Abstract
Chromatin remodelers act to regulate multiple cellular processes, such as transcription and DNA repair, by controlling access to genomic DNA. Four families of chromatin remodelers have been identified in yeast, each with non-redundant roles within the cell. There has been a recent surge in structural models of chromatin remodelers in complex with their nucleosomal substrate. These structural studies provide new insight into the mechanism of action for individual chromatin remodelers. In this review, we summarize available data for the structure and mechanism of action of the four chromatin remodeling complex families.
Keywords: Chromatin remodeling, ISWI, SWI/SNF, INO80, CHD
Introduction
Essential processes such as transcription, DNA replication, and repair depend on DNA being accessible to the protein complexes that initiate and conduct their respective processes. A group of ATP-dependent chromatin remodelers serves to regulate DNA accessibility by repositioning, ejecting, or modifying nucleosomes. This important class of enzymes ensures the proper positioning of nucleosomes to allow DNA-centric processes to occur and represent another level of regulation. Eukaryotic cells contain four families of chromatin remodelers, which are categorized based on the similarities and differences of the ATPase subunits [1], including switch/sucrose non-fermentable (SWI/SNF), imitation switch (ISWI), chromodomain helicase DNA-binding (CHD), and INOsitol requiring 80 (INO80).
Each family of chromatin remodelers carries out specialized functions within the cell. SWI/SNF remodelers, which include the SWI/SNF and RSC complexes, establish nucleosome depleted regions [2] as well as position the +1 nucleosome for transcription initiation through nucleosome sliding and ejection [3, 4]. ISWI remodelers, Isw1a, Isw1b, and Isw2 complexes, and CHD function in nucleosome maturation and spacing to create nucleosomal arrays with fixed distances [5]. INO80 family chromatin remodelers, INO80 and SWR1, play a role in nucleosome editing by exchanging histone variants [6, 7].
In recent years a considerable amount of biochemical, biophysical, and genomic work has been done to determine the mechanism of chromatin remodeling and how remodelers serve to regulate cellular processes. All four families of remodelers are highly conserved throughout evolution, but we will focus mainly on the yeast chromatin remodelers. In this review, we will focus on the most recent high-resolution cryo-electron microscopy (cryo-EM) structures of the various remodelers and the current mechanistic understanding of remodeler function.
Structure of chromatin remodeling complexes
Recent cryo-EM studies have provided high-resolution information about the interaction of chromatin remodeler complexes with their nucleosomal substrate. This can provide insight into the organization and mechanism of action of the different families of chromatin remodelers. Here we review the currently available structural models of yeast chromatin remodelers.
SWI/SNF
The SWI/SNF complex can be divided into three main modules – the ATPase, Actin-Related Protein (ARP), and Body modules (Figure 1A, Table 1) [8]. The ATPase module includes Snf2 and, as the name suggests, is responsible for coupling ATP hydrolysis to DNA translocation. Similar to other remodelers, the ATPase domain of Snf2 is made up of two RecA-like domains connected by a pair of brace helices [8, 9]. Snf2 interacts with the exposed surface of nucleosomal DNA at superhelical location 2 (SHL 2) through the cleft formed by the RecA-like domains (Figure 1A). Basic residues of the DNA binding cleft interact with the phosphate backbone of DNA, which can explain its ability to bind in a sequence-independent manner [10]. This mode of Snf2 binding to the nucleosome is similar in structures of Snf2 alone and in the context of the full SWI/SNF complex [8,9,11]. In addition to the ATPase domain, Snf2 also harbors a C-terminal bromodomain that increases binding affinity to acetylated nucleosomes [12, 13]. Previous studies of the homologous human BRG1 showed a canonical four-helix bundle bromodomain with moderate affinity for acetylated histone H3 and H4 tails [14]. The Snf2 bromodomain is unresolved in SWI/SNF cryo-EM structures, likely due to flexibility of histone tails or heterogenous post-translational modifications of the nucleosome substrate used in most structural studies to date.
Figure 1.
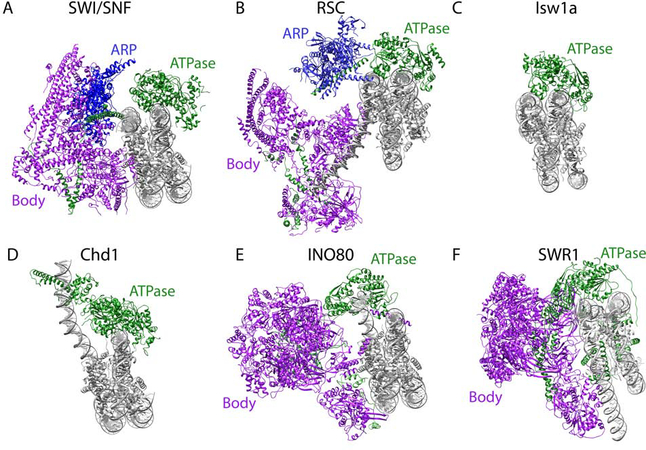
Architecture of chromatin remodelers. Structural models of A) SWI/SNF (PDB 6UXW), B) RSC (PSB 6TDA) C) Isw1a (PDB 6JYL), D) Chd1 (PDB 5O9G), E) INO80 (PDB 6FML), and F) SWR1 (PDB 6GEN). Each structural model is colored to denote structural features – nucleosome (grey), ATPase module (green), ARP module (blue), Body module (purple).
Table 1.
Composition of chromatin remodelers. Each remodeler is organized into their respective family and broken down into the modules highlighted in Fig 1. Each protein within a complex is organized into the module(s) that it is most commonly associated with, even though some proteins span multiple modules.
| Family | Complex | Module | Protein | ||||||
|---|---|---|---|---|---|---|---|---|---|
| SWI/SNF | SWI/SNF | ATPase | Snf2 | ||||||
| ARP | Snf2 | Rtt102 | Arp7 | Arp9 | |||||
| Body | Swi1 | Snf5 | Swi3 | Snf12/Swp73 | Snf6 | Swp82 | |||
| RSC | ATPase | Sth1 | |||||||
| ARP | Sth1 | Rtt102 | Arp7 | Arp9 | |||||
| Body | Sfh1 | Rsc7 | Rsc8a/b | Rsc9 | Rsc58 | Rsc6 | Rsc4 | ||
| Rsc2 | Rsc3 | Rsc30 | Htl1 | ||||||
| ISWI | ISW1a | ATPase | Isw1 | ||||||
| Body | Ioc3 | ||||||||
| ISW1b | ATPase | Isw1 | |||||||
| Body | Ioc2 | Ioc4 | |||||||
| ISW2 | ATPase | Isw2 | |||||||
| Body | Itc1 | ||||||||
| CHD | CHD1 | ATPase | Chd1 | ||||||
| INO80 | ATPase | Ino80 | Ies2 | ||||||
| ARP | Ino80 | Arp4 | Arp8 | N-actin | Nhp10 | ||||
| Body | Ruvb1 | Ruvb2 | Arp5 | Ies2 | Ies1 | Ies3 | Ies4 | ||
| Ies5 | Ies6 | Taf14 | |||||||
| SWR1 | ATPase | Swr1 | |||||||
| ARP | Swr1 | Arp4 | N-actin | Swc4 | Swc7 | Yaf9 | Bdf1 | ||
| Body | Arp6 | Ruvbl1 | Ruvbl2 | Swc2 | Swc3 | Swc5 | Swc6 | ||
The ARP module includes the Snf2 HSA (helicase-SANT associated) domain, Rtt102, Arp7, and Arp9, and acts to support and coordinate the ATPase module with the Body module (Table 1). The Snf2 HSA domain forms a long, single helix between the ATPase domain and the N-terminus of Snf2, which is within the Body module (Figure 1A). Arp7 and Arp9 pack against and straddle the Snf2 HSA domain. Rtt102 binds to one side of the HSA-Arp7/9 subcomplex to further stabilize the ARP module [15].
The Body module of SWI/SNF consists of Swi1, Snf5, Swi3, Snf12/Swp73, Snf6, and Swp82 [Table 1]. The Body is the largest module of the SWI/SNF complex and packs against the opposite face of the nucleosome from the ATPase module, creating additional nucleosome interactions with histone- and DNA-interacting subunits. These additional nucleosome interactions are suggested to play an important role in substrate recognition and anchoring the octamer during DNA translocation. One essential histone binding protein is Snf5, which interacts with the acidic patch of the histone octamer. Loss of Snf5 uncouples ATP hydrolysis from DNA translocation [16], indicating an important role in anchoring the octamer as DNA slides around the nucleosome. Swi1, which acts as a scaffold for the Body module, also includes an AT-rich interaction domain (ARID). Previous work has shown that Swi1 ARID interacts with AT-rich DNA sequences [17], and structural studies of this domain in humans revealed a helix-turn-helix motif that interacts with the major groove of DNA [18]. Swi1 ARID is unresolved in the cryo-EM structure, likely due to the conformational flexibility of its interaction with DNA, and it is unknown how this domain engages DNA when bound to the nucleosome. Other subunits work to act as a scaffold for assembly or further stabilize the Body module. Two long helices of Snf12 and the coiled-coil domains of Swi3 form a four-helix bundle within the Body, likely playing a role in early complex assembly and transcription regulation [8, 19, 20]. Swi3 also contains two SANT domains that anchor and stabilize Snf2 within the complex. Snf6 and Swp82 are yeast specific subunits that occupy peripheral regions of the Body module and are believed to further stabilize the complex [8].
RSC
The cryo-EM structure of RSC, another member of the SWI/SNF family, bound to the nucleosome reveals a similar overall architecture when compared to SWI/SNF containing corresponding ATPase, ARP, and Body modules [21–23] (Figure 1B, Table 1). Sth1, an Snf2 paralog, forms the ATPase module with C-terminal RecA-like domains and a classical bromodomain that interacts with acetylated histone peptides [1,24]. Similar to Snf2, Sth1 engages its nucleosome substrate at SHL 2. However, one of the ATPase lobes of Sth1 also engages the second DNA gyre near SHL 6 [22].
Remarkably similar to the SWI/SNF ARP module, the Sth1 HSA helix, Arp9, Arp7, and Rtt102 form the RSC ARP module. The ARP module bridges the ATPase and Body modules, allowing for coupling and regulation of ATP hydrolysis and DNA translocation [22, 25]. Very similar to the SWI/SNF ARP module, Arp7 and Arp9 pack against the HSA helix of Sth1 with Rtt102 wrapping around one face of Arp 7 and Arp9. A recent study suggests that binding of Arp7 and Arp9 to the Sth1 HSA domain induces proper folding of HSA [22].
The Body module of RSC consists of Sfh1 (a Snf5 paralog), Rsc7, Rsc8a/b (a Swi3 paralog), Rsc9, Rsc58, Rsc6 (a Swp73 paralog), Rsc2, Rsc4, Rsc3, Rsc30, and Htl1 (Table 1], which interacts with the face of the nucleosome opposite from the ATPase module. In the Body module, Sfh1 interacts directly with the H2A/H2B acidic patch anchoring the Body to the core histone octamer, serving a similar function as Snf5 in the SWI/SNF complex. The RSC Body module harbors histone tail binding domains, including bromodomains in Rsc2 (known to interact with H3 tails), Rsc4 (which binds acetylated H3 tails), and Rsc58. The Body module also contains various DNA-binding domains, including zinc clusters in Rsc3/30 that are unresolved in the cryo-EM structure, an RFX domain in Rsc9, and a zinc-finger domain in Rsc8 [26–28]. However, it is not fully understood how these histone tail and DNA binding domains contribute to the positioning of the RSC complex and sensing post-translational modifications on the nucleosome. A major difference between the SWI/SNF and RSC Body modules is that RSC is also shown to bind DNA that has exited the nucleosome (Figure 1A, B). Rsc3/30, which are known to recognize DNA elements within promoters, are included in the region of the Body that binds DNA upstream of SHL 7 [29]. This DNA binding site likely contributes to the high affinity that RSC has for nucleosomes flanking nucleosome depleted regions (NDRs).
ISWI
Currently, high-resolution cryo-EM structures of ISWI complexes bound to nucleosome are limited to the Iswi1 ATPase subunit (Figure 1C, Table 1). The catalytic subunits of the ISWI family of remodelers, Isw1 or 2, contain N-terminal RecA-like helicase domains, a C-terminal HAND-SANT-SLIDE (HSS) domain, and a regulatory auto-inhibition domain. The RecA-like domains pack together to form the ATPase domain, as seen in other chromatin remodeling complexes. The cryo-EM structure of yeast Isw1 shows that the ATPase domain engages nucleosomes at SHL 2 in a similar manner to Snf2 (Figure 1C) [30,31]. The C-terminus of Isw1 harbors a four-helix HAND domain followed by a SANT domain and SLIDE domain, referred to as the HSS domain, that is involved in nucleosome substrate binding. THE SANT and SLIDE domains are known to bind DNA. Furthermore, deletion of the SANT-SLIDE domains leads to decreased remodeling by Isw1 [32]. This effect may be due to the HSS-DNA interaction preventing the C-terminal negative regulator (NegC) from folding back on the helicase domain.
A comparison of the structures of Isw1 alone or in complex with the histone H4 tail demonstrates the mechanism of autoinhibition [33]. The N-terminal auto inhibition domain (AutoN) binds to RecA-like helicase in the absence of nucleosomal substrate, locking it into an inactive conformation. Histone H4 tails compete with AutoN to bind to the negatively charged surface of the second lobe of the RecA-like ATPase [34]. Additionally, NegC binds to one lobe of Isw1, preventing the helicase domain from coming together into the active conformation. When Isw1 is bound to DNA, NegC is prevented from folding back and inhibiting the activation of the helicase domain. Together these suggest a mechanism whereby Isw1 is inhibited until it binds to its nucleosomal substrate. The H4 tails and DNA compete with autoinhibitory domains, allowing Isw1 to adopt its active conformation.
Individual ISWI complexes are composed of a catalytic subunit in addition to accessory proteins that affect their function [35] (Figure 1C, Table 1). Isw2 forms a single complex consisting of Isw2 and Itc1. Isw1 is included in two different complexes, Isw1a and Isw1b, that are differentiated by their accessory subunits. Isw1a includes Ioc3, and Isw1b includes Ioc2 and Ioc4. A previous study showed that the Isw1 HSS domain interacts with Ioc3, revealing an extensive interface between a hydrophobic pocket of Ioc3 and the HSS SLIDE domain [32]. Low-resolution cryo-EM studies indicate that Isw1 HSS and Ioc3 interact with the nucleosome dyad within mono-nucleosomes. A recent study revealed that Isw1a preferentially binds and more efficiently remodels dinucleosomes, likely due to Ioc3 not binding the dyads of dinucleosomes [36]. When Isw1a binds mono-nucleosomes, Ioc3 extensively binds to the lateral face and acidic patch of the nucleosome. However, when binding di-nucleosomes, it is Isw1 that binds the lateral face and acidic patch. The Isw1b auxiliary subunits, Ioc4 and Ioc2, contribute to nucleosome binding through interactions with methylated histones [37,38]. Further work is required to better understand how Ioc3, Itc1, Ioc2, and Ioc4 contribute to complex assembly, substrate binding, and DNA translocation in the context of nucleosome arrays.
CHD
CHD chromatin remodelers are known to interact with elongation and chromatin-modifying factors, such as Paf1, FACT, and SAGA [39–41]. Whereas the previously reviewed chromatin remodelers form their own multi-subunit complexes, CHD proteins appear to coordinate with multiple chromatin-modifying complexes. The details of CHD protein interactions with these complexes remain unclear. However, recent cryo-EM studies provide insight into the interaction between Chd1, the only CHD protein in S. cerevisiae, and its nucleosome substrate [42,43].
The Chd1 ATPase interacts with the nucleosome between DNA gyres at SHL 2 in a similar manner as Isw1 and Snf2 (Figure 1D, Table 1). Additional DNA contacts occur involving the N-terminal double chromodomains and the C-terminal SANT-SLIDE domains with SHL 1 and extranucleosomal DNA, respectively, introducing a kink in the DNA trajectory and unraveling DNA from the octamer [42,43]. Minimal engagement occurs between Chd1 and histones, although the ATPase domain of Chd1 does interact with the first alpha-helix of histone H3 and the histone H4 tail. It is likely that subunits of associated factors (i.e., FACT, SAGA, etc.) engage in extensive contact with the histone octamer and assist in DNA translocation. Further studies of Chd1 potential interactions with these factors are necessary to understand the underlying mechanism by which they cooperate.
INO80
The cryo-EM structure of the C. thermophilum INO80 complex was solved in complex with nucleosome [44] (Figure 1E) and displays a unique architecture when compared to SWI/SNF and RSC chromatin remodelers. Like SWI/SNF and RSC, INO80 can be organized into the ATPase, ARP, and Body modules (Table 1). Although the Ino80 ATPase domain shares a similar structure with the ATPase domains discussed above, it interacts with the nucleosome at a unique position, SHL 6. Ies2 is also part of the INO80 ATPase module and wraps around nucleosomal DNA to bind SHL 2, providing a more extensive DNA binding interface than is observed in SWI/SNF and RSC. The INO80 family of remodelers is also known for a long linker, often referred to as the INO80 insertion, between the ATPase lobes of the catalytic subunits. The Ino80 insertion spans most of the complex, engaging in specific interactions with Ruvb1/Ruvb2, Ies2, Arp5, and Ies6 [44].
The ARP module of INO80 consists of Ino80 HSA domain, Arp4, Arp8, Nhp10, and N-actin. This module is unresolved within the cryo-EM structure of the INO80-nucleosome complex, although additional density suggests it binds to extranucleosomal DNA [44]. The crystal structure of the ARP module alone contains a long, helical HSA domain providing a binding scaffold for additional subunits of the module [45]. It was further reported that the ARP module preferentially binds to extranucleosomal DNA and that this interaction is mediated by basic residues of the HSA helix along with the N-terminus of Arp8 and the C-terminus of Arp4 [46]. This contrasts with SWI/SNF and RSC ARP modules, which do not appear to interact with extranucleosomal DNA and are positioned away from the nucleosome dyad. Arp8 shows a strong preference for binding H3-H4 tetramers suggesting that Arp8 plays a role in maintaining nucleosome recognition [47].
Ruvb1/Ruvb2 make up the core of the Body module, which also includes Arp5, Ies2, Ies4, Ies6, and Taf14. The yeast INO80 complex also consists of Ies1, 3, and 5. Arp5 binds the H2A/H2B acidic patch on the face of the nucleosome proximal to the Body module, while Ies2 wraps around the nucleosome to bind the distal H2A/H2B acidic patch [44]. Additional contacts occur between Ies6 and the H2B C-terminal helix. Arp5 and Ies6 also bind DNA at SHL 2 and 3, respectively, forming an interface on the opposite side of the H2A/H2B dimer from the ATPase domain. This unique mode of binding to a nucleosomal substrate is more extensive than that found in SWI/SNF and RSC, suggesting a distinct mechanism of chromatin remodeling.
SWR1
SWR1, a member of the INO80 family, is remarkably similar to the INO80 complex in its overall architecture and can also be organized into the ATPase, ARP, and Body modules [48] (Figure 1F, Table 1). The ATPase module includes Swr1, which consists of an ATPase domain, an HSA domain, and an insert between the two ATPase lobes that is characteristic of INO80 remodelers. Unlike Ino80, but similar to other ATPase subunits discussed above, the Swr1 ATPase lobes bind nucleosomes at SHL 2. A unique feature of Swr1 binding at SHL 2 involves a displacement of the bound DNA that results in a significant rotation of Swr1, allowing the first ATPase lobe to also make extensive contacts with the second DNA gyre. Binding of the second ATPase lobe is also associated with significant DNA distortion due to alpha helices within the second lobe being pushed against the bound DNA [48]. Unlike other remodelers, Swr1 only translocates DNA near SHL 2 and is unable to further propagate DNA to reposition nucleosomes, likely due to the tighter interactions made by the first ATPase lobe [49,50].
Although the densities of the ARP module are disordered in the SWR1-nucleosome cryo-EM structure, the ARP module consists of the N-terminus of Swr1, N-actin, Arp4, Swc4, Swc7, Yaf9, and Bdf1. The Swr1 N-terminus is necessary to direct the association of Arp4, Swc4, Swc7, Yaf9, and Bdf1, suggesting that it is the primary scaffold within the ARP module [51]. Bdf1 is known to bind acetylated lysines on the histone H4 tail, possibly contributing to the recruitment of SWR1 to +1 nucleosomes [52,53]. The N-terminus of Swr1 also contains a H2AZ-H2B binding site, which likely plays a role in regulating SWR1 histone exchange activity.
The Body module of SWR1 is very similar to INO80 and consists of Ruvb1, Ruvb2, Swc2, Arp6, Swc3, Swc5, and Swc6 [48] (Figure 1E, F). Arp6 contains an actin fold that interacts with Swc6 to form a heterodimer very similar to the Arp5/Ies6 dimer formed in INO80. The Arp6/Swc6 heterodimer connects to H2A and linker DNA on the opposite side of the Swr1 nucleosome binding site (Figure 1F). Swc6 also contains a hydrophobic core that allows for further interactions between Swc6 and H2A. Swc2 spans a large part of the Body and extends into the ATPase module, similar to Ies2 in INO80, creating an extensive interface for connections within the SWR1 complex as well as additional binding to the nucleosome acidic patch and linker DNA [48,54]. The acidic N-terminus of Swc5 binds H2A-H2B dimers, likely contributing to nucleosome recognition and regulation of histone exchange [55]. The RuvBL ATPase ring is believed to mainly play an architectural role within the SWR1 complex, as it does in INO80, although the ATP binding sites within the RuvBL ring are occupied by nucleotides in the cryo-EM structure. The structural similarities among the various remodelers, especially within the ATPase modules, help explain the common DNA translocating mechanism shared across the families. While the structural differences, especially within the ARP and Body modules, shed some light on the differences in overall function among remodeler families.
Mechanism of Chromatin Remodeling
Cryo-EM structures of Snf2, the ATPase in SWI/SNF, provide insight into the mechanism by which DNA is translocated around the histone octamer [9, 11]. The shared mechanism of DNA translocation by ATP-dependent chromatin remodelers has been extensively reviewed and illustrated previously [56–58]. Data suggest that the ATPase subunit induces a ~1 base pair bulge at SHL 2 on the tracking strand of DNA in the absence of nucleotide or with ADP. This advances the tracking strand of DNA (oriented 5’ to 3’ towards the dyad) forward in relation to the histone octamer while the rest of the DNA remains in position. The addition of a stable ATP analog leads to a closed state of the ATPase domain where DNA is not strained by a 1bp bulge. This suggests a conformational cycle whereby SWI/SNF remodelers slide a DNA wave that is propagated along the histone octamer. In this model, Snf2-like ATPases engage nucleosomal DNA at SHL 2 and induce a 1base pair bulge in the DNA binding cleft. A conformational change occurs upon ATP binding where the ATPase domain grabs and pushes the bulging DNA strand translocating DNA one base pair toward the exit site of the nucleosome. ATP hydrolysis resets the ATPase domain to release the DNA strand and introduce another one base pair DNA bulge at SHL 2. ADP is then released and the cycle is reset. This model is consistent with smFRET studies of the RSC complex that shows the DNA at both the nucleosome entry and exit sites do not significantly lift from the histone octamer, further supporting the creation of a bulge within the nucleosome and the observation that the RSC ATPase remains at a fixed site on the nucleosome during remodeling [59,60]. This mechanism would keep most of the nucleosomal DNA along its canonical path around the histone octamer, which can help the H2A-H2B dimer to remain mostly undisturbed and keep the octamer intact [59]. However, SWI/SNF remodelers are also able to eject H2A-H2B dimers or the full octamer [61,62]. H2A-H2B dimer ejection can potentially act as a means for increasing DNA accessibility while maintaining regulation offered by the presence of a partial nucleosome. Full octamer ejection is likely a more efficient means of nucleosome reorganization at sites, such as promoters, that would require substantial sliding to achieve adequate nucleosome depletion [62,63].
Cryo-EM structures show a conserved mode of binding between the SWI/SNF complex and the ATPase domains of Isw1, Chd1, Sth1, and Swr1 [8, 30, 43]. Furthermore, the same conformational cycle of DNA distortion is observed in the structure of Isw1, wherein a one base pair bulge is introduced in the ADP bound state but not present in the ADP-BeFx structure [31]. Although a high-resolution structure of Isw2 is not available, biochemical evidence supports a similar mode of nucleosome binding and cyclical ATP-dependent DNA translocation mechanism described above [64–66]. Chd1 ATPase also adopts a closed conformation upon the addition of a non-hydrolyzable ATP analog [43], although currently there is no high-resolution structure available for the ADP or nucleotide-free states. However, chemical crosslinking experiments of Chd1 identified a similar one base pair step DNA translocation cycle [67]. smFRET experiments of Chd1 and Isw1a also showed that movement first occurs at the entry-side DNA with a delay of movement at the exit-side DNA [68]. This suggests a similar mechanism of DNA translocation among these chromatin remodelers as found in the SWI/SNF family. ISWI and CHD remodelers are known for globally establishing equally spaced nucleosome arrays that heavily rely on the ability to bind extranucleosomal DNA [35,69–72]. In vitro studies have shown that sensing extranucleosomal DNA is critical for the ability of ISWI and CHD remodelers to bind and reposition nucleosomes [71,73–75]. Adjacent nucleosomes move closer together, as the ISWI and CHD ATPases translocate DNA around the histone octamer. Eventually, the extranucleosomal DNA between adjacent nucleosomes becomes too short for ISWI and CHD to bind, resulting in equally spaced nucleosomes.
As stated previously, INO80 has a unique binding mode with nucleosomes when compared to other chromatin remodelers. A similar cyclic mechanism of ATP binding and hydrolysis is used as revealed by cryo-EM structures of the yeast, fungus, and human INO80 complexes showing similar conformational states for the nucleotide-free and ATP analog bound complexes when compared to other remodelers [44,76]. The unique interaction of INO80 with the nucleosome instead affects the mechanism of DNA translocation and H2A-H2B dimer exchange [6,59,77]. Unlike the SWI/SNF ATPases, Ino80 induces unwrapping of nucleosomal DNA very close to the nucleosome entry site and disrupts DNA-histone interactions to partially expose H2AZ-H2B dimers [44,45]. Although it has not been directly observed, the INO80-nucleosome structure suggests that DNA could be looped out in a ratchet-like manner during DNA translocation and to mediate histone exchange. Binding sites at SHL 2 and SHL 6 would allow for the ATPase to pump DNA across the octamer toward Arp5 without DNA moving on the exit side. This loop could facilitate H2A-H2B exchange by transiently exposing the H2AZ-H2B dimer prior to the release of DNA by Arp5. A large loop of DNA within the nucleosome would require the Ino80 motor to remain in place while DNA is being looped. The Ino80 motor is anchored by Ies2, Ruvb1/Ruvb2, Arp5, and Ies6, which bind the H2A-H2B acidic patch, SHL 2, and the Ino80 insertion, as DNA is pumped into the nucleosome [44]. Arp5 undergoes a conformational change that is believed to block DNA from exiting the nucleosome, likely playing a role in regulating histone exchange. Ies2 and Arp5 bind the nucleosome acidic patch at opposite sides of the nucleosome to stabilize the H2A histones as the entry DNA is being unwrapped and looped within the nucleosome. Binding of Arp5 to the nucleosome acidic patch is partially regulated by Arp8 and Arp4, which have been shown to be critical in coupling the sensing of extranucleosomal DNA to nucleosome positioning [45,46]. Notably, the INO80 complex is recruited to and helps position the +1 nucleosome [78,79]. The DNA translocation step is completed upon release of DNA by Arp5, allowing the DNA loop to exit and reposition the nucleosome in a relatively large step, ~15 bp, when compared to other chromatin remodelers. [44,77].
The ATPase subunit of SWR1 binds to nucleosome in a similar manner as Snf2, Sth1, Isw1, and Chd1[8,21,22,30,42,43]. However, the mechanism of DNA distortion within the nucleosome caused by Swr1 is slightly different and does not result in net DNA translocation. While Swr1 binds the nucleosome at SHL 2, the binding is tighter due to a rotation in the first ATPase lobe allowing Swr1 to interact with both DNA gyres. The second ATPase lope causes DNA distortion at SHL 2, like the subunits mentioned above, but the distortion is caused by an alpha helix pushing against the DNA and significantly changes the path of the DNA when compared to the Chd1 structure. Similar to the SWI/SNF, ISWI, and CHD remodelers, the ability of SWR1 to translocate DNA within the nucleosome is dependent on ATP binding [48]. The differences in binding and DNA distortion between SWR1 and the other families of remodelers likely play a role in histone exchange and preventing significant DNA translocation out of the nucleosome. Unlike INO80, SWR1 is able to function as a monomer. This ability is potentially due to an H2A.Z binding site being present at the N-terminal domains of Swr1 and Swc2 while Arp6 and Swc6 bind H2A-H2B, allowing for one SWR1 complex to differentiate and bind both H2A and H2A.Z [48,80]. It is known that SWR1 exchanges H2A-H2B for H2A.Z-H2B dimers in a stepwise fashion, and the ATPase activity of SWR1 is enhanced by the presence of H2A containing nucleosomes and free H2A.Z-H2B dimers [48,81]. Although the exact mechanism of histone exchange by SWR1 is not fully understood, a model can be proposed based on recent biophysical and biochemical studies. An SWR1 monomer binds H2A containing nucleosome at SHL 2, and ATP binding causes DNA distortion and some translocation around SHL 2 [48,49]. Binding of H2A and free H2A.Z-H2B dimers enhance the ATPase activity of Swr1, and ATP hydrolysis leads to the exchange of one H2A-H2B dimer for H2A.Z-H2B. A second round of SWR1 binding and ATP hydrolysis on the heterotypic nucleosome result in the other H2A-H2B dimer being exchanged, creating a homotypic H2A.Z nucleosome [81].
Cryo-EM and single-molecule studies support a common mechanism of DNA translocation by most ATPase domains of chromatin remodelers, despite differences in how they engage the nucleosome. A unique translocation mechanism by INO80 is likely due to the difference in ATPase binding and regulation by accessory proteins within the Body module. Additional investigation of the Arp and Body modules is needed to understand the mechanism by which ISWI, CHD, and INO80 complexes display novel remodeling activities, such as maintaining nucleosome spacing and exchanging histones.
Perspective
The recent influx of cryo-EM structures of chromatin remodelers has provided crucial insight into their function and mechanism of action. Although models are available for their ATPase domains on the nucleosome, we currently lack high-resolution structures of ISWI family remodelers. More complete models of the INO80 remodelers that include high-resolution density for the Arp modules are also needed. Future studies of the fully intact complexes can explain the role of accessory proteins in remodeling complex function. This can help to explain the unique functions of ISWI, CHD, and INO80 complexes within the cell.
Previous structural and single-molecule studies have presented a unified mechanism of DNA translocation by the ATPase domain in chromatin remodeler complexes. The differences in complex localization and function are instead determined by additional non-catalytic subunits. This raises fundamental questions such as: How do chromatin remodelers sense DNA linker length? What is the molecular mechanism by which nucleosome spacing is maintained? How do histone chaperones cooperate with chromatin remodelers to accomplish specific histone exchange?
Research Highlights.
High-resolution structures of chromatin remodelers explain the mechanism by which individual remodelers interact with the nucleosome and translocate DNA. There are shared features across families of chromatin remodelers, such as the structure of the RecA-like ATPase domain and key contacts between accessory proteins and the H2A-H2B acidic patch.
The ATPase domain of chromatin remodelers displays a shared mechanism of DNA translocation throughout different families. A cyclical conformational change occurs, which introduces a one base pair bulge in nucleosomal DNA and induces a DNA wave that translocates DNA across the nucleosome.
Acknowledgments:
This work was supported by R01GM135651, P01CA092584, and U54CA193419 from NIH. A. A. Reyes is supported by the Molecular Biophysics Training Program from NIGMS/NIH (5T32 GM008382).
Abbreviations
- SWI/SNF
Switch/Sucrose non-fermentable
- RSC
Remodeling Structure of Chromatin
- ISWI
Imitation Switch
- CHD
Chromodomain Helicase DNA-binding
- INO80
INOsitol Requiring 80
- SWR1
SWI2/SNF2-Related 1 Chromatin Remodeling
- Cryo-EM
Cryogenic Electron Microscopy
- ARP
actin-related protein
- AutoN
auto inhibition domain
- NegC
C-terminal negative regulator
- HSA
helicase-SNAT associated
- TSS
transcription start site
- NDR
nucleosome depleted region
Footnotes
Conflicts of Interest: The authors declare no conflict of interest.
Declaration of interests
☒ The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Flaus A, Martin DM, Barton GJ, Owen-Hughes T. Identification of multiple distinct Snf2 subfamilies with conserved structural motifs. Nucleic Acids Res 2006;34:2887–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Sudarsanam P, Winston F. The Swi/Snf family nucleosome-remodeling complexes and transcriptional control. Trends Genet 2000;16:345–51. [DOI] [PubMed] [Google Scholar]
- [3].Kubik S, Bruzzone MJ, Challal D, Dreos R, Mattarocci S, Bucher P, et al. Opposing chromatin remodelers control transcription initiation frequency and start site selection. Nat Struct Mol Biol 2019;26:744–54. [DOI] [PubMed] [Google Scholar]
- [4].Rawal Y, Chereji RV, Qiu H, Ananthakrishnan S, Govind CK, Clark DJ, et al. SWI/SNF and RSC cooperate to reposition and evict promoter nucleosomes at highly expressed genes in yeast. Genes Dev 2018;32:695–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Clapier CR, Iwasa J, Cairns BR, Peterson CL. Mechanisms of action and regulation of ATP-dependent chromatin-remodelling complexes. Nat Rev Mol Cell Biol 2017;18:407–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Brahma S, Udugama MI, Kim J, Hada A, Bhardwaj SK, Hailu SG, et al. INO80 exchanges H2A.Z for H2A by translocating on DNA proximal to histone dimers. Nat Commun 2017;8:15616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Mizuguchi G, Shen X, Landry J, Wu WH, Sen S, Wu C. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science. 2004;303:343–8. [DOI] [PubMed] [Google Scholar]
- [8].Han Y, Reyes AA, Malik S, He Y. Cryo-EM structure of SWI/SNF complex bound to a nucleosome. Nature. 2020;579:452–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Liu X, Li M, Xia X, Li X, Chen Z. Mechanism of chromatin remodelling revealed by the Snf2-nucleosome structure. Nature. 2017;544:440–5. [DOI] [PubMed] [Google Scholar]
- [10].Quinn J, Fyrberg AM, Ganster RW, Schmidt MC, Peterson CL. DNA-binding properties of the yeast SWI/SNF complex. Nature. 1996. February 29;379(6568):844–7. [DOI] [PubMed] [Google Scholar]
- [11].Li M, Xia X, Tian Y, Jia Q, Liu X, Lu Y, et al. Mechanism of DNA translocation underlying chromatin remodelling by Snf2. Nature. 2019;567:409–13. [DOI] [PubMed] [Google Scholar]
- [12].Sen P, Ghosh S, Pugh BF, Bartholomew B. A new, highly conserved domain in Swi2/Snf2 is required for SWI/SNF remodeling. Nucleic Acids Res 2011;39:9155–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Hassan AH, Awad S, Prochasson P. The Swi2/Snf2 bromodomain is required for the displacement of SAGA and the octamer transfer of SAGA-acetylated nucleosomes. J Biol Chem 2006;281:18126–34. [DOI] [PubMed] [Google Scholar]
- [14].Shen W, Xu C, Huang W, Zhang J, Carlson JE, Tu X, et al. Solution structure of human Brg1 bromodomain and its specific binding to acetylated histone tails. Biochemistry. 2007;46:2100–10. [DOI] [PubMed] [Google Scholar]
- [15].Schubert HL, Wittmeyer J, Kasten MM, Hinata K, Rawling DC, et al. Structure of an actin-related subcomplex of the SWI/SNF chromatin remodeler. PNAS. 2013;110:3345–3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Sen P, Luo J, Hada A, Hailu SG, Dechassa ML, Persinger J, et al. Loss of Snf5 Induces Formation of an Aberrant SWI/SNF Complex. Cell Rep 2017;18:2135–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Wilsker D, Patsialou A, Zumbrun SD, Kim S, Chen Y, Dallas PB, et al. The DNA-binding properties of the ARID-containing subunits of yeast and mammalian SWI/SNF complexes. Nucleic Acids Res 2004;32:1345–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Kim S, Zhang Z, Upchurch S, Isern N, Chen Y. Structure and DNA-binding sites of the SWI1 AT-rich interaction domain (ARID) suggest determinants for sequence-specific DNA recognition. J Biol Chem 2004;279:16670–6. [DOI] [PubMed] [Google Scholar]
- [19].Cairns BR, Lorch Y, Li Y, Zhang M, Lacomis L, Erdjument-Bromage H, Tempst P, Du J, Laurent B, Kornberg RD. RSC, an essential, abundant chromatin-remodeling complex. Cell. 1996. December 27;87(7):1249–60. [DOI] [PubMed] [Google Scholar]
- [20].Mashtalir N, D’Avino AR, Michel BC, Luo J, Pan J, Otto JE, Zullow HJ, McKenzie ZM, Kubiak RL, St Pierre R, Valencia AM, Poynter SJ, Cassel SH, Ranish JA, Kadoch C. Modular Organization and Assembly of SWI/SNF Family Chromatin Remodeling Complexes. Cell. 2018. November 15;175(5):1272–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Ye Y, Wu H, Chen K, Clapier CR, Verma N, Zhang W, et al. Structure of the RSC complex bound to the nucleosome. Science. 2019;366:838–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Wagner FR, Dienemann C, Wang H, Stutzer A, Tegunov D, Urlaub H, et al. Structure of SWI/SNF chromatin remodeller RSC bound to a nucleosome. Nature. 2020;579:448–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Patel AB, Moore CM, Greber BJ, Luo J, Zukin SA, Ranish J, et al. Architecture of the chromatin remodeler RSC and insights into its nucleosome engagement. Elife. 2019;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Chen G, Li W, Yan F, Wang D, Chen Y. The Structural Basis for Specific Recognition of H3K14 Acetylation by Sth1 in the RSC Chromatin Remodeling Complex. Structure. 2020;28:111–8 e3. [DOI] [PubMed] [Google Scholar]
- [25].Baker RW, Reimer JM, Carman PJ, Turegun B, Arakawa T, Dominguez R, Leschziner AE. Structural insights into assembly and function of the RSC chromatin remodeling complex. Nat Struct Mol Biol 2021. January;28(1):71–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Chambers AL, Pearl LH, Oliver AW, Downs JA. The BAH domain of Rsc2 is a histone H3 binding domain. Nucleic Acids Res 2013;41:9168–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].VanDemark AP, Kasten MM, Ferris E, Heroux A, Hill CP, Cairns BR. Autoregulation of the rsc4 tandem bromodomain by gcn5 acetylation. Mol Cell. 2007;27:817–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Angus-Hill ML, Schlichter A, Roberts D, Erdjument-Bromage H, Tempst P, Cairns BR. A Rsc3/Rsc30 zinc cluster dimer reveals novel roles for the chromatin remodeler RSC in gene expression and cell cycle control. Mol Cell. 2001;7:741–51. [DOI] [PubMed] [Google Scholar]
- [29].Badis G, Chan ET, Bakel H, Pena-Castillo L, Tillo D, Tsui K, et al. A Library of Yeast Transcription Factor Motifs Reveals a Widespread Function for Rsc3 in Targeting Nucleosome Exclusion at Promoters. Mol Cell. 2008;32:878–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Chittori S, Hong J, Bai Y, Subramaniam S. Structure of the primed state of the ATPase domain of chromatin remodeling factor ISWI bound to the nucleosome. Nucleic Acids Res 2019;47:9400–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Yan L, Wu H, Li X, Gao N, Chen Z. Structures of the ISWI-nucleosome complex reveal a conserved mechanism of chromatin remodeling. Nat Struct Mol Biol 2019;26:258–66. [DOI] [PubMed] [Google Scholar]
- [32].Yamada K, Frouws TD, Angst B, Fitzgerald DJ, DeLuca C, Schimmele K, et al. Structure and mechanism of the chromatin remodelling factor ISW1a. Nature. 2011;472:448–53. [DOI] [PubMed] [Google Scholar]
- [33].Yan L, Wang L, Tian Y, Xia X, Chen Z. Structure and regulation of the chromatin remodeller ISWI. Nature. 2016;540:466–9. [DOI] [PubMed] [Google Scholar]
- [34].Ludwigsen J, Pfennig S, Singh AK, Schindler C, Harrer N, Forne I, et al. Concerted regulation of ISWI by an autoinhibitory domain and the H4 N-terminal tail. Elife. 2017;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Vary JC Jr., Gangaraju VK, Qin J, Landel CC, Kooperberg C, Bartholomew B, et al. Yeast Isw1p forms two separable complexes in vivo. Mol Cell Biol 2003;23:80–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Bhardwaj SK, Hailu SG, Olufemi L, Brahma S, Kundu S, Hota SK, Persinger J, Bartholomew B. Dinucleosome specificity and allosteric switch of the ISW1a ATP-dependent chromatin remodeler in transcription regulation. Nat Commun 2020. November 20;11(1):5913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Maltby VE, Martin BJ, Schulze JM, Johnson I, Hentrich T, Sharma A, Kobor MS, Howe L. Histone H3 lysine 36 methylation targets the Isw1b remodeling complex to chromatin. Mol Cell Biol 2012. September;32(17):3479–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Smolle M, Venkatesh S, Gogol MM, Li H, Zhang Y, Florens L, Washburn MP, Workman JL. Chromatin remodelers Isw1 and Chd1 maintain chromatin structure during transcription by preventing histone exchange. Nat Struct Mol Biol 2012. September;19(9):884–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Simic R, Lindstrom DL, Tran HG, Roinick KL, Costa PJ, Johnson AD, et al. Chromatin remodeling protein Chd1 interacts with transcription elongation factors and localizes to transcribed genes. EMBO J 2003;22:1846–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Krogan NJ, Kim M, Ahn SH, Zhong G, Kobor MS, Cagney G, et al. RNA polymerase II elongation factors of Saccharomyces cerevisiae: a targeted proteomics approach. Mol Cell Biol 2002;22:6979–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Pray-Grant MG, Daniel JA, Schieltz D, Yates JR 3rd, Grant PA. Chd1 chromodomain links histone H3 methylation with SAGA- and SLIK-dependent acetylation. Nature. 2005;433:434–8. [DOI] [PubMed] [Google Scholar]
- [42].Sundaramoorthy R, Hughes AL, El-Mkami H, Norman DG, Ferreira H, Owen-Hughes T. Structure of the chromatin remodelling enzyme Chd1 bound to a ubiquitinylated nucleosome. Elife. 2018;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Farnung L, Vos SM, Wigge C, Cramer P. Nucleosome-Chd1 structure and implications for chromatin remodelling. Nature. 2017;550:539–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Eustermann S, Schall K, Kostrewa D, Lakomek K, Strauss M, Moldt M, et al. Structural basis for ATP-dependent chromatin remodelling by the INO80 complex. Nature. 2018;556:386–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Knoll KR, Eustermann S, Niebauer V, Oberbeckmann E, Stoehr G, Schall K, et al. The nuclear actin-containing Arp8 module is a linker DNA sensor driving INO80 chromatin remodeling. Nat Struct Mol Biol 2018;25:823–32. [DOI] [PubMed] [Google Scholar]
- [46].Brahma S, Ngubo M, Paul S, Udugama M, Bartholomew B. The Arp8 and Arp4 module acts as a DNA sensor controlling INO80 chromatin remodeling. Nat Commun 2018. August 17;9(1):3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Gerhold CB, Winkler DD, Lakomek K, et al. Structure of Actin-related protein 8 and its contribution to nucleosome binding. Nucleic Acids Res 2012;40(21):11036–11046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Willhoft O, Ghoneim M, Lin CL, Chua EYD, Wilkinson M, Chaban Y, Ayala R, McCormack EA, Ocloo L, Rueda DS, Wigley DB. Structure and dynamics of the yeast SWR1-nucleosome complex. Science. 2018. October 12;362(6411):eaat7716. [DOI] [PubMed] [Google Scholar]
- [49].Singh RK, Fan J, Gioacchini N, Watanabe S, Bilsel O, Peterson CL. Transient Kinetic Analysis of SWR1C-Catalyzed H2A.Z Deposition Unravels the Impact of Nucleosome Dynamics and the Asymmetry of Histone Exchange. Cell Rep 2019;27(2):374–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Ranjan A, Wang F, Mizuguchi G, Wei D, Huang Y, Wu C. H2A histone-fold and DNA elements in nucleosome activate SWR1-mediated H2A.Z replacement in budding yeast. Elife. 2015. June 27;4:e06845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Wu WH, Wu CH, Ladurner A, Mizuguchi G, Wei D, Xiao H, Luk E, Ranjan A, Wu C. N terminus of Swr1 binds to histone H2AZ and provides a platform for subunit assembly in the chromatin remodeling complex. J Biol Chem 2009. March 6;284(10):6200–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Matangkasombut O, Buratowski S. Different sensitivities of bromodomain factors 1 and 2 to histone H4 acetylation. Mol Cell. 2003. February;11(2):353–63. [DOI] [PubMed] [Google Scholar]
- [53].Koerber RT, Rhee HS, Jiang C, Pugh BF. Interaction of transcriptional regulators with specific nucleosomes across the Saccharomyces genome. Mol Cell. 2009;35(6):889–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Ranjan A, Mizuguchi G, FitzGerald PC, Wei D, Wang F, Huang Y, Luk E, Woodcock CL, Wu C. Nucleosome-free region dominates histone acetylation in targeting SWR1 to promoters for H2A.Z replacement. Cell. 2013. September 12;154(6):1232–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Sun L, Luk E. Dual function of Swc5 in SWR remodeling ATPase activation and histone H2A eviction. Nucleic Acids Res 2017;45(17):9931–9946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Clapier CR, Iwasa J, Cairns BR, Peterson CL. Mechanisms of action and regulation of ATP-dependent chromatin-remodelling complexes. Nat Rev Mol Cell Biol 2017. July;18(7):407–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Yan L, Chen Z. A Unifying Mechanism of DNA Translocation Underlying Chromatin Remodeling. Trends Biochem Sci 2020. March;45(3):217–227. [DOI] [PubMed] [Google Scholar]
- [58].Jungblut A, Hopfner KP, Eustermann S. Megadalton chromatin remodelers: common principles for versatile functions. Curr Opin Struct Biol 2020. October;64:134–144. [DOI] [PubMed] [Google Scholar]
- [59].Harada BT, Hwang WL, Deindl S, Chatterjee N, Bartholomew B, Zhuang X. Stepwise nucleosome translocation by RSC remodeling complexes. Elife. 2016;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Saha A, Wittmeyer J, Cairns BR. Chromatin remodeling through directional DNA translocation from an internal nucleosomal site. Nat Struct Mol Biol 2005;12:747–55. [DOI] [PubMed] [Google Scholar]
- [61].Bash R, Wang H, Anderson C, Yodh J, Hager G, Lindsay SM, Lohr D. AFM imaging of protein movements: histone H2A-H2B release during nucleosome remodeling. FEBS Lett. 2006. August 21;580(19):4757–61. [DOI] [PubMed] [Google Scholar]
- [62].Boeger H, Griesenbeck J, Strattan JS, Kornberg RD. Removal of promoter nucleosomes by disassembly rather than sliding in vivo. Mol Cell. 2004. June 4;14(5):667–73. [DOI] [PubMed] [Google Scholar]
- [63].Brown CR, Mao C, Falkovskaia E, Law JK, Boeger H. In vivo role for the chromatin-remodeling enzyme SWI/SNF in the removal of promoter nucleosomes by disassembly rather than sliding. J Biol Chem 2011. November 25;286(47):40556–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Kagalwala MN, Glaus BJ, Dang W, Zofall M, Bartholomew B. Topography of the ISW2-nucleosome complex: insights into nucleosome spacing and chromatin remodeling. EMBO J 2004. May 19;23(10):2092–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Fitzgerald DJ, DeLuca C, Berger I, et al. Reaction cycle of the yeast Isw2 chromatin remodeling complex. EMBO J 2004;23(19):3836–3843. doi: 10.1038/sj.emboj.7600364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Zofall M, Persinger J, Kassabov SR, Bartholomew B. Chromatin remodeling by ISW2 and SWI/SNF requires DNA translocation inside the nucleosome. Nat Struct Mol Biol 2006. April;13(4):339–46. [DOI] [PubMed] [Google Scholar]
- [67].Winger J, Nodelman IM, Levendosky RF, Bowman GD. A twist defect mechanism for ATP-dependent translocation of nucleosomal DNA. Elife. 2018;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Sabantsev A, Levendosky RF, Zhuang X, Bowman GD, Deindl S. Direct observation of coordinated DNA movements on the nucleosome during chromatin remodelling. Nat Commun 2019;10:1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Tsukiyama T, Palmer J, Landel CC, Shiloach J, Wu C. Characterization of the imitation switch subfamily of ATP-dependent chromatin-remodeling factors in Saccharomyces cerevisiae. Genes Dev 1999. March 15;13(6):686–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Lusser A, Urwin DL, Kadonaga JT. Distinct activities of CHD1 and ACF in ATP-dependent chromatin assembly. Nat Struct Mol Biol 2005. February;12(2):160–6. [DOI] [PubMed] [Google Scholar]
- [71].McKnight JN, Jenkins KR, Nodelman IM, Escobar T, Bowman GD. Extranucleosomal DNA binding directs nucleosome sliding by Chd1. Mol Cell Biol. 2011. December;31(23):4746–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Eriksson PR, Clark DJ. The yeast ISW1b ATP-dependent chromatin remodeler is critical for nucleosome spacing and dinucleosome resolution. Sci Rep 2021;11(1):4195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Gangaraju VK, Bartholomew B. Dependency of ISW1a chromatin remodeling on extranucleosomal DNA. Mol Cell Biol 2007. April;27(8):3217–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Zofall M, Persinger J, Bartholomew B. Functional role of extranucleosomal DNA and the entry site of the nucleosome in chromatin remodeling by ISW2. Mol Cell Biol. 2004. November;24(22):10047–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Nodelman IM, Horvath KC, Levendosky RF, Winger J, Ren R, Patel A, Li M, Wang MD, Roberts E, Bowman GD. The Chd1 chromatin remodeler can sense both entry and exit sides of the nucleosome. Nucleic Acids Res 2016. September 19;44(16):7580–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Ayala R, Willhoft O, Aramayo RJ, Wilkinson M, McCormack EA, Ocloo L, et al. Structure and regulation of the human INO80-nucleosome complex. Nature. 2018;556:391–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Brahma S, Udugama MI, Kim J, Hada A, Bhardwaj SK, Hailu SG, Lee TH, Bartholomew B. INO80 exchanges H2A.Z for H2A by translocating on DNA proximal to histone dimers. Nat Commun 2017. June 12;8:15616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Yen K, Vinayachandran V, Pugh BF. SWR-C and INO80 chromatin remodelers recognize nucleosome-free regions near +1 nucleosomes. Cell. 2013. September 12;154(6):1246–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Krietenstein N, Wal M, Watanabe S, Park B, Peterson CL, Pugh BF, Korber P. Genomic Nucleosome Organization Reconstituted with Pure Proteins. Cell. 2016. October 20;167(3):709–721.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Wu WH, Alami S, Luk E, Wu CH, Sen S, Mizuguchi G, Wei D, Wu C. Swc2 is a widely conserved H2AZ-binding module essential for ATP-dependent histone exchange. Nat Struct Mol Biol 2005. December;12(12):1064–71. [DOI] [PubMed] [Google Scholar]
- [81].Luk E, Ranjan A, Fitzgerald PC, Mizuguchi G, Huang Y, Wei D, Wu C. Stepwise histone replacement by SWR1 requires dual activation with histone H2A.Z and canonical nucleosome. Cell. 2010. November 24;143(5):725–36. [DOI] [PMC free article] [PubMed] [Google Scholar]


