Abstract
Background
Chemoresistance is one of the major factors for treatment failure in OSCC. Identifying key resistance triggering molecules will be useful strategy for developing novel treatment methods.
Methods
To identify the causative factors of chemoresistance, we performed RNA sequencing and global proteomic profiling of human OSCC lines presenting with sensitive, early and late cisplatin-resistance patterns.
Results
From the common set of dysregulated genes from both the analysis, RRBP1 was identified to be upregulated in both early and late cisplatin-resistant cells with respect to the sensitive counterpart. Analysis of OSCC patient sample indicates that RRBP1 expression is upregulated in chemotherapy-non-responder tumours as compared to chemotherapy-responder tumours. Genetic (knockout) or pharmacological (Radezolid, represses expression of RRBP1) inhibition of RRBP1 restores cisplatin-mediated cell death in chemo-resistant OSCC. Mechanistically, RRBP1 regulates Yes-associated protein1 (YAP1), a key protein in the Hippo pathway to induce chemoresistance. The PDC xenograft data suggests that knockout of RRBP1 induces cisplatin-mediated cell death and facilitates a significant reduction of tumour burden.
Conclusion
Overall, our data suggests that (I) RRBP1 is a major driver of cisplatin-resistance in OSCC, (II) RRBP1 regulates YAP1 expression to mediate cisplatin-resistance, (III) Radezolid represses RRBP1 expression and (IV) targeting RRBP1 reverses cisplatin-induced chemoresistance in advanced OSCC.
Subject terms: Oral cancer, Drug regulation
Background
HNSCC is the sixth most common cancer globally and most prevalent in developing countries. OSCC is an aggressive form of HNSCC and is the most common cancer among Indian males.1 The traditional treatment modalities for advanced OSCC comprises of surgical removal of primary tumours followed by concomitant adjuvant chemoradiotherapy.2 In addition, neoadjuvant chemotherapy is frequently recommended for surgically unresectable OSCC tumours. One of the emerging therapeutic regimens is RADPLAT, where intra-arterial delivery of cisplatin is done followed by systemic neutralisation by sodium thiosulphate with concomitant radiotherapy.3 Despite having these modalities, the 5 years survival rate of advance tongue OSCC is ~50%, indicating a possible resistance to currently available therapeutics. Chemoresistance is one of the important factors for treatment failure in OSCC.4 Cisplatin alone or in combination with 5- fluorouracil and taxane/docetaxel (TPF) are generally used as chemotherapy regimen for OSCC.5 But due to chemoresistance development, patients experience relapse, which leads to continued tumour growth and metastasis. The chemo-resistant properties could be attributed to enhanced cancer stem cell population, decreased drug accumulation, reduced drug-target interaction, reduced apoptotic response and enhanced autophagic activities.6 These hallmarks present the endpoint events in cancer cell with acquired chemoresistance. Till date, the causative factors responsible for acquiring chemoresistance in cells are yet to be explored.
In the present study, we aimed to identify the causative factors, dysregulated molecules, responsible for cisplatin-resistance development in OSCC by employing global quantitative proteomics and transcriptome tools. RRBP1 was found to be one of the important proteins, identified from the common list of dysregulated molecules from proteomics and RNA-seq analyses. RRBP1 plays critical role in translation, transportation and release of secretory proteins, anchors to the rough endoplasmic reticulum (ER) and present in both cytoplasm and nucleus.7,8 RRBP1 plays an important role in augmenting collagen synthesis and its secretion. Knocking down RRBP1 in human fibroblast resulted in a significant reduction of secreted collagen.9
In this study, we observed that knocking out of RRBP1 protein in cisplatin-resistant cells restored drug effect. The patient-derived cell (PDC) xenograft experiment suggests that knocking out of RRBP1 induces cisplatin-mediated cell death and facilitate significant reduction of tumour burden. Mechanistically, RRBP1 regulates hippo signalling and activates YAP1 target genes, which principally confers chemoresistance in OSCC. An oxazolidinone group of antibiotics, Radezolid, was found to reduce the expression of the RRBP1 and reverses drug resistance in OSCC chemo-resistant cell lines. This preclinical study highlights the importance of RRBP1 as a putative cancer marker explaining cisplatin-resistant development in OSCC cells.
Methods
OSCC patient sample
Human Ethics Committee (HEC) of the Institute of Life Sciences approved all patient-related studies, and written informed consent was obtained from all patients participated in this study. Subject details of recruited patients with complete treatment modalities are presented in Supplementary table 1 and 2. All the samples collected are locoregionally advanced OSCC. Neoadjuvant chemotherapy has been prescribed before surgery and/or radiotherapy. The three-drug combination (TPF) showed highest response (TAX 324). However, some cases had poor ECOG (Eastern Cooperative Oncology Group) performance status, two drugs (TP) were prescribed instead of TPF. After chemotherapy (CT) the response is evaluated as per RECIST (Response evaluation criteria in solid tumours) criteria by clinical and radiological evaluation. If there is no evidence of malignancy, then it is diagnosed as Complete Response (CR). With a decrease of target lesions ≥30% of the sum of the longest diameter, then subjects were grouped as Partial Response (PR). Subjects without sign of CR or PR were grouped as stable disease but with a ≥20% of the sum of the longest diameter were grouped as PD (Progressive Disease). Patients of CR and PR groups responded to the CT and categorised as Responders and rest of the patients with stable disease or PD with almost no response to CT were categorised as Non-Responders. Primary patient-derived cells (PDC) were isolated from harvested tissues of patients not responding to treatment and cultured.10
Cell culture
The human tongue OSCC cell lines (H357, SCC-9 and SCC-4) were obtained from Sigma–Aldrich, sourced from European collection of authenticated cell culture. PCR fingerprinting to establish the cell line authentication were done by the provider. All OSCC cell lines were cultured and maintained as described earlier.11
Generation of early and late cisplatin-resistance cell lines
To establish a chemo-resistant cell model system, OSCC cell lines (H357, SCC-9 and SCC-4) were initially treated with low dose of cisplatin (1 μM) for a week and gradually increased 15 μM (IC50 value) by 3 months and subsequently grown at IC50 until 8 months. Here, drugs efficiently eliminated the rapidly dividing cancer cells by inducing cell death, but poorly targeted the slowly dividing cells. Gradually, the poorly sensitive cells regained the normal growth cycle. Cells at the starting time were grouped as sensitive (CisS) and at 4 and 8 months of treatment were termed as early (Cis EarlyR) and late resistant (CisR Late R) cells, respectively.
iTRAQ based proteomics analysis
Harvested cells (5 × 106), from three time points (0 M: H357CisS, 4 M: H357CisR4M and 8 M: H357 CisR8M) were treated with RIPA buffers (Thermo Scientific, Cat #88665) and inhibitors (Sigma, Cat # P0044,). Extracted proteins (100 μg) from study samples were trypsinised and isobaric tags were added following manufacturer’s instructions (AB Sciex, USA). Tagged tryptic peptides (~250 μg) were subjected to fractionation using a strong cation exchange chromatography before mass spectrometry data acquisition. Earlier published methods without any modifications were used for mass spectrometry data acquisition and for protein identification and quantification.12 The false discovery rate (FDR) at both protein and peptide level was calculated at 5%. Proteins and fold change values were selected for principal component analysis (PCA) using MetaboAnalyst 3.0. Ingenuity Pathway Analysis (IPA) was used for functional analysis with the identified protein sets. All the mass spectrometry data files are available in the ProteomeXchange Consortium (PXD0016977).
Library preparation and RNA sequencing
The RNA-seq library was prepared using H357CisS, H357CisR4M (early resistance), H357 CisR8M (Late resistance), H357CisR NtSh and H357CisR RRBP1sh groups. Two independent biological replicates from each study groups were included. For RNA-seq library preparation 2 μg of total RNA was used to isolate mRNA through magnetic beads using mRNA isolation kit (PolyA mRNA isolation Module, NEB) followed by RNA-seq library preparation using mRNA library preparation kit (NEB) strictly following the vendor recommended protocol. After library preparation, sample concentrations were estimated using Qubit 2.0 (Invitrogen) and the expected fragmentation sizes were confirmed by Bio-analyzer (Agilent, USA).
Transcriptome analysis
For RNA-seq sequencing, the library was prepared following the manufacturer’s recommendation of NEBNext RNA-kit (Cat No. E7335L). The paired-end sequencing was performed using Illumina Next550 sequencer. The BCL files were obtained from the sequencer and demultiplexed to generate fastq files through the bcl2fastq tool using 5’ index sequences. Adaptor sequences were removed during the demultiplexing. The read quality of each sample was assessed through fastqc. Next, the alignment was performed by HISAT2 using the GRch38 human reference genome. Aligned reads were processed in SAMtools to make the index. Both paired-end aligned sequences were mapped with human GENCODEv35 (GTF) to quantify the read count abundance through featureCounts2.0.1. The biological replicates were used for PCA analysis. Additionally, the raw read count was normalised to read per million (RPM) for quantitative representation. Differential Gene Expression (DGEs) was performed using DESeq2 in R 3.0.1/BioConductor. Genes were considered differentially expressed if |log2FC > 1 and p-value < 0.05 | . The heatmap for each individual study was plotted by complexheatmap library.
Pathway analysis
The Ingenuity Pathway Analysis (IPA) was used to check enriched biological pathways among the differentially expressed proteins or genes. The significant pathways were selected based on the p-value < 0.05.
Lentivirus production and generation of stable Cas9 overexpressing chemo-resistant lines
For Cas9 lentivirus generation, lentiCas9-Blast (Addgene, Cat#52962) was transfected along with its packaging psPAX2 and envelop pMD2G in HEK293T cells as lentiviral particles were generated following protocol described in Shriwas et al.13 Chemo-resistant cells were infected with lento Cas9 particles and treated with blastidicines hydrochloride (5 µg/ml, MP biomedical, Cat# 2150477). Single clones were selected, and Cas9 overexpression was confirmed by western blot using anti-Cas9 antibody (CST Cat #14697). The lentiCas9-Blast vector was kindly deposited to Addgene by Feng Zhang lab.14
Generation of CRISPR based RRBP1 KO cell line
For generation of RRBP1 knockout cells, lentiviral vector expressing RRBP1 sgRNA (GGCGTTTCAGAATCGCCACA) was procured from Addgene (Cat# 92157). This lentiGuide-RRBP1-2 vector was kindly deposited by Alice Ting Lab.15 Lentiviral particles were generated as described above using HEK293T cells. Stable clones of Cas9 overexpressing chemo-resistant cells (H357CisR, SCC-9 CisR and patient-derived cells PDC-1) were infected with lentiGuide-RRBP1-2 for 48 h in presence of polybrene (8 µg/ml), after which cells were treated with puromycin (2 μg/ml, Invitrogen, USA, Cat #A11138-03) for 7 days. Single clones were selected, and RRBP1 knockout was confirmed by western blot using anti-RRBP1 antibody (Abcam, USA, Cat # ab95983). The RRBP1 KO clones were confirmed by cleavage detection assay. The genomic cleavage efficiency was measured by the GeneArt® Genomic Cleavage Detection kit (Thermo Fisher Scientific, Cat # A24372) according to manufacturer’s protocol. Details of oligos used in this study are presented in supplementary table 3B.
Generation of lentivirus-based RRBP1 shRNA and overexpression of YAP1
ShRNAs targeting RRBP1 were cloned into pLKO.1 vector following the protocol shared by the manufacturer (addgene). Cloning of shRNAs were confirmed by sequencing. Lentivirus was produced by transfection of pLKO.1-shRNA plasmid along with packaging plasmid psPAX2 and envelop plasmid pMD2G into HEK293T cells. Lentivirus infected cells were incubated with puromycin up to 5 µg/ml for two weeks, stable clones were picked and confirmed by immunoblotting. All shRNA sequences used in this study are presented in Supplementary table 3C. For overexpression of YAP1, similar lentivirus-based protocol was followed using pQCXIH-Flag-YAP-S127A plasmid along with packaging and envelop. pQCXIH-Flag-YAP-S127A (Addgene #33092) deposited by Dr. Guan lab.16
Transient transfection with plasmids
For transient expression, H357 and SCC-9 cells were transfected with RRBP1 pcDNA4 HisMax-V5-GFP-RRBP1(Addgene: Cat#92150) and YAP1, pQCXIH-Flag-YAP-S127A (Addgene Cat #33092) overexpression plasmids using the ViaFect transfection reagent (Promega Cat# E4982). The transfection efficiency was determined by immunoblotting with Anti GFP (Abcam, USA, Cat # ab6556). pcDNA4 HisMax-V5-GFP-RRBP1 was kindly deposited by Alice Ting Lab.15
Organoid culture
Following earlier published methods with minor modification, chemo-resistant lines (H357CisR, SCC-9 CisR) and PDC-1 were used for developing 3D organoid.17 Organoid formation rate was defined as the average number of 50-mm spherical structures at day 14 that was divided by the total number of seeded cells in each well at day 0. During this experiment, at day 8 from establishing organoid culture, cisplatin (10 μM) or DMSO (vehicle control) was used for treatment.
Immunoblotting of targeted proteins
Cell lysates were used for immunoblotting experiments as described earlier.18 For this study, we used antibody against anti β-actin (Abcam, USA,Cat#A2066), Anti-RRBP1 (Sigma, Cat# HPA011924, Abcam USA, Cat# ab95983), Anti YAP1 (CST, Cat # 14074), Anti-YAP1s-397 (CST, Cat # 13619 S), Anti-YAP1s-127 (CST, Cat # 13008), Anti PARP (CST, Cat #9542 L), Anti ps-139-H2AX (CST, Cat # 9718 S), Anti GFP (Abcam, USA, Cat # ab6556) and YAP/TAZ Transcriptional Targets Antibody Sampler Kit (CST, Cat#56674), anti-Cas9 (CST, Cat # 14697). Nuclear and cytoplasmic fractions were separated using the NE-PER® nuclear and cytoplasmic extraction kit (Thermo Scientific Massachusetts, USA-78833), following the manufacturer’s instructions.
Patient-derived xenograft
BALB/C-nude mice (6–8 weeks, male, NCr-Foxn1nu athymic) were procured from Vivo biotech Ltd (Secunderabad, India) and maintained under pathogen-free conditions in the animal house of Institute of Life Sciences Bhubaneswar. Xenograft experiment were carried out using PDC-1 of chemo non-responders.10 The patient (PDC#1) was treated with TP without any response. Cells (1 × 106) were suspended in phosphate-buffered solution-Matrigel (1:1, 100 μl) and transplanted into upper flank of mice. The PDC1RRBP1KO cells were injected in the left upper flank and PDC-1 WT cells were injected in right upper flank of same mice. After the tumour volume reached to 50 mm,3 these mice were randomly divided into two groups for treatment with vehicle and cisplatin (3 mg/Kg) intraperitonially twice a week. At the end of experiment, mice were killed by CO2 asphyxiation and tumour were isolated.
RNA isolation and quantitative real‐time PCR
Total RNA of experimental samples were isolated using RNA mini kit (Himedia, Cat# MB602) following manufacturer’s instruction and yield was quantified by Nanodrop. RNA (300 ng) was used for reverse transcription by using first cDNA synthesis (VERSO CDNA KIT Thermo Fisher Scientific, Cat # AB1453 A) and qRT-PCR was carried out using SYBR Green master mix (Thermo Fisher scientific Cat # 4367659). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as a loading control and complete primer details used in this article are presented in supplementary table 3A.
Immunohistochemistry
Tumour from OSCC patients and mice models were isolated for paraffin embedding for immunohistochemistry analysis following earlier published method.13 Antibodies for RRBP1 (Sigma, cat# HPA011924), Ki67 (Vector, Cat# VP-RM04), cleaved caspase (CST, cat # 9661 S), CTGF (Santa Cruz Cat# SC 25440), Survivin (CST Cat# 2808) were used for immunohistochemistry. Images were captured using the Leica DM500 microscope. Q-score was determined by multiplication of the percentage of positive cells (P) and intensity (I). ‘P’ was determined by the presence of positive cells in a slide. ‘I’ was determined by the level of staining in the slide i.e. negative (value = 0), weak (value = 1), intermediate (value = 2) and strong (value = 3) respectively. Detailed Q-score protocol used in this study was followed from Maji et al.10
Annexin V PE/7-ADD assay
Apoptosis and cell death of experimental samples were measured using Annexin V Apoptosis Detection Kit PE (eBioscience™, USA, Cat # 88-8102-74).10
Immunofluorescence
Cells (103) were seeded on coverslip and allowed to adhere to the surface. The adhered cells were fixed in formaldehyde (4%) for 15 min and permeabilise with permeabilisation buffer (eBioscience™, USA, Cat # 00-8333-56) followed by blocking with bovine serum albumin (BSA,3 %) for 1 h at room temperature. Then cells were incubated with primary antibody (RRBP1: Abcam, USA, Cat # ab95983 and YAP: CST cat # 14074) overnight at 4 °C, washed thrice with PBST followed by 1 h incubation with Alexa fluor conjugated secondary antibody (Thermo Fisher Scientific Cat # 11008, 21244) then again washed thrice with PBST and coverslips were mounted with DAPI (Slow Fade ® GOLD Antifade, Thermo Fisher Scientific, Cat # S36938). Images were captured using a confocal microscopy (LEICA TCS-SP5).
Clonogenic assay
For Clonogenic assay, cisplatin-resistance cell lines (H357, SCC-9 and SCC-4) were seeded in six well plate and treated with DMSO, Cisplatin (cis-diammineplatinum (II) dichloride, Sigma, Cat# 479306), Radezolid (MedChemExpress USA, Cat# HY-14800) or in combination then allowed for 10 days to grow. The colonies were stained with 0.5% crystal violet and counted by ImageJ software.
Validation of relative expression levels of YAP1 target genes with RRBP1
The relative expression level of YAP1 target genes with RRBP1 in HNSCC patient tumours were validated using the online analysis software GEPIA platform (http://gepia.cancer-pku.cn/detail.php?gene=RRBP1).19
Insilco molecular docking
Structure of Radezolid was obtained from DrugBank (https://www.drugbank.ca/). The ligand was prepared by LigPrep module in Schrodinger molecular modelling software. Since crystal structure of the target protein RRBP1 in Protein Databank (PDB) is not available, a homology model of RRBP1 was built using Modller9.22 software. The protein template for homology modelling was selected using DELTA-BLAST. Two templates (PDB ID 6FSA, and 5TBY) were selected and multiple sequence alignment with RRBP1 sequence was done for the modelling. Ten models were built using Modeller9.22 and based on MolPDF score the best model was chosen. The selected model was imported in the Maestro module of Schrodinger software for molecular docking. The hydrogens were added, bond orders and ionisation states were assigned, and charges were calculated for the atoms of the RRBP1 protein. The active site of RRBP1 protein was identified by SiteMap and a cavity with 772.7 A3 was selected for docking. The docking study was done using the Glide module with extra precision (XP) mode.
Statistical analysis
All data points are presented as mean and standard deviation and Graph Pad Prism 5.0 was used for calculation. The statistical significance was calculated by one/two-way variance (one/two-way ANOVA) and considered significance at P ≤ 0.05.
Results
Establishment of chemoresistance OSCC cells
To identify the key resistance triggering molecules, cisplatin-resistant cells were established by prolonged treatment of cisplatin to OSCC cell lines as described in the method section. Monitoring cisplatin-induced cell death in three stages (CisS, Cis EarlyR and Cis LateR) of H357, SCC-9 and SCC-4 cells by flow cytometry assay showed Cis LateR achieved complete acquired resistance and Cis EarlyR achieved partial resistance (Supplementary Fig. 1A, B). In addition to this, our flow cytometry data suggest that CD44 expression is higher in early and late resistant cells as compared to sensitive counterpart indicating enhanced population of cancer stem like cells (CSCs) (Supplementary Fig. 1C).
RRBP1 expression is elevated in chemo-resistant OSCC
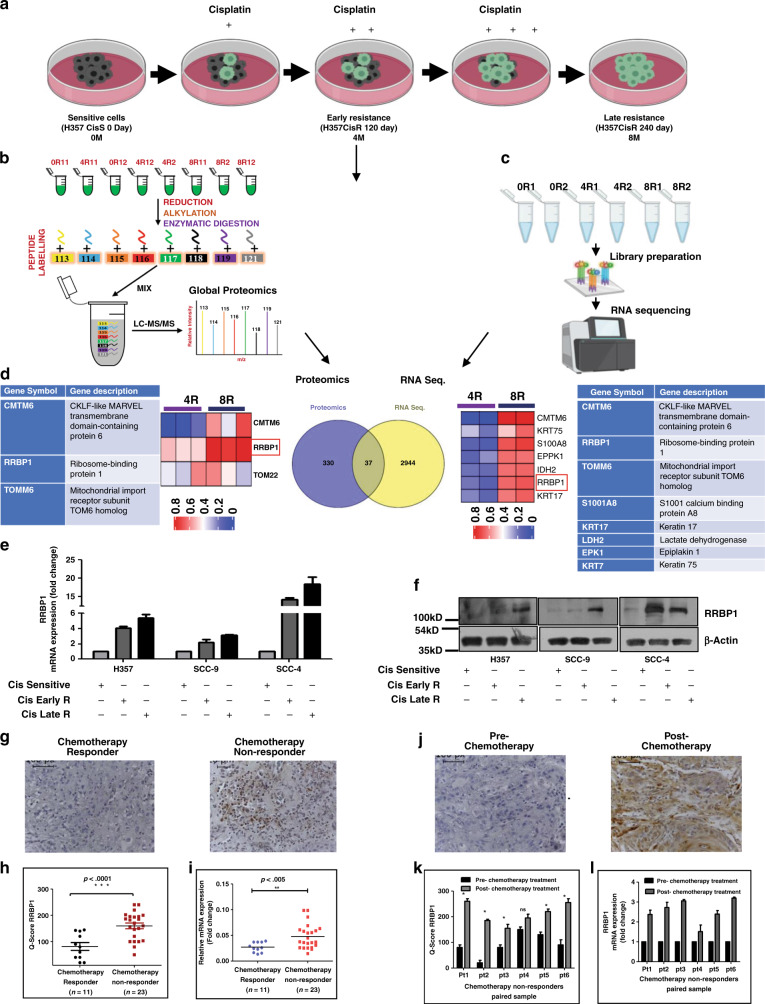
To identify the causative factors responsible for acquired cisplatin resistance in OSCC, an unbiased global proteomic profiling as well as RNA sequencing of H357CisS, H357CisR4M and H357 CisR8M cells were performed (Fig. 1a–c). A set of 37 genes were found to be commonly deregulated in both proteomics (367 proteins) and RNA-Seq (2981 genes) analyses (Fig. 1d and Supplementary Table 4). PCA analysis of all the identified proteins grouped these groups into distinctly different clusters (Supplementary Fig. 2A, B). Out of these 37 dysregulated genes, a novel and less studied oncogenic protein i.e. RRBP1 was selected for further validation. RRBP1 expression was elevated during the development of cisplatin resistance (Supplementary Fig. 2C, D). The fragmentation spectra of ALNQATSQVES peptide used for RRBP1 protein identification and quantification was manually checked and confirmed (Supplementary Fig. 2E). Based on these observations, we focused on RRBP1 to elucidate its potential role in acquired cisplatin resistance. RRBP1 expression was monitored in H357, SCC-9 and SCC-4 OSCC cell lines showing different acquired chemo-resistant patterns. Expression of RRBP1 at protein and mRNA levels were found to be upregulated in CisR4M and CisR8M cells with respect to CisS (cisplatin sensitive) counterparts (Fig. 1e, f). A similar profile of RRBP1 was also observed in tumour sections isolated from chemotherapy responder and patients not responding to neoadjuvant chemotherapy (TPF) (Fig. 1g–i). Similarly, RRBP1 expression was also monitored in chemotherapy naive and post-chemotherapy-non-responder paired tumour samples of OSCC patients, and a higher RRBP1 expression was observed in tumour samples post-chemotherapy (Fig. 1j–l). Here onwards, we indicate CisR8M cells as CisR for convenience.
Fig. 1. Global proteomics and transcriptomics data revealed RRBP1 is upregulated in OSCC chemo-resistant cells.
a Schematic representation of sensitive, early and late cisplatin-resistant OSCC line for global and proteome and RNA-seq profiling. The establishment of sensitive, early and late resistant cells is described in the materials and method section. b The lysates were isolated from parental sensitive (H3457CisS), early (H357Cis Early R) and late (H357Cis Late R) cisplatin-resistant cells and subjected to global proteomic profiling. The schematic diagram depicts the iTRAQ labelling strategy for proteomic analysis. 0R11 and 0R12 are technical replicates of H357CisS group, 4R11: 4R12 are technical and 4R2 is biological replicates of H357Cis earlyR group, 8R11: 8R12 are technical and 8R2 is biological replicates of H357Cis lateR group (0- not treated cisplatin, 4-Cisplatin treated 4 month, 8-Cispaltin treated 8 month). c Total RNA was isolated from (H3457CisS), early (H357Cis Early R) and late (H357Cis Late R) cisplatin-resistant cells Schematic representation of workflow for RNA Sequencing. 0R1 and 0R2 are technical replicates of H357CisS group, 4R1: 4R2 are technical replicates of H357Cis Early R group, 8R1: 8R2 are technical replicates of H357Cis LateR group. d Overlapping of differentially expressed genes from proteomics and RNA-seq dataset, left panel: The heatmap and table represents the deregulated genes from proteomic analysis (from early to late resistance normalised with sensitive). Right panel: heatmap and table represents the deregulated genes from RNA-seq analysis (from early to late resistance normalised with sensitive). e Relative mRNA (fold change) expression of RRBP1 was analysed by qRT-PCR in indicated acquired chemo-resistant OSCC cells as compared to the sensitive counterpart (mean ± SEM, n = 2). GAPDH was used as an internal control. f Cell lysates from indicated resistant and sensitive OSCC cells were isolated and subjected to immunoblotting against RRBP1 and β-actin antibodies. g Protein expression of RRBP1 was analysed by immunohistochemistry (IHC) in chemotherapy-responder and chemotherapy-non-responder OSCC tumours. h Representative IHC Scoring for RRBP1 from panel G (Q Score = Staining Intensity × % of Staining), (Median, n = 11 for chemotherapy-responder and n = 23 for chemotherapy-non-responder) *P < 0.05. i Relative mRNA expression of RRBP1 was analysed by qRT-PCR in different chemotherapy-non-responder OSCC tumours as compared to chemotherapy-responder tumours (Median, n = 11 for chemotherapy-responder and n = 23 for chemotherapy-non-responder). *P < 0.05. j Protein expression of RRBP1 was analysed by immunohistochemistry (IHC) in pre- and post-TPF treated paired tumour samples for chemotherapy-non-responder patients. k IHC Scoring for RRBP1 from panel J (Q Score = Staining Intensity × % of Staining), n = 6. l Relative mRNA expression of RRBP1 was analysed by qRT-PCR in pre- and post-TPF treated paired tumour samples for chemotherapy-non-responder patients (n = 6). Pt represents each patient.
RRBP1 dependency in chemo-resistant cell lines and Cancer Associated Fibroblast (CAF)
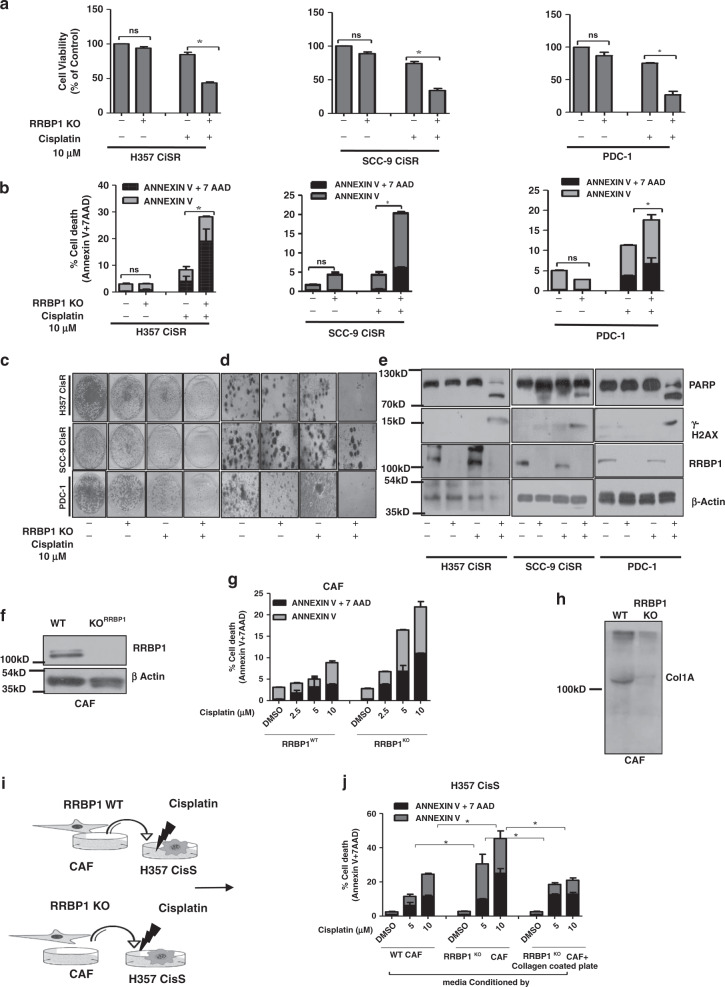
Further, we generated RRBP1 knockout clones in cell line (H357CisR and SCC-9 CisR) and in patient-derived tumour primary cells not responding to TP, which was confirmed by cleavage detection assay (Supplementary Fig. 3A–D). Cell viability and cell death assay data suggest that knockout of RRBP1 significantly sensitised chemo-resistant lines to cisplatin (Fig. 2a, b). The anchorage-dependent colony forming assay and 3D organoid data suggest that the viability and organoid formation efficiency is significantly reduced in RRBP1 KO cells compared with WT (Fig. 2c, d). The immunoblotting data suggests that knockout RRBP1 in chemo-resistant cells results in significant increase in cisplatin-induced cleaved PARP and γ-H2AX expression (Fig. 2e). In addition to this, transient overexpression of RRBP1 in H357CisS and SCC-9 CisS cells using overexpression construct (pcDNA4 HisMax-V5-GFP-RRBP1), showed the development of cisplatin resistance (Supplementary Fig. 4 A-F). The extra-cellular matrix (ECM) plays a key role in cancer progression. The collagen induced by CAFs is an important component of ECM. Earlier, it is established that RRBP1 regulates collagen secretion in fibroblast and knockdown of RRBP1 in fibroblast resulted in depleted collagen secretion.9 Henceforth, to investigate the role of RRBP1 in CAF to promote chemoresistance, we isolated cancer associated fibroblast (CAF) and epithelial cancer cells (PDC) from neoadjuvant treated OSCC patients that did not respond to TP (Supplementary Fig. 5A). To characterise the isolated CAF and PDC, we stained pan-cytokeratin for epithelial and FSP1(fibroblast specific protein 1) for CAF (Supplementary Fig. 5B). The data suggest that FSP1 is expressed in CAF, whereas pan-cytokeratin is expressed in epithelial cells. Further, immunoblotting and cell death data suggest that knockout of RRBP1 in CAF resulted in enhanced cell death against cisplatin (Fig. 2f, g). Here, corroborating with earlier study we also found reduced secretion of collagen1A in RRBP1 KO CAF (Fig. 2h). To investigate possible RRBP1 mediated secretary function, we treated the condition media from WT CAF and RRBP1 KO CAF cells (CM) to H357CisS culture and measured their effects on the cisplatin-induced apoptotic response. Interestingly, we found that WT CAF-CM promoted resistance against cisplatin but not the RRBP1 KO CAF-CM (Fig. 2i, j).
Fig. 2. RRBP1 knockout sensitised chemo-resistant resistant cells to cisplatin.
a RRBP1 KO cells in human OSCC line H357CisR, SCC-9 CisR and PDC-1 were generated using a lentiviral approach as described in materials and methods. RRBP1 KO and RRBP1 WT cells of H357CisR, SCC-9 CisR and PDC-1 were treated with cisplatin (10 μM) for 48 h and cell viability was determined using MTT assay (Mean ± SEM, n = 3) *P < 0.05. b RRBP1 KO and WT cells of H357CisR, SCC-9 CisR and PDC-1 were treated with cisplatin (10 μM) for 48 h, after which cell death was determined by annexin V/7AAD assay using flow cytometer. Bar diagrams indicate the percentage of cell death with respective treated groups. c RRBP1 KO and RRBP1 WT cells of H357CisR, SCC-9 CisR and PDC-1 were treated with cisplatin (10 μM) and anchorage-dependent colony forming assay was performed as described in materials methods. d RBP1KO and WT cells were treated with cisplatin (10 μM) and 3D organoid assay was performed as described in materials and methods. e RRBP1 KO and WT cells were treated with cisplatin (10 μM) for 48 h and immunoblotting was performed with indicated antibodies. f Confirmation of RRBP1 KO in CAF by immunoblotting. g RRBP1 KO and WT CAF of OSCC, were treated with cisplatin up to 10 μM for 48 h, after which cell death was determined by annexin V/7AAD assay using flow cytometer. h Immunoblotting was performed against collagen 1 antibody in condition media of WT and RRBP1 KO CAF. i Schematic diagram of approach to evaluate the effects of conditioned media by WT RRBP1 and KO RRBP1 CAF on cell death of H357CisS. j Conditioned media of RRBP1 KO and WT CAF collected and supplemented in H357CisS cultured in with or without collagen coated plates then treated with cisplatin up to 10 μM for 48 h, after which cell death was determined by annexin V/7AAD assay using flow cytometer.
Transcriptome analysis revealed RRBP1 as potential modulator of YAP1
The deregulated proteins, identified from global proteomics analysis, were converted to gene list and a functional analysis was carried out using Ingenuity Pathway Analysis (IPA). IPA analysis of functional pathways in acquired chemoresistance cells showed highest downregulation of Hippo signalling (Supplementary Fig. 6A). Hippo pathway negatively regulates the activity of its downstream transcriptional co-activators, Yes-associated protein 1 (YAP1) and Transcriptional coactivator with PDZ-binding motif (TAZ). The active YAP1/TAZ translocate to the nucleus and binds with TEA domain family member (TEAD), which in turn transcribes genes that promotes cell proliferation and inhibit apoptosis. We also performed the RNA sequencing of H357CisR RRBP1KD along with H357CiSR Nt Sh cells and compared with RNA-seq dataset of H357CisR vs H357CisS cells. The efficacy of RRBP1 ShRNA is described in Supplementary Fig. 7A–C. Total 1456 and 2534 no of genes are found to be dysregulated between H357CisR vs RRBP1KD and H357CisR vs H357CisS, respectively (Supplementary Fig. 6B). After overlapping the two datasets from Supplementary Fig. 6C, we found 810 common gene were dysregulated. When we analysed the common set of deregulated genes between these two groups (H357CisR vs RRBP1KD and H357CisR vs H357CisS), it was found that YAP1 target genes (CCN1 and CCN2) were downregulated in H357CisS and RRBP1KD cells (Supplementary Fig. 6 D, E). Volcano plot yielded similar observation (Supplementary Fig. 6F, G).
RRBP1 regulates YAP1 expression in chemo-resistant cells
In line with RNA-seq data, the expression of YAP1 and its target genes (CYR61, CTGF, Jagged 1, AXL, Integrin β2, IGFBP3 and Laminin B2) were found to be significantly downregulated both at mRNA and protein levels in RRBP1 KO cells with respect to controls (Fig. 3a, b). Confocal microscopy and immunoblotting data showed significantly reduced nuclear YAP1 expression in RRBP1 KO cells as compared to control cells (Fig. 3c, d). Further, constitutive overexpression of s-127 Yap1 in chemo-sensitive line rescued the resistance phenotype as well as all the target genes expression (Supplementary Fig. 8A–C). Hippo signalling is regulated by MST1/2 and LATS1/2, which phosphorylates the YAP1 at Ser-127 (Hippo on) resulting in its cytoplasmic retention and proteasome-mediated degradation. Similarly, phosphorylation at Ser-397 of YAP1 by LATS1/2 creates a phosphodegron motif for β‐TrCP binding followed by proteasomal degradation.20 We did not find any increase in phosphorylated YAP1, rather, lower expression of p-YAP (at Ser-127 and Ser-397) were observed in RRBP1 KO as compared to WT cells (Fig. 3e). Further, association analysis of RRBP1 mRNA levels with YAP1 target genes (CTGF, CYR61, Jagged 1, AXL, TEAD1, COL1A1, DCN, NUAK1, FOXO1, AMOTL2, PCDH7, MYOF, JAK1, SEMAC3 and RUNX1) in the The Cancer Genome Atlas (TCGA) HNSCC cohort using GEPIA showed a positive correlation (r ≥ 0.3) (Fig. 3f).
Fig. 3. RRBP1 regulates YAP1 in chemo-resistant OSCC.
a Relative mRNA expression of indicated genes was analysed by qRT-PCR in indicated RRBP1 KO and RRBP1 WT chemo-resistant cells (mean ± SEM, n = 3). GAPDH was used as an internal control. b Cell lysates from indicated RRBP1 KO and WT cells were isolated and subjected to immunoblotting against indicated antibodies. c H357CiSR RRBP1 KO and H357CisR RRBP1 WT cells were cultured and immunostaining were performed using the anti-YAP1 antibody as described in materials and methods. Images were acquired using confocal microscopy (LEICA TCS-SP8). d Lysates from cytoplasm and nucleus were separated and immunoblot was performed against indicated antibodies. e Cell lysates from indicated RRBP1 KO and WT cells were isolated and subjected to immunoblotting against indicated antibodies. f Expression correlation of RRBP1 and indicated YAP1 target genes (mRNA) analysed in the HNSCC TCGA cohort. Correlation was analysed using Spearman’s correlation coefficient test, n = 520. The analysis was performed in Gene Expression Profiling Interactive Analysis (GEPIA) platform.
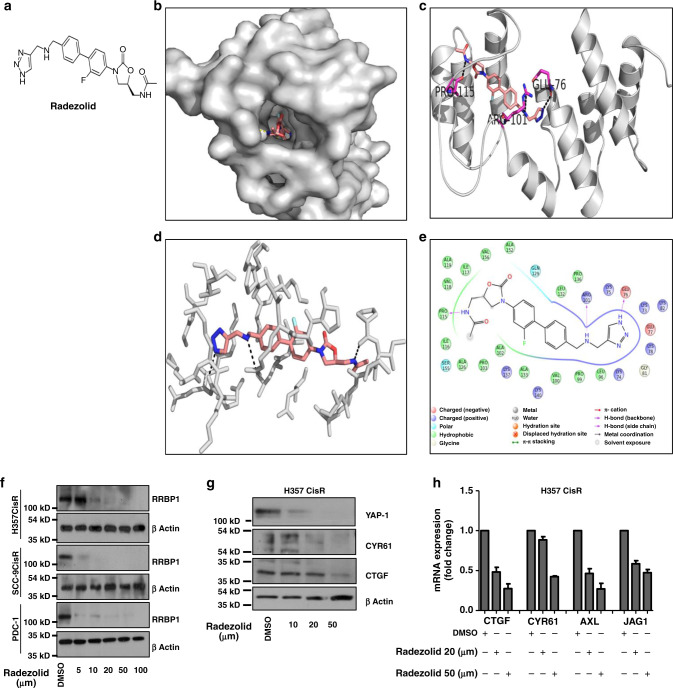
Radezolid, a potential inhibitor of RRBP1, restores cisplatin-induced cell death in chemo-resistant OSCC
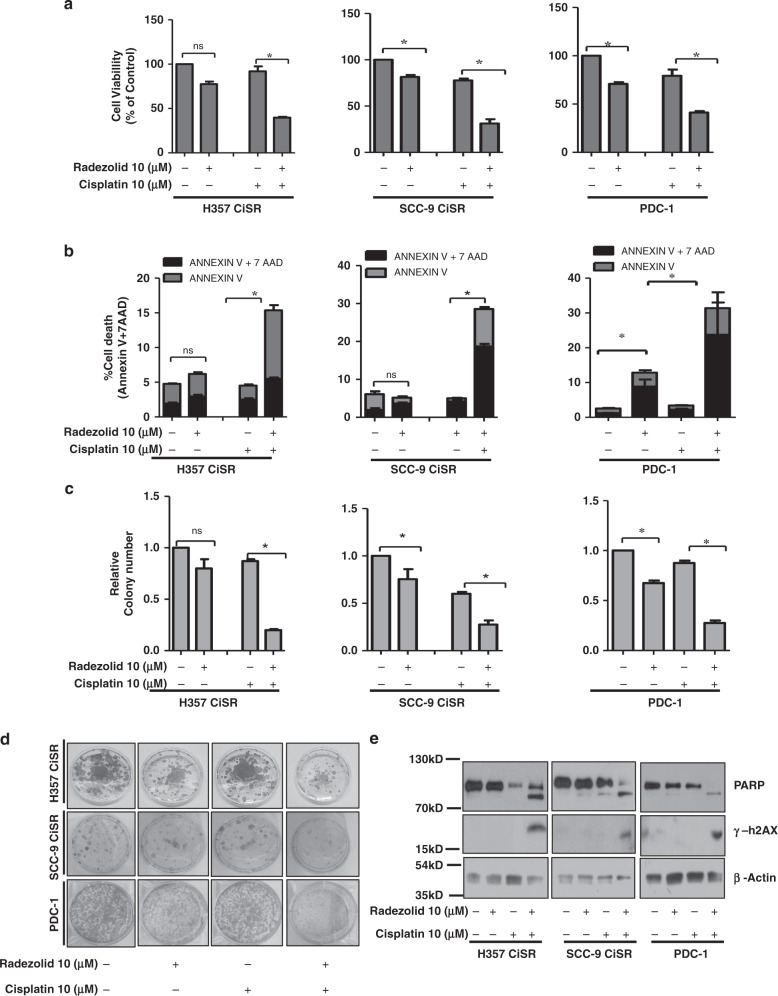
Limited information on the potential inhibitors of RRBP1 are available in the literature. From PubChem and drugbank database search, it seemed that Radezolid, a second generation oxazolidinone antibiotic, may be used as a potential candidate to target RRBP1 (Fig. 4a). The Insilco molecular docking study suggested that Radezolid docks well at the active site of RRBP1 (Fig. 4b, c) with three hydrogen bonds and a docking score of −8.0 indicating a high affinity for the target protein. The first hydrogen bond with GLU78 while the nearby amide makes a hydrogen bond with PRO115. The other amide makes a hydrogen bond interaction with ARG101 of RRBP1 (Fig. 4 d, e). Next, RRBP1 expression was monitored in chemo-resistant cells (H357CisR, SCC-9 CisR and PDC-1) treated with Radezolid. We observed a dose dependent (≥5 μM) lowering of RRBP1 protein expression with treatment of Radezolid (Fig. 4f). However, Radezolid treatment significantly reduced the expression of YAP1 target genes (both at protein and mRNA level) in H357CisR and SCC-9 CisR cells (Fig. 4g, h). Further, we evaluated if treatment of Radezolid can overcome cisplatin resistance in OSCC. Upon combinatorial treatment of radezolid and cisplatin in chemo-resistant OSCC cells and PDC-1, we observed a reversal of chemoresistance (Fig. 5 a–d). Apoptosis induced by this combinatorial effect was confirmed from a significant increase in cleaved PARP and increased γH2AX level (Fig. 5 e).
Fig. 4. Radezolid represses RRBP1 protein expression and regulate YAP1 target genes in chemo-resistant OSCC.
a Structure of Radezolid. b Molecular docking of RRBP1 with Radezolid compounds carried out in Glide and representation in surface view receptor of drug interaction. c Ribbon model structure of RRBP1 showing the hydrogen bonding interaction with Radezolid at PRO115, ARG101 and GLU-76. Hydrogen bonds are shown as dashed lines. d Active site residues within 5 Angstrom of RRBP1 and Radezolid interactions. e Ligand plot showing interaction of the Radezolid interaction with different residues of RRBP1 within 5 angstrom distance. f Indicated chemo-resistant OSCC cells were treated with Radezolid in a dose dependent manner for 48 h, after which lysates were isolated to perform immunoblotting against β-actin and RRBP1. g H357CisR cells were treated with Radezolid in a dose dependent manner for 48 h, after which lysates were isolated to perform immunoblotting against indicated antibody. h H357CisR cells were treated with Radezolid in a dose dependent manner for 48 h, after qRT-PCR was performed to determine the relative mRNA expression (fold change) of indicated genes. GAPDH was used as an internal control.
Fig. 5. Radezolid (oxazolidinone group antibiotic) restored cisplatin-induced cell death in chemo-resistant OSCC.
a Cisplatin-resistant OSCC line H357CisR, SCC-9 CisR and PDC-1 cells were treated with Radezolid (10 µM) and cisplatin(10 µM) alone or in combination for 48 h and cell viability was determined using MTT assay (Mean ± SEM, n = 3) *P < 0.05. b H357CisR, SCC-9 CisR and PDC-1 cells were treated with Radezolid (10 µM) and cisplatin(10 µM) for 48 h, after which cell death was determined by annexin V/7AAD assay using flow cytometer. Bar diagrams indicate the percentage of cell death from a panel with respective treated groups (Mean ± SEM, n = 3). c H357CisR, SCC-9 CisR and PDC-1 cells were treated with Radezolid (10 µM) and cisplatin(10 µM) after which anchorage-dependent colony forming assay was performed as described in materials methods. The bar describes the colony number as compared to vehicle treated H357CisR (cells (Mean ± SEM, n = 3) *P < 0.05. d Representative Images of the anchorage-dependent colony forming assay described in c. e H357CisR, SCC-9 CisR and PDC-1 cells were treated with Radezolid (10 µM) and cisplatin (10 µM) for 48 h, after which immunoblotting was performed with indicated antibodies.
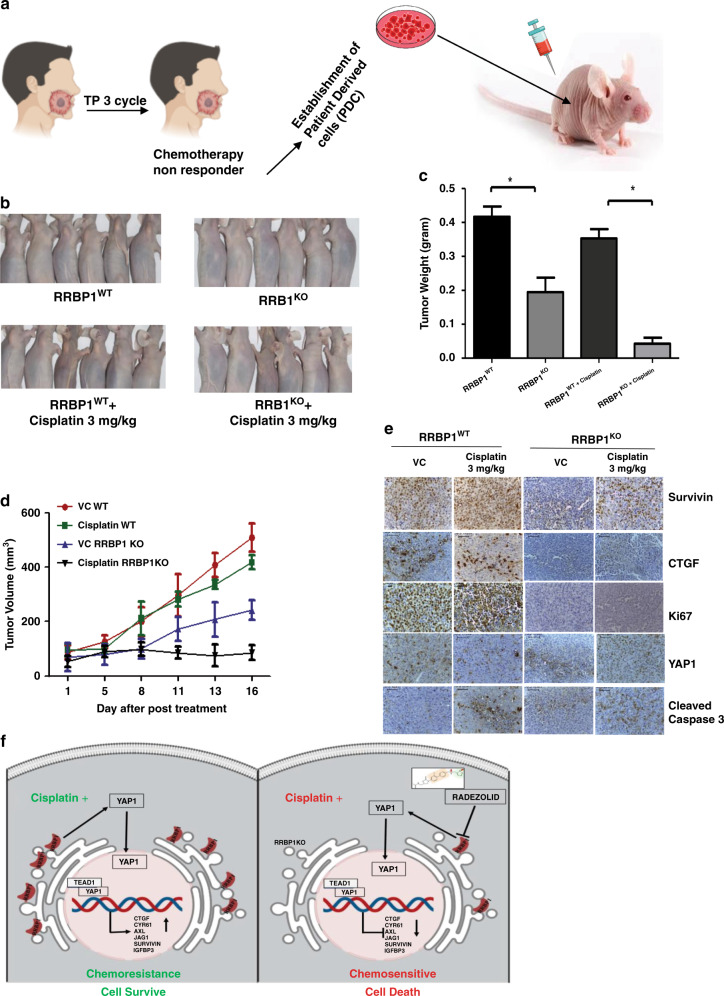
Knockout of RRBP1 significantly induced cisplatin-mediated apoptosis in chemo-resistant patient-derived xenograft
To evaluate in vivo efficacy of knocking out RRBP1 in restoring cisplatin-induced cell death in chemo-resistant OSCC, we established xenograft tumours in nude mice using PDC isolated from tumours of a chemotherapy-non-responder patient (Fig. 6a) (patient# 1, Supplementary Table 2). We observed a reduction in tumour growth and size in RRBP1 KO group as compared to WT PDC-1. Treating with cisplatin (3 mg/kg) significantly reduced the tumour burden in case of RRBP1 KO group (Fig. 6 b–d). Immunohistochemistry analysis of harvested tumours showed significantly higher apoptosis levels, reduced expression of YAP1 and its target genes in RBBP1 KO groups treated with cisplatin (Fig. 6 e).
Fig. 6. Knockout of RRBP1 restores cisplatin-induced cell death in chemo-resistant xenografts.
a Schematic representation of establishment of PDX, using patient-derived cells isolated from chemotherapy-non-responder patient. b RRBP1 WT cells were injected to the right flank of athymic nude mice and PDC1RRBP1KO cells were injected to the left flank of same mice as described in materials and methods. After which mice were treated with either vehicle control or 3 mg/Kg of cisplatin in two different groups (twice a week). At the end of the experiment’s mice were sacrificed and images were captured (n = 6). c At the end of experiments, tumour weight was measured and represented in bar diagram (mean ± SEM, **P < 0.05, n = 6). d Tumour growth was measured in the indicated time point using digital slide calliper and plotted as a graph (mean ± SEM, n = 6). e After completion of treatment, tumours from each group were fixed with formalin, and paraffin-embedded sections were prepared to perform immunohistochemistry with indicated antibodies. f Schematic presentation of the mechanism by which RRBP1 mediates chemoresistance in OSCC.
Discussion
Earlier, cisplatin-resistance models have been successfully established by prolonged treatment of drugs to cancer cell lines representing various neoplasms.21 These models can be broadly divided into two groups: (I) clinically relevant model, where cells were grown with lower doses of drug adopting a pulse treatment strategy,22,23 (II) High-level laboratory models, where cells are continually grown in presence of drugs with dosage escalation from lower dose to IC50.24,25 The advantage of clinically relevant models is that it mimics the chemotherapy strategies in patients; on the other hand, its resistance pattern is very inconsistent. High-level laboratory models showed consistent resistant pattern and are generally preferred to study the mechanism of chemoresistance in cancer cells. All these studies engage the parental sensitive cells and late drug resistant cells to understand the molecular mechanism for chemoresistance. Here, to explore the causative factors of chemoresistance in OSCC, we have established and characterised sensitive, early and late cisplatin-resistant cells. Using global proteomic and genomic profiling of oral cancer cells with different grades of resistance to cisplatin, we have identified and validated that Ribosomal binding protein 1 (RRBP1) is one of the critical proteins responsible for resistance development. RRBP1 is localised in the rough endoplasmic reticulum (rER) and supposed to play a role in secretion of newly synthesised protein.26 RRBP1 is reported to be over expressed in breast,27 lung,26 colorectal,28 oesophageal,29 endometrial,30 prostate31 and ovarian cancer32 patient tissues. RRBP1 expression is associated with the disease progression and also envisaged as an unfavourable post-operative prognosis.28 Here, we established that inhibiting RRBP1 has potential to rewire cisplatin-resistance development and making it susceptible to cisplatin. To the best of our knowledge, this is the first study to demonstrate that elevated RRBP1 in cisplatin-resistant OSCC cancer cells could be reversed to make them susceptible to cisplatin treatment.
In this study, we also observed a significant reduction of YAP1 and its target genes in RRBP1 knockout cells, limited YAP1 phosphorylation, indicating that it is not degraded by proteasomal degradation. Our results indicate that RRBP1 might be playing a role in the translation of YAP1 mRNA. In absence of RRBP1, the translation of YAP1 is partially blocked (Fig. 6F). It is well established that dysregulated Hippo signalling promotes malignancy in cancer cells.33,34 Recently, Hippo signalling has also been correlated to mediate chemoresistance in different neoplasms to several chemotherapeutic drugs.35 Similarly, phosphorylation at S397 of YAP1 by LATS1/2 creates a phospho‐degron motif for β‐TrCP binding followed by proteasomal degradation.20 Overall, dephosphorylated YAP1 (Hippo off) translocate to the nucleus to transcribe the YAP1 target genes.
We additionally provide evidence that a second generation Oxazolidinones i.e. Radezolid is a synthetic antibiotic affecting the initiation phase of bacterial protein synthesis (under development by Melinta Therapeutics Inc) represses RRBP1 in chemo-resistant cells. Further, preclinical studies are underway to determine the toxicity profile, pharmacokinetic properties of Radezolid. We are in the process of evaluating the in vivo efficacy of Radezolid to understand its role in reversing the cisplatin resistance in OSCC.
In conclusion, our study suggests that RRBP1 mediates cisplatin resistance in OSCC by modulating hippo signalling. Knocking out RRBP1 restores cisplatin-mediated cell death in chemo-resistant OSCC cells. We further demonstrated that Radezolid (which inhibits RRBP1) could be useful to reverse cisplatin resistance in acquired chemo-resistant lines and PDX models. Thus, this unique combinatorial approach needs further clinical investigation to target advanced OSCC.
Supplementary information
Supplementary Fig 1: Characterization of sensitive, early and late OSCC cisplatin-resistant lines
Supplementary Fig 2: PCA plot and fragmentation pattern of the RRBP1 protein
Supplementary figure 3: The confirmation of RRBP1 KO in OSCC Chemoresistance cell line done by using GeneArt®Genomic Cleavage Detection (GCD) Kit (Life technology) with PCR primers listed in supplem
Supplementary Figure 4: Overexpression of RRBP1 in human OSCC lines results in development of cisplatin resistance
Supplementary Fig 5: PDC and CAF isolation from primary chemotherapy non-responder tumor
Supplementary Figure 6: RNA Sequencing revealed YAP1 target genes modulated by RRBP1.
Supplementary Fig 7: Characterisation of RRBP1 knockdown in H357CisR cells
Supplementary Figure 8: Constitutive overexpression of s-127 rescue the resistance property in sensitive cells
Acknowledgements
O.S. is a UGC-SRF, S.M. is UGC-JRF, P.M. is CSIR SRF. S.R.K. and F.P. received fellowship from Department of Biotechnology. K.C.M. received institutional fellowship.
Author contributions
O.S., S.M., R.A., P.M., S.R.K., F.P., S.K., P.P. and K.C.M. performed experiments, and analysed the data, under the direction of R.D. and R.K.N. R.R., D.K.M., A.D. and S.K.M. performed part of experiments. O.S. and R.D. designed experiments and supervised the study. O.S., R.D. and R.K.N. wrote the manuscript. All authors approved the final version.
Ethics approval and consent to participate
This study was approved by Human Ethics committees (HEC) of Institute of Life Sciences, (84/HEC/18) and All India Institute of Medical Sciences (AIIMS), (T/EMF/Surg.Onco/19/03). The animal-related experiments were approved by Institutional Animal Ethics Committee of Institute of Life Sciences, (ILS/IAEC-147-AH/FEB-19).
Data availability
All the mass spectrometry data files (. raw and. mgf) with result files were deposited in the ProteomeXchange Consortium (PXD0016977) and all the RNA-seq data file were deposited in ArrayExpress (E-MTAB-9697).
Competing interests
The authors declare no competing interests.
Funding information
Grant support: This work is supported by Institute of Life Sciences, Bhubaneswar intramural support, ICMR (5/13/9/2019-NCD-III) and DBT BT/INF/22/SP28293/2018 (for imaging facility). Core support from International Centre for Genetic Engineering and Biotechnology to R.K.N. is highly acknowledged.
Footnotes
The original online version of this article was revised: During the proof process the authors unfortunately removed of one the co-corresponding authors. The correct version should read: Correspondence: Rupesh Dash (rupesh.dash@gmail.com) or Ranjan K. Nanda (HYPERLINK “mailto:ranjan@icgeb.res.in” ranjan@icgeb.res.in).
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
5/7/2021
A Correction to this paper has been published: 10.1038/s41416-021-01414-w
Contributor Information
Ranjan K. Nanda, Email: ranjan@icgeb.res.in
Rupesh Dash, Email: rupesh.dash@gmail.com.
Supplementary information
The online version contains supplementary material available at 10.1038/s41416-021-01336-7.
References
- 1.Dikshit R, Gupta PC, Ramasundarahettige C, Gajalakshmi V, Aleksandrowicz L, Badwe R, et al. Cancer mortality in India: a nationally representative survey. Lancet. 2012;379:1807–1816. doi: 10.1016/S0140-6736(12)60358-4. [DOI] [PubMed] [Google Scholar]
- 2.Huang SH, O’Sullivan B. Oral cancer: current role of radiotherapy and chemotherapy. Med. Oral. Patol. Oral. Cir. Bucal. 2013;18:e233–e240. doi: 10.4317/medoral.18772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ackerstaff AH, Rasch CR, Balm AJ, de Boer JP, Wiggenraad R, Rietveld DH, et al. Five-year quality of life results of the randomized clinical phase III (RADPLAT) trial, comparing concomitant intra-arterial versus intravenous chemoradiotherapy in locally advanced head and neck cancer. Head Neck. 2012;34:974–980. doi: 10.1002/hed.21851. [DOI] [PubMed] [Google Scholar]
- 4.Wang C, Liu XQ, Hou JS, Wang JN, Huang HZ. Molecular mechanisms of chemoresistance in oral cancer. Chin. J. Dent. Res. 2016;19:25–33. doi: 10.3290/j.cjdr.a35694. [DOI] [PubMed] [Google Scholar]
- 5.Vishak S, Rangarajan B, Kekatpure VD. Neoadjuvant chemotherapy in oral cancers: selecting the right patients. Indian J. Med. Paediatr. Oncol. 2015;36:148–153. doi: 10.4103/0971-5851.166716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maji S, Panda S, Samal SK, Shriwas O, Rath R, Pellecchia M, et al. Bcl-2 antiapoptotic family proteins and chemoresistance in cancer. Adv. Cancer Res. 2018;137:37–75. doi: 10.1016/bs.acr.2017.11.001. [DOI] [PubMed] [Google Scholar]
- 7.Wanker EE, Sun Y, Savitz AJ, Meyer DI. Functional characterization of the 180-kD ribosome receptor in vivo. J. Cell Biol. 1995;130:29–39. doi: 10.1083/jcb.130.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Savitz AJ, Meyer DI. 180-kD ribosome receptor is essential for both ribosome binding and protein translocation. J. Cell Biol. 1993;120:853–863. doi: 10.1083/jcb.120.4.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ueno T, Tanaka K, Kaneko K, Taga Y, Sata T, Irie S, et al. Enhancement of procollagen biosynthesis by p180 through augmented ribosome association on the endoplasmic reticulum in response to stimulated secretion. J. Biol. Chem. 2010;285:29941–29950. doi: 10.1074/jbc.M109.094607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maji S, Shriwas O, Samal SK, Priyadarshini M, Rath R, Panda S, et al. STAT3- and GSK3beta-mediated Mcl-1 regulation modulates TPF resistance in oral squamous cell carcinoma. Carcinogenesis. 2019;40:173–183. doi: 10.1093/carcin/bgy135. [DOI] [PubMed] [Google Scholar]
- 11.Samal SK, Routray S, Veeramachaneni GK, Dash R, Botlagunta M. Ketorolac salt is a newly discovered DDX3 inhibitor to treat oral cancer. Sci. Rep. 2015;5:9982. doi: 10.1038/srep09982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arya R, Dabral D, Faruquee HM, Mazumdar H, Patgiri SJ, Deka T, et al. Serum small extracellular vesicles proteome of tuberculosis patients demonstrated deregulated immune response. Proteom. Clin. Appl. 2020;14:e1900062. doi: 10.1002/prca.201900062. [DOI] [PubMed] [Google Scholar]
- 13.Shriwas, O., Priyadarshini, M., Samal, S. K., Rath, R., Panda, S., Das Majumdar, S. K. et al. DDX3 modulates cisplatin resistance in OSCC through ALKBH5-mediated m(6)A-demethylation of FOXM1 and NANOG. Apoptosis10.1007/s10495-020-01591-8 (2020). [DOI] [PubMed]
- 14.Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hung, V., Lam, S. S., Udeshi, N. D., Svinkina, T., Guzman, G., Mootha, V. K. et al. Proteomic mapping of cytosol-facing outer mitochondrial and ER membranes in living human cells by proximity biotinylation. elife10.7554/eLife.24463 (2017). [DOI] [PMC free article] [PubMed]
- 16.Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim J, et al. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 2007;21:2747–2761. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kijima T, Nakagawa H, Shimonosono M, Chandramouleeswaran PM, Hara T, Sahu V, et al. Three-dimensional organoids reveal therapy resistance of esophageal and oropharyngeal squamous cell carcinoma cells. Cell Mol. Gastroenterol. Hepatol. 2019;7:73–91. doi: 10.1016/j.jcmgh.2018.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maji S, Samal SK, Pattanaik L, Panda S, Quinn BA, Das SK, et al. Mcl-1 is an important therapeutic target for oral squamous cell carcinomas. Oncotarget. 2015;6:16623–16637. doi: 10.18632/oncotarget.3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang Z, Kang B, Li C, Chen T, Zhang Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47:W556–W560. doi: 10.1093/nar/gkz430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rozengurt E, Sinnett-Smith J, Eibl G. Yes-associated protein (YAP) in pancreatic cancer: at the epicenter of a targetable signaling network associated with patient survival. Signal Transduct. Target Ther. 2018;3:11. doi: 10.1038/s41392-017-0005-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McDermott M, Eustace AJ, Busschots S, Breen L, Crown J, Clynes M, et al. In vitro development of chemotherapy and targeted therapy drug-resistant cancer cell lines: a practical guide with case studies. Front. Oncol. 2014;4:40. doi: 10.3389/fonc.2014.00040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stordal BK, Davey MW, Davey RA. Oxaliplatin induces drug resistance more rapidly than cisplatin in H69 small cell lung cancer cells. Cancer Chemother. Pharmacol. 2006;58:256–265. doi: 10.1007/s00280-005-0148-7. [DOI] [PubMed] [Google Scholar]
- 23.Ma J, Maliepaard M, Kolker HJ, Verweij J, Schellens JH. Abrogated energy-dependent uptake of cisplatin in a cisplatin-resistant subline of the human ovarian cancer cell line IGROV-1. Cancer Chemother. Pharmacol. 1998;41:186–192. doi: 10.1007/s002800050727. [DOI] [PubMed] [Google Scholar]
- 24.Shen DW, Akiyama S, Schoenlein P, Pastan I, Gottesman MM. Characterisation of high-level cisplatin-resistant cell lines established from a human hepatoma cell line and human KB adenocarcinoma cells: cross-resistance and protein changes. Br. J. Cancer. 1995;71:676–683. doi: 10.1038/bjc.1995.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liang XJ, Shen DW, Garfield S, Gottesman MM. Mislocalization of membrane proteins associated with multidrug resistance in cisplatin-resistant cancer cell lines. Cancer Res. 2003;63:5909–5916. [PubMed] [Google Scholar]
- 26.Tsai HY, Yang YF, Wu AT, Yang CJ, Liu YP, Jan YH, et al. Endoplasmic reticulum ribosome-binding protein 1 (RRBP1) overexpression is frequently found in lung cancer patients and alleviates intracellular stress-induced apoptosis through the enhancement of GRP78. Oncogene. 2013;32:4921–4931. doi: 10.1038/onc.2012.514. [DOI] [PubMed] [Google Scholar]
- 27.Telikicherla D, Marimuthu A, Kashyap MK, Ramachandra YL, Mohan S, Roa JC, et al. Overexpression of ribosome binding protein 1 (RRBP1) in breast cancer. Clin. Proteomics. 2012;9:7. doi: 10.1186/1559-0275-9-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pan Y, Cao F, Guo A, Chang W, Chen X, Ma W, et al. Endoplasmic reticulum ribosome-binding protein 1, RRBP1, promotes progression of colorectal cancer and predicts an unfavourable prognosis. Br. J. Cancer. 2015;113:763–772. doi: 10.1038/bjc.2015.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang L, Wang M, Zhang M, Li X, Zhu Z, Wang H. Expression and significance of RRBP1 in esophageal carcinoma. Cancer Manag. Res. 2018;10:1243–1249. doi: 10.2147/CMAR.S158013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu S, Lin M, Ji H, Ding J, Zhu J, Ma R, et al. RRBP1 overexpression is associated with progression and prognosis in endometrial endometrioid adenocarcinoma. Diagn. Pathol. 2019;14:7. doi: 10.1186/s13000-019-0784-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li T, Wang Q, Hong X, Li H, Yang K, Li J, et al. RRBP1 is highly expressed in prostate cancer and correlates with prognosis. Cancer Manag. Res. 2019;11:3021–3027. doi: 10.2147/CMAR.S186632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma, J., Ren, S., Ding, J., Liu, S., Zhu, J., Ma, R. et al. Expression of RRBP1 in epithelial ovarian cancer and its clinical significance. Biosci. Rep.10.1042/BSR20190656 (2019). [DOI] [PMC free article] [PubMed]
- 33.Han Y. Analysis of the role of the Hippo pathway in cancer. J. Transl. Med. 2019;17:116. doi: 10.1186/s12967-019-1869-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Misra JR, Irvine KD. The Hippo signaling network and its biological functions. Annu. Rev. Genet. 2018;52:65–87. doi: 10.1146/annurev-genet-120417-031621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gujral TS, Kirschner MW. Hippo pathway mediates resistance to cytotoxic drugs. Proc. Natl Acad. Sci. USA. 2017;114:E3729–E3738. doi: 10.1073/pnas.1703096114. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig 1: Characterization of sensitive, early and late OSCC cisplatin-resistant lines
Supplementary Fig 2: PCA plot and fragmentation pattern of the RRBP1 protein
Supplementary figure 3: The confirmation of RRBP1 KO in OSCC Chemoresistance cell line done by using GeneArt®Genomic Cleavage Detection (GCD) Kit (Life technology) with PCR primers listed in supplem
Supplementary Figure 4: Overexpression of RRBP1 in human OSCC lines results in development of cisplatin resistance
Supplementary Fig 5: PDC and CAF isolation from primary chemotherapy non-responder tumor
Supplementary Figure 6: RNA Sequencing revealed YAP1 target genes modulated by RRBP1.
Supplementary Fig 7: Characterisation of RRBP1 knockdown in H357CisR cells
Supplementary Figure 8: Constitutive overexpression of s-127 rescue the resistance property in sensitive cells
Data Availability Statement
All the mass spectrometry data files (. raw and. mgf) with result files were deposited in the ProteomeXchange Consortium (PXD0016977) and all the RNA-seq data file were deposited in ArrayExpress (E-MTAB-9697).