Abstract
Background
GNG11 (G protein subunit gamma 11) is a member of guanine nucleotide-binding protein (G protein) gamma family. Few studies elucidated the role of GNG11 in human disease, especially in tumors. The present study initially analyzed the function of GNG11 in ovarian serous cystadenocarcinoma.
Methods
The differential expression of GNG11 mRNA in ovarian cancer and normal tissues was evaluated through Oncomine, CCLE, Gepia, UCSC Xena and UALCAN databases. The protein expression of GNG11 was assessed via HPA database. Prognosis analysis was performed by Kaplan–Meier Plotter. Restrict survival analysis to subtypes including tumor grade, cancer stage and TP53 mutation status was then carried out. GSEA enrichment analysis was performed to explore the significant pathways associated with GNG11 in ovarian cancer. Finally, the upstream miRNAs of GNG11 were predicted by DIANA, Target Scan, miRDB and miRWalk databases, and the potential key KEGG pathways were subsequently determined by DIANA.
Results
The mRNA expression of GNG11 was down-regulated in ovarian cancer patients (P<0.05). The cancer stage of patients correlated with the expression of GNG11 (P<0.05). Survival analysis indicated that GNG11 high expression statistically shortened the overall survival time of patients (HR=1.26, P=0.0043) compared with low expression group, especially for the patients with earlier stage (HR=2.48, P=0.035) and lower grade (HR=1.72, P=0.0016). Subsequently, the consistent upstream miRNA of GNG11, hsa-miR-22-5p, was predicted from 4 databases. The differential expression profile of hsa-miR-22-5p in blood was observed in ovarian cancer patients. According to the GSEA analysis on GNG11 and KEGG analysis on hsa-miR-22-5p, the consistent pathway of ECM-receptor interaction was observed (all P<0.01). ECM-receptor interaction pathway and differential expression of hsa-miR-22-5p in blood suggested the migration risk of ovarian cancer.
Conclusion
High expression of GNG11 indicated the poor prognosis of ovarian cancer patients. GNG11 might play a crucial role in the biological process of ovarian serous cystadenocarcinoma by ECM-receptor interaction pathway, thus affecting the prognosis of patients.
Keywords: GNG11, ovarian serous cystadenocarcinoma, poor prognosis, ECM-receptor interaction
Introduction
Ovarian cancer has became one of the most common gynecological malignancies worldwide. Several risk factors were associated with ovarian cancer occurrence, such as a family history of ovarian cancer, excess body weight, smoking and not giving birth.1 Ovarian serous cystadenocarcinoma (OSC), the most common subtype of ovarian cancer, constituted 60–80% of ovarian epithelial neoplasms.2,3 TP53 mutation was closely related to the ovarian serous cancer. Mutation in the tumor suppressor gene TP53 was an early event in the development of high-grade serous ovarian cancer.4 The Cancer Genome Atlas has reported that 96% of ovarian high-grade serous carcinomas have TP53 somatic mutations, suggesting that mutation of this gene was a defining feature of this neoplasm.5 Genetic mutation was only one of the factors of cancer pathogenesis. Owing to the lack of effective detection methods or early screening, approximately 75% of women have an advanced stage of the disease at diagnosis, which was consequently associated with a poor clinical outcome6 and heavy tumor burden.7 In most countries, the 5-year survival rate of ovarian cancer was lower than 40%.8 Ovarian cancer has ranked first among gynecologic malignant tumors.9 Therefore, it was urgently necessary to identify effective predictive biomarkers and molecular mechanisms involved in the progression of OSC, which may help find better predictive and therapeutic targets.
G Protein Subunit Gamma 11 (GNG11), is a member of the guanine nucleotide-binding protein (G protein) gamma family and encodes a lipid-anchored, cell membrane protein. As a member of the heterotrimeric G protein complex, this protein plays a vital role in the transmembrane signaling system.10 The diseases closely associated with GNG11 included splenic marginal zone lymphoma.11 Previous study has shown that over-expression of GNG11 immediately induced the cellular senescence in normal human fibroblasts.12 Further study examined the effect of the expression of GNG11 on the growth of immortalized human cell lines, and found that it suppressed the growth of SUSM-1 cells with induction of reactive oxygen species and abnormal nuclear morphology.13 Until now, there was fewer studies elucidating the role of GNG11 in human disease, especially in tumors, and its function has been largely unidentified.
On the basis of various bioinformatics databases, the present study comprehensively and objectively analyzed the role of GNG11 in ovarian serous cystadenocarcinoma. We initially revealed the expression distribution, prognostic value, biological function of GNG11 in ovarian cancer, as well as the prospective pathway involved in ovarian cancer. In addition, the upstream miRNAs of GNG11 was predicted and potential regulatory pathway was explored.
Methods
Expression Analysis
We first explored the mRNA expression of GNG11 in human cancers and various cancer cell lines through the Oncomine (https://www.oncomine.org/) and Cancer Cell Line Encyclopedia (CCLE) databases (https://portals.broadinstitute.org/ccle/). The quantitative analysis on mRNA expression of GNG11 was also assessed through Gene Expression Profiling Interactive Analysis (Gepia) (http://gepia.cancer-pku.cn/), UCSC (https://xenabrowser.net/) Xena and Oncomine databases. The filter criteria on GNG11 expression was set as follows: Oncomine (P value<0.01, fold change>2 and gene rank=top 10%), Gepia (|Log2FC| Cutoff=1, P value Cutoff=0.01). In addition, the expression of GNG11 based on clinical characteristics in patients with ovarian serous cystadenocarcinoma was evaluated via UALCAN database (http://ualcan.path.uab.edu/). The clinical characteristics included age (21–40, 41–60, 61–80, 81–100), tumor grade (1, 2, 3, 4), cancer stage (I, II, III, IV) and TP53 mutation status (TP53-mutant, TP53-nonmutant). Subtype descriptions for tumor grade: grade 1-well differentiated (low grade), grade 2-moderately differentiated (intermediate grade), grade 3-poorly differentiated (high grade), grade 4-undifferentiated (high grade). Moreover, the protein expression of GNG11 in ovarian cancer and normal tissues with immunohistochemistry was assessed via the Human Protein Atlas (HPA) database (https://www.proteinatlas.org/).
Survival Analysis
The association of GNG11 expression with prognosis of patients with ovarian serous cystadenocarcinoma was evaluated by Kaplan-Meier Plotter (http://kmplot.com/). The survival in terms of overall survival (OS), progression free survival (PFS) and post progression survival (PPS) was analyzed between high and low expression groups. In this study, all the patients were split by automatically selecting best cutoff by system. Then, we performed restrict analysis to subtypes including tumor grade (1+2 vs 3), cancer stage (1+2 vs 3+4) and TP53 mutation status (mutated vs wild type). Another filter conditions were as follows: follow-up threshold: all, and log-rank P value<0.05 was regarded as statistically significant difference.
GSEA Analysis
We performed the gene set enrichment analysis (GSEA) to investigate the potential function of GNG11 in ovarian cancer. To obtain normalized enrichment scores (NES), the nominal P‑value and false discovery rate (FDR) q‑value were determined. The number of gene set permutations for each analysis was set at 1000, and the phenotype label was the expression level of GNG11. Gene sets with a nominal P‑value<0.05 and an FDR q‑value<0.25 were considered significantly enriched.
Biological Function Analysis
To elucidate the biological functions of GNG11, we performed Gene Ontology (GO) annotation analysis including biological process (BP), molecular function (MF) and cellular component (CC). The co-expressed genes were firstly explored through LinkedOmics (http://www.linkedomics.org/). The filter criteria were as follows: cancer cohort: TCGA_OV; dataset: TCGA_OV (RNAseq); dataset attribute: GNG11; target dataset: TCGA_OV (RNAseq); statistical method: Pearson correlation test. The co-expressed genes positively and negatively associated with GNG11 were presented with volcano plot and heat map. The identified genes with P value<0.001 and FDR<0.001 were selected for the further GO annotation analysis. In addition, we used GeneMANIA (https://genemania.org/) to find other important genes that were related to GNG11, using a very large set of functional association data. Association data included protein and genetic interactions, pathways, co-expression, co-localization and protein domain similarity.
Identification of Upstream miRNAs
In this study, we also predicted the target upstream miRNAs of GNG11. Four databases including DIANA (http://diana.imis.athena-innovation.gr/DianaTools/), Target Scan (http://www.targetscan.org/), miRDB (http://mirdb.org/) and miRWalk (http://mirwalk.umm.uni-heidelberg.de/) were used to explore the targeted miRNAs. The consistent miRNA, namely hsa-miR-22-5p, among 4 databases was determined by Venn analysis (http://bioinformatics.psb.ugent.be/webtools/Venn/). Subsequently, the association of hsa-miR-22-5p expression with overall survival of ovarian cancer patients was evaluated through Kaplan-Meier Potter (http://kmplot.com/). The differential expression profile of hsa-miR-22-5p was explored via the Database of Differentially Expressed miRNAs in Human Cancers (dbDEMC) (https://www.picb.ac.cn/dbDEMC/). Finally, the significant biological pathway associated with hsa-miR-22-5p in ovarian cancer was analyzed by DIANA database.
Results
The Expression Analysis of GNG11
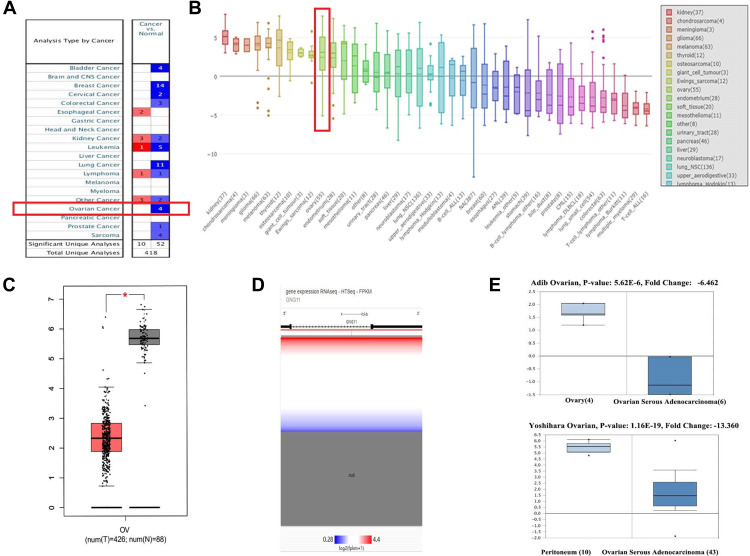
As shown in Figure 1A, a total of 10 cases indicated the up-regulation of GNG11 in human cancers and 52 showed down-regulation. Four researches reported that GNG11 was down-regulated in ovarian cancer compared with normal tissue. Figure 1B shows that expression of GNG11 was relatively higher in ovarian cancer cell among various human cancer cell lines. Figure 1C–E presented the down-regulation of GNG11 in ovarian cancer from different databases through quantitative analysis.
Figure 1.
The mRNA expression analysis of GNG11. (A) Disease summary. The study with red box indicated the research associated with ovarian cancer. (B) GNG11 expression in various human cancer cell lines. The term with red box indicated the ovarian cancer cell lines. Differential expression analysis between ovarian cancer and normal tissues in (C) Gepia (*P<0.05); (D) UCSC Xena; (E) Oncomine.
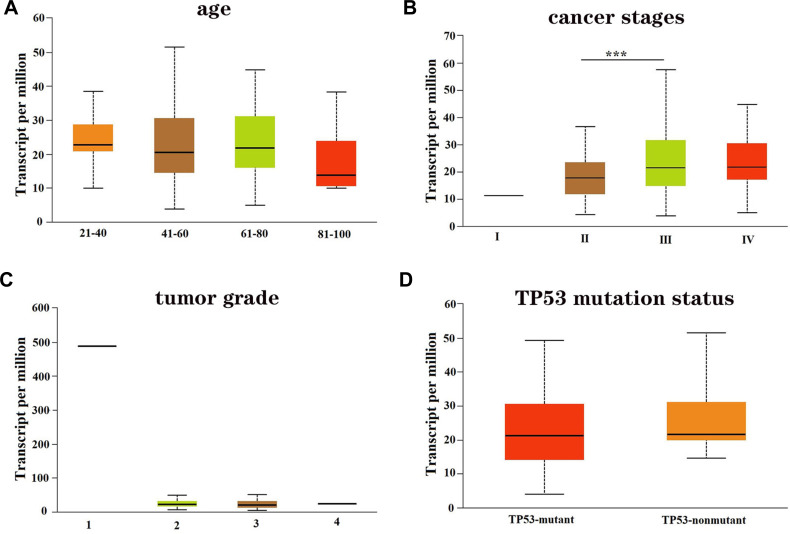
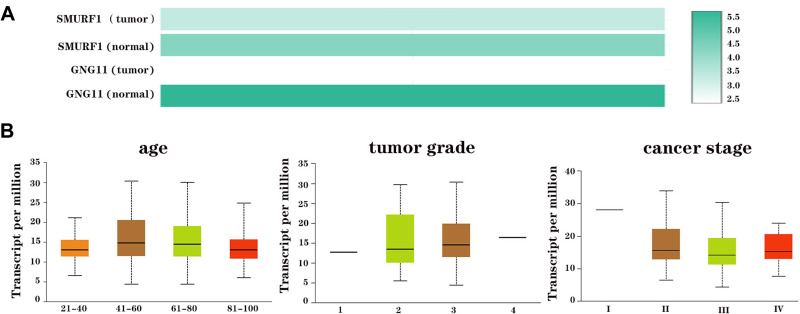
We then evaluated the association of GNG11 mRNA expression with the clinical characteristics of patients with ovarian serous cystadenocarcinoma. The age of patients did not correlate with the GNG11 expression (Figure 2A). The cancer stage of patients influenced the expression of GNG11, and patients in stage III showed higher mRNA expression than in other stages (Figure 2B). There was no difference of GNG11 expression in different tumor grade (Figure 2C). TP53 mutation status was not associated with the expression of GNG11 as well (Figure 2D).
Figure 2.
The association of GNG11 mRNA expression with clinical characteristics of patients with ovarian serous cystadenocarcinoma. (A) Age; (B) cancer stage; (C) tumor grade; (D) TP53 mutation status. ***P<0.001.
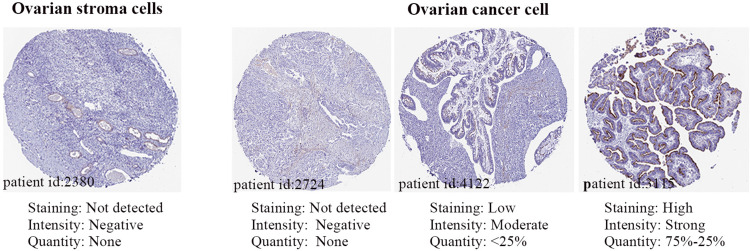
Figure 3 shows the protein expression of GNG11 in normal and cancer tissues. There was no protein expression detected in normal tissue (id:2380). However, it was observed that the GNG11 protein expression differed for each individual in ovarian cancer. The intensity of GNG11 protein expression appeared negative (id:2724) or moderate (id: 4122) or strong (id:3115) in cancer patients. It seemed that protein expression of GNG11 did not correspond with its mRNA expression.
Figure 3.
The protein expression of GNG11 in normal and ovarian cancer tissues.
The Association of GNG11 Expression with Prognosis
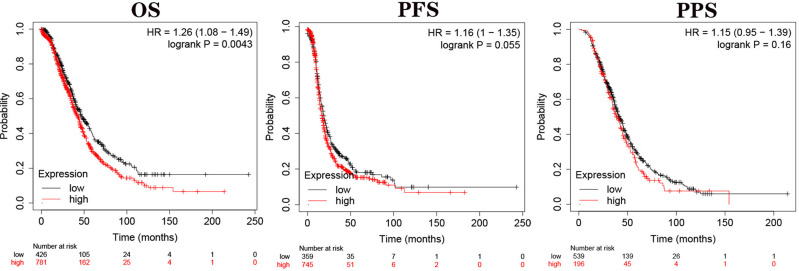
We used Kaplan-Meier Plotter database to explore the influence of GNG1 expression on the prognosis of patients with ovarian cancer. As shown in Figure 4, high expression of GNG11 shortened the overall survival time of patients compared with low expression group (HR=1.26, P=0.0043). The median survival time of patients in high and low expression cohorts was 42.17 and 46.60 months, respectively. However, the progression free survival and post progression survival of patients were not affected by the expression level of GNG11 (all P>0.05).
Figure 4.
The association of GNG11 mRNA expression with prognosis of patients with ovarian serous cystadenocarcinoma.
Abbreviations: OS, overall survival; PFS, progression-free survival; PPS, post progression survival.
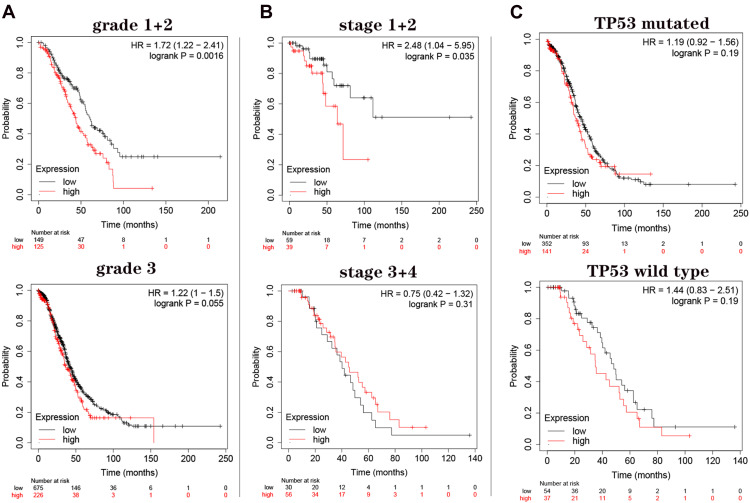
Due to the significance of GNG11 mRNA expression on OS of patients, we subsequently performed restrict survival analysis to subtypes in terms of tumor grade (1+2 vs 3), cancer stage (1+2 vs 3+4) and TP53 mutation status (mutated vs wild type). The survival analysis regarding grade 4 was missing because the sample number was too low for meaningful analysis. Survival analysis indicated that high expression of GNG11 mRNA caused the shorter overall survival time of patients in tumor grade (1+2) compared with low expression group (Figure 5A, HR=1.72, P=0.0016). The survival of patients in grade 3 was not associated with the expression level of GNG11 (P=0.055). For patients in stage (1+2), GNG11 high expression was not conducive to their prognosis compared with low expression (Figure 5B, HR=2.48, P=0.035). But the survival time of patients in stage (3+4) showed no difference between high and low expression groups (P=0.031). In addition, TP53 mutation status was not associated with the survival of patients with ovarian cancer (Figure 5C, all P>0.05).
Figure 5.
Restrict overall survival analysis on GNG11 mRNA expression in patients with ovarian serous cystadenocarcinoma. (A) Tumor grade; (B) cancer stage; (C) TP53 mutation status.
Enrichment Analysis on GNG11
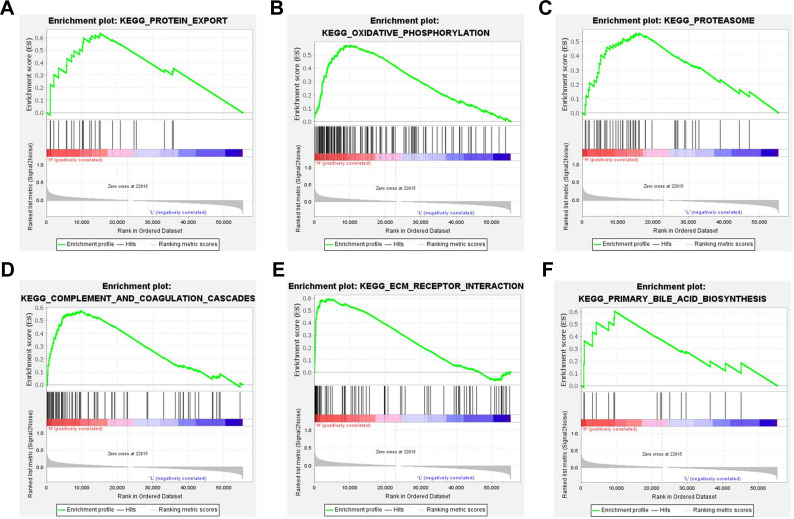
We then explored the biological pathogenesis of GNG11 in ovarian cancer through GSEA analysis. According to the screening criteria of normal P value<0.05 and FDR<0.25, there was no down-regulated pathways appeared, and 24 up-regulated pathways were observed under this threshold criterion. GSEA analysis indicated that 6 significant pathways (Figure 6, Table 1), including Protein export, Oxidative phosphorylation, Proteasome, Complement and coagulation cascades, ECM receptor interaction and Primary bile acid biosynthesis, were activated with GNG11 high expression. The results suggested that these biological pathways might play a vital role in the progression of ovarian cancer.
Figure 6.
Enrichment plots from GSEA analysis on GNG11 in ovarian cancer. (A) Protein export. (B) Oxidative phosphorylation. (C) Proteasome. (D) Complement and coagulation cascades. (E) ECM receptor interaction. (F) Primary bile acid biosynthesis.
Abbreviation: GSEA, gene set enrichment analysis.
Table 1.
GSEA Analysis on GNG11 in Ovarian Cancer
| KEGG Name | ES | NES | Nominal p-value | FDR q-value |
|---|---|---|---|---|
| Protein export | 0.634032 | 2.0384740 | 3.97E-03 | 0.037414 |
| Oxidative phosphorylation | 0.571145 | 1.9287751 | 2.55E-02 | 0.067186 |
| Proteasome | 0.558876 | 1.8173838 | 5.44E-02 | 0.123006 |
| Complement and coagulation cascades | 0.575297 | 1.7534364 | 1.19E-02 | 0.145108 |
| ECM receptor interaction | 0.593909 | 1.7346296 | 1.84E-02 | 0.135957 |
| Primary bile acid biosynthesis | 0.605069 | 1.7007636 | 8.16E-03 | 0.155467 |
Abbreviations: GSEA, gene set enrichment analysis; ES, Enrichment Score; NES, Normalized Enrichment Score; FDR, false discovery rate.
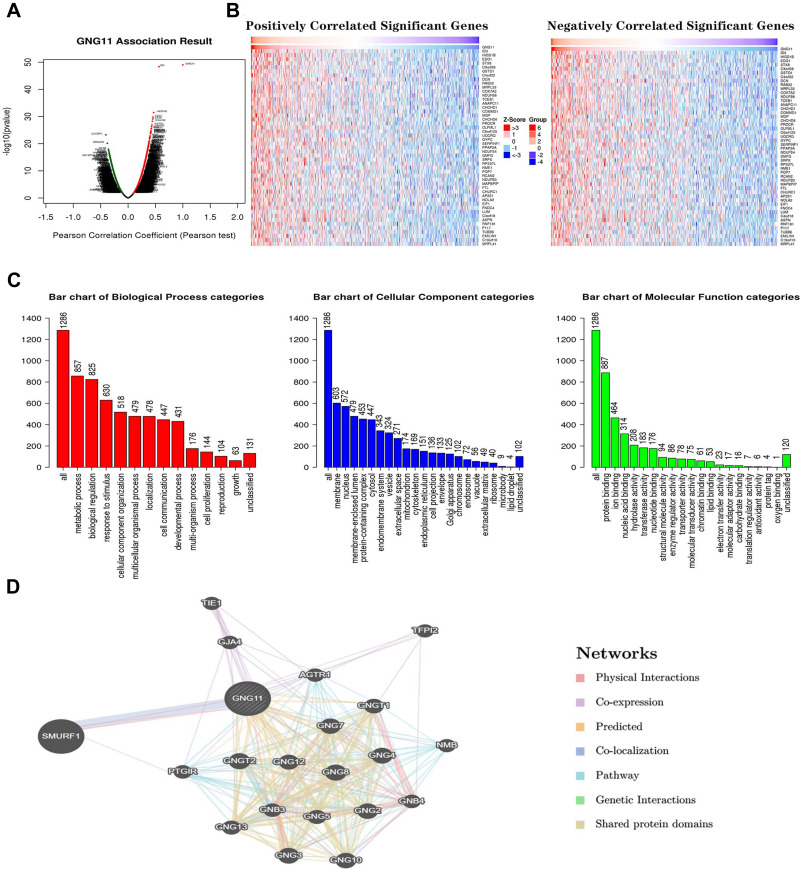
In order to disclose the biological function of GNG11 in ovarian cancer, the co-expressed genes closely correlated with GNG11 in ovarian serous cystadenocarcinoma were determined through LinkedOmics. Figure 7A displays the related co-expressed genes with GNG11 via volcano map. Figure 7B presents the top 50 co-expressed genes that were positively and negatively associated with GNG11 in ovarian cancer. The associated genes meeting the threshold of P<0.001 and FDR<0.001 were selected for further GO annotation analysis. Figure 7C indicates that cellular component were mainly enriched in membrane, nucleus and membrane-enclosed lumen. The biological processes that GNG11 participated in included metabolic process, biological regulation and response to stimulus. The molecular function of GNG11 were mainly enriched in protein binding, ion binding and nucleic acid binding. Figure 7D presents that SMURF1 was closely associated with GNG11.
Figure 7.
Co-expressed genes analysis associated with GNG11. (A) Volcano plot of co-expressed genes with GNG11. (B) The top 50 co-expressed genes positively and negatively associated with GNG11 in ovarian cancer. (C) GO annotation analysis. (D) Interaction analysis of GNG11 with other related genes.
Upstream miRNA Identification of GNG11
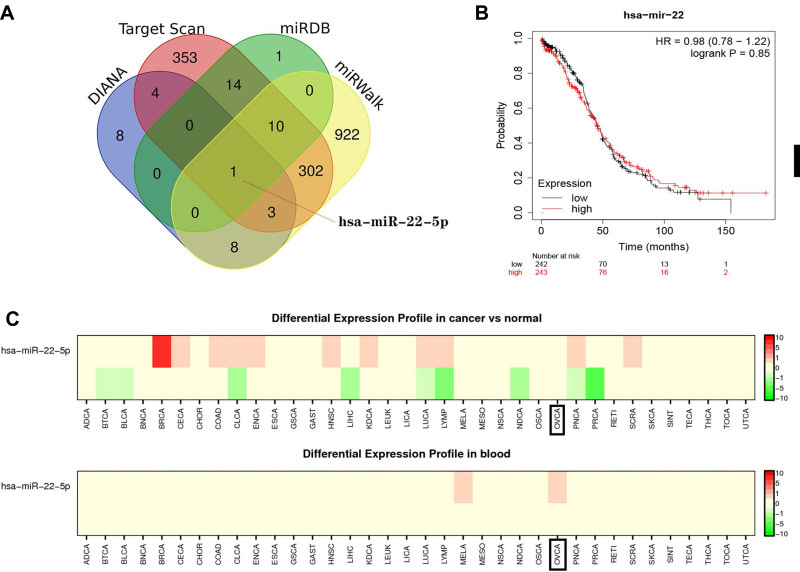
This study also explored the potential upstream miRNAs of GNG11 from 4 databases. There were 23, 26, 1676 and 842 miRNAs predicted in DIANA, miRDB, miRWalk and Target Scan databases, respectively. At last, one consistent miRNA, hsa-miR-22-5p, was screened by venn analysis (Figure 8A). Kaplan-Meier Plotter analysis indicated that expression of hsa-miR-22 did not influence the prognosis of patients with ovarian cancer (Figure 8B). Differential expression analysis (Figure 8C) on hsa-miR-22 found no difference between normal and ovarian cancer patients. However, differential expression profile in blood was observed in ovarian cancer patients.
Figure 8.
The upstream miRNAs analysis of GNG11. (A) hsa-miR-22-5p identification by venn analysis. (B) Survival analysis on hsa-miR-22-5p in ovarian serous cystadenocarcinoma. (C) Differential expression analysis on hsa-miR-22-5p.
The possible significant pathway related with miR-22-5p was investigated through DIANA database. The results indicated that hsa-miR-22-5p was significantly associated with Adherens junction, Proteoglycans in cancer, Lysine degradation, Estrogen signaling pathway and ECM-receptor interaction pathways (Table 2).
Table 2.
The KEGG Pathway Analysis on hsa-miR-22-5p
| Pathway ID | KEGG Terms | P-value |
|---|---|---|
| hsa04520 | Adherens junction | 7.01E-10 |
| hsa05205 | Proteoglycans in cancer | 2.04E-08 |
| hsa00310 | Lysine degradation | 8.69E-04 |
| hsa04915 | Estrogen signaling pathway | 8.91E-04 |
| hsa04512 | ECM-receptor interaction | 5.08E-03 |
Expression and Pathway Analysis on SMURF1
According to the results mentioned above, SMURF1 was closely related to GNG11. Further study found that mRNA expression of SMURF1 was higher in normal tissue than in ovarian cancer tissue. In addition, the mRNA expression of SMURF1 was higher than GNG11 in ovarian cancer, but lower in normal samples (Figure 9A). Association analysis showed that SMURF1 mRNA expression did not correlate with the clinical characteristics of ovarian cancer patients (Figure 9B, all P>0.05).
Figure 9.
The mRNA expression of SMURF1 in ovarian serous cystadenocarcinoma. (A) mRNA expression comparison in ovarian cancer and normal tissues. (B) Association analysis on SMURF1 mRNA expression with clinical phenotypes of patients with ovarian serous cystadenocarcinoma.
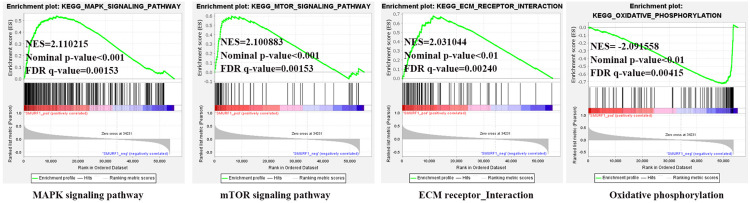
According to the screening criteria of normal P value<0.05 and FDR<0.25, a total of 8 down-regulated and 83 up-regulated pathways were observed associated with SMURF1 in ovarian cancer. GSEA analysis (Figure 10) showed that SMURF1 was positively related to MAPK signaling pathway, mTOR signaling pathway and ECM receptor interaction pathway, as well as negatively correlated with Oxidative phosphorylation pathway.
Figure 10.
Enrichment plots of SMURF1 from GSEA analysis in ovarian cancer.
Combining the GSEA analysis on GNG11 and SMURF1, as well as KEGG analysis on hsa-miR-22-5p, the significant pathway of ECM-receptor interaction was identified consistently. As the upstream of GNG11, hsa-miR-22-5p might regulate the expression of GNG11 via participating in ECM-receptor interaction pathway. In addition, GNG11 acted as the downstream of hsa-miR-22-5p, might influence the progression of ovarian cancer through ECM-receptor interaction pathway. It can be seen that ECM-receptor interaction pathway might play an important role in the progression of ovarian cancer. The detailed information of ECM-receptor interaction pathway is presented in Figure 11.
Figure 11.
ECM-receptor interaction pathway overview.
Dicussion
GNG11, a key member of G protein, plays a vital role in transmembrane system. Previous study has demonstrated the potential role of GNG11 in cancers. It has been identified as the hub gene and potential biomarker for lung adenocarcinoma,10 colon cancer14 and breast cancer.15 In addition, GNG11 has been demonstrated as a hub gene associated with ovarian cancer prognosis (HR=1.30).16 However, the potential function of GNG11 in ovarian serous cystadenocarcinoma has not been elucidated yet. Hence, the present study initially investigated the potential role of GNG11 in ovarian cancer. We assessed the expression distribution of GNG11 in ovarian serous cystadenocarcinoma, performed the deeper survival analysis, and explored the significant pathways and biological functions associated with GNG11 in ovarian cancer. In addition, the upstream miRNAs of GNG11 and potential regulatory mechanisms were discovered as well.
In this study, we disclosed the down-regulation of GNG11 mRNA in ovarian serous cystadenocarcinoma. Age, tumor grade and TP53 mutation status of patients were not associated with the GNG11 mRNA expression. Cancer stage was related with the expression of GNG11, and relatively higher expression was observed in grade 3 patients. However, the protein expression of GNG11 did not correspond with its mRNA expression, and we speculated that the desynchrony might be attributed to several reasons. The half-life period is different between mRNA and protein. The mRNA is easy to degrade and exists in tissues for a short time, while its properties are relatively stable after being translated into protein. In addition, after finishing transcription, there is post-transcriptional processing, degradation of transcription product, translation, post-translation processing and modification. Therefore, transcription level is not exactly the same as the translation level, and this regulation differs between different proteins. Our results indicated the difference between transcription and translation level of GNG11, which needed to be studied further.
The study then predicted the effects of GNG11 expression on the prognosis of patients. Survival analysis indicated that GNG11 high expression was unfavorable for the overall survival time of patients with ovarian serous cystadenocarcinoma, especially for the subjects in grade (1+2) and stage (1+2). Clinicopathologic factors associated with long-term survival in ovarian cancer included younger age at diagnosis, lower grade, earlier stage, absence of ascites, primary debulking surgery and microscopic disease after cytoreductive surgery.17,18 The advanced stage, platinum resistance, and recurrence might be the negative prognostic factors of ovarian cancer patients.19,20 The present study also indicated that the survival of patients did not correlate with the advanced stage (3+4). From another perspective, GNG11 low expression was conducive to the patient survival, which reminded us that controlling the expression of GNG11 within a relatively low state might be able to improve the clinical outcome of ovarian cancer patients.
It should be noted that the expression and prognostic impacts of GNG11 varies with different cancer types. Most of researches associated with GNG11 focused on the lung cancer-related studies. Down regulation of GNG11 was observed in lung cancer, and low expression of GNG11 was associated with worse OS for the female lung cancer patients who never smoked.21 GNG11 has been identified as a core gene associated with triple-negative breast cancer, and was down-regulated in tumor tissue.22 In addition, GNG11 was also screened as the most valuable protein involved in cervical cancer with down-regulation.23 Our study also found the high expression of GNG11 in some cancers such as kidney cancer and esophageal cancer. It followed that GNG11 might act as tumor suppressor gene or an oncogene, and these differences were important for revealing pathogenesis of cancer. However, few studies revealed the expression and prognosis influence of GNG11 in human cancers up to now, and more future researches needed to be done to investigate the significance of GNG11 in cancers.
Moreover, we explored the significant pathways associated with GNG11 in ovarian cancer. GSEA analysis in this study indicated that GNG11 mainly participated in the up-regulated pathways such as oxidative phosphorylation, complement and coagulation cascades, and ECM receptor interaction. These pathways could be activated under the condition of GNG11 high expression, which might be closely related with the progression and poor prognosis of ovarian cancer patients. The upstream miRNA of GNG11, hsa-miR-22-5p, was determined as well, and DIANA analysis was used to predict the key KEGG pathway about hsa-miR-22-5p. The results suggested that hsa-miR-22-5p was possibly involved in adherens junction, estrogen signaling pathway and ECM-receptor interaction. In addition, SMURF1 was closely associated with GNG11. GSEA analysis also predicted the positive pathways associated with SMURF1 in ovarian cancer, such as ECM receptor interaction pathway and oxidative phosphorylation pathway. It followed that ECM-receptor interaction was the consistent pathway associated with GNG11, its upstream and GNG11-related gene.
The regulation of ECM-receptor interaction pathway was significantly associated with the metastasis of cancers such as colorectal cancer,24 kidney cancer25 and breast cancer.26 Our results showed that ECM-receptor interaction was enriched in high expression group of GNG11, which indicating the migration risk of ovarian cancer. The dbDEMC analysis indicated that differential expression profile of hsa-miR-22-5p in blood was observed in ovarian cancer patients, suggesting that the tumor cells secreted the hsa-miR-22-5p into blood, and possibly caused the risk of distant metastasis and absorption by other organs. These results reminded us that the invasion and migration risk of ovarian cancer patients with GNG11 high expression should not be ignored. Other study has identified the hsa-miR-22-5p as potential diagnostic biomarkers in gastric cardia adenocarcinoma.27 The significance of hsa-miR-22-5p in the progression of other diseases deserved further investigation.
The present study initially explored the function of GNG11 in ovarian cancer, and provided a novel therapeutic target for the treatment of ovarian cancer. However, we should concede that there were several imperfections in this study. The potential difference on GNG11 protein expression in ovarian cancer tissues should be confirmed further. The related pathway about ECM-receptor interaction analyzed in this study needed further investigation in future work.
Conclusions
Collectively, we adopted the bioinformatic method to comprehensively analyze the function of GNG11 in ovarian serous cystadenocarcinoma. The results presented here suggested that GNG11 high expression was unfavorable for the overall survival of patients with ovarian cancer, especially for the patients in early stage and lower grade. Cancer stage was associated with the GNG11 expression level. GNG11 might play an important role in the progression of ovarian serous cystadenocarcinoma through ECM-receptor interaction pathway. However, further studies in vitro and in vivo were still needed on the pathogenesis to validate the role of GNG11-regulated molecular networks in ovarian cancer.
Funding Statement
There is no funding to report.
Data Sharing Statement
The datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request.
Author Contributions
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agreed to be accountable for all aspects of the work.
Disclosure
The authors report no competing interests in this work.
References
- 1.Torre LA, Islami F, Siegel RL, Ward EM, Jemal A. Global cancer in women: burden and trends. Cancer Epidemiol Biomarkers Prev. 2017;26(4):444–457. doi: 10.1158/1055-9965.EPI-16-0858 [DOI] [PubMed] [Google Scholar]
- 2.Li J, Fadare O, Xiang L, Kong B, Zheng W. Ovarian serous carcinoma: recent concepts on its origin and carcinogenesis. J Hematol Oncol. 2012;5(1):8. doi: 10.1186/1756-8722-5-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Slomovitz B, Gourley C, Carey MS, et al. Low-grade serous ovarian cancer: state of the science. Gynecol Oncol. 2020;156(3):715–725. doi: 10.1016/j.ygyno.2019.12.033 [DOI] [PubMed] [Google Scholar]
- 4.Fransson A, Glaessgen D, Alfredsson J, Wiman KG, Bajalica-Lagercrantz S, Mohell N. Strong synergy with APR-246 and DNA-damaging drugs in primary cancer cells from patients with TP53 mutant high-grade serous ovarian cancer. J Ovarian Res. 2016;9(1):27. doi: 10.1186/s13048-016-0239-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vang R, Levine DA, Soslow RA, Zaloudek C, Shih Ie M, Kurman RJ. Molecular alterations of TP53 are a defining feature of ovarian high-grade serous carcinoma: a rereview of cases lacking TP53 mutations in the Cancer Genome Atlas Ovarian Study. Int J Gynecol Pathol. 2016;35(1):48–55. doi: 10.1097/PGP.0000000000000207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shapira I, Oswald M, Lovecchio J, et al. Circulating biomarkers for detection of ovarian cancer and predicting cancer outcomes. Br J Cancer. 2014;110(4):976–983. doi: 10.1038/bjc.2013.795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bachmayr-Heyda A, Aust S, Auer K, et al. Integrative systemic and local metabolomics with impact on survival in high-grade serous ovarian cancer. Clin Cancer Res. 2017;23(8):2081–2092. doi: 10.1158/1078-0432.CCR-16-1647 [DOI] [PubMed] [Google Scholar]
- 8.Allemani C, Weir HK, Carreira H, et al. Global surveillance of cancer survival 1995–2009: analysis of individual data for 25,676,887 patients from 279 population-based registries in 67 countries (CONCORD-2). Lancet. 2015;385(9972):977–1010. doi: 10.1016/S0140-6736(14)62038-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu M, Sun Y, Wu J, Liu G. Identification of hub genes in high-grade serous ovarian cancer using weighted gene co-expression network analysis. Med Sci Monit. 2020;26:e922107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hua P, Zhang Y, Jin C, Zhang G, Wang B. Integration of gene profile to explore the hub genes of lung adenocarcinoma: a Quasi-Experimental Study. Medicine (Baltimore). 2020;99(43):e22727. doi: 10.1097/MD.0000000000022727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ruiz-Ballesteros E, Mollejo M, Rodriguez A, et al. Splenic marginal zone lymphoma: proposal of new diagnostic and prognostic markers identified after tissue and cDNA microarray analysis. Blood. 2005;106(5):1831–1838. doi: 10.1182/blood-2004-10-3898 [DOI] [PubMed] [Google Scholar]
- 12.Hossain MN, Sakemura R, Fujii M, Ayusawa D. G-protein gamma subunit GNG11 strongly regulates cellular senescence. Biochem Biophys Res Commun. 2006;351(3):645–650. doi: 10.1016/j.bbrc.2006.10.112 [DOI] [PubMed] [Google Scholar]
- 13.Takauji Y, Kudo I, En A, et al. GNG11 (G-protein subunit gamma 11) suppresses cell growth with induction of reactive oxygen species and abnormal nuclear morphology in human SUSM-1 cells. Biochem Cell Biol. 2017;95(4):517–523. doi: 10.1139/bcb-2016-0248 [DOI] [PubMed] [Google Scholar]
- 14.Xing S, Wang Y, Hu K, Wang F, Sun T, Li Q. WGCNA reveals key gene modules regulated by the combined treatment of colon cancer with PHY906 and CPT11. Biosci Rep. 2020;40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mehrpour Layeghi S, Arabpour M, Esmaeili R, Naghizadeh MM, Tavakkoly Bazzaz J, Shakoori A. Evaluation of the potential role of long non-coding RNA LINC00961 in luminal breast cancer: a case-control and systems biology study. Cancer Cell Int. 2020;20(1):478. doi: 10.1186/s12935-020-01569-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng MJ, Li X, Hu YX, et al. Identification of molecular marker associated with ovarian cancer prognosis using bioinformatics analysis and experiments. J Cell Physiol. 2019;234(7):11023–11036. doi: 10.1002/jcp.27926 [DOI] [PubMed] [Google Scholar]
- 17.Cress RD, Chen YS, Morris CR, Petersen M, Leiserowitz GS. Characteristics of long-term survivors of epithelial ovarian cancer. Obstet Gynecol. 2015;126(3):491–497. doi: 10.1097/AOG.0000000000000981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaern J, Aghmesheh M, Nesland JM, et al. Prognostic factors in ovarian carcinoma stage III patients. Can biomarkers improve the prediction of short- and long-term survivors? Int J Gynecol Cancer. 2005;15(6):1014–1022. doi: 10.1111/j.1525-1438.2005.00185.x [DOI] [PubMed] [Google Scholar]
- 19.Petrillo M, Fagotti A, Ferrandina G, et al. Ovarian cancer patients with localized relapse: clinical outcome and prognostic factors. Gynecol Oncol. 2013;131(1):36–41. doi: 10.1016/j.ygyno.2013.06.020 [DOI] [PubMed] [Google Scholar]
- 20.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34. doi: 10.3322/caac.21551 [DOI] [PubMed] [Google Scholar]
- 21.Shi K, Li N, Yang M, Li W. Identification of key genes and pathways in female lung cancer patients who never smoked by a bioinformatics analysis. J Cancer. 2019;10(1):51–60. doi: 10.7150/jca.26908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang X, Kang X, Jin L, et al. ABCC9, NKAPL, and TMEM132C are potential diagnostic and prognostic markers in triple-negative breast cancer. Cell Biol Int. 2020;44(10):2002–2010. doi: 10.1002/cbin.11406 [DOI] [PubMed] [Google Scholar]
- 23.Liu J, Li S, Lin L, et al. Co-expression network analysis identified atypical chemokine receptor 1 (ACKR1) association with lymph node metastasis and prognosis in cervical cancer. Cancer Biomark. 2020;27(2):213–223. doi: 10.3233/CBM-190533 [DOI] [PubMed] [Google Scholar]
- 24.Yuan J, Xie A, Cao Q, Li X, Chen J, Fang Y. INHBB is a novel prognostic biomarker associated with cancer-promoting pathways in colorectal cancer. Biomed Res Int. 2020;2020:6909672. doi: 10.1155/2020/6909672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang HJ, Tao J, Sheng L, et al. Twist2 promotes kidney cancer cell proliferation and invasion by regulating ITGA6 and CD44 expression in the ECM-receptor interaction pathway. Onco Targets Ther. 2016;9:1801–1812. doi: 10.2147/OTT.S96535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peng W, Li JD, Zeng JJ, et al. Clinical value and potential mechanisms of COL8A1 upregulation in breast cancer: a comprehensive analysis. Cancer Cell Int. 2020;20(1):392. doi: 10.1186/s12935-020-01465-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang J, Zhang H, Zhou X, et al. Five serum-based miRNAs were identified as potential diagnostic biomarkers in gastric cardia adenocarcinoma. Cancer Biomark. 2018;23(2):193–203. doi: 10.3233/CBM-181258 [DOI] [PubMed] [Google Scholar]