Abstract
Cancer cells have the plasticity to adjust their metabolic phenotypes for survival and metastasis. A developmental programme known as epithelial-to-mesenchymal transition (EMT) plays a critical role during metastasis, promoting the loss of polarity and cell–cell adhesion and the acquisition of motile, stem-cell characteristics. Cells undergoing EMT or the reverse mesenchymal-to-epithelial transition (MET) are often associated with metabolic changes, as the change in phenotype often correlates with a different balance of proliferation versus energy-intensive migration. Extensive crosstalk occurs between metabolism and EMT, but how this crosstalk leads to coordinated physiological changes is still uncertain. The elusive connection between metabolism and EMT compromises the efficacy of metabolic therapies targeting metastasis. In this review, we aim to clarify the causation between metabolism and EMT on the basis of experimental studies, and propose integrated theoretical–experimental efforts to better understand the coupled decision-making of metabolism and EMT.
Subject terms: Metastasis, Regulatory networks, Cancer metabolism
Background
Metastasis remains the leading cause of cancer-related death, and efforts to interfere with this process—and the associated emergence of drug resistance—therefore remain a priority. A developmental programme referred to as epithelial-to-mesenchymal transition (EMT) is often implicated in metastasis and also in the acquisition of stemness, which is typically associated with drug resistance.1 During EMT, cobblestone-shaped epithelial cells lose their apical–basal polarity and cell–cell adhesion and become spindle-shaped, with increased motility and invasiveness.1 Crucially, cells undergoing EMT typically become more resilient in the face of stress that arises either from applied therapeutics and/or from being faced with a new microenvironment in a distant metastatic niche. Several review articles summarise the state-of-affairs concerning EMT phenomenology and EMT systems-biology modelling.2,3
Metabolic reprogramming is an emerging hallmark of cancer.4 Whereas normal cells mainly use oxidative phosphorylation (OXPHOS) for ATP production, cancer cells have often been observed to rely primarily or partially on glycolysis, irrespective of the presence of oxygen, in a process referred to as the Warburg effect or aerobic glycolysis, which takes place in the cytosol. The increased glycolytic activity in cancer cells was originally hypothesised to be the result of their defective mitochondria,5 but this has been proven not to be the case. The proposed benefits of the Warburg effect include the rapid production of ATP, biomass synthesis and balanced levels of reactive oxygen species (ROS).6 Notably, several types of non-cancer cell also exhibit aerobic glycolysis. For example, the adult stem cells that reside in hypoxic niches use glycolysis to maintain their self-renewal capacity,7 and neural crest cells use aerobic glycolysis when undergoing EMT for migration throughout the embryo,8 supporting the hypothesis that aerobic glycolysis is a physiological adaptive developmental programme that can be anomalously activated in cancer.
Although the Warburg effect is a common phenomenon in cancer, the role of mitochondria in cancer cells cannot be ignored. Increasing glycolytic activity in the cells of the primary tumour relative to normal cells does not necessarily mean their mitochondrial activity has to be suppressed. As one example, PANC-1 pancreatic cells exhibit significantly higher glucose oxidation activity relative to healthy pancreatic epithelial cells.9 Furthermore, a meta-analysis of 31 cancer cell lines shows that the contribution of OXPHOS to ATP production ranges from 36% to 99% across cancer cell lines.10 Moreover, increased mitochondrial mass and activity can confer stem-like traits on cancer cells and promote their resistance to chemotherapy.11 In short, mitochondrial activity has a critical role in cancer cells, especially in drug resistance and metastasis.12–15
Studies carried out over the past decade have witnessed significant advances in characterising EMT and metabolic plasticity. It has been convincingly shown that neither EMT nor metabolic reprogramming is a binary decision-making process in which cells can be only epithelial or mesenchymal, or can use only glycolysis or OXPHOS. Instead, a more nuanced set of pictures has emerged. Cancer cells can exist along a spectrum of EMT states that are characterised by varying proportions of epithelial and mesenchymal traits.16–18 Stable hybrid epithelial/mesenchymal (E/M) phenotypes that co-express E-cadherin and vimentin and exhibit collective migratory behaviour have been identified at the single-cell level and have been argued to be of critical importance for metastasis and/or drug resistance.18,19 In addition, many researchers have shown that cancer cells can mix and match different aspects of energy production and resource utilisation and acquire a mixed metabolic phenotype that is characterised by high rates of both glycolysis and OXPHOS.11,13,14,20 Moreover, cancer cell populations can exhibit metabolic coordination as shown during collective invasion wherein leader cells use more OXPHOS and the follower cells use more glycolysis.21 This flexibility has proven problematic for the development of effective drug treatments, as tumour cells can readily adapt and continue to flourish even when putatively crucial pathways involved in EMT and/or metabolic reprogramming are blocked.
It is reasonable to expect that changing motility phenotypes would necessitate altered cellular bioenergetics and, thereby, altered metabolism and, indeed, extensive regulatory crosstalk between EMT and metabolic reprogramming has been demonstrated.22 However, the cause-and-effect relationship between EMT and metabolic reprogramming remains elusive. In this article, we focus on elucidating the EMT–metabolic reprogramming causation, exploring how EMT can affect cancer cell metabolism and how cancer cell metabolism influences EMT, and discuss how systems-biology approaches can be developed to rationalise the connection between EMT and metabolic reprogramming.
How does EMT affect metabolism?
EMT is a multidimensional transformation process that involves changes in cellular mechanics and biochemical signalling that are fine-tuned by the underlying epigenetic landscape.23 These changes can be instigated by signals from the microenvironment, such as transforming growth factor (TGF)-β, Notch ligands such as Delta and Jagged, cytokines such as interleukin (IL)-6 and tumour necrosis factor (TNF)-α (acting through nuclear factor (NF)-κB), interactions with the extracellular matrix (ECM), and possibly by mutational events. These microenvironmental signals eventually impinge on a network involving EMT-inducing transcription factor (EMT-TF) families such as SNAIL and ZEB, and microRNA (miRNA) families such as miR-200 and miR-34. Surprisingly, the details of the interactions between EMT-TFs and miRNAs seem to be context-dependent, indicating an elaboration of possible mechanisms en route to a relatively conserved set of phenotypic changes. Of note, the EMT-inducing signals and EMT-TFs also play critical roles in regulating cancer invasiveness in non-carcinoma contexts such as glioblastoma.24 Below, we discuss how these most important nodes of EMT regulation impinge upon metabolic reprogramming.
EMT-inducing signals and metabolism
Multiple microenvironmental signals that can trigger EMT can also reprogramme metabolism. The EMT-inducing signal that best exemplifies such a dual effect is TGF-β. TGF-β is widely used to induce EMT in a plethora of cancer types;23 it does so through a variety of mechanisms.23 In terms of metabolism, TGF-β can promote glycolysis24 as well as upregulating fatty acid β-oxidation (FAO) to meet the energy needs associated with EMT and motility.22,25
TGF-β signalling and glycolysis
TGF-β signalling upregulates glycolysis by increasing the expression of glucose transporters and glycolytic enzymes (Fig. 1). For example, TGF-β can increase the mRNA levels of glucose transporter 1 (GLUT1), hexokinase 2 (HK2) and lactate dehydrogenase A (LDHA) in glioblastoma.24 The glycolysis-enhancing effect of TGF-β has also been reported in PANC-1 cells, which exhibit increased expression of 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 (PFKFB-3), an allosteric activator of the glycolytic enzyme phosphofructokinase (PFK)-1, followed by increased glucose uptake and lactate production upon TGF-β treatment.26 Intriguingly, OXPHOS activity remains significant in PANC-1 cells,27 indicating the acquisition of a hybrid metabolic phenotype. It is possible that the increase in glycolysis during TGF-β induced EMT is connected to the established relationship between EMT and stemness28,29 and the well-known tendency for stem cells to use this form of energy production.7 This putative link then leads to the question of whether these metabolic changes are, in fact, necessary for EMT. At least in some cases, the answer appears to be yes. As one example, PFKFB-3 knockdown suppresses SNAIL and reduces the EMT-dependent invasiveness of PANC-1 cells.26 A detailed discussion of the effects of induced metabolic changes on EMT is presented below.
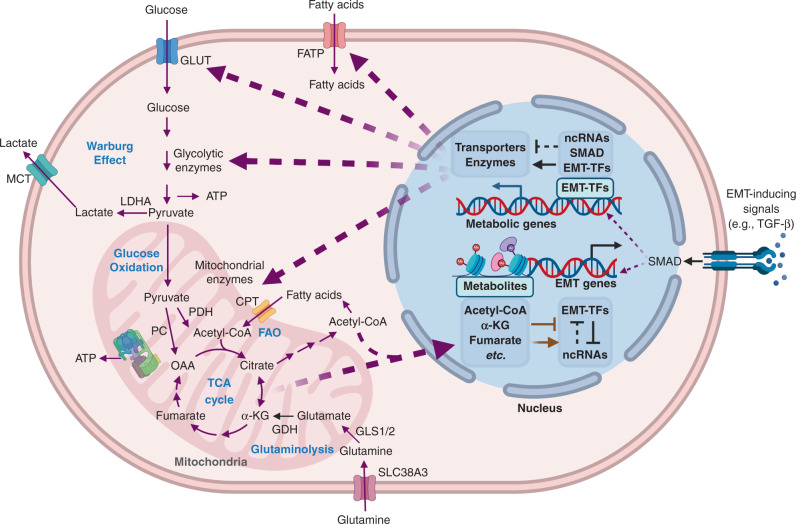
Fig. 1. Crosstalk between EMT and metabolism in cancer.
The purple arrows represent metabolic fluxes. The black arrows/bar-headed arrows represent transcriptional regulation. The black dotted bar-headed arrows represent ncRNA-mediated regulation. The brown arrows/bar-headed arrows represent epigenetic regulations. The purple dotted arrows represent the transportation of molecules that mediate the interaction between EMT and metabolic reprogramming. EMT-inducing signals, EMT-TFs and EMT-associated ncRNAs can directly regulate the transcription or translation of metabolic enzymes and transporters. In turn, the metabolic intermediates can facilitate epigenetic modification of EMT-associated genes or proteins. For example, the EMT-TFs Slug/Twist can repress mitochondrial respiration via suppression of SDH. SDH suppression leads to accumulation of succinate. Succinate accumulation can cause DNA hypermethylation by inhibiting TET2, and subsequently promotes EMT. More details about the EMT-metabolism crosstalk can be found in sections “how does EMT affect metabolism?” and “how does cancer metabolism affect EMT”. GLUT glucose transporter, FATP fatty acid transporter protein, MCT monocarboxylate transporter, LDHA lactate dehydrogenase A, PDH pyruvate dehydrogenase, PC pyruvate carboxylase, OAA oxaloacetic acid, α-KG α-ketoglutarate, CPT carnitine palmitoyltransferase, GDH glutamate dehydrogenase, GLS glutaminase, ncRNA non-coding RNA, EMT-TF EMT-inducing transcription factor. The Figure was created by BioRender.com.
TGF-β signalling and FAO
Aside from upregulating glycolysis, TGF-β signalling can promote FAO, which will increase the rate of energy production compared with the pre-EMT baseline. The non-small cell lung cancer (NSCLC) cell line A549, which uses basal glycolysis and OXPHOS,30 exhibits increased FAO along with EMT upon TGF-β treatment.22 TGF-β treatment decreases the master lipogenic regulator carbohydrate-responsive element-binding protein (ChREBP), which consequently decreases the expression of fatty acid synthase and induces FAO. Again, the connection is two-sided; fatty acid synthase knockdown is sufficient to induce EMT in vitro and can promote metastasis in vivo.22 TGF-β signalling can also promote FAO by increasing fatty acid uptake via the membrane fatty acid transporter CD36.25 Interestingly, enhanced FAO enriches the cellular pool of acetyl coenzyme A (acetyl-CoA), which can increase the acetylation of Smad2, a component of TGF-β signalling, thereby strengthening the TGF-β signalling pathway. A mutual excitatory feedback loop therefore operates between TGF-β signalling and FAO. Interestingly, in renal tubular epithelial cells, TGF-β signalling suppresses FAO via peroxisome proliferator-activated receptor (PPAR) γ coactivator 1α (PGC-1α) and Smad3, and inhibiting FAO leads to fibrosis,31 indicating potential distinct effects of TGF-β on FAO in cancer and non-cancer contexts.
Along with EMT induction, TGF-β therefore has the potential to induce cells to increase glycolysis or FAO, or both. Depending on how significant the increases in glycolysis and FAO are, cells can exhibit mostly glycolytic activity or mostly OXPHOS activity. Therefore, the net effect of the coupling between EMT and metabolic reprogramming due to TGF-β stimulation can be context-dependent. The dual effect of TGF-β on EMT and glycolysis is perhaps analogous to the dual role played by hypoxia in tumours. Hypoxic conditions in cancer cells can stabilise the transcription factor hypoxia-inducible factor (HIF)-1, which can then induce EMT by transcriptionally activating SNAIL as well as promoting glycolysis by acting on multiple glycolytic enzymes,11 thereby associating increased glycolysis with EMT.
EMT-TFs and metabolism
In addition to investigating how external EMT-inducing signals couple to metabolic reprogramming, how specific EMT-TFs (SNAIL, SLUG, TWIST, ZEB, etc.) cause specific metabolic changes independent of whether they were stimulated by TGF-β or by other pathways (Fig. 1) can also be studied. Characterizing the activity of EMT-TFs and their roles in metabolism allows for a better understanding of the EMT status of a given cell line and its expected metabolism.
SNAIL
SNAIL can downregulate OXPHOS and upregulate glycolysis. In a panel of basal-like breast cancer cell lines, SNAIL directly represses fructose-1,6-bisphosphatase (FBP) 1, a rate-limiting enzyme in gluconeogenesis.32 FBP1 repression leads to a reduction in both oxygen consumption and the levels of ROS, and an increase in glycolysis and biomass synthesis. The repression of FBP1 is required for SNAIL-induced EMT, as ectopic expression of FBP1 abrogates the decrease in E-cadherin and EMT-related morphological change upon SNAIL induction. Conversely, knockdown of SNAIL can increase FBP1 expression.
SLUG and TWIST
SLUG and TWIST can also inhibit mitochondrial respiration and activate glycolysis. Overexpression of SLUG or TWIST in the luminal A breast cancer MCF7 cell line and in immortalised but non-tumorigenic MCF10A cells leads to decreased mitochondrial respiration and reduced mitochondrial mass, owing to the suppression of succinate dehydrogenase (SDH), an enzyme involved in the TCA cycle and the electron transport chain.33 Notably, both the MCF7 and the MCF10A cells exhibit active basal OXPHOS.34 An alternative mechanism underlying TWIST-mediated metabolic change in MCF10A cells depends on the phosphatidylinositol 3-kinase (PI3K)–AKT–mammalian target of rapamycin (mTOR) signalling pathway, which leads to an increase in the levels of pyruvate kinase M2 and lactate dehydrogenase A,35 ultimately promoting glycolysis.
ZEB1
ZEB1, an EMT-TF considered to be more critical for a sustained EMT response relative to SNAIL/TWIST in certain scenarios,36,37 can also be crucial for cancer metabolic plasticity.36 In the KPC mouse model of pancreatic cancer, ZEB1-knockout KPC cells fail to upregulate their glycolytic activity to compensate for the decreased OXPHOS upon treatment with the ATP synthase inhibitor oligomycin.36 ZEB1 can promote glucose uptake by transcriptionally activating the glucose transporter 3 (GLUT3) gene during EMT.38
Beyond increased glycolysis and decreased OXPHOS
Thus, the EMT-TFs (SNAIL, SLUG, TWIST and ZEB1) exhibit consistent activation of glycolysis and inhibition of glucose-based OXPHOS through the altered expression of genes that encode enzymes involved in energy metabolism. Cells might or might not compensate for this rerouting of glucose by increasing FAO. However, there do appear to be exceptions to this general behaviour. Under conditions of oxidative stress, SNAIL can suppress glycolysis in MCF7 cells and in triple negative breast cancer (TNBC) MDA-MB-231 cells by transcriptionally inhibiting the platelet isoform of the phosphofructokinase (PFKP) glycolytic enzyme.39 Downregulation of PFKP diverts the glucose flux from glycolysis to the pentose phosphate pathway to generate NADPH for survival under oxidative stress. As another example, circulating tumour cells (CTCs) formed in the 4T1 mouse breast cancer model have undergone EMT but exhibit increased levels of OXPHOS and ATP production, mediated by PGC-1α; however, these cells retain a level of glycolytic activity that is similar to levels seen in both the primary tumour and lung metastases.13 It would be interesting to investigate whether the increased OXPHOS in 4T1-CTCs is due to increased FAO. We will return to this example below.
EMT-TFs can also regulate other aspects of metabolism in addition to glycolysis and OXPHOS. For example, ZEB1 can promote the synthesis of long-chain polyunsaturated fatty acids, which become substrates for lipid peroxidation and can lead to ferroptosis.40 ZEB1 can also promote gangliosides synthesis by upregulating the enzyme GM3 synthase (GM3S) both directly (as a transcription factor) and indirectly (by repressing miRNAs that inhibit GM3S).41 Indeed, GM3S is required for EMT as repression of GM3S is sufficient to increase epithelial cell adhesion. Discussing all such effects are beyond the scope of this review, but are described in.40–43
EMT-associated non-coding RNAs and metabolism
Non-coding RNAs, which include miRNAs (e.g. miR-200 and miR-34) and long non-coding RNAs (lncRNAs; e.g. NEAT1 and ANRIL) comprise an important layer in the post-transcriptional regulation of EMT. Both miRNAs and lncRNAs can function either as inducers of EMT (such as miR-10b44 and HOTAIR45) or suppressors of EMT (such as miR-20046 and TUSC747).
EMT-associated miRNAs and metabolism
Generally speaking, the miRNAs that maintain an epithelial phenotype tend to repress glycolysis; conversely, the miRNAs that repress EMT can repress FAO as well. For example, the miR-200 family and the miR-34 family function as critical gatekeepers of the epithelial phenotype.46 Members of the miR-200 family directly targeting and silencing ZEB, and miR-34 family members directly target SNAIL. Both miR-200 and miR-34 can target lactate dehydrogenase A to repress glycolysis.48,49 miR-33, which inhibits EMT by targeting SNAIL and ZEB,50 can inhibit FAO by targeting the genes encoding carnitine palmitoyltransferase IA (CPT1A) and 5’ AMP-activated protein kinase (AMPK) α.51 As expected, the effects are directly opposite to those of TGF-β on both glycolysis and FAO upon induction of EMT.
EMT-associated lncRNAs and metabolism
In general, EMT-promoting lncRNAs can upregulate glycolysis and FAO. For example, HOTAIR, which can induce EMT by upregulating the transcriptional activator EZH2 and, consequently, EMT-TFs (SNAIL, ZEB and TWIST),45 can enhance glycolysis by increasing the expression of GLUT1 by activating mTOR signalling.52 Another example is the EMT-promoting lncRNA NEAT1, which can promote FAO through the activation of adipose triglyceride lipase (ATGL)–PPARα signalling.53 Again, the dual nature of the EMT effect on metabolism—increasing both glycolysis and FAO with an adjustable balance to meet the cell’s new requirements—is apparent.
In summary, we have described how EMT-inducing signals, EMT-TFs and EMT-promoting lncRNAs can enhance both glycolysis and FAO, and suppress glucose-based OXPHOS. We have also described how EMT-suppressing miRNAs can repress both glycolysis and FAO by altering the expression of metabolic transporters and enzymes. The dual effect of EMT on glucose-based oxidation repression and FAO activation might reconcile contradictory conclusions in the literature where either decreased OXPHOS or increased OXPHOS have been associated with EMT.54 We have also mentioned several reciprocal cases where metabolic changes act as a causative agent for promoting or inhibiting EMT. We look at this question more systematically in the following section.
How does cancer metabolism affect EMT?
Cancer cells are endowed with the ability to adjust their metabolic activity—through glycolysis, mitochondrial respiration, fatty acid metabolism and glutamine metabolism11—to exploit the surrounding nutrients.11 In this section, we will discuss how each of these processes can shape EMT (Fig. 1).
Glycolysis can promote EMT
Emerging evidence supports the notion that increased glycolysis can facilitate EMT. Cancer cells exhibit elevated glucose uptake relative to normal cells, which can be achieved via the overexpression of GLUT1; increased GLUT1 expression has been reported to increase the expression of matrix metalloproteinase 2, which is overexpressed in many cancer types and associated with increased invasiveness.55 Upregulation of HK2, which is involved in the first rate-limiting step of glycolysis, can increase glycolysis and enhance the metastasis of PANC-1 cells in a lactate-dependent manner.56 Consistently, downregulation of HK2 results in decreased glycolysis and suppressed EMT.57 The enzyme glucose-6-phosphate isomerase, as well as converting glucose 6-phosphate into fructose 6-phosphate, can act as a cytokine. When secreted by tumour cells, glucose-6-phosphate isomerase is often referred to as an autocrine motility factor that can induce EMT via ZEB1/2 in an NF-κB-dependent manner.58 Inhibition of either PFKFB-3 or glyceraldehyde-3-phosphate dehydrogenase can inhibit EMT via the downregulation of SNAIL.26,59 Pyruvate kinase M2 (PKM2) can induce EMT via its nuclear translocation and subsequent transcriptional suppression of the gene that encodes E-cadherin (CDH1).60 Intriguingly, nuclear PKM2 can also activate β-catenin, which enhances MYC activity and results in the increased expression of PKM2, LDHA and GLUT1, thus forming a self-reinforcing feedback loop to strengthen the connection between glycolysis and EMT.61
PDK1, the enzyme that inactivates pyruvate dehydrogenase (PDH) to inhibit the conversion of pyruvate into acetyl-CoA to fuel the TCA cycle, promotes EMT by activating NF-κB signalling in gastric cancer.62 In ovarian cancer, downregulation of PDK1 reverts EMT and eliminates cisplatin resistance, a common EMT-associated trait.63 Interestingly, another isozyme of PDK—PDK4—exhibits the opposite effect on EMT. PDK4 overexpression partially blocks TGF-β-induced EMT in lung cancer.64 PDK4 inhibition is sufficient to induce EMT and enables resistance to erlotinib. This result is reminiscent of the anti-metastasis effect seen in breast cancer by suppressing pyruvate carboxylase, which catalyses the carboxylation of pyruvate to oxaloacetate to replenish the TCA cycle.65 These results suggest that lung and breast cancer cells that have undergone EMT might still need the diversion of some glucose to the TCA cycle. This diversion might reflect the need for the TCA activity to provide citrate to enrich the cellular pool of acetyl-CoA, as discussed in the next section. It would be interesting to investigate the dependence of this finding on the transport of glutamine, which can act as an alternative source of acetyl-CoA.
Finally, LDHA, which converts pyruvate into lactate, the end product of glycolysis, can promote EMT by upregulating ZEB2.66 In addition, the lactate produced and secreted by tumour cells can lower the extracellular pH and convert inactive extracellular TGF-β into its active form, thus promoting EMT.67 Meanwhile, the secretion of lactate from cells elevates the intracellular pH, which activates Wnt signalling, potentially leading to EMT.68 In summary, glucose transporters, most glycolytic enzymes (except for PDK4), and lactate accumulation all consistently promote EMT across cancer types.
FAO can promote EMT
In addition to glycolysis, the uptake of fatty acids and subsequent FAO can induce EMT/metastasis. The Cancer Genome Atlas (TGCA) pan-cancer analysis has revealed that samples with a higher EMT score have a higher expression of the genes encoding caveolin 1, a major component of caveolae, and the scavenger receptor CD36, both of which are involved in fatty acid uptake.69 A follow-up study showed that elevated fatty acid uptake via CD36 activates TGF-β and Wnt signalling pathways and thereby leads to EMT induction.70 Enhanced FAO can promote metastasis by activating oncogenic pathways such as Src15 to promote EMT.71 Consequently, blocking FAO by silencing CPT1A represses metastasis.15
Amino acid metabolism and EMT
As well as glycolysis and FAO, changes in glutamine metabolism have emerged as another hallmark of EMT.72 Among the enzymes involved in glutaminolysis (the process by which glutamine is converted into TCA cycle metabolites), the role of glutaminase (GLS) in the regulation of EMT has been the most widely reported. Two isoforms of GLS exist—the kidney-type, GLS1, and the liver-type, GLS2. Both isoforms can convert glutamine into glutamate; however, GLS1 functions as an EMT inducer while GLS2 functions as an EMT suppressor. GLS1, when knocked out, inhibits TGF-β-induced EMT in MCF7 cells and represses metastasis in vivo.73 GLS2 can bind to and stabilise Dicer to increase the expression of miR-34 and thereby repress EMT.74 Accordingly, benign breast HMLE cells undergoing EMT exhibit increased levels of GLS1 and decreased levels of GLS2.75 As well as GLS, glutamine dehydrogenase (GDH), which converts glutamate into α-ketoglutarate (α-KG), potentially promotes EMT via activation of signal transducer and activator of transcription (STAT) 3 signalling and consequently upregulation of TWIST and ZEB.76 Interestingly, a deficiency in intracellular glutamine as a result of increased export by the glutamine transporter SLC38A3, the expression of which is upregulated in metastatic NSCLC cells and correlates with poor prognosis, can promote EMT by activating the PDK1–AKT signalling pathway.77
Of course, the possible effects of other aspects of amino acid metabolism should not be overlooked. One example concerns branched-chain α-keto acid dehydrogenase kinase (BCKDK), the key enzyme for the catabolism of branched-chain amino acids such as leucine, valine and isoleucine, which has been reported to promote EMT in metastatic colorectal cancer. In this context, EMT is facilitated by decreasing the expression of E-cadherin and increasing the expression of the well-known mesenchymal markers N-cadherin and vimentin.78
Mitochondrial regulation of EMT epigenetics
Mitochondrial enzymes and metabolites can promote EMT via their effects on epigenetic modifications. Cancer cells undergoing EMT exhibit epigenetic plasticity involving both DNA and histone modifications.23 Modification enzymes are mainly classified into three categories: ‘writers’ (e.g. histone acetyltransferases (HATs)), which attach a molecular modification to chromatin or DNA; ‘erasers’ (e.g. DNA demethylation ten eleven translocation hydroxylase (TET) enzymes), which remove the modifications; and ‘readers’ (e.g. bromodomain-containing protein 4 (BRD4)), which recognise acetylated or methylated residues.79 Metabolites—acetyl-CoA, α-KG, 2-HG, succinate, fumarate, and so on—generated in mitochondria can serve as crucial cofactors or substrates (referred to as ‘ink’) that directly impact the activity of histone modification enzymes.80 Thus, it is not surprising that many studies have reported that mitochondrial dysfunction or mitochondrial retrograde signalling can regulate EMT-related epigenetics in cancer.81
We note also that non-epigenetic pathways exist by which mitochondrial dysfunction can affect EMT—for example, by altering mitophagy or regulating the distribution of ROS in the cell. Mitophagy, a specialized form of autophagy, leads to mtDNA depletion, which is known to induce EMT in mammary epithelial cells.82 On the other hand, ROS interacts with EMT in a complex manner; for a review of the various mechanisms underlying this interaction see ref. 83
Acetyl-CoA
Acetyl-CoA is a substrate that HATs use to carry out the process of acetylation, which generates an euchromatin state of the target genes that encode EMT-TFs and thereby induces EMT. The accumulation of acetyl-CoA can increase H3 acetylation in the promoter region of TWIST2, consequently inducing TWIST2 expression and promoting EMT, as shown in hepatocellular carcinoma.84 Acetyl-CoA can also function as a key cofactor for the post-translational modification of key proteins involved in EMT. For example, a high level of histone acetylation on Snail enables BRD4 to bind to this protein and prevent its proteasomal degradation, consequently promoting EMT, as shown in gastric cancer.85 Elevated acetyl-CoA levels can also increase Smad2 acetylation to facilitate EMT, as demonstrated in breast cancer.25
α-KG and α-KG-dependent demethylases
α-KG, which is generated from citrate during the TCA cycle or by glutaminolysis, is a cofactor for the ‘eraser’ TETs.86 The accumulation of α-KG can increase TET activity to cause DNA demethylation of miR-200 and, as a result, miR-200 is activated and EMT is inhibited.87 Due to their structural similarity to α-KG, 2-HG, succinate and fumarate function as competitive inhibitors of α-KG-dependent demethylases such as TET- and Jumonji-family proteins.88–90 In various human cancer types, gain-of-function mutations in isocitrate dehydrogenase 1 (IDH1; in the cytosol) and/or IDH2 (in mitochondria) are frequently observed. As a result, 2-HG accumulates and can promote EMT by increasing H3K4 methylation (by inhibiting Jumonji-family histone demethylases) in the promoter region of ZEB1.91 Increased levels of succinate and fumarate in cancer often result from loss-of-function mutations in succinate dehydrogenase (SDH) and fumarate hydratase (FH), respectively. Succinate accumulation can cause DNA hypermethylation by inhibiting TET2, and subsequently promotes EMT.92 Fumarate accumulation inhibits TET-mediated DNA demethylation of the miR-200ba429 locus and, consequently, the repression of ZEB by miR-200 is relieved and EMT is promoted.93
Complexity of EMT–metabolic reprogramming connection
Our focus in this review has been on how cancer metabolic activity, through glycolysis, mitochondrial respiration, fatty acid metabolism and glutamine metabolism, can enable or disable EMT. Notably, there have been studies elucidating the connection between metabolism and EMT via additional metabolic pathways—see Shaul et al.94 for the role of dihydropyrimidine dehydrogenase (DPYD) and Schwab et al.95 for a discussion of the polyol pathway, which converts excess intracellular glucose into fructose. These examples indicate the complex nature of the connection between EMT and the overall metabolic system of a cell. Although certain trends are more common, it should be acknowledged that no simplistic rule exists for how different EMT stages are coordinated with various metabolism phenotypes in a specific cell line or specific patient sample. Interestingly, this finding is analogous to the connection between EMT and stemness, where it has also been realised that cells at various stages of EMT can all become stem-like;29 although a most common behaviour exists (namely that hybrid E/M cells are more conducive to de-differentiation), the detailed behaviour can be cell-line- and patient-sample-dependent. It is worth noting that, as the role of metabolism in EMT regulation is gradually revealed, metabolic inhibitors might play an increasing role in targeting EMT.96
Metabolic correlates to the hybrid E/M phenotype
Let us now step back from the detailed discussion of molecular pathways and focus on energy production. From a more general perspective, we have seen that cells fulfil their bioenergetic needs in variable ways and, similarly, cells can choose to operate at various locations along the E/M phenotypic axis. Interestingly, hybrid E/M phenotypes and mixed metabolic states have both been proposed to account for high metastatic potential and cancer stemness. There then ensues an interesting question: does the hybrid metabolic phenotype typically characterise the hybrid E/M phenotype? Preliminary evidence suggests that the answer might be yes.
Evidence from 4T1-CTCs and breast cancer stem cells
Returning to the example mentioned above of CTCs identified in the 4T1 mouse breast cancer model, these CTCs exhibit a higher level of OXPHOS and a similar level of glycolysis relative to both the primary tumours and lung metastases formed by 4T1 cells, indicating the acquisition of a mixed metabolic phenotype.13 At the same time, 4T1 tumours have a significant percentage of hybrid E/M cells, suggesting that the 4T1-CTCs are likely to consist largely of clusters of CTCs with dual hybrid characteristics. Supporting increased OXPHOS in CTCs, levels of the antioxidation regulator nuclear factor erythroid 2-related factor 2 (NRF2) reach their maximum in the hybrid E/M phenotype relative to epithelial or mesenchymal entities.97
Similar observations are emerging from the consideration of breast cancer stem cells (BCSCs). BCSCs contain two subpopulations: the CD44high/CD24low more-mesenchymal-like BCSCs (M-BCSCs) and the aldehyde dehydrogenase (ALDH)+ hybrid E/M-BCSCs (sometimes referred to as the more-epithelial-like BCSCs).98,99 Relative to M-BCSCs, the hybrid E/M-BCSCs demonstrate higher OXPHOS and similar glycolysis—that is, they have acquired a mixed metabolic phenotype—and their proliferation can drive metastatic growth. The quiescent M-phenotype is the most stem-like state, consistent with the notion that enhanced glycolysis is useful for enabling cellular plasticity7 (Fig. 2). These concepts could offer a path forward for targeting dormant M-BCSCs.
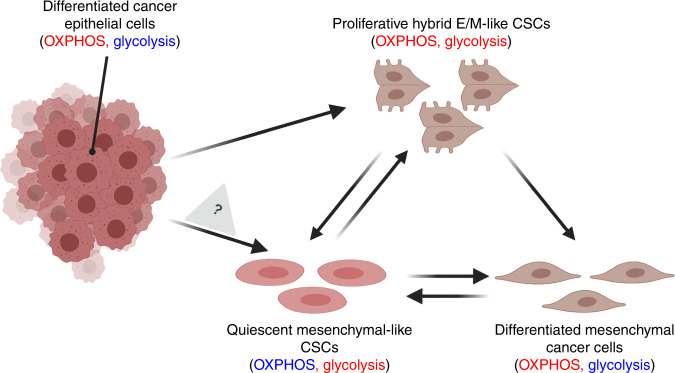
Fig. 2. Hypothetical coupling of EMT and metabolic reprogramming during the acquisition of stemness.
Differentiated epithelial cancer cells depend on oxidative phosphorylation (OXPHOS).9,10 Upon undergoing EMT, these cells can increase their glycolytic activity as needed to acquire stemness and become hybrid E/M-like cancer stem cells (CSCs; OXPHOShigh/glycolysishigh, proliferative). The hybrid E/M-like CSCs can either decrease their OXPHOS activity to transition into mesenchymal-like CSCs (glycolysishigh, quiescent)98,99 or decrease their glycolytic activity and lose stemness to transition into the differentiated mesenchymal cancer cells (OXPHOShigh). Note that being labelled OXPHOShigh does not determine the extent to which fatty acid oxidation is operative, which could differ between different types of differentiated cancer cell. Quiescent mesenchymal-like CSCs (glycolysishigh) can either increase their OXPHOS activity to become hybrid E/M-like CSCs99 or switch from glycolysis to OXPHOS and differentiate into mesenchymal cancer cells; these cells might undergo dedifferentiation to become mesenchymal-like CSCs. It will be interesting to investigate whether differentiated epithelial cancer cells can directly transition into mesenchymal-like CSCs without passing through hybrid E/M phenotypes. The Figure was created by BioRender.com.
The evidence outlined above supporting a connection between the hybrid E/M phenotype and a hybrid metabolism is fairly preliminary, with two key questions still unresolved. First, it remains to be determined if the observed hybrid metabolic behaviour in the cases of 4T1-CTCs and BCSCs results from a hybrid metabolic character at the single-cell level or is simply a consequence of different cells in the population exhibiting different metabolic behaviours. Technical advances in single-cell metabolomics should help to answer this question. Second, and more importantly, detailed regulatory connections between the hybrid E/M state and hybrid metabolism have not yet been established. Experiments that directly probe the metabolic character of hybrid E/M cells are needed to determine whether or not hybrid E/M cells are more likely to exhibit hybrid metabolism compared with epithelial or mesenchymal cells and, conversely, whether or not the maintenance of a mixed set of metabolic processes is necessary for the maintenance of a hybrid E/M state.
Using systems biology to solve the EMT–metabolic reprogramming puzzle
In the face of increasing sophisticated data regarding the various links between EMT and the metabolic state, how can further progress in elucidating the association between EMT and metabolic reprogramming be made? We believe it will be necessary to systematically investigate the quantitative dynamics of the coupled decision-making networks of EMT and metabolism and, to that end, systems-biology approaches will need to be developed and applied (Fig. 3).
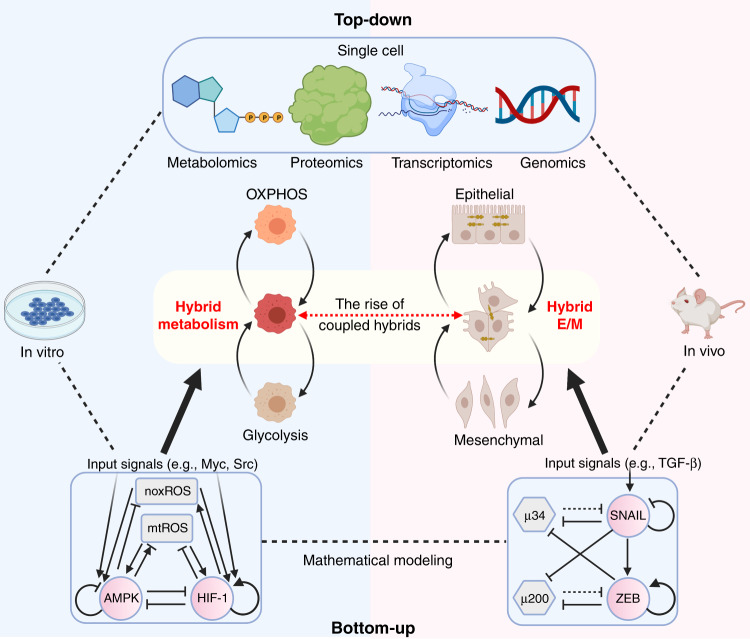
Fig. 3. A systematic pipeline to elucidate the connection between EMT and metabolic reprogramming.
Mathematical modelling approaches have provided critical insights into the dynamics of EMT and metabolism and predicted the acquisition of stable hybrid phenotypes by cancer cells. The AMPK/HIF-1/ROS model (bottom left) is an initial effort to understand how genetic regulation interacts with metabolic fluxes.104 The miR-34/SNAIL/miR-200/ZEB model (bottom right) was used in the initial studies that proposed that hybrid epithelial/mesenchymal (E/M) phenotypes can constitute an end point of EMT.37 The predicted hybrid metabolic phenotype and hybrid E/M phenotype have been validated by several in vitro and in vivo studies, respectively.16,19,20 With further advances in single-cell multi-omics, acquisition of the transcriptomics and metabolomics profiles of the same single cell can facilitate the understanding of EMT–metabolism coupling. Through the integration of mathematical modelling, data analysis and experimental studies, the nuanced EMT–metabolism connection can gradually be elucidated. The Figure was created by BioRender.com.
Two main categories of systems-biology model exist: bottom-up and top-down. The bottom-up approach often starts with a relatively well-defined molecular network and formulates mathematical models to simulate its dynamics; the goal is to better explain observed experimental results and to generate predictions to guide new experiments. The top-down approach often starts from omics data from which phenomenological models are inferred to capture the data. Both bottom-up and top-down approaches have been applied to EMT and metabolism, albeit independently. To gain an integrated view of EMT–metabolic reprogramming coupling, however, these separate studies need to be combined.
Models of EMT and metabolism
To simulate EMT, mechanism-based mathematical models have been developed to analyse the dynamics of the proposed regulatory networks. These models consist of two mutually inhibitory feedback loops, SNAIL/miR-34 and ZEB/miR-200,37 which predict the possible acquisition of stable hybrid E/M states as a possible end point of EMT. A detailed review of various modelling approaches to EMT dynamics can be found in refs. 2,3
A detailed review of traditional mathematical models of metabolism can be found in ref. 100 Constraint-based methods, including flux-balance analysis (FBA) based on mass conservation, constitute the most widely used approach.101 FBA is well suited to quantify the steady-state metabolic flux distribution and can be adapted to characterise the effects of perturbations in a computationally inexpensive way. However, in its basic form, FBA does not incorporate genetic regulation, and cannot determine metabolite concentrations.101 To begin to investigate crosstalk between genetic regulation and metabolic pathways, a mechanism-based mathematical model coupling the metabolic fluxes (glucose oxidation, glycolysis and FAO) with the AMPK/HIF-1 genetic circuit has been proposed.20 This mathematical model reveals a direct association between AMPK/HIF-1 and OXPHOS/glycolysis, illustrates how cancer cells switch between different metabolic phenotypes, and predicts the existence of a stable hybrid metabolic phenotype and a metabolic inactivity phenotype, which have been confirmed in TNBC cells and melanoma cells, respectively.20,102 Future progress using computational models to elucidate the coupled decision-making in the EMT–metabolic reprogramming connection must focus on extending our assessment of this type of crosstalk and applying it to the EMT circuit.
Coupling EMT and metabolic plasticity
One attempt to achieve a better understanding of the correlated temporal changes in EMT status and metabolism relies on computing a probabilistic landscape for a regulatory network integrating the decision-making modules of metabolism, EMT and metastasis. Given a regulatory network, the landscape approach can characterise the basins of attractions corresponding to stable cell phenotypes and approximate transition paths to predict the temporal orders of events. With that in mind, Kang et al.103 proposed a path to metastasis in which epithelial cells first reprogramme their metabolism by upregulating HIF-1, then transition to mesenchymal cells by upregulating ZEB, which is followed by transitioning into a metastatic state upon the upregulation of BACH1, a transcription factor regulating multiple metastasis-related genes.103 The result indicates that, as we have argued, metabolic reprogramming is necessary for EMT, and interfering with these metabolic changes can intercept EMT at a very early stage. Hopefully, this early attempt will spur the systems-biology community to stop viewing EMT and metabolic plasticity as uncoupled modules — as we have seen, they are strongly intertwined.
Concluding remarks
As both EMT and metabolism are multidimensional processes that involve stable hybrid phenotypes, it can be misleading when considering the connection between EMT and metabolic reprogramming if we simply classify a cancer cell line as purely epithelial or mesenchymal, or imagine that genetic or pharmacological perturbations have an all-or-nothing EMT-related consequence. Similarly, we cannot label a cell’s energy metabolism as being glycolytic or OXPHOS without having a detailed quantitative evaluation of both processes. Therefore, quantifying both EMT and metabolism is critical for probing the connection between EMT and metabolic reprogramming. It is probable that, as EMT proceeds through different stages, different metabolic processes predominate accordingly. It is possible that metabolic changes that accompany EMT precede some of the more obvious indicators of becoming more mesenchymal and indeed actively contribute to the activation of these indicators. By coupling the decision-making networks of EMT and metabolism, systems-biology approaches can help to identify the association of specific EMT states and metabolic states and the conditions that enable that association. The predicted connections between EMT and metabolic reprogramming can ideally be tested by simultaneously performing transcriptomics and metabolomics analyses of the same single cells and by experiments that perturb EMT to identify metabolic change and vice versa. A synergistic approach of theoretical and experimental efforts should be undertaken to delineate the dynamic connection map between EMT and metabolic reprogramming.
Acknowledgements
Not applicable.
Author contributions
D.J., B.A.K., J.N.O. and H.L. conceptualized the review. D.J., J.H.P., H.K., K.W.J., S.Y., S.T., M.G., Y.D. and M.K.J. contributed to writing early drafts of the paper. D.J., B.A.K., J.N.O. and H.L. revised the final version of the paper.
Ethics approval and consent to participate
Not applicable.
Consent to publish
Not applicable.
Data availability
Not applicable.
Competing interests
The authors declare no competing interests.
Funding information
This work is supported by the National Science Foundation (NSF) Center for Theoretical Biological Physics (NSF PHY-2019745) and NSF grants nos. PHY-1605817, PHY-1522550, and CHEM-1614101. J.N.O. is a CPRIT Scholar in Cancer Research. D.J. is supported by a training fellowship from the Gulf Coast Consortia, on the Computational Cancer Biology Training Program (CPRIT grant no. RP170593). M.G. is supported by the NSF GRFP no. 1842494. B.A.K. is supported by NIH grants nos. CA253445, CA234479, DK117001 and CA235113 and DOD grant no. W81XWH-18-1-0714. M.K.J. is supported by Ramanujan Fellowship awarded by Science and Engineering Research Board (SERB), Department of Science and Technology, Government of India (SB/S2/RJN-049/2018).
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Dongya Jia, Email: dyajia@gmail.com.
Benny Abraham Kaipparettu, Email: kaippare@bcm.edu.
José N. Onuchic, Email: jonuchic@rice.edu
Herbert Levine, Email: h.levine@northeastern.edu.
References
- 1.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J. Clin. Invest. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burger GA, Danen EHJ, Beltman JB. Deciphering epithelial-mesenchymal transition regulatory networks in cancer through computational approaches. Front. Oncol. 2017;7:162. doi: 10.3389/fonc.2017.00162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tripathi S, Levine H, Jolly MK. The physics of cellular decision making during epithelial-mesenchymal transition. Annu. Rev. Biophys. 2020;49:1–18. doi: 10.1146/annurev-biophys-121219-081557. [DOI] [PubMed] [Google Scholar]
- 4.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 5.Weinhouse S, Warburg O, Burk D, Schade AL. On respiratory impairment in cancer cells. Science. 1956;124:267–272. doi: 10.1126/science.124.3215.267. [DOI] [PubMed] [Google Scholar]
- 6.Liberti MV, Locasale JW. The Warburg effect: how does it benefit cancer cells? Trends Biochem. Sci. 2016;41:211–218. doi: 10.1016/j.tibs.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shyh-Chang N, Ng H-H. The metabolic programming of stem cells. Genes Dev. 2017;31:336–346. doi: 10.1101/gad.293167.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bhattacharya D, Azambuja AP, Simoes-Costa M. Metabolic reprogramming promotes neural crest migration via yap/tead signaling. Dev. Cell. 2020;53:199–211.e6. doi: 10.1016/j.devcel.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krapf SA, Lund J, Lundkvist M, Dale MG, Nyman TA, Thoresen GH, et al. Pancreatic cancer cells show lower oleic acid oxidation and their conditioned medium inhibits oleic acid oxidation in human myotubes. Pancreatology. 2020;20:676–682. doi: 10.1016/j.pan.2020.04.014. [DOI] [PubMed] [Google Scholar]
- 10.Zu XL, Guppy M. Cancer metabolism: facts, fantasy, and fiction. Biochem. Biophys. Res. Commun. 2004;313:459–465. doi: 10.1016/j.bbrc.2003.11.136. [DOI] [PubMed] [Google Scholar]
- 11.Jia, D., Park, J. H., Jung, K. H., Levine, H. & Kaipparettu, B. A. Elucidating the metabolic plasticity of cancer: mitochondrial reprogramming and hybrid metabolic states. Cells7, 21 (2018). [DOI] [PMC free article] [PubMed]
- 12.Porporato PE, Payen VL, Pérez-Escuredo J, De Saedeleer CJ, Danhier P, Copetti T, et al. A mitochondrial switch promotes tumor metastasis. Cell Rep. 2014;8:754–766. doi: 10.1016/j.celrep.2014.06.043. [DOI] [PubMed] [Google Scholar]
- 13.LeBleu VS, O’Connell JT, Gonzalez Herrera KN, Wikman H, Pantel K, Haigis MC, et al. PGC-1α mediates mitochondrial biogenesis and oxidative phosphorylation in cancer cells to promote metastasis. Nat. Cell Biol. 2014;16:992–1003. doi: 10.1038/ncb3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dupuy F, Tabariès S, Andrzejewski S, Dong Z, Blagih J, Annis MG, et al. PDK1-dependent metabolic reprogramming dictates metastatic potential in breast cancer. Cell Metab. 2015;22:577–589. doi: 10.1016/j.cmet.2015.08.007. [DOI] [PubMed] [Google Scholar]
- 15.Park JH, Vithayathil S, Kumar S, Sung P-L, Dobrolecki LE, Putluri V, et al. Fatty acid oxidation-driven Src links mitochondrial energy reprogramming and oncogenic properties in triple-negative breast cancer. Cell Rep. 2016;14:2154–2165. doi: 10.1016/j.celrep.2016.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pastushenko I, Brisebarre A, Sifrim A, Fioramonti M, Revenco T, Boumahdi S, et al. Identification of the tumour transition states occurring during EMT. Nature. 2018;556:463–468. doi: 10.1038/s41586-018-0040-3. [DOI] [PubMed] [Google Scholar]
- 17.Lu M, Jolly MK, Levine H, Onuchic JN, Ben-Jacob E. MicroRNA-based regulation of epithelial-hybrid-mesenchymal fate determination. Proc. Natl. Acad. Sci. USA. 2013;110:18144–18149. doi: 10.1073/pnas.1318192110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jolly MK, Boareto M, Huang B, Jia D, Lu M, Ben-Jacob E, et al. Implications of the hybrid epithelial/mesenchymal phenotype in metastasis. Front. Oncol. 2015;5:155. doi: 10.3389/fonc.2015.00155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jolly MK, Tripathi SC, Jia D, Mooney SM, Celiktas M, Hanash SM, et al. Stability of the hybrid epithelial/mesenchymal phenotype. Oncotarget. 2016;7:27067–27084. doi: 10.18632/oncotarget.8166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jia D, Lu M, Jung KH, Park JH, Yu L, Onuchic JN, et al. Elucidating cancer metabolic plasticity by coupling gene regulation with metabolic pathways. Proc. Natl Acade. Sci. USA. 2019;116:3909–3918. doi: 10.1073/pnas.1816391116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Commander R, Wei C, Sharma A, Mouw JK, Burton LJ, Summerbell E, et al. Subpopulation targeting of pyruvate dehydrogenase and GLUT1 decouples metabolic heterogeneity during collective cancer cell invasion. Nat. Commun. 2020;11:1533. doi: 10.1038/s41467-020-15219-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang L, Xiao L, Sugiura H, Huang X, Ali A, Kuro-o M, et al. Metabolic reprogramming during TGFβ1-induced epithelial-to-mesenchymal transition. Oncogene. 2015;34:3908–3916. doi: 10.1038/onc.2014.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tam WL, Weinberg RA. The epigenetics of epithelial-mesenchymal plasticity in cancer. Nature Medicine. 2013;19:1438–1449. doi: 10.1038/nm.3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rodríguez-García A, Samsó P, Fontova P, Simon-Molas H, Manzano A, Castaño E, et al. TGF-β1 targets Smad, p38 MAPK, and PI3K/Akt signaling pathways to induce PFKFB3 gene expression and glycolysis in glioblastoma cells. FEBS J. 2017;284:3437–3454. doi: 10.1111/febs.14201. [DOI] [PubMed] [Google Scholar]
- 25.Corbet C, Bastien E, Santiago de Jesus JP, Dierge E, Martherus R, Vander Linden C, et al. TGFβ2-induced formation of lipid droplets supports acidosis-driven EMT and the metastatic spreading of cancer cells. Nat. Commun. 2020;11:454. doi: 10.1038/s41467-019-14262-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yalcin A, Solakoglu TH, Ozcan SC, Guzel S, Peker S, Celikler S, et al. 6-phosphofructo-2-kinase/fructose 2,6-bisphosphatase-3 is required for transforming growth factor β1-enhanced invasion of Panc1 cells in vitro. Biochem. Biophys. Res. Commun. 2017;484:687–693. doi: 10.1016/j.bbrc.2017.01.178. [DOI] [PubMed] [Google Scholar]
- 27.Liu M, Quek L-E, Sultani G, Turner N. Epithelial-mesenchymal transition induction is associated with augmented glucose uptake and lactate production in pancreatic ductal adenocarcinoma. Cancer Metab. 2016;4:19. doi: 10.1186/s40170-016-0160-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mani SA, Guo W, Liao M-J, Eaton EN, Ayyanan A, Zhou AY, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jolly MK, Huang B, Lu M, Mani SA, Levine H, Ben-Jacob E, et al. Towards elucidating the connection between epithelial–mesenchymal transitions and stemness. J. R. Soc. Interface. 2014;11:20140962. doi: 10.1098/rsif.2014.0962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee S, Lee J-S, Seo J, Lee S-H, Kang JH, Song J, et al. Targeting mitochondrial oxidative phosphorylation abrogated irinotecan resistance in NSCLC. Sci. Rep. 2018;8:15707. doi: 10.1038/s41598-018-33667-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kang HM, Ahn SH, Choi P, Ko Y-A, Han SH, Chinga F, et al. Defective fatty acid oxidation in renal tubular epithelial cells has a key role in kidney fibrosis development. Nat. Med. 2015;21:37–46. doi: 10.1038/nm.3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dong C, Yuan T, Wu Y, Wang Y, Fan TWM, Miriyala S, et al. Loss of FBP1 by Snail-mediated repression provides metabolic advantages in basal-like breast cancer. Cancer Cell. 2013;23:316–331. doi: 10.1016/j.ccr.2013.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Røsland GV, Dyrstad SE, Tusubira D, Helwa R, Tan TZ, Lotsberg ML, et al. Epithelial to mesenchymal transition (EMT) is associated with attenuation of succinate dehydrogenase (SDH) in breast cancer through reduced expression of. Cancer Metab. 2019;7:6. doi: 10.1186/s40170-019-0197-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zancan, P., Sola-Penna, M. & Furtado, C. M. & Da Silva, D. Differential expression of phosphofructokinase-1 isoforms correlates with the glycolytic efficiency of breast cancer cells. Mol. Genet. Metab.100, 372–378 (2010).. [DOI] [PubMed]
- 35.Yang L, Hou Y, Yuan J, Tang S, Zhang H, Zhu Q, et al. Twist promotes reprogramming of glucose metabolism in breast cancer cells through PI3K/AKT and p53 signaling pathways. Oncotarget. 2015;6:25755–25769. doi: 10.18632/oncotarget.4697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krebs AM, Mitschke J, Lasierra Losada M, Schmalhofer O, Boerries M, Busch H, et al. The EMT-activator Zeb1 is a key factor for cell plasticity and promotes metastasis in pancreatic cancer. Nat. Cell Biol. 2017;19:518–529. doi: 10.1038/ncb3513. [DOI] [PubMed] [Google Scholar]
- 37.Jia D, Jolly MK, Tripathi SC, Den Hollander P, Huang B, Lu M, et al. Distinguishing mechanisms underlying EMT tristability. Cancer Converg. 2017;1:2. doi: 10.1186/s41236-017-0005-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Masin M, Vazquez J, Rossi S, Groeneveld S, Samson N, Schwalie PC, et al. GLUT3 is induced during epithelial-mesenchymal transition and promotes tumor cell proliferation in non-small cell lung cancer. Cancer Metab. 2014;2:11. doi: 10.1186/2049-3002-2-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim NH, Cha YH, Lee J, Lee S-H, Yang JH, Yun JS, et al. Snail reprograms glucose metabolism by repressing phosphofructokinase PFKP allowing cancer cell survival under metabolic stress. Nat. Commun. 2017;8:14374. doi: 10.1038/ncomms14374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Viswanathan VS, Ryan MJ, Dhruv HD, Gill S, Eichhoff OM, Seashore-Ludlow B, et al. Dependency of a therapy-resistant state of cancer cells on a lipid peroxidase pathway. Nature. 2017;547:453–457. doi: 10.1038/nature23007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mathow D, Chessa F, Rabionet M, Kaden S, Jennemann R, Sandhoff R, et al. Zeb1 affects epithelial cell adhesion by diverting glycosphingolipid metabolism. EMBO Rep. 2015;16:321–331. doi: 10.15252/embr.201439333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Siddiqui A, Vazakidou ME, Schwab A, Napoli F, Fernandez-Molina C, Rapa I, et al. Thymidylate synthase is functionally associated with ZEB1 and contributes to the epithelial-to-mesenchymal transition of cancer cells. J. Pathol. 2017;242:221–233. doi: 10.1002/path.4897. [DOI] [PubMed] [Google Scholar]
- 43.Lee J, You JH, Kim M-S, Roh J-L. Epigenetic reprogramming of epithelial-mesenchymal transition promotes ferroptosis of head and neck cancer. Redox Biol. 2020;37:101697. doi: 10.1016/j.redox.2020.101697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–688. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- 45.Battistelli C, Cicchini C, Santangelo L, Tramontano A, Grassi L, Gonzalez FJ, et al. The Snail repressor recruits EZH2 to specific genomic sites through the enrollment of the lncRNA HOTAIR in epithelial-to-mesenchymal transition. Oncogene. 2017;36:942–955. doi: 10.1038/onc.2016.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Korpal M, Lee ES, Hu G, Kang Y. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J. Biol. Chem. 2008;283:14910–14914. doi: 10.1074/jbc.C800074200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Y, Liu Z, Yao B, Dou C, Xu M, Xue Y, et al. Long non-coding RNA TUSC7 acts a molecular sponge for miR-10a and suppresses EMT in hepatocellular carcinoma. Tumor Biol. 2016;37:11429–11441. doi: 10.1007/s13277-016-4892-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kaller M, Liffers S-T, Oeljeklaus S, Kuhlmann K, Röh S, Hoffmann R, et al. Genome-wide characterization of miR-34a induced changes in protein and mRNA expression by a combined pulsed SILAC and microarray analysis. Mol. Cell. Proteomics. 2011;10:M111.010462. doi: 10.1074/mcp.M111.010462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yuan D, Zheng S, Wang L, Li J, Yang J, Wang B, et al. MiR-200c inhibits bladder cancer progression by targeting lactate dehydrogenase A. Oncotarget. 2017;8:67663–67669. doi: 10.18632/oncotarget.18801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gao C, Wei J, Tang T, Huang Z. Role of microRNA‑33a in malignant cells (Review) Oncol. Lett. 2020;20:2537–2556. doi: 10.3892/ol.2020.11835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dávalos A, Goedeke L, Smibert P, Ramírez CM, Warrier NP, Andreo U, et al. miR-33a/b contribute to the regulation of fatty acid metabolism and insulin signaling. Proc. Natl. Acad. Sci. USA. 2011;108:9232–9237. doi: 10.1073/pnas.1102281108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wei S, Fan Q, Yang L, Zhang X, Ma Y, Zong Z, et al. Promotion of glycolysis by HOTAIR through GLUT1 upregulation via mTOR signaling. Oncol. Rep. 2017;38:1902–1908. doi: 10.3892/or.2017.5840. [DOI] [PubMed] [Google Scholar]
- 53.Liu X, Liang Y, Song R, Yang G, Han J, Lan Y, et al. Long non-coding RNA NEAT1-modulated abnormal lipolysis via ATGL drives hepatocellular carcinoma proliferation. Mol. Cancer. 2018;17:90. doi: 10.1186/s12943-018-0838-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gaude E, Frezza C. Tissue-specific and convergent metabolic transformation of cancer correlates with metastatic potential and patient survival. Nat. Commun. 2016;7:13041. doi: 10.1038/ncomms13041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ito S, Fukusato T, Nemoto T, Sekihara H, Seyama Y, Kubota S. Coexpression of glucose transporter 1 and matrix metalloproteinase-2 in human cancers. J. Natl. Cancer Inst. 2002;94:1080–1091. doi: 10.1093/jnci/94.14.1080. [DOI] [PubMed] [Google Scholar]
- 56.Anderson M, Marayati R, Moffitt R, Yeh JJ. Hexokinase 2 promotes tumor growth and metastasis by regulating lactate production in pancreatic cancer. Oncotarget. 2017;8:56081–56094. doi: 10.18632/oncotarget.9760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen G, Zhang Y, Liang J, Li W, Zhu Y, Zhang M, et al. Deregulation of Hexokinase II Is Associated with Glycolysis, Autophagy, and the Epithelial-Mesenchymal Transition in Tongue Squamous Cell Carcinoma under Hypoxia. BioMed Res. Int. 2018;2018:1–15. doi: 10.1155/2018/8480762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ahmad A, Aboukameel A, Kong D, Wang Z, Sethi S, Chen W, et al. Phosphoglucose isomerase/autocrine motility factor mediates epithelial-mesenchymal transition regulated by miR-200 in breast cancer cells. Cancer Res. 2011;71:3400–3409. doi: 10.1158/0008-5472.CAN-10-0965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu K, Tang Z, Huang A, Chen P, Liu P, Yang J, et al. Glyceraldehyde-3-phosphate dehydrogenase promotes cancer growth and metastasis through upregulation of SNAIL expression. Int. J. Oncol. 2017;50:252–262. doi: 10.3892/ijo.2016.3774. [DOI] [PubMed] [Google Scholar]
- 60.Hamabe A, Konno M, Tanuma N, Shima H, Tsunekuni K, Kawamoto K, et al. Role of pyruvate kinase M2 in transcriptional regulation leading to epithelial-mesenchymal transition. Proc. Natl. Acad. Sci. USA. 2014;111:15526–15531. doi: 10.1073/pnas.1407717111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tanaka F, Yoshimoto S, Okamura K, Ikebe T, Hashimoto S. Nuclear PKM2 promotes the progression of oral squamous cell carcinoma by inducing EMT and post-translationally repressing TGIF2. Oncotarget. 2018;9:33745–33761. doi: 10.18632/oncotarget.25850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu N, He C, Zhu B, Jiang J, Chen Y, Ma T. 3-Phosphoinositide Dependent Protein Kinase-1 (PDK-1) Promotes Migration and Invasion in Gastric Cancer Cells Through Activating the NF-κB Pathway. Oncol. Res. 2017;25:1153–1159. doi: 10.3727/096504017X14845839228545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang M, Cong Q, Zhang X-Y, Zhang M-X, Lu Y-Y, Xu C-J. Pyruvate dehydrogenase kinase 1 contributes to cisplatin resistance of ovarian cancer through EGFR activation. J. Cell. Physiol. 2019;234:6361–6370. doi: 10.1002/jcp.27369. [DOI] [PubMed] [Google Scholar]
- 64.Sun Y, Daemen A, Hatzivassiliou G, Arnott D, Wilson C, Zhuang G, et al. Metabolic and transcriptional profiling reveals pyruvate dehydrogenase kinase 4 as a mediator of epithelial-mesenchymal transition and drug resistance in tumor cells. Cancer Metab. 2014;2:20. doi: 10.1186/2049-3002-2-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lin Q, He Y, Wang X, Zhang Y, Hu M, Guo W, et al. Targeting pyruvate carboxylase by a small molecule suppresses breast cancer progression. Adv. Sci. 2020;7:1903483. doi: 10.1002/advs.201903483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang Y, Lin S, Chen Y, Yang F, Liu S. LDH-A promotes epithelial–mesenchymal transition by upregulating ZEB2 in intestinal-type gastric cancer. Onco. Targets Ther. 2018;11:2363–2373. doi: 10.2147/OTT.S163570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li, X., Zhang, Z., Zhang, Y., Cao, Y., Wei, H. Wu, Z. Upregulation of lactate-inducible snail protein suppresses oncogene-mediated senescence through p16INK4a inactivation. J. Exp. Clin. Cancer Res.37, 39 (2018). [DOI] [PMC free article] [PubMed]
- 68.Oginuma M, Harima Y, Tarazona OA, Diaz-Cuadros M, Michaut A, Ishitani T, et al. Intracellular pH controls WNT downstream of glycolysis in amniote embryos. Nature. 2020;584:98–101. doi: 10.1038/s41586-020-2428-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nath A, Chan C. Genetic alterations in fatty acid transport and metabolism genes are associated with metastatic progression and poor prognosis of human cancers. Sci. Rep. 2016;6:18669. doi: 10.1038/srep18669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nath A, Li I, Roberts LR, Chan C. Elevated free fatty acid uptake via CD36 promotes epithelial-mesenchymal transition in hepatocellular carcinoma. Sci. Rep. 2015;5:14752. doi: 10.1038/srep14752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Patel A, Sabbineni H, Clarke A, Somanath PR. Novel roles of Src in cancer cell epithelial-to-mesenchymal transition, vascular permeability, microinvasion and metastasis. Life Sci. 2016;157:52–61. doi: 10.1016/j.lfs.2016.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Altman BJ, Stine ZE, Dang CV. From Krebs to clinic: glutamine metabolism to cancer therapy. Nat. Rev. Cancer. 2016;16:749. doi: 10.1038/nrc.2016.114. [DOI] [PubMed] [Google Scholar]
- 73.Lee SY, Jeon HM, Ju MK, Jeong EK, Kim CH, Park HG, et al. Dlx-2 and glutaminase upregulate epithelial-mesenchymal transition and glycolytic switch. Oncotarget. 2016;7:7925–7939. doi: 10.18632/oncotarget.6879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kuo T-C, Chen C-K, Hua K-T, Yu P, Lee W-J, Chen M-W, et al. Glutaminase 2 stabilizes Dicer to repress Snail and metastasis in hepatocellular carcinoma cells. Cancer Letters. 2016;383:282–294. doi: 10.1016/j.canlet.2016.10.012. [DOI] [PubMed] [Google Scholar]
- 75.Ramirez-Peña E, Arnold J, Shivakumar V, Joseph R, Vijay GV, den Hollander P, et al. The epithelial to mesenchymal transition promotes glutamine independence by suppressing GLS2 expression. Cancers. 2019;11:1610. doi: 10.3390/cancers11101610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liu, G., Zhu, J., Yu, M., Cai, C., Zhou, Y., Yu, M. et al. Glutamate dehydrogenase is a novel prognostic marker and predicts metastases in colorectal cancer patients. J. Transl. Med.13, 144 (2015). [DOI] [PMC free article] [PubMed]
- 77.Wang Y, Fu L, Cui M, Wang Y, Xu Y, Li M, et al. Amino acid transporter SLC38A3 promotes metastasis of non-small cell lung cancer cells by activating PDK1. Cancer Lett. 2017;393:8–15. doi: 10.1016/j.canlet.2017.01.036. [DOI] [PubMed] [Google Scholar]
- 78.Tian Q, Yuan P, Quan C, Li M, Xiao J, Zhang L, et al. Phosphorylation of BCKDK of BCAA catabolism at Y246 by Src promotes metastasis of colorectal cancer. Oncogene. 2020;39:3980–3996. doi: 10.1038/s41388-020-1262-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bates SE. Epigenetic therapies for cancer. N. Engl. J. Med. 2020;383:650–663. doi: 10.1056/NEJMra1805035. [DOI] [PubMed] [Google Scholar]
- 80.Kinnaird A, Zhao S, Wellen KE, Michelakis ED. Metabolic control of epigenetics in cancer. Nat. Rev. Cancer. 2016;16:694–707. doi: 10.1038/nrc.2016.82. [DOI] [PubMed] [Google Scholar]
- 81.Guerra F, Guaragnella N, Arbini AA, Bucci C, Giannattasio S, Moro L. Mitochondrial dysfunction: a novel potential driver of epithelial-to-mesenchymal transition in cancer. Front. Oncol. 2017;7:295. doi: 10.3389/fonc.2017.00295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Guha M, Srinivasan S, Ruthel G, Kashina AK, Carstens RP, Mendoza A, et al. Mitochondrial retrograde signaling induces epithelial-mesenchymal transition and generates breast cancer stem cells. Oncogene. 2014;33:5238–5250. doi: 10.1038/onc.2013.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wu W-S. The signaling mechanism of ROS in tumor progression. Cancer Metastasis Rev. 2006;25:695–705. doi: 10.1007/s10555-006-9037-8. [DOI] [PubMed] [Google Scholar]
- 84.Lu M, Zhu W-W, Wang X, Tang J-J, Zhang K-L, Yu G-Y, et al. ACOT12-Dependent Alteration of Acetyl-CoA Drives Hepatocellular Carcinoma Metastasis by Epigenetic Induction of Epithelial-Mesenchymal Transition. Cell Metabolism. 2019;29:886–900.e5. doi: 10.1016/j.cmet.2018.12.019. [DOI] [PubMed] [Google Scholar]
- 85.Qin Z-Y, Wang T, Su S, Shen L-T, Zhu G-X, Liu Q. BRD4 promotes gastric cancer progression and metastasis through acetylation-dependent stabilization of Snail. Cancer Res. 2019;79:4869–4881. doi: 10.1158/0008-5472.CAN-19-0442. [DOI] [PubMed] [Google Scholar]
- 86.Islam MS, Leissing TM, Chowdhury R, Hopkinson RJ, Schofield CJ. 2-Oxoglutarate-dependent oxygenases. Annu. Rev. Biochem. 2018;87:585–620. doi: 10.1146/annurev-biochem-061516-044724. [DOI] [PubMed] [Google Scholar]
- 87.Atlante S, Visintin A, Marini E, Savoia M, Dianzani C, Giorgis M, et al. α-ketoglutarate dehydrogenase inhibition counteracts breast cancer-associated lung metastasis. Cell Death Dis. 2018;9:756. doi: 10.1038/s41419-018-0802-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim S-H, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of α-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011;19:17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yang M, Pollard PJ. Succinate: a new epigenetic hacker. Cancer cell. 2013;23:709–711. doi: 10.1016/j.ccr.2013.05.015. [DOI] [PubMed] [Google Scholar]
- 90.Fuhler GM, Eppinga H, Peppelenbosch MP. Fumarates and cancer. Trends Mol. Med. 2017;23:3–5. doi: 10.1016/j.molmed.2016.12.001. [DOI] [PubMed] [Google Scholar]
- 91.Colvin, H., Nishida, N., Konno, M., Haraguchi, N., Takahashi, H., Nishimura, J. et al. Oncometabolite D-2-hydroxyglurate directly induces epithelial-mesenchymal transition and is associated with distant metastasis in colorectal cancer. Sci. Rep. 6, 36289 (2016). [DOI] [PMC free article] [PubMed]
- 92.Letouzé E, Martinelli C, Loriot C, Burnichon N, Abermil N, Ottolenghi C, et al. SDH mutations establish a hypermethylator phenotype in paraganglioma. Cancer Cell. 2013;23:739–752. doi: 10.1016/j.ccr.2013.04.018. [DOI] [PubMed] [Google Scholar]
- 93.Sciacovelli M, Gonçalves E, Johnson TI, Zecchini VR, da Costa ASH, Gaude E, et al. Fumarate is an epigenetic modifier that elicits epithelial-to-mesenchymal transition. Nature. 2016;537:544–547. doi: 10.1038/nature19353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Shaul YD, Freinkman E, Comb WC, Cantor JR, Tam WL, Thiru P, et al. Dihydropyrimidine accumulation is required for the epithelial-mesenchymal transition. Cell. 2014;158:1094–1109. doi: 10.1016/j.cell.2014.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Schwab A, Siddiqui A, Vazakidou ME, Napoli F, Böttcher M, Menchicchi B, et al. Polyol pathway links glucose metabolism to the aggressiveness of cancer cells. Cancer Res. 2018;78:1604–1618. doi: 10.1158/0008-5472.CAN-17-2834. [DOI] [PubMed] [Google Scholar]
- 96.Ramesh V, Brabletz T, Ceppi P, Targeting EMT. Cancer with repurposed metabolic inhibitors. Trends Cancer Res. 2020;6:942–950. doi: 10.1016/j.trecan.2020.06.005. [DOI] [PubMed] [Google Scholar]
- 97.Bocci F, Tripathi SC, Vilchez Mercedes SA, George JT, Casabar JP, Wong PK, et al. NRF2 activates a partial epithelial-mesenchymal transition and is maximally present in a hybrid epithelial/mesenchymal phenotype. Integr. Biol. 2019;11:251–263. doi: 10.1093/intbio/zyz021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Colacino JA, Azizi E, Brooks MD, Harouaka R, Fouladdel S, McDermott SP, et al. Heterogeneity of human breast stem and progenitor cells as revealed by transcriptional profiling. Stem Cell Reports. 2018;10:1596–1609. doi: 10.1016/j.stemcr.2018.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Luo M, Shang L, Brooks MD, Jiagge E, Zhu Y, Buschhaus JM, et al. Targeting breast cancer stem cell state equilibrium through modulation of redox signaling. Cell Metab. 2018;28:69–86.e6. doi: 10.1016/j.cmet.2018.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Medina MÁ. Mathematical modeling of cancer metabolism. Crit. Rev. Oncol. Hematol. 2018;124:37–40. doi: 10.1016/j.critrevonc.2018.02.004. [DOI] [PubMed] [Google Scholar]
- 101.Orth JD, Thiele I, Palsson BØ. What is flux balance analysis? Nat. Biotechnol. 2010;28:245–248. doi: 10.1038/nbt.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Jia D, Paudel BB, Hayford CE, Hardeman KN, Levine H, Onuchic JN, et al. Drug-tolerant idling melanoma cells exhibit theory-predicted metabolic low-low phenotype. Front. Oncol. 2020;10:1426. doi: 10.3389/fonc.2020.01426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kang X, Wang J, Li C. Exposing the underlying relationship of cancer metastasis to metabolism and epithelial-mesenchymal transitions. iScience. 2019;21:754–772. doi: 10.1016/j.isci.2019.10.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yu L, Lu M, Jia D, Ma J, Ben-Jacob E, Levine H, et al. Modeling the genetic regulation of cancer metabolism: interplay between glycolysis and oxidative phosphorylation. Cancer Res. 2017;77:1564–1574. doi: 10.1158/0008-5472.CAN-16-2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.