Abstract
Upregulation of programmed death ligand 1 (PD-L1) helps tumor cells escape from immune surveillance, and therapeutic antibodies targeting PD-1/PD-L1 have shown better patient outcomes only in several types of malignancies. Recent studies suggest that the clinical efficacy of anti-PD-1/PD-L1 treatments is associated with PD-L1 levels; however, the underlying mechanism of high PD-L1 protein levels in cancers is not well defined. Here, we report that the deubiquitinase OTUB1 positively regulates PD-L1 stability and mediates cancer immune responses through the PD-1/PD-L1 axis. Mechanistically, we demonstrate that OTUB1 interacts with and removes K48-linked ubiquitin chains from the PD-L1 intracellular domain in a manner dependent on its deubiquitinase activity to hinder the degradation of PD-L1 through the ERAD pathway. Functionally, depletion of OTUB1 markedly decreases PD-L1 abundance, reduces PD-1 protein binding to the tumor cell surface, and causes increased tumor cell sensitivity to human peripheral blood mononuclear cells (PBMCs)-mediated cytotoxicity. Meanwhile, OTUB1 ablation-induced PD-L1 destabilization facilitates more CD8+ T cells infiltration and increases the level of IFN-γ in serum to enhance antitumor immunity in mice, and the tumor growth suppression by OTUB1 silencing could be reversed by PD-L1 overexpression. Furthermore, we observe a significant correlation between PD-L1 abundance and OTUB1 expression in human breast carcinoma. Our study reveals OTUB1 as a deubiquitinating enzyme that influences cancer immunosuppression via regulation of PD-L1 stability and may be a potential therapeutic target for cancer immunotherapy.
Subject terms: Proteins, Immune evasion
Introduction
Programmed death ligand 1 (PD-L1) and its receptor PD-1 are important targets for immune checkpoint blockade therapy [1–3], as high expression of PD-L1 on tumor cells leads to T cell inhibition through PD-1 engagement [4–7]. Hence, antibodies targeting PD-L1/PD-1 interaction can impede immune evasion of tumor cells. This strategy has shown promise for the treatment of several cancer types, including melanoma, renal cell carcinoma, Hodgkin’s lymphoma, and non-small cell lung cancer [8–11]. However, patients with other types of cancer exhibit reduced responses to PD-1/PD-L1 blockade therapy, and thus it is necessary to uncover the molecular mechanisms regulating PD-L1 and further provide hopeful strategies to enhance the clinical efficacy of anti-PD-l/PD-L1 therapy [12, 13].
Recent studies have revealed the critical roles of post-translational modifications (PTMs) in controlling PD-L1 protein levels and regulating cancer cell immunosuppression [14, 15]. N-glycosylation promotes PD-L1 protein stability and inhibits antitumor T cell immunity via the IL-6/JAK1/PD-L1 Y112 phosphorylation/STT3A signaling axis [16]. Glycogen synthase kinase 3β (GSK3β) is regarded as a central node of PD-L1 regulation, as it stringently phosphorylates non-glycosylated PD-L1 and results in downregulation of PD-L1 [17]. The ubiquitin E3 ligases SPOP and β-TrCP are responsible for the polyubiquitination of PD-L1 and target PD-L1 to the proteasome degradation pathway [17, 18], meanwhile, PD-L1 level is also reported to be regulated by the reversed deubiquitination process [19–22]. On the basis of these findings, targeting the PTMs of PD-L1 may be a promising antitumor therapeutic strategy. Thus identification of PTM regulators of PD-L1 expression in response to different stimuli in cells is of great importance.
OTUB1 (OTU domain-containing ubiquitin aldehyde-binding protein 1) belongs to the ovarian tumor domain protease (OTU) subfamily of deubiquitinases (DUBs) and negatively regulates ubiquitination to control protein stability and activity [23–25]. Accumulating evidence suggests that OTUB1 is a critical regulator in DNA damage response, cell apoptosis, proliferation, and cancer development [26–28]. Many important proteins, such as p53, SMAD2/3, and TRAF3, are targets of OTUB1 [29–31]. A recent study found that OTUB1 controls IL-15-stimulated activation of CD8+ T cells and NK cells [32]. Deletion of OTUB1 profoundly promotes antitumor immunity, suggesting that OTUB1 acts as a checkpoint during T cell-mediated immune responses. As OTUB1 modulates T cell function, the possible role of highly expressed OTUB1 in cancer cells to regulate immune evasion is worthy to be explored.
Here, we identify OTUB1 as a deubiquitinase that stabilizes PD-L1. Knockdown of OTUB1 leads to increased ubiquitination and degradation of PD-L1 through the proteasome-dependent ERAD pathway. OTUB1 deficiency causes cancer cells to become more sensitive to T cell-mediated cytotoxicity. Moreover, OTUB1 depletion leads to the inhibitory effect on tumor growth and enhances antitumor immunity through regulating PD-L1 in mice. OTUB1 is positively correlated with PD-L1 abundance in human breast cancer specimens. Together, our results reveal a new role for OTUB1 in mediating cancer immunosuppression via regulation of PD-L1 abundance and suggest that inhibition of OTUB1 may offer substantial clinical benefit for patients.
Materials and methods
Clinical samples and Ethics statement
Peripheral blood samples from healthy individuals (20–35 years of age) were collected at Peking University Hospital according to the guidelines of the Ethics Committee of Peking University (IRB00001052-16020). All samples were obtained with patients’ informed contents. This study was performed according to the ethical guidelines of the Declaration of Helsinki. Human peripheral blood mononuclear cells (PBMCs) were purified by Histopaque-1077 (Sigma-Aldrich, USA) density-gradient centrifugation and maintained in phosphate buffered saline (PBS).
Animal models
All animals were handled following the ‘Principles for the Utilization and Care of Vertebrate Animals’ and the ‘Guide for the Care and Use of Laboratory Animals’. Animal studies were approved by the IACUC of the Center for Experimental Animal Research (China) and Peking University Laboratory Animal Center (IACUC No. LSC-ZhengX-2-1). The number of production license for laboratory animals is SYXK-2019-0032. Female BALB/c normal mice (6–8 weeks of age) were purchased from Beijing Vital River Laboratory Animal Technology. The 4T1 murine breast cancer cells (stable clones expressing shOTUB1, PD-L1, shOTUB1/PD-L1, and control plasmid) were injected subcutaneously into the right armpits of BALB/c mice (32 mice were divided into four groups randomly, 1 × 106 cells per mouse). Tumor size was measured every 2 days using a caliper, and tumor volume was calculated using the formula: volume = length × width2/2. Mice were sacrificed when the sizes of tumors reached 1000 mm3, or when ulceration is found. Tumor tissues were analyzed by immunohistochemistry to determine PD-L1 expression, tumor cell proliferation (Ki67 staining), and infiltration of CD8+ cytotoxic T cells. ELISA assays were performed to detect the abundance of IFN-γ in mice serum.
Cell culture
HEK293T, MDA-MB-231, HepG2, A549, and MCF-7 cells were cultured in DMEM supplemented with 10% fetal bovine serum (Gibco, USA). BT-549 and RKO cells were maintained in RPMI 1640 (Gibco, USA). MDA-MB-436, HCC1954, HCC1937, T47D, Hs578T, ZR-75-1, SKBR3, B16-F10, CT-26, and BT20 cell lines were from the American Type Culture Collection (ATCC) and maintained in DMEM media. 4T1 murine cell line was a kind gift from Dr. Jiadong Wang (Peking University). The identities of cell lines were authenticated by short tandem repeat analysis. Cell lines were confirmed to have no mycoplasma contamination by PCR analysis.
Antibodies, reagents, and plasmids
The antibodies and reagents used in this study are listed in the Supplementary Materials. Plasmids expressing human deubiquitinase OTU subfamily members were kindly provided by Prof. Lingqiang Zhang from the Beijing Institute of Radiation Medicine. Signal peptide (SP)-HA-PD-L1 plasmid, in which the HA tag was inserted after the signal peptide sequence, was provided by Prof. Jinfang Zhang from Wuhan University [33], and the SP-HA-ΔC-tail truncation and SP-HA-5KR mutant of PD-L1 were constructed in our laboratory. All SP-HA-PD-L1 plasmids used in the article were represented by HA-PD-L1, HA-PD-L1-ΔC-tail, and HA-PD-L1-5KR. Mouse PD-L1/CD274 cDNA clone (MC201908) was purchased from OriGene (USA). Full-length, 1-85 aa, 47-271 aa, C91S, D88A, and ASA (D88A/C91S/H265A) OTUB1 were cloned into the pcDNA3.0-3flag vector. His-tagged OTUB1 was cloned into the pET28a vector. All constructs were confirmed by DNA sequencing.
In vitro deubiquitination assays
HEK293T cells were co-transfected with K48- or K63- specific linkage His-ubiquitin and HA-tagged PD-L1. Before harvest, cells were treated with MG132 to increase the level of ubiquitinated PD-L1. Subsequently, immunoprecipitation was performed to obtain ubiquitinated PD-L1, which was enriched using anti-HA antibodies and protein G beads. The beads were washed three times using deubiquitinating buffer (60 mM HEPES, 5 mM MgCl2, 4% glycerol, pH 7.6) and incubated with recombinant His-OTUB1 proteins (purified from E. coli) at 30 °C for 4 h. The mixtures were boiled in 2× SDS loading buffer and analyzed by immunoblot analysis.
Tandem ubiquitin-binding entity (TUBE) pull-down analysis
To enrich endogenous ubiquitinated proteins, we performed TUBE pull-down analyses using agarose-TUBEs (UM402) and K48-TUBE-HF (UM607) (Life Sensors). Briefly, cells were treated with MG132 (10 µM) overnight and harvested in TUBE lysis buffer (50 mM Tris HCl, 0.15 M NaCl, 1 mM EDTA, 1% NP-40, 10% glycerol, pH 7.5) or K48-TUBE lysis buffer (100 mM Tris HCl, 0.15 M NaCl, 5 mM EDTA, 1% NP-40, pH 8.0) containing additional inhibitors, 1,10-phenanthroline (o-PA) (5 mM), N-Ethylmaleimide (NEM) (5 mM), and PR-619 (100 µM). Cell lysates were then centrifuged to remove cell debris and incubated with agarose-TUBEs or K48-TUBE-FLAG at 4 °C for 2 h. The agarose-TUBEs beads were washed with TBST three times and boiled with 2×SDS loading buffer. To enrich FLAG-tagged TUBEs, we next added clarified cell lysate to equilibrated FLAG M2 Affinity Resin (A2220, Sigma-Aldrich) and incubated for 4 h with rotation. Flag M2 beads were collected and washed with Wash Buffer 1 and 2 and then boiled with 2×SDS loading buffer. All samples were analyzed by immunoblotting and visualized using the Odyssey Infrared Imaging System (LI-COR Biosciences, USA).
Duolink proximity ligation assay (PLA)
To detect the interaction between OTUB1 and PD-L1, we used the Duolink® In Situ PLA® kit (DUO92101, Sigma-Aldrich). MDA-MB-231 cells were fixed in 4% paraformaldehyde for 10 min at room temperature and subsequently blocked using 1× blocking solution. Cells were then incubated with primary antibodies targeting OTUB1 and PD-L1 at 4 °C overnight, followed by incubation with PLA probes at 37 °C for 1 h. After washing three times, the ligation-ligase solution was added and incubated at 37 °C for 30 min. Next, slides were incubated with amplification-polymerase solution at 37 °C in the dark for 100 min. Finally, cells were stained with Mounting Medium containing DAPI. Fluorescence images were obtained under a confocal laser scanning microscope (Zeiss LSM 710) using a 63× oil objective lens.
Endoplasmic reticulum enrichment and cell fractionation
The extraction of ER was performed by using an ER Enrichment kit (NBP2-29482, Novus Biologicals). Briefly, cells were washed with ice-cold PBS and harvested in a centrifuge tube with isosmotic homogenization buffer containing protease inhibitors. The cell extracts were lysed in the supplied buffer using a glass Dounce homogenizer with 30 strokes and centrifuged at 1000 g for 10 min to remove nuclei and cell debris. The supernatant was further centrifuged at 12000 g for 15 min to discard mitochondria, and the rough ER fraction was then precipitated in CaCl2 buffer and obtained by centrifugation of 8000 g for 10 min. The fractionation of MDA-MB-231 cells was performed using the Cell Fractionation Kit (#9038, Cell Signaling Technology). Typically, approximately 5 × 106 MDA-MB-231 cells were collocated in 0.5 mL PBS, and 100 µL of the cell suspension was used to prepare the Whole Cell Lysate (WCL). The remaining 400 µL were centrifuged, and the cell pellet was resuspended in 500 µL of CIB Buffer and vortexed before further centrifugation. The supernatant was saved as the Cytoplasmic Fraction, and the pellet was resuspended in 500 µL of MIB buffer to separate the Membrane Fraction. The Nuclear Fraction was obtained by addition of 250 µL CyNIB Buffer. All cell fractions were boiled with 2×SDS loading buffer and analyzed by immunoblotting.
PD-1 binding assay
The binding of PD-1 to PD-L1 was measured following procedures described previously [17]. MDA-MB-231 cells stably expressing shControl and shOTUB1 were seeded into 6-well plates and then fixed in 4% paraformaldehyde, followed by incubation with 5 µg/ml recombinant human PD-1 Fc chimera protein (1:100, R&D Systems). After washing with PBS, cells were subsequently incubated with anti-human IgG/Alexa Fluor 488 dye (1:100, Bioss) at room temperature for 1 h and stained with Mounting Medium containing DAPI (Zsbio). The green fluorescent signal was visualized using a confocal microscope (Zeiss LSM-710 NLO and DuoScan, Germany), and signal intensity was calculated using the ImageJ software.
T cell-mediated tumor cell killing
The procedure was performed as previously described [19, 34]. Briefly, MDA-MB-231 cells stably expressing shControl or shOTUB1 were seeded into 12-well plates at a density of 105 cells per well. Human PBMCs were activated in the presence of 2 µg/ml CD3 antibody and 1 µg/ml CD28 antibody (BioLegend) for 24 h. Then, activated PBMCs were added to the culture medium of MDA-MB-231 cells at a ratio of 5:1. At 72 h after co-incubation, tumor cells were collected and stained with Annexin V and propidium iodide (Beyotime) for FACS analysis of apoptotic rate.
Ki67 staining and TUNEL fluorescence assay
After co-culture with MDA-MB-231 shControl or shOTUB1 cells, PBMCs were collected and adhered to glass slides using a Thermo Scientific Cytospin 4 Cytocentrifuge, followed by incubation with Ki67 antibody or TUNEL reaction mixture (C1086, Beyotime). Nuclear DNA was stained with DAPI. Images were visualized with a confocal microscope using a × 63 oil objective lens. The percentage of Ki67 or TUNEL positive staining cells were analyzed using ImageJ software.
Immunohistochemistry (IHC) and histopathological analyses
Human breast cancer tissue arrays containing 90 cancer specimens were purchased from Shanghai Biochip Company Ltd. (Shanghai, China). The slides were stained with rabbit anti-PD-L1 (1:100) and mouse anti-OTUB1 (1:200) at 4 °C overnight, followed by incubation with secondary antibodies and visualization using 3′3′-diaminobenzidine tetrahydrochloride as the substrate. The negative control was prepared identically without the primary antibody. All immunostainings were evaluated blindly by pathologists from Peking University Health Science Center based on the histochemical score. The intensities of PD-L1 and OTUB1 were classified as follows: 0, no staining; 1, weak reactivity; 2, moderate reactivity; 3, strong reactivity; 4, very strong reactivity. Images were obtained using a Leica DM IRE2 microscope.
Bioinformatic analyses of PD-L1 and OTUB1 expression
To analyze the expression of PD-L1 and OTUB1 in human breast cancers, we downloaded an expression dataset of TCGA BRCA cancer type, based on the gencode v23 gene model, from UCSC Xena (http://xena.ucsc.edu/). The clinical data was downloaded from the GDC Data Portal (https://gdc-portal.nci.nih.gov/). The expression unit was TPM (transcript per million). We applied one-way analysis of variance (ANOVA) using disease state (Tumor or Normal) as the variable for calculating differential expression. The expression data for differential analysis were first log2(TPM+1) transformed. We defined the differentially expressed genes as those satisfying following thresholds: p (disease state) < 0.01.
Statistical analyses
All statistical results are reported as the mean ± SEM of three or more independent biological replicates. Unless stated otherwise, comparisons were performed with a two-tailed Student’s t test, and P values < 0.05 were considered to be statistically significant (variance is similar between the groups) (*P < 0.05, **P < 0.01, ***P < 0.001). Correlations were performed using the Pearson correlation test. The overall survival curves of patients were drawn by the Kaplan–Meier method and the difference was determined using a log-rank test. For every figure, statistical tests are justified as appropriate. Analyses and graphical presentation were performed using the GraphPad Prism 8.0 software.
Results
OTUB1 maintains PD-L1 protein stability
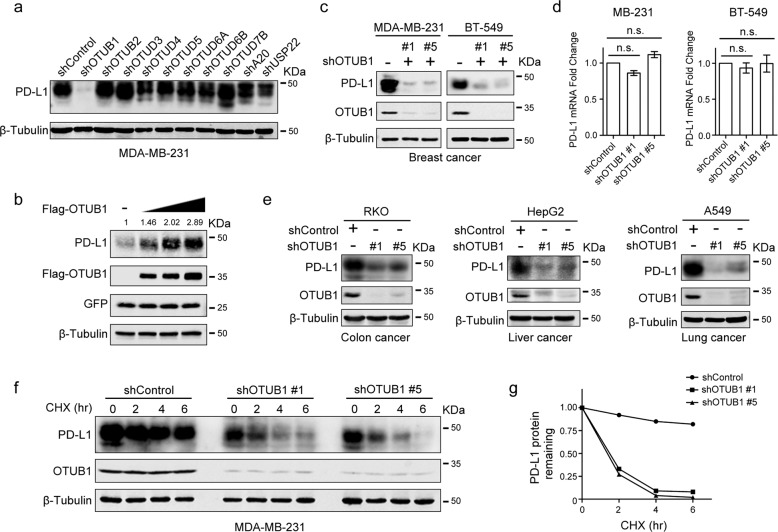
PD-L1 levels are commonly regulated by the ubiquitin proteasome system, we wanted to identify OTU family deubiquitinases (DUBs) that target PD-L1. MDA-MB-231 cells were infected with viruses harboring distinct shDUBs to examine whether the PD-L1 protein level could be affected by DUB depletion. As shown in Fig. 1a, OTUB1, but not other members of the indicated OTU subfamily, was able to positively regulate PD-L1, but shRNAs targeting some USP family members had no effect on PD-L1 abundance (Fig. S1).
Fig. 1. OTUB1 maintains PD-L1 protein stability.
a Screening of OTU family members that regulate PD-L1 protein level. MDA-MB-231 cells individually were infected with shRNAs targeting different OTU subfamily DUBs, and the expression of PD-L1 was detected. b Immunoblotting of PD-L1 expression in HEK293T cells transfected with increasing amounts of Flag-OTUB1 together with a constant amount of HA-PD-L1 plasmid in each group. GFP expression was used to confirm the constant transfection efficiency across the four experimental groups. c Immunoblot analysis of PD-L1 in MDA-MB-231 and BT-549 cells infected with shOTUB1- or shControl-encoding lentivirus, respectively. d The qRT-PCR analyses of PD-L1 mRNA expression in MB-MDA-231 and BT-549 cells stably expressing shControl or shOTUB1. Data are shown as the mean ± SEM (n = 3). Statistical analysis was performed using Student’s t test. n.s., not significant. e Immunoblotting of PD-L1 expression in RKO, HepG2, and A549 cells after introducing shControl or shOTUB1 lentiviruses. f Half-life analysis of PD-L1 in shControl or shOTUB1 MDA-MB-231 cells treated with 50 µg/mL cycloheximide for the indicated times before harvesting. g Semi-quantification of PD-L1 levels, with β-tubulin used as a loading control. Relative PD-L1 levels at time 0 were set as 1.
We thus further investigated the relationship of the deubiquitinase OTUB1 with PD-L1. Overexpression of OTUB1 significantly increased PD-L1 abundance in a dose-dependent manner (Fig. 1b), while OTUB1 knockdown strongly decreased PD-L1 protein levels in both MDA-MB-231 and BT-549 cells (Fig. 1c). Additionally, OTUB1 depletion did not affect the abundance of PD-L1 mRNA (Fig. 1d). The effects of OTUB1 on PD-L1 protein levels were also found in other human tumor cell lines such as colon cancer cell line RKO, liver cancer cell line HepG2, and lung cancer cell line A549 (Fig. 1e). Next, we performed half-life analysis to validate the role of OTUB1 in the regulation of PD-L1 stability and found that the half-life of PD-L1 was shortened in OTUB1-depleted cells (Fig. 1f, g). Together, we identify OTUB1 as a specific positive regulator to maintain high PD-L1 levels in different cancer cells. As a deubiquitinase, OTUB1 likely stabilizes PD-L1 through post-translational modification.
OTUB1 specifically interacts with PD-L1 in vivo and in vitro
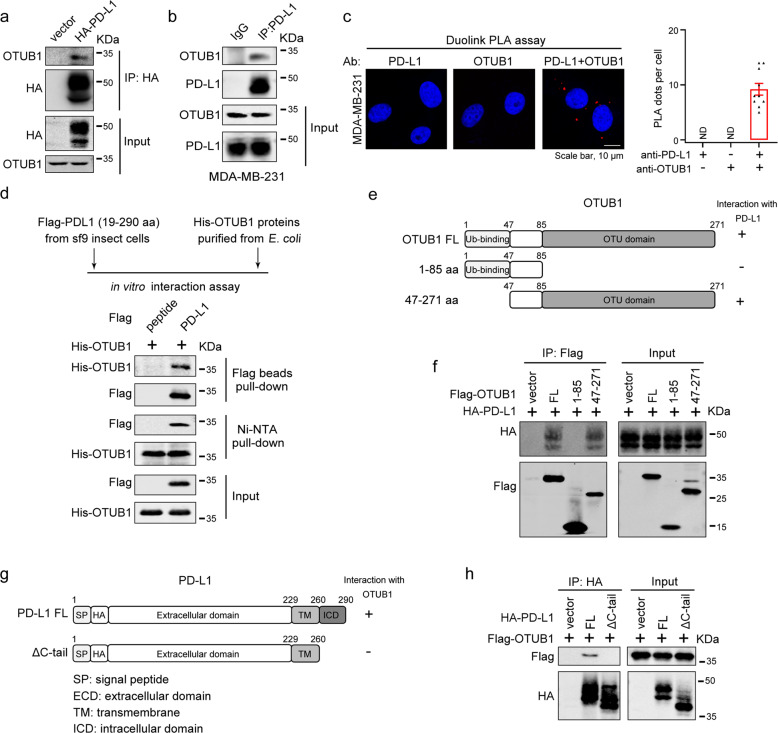
To further illustrate the mechanism underlying OTUB1-mediated PD-L1 regulation, we utilized HA-PD-L1 plasmid with signal peptide inserted before HA-tag that has been proved to localize correctly to the plasma membrane (Fig. S2) to perform co-IP assays. The result showed that endogenous OTUB1 could bind to exogenously expressed HA-PD-L1 (Fig. 2a). Moreover, their fully endogenous association was confirmed in MDA-MB-231 cells under normal physiological conditions, and the in situ PD-L1-OTUB1 interaction was observed using a Duolink PLA assay (Fig. 2b, c). We also demonstrated a direct association between recombinant Flag-PD-L1 19-290 aa and His-OTUB1 proteins in vitro under cell-free conditions (Fig. 2d).
Fig. 2. OTUB1 specifically interacts with PD-L1 in vivo and in vitro.
a HEK293T cells were transfected with HA-PD-L1 or HA vector, and immunoprecipitation was performed with anti-HA antibody to examine the interaction between HA-PD-L1 and endogenous OTUB1. b Association between endogenous OTUB1 and PD-L1 in MDA-MB-231 cell lines was detected by Co-IP assays using IgG or PD-L1 antibodies. c In situ interaction between OTUB1 and PD-L1. Cells were fixed with 4% paraformaldehyde, immunostained with OTUB1 and PD-L1 antibodies and then assessed using the Duolink PLA assay. Scale bar, 10 µm. Quantification of the PLA dots indicating PD-L1-OTUB1 interactions was shown as mean ± SEM. d The direct interaction between recombinant Flag-PD-L1 19-290 aa proteins and His-OTUB1 proteins examined by in vitro pull-down assays. e Schematic representation of various OTUB1 truncations. f Mapping of OTUB1 domains critical for PD-L1 binding. HEK293T cells were transfected with different OTUB1 truncations, and cell lysates were immunoprecipitated with anti-Flag antibody to detect their PD-L1 binding ability. g Schematic representation of PD-L1 full-length and ΔC-tail constructs. h The interaction between Flag-OTUB1 and PD-L1 full-length or ΔC-tail detected by Co-IP assays.
Based on the above observations, we sought to identify which OTUB1 regions are critically required for its interaction with PD-L1. We generated two OTUB1 truncations (Fig. 2e), and through Co-IP assays, we found that the C-terminal OTU domain of OTUB1 mediated its association with PD-L1 (Fig. 2f). On the other hand, we constructed a plasmid expressing PD-L1 lacking its C-terminal tail (ΔC-tail) (Fig. 2g) and found that deletion of the C-tail of PD-L1 disrupted its binding to OTUB1, suggesting that the intracellular region of PD-L1 was necessary for its binding to OTUB1 (Fig. 2h). Collectively, our data show that OTUB1 positively regulates PD-L1 stability through direct interaction with PD-L1.
OTUB1 stabilizes PD-L1 through cleaving K48-linked poly-ubiquitin chains of PD-L1
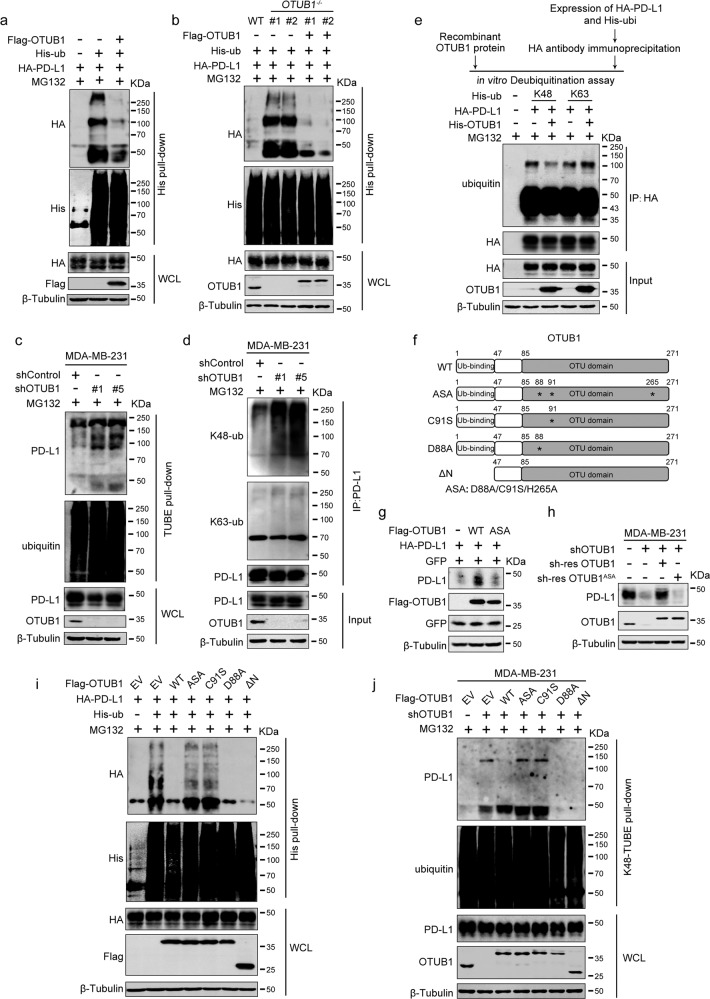
To determine if OTUB1 stabilized PD-L1 via deubiquitination, we performed His-ubiquitin pull-down to examine the level of PD-L1 ubiquitination. As expected, OTUB1 overexpression remarkably reduced PD-L1 ubiquitination in vivo (Fig. 3a). Subsequently, we generated OTUB1−/− HEK293T cells (Fig. S3a, b) and assessed the effect of OTUB1 deletion on PD-L1 ubiquitination. Compared to wild-type OTUB1 cells, PD-L1 polyubiquitination dramatically increased in OTUB1−/− cell. Consistently, re-expression of Flag-OTUB1 recovered its inhibitory effect on PD-L1 ubiquitination levels (Fig. 3b). To enrich endogenous ubiquitinated protein, we carried out TUBE pull-down experiments and confirmed the effect of depleted OTUB1 on the level of endogenous PD-L1 ubiquitination (Fig. 3c). These results demonstrated that OTUB1 specifically deubiquitinates PD-L1 in vivo.
Fig. 3. OTUB1 stabilizes PD-L1 through cleaving K48-linked poly-ubiquitin chains of PD-L1.
a HA-PD-L1 ubiquitination levels in HEK293T cells transfected with Flag-vector or Flag-OTUB1 were analyzed by His pull-down assay. b The effect of PD-L1 ubiquitination by depleted OTUB1 was examined in OTUB1WT, OTUB1-/- and re-expressed OTUB1 HEK293T cells by His pull-down assay. Two OTUB1-/- strains were obtained through CRISPR/Cas9 genome editing. c The shControl- and shOTUB1- MDA-MB-231 cells were subjected to TUBE pull-down assays to examine the effect of depleted OTUB1 on endogenous ubiquitination levels of PD-L1. d The influence of OTUB1 on specific ubiquitination types of endogenous PD-L1 was determined by immunoblotting analysis using the K48- or K63- ubiquitin linkage-specific antibodies. MDA-MB-231 cells infected with shControl- or shOTUB1-encoding lentiviruses were treated with MG132 overnight and then subjected to immunoprecipitation. e In vitro deubiquitination assays of recombinant OTUB1 proteins and enriched K48-linked or K63-linked ubiquitinated PD-L1 from cell extracts. The mixture was incubated at 30 °C for 4 h and then analyzed by immunoblotting. f Schematic representation of the OTUB1 C91S, D88A, ASA (D88A/C91S/H265A) mutants and ΔN truncation. Stars represent mutated amino acids in the OTU domain. g Examination of the effect of wild-type OTUB1 or ASA mutant on PD-L1 abundance. GFP expression was used to confirm constant transfection efficiency across the three experimental groups. h Determination of PD-L1 levels after re-expression of the indicated OTUB1 mutants. The shOTUB1 MDA-MB-231 cells were infected with lentiviruses stably expressing shRNA-resistant wild-type OTUB1 or ASA mutant and then subjected to immunoblotting of PD-L1. i Immunoblot analysis of PD-L1 ubiquitination affected by wild-type OTUB1, ASA, C91S, D88A, or ΔN truncation mutants. HEK293T cells were transfected with HA-tagged PD-L1 and His-ubiquitin along with different OTUB1 mutants. Following MG132 treatment, cell extracts were enriched by Ni+-NTA agarose and then prepared for immunoblotting with the indicated antibodies. j Detection of endogenous K48-linked polyubiquitination of PD-L1 affected by OTUB1. MDA-MB-231 cells with shControl or shOTUB1 and OTUB1-depleted cells with re-overexpressed wild-type or mutation constructs of Flag-OTUB1 were subjected to K48-TUBE pull-down analyses.
Next, we wanted to determine whether OTUB1 could remove K48-linked ubiquitin chains of PD-L1. OTUB1 depletion in MDA-MB-231 cells promoted endogenous K48-linked ubiquitination of PD-L1, but had no effect on K63-linked ubiquitination (Fig. 3d). Moreover, in vitro deubiquitination assay in a cell-free system further confirmed that OTUB1 was able to remove K48-linked ubiquitin chains from PD-L1 directly (Fig. 3e). These data indicate that OTUB1 stabilizes PD-L1 by directly cleaving K48-linked poly-ubiquitin chains and protecting it from proteolytic degradation.
OTUB1 catalytic activity is required for its regulation of PD-L1
Generally, OTUB1 mediates the deubiquitination process via two different mechanisms. OTUB’s N-terminal ubiquitin-binding motif interacts with and inhibits E2 ubiquitin-conjugating enzymes, and the C-terminal OTU domain is responsible for its catalytic deubiquitinase activity [35]. The catalytic triad of OTUB1 is composed of the D88, C91, and H265 residues in the OTU domain [36], however, recent studies have shown that the C91S alone is a catalytically dead mutant, while the D88A mutation alters the E2 suppressing activity of OTUB1 [37, 38]. To uncover how OTUB1 deubiquitinated PD-L1, we constructed the catalytically inactive OTUB1 C91S and ASA (D88A/C91S/H265A) mutants, the non-canonical activity mutant D88A, and an OTUB1 ΔN truncation in which the ubiquitin-binding motif was deleted (Fig. 3f). We first observed that PD-L1 expression was affected by wild-type OTUB1, while the ASA mutant had no obvious effect (Fig. 3g). Next, the results of rescue experiments revealed that the expression of shRNA-resistant wild-type OTUB1 but not the ASA mutant restored PD-L1 expression (Fig. 3h).
To ensure that the inability of the catalytically inactive OTUB1 mutants to enhance PD-L1 expression was due to its impaired deubiquitination capability, we performed His-ubiquitin pull-down assays and found that overexpression of wild-type OTUB1 largely abolished the ubiquitination of PD-L1, similar to the effect observed for the OTUB1 D88A and ΔN truncation, while the catalytically inactive mutants, C91S and ASA, failed to remove ubiquitin chains from PD-L1 (Fig. 3i). Furthermore, K48-TUBE pull-down assays verified that the catalytic activity of OTUB1 is necessary to decrease endogenous K48-linked ubiquitin chains of PD-L1 (Fig. 3j). In summary, our results indicate that OTUB1 relies on its catalytic activity to regulate PD-L1 abundance.
OTUB1 regulates PD-L1 abundance through the ERAD pathway
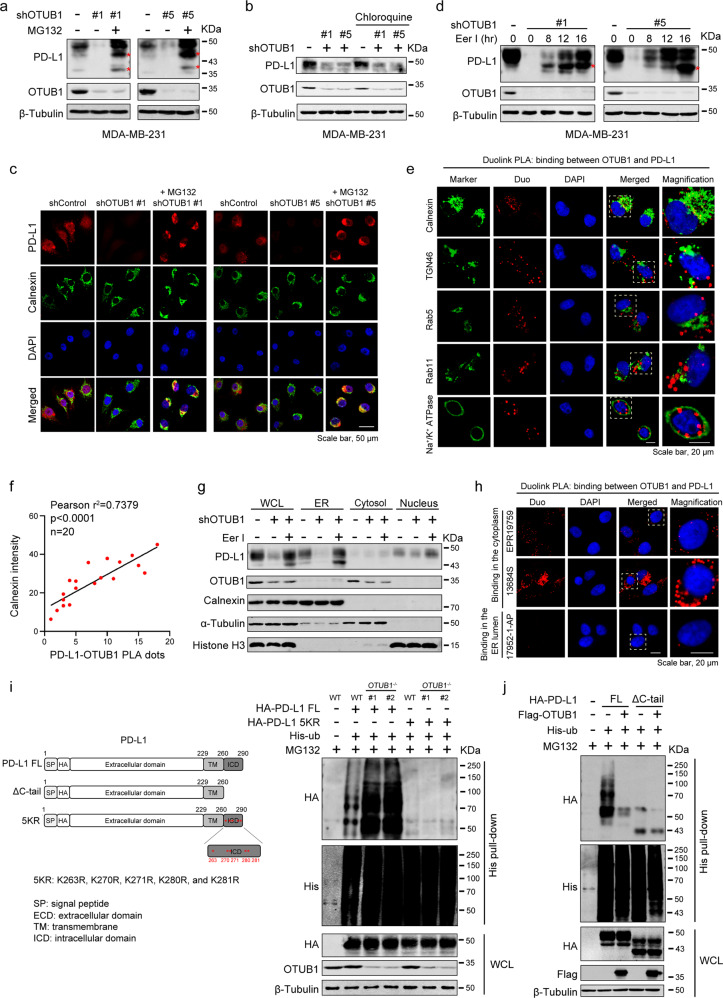
Membrane proteins in eukaryotes are mainly degraded via the following three distinct pathways: ERAD (endoplasmic reticulum-associated degradation) through the proteasome, ESCRT (endosomal sorting complexes required for transport) and EGAD (Golgi-associated degradation) through the lysosome [39]. As described above, OTUB1 promotes PD-L1 stability by regulating its K48-linked ubiquitination. To further explore the cellular location where OTUB1 impeded PD-L1 degradation, we treated OTUB1 knockdown cells with the proteasome inhibitor MG132 for a short period of time (10 µM, 4 h) and found that inhibition of PD-L1 stability by OTUB1 depletion was clearly blocked by MG132 treatment (Fig. 4a). Meanwhile, we observed bands for incomplete glycosylated PD-L1 lower than 50 KDa in the presence of MG132, whereas in cells treated with the lysosomal inhibitor chloroquine, PD-L1 abundance remained unchanged (Fig. 4b), indicating that OTUB1 regulates PD-L1 protein levels in the early stage of protein synthesis through the proteasome degradation pathway. By immunofluorescence experiments, we also found that the restored PD-L1 after MG132 treatment accumulated in the cytoplasm, which is colocalized with the endoplasmic reticulum (ER) membrane protein calnexin (Fig. 4c). Consistently, addition of eeyarestatin I (Eer I), a specific inhibitor of ERAD [40], to shOTUB1 MDA-MB-231 cells significantly replenished PD-L1 expression (Fig. 4d), suggesting that OTUB1 regulates PD-L1 in the ER.
Fig. 4. OTUB1 regulates PD-L1 abundance through ERAD pathway.
a The MDA-MB-231 cells harboring shControl or shOTUB1 treated with or without 10 µM MG132 for 4 h were collected to analyze PD-L1 expression. Red stars represent incomplete glycosylated PD-L1. b Immunoblot analysis of PD-L1 in shControl or shOTUB1 MDA-MB-231 cells treated with or without the lysosomal inhibitor chloroquine (20 µM). c MDA-MB-231 cells with shControl and shOTUB1 treated with MG132 were immunostained with a PD-L1 antibody (red), a calnexin antibody (green), and a DAPI antibody (blue). Scale bar, 50 µm. d The shOTUB1 MDA-MB-231 cells were treated with the ERAD pathway inhibitor Eer I (10 µM) at the indicated times for immunoblot detection of PD-L1 expression. Red stars represent incomplete glycosylated PD-L1. e MDA-MB-231 cells were subjected to immunofluorescence assays with calnexin (ER), TGN46 (Golgi), Rab5 (endosome), Rab11 (recycling endosome), and Na+/K+ ATPase (plasma membrane) antibodies following Duolink PLA to identify the location of OTUB1 interaction with PD-L1. f The positive correlation between PD-L1-OTUB1 PLA dots and the fluorescence intensity of ER marker calnexin was shown in scatterplot. Pearson’s coefficient tests were performed to assess statistical significance (Pearson r2 = 07379, p < 0.001, n = 20). g The presence of PD-L1 and OTUB1 in different fractions was analyzed by immunoblotting, using antibodies against PD-L1 and OTUB1, the ER protein calnexin, cytosolic α-Tubulin, as well as the nuclear protein histone H3. h The in situ binding between PD-L1 and OTUB1 was detected by Duolink PLA immunofluorescence using two antibodies specific for the PD-L1 intracellular domain (ICD) (EPR19759 and 13684 S) and extracellular domain (ECD) (17952-1-AP). i Schematic representation of HA-PD-L1 ΔC-tail and 5KR constructs. OTUB1WT and OTUB1-/- HEK293T cells transfected with PD-L1 full-length or the 5KR mutant were subjected to His pull-down and SDS-PAGE analyses. j The effect of OTUB1 on the ubiquitination of full-length PD-L1 or ΔC-tail truncation was monitored by His-ubiquitin pull-down assay.
To confirm these results, we performed IF following Duolink PLA assays using antibodies targeting the ER protein calnexin, the Golgi marker TGN46, the endosome marker Rab5, the recycling endosome protein Rab11, and the cell membrane marker Na+/K+ ATPase to explore the subcellular location of OTUB1-PD-L1 binding. As shown in Fig. 4e, the association of OTUB1 with PD-L1 (red signals) was mainly colocalized with the ER marker calnexin. The number of PD-L1-OTUB1 PLA dots positively correlated with the fluorescence intensity of calnexin (Fig. 4f), while Pearson correlation analysis between PLA dots with other cell structure markers showed no significance (Fig. S4). In addition, the results of cell fractionation showed that Eer I treatment indeed rescued PD-L1 levels in the ER (Fig. 4g).
OTUB1 proteins were found to be mainly located in the cytosol (Fig. 4g). By performing PLA Duolink assay using antibodies specific for the extracellular domain (ECD) and the intracellular domain (ICD) of PD-L1, we found that OTUB in situ binds to the ICD of PD-L1 (Fig. 4h). PD-L1 is a transmembrane protein, and only the intracellular domain of ER-associated PD-L1 exposes to the cytosol, as OTUB1 interacts with the C-tail of PD-L1 (Fig. 2h). Therefore, we speculated that OTUB1 removes ubiquitin chains from the ICD region of PD-L1. We constructed a HA-PD-L1 5KR mutant to examine whether OTUB1 can still affect HA-PD-L1 5KR ubiquitination. In OTUB1-/- HEK293T cells, ubiquitination of wild-type HA-PD-L1 significantly increased while the 5KR mutant displayed almost no ubiquitination (Fig. 4i). Furthermore, we found that compared with the full-length PD-L1, the PD-L1 ΔC-tail was barely modified by ubiquitination, and OTUB1 no longer affected the ubiquitination of the PD-L1 ΔC-tail mutant (Fig. 4j). Taken together, these findings indicate that OTUB1 in the cytosol promotes PD-L1 protein stability by decreasing the K48-linked polyubiquitination of the PD-L1 intracellular domain and thus prevents its degradation through the ERAD pathway.
Depletion of OTUB1 impairs PD-L1 function in T cell-mediated cancer cell killing
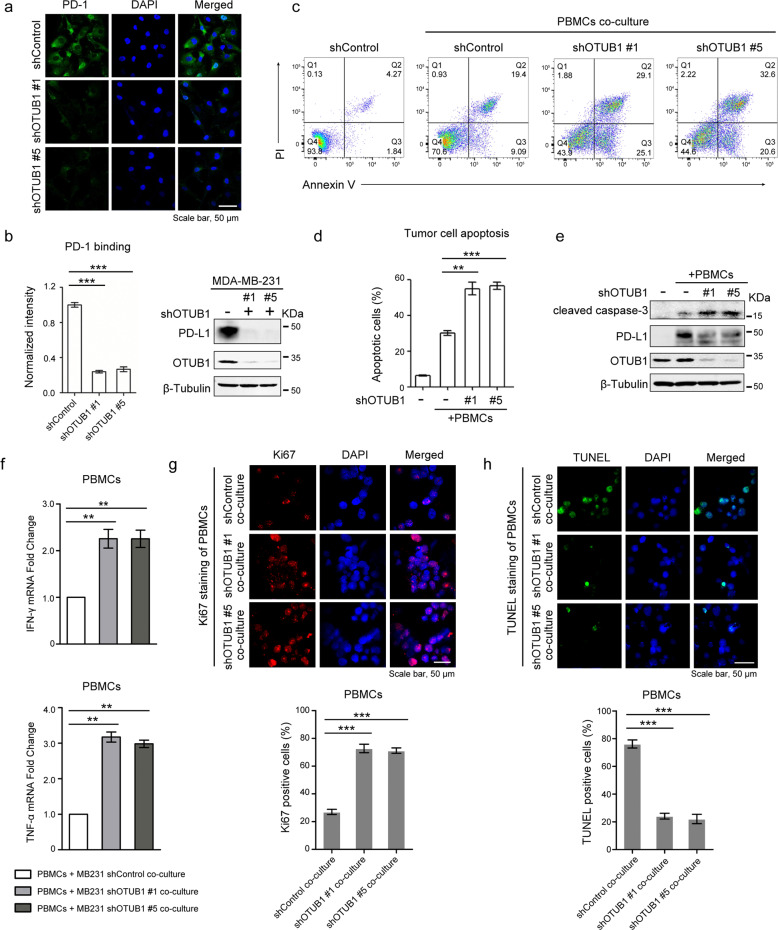
PD-L1 is known to regulate T cell tolerance through interaction with its receptor PD-1 [41]. As OTUB1 stabilizes PD-L1, we further tested whether OTUB1 mediates cancer immunosuppression via regulation of PD-L1. Immunofluorescence assays revealed that depletion of OTUB1 reduced PD-1 protein binding intensity to the tumor cell surface (Fig. 5a, b).
Fig. 5. Depletion of OTUB1 impairs PD-L1 function in T cell-mediated cancer cell killing.
a Immunofluorescence assays to detect PD-1 binding intensity. MDA-MB-231 cells stably expressing shControl or shOTUB1 were incubated with recombinant human PD-1 Fc protein and then anti-human Alexa Fluor 488 dye. Scale bar, 50 µm. b Normalized intensity of PD-1 binding and immunoblotting of OTUB1 expression to show knockdown efficiency. c FACS analysis of PBMCs-mediated killing of tumor cells using Annexin V and propidium iodide (PI) double staining. Healthy cells are negative for both stains in the Q4 quadrant. The Q3 quadrant shows Annexin V-positive cells, which are in the early stage of apoptosis. The Q2 quadrant shows cells that are both Annexin V- and PI-positive, which are in the late stage of apoptosis. d Quantitative apoptotic measurement of Q2 and Q3 quadrants. Greater than 104 cells were counted for each group. e Immunoblotting of cleaved caspase-3 levels in shControl or shOTUB1 MDA-MB-231 cells after incubation with activated PBMCs. f The qRT-PCR analyses of IFN-γ and TNF-α mRNA expressions in PBMCs after co-culture with MDA-MB-231 shControl or shOTUB1 cells. g, h The Ki67 or TUNEL staining of PBMCs after co-culture with MDA-MB-231 shControl or shOTUB1 cells. The percentage of Ki67 or TUNEL positive staining cells were analyzed using the ImageJ software. The results of b, d, f, g and h are shown as the mean ± SEM (n = 3). Statistical analysis was performed using Student’s t test. **p < 0.01, ***p < 0.001.
We also performed T cell cytotoxicity assays to further assess whether OTUB1 influences the function of PD-L1 in T cell-mediated cancer cell killing. As shown in Fig. 5c, d, OTUB1 knockdown increased the apoptotic ratio and caused cells to become more sensitive to human PBMCs-mediated cytolysis. To verify this result, we harvested a small fraction of cell lysates for immunoblotting and found that OTUB1-depleted cells exhibited higher expression of active caspase-3 proteins (Fig. 5e). Moreover, co-culture with PBMCs increased PD-L1 expression, which was significantly reduced in the shOTUB1-treated groups (Fig. 5e). To confirm the immune inhibition function of PD-L1 affected by OTUB1, we next compared the immunosuppressive effects of shOTUB1 tumor cells on PBMCs with that of shControl cells. As shown in Fig. 5f, OTUB1 ablation attenuated the tumor cell-induced immune suppression activity, as the levels of IFN-γ and TNF-α expression increased significantly. Meanwhile, co-culture of PBMCs with MDA-MB-231 shOTUB1 cells resulted in higher Ki67 staining (Fig. 5g) and lower TUNEL ratio (Fig. 5h) of PBMCs, suggesting that OTUB1 in tumor cells promotes the inhibition of immune cells. Taken together, these results indicate that OTUB1 depletion-mediated PD-L1 destabilization reduces the immunosuppressive function of PD-L1.
OTUB1 silencing-mediated downregulation of PD-L1 promotes antitumor immunity
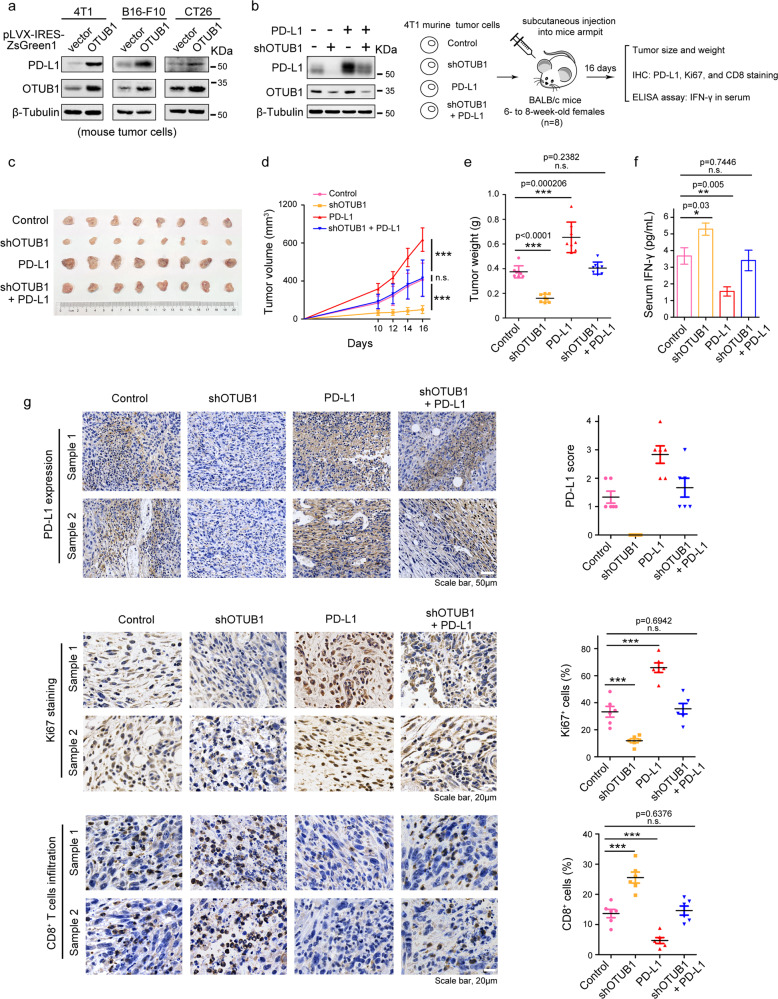
To further elucidate the effect of OTUB1 on PD-L1 in mice, we generated 4T1 murine breast cancer cells stably expressing pLVX-IRES-ZsGreen1-OTUB1 and found that OTUB1 substantially elevated PD-L1 protein expression, as well as in murine B16-F10 melanoma cells, and CT26 colon cancer cells (Fig. 6a).
Fig. 6. OTUB1 silencing-mediated downregulation of PD-L1 promotes antitumor immunity.
a Immunoblot analysis of PD-L1 in murine 4T1, B16-F10, and CT26 tumor cells with overexpressed OTUB1. b Experimental design for the in vivo tumorigenesis assay and the expression levels of OTUB1 and PD-L1 in four different 4T1 cell types (shOTUB1, PD-L1, shOTUB1/PD-L1, and control) were confirmed by immunoblot analysis. c Photographs of mice tumors of each group (n = 8) at the end of the experiment. d The curve graph exhibited the tumor growth measured at different time-points after inoculation (n = 8). e The image shows the tumor weight for each group (n = 8). f The abundance of IFN-γ (pg/mL) in mice serum (n = 5) detected by ELISA assays. g IHC of PD-L1 expression (Scale bar, 50 µm), Ki67 staining (Scale bar, 20 µm), and CD8+ cell infiltration (Scale bar, 20 µm) in tumor sections (n = 6 for each group). Statistical results are quantified by ImageJ software. Data in d, e, f, and g are presented as the mean ± SEM in each group, and statistical analysis was performed using Student’s t test. n.s., not significant, *p < 0.05, **p < 0.01, ***p < 0.001.
To clarify the role of OTUB1 in cancer cell immunosuppression through regulation of PD-L1 in vivo, 4T1 cells with stably expressed shOTUB1, PD-L1, shOTUB1/PD-L1, and control plasmids, as confirmed in Fig. 6b, were injected subcutaneously into the right armpits of BALB/c mice, respectively (8 mice each group, Fig. 6b). At Day10 after inoculation, we started to measure tumor volume every 2 days until the sizes of tumors approached 1000 mm3 (Day16). As expected, in comparison to the control group, overexpressed PD-L1 resulted in a substantial increase of tumor growth, whereas depletion of endogenous OTUB1 significantly attenuated both the tumor size and tumor weight in vivo. We also found that the knockdown effect of OTUB1 could be compensated for PD-L1 expression (Fig. 6c–e). To further confirm that the inhibition of tumor growth is attributed to enhanced T-cell activation affected by depleted OTUB1, we collected mice serum to detect the abundance of IFN-γ by ELISA assays. As shown in Fig. 6f, IFN-γ, an indicator of activated immunity that is primarily secreted by T cells, was significantly increased in serum of mice with depleted OTUB1. To determine the expression of PD-L1, Ki67, and CD8 in tumor specimens, tumor samples from six mice in each group were collected and IHC staining was performed. Consistently, the tumor-bearing mice induced with depleted OTUB1 showed great reductions of cell proliferation marker Ki67 and increased numbers of CD8+ tumor-infiltrating lymphocytes. In contrast, a rapid elevation of the positive Ki67 staining cells was found in tumors receiving overexpressed PD-L1, and a significant decrease in infiltration of CD8+ cells was also observed (Fig. 6g). These results indicate that OTUB1 knockdown causes PD-L1 degradation and thus activates cytotoxic T lymphocytes to promote antitumor effect. In the control and rescue (shOTUB1/PD-L1) groups, mice displayed the same percentage of positive Ki67 and CD8 staining. These results support the conclusion that shOTUB1 mediates the inhibitory effect on tumor growth and enhances antitumor immunity through regulating PD-L1 stability.
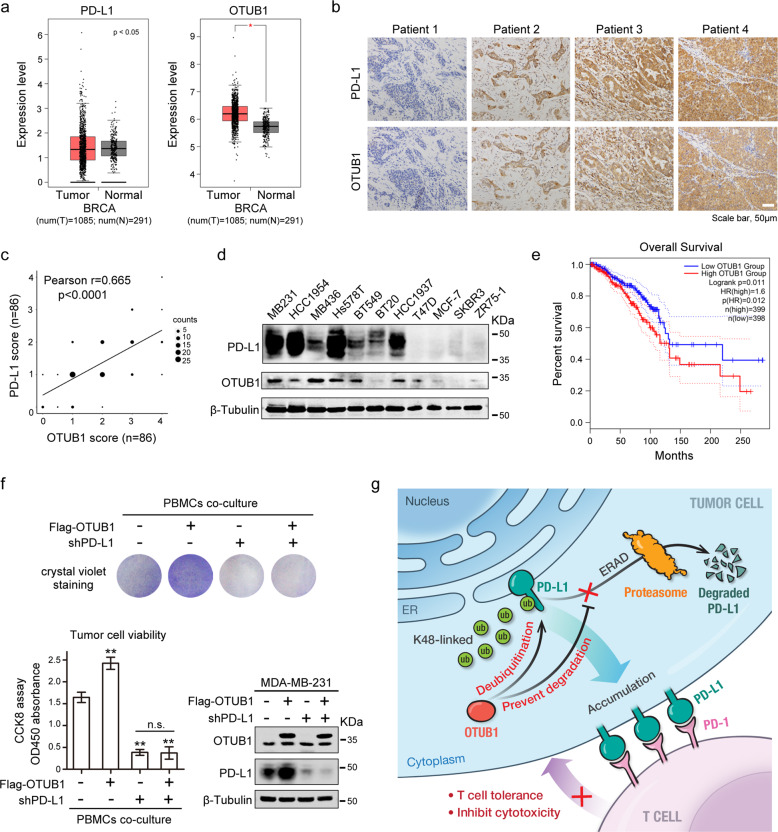
OTUB1 correlates with PD-L1 abundance in breast carcinoma
To further investigate the pathological relevance of OTUB1 and PD-L1 in breast cancer, we first extracted BRCA samples from TCGA dataset and compared the differential expressions of OTUB1 and PD-L1 between the tumor group and adjacent normal group. These bioinformatics analyses revealed that OTUB1 was more highly expressed in breast tumor tissue than in normal tissue while the expression level of PD-L1 showed no significant difference (Fig. 7a). We next assessed PD-L1 and OTUB1 protein expressions in human BRCA specimens by IHC. Representative images of low/no and high staining of PD-L1 and OTUB1 were shown in Fig. 7b. A quantitative analysis showed that PD-L1 protein levels were positively correlated with OTUB1 (Pearson r = 0.665, p < 0.0001) (Fig. 7c). Moreover, we measured PD-L1 and OTUB1 expression in a panel of human breast cancer cell lines and found that PD-L1 expression indeed displayed a relationship with OTUB1 abundance (Fig. 7d). We also performed overall survival analysis of patient samples in the TCGA BRCA database and found that patients with high levels of OTUB1 exhibited poor overall survival (Fig. 7e), suggesting that OTUB1 has crucial clinical significance and could be served as a biomarker for cancer diagnosis.
Fig. 7. OTUB1 correlates with PD-L1 abundance in breast carcinoma.
a Bioinformatic analyses of PD-L1 and OTUB1 expression in breast invasive carcinoma (BRCA) patients (n = 1085) relative to those of healthy controls (n = 291). b Representative images of IHC staining of PD-L1 and OTUB1 in human BRCA specimens. Scale bars, 50 µm. c Pearson correlation analysis to determine the degree of association between OTUB1 and PD-L1 by IHC staining (n = 86). P-value was obtained by Student’s t test. d Immunoblotting of PD-L1 and OTUB1 expression in the indicated human breast cancer cell lines. e Kaplan–Meier curve of overall survival of patients in the TCGA BRCA database. Patients were divided into the high OTUB1 group (n = 399) and the low OTUB1 group (n = 398). The difference in the overall survival between these two groups was determined using a log-rank test. f Crystal violet staining and CCK8 analysis to detect tumor cell viability after incubation with activated PBMCs. The results of CCK8 assay are shown as the mean ± SEM (n = 3) and statistical analysis was performed using Student’s t test (**p < 0.01). The expression levels of OTUB1 and PD-L1 in different MDA-MB-231 cell types were confirmed by immunoblot analysis. g Working model for regulation of PD-L1 stability and cancer cell immunosuppression by OTUB1.
Because OTUB1 depletion rendered tumor cells hyper-sensitive to human PBMCs-mediated killing (Fig. 5), we wanted to further confirm that OTUB1 modulates cancer cell immunosuppression through regulation of PD-L1. As shown in Fig. 7f, OTUB1 overexpression significantly enhanced tumor cell resistance to PBMCs-mediated cytotoxicity, but upon PD-L1 knockdown, we did not observe any appreciable differences between wild-type and OTUB1-overexpressing cells (Fig. 7f), suggesting that OTUB1 serves as an upstream regulator that influences cancer cell immune evasion via targeting PD-L1.
In summary, the positive correlation between OTUB1 and PD-L1 expression in clinical samples is consistent with our finding that OTUB1 functions as a deubiquitinating enzyme to maintain PD-L1 protein levels. In addition, we confirmed that OTUB1 mediates immune evasion via regulation of PD-L1.
Discussion
The levels of PD-L1 expression on tumor cells likely correlate with the clinical response and efficacy of anti-PD-1/PD-L1 therapies [42]. Hence, there is a significant need to understand the molecular mechanisms underlying regulation of PD-L1 expression. Recent reports have described different mechanisms to control PD-L1 abundance at the post-translational level, such as proteasome-mediated or lysosomal-mediated degradation [43, 44], while regulation of PD-L1 stabilization by the ubiquitin proteasome system is not fully understood. Here we found that OTUB1, a member of the OTU subfamily of deubiquitinases, stabilizes PD-L1 and consequently impairs cancer cell immunity. OTUB1 specifically interacts with PD-L1, directly cleaves the Lys 48-linked poly-ubiquitin chains on the ICD region, and thus prevents proteasomal degradation of ER-associated PD-L1. Stabilized PD-L1 proteins accumulate on the membrane of tumor cells, where they interact with the receptor PD-1 on T cell membranes. The PD-L1/PD-1 axis provides signals to inhibit T cell activity and thus contributes to cancer cell evasion of immune surveillance (Fig. 7g).
Previous studies demonstrated that USP9X [22] and USP22 [21] influence PD-L1 in oral squamous cell carcinoma or liver cancer, respectively, whereas our results proved that OTUB1-mediated PD-L1 regulation is not just working in breast cancer, the same effects are observed in RKO (colon cancer), HepG2 (liver cancer), and A549 (lung cancer) cells, suggesting that OTUB1 possesses more general roles in regulation of PD-L1 in various tumor cells. Meanwhile, knockdown of USP9X or USP22 by two different shRNAs in breast cancer only mildly reduces PD-L1 abundance, while OTUB1 depletion has the most significant impact (Fig. S5a). Previous study showed that CSN5 promotes PD-L1 in response to TNF-α or CCL-5 stimuli via the NF-κB pathway [19, 20]. In this study, we found that TNF-α treatment significantly promotes PD-L1 expression (Fig. S5b), which is consistent with previous findings; however, the OTUB1 protein level remains unchanged after incubation with TNF-α. In OTUB1 knockdown cells, TNF-α treatment still enhances PD-L1 expression levels, suggesting that OTUB1 functions differently from CSN5 as a novel deubiquitinase of PD-L1. The OTUB1 protein is frequently upregulated in many tumors [26] and the upstream signaling pathway regulating OTUB1-modulated PD-L1 abundance needs to be studied in the future.
So far, the effects of reported DUBs on PD-L1 expression and function are well characterized, but little is known about where these DUBs function in cells. The biogenesis of membrane proteins involves multiple transportation and modification steps along the ER-Golgi-plasma membrane axis [45]; the synthesized proteins can also be targeted for degradation at each step [39, 46]. Our findings here reveal that OTUB1 inhibits PD-L1 degradation through the ERAD pathway before it fully matures (Fig. 4a–c), which is different from CMTM6 that maintains PD-L1 protein at the plasma membrane through preventing recycling endosome- and lysosome- mediated pathway [43]. Consistent with previous studies that OTUB1 primarily localizes in the cytosol, we found that OTUB1 binds to and deubiquitinates the ICD region of ER-associated PD-L1 that is exposed to the cytosol (Figs. 3h and 4i, j). Recent studies have shown that mutation of PD-L1 glycosylation sites significantly increases its ubiquitination and speeds up degradation [17], indicating that there is an antagonistic relationship between glycosylation and ubiquitination of PD-L1. To investigate the physiological significance of OTUB1 in regulating the ER-associated PD-L1 protein level, we treated cells using tunicamycin, a glycosylation inhibitor, and found that tunicamycin promoted the binding of PD-L1 to OTUB1 (Fig. S6), suggesting that OTUB1 protects PD-L1 protein from degradation potentially before glycosylation. This fundamental mechanism eventually regulates the level of PD-L1 on the cell surface and maintains the immunosuppressive ability of cancer cells. A recent study has shown that AMPK-mediated PD-L1 phosphorylation causes its erroneous glycosylation and promotes HRD1, the E3 ubiquitin ligase located in the ER, to ubiquitinate and degrade PD-L1 [47]. The ERAD pathway not only controls the quality of misfolded or mismodified ER proteins but also regulates the physiological quantity of membrane-associated proteins [48, 49]. For instance, ERAD contributes to the regulation of plasma membrane-localized signaling receptors, like the transmembrane ErbB3 receptor tyrosine kinase [50, 51]. In this study, we demonstrate that OTUB1 is likely to keep newly synthesized PD-L1 proteins in a less ubiquitinated state, which saves PD-L1 from ERAD degradation, facilitates its glycosylation, and thus promotes high levels of PD-L1 in different cancers through modulating ER control of protein levels. However, the ER-associated E3 ubiquitin ligase that regulates the ubiquitination and quantity of PD-L1 remains unknown, and identification of the PD-L1-associated E3 ubiquitin ligase responsible for ERAD quantity control will aid a deep understanding of the regulatory mechanism of PD-L1 abundance.
Collectively, our study identifies a new target of deubiquitinase OTUB1, the immune checkpoint protein PD-L1, and establishes the association between OTUB1 and cancer immune response. We elucidate the regulatory mechanism of OTUB1-mediated PD-L1 stability via inhibition of the ERAD pathway and reveal the function of OTUB1 in promoting tumor growth in vivo by PD-L1-mediated immune evasion. Therefore, targeting the deubiquitinase OTUB1 may potentially improve patient outcomes through influencing PD-L1 expression in cancer immunotherapy.
Supplementary information
Acknowledgements
The authors sincerely thank Prof. Lingqiang Zhang for providing human deubiquitinase OTU subfamily plasmids and several shDUBs vectors, Profs. Jiadong Wang and Ceshi Chen for providing 4T1 cells, and Prof. Wensheng Wei for providing the CRISPR/Cas9 related plasmids. We thank Dr. Weicheng Zang for helpful discussion, and the National Center for Protein Sciences at Peking University, particularly Liying Du and Hongxia Lv for technical help. We also appreciate the assistance of Xiaochen Li from the Core Facilities of Life Sciences at Peking University for their assistance with microscopic imaging. We thank the Human Genetic Resource Core Facility at Peking University for providing peripheral blood samples from Peking University Hospital. This work was supported by the National Key Research and Development Program of China (2016YFC1302401) and the National Natural Science Foundation of China (81730080, 31670786, 32000917).
Author contributions
D.Z. designed and performed the experiments, analyzed the data, and wrote the manuscript. R.X., X.H., and J.Z. performed the experiments. Z.T. performed the bioinformatics analysis. Y.T. performed the cancer cell injection into mice. X.Z. supervised this study and wrote the manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All experiments using human samples and mice were approved by the Ethics Committee of Peking University and complied with all relevant ethical guidelines.
Footnotes
Edited by X. Lu
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version of this article (10.1038/s41418-020-00700-z) contains supplementary material, which is available to authorized users.
References
- 1.Zou W, Wolchok JD, Chen L. PD-L1 (B7-H1) and PD-1 pathway blockade for cancer therapy: Mechanisms, response biomarkers, and combinations. Sci Transl Med. 2016;8:328rv324. doi: 10.1126/scitranslmed.aad7118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen L, Han X. Anti-PD-1/PD-L1 therapy of human cancer: past, present, and future. J Clin Invest. 2015;125:3384–91. doi: 10.1172/JCI80011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bardhan K, Anagnostou T, Boussiotis VA. The PD1:PD-L1/2 pathway from discovery to clinical implementation. Front Immunol. 2016;7:550. doi: 10.3389/fimmu.2016.00550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Freeman GJ, Long AJ, Iwai Y, Bourque K, Chernova T, Nishimura H, et al. Engagement of the PD-1 immunoinhibitory receptor by a novel B7 family member leads to negative regulation of lymphocyte activation. J Exp Med. 2000;192:1027–34. doi: 10.1084/jem.192.7.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Iwai Y, Hamanishi J, Chamoto K, Honjo T. Cancer immunotherapies targeting the PD-1 signaling pathway. J Biomed Sci. 2017;24:26. doi: 10.1186/s12929-017-0329-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dong H, Strome SE, Salomao DR, Tamura H, Hirano F, Flies DB, et al. Tumor-associated B7-H1 promotes T-cell apoptosis: a potential mechanism of immune evasion. Nat Med. 2002;8:793–800. doi: 10.1038/nm730. [DOI] [PubMed] [Google Scholar]
- 7.Zou W, Chen L. Inhibitory B7-family molecules in the tumour microenvironment. Nat Rev Immunol. 2008;8:467–77. doi: 10.1038/nri2326. [DOI] [PubMed] [Google Scholar]
- 8.La-Beck NM, Jean GW, Huynh C, Alzghari SK, Lowe DB. Immune checkpoint inhibitors: new insights and current place in cancer therapy. Pharmacotherapy. 2015;35:963–76. doi: 10.1002/phar.1643. [DOI] [PubMed] [Google Scholar]
- 9.Sharma P, Allison JP. Immune checkpoint targeting in cancer therapy: toward combination strategies with curative potential. Cell. 2015;161:205–14. doi: 10.1016/j.cell.2015.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun C, Mezzadra R, Schumacher TN. Regulation and function of the PD-L1 Checkpoint. Immunity. 2018;48:434–52. doi: 10.1016/j.immuni.2018.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen J, Jiang CC, Jin L, Zhang XD. Regulation of PD-L1: a novel role of pro-survival signalling in cancer. Ann Oncol. 2016;27:409–16. doi: 10.1093/annonc/mdv615. [DOI] [PubMed] [Google Scholar]
- 12.Gotwals P, Cameron S, Cipolletta D, Cremasco V, Crystal A, Hewes B, et al. Prospects for combining targeted and conventional cancer therapy with immunotherapy. Nat Rev Cancer. 2017;17:286–301. doi: 10.1038/nrc.2017.17. [DOI] [PubMed] [Google Scholar]
- 13.Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer. 2012;12:252–64. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hsu JM, Li CW, Lai YJ, Hung MC. Posttranslational modifications of PD-L1 and their applications in cancer therapy. Cancer Res. 2018;78:6349–53. doi: 10.1158/0008-5472.CAN-18-1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang J, Dang F, Ren J, Wei W. Biochemical aspects of PD-L1 regulation in cancer immunotherapy. Trends Biochem Sci. 2018;43:1014–32. doi: 10.1016/j.tibs.2018.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chan LC, Li CW, Xia W, Hsu JM, Lee HH, Cha JH, et al. IL-6/JAK1 pathway drives PD-L1 Y112 phosphorylation to promote cancer immune evasion. J Clin Invest. 2019;129:3324–38. [DOI] [PMC free article] [PubMed]
- 17.Li CW, Lim SO, Xia W, Lee HH, Chan LC, Kuo CW, et al. Glycosylation and stabilization of programmed death ligand-1 suppresses T-cell activity. Nat Commun. 2016;7:12632. doi: 10.1038/ncomms12632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang J, Bu X, Wang H, Zhu Y, Geng Y, Nihira NT, et al. Cyclin D-CDK4 kinase destabilizes PD-L1 via cullin 3-SPOP to control cancer immune surveillance. Nature. 2018;553:91–95. doi: 10.1038/nature25015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lim SO, Li CW, Xia W, Cha JH, Chan LC, Wu Y, et al. Deubiquitination and Stabilization of PD-L1 by CSN5. Cancer Cell. 2016;30:925–39. doi: 10.1016/j.ccell.2016.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu C, Yao Z, Wang J, Zhang W, Yang Y, Zhang Y, et al. Macrophage-derived CCL5 facilitates immune escape of colorectal cancer cells via the p65/STAT3-CSN5-PD-L1 pathway. Cell Death Differ. 2020;27:1765–81. [DOI] [PMC free article] [PubMed]
- 21.Huang X, Zhang Q, Lou Y, Wang J, Zhao X, Wang L, et al. USP22 deubiquitinates CD274 to suppress anticancer immunity. Cancer Immunol Res. 2019;7:1580–90. doi: 10.1158/2326-6066.CIR-18-0910. [DOI] [PubMed] [Google Scholar]
- 22.Jingjing W, Wenzheng G, Donghua W, Guangyu H, Aiping Z, Wenjuan W. Deubiquitination and stabilization of programmed cell death ligand 1 by ubiquitin-specific peptidase 9, X-linked in oral squamous cell carcinoma. Cancer Med. 2018;7:4004–11. doi: 10.1002/cam4.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Komander D, Clague MJ, Urbe S. Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol. 2009;10:550–63. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- 24.Edelmann MJ, Iphofer A, Akutsu M, Altun M, di Gleria K, Kramer HB, et al. Structural basis and specificity of human otubain 1-mediated deubiquitination. Biochem J. 2009;418:379–90. doi: 10.1042/BJ20081318. [DOI] [PubMed] [Google Scholar]
- 25.He M, Zhou Z, Wu G, Chen Q, Wan Y. Emerging role of DUBs in tumor metastasis and apoptosis: Therapeutic implication. Pharm Ther. 2017;177:96–107. doi: 10.1016/j.pharmthera.2017.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saldana M, VanderVorst K, Berg AL, Lee H, Carraway KL. Otubain 1: a non-canonical deubiquitinase with an emerging role in cancer. Endocr Relat Cancer. 2019;26:R1–R14. doi: 10.1530/ERC-18-0264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baietti MF, Simicek M, Abbasi Asbagh L, Radaelli E, Lievens S, Crowther J, et al. OTUB1 triggers lung cancer development by inhibiting RAS monoubiquitination. EMBO Mol Med. 2016;8:288–303. doi: 10.15252/emmm.201505972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakada S, Tai I, Panier S, Al-Hakim A, Iemura S, Juang YC, et al. Non-canonical inhibition of DNA damage-dependent ubiquitination by OTUB1. Nature. 2010;466:941–6. doi: 10.1038/nature09297. [DOI] [PubMed] [Google Scholar]
- 29.Herhaus L, Al-Salihi M, Macartney T, Weidlich S, Sapkota GP. OTUB1 enhances TGFbeta signalling by inhibiting the ubiquitylation and degradation of active SMAD2/3. Nat Commun. 2013;4:2519. doi: 10.1038/ncomms3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li S, Zheng H, Mao AP, Zhong B, Li Y, Liu Y, et al. Regulation of virus-triggered signaling by OTUB1- and OTUB2-mediated deubiquitination of TRAF3 and TRAF6. J Biol Chem. 2010;285:4291–7. doi: 10.1074/jbc.M109.074971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Y, Sun XX, Elferich J, Shinde U, David LL, Dai MS. Monoubiquitination is critical for ovarian tumor domain-containing ubiquitin aldehyde binding protein 1 (Otub1) to suppress UbcH5 enzyme and stabilize p53 protein. J Biol Chem. 2014;289:5097–108. doi: 10.1074/jbc.M113.533109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou X, Yu J, Cheng X, Zhao B, Manyam GC, Zhang L, et al. The deubiquitinase Otub1 controls the activation of CD8(+) T cells and NK cells by regulating IL-15-mediated priming. Nat Immunol. 2019;20:879–89. doi: 10.1038/s41590-019-0405-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gao Y, Nihira NT, Bu X, Chu C, Zhang J, Kolodziejczyk A, et al. Acetylation-dependent regulation of PD-L1 nuclear translocation dictates the efficacy of anti-PD-1 immunotherapy. Nat Cell Biol. 2020;22:1064–75. doi: 10.1038/s41556-020-0562-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jiao S, Xia W, Yamaguchi H, Wei Y, Chen MK, Hsu JM, et al. PARP Inhibitor Upregulates PD-L1 Expression and Enhances Cancer-Associated Immunosuppression. Clin Cancer Res. 2017;23:3711–20. doi: 10.1158/1078-0432.CCR-16-3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wiener R, Zhang X, Wang T, Wolberger C. The mechanism of OTUB1-mediated inhibition of ubiquitination. Nature. 2012;483:618–22. doi: 10.1038/nature10911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Balakirev MY, Tcherniuk SO, Jaquinod M, Chroboczek J. Otubains: a new family of cysteine proteases in the ubiquitin pathway. EMBO Rep. 2003;4:517–22. doi: 10.1038/sj.embor.embor824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu T, Jiang L, Tavana O, Gu W. The deubiquitylase OTUB1 mediates ferroptosis via stabilization of SLC7A11. Cancer Res. 2019;79:1913–24. doi: 10.1158/0008-5472.CAN-18-3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jahan AS, Biquand E, Munoz-Moreno R, Le Quang A, Mok CK, Wong HH, et al. OTUB1 is a key regulator of RIG-I-dependent immune signaling and is targeted for proteasomal degradation by influenza A NS1. Cell Rep. 2020;30:1570–84 e1576. doi: 10.1016/j.celrep.2020.01.015. [DOI] [PubMed] [Google Scholar]
- 39.Schmidt O, Weyer Y, Baumann V, Widerin MA, Eising S, Angelova M, et al. Endosome and Golgi-associated degradation (EGAD) of membrane proteins regulates sphingolipid metabolism. EMBO J. 2019;38:e101433. doi: 10.15252/embj.2018101433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Q, Shinkre BA, Lee JG, Weniger MA, Liu Y, Chen W, et al. The ERAD inhibitor Eeyarestatin I is a bifunctional compound with a membrane-binding domain and a p97/VCP inhibitory group. PLoS One. 2010;5:e15479. doi: 10.1371/journal.pone.0015479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zerdes I, Matikas A, Bergh J, Rassidakis GZ, Foukakis T. Genetic, transcriptional and post-translational regulation of the programmed death protein ligand 1 in cancer: biology and clinical correlations. Oncogene. 2018;37:4639–61. doi: 10.1038/s41388-018-0303-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Patel SP, Kurzrock R. PD-L1 expression as a predictive biomarker in cancer immunotherapy. Mol Cancer Ther. 2015;14:847–56. doi: 10.1158/1535-7163.MCT-14-0983. [DOI] [PubMed] [Google Scholar]
- 43.Burr ML, Sparbier CE, Chan YC, Williamson JC, Woods K, Beavis PA, et al. CMTM6 maintains the expression of PD-L1 and regulates anti-tumour immunity. Nature. 2017;549:101–5. doi: 10.1038/nature23643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mezzadra R, Sun C, Jae LT, Gomez-Eerland R, de Vries E, Wu W, et al. Identification of CMTM6 and CMTM4 as PD-L1 protein regulators. Nature. 2017;549:106–10. doi: 10.1038/nature23669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guna A, Hegde RS. Transmembrane domain recognition during membrane protein biogenesis and quality control. Curr Biol. 2018;28:R498–R511. doi: 10.1016/j.cub.2018.02.004. [DOI] [PubMed] [Google Scholar]
- 46.Avci D, Lemberg MK. Clipping or extracting: two ways to membrane protein degradation. Trends Cell Biol. 2015;25:611–22. doi: 10.1016/j.tcb.2015.07.003. [DOI] [PubMed] [Google Scholar]
- 47.Cha JH, Yang WH, Xia W, Wei Y, Chan LC, Lim SO, et al. Metformin promotes antitumor immunity via endoplasmic-reticulum-associated degradation of PD-L1. Mol Cell. 2018;71:606–20 e607. doi: 10.1016/j.molcel.2018.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Printsev I, Curiel D, Carraway KL., 3rd Membrane protein quantity control at the endoplasmic reticulum. J Membr Biol. 2017;250:379–92. doi: 10.1007/s00232-016-9931-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hegde RS, Ploegh HL. Quality and quantity control at the endoplasmic reticulum. Curr Opin Cell Biol. 2010;22:437–46. doi: 10.1016/j.ceb.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yen L, Cao Z, Wu X, Ingalla ER, Baron C, Young LJ, et al. Loss of Nrdp1 enhances ErbB2/ErbB3-dependent breast tumor cell growth. Cancer Res. 2006;66:11279–86. doi: 10.1158/0008-5472.CAN-06-2319. [DOI] [PubMed] [Google Scholar]
- 51.Fry WH, Simion C, Sweeney C, Carraway KL., 3rd Quantity control of the ErbB3 receptor tyrosine kinase at the endoplasmic reticulum. Mol Cell Biol. 2011;31:3009–18. doi: 10.1128/MCB.05105-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.