Abstract
Purpose:
To (1) use All of Us (AoU) data to validate a previously published single-center model predicting need for surgery among individuals with glaucoma, (2) train new models using AoU data, and (3) share insights regarding this novel data source for ophthalmic research.
Design:
Development and evaluation of machine learning models.
Methods:
Electronic health record data were extracted from AoU for 1231 adults diagnosed with primary open-angle glaucoma. The single-center model was applied to AoU data for external validation. AoU data were then used to train new models for predicting need for glaucoma surgery using multivariable logistic regression, artificial neural networks, and random forests. Five-fold cross-validation was performed. Model performance was evaluated based on area under the receiver operating characteristic curve (AUC), accuracy, precision and recall.
Results:
The mean (standard deviation) age of the All of Us cohort was 69.1 (10.5) years, with 57.3% women and 33.5% Black, significantly exceeding representation in the single-center cohort (p=0.04 and p<0.001, respectively). Of 1231 participants, 286 (23.2%) needed glaucoma surgery. When applying the single-center model to AoU data, accuracy was 0.69, and AUC was only 0.49. Using AoU data to train new models resulted in superior performance: AUCs ranged from 0.80 (logistic regression) to 0.99 (random forests).
Conclusions:
Models trained with national AoU data achieved superior performance compared to using single-center data. Although AoU does not currently include ophthalmic imaging, it offers several strengths over similar big-data sources such as claims data. AoU is a promising new data source for ophthalmic research.
INTRODUCTION
The digitization of healthcare data offers an outstanding opportunity to better understand the complex relationships between systemic disease and glaucoma, the leading cause of irreversible blindness globally.1,2 Understanding these relationships is critical to enabling precision management of glaucoma patients, who are often elderly and have co-morbid conditions such as hypertension and diabetes.3 Several studies suggested that systemic diseases and medications may influence the development or progression of glaucoma.4–8 Electronic health records (EHRs) contain a vast amount of clinical data that may help further our understanding of these associations. EHR data have been employed to develop predictive models in a wide range of clinical applications.9,10 Within ophthalmology, several models have been developed to predict glaucoma onset and progression based on structural and functional data related to the eye,11–20 but few have used systemic data from the EHR. To address this gap, we previously developed a single-center model predicting glaucoma progression using systemic data from the EHR at our institution.21 Its predictive performance suggested that systemic EHR data could help predict patients at risk for progressing into glaucoma surgery, even in the absence of ophthalmic data. The results of that study also provided the rationale for further investigation and model refinement, particularly since the initial model was derived from a small sample.
To test generalizability, in this study we leveraged nationwide data from the All of Us Research Program (“All of Us”) to further examine the utility of systemic EHR data for prediction of glaucoma progression. Motivated by the success of other large-scale national-level cohort studies such as the UK Biobank,22 the Million Veteran Program,23 and the China Kadoorie Biobank,24 All of Us aims to collect data for 1 million persons living in the United States to advance precision medicine.25 Data collected include health questionnaires, EHRs, physical measurements, and data derived from digital health technology. The program also performs collection and analysis of biospecimens. The program emphasizes enrollment of diverse participants traditionally underrepresented in biomedical research. Enrollment opened in May 2018; as of June 2020, All of Us had enrolled >345,000 participants from both clinic-based and community-based recruitment sites.26 In October 2019, the program began an alpha demonstration phase for the Researcher Workbench, allowing selected research teams to have access to some participant data within the All of Us Researcher Workbench as the Workbench was being refined. Ours was the only alpha demonstration project related to ophthalmology.
The aims of our study were to: (1) externally validate our single-center model’s performance with All of Us data, (2) develop models trained by the All of Us data and compare their performance to our single-center model, and (3) share insights from our experience using All of Us data and the Researcher Workbench with other ophthalmology researchers who may be interested in using this novel data source.
METHODS
Study Population, Data Source, and Demonstration Project Information
The methods underlying our initial single-center model were described in detail previously.21 In short, we examined data from a cohort of adult participants from a single academic center with primary open-angle glaucoma over a 5-year period; we extracted their systemic EHR data from our institution’s clinical data warehouse, and developed predictive models using multivariable logistic regression, random forests, and artificial neural networks (ANNs).
The goals, recruitment methods and sites, and scientific rationale for All of Us have been described previously.25 Demonstration projects were designed to describe the cohort, replicate previous findings for validation, and avoid novel discovery in line with the program value to ensure equal access by researchers to the data.26 The work described here was proposed by Consortium members, reviewed and overseen by the program’s Science Committee, and was confirmed as meeting criteria for non-human subjects research by the All of Us Institutional Review Board. The initial release of data and tools used in this work was published recently.26 Results reported are in compliance with the All of Us Data and Statistics Dissemination Policy disallowing disclosure of group counts under 20.
This work was performed on data collected by the previously described26 All of Us Research Program using the All of Us Researcher Workbench, a cloud-based platform where approved researchers can access and analyze All of Us data. The All of Us data currently includes surveys, EHRs, and physical measurements (PM). The details of the surveys are available in the Survey Explorer found in the Research Hub, a website designed to support researchers.27 Each survey includes branching logic and all questions are optional and may be skipped by the participant. PM recorded at enrollment include systolic and diastolic blood pressure, height, weight, heart rate, waist and hip measurement, wheelchair use, and current pregnancy status. EHR data was linked for those consented participants. All three datatypes (survey, PM, and EHR) are mapped to the Observational Health and Medicines Outcomes Partnership (OMOP) common data model v 5.2 maintained by the Observational Health and Data Sciences Initiative (OHDSI) collaborative. To protect participant privacy, a series of data transformations were applied. These included data suppression of codes with a high risk of identification such as military status; generalization of categories, including age, sex at birth, gender identity, sexual orientation, and race; and date shifting by a random (less than one year) number of days, implemented consistently across each participant record. Documentation on privacy implementation and creation of the Curated Data Repository (CDR) is available in the All of Us Registered Tier CDR Data Dictionary.28 The Researcher Workbench currently offers tools with a user interface built for selecting groups of participants (Cohort Builder), creating datasets for analysis (Dataset Builder), and Workspaces with Jupyter Notebooks (Notebooks) to analyze data. The Notebooks enable use of saved datasets and direct query using R and Python 3 programming languages.
Figure 1 depicts our study workflow. At the time of the alpha demonstration phase, there were 242,070 adult participants in All of Us. Queries using Systematized Nomenclature of Medicine (SNOMED) codes for “Glaucoma” and subsequently “Primary open-angle glaucoma” narrowed the cohort. Finally, to maintain consistency in cohort definition with the initial single-center model, our final study cohort consisted of adult (age 18 years and above) participants with International Classification of Diseases (ICD)-9 code 365.11 or any variants of ICD-10 code H40.11, derived from 26 distinct enrollment sites.
Figure 1.
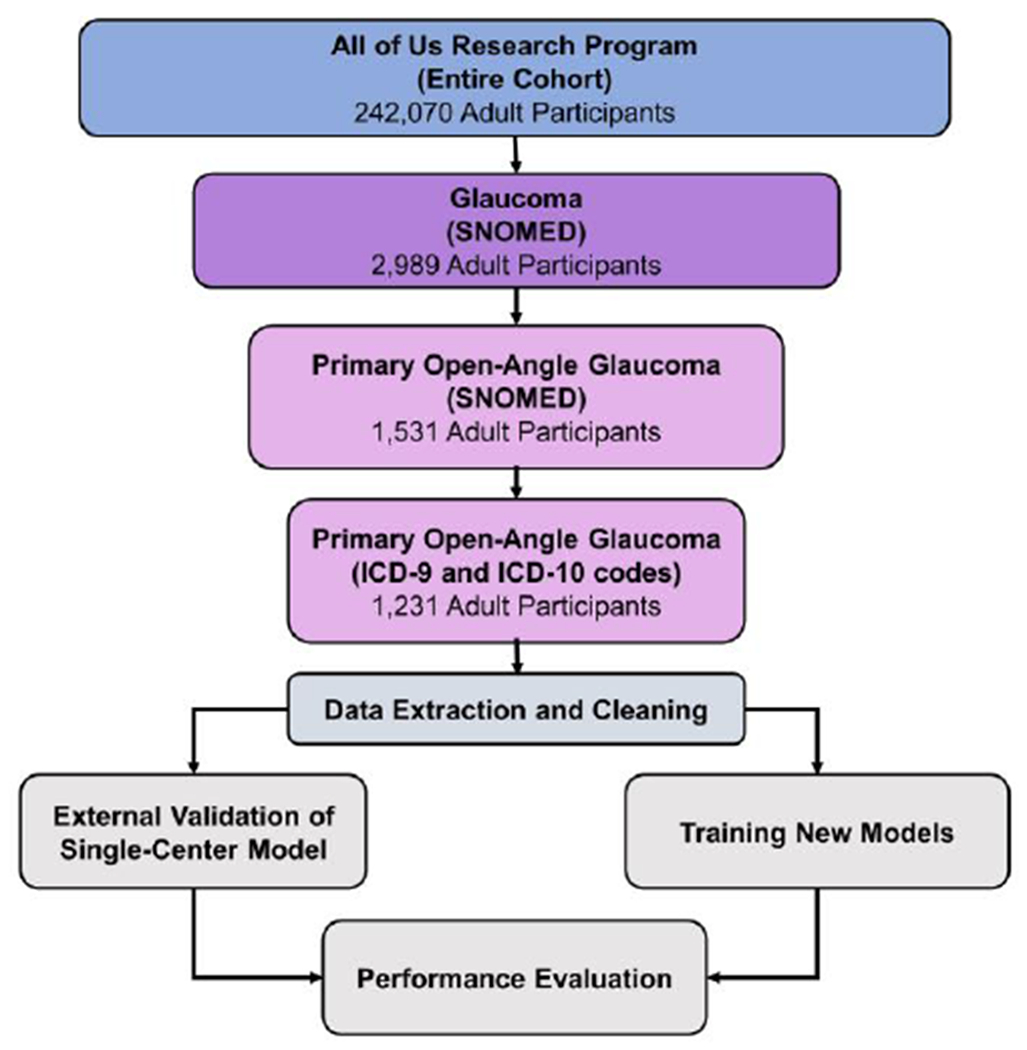
Study workflow and cohort definition for evaluating predictive models for participants with primary open-angle glaucoma in the All of Us Research Program.
Data Processing
We used the Researcher Workbench to extract relevant data for the analysis. Within the Workbench, we first defined the cohort according to the criteria above. Next, we built concept sets for the outcome and each predictor in the Workbench by selecting relevant codes (e.g. ICD and/or SNOMED codes for conditions, Logical Observation Identifiers Names and Codes [LOINC] for measurements and observations, RxNorm codes for medications, and Current Procedure Terminology [CPT] codes for procedures). Because a key aim was to validate the prior single-center model, we extracted the same data types for this cohort. For example, we defined the outcome of interest equivalently to the initial single-center model (i.e., need for glaucoma procedural intervention – laser or surgery – within 6 months of diagnosis) based on qualifying CPT codes.21 The rationale for this was that need for surgery serves as a surrogate for advancing/progressive disease. This approach was also used by Zheng et al. in examining associations between systemic medications and glaucoma.5 We then built concept sets for all predictors, such as vital signs (pulse, blood pressure), physical measurements (height, weight, body mass index [BMI]), and co-morbid conditions (ICD and SNOMED codes in categories included in the prior single-center model). These concept sets were linked to the study cohort to create “datasets,” or analysis-ready tables linking participants with the values of the selected concept sets. For participants who underwent glaucoma surgery intervention, data regarding predictors were restricted to the time period before the occurrence of the qualifying procedure code. In other words, all data with a timestamp occurring after the glaucoma procedure were censored and not included in modeling procedures in order to establish an appropriate temporal relationship between predictors and the outcome. We then exported these datasets to a Python 3.0 notebook within the All of Us Workbench environment to conduct the analyses. During the export process, the Workbench generated structured query language (SQL) codes to extract the data of interest for the selected cohort and populated these directly into the notebook. All data extraction and cleaning procedures can be found in the referenced Python 3.0 notebook29 in our publicly available workspace.30
Of note, the OMOP Visits table was not included in the Workbench interface during the alpha demonstration phase. Therefore, to extract data on predictors related to visits (e.g. total days of contact with the healthcare system, which was a predictor in our original model), we manually constructed a custom SQL query within the notebook. Similarly, we performed a custom SQL query to extract all medications in order to group them within therapeutic classes that we used in the original model. The Workbench interface allowed search and selection of individual medications, but not of medication classes.
The original single-center model did not include ophthalmic data, as the focus was on examining the predictive value of systemic data alone. Here, we also did not include ophthalmic data. One reason was to maintain the same data structure as the original model. Another was because ophthalmic data (beyond diagnosis codes) were sparse in the current All of Us data repository. As an illustration, despite 6665 All of Us participants having a SNOMED code including “glaucoma,” structured intraocular pressure (IOP) data (LOINC code 56844-4) were available for less than 20 participants. Ophthalmic data coverage in All of Us is detailed in Supplemental Table 1.
Table 1.
Comparison of demographic characteristics and primary outcome between cohorts of adults with primary open-angle glaucoma derived from a single academic center and derived from the All of Us data repository.
| Single-center Cohort (N=385) | All of Us Cohort (N=1231) | p-value | |
|---|---|---|---|
| Mean (SD) Age | 73.1 (12.2) years | 69.1 (10.5) years | <0.001 |
| Female | 198 (51.4%) | 705 (57.3%) | 0.04 |
| Self-Reported Race | <0.001 | ||
| White | 214 (55.6%) | 508 (41.3%) | |
| Black or African American Asian | 23 (6.0%) | 412 (33.5%) | |
| Asian | 49 (12.7%) | 27 (2.2%) | |
| Other Race or Mixed Race | 70 (18.2%) | 21 (1.7%) | |
| None or Skipped | 29 (7.5%) | 263 (21.4%) | |
| Participants who underwent glaucoma surgery | 174 (45.2%) | 286 (23.2%) | <0.001 |
Data Analysis and Modeling
General cohort characteristics
We generated descriptive statistics of the All of Us study cohort for age, gender, race, and need for glaucoma surgery (Table 1). These characteristics were compared to the initial single-center cohort using t-tests for continuous variables and Chi-squared tests for categorical variables. We considered p-values < 0.05 as statistically significant.
Using All of Us Data to externally validate the published single-center model
Out of the models developed in the initial study, the best-performing and most interpretable model was the multivariable logistic regression model (Supplemental Table 2).21 We therefore developed a dataset from All of Us with the top 15 predictors from the single-center model and used it as an external validation set for this regression model. To evaluate performance, we measured area under the receiver operating characteristic curve (AUC) as well as accuracy, sensitivity, and specificity. These performance measures were compared to the performance measures achieved in the original analysis of the single-center datasets, which were generated with leave-one-out cross-validation. This validation was performed in R in the referenced notebook31 using tidyverse and dplyr.
Development of new models trained by All of Us data
Next, we developed a broader dataset from All of Us using 56 predictors, rather than limiting to only the 15 predictors included in the single-center model. This dataset was used to train multiple models, including multivariable logistic regression, random forests, and ANNs using the scikit-learn and keras libraries in Python.32,33 Data were split into 80% for training and 20% for testing. The test dataset was separated prior to any training procedures, and 5-fold cross-validation was used for all modeling approaches to reduce the risk of overfitting. We evaluated various feature selection techniques such as using Pearson correlation, backward elimination, recursive feature elimination, and Lasso regularization. Because the feature selection methods did not improve performance, the full complement of features were used for subsequent modeling. Using the test dataset, we evaluated performance metrics such as AUC, accuracy, precision (also known as positive predictive value), and recall (also known as sensitivity). For ANNs, we compared a variety of neural network architectures (e.g., varying the number of epochs, the number of hidden layers, and the number of nodes within each layer) using a grid search method. Because of class imbalance of the outcome label in the dataset (286 participants with surgery out of 1231 total in the cohort), we also evaluated models incorporating minority upsampling or majority downsampling for balancing the classes. Variables of importance were evaluated for the best-performing random forests model.
We also evaluated models trained with data from All of Us that were limited to only the 15 predictors available in the original single-center model. Again, we developed models using multivariable logistic regression, random forests, and ANNs using an 80%/20% training/testing split and 5-fold cross-validation. The same ANN architecture was used as the best-performing ANN developed using the broader dataset. Details of all modeling procedures and execution are described in the referenced Python 3.0 notebook.34
RESULTS
General Cohort Characteristics
There were significant differences between the cohorts of participants with glaucoma from the initial single-center study with that using All of Us (Table 1). The All of Us cohort was over triple the size of the single-center cohort and consisted of EHR data derived from 26 distinct enrollment sites. The mean (standard deviation, SD) age was 69.1 (10.5) years, which was significantly younger than the single-center cohort, where the mean (SD) age was 73.1 (12.2) years (p<0.001). Participants identifying as female comprised more than half of both cohorts and were significantly better-represented in the All of Us cohort (57.3% vs. 51.4% in the single-center cohort, p=0.04). The proportion of participants identifying as Black or African American was 33.5% in the All of Us cohort, over five times that in the single-center cohort. However, the single-center cohort had a much higher proportion of Asian participants (12.7% compared to 2.2% in All of Us). In both cohorts, about a quarter of participants did not indicate a single racial category, e.g. answering “other” or “mixed” or not providing any answer. In total, 286 (23.2%) participants in the All of Us cohort underwent some kind of glaucoma procedure. This was a significantly lower percentage than the 45.2% of participants in the single-center cohort (p<0.001).
External Validation of the Single-Center Model
To externally validate a previously published single-center model,21 we used data from All of Us as an independent test set using the same coefficients included in the initial model (Supplemental Table 2). The overall accuracy of the model when validated on All of Us data was 0.69, exceeding the accuracy of the single-center model when using leave-one-out cross-validation (0.62). The overall discriminative ability (demonstrated by the AUC) of the single-center model when applied to the All of Us cohort was only 0.49, indicating no discrimination between those who progressed and did not progress to surgery. This was lower than the AUC of the single-center model when previously validated with data from the same center (0.67).
Development of New Models using All of Us Data
We then used All of Us data with an expanded set of predictors to train new models predicting need for glaucoma surgery using multivariable logistic regression, random forests, and ANNs. The predictive performance of multivariable logistic regression models trained with All of Us data was better than the performance of the model trained with single-center data, with the best-performing logistic regression model trained with All of Us achieving an AUC of 0.80 and accuracy of 0.87, although recall was only 0.51 (Table 2). Variables in the logistic regression model are detailed in Supplemental Table 3. The best-performing ANN was a deep learning network consisting of 4 dense layers trained over 25 epochs on a minority upsampled dataset, resulting in an AUC of 0.93.
Table 2.
Predictive performance of models predicting need for glaucoma surgery trained with data from All of Us.
| AUC | Precision | Recall | Accuracy | |
|---|---|---|---|---|
| Models trained with dataset containing broad set of predictor variables (56 predictors) | ||||
| Multivariable Logistic Regression | 0.80 | 0.90 | 0.51 | 0.87 |
| Artificial Neural Networks | 0.93 | 0.83 | 0.84 | 0.92 |
| Random Forests | 0.99 | 1.00 | 0.88 | 0.97 |
| Models trained with dataset containing predictor variables restricted to those from the original single-center model (15 predictors) | ||||
| Multivariable Logistic Regression | 0.81 | 0.40 | 0.75 | 0.68 |
| Artificial Neural Networks | 0.94 | 0.84 | 0.74 | 0.91 |
| Random Forests | 0.97 | 0.91 | 0.75 | 0.93 |
Predictive modeling using random forests yielded the best performance. With minority upsampling as a class-balancing procedure on the training dataset, random forests achieved an AUC of 0.99 in identifying participants who needed glaucoma surgery (Figure 2). In addition, the random forests model had excellent accuracy, precision, and recall (Table 2).
Figure 2.
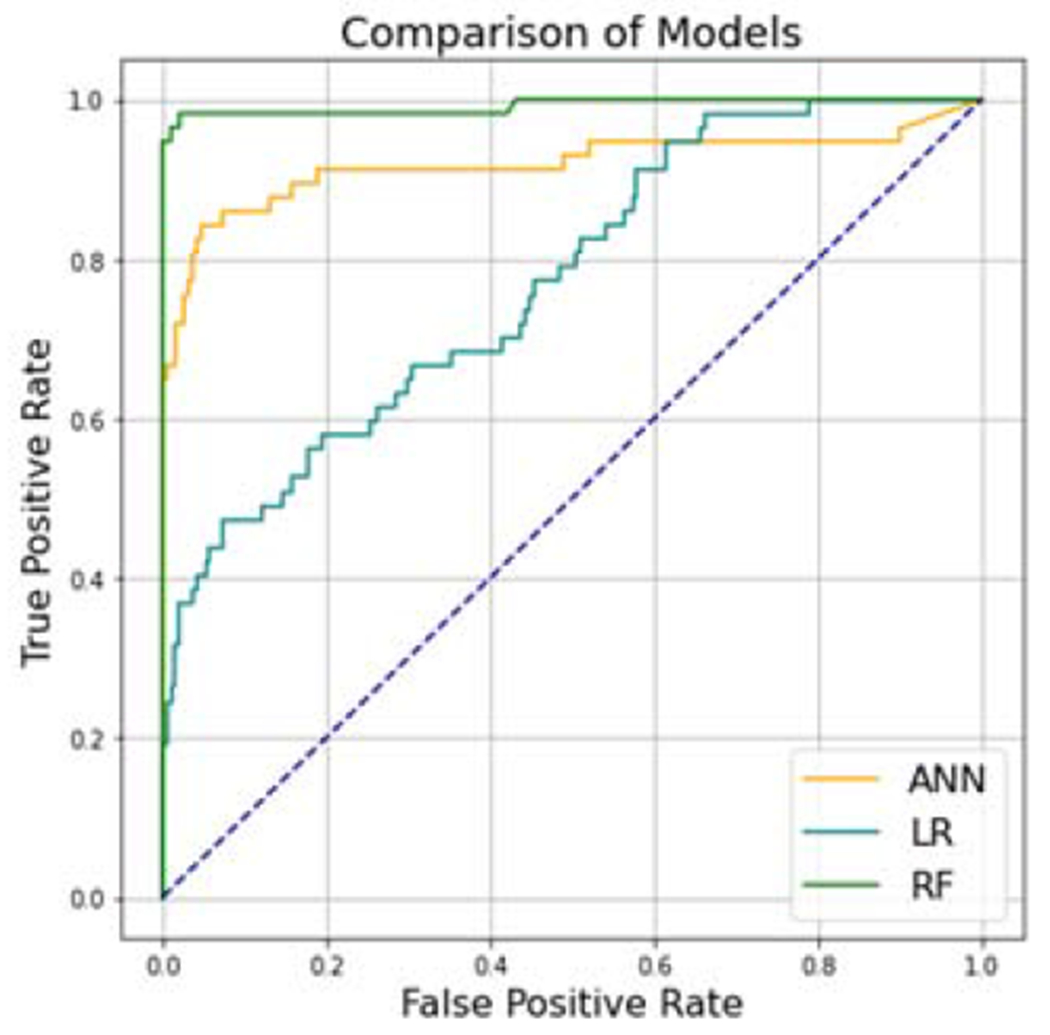
Composite receiver operating characteristic (ROC) curves for multivariable Logistic Regression (LR) model with area under the curve (AUC) of 0.80, Artificial Neural Network (ANN) model with AUC of 0.93, and Random Forests (RF) model with AUC of 0.99 predicting need for surgery among All of Us research participants with glaucoma.
Based on an analysis of feature importance in the random forests model, predictors related to days of contact with the healthcare system, systolic blood pressure, diastolic blood pressure, pulse, and body measurements (e.g. BMI) were of highest relative importance (Supplemental Table 4). These carried more importance in predictions than co-morbid conditions or medications.
To evaluate whether predictive performance could be maintained with a narrower set of predictor variables, we also trained models using data from All of Us using only the 15 significant predictor variables from the original single-center model. Even with a narrower training dataset, these models achieved comparable AUCs (0.81 for logistic regression, 0.94 for ANNs, and 0.97 for random forests). However, their precision, recall, and accuracy were generally not as strong as the models trained with the broader dataset (Table 2).
DISCUSSION
Here, we report findings from a demonstration project leveraging early access to the All of Us Research Program, a large-scale prospective nationwide cohort study, centered on predictive modeling of need for glaucoma surgery among adults with primary open-angle glaucoma. This is the first analysis of data related to ophthalmology from All of Us.
Our first key finding was that All of Us offered a larger number and generally greater diversity of participants compared to our cohort for the original model, which was derived from a single academic center. Notably, the representation of women and of participants identifying as Black or African American was significantly higher. This is highly relevant for glaucoma, since women and minorities – and particularly individuals of African descent – bear a disproportionate burden of disease and blindness.34–37 However, about a quarter of each cohort (for both the single-center and All of Us) had incomplete race information, i.e. “other race” or not indicated/completed. Missing data in EHRs is a well-known limitation.9 For studies aimed at understanding healthcare disparities or differential risk based on race, excluding individuals with incomplete race information may negatively affect the generalizability of results. The inclusion of genomic data in All of Us, anticipated for release at a future date, will enable determination of racial admixtures via sequencing analyses rather than by self-report and may help address some of these gaps.
The initial model trained with data from a single academic center demonstrated weaker performance when using All of Us data for external validation, compared to the internal validation.21 This demonstrated that the model developed with data sampled from one population could not necessarily be applied to patients from another population. This was not surprising for several reasons. First, the initial model was based on a relatively small number of participants originating from a single center. Second, comparing the two cohorts revealed significant differences in demographics and incidence of glaucoma surgery. The single-center cohort was older and had higher rates of surgery, which was expected for a clinic-based population at a tertiary referral center. In contrast, All of Us includes community-based recruitment sites in addition to clinic-based sites, reflecting an overall younger and healthier population. Using data from this population may therefore offer additional insights for primary prevention efforts, by including individuals beyond clinic- or hospital-based populations alone.
Finally, decreased performance of predictive models on external validation is not uncommon, especially when the sample size for developing the model is small.38 However, a systematic review of 120 clinical risk prediction models found that there is a relative dearth of well-conducted and clearly reported external validation studies on independent data, which limits the translation of predictive models to clinical guidelines and practice.38 All of Us can serve as a data source for external validation studies to help address this issue.
Models trained with All of Us data achieved superior predictive performance than the single-center models, even when trained with data limited to only 15 predictor variables from the original single-center model. The best-performing model overall was the random forests model trained with the broader All of Us dataset, achieving an AUC of 0.99. Variables in this model with the highest relative importance for driving predictions included measurements related to blood pressure. This supports findings from the initial single-center model, where predictions were also heavily influenced by blood pressure measurements. Several prior studies have demonstrated relationships between blood pressure, intraocular pressure, and glaucoma.8,39,40 However, these relationships are complex and not completely understood. With rapidly evolving blood pressure management guidelines recommending more aggressive blood pressure reduction,41 and the potential for increased glaucoma progression events secondary to optic nerve hypoperfusion,3 improving understanding of these relationships is critically important. Our study results provide additional support for further investigation in this area.
Another advantage of models such as these is that they facilitate integration of systemic data into clinical decision-making. Ophthalmology is known to be a high-volume specialty that demands high efficiency during patient encounters.42 As such, time spent in reviewing the medical record for each patient is minimal, which has been demonstrated in several prior studies.43,44 In light of these time pressures and the lack of detailed medical record review, clinical decision support (CDS) tools that are embedded into EHR systems and have programmed logic to automatically extract relevant data elements (such as medications, comorbidities, and blood pressure measurements) and calculate risk scores directly for ophthalmologists to view would enable these important factors to be considered in the assessment and management of patients. Therefore, ophthalmologists would not need to extract these data elements themselves or perform any manual calculations. For a given patient, they could theoretically view the risk calculated by the model based on these systemic factors in the EHR and incorporate that information into their overall assessment of the patient. In general, CDS tools that calculate risk scores in real-time within EHRs to enable predictive analytics are increasingly common,45 although several implementation challenges are present.46,47 Moreover, a recent systematic review did not find any publications describing predictive models related to ophthalmology that had been embedded into EHRs for real-time use.48 Therefore, developing best practices for implementing these models for routine use by ophthalmologists is a nascent area ripe for ongoing investigation.
Although we have highlighted some of the strengths of All of Us data in the context of our study findings above, there are certainly limitations as well. Data on ophthalmic observations (e.g., intraocular pressure) are sparse (Supplemental Table 1), and this could be secondary to many ophthalmology observations being captured in clinical notes rather than in discrete data fields mapped to LOINC codes. Although free-text clinical notes are not currently available in All of Us, future inclusion of these notes and the use of natural language processing techniques may allow more ophthalmic data to be extracted and analyzed. Although All of Us includes procedure codes for imaging and visual fields, the data repository does not currently include images or visual fields themselves. Figure 3 delineates current strengths and limitations of All of Us data for ophthalmic research in greater detail.
Figure 3.
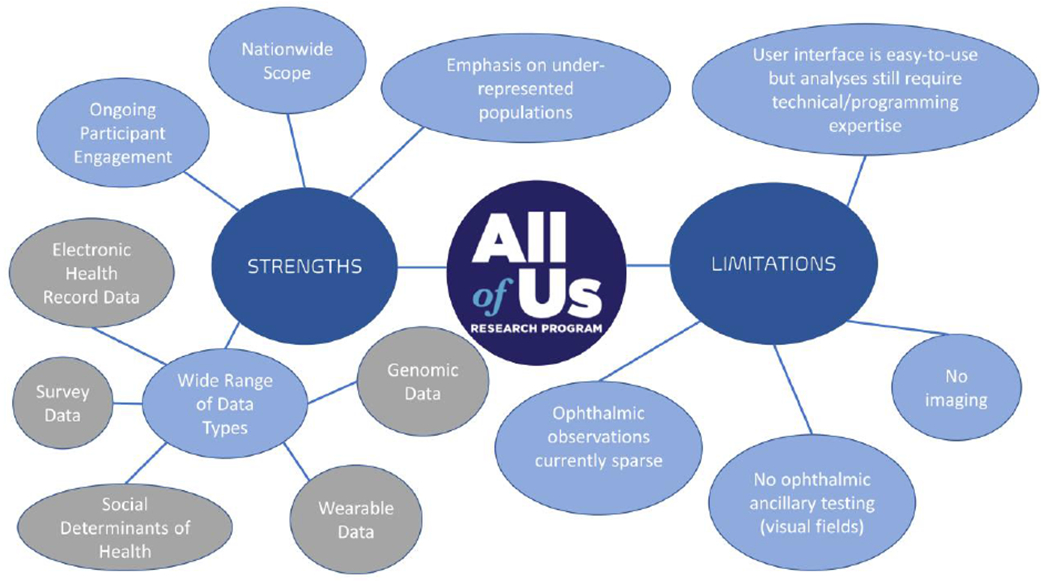
Strengths and limitations of All of Us data for ophthalmic research.
Regarding limitations of our specific study, we recognize that the outcome of glaucoma surgery is a crude proxy of glaucoma progression, and that other factors (e.g. surgeon preference) may factor into the decision to pursue surgery other than disease severity alone. The lack of visual field and imaging data precluded a more precise measure of glaucoma progression.
Overall, All of Us offers many opportunities for ophthalmic research. Despite the scarcity of ophthalmic observations and lack of imaging, many high-quality studies in ophthalmology have been performed using claims data or data from other cohorts which also lacked this information. Furthermore, this data repository offers a much wider array of data types than claims data. All of Us will be of particular benefit to ophthalmic researchers interested in studying interactions between systemic disease and eye conditions, traditionally underrepresented populations, social determinants of health, and those using big-data analytic techniques. This demonstration project illustrated the diversity of the participants, the wealth of data, and the improved performance of predictive models using national-level diverse data.
Table of Contents Statement
This study describes an alpha phase demonstration project using data from the nationwide All of Us Research Program to validate and develop machine learning models predicting need for glaucoma surgical intervention in adults with primary open angle glaucoma. Models developed using national data from diverse participants performed better than those previously trained with data from a single academic medical center. All of Us is a novel data source offering exciting opportunities for ophthalmic research.
Supplementary Material
APPENDIX 1:
All of Us Research Program Investigators
| First Name | Last Name | Degrees | Institutional Affiliation |
|---|---|---|---|
| Confidence | Achilike | MBBS, MPH | Boston University School of Medicine, Department of Obstetrics and Gynecology, Boston, MA, USA |
| Brian | Ahmedani | PhD | Henry Ford Health System, Duluth, MN, USA |
| Habibul | Ahsan | MD, MMedSc |
Institute for Population and Precision Health, University of Chicago, Chicago, IL, USA |
| Toluwalase (Lase) |
Ajayi | MD | The Participant Center / Scripps Research, La Jolla, CA, USA |
| Alvaro | Alonso | MD, PhD | Department of Epidemiology, Rollins School of Public Health, Emory University, Atlanta, GA, USA |
| Amit | Arora | MPH | University of Arizona, Mel and Enid Zuckerman College of Public Health, Tucson, AZ, USA |
| Briseis | Aschebrook-Kilfoy | PhD, MPH |
Department of Public Health Sciences, University of Chicago, Chicago, IL, USA |
| Sally L. | Baxter | MD, MSc | University of California San Diego Viterbi Family Department of Ophthalmology and Shiley Eye Institute; University of California San Diego Health Department of Biomedical Informatics, La Jolla, CA, USA |
| Dean | Billheimer | PhD | Department of Epidemiology and Biostatistics / University of Arizona, Tucson, AZ, USA |
| Eugene R. | Bleeker | MD | Department of Medicine / University of Arizona Health Sciences, Tucson, AZ, USA |
| Luca | Bonomi | PhD | University of California San Diego Health Department of Biomedical Informatics, La Jolla, CA, USA |
| Olveen | Carrasquillo | MD, MPH | University of Miami, Department of Medicine, Miami, FL, USA |
| Paulette | Chandler | MD | Brigham & Women’s Hospital, Department of Medicine, Boston, MA, USA |
| Qingxia | Chen | PhD | Vanderbilt University School of Medicine, Department of Biostatistics, Nashville, TN, USA |
| Dave | Chesla | Spectrum Health, Temple, TX, USA | |
| Cheryl | Clark | MD, ScD | Brigham & Women’s Hospital, Department of Medicine, Boston, MA, USA |
| Andrew | Craver | MPH, MS | Institute for Population and Precision Health, University of Chicago, Chicago, IL, USA |
| Robert | Cronin | MD, MS | Vanderbilt University School of Medicine, Department of Biomedical Informatics, Internal Medicine, and Pediatrics, Nashville, TN, USA |
| Zubin | Dastur | MS, MPH | Stanford University School of Medicine, Department of Obstetrics and Gynecology, Palo Alto, CA, USA |
| The PRIDE Study/PRIDEnet, Palo Alto, CA, USA | |||
| John | Ehiri | PhD | Mel and Enid Zuckerman College of Public Health, University of Arizona, Tucson, AZ, USA |
| Mara M. | Epstein | ScD | Department of Medicine, University of Massachusetts Medical School; The Meyers Primary Care Institute, a joint venture of Reliant Medical Group, Fallon Health, and the University of Massachusetts Medical School, Worcester, MA, USA |
| Xiaoke | Feng | MS | Vanderbilt University Medical Center, Department of Biostatistics, Nashville, TN, USA |
| Annesa | Flentje | PhD | University of California, San Francisco, School of Nursing, Department of Community Health Systems, San Francisco, CA, USA |
| Alliance Health Project, Department of Psychiatry and Behavioral Sciences, School of Medicine, University of California, San Francisco, San Francisco, CA, USA | |||
| The PRIDE Study/PRIDEnet, Palo Alto, CA, USA | |||
| Lawrence | Garber | MD | Reliant Medical Group, Worcester, MA, USA |
| Nicholas | Giangreco | MA, MPhil |
Columbia University Irving Medical Center, New York, NY, USA |
| Yi | Guo | PhD | University of Florida, Gainesville, FL, USA |
| Alese | Halvorson | MS | Vanderbilt University School of Medicine, Department of Biostatistics, Nashville, TN, USA |
| Robert A. | Hiatt | MD, PhD | University of California, San Francisco, Department of Epidemiology & Biostatistics, San Francisco, CA, USA |
| Kai Yin | Ho | MPhil | Northwestern University, Department of Preventive Medicine, Chicago, IL, USA |
| Joyce | Ho | PhD | Northwestern University, Department of Preventive Medicine, Chicago, IL, USA |
| William | Hogan | MD, MS | University of Florida, Gainesville, FL, USA |
| George | Hripcsak | MD, MS | Columbia University Irving Medical Center, New York, NY, USA |
| Carolyn | Hunt | MPA | University of California, San Francisco, School of Nursing, Department of Community Health Systems, San Francisco, CA, USA |
| The PRIDE Study/PRIDEnet, Palo Alto, CA, USA | |||
| Rosario | Isasi | JD, MPH | John P. Hussman Institute for Human Genomics, Miller School of Medicine, University of Miami; The Dr. John T. Macdonald Foundation Department of Human Genetics, Miller School of Medicine, University of Miami, Miami, FL, USA |
| Xinzhuo | Jiang | MS | Department of Biomedical Informatics, Columbia University Medical Center, New York, NY, USA |
| Christine C. | Johnson | PhD | Henry Ford Health System, Public Health Sciences, Detroit, MI, USA |
| Christina D. | Jordan | PhD | All-of-Us Research Program Director of Operations, Biobank, The University of Mississippi Medical Center, Jackson, MS, USA |
| King | Jordan | PhD | School of Biological Sciences, Georgia Institute of Technology, Atlanta, GA, USA |
| Christine LM | Joseph | PhD | Henry Ford Health System, Detroit, MI, USA |
| Hooman | Kamel | MS, MD | Weill Cornell Medicine, Neurology & BMRI, New York, NY, USA |
| Elizabeth | Karlson | MD, MS | Brigham & Women’s Hospital, Department of Medicine, Boston, MA, USA |
| Jason H. | Karnes | PharmD, PhD | University of Arizona College of Pharmacy, Department of Pharmacy Practice and Science, Tucson, AZ, USA |
| Theresa H. | Keegan | PhD | University of California Davis Comprehensive Cancer Center, Sacramento, CA, USA |
| Karen | Kim | MD, MS | Illinois Precision Medicine: University of Chicago, Chicago, IL, USA |
| Katherine K. | Kim | PhD, MPH, MBA |
University of California Davis, Betty Irene Moore School of Nursing, Sacramento, CA, USA |
| Jihoon | Kim | MS | University of California San Diego Health Department of Biomedical Informatics, La Jolla, CA, USA |
| Paula | King | MPH | The Participant Center / Scripps Research, La Jolla, CA, USA |
| Yann C. | Klimentidis | PhD | University of Arizona, Mel and Enid Zuckerman College of Public Health, Tucson, AZ, USA |
| Irving L. | Kron | MD | University of Arizona Health Sciences, Tucson, AZ, USA |
| Tsung-Ting | Kuo | PhD | University of California San Diego Health Department of Biomedical Informatics, La Jolla, CA, USA |
| Helen | Lam | PhD, RN | Illinois Precision Medicine: University of Chicago, Chicago, IL, USA |
| James P. | Lash | MD | University of Illinois at Chicago, Department of Medicine, Chicago, IL, USA |
| Micah E. | Lubensky | PhD | University of California, San Francisco, School of Nursing, Department of Community Health Systems, San Francisco, CA, USA |
| The PRIDE Study/PRIDEnet, Palo Alto, CA, USA | |||
| Mitchell R. | Lunn | MD, MAS | Stanford University School of Medicine, Department of Medicine, Division of Nephrology, Palo Alto, CA, USA |
| The PRIDE Study/PRIDEnet, Palo Alto, CA, USA | |||
| Yves A. | Lussier | MD | The University of Arizona Health Sciences, Tucson, AZ, USA |
| Jacob L. | McCauley | PhD | John P. Hussman Institute for Human Genomics, Miller School of Medicine, University of Miami; The Dr. John T. Macdonald Foundation Department of Human Genetics, Miller School of Medicine, University of Miami, Miami, FL, USA |
| Robert | Meller | D.Phil | Neuroscience Institute, Morehouse School of Medicine, Atlanta, GA, USA |
| Lizette | Mendez | MPH | Boston University School of Medicine, Department of Obstetrics and Gynecology, Boston, MA, USA |
| Deborah A. | Meyers | PhD | Department of Medicine / University of Arizona Health Sciences, Tucson, AZ, USA |
| Raul A. | Montanez Valverde |
University of Miami, Department of Medicine, Miami, FL, USA | |
| Julia L. | Moore Vogel |
PhD | The Participant Center / Scripps Research, La Jolla, CA, USA |
| Shashwat D. | Nagar | MS | School of Biological Sciences, Georgia Institute of Technology, Atlanta, GA, USA |
| Kartnik | Natarajan | PhD | Columbia University Irving Medical Center, New York, NY, USA |
| Nyia L. | Noel | MD, MPH | Boston University School of Medicine, Department of Obstetrics and Gynecology, Boston, MA, USA |
| Juno | Obedin-Maliver | MD, MPH, MAS |
Stanford University School of Medicine, Department of Obstetrics and Gynecology, Palo Alto, CA, USA |
| The PRIDE Study/PRIDEnet, Palo Alto, CA, USA | |||
| Lucila | Ohno-Machado | MD, PhD | University of California San Diego Health Department of Biomedical Informatics, La Jolla, CA, USA |
| Paulina | Paul | MS | University of California San Diego Health Department of Biomedical Informatics, La Jolla, CA, USA |
| Pamala A. | Pawloski | Pharm.D. | Health Partners Institute, La Jolla, CA, USA |
| Cathryn | Peltz-Rauchman | PhD | Henry Ford Health System, Public Health Sciences, Detroit, MI, USA |
| Priscilla | Pemu | MD | Morehouse School of Medicine, Atlanta, GA, USA |
| Fornessa T. | Randal | Asian Health Coalition, Chicago, IL, USA | |
| Ana | Rescate | MBA | Stanford University School of Medicine, Department of Obstetrics and Gynecology, Palo Alto, CA, USA |
| Ana C. | Ricardo | MD, MPH, MS |
University of Illinois at Chicago, Department of Medicine, Chicago, IL, USA |
| M. Elizabeth | Ross | MD, PhD | Weill Cornell Medicine, Brain & Mind Research Institute, New York, NY, USA |
| Brittney | Roth-Manning | MPH | University of Florida, Gainesville, FL, USA |
| Madhi | Saranadasa | University of Illinois at Chicago, School of Public Health, Chicago, IL, USA | |
| Sheri D. | Schully | PhD | National Institutes of Health, All of Us Research Program, Bethesda, MD, USA |
| Ning | Shang | PhD | Columbia University Irving Medical Center, New York, NY, USA |
| Emily G. | Spencer | PhD | The Participant Center / Scripps Research, La Jolla, CA, USA |
| Cassie | Springer | MPH | Vanderbilt University Medical Center, Nashville, TN, USA |
| Alan | Stevens | PhD | Baylor Scott & White Research Institute, Dallas, TX, USA |
| Vignesh | Subbian | PhD | College of Engineering, University of Arizona, Tucson, AZ, USA |
| Amy | Tang | PhD | Henry Ford Health System, Public Health Sciences, Detroit, MI, USA |
| Rhonda K. | Trousdale | MD | NYC Health + Hospital/Harlem, Department of Medicine Columbia University Vagelos College of Physicians & Surgeons, Department of Medicine, New York, NY, USA |
| Jill | Waalen | MD, MPH | The Participant Center / Scripps Research, La Jolla, CA, USA |
| Stephen | Waring | DVM, PhD |
Essentia Institute of Rural Health, Duluth, MN, USA |
| Chunhua | Weng | PhD | Columbia University Irving Medical Center, New York, NY, USA |
| Lisa | White | MS | BI05 Institute / University of Arizona, Tucson, AZ, USA |
| Sonya | White | PMP | University of Florida, Gainesville, FL, USA |
| Nathan E. | Wineinger | PhD | Scripps Research Translational Institute, La Jolla, CA, USA |
| Chen | Yeh | MS | Northwestern University, Department of Preventive Medicine, Division of Biostatistics, Chicago, IL, USA |
| Hsueh-Han | Yeh | PhD | Henry Ford Health System, Bloomington, MN, USA |
| Paul | Zakin | Institute for Population and Precision Health, University of Chicago, Chicago, IL, USA | |
| Guohai | Zhou | PhD | Brigham & Women’s Hospital, Center for Clinical Investigation, Boston, MA, USA |
| Yanhua | Zhou | MS | The Meyers Primary Care Institute, a joint venture of Reliant Medical Group, Fallon Health, and the University of Massachusetts Medical School, Worcester, MA, USA |
| Stephan | Zuchner | MD, PhD | John P. Hussman Institute for Human Genomics, Miller School of Medicine, University of Miami; The Dr. John T. Macdonald Foundation Department of Human Genetics, Miller School of Medicine, University of Miami, Miami, FL, USA |
ACKNOWLEDGMENTS
Funding/Support:
This study was supported by National Institutes of Health grants T15LM011271, 1DP5OD029610, and OT2OD026552. The All of Us Research Program is supported (or funded) by grants through the National Institutes of Health, Office of the Director: Regional Medical Centers: 1 OT2 OD026549; 1 OT2 OD026554; 1 OT2 OD026557; 1 OT2 OD026556; 1 OT2 OD026550; 1 OT2 OD 026552; 1 OT2 OD026553; 1 OT2 OD026548; 1 OT2 OD026551; 1 OT2 OD026555; IAA #: AOD 16037; Federally Qualified Health Centers: HHSN 263201600085U; Data and Research Center: 5 U2C OD023196; Biobank: 1 U24 OD023121; The Participant Center: U24 OD023176; Participant Technology Systems Center: 1 U24 OD023163; Communications and Engagement: 3 OT2 OD023205; 3 OT2 OD023206; and Community Partners: 1 OT2 OD025277; 3 OT2 OD025315; 1 OT2 OD025337; 1 OT2 OD025276. In addition to the funded partners, the All of Us Research Program would not be possible without the contributions made by its participants.
Financial Disclosures:
No financial disclosures.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Meeting Presentation: None previously. An abstract based on this work has been submitted to the 2021 Association for Research in Vision and Ophthalmology (ARVO) Annual Meeting.
Supplemental Material: Supplemental Material available at AJO.com.
REFERENCES
- 1.Tham Y-C, Li X, Wong TY, Quigley HA, Aung T, Cheng C-Y. Global prevalence of glaucoma and projections of glaucoma burden through 2040: a systematic review and meta-analysis. Ophthalmology . 2014;121(11):2081–2090. doi: 10.1016/j.ophtha.2014.05.013 [DOI] [PubMed] [Google Scholar]
- 2.Weinreb RN, Aung T, Medeiros FA. The pathophysiology and treatment of glaucoma: a review. JAMA . 2014;311(18):1901–1911. doi: 10.1001/jama.2014.3192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Moraes CG, Cioffi GA, Weinreb RN, Liebmann JM. New Recommendations for the Treatment of Systemic Hypertension and their Potential Implications for Glaucoma Management. J Glaucoma. 2018;27(7):567–571. doi: 10.1097/IJG.0000000000000981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin H-C, Stein JD, Nan B, et al. Association of Geroprotective Effects of Metformin and Risk of Open-Angle Glaucoma in Persons With Diabetes Mellitus. JAMA Ophthalmol. 2015;133(8):915–923. doi: 10.1001/jamaophthalmol.2015.1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zheng W, Dryja TP, Wei Z, et al. Systemic Medication Associations with Presumed Advanced or Uncontrolled Primary Open-Angle Glaucoma. Ophthalmology. 2018;125(7):984–993. doi: 10.1016/j.ophtha.2018.01.007 [DOI] [PubMed] [Google Scholar]
- 6.Chung HJ, Hwang HB, Lee NY. The Association between Primary Open-Angle Glaucoma and Blood Pressure: Two Aspects of Hypertension and Hypotension. Biomed Res Int. 2015;2015. doi: 10.1155/2015/827516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Talwar N, Musch DC, Stein JD. Association of Daily Dosage and Type of Statin Agent With Risk of Open-Angle Glaucoma. JAMA Ophthalmol. 2017;135(3):263–267. doi: 10.1001/jamaophthalmol.2016.5406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dielemans I, Vingerling JR, Algra D, Hofman A, Grobbee DE, de Jong PT. Primary open-angle glaucoma, intraocular pressure, and systemic blood pressure in the general elderly population. The Rotterdam Study. Ophthalmology. 1995;102(1):54–60. [DOI] [PubMed] [Google Scholar]
- 9.Goldstein BA, Navar AM, Pencina MJ, Ioannidis JPA. Opportunities and challenges in developing risk prediction models with electronic health records data: a systematic review. Journal of the American Medical Informatics Association. 2017;24(1):198–208. doi: 10.1093/jamia/ocw042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Richter AN, Khoshgoftaar TM Efficient learning from big data for cancer risk modeling: A case study with melanoma. Comput Biol Med. 2019;110((Richter AN, arichter@fau.edu; Khoshgoftaar TM, khoshgof@fau.edu) Department of Computer&Electrical Engineering and Computer Science College of Engineering and Computer Science, Florida Atlantic University, 777 Glades Road EE 403, Boca Raton, FL, United States: ):29–39. doi: 10.1016/j.compbiomed.2019.04.039 [DOI] [PubMed] [Google Scholar]
- 11.Gordon MO, Beiser JA, Brandt JD, et al. The Ocular Hypertension Treatment Study: Baseline Factors That Predict the Onset of Primary Open-Angle Glaucoma. Arch Ophthalmol. 2002;120(6):714–720. doi: 10.1001/archopht.120.6.714 [DOI] [PubMed] [Google Scholar]
- 12.Bowd C, Goldbaum MH. Machine learning classifiers in glaucoma. Optom Vis Sci. 2008;85(6):396–405. doi: 10.1097/OPX.0b013e3181783ab6 [DOI] [PubMed] [Google Scholar]
- 13.Christopher M, Belghith A, Weinreb RN, et al. Retinal Nerve Fiber Layer Features Identified by Unsupervised Machine Learning on Optical Coherence Tomography Scans Predict Glaucoma Progression. Invest Ophthalmol Vis Sci. 2018;59(7):2748–2756. doi: 10.1167/iovs.17-23387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Medeiros FA, Weinreb RN. Predictive models to estimate the risk of glaucoma development and progression. In: Progress in Brain Research. Vol 173. Elsevier; 2008:15–24. doi: 10.1016/S0079-6123(08)01102-3 [DOI] [PubMed] [Google Scholar]
- 15.Medeiros FA, Zangwill LM, Girkin CA, Liebmann JM, Weinreb RN. Combining Structural and Functional Measurements to Improve Estimates of Rates of Glaucomatous Progression. Am J Ophthalmol. 2012;153(6):1197–205. et al. doi: 10.1016/j.ajo.2011.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Medeiros FA, Weinreb RN, Sample PA, et al. Validation of a predictive model to estimate the risk of conversion from ocular hypertension to glaucoma. Arch Ophthalmol. 2005;123(10):1351–1360. doi: 10.1001/archopht.123.10.1351 [DOI] [PubMed] [Google Scholar]
- 17.Medeiros FA, Zangwill LM, Alencar LM, et al. Detection of Glaucoma Progression with Stratus OCT Retinal Nerve Fiber Layer, Optic Nerve Head, and Macular Thickness Measurements. Invest Ophthalmol Vis Sci. 2009;50(12):5741–5748. doi: 10.1167/iovs.09-3715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Moraes CG, Sehi M, Greenfield DS, Chung YS, Ritch R, Liebmann JM. A Validated Risk Calculator to Assess Risk and Rate of Visual Field Progression in Treated Glaucoma Patients. Invest Ophthalmol Vis Sci. 2012;53(6):2702–2707. doi: 10.1167/iovs.11-7900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oh E, Yoo TK, Hong S. Artificial Neural Network Approach for Differentiating Open-Angle Glaucoma From Glaucoma Suspect Without a Visual Field Test. Invest Ophthalmol Vis Sci. 2015;56(6):3957–3966. doi: 10.1167/iovs.15-16805 [DOI] [PubMed] [Google Scholar]
- 20.Validated Prediction Model for the Development of Primary Open-Angle Glaucoma in Individuals with Ocular Hypertension. Ophthalmology. 2007;114(1):10–19.e2. doi: 10.1016/j.ophtha.2006.08.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baxter SL, Marks C, Kuo T-T, Ohno-Machado L, Weinreb RN. Machine Learning-Based Predictive Modeling of Surgical Intervention in Glaucoma Using Systemic Data From Electronic Health Records. American Journal of Ophthalmology. 2019;208:30–40. doi: 10.1016/j.ajo.2019.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sudlow C, Gallacher J, Allen N, et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12(3):e1001779. doi: 10.1371/journal.pmed.1001779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gaziano JM, Concato J, Brophy M, et al. Million Veteran Program: A mega-biobank to study genetic influences on health and disease. J Clin Epidemiol. 2016;70:214–223. doi: 10.1016/j.jclinepi.2015.09.016 [DOI] [PubMed] [Google Scholar]
- 24.Chen Z, Chen J, Collins R, et al. China Kadoorie Biobank of 0.5 million people: survey methods, baseline characteristics and long-term follow-up. Int J Epidemiol. 2011. ;40(6):1652–1666. doi: 10.1093/ije/dyr120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.The All of Us Research Program Investigators. The “All of Us” Research Program. N Engl J Med. 2019;381 (7):668–676. doi: 10.1056/NEJMsr1809937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramirez AH, Sulieman L, Schlueter DJ, et al. The All of Us Research Program: data quality, utility, and diversity. medRxiv. Published online June 3, 2020:2020.05.29.20116905. doi: 10.1101/2020.05.29.20116905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.All of Us Research Hub. Accessed May 20, 2020. https://www.researchallofus.org/
- 28.Methods – All of Us Research Hub. Accessed May 20, 2020. https://www.researchallofus.org/methods/
- 29.Baxter S Data Extraction and Cleaning Notebook. Systemic Disease and Glaucoma Workspace, https://workbench.researchallofus.org/workspaces/aou-rw-1c4fddc2/duplicateofsystemicdiseaseandglaucoma/notebooks/preview/1_Data%20Extraction%20and%20Cleaning.ipynb [Google Scholar]
- 30.Systemic Disease and Glaucoma Workspace. https://workbench.researchallofus.org/workspaces/aou-rw-1c4fddc2/duplicateofsystemicdiseaseandglaucoma/data
- 31.Baxter S Validation of Single-Center Model Notebook. Systemic Disease and Glaucoma Workspace, https://workbench.researchallofus.org/workspaces/aou-rw-1c4fddc2/duplicateofsystemicdiseaseandglaucoma/notebooks/preview/2_Validation%20of%20Single-Center%20Model.ipynb [Google Scholar]
- 32.Pedregosa F, Varoquaux G, Gramfort A, et al. Scikit-learn: Machine Learning in Python. Journal of Machine Learning Research. 2011; 12:2825–2830. [Google Scholar]
- 33.Chollet F, others. Keras.; 2015. https://keras.io [Google Scholar]
- 34.Glaucoma, Open-angle | National Eye Institute. Accessed March 2, 2019. https://www.nei.nih.gov/eyedata/glaucoma
- 35.Tielsch JM, Sommer A, Katz J, Royall RM, Quigley HA, Javitt J. Racial variations in the prevalence of primary open-angle glaucoma. The Baltimore Eye Survey. JAMA. 1991. ;266(3):369–374. [PubMed] [Google Scholar]
- 36.Varma R, Ying-Lai M, Francis BA, et al. Prevalence of open-angle glaucoma and ocular hypertension in Latinos: the Los Angeles Latino Eye Study. Ophthalmology. 2004;111(8): 1439–1448. doi: 10.1016/j.ophtha.2004.01.025 [DOI] [PubMed] [Google Scholar]
- 37.Leske MC, Connell AMS, Schachat AP, Hyman L. The Barbados Eye Study: Prevalence of Open Angle Glaucoma. Arch Ophthalmol. 1994;112(6):821–829. doi: 10.1001/archopht.1994.01090180121046 [DOI] [PubMed] [Google Scholar]
- 38.Collins GS, de Groot JA, Dutton S, et al. External validation of multivariable prediction models: a systematic review of methodological conduct and reporting. BMC Medical Research Methodology. 2014;14(1):40. doi: 10.1186/1471-2288-14-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mitchell P, Lee AJ, Rochtchina E, Wang JJ. Open-angle glaucoma and systemic hypertension: the blue mountains eye study. J Glaucoma. 2004;13(4):319–326. [DOI] [PubMed] [Google Scholar]
- 40.Leske MC, Wu S-Y, Hennis A, Honkanen R, Nemesure B, BESs Study Group. Risk factors for incident open-angle glaucoma: the Barbados Eye Studies. Ophthalmology. 2008;115(1):85–93. doi: 10.1016/j.ophtha.2007.03.017 [DOI] [PubMed] [Google Scholar]
- 41.The SPRINT Research Group. A Randomized Trial of Intensive versus Standard Blood-Pressure Control. New England Journal of Medicine. 2015;373(22):2103–2116. doi: 10.1056/NEJMoa1511939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chiang MF, Boland MV, Brewer A, et al. Special requirements for electronic health record systems in ophthalmology. Ophthalmology. 2011;118(8):1681–1687. doi: 10.1016/j.ophtha.2011.04.015 [DOI] [PubMed] [Google Scholar]
- 43.Read-Brown S, Hribar MR, Reznick LG, et al. Time Requirements for Electronic Health Record Use in an Academic Ophthalmology Center. JAMA Ophthalmol. 2017;135(11):1250–1257. doi: 10.1001/jamaophthalmol.2017.4187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baxter SL, Gali HE, Mehta MC, et al. Multi-center analysis of electronic health record use among ophthalmologists. Ophthalmology. Published online June 2020:S0161642020305236. doi: 10.1016/j.ophtha.2020.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parikh RB, Kakad M, Bates DW. Integrating Predictive Analytics Into High-Value Care: The Dawn of Precision Delivery. JAMA. 2016;315(7):651–652. doi: 10.1001/jama.2015.19417 [DOI] [PubMed] [Google Scholar]
- 46.Shaw J, Rudzicz F, Jamieson T, Goldfarb A. Artificial Intelligence and the Implementation Challenge. J Med Internet Res. 2019;21(7). doi: 10.2196/13659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.He J, Baxter SL, Xu J, Xu J, Zhou X, Zhang K. The practical implementation of artificial intelligence technologies in medicine. Nat Med. 2019;25(1):30–36. doi: 10.1038/s41591-018-0307-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee TC, Shah NU, Haack A, Baxter SL. Clinical Implementation of Predictive Models Embedded within Electronic Health Record Systems: A Systematic Review. Informatics. 2020;7(3):25. doi: 10.3390/informatics7030025 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


