Summary
The impact of systemic therapy on the tumor microenvironment has been difficult to study in human solid tumors. Our protocol describes steps for establishing slice cultures to investigate response to chemotherapies, immunotherapies, or adoptive cell therapies. Endpoints include changes in viability, histology, live-cell imaging, and multi-omics analyses. The protocol has been applied to a broad array of gastrointestinal malignancies. Culture conditions and treatment parameters can be modified for specific experiments. The platform is highly flexible and easy to manipulate.
For complete details on the use and execution of this protocol, please refer to Kenerson et al. (2020), Jabbari et al. (2020), Brempelis et al. (2020), and Jiang et al. (2017).
Subject areas: Cell culture, Cell-based Assays, Cancer, High Throughput Screening, Immunology
Graphical abstract
Highlights
-
•
Organotypic tumor slice cultures (TSCs) can be utilized to study human cancer
-
•
TSCs provide a model to study effects of chemo-, immuno-, and cell-based therapies
-
•
Tumor response to treatment can be assessed using multiple readouts
The impact of systemic therapy on the tumor microenvironment has been difficult to study in human solid tumors. Our protocol describes steps for establishing slice cultures to investigate response to chemotherapies, immunotherapies, or adoptive cell therapies. Endpoints include changes in viability, histology, live-cell imaging, and multi-omics analyses. The protocol has been applied to a broad array of gastrointestinal malignancies. Culture conditions and treatment parameters can be modified for specific experiments. The platform is highly flexible and easy to manipulate.
Before you begin
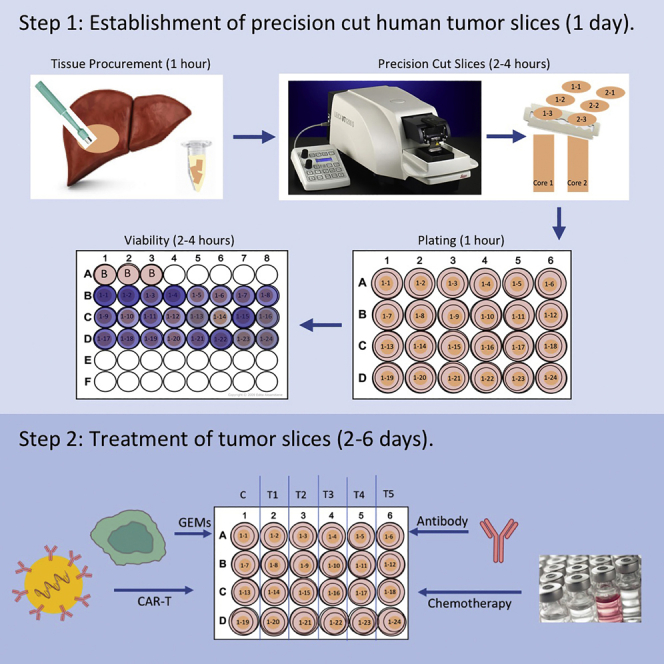
Organotypic precision-cut slices from solid tumors are an effective in vitro model for studying human solid cancer that preserves the tumor microenvironment, including immune components. Tumor slice cultures (TSC) can be established from most tumors within a day. However, they have a finite lifespan and are non-renewable. TSC are most suitable for short-term studies including experiments using drugs and cell-based therapies. Response to treatment can be assessed using multiple readouts including tissue viability, RNA and protein analyses, histology, and live-cell imaging.
Identification of potential research tissue and patient consent
Timing: 1 week
-
1.
Patients undergoing elective surgical resection are reviewed for potential tissue collection.
-
2.The following criteria for research collection must be met:
-
a.Patient has consented to participate in the study.
-
b.Tumors are greater than 2 cm in diameter.
-
c.Pathologic assessment will not be compromised from the tissue collection.
-
a.
CRITICAL: Tissue is only collected if the patient consents to the research study.
Note: Cases that are treatment -naïve or have progressed despite treatment are better suited for slice culture experiments given the likelihood of viable tumor.
Preparation of reagents prior to tissue collection
Timing: 30 min–1 h
-
3.
Prepare 5 mL Eppendorf tubes with 3 mL of collection buffer. Keep on ice. Here we use Belzer UW cold Storage Solution, but PBS or media could be used.
-
4.
Prepare cutting buffer. Keep on ice or at 4°C until use. Here we use William’s E Media with 1% Penicillin Streptomycin, but another appropriate media or solution could be used.
-
5.
Prepare wash/growth media. Here we use modified William’s E Media (Kenerson et al., 2020), but any other appropriate media can be used as well.
Note: The media used to grow tumor slices or non-tumor liver slices in culture should be optimized for specific tissue and endpoint assays.
-
6.
Transfer 400 μL wash buffer into each well of a 48-well plate(s), depending on the number of slices desired, and place in tissue culture incubator.
-
7.Prepare 2% agarose solution and place in 37°C water bath.
-
a.Add 1 gram of low- melting point agarose to 25 mL of sterile dPBS in a sterile 250 mL Erlenmeyer flask.
-
i.Microwave on low power until agarose has completely dissolved.
-
i.
-
b.Stop microwave and swirl liquid every 20 s so that the contents do not boil over.
-
c.Allow the liquid to cool down slightly in 37°C water bath and then add 25 mL of pre-warmed William’s Medium E or base media of choice. Leave in 37°C water bath until ready to use.
-
a.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Ki67 | Agilent | Cat# M7249; RRID:AB_2250503 |
| Alexa 647 EpCAM (9C4) | BioLegend | Cat#324228; RRID:AB_2563209 |
| Biological samples | ||
| Human tumor tissue (CRCLM, PDA, HCC) | University of Washington Medical Center | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| HEPES | Gibco | Cat #15630080 |
| Sodium bicarbonate | Gibco | Cat #25080-094 |
| Sodium pyruvate | Gibco | Cat #11360070 |
| L-Glutamine | Gibco | Cat #25030081 |
| Penicillin-Streptomycin | Gibco | Cat #15140-122 |
| Nicotinamide | Sigma-Aldrich | Cat #N-0636 CAS: 98-92-0 |
| L-Ascorbic acid 2-phosphate | Sigma-Aldrich | Cat #A8960-5G CAS: 113170-55-1 |
| D-(+)-Glucose | Sigma-Aldrich | Cat #G5767 CAS: 50-99-7 |
| hEGF | Fisher Scientific | Cat #354052 |
| Corning ITS + premix supplement | Fisher Scientific | Cat #354352 |
| William’s E Medium, no glutamine | Gibco | Cat #12551032 |
| PBS | Gibco | Cat #20012027 |
| DMSO | Sigma-Aldrich | Cat #D2650 CAS: 67-68-5 |
| Fluorouracil (5-FU) | Selleck | Cat #S1209 CAS: 51-21-8 |
| Oxaliplatin | Selleck | Cat #S1224 CAS: 61825-94-3 |
| Irinotecan | Selleck | Cat #S2217 CAS: 136572-09-3 |
| RNA Later | Invitrogen | Cat #AM7020 |
| 10% Neutral buffered formalin | Fisher Scientific | Cat #23-245684 CAS: 50-00-0 |
| UltraPure Low Melting Point Agarose | Sigma-Aldrich | Cat#16520100 |
| Carboxyfluorescein succinimidyl ester (CFSE) | BioLegend | Cat#423801 |
| Hoechst 33342 | ImmunoChemistry Technologies | Cat#639 |
| 8-Well culture slide | ibidi | Cat#80806 |
| Critical commercial assays | ||
| CellTiter 96®AQueous One Solution Cell Proliferation Assay | Promega | Cat #G3582 |
| SR-FLICA | ImmunohistoChemistry Technologies | Cat#932 |
| Experimental models: Cell lines | ||
| Genetically engineered Macrophages | Brempelis et al., 2020, Moyes et al., 2017 | N/A |
| Software and algorithms | ||
| Leica LAS X software | Lecia Microsystems | https://www.leica-microsystems.com/products/microscope-software/p/leica-las-x-ls/ |
| Imaris | Bitplane | https://imaris.oxinst.com/ |
| Other | ||
| Integra™ Miltex™ Standard Biopsy Punches (6 mm) | Fisher Scientific | Cat #12-460-412 |
| Leica VT 1200S vibrating microtome | Leica Biosystems | https://www.leicabiosystems.com/histology-equipment/sliding-and-vibrating-blade-microtomes/vibrating-blade-microtomes/leica-vt1200-s/ |
| Blades (double edge, PTFE coated) | Ted Pella | Cat #121-6 |
| Adhesive | Ted Pella | Cat #7085-85-0 |
| Millicell Cell Culture Insert, 12 mm, hydrophilic PTFE, 0.4 μm | Millipore | Cat #PICM01250 |
| Microplate reader | Molecular Devices Optimax | N/A |
| Confocal microscope | Leica Biosystems | Cat#SP8x |
Materials and equipment
Alternatives: This protocol requires a precision cutting instrument, such as a vibrating blade microtome, in order to generate thin slices of tumor tissue for in vitro culturing. The vibrating blade microtome aids in maintaining the morphology and cell viability of the tissue. The use of a microtome for slicing fresh tissue also minimizes artifacts, compression distortion, cell destruction and other inherent deleterious effects of sectioning. In this protocol we used a Leica VT 1200S vibrating blade microtome to generate 250 μm sections of fresh tissue. Detailed description of this instrument can be found on manufacturer’s website (www.leica.com). Similar instruments are available from other manufacturers.
Alternatives: When working with fresh tissue cores, an ice-cold collection buffer is used to submerge tissue upon procurement to aid in maintaining the vitality of the tissue. For our purposes we used UW Belzer cold storage solution. Alternatively, tissue could be collected in ice-cold PBS or media. When tissue is being sectioned into thin slices it is submerged in an ice-cold cutting buffer. We use William’s E Media supplemented with 1% Pen/Strep. However one could substitute this with another buffer such as ice-cold UW Belzer solution, Krebs-Henseleit buffer (KHB) or base media of choice.
Alternatives: Here we use a modified William’s E Media without human EGF for wash buffer. When slices are plated and cultured we add human EGF (200 ng/mL) to the modified Williams E media. Wash/growth buffer should be optimized by user depending on tissue type and desired endpoints (Jiang et al., 2017, Wu et al., 2018, Wu et al., 2020).
Modified William’s E Media
| Reagent | Final concentration | Stock concentration | Add to 500 mL |
|---|---|---|---|
| Nicotinamide | 12 mM | 100 mM | 6 mL |
| L-Ascorbic Acid 2-phosphate | 175 μM | 14.5 mM | 6 mL |
| Sodium Bicarbonate | 0.225% (w/v) | 7.5% (w/v) | 15 mL |
| HEPES | 20 mM | 1 M | 10 mL |
| D-(+)-Glucose | 0.5% (w/v) | 25% (w/v) | 10 mL |
| Sodium Pyruvate | 1 mM | 100 mM | 5 mL |
| L-Glutamine | 2 mM | 200 mM | 5 mL |
| Penicillin Streptomycin | 0.4% | N/A | 2 mL |
| ITS + Premix | 1% | N/A | 5 mL |
| Williams’ Media E | 436 mL |
Modified Williams E media can be stored for 4 weeks at 4°C. Before use as growth media, human EGF is added fresh to small aliquots to a final concentration of 200 ng/mL. Once human EGF is added aliquot should be used within 3–5 days.
Alternatives: In order to assess tissue viability we use the CellTiter 96® AQueous One Solution Cell Proliferation Assay (MTS) from Promega. The MTS reagent is converted by cells into a soluble colored formazan product, which uses absorbance as a read out. There are different assays available that could be used to assess tissue viability, such as Real Time Glo (Promega) (Sivakumar et al., 2019).
Alternatives: Tissue must be placed on a membrane in order for it to survive in culture. Here we use cell culture inserts from Millipore which consist of a hydrophilic polytetrafluoroethylene (PTFE) membrane with pore size of 0.4 μm. Alternative cell culture inserts consisting of other types of membrane material, such as polycarbonate or PET, or different pores sizes are available. However, we have not performed a direct comparison of these products to the one used here.
Step-by-step method details
Sterile collection of human tissue
Timing: 1 h
Sterile collection of 6 mm biopsy cores from human solid tumors for the purpose of establishing an in vitro model for testing/manipulation.
-
1.
Research representative and pathology representative are called to the operating room when resection is near complete.
-
2.
Once the specimen is handed off the operative field, it is kept sterile for immediate tissue procurement.
CRITICAL: The specimen must remain in the sterile field and manipulated with sterile gloves and instruments only.
-
3.
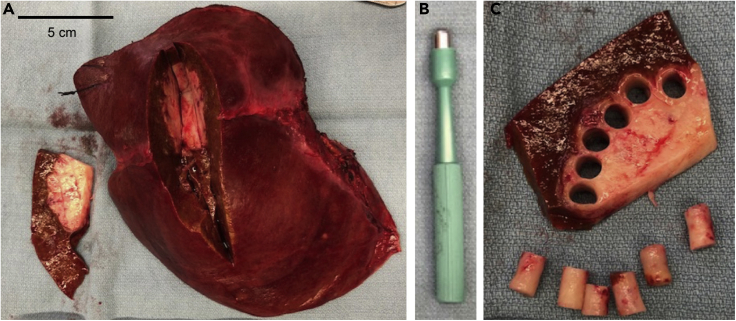
Under the observation of a pathologist, a wedge of tumor tissue ∼ 1 cm thick is removed using sterile surgical scalpel or scissors (Figure 1A).
CRITICAL: Tissue collection is done in the presence of a pathology representative to ensure that a complete and proper evaluation of the specimen, including tumor margins and orientation will be maintained. Tissue should not be removed if it will compromise the pathological evaluation.
-
4.
Using a 6-mm biopsy punch, cores of tumor tissue are obtained from the wedge (Figures 1B and 1C). Samples are placed directly in 3 mL of ice-cold Belzer UW Cold Storage Solution in 5 mL Eppendorf tubes (sterile), placed on ice, and transported to research lab.
Note: To maximize tissue viability, cores are taken from the periphery of the tumor to avoid areas of central necrosis (Figure 1C). There is significant heterogeneity among cores taken from different regions of a tumor (Kenerson et al., 2020), and one needs to consider this issue when designing the experiment.
CRITICAL: The time between the completion of surgical resection and tissue procurement (i.e., warm ischemic time) should be kept to a minimum – ideally under 10 min. If time between collection and tissue procurement exceeds 45 min viability of slices should be tested prior to proceeding with an experiment. Once tissue is procured it is transported on ice immediately to the research lab for the generation of precision cut slices.
Figure 1.
Sterile tumor tissue procurement and tumor core punches
(A) Upon completion of liver resection, specimen is placed on a sterile field. With guidance of pathologist a 1 cm thick wedge of tissue is removed from specimen to access tumor tissue. Scale bar is equal to 5 cm.
(B) A 6 mm biopsy punch is used to core tumor tissue for slice protocol.
(C) Samples are taken from tumor periphery to avoid areas of central necrosis.
Precision cut slices
Timing: 2–4 h
Note: The following procedure is described for a vibrating blade microtome instrument such as the Leica VT 1200S vibratome.
-
5.
Remove tissue cores from UW solution with sterile forceps and place in a sterile petri dish.
-
6.
Dab excess liquid on sterile gauze (Figure 2A).
-
7.Embed cores in 2% agarose for slicing.
-
a.Individual cores can be positioned in the center of one well of a sterile 24-well plate.
-
b.Pipette warm (37°C), 2% agarose solution over top of core(s) in each well until tissue is entirely covered, cover plate, and place on ice (Figure 2B).
-
i.Agarose should solidify in 5 min.
-
i.
-
a.
Note: Multiple cores (up to 5) can also be embedded in agarose using one well of a 6-well plate instead of in individual wells of a 24-well dish (Figure 2C).
CRITICAL: Embedding cores in agarose is important for tissue cores with a softer consistency.
Note: Tissue cores can also be directly glued to the specimen disc with an adhesive for slicing and longer cores can be cut in half with sterile scalpel if desired for greater stability.
-
8.
Once the agarose has solidified, use a sterile scalpel or spatula to cut along the edge of the agarose in the well. With a scooping motion pop the embedded tissue out of the well and onto a sterile petri dish.
-
9.Using a drop of superglue, glue the face of the embedded core that was on the bottom of the well directly to the specimen disc. The top of the agarose will have a concave shape.
-
a.Up to 4 cores in individual agarose from 24-well or 6 cores not in agarose can be glued to the specimen disc at one time for efficient slicing. If cores were embedded in a 6-well plate, one well (containing up to 5 cores) can be adhered to specimen disc (Figures 2D and 2E).
-
a.
CRITICAL: Ensure that both the tissue and agarose are in contact with the adhesive.
-
10.
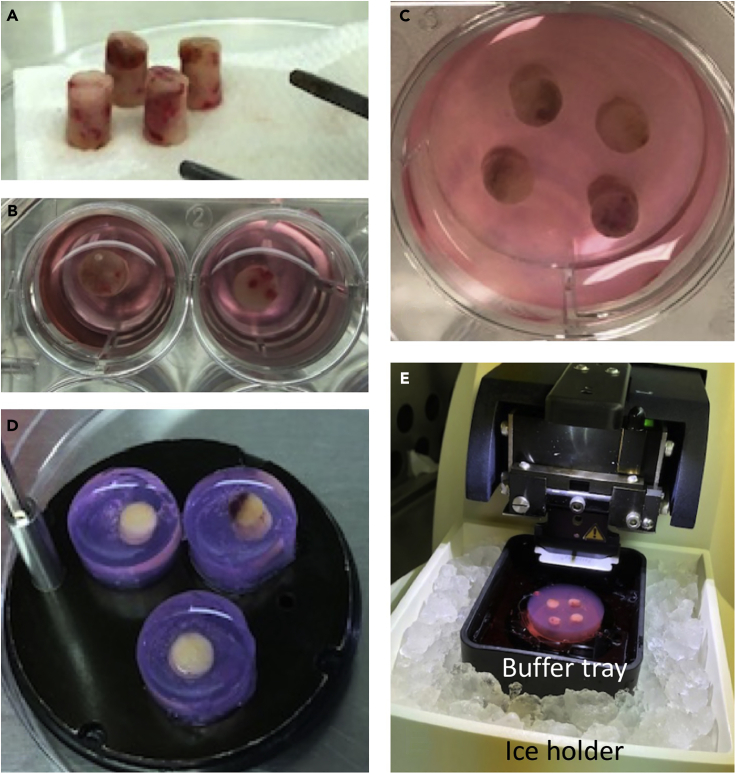
Place the buffer tray (Figure 2E) in ice holder and cover. Add ice around the buffer tray.
-
11.
The specimen disc is then placed in the buffer tray and the buffer tray filled with ice-cold cutting buffer (Figure 2E).
CRITICAL: The vibratome should be located in a BSL-2-approved biosafety cabinet for slicing human tissue. Appropriate PPE should be worn while working in a BSL-2 area and when working with human tissue.
-
12.
Slide the ice tray onto the vibratome stand and lock the tray in place by moving the lever to the right.
-
13.
Secure blade into blade holder with an Allen wrench.
-
14.
Using the Allen wrench rotate the blade into cutting position 1, 2, or 3.
-
15.
Raise the stage up using the control pad until the blade touches the liquid in the buffer tray.
-
16.
Set cutting window using the control pad.
-
17.
Once the window is set, using the run feature step down 250 μm at a time at 1.5 mm/s until blade reaches the tallest tissue core.
-
18.
Adjust settings on the vibratome depending on the consistency and integrity of the tissue and the number of cores that are being sliced at one time, (amplitude 2–3, speed 0.5–1.5 mm/s). Troubleshooting
-
19.
Once uniform 250 μm slices are obtained, gently scoop them out of the bath using sterile spatula or forceps.
-
20.
Place each slice in one well of the 48-well dish containing prewarmed modified WME or media of choice.
-
21.
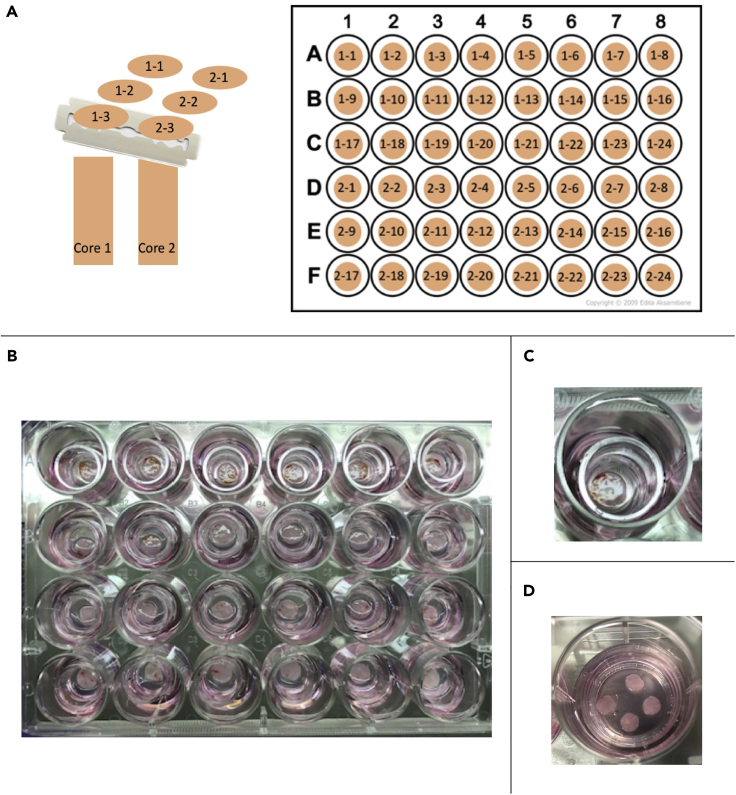
Each core and slice are assigned numerical values so that sequence and orientation of slices can be tracked and documented (Figure 3A).
Note: Depending on core length and tissue consistency the number of slices obtained from a core can vary. On average from a 1 cm long core, 24 – 32 slices can be generated. When tissue consistency is too firm or too soft the blade may run over the tissue.
Note: Depending on specific experimental needs, slice thickness can be adjusted accordingly. In our experience, 250 μm slices are a good compromise between ease of tissue slicing/handling and diffusion/penetration of nutrients and drugs.
-
22.
Once a 48-well plate is filled with slices, place in incubator on a rocker (37°C , ∼ 20 rocks/min) for 1 – 4 h for washing until all slicing is complete.
Note: Before plating slices in cell culture inserts it is recommended to assess tissue viability on a small sample of slices to ensure tissue is viable before experiments commence.
Figure 2.
Tissue cores embedded in agarose
(A) Cores are removed from transport solution and excess liquid removed.
(B and C) (B) Cores are embedded in agarose individually in wells of a 24-well plate or (C) grouped in a well of a 6-well plate.
(D) Agarose and tissue are adhered to specimen disc.
(E) Specimen disc is placed in buffer tray with cutting solution on ice for slicing.
Figure 3.
Plating tissue slices
(A) Tumor slices are placed in wash buffer in a 48-well dish in order of core and slice number.
(B) Slices are then transferred onto individual Millicell culture inserts placed in a 24-well plate with 450 μL of media.
(C) One tumor slice plated on a 12 mm insert in a 24-well dish.
(D) 4 tumor slices plated on a 30 mm insert in a 6-well dish.
Plating slices for culture
Timing: 1 h
-
23.
Place one Millicell cell culture insert into each well of a 24-well tissue culture plate using sterile forceps for each slice that will be plated.
-
24.
Pipette 450 μL of growth media, modified William’s media E supplemented with hEGF (20 ng/mL), into each well, ensuring that at least 100 μL of media is placed in the insert.
CRITICAL: Tissue slices must be plated on cell culture inserts to maintain viability.
Note: Media will eventually equilibrate on its own, but in order to plate slices without damaging softer tissue, at least 100 μL of media is needed in each insert.
-
25.
Using a 1000 μL pipette tip or sterile forceps gently transfer one slice into each insert. Use the liquid in the insert to help remove slice from tip. Once transferred, use the pipette tip to gently move the floating slice so that it is completely flat (some slices will fold during transfer). Remove media in the insert to the outside of insert to ensure the slice is flat on the cell insert membrane, otherwise the slice may fold or bend if media is allowed to equilibrate out of the insert (Figures 3B and 3C). Some media will eventually equilibrate back into the insert.
Note: If desired, 3 to 4 slices can be placed on a 30 mm Millicell cell culture insert that fits in a 6-well tissue culture dish with 1.25 mL of media (Figure 3D).
-
26.
Once slices are plated, place plate on a rocker (20 rocks/min) at 37°C, 5% CO2. Rocking allows the tissue interface to be exposed to media and air intermittently. (Leeman, et al., 1995)
Note: Rocking is optional.
Assessment of tissue viability (MTS assay) or pre-treatment MTS
Timing: 2–4 h
Assessment of initial tumor viability can be performed on the day of slicing or the after one day in culture depending on timing and treatment scheme. This can be done by performing a cell viability/proliferation assay on at least 3 slices from different areas of the tumor. The more slices that are cultured from different cores or areas of tumor, the more slices should be assessed and the slices should be dispersed throughout the sampling of the tumor. This will ensure that the tissue that you are working with is viable.
We have chosen to asses viability by using the CellTiter 96® AQueous One Solution Cell Proliferation Assay (MTS) from Promega which does not require solubilization. We have used MTS to assess viability of slices before and after drug treatment to calculate a percent change in viability for each slice. This method provides the ultimate level of control since you are able to quantify the change in viability for each individual slice. We have had success using this method on tumor slices that are highly robust, such as treatment naïve colorectal liver metastases. However, the MTS reagent/process can have deleterious effects in some tissue slices, which can negatively affect tissue growth in culture. Since one is unable to predict how the tissue will behave in vitro, we have largely relied on a single MTS readout after drug treatment.
-
27.
Thaw 5 mL aliquots of CellTiter 96 Aqueous One Solution (MTS reagent) in 37°C water bath.
-
28.
Place 400 μL of media into each well of a 48-well dish
-
29.
Transfer slices from the Millicell insert to individual wells. Keeping track of slice identification number. Keep 3 wells without slices for blanks.
-
30.
Add 80 μL of MTS reagent to each well of the plate.
Note: MTS reagent is light sensitive, turn off lights in Biosafety Cabinet when using.
-
31.Incubate tissue with MTS reagent for approximately 3 h.
-
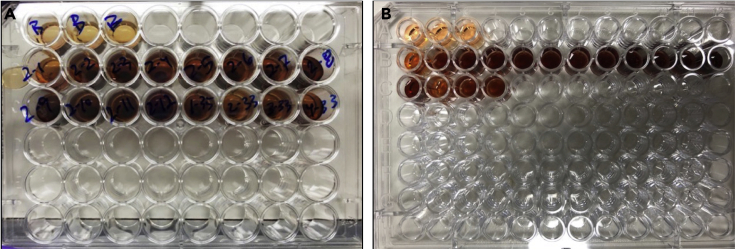
a.Media and slice will change to a purple color (Figure 4A). Slices can be incubated with MTS reagent from 1 – 4 ho depending on color change that occurs. In our experience we incubate most tissue for 3 h.
-
a.
-
32.
Once incubation is finished, transfer 200 μL of each blank well and each slice well to a 96-well assay plate (Figure 4B).
-
33.
Read absorbance at 490 nm, blanks are averaged and subtracted from the values of the wells containing slices. Troubleshooting
-
34.
At this point slices can be fixed in 10% formalin for histology, frozen for protein analysis, or transferred back on initial Millicell insert for drug testing and further culturing.
CRITICAL: It is important to keep track of every slice when moving from insert to 48-well dish for MTS so that the pre and post MTS can be used to calculate change in viability for each slice.
Figure 4.
MTS assay on tissue slices
(A) Tissue slices are transferred to individual wells of a 48-well dish containing 400 μL of media. 80 μL of MTS reagent is added. Plate is incubated at 37°C and 5% CO2 for 3 h.
(B) 200 μL of media is transferred to 96-well assay plate and absorbance read at 490 nm.
Example of treatment of slices: Chemotherapy/immunotherapy
Timing: 2–6 days
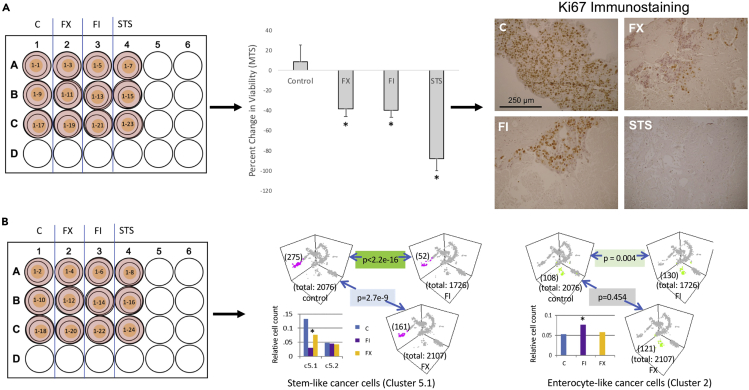
The tumor slice culture method can be used to study tumor response to chemotherapies, immunotherapies, or combined chemo-immunotherapies. In previously published work (Jabbari et al., 2020) the response of human colorectal liver metastases (CRLM) treated with chemotherapy and immunotherapy combinations was evaluated utilizing tissue slice cultures. Here we show an example of a case of CRLM treated with two different chemotherapy combinations with endpoints for tumor response including viability (based on percent change in MTS absorbance), histology including immunohistochemistry, and single-cell RNA sequencing.
CRITICAL: In our experience, slices that are generated from tumors that have undergone treatments such as chemotherapy or radiation may not grow well in the in vitro system. Untreated tumors are ideal for drug treatment experiments since they are treatment naïve. Tumors that are progressing on treatment are also likely to grow well in vitro.
-
35.Tumor slices are prepared as described above and incubated for 12 to 18 h.
-
a.In this example (Figure 5), a pre-treatment MTS is performed on half of the slices before plating in order to determine change in viability with treatment.
-
a.
-
36.The following day working solutions of chemotherapies or immunotherapies are prepared fresh by diluting stock solutions of each drug in media.
-
a.In this example n of 3 slices were treated with the following
-
i.Control - 0.2% DMSO (vehicle)
-
ii.FX - 1 μg/mL 5-fluorouracil + 1 μg/mL oxaliplatin
-
iii.FI - 1 μg/mL 5-fluorouracil + 2 μg/mL irinotecan
-
iv.10 μM Staurosporine - positive control
-
i.
-
a.
CRITICAL: Since consecutive slices are more biologically identical and have been shown to have comparable MTS readouts, they are purposely dispersed across treatment groups when comparing different treatments. See Figures 5A and 5B for example of slice dispersion.
Note: If slices are treated with immunotherapies or combined chemo-immunotherapies (Jabbari et al., 2020) use appropriate IgG antibody for negative control.
-
37.Slices are incubated at 37°C and 5% CO2 with chemotherapy or immunotherapy for desired treatment period. Adding fresh drug every 1 to 2 days depending on treatment duration.
-
a.In Figure 5, tissue slices were treated with 3 consecutive days adding fresh media with drug daily.
-
a.
Note: Depending on chosen experimental chemotherapies and immunotherapies, length and dosing of treatments will vary. For FDA-approved drugs, we use drug concentrations that are achievable in serum based on available clinical data. In general, we treat slices for 3 to 5 days depending on desired endpoints. We add media with fresh drugs every one to two days.
-
38.At the end of the treatment downstream assays are performed.
-
a.In Figure 5A, one set of slices were subject to a post-treatment MTS followed by fixation in 10% formalin for histological analysis.
-
i.Percent change in viability was calculated for each slice using the following equation. Percent change in viability = ((post-treatment MTS – pre-treatment MTS)/pre-treatment MTS Absorbance))∗100. The percent change in viability is averaged for each treatment group (Figure 5A). P-value was calculated using Students t-test. Troubleshooting
-
i.
-
b.Additional treated slices are placed in RNAlater for RNAseq analysis (Figure 5B).
-
a.
CRITICAL: It is necessary to keep track of the identification number of each slice and its corresponding treatment group. This will be imperative when calculating percent change in viability from an MTS absorbance obtained on each slice before and after treatment.
Note: Slices that have been exposed to MTS reagent can also be fixed for histological analysis or frozen for protein analysis, such as Western Blot. This is beneficial since you can achieve multiple readouts on the same tissue slice and streamline experiments.
CRITICAL: If slices will be used for RNA analysis or live imaging they should not be exposed to MTS reagent and need to have parallel treatment replicates for these endpoints.
Figure 5.
Tumor treatment scheme with multiple readouts
Tumor slices from a CRLM were treated with vehicle control (0.2% DMSO), FX (1 μg/mL 5FU + 1 μg/mL Oxaliplatin), FI (1 μg/mL 5FU + 2 μg/mL Irinotecan, or STS (10 μm Staurosporine, to serve as a positive control) two times over 4 days.
(A) One set of treated slices were used to calculate percent change in viability using pre-treatment and post-treatment MTS values and subsequently fixed for histology. Data are represented as mean ± standard deviation. Fixed slices were processed for immunohistochemistry staining with Ki67 (Dako, 1:200). Scale bar is equal to 250 μm.
(B) Another set of treated slices were used to perform single-cell RNA sequencing. Figure reprinted with permission from Jabbari et al., 2020. Note how slices are dispersed over the treatment and endpoint assay.
Example of treatment of slices: Adoptive cellular therapy
Timing: 3–6 days
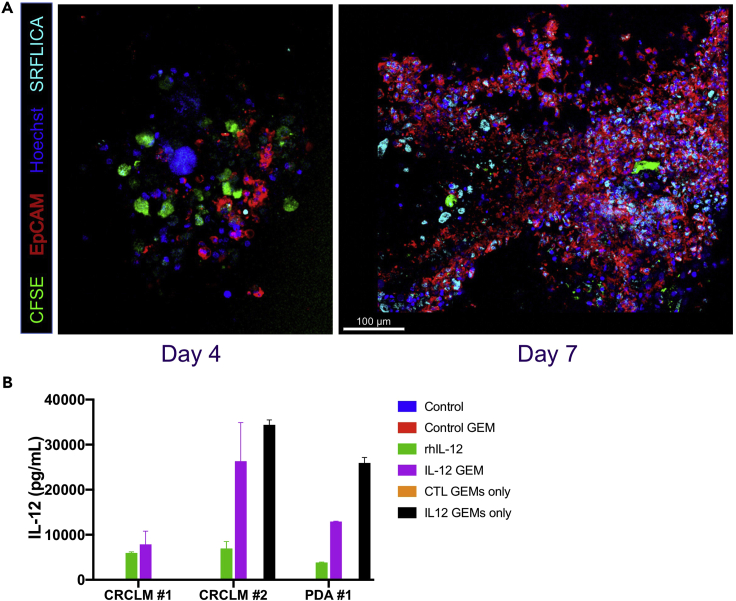
The tumor slice culture system can be used to evaluate adoptive cellular therapy. In previously published work (Brempelis et al., 2020) the therapeutic efficacy of genetically engineered macrophages (GEMs) were evaluated in tumor slice culture. The GEMs infiltrate and persist in tumor slice for at least 1 week in culture (Figure 6). Downstream assays of tumor cell death, cytokine production and modification of tumor immune microenvironment were conducted. Our lab has also treated slices with CAR-T cells in order to evaluate the effect of modulation of the tumor microenvironment on CAR-T anti-tumor immunity.
-
39.
GEMs are prepared. See previously published work for details (Brempelis et al., 2020, Moyes et al., 2017). Alternatively, CAR-T cells can be utilized and thawed from frozen aliquots at the time of the experiment.
-
40.
Tumors slices are prepared as described above.
-
41.
Slices are incubated 12 – 18 h.
-
42.The day after slicing, cells are prepared as follows:
-
a.GEMs or CAR-T cells are stained with Carboxyfluorescein succinimidyl ester (CFSE) per manufacturer’s protocols.
-
b.GEMs or CAR-T cells are resuspended in cell culture media to a predetermined cell number and concentration desired for the experiment (we have used CAR-T cells at a count of 105–106 per slice).
-
a.
-
43.
Previously untreated slice culture media was removed and replaced with 400 μL of slice culture media with any additional treatment at appropriate concentration or controls (for example, cytokine blockade).
-
44.
Cellular therapy was added to each slice to achieve a final volume of 450 μL.
-
45.
Slices with GEMs or CAR-T cells are incubated at 37°C and 5% CO2 for the desired treatment period.
-
46.Downstream assays performed
-
a.Four and seven days after GEM addition, slices were fixed in formalin for histology and immunohistochemistry, stored in RNAlater for transcriptomic analyses, and/or Live Cell Imaging performed as well as quantification of IL-12 production via ELISA (Figure 6).
-
a.
Figure 6.
GEMS infiltrate, persist, and function in tumor slice culture
(A) Confocal fluorescent microscopic images of colorectal cancer liver metastasis slice four (left panel) and seven days (right panel) after addition of GEMs. Scale bar is equal to 100 μm.
(B) IL-12 production quantified by ELISA in 3 slice culture experiments, 2 colorectal cancer liver metastasis (CRCLM) and 1 pancreatic ductal adenocarcinoma (PDA). Columns represent mean of 3 biologic replicates ± standard deviation.
Live cell imaging
Timing: 5–8 h
We have used fluorescent staining and confocal microscopic imaging of live slices to evaluate for evidence of anti-tumor immunity as a result of manipulation of the tumor microenvironment (Figure 6 and Methods video S1). In the above protocol, we stain the adaptive cellular therapy (for example, CAR-T cells) with CFSE for imaging. We also use fluorescently labeled antibodies to stain and identify tumor cells, for example those which express the EpCAM antigen. In addition, we labeled apoptotic cells with SR-FLICA reagent, which stains activated cleaved-caspase-3.
-
47.
Tumor slices are transferred from the original media which is set aside to a 48-well plate with 500 μL of fresh media with 10 μg/mL Alexa 647 EpCAM antibody and SR-FLICA reagent per the manufacturer’s instructions.
-
48.
The slices are incubated with the primarily conjugated fluorescent antibodies for 3 h at 37°C
-
49.
The slices are stained for nuclei using 10 μg/mL Hoechst 33342 (ImmunoChemistry Technologies) for 10 min.
-
50.
The slices are washed of the fluorescent staining antibodies by sequentially transferring twice to separate wells containing PBS, and returned with their original media containing the treatment to an 8-well chamber slides with a coverslip bottom (Ibidi) for imaging.
-
51.
The slices are maintained at 37°C using a covered stage (PeCon, Erbach, Germany) while flushing warmed, humidified CO2 through the enclosure in order to maintain culture conditions during the duration of the imaging.
-
52.
The slices are imaged using a confocal microscope (for example, Leica SP8X, Leica Microsystems) at 20× magnification. Each treated slice was imaged for 1 h at multiple positions throughout the slice with a z-stack of 20 mm (see Methods video S1 for live-imaging video of tissue slice culture).
-
53.
Image processing and data analysis were performed on Leica LAS X software (Leica Microsystems) and Imaris software (Bitplane).
-
54.
Apoptotic tumor cells that are EpCAM+ SR-FLICA+ and CFSE+ CAR-T cells can be counted to quantify cell populations. Imaris also has the capability to determine proximity analysis amongst other analyses.
EpCAM tumor cell (red) and immune cell (green) in HCC tumor slice. Video demonstrating cell movement during live microscopy in a tumor slice culture of hepatocellular carcinoma. An immune cell labelled with Alexa 488 CD45 migrates toward an Alexa 647 EpCAM tumor cell over the course of 1 h of imaging at 20× magnification.
Expected outcomes
Based on our experience, one can expect to generate 24-32 precision-cut (250μm) slices from a 1-cm long tissue core of 6 mm diameter. Once placed in culture, one can expect these short-lived human tumor slices to survive at least 5 days, depending on the state of the primary tissue (e.g., prior treatment effects, warm ischemic time, etc.). During this time, they can undergo many different types of manipulation and analyses, from global viability assessment to single-cell transcriptome analyses, as well as live imaging of immune cells. Examples of these outcomes are shown in Figures 4 – 6 and Video 1.
Limitations
The tumor slice culture system is a non-renewable system and is best suited for short-term studies including experiments using drugs and cell-based therapies. Due to the inherent nature of the tissue slice culture being derived from individual patients not all tumors behave the same. In our experience CRLM is the most reliably grown tumor in vitro unless there is extensive necrosis due to prior chemotherapy or radiation. On the other hand, tumors such as hepatocellular carcinoma are more variable in their ability to remain viable long-term in culture. For any particular sample, there remain many conditions that can be fine-tuned for optimal growth including, but not limited to, components of the media, thickness of the slice, ambient oxygen level, and the extent of physical motion. Our protocol provides a starting point for anyone who wants to explore this method in their studies. A critical component of the protocol is the access to fresh, sterile tissue specimens.
Troubleshooting
Problem 1
Difficulty slicing tissue
Potential solution
Tissue consistency plays an important role in the ability to slice tissue. If tissue is too hard then the blade has a tendency to run over the tissue causing inconsistent tissue thickness between slices. If the tissue is too soft it lacks stability to hold up to the blade when slicing. Adjust speed and amplitude settings on vibratome. Harder tissues cut best when blade is in position 3 (more angled). If still having trouble thicker tumor slices may be achievable. Embed softer tissue cores in agarose to provide stability.
Problem 2
Initial tumor viability low
Potential solution
An MTS assay readout with an average absorbance of 0.75 or less after 3 h of incubation with MTS reagent is considered a low viability and slices are not suitable for treatment studies. Ensure that tissue cores are taken from periphery of tumor to avoid central necrosis. When selecting possible research tissue avoid cases that are heavily pre-treated or had a near complete response to treatment for tissue slice experiments.
Problem 3
Absorbance reading from MTS viability assay is too high
Potential solution
Check the color change of the media by naked eye every 30 min to determine the appropriate incubation of slices with MTS reagent. Ideally the absorbance at 490 nm should be between 1 and 2, but can range from 0 – 3.5 as read on a Molecular Devices Optimax plate reader used in our lab. Check absorbance reading of an extra slice(s) during the MTS assay (1–2 h) to assess how the purple color change correlates to the absorbance reading in order to determine an appropriate incubation time for specific tissue.
Problem 4
Large percent change in viability in control slices
Potential solution
A calculated percent change in viability of - 50% in control slices or lower indicates that the tissue may have been affected by the MTS reagent. Perform a post-treatment MTS only as to not expose tissue slice to MTS reagent prior to treatment.
Problem 5
Low protein or RNA yield.
Potential solution
Pool more slices from each treatment group to increase yield.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Raymond Yeung (ryeung@uw.edu).
Materials availability
This protocol relies on fresh primary tissues. There is no cell line or vector generated in this study.
Data and code availability
Not applicable.
Acknowledgments
The authors thank Nathaniel Peters at the University of Washington W.M. Keck Microscopy Center for assistance with live image confocal microscopy and Dr. Alan F. Utria for critical reading of the protocol. This work was supported by the DoD grant W81XWH-16-1-0149.
Author contributions
H.L.K., K.P.L., and K.M.S. designed and performed experiments and analyzed and interpreted data. R.S.Y. and V.G.P. designed experiments, interpreted data, and provided funding and resources. H.L.K., K.P.L., and K.M.S. contributed protocols for the manuscript with input from R.S.Y. All authors contributed feedback for the final manuscript.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xpro.2021.100574.
Contributor Information
Heidi L. Kenerson, Email: kenerson@uw.edu.
Raymond S. Yeung, Email: ryeung@uw.edu.
References
- Brempelis K., Cowan C., Kreuser S., Labadie K., Prieskorn B., Lieberman N., Ene C., Moyes K., Chinn H., DeGolier K. Genetically engineered macrophages persist in solid tumors and locally deliver therapeutic proteins to activate immune responses. J. ImmunoTher. Cancer. 2020;8:e001356. doi: 10.1136/jitc-2020-001356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabbari N., Kenerson H., Lausted C., Yan X., Meng C., Sullivan K., Baloni P., Bergey D., Pillarisetty V., Hood L. Modulation of immune checkpoints by chemotherapy in human colorectal liver metastases. Cell Rep. Med. 2020;1:100160. doi: 10.1016/j.xcrm.2020.100160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X., Seo Y., Chang J., Coveler A., Nigjeh E., Pan S., Jalikis F., Yeung R., Crispe I., Pillarisetty V. Long-lived pancreatic ductal adenocarcinoma slice cultures enable precise study of the immune microenvironment. OncoImmunology. 2017;6:e1333210. doi: 10.1080/2162402X.2017.1333210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenerson H., Sullivan K., Seo Y., Stadeli K., Ussakli C., Yan X., Lausted C., Pillarisetty V., Park J., Riehle K. Tumor slice culture as a biologic surrogate of human cancer. Ann. Transl. Med. 2020;8:114. doi: 10.21037/atm.2019.12.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeman W.R., van de Gevel I.A., Rutten A.A. Cytotoxicity of retinoic acid, menadione and aflatoxin B(1) in rat liver slices using Netwell inserts as a new culture system. Toxicol. In Vitro. 1995;9:291–298. doi: 10.1016/0887-2333(95)00008-v. [DOI] [PubMed] [Google Scholar]
- Moyes K., Lieberman N., Kreuser S., Chinn H., Winter C., Deutsch G., Hoglund V., Watson R., Crane C. Genetically engineered macrophages: a potential platform for cancer immunotherapy. Human Gene Therapy. 2017;28:200–215. doi: 10.1089/hum.2016.060. [DOI] [PubMed] [Google Scholar]
- Sivakumar R., Chan M., Shin J., Nishida-Aoki N., Kenerson H., Elemento O., Beltran H., Yeung R., Gujral T. Organotypic tumor slice cultures provide a versatile platform for immuno-oncology and drug discovery. OncoImmunology. 2019;8:e1670019. doi: 10.1080/2162402X.2019.1670019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Roberto J., Knupp A., Kenerson H., Truong C., Yuen S., Brempelis K., Tuefferd M., Chen A., Horton H. Precision-cut human liver slice cultures as an immunological platform. J. Immunol. Methods. 2018;455:71–79. doi: 10.1016/j.jim.2018.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Hollingshead N., Roberto J., Knupp A., Kenerson H., Chen A., Strickland I., Horton H., Yeung R., Soysa R. Human liver macrophage subsets defined by CD32. Front. Immunol. 2020;11:2108. doi: 10.3389/fimmu.2020.02108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
EpCAM tumor cell (red) and immune cell (green) in HCC tumor slice. Video demonstrating cell movement during live microscopy in a tumor slice culture of hepatocellular carcinoma. An immune cell labelled with Alexa 488 CD45 migrates toward an Alexa 647 EpCAM tumor cell over the course of 1 h of imaging at 20× magnification.
Data Availability Statement
Not applicable.