Abstract
Metabolism influences locomotor behaviors, but the understanding of neural curcuit control for that is limited. Under standard light-dark cycles, Drosophila exhibits bimodal morning (M) and evening (E) locomotor activities that are controlled by clock neurons. Here, we showed that a high-nutrient diet progressively extended M activity but not E activity. Drosophila tachykinin (DTk) and Tachykinin-like receptor at 86C (TkR86C)-mediated signaling was required for the extension of M activity. DTk neurons were anatomically and functionally connected to the posterior dorsal neuron 1s (DN1ps) in the clock neuronal network. The activation of DTk neurons reduced intracellular Ca2+ levels in DN1ps suggesting an inhibitory connection. The contacts between DN1ps and DTk neurons increased gradually over time in flies fed a high-sucrose diet, consistent with the locomotor behavior. DN1ps have been implicated in integrating environmental sensory inputs (e.g., light and temperature) to control daily locomotor behavior. This study revealed that DN1ps also coordinated nutrient information through DTk signaling to shape daily locomotor behavior.
Subject terms: Behavioural genetics, Circadian regulation
Lee and colleagues report the effect of a high-sucrose diet on Drosophila locomotor activity via DTk-TkR86C neuropeptide signalling. This signalling pattern appears to involve a circadian element, with pacemaker neuron involvement having a possible time-of-day effect on locomotor behaviour.
Introduction
The circadian clock system allows living organisms to anticipate environmental changes that are driven by the earth’s daily rotation, resulting in ~24-h rhythms in behavior and physiology. In animals, the cell-autonomous circadian clocks are organized into the master clock, residing in the brain, and peripheral clocks located throughout the body. The master clock is reset by external timing signals called zeitgebers, which in turn synchronize peripheral clocks through innervation and humoral signals1,2. The molecular mechanism controlling the circadian clock is a cell-autonomous transcriptional–translational feedback loop comprising the core clock genes3,4.
While the most potent zeitgeber is light, food also influences the circadian clock system5–7. Notably, timed-restricted feeding drives animal’s food-anticipatory activity8 and resets peripheral clocks, independent of master clocks6,9. Also, food content modulates rhythmic behaviors. In mice, a high-fat diet (HFD) reduces the rhythmicity and lengthens the periods of activity10, and a high-fat /high-salt diet reduces locomotor activity11. While metabolic control of the cell’s molecular clock has been extensively studied12, the effect of food on the neural circuit control of daily locomotor activity is not well understood. The fruit fly, Drosophila melanogaster, provides a genetically tractable model system to study fundamental aspects of the circadian clock and metabolism that are shared with mammals3,13,14.
In light–dark cycle, Drosophila exhibits bimodal patterns of locomotor activity with morning (M) peak and evening (E) peak, separated by a siesta. The flies' rhythmic locomotor activity profile is determined by the circadian neuron network located in the lateral and dorsal regions of the brain15. Large and small lateral ventral neurons (lLNv and sLNv) called M oscillators control M activity, and lateral dorsal neurons (LNd) and a fifth sLNv neuron called E oscillators control E activity16–18. Posterior dorsal neuron 1s (DN1ps) can control both M and E activities and integrate environmental stimuli, such as light and temperature, for locomotor regulation19–23.
Neuropeptides which control many aspects of behavior and physiology24,25 play a role in rhythmic locomotor activity. The pigment dispersing factor (PDF) released by LNvs synchronizes the clock neuron network and determines the anticipatory M activity and the phase of E activity. In addition, neuropeptide F (NPF), short neuropeptide F (sNPF), and the ion transport peptide (ITP) contribute to rhythmicity26–28. Diuretic hormone 31 (DH31) awakens flies in the early morning29, while Diuretic hormone 44 (DH44), expressed in the cells of neuroendocrine pars intercerebralis (PI), functions as an output molecule to communicate with the downstream locomotor center30. Neuropeptide leucokinin (LK), expressed in the lateral horn (LHLK), controls rhythmicity and levels of locomotor activity31. Some neuropeptides regulate both locomotor activity and metabolism; sNPF and NPF promote feeding and sleep32,33. After nutrient depletion, NPF signaling promotes feeding and suppresses sleep via independent circuits34. Conversely, allatostatin A suppresses feeding but promotes sleep35. Insulin-like hormone peptides (Ilps, an ortholog of mammalian insulin and insulin-like growth factor), which control metabolic homeostasis, regulate age-dependent sleep fragmentation36 and sleep depth in starved animals37. The neuropeptide SIFamide that is expressed in a subset of PI cells is required for rhythms of both locomotor activity and feeding/fasting30,38.
How dietary nutrient affects Drosophila locomotor behavior has been studied. A high-sucrose diet (HSD) reduces total sleep39 or alters the timing of sleep40. Siesta begins slightly later in Drosophila fed a HSD than in flies fed a low-sucrose diet (LSD)40. Sleep analysis in flies is based on measuring the duration of inactivity hence the delayed siesta onset associated with a HSD could instead reflect a lengthened period of M activity, but Linford et al. did not discuss M activity in detail40. Flies reared on a HFD increase their total sleep and bouts of sleep, together with reduced lifespan and fecundity mediated by increased expression of adipokinetic hormone (AKH)41. Although the locomotor assay is used for sleep studies, the molecular and neural mechanisms that control sleep versus activity are clearly separable. However, the effects of diet on daily locomotor activity and their underlying neural mechanisms are unknown.
In this study, we examined D. melanogaster’s daily locomotor activity in high-nutrient conditions and investigated underlying neuropeptidergic control mechanisms. A HSD or a HFD extended M activity, but not E activity. DTk- and TkR86C-mediated signaling were required for the extended M activity. DTk-expressing neurons were anatomically and functionally connected to DN1ps. The contacts between DN1ps- and DTk-expressing neurons gradually increased over time in flies fed a HSD, which is consistent with the locomotor activity behavior. Collectively, these results indicated that in addition to the role of integrating temperature signals into the circadian clock, DN1ps integrated nutrient information through DTk signaling and controlled Drosophila locomotor behavior in a nutrient state-dependent manner.
Results
HSD extended morning activity but not evening activity
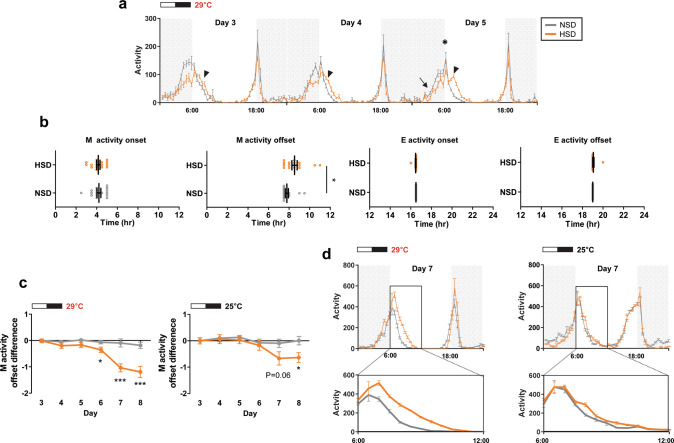
To evaluate how nutrient concentration affected fly daily locomotor behavior, we analyzed Drosophila melanogaster activity in normal concentration (5% sucrose, normal sucrose diet, NSD) or high concentration (30% sucrose, high sucrose diet, HSD) sucrose-containing food in a 12-h light/12-h dark (12L:12D) condition at 29 °C. Flies exhibited bimodal M and E activities around light on/off transition. The control w1118 flies fed on a NSD or a HSD showed differences in the M activity but not the E activity (Fig. 1a). Anticipatory activities controlled by the circadian clock (arrow, Fig. 1a) and the startle responses that occurred immediately after the light on/off transitions (asterisk, Fig. 1a) were not largely different under the two diet conditions42. However, while M activity in NSD was sharply reduced following the startle response M activity was extended in HSD (arrowhead, Fig. 1a). To quantitate this behavior for the two diet conditions, we compared locomotor activity onset and offset times (Fig. 1b). Activity onset indicated the largest 1-h increase before the light transition, while activity offset indicated the largest 1-h decrease after the light transition. As expected from the activity pattern (Fig. 1a), while the M activity onset, the E activity onset, and E activity offset were the same between the two diets, the M activity offset was delayed about 1 h, on average, in flies fed a HSD on day 5 compared to the M activity offset in flies fed a NSD (Fig. 1b). To test whether this effect was specific to sucrose, we measured fly locomotor activity in HFD containing 20% coconut oil with NSD. Because coconut oil melted at high temperatures, we performed the behavior analysis at 25 °C. Compared with the NSD, M activity was extended after the startle response in both the HSD and HFD, but E activity was not different in any diets (Supplementary Fig. 1a). Activity offset was delayed only in the mornings in HFD and HSD conditions (Supplementary Fig. 1b). We also noted that the effect of a high-nutrient diet on M activity was enhanced over time (Fig. 1a and Supplementary Fig. 2). M activity offset showed a delay each day until day 7 after which the delay was maintained (Fig. 1c, d). The HSD-induced M activity offset delay was somewhat reduced at 25 °C compared to 29 °C (Fig. 1d and Supplementary Fig. 2). Collectively, M activity was extended after the startle response when the nutrient content was high, and high-temperature augmented this effect.
Fig. 1. Fly M activity but not E activity offset was extended in HSD.
a, b w1118 fly locomotor activity was analyzed in normal sucrose diet (NSD) and high-sucrose diet (HSD) under a 12L:12D cycle at 29 °C. a Daily activity profiles from day 3 to day 5 are shown. Arrow indicates anticipatory activity and asterisk indicates the startle response. M activity, but not E activity, was extended in flies fed a HSD (arrowhead). b M and E activity onset and offset times for individual w1118 flies on day 5 are shown. Bars indicate mean ± SEM values (n = 28–31). Statistically significant differences in the onset or offset between NSD and HSD (independent t test): *P < 0.05. c, d w1118 fly locomotor activity was analyzed in NSD and HSD under 12L:12D cycle at 29 °C and 25 °C. Daily activity profiles for days 3–8 are in Supplementary Fig. 2. c M activity offsets of individual w1118 flies were obtained and the differences versus day 3 are shown. The M activity offset in HSD was progressively delayed at 29 °C and 25 °C. Values indicate mean ± SEM (n = 30–37). Statistically significant differences in the M activity offset between NSD and HSD at each day (independent t test): *P < 0.05; ***P < 0.001. d Daily activity profiles on day 7 at 29 °C and 25 °C are shown. Lower panels show a magnified image of the boxed region in the upper panel. The extent of M activity offset delay was greater at 29 °C than at 25 °C.
Since only the morning locomotor activity was extended in flies fed a HSD, we attempted to increase the phase relationship between the M and E activities by exposing flies to a long-day photoperiod, 16L:8D43. Extended M activity after the startle response was more prominent in the 16L:8D compared to 12L:12D (Figs. 1a and 2a). The extent of M activity offset delay was greater in the 16L:8D cycle compared to the 12L:12D, but M activity onset, E activity onset, and E activity offset were not altered in HSD (Fig. 2b). Therefore, we used a 16L:8D condition for the subsequent experiments and analyzed the activity profile at day 7 if not mentioned otherwise.
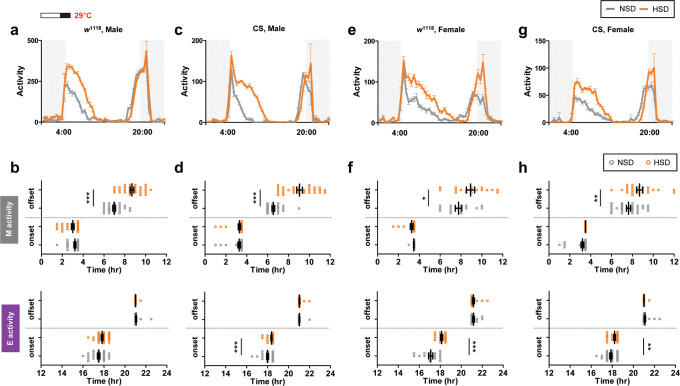
Fig. 2. The effect of a HSD on M activity offset was observed in other genotypes of control flies and in females.
a, c, e, g Locomotor activities of w1118 male (a) and female (e), Canton S (CS) male (c), and female (g) flies locomotor activities were analyzed in NSD and HSD conditions under a 16L:8D cycle at 29 °C. Daily activity profiles of given genotypes of flies (denoted on top) on day 7 are shown. b, d, f, h M and E activity onset/offset of w1118 male (b), w1118 female (f), CS male (d), and CS female (h) flies on day 7 are plotted. Bars indicate mean ± SEM (n = 15–32). Statistically significant differences in the onset or offset between NSD and HSD conditions (independent t test): *P < 0.05, **P < 0.01, ***P < 0.001.
Wild-type flies, Canton S, also had extended M activity (Fig. 2c) and showed an M offset delay in a HSD (Fig. 2d). Next, to examine for gender differences, we analyzed female locomotor behavior and found the same HSD effect on M activity for both male and female flies (Fig. 2e–h). In some instances, the E activity onset was slightly delayed but not as consistently as the M activity offset. Taken together, these results indicated that M activity extension in high-nutrient conditions is a universal behavioral response of Drosophila.
Neuropeptide tachykinin was required for M activity extension in flies on a HSD
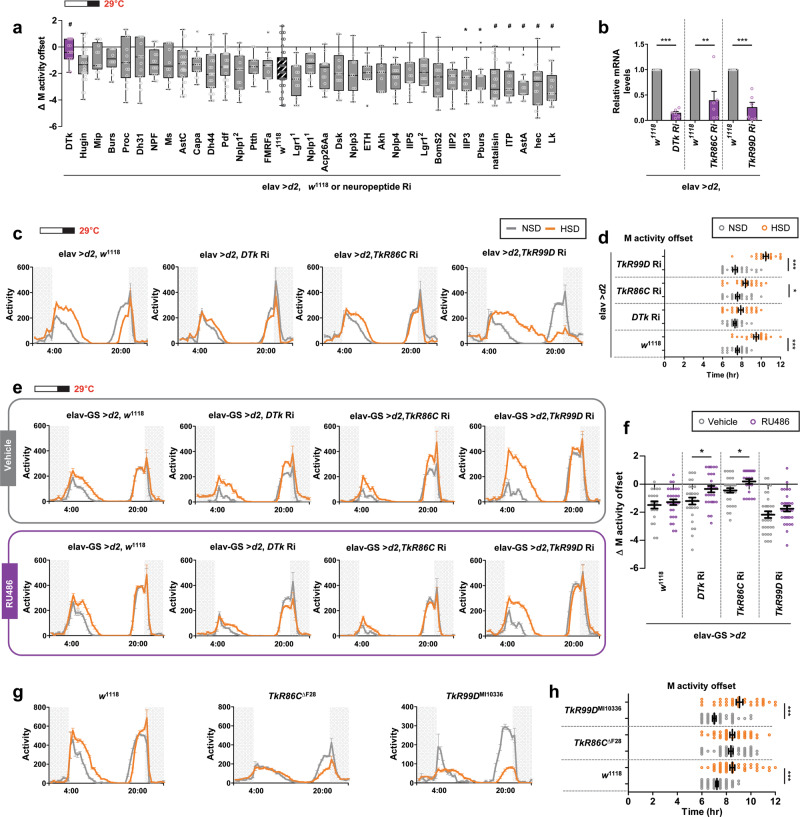
Neuropeptides are small proteins that modulate many aspects of physiology and behavior, such as feeding and rhythmic locomotor behavior24,25. We searched for neuropeptides mediating the HSD effect on locomotor behavior using an RNAi screen via a binary Gal4/UAS system44. Neuron-specific elav-Gal4 driver flies were crossed with w1118 or UAS-neuropeptide RNAi flies, and the offspring were used as a control or knockdown flies, respectively. Thirty-four neuropeptide genes were tested, and the ΔM activity offset on day 7 was determined and compared to the control (elav > dcr2, w1118) (Fig. 3a). Eight neuropeptide knockdown flies had an enhanced HSD-associated response. The DTk knockdown flies showed only a reduced HSD-associated response. Downregulation of DTk mRNA in the heads of the knockdown flies was confirmed by qRT-PCR (Fig. 3b). Daily activity profiles and comparisons of the M activity offset in individual flies indicated that pan-neuronal knockdown of DTk reduced the HSD-associated effects on M activity (Fig. 3c, d and Supplementary Fig. 3). Two DTk receptor isoforms, TkR86C (CG6515; neurokinin receptor from Drosophila, NKD) and TkR99D (CG7887; Drosophila tachykinin receptor, DTkR) have been cloned in Drosophila45–48. Pan-neuronal knockdown of TkR86C but not TkR99D, confirmed by qRT-PCR (Fig. 3b), diminished the M activity extension in flies fed a HSD (Fig. 3c, d). Knockdown of DTk also diminished the effects of a HSD in standard 12L:12D conditions at 25 °C (Supplementary Fig. 4). We then used the drug-inducible pan-neuronal elav-GeneSwitch driver to determine whether knockdown of DTk or TkR in adults diminished the HSD effect49. Flies in which DTk or TkR86C expression was downregulated by ingestion of RU486 showed less effect of a HSD compared with the control vehicle-treated flies (Fig. 3e, f). Flies in which most of the first exon of TkR86C was deleted (TkR86CΔF28) did not show M activity extension in a HSD (Figs. 3g, h)50. In contrast, a putative loss-of-function insertion mutation of TkR99D, TkR99DMI10336, did not affect the HSD-induced M activity offset delay (Fig. 3g, h). Interestingly, flies with loss-of-function mutations in TkR86C or TkR99D showed reduced E activity in HSD, suggesting that there might be a common regulatory role for these two receptors on E activity in flies fed a HSD.
Fig. 3. Neuropeptide DTk and the DTk receptor TkR86C were required for M activity extension in HSD.
a–d w1118 flies or UAS-neuropeptide RNAi (denoted on bottom) flies were crossed with elav-Gal4, UAS-dcr2 (elav > d2). a The locomotor activities of offspring were analyzed in NSD or a HSD under a 16L:8D cycle at 29 °C. Differences in average M activity offset between NSD and HSD (ΔM activity offset) on day 7 are shown. Control flies (hatched box;elav > d2, w1118) showed delayed M activity offset in HSD. The DTk knockdown flies (purple box; elav > d2, DTk Ri) showed little difference with a NSD versus a HSD (n = 9–16). Statistically significant differences in ΔM activity offset between the control and knockdown flies (independent t test): *P < 0.05, #P < 0.01. b Flies with the indicated genotypes fed a NSD on a 16L:8D cycle at 29 °C were collected at ZT2. DTk, TkR86C, and TkR99D mRNA levels were quantified by qRT-PCR. The mRNA levels in the knockdown flies were normalized to the control (elav > d2, w1118) flies. Values indicate mean ± SEM from six independent experiments. Statistically significant differences in mRNA levels between control and knockdown flies (independent t test): **P < 0.01, ***P < 0.001. c–h The locomotor activities of each fly genotype (denoted above each graph) were analyzed in NSD and HSD under a 16L:8D cycle at 29 °C. c, e, g Daily activity profiles of flies on day 7 are shown. d, h The M activity offsets of individual flies on day 7 are shown. Bars indicate mean ± SEM (n = 21–62). Statistically significant differences in the average time between NSD and HSD (independent t test): *P < 0.05, ***P < 0.001. f ΔM activity offset on day 7 are shown. Statistically significant differences in ΔM activity offset between vehicle- and RU486-treated groups for each genotype of flies (independent t test): *P < 0.05.
Tk includes an evolutionarily well-conserved family of brain/gut neuropeptides that function as important neuromodulators in the central and peripheral nervous systems (reviewed in ref. 51). DTks are also involved in various aspects of behavior and physiology, including locomotion52,53 and food-seeking behavior50. We tested the possibility that the downregulation of DTk or TkR86C affects feeding, thereby contributing to the reduced response in flies fed a HSD; however, food intake was similar in control, DTk and TkR86C knockdown flies consuming either diet (Supplementary Fig. 5). We also tested whether DTk signaling affects circadian rhythmicity behavior. Flies were entrained under the 12L:12D cycle followed by constant darkness, and their circadian locomotor behavior was analyzed in NSD or HSD (Supplementary Fig. 6). elav > d2, DTk Ri, elav > d2, TkR86C Ri, and elav > d2, TkR99D Ri flies exhibited a similar period and rhythmicity compared to control flies, indicating DTk signaling is not involved in regulating general circadian locomotor behavior. Nevertheless, all the fly genotypes showed a tendency toward a lengthened period and reduced robustness of rhythm in HSD, consistent with the previous mammalian study conducted in HFD50. In this behavior analysis, we also noted the M activity extension and offset delay in constant darkness; however different from LD cycle, E activity onset was slightly delayed in HSD. In constant darkness, the duration of the siesta decreased due to the absence of a strong, light-driven paradoxical masking effect54–56. Thus, the homeostatic drive to maintain a critical length of siesta might delay E activity onset. Taken together, these results revealed that neuropeptide DTk signaling via TkR86C specifically mediated the HSD-induced M activity extension in Drosophila.
DTk levels were upregulated in DTk neurons in flies on a HSD
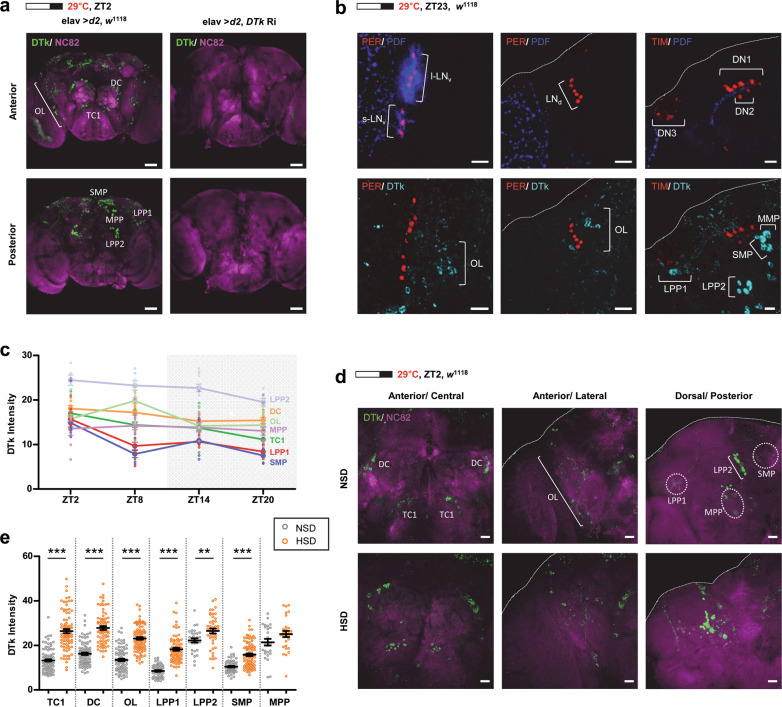
To examine how DTk mediated HSD affects on M activity, we immunostained fly brains with newly raised DTk antibodies. Immunostaining of control fly brains revealed DTk-positive clusters consistent with previous study51,57. In the anterior region, deutocerebrum (DC), tritocerebrum (TC1), and optic lobe (OL) clusters were DTk-positive. In the posterior region, superior median protocerebrum (SMP), lateral posterior protocerebrum 1 (LPP1), lateral posterior protocerebrum 2 (LPP2), and median posterior protocerebrum (MPP) clusters were DTk-positive (Fig. 4a). elav-Gal4 driving DTk knockdown flies did not exhibit DTk staining, which verified the antibody specificity and the downregulation of DTk in the knockdown flies. Because the DTk-positive neurons were in the lateral and dorsal areas where clock neurons are located, we determined whether the DTk-positive neurons were clock neurons. Clock neurons (e.g., LNvs, LNds, and DNs) labeled with anti-PERIOD (PER) or anti-TIMELESS (TIM) antibodies were contiguous with DTk-labeled neurons but did not overlap (Fig. 4b).
Fig. 4. DTk levels were increased in DTk neurons in flies fed a HSD.
a Flies of the indicated genotypes were maintained under 16L:8D cycle at 29 °C. Brains were dissected at ZT2 and stained with anti-DTk (green) and anti-NC82 (magenta) antibodies. In control flies (elav > d2, w1118), DTk-positive clusters were observed in DC, TC1, and in the OL on the anterior side. On the posterior side, DTk-positive clusters were observed in the SMP, LPP1, LPP2, and the MPP. DTk-positive clusters were absent in pan-neuronal DTk knockdown flies (elav > d2, DTk Ri). All scale bars represented 50 μm. b w1118 flies were maintained under a 16L:8D cycle at 29 °C. Brains were dissected at ZT23 and stained with anti-PER (red), anti-TIM (red), anti-PDF (blue), and anti-DTk (cyan blue) antibodies. DTk-positive neurons and clock neurons did not overlap but were in close proximity. All scale bars represented 20 μm. c w1118 flies were maintained with a NSD and 12L:12D cycle at 29 °C. Brains were dissected at each indicated time and stained with anti-DTk antibodies. DTk intensities of each cluster were quantified using ImageJ software and are shown. d, e w1118 flies were maintained with a NSD or HSD under a 16L:8D photoperiod at 29 °C. Brains were dissected on day 7, ZT2, and stained with anti-DTk (cyan blue) and anti-NC82 (magenta) antibodies. All scale bars represented 50 μm. e DTk intensities in each DTk-positive cluster were quantified using ImageJ software. Bars indicate mean ± SEM (n = 26–98). Statistically significant differences in the average intensity value between NSD and HSD (independent t test): **P < 0.01, ***P < 0.001.
We then assessed DTk levels under different diets. We first examined DTk levels throughout the day in flies fed a NSD and found that levels were highest early in the morning (e.g., ZT2) in most DTk neuronal clusters (Fig. 4c). Flies were maintained in a NSD or HSD, and on day 7 brains were dissected at ZT2 and immunostained with DTk antibody (Fig. 4d). DTk staining intensities were higher in every DTk neuron in the brains from the flies fed a HSD compared with the NSD (Fig. 4e). The DTk signal increase was observed in the soma and the knockdown of DTk or TkR using RNAi attenuated the HSD effects; therefore, the enhanced staining intensity likely resulted from increased expression rather than inhibition of DTk release in HSD.
DTk neurons and DN1ps were anatomically and functionally connected
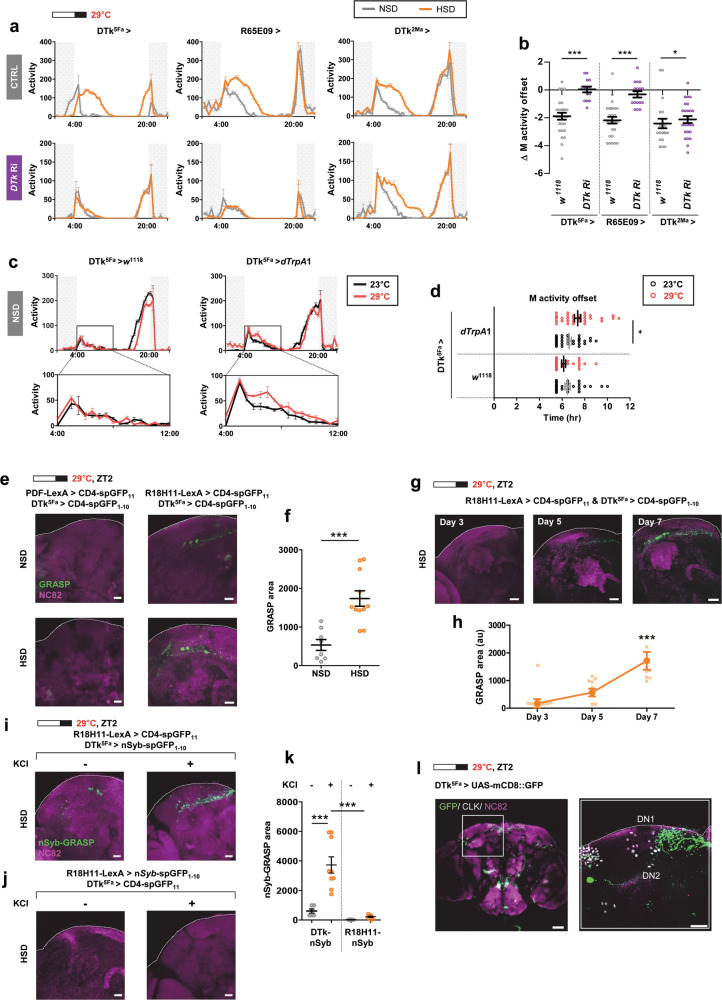
Instead of using elav-Gal4 to knock down DTk in a pan-neuronal manner, DTk was downregulated in limited groups of cells using three Gal4 lines, DTk5Fa-Gal4, DTk2Ma-Gal4, and R65E09-Gal4. DTk5Fa-Gal4 and DTk2Ma-Gal4 were generated in the collection of neuropeptide promoter-GAL4 strains58,59. R65E09-Gal4, a Janelia Gal4 line, is associated with the DTk promoter region60,61. While three DTk-Gal4 control flies exhibited M activity extension (Fig. 5a) and prominent M offset delays (Fig. 5b) in HSD, the knockdown of DTk using DTk5Fa-Gal4 or R65E09-Gal4, but not DTk2Ma-Gal4, abolished the HSD effects. Thus, DTk5Fa and R65E09-Gal4 neurons mediated the HSD effects on M activity.
Fig. 5. DTk neurons and DN1Ps were anatomically and functionally connected.
a, b Fly locomotor activity for w1118 (control) or UAS-DTk Ri (DTk Ri) driven by different DTk-Gal4 (DTk5Fa-Gal4, DTk2Ma-Gal4, and R65E09-Gal4) was analyzed in NSD and HSD under a 16L:8D cycle at 29 °C. a Daily activity profiles of flies on day 7 are shown. b ΔM activity offsets on day 7 are shown. DTk knockdown in DTk5Fa-Gal4, and 65E09-Gal4 active cells abolished the HSD effect, but not in the DTk2Ma-Gal4 active cells. (n = 14–26). Statistically significant differences in ΔM activity between control and DTk knockdown flies (independent t test): *P < 0.05, ***P < 0.001. c, d Flies were entrained in 16L:8D cycle at 23 °C for 7 days in NSD. The temperature was then elevated to 29 °C for 2 more days. c Daily activity profiles of flies on the last day at 23 °C (before activation) and on the 2nd day at 29 °C were overlaid. d M activity offset of individual flies on the last day at 23 °C and on the 2nd day after the temperature elevation to 29 °C to avoid temperature change-induced strong startle activity are shown. Bars indicate mean ± SEM (n = 32). Statistically significant differences in the average time between 23 and 29 °C (independent t test): *P < 0.05. e–l Flies of the indicated genotypes were maintained on a 16L:8D cycle at 29 °C. Brains were dissected at ZT2. e, f On day 7, GRASP-positive signals were produced between DN1ps in R18H11-LexA > LexAop-CD4-spGFP11 and DTk neurons in DTk5Fa > UAS-CD4-spGFP1-10, but not between LNvs in PDF-LexA > LexAop-CD4-spGFP11 and DTk neurons in DTk5Fa > UAS-CD4-spGFP1-10. GRASP signals were detected more broadly in flies fed a HSD. All scale bars represented 20 μm. f The areas showing GRASP signals were quantified using ImageJ software (n = 8–11). Statistically significant differences in GRASP area between NSD and HSD groups (independent t test): ***P < 0.001. g, h GRASP signals between DTk neurons in DTk5Fa > UAS-CD4-spGFP1-10 and DN1ps in R18H11-LexA > LexAop-CD4-spGFP11 were analyzed on days 3, 5, and 7. All scale bars represented 20 μm. h The areas showing GRASP signals were quantified using ImageJ software. The GRASP areas were progressively increased over time in flies fed a HSD. Values indicate mean ± SEM (n = 8–9). Statistically significant differences in GRASP area (one-way ANOVA): ***P 0.001. i–k Flies of the indicated genotypes were maintained on a 16L:8D cycle at 29 °C. Brains were dissected at ZT2. On day 7, nSyb-GRASP-positive signals were produced between DN1ps in R18H11-LexA > LexAop-CD4-spGFP11 and DTk neurons in DTk5Fa > UAS-nSyb-spGFP1-10 (i), but not between DN1ps in R18H11-LexA > LexAop-nSyb-spGFP1-10 and DTk neurons in DTk5Fa > UAS-CD4-spGFP11 (j). Stronger nSyb-GRASP signals were detected when brains were exposed to KCl (final 70 mM, +) than to AHL (−). All GRASP- and nSyb-GRASP-positive signals represented endogenous GFP fluorescence. Brains were counter-stained with anti-NC82 (magenta) antibodies. All scale bars represented 20 μm. k The areas showing nSyb-GRASP-positive signals were quantified using ImageJ software (n = 6–9). Statistically significant differences in nSyb-GRASP area between NSD and HSD groups (independent t test): ***P < 0.001. l Flies of the indicated genotypes (denoted on top) were maintained on a 16L:8D cycle at 29 °C. Brains were dissected at ZT2 and stained with anti-GFP (green), anti-CLK (gray), and anti-NC82 (magenta) antibodies. The right panel shows magnified images of the boxed regions in the left panel. All scale bars represented 20 μm.
Next, we investigated whether acute manipulation of DTk neuronal activity affected M activity in NSD. We activated DTk neuron subsets by expressing warmth-activated cation channel, dTrpA1, which is inactive below 25 °C62. Because DTk5Fa-Gal4-driven DTk RNAi successfully suppressed the M activity extension, we used DTk5Fa-Gal4 for neuronal activity manipulation. Flies were entrained in a 16L:8D cycle at the non-permissive temperature of 23 °C for 7 days in NSD, and then the temperature was elevated to the permissive temperature of 29 °C for 2 more days. Locomotor activities and M activity offsets were compared on the last day at 23 °C and on the 2nd day after temperature elevation to avoid temperature change-induced strong startle activity. M activity in control flies (DTk5Fa > w1118) was the same at 23 °C or 29 °C, indicating that the temperature increase alone did not affect M activity significantly. On the other hand, flies expressing dTrpA1 (DTk5Fa > dTrpA1) exhibited a small but obvious M activity increase at 29 °C (Fig. 5c). In control flies, temperature elevation alone induced M activity offset differences were not evident. However, for flies expressing dTrpA1 (i.e., DTk neurons were activated by temperature elevation) there was a slight but statistically significant M activity offset delay (Fig. 5d). On the other hand, the warmth-induced activation of DTk2Ma–Gal4, which did not affect the HSD-associated behavior (Fig. 5a), did not increase M activity (Supplementary Fig. 7). Collectively, DTk neuron subgroup’s activity was involved in the HSD mediated M activity extension.
The M activity is controlled by sLNvs and DN1ps19,22,30,63, suggesting that DTk neurons might affect M activity by communicating with either sLNvs or DN1ps. To test this idea, we performed a GFP Reconstitution Across Synaptic Partners (GRASP) experiment to examine synaptic connections between two cells64,65 (Fig. 5e). We paired the Pdf-LexA (LNvs driver)66 or the R18H11-LexA (DN1ps driver)29 with DTk5Fa-Gal4 to express the split-GFP fragments, UAS-CD4::spGFP1-10 and LexAop-CD4::spGFP11. When the split-GFP fragments were in LNvs and DTk cells, no GFP signal was reconstituted. In contrast, high GFP signals were observed when split-GFP fragments were expressed by R18H11-LexA and DTk5Fa-Gal4. We found reconstituted GFP signals in the soma and nearby neurites in the DN1 region, indicating that DTk neurons were in physical contact with DN1ps but not with LNvs. We also found that reconstituted GFP-labeled neurites were greatly increased in HSD. We quantified this by measuring the GRASP signal area, and the results indicated that physical contacts increased between DTk neurons and DN1ps in flies fed a HSD (Fig. 5f). The M activity extension behavior was progressively enhanced over time (Fig. 1c and Supplementary Fig. 2). Intriguingly, the GRASP area between DTk neurons and DN1ps gradually increased over time for flies fed a HSD (Fig. 5g, h). These results support the idea that the HSD-induced M activity extension required plasticity in synapses between the DTk neurons and DN1ps.
Next, we used a modified GRASP technique to determine whether GRASP signals between DN1ps and DTk neurons resulted from active synapses. We used a neuronal-synaptobrevin-spGFP1-10 chimera (nSyb-spGFP1-10) instead of CD4-spGFP1-10 (Fig. 5i). nSyb-spGFP1-10 is exposed only after presynaptic neuronal activation and, therefore, preferentially labels active synapses67. The freshly isolated live brain was exposed to KCl (three times, 5 s each time) to induce depolarization67. When DTk5Fa-Gal4 drove the expression of nSyb-spGFP1-10 and R18H11-LexA drove the expression of spGFP11, nSyb-GRASP signal was produced (Fig. 5i, −). Application of KCl enlarged this nSyb-GRASP signal (Fig. 5i, + and Fig. 5k, +). When reciprocal nSyb-GRASP partners were used, no GFP signal appeared (Fig. 5j). These results further indicated that the DTk neurons were presynaptically innervated the DN1ps. Indeed, DTk5Fa-Gal4-driven CD8::GFP reporter revealed that dCLK labeled DN1ps contacted by the neurites of DTk neurons (Fig. 5l).
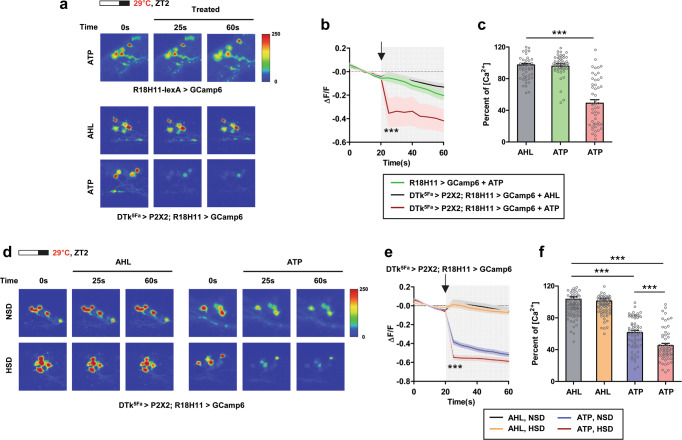
To further examine the functionality of this connection, we expressed P2X2, a mammalian ATP receptor, in DTk5Fa-Gal4 cells, and GCaMP6, a fluorescent Ca2+ sensor, in R18H11-DN1ps. While R18H11 cells did not respond to ATP addition, R18H11 cells with DTk5Fa–Gal4 driving P2X2 expression showed a 60% decrease in Ca2+ levels after the addition of ATP compared to the AHL treated controls (Fig. 6a–c). These results indicated an inhibitory connection between in DTk5Fa and R18H11-DN1ps. We next examined whether diets affected Ca2+ response. Consistent with the increase in synaptic contacts in flies fed a HSD, these flies showed a much greater decrease in intracellular Ca2+ levels in R18H11-DN1ps compared to the flies fed a NSD (Fig. 6d–f). We also noted the ATP response was observed in all GCaMP6-positive cells, indicating that DTk neurons innervated most if not all R18H11-DN1ps. This structural and physiological plasticity of the DTk and DN1p circuit in flies fed a HSD was not unique to the high-temperature and long-photoperiod condition and were observed in flies maintained under the standard 12L:12D cycle at 25 °C conditions (Supplementary Fig. 8). Collectively, our results imply that the suppression of R18H11-DN1ps by DTk neurons extended M activity. Our results are consistent with a previous report that the optogenetic inhibition of R18H11 from the midday extended the E activity, but the timing is different68. In addition, the thermogenetic activation of R18H11-DN1ps promoted activity around dawn followed by siesta29,68. Thus, we think that DTk neurons regulate R18H11-DN1ps activity in a temporally gated manner in a way to suppress sleep-promoting output from DN1ps.
Fig. 6. DTk neuron reduced intracellular Ca2+ levels in DN1ps.
a–f Flies of the indicated genotypes were maintained on a 16L:8D cycle at 29 °C. On day 7, brains were dissected at ZT2 ~ 4. a, d Images with GCaMP-positive DN1ps following application of AHL or ATP. b, e ΔF/F values over time following AHL or ATP application (arrow) are shown. c, f Relative fold changes of intracellular Ca2+ levels. c ΔF/F values were normalized to AHL applied DTk5Fa > P2X2; R18H11 > GCamp6 flies at 25 s. Bars indicate mean ± SEM (n = 45–51). Statistically significant differences between AHL and ATP treated groups (independent t test): ***P < 0.001. f ΔF/F values were normalized to AHL applied DTk5Fa > P2X2; R18H11 > GCamp6 flies fed with a NSD at 25 s. Bars indicate mean ± SEM (n = 61–89). Statistically significant differences between AHL and ATP treated groups or between NSD and HSD condition (independent t test): ***P < 0.001.
Subsets of DN1ps were TkR86C-positive and required for M activity extension in HSD
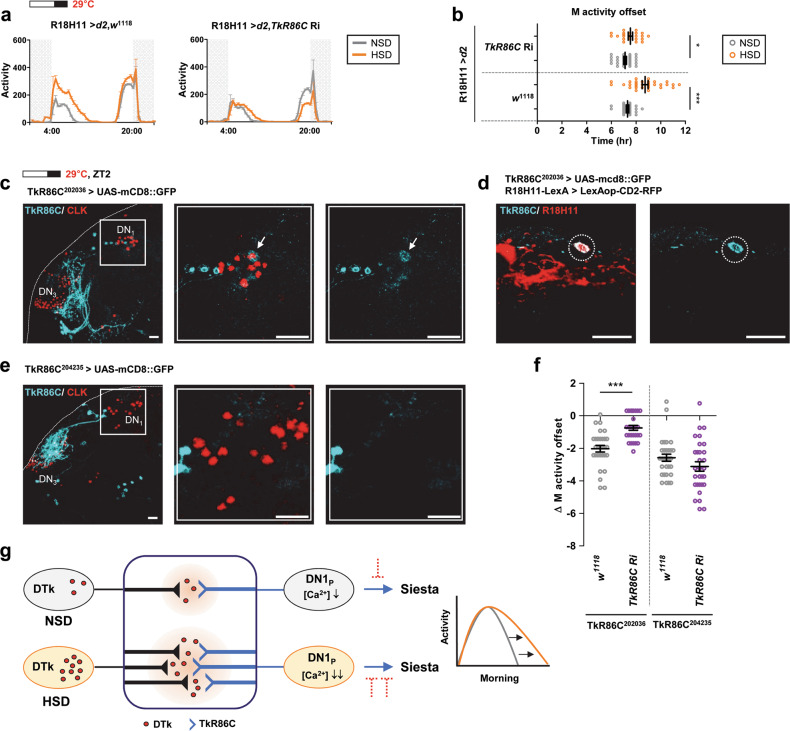
To further demonstrate that DN1ps innervated by DTk neurons mediated the M activity extension in HSD, we knocked down TkR86C using R18H11-Gal4. Compared with the control flies, the R18H11-Gal4-driven TkR86C knockdown abolished the effect of a HSD on M activity (Fig. 7a, b). To determine whether DN1ps expressed TkR86C, TkR86C-expressing cells were marked by TkR86C-Gal4-driven mCD8::GFP reporter and DN1ps were visualized using dCLK immunostaining. A single DN1p pair was positive for both TkR86C202036-Gal4 (Fig. 7c) and R18H11-LexA (Fig. 7d). Another TkR86C-Gal4 line, TkR86C204235-Gal4, expressed the mCD8::GFP reporter similarly to TkR86C202036-Gal4 in the lateral dorsal brain region, but no DN1ps were positive (Fig. 7e). When TkR86C expression was downregulated using TkR86C202036-Gal4, the M activity offset delay in flies fed a HSD was mitigated compared with the control, but not completely suppressed (Fig. 7f and Supplementary Fig. 9a). Since the DTk neuron-dependent Ca2+ signal was observed in most of R18H11-DN1ps (Fig. 6), it seems likely that TkR86C202036-Gal4 did not target all the TkR86C-positive DN1ps. Nevertheless, TkR86C204235-Gal4-driven downregulation of TkR86C did not affect the HSD-induced M activity offset delay (Fig. 7f and Supplementary Fig. 9b).
Fig. 7. Subsets of DN1ps were TkR86C-positive and were required for M activity extension in HSD.
a, b Locomotor activities of given genotypes of flies (denoted on top) were analyzed in NSD and HSD on a 16L:8D cycle at 29 °C. Daily activity profiles of flies on day 7 are shown. b M activity offset of individual flies on day 7 is shown. Bars indicate mean ± SEM values (n = 22–30). Statistically significant differences in M activity offset between control (R18H11 > d2, w1118) and TkR86C knockdown flies (R18H11 > d2, TkR86C Ri) (independent t test): *P < 0.05, ***P < 0.001. c, e Flies of the indicated genotypes (denoted on top) were maintained on a 16L:8D cycle at 29 °C. Brains were dissected at ZT2 and stained with anti-GFP (cyan blue) and anti-CLK (red) antibodies. The middle and right panels show a magnified image of the boxed region in the left panel. Arrow indicates a TkR86C-positive DN1p. All scale bars represented 20 μm. d Brains were stained with anti-GFP (cyan blue) and anti-RFP (red) antibodies. Dashed circle marks R18H11 and TkR86C positive cells in DN1p region. All scale bars represented 20 μm. f Differences in M activity offset on day 7 between NSD and HSD groups (ΔM activity offset) for given genotypes of flies are shown (n = 26–31). Statistically significant differences in ΔM activity offset between control (TkR86C202036 > w1118 or TkR86C204235 > w1118) and TkR86C knockdown flies (TkR86C202036 > TkR86C Ri or TkR86C204235 > TkR86C Ri) (independent t test): ***P < 0.001. g Schematic of our model for a HSD-induced M activity extension in flies. DTk signaling is transmitted via TkR86C receptors onto postsynaptic DN1ps. The activation of DTk neurons reduced intracellular Ca2+ levels in DN1ps indicating the inhibitory connection between two neurons. A HSD increased the connections between DTk and DN1ps anatomically and physiologically. DN1ps promote activity at dawn and sleep at midday, we hypothesized that DTk modulates DN1ps activity in a time-gated manner to inhibit siesta, leading to the M activity extension (marked as a dashed line, because it was not proven in our study).
Taken all together, this study revealed that DTk signaling onto DN1ps via TkR86C was required for HSD-induced M activity extension. A HSD augmented the inhibitory synaptic connection between DTk neuron and R18H11-DN1ps, likely suppressing the siesta-promoting activity of DN1ps (Fig. 7g).
Discussion
In this study, we found that D. melanogaster had extended the M activity without much effect on E activity in a high-nutrient condition. DTk signaling onto DN1ps via TkR86C mediated this behavior with a concomitant increase in anatomical and physiological synaptic contacts between DTk neurons and DN1ps. It is known that DN1ps integrates environmental stimuli such as light and temperature for daily locomotor activity and sleep regulation19–23, and our results further indicated that DN1ps also coordinated the metabolic input via DTk signaling which shaped daily locomotor behavior.
Tk constitutes an evolutionarily well-conserved family of brain/gut neuropeptides that function as important neuromodulators in the central and peripheral nervous systems (reviewed in ref. 51). The mammalian Tk family members are substance P (SP), neurokinin A, and neurokinin B, which are produced from the pre-protachykinin-A gene. SP plays important modulatory roles in many processes (e.g., sensory processing, pain transmission, neurogenic inflammation, and stress) and has been implicated in the regulation of the circadian timing system. In the photic entrainment pathway, glutamatergic signals following photoreception are transmitted to the suprachiasmatic nucleus master clock of the mammalian circadian timing system. This process is enhanced by SP via the NK1 receptor (NK1R)69–71. Interestingly, decreasing NK1R by an antagonist attenuates the light pulse-induced phase shift during the late-night but not during the early night72. SP enhances acetylcholine release in the limbic/prefrontal area during the morning, but not during the afternoon73. These findings suggest that SP affects the circadian timing system during a time-restricted window, which is consistent with our results showing DTk-associated morning-restricted effects on locomotor behavior in flies on a HSD. In addition, our study suggested that SP might also be implicated to signal metabolic input in mammals.
DTk and two TkRs, TkR86C and TkR99D, are homologs of SP and its receptor NK1R, respectively46,47. Similar to mammalian Tk, the DTk gene encodes a pre-protachykinin that is processed into Tk-1–657,74. In the CNS, DTk sensitizes sensory processing52 and participates in regulating systemic responses, including locomotor activity52,53, metabolic stress resistance75, and aggressive behavior50. While pan-neuronal or pontine neuronal knockdown of DTk increases activity or rest-activity bouts, respectively52,53, we did not observe an obvious change in locomotor activity in DTk knockdown flies fed a NSD (Fig. 3c). It appears that the effect of DTk on the regulation of general locomotor activity is not substantial. However, we found that DTk controlled the daily locomotor activity profile depending on the flies’ nutritional status, providing a novel neuromodulatory role for DTk in the CNS. A previous report that DTk expressed in five pairs of large protocerebral neurosecretory cells (designated ipc-1 and ipc-2a) regulates metabolic stress responses further supports the role of DTk in the regulation of metabolism in the CNS75. We found that in flies fed a HSD, there were increased levels of intracellular DTk in the brain. In the midgut, starvation promotes intracellular DTk production, but only amino acids, not sucrose or coconut oil, affect DTk76, suggesting that the metabolic stimuli that induce DTk production might be different in the CNS versus the peripheral nervous system.
In this study, we found that DN1ps coordinated the metabolic input via DTk signaling and extend M activity. How DN1ps control this behavior? The activation of R18H11-DN1ps promotes activity around dawn29 but promotes midday sleep68. Neural circuits from DN1ps to drive activity and sleep have been identified. DN1ps promoting wakefulness project to the dorsomedial protocerebrum, pars intercerebralis (PI) region19,29,30. DN1ps also targets the ellipsoid body (EB) region via a subgroup of tubercular-bulbar (TuBu) neurons in the anterior region. This circuit appeared to be sleep promoting in one study77 and wake promoting in the other study78. Our internal Ca2+ measurements following DTk neuron activation showed that connections between DTk neurons and R18H11-DN1ps are inhibitory. Given that the enhancement of DTk signaling onto DN1ps extended M activity without affecting M activity onset, we hypothesized that DTk suppresses the siesta-promoting DN1p circuit thereby extend the M activity in the temporally gated manner (Fig. 7g). Interestingly, optogenetic inhibition of R18H11 from the midday extended the E activity and that is consistent with our idea, yet the timing is different68. High nutrition impacted the flies’ locomotor activity largely in the morning. Our immunostaining data showed that DTk levels were higher in the morning in some DTk-expressing nuclei in the brain such as LPP1, LPP2, or SMP (Fig. 4c). The rhythmic oscillation of DTk in the specific nucleus mediating the HSD effect might be the underlying mechanism for DTk-associated morning-restricted HSD effects on locomotor behavior. The rhythmic presentation of TkR86C on DN1ps might cause phase-specific effects as well, which require further study. Given DN1ps excitability is maximal in the morning79, the inhibitory inputs from DTk neurons might have the strongest impact on DN1ps in the morning.
The effect of a HSD on M activity increased in an environment of a summer-like high temperature and a long photoperiod (Supplementary Fig. 1 and Fig. 2); however, the effect of HSD on structural and physiological connections between DTk5Fa neurons and R18H11-DN1ps were similar in flies reared in either 25 °C 12L:12D or 29 °C 16L:8D. These results suggested that temperature and photoperiod might have additive effects with the HSD on M activity in flies. Intriguingly, in vivo Ca2+ imaging indicates that DN1ps are inhibited by heating80 supporting our hypothesis of the additive effect of high temperature and DTk-mediated signaling both decreasing internal Ca2+. There is a previous report that temperature elevation to high levels (>30 °C) prolong morning activity and delay midday sleep onset, which is similar to the effect of a HSD in our study except prolonged morning activity is observed only in male flies78. The temperature information that affected sleep was transmitted to DN1ps also via two neuronal groups expressing TrpA1 (i.e., TrpA1[SH]-Gal4- and ppk-Gal4-active cells). The separate circuits appear to converge onto DN1ps to deliver temperature and high-nutrient information. The morning activity of the fly comprises the lights-on startle component, which is the sharp increase in activity and an endogenous circadian component, the morning peak43. Under the standard 12L:12D cycle, the two components are not separable because the circadian component is largely masked by the startle response. With a long photoperiod, circadian activity appears separately after the startle response. Therefore, we think that photoperiodic gating of circadian M activity is timely followed by DTk signaling, leading to enhancement of the M activity extension in long photoperiod.
Tachykinin receptors are G-protein-coupled receptors, and NK1R is usually coupled to the Gq/11 cascade, leading to an increase in internal Ca2+ (reviewed in ref. 81). However, interaction with other G proteins and diverse downstream signaling pathways had been also known in different tissues (reviewed in ref. 82). In Drosophila, DTk also increases Ca2+ levels in TkR99D-transfected HEK293 cells suggesting an increase in neural activity45,83. On the other hand, olfactory receptor neurons expressing TkR99D are suppressed by DTk, suggesting that DTk mediates inhibitory neuromodulation84,85. While the TkR86C downstream intracellular signaling pathway in Drosophila is unknown, in this study we showed that DTk5Fa neuron activation reduced intracellular Ca2+ levels of R18H11-DN1ps possibly via TkR86C (Figs. 6 and 7).
Mice fed a HFD exhibit reduced rhythmicity and lengthened periods of activity10 with slower responses to light86; however, whether a HFD affects the locomotor activity profile of mice as it does in flies is unknown. A hypocaloric diet with restricted feeding advances mouse activity onset without a change of period87, which is comparable to the delayed M activity offset of flies in HSD in the opposite direction87. The question remains, why did flies in a high-nutrient diet show extended locomotor activity only in the morning? The total activity was generally higher in flies fed a HSD than a NSD, but the increase for the w1118 flies was not significant. Thus, it is conceivable that flies may increase their locomotor activity to balance energy input and expenditure but may restrict this change to the morning phase not to compromise a deep sleep during the night phase88,89.
Methods
Fly stocks
TkR86C▵F28 flies50 were provided by David Anderson (California Institute of Technology, USA). UAS-mCD8::GFP;lexAop-CD2 RFP flies were provided by Seok Jun Moon (Yonsei University, Republic of Korea). UAS-CD4-spGFP1-10;lexAop-CD4-spGFP11 (BL58755), UAS-nSyb-spGFP1-10;lexAop-CD4-spGFP11 (BL64314), and lexAop-nSyb-spGFP1-10;UAS-CD4-spGFP11 (BL64315) flies were provided by Chunghun Lim (UNIST, Republic of Korea). The pdf-Gal490 flies were a gift from Jae H. Park (University of Tennessee, USA). The following lines were obtained from the Bloomington Drosophila Stock Center: w1118(BL5905), elav-Gal4C155 (BL458), elav-GS-Gal4 (BL43642), DTk2Ma-Gal4 (BL51973), DTk5Fa-Gal4 (BL51975), R65E09-Gal4 (BL39358), UAS-TkR99D RNAi (BL55732), UAS-GFP.nls (BL4776), 10XUAS-IVS-mCD8::GFP (BL32187), UAS-dTrpA1(BL26263), R18H11-Gal4 (BL48832), R18H11-lexA (BL52535), pdf-lexA (BL52685), UAS-P2X2 (BL91222), 13XLexAop2-IVS-GCaMP6m (BL44276). The following fly stocks were obtained from the Vienna Drosophila Resource Center: UAS-DTk RNAi (V103662), UAS-TkR86C RNAi (V13392), TkR86C039622-Gal4 (V202036), TkR86C039625-Gal4 (V204235). Neuropeptide RNAi lines: DTk(V103662), Dh44(V108473), Ptth(V102043), FMRFa(V103981), Nplp11(V107116), Capa(V101705), Burs(V102204), Mip(V106076), Nplp12(V14035), NPF(V108772), Proc(V102488), Nplp3(V105584), Akh(V105063), AstA(V103215), Dh31(V50295), Nplp4(V104662), Ms(V108760), Pdf(BL25802), Dsk(V14201), AstC(V102735), Ilp3(V106512), Ilp5(V105004), Acp26Aa(V41193), Pburs(V102690), Ilp2(V44761), Hug(V107771), hec(V7223), BomS2(V10586), Lgr11(V104877), ETH(V18825), Lk(V14091), natalisin(V19547), Lgr12(V13566), ITP(V43848). elav-Gal4, elav-GS-Gal4, and pdf-Gal4 were crossed to UAS-dicer2/CyO to generate dcr2;elav-Gal4, dcr2;elav-GS-Gal4, and dcr2;pdf-Gal4 and used as driver flies for knockdown of expression. w1118 (BL5905) flies were used as a background strain in this study. UAS-DTk RNAi and UAS-TkR86C RNAi were outcrossed to w1118 (BL5905) for six generations.
Locomotor behavior analysis
Locomotor activity of individual flies was determined using the Drosophila Activity Monitoring System 3 software (Trikinetics, version 1.02). Young male flies were used for the analysis and maintained in glass tubes containing 2% agar and 5% sucrose (normal sucrose diet, NSD) or 30% sucrose (high-sucrose diet, HSD) or 5% sucrose-containing 20% coconut oil (Nutiva) (high-fat diet, HFD). Flies were kept in incubators at the indicated temperature (25 °C or 29 °C) and were exposed to a 12L:12D or 16L:8D cycle for the indicated number of days of the experiment. Averaged fly locomotor activity profiles were plotted using GraphPad Prism5 software. To obtain the M and E activity phases, the onset/offset formula [(An+2 + An+1) – (An-1 + An-2) = ∆Activity] was used91. For M and E onset assessments, the largest 1-h increase in the activity window before light-on (M) or light-off (E) transitions, respectively, was used. For M and E offset assessments, the largest 1-h decrease in the activity window after light-on (M) or light-off (E) transitions, respectively, was used. Activities during the first 30 min after the light-on/off transition were removed to minimize the light-induced startle response.
Food intake assays
To quantify the food intake of the flies, the absorbance of ingested dye was measured following method with slight modification92. Flies were maintained at 16L:8D cycle at 29 °C. Flies in groups of 16 were collected at ZT2 and starved for 18 h in 2% agar. Then flies were allowed to feed on 5% sucrose in 2% agarose for 20 min, transferred to new vials containing 1% blue dye (McCormick), and left to feed for another 15 min. Flies were homogenized in PBS, centrifuged for 3 min, and the absorbance of the blue dye in the supernatant was measured at 620 nm.
Antibody production
We raised guinea pig anti-DTk antiserum (DTk-gp2) using the full-length protein as the antigen (Young In Frontier, Korea). We raised guinea pig anti-CLK antiserum (CLK-gp2) using the C-terminal 1138–3081 amino acids of the protein as the antigen (Young In Frontier, Korea). Antibody was purified from the antiserum with antigens immobilized on PVDF membranes. The antibody was dialyzed in PBS; glycerol (final 30%, v/v) was added as a stabilizer.
Immunohistochemistry and confocal imaging
Immunostaining was performed as described previously with minor modifications93. Fly heads were cut open, fixed in 2% formaldehyde, and washed with 0.5% PAXD buffer (1× PBS, 5% BSA, 0.03% sodium deoxycholate, 0.03% Triton X-100)94. The fixed heads were dissected, and the isolated brains were permeabilized in 1% PBT for 20 min and then blocked in 0.5% PAXD containing 5% horse serum for 1 h. The following primary antibodies were diluted 1:200 and added directly to the mixtures: anti-DTk antibody (Gp2), anti-PDF antibody (C7) (DSHB), anti-PER antibody (Rb1)95, anti-TIM antibody (Rb1)93, anti-CLK antibody (Gp2), anti-GFP antibody (MBL International), anti-RFP antibody (MBL International), and anti-NC82 antibody (DSHB). The brains were washed with PAXD and incubated overnight with secondary antibodies in a blocking solution at 4 °C. The following secondary antibodies were used at a 1:200 dilution: goat anti-rabbit Alexa-488 (Thermo Fisher Scientific), goat anti-guinea pig Alexa-555 (Thermo Fisher Scientific), goat anti-mouse Alexa-555 (Thermo Fisher Scientific), and goat anti-mouse Alexa-633 (Thermo Fisher Scientific). Stained brain samples were washed with PAXD, incubated in 0.1 M phosphate buffer containing 50% glycerol for 30 min, and mounted using a mounting medium. Confocal images were obtained using an LSM 800 confocal microscope (Carl Zeiss) and were processed using Zen software (ZEN Digital Imaging for Light Microscopy, Carl Zeiss, version 3.1). For signal quantification, the pixel intensity of each cell was determined using ImageJ software. The intensity was the average of at least eight brains for each genotype.
GFP reconstitution across synaptic partners analysis
GFP Reconstitution Across Synaptic Partners (GRASP) was performed to detect membrane contacts between flies expressing the CD4::spGFP1-10 fragment in one neuronal type and the CD4::spGFP11 fragment in the other neuronal type using the GAL4/UAS and LexA/lexAop systems, respectively64,65. pdf-LexA or R18H11-LexA drivers were used to express CD4::spGFP11 in LNvs or DN1ps, respectively. CD4::spGFP1-10 was expressed in DTk neurons using the DTk5Fa-Gal4 driver. A modified GRASP (i.e., nSyb-GRASP) analysis was performed to determine whether the contacts between two neuronal groups were active synapses67. In the nSyb-GRASP system, neuronal synaptobrevin fused to spGFP1-10 (nSyb::spGFP1-10) fragment is expressed in one neuronal type. spGFP1-10 is exposed to the extracellular space following neuronal activation because n-Syb is a component of the synaptic vesicle membrane. R18H11-LexA and DTk5Fa-Gal4 drivers were crossed with either UAS-nSyb-spGFP1–10, lexAop-CD4-spGFP11 or UAS-CD4-spGFP11, lexAop-nSyb-spGFP1-10. To apply KCl to evoke neuronal activation, flies were anesthetized on ice, and their brains were dissected in adult hemolymph (AHL, containing 108 mM NaCl, 5 mM KCl, 4 mM NaHCO3, 1 mM NaH2PO4, 15 mM sucrose, 5 mM HEPES, 8.2 mM MgCl2, 2 mM CaCl2, pH 7.4). Dissected brains were rinsed quickly (5 s) three times with 70 mM KCl in AHL, and then imaged in AHL 20 min after KCl application. Control flies were rinsed in AHL containing no additional KCl and imaged the same way. GRASP Area was determined using the ImageJ software. A GFP (positive GRASP) signal above background levels was selected by adjusting the color threshold and the area of the GFP signal was obtained from ImageJ software (version 1.53c).
GCaMP imaging and analysis
Adult male flies were entrained for 7 days in incubators at the indicated temperature and light cycle. From ZT2 to ZT4, flies were dissected in adult hemolymph-like buffer (AHL, 108 mM NaCl, 8.2 mM MgCl2, 4 mM NaHCO3, 1 mM NaH2PO4, 2 mM CaCl2, 5 mM KCl, 5 mM HEPES, 80 mM sucrose)96. Dissected Drosophila brains were rapidly mounted on a cover glass and sprayed with 20 μl of AHL buffer to prevent the brain from drying out. After stabilizing the samples for 3 min in AHL buffer, confocal imaging was performed to determine the baseline Ca2+ levels. ATP, at a concentration of 2.5 mM, or AHL (control) were applied directly to the AHL buffer covering the brain, and imaging was performed. The Z stack images were taken (three layers) every 5 seconds to measure all the DN1ps. Image processing and measurement of fluorescence intensity were performed in ZEN (black edition) and ImageJ programs. A sum-intensity Z-projection of each time interval was measured after combining the images using the ZEN program (orthogonal projection). GCaMP-positive regions of interest (DN1pS cells) were manually drawn and mean intensities were measured at each time interval using the ImageJ program. The ratio changes were calculated using the following formula: ΔF/F = (Fn − F0)/F0, where Fn was the mean intensity of GCaMP-positive cells, F0 was the average baseline intensity. Brains with cells that had unstable baselines were not used.
qRT-PCR
The total RNA was extracted from fly heads using QIAzol reagent (QIAGEN). The total RNA (1 μg) was reverse transcribed using an oligo(dT)20 primer (for mRNA) and PrimeScript RTase (TaKaRa). Quantitative, real-time PCR (qPCR) was performed using Rotor Gene 6000 (QIAGEN) with TB Green Premix Ex Taq (Tli RNaseH Plus, TaKaRa). The following primers were used: DTk forward, 5′-CGGTCAATTCCTTTGTGGG-3′; DTk reverse, 5′-ATTCGGAGAGAGCTGCAC-3′; TkR86C forward, 5′-GACCAAGCACTATTACAATGG-3′; TkR86C reverse, 5′-GCCATAGAAGTGGGATATCG-3′; TkR99D forward, 5′-GTGGAGAATGTGCGGAGTAAG-3′; and TkR99D reverse, 5′-CGGGTAGCAGGATGTGATTATG-3′. Noncycling mRNA encoding cbp20 was used to normalize gene expression with the primers cbp20 forward, 5′-GTATAAGAAGACGCCCTGC-3′; and cbp20 reverse, 5′-TTCACAAATCTCATGGCCG-3′. The data were analyzed using Rotor Gene Q- Pure Detection software (version 2.2.3), and the relative mRNA levels were quantified using the 2−∆∆Ct method in which ∆∆Ct = [(Ct target − Ct cbp20) of the experimental group] − [(Ct target − Ct cbp20) of control group].
Statistics and reproducibility
GraphPad Prism5 software was used for the statistical analysis. All population assays were performed with the experimental and control genotypes in parallel and with more than n = 16 flies per genotype. All data represented multiple independent experiments. Nonparametric t test statistics were used unless otherwise indicated.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
We thank David Anderson (California Institute of Technology, USA), Jae H. Park (University of Tennessee, USA), Seok Joon Moon (Yonsei University, Republic of Korea), and Chunghun Lim (UNIST, Republic of Korea) for sharing fly lines. This research was supported by the National Research Foundation of Korea (NRF) grants funded by the Korean government (Ministry of Science and ICT; grant numbers 2019M3C7A1031905, 2019R1A5A2026045, and 2020R1A2C2007158) and intramural research fund of Ajou University Medical Center to Eun Young Kim.
Author contributions
Conceptualization of the work by E.C. and E.Y.K.; design of the work by S.H.L., E.C., and E.Y.K.; acquisition and analysis of data by S.H.L.; interpretation of the data by S.H.L., E.C., and E.Y.K.; resources by S.Y. and Y.K.; writing original draft and revision by S.H.L. and E.Y.K.
Data availability
The source data underlying the graphs are shown as Supplementary Data 1. All other data supporting the findings of this study are available from the corresponding author upon reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Peer review information Communications Biology thanks the anonymous reviewers for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s42003-021-02219-6.
References
- 1.Schibler U, et al. Clock-talk: interactions between central and peripheral circadian oscillators in mammals. Cold Spring Harb. Symp. Quant. Biol. 2015;80:223–232. doi: 10.1101/sqb.2015.80.027490. [DOI] [PubMed] [Google Scholar]
- 2.Mohawk JA, Green CB, Takahashi JS. Central and peripheral circadian clocks in mammals. Annu Rev. Neurosci. 2012;35:445–462. doi: 10.1146/annurev-neuro-060909-153128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bell-Pedersen D, et al. Circadian rhythms from multiple oscillators: lessons from diverse organisms. Nat. Rev. Genet. 2005;6:544–556. doi: 10.1038/nrg1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takahashi JS. Transcriptional architecture of the mammalian circadian clock. Nat. Rev. Genet. 2017;18:164–179. doi: 10.1038/nrg.2016.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Challet E. Circadian clocks, food intake, and metabolism. Prog. Mol. Biol. Transl. Sci. 2013;119:105–135. doi: 10.1016/B978-0-12-396971-2.00005-1. [DOI] [PubMed] [Google Scholar]
- 6.Damiola F, et al. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev. 2000;14:2950–2961. doi: 10.1101/gad.183500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hara R, et al. Restricted feeding entrains liver clock without participation of the suprachiasmatic nucleus. Genes Cells. 2001;6:269–278. doi: 10.1046/j.1365-2443.2001.00419.x. [DOI] [PubMed] [Google Scholar]
- 8.Mistlberger RE. Neurobiology of food anticipatory circadian rhythms. Physiol. Behav. 2011;104:535–545. doi: 10.1016/j.physbeh.2011.04.015. [DOI] [PubMed] [Google Scholar]
- 9.Stokkan KA, Yamazaki S, Tei H, Sakaki Y, Menaker M. Entrainment of the circadian clock in the liver by feeding. Science. 2001;291:490–493. doi: 10.1126/science.291.5503.490. [DOI] [PubMed] [Google Scholar]
- 10.Kohsaka A, et al. High-fat diet disrupts behavioral and molecular circadian rhythms in mice. Cell Metab. 2007;6:414–421. doi: 10.1016/j.cmet.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 11.Yokoyama Y, et al. A high-salt/high fat diet alters circadian locomotor activity and glucocorticoid synthesis in mice. PLoS ONE. 2020;15:e0233386. doi: 10.1371/journal.pone.0233386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brown SA. Circadian metabolism: from mechanisms to metabolomics and medicine. Trends Endocrinol. Metab. 2016;27:415–426. doi: 10.1016/j.tem.2016.03.015. [DOI] [PubMed] [Google Scholar]
- 13.King AN, Sehgal A. Molecular and circuit mechanisms mediating circadian clock output in the Drosophila brain. Eur. J. Neurosci. 2020;51:268–281. doi: 10.1111/ejn.14092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rajan A, Perrimon N. Of flies and men: insights on organismal metabolism from fruit flies. BMC Biol. 2013;11:38. doi: 10.1186/1741-7007-11-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Allada R, Chung BY. Circadian organization of behavior and physiology in Drosophila. Annu Rev. Physiol. 2010;72:605–624. doi: 10.1146/annurev-physiol-021909-135815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grima B, Chelot E, Xia R, Rouyer F. Morning and evening peaks of activity rely on different clock neurons of the Drosophila brain. Nature. 2004;431:869–873. doi: 10.1038/nature02935. [DOI] [PubMed] [Google Scholar]
- 17.Rieger D, Shafer OT, Tomioka K, Helfrich-Forster C. Functional analysis of circadian pacemaker neurons in Drosophila melanogaster. J. Neurosci. 2006;26:2531–2543. doi: 10.1523/JNEUROSCI.1234-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stoleru D, Peng Y, Agosto J, Rosbash M. Coupled oscillators control morning and evening locomotor behaviour of Drosophila. Nature. 2004;431:862–868. doi: 10.1038/nature02926. [DOI] [PubMed] [Google Scholar]
- 19.Chatterjee A, et al. Reconfiguration of a multi-oscillator network by light in the Drosophila circadian clock. Curr. Biol. 2018;28:2007–2017. doi: 10.1016/j.cub.2018.04.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen C, Xu M, Anantaprakorn Y, Rosing M, Stanewsky R. nocte is required for integrating light and temperature inputs in circadian clock neurons of Drosophila. Curr. Biol. 2018;28:1595–1605. doi: 10.1016/j.cub.2018.04.001. [DOI] [PubMed] [Google Scholar]
- 21.Lamaze A, et al. Regulation of sleep plasticity by a thermo-sensitive circuit in Drosophila. Sci. Rep. 2017;7:40304. doi: 10.1038/srep40304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang L, et al. DN1(p) circadian neurons coordinate acute light and PDF inputs to produce robust daily behavior in Drosophila. Curr. Biol. 2010;20:591–599. doi: 10.1016/j.cub.2010.02.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Y, Liu Y, Bilodeau-Wentworth D, Hardin PE, Emery P. Light and temperature control the contribution of specific DN1 neurons to Drosophila circadian behavior. Curr. Biol. 2010;20:600–605. doi: 10.1016/j.cub.2010.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nassel DR, Winther AM. Drosophila neuropeptides in regulation of physiology and behavior. Prog. Neurobiol. 2010;92:42–104. doi: 10.1016/j.pneurobio.2010.04.010. [DOI] [PubMed] [Google Scholar]
- 25.Taghert PH, Nitabach MN. Peptide neuromodulation in invertebrate model systems. Neuron. 2012;76:82–97. doi: 10.1016/j.neuron.2012.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hermann C, Yoshii T, Dusik V, Helfrich-Forster C. Neuropeptide F immunoreactive clock neurons modify evening locomotor activity and free-running period in Drosophila melanogaster. J. Comp. Neurol. 2012;520:970–987. doi: 10.1002/cne.22742. [DOI] [PubMed] [Google Scholar]
- 27.Hermann-Luibl C, Yoshii T, Senthilan PR, Dircksen H, Helfrich-Forster C. The ion transport peptide is a new functional clock neuropeptide in the fruit fly Drosophila melanogaster. J. Neurosci. 2014;34:9522–9536. doi: 10.1523/JNEUROSCI.0111-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johard HA, et al. Peptidergic clock neurons in Drosophila: ion transport peptide and short neuropeptide F in subsets of dorsal and ventral lateral neurons. J. Comp. Neurol. 2009;516:59–73. doi: 10.1002/cne.22099. [DOI] [PubMed] [Google Scholar]
- 29.Kunst M, et al. Calcitonin gene-related peptide neurons mediate sleep-specific circadian output in Drosophila. Curr. Biol. 2014;24:2652–2664. doi: 10.1016/j.cub.2014.09.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cavanaugh DJ, et al. Identification of a circadian output circuit for rest:activity rhythms in Drosophila. Cell. 2014;157:689–701. doi: 10.1016/j.cell.2014.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cavey M, Collins B, Bertet C, Blau J. Circadian rhythms in neuronal activity propagate through output circuits. Nat. Neurosci. 2016;19:587–595. doi: 10.1038/nn.4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.He C, Yang Y, Zhang M, Price JL, Zhao Z. Regulation of sleep by neuropeptide Y-like system in Drosophila melanogaster. PLoS ONE. 2013;8:e74237. doi: 10.1371/journal.pone.0074237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shang Y, et al. Short neuropeptide F is a sleep-promoting inhibitory modulator. Neuron. 2013;80:171–183. doi: 10.1016/j.neuron.2013.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chung BY, et al. Drosophila neuropeptide F signaling independently regulates feeding and sleep-wake behavior. Cell Rep. 2017;19:2441–2450. doi: 10.1016/j.celrep.2017.05.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen J, et al. Allatostatin a signalling in Drosophila regulates feeding and sleep and is modulated by PDF. PLoS Genet. 2016;12:e1006346. doi: 10.1371/journal.pgen.1006346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Metaxakis A, et al. Lowered insulin signalling ameliorates age-related sleep fragmentation in Drosophila. PLoS Biol. 2014;12:e1001824. doi: 10.1371/journal.pbio.1001824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brown EB, Shah KD, Faville R, Kottler B, Keene AC. Drosophila insulin-like peptide 2 mediates dietary regulation of sleep intensity. PLoS Genet. 2020;16:e1008270. doi: 10.1371/journal.pgen.1008270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dreyer AP, et al. A circadian output center controlling feeding:fasting rhythms in Drosophila. PLoS Genet. 2019;15:e1008478. doi: 10.1371/journal.pgen.1008478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Catterson JH, et al. Dietary modulation of Drosophila sleep-wake behaviour. PLoS ONE. 2010;5:e12062. doi: 10.1371/journal.pone.0012062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Linford NJ, Chan TP, Pletcher SD. Re-patterning sleep architecture in Drosophila through gustatory perception and nutritional quality. PLoS Genet. 2012;8:e1002668. doi: 10.1371/journal.pgen.1002668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liao, S., Amcoff, M. & Nassel, D. R. Impact of high-fat diet on lifespan, metabolism, fecundity and behavioral senescence in Drosophila. Insect Biochem. Mol. Biol. 103495, 10.1016/j.ibmb.2020.103495 (2020). [DOI] [PubMed]
- 42.Potdar S, Sheeba V. Large ventral lateral neurons determine the phase of evening activity peak across photoperiods in Drosophila melanogaster. J. Biol. Rhythms. 2012;27:267–279. doi: 10.1177/0748730412449820. [DOI] [PubMed] [Google Scholar]
- 43.Rieger D, Stanewsky R, Helfrich-Forster C. Cryptochrome, compound eyes, Hofbauer-Buchner eyelets, and ocelli play different roles in the entrainment and masking pathway of the locomotor activity rhythm in the fruit fly Drosophila melanogaster. J. Biol. Rhythms. 2003;18:377–391. doi: 10.1177/0748730403256997. [DOI] [PubMed] [Google Scholar]
- 44.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 45.Birse RT, Johnson EC, Taghert PH, Nassel DR. Widely distributed Drosophila G-protein-coupled receptor (CG7887) is activated by endogenous tachykinin-related peptides. J. Neurobiol. 2006;66:33–46. doi: 10.1002/neu.20189. [DOI] [PubMed] [Google Scholar]
- 46.Li XJ, Wolfgang W, Wu YN, North RA, Forte M. Cloning, heterologous expression and developmental regulation of a Drosophila receptor for tachykinin-like peptides. EMBO J. 1991;10:3221–3229. doi: 10.1002/j.1460-2075.1991.tb04885.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Monnier D, et al. NKD, a developmentally regulated tachykinin receptor in Drosophila. J. Biol. Chem. 1992;267:1298–1302. doi: 10.1016/S0021-9258(18)48429-3. [DOI] [PubMed] [Google Scholar]
- 48.Poels J, et al. Characterization and distribution of NKD, a receptor for Drosophila tachykinin-related peptide 6. Peptides. 2009;30:545–556. doi: 10.1016/j.peptides.2008.10.012. [DOI] [PubMed] [Google Scholar]
- 49.Osterwalder T, Yoon KS, White BH, Keshishian H. A conditional tissue-specific transgene expression system using inducible GAL4. Proc. Natl Acad. Sci. USA. 2001;98:12596–12601. doi: 10.1073/pnas.221303298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Asahina K, et al. Tachykinin-expressing neurons control male-specific aggressive arousal in Drosophila. Cell. 2014;156:221–235. doi: 10.1016/j.cell.2013.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nassel DR, Zandawala M, Kawada T, Satake H. Tachykinins: neuropeptides that are ancient, diverse, widespread and functionally pleiotropic. Front. Neurosci. 2019;13:1262. doi: 10.3389/fnins.2019.01262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Winther AM, Acebes A, Ferrus A. Tachykinin-related peptides modulate odor perception and locomotor activity in Drosophila. Mol. Cell Neurosci. 2006;31:399–406. doi: 10.1016/j.mcn.2005.10.010. [DOI] [PubMed] [Google Scholar]
- 53.Kahsai L, Martin JR, Winther AM. Neuropeptides in the Drosophila central complex in modulation of locomotor behavior. J. Exp. Biol. 2010;213:2256–2265. doi: 10.1242/jeb.043190. [DOI] [PubMed] [Google Scholar]
- 54.Mrosovsky N. Masking: history, definitions, and measurement. Chronobiol. Int. 1999;16:415–429. doi: 10.3109/07420529908998717. [DOI] [PubMed] [Google Scholar]
- 55.Lu B, Liu W, Guo F, Guo A. Circadian modulation of light-induced locomotion responses in Drosophila melanogaster. Genes Brain Behav. 2008;7:730–739. doi: 10.1111/j.1601-183X.2008.00411.x. [DOI] [PubMed] [Google Scholar]
- 56.Matsumoto A, Matsumoto N, Harui Y, Sakamoto M, Tomioka K. Light and temperature cooperate to regulate the circadian locomotor rhythm of wild type and period mutants of Drosophila melanogaster. J. Insect Physiol. 1998;44:587–596. doi: 10.1016/S0022-1910(98)00046-8. [DOI] [PubMed] [Google Scholar]
- 57.Winther AM, Siviter RJ, Isaac RE, Predel R, Nassel DR. Neuronal expression of tachykinin-related peptides and gene transcript during postembryonic development of Drosophila. J. Comp. Neurol. 2003;464:180–196. doi: 10.1002/cne.10790. [DOI] [PubMed] [Google Scholar]
- 58.Hergarden AC, Tayler TD, Anderson DJ. Allatostatin-A neurons inhibit feeding behavior in adult Drosophila. Proc. Natl Acad. Sci. USA. 2012;109:3967–3972. doi: 10.1073/pnas.1200778109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tayler TD, Pacheco DA, Hergarden AC, Murthy M, Anderson DJ. A neuropeptide circuit that coordinates sperm transfer and copulation duration in Drosophila. Proc. Natl Acad. Sci. USA. 2012;109:20697–20702. doi: 10.1073/pnas.1218246109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jenett A, et al. A GAL4-driver line resource for Drosophila neurobiology. Cell Rep. 2012;2:991–1001. doi: 10.1016/j.celrep.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pfeiffer BD, et al. Tools for neuroanatomy and neurogenetics in Drosophila. Proc. Natl Acad. Sci. USA. 2008;105:9715–9720. doi: 10.1073/pnas.0803697105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pulver SR, Pashkovski SL, Hornstein NJ, Garrity PA, Griffith LC. Temporal dynamics of neuronal activation by Channelrhodopsin-2 and TRPA1 determine behavioral output in Drosophila larvae. J. Neurophysiol. 2009;101:3075–3088. doi: 10.1152/jn.00071.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Choi C, et al. Autoreceptor control of peptide/neurotransmitter corelease from PDF neurons determines allocation of circadian activity in drosophila. Cell Rep. 2012;2:332–344. doi: 10.1016/j.celrep.2012.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Feinberg EH, et al. GFP reconstitution across synaptic partners (GRASP) defines cell contacts and synapses in living nervous systems. Neuron. 2008;57:353–363. doi: 10.1016/j.neuron.2007.11.030. [DOI] [PubMed] [Google Scholar]
- 65.Gordon MD, Scott K. Motor control in a Drosophila taste circuit. Neuron. 2009;61:373–384. doi: 10.1016/j.neuron.2008.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pfeiffer BD, et al. Refinement of tools for targeted gene expression in Drosophila. Genetics. 2010;186:735–755. doi: 10.1534/genetics.110.119917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Macpherson LJ, et al. Dynamic labelling of neural connections in multiple colours by trans-synaptic fluorescence complementation. Nat. Commun. 2015;6:10024. doi: 10.1038/ncomms10024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Guo F, et al. Circadian neuron feedback controls the Drosophila sleep-activity profile. Nature. 2016;536:292–297. doi: 10.1038/nature19097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hamada T, Yamanouchi S, Watanabe A, Shibata S, Watanabe S. Involvement of glutamate release in substance P-induced phase delays of suprachiasmatic neuron activity rhythm in vitro. Brain Res. 1999;836:190–193. doi: 10.1016/S0006-8993(99)01565-6. [DOI] [PubMed] [Google Scholar]
- 70.Shibata S, Tsuneyoshi A, Hamada T, Tominaga K, Watanabe S. Effect of substance P on circadian rhythms of firing activity and the 2-deoxyglucose uptake in the rat suprachiasmatic nucleus in vitro. Brain Res. 1992;597:257–263. doi: 10.1016/0006-8993(92)91482-T. [DOI] [PubMed] [Google Scholar]
- 71.Shirakawa T, Moore RY. Responses of rat suprachiasmatic nucleus neurons to substance P and glutamate in vitro. Brain Res. 1994;642:213–220. doi: 10.1016/0006-8993(94)90924-5. [DOI] [PubMed] [Google Scholar]
- 72.Challet E, Naylor E, Metzger JM, MacIntyre DE, Turek FW. An NK1 receptor antagonist affects the circadian regulation of locomotor activity in golden hamsters. Brain Res. 1998;800:32–39. doi: 10.1016/S0006-8993(98)00491-0. [DOI] [PubMed] [Google Scholar]
- 73.Perez S, Tierney A, Deniau JM, Kemel ML. Tachykinin regulation of cholinergic transmission in the limbic/prefrontal territory of the rat dorsal striatum: implication of new neurokinine 1-sensitive receptor binding site and interaction with enkephalin/mu opioid receptor transmission. J. Neurochem. 2007;103:2153–2163. doi: 10.1111/j.1471-4159.2007.04944.x. [DOI] [PubMed] [Google Scholar]
- 74.Siviter RJ, et al. Expression and functional characterization of a Drosophila neuropeptide precursor with homology to mammalian preprotachykinin A. J. Biol. Chem. 2000;275:23273–23280. doi: 10.1074/jbc.M002875200. [DOI] [PubMed] [Google Scholar]
- 75.Kahsai L, Kapan N, Dircksen H, Winther AM, Nassel DR. Metabolic stress responses in Drosophila are modulated by brain neurosecretory cells that produce multiple neuropeptides. PLoS ONE. 2010;5:e11480. doi: 10.1371/journal.pone.0011480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Song W, Veenstra JA, Perrimon N. Control of lipid metabolism by tachykinin in Drosophila. Cell Rep. 2014;9:40–47. doi: 10.1016/j.celrep.2014.08.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Guo F, Holla M, Diaz MM, Rosbash M. A circadian output circuit controls sleep-wake arousal in Drosophila. Neuron. 2018;100:624–635. doi: 10.1016/j.neuron.2018.09.002. [DOI] [PubMed] [Google Scholar]
- 78.Lamaze A, Kratschmer P, Chen KF, Lowe S, Jepson JEC. A wake-promoting circadian output circuit in Drosophila. Curr. Biol. 2018;28:3098–3105. doi: 10.1016/j.cub.2018.07.024. [DOI] [PubMed] [Google Scholar]
- 79.Flourakis M, et al. A conserved bicycle model for circadian clock control of membrane excitability. Cell. 2015;162:836–848. doi: 10.1016/j.cell.2015.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yadlapalli S, et al. Circadian clock neurons constantly monitor environmental temperature to set sleep timing. Nature. 2018;555:98–102. doi: 10.1038/nature25740. [DOI] [PubMed] [Google Scholar]
- 81.Garcia-Recio, S. & Gascon, P. Biological and pharmacological aspects of the NK1-receptor. Biomed. Res. Int.2015, 495704 (2015). [DOI] [PMC free article] [PubMed]
- 82.Chang, C. T., Jiang, B. Y. & Chen, C. C. Ion channels involved in substance P-mediated nociception and antinociception. Int. J. Mol. Sci.20, 10.3390/ijms20071596 (2019). [DOI] [PMC free article] [PubMed]
- 83.Im SH, et al. Tachykinin acts upstream of autocrine Hedgehog signaling during nociceptive sensitization in Drosophila. eLife. 2015;4:e10735. doi: 10.7554/eLife.10735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ignell R, et al. Presynaptic peptidergic modulation of olfactory receptor neurons in Drosophila. Proc. Natl Acad. Sci. USA. 2009;106:13070–13075. doi: 10.1073/pnas.0813004106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ko, K. I. et al. Starvation promotes concerted modulation of appetitive olfactory behavior via parallel neuromodulatory circuits. eLife4, 10.7554/eLife.08298 (2015). [DOI] [PMC free article] [PubMed]
- 86.Mendoza J, Pevet P, Challet E. High-fat feeding alters the clock synchronization to light. J. Physiol. 2008;586:5901–5910. doi: 10.1113/jphysiol.2008.159566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Challet E, Solberg LC, Turek FW. Entrainment in calorie-restricted mice: conflicting zeitgebers and free-running conditions. Am. J. Physiol. 1998;274:R1751–R1761. doi: 10.1152/ajpregu.1998.274.6.R1751. [DOI] [PubMed] [Google Scholar]
- 88.Ishimoto H, Lark A, Kitamoto T. Factors that differentially affect daytime and nighttime sleep in Drosophila melanogaster. Front. Neurol. 2012;3:24. doi: 10.3389/fneur.2012.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.van Alphen B, Yap MH, Kirszenblat L, Kottler B, van Swinderen B. A dynamic deep sleep stage in Drosophila. J. Neurosci. 2013;33:6917–6927. doi: 10.1523/JNEUROSCI.0061-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Park JH, et al. Differential regulation of circadian pacemaker output by separate clock genes in Drosophila. Proc. Natl Acad. Sci. USA. 2000;97:3608–3613. doi: 10.1073/pnas.97.7.3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kula-Eversole E, et al. Phosphatase of regenerating liver-1 selectively times circadian behavior in darkness via function in PDF neurons and dephosphorylation of TIMELESS. Curr. Biol. 2021;31:138–149. doi: 10.1016/j.cub.2020.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Soderberg JA, Carlsson MA, Nassel DR. Insulin-producing cells in the Drosophila brain also express satiety-inducing cholecystokinin-like peptide, drosulfakinin. Front. Endocrinol. 2012;3:109. doi: 10.3389/fendo.2012.00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lee E, et al. Pacemaker-neuron-dependent disturbance of the molecular clockwork by a Drosophila CLOCK mutant homologous to the mouse Clock mutation. Proc. Natl Acad. Sci. USA. 2016;113:E4904–E4913. doi: 10.1073/pnas.1523494113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gunawardhana KL, Hardin PE. VRILLE controls PDF neuropeptide accumulation and arborization rhythms in small ventrolateral neurons to drive rhythmic behavior in Drosophila. Curr. Biol. 2017;27:3442–3453. doi: 10.1016/j.cub.2017.10.010. [DOI] [PubMed] [Google Scholar]
- 95.Kim EY, et al. A role for O-GlcNAcylation in setting circadian clock speed. Genes Dev. 2012;26:490–502. doi: 10.1101/gad.182378.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yang Z, et al. A post-ingestive amino acid sensor promotes food consumption in Drosophila. Cell Res. 2018;28:1013–1025. doi: 10.1038/s41422-018-0084-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The source data underlying the graphs are shown as Supplementary Data 1. All other data supporting the findings of this study are available from the corresponding author upon reasonable request.