Portz B, Lu F, Gibbs EB, Mayfield JE, Rachel Mehaffey M, Zhang YJ, Brodbelt JS, Showalter SA, Gilmour DS, Structural heterogeneity in the intrinsically disordered RNA polymerase II C-terminal domain, Nat. Commun. 8 (2017) 15231. 10.1038/ncomms15231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Mayfield JE, Robinson MR, Cotham VC, Irani S, Matthews WL, Ram A, Gilmour DS, Cannon JR, Zhang YJ, Brodbelt JS, Mapping the Phosphorylation Pattern of Drosophila melanogaster RNA Polymerase II Carboxyl-Terminal Domain Using Ultraviolet Photodissociation Mass Spectrometry, ACS Chem. Biol. 12 (2017) 153–162. 10.1021/acschembio.6b00729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Burkholder NT, Sipe SN, Escobar EE, Venkatramani M, Irani S, Yang W, Wu H, Matthews WM, Brodbelt JS, Zhang Y, Mapping RNAPII CTD Phosphorylation Reveals That the Identity and Modification of Seventh Heptad Residues Direct Tyr1 Phosphorylation, ACS Chem. Biol. 14 (2019) 2264–2275. 10.1021/acschembio.9b00610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Mayfield JE, Irani S, Escobar EE, Zhang Z, Burkholder NT, Robinson MR, Mehaffey MR, Sipe SN, Yang W, Prescott NA, Kathuria KR, Liu Z, Brodbelt JS, Zhang Y, Tyr1 phosphorylation promotes phosphorylation of Ser2 on the C-terminal domain of eukaryotic RNA polymerase II by P-TEFb, ELife. 8 (2019) e48725. 10.7554/eLife.48725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Brodbelt JS, Photodissociation mass spectrometry: new tools for characterization of biological molecules, Chem. Soc. Rev. 43 (2014) 2757–2783. 10.1039/C3CS60444F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Madsen JA, Xu H, Robinson MR, Horton AP, Shaw JB, Giles DK, Kaoud TS, Dalby KN, Trent MS, Brodbelt JS, High-throughput Database Search and Large-scale Negative Polarity Liquid Chromatography–Tandem Mass Spectrometry with Ultraviolet Photodissociation for Complex Proteomic Samples, Mol. Cell. Proteomics. 12 (2013) 2604–2614. 10.1074/mcp.O113.028258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Madsen JA, Kaoud TS, Dalby KN, Brodbelt JS, 193-nm photodissociation of singly and multiply charged peptide anions for acidic proteome characterization, PROTEOMICS. 11 (2011) 1329–1334. 10.1002/pmic.201000565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Han S-W, Lee S-W, Bahar O, Schwessinger B, Robinson MR, Shaw JB, Madsen JA, Brodbelt JS, Ronald PC, Tyrosine sulfation in a Gram-negative bacterium, Nat. Commun. 3 (2012) 1153. 10.1038/ncomms2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Greer SM, Cannon JR, Brodbelt JS, Improvement of Shotgun Proteomics in the Negative Mode by Carbamylation of Peptides and Ultraviolet Photodissociation Mass Spectrometry, Anal. Chem. 86 (2014) 12285–12290. 10.1021/ac5035314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Madsen JA, Ko BJ, Xu H, Iwashkiw JA, Robotham SA, Shaw JB, Feldman MF, Brodbelt JS, Concurrent Automated Sequencing of the Glycan and Peptide Portions of O-Linked Glycopeptide Anions by Ultraviolet Photodissociation Mass Spectrometry, Anal. Chem. 85 (2013) 9253–9261. 10.1021/ac4021177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Robinson MR, Moore KL, Brodbelt JS, Direct Identification of Tyrosine Sulfation by using Ultraviolet Photodissociation Mass Spectrometry, J. Am. Soc. Mass Spectrom. 25 (2014) 1461–1471. 10.1007/s13361-014-0910-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Robinson MR, Brodbelt JS, Integrating Weak Anion Exchange and Ultraviolet Photodissociation Mass Spectrometry with Strategic Modulation of Peptide Basicity for the Enrichment of Sulfopeptides, Anal. Chem. 88 (2016) 11037–11045. 10.1021/acs.analchem.6b02899. [DOI] [PubMed] [Google Scholar]
- [43].Lee J-S, Shukla A, Schneider J, Swanson SK, Washburn MP, Florens L, Bhaumik SR, Shilatifard A, Histone Cross-talk between H2B Monoubiquitination and H3 Methylation Mediated by COMPASS, Cell. 131 (2007) 1084–1096. 10.1016/j.cell.2007.09.046. [DOI] [PubMed] [Google Scholar]
- [44].Chen S, Li J, Wang D-L, Sun F-L, Histone H2B lysine 120 monoubiquitination is required for embryonic stem cell differentiation, Cell Res. 22 (2012) 1402–1405. 10.1038/cr.2012.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Yuan W, Xu M, Huang C, Liu N, Chen S, Zhu B, H3K36 Methylation Antagonizes PRC2-mediated H3K27 Methylation, J. Biol. Chem. 286 (2011) 7983–7989. 10.1074/jbc.M110.194027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Tamburri S, Lavarone E, Fernández-Pérez D, Conway E, Zanotti M, Manganaro D, Pasini D, Histone H2AK119 Mono-Ubiquitination Is Essential for Polycomb-Mediated Transcriptional Repression, Mol. Cell. 77 (2020) 840-856.e5. 10.1016/j.molcel.2019.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Stiller JW, Cook MS, Functional Unit of the RNA Polymerase II C-Terminal Domain Lies within Heptapeptide Pairs, Eukaryot. Cell. 3 (2004) 735–740. 10.1128/EC.3.3.735740.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Lu KP, Liou Y-C, Zhou XZ, Pinning down proline-directed phosphorylation signaling, Trends Cell Biol. 12 (2002) 164–172. 10.1016/S0962-8924(02)02253-5. [DOI] [PubMed] [Google Scholar]
- [49].Zhang Y, Daum S, Wildemann D, Zhou XZ, Verdecia MA, Bowman ME, Lücke C, Hunter T, Lu K-P, Fischer G, Noel JP, Structural Basis for High-Affinity Peptide Inhibition of Human Pin1, ACS Chem. Biol. 2 (2007) 320–328. 10.1021/cb7000044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Ping Lu K, Hanes SD, Hunter T, A human peptidyl–prolyl isomerase essential for regulation of mitosis, nature. 380 (1996) 544–547. 10.1038/380544a0. [DOI] [PubMed] [Google Scholar]
- [51].Xiang K, Nagaike T, Xiang S, Kilic T, Beh MM, Manley JL, Tong L, Crystal structure of the human symplekin–Ssu72–CTD phosphopeptide complex, nature. 467 (2010) 729–733. 10.1038/nature09391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Werner-Allen JW, Lee C-J, Liu P, Nicely NI, Wang S, Greenleaf AL, Zhou P, cis-Proline-mediated Ser(P)5 Dephosphorylation by the RNA Polymerase II C-terminal Domain Phosphatase Ssu72, J. Biol. Chem. 286 (2011) 5717–5726. 10.1074/jbc.M110.197129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Luo Y, Yogesha SD, Cannon JR, Yan W, Ellington AD, Brodbelt JS, Zhang Y, Novel Modifications on C-terminal Domain of RNA Polymerase II Can Fine-tune the Phosphatase Activity of Ssu72, ACS Chem. Biol. 8 (2013) 2042–2052. 10.1021/cb400229c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Mayfield JE, Fan S, Wei S, Zhang M, Li B, Ellington AD, Etzkorn FA, Zhang YJ, Chemical Tools To Decipher Regulation of Phosphatases by Proline Isomerization on Eukaryotic RNA Polymerase II, ACS Chem. Biol. 10 (2015) 2405–2414. 10.1021/acschembio.5b00296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Noble CG, Hollingworth D, Martin SR, Ennis-Adeniran V, Smerdon SJ, Kelly G, Taylor IA, Ramos A, Key features of the interaction between Pcf11 CID and RNA polymerase II CTD, Nat. Struct. Mol. Biol. 12 (2005) 144–151. 10.1038/nsmb887. [DOI] [PubMed] [Google Scholar]
- [56].Irani S, Sipe SN, Yang W, Burkholder NT, Lin B, Sim K, Matthews WL, Brodbelt JS, Zhang Y, Structural determinants for accurate dephosphorylation of RNA polymerase II by its cognate CTD phosphatase during eukaryotic transcription, J. Biol. Chem. (2019) jbc.RA119.007697. 10.1074/jbc.RA119.007697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Spector BM, Turek ME, Price DH, Functional interaction of human Ssu72 with RNA polymerase II complexes, PLOS ONE. 14 (2019) e0213598. 10.1371/journal.pone.0213598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Czudnochowski N, Bösken CA, Geyer M, Serine-7 but not serine-5 phosphorylation primes RNA polymerase II CTD for P-TEFb recognition, Nat. Commun. 3 (2012) 842. 10.1038/ncomms1846. [DOI] [PubMed] [Google Scholar]
- [59].Missra A, Gilmour DS, Interactions between DSIF (DRB sensitivity inducing factor), NELF (negative elongation factor), and the Drosophila RNA polymerase II transcription elongation complex, Proc. Natl. Acad. Sci. 107 (2010) 11301–11306. 10.1073/pnas.1000681107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Bartkowiak B, Liu P, Phatnani HP, Fuda NJ, Cooper JJ, Price DH, Adelman K, Lis JT, Greenleaf AL, CDK12 is a transcription elongation-associated CTD kinase, the metazoan ortholog of yeast Ctk1, Genes Dev. 24 (2010) 2303–2316. 10.1101/gad.1968210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Blazek D, Kohoutek J, Bartholomeeusen K, Johansen E, Hulinkova P, Luo Z, Cimermancic P, Ule J, Peterlin BM, The Cyclin K/Cdk12 complex maintains genomic stability via regulation of expression of DNA damage response genes, Genes Dev. 25 (2011) 2158–2172. 10.1101/gad.16962311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Chi A, Huttenhower C, Geer LY, Coon JJ, Syka JEP, Bai DL, Shabanowitz J, Burke DJ, Troyanskaya OG, Hunt DF, Analysis of phosphorylation sites on proteins from Saccharomyces cerevisiae by electron transfer dissociation (ETD) mass spectrometry, Proc. Natl. Acad. Sci. 104 (2007) 2193–2198. 10.1073/pnas.0607084104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Modesti A, Bini L, Carraresi L, Magherini F, Liberatori S, Pallini V, Manao G, Pinna LA, Raugei G, Ramponi G, Expression of the small tyrosine phosphatase (Stp1) in Saccharomyces cerevisiae: A study on protein tyrosine phosphorylation, ELECTROPHORESIS. 22 (2001) 576–585. [DOI] [PubMed] [Google Scholar]
- [64].Heidemann M, Eick D, Tyrosine-1 and threonine-4 phosphorylation marks complete the RNA polymerase II CTD phospho-code, RNA Biol. 9 (2012) 1144–1146. 10.4161/rna.21726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Hsin J-P, Li W, Hoque M, Tian B, Manley JL, RNAP II CTD tyrosine 1 performs diverse functions in vertebrate cells, ELife. 3 (2014) e02112. 10.7554/eLife.02112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Yankulov KY, Bentley DL, Regulation of CDK7 substrate specificity by MAT1 and TFIIH, EMBO J. 16 (1997) 1638–1646. 10.1093/emboj/16.7.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Greber BJ, Perez-Bertoldi JM, Lim K, Iavarone AT, Toso DB, Nogales E, The cryoelectron microscopy structure of the human CDK-activating kinase, Proc. Natl. Acad. Sci. 117 (2020) 22849–22857. 10.1073/pnas.2009627117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Peissert S, Schlosser A, Kendel R, Kuper J, Kisker C, Structural basis for CDK7 activation by MAT1 and Cyclin H, Proc. Natl. Acad. Sci. 117 (2020) 26739–26748. 10.1073/pnas.2010885117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Wong KH, Jin Y, Struhl K, TFIIH Phosphorylation of the Pol II CTD Stimulates Mediator Dissociation from the Preinitiation Complex and Promoter Escape, Mol. Cell. 54 (2014) 601–612. 10.1016/j.molcel.2014.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Robinson PJJ, Bushnell DA, Trnka MJ, Burlingame AL, Kornberg RD, Structure of the Mediator Head module bound to the carboxy-terminal domain of RNA polymerase II, Proc. Natl. Acad. Sci. 109 (2012) 17931–17935. 10.1073/pnas.1215241109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Ghosh A, Shuman S, Lima CD, Structural Insights to How Mammalian Capping Enzyme Reads the CTD Code, Mol. Cell. 43 (2011) 299–310. 10.1016/j.molcel.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Schwer B, Shuman S, Deciphering the RNA Polymerase II CTD Code in Fission Yeast, Mol. Cell. 43 (2011) 311–318. 10.1016/j.molcel.2011.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Boeing S, Rigault C, Heidemann M, Eick D, Meisterernst M, RNA Polymerase II C-terminal Heptarepeat Domain Ser-7 Phosphorylation Is Established in a Mediator-dependent Fashion, J. Biol. Chem. 285 (2010) 188–196. 10.1074/jbc.M109.046565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Akhtar Md.S., Heidemann M, Tietjen JR, Zhang DW, Chapman RD, Eick D, Ansari AZ, TFIIH Kinase Places Bivalent Marks on the Carboxy-Terminal Domain of RNA Polymerase II, Mol. Cell. 34 (2009) 387–393. 10.1016/j.molcel.2009.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Glover-Cutter K, Larochelle S, Erickson B, Zhang C, Shokat K, Fisher RP, Bentley DL, TFIIH-associated Cdk7 kinase functions in phosphorylation of C-terminal domain Ser7 residues, promoter-proximal pausing, and termination by RNA polymerase II, Mol. Cell. Biol. 29 (2009) 5455–5464. 10.1128/MCB.00637-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Kelso TWR, Baumgart K, Eickhoff J, Albert T, Antrecht C, Lemcke S, Klebl B, Meisterernst M, Cyclin-Dependent Kinase 7 Controls mRNA Synthesis by Affecting Stability of Preinitiation Complexes, Leading to Altered Gene Expression, Cell Cycle Progression, and Survival of Tumor Cells, Mol. Cell. Biol. 34 (2014) 3675–3688. 10.1128/MCB.00595-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Egloff S, O’Reilly D, Chapman RD, Taylor A, Tanzhaus K, Pitts L, Eick D, Murphy S, Serine-7 of the RNA Polymerase II CTD Is Specifically Required for snRNA Gene Expression, Science. 318 (2007) 1777–1779. 10.1126/science.1145989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Egloff S, Zaborowska J, Laitem C, Kiss T, Murphy S, Ser7 Phosphorylation of the CTD Recruits the RPAP2 Ser5 Phosphatase to snRNA Genes, Mol. Cell. 45 (2012) 111–122. 10.1016/j.molcel.2011.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Reyes-Reyes M, Hampsey M, Role for the Ssu72 C-Terminal Domain Phosphatase in RNA Polymerase II Transcription Elongation, Mol. Cell. Biol. 27 (2007) 926–936. 10.1128/MCB.01361-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Myers LC, Lacomis L, Erdjument-Bromage H, Tempst P, The Yeast Capping Enzyme Represses RNA Polymerase II Transcription, Mol. Cell. 10 (2002) 883–894. 10.1016/S1097-2765(02)00644-5. [DOI] [PubMed] [Google Scholar]
- [81].Egloff S, Szczepaniak SA, Dienstbier M, Taylor A, Knight S, Murphy S, The Integrator Complex Recognizes a New Double Mark on the RNA Polymerase II Carboxyl-terminal Domain, J. Biol. Chem. 285 (2010) 20564–20569. 10.1074/jbc.M110.132530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Lu F, Portz B, Gilmour DS, The C-Terminal Domain of RNA Polymerase II Is a Multivalent Targeting Sequence that Supports Drosophila Development with Only Consensus Heptads, Mol. Cell. 73 (2019) 1232–1242.e4. 10.1016/j.molcel.2019.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Gibbs EB, Lu F, Portz B, Fisher MJ, Medellin BP, Laremore TN, Zhang YJ, Gilmour DS, Showalter SA, Phosphorylation induces sequence-specific conformational switches in the RNA polymerase II C-terminal domain, Nat. Commun. 8 (2017) 15233. 10.1038/ncomms15233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Voss K, Forné I, Descostes N, Hintermair C, Schüller R, Maqbool MA, Heidemann M, Flatley A, Imhof A, Gut M, Gut I, Kremmer E, Andrau J-C, Eick D, Site-specific methylation and acetylation of lysine residues in the C-terminal domain (CTD) of RNA polymerase II, Transcription. 6 (2015) 91–101. 10.1080/21541264.2015.1114983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Baskaran R, Chiang GG, Wang JY, Identification of a binding site in c-Ab1 tyrosine kinase for the C-terminal repeated domain of RNA polymerase II., Mol. Cell. Biol. 16 (1996) 3361–3369. 10.1128/MCB.16.7.3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Baskaran R, Chiang GG, Mysliwiec T, Kruh GD, Wang JYJ, Tyrosine Phosphorylation of RNA Polymerase II Carboxyl-terminal Domain by the Abl-related Gene Product, J. Biol. Chem. 272 (1997) 18905–18909. 10.1074/jbc.272.30.18905. [DOI] [PubMed] [Google Scholar]
- [87].Sharma P, Lioutas A, Fernandez-Fuentes N, Quilez J, Carbonell-Caballero J, Wright RHG, Di Vona C, Le Dily F, Schüller R, Eick D, Oliva B, Beato M, Arginine Citrullination at the C-Terminal Domain Controls RNA Polymerase II Transcription, Mol. Cell. 73 (2019) 84–96.e7. 10.1016/j.molcel.2018.10.016. [DOI] [PubMed] [Google Scholar]
- [88].McNeil JB, Agah H, Bentley D, Activated transcription independent of the RNA polymerase II holoenzyme in budding yeast, Genes Dev. 12 (1998) 2510–2521. 10.1101/gad.12.16.2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Kaneko S, Chu C, Shatkin AJ, Manley JL, Human capping enzyme promotes formation of transcriptional R loops in vitro, Proc. Natl. Acad. Sci. 104 (2007) 17620–17625. 10.1073/pnas.0708866104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].BURATOWSKIt S, Sharp PA, Transcription Initiation Complexes and Upstream Activation with RNA Polymerase II Lacking the C-Terminal Domain of the Largest Subunit, (n.d.) 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Yang Z, Yik JHN, Chen R, He N, Jang MK, Ozato K, Zhou Q, Recruitment of P-TEFb for Stimulation of Transcriptional Elongation by the Bromodomain Protein Brd4, Mol. Cell. 19 (2005) 535–545. 10.1016/j.molcel.2005.06.029. [DOI] [PubMed] [Google Scholar]
- [92].Bhagwat AS, Roe J-S, Mok BYL, Hohmann AF, Shi J, Vakoc CR, BET Bromodomain Inhibition Releases the Mediator Complex from Select cis -Regulatory Elements, Cell Rep. 15 (2016) 519–530. 10.1016/j.celrep.2016.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Anand P, Brown JD, Lin CY, Qi J, Zhang R, Artero PC, Alaiti MA, Bullard J, Alazem K, Margulies KB, Cappola TP, Lemieux M, Plutzky J, Bradner JE, Haldar SM, BET Bromodomains Mediate Transcriptional Pause Release in Heart Failure, Cell. 154 (2013) 569–582. 10.1016/j.cell.2013.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Stewart HJS, Horne GA, Bastow S, Chevassut TJT, BRD4 associates with p53 in DNMT3A-mutated leukemia cells and is implicated in apoptosis by the bromodomain inhibitor JQ1, Cancer Med. 2 (2013) 826–835. 10.1002/cam4.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Sansó M, Parua PK, Pinto D, Svensson JP, Pagé V, Bitton DA, MacKinnon S, Garcia P, Hidalgo E, Bähler J, Tanny JC, Fisher RP, Cdk9 and H2Bub1 signal to Clr6-CII/Rpd3S to suppress aberrant antisense transcription, Nucleic Acids Res. (2020) gkaa474. 10.1093/nar/gkaa474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Sun Z-W, Allis CD, Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast, nature. 418 (2002) 104–108. 10.1038/nature00883. [DOI] [PubMed] [Google Scholar]
- [97].Briggs SD, Xiao T, Sun Z-W, Caldwell JA, Shabanowitz J, Hunt DF, Allis CD, Strahl BD, Trans -histone regulatory pathway in chromatin, nature. 418 (2002) 498–498. 10.1038/nature00970. [DOI] [PubMed] [Google Scholar]
- [98].Wojcik F, Dann GP, Beh LY, Debelouchina GT, Hofmann R, Muir TW, Functional cross-talk between histone H2B ubiquitylation and H2A modifications and variants, Nat. Commun. 9 (2018) 1394. 10.1038/s41467-018-03895-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Worden EJ, Hoffmann NA, Hicks CW, Wolberger C, Mechanism of Crosstalk between H2B Ubiquitination and H3 Methylation by Dot1L, Cell. 176 (2019) 1490–1501.e12. 10.1016/j.cell.2019.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Worden EJ, Zhang X, Wolberger C, Structural basis for COMPASS recognition of an H2B-ubiquitinated nucleosome, ELife. 9 (2020) e53199. 10.7554/eLife.53199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, Wang W, Weng Z, Green RD, Crawford GE, Ren B, Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome, Nat. Genet. 39 (2007) 311–318. 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- [102].Schneider R, Bannister AJ, Myers FA, Thorne AW, Crane-Robinson C, Kouzarides T, Histone H3 lysine 4 methylation patterns in higher eukaryotic genes, Nat. Cell Biol. 6 (2004) 73–77. 10.1038/ncb1076. [DOI] [PubMed] [Google Scholar]
- [103].Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, Huebert DJ, McMahon S, Karlsson EK, Kulbokas EJ, Gingeras TR, Schreiber SL, Lander ES, Genomic Maps and Comparative Analysis of Histone Modifications in Human and Mouse, Cell. 120 (2005) 169–181. 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- [104].Ebmeier CC, Erickson B, Allen BL, Allen MA, Kim H, Fong N, Jacobsen JR, Liang K, Shilatifard A, Dowell RD, Old WM, Bentley DL, Taatjes DJ, Human TFIIH Kinase CDK7 Regulates Transcription-Associated Chromatin Modifications, Cell Rep. 20 (2017) 1173–1186. 10.1016/j.celrep.2017.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Bae HJ, Dubarry M, Jeon J, Soares LM, Dargemont C, Kim J, Geli V, Buratowski S, The Set1 N-terminal domain and Swd2 interact with RNA polymerase II CTD to recruit COMPASS, Nat. Commun. 11 (2020) 2181. 10.1038/s41467-020-16082-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Kizer KO, Phatnani HP, Shibata Y, Hall H, Greenleaf AL, Strahl BD, A Novel Domain in Set2 Mediates RNA Polymerase II Interaction and Couples Histone H3 K36 Methylation with Transcript Elongation, Mol. Cell. Biol. 25 (2005) 3305–3316. 10.1128/MCB.25.8.3305-3316.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Krogan NJ, Kim M, Tong A, Golshani A, Cagney G, Canadien V, Richards DP, Beattie BK, Emili A, Boone C, Shilatifard A, Buratowski S, Greenblatt J, Methylation of Histone H3 by Set2 in Saccharomyces cerevisiae Is Linked to Transcriptional Elongation by RNA Polymerase II, Mol. Cell. Biol. 23 (2003) 4207–4218. 10.1128/MCB.23.12.4207-4218.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Li M, Phatnani HP, Guan Z, Sage H, Greenleaf AL, Zhou P, Solution structure of the Set2–Rpb1 interacting domain of human Set2 and its interaction with the hyperphosphorylated C-terminal domain of Rpb1, Proc. Natl. Acad. Sci. 102 (2005) 17636–17641. 10.1073/pnas.0506350102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Vojnic E, Simon B, Strahl BD, Sattler M, Cramer P, Structure and Carboxyl-terminal Domain (CTD) Binding of the Set2 SRI Domain That Couples Histone H3 Lys36 Methylation to Transcription, J. Biol. Chem. 281 (2006) 13–15. 10.1074/jbc.C500423200. [DOI] [PubMed] [Google Scholar]
- [110].Govind CK, Zhang F, Qiu H, Hofmeyer K, Hinnebusch AG, Gcn5 Promotes Acetylation, Eviction, and Methylation of Nucleosomes in Transcribed Coding Regions, Mol. Cell. 25 (2007) 31–42. 10.1016/j.molcel.2006.11.020. [DOI] [PubMed] [Google Scholar]
- [111].Meyer KD, Donner AJ, Knuesel MT, York AG, Espinosa JM, and Taatjes DJ, Cooperative activity of cdk8 and GCN5L within Mediator directs tandem phosphoacetylation of histone H3, EMBO J. (2008). 10.1038/emboj.2008.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Kim T, Buratowski S, Dimethylation of H3K4 by Set1 Recruits the Set3 Histone Deacetylase Complex to 5′ Transcribed Regions, Cell. 137 (2009) 259–272. 10.1016/j.cell.2009.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Govind CK, Qiu H, Ginsburg DS, Ruan C, Hofmeyer K, Hu C, Swaminathan V, Workman JL, Li B, Hinnebusch AG, Phosphorylated Pol II CTD Recruits Multiple HDACs, Including Rpd3C(S), for Methylation-Dependent Deacetylation of ORF Nucleosomes, Mol. Cell. 39 (2010) 234–246. 10.1016/j.molcel.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Mayer A, Heidemann M, Lidschreiber M, Schreieck A, Sun M, Hintermair C, Kremmer E, Eick D, Cramer P, CTD Tyrosine Phosphorylation Impairs Termination Factor Recruitment to RNA Polymerase II, Science. 336 (2012) 1723–1725. 10.1126/science.1219651. [DOI] [PubMed] [Google Scholar]
- [115].Lunde BM, Reichow SL, Kim M, Suh H, Leeper TC, Yang F, Mutschler H, Buratowski S, Meinhart A, Varani G, Cooperative interaction of transcription termination factors with the RNA polymerase II C-terminal domain, Nat. Struct. Mol. Biol. 17 (2010) 1195–1201. 10.1038/nsmb.1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Kecman T, Kuś K, Heo D-H, Duckett K, Birot A, Liberatori S, Mohammed S, Geis-Asteggiante L, Robinson CV, Vasiljeva L, Elongation/Termination Factor Exchange Mediated by PP1 Phosphatase Orchestrates Transcription Termination, Cell Rep. 25 (2018) 259–269.e5. 10.1016/j.celrep.2018.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Jasnovidova O, Krejcikova M, Kubicek K, Stefl R, Structural insight into recognition of phosphorylated threonine-4 of RNA polymerase II C-terminal domain by Rtt103p, EMBO Rep. 18 (2017) 906–913. 10.15252/embr.201643723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Nemec CM, Yang F, Gilmore JM, Hintermair C, Ho Y-H, Tseng SC, Heidemann M, Zhang Y, Florens L, Gasch AP, Eick D, Washburn MP, Varani G, Ansari AZ, Different phosphoisoforms of RNA polymerase II engage the Rtt103 termination factor in a structurally analogous manner, Proc. Natl. Acad. Sci. 114 (2017) E3944–E3953. 10.1073/pnas.1700128114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Brázda P, Krejčíková M, Kasiliauskaite A, Šmiřáková E, Klumpler T, Vácha R, Kubíček K, Štefl R, Yeast Spt6 Reads Multiple Phosphorylation Patterns of RNA Polymerase II C-Terminal Domain In Vitro, J. Mol. Biol. 432 (2020) 4092–4107. 10.1016/j.jmb.2020.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Vasiljeva L, Kim M, Mutschler H, Buratowski S, Meinhart A, The Nrd1–Nab3–Sen1 termination complex interacts with the Ser5-phosphorylated RNA polymerase II C-terminal domain, Nat. Struct. Mol. Biol. 15 (2008) 795–804. 10.1038/nsmb.1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Kubicek K, Cerna H, Holub P, Pasulka J, Hrossova D, Loehr F, Hofr C, Vanacova S, Stefl R, Serine phosphorylation and proline isomerization in RNAP II CTD control recruitment of Nrd1, Genes Dev. 26 (2012) 1891–1896. 10.1101/gad.192781.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Lukarska M, Fournier G, Pflug A, Resa-Infante P, Reich S, Naffakh N, Cusack S, Structural basis of an essential interaction between influenza polymerase and Pol II CTD, Nature. 541 (2017) 117–121. 10.1038/nature20594. [DOI] [PubMed] [Google Scholar]
- [123].Gemmill TR, Wu X, Hanes SD, Vanishingly Low Levels of Ess1 Prolyl-isomerase Activity Are Sufficient for Growth in Saccharomyces cerevisiae, J. Biol. Chem. 280 (2005) 15510–15517. 10.1074/jbc.M412172200. [DOI] [PubMed] [Google Scholar]
- [124].Atencio D, Barnes C, Duncan TM, Willis IM, Hanes SD, The Yeast Ess1 Prolyl Isomerase Controls Swi6 and Whi5 Nuclear Localization, G3amp58 GenesGenomesGenetics. 4 (2014) 523–537. 10.1534/g3.113.008763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Myers JK, Morris DP, Greenleaf AL, Oas TG, Phosphorylation of RNA Polymerase II CTD Fragments Results in Tight Binding to the WW Domain from the Yeast Prolyl Isomerase Ess1 †, Biochemistry. 40 (2001) 8479–8486. 10.1021/bi0027884. [DOI] [PubMed] [Google Scholar]
- [126].Verdecia MA, Bowman ME, Ping K, Hunter T, Noel JP, Structural basis for phosphoserine-proline recognition by group IV WW domains, Nat. Struct. Biol. 7 (2000) 5. [DOI] [PubMed] [Google Scholar]
- [127].Doamekpor SK, Sanchez AM, Schwer B, Shuman S, Lima CD, How an mRNA capping enzyme reads distinct RNA polymerase II and Spt5 CTD phosphorylation codes, Genes Dev. 28 (2014) 1323–1336. 10.1101/gad.242768.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Bharati AP, Singh N, Kumar V, Kashif M, Singh AK, Singh P, Singh SK, Siddiqi MI, Tripathi T, Akhtar MS, The mRNA capping enzyme of Saccharomyces cerevisiae has dual specificity to interact with CTD of RNA Polymerase II, Sci. Rep. 6 (2016) 31294. 10.1038/srep31294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Becker R, Loll B, Meinhart A, Snapshots of the RNA Processing Factor SCAF8 Bound to Different Phosphorylated Forms of the Carboxyl-terminal Domain of RNA Polymerase II, J. Biol. Chem. 283 (2008) 22659–22669. 10.1074/jbc.M803540200. [DOI] [PubMed] [Google Scholar]
- [130].Sikorsky T, Hobor F, Krizanova E, Pasulka J, Kubicek K, Stefl R, Recognition of asymmetrically dimethylated arginine by TDRD3, Nucleic Acids Res. 40 (2012) 11748–11755. 10.1093/nar/gks929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Appel L-M, Franke V, Bruno M, Grishkovskaya I, Kasiliauskaite A, Schoeberl UE, Puchinger MG, Kostrhon S, Beltzung E, Mechtler K, Lin G, Vlasova A, Leeb M, Pavri R, Stark A, Akalin A, Stefl R, Bernecky C, Djinovic-Carugo K, Slade D, PHF3 regulates neuronal gene expression through the new Pol II CTD reader domain SPOC, BioRxiv. (2020) 2020.02.11.943159. 10.1101/2020.02.11.943159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Ni Z, Xu C, Guo X, Hunter GO, Kuznetsova OV, Tempel W, Marcon E, Zhong G, Guo H, Kuo W-HW, Li J, Young P, Olsen JB, Wan C, Loppnau P, El Bakkouri M, Senisterra GA, He H, Huang H, Sidhu SS, Emili A, Murphy S, Mosley AL, Arrowsmith CH, Min J, Greenblatt JF, RPRD1A and RPRD1B are human RNA polymerase II C-terminal domain scaffolds for Ser5 dephosphorylation, Nat. Struct. Mol. Biol. 21 (2014) 686–695. 10.1038/nsmb.2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Mei K, Jin Z, Ren F, Wang Y, Chang Z, Wang X, Structural basis for the recognition of RNA polymerase II C-terminal domain by CREPT and p15RS, Sci. China Life Sci. 57 (2014) 97–106. 10.1007/s11427-013-4589-7. [DOI] [PubMed] [Google Scholar]
- [134].Liu J, Fan S, Lee C-J, Greenleaf AL, Zhou P, Specific Interaction of the Transcription Elongation Regulator TCERG1 with RNA Polymerase II Requires Simultaneous Phosphorylation at Ser2, Ser5, and Ser7 within the Carboxyl-terminal Domain Repeat, J. Biol. Chem. 288 (2013) 10890–10901. 10.1074/jbc.M113.460238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Liu H, Ramachandran S, Fong N, Phang T, Lee S, Parsa P, Liu X, Harmacek L, Danhorn T, Song T, Oh S, Zhang Q, Chen Z, Zhang Q, Tu T-H, Happoldt C, O’Conner B, Janknecht R, Li C-Y, Marrack P, Kappler J, Leach S, Zhang G, JMJD5 couples with CDK9 to release the paused RNA polymerase II, Proc. Natl. Acad. Sci. 117 (2020) 19888–19895. 10.1073/pnas.2005745117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Yanling Zhao D, Gish G, Braunschweig U, Li Y, Ni Z, Schmitges FW, Zhong G, Liu K, Li W, Moffat J, Vedadi M, Min J, Pawson TJ, Blencowe BJ, Greenblatt JF, SMN and symmetric arginine dimethylation of RNA polymerase II C-terminal domain control termination, nature. 529 (2016) 48–53. 10.1038/nature16469. [DOI] [PubMed] [Google Scholar]