Highlights
-
•
A CT imaging dataset of 1222 lung adenocarcinoma patients from 3 medical centers was used to construct the classification model.
-
•
The proposed algorithms achieved encouraging performance in the differentiation of lung adenocarcinoma subtypes.
-
•
The C-index of prognosis model based on deep radiomics combined classifier was 0.892(95% confidence Intervals: 0.846–0.937) in internal validation set.
Keywords: Deep learning, Computed tomography, Lung adenocarcinoma, Subtype
Abstract
Objectives
The subtype classification of lung adenocarcinoma is important for treatment decision. This study aimed to investigate the deep learning and radiomics networks for predicting histologic subtype classification and survival of lung adenocarcinoma diagnosed through computed tomography (CT) images.
Methods
A dataset of 1222 patients with lung adenocarcinoma were retrospectively enrolled from three medical institutions. The anonymised preoperative CT images and pathological labels of atypical adenomatous hyperplasia, adenocarcinoma in situ, minimally invasive adenocarcinoma, invasive adenocarcinoma (IAC) with five predominant components were obtained. These pathological labels were divided into 2-category classification (IAC; non-IAC), 3-category and 8-category. We modeled the classification task of histological subtypes based on modified ResNet-34 deep learning network, radiomics strategies and deep radiomics combined algorithm. Then we established the prognostic models in lung adenocarcinoma patients with survival outcomes. The accuracy (ACC), area under ROC curves (AUCs) and C-index were primarily performed to evaluate the algorithms.
Results
This study included a training set (n = 802) and two validation cohorts (internal, n = 196; external, n = 224). The ACC of deep radiomics algorithm in internal validation achieved 0.8776, 0.8061 in the 2-category, 3-category classification, respectively. Even in 8 classifications, the AUC ranged from 0.739 to 0.940 in internal set. Further, we constructed a prognosis model that C-index was 0.892(95% CI: 0.846–0.937) in internal validation set.
Conclusions
The automated deep radiomics based triage system has achieved the great performance in the subtype classification and survival predictability in patients with CT-detected lung adenocarcinoma nodules, providing the clinical guide for treatment strategies.
Introduction
During the past few decades, computed tomography (CT) has been widely used in clinical examinations as an effective tool for lung cancer screening, seriously reducing about 20% mortality of the whole patient population according to the U.S. National Lung Screening Trial (NLST) and European Nederlands–Leuvens Longkanker Screenings Onderzoek (NELSON) study [1, 2]. However, lung cancer remains the leading cause of cancer-related mortality (18.0% of the total cancer deaths) with estimated 1.8 million deaths globally and the second common type of malignancies (11.4% of new cancer cases) in 2020 [3].
As the most prevalent subtype, lung adenocarcinoma accounts for approximately more than half of all lung cancer cases, which displays as pulmonary nodules in the CT images [4]. It is also worth mentioning that the prognosis of adenocarcinoma varies widely among the different subtypes [5, 6]. For example, the patients with preinvasive lesions pathologically diagnosed with atypical adenomatous hyperplasia (AAH), adenocarcinoma in situ (AIS), or minimally invasive adenocarcinoma (MIA), manifesting as indolent nodules with ground glass opacities will have almost 100% 5-year disease-free survival rate after complete resection, and have no need of adjuvant chemotherapy [7, 8]. Traditionally, the specific subtype of adenocarcinoma needs to be determined once surgically resected, which may expose patients to unnecessary investigations, and create a drain on health-care resources [5, 9]. In contrast, hispathological micropapillary or solid patterns with solid aggressive nodules in CT images, even less than 5% of the entire tumor, are associated with unfavorable prognosis, and these patients usually benefit from adjuvant chemotherapy [10], [11], [12]. What's more, patients with comorbidities or the elderly may make the complete surgical intervention a big risky endeavor. Therefore, novel non-invasive techniques should be developed to predict the histologic subtypes precisely in CT-detected lung adenocarcinoma nodules.
Recently, machine learning provided decision support in the diagnosis of lung nodules via CT images, particularly at centers where adequate expert thoracic radiologists are unavailable [13, 14]. Deep learning, a subset of machine learning, directly sends image into a convolutional neural network (CNN) and works as a “black box”. It has achieved impressive results with robust evidence in several medical image classification tasks, including grading of diabetic retinopathy, classification of skin lesions, and prediction of the malignancy risk of lung cancer screening with good accuracy equivalent to specialist physicians [15], [16], [17]. Another critical machine learning method is radiomics, which refers to the manual extraction and analysis of advanced quantitative imaging features and provides predictive models relating image features to tumor phenotypes [18, 19]. In addition, previous studies have demonstrated that deep learning or radiomics system could distinguish AAH-AIS, MIA and invasive adenocarcinoma (IAC), and achieved better classification performance than the radiologists [20], [21], [22]. However, there was very little literature to predict the specific pathological subtype categories of lung adenocarcinoma using deep learning or radiomics methods and further to help with the precision of survival estimations.
In the current study, we investigated radiomics and deep learning methods to predict the histologic subtypes of lung adenocarcinoma and evaluated their prognostic value through analyses of non-invasive CT images.
Methods
Patient inclusion
A total of 1222 patients with lung adenocarcinoma who had went through resection at West China Hospital of Sichuan University, Suining Central Hospital and Chengdu Shangjin Nanfu Hospital, were enrolled in this study before December 2017. Our inclusion criteria were (1) pathologically diagnosed with lung adenocarcinoma according to the 2015 WHO Classification [23]; (2) complete surgery specimen with clear pathological pattern; (3) having not gone through radiotherapy or chemotherapy when received imaging screening; (4) having preoperative chest CT images within one month before surgery. On the other hand, our exclusion criteria were: (1) having tumors with metastases to extrapulmonary tissues; (2) multi-focal lung cancers; (3) diagnosed with adenosquamous carcinoma or other types of lung cancer. Patients with complete 5-year follow-up were enrolled in the prognosis estimation analysis.
All patients were followed up until December 2020. Clinical parameters including but not limited to sex, age at diagnosis and smoking status, and corresponding electric histopathologic reports with the evaluation and verification of senior pathologists served as the gold standard during the algorithm development process, were obtained through the Hospital Information System (HIS). In this study, lung adenocarcinomas were classified into AAH, AIS, MIA and IAC, and IACs were further subdivided into predominant lepidic adenocarcinoma (pLAC), predominant acinar adenocarcinoma (pAAC), predominant papillary adenocarcinoma(pPAC), predominant micropapillary adenocarcinoma (pMAC), and predominant solid adenocarcinoma (pSAC) on the basis of the WHO 2015 standards [23]. CT DICOM images within one month before operation were extracted from Picture Archiving and Communication System (PACS), with duplicate or incomplete images were removed. The natural language processing (NLP) algorithm was used to detect pathological subtype patterns from clinical histopathologic reports, which had been reviewed by the senior pathologist. This study was approved by the Ethics Committee of the participating institutions .
Dataset preparation
We randomly allocated a dataset of 998 patients with lung adenocarcinoma from West China hospital into a training set (n = 802) and an internal validation cohort (n = 196), while 224 patients from another two independent cohorts constituted the external validation group. The three subsets were mutually exclusive. In order to ensure the average distribution of the data of each category, 200 patients were randomly selected for each category except for the extremely rare AAH and pMAC. In our research, the region of interest (ROI) detection of lung nodules were built on a deep residual network, which is able to detect target ROI automatically [24]. Then two senior radiologists (with 10 years of work experience in thoracic imaging diagnosis) blindly and independently assessed the mask of all images, and any disagreements between two observers were resolved by discussion until a consensus was reached. And the external test set from two independent institutions was obtained to evaluate the robustness and generalizability of this model.
Deep learning methods
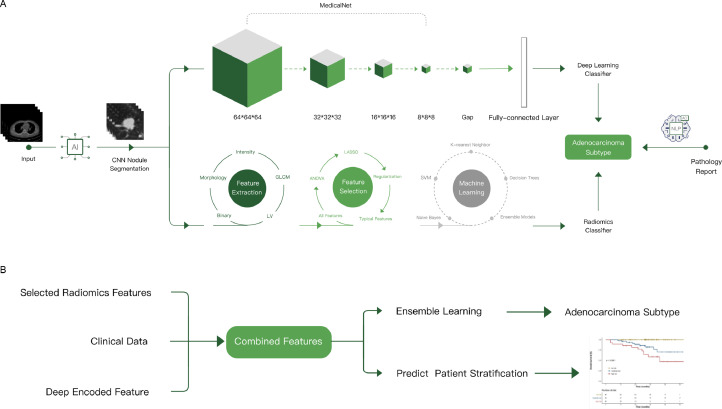
The deep learning model(Fig. 1) was trained from MedicalNet, which was a series of publicly released pre-trained neural networks [25]. Before our study, the model was trained on 8 public datasets of radiologic images from diverse modalities, target organs, and pathologies, which showed marvelous adaptability in the medical domain. Among the pre-trained models released in the MedicalNet, 3D-ResNet-34 was chosen as the backbone in this study.
Fig. 1.
Overall modeling framework: (A) classification algorithm based on deep learning and radiomics; (B) deep radiomics combined classifier and prognosis models.
We made some modifications on model structures of the baseline model, to get better performance generalization abilities. The modification details in structure included: (1) The Flatten operation after the last convolutional layer was replaced by a Global Average Pooling (GAP) layer, which led to a better generalization ability; (2) Information regarding the proportional size of lesions in transverse, coronal and sagittal planes was extracted to represent the actual size of the lesion. The 3 proportional size features were set as input to the fully connected layer after GAP layer in the model; (3) Information regarding the lesion attenuation (solid, subsolid, ground glass, calcification), margin (smooth or non-smooth), spiculation, lobulation, pleural indentation, vascular bundle sign, vacuole sign, and air bronchogram were collected and determined by experienced physicians. These features were also set as input to the fully connected layer after GAP layer in the model. Therefore, concatenating all these 11 features mentioned above with 512 features encoded from 3D cropped image, 523 features were finally collected in the fully connection layer; (4) To avoid overfitting, a dropout strategy with 0.5 probability was applied to the last two fully connected layers. In terms of output, we computed 8 softmax outputs as probabilities over classes. In order to obtain better spatial interpretability of the model, GradCAM method was used to visualize. Also, the 512 features were extracted as encoded image features, to help build prognosis prediction models.
We also sought for innovations in the training strategies: (1) Online data augmentation was conducted, including but not limited to techniques such as image flipping, rotation, transposing, size scaling, translation in 3 axes, Gaussian blur, linear contrast augmentation and gray value scaling. (2) The sample sizes for each pathological category in each training epoch were balanced after augmentation. (3) The optimizer adopted SGD with momentum, and with weight decay coefficient of 0.01. With the initial learning rate of shallow parameters to be 0.001 and the initial learning rate of the deep parameter to be 0.005, the learning rate of all parameters were decayed exponentially by 0.95 along all epochs, The batch size was set to 4.
Radiomics explorations
At the same time, we explored the radiomics model to classify various subtypes of adenocarcinoma (Fig. 1). Due to the high dimensionality of the radiomics feature space, the similarity of each feature pair was compared. If the Pearson's correlation coefficient of a feature pair is >0.9, one pair is removed. Then, we trained a penalized multinomial logistic regression model with least absolute shrinkage and selection operator (LASSO) using glmnet package. To avoid overfitting, resampling of training set was done through 5-fold cross validation. The model with the least misclassification error was chosen as optimal, and it also contained the best set of features (details in Supplementary Methods).
Deep radiomic combined classifier algorithm
Before building the model, we used stepwise multinomial logistic regression model to reduce the dimensionality of 512-dimensional features generated by deep learning model. Then we combined the features left by the stepwise regression, the features from the radiomic results and the features from clinical results (such as sex, age, smoking history), using optimal subset method and Akaike information criterion (AIC) to select the best subset combination. The optimal subset method provides the corresponding chi-square value for various feature number combinations. However, it could not select the best combination. Therefore, the corresponding AIC values under various combinations can be calculated to find the smallest corresponding value. The final combined model was constructed using the combination of features under the minimum AIC value with multinomial logistic regression algorithm (Fig. 1).
Statistical analysis
To compare the clinical characteristics of patients among three datasets, continuous and categorical variables were evaluated by ANOVA and Chi-square test, respectively. In addition, the area under ROC curves (AUCs) of model designed for the multiclass problems were calculated using the prediction performance of the positive class (ie, the respective metastatic tumor type) versus all other classes. ROC curves based on means were estimated from all cross-validation sets. Confidence intervals (CIs) for sensitivities and specificities were derived from 2000 bootstrap replicates using pROC, version 1.10 and qwraps2, version 0.3.0, R-packages. And exemplified classifier cutoff values for further sensitivity and specificity analyses were determined according to minimum-distance cutoff points.
The probability scores for all eight subtypes of adenocarcinoma were calculated by putting them through the deep learning and radiomics model, and heatmaps were generated for all models. We used these probability scores to build independent Cox proportional hazard models. And we used Kaplan-Meier plots to visualize the survival curves of different subtypes and stratify three risk levels. X-tile plots were used to generate two optimal cut-off values to classify patients into low-risk, medium-risk and high-risk groups [26].
Results
Clinical characteristics of patients
Baseline characteristics of 1222 lung adenocarcinoma patients were delineated in Table 1. The enrolled patients were allocated into a training set (n = 802), an internal validation set (n = 196), and an external validation test set (n = 224 patients from two independent medical centers). The median age of training set was 58.78 years with more than 69% being no-smokers and females accounting for the majority. There was no statistical significance among those cohorts in terms of age (p = 0.09), smoking status (p = 0.098) and overall survival status (p = 0.392), while the sex, and pathological classification showed the statistical difference. According to the degree of invasion, adenocarcinoma was divided into 2 categories (IAC; non-IAC including AAH, AIS and MIA). Regarding that micropapillary and solid components could significantly affect the prognosis of patients [10, 11, 27, 28], then we divided all adenocarcinoma cases into 3 groups: a group without micropapillary or solid components (non-MAC/SAC), a group without micropapillary or solid components(non-pMAC/pSAC), and a group with predominant micropapillary and solid patterns (pMAC+pSAC). Also, we also merged some categories to form a 6-category classification (AAH+AIS, MIA, pLAC, pAAC, pPAC, pMAC+pSAC) referred to the 8-category classification.
Table 1.
Clinical characteristics of patients.
| Variable | Level | Training set | Internal Validation set | External Validation set | P value |
|---|---|---|---|---|---|
| N | N = 802 | N = 196 | N = 224 | ||
| Age (mean (SD)) | 58.78 (10.09) | 57.79 (11.06) | 57.17 (10.74) | 0.09 | |
| Sex (%) | Male | 342 (42.6) | 88 (44.9) | 77 (34.4) | 0.049 |
| Female | 460 (57.4) | 108 (55.1) | 147 (65.6) | ||
| Smoke (%) | No smoke | 556 (69.3) | 146 (74.5) | 170 (75.9) | 0.098 |
| Smoke | 229 (28.6) | 49 (25.0) | 54 (24.1) | ||
| Unknown | 17 (2.1) | 1 (0.5) | 0 (0.0) | ||
| 2-category classification (%) | Non-IAC | 182 (22.7) | 57 (29.1) | 19 (8.5) | <0.001 |
| IAC | 620 (77.3) | 139 (70.9) | 205 (91.5) | ||
| 3-category classification (%) | Non-MAC/SAC | 561 (70.0) | 148 (75.5) | 164 (73.2) | 0.025 |
| Non-pMAC/pSAC | 103 (12.8) | 17 (8.7) | 37 (16.5) | ||
| pMAC+pSAC | 138 (17.2) | 31 (15.8) | 23 (10.3) | ||
| 6-category classification (%) | AAH+AIS | 68 (8.5) | 30 (15.3) | 5 (2.2) | <0.001 |
| MIA | 114 (14.2) | 27 (13.8) | 14 (6.2) | ||
| pLAC | 154 (19.2) | 38 (19.4) | 30 (13.4) | ||
| pAAC | 173 (21.6) | 36 (18.4) | 116 (51.8) | ||
| pPAC | 155 (19.3) | 34 (17.3) | 36 (16.1) | ||
| pMAC+pSAC | 138 (17.2) | 31 (15.8) | 23 (10.3) | ||
| 8-category classification (%) | AAH | 10 (1.2) | 6 (3.1) | 1 (0.4) | <0.001 |
| AIS | 58 (7.2) | 24 (12.2) | 4 (1.8) | ||
| MIA | 114 (14.2) | 27 (13.8) | 14 (6.2) | ||
| pLAC | 154 (19.2) | 38 (19.4) | 30 (13.4) | ||
| pAAC | 173 (21.6) | 36 (18.4) | 116 (51.8) | ||
| pPAC | 155 (19.3) | 34 (17.3) | 36 (16.1) | ||
| pMAC | 16 (2.0) | 3 (1.5) | 0 (0.0) | ||
| pSAC | 122 (15.2) | 28 (14.3) | 23 (10.3) | ||
| OS_Status (%) | Survival | 720 (89.8) | 180 (91.8) | 139 (87.4)* | 0.392 |
| Death | 82 (10.2) | 16 (8.2) | 20 (12.6)* | ||
| OS_month (mean (SD)) | 44.51 (13.32) | 44.43 (13.01) | 33.16 (12.96) | <0.001 |
Abbreviations.
SD, Standard Deviation; IAC, invasive adenocarcinoma; pMAC, predominant micropapillary adenocarcinoma; pSAC, predominant solid adenocarcinoma; AAH, atypical adenomatous hyperplasia; AIS, adenocarcinoma in situ; MIA, minimally invasive adenocarcinoma; pLAC, predominant lepidic adenocarcinoma; pAAC, predominant acinar adenocarcinoma, pPAC, predominant papillary adenocarcinoma, OS, Overall Survival. *159 patients with complete follow-up data in external validation set. ⁎⁎Statistical significance in training and internal validation set.
Deep radiomics classification evaluation
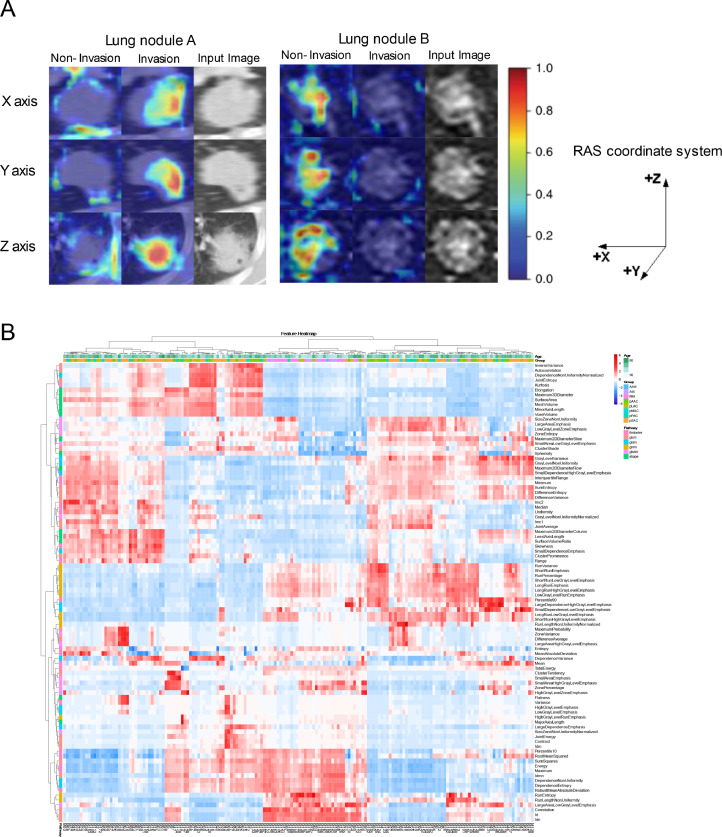
Deep learning model achieved great performance in the adenocarcinoma classification task (Table 2, Supplementary Figure 1). The performance of 3-category classification based on deep learning achieved better than the performance of other models, which the accuracy (ACC) reached 0.8112, 0.7366 in internal and external validation set respectively. Moreover, we used a deep learning visualization method to find the tumor region related to pathological subtypes. Fig. 2(A) depicted the attention map of the deep learning model. Dark colors represented high-response area, also called suspicious tumor area. The suspicious area in lung nodule A was the tumor edge and tissue between tumor and pleura, which showed that the tumor had a high tendency to spread or invade. Meanwhile, the deep learning model focused on the tumor edge in lung nodule B and predicted them to be non-invasive.
Table 2.
Predictive performance of three machine learning models.
| ACC of Classification | Validation Set | Deep Radiomics | Deep Learning | Radiomics |
|---|---|---|---|---|
| 2-category | Internal | 0.8776 | 0.8622 | 0.8827 |
| External | 0.9063 | 0.8527 | 0.8973 | |
| 3-category | Internal | 0.8061 | 0.8112 | 0.7908 |
| External | 0.7143 | 0.7366 | 0.7054 | |
| 6-category | Internal | 0.4796 | 0.4031 | 0.3724 |
| External | 0.4554 | 0.4241 | 0.4375 | |
| 8-category | Internal | 0.4643 | 0.4235 | 0.3520 |
| External | 0.4643 | 0.4286 | 0.4464 | |
| C-index of prognosis models (95%CI) | Internal | 0.892(0.846–0.937) | 0.836(0.765–0.906) | 0.854(0.792–0.917) |
| External | 0.835(0.774–0.896) | 0.800(0.709–0.891) | 0.840(0.786–0.894) |
Abbreviations.
ACC, accuracy; CI, Confidence Interval.
Fig. 2.
Deep learning and radiomics feature analysis. (A)Attention map of the deep learning model (RAS coordinate system was adopted in the project). (B)Unsupervised clustering of lung adenocarcinoma patients on the horizontal axis vertical axis and radiomics feature expression on the vertical axis.
The performance of radiomics was almost as great as other models. The 2-category classification had excellent accuracy with 0.8827, 0.8973 in internal validation cohort and external validation set respectively (Table 2, Supplementary Figure 2). Focus on six and eight classifications, the AUCs of each subtype were above 0.7 in internal validation set (Supplementary Figure 3). Furthermore, the cluster graph proved the relative correlation between machine learning features and pathological types in Fig. 2(B).
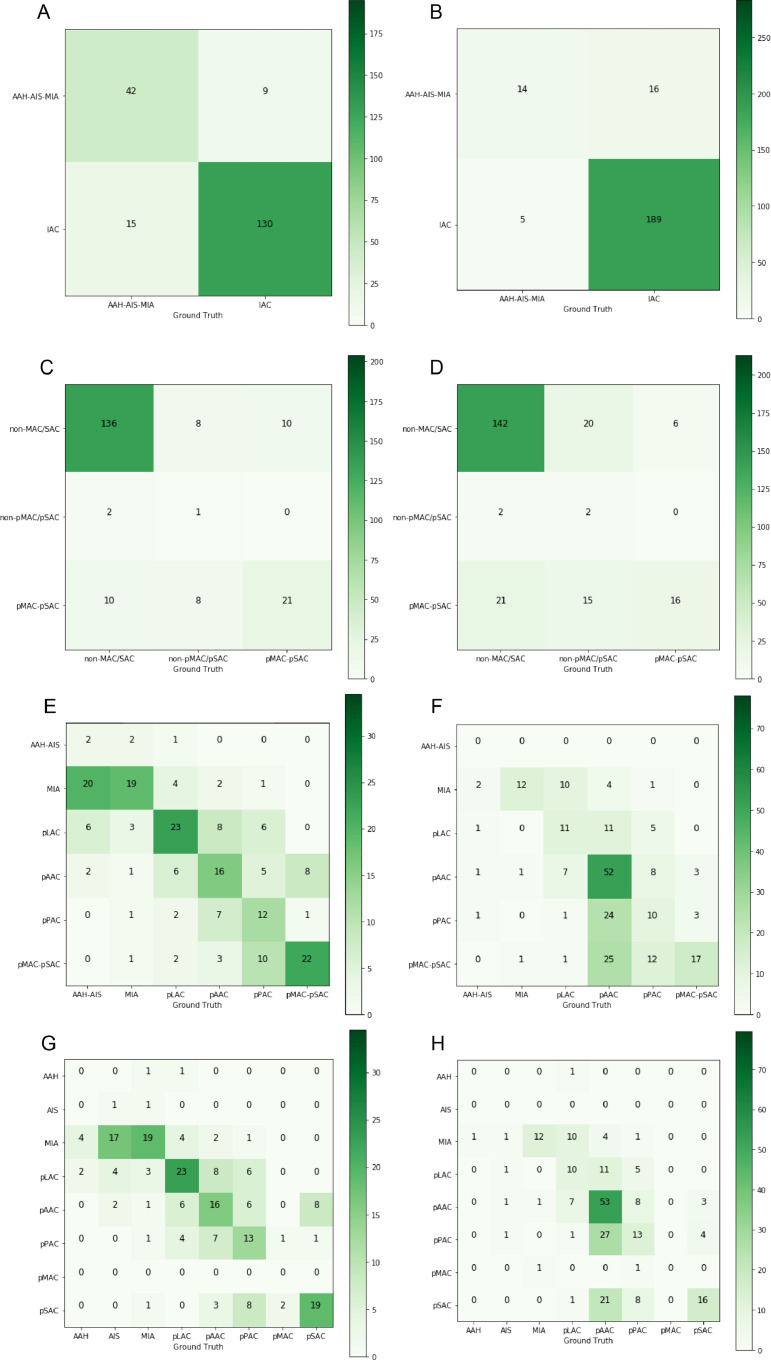
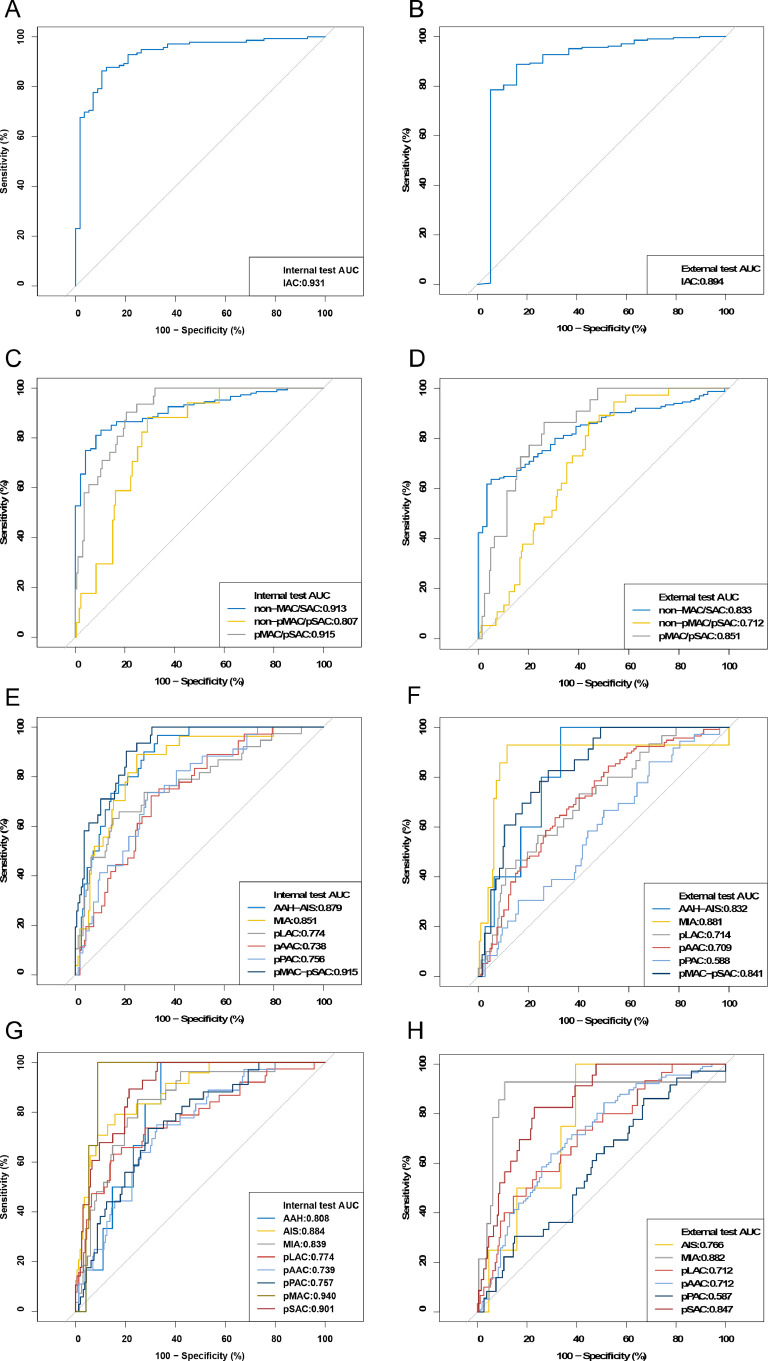
Significantly, the deep radiomics combined model accurately classified different subtypes of adenocarcinoma (Table 2). Using the histopathologic reports as the reference standard, ACC scores of 2-category classification achieved 0.8776, 0.9063 in internal and external validation set respectively. When we evaluated according to three categories, the performance of this model still was promising (internal validation set, ACC=0.8061; external validation set, ACC=0.7143). The confusion matrix was shown in Fig. 3, suggesting that this model was not prone to make errors and implicitly learned the relationship among these categories. Even in 6 or 8 categories, the ACC of both internal and external validation sets was more than 0.45. Most subtypes in 8 categories were well identified such as pMAC (AUC=0.940, 95% CI: 0.898–0.981), pSAC (AUC=0.901, 95% CI: 0.854–0.948), while the minimum AUC was at 0.739 (95% CI: 0.657–0.822) for pAAC in internal test (Fig. 4).
Fig. 3.
Confusion matrix on deep radiomics combined model: the 2-category classification in (A) internal validation set and (B) external validation set; 3-category classification in (C) internal validation set and (D) external validation set; 6-category classification in (E) internal validation set and (F) external validation set; 8-category classification in (G) internal validation set and (H) external validation set.
Fig. 4.
ROC curve on deep radiomics combined model: the 2-category classification in (A) internal validation set and (B) external validation set; 3-category classification in (C) internal validation set and (D) external validation set; 6-category classification in (E) internal validation set and (F) external validation set; 8-category classification in (G) internal validation set and (H) external validation set.
Survival analysis
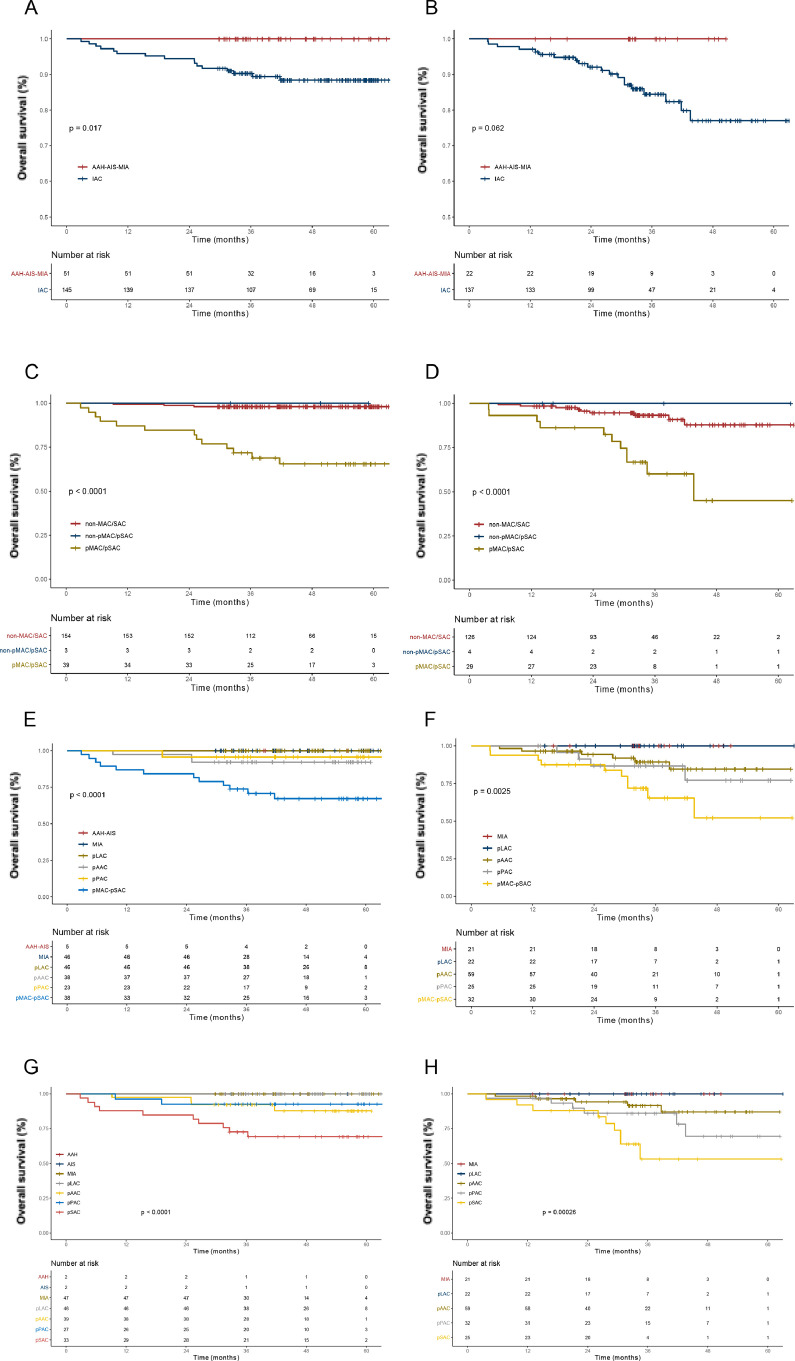
On the basis of selected deep radiomics features, we constructed prognosis models that C-indexes were 0.892(95% CI: 0.846–0.937) in internal validation set and 0.835(95% CI: 0.774–0.896) in external validation set (Table 2). There were significant differences of survival among all category groups of internal validation set (all p<0.05) (Fig. 5). Based on training dataset, two optimal cut-off values (0.62 and 1.38) were used to separate the dataset into low-, medium- and high-risk groups (Supplementary Table 1). Also, the algorithm provided prognostic discrimination among them in internal and external validation set (both p<0.0001) (Supplementary Figure 4).
Fig. 5.
Survival curves based on deep radiomics combined model: the 2-category classification in (A) internal validation set and (B) external validation set; 3-category classification in (C) internal validation set and (D) external validation set; 6-category classification in (E) internal validation set and (F) external validation set; 8-category classification in (G) internal validation set and (H) external validation set.
The prognostic model on deep learning revealed great performance (internal validation set, C-index 0.836, 95% CI: 0.765–0.906; external validation set, C-index 0.800, 95% CI: 0.709–0.891). The C-indexes of prognosis prediction model on radiomics were 0.854(95% CI: 0.792–0.917), 0.840(95% CI: 0.786–0.894), in internal and external validation set respectively (Table 2).
Discussion
In this study, we developed the machine learning methods including ResNet-34 deep learning network and the radiomics which could successfully classify the subtypes and predict the survival outcome of lung adenocarcinoma using non-invasive CT images. To our knowledge, this is the first study with the largest dataset up to date to develop a deep learning algorithm that had a promising performance to automatically make the histologic subtype classification and survival prediction of lung adenocarcinoma patients, which would dramatically help with the clinical efficiency and guide for physicians in decision making.
As is known, the pathological classification of lung adenocarcinoma serves as a vital reference standard in the modality selection of treatment strategy, and it is usually determined according to pathologic results after complete surgical resection [27, 29]. However, in spite of the wide adoption of those invasive modalities, attempts to find alternative non-invasive methods never cease. Compared with the radiologists in the former study, the deep learning model had higher accuracy and better classification performance in predicting tumor invasiveness of sub-centimeter pulmonary adenocarcinomas [20]. What was pioneering for this research was that we proposed the first machine learning system with a comprehensive coverage of almost all common histopathologic subtypes including AAH, AIS, MIA and IAC of lung adenocarcinoma. And IAC was further subclassified into finer divisions(predominant lepidic, acinar, papillary, micropapillary and solid patterns).
But interestingly, in the current study, the performance of the six or eight classification model was inferior to the other classification models. It was supposed that some mixed components among the complicated pathological constitution might affect the imaging features even though they were not high in proportion. To estimate our conjecture, we evaluated the three classification models in cases with different micropapillary or and solid components and figured out a superior model. This classification method had also been supported by previous research [12, 30, 31]. NCCN guidelines recommend that patients with non-invasive adenocarcinoma such as AIS, MIA or IAC at early stage have no need for chemotherapy, and adjuvant chemotherapy maybe the appropriate choice for invasive adenocarcinoma especially for pMAC or pSAC, which will lead to the more recurrence or metastasis [32]. Our results suggest that the proposed deep learning and radiomics algorithm can potentially select appropriate patients with pMAC or pSAC with potentially poorer prognosis, who would be the ideal candidates for systemic therapy, making the best opportunity for surgery.
Also, we established a prognosis model based on imaging semantic features, and stratified the risk of prognosis. A strong performance of the prognosis prediction model was shown in the external validation set with two independent cohorts. Therefore, it was conjectured that the imaging features involved in pathological classification could also play a central role in predicting the prognosis of lung cancer patients [33]. At the same time, there were many factors determining the choice of adjuvant vs non-adjuvant treatment such as tumor stage. The clinical model also could predict the prognosis, although it needed more evaluation of pulmonary nodules, lymph nodes and organs of the whole body. In practice, the machine learning method with distinct features in various dimensions of lung nodules based on CT scans could help make better-individualized management decisions. Also, it may be of great value to explore this technique in predicting recurrence or metastasis after surgery, which lead to clinical utility for the choice of any adjuvant or neoadjuvant medical therapy.
Diverse features and spheres of the application were seen between the deep learning and radiomics model. The deep learning system, only requiring user-defined ROI as input, could discover the underlying hierarchical features, so we used GradCAM to explain the decision-making process of deep neural network. Relatively, radiomics has better interpretability, which was based on feature engineering, comprising of complex procedures including image segmentation, feature extraction, feature selection and model building[34]. In addition, deep learning methods generally require a lot of data, but prognostic information is difficult to track [35]. Therefore, due to the small number of cases, the deep learning method did not display a comparably favorable prognostic performance as radiomics. In contrast, massive information regarding pathological and imaging characteristics were convenient to obtain. With these large amount of data, further application of artificial intelligence in precise diagnosis of lung cancer and prognosis prediction were thus possible.
It has been a significant research field in recent years that using radiographic features to predict pathological classification or prognosis. And a host of studies have confirmed that model construction based on radiomics can achieve good performance even with a small sample size. However, CT radiomic features vary according to the reconstruction kernel used for image generation [36]. To improve the reproducibility of imaging features, we thereby choose to finely segment the nodules through deep learning. However, this may lead to the model focus on the heterogeneity within the tumor instead of tumor microenvironment. But the deep learning model can make up for this shortcoming. It can be seen from the attention map that the deep-learning–based model tends to focus on the area adjacent to the nodules and pleura to predict the pathological type of adenocarcinoma, yet from where features cannot be extracted by the radiomics-based model. In addition, with the increase of data samples, deep learning may perform a better performance and our experimental results show that the model ensemble deep learning and radiomics may bring better performance.
There still were several limitations of the current study. Firstly, it was a multicenter retrospective study based on the real-world clinical practice, and the number of samples and the clinical features of adenocarcinoma were heterogeneous depending on the different medical centers. The number of patients in the validation cohort was relatively small. Secondly, it may be challenging to provide the treatment modalities through AI-based classification on non-invasive CT scans with comprehensive histologic evaluation for lung adenocarcinoma patients in the context of the numerous treatment modalities currently available based on histopathological results. Thirdly, patients with lung cancer undergoing surgery were selected, most of whom were at an early stage. Lung adenocarcinoma consisted of mixtures of several histologic subtypes and pathological subtypes such as AAH and micropapillary patterns were rare. Thus, might not be fully learned by the deep neural networks and the population we selected may not represent the whole population. All factors above may cause the model deviation and affect the robustness of our model.
In summary, deep learning makes it possible to determine adenocarcinoma subtypes without invasive complete surgery, which is a powerful and influential tool to automatically predict prognosticate prognosis of lung cancer patients, reforming the selection strategies of the adjuvant treatment. With competent prediction accuracy, our diagnostic and prognostic models of lung adenocarcinoma based on deep learning methods could enhance the efficiency in daily medical practice. Further, to improve the practicability and generalizability of this model, we will seek to develop a more comprehensive integration of imaging, pathological and biomarker features based on the combination of deep learning and radiomics modalities in the foreseeable future.
Contributors
WML conceived and supervised the project. CDW, JS, JWL and DL recruited the patients, collected and analyzed the data. CDW, JWL, YDC, CNZ, WS and LS contributed to acquisition, analysis, or interpretation of data. All authors discussed the results, wrote and reviewed the manuscript, agreed to its submission.
Ethical approval
This study was conducted in accordance with the Declaration of Helsinki and approved by the institutional ethics committee of participating institutions.
Declaration of Competing Interest
The authors declare no conflict of interest.
Acknowledgments
This study was supported by grants 91859203, 81871890 from National Natural Science Foundation of China, grant 2017-CY02–00030-GX from the Science and Technology Project of Chengdu, grant 2020YFG0473 from the Science and Technology Project of Sichuan, 2021SCU12018 from Postdoctoral Program of Sichuan University, and 2020HXBH084 from Postdoctoral Program of West China Hospital, Sichuan University.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2021.101141.
Contributor Information
Dan Liu, Email: liudanscu@qq.com.
Weimin Li, Email: weimi003@scu.edu.cn.
Appendix. Supplementary materials
References
- 1.Aberle D.R., Adams A.M., Berg C.D., Black W.C., Clapp J.D., Fagerstrom R.M., Gareen I.F., Gatsonis C., Marcus P.M., Sicks J.D. Reduced lung-cancer mortality with low-dose computed tomographic screening. N. Engl. J. Med. 2011;365(5):395–409. doi: 10.1056/NEJMoa1102873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Koning H.J., van der Aalst C.M., de Jong P.A., Scholten E.T., Nackaerts K., Heuvelmans M.A., Lammers J.J., Weenink C., Yousaf-Khan U., Horeweg N., van 't Westeinde S., Prokop M., Mali W.P., Mohamed Hoesein F.A.A., van Ooijen P.M.A., Aerts J., den Bakker M.A., Thunnissen E., Verschakelen J., Vliegenthart R., Walter J.E., Ten Haaf K., Groen H.J.M., Oudkerk M. Reduced lung-cancer mortality with volume CT screening in a randomized trial. N. Engl. J. Med. 2020;382(6):503–513. doi: 10.1056/NEJMoa1911793. [DOI] [PubMed] [Google Scholar]
- 3.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021 doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 4.Shi J.F., Wang L., Wu N., Li J.L., Hui Z.G., Liu S.M., Yang B.Y., Gao S.G., Ren J.S., Huang H.Y., Zhu J., Liu C.C., Fan J.H., Zhao S.J., Xing P.Y., Zhang Y., Li N., Lei W.D., Wang D.B., Huang Y.C., Liao X.Z., Xing X.J., Du L.B., Yang L., Liu Y.Q., Zhang Y.Z., Zhang K., Qiao Y.L., He J., Dai M. Clinical characteristics and medical service utilization of lung cancer in China, 2005-2014: overall design and results from a multicenter retrospective epidemiologic survey. Lung Cancer. 2019;128:91–100. doi: 10.1016/j.lungcan.2018.11.031. [DOI] [PubMed] [Google Scholar]
- 5.Travis W.D., Brambilla E., Noguchi M., Nicholson A.G., Geisinger K.R., Yatabe Y., Beer D.G., Powell C.A., Riely G.J., Van Schil P.E., Garg K., Austin J.H., Asamura H., Rusch V.W., Hirsch F.R., Scagliotti G., Mitsudomi T., Huber R.M., Ishikawa Y., Jett J., Sanchez-Cespedes M., Sculier J.P., Takahashi T., Tsuboi M., Vansteenkiste J., Wistuba I., Yang P.C., Aberle D., Brambilla C., Flieder D., Franklin W., Gazdar A., Gould M., Hasleton P., Henderson D., Johnson B., Johnson D., Kerr K., Kuriyama K., Lee J.S., Miller V.A., Petersen I., Roggli V., Rosell R., Saijo N., Thunnissen E., Tsao M., Yankelewitz D. International association for the study of lung cancer/american thoracic society/european respiratory society international multidisciplinary classification of lung adenocarcinoma. J. Thorac. Oncol. 2011;6(2):244–285. doi: 10.1097/JTO.0b013e318206a221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Russell P.A., Wainer Z., Wright G.M., Daniels M., Conron M., Williams R.A. Does lung adenocarcinoma subtype predict patient survival?: a clinicopathologic study based on the new International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society international multidisciplinary lung adenocarcinoma classification. J. Thorac. Oncol. 2011;6(9):1496–1504. doi: 10.1097/JTO.0b013e318221f701. [DOI] [PubMed] [Google Scholar]
- 7.Murakami S., Ito H., Tsubokawa N., Mimae T., Sasada S., Yoshiya T., Miyata Y., Yokose T., Okada M., Nakayama H. Prognostic value of the new IASLC/ATS/ERS classification of clinical stage IA lung adenocarcinoma. Lung Cancer. 2015;90(2):199–204. doi: 10.1016/j.lungcan.2015.06.022. [DOI] [PubMed] [Google Scholar]
- 8.Wang C., Wu Y., Shao J., Liu D., Li W. Clinicopathological variables influencing overall survival, recurrence and post-recurrence survival in resected stage I non-small-cell lung cancer. BMC Cancer. 2020;20(1):150. doi: 10.1186/s12885-020-6621-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Van Schil P.E., Asamura H., Rusch V.W., Mitsudomi T., Tsuboi M., Brambilla E., Travis W.D. Surgical implications of the new IASLC/ATS/ERS adenocarcinoma classification. Eur. Respir. J. 2012;39(2):478–486. doi: 10.1183/09031936.00027511. [DOI] [PubMed] [Google Scholar]
- 10.Wang W., Hu Z., Zhao J., Huang Y., Rao S., Yang J., Xiao S., Cao R., Ye L. Both the presence of a micropapillary component and the micropapillary predominant subtype predict poor prognosis after lung adenocarcinoma resection: a meta-analysis. J. Cardiothorac. Surg. 2020;15(1):154. doi: 10.1186/s13019-020-01199-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miyahara N., Nii K., Benazzo A., Hoda M.A., Iwasaki A., Klepetko W., Klikovits T., Hoetzenecker K. Solid predominant subtype in lung adenocarcinoma is related to poor prognosis after surgical resection: a systematic review and meta-analysis, European journal of surgical oncology: the journal of the. Eur. Soc. Surg. Oncol. Br. Assoc. Surg. Oncol. 2019;45(7):1156–1162. doi: 10.1016/j.ejso.2019.01.220. [DOI] [PubMed] [Google Scholar]
- 12.Song S.H., Park H., Lee G., Lee H.Y., Sohn I., Kim H.S., Lee S.H., Jeong J.Y., Kim J., Lee K.S., Shim Y.M. Imaging phenotyping using radiomics to predict micropapillary pattern within lung adenocarcinoma. J. Thorac. Oncol. 2017;12(4):624–632. doi: 10.1016/j.jtho.2016.11.2230. [DOI] [PubMed] [Google Scholar]
- 13.MacMahon H., Naidich D.P., Goo J.M., Lee K.S., Leung A.N.C., Mayo J.R., Mehta A.C., Ohno Y., Powell C.A., Prokop M., Rubin G.D., Schaefer-Prokop C.M., Travis W.D., Van Schil P.E., Bankier A.A. Guidelines for management of incidental pulmonary nodules detected on CT images: from the Fleischner society 2017. Radiology. 2017;284(1):228–243. doi: 10.1148/radiol.2017161659. [DOI] [PubMed] [Google Scholar]
- 14.Ettinger D.S., Wood D.E., Aisner D.L., Akerley W., Bauman J., Chirieac L.R., D'Amico T.A., DeCamp M.M., Dilling T.J., Dobelbower M., Doebele R.C., Govindan R., Gubens M.A., Hennon M., Horn L., Komaki R., Lackner R.P., Lanuti M., Leal T.A., Leisch L.J., Lilenbaum R., Lin J., Loo B.W., Jr., Martins R., Otterson G.A., Reckamp K., Riely G.J., Schild S.E., Shapiro T.A., Stevenson J., Swanson S.J., Tauer K., Yang S.C., Gregory K., Hughes M. Non-small cell lung cancer, version 5.2017, NCCN clinical practice guidelines in oncology. J. Natl. Compr. Canc. Netw. 2017;15(4):504–535. doi: 10.6004/jnccn.2017.0050. [DOI] [PubMed] [Google Scholar]
- 15.Esteva A., Kuprel B., Novoa R.A., Ko J., Swetter S.M., Blau H.M., Thrun S. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542(7639):115–118. doi: 10.1038/nature21056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gulshan V., Peng L., Coram M., Stumpe M.C., Wu D., Narayanaswamy A., Venugopalan S., Widner K., Madams T., Cuadros J., Kim R., Raman R., Nelson P.C., Mega J.L., Webster D.R. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA. 2016;316(22):2402–2410. doi: 10.1001/jama.2016.17216. [DOI] [PubMed] [Google Scholar]
- 17.Ardila D., Kiraly A.P., Bharadwaj S., Choi B., Reicher J.J., Peng L., Tse D., Etemadi M., Ye W., Corrado G., Naidich D.P., Shetty S. End-to-end lung cancer screening with three-dimensional deep learning on low-dose chest computed tomography. Nat. Med. 2019;25(6):954–961. doi: 10.1038/s41591-019-0447-x. [DOI] [PubMed] [Google Scholar]
- 18.Limkin E.J., Sun R., Dercle L., Zacharaki E.I., Robert C., Reuzé S., Schernberg A., Paragios N., Deutsch E., Ferté C. Promises and challenges for the implementation of computational medical imaging (radiomics) in oncology. Ann. Oncol. 2017;28(6):1191–1206. doi: 10.1093/annonc/mdx034. [DOI] [PubMed] [Google Scholar]
- 19.Zhou Y., Xu X., Song L., Wang C., Guo J., Yi Z., Li W. The application of artificial intelligence and radiomics in lung cancer. Precis. Clin. Med. 2020;3(3):214–227. doi: 10.1093/pcmedi/pbaa028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao W., Yang J., Sun Y., Li C., Wu W., Jin L., Yang Z., Ni B., Gao P., Wang P., Hua Y., Li M. 3D deep learning from CT scans predicts tumor invasiveness of subcentimeter pulmonary adenocarcinomas. Cancer Res. 2018;78(24):6881–6889. doi: 10.1158/0008-5472.CAN-18-0696. [DOI] [PubMed] [Google Scholar]
- 21.Yanagawa M., Niioka H., Kusumoto M., Awai K., Tsubamoto M., Satoh Y., Miyata T., Yoshida Y., Kikuchi N., Hata A., Yamasaki S., Kido S., Nagahara H., Miyake J., Tomiyama N. Diagnostic performance for pulmonary adenocarcinoma on CT: comparison of radiologists with and without three-dimensional convolutional neural network. Eur. Radiol. 2020 doi: 10.1007/s00330-020-07339-x. [DOI] [PubMed] [Google Scholar]
- 22.Wang X., Li Q., Cai J., Wang W., Xu P., Zhang Y., Fang Q., Fu C., Fan L., Xiao Y., Liu S. Predicting the invasiveness of lung adenocarcinomas appearing as ground-glass nodule on CT scan using multi-task learning and deep radiomics. Transl. Lung Cancer Res. 2020;9(4):1397–1406. doi: 10.21037/tlcr-20-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Travis W.D., Brambilla E., Nicholson A.G., Yatabe Y., Austin J.H.M., Beasley M.B., Chirieac L.R., Dacic S., Duhig E., Flieder D.B., Geisinger K., Hirsch F.R., Ishikawa Y., Kerr K.M., Noguchi M., Pelosi G., Powell C.A., Tsao M.S., Wistuba I. The 2015 world health organization classification of lung tumors: impact of genetic, clinical and radiologic advances since the 2004 classification. J. Thorac. Oncol. 2015;10(9):1243–1260. doi: 10.1097/JTO.0000000000000630. [DOI] [PubMed] [Google Scholar]
- 24.Cui S., Ming S., Lin Y., Chen F., Shen Q., Li H., Chen G., Gong X., Wang H. Development and clinical application of deep learning model for lung nodules screening on CT images. Sci. Rep. 2020;10(1):13657. doi: 10.1038/s41598-020-70629-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.S. Chen, K. Ma, Y. Zheng, Med3D: transfer learning for 3D medical image analysis, ArXiv abs/1904.00625 (2019).
- 26.Camp L.R. X-tile: a new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin. Cancer Res. 2004;10(21):7252–7259. doi: 10.1158/1078-0432.CCR-04-0713. [DOI] [PubMed] [Google Scholar]
- 27.Tsao M.S., Marguet S., Teuff G.Le, Lantuejoul S., Shepherd F.A., Seymour L., Kratzke R., Graziano S.L., Popper H.H., Rosell R., Douillard J.Y., Le-Chevalier T., Pignon J.P., Soria J.C., Brambilla E.M. Subtype classification of lung adenocarcinoma predicts benefit from adjuvant chemotherapy in patients undergoing complete resection. J. Clin. Oncol. 2015;33(30):3439–3446. doi: 10.1200/JCO.2014.58.8335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suh Y.J., Lee H.J., Kim Y.T., Kang C.H., Park I.K., Jeon Y.K., Chung D.H. Added prognostic value of CT characteristics and IASLC/ATS/ERS histologic subtype in surgically resected lung adenocarcinomas. Lung Cancer. 2018;120:130–136. doi: 10.1016/j.lungcan.2018.04.007. [DOI] [PubMed] [Google Scholar]
- 29.Cruz V.Da, Yvorel V., Casteillo F., Tissot C., Luchez A., Bayle-Bleuez S., Fournel P., Tiffet O., Péoc'h M., Forest F. Histopathological subtyping is a prognostic factor in stage IV lung adenocarcinoma. Lung Cancer. 2020;147:77–82. doi: 10.1016/j.lungcan.2020.07.010. [DOI] [PubMed] [Google Scholar]
- 30.Su H., Xie H., Dai C., Zhao S., Xie D., She Y., Ren Y., Zhang L., Fan Z., Chen D., Jiang F., Liu J., Zhu Q., Yao J., Ke H., Zhang L., Wu C., Jiang G., Chen C. Procedure-specific prognostic impact of micropapillary subtype may guide resection strategy in small-sized lung adenocarcinomas: a multicenter study. Ther. Adv. Med Oncol. 2020;12 doi: 10.1177/1758835920937893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ujiie H., Kadota K., Chaft J.E., Buitrago D., Sima C.S., Lee M.C., Huang J., Travis W.D., Rizk N.P., Rudin C.M., Jones D.R., Adusumilli P.S. Solid predominant histologic subtype in resected stage I lung adenocarcinoma is an independent predictor of early, extrathoracic, multisite recurrence and of poor postrecurrence survival. J. Clin. Oncol. 2015;33(26):2877–2884. doi: 10.1200/JCO.2015.60.9818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ettinger D.S., Wood D.E., Aggarwal C., Aisner D.L., Akerley W., Bauman J.R., Bharat A., Bruno D.S., Chang J.Y., Chirieac L.R., D'Amico T.A., Dilling T.J., Dobelbower M., Gettinger S., Govindan R., Gubens M.A., Hennon M., Horn L., Lackner R.P., Lanuti M., Leal T.A., Lin J., Loo B.W., Jr., Martins R.G., Otterson G.A., Patel S.P., Reckamp K.L., Riely G.J., Schild S.E., Shapiro T.A., Stevenson J., Swanson S.J., Tauer K.W., Yang S.C., Gregory K., Hughes M. NCCN guidelines insights: non-small cell lung cancer, version 1.2020. J. Natl. Compr. Canc. Netw. 2019;17(12):1464–1472. doi: 10.6004/jnccn.2019.0059. [DOI] [PubMed] [Google Scholar]
- 33.Varghese C., Rajagopalan S., Karwoski R.A., Bartholmai B.J., Maldonado F., Boland J.M., Peikert T. Computed tomography-based score indicative of lung cancer aggression (SILA) predicts the degree of histologic tissue invasion and patient survival in lung adenocarcinoma spectrum. J. Thorac. Oncol. 2019;14(8):1419–1429. doi: 10.1016/j.jtho.2019.04.022. [DOI] [PubMed] [Google Scholar]
- 34.Gillies R.J., Kinahan P.E., Hricak H. Radiomics: images are more than pictures, they are data. Radiology. 2016;278(2):563–577. doi: 10.1148/radiol.2015151169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.He J., Baxter S.L., Xu J., Xu J., Zhou X., Zhang K. The practical implementation of artificial intelligence technologies in medicine. Nat. Med. 2019;25(1):30–36. doi: 10.1038/s41591-018-0307-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Choe J., Lee S.M., Do K.H., Lee G., Lee J.G., Lee S.M., Seo J.B. Deep learning-based image conversion of CT reconstruction kernels improves radiomics reproducibility for pulmonary nodules or masses. Radiology. 2019;292(2):365–373. doi: 10.1148/radiol.2019181960. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.