Abstract
Structural magnetic resonance imaging (MRI) has shown great clinical and practical values in computer-aided brain disorder identification. Multi-site MRI data increase sample size and statistical power, but are susceptible to inter-site heterogeneity caused by different scanners, scanning protocols, and subject cohorts. Multi-site MRI harmonization (MMH) helps alleviate the inter-site difference for subsequent analysis. Some MMH methods performed at imaging level or feature extraction level are concise but lack robustness and flexibility to some extent. Even though several machine/deep learning-based methods have been proposed for MMH, some of them require a portion of labeled data in the to-be-analyzed target domain or ignore the potential contributions of different brain regions to the identification of brain disorders. In this work, we propose an attention-guided deep domain adaptation (AD2A) framework for MMH and apply it to automated brain disorder identification with multi-site MRIs. The proposed framework does not need any category label information of target data, and can also automatically identify discriminative regions in whole-brain MR images. Specifically, the proposed AD2A is composed of three key modules: 1) an MRI feature encoding module to extract representations of input MRIs, 2) an attention discovery module to automatically locate discriminative dementia-related regions in each whole-brain MRI scan, and 3) a domain transfer module trained with adversarial learning for knowledge transfer between the source and target domains. Experiments have been performed on 2, 572 subjects from four benchmark datasets with T1-weighted structural MRIs, with results demonstrating the effectiveness of the proposed method in both tasks of brain disorder identification and disease progression prediction.
Keywords: Brain disorder, structural MRI, harmonization, domain adaptation, attention
1. Introduction
Structural magnetic resonance imaging (MRI) has shown great clinical and practical values in computer-aided brain disorder identification, such as Alzheimers disease (AD) and its early stage, i.e., Mild Cognitive Impairment (MCI), is of great clinical value (Brookmeyer et al., 2007; Alzheimer’s Association, 2019). With MRI data acquired from multiple neuroimaging centers/sites (Frisoni et al., 2010), numerous learning-based learning methods have been proposed to tackle the problem of brain disorder identification (Falahati et al., 2014; Cuingnet et al., 2011). Among these methods, deep learning (LeCun et al., 2015), e.g., convolutional neural networks (CNNs) (Krizhevsky et al., 2012), has recently demonstrated its advantages over traditional machine learning methods in neuroimaging-based diagnosis and prognosis of brain dementia (Liu et al., 2018, 2020).
Multi-site MRI data help increase sample size and statistical power but maybe susceptible to inter-site heterogeneity caused for instance, by different scanners, scanning protocols, and subject cohorts. Previous studies typically assume that multi-site neuroimaging data are sampled from the identical distribution (Valiant, 1984; Lian et al., 2020), and directly apply a model (trained on source domain) to target data. However, such an assumption is too strong and may not hold in real-world applications due to the inter-site heterogeneity (Quionero-Candela et al., 2009). Multi-site MRI harmonization (MMH) helps alleviate the inter-site difference for subsequent analysis. Failure to perform MMH will cause biased results and erroneous conclusions that can potentially mislead future scientific endeavors. To deal with this problem, some methods facilitate MMH at the imaging level through hardware and software tuning (Clarke et al., 2020). Some methods adopt statistical techniques at the feature extraction level for MMH. Pomponio et al. (Pomponio et al., 2020) estimate the location and scale differences in ROI volumes across sites, and then remove these effects to achieve standardized ROI volumes for feature extraction. Wrobel et al. (Wrobel et al., 2020) adopt nonlinear transformations which are calculated by aligning distribution functions of intensity values to facilitate MMH. These methods are concise and effective to some extent, but often rely on some prior knowledge and assumptions which limit their robustness and flexibility. A more promising solution for MMH is to use domain adaptation methods to improve the transferability of models across multi-site data (Cheng et al., 2015; Madani et al., 2018), thereby generating a model that can work well on both source and target domains.
Existing domain adaptation methods can be generally divided into two categories: 1) feature transfer and 2) model transfer approaches. The first category aims to learn transferable features through deep learning techniques. It has been revealed that deep convolutional networks (CNNs) can be used to learn discriminative and transferable features across different domains (Oquab et al., 2014; Zeiler and Fergus, 2014). Based on this finding, CNN has been introduced to deal with various tasks of brain dementia classification, aiming to achieve higher transferability across different sites (Korolev et al., 2017; Lian et al., 2020). These methods do not use target samples during the learning process, which may limit their generalizability to the target data. The second category aims to learn transferable models by fine-tuning a pretrained model using samples in the target domain (Khan et al., 2019; Hosseini-Asl et al., 2016; Cheng et al., 2015). Taking the domain heterogeneity into consideration during the learning process, these methods tend to show higher generalizability. However, these methods often suffer from the following limitations. First, many of them require a part of labeled target data for model fine-tuning, thus greatly limiting their applications to unsupervised scenarios where no labeled target data are available. Note that labeling MRIs is a tedious and time-consuming task that requires the participation of experienced radiologists. Second, most existing methods equally treat all voxels in the whole-brain MRI, ignoring the potential different contributions of different regions to brain disorder identification, resulting in less robust models. It has been revealed that different brain regions have different effects on brain disorders (Mu and Gage, 2011; Ott et al., 2010; Lian et al., 2020). Intuitively, incorporating such prior knowledge into the training process of domain adaptation models will improve the performance of brain disorder identification.
In this work, we propose an attention-guided deep domain adaptation (AD2A) framework for MMH and apply it to the automated identification of brain disorders. The proposed AD2A method leverages domain adaptation to overcome the shortage of labeled target data for model fine-tuning (transferability enhancement) via adversarial learning (Goodfellow et al., 2014; Ganin and Lempitsky, 2015) and also can locate disease-related brain areas shared by cross-domain MRIs via an attention mechanism (Zhou et al., 2016; Woo et al., 2018). As shown in Fig. 1, our AD2A framework consists of three key components: 1) an MRI feature encoding module that extracts hierarchical feature representations of the input brain MRIs in both source and target domains, 2) an attention discovery module that automatically locates disease-related regions in whole-brain MRIs, and 3) a domain transfer module with adversarial learning that transfers knowledge between the source and target domains. In the experiments, the proposed AD2A method is evaluated on four independent datasets (i.e., ADNI-1 (Jack Jr et al., 2008), ADNI-2, ADNI-3, and AIBL (Ellis et al., 2009)) for multiple AD-related diagnosis tasks. Experimental results demonstrate that AD2A can yield superior cross-domain diagnostic performance compared with the state-of-the-art methods, and also effectively identify AD-related discriminative atrophy locations in MRIs.
Figure 1:
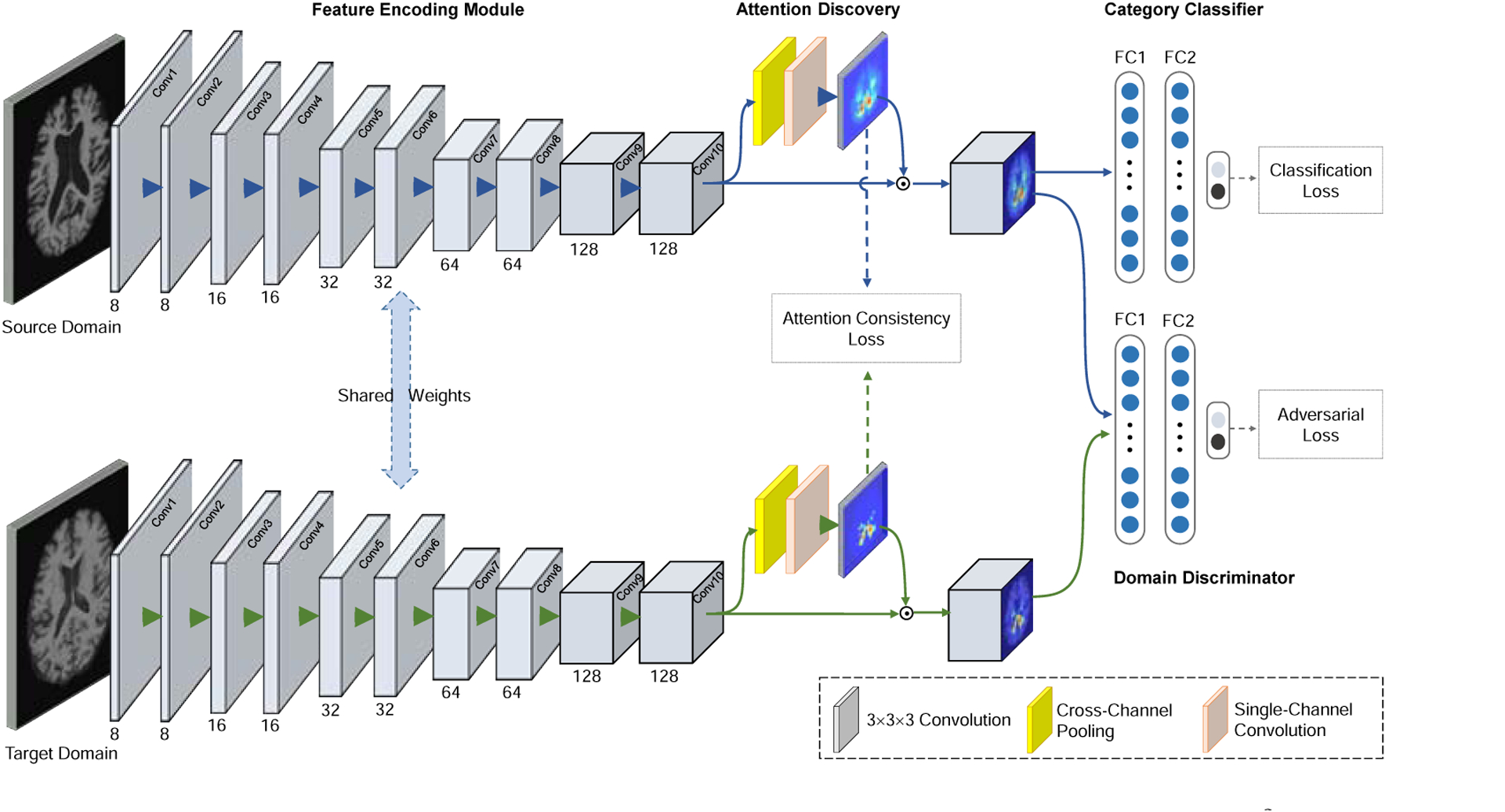
Illustration of the proposed attention-guided deep domain adaptation (AD2A) framework for MRI-based dementia identification. There are three main components: 1) a feature encoding module, 2) an attention discovery module, and 3) a domain transfer module with adversarial learning for knowledge transfer between the source and target domains.
The major contributions of this work can be summarized as follows. First, an unsupervised MMH framework is proposed for MRI-based brain disorder identification without requiring any label information of target data. Second, we propose to incorporate discriminative brain region localization into the model learning process for domain adaptation, which can reduce the negative influence of brain regions that are uninformative for prognosis. Besides, extensive experiments have been performed on 2, 572 subjects from four benchmark datasets with multi-site structural MRI scans.
The remainder of this paper is organized as follows. We first review relevant studies in Section 2. Section 3 introduces the materials used in this work and the details of the proposed method. In Section 4, we present the experimental settings, evaluation metrics, and experimental results. We further analyze the influence of several key components of the proposed method and discuss the limitations of the current work and future work in Section 5. The paper is finally concluded in Section 6.
2. Related Works
2.1. MRI-based Brain Disorder Analysis
Structural MRI data have been widely used in the computer-aided systems for brain disorder diagnosis and prognosis. Conventional methods usually extract hand-crafted MRI features and enhance robustness through feature fusion or selection (Falahati et al., 2014; Cuingnet et al., 2011; Shi et al., 2014; Zhu et al., 2014; Rathore et al., 2017). (Klöppel et al., 2008) extracted the grey matter density map of the entire brain MRI to train a support vector machine (SVM) for AD classification. (Cho et al., 2012) used converted thickness features with an incremental learning-based LDA for AD classification. (Chincarini et al., 2011) proposed to extract statistical and textural features of predefined brain regions, and subsequently trained an SVM for MCI conversion prediction.
In recent years, deep learning (e.g., CNN) has achieved promising results in computer vision (Krizhevsky et al., 2012; He et al., 2016) and neuroimaging analysis (Shen et al., 2017). (Gupta et al., 2013) used a sparse auto-encoder to extract features from brain MRIs, followed by a 2D CNN for AD classification. (Suk et al., 2014) proposed to use Deep Boltzmann Machine (DBM) trained with multi-modal images, i.e., MRI and positron emission tomography (PET), for automated AD classification. (Korolev et al., 2017) proposed VoxCNN, a 3D VGG-like CNN, for brain MRI classification. (Parisot et al., 2018) proposed to adopt graph convolutional networks (GCN) for the task of MCI conversion prediction.
These methods have shown promising performance for the task of brain dementia identification. However, they only rely on source data for model learning and ignore the distribution of target data, which may limit their transferability.
2.2. Domain Adaptation for Medical Image Analysis
Conventional machine learning methods typically assume that the training/source MRI data and test/target MRI data have identical distribution. However, this assumption does not always hold in real-world applications. For example, domain distributional heterogeneity (i.e., domain shift) is widespread among multi-site MRI datasets caused by different scanners, scanning parameters, and subject populations. Therefore, multi-site MRI harmonization (MMH) is essential to avoid biased results and erroneous conclusions by alleviating the inter-site difference.
As a promising solution to MMH, domain adaptation has attracted increasing attention in the filed (Kouw and Loog, 2019; Wang and Deng, 2018; Csurka et al., 2017; Pan and Yang, 2010). (Wachinger and Reuter, 2016) computed thickness and shape features from brain MRIs, then trained an elastic-net regression model based on instance weighting strategy to alleviate domain shift. (Moradi et al., 2014) proposed to utilize a transductive support vector machine (TSVM) for domain adaptation, based on gray matter density features of brain MRIs. (Li et al., 2019) adopted subspace alignment to reduce domain boundaries and trained a discriminative analysis classifier for AD identification. (Cheng et al., 2015) proposed a feature selection method based on gray matter tissue volumes of predefined regions-of-interest (ROIs), followed by a TSVM for MCI conversion prediction. (Madani et al., 2018) proposed a semi-supervised generative adversarial network (GAN) model for chest X-ray classification, which can incorporate unlabeled target data into network training to enhance the model transferability. (Zhang et al., 2019b) proposed a noise GAN model, an image-to-image translation GAN, which can map source samples to the target domain to alleviate the domain shift. (Ahn et al., 2020) proposed a zero-bias convolutional auto-encoder to learn features of target samples in an unsupervised manner. (Khan et al., 2019) first pretrained a VGG network on natural images, then performed layer-wise fine-tuning with MRIs for AD classification. (Hosseini-Asl et al., 2016) proposed an adaptive 3D CNN that was pretrained on MRIs in the source domain, and then fine-tuned task-specific layers on MRIs in the target domain. (Zhang et al., 2019a) developed an unsupervised conditional adversarial network for brain disease identification, by learning both domain-invariant and domain-specific features of structural MRI scans. However, existing methods rarely exploit the unique characteristics of brain images; that is, different brain regions may have different contributions to the recognition of specific brain diseases.
3. Materials and Methodology
3.1. Materials and MRI Preprocessing
Four benchmark datasets with baseline MRIs are used in this work, including 1) Alzheimer’s Disease Neuroimaging Initiative (ADNI-1) (Jack Jr et al., 2008), 2) ADNI-2, 3) ADNI-3, and 4) Australian Imaging Biomarkers and Lifestyle Study of Aging database (AIBL) (Ellis et al., 2009). Subjects that simultaneously appear in ADNI-1, ADNI-2 and ADNI-3 are removed from ADNI-2 and ADNI-3 for the sake of independent evaluation. Specifically, ADNI-1 consists of 748 subjects with 1.5T T1-weighted structural MRIs, including 205 AD, 231 cognitively normal (CN), 165 progressive MCI (pMCI) and 147 stable MCI (sMCI) subjects. ADNI-2 contains 708 subjects with 3T T1-weighted structural MRIs, including 162 AD, 205 CN, 88 pMCI and 253 sMCI subjects. ADNI-3 involves 567 subjects with 3T T1-weighted structural MRIs, including 60 AD, 329 CN, 178 MCI subjects. Note that there are no pMCI and sMCI labels for the MCI subjects in ADNI-3. Besides, AIBL has structural MRIs acquired from 549 subjects, including 71 AD, 447 CN, 11 pMCI and 20 sMCI subjects. The demographic and clinical information of studied subjects can be found in Table 1.
Table 1:
Demographic and clinical information of subjects included in four benchmark datasets (i.e., ADNI-1, ADNI-2, ADNI-3, and AIBL). The gender is presented as male/female. The age, education years, and mini-mental state examination (MMSE) scores are presented as mean ± standard deviation (std).
| Datasets | Category | Gender | Age | Education | MMSE |
|---|---|---|---|---|---|
| ADNI-1 | NC | 119/112 | 76.0 ± 5.0 | 15.9 ± 4.1 | 28.5 ± 2.6 |
| sMCI | 101/46 | 74.6 ± 7.7 | 15.6 ± 3.0 | 27.1 ± 1.5 | |
| pMCI | 101/64 | 74.8 ± 6.8 | 15.4 ± 3.5 | 26.5 ± 1.1 | |
| AD | 106/99 | 75.7 ± 7.6 | 13.1 ± 6.8 | 24.1 ± 1.4 | |
| ADNI-2 | NC | 110/95 | 73.2 ± 6.4 | 16.5 ± 2.5 | 26.5 ± 1.3 |
| sMCI | 146/107 | 71.0 ± 7.4 | 16.2 ± 2.1 | 27.2 ± 1.5 | |
| pMCI | 52/36 | 73.1 ± 7.0 | 16.0 ± 2.5 | 27.0 ± 2.0 | |
| AD | 95/67 | 74.2 ± 8.0 | 15.9 ± 2.6 | 24.0 ± 1.2 | |
| ADNI-3 | NC | 118/211 | 70.4 ± 7.5 | 15.7 ± 2.8 | 29.1 ± 1.1 |
| MCI | 100/78 | 72.4 ± 7.7 | 16.2 ± 2.7 | 27.8 ± 2.1 | |
| AD | 37/23 | 74.1 ± 12.7 | 15.9 ± 2.6 | 23.9 ± 2.8 | |
| AIBL | NC | 192/255 | 72.8 ± 6.6 | – | 28.7 ± 1.2 |
| AD | 30/41 | 73.4 ± 7.8 | – | 20.5 ± 5.7 |
All brain MR images were preprocessed through a standard pipeline, including skull stripping, intensity correction, and spatial normalization to Automated Anatomical Labeling (AAL) template. To avoid losing useful information, we followed the requirement that all brain tissues should be completely preserved.
3.2. Problem Setting
We focus on the problem of unsupervised domain adaptation for MRI-based brain disorder classification. Let represent the joint space of samples (subjects) and the corresponding category labels. A source domain S and a target domain T are defined on the joint space, with unknown distributions P and Q (P ≠ Q), respectively. Suppose Ns samples are provided with category labels in the source domain, i.e., . Also, we have Nt samples in the target domain but without category labels, i.e., . These two domains are assumed to share the same set of category labels. Our goal is to design an unsupervised learning model, which is constructed on labeled source samples and can accurately predict the labels of subjects in the target domain without any help of label information of target samples.
There are two important concepts for understanding the problem in this work: 1) category label and 2) domain label. Specifically, the category label indicates the category of a subject (e.g., AD, CN, and MCI). The term “label” or “label information” refers to the category label in this paper. The domain label indicates the domain to which the subject belongs. For example, “1” indicates the source domain, and “0” indicates the target domain. It should be noted that the domain label is determined by the model setting for a specific task.
3.3. Proposed Method
As shown in Fig. 1, the proposed attention-guided deep domain adaptation (AD2A) framework consists of three main components: 1) a feature encoding module, 2) an attention discovery module, and 3) a domain transfer module. We now introduce the details of each component as follows.
3.3.1. MRI Feature Encoding
We design a 3D CNN to extract features of brain MR images both in both source and target domains. As illustrated in the left panel of Fig. 1, the feature encoding module contains ten 3×3×3 convolution (Conv) layers, with the channel numbers of 8, 8, 16, 16, 32, 32, 64, 64, 128, and 128, respectively. Each Conv layer is followed by batch normalization (BN) and a rectified linear unit (ReLU). To avoid overfitting and enlarge receptive fields, down-sampling operations (stride: 2×2×2) are added to the Conv2, Conv4, Conv6, Conv8 and Conv10, respectively.
3.3.2. Dementia Attention Discovery
Previous studies have revealed that brain disorders are highly associated with certain regions in the brain (Mu and Gage, 2011; Ott et al., 2010; Woo et al., 2018; Lian et al., 2020). In addition, we also find in our experiments that locating disease-related areas can improve the transferability of the learning model. Based on these motivations, we design a trainable attention discovery module to automatically identify essential brain regions that are more closely linked to subject-specific abnormal status in brain MR images.
As illustrated in the middle part of Fig. 1, the feature maps generated by the Conv10 layer of feature encoder is used as the input of the proposed attention discovery module. Let M = [M1, … , MC] denote the input feature map, where is the feature map at the ith channel and C represents the number of channels. Cross-channel average pooling and max-pooling are then performed on M to generate two feature maps, i.e., Mavg and Mmax, respectively. We concatenate these two feature maps and send them to a Conv layer (i.e., Conv 11 with only one channel) to produce a spatial attention map. The sigmoid function is then used as the nonlinear activation to calculate the final attention map A. The role of the sigmoid function is to constrain each element in A within the range of [0, 1], which can reflect the importance of different areas in the MRI feature map. That is, important brain areas in the feature map would be assigned larger weights, while less important ones would be assigned smaller weights. Mathematically, the attention map is defined as:
| (1) |
where σ represents the sigmoid function and f3×3×3 denotes a convolution operator with a 3 × 3 × 3 kernel.
As shown in Fig 1, the proposed AD2A has two parallel branches corresponding to the source and target domains, respectively. Each of the branches follows the same pipeline to generate the attention maps as presented above. Let As and At denote the attention maps for source and target domains, respectively. To encourage the attention consistency and transfer semantic information from the source domain to target domain, we design an attention consistency loss in AD2A, which is defined as the mean square difference between As and At as follows:
| (2) |
where N is the number of samples.
Besides the attention consistency loss, our attention module also leverages both image-level category labels and domain labels as supervision for end-to-end training (see Fig. 1). This is the main difference between our model and previous deep learning models (e.g., localized class activation maps (Zhou et al., 2016)) that are trained by using only category labels as supervision. Therefore, our attention module helps highlight discriminative regions across different domains, while others can only focus on a single domain.
3.3.3. Domain Transfer via Adversarial Learning
Due to the data heterogeneity and distribution difference between the source and target domains, a model that is well-trained on a source domain may have degraded performance when directly applied to the target domain. It is especially challenging when there is no label information offered in the target domain for model fine-tuning. Thus our goal is to build a robust learning model based on only labeled source data. To this end, we develop a domain transfer module in the proposed AD2A (see the right panel of Fig. 1). This module is trained in an adversarial learning manner to balance the classification performance and generalization ability. More importantly, it does not require any label information of target samples.
Specifically, the proposed transfer module consists of a category classifier for classification and a domain discriminator/classifier for telling whether an input sample is from the source or target domain. Through co-training of these two classifiers, the proposed AD2A is encouraged to not only achieve good classification performance on source data but also learn domain-invariant features for both domains. In this way, we could improve the robustness of the learned model when applying it to the target domain.
Category Classifier.
The category classifier is built to estimate the labels of input MRI samples. Since no labeled data are available for the target domain, we can only train this classifier using labeled data in the source domain. Using the feature map generated by the feature encoder and weighted by the attention map as input, we employ three fully-connected layers with 128, 64 and 2 units in the category classifier for classification, with the loss defined as:
| (3) |
where L(·) denotes the cross-entropy loss.
Domain Discriminator.
The domain discriminator is designed to distinguish MRI features from different domains. It is trained by adversarial learning in which it serves as a player (the other one is the feature encoding module) in a min-max game. In this game, we try to maximize the loss of the domain classifier; thus the feature encoding module is encouraged to learn domain-invariant MRI features for both source and target data. To this end, three successive fully-connected layers with 128, 64 and 2 units are added in the domain discriminator. For network training, a training set with N samples is formed, where indicates that xi comes from the source domain and denotes that xi is from the target domain. In each batch, we select equal numbers of training samples from both the source domain and target domain to avoid bias towards either of them. Then the domain discriminator is trained by minimizing the following loss:
| (4) |
where L(·) denotes the cross-entropy loss and is the domain label.
The final goal of our system is to learn domain-invariant and disease-related features across the source and target domains. To achieve this, the task can be performed by learning a model that is capable of predicting category labels correctly without any domain cues. In this work, we jointly minimize the category classification loss in Eq. (3), minimize the attention consistency loss in Eq. (2), and maximize the domain classification loss in Eq. (4). The overall objective function of AD2A is defined as follows:
| (5) |
where α and β are the hyperparameters used to control the contributions of three terms. The proposed method can be used in various applications where the to-be-analyzed domain has no labeled data, especially for problems with few or even no labeled target data.
3.3.4. Implementation
The proposed AD2A model was implemented using Python based on PyTorch. The network was trained for 100 epochs. The Adam solver (Kingma and Ba, 2015) was used as the optimizer with a learning rate of 1 × 10−4 and a batch size of 2. The dropout operation with a rate of 0.5 was used to prevent over-fitting. We empirically set the parameter α and β in Eq. (5) to be 0.5 and 0.1, respectively. In the training process, we first pretrain the feature encoding network and the attention discovery module for classification according to Eq. (3) for 30 epochs. Then, these modules were further fine-tuned and co-trained with both the domain discriminator and category classifier via Eq. (5).
4. Experiments
4.1. Experimental Setup
We conduct four groups of experiments, including: 1) AD identification (i.e., AD vs. CN classification), 2) MCI conversion prediction (i.e., pMCI vs. sMCI classification), 3) AD vs. MCI classification, and 4) MCI vs. CN classification.
For AD identification, six transfer learning settings are considered: 1) “ADNI-1→ADNI-2” with ADNI-1 as the source domain and ADNI-2 as the target domain; 2) “ADNI-2→ADNI-1” with ADNI-2 and ADNI-1 as the source and target domains, respectively; 3) “ADNI-1→ADNI-3” with ADNI-1 and ADNI-3 as the source and target domains, respectively; 4) “ADNI-1+ADNI-2 → ADNI-3” with the combination of ADNI-1 and ADNI-2 as the source domain and ADNI-3 as the target domain; 5) “ADNI-1→AIBL” with ADNI-1 and AIBL as the source and target domains, respectively; and 6) “ADNI-1+ADNI-2 → AIBL” with the combination of ADNI-1 and ADNI-2 as the source domain and AIBL as the target domain. Since the number of MCI subjects in AIBL is small (i.e., 32) and there is no pMCI and sMCI labels in ADNI-3, we only evaluate the performance of MCI conversion prediction on two transfer learning settings: 1) “ADNI-1→ADNI-2”; and 2) “ADNI-2→ADNI-1”. For AD vs. MCI classification, four transfer learning settings are considered: 1) “ADNI-1→ADNI-2”; 2) “ADNI-2→ADNI-1”; 3) “ADNI-1→ADNI-3”; and 4) “ADNI-1+ADNI-2→ADNI-3”. For MCI vs. CN classification, four transfer learning settings are considered: 1) “ADNI-1→ADNI-2”; 2) “ADNI-2→ADNI-1”; 3) “ADNI-1→ADNI-3”; and 4) “ADNI-1+ADNI-2→ADNI-3”.
Four metrics were employed for performance evaluation in the experiments, i.e., classification accuracy (ACC), sensitivity (SEN), specificity (SPE), and area under the receiver operating characteristic curve (AUC). Denote TP, TN, FP, FN as the true positive, true negative, false positive and false negative, respectively. Then, these four evaluation metrics can be defined as , , and . For each metric, a higher value indicates better classification performance.
4.2. Competing Methods
In our experiments, we compared the proposed AD2A with four hand-crafted feature-based domain adaptation methods, including 1) Transfer Component Analysis (TCA) (Pan et al., 2010), 2) Subspace Alignment (SA) (Fernando et al., 2013), 3) Geodesic Flow Kernel (GFK) (Gong et al., 2012), and 4) Correlation Alignment (CORAL) (Sun et al., 2017; Kumar et al., 2017). We also compare AD2A with two state-of-the-art deep learning methods, including 1) VoxCNN (Korolev et al., 2017), and 2) Domain-Adversarial Training of Neural Network (DANN) (Ganin and Lempitsky, 2015). The four hand-crafted feature-based methods use gray matter volumes of 90 regions defined in the AAL template as the feature representation of MRIs, and logistic regression as the classifier. The deep learning methods (i.e., VoxCNN, DANN and our AD2A) learn MRI feature representations automatically from data in an end-to-end manner. We briefly introduce these competing methods as follows.
TCA (Pan et al., 2010). In the TCA method, several transfer components are learned based on the MR image features in different domains. Then, Maximum Mean Discrepancy (MMD) is utilized in the Reproducing Kernel Hilbert Space to make the distribution of multiple domains close to each other. In the experiments, we use a linear kernel for feature learning in TCA. We set the four key parameters of TCA as σ = 2, μ = 1, λ = 0, γ = 0.1, respectively.
SA (Fernando et al., 2013). In the SA method, MRI features of source and target domains are represented by a subspace spanned by eigenvectors. Then a mapping function is learned to align the subspace representations by minimizing the Bregman matrix divergence. The parameter for the new feature dimension in SA is set to 20.
GFK (Gong et al., 2012). In the GFK method, low dimensional representations of the MRIs from the source and target domain are learned. The data distribution difference is reduced by exploring the low-dimensional data structures that are domain-invariant. The parameter of the subspace dimension in GFK is set to 20.
CORAL (Sun et al., 2017). In the CORAL method, domain shift is minimized by aligning the second-order statistics of source and target distributions. CORAL needs to compute the covariance of the source and target features without extra parameters.
VoxCNN (Korolev et al., 2017). VoxCNN is a deliberately designed CNN model for MRI-based dementia classification. It contains ten 3 × 3 × 3 Conv layers (with the channel numbers of 8, 8, 16, 16, 32, 32, 32, 64, 64, and 64, respectively) for feature learning, and two fully-connected layers for classification. Note that this VoxCNN method does not include any data adaptation process, since the model is trained on the source domain and directly applied to the target domain.
DANN (Ganin and Lempitsky, 2015). DANN is a state-of-the-art adversarial learning-based domain adaptation method that has been widely used in modern medical imaging tasks (Yang et al., 2019; Kamnitsas et al., 2017; Javanmardi and Tasdizen, 2018). It adopts AlexNet for feature learning and a domain classifier for domain adaptation. Different from our model, it only aligns feature distributions of source and target domains in the top fully-connected layers, whereas our method also aligns the attention maps learned from convolution layers (with more spatial information).
4.3. Results of Cross-Domain Classification
We first evaluate the proposed AD2A and the competing methods in cross-domain problems, with one dataset used as the source domain and the other as the target domain. In this group of experiments, 80% source samples are used for training and the remaining 20% source samples are used for validation. Target samples (with an equal number of the training source data) are used for model training, and these target samples have no label information. More discussions on the number of target samples are reported in the Supplementary Materials.
4.3.1. AD vs. CN Classification
Table 2 reports the results achieved by different methods in the task of AD identification. From Table 2, one can observe that the proposed AD2A consistently outperforms the conventional hand-crafted feature based methods and the deep learning method in six transfer learning settings. Besides, our AD2A achieves overall better performance in the setting of “ADNI-1+ADNI-2 → AIBL” than “ADNI-1 → AIBL”. This implies that training with more diverse data in the source domain may enhance the robustness of learned models when applied to the target domain.
Table 2:
Results of seven methods in AD identification (i.e., AD vs. CN classification) in six different transfer learning settings.
| Source Domain→Target Domain | Method | ACC (%) | SEN (%) | SPE (%) | AUC (%) |
|---|---|---|---|---|---|
| ADNI-1→ADNI-2 | TCA | 74.39 | 56.79 | 88.29 | 80.55 |
| SA | 74.65 | 61.72 | 84.88 | 80.10 | |
| GFK | 65.40 | 55.56 | 73.17 | 70.33 | |
| CORAL | 77.65 | 82.71 | 73.65 | 85.16 | |
| VoxCNN | 83.65 | 85.80 | 81.95 | 90.43 | |
| DANN | 87.19 | 83.33 | 90.24 | 90.77 | |
| AD2A (Ours) | 89.92 | 87.65 | 91.70 | 94.01 | |
| ADNI-2→ADNI-1 | TCA | 73.62 | 66.83 | 79.65 | 80.61 |
| SA | 73.39 | 59.51 | 85.71 | 80.12 | |
| GFK | 62.16 | 49.27 | 73.59 | 65.05 | |
| CORAL | 76.38 | 72.20 | 80.09 | 84.00 | |
| VoxCNN | 82.33 | 70.24 | 93.07 | 89.94 | |
| DANN | 84.17 | 79.51 | 88.31 | 90.01 | |
| AD2A (Ours) | 87.84 | 86.83 | 88.74 | 92.07 | |
| ADNI-1→ADNI-3 | TCA | 76.35 | 60.00 | 79.33 | 80.15 |
| SA | 77.89 | 56.67 | 81.76 | 80.33 | |
| GFK | 74.55 | 61.67 | 76.90 | 72.22 | |
| CORAL | 71.21 | 58.33 | 73.56 | 84.50 | |
| VoxCNN | 86.11 | 61.67 | 90.57 | 89.07 | |
| DANN | 88.17 | 61.67 | 93.00 | 92.46 | |
| AD2A (Ours) | 92.03 | 66.67 | 96.65 | 95.01 | |
| ADNI-1+ADNI-2→ADNI-3 | TCA | 80.21 | 63.33 | 83.28 | 83.00 |
| SA | 83.55 | 56.67 | 88.45 | 85.29 | |
| GFK | 75.32 | 65.00 | 77.20 | 76.10 | |
| CORAL | 76.35 | 63.33 | 78.72 | 85.87 | |
| VoxCNN | 87.66 | 66.67 | 91.18 | 90.74 | |
| DANN | 88.69 | 71.67 | 91.79 | 92.82 | |
| AD2A (Ours) | 92.54 | 75.00 | 95.74 | 95.66 | |
| ADNI-1→AIBL | TCA | 68.34 | 32.39 | 74.05 | 50.92 |
| SA | 69.69 | 36.62 | 74.94 | 51.37 | |
| GFK | 59.85 | 46.48 | 61.97 | 50.25 | |
| CORAL | 54.44 | 45.07 | 55.93 | 54.48 | |
| VoxCNN | 85.91 | 66.20 | 89.04 | 86.06 | |
| DANN | 86.49 | 73.24 | 88.59 | 90.10 | |
| AD2A (Ours) | 88.80 | 85.92 | 89.26 | 92.73 | |
| ADNI-1+ADNI-2→AIBL | TCA | 69.31 | 30.99 | 75.39 | 51.69 |
| SA | 74.02 | 26.46 | 85.84 | 52.33 | |
| GFK | 63.71 | 36.62 | 68.00 | 50.90 | |
| CORAL | 57.72 | 50.70 | 58.84 | 57.24 | |
| VoxCNN | 87.07 | 83.10 | 87.70 | 92.28 | |
| DANN | 88.03 | 80.28 | 89.26 | 93.05 | |
| AD2A (Ours) | 90.35 | 87.32 | 90.83 | 95.37 |
4.3.2. MCI Conversion Prediction
Table 3 reports the results achieved by different methods in the task of MCI conversion prediction. From Table 3, we can see that the performance of seven methods in “ADNI-2 → ADNI-1” is usually worse than “ADNI-1 → ADNI-2”. This result could be caused by imbalanced pMCI (i.e., 88) and sMCI (i.e., 253) subjects in ADNI-2. Also, results in Tables 2–3 show that accurately predicting the future conversion of MCI subjects is more challenging than the task of AD identification, while our AD2A still achieves the overall best performance.
Table 3:
Results of seven methods in MCI conversion prediction (i.e., pMCI vs. sMCI classification) in two different transfer learning settings.
| Source Domain→Target Domain | Method | ACC (%) | SEN (%) | SPE (%) | AUC (%) |
|---|---|---|---|---|---|
| ADNI-1→ADNI-2 | TCA | 70.09 | 43.18 | 79.45 | 61.33 |
| SA | 67.44 | 34.09 | 79.05 | 58.29 | |
| GFK | 69.20 | 42.05 | 78.66 | 55.17 | |
| CORAL | 68.91 | 50.00 | 75.49 | 67.57 | |
| VoxCNN | 73.21 | 34.09 | 86.96 | 74.56 | |
| DANN | 75.07 | 52.27 | 83.00 | 76.01 | |
| AD2A (Ours) | 78.01 | 53.41 | 86.56 | 78.82 | |
| ADNI-2→ADNI-1 | TCA | 58.33 | 53.94 | 63.27 | 60.11 |
| SA | 58.65 | 57.58 | 59.86 | 57.33 | |
| GFK | 54.17 | 47.88 | 61.22 | 51.00 | |
| CORAL | 59.61 | 40.61 | 80.95 | 58.45 | |
| VoxCNN | 63.14 | 64.24 | 61.90 | 66.42 | |
| DANN | 66.99 | 60.61 | 74.14 | 67.87 | |
| AD2A (Ours) | 69.88 | 65.45 | 74.82 | 71.41 |
4.3.3. AD vs. MCI Classification
Results achieved by different methods in the task of AD vs. MCI classification are shown in Table 4. From Table 4, we can see that the proposed AD2A still achieves the best performance among the conventional and deep learning methods. In addition, our AD2A yields overall better results in the setting of “ADNI-1+ADNI-2 → ADNI-3” than “ADNI-1 → ADNI-3”. This again validates that using more diverse training data helps produce models with higher transferability for multi-site MRI harmonization.
Table 4:
Results of seven methods in AD vs. MCI classification in four different transfer learning settings.
| Source Domain→Target Domain | Method | ACC (%) | SEN (%) | SPE (%) | AUC (%) |
|---|---|---|---|---|---|
| ADNI-1→ADNI-2 | TCA | 63.62 | 22.22 | 83.28 | 58.76 |
| SA | 64.02 | 25.31 | 82.40 | 61.25 | |
| GFK | 65.01 | 35.80 | 78.89 | 64.10 | |
| CORAL | 67.99 | 29.01 | 86.51 | 67.40 | |
| VoxCNN | 69.58 | 46.91 | 80.35 | 70.79 | |
| DANN | 73.76 | 41.98 | 88.86 | 75.02 | |
| AD2A (Ours) | 75.55 | 46.29 | 89.44 | 77.67 | |
| ADNI-2→ADNI-1 | TCA | 53.57 | 22.44 | 74.04 | 58.21 |
| SA | 53.80 | 21.95 | 74.68 | 60.12 | |
| GFK | 60.35 | 43.41 | 71.47 | 63.65 | |
| CORAL | 67.70 | 41.46 | 84.93 | 66.79 | |
| VoxCNN | 68.09 | 41.95 | 85.26 | 68.96 | |
| DANN | 68.47 | 42.43 | 85.57 | 69.65 | |
| AD2A (Ours) | 70.41 | 43.90 | 87.82 | 71.99 | |
| ADNI-1→ADNI-3 | TCA | 71.84 | 46.67 | 80.90 | 60.48 |
| SA | 73.11 | 51.67 | 80.33 | 59.30 | |
| GFK | 64.29 | 50.00 | 69.10 | 51.75 | |
| CORAL | 67.23 | 51.67 | 72.47 | 69.67 | |
| VoxCNN | 76.89 | 51.67 | 85.39 | 70.15 | |
| DANN | 79.83 | 53.33 | 88.76 | 72.90 | |
| AD2A (Ours) | 81.51 | 55.00 | 90.45 | 75.88 | |
| ADNI-1+ADNI-2→ADNI-3 | TCA | 72.68 | 40.00 | 83.71 | 67.32 |
| SA | 73.95 | 41.67 | 84.83 | 64.11 | |
| GFK | 65.13 | 38.33 | 74.16 | 56.80 | |
| CORAL | 68.49 | 50.00 | 74.72 | 69.85 | |
| VoxCNN | 78.57 | 45.00 | 89.89 | 74.74 | |
| DANN | 81.09 | 48.33 | 92.13 | 75.96 | |
| AD2A (Ours) | 82.35 | 50.00 | 93.25 | 77.61 |
4.3.4. MCI vs. CN Classification
Table 5 reports the results achieved by different methods in the task of MCI vs. CN classification. From Table 5, we can see that the results of all methods are worse than those in Tables 2–4, suggesting that the task of MCI vs. CN classification is quite challenging. This can be attributed to that the MCI and CN subjects are relatively closer in the MRI feature space, since only very subtle structural changes occur in brain MRIs of MCI subjects.
Table 5:
Results of seven methods in MCI vs. CN classification in four different transfer learning settings.
| Source Domain→Target Domain | Method | ACC (%) | SEN (%) | SPE (%) | AUC (%) |
|---|---|---|---|---|---|
| ADNI-1→ADNI-2 | TCA | 57.88 | 53.96 | 64.39 | 58.19 |
| SA | 58.05 | 54.25 | 64.39 | 55.62 | |
| GFK | 59.52 | 54.55 | 67.80 | 58.14 | |
| CORAL | 60.44 | 62.46 | 57.07 | 63.60 | |
| VoxCNN | 61.72 | 58.65 | 66.83 | 63.90 | |
| DANN | 64.10 | 59.53 | 71.71 | 69.58 | |
| AD2A (Ours) | 67.03 | 63.64 | 72.68 | 70.33 | |
| ADNI-2→ADNI-1 | TCA | 53.22 | 48.08 | 60.17 | 54.25 |
| SA | 54.14 | 47.76 | 62.77 | 54.72 | |
| GFK | 55.25 | 47.44 | 65.80 | 55.18 | |
| CORAL | 60.41 | 55.77 | 66.67 | 60.05 | |
| VoxCNN | 60.77 | 56.09 | 67.10 | 65.12 | |
| DANN | 63.90 | 60.90 | 67.97 | 67.55 | |
| AD2A (Ours) | 65.19 | 61.53 | 70.13 | 69.96 | |
| ADNI-1→ADNI-3 | TCA | 59.17 | 38.20 | 70.51 | 58.68 |
| SA | 54.24 | 22.47 | 71.43 | 58.12 | |
| GFK | 52.46 | 23.03 | 68.39 | 52.51 | |
| CORAL | 60.35 | 28.65 | 77.51 | 52.67 | |
| VoxCNN | 62.92 | 42.13 | 74.16 | 59.06 | |
| DANN | 68.84 | 41.57 | 83.59 | 64.90 | |
| AD2A (Ours) | 70.22 | 42.70 | 85.11 | 67.22 | |
| ADNI-1+ADNI-2→ADNI-3 | TCA | 61.14 | 33.71 | 75.99 | 60.05 |
| SA | 60.16 | 31.46 | 75.68 | 58.55 | |
| GFK | 57.99 | 39.89 | 67.78 | 56.72 | |
| CORAL | 60.95 | 40.45 | 72.04 | 60.52 | |
| VoxCNN | 65.48 | 41.01 | 78.72 | 63.11 | |
| DANN | 69.82 | 41.57 | 85.11 | 65.00 | |
| AD2A (Ours) | 71.60 | 44.94 | 86.02 | 69.23 |
4.4. Results of Within-Domain Classification
We further evaluate the performance of different methods for within-domain classification by using the mixed data from ADNI-1, ADNI-2, ADNI-3 and AIBL. A 5-fold cross-validation strategy is used here. That is, we first randomly partitioned all AD and CN subjects from these four datasets into five folds. One of these five folds is used as the testing set (target domain) alliteratively, while the remaining four folds are used as the training set (source domain). The AD vs. CN classification results of different methods in each fold are listed in Table 6.
Table 6:
Results of AD vs. CN classification achieved by the proposed method and six competing methods on the mixed data from ADNI-1, ADNI-2, ADNI-3 and AIBL using 5-fold cross validation.
| Method | Fold #1 | Fold #2 | Fold #3 | Fold #4 | Fold #5 | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| ACC (%) | AUC (%) | ACC (%) | AUC (%) | ACC (%) | AUC (%) | ACC (%) | AUC (%) | ACC (%) | AUC (%) | |
| TCA | 73.97 | 79.88 | 74.56 | 80.23 | 73.39 | 77.32 | 74.85 | 80.98 | 74.27 | 79.11 |
| SA | 75.14 | 80.76 | 74.26 | 77.28 | 73.10 | 76.09 | 73.39 | 77.89 | 75.43 | 81.42 |
| GFK | 71.64 | 70.22 | 70.17 | 70.05 | 69.88 | 70.30 | 69.29 | 70.11 | 70.46 | 70.50 |
| CORAL | 73.10 | 83.79 | 76.02 | 84.71 | 77.20 | 84.85 | 78.95 | 87.75 | 77.78 | 85.31 |
| VoxCNN | 83.33 | 89.02 | 84.80 | 90.05 | 86.25 | 88.14 | 85.09 | 90.12 | 84.50 | 89.03 |
| DANN | 90.64 | 90.02 | 90.35 | 90.44 | 88.89 | 90.00 | 90.05 | 91.75 | 90.93 | 91.86 |
| AD2A (Ours) | 92.40 | 94.76 | 90.64 | 90.86 | 90.93 | 91.06 | 92.98 | 94.31 | 93.57 | 94.98 |
Table 6 suggests that the proposed AD2A can achieve the overall superior performance in terms of both ACC and AUC values, compared with those in Table 2. This can be attributed to the decrease in distribution difference between the source and target domains when performing cross-validation on mixed data of four datasets.
5. Discussion
In this section, we will investigate several major components in the proposed AD2A, analyze the influences of parameters, and present the limitations of the current work. Besides, we study the influence of a fine-tuning strategy (i.e., using a part of labeled target data for network refinement), and report the experimental results in the Supplementary Materials.
5.1. Ablation Study
The proposed AD2A consists of two key components, i.e., the attention discovery module and the 2) domain discriminator. To evaluate their contribution, we compare AD2A with its three variants for ablation analysis. These variants include: 1) ADN that only contains the feature encoding module and the category classifier in Fig. 1; 2) ADN-T that contains the feature encoding module, attention discovery module, and the category classifier; and 3) AD2A-S that includes the feature encoding and domain transfer modules. Note that ADN and ADN-T do not have domain adaptation modules. That is, these two models are firstly trained on source data and then directly applied to target data. Fig. 2 shows the AUC results achieved by AD2A and its three variants in four cross-domain tasks.
Figure 2:
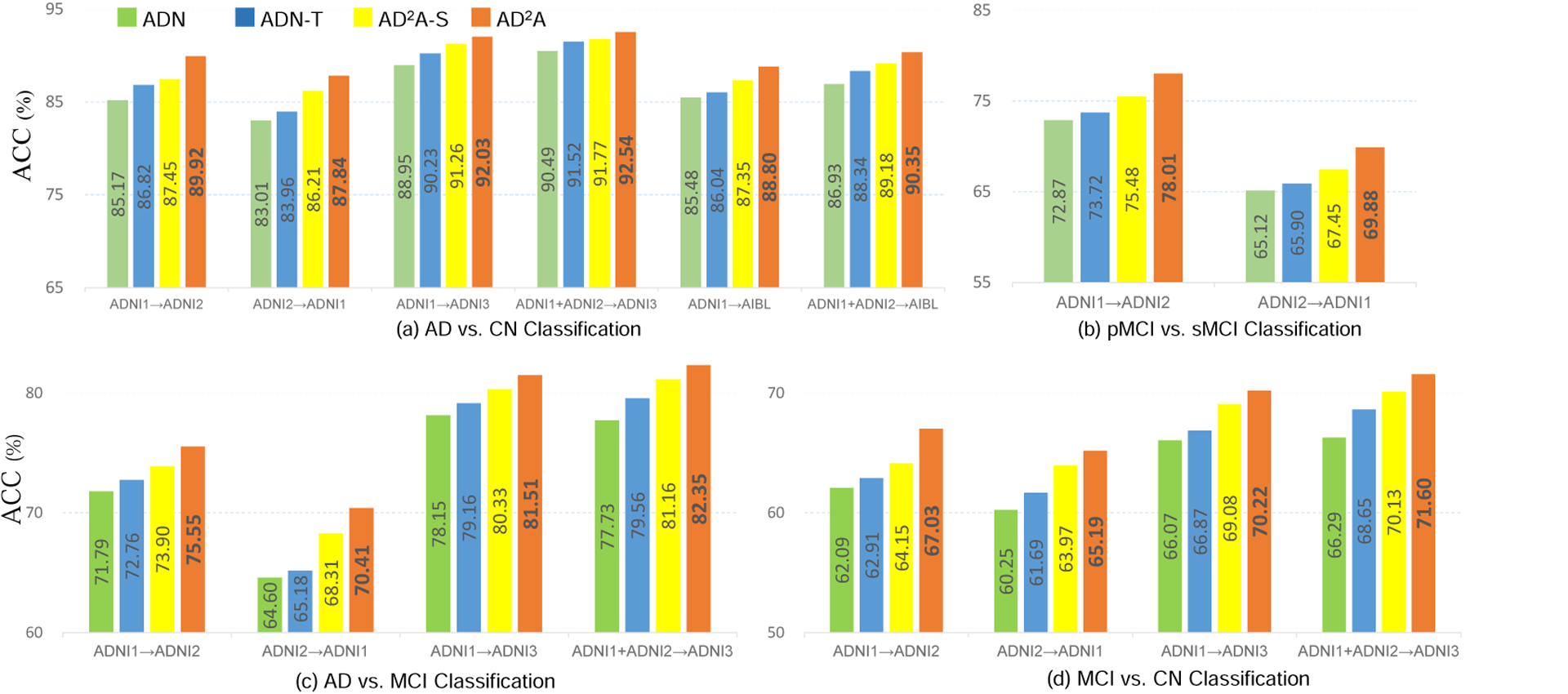
Ablation study for verifying the effectiveness of different components in AD2A.
From Fig. 2, we can derive the following observations. First, ADN (without the attention discovery module and domain discriminator) yields the worst performance in four classification tasks. Second, the results of AD2A-S (without the attention discovery module) and ADN-T (without the domain discriminator) are generally inferior to AD2A. These results suggest that both the attention discovery module and the domain discriminator are useful in boosting the learning performance. The underlying reason could be that the attention mechanism plays a role in feature selection that enables the model to focus on the discriminative patterns across domains for dementia identification. And the proposed domain discriminator helps extract domain-invariant features that are robust for cross-domain classification.
5.2. Parameter Analysis
The parameters α and β in Eq. (5) play important roles in balancing the contributions of the attention alignment and domain discriminator. To study their influence on the proposed AD2A model, we vary their values within the range of [0, 0.01, 0.02, 0.05, 0.1, 0.2, 0.5, 1], and report the corresponding AUC values. Fig. 3 reports the experimental results of AD2A with different values of α and β in MCI conversion prediction in the setting of “ADNI-1 → ADNI-2”. From Fig. 3, we can see that AD2A can yield good performance with α ∈ [0.05, 0.5] and β ∈ [0.01, 0.2]. Also, with α = 0 or β = 0, the AUC values of AD2A are not that good. This suggests the attention discovery module and the domain discriminator have positive complementary effects on enhancing transferability.
Figure 3:
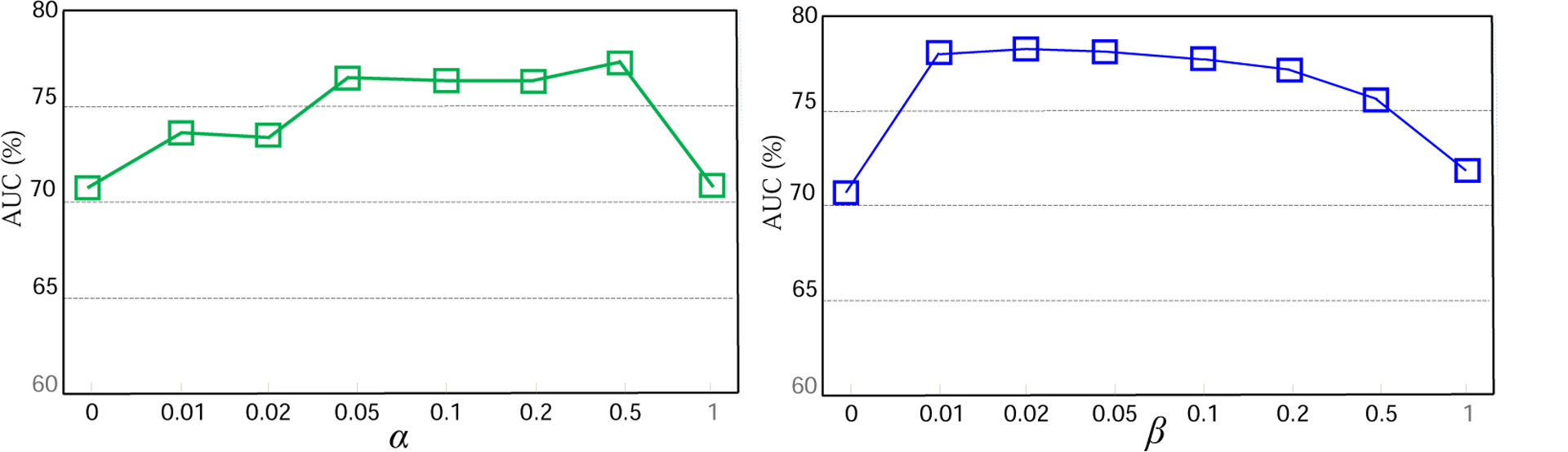
Impact of two parameters, i.e., (top) α (with β = 0) and (bottom) β (with α = 0), on the proposed method in MCI conversion prediction.
5.3. Learned Attention Maps
We further visualize the generated attention maps for eight subjects from ADNI-1 and ADNI-2, as shown in Fig. 4. From the visualization results, we can see that the most discriminative areas (denoted by red) for dementia prognosis mainly located in the hippocampus (Mu and Gage, 2011) and ventricles (Ott et al., 2010). Besides, it can be observed that the discriminative regions within AD subjects are more distinct than those of MCI (i.e., pMCI and sMCI) subjects. Considering the fact that structural changes caused by AD are relatively easier to be detected than MCI, these results suggest that the learned attention maps of the proposed AD2A are reasonable.
Figure 4:
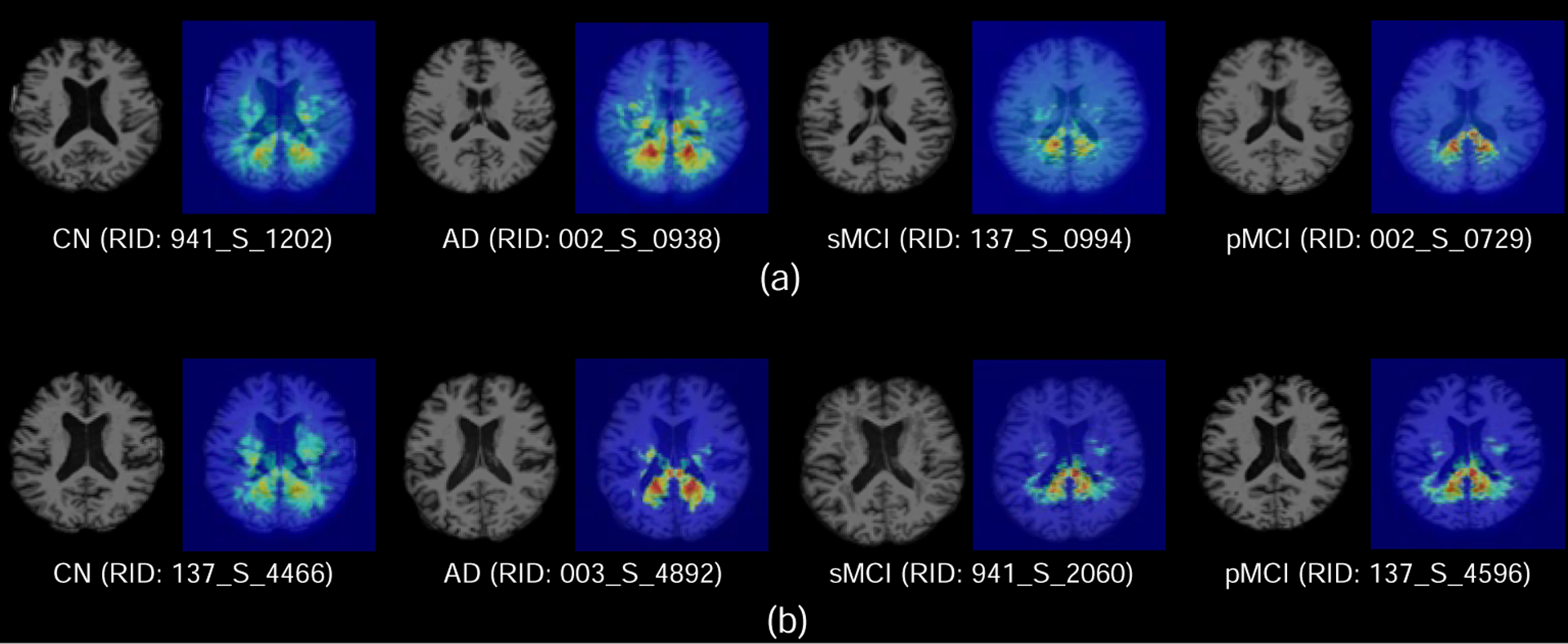
Attention maps generated by our AD2A for eight typical subjects from ADNI-1 (a) and ADNI-2 (b). The red and blue denote the high and low discriminative capability of brain regions in disease identification, respectively.
5.4. Visualization of Distribution after Adaptation
To intuitively illustrate the effectiveness of the proposed method, we visualize the data distribution of two datasets (i.e., ADNI-1, ADNI-2) before and after domain adaptation (via our AD2A). To visualize the domain heterogeneity before adaptation, we randomly selected 200 subjects (AD and CN) from ADNI-1, ADNI-2, and extracted gray matter volumes of 90 regions defined in the AAL template as feature representation of brain MRIs. Then, we use the t-SNE algorithm (Maaten and Hinton, 2008) to visualize their data distributions in Fig. 5 (a), from which we can observe that there is a significant domain shift between ADNI-1 and ADNI-2. After adaptation via AD2A, we use the trained network to extract features of samples from these two datasets, and then use t-SNE to plot their distribution as shown in Fig. 5 (b). From Fig. 5, we can see that the domain shift has been largely reduced, suggesting the effectiveness of the proposed method.
Figure 5:
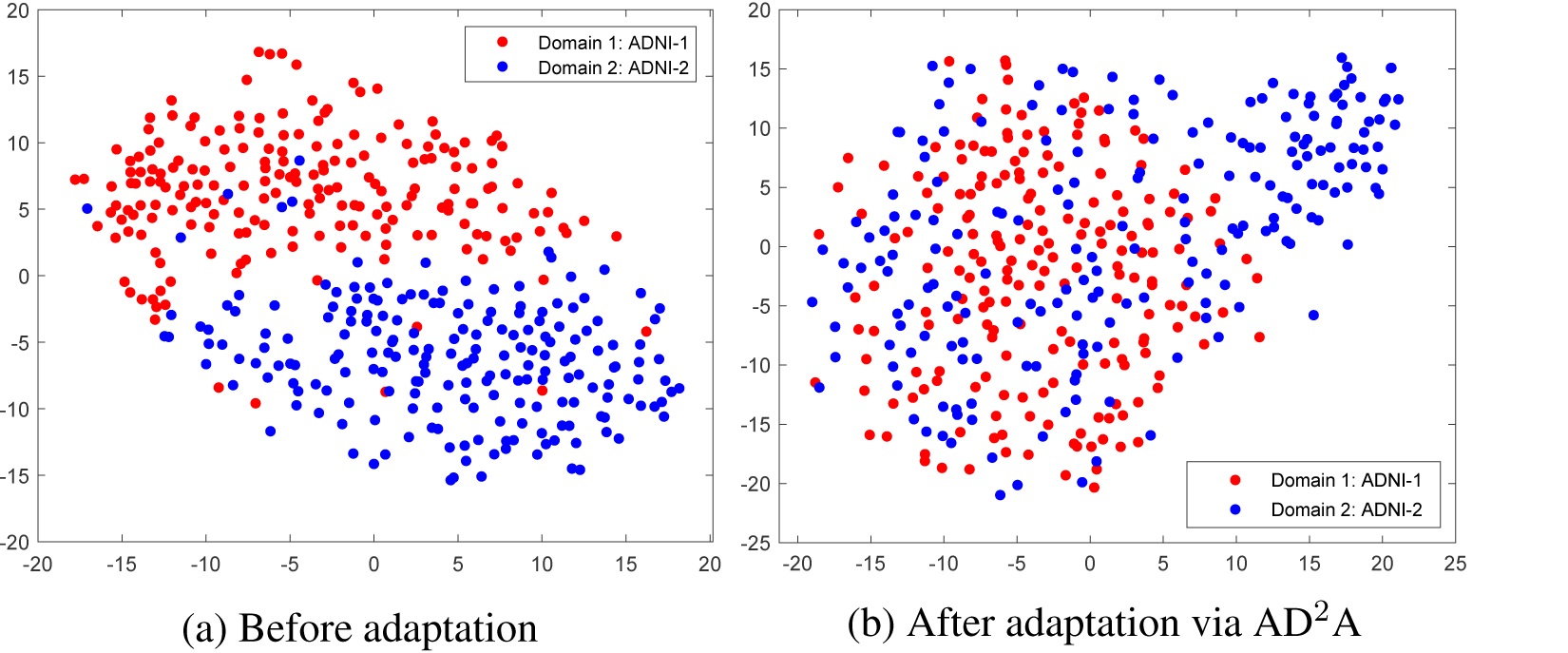
Visualization of (a) the original distribution and (b) the distribution after adaptation via our proposed AD2A for two structural MRI datasets (i.e., ADNI-1, ADNI-2).
5.5. Computational Cost
We now analyze the computational cost of the proposed AD2A model. Since the training process is conducted in an off-line manner, we only analyze the computational cost for the online test stage for new test MR images. The proposed network was implemented in PyTorch on a workstation equipped with a GPU (TI-TANX, 12G), and it took about 0.08 s to predict an input MRI scan. This result indicates that our AD2A method can perform real-time diagnosis of brain diseases, which is very useful in real-world applications.
5.6. Limitations and Future Work
Although the proposed AD2A model has obtained good performance in brain dementia identification, there are still some limitations that need to be addressed in the future.
First, the feature encoding network is trained from scratch in the current work. It is interesting to pretrain existing 3D CNNs on the other large-scale 3D medical image datasets and fine-tune them on the dementia dataset to further improve the classification performance. Second, only neuroimaging data are considered in our current work, while demographic information (e.g., age (Peters, 2006)) may also play a role in brain dementia prediction. It is interesting to incorporate some demographic information to improve the classification results. Besides, our current model is mainly trained on one domain and transferred to other domains. As future work, one can study how to leverage multi-source domain learning (Zhao et al., 2020) to incorporate more diverse training sets into the whole learning process to further enhance the robustness and transferability. Furthermore, the size of the training samples is still relatively small. It is desired to collect more neuroimaging data from multi-site MRI studies and use generative models (e.g., generative adversarial network (Yi et al., 2019)) to augment the training samples.
6. Conclusion
In this paper, we proposed an attention-guided deep domain adaptation (AD2A) framework for multi-site MRI harmonization and applied it to automated brain disorder identification. Specifically, the proposed AD2A consists of three main components, i.e., a feature encoding model for MRI feature extraction, an attention discovery module to locate disease-related regions in brain MRIs, and a domain transfer module for knowledge transfer between the source and target domains. We evaluated the AD2A model on four benchmark datasets with T1-weighted structural MRIs acquired from multiple imaging centers. Experimental results show that this method is effective in identifying brain diseases compared to several state-of-the-art methods.
Supplementary Material
Highlights.
An unsupervised domain adaptation framework for brain disorder identification.
Avoid the demand for labeled target data for training.
Automatically locate disease-related brain areas.
Extensive experiments on multi-site neuroimaging datasets.
Acknowledgment
This work was partly supported by NIH grants (Nos. AG041721, AG053867, and MH108560). Part of the data used in this paper were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database. The investigators within the ADNI contributed to the design and implementation of ADNI and provided data but did not participate in analysis or writing of this article.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Ahn E, Kumar A, Fulham M, Feng D, Kim J, 2020. Unsupervised domain adaptation to classify medical images using zero-bias convolutional autoencoders and context-based feature augmentation. IEEE Transactions on Medical Imaging 39, 2385–2394. [DOI] [PubMed] [Google Scholar]
- Alzheimer’s Association, 2019. Alzheimer’s disease facts and figures. Alzheimer’s & Dementia 15, 321–387. [DOI] [PubMed] [Google Scholar]
- Brookmeyer R, Johnson E, Ziegler-Graham K, Arrighi HM, 2007. Forecasting the global burden of Alzheimer’s disease. Alzheimer’s & Dementia 3, 186–191. [DOI] [PubMed] [Google Scholar]
- Cheng B, Liu M, Zhang D, Munsell BC, Shen D, 2015. Domain transfer learning for MCI conversion prediction. IEEE Transactions on Biomedical Engineering 62, 1805–1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chincarini A, Bosco P, Calvini P, Gemme G, et al. , 2011. Local MRI analysis approach in the diagnosis of early and prodromal Alzheimer’s disease. NeuroImage 58, 469–480. [DOI] [PubMed] [Google Scholar]
- Cho Y, Seong JK, Jeong Y, Shin SY, 2012. Individual subject classification for alzheimer’s disease based on incremental learning using a spatial frequency representation of cortical thickness data. NeuroImage 59, 2217–2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke WT, Mougin O, Driver ID, Rua C, et al. , 2020. Multi-site harmonization of 7 tesla MRI neuroimaging protocols. NeuroImage 206, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csurka G, et al. , 2017. Domain adaptation in computer vision applications. Springer. [Google Scholar]
- Cuingnet R, Gerardin E, Tessieras J, et al. , 2011. Automatic classification of patients with Alzheimer’s disease from structural MRI: A comparison of ten methods using the ADNI database. NeuroImage 56, 766–781. [DOI] [PubMed] [Google Scholar]
- Ellis KA, Bush AI, Darby D, et al. , 2009. The Australian Imaging, Biomarkers and lifestyle (AIBL) study of aging: Methodology and baseline characteristics of 1112 individuals recruited for a longitudinal study of Alzheimer’s disease. International Psychogeriatrics 21, 672–687. [DOI] [PubMed] [Google Scholar]
- Falahati F, Westman E, Simmons A, 2014. Multivariate data analysis and machine learning in Alzheimer’s disease with a focus on structural magnetic resonance imaging. Journal of Alzheimer’s Disease 41, 685–708. [DOI] [PubMed] [Google Scholar]
- Fernando B, Habrard A, Sebban M, Tuytelaars T, 2013. Unsupervised visual domain adaptation using subspace alignment, in: ICCV, pp. 2960–2967. [Google Scholar]
- Frisoni GB, et al. , 2010. The clinical use of structural MRI in Alzheimer disease. Nature Reviews Neurology 6, 67–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganin Y, Lempitsky V, 2015. Unsupervised domain adaptation by backpropagation, in: International Conference on Machine Learning (ICML), pp. 1180–1189. [Google Scholar]
- Gong B, Shi Y, Sha F, Grauman K, 2012. Geodesic flow kernel for unsupervised domain adaptation, in: CVPR, pp. 2066–2073. [Google Scholar]
- Goodfellow I, Pouget-Abadie J, Mirza M, Xu B, et al. , 2014. Generative adversarial nets, in: NeurIPS, pp. 2672–2680. [Google Scholar]
- Gupta A, Ayhan M, Maida A, 2013. Natural image bases to represent neuroimaging data, in: International Conference on Machine Learning (ICML), pp. 987–994. [Google Scholar]
- He K, Zhang X, Ren S, Sun J, 2016. Deep residual learning for image recognition, in: CVPR, pp. 770–778. [Google Scholar]
- Hosseini-Asl E, Keynton R, El-Baz A, 2016. Alzheimer’s disease diagnostics by adaptation of 3D convolutional network, in: ICIP, pp. 126–130. [Google Scholar]
- Jack CR Jr, Bernstein MA, Fox NC, et al. , 2008. The Alzheimer’s disease neuroimaging initiative (ADNI): MRI methods. Journal of Magnetic Resonance Imaging 27, 685–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javanmardi M, Tasdizen T, 2018. Domain adaptation for biomedical image segmentation using adversarial training, in: IEEE 15th International Symposium on Biomedical Imaging (ISBI), pp. 554–558. [Google Scholar]
- Kamnitsas K, Baumgartner C, Ledig C, Newcombe V, Simpson J, et al. , 2017. Unsupervised domain adaptation in brain lesion segmentation with adversarial networks, in: International Conference on Information Processing in Medical Imaging, pp. 597–609. [Google Scholar]
- Khan NM, Abraham N, Hon M, 2019. Transfer learning with intelligent training data selection for prediction of Alzheimer’s disease. IEEE Access 7, 72726–72735. [Google Scholar]
- Kingma DP, Ba J, 2015. Adam: a method for stochastic optimization, in: ICLR. [Google Scholar]
- Klöppel S, Stonnington CM, Chu C, Draganski B, et al. , 2008. Automatic classification of MR scans in Alzheimer’s disease. Brain 131, 681–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korolev S, Safiullin A, Belyaev M, Dodonova Y, 2017. Residual and plain convolutional neural networks for 3D brain MRI classification, in: IEEE International Symposium on Biomedical Imaging (ISBI), pp. 835–838. [Google Scholar]
- Kouw WM, Loog M, 2019. A review of domain adaptation without target labels. IEEE Transactions on Pattern Analysis and Machine Intelligence. [DOI] [PubMed] [Google Scholar]
- Krizhevsky A, Sutskever I, Hinton GE, 2012. Imagenet classification with deep convolutional neural networks, in: NeurIPS, pp. 1097–1105. [Google Scholar]
- Kumar D, Kumar C, Shao M, 2017. Cross-database mammographic image analysis through unsupervised domain adaptation, in: IEEE International Conference on Big Data, pp. 4035–4042. [Google Scholar]
- LeCun Y, Bengio Y, Hinton G, 2015. Deep learning. Nature 521, 436–444. [DOI] [PubMed] [Google Scholar]
- Li W, Zhao Y, Chen X, Xiao Y, Qin Y, 2019. Detecting Alzheimer’s disease on small dataset: A knowledge transfer perspective. IEEE Journal of Biomedical and Health Informatics 23, 1234–1242. [DOI] [PubMed] [Google Scholar]
- Lian C, Liu M, Zhang J, Shen D, 2020. Hierarchical fully convolutional network for joint atrophy localization and Alzheimer’s disease diagnosis using structural MRI. IEEE Transactions on Pattern Analysis and Machine Intelligence 42, 880–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Zhang J, Adeli E, Shen D, 2018. Joint classification and regression via deep multi-task multi-channel learning for Alzheimer’s disease diagnosis. IEEE Transactions on Biomedical Engineering 66, 1195–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Zhang J, Lian C, Shen D, 2020. Weakly supervised deep learning for brain disease prognosis using MRI and incomplete clinical scores. IEEE Transactions on Cybernetics 50, 3381–3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maaten LVD, Hinton G, 2008. Visualizing data using t-SNE. Journal of Machine Learning Research 9, 2579–2605. [Google Scholar]
- Madani A, Moradi M, Karargyris A, Syeda-Mahmood T, 2018. Semi-supervised learning with generative adversarial networks for chest X-ray classification with ability of data domain adaptation, in: IEEE International Symposium on Biomedical Imaging (ISBI), pp. 1038–1042. [Google Scholar]
- Moradi E, Gaser C, Huttunen H, Tohka J, 2014. MRI based dementia classification using semi-supervised learning and domain adaptation, in: MICCAI Workshop Proceedings, Challenge on Computer-Aided Diagnosis of Dementia, based on Structural MRI Data. [Google Scholar]
- Mu Y, Gage FH, 2011. Adult hippocampal neurogenesis and its role in Alzheimer’s disease. Molecular Neurodegeneration 6, 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oquab M, Bottou L, Laptev I, Sivic J, 2014. Learning and transferring mid-level image representations using convolutional neural networks, in: CVPR, pp. 1717–1724. [Google Scholar]
- Ott BR, Cohen RA, Gongvatana A, et al. , 2010. Brain ventricular volume and cerebrospinal fluid biomarkers of Alzheimer’s disease. Journal of Alzheimer’s Disease 20, 647–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan SJ, Tsang IW, Kwok JT, Yang Q, 2010. Domain adaptation via transfer component analysis. IEEE Transactions on Neural Networks 22, 199–210. [DOI] [PubMed] [Google Scholar]
- Pan SJ, Yang Q, 2010. A survey on transfer learning. IEEE Transactions on Knowledge and Data Engineering 22, 1345–1359. [Google Scholar]
- Parisot S, Ktena SI, Ferrante E, Lee M, et al. , 2018. Disease prediction using graph convolutional networks: Application to autism spectrum disorder and alzheimer’s disease. Medical Image Analysis 48, 117–130. [DOI] [PubMed] [Google Scholar]
- Peters R, 2006. Ageing and the brain. Postgraduate Medical Journal 82, 84–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomponio R, Erus G, Habes M, Doshi J, et al. , 2020. Harmonization of large MRI datasets for the analysis of brain imaging patterns throughout the lifespan. NeuroImage 208, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quionero-Candela J, Sugiyama M, Schwaighofer A, Lawrence ND, 2009. Dataset Shift in Machine Learning. The MIT Press. [Google Scholar]
- Rathore S, Habes M, Iftikhar MA, Shacklett A, Davatzikos C, 2017. A review on neuroimaging-based classification studies and associated feature extraction methods for Alzheimer’s disease and its prodromal stages. NeuroImage 155, 530–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen D, Wu G, Suk HI, 2017. Deep learning in medical image analysis. Annual Review of Biomedical Engineering 19, 221–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Suk HI, Gao Y, Shen D, 2014. Joint coupled-feature representation and coupled boosting for AD diagnosis, in: CVPR, pp. 2721–2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suk HI, Lee SW, Shen D, 2014. Hierarchical feature representation and multimodal fusion with deep learning for AD/MCI diagnosis. NeuroImage 101, 569–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun B, Feng J, Saenko K, 2017. Return of frustratingly easy domain adaptation, in: Domain Adaptation in Computer Vision Applications, pp. 153–171. [Google Scholar]
- Valiant LG, 1984. A theory of the learnable. Communications of the ACM 27, 1134–1142. [Google Scholar]
- Wachinger C, Reuter M, 2016. Domain adaptation for Alzheimer’s disease diagnostics. NeuroImage 139, 470–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Deng W, 2018. Deep visual domain adaptation: A survey. Neurocomputing 312, 135–153. [Google Scholar]
- Woo S, Park J, Lee JY, So Kweon I, 2018. CBAM: Convolutional block attention module, in: ECCV, pp. 3–19. [Google Scholar]
- Wrobel J, Martin ML, Bakshi R, Calabresi PA, et al. , 2020. Intensity warping for multisite MRI harmonization. NeuroImage 223, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Vetterli T, Balte PP, Barr RG, Laine AF, Angelini ED, 2019. Unsupervised domain adaption with adversarial learning (UDAA) for emphysema subtyping on cardiac CT scans: The mesa study, in: IEEE 16th International Symposium on Biomedical Imaging (ISBI), pp. 289–293. [Google Scholar]
- Yi X, Walia E, Babyn P, 2019. Generative adversarial network in medical imaging: A review. Medical Image Analysis 58, 101552. [DOI] [PubMed] [Google Scholar]
- Zeiler MD, Fergus R, 2014. Visualizing and understanding convolutional networks, in: ECCV, pp. 818–833. [Google Scholar]
- Zhang J, Liu M, Pan Y, Shen D, 2019a.. Unsupervised conditional consensus adversarial network for brain disease identification with structural MRI, in: International Workshop on Machine Learning in Medical Imaging, pp. 391–399. [Google Scholar]
- Zhang T, Cheng J, Fu H, Gu Z, et al. , 2019b. Noise adaptation generative adversarial network for medical image analysis. IEEE Transactions on Medical Imaging 39, 1149–1159. [DOI] [PubMed] [Google Scholar]
- Zhao S, Wang G, Zhang S, Yang G, Li Y, Song Z, Xu P, Hu R, Chai H, Keutzer K, 2020. Multi-source distilling domain adaptation, in: AAAI. [Google Scholar]
- Zhou B, Khosla A, Lapedriza A, Oliva A, Torralba A, 2016. Learning deep features for discriminative localization, in: CVPR, pp. 2921–2929. [Google Scholar]
- Zhu X, Suk HI, Shen D, 2014. Matrix-similarity based loss function and feature selection for Alzheimer’s disease diagnosis, in: CVPR, pp. 3089–3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


