Abstract
Nucleic acid testing and antibody testing data from 143 recovered COVID-19 patients during the convalescent phase were retrospectively analyzed. A total of 23 (16.1%) recovered patients re-tested positive for SARS-CoV-2 RNA by RT-PCR. Three months after symptom onset, 100% and 99.3% of the patients remained positive for total and IgG antibodies, and the antibody levels remained high. IgM antibodies declined rapidly, with a median time to seroconversion of 67 (95% CI: 59, 75) days after onset. Approximately 25% of patients were seronegative for IgA antibodies at three months after onset. There was no statistically significant difference in antibody kinetics between patients with and without re-positive RT-PCR results during the convalescent phase.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00705-021-05132-9.
Coronavirus disease 2019 (COVID-19) is now pandemic globally, spreading rapidly to over 200 countries, with over 117 million confirmed cases and 2.6 million deaths up to March 8, 2021 [1, 2]. After discharge from the hospital, most recovered COVID-19 patients remain negative (non-re-positive) for novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNA by RT-PCR, while a proportion of recovered COVID-19 patients test positive again (re-positive) [3–5]. Young and mild-COVID-19 patients seemed to have a higher risk of having re-positive RT-PCR results during the convalescent phase [3, 6]. The antibody level tends to be similar in the re-positive and non-re-positive groups [3, 6]. However, these studies included only one antibody test per patient and did not adjust for the time of onset, which was significantly correlated with antibody level. It remains unclear whether the dynamics of the antibody response are different in re-positive and non-re-positive patients. This study analyzed the dynamics of total, IgA, IgM, and IgG antibodies in COVID-19 patients during the convalescent phase to understand the kinetics of antibody response in recovered patients.
In Shenzhen, all COVID-19 patients were treated in a designated hospital (the Shenzhen Third People’s Hospital). Patients were discharged if they met the following criteria: a) normal body temperature for more than three days, b) significant improvement in respiratory symptoms, c) significant improvement in absorption of acute exudative lesions on lung imaging, and d) negative nucleic acid testing of two consecutive respiratory specimens (at least one day between samplings). After discharge from the designated hospital, the recovered patients were also required to be quarantined in the Shenzhen Sami Medical Center for 14 days of medical observation. Considering the possibility of extended duration of viral shedding in feces [7], nasopharyngeal and anal swabs were collected for SARS-CoV-2 RT-PCR on days 7 and 14 during the 14-day quarantine period. Serum was collected at least one time for antibody testing. If patients remained negative (non-re-positive) by RT-PCR during the 14-quarantine period, they were discharged. If the patient tested positive again by RT-PCR (re-positive), they were re-admitted to the designated hospital. After discharge, all patients were also invited to make semi-monthly or monthly follow-up visits for collection of blood samples for antibody testing. All participants provided written informed consent. This study retrospectively reviewed and analyzed the medical records of recovered COVID-19 patients in the Shenzhen Sami Medical Center. A total of 143 recovered COVID-19 patients with an exact date of symptom onset available were included in this study. This study was approved by Medical Ethical Committee of Shenzhen Sami Medical Center.
Nasopharyngeal and anal swabs were sent to the Shenzhen Center for Disease Control and Prevention (CDC) for RT-PCR testing as described previously [6]. Briefly, nucleic acids were extracted using a High Pure Viral RNA Kit (Roche, Mannheim, Germany). Nucleic acid amplification and identification were performed using a commercial RT-PCR assay targeting the ORF1ab and N genes of SARS-CoV-2 (Bio-Germ, Shanghai, China). The assay was approved by China's National Medical Products Administration. Total, IgA, IgM, and IgG (all IgG subclasses) antibodies against the receptor-binding domain (RBD) of the SARS-CoV-2 spike protein in the sera of COVID-19 patients were detected using a commercial chemiluminescence microparticle immunoassay (CMIA, Xiamen InnoDx Biotech Co., Ltd., China) as described previously [8]. Briefly, total antibody detection was based on a double-antigen sandwich method. A capture method was applied to detect the IgM antibodies. For the detection of IgG and IgA, an indirect method was used. The test procedure for all assays was performed following the manufacturer's instructions. Antibody levels were expressed as relative binding signals compared to the cutoff value of each assay (S/CO). The sensitivity of detection of total, IgM, IgG, and IgA antibodies was 96.3%, 86.3%, 99.6% and 91.9%, respectively. The specificity of detection of total, IgM, IgG, and IgA antibodies was 99.3%, 99.3%, 99.0%, and 98.6%, respectively (unpublished data for IgG and IgA) [8].
Geometric mean titers (GMTs) with 95% confidence interval were calculated for total, IgA, IgM, and IgG antibodies by days post-symptom-onset. Multivariable log-binomial regression models with generalized estimating equations were used to compare the antibody level between the re-positive and non-re-positive groups, adjusted for age and days post-onset. The probability of seropositivity and median time to seronegativity were calculated using the Kaplan-Meier method and compared using the log-rank test. All analyses were performed using SAS software (version 9.4), and a p-value less than 0.05 was considered statistically significant.
From March 4 to April 29, 2020, 306 recovered COVID-19 patients were under medical observation at the Shenzhen Sami Medical Center. From among these patients, 143 (46.7%) with an exact symptom onset date available were included in this study. During the 14-day medical observation period, a total of 23 (16.1%) recovered patients tested positive again by RT-PCR: 14 (60.9%, 14/23) on day 7 and 9 (39.1%) on day 14. As shown in Table 1, the median duration of treatment at the designated hospital was 31 days (IQR: 23-39). The median age (IQR) of the re-positive and non-re-positive patients was 50 (34, 58) and 50 (37, 61), respectively. Sixty-seven patients (46.9%) were male. The median follow-up was 45 days (IQR: 40-70, range: 26-96) post-onset for all patients. Patients with re-positive RT-PCR results were followed up longer than those without (72 days vs. 44 days post-onset).
Table 1.
Baseline characteristics of patients
| Variable | Re-positive | Non-re-positive | Total |
|---|---|---|---|
| Number | 23a | 120 | 143 |
| Age (median, IQR) | 50 (34, 58) | 50 (37, 61) | 50 (37, 61) |
| Gender (%) | |||
| Male | 7 (30.4) | 60 (50.0) | 67 (46.9) |
| Female | 16 (69.6) | 60 (50.0) | 76 (53.1) |
| Duration of treatment in the designated hospital | |||
| Median (IQR) | 35 (27, 49) | 30 (22, 39) | 31 (23, 39) |
| No. of sera samples collected | |||
| Of each case, median (IQR) | 4 (2, 5) | 1 (1, 1) | 1 (1, 3) |
| Total | 85 | 190 | 275 |
| Duration of serological follow-up (days after discharge from the designated hospital) | |||
| Median (IQR) | 27 (21, 36) | 10 (5, 14) | 13 (5, 15) |
| Duration of clinical follow-up (days after symptom onset) | |||
| Median (IQR) | 72 (62, 80) | 44 (39.5, 62) | 45 (40, 70) |
| Range | 40, 91 | 26, 96 | 26, 96 |
a19 (82.6%) patients tested positive only with nasopharyngeal swabs, 4 (17.4%) only with anal swabs.
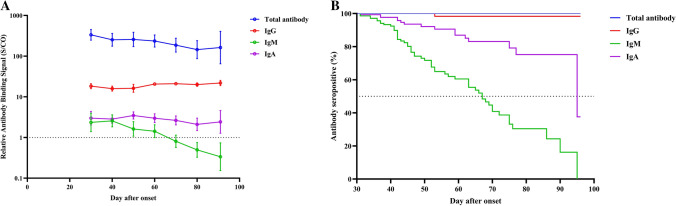
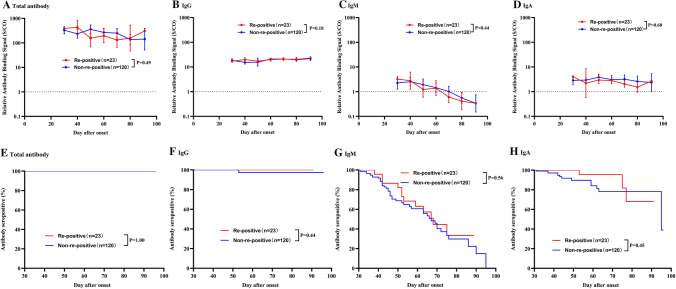
A total of 275 blood samples were collected from 143 patients, with a median of 4 and 1 samples from the re-positive and non-re-positive group, respectively. All recovered patients were seropositive for total antibodies in their first sample taken at the medical center. As shown in Fig. 1, total antibody levels slowly decreased and remained at high S/CO values around three months after onset (GMT = 162.5, 95%CI: 64.7-408.0). No patient was seronegative at the last sampling time. For IgG antibodies, the levels remained at a plateau from 1 month to 3 months after onset. Only one patient was negative for IgG antibody at 53 days after onset. IgM antibody levels declined faster than those of other antibodies, with a median time to seroconversion of 67 (95% CI: 59, 75) days after onset. The rate of decline of IgA antibodies was intermediate between those of IgM and IgG. Approximately 25% of patients were seronegative for IgA antibodies at three months post-onset. The antibody levels and the seropositivity rate for total, IgG, IgM, and IgA were similar among patients with and without re-positive RT-PCR (all p-values higher than 0.05, Fig. 2 and Supplementary Fig. S1).
Fig. 1.
Antibody kinetics of all COVID-19 patients during the convalescent phases (n = 143). A Changes in levels of total, IgG, IgM and IgA antibodies. B Kaplan-Meier estimates of the probability of seropositivity for total, IgG, IgM, and IgA antibodies
Fig. 2.
Comparison of antibody kinetics in COVID-19 patient with and without re-positive RT-PCR results during the convalescent phase. Comparison of antibody levels: A total antibody, B IgG antibody, C IgM antibody, D IgA antibody. P-values were estimated using multivariable log-binomial regression models with generalized estimating equations adjusted for age and days after symptom onset. Kaplan-Meier curves comparing the prevalence of seropositivity: E total antibody, F IgG antibody, G IgM antibody, H IgA antibody. P-values were estimated using the log-rank test
This study confirmed that IgG antibodies remained stable for three months, consistent with other current studies [9, 10]. The antibody response to SARS-CoV-2 infection may be similar to that of SARS-CoV-1 or other viral infections [11]. The IgM antibody response was transient, and in half of the patients, IgM antibodies were undetectable after about two months. Therefore, similar to other viral infections [12], IgM antibodies against SARS-CoV-2 may be a marker of acute SARS-CoV-2 infection.
Antibody levels and seropositivity rates for total, IgG, IgM, and IgA antibodies were similar in patients with and without re-positive RT-PCR results. Studies have shown that the SARS-CoV-2 nucleic acids can be detected in lower-respiratory-tract specimens and feces for up to 50 days [5, 7]. In addition, the sample quality, sensitivity, and specificity of commercial test kits may result in false-negative results, which to some extent explains why some patients become positive again after a negative result [3]. One study showed that IgG and IgM levels were similar in re-positive and non-re-positive patients, which is in agreement with this study [3]. Another preprint study showed that IgG and IgM antibody levels did not correlate with the clinical course of disease and therefore could not be used as a predictor of disease progression.
There are some limitations of this study. The convenience of collection of blood samples resulted in an uneven distribution of sample size over time. Most non-re-positive patients provided only one blood sample during the 14-day quarantine period. They were reluctant to come back to provide blood samples after being discharged. Also, the duration of the follow-up was relatively short.
In conclusion, this study confirmed that high antibody titers for total and IgG antibodies persisted for three months in recovered COVID-19 patients. IgM antibodies declined rapidly, with a median time to seroconversion of about two months. IgA antibodies declined more slowly than IgM but more rapidly than IgG. Antibody responses were similar between patients with and without re-positive RT-PCR during the convalescent phase.
Supplementary Information
Below is the link to the electronic supplementary material.
Author contributions
Conceptualization was performed by JP and C-QS. Investigation was performed by JP, Z-YL, X-YC, KZ and YL. Data curation was performed by Z-YL, X-JY, X-YC, KZ, YL, and C-QS. Methodology was performed by Z-YL, Y-YS and C-QS. Formal analysis was performed by X-JY and Y-YS. Writing of the original draft was performed by JP, Z-YL, X-JY and Y-YS. Reviewing and editing were performed by Y-YS and C-QS. All authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Availability of data and material
The data will be available beginning 9 months and ending 36 months following article publication. The data will be shared with investigators whose proposed use of the data has been approved by an independent review committee identified for individual participant data meta-analysis. Proposals should be directed to yingyingsu@xmu.edu.cn or sunchangqing@ssmc-sz.com. To gain access, data requestors will need to sign a data access agreement.
Code availability
Software application.
Declarations
Conflict of interest
The authors have declared that no conflict of interest exists.
Ethical approval
This study was approved by Medical Ethical Committee of Shenzhen Sami Medical Center.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Jing Peng, Zhi-Yong Liu, Xiao-Juan Yu contributed equally to this work.
Contributor Information
Ying-Ying Su, Email: yingyingsu@xmu.edu.cn.
Chang-Qing Sun, Email: sunchangqing@ssmc-sz.com.
References
- 1.Word Health Organization (2021) Coronavirus disease (COVID-19) situation report. https://www.who.int/publications/m/item/weekly-epidemiological-update---12-january-2021. Accessed 13 Jan 2021
- 2.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W, I. China Novel Coronavirus and T. Research A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.An JH, Liao XJ, Xiao TY, Qian S, Yuan J, Ye HC, Qi FR, Shen CG, Liu Y, Wang LF, Cheng XY, Li N, Cai QX, Wang F, Chen J, Liu YX, Wang YF, Zhang F, Fu Y, Tan XH, Chen J, Liu YX, Wang YF, Zhang F, Fu Y, Tan XH, Liu L, Zhang Z. Clinical characteristics of the recovered COVID-19 patients with re-detectable positive RNA test. Ann Transl Med. 2020;8(17):1084. doi: 10.21037/atm-20-5602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lan L, Xu D, Ye GM, Xia C, Wang SK, Li YR, Xu HB. Positive RT-PCR test results in patients recovered from COVID-19. JAMA. 2020;323(15):1502–1503. doi: 10.1001/jama.2020.2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yao XH, He ZC, Li TY, Zhang HR, Wang Y, Mou H, Guo Q, Yu SC, Ding Y, Liu X, Ping YF, Bian XW. Pathological evidence for residual SARS-CoV-2 in pulmonary tissues of a ready-for-discharge patient. Cell Res. 2020;30(6):541–543. doi: 10.1038/s41422-020-0318-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yuan B, Liu HQ, Yang ZR, Chen YX, Liu ZY, Zhang K, Wang C, Li WX, An YW, Wang JC, Song S. Recurrence of positive SARS-CoV-2 viral RNA in recovered COVID-19 patients during medical isolation observation. Sci Rep. 2020;10(1):11887. doi: 10.1038/s41598-020-68782-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu Y, Guo C, Tang L, Hong Z, Zhou J, Dong X, Yin H, Xiao Q, Tang Y, Qu X, Kuang L, Fang X, Mishra N, Lu J, Shan H, Jiang G, Huang X. Prolonged presence of SARS-CoV-2 viral RNA in faecal samples. Lancet Gastroenterol Hepatol. 2020;5(5):434–435. doi: 10.1016/s2468-1253(20)30083-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lou B, Li TD, Zheng SF, Su YY, Li ZY, Liu W, Yu F, Ge SX, Zou QD, Yuan Q, Lin S, Hong CM, Yao XY, Zhang XJ, Wu DH, Zhou GL, Hou WH, Li TT, Zhang YL, Zhang SY, Fan J, Zhang J, Xia NS, Chen Y. Serology characteristics of SARS-CoV-2 infection after exposure and post-symptom onset. Eur Respir J. 2020 doi: 10.1183/13993003.00763-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gudbjartsson DF, Norddahl GL, Melsted P, Gunnarsdottir K, Holm H, Eythorsson E, Arnthorsson AO, Helgason D, Bjarnadottir K, Ingvarsson RF, Thorsteinsdottir B, Kristjansdottir S, Birgisdottir K, Kristinsdottir AM, Sigurdsson MI, Arnadottir GA, Ivarsdottir EV, Andresdottir M, Jonsson F, Agustsdottir AB, Berglund J, Eiriksdottir B, Fridriksdottir R, Gardarsdottir EE, Gottfredsson M, Gretarsdottir OS, Gudmundsdottir S, Gudmundsson KR, Gunnarsdottir TR, Gylfason A, Helgason A, Jensson BO, Jonasdottir A, Jonsson H, Kristjansson T, Kristinsson KG, Magnusdottir DN, Magnusson OT, Olafsdottir LB, Rognvaldsson S, le Roux L, Sigmundsdottir G, Sigurdsson A, Sveinbjornsson G, Sveinsdottir KE, Sveinsdottir M, Thorarensen EA, Thorbjornsson B, Thordardottir M, Saemundsdottir J, Kristjansson SH, Josefsdottir KS, Masson G, Georgsson G, Kristjansson M, Moller A, Palsson R, Gudnason T, Thorsteinsdottir U, Jonsdottir I, Sulem P, Stefansson K. Humoral immune response to SARS-CoV-2 in Iceland. N Engl J Med. 2020;383(18):1724–1734. doi: 10.1056/NEJMoa2026116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seow J, Graham C, Merrick B, Acors S, Steel KJA, Hemmings O, O'Bryne A, Kouphou N, Pickering S, Galao R, Betancor G, Wilson HD, Signell AW, Winstone H, Kerridge C, Temperton N, Snell L, Bisnauthsing K, Moore A, Green A, Martinez L, Stokes B, Honey J, Izquierdo-Barras A, Arbane G, Patel A, L. OConnell, G. O Hara, E. MacMahon, S. Douthwaite, G. Nebbia, R. Batra, R. Martinez-Nunez, J. D. Edgeworth, S. J. D. Neil, M. H. Malim and K. Doores, Longitudinal observation and decline of neutralizing antibody responses in the three months following SARS-CoV-2 infection in humans. Nat Microbiol. 2020;5:1598–1607. doi: 10.1038/s41564-020-00813-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alter G, Seder R. The power of antibody-based surveillance. N Engl J Med. 2020;383(18):1782–1784. doi: 10.1056/NEJMe2028079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ponde RAA. The serological markers of acute infection with hepatitis A, B, C, D, E and G viruses revisited. Arch Virol. 2017;162(12):3587–3602. doi: 10.1007/s00705-017-3538-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data will be available beginning 9 months and ending 36 months following article publication. The data will be shared with investigators whose proposed use of the data has been approved by an independent review committee identified for individual participant data meta-analysis. Proposals should be directed to yingyingsu@xmu.edu.cn or sunchangqing@ssmc-sz.com. To gain access, data requestors will need to sign a data access agreement.
Software application.