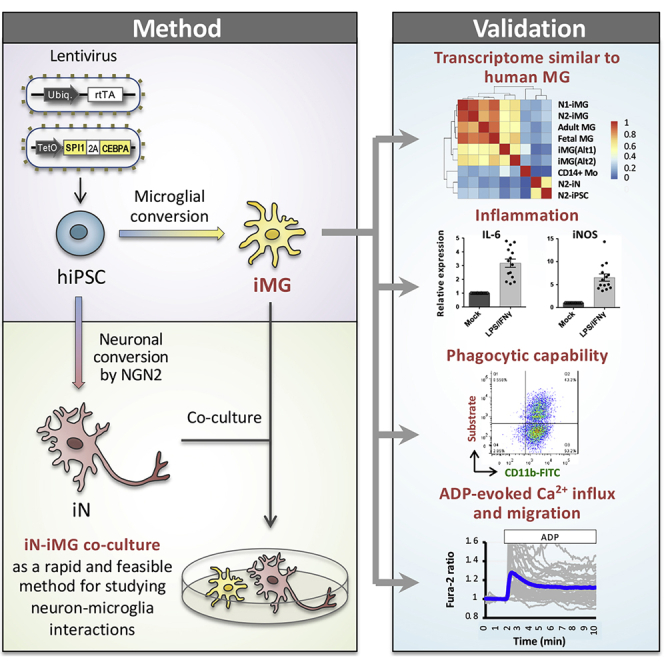
Summary
Microglia, the immune cells of the central nervous system, play critical roles in brain physiology and pathology. We report a novel approach that produces, within 10 days, the differentiation of human induced pluripotent stem cells (hiPSCs) into microglia (iMG) by forced expression of both SPI1 and CEBPA. High-level expression of the main microglial markers and the purity of the iMG cells were confirmed by RT-qPCR, immunostaining, and flow cytometry analyses. Whole-transcriptome analysis demonstrated that these iMGs resemble human fetal/adult microglia but not human monocytes. Moreover, these iMGs exhibited appropriate physiological functions, including various inflammatory responses, ADP/ATP-evoked migration, and phagocytic ability. When co-cultured with hiPSC-derived neurons, the iMGs respond and migrate toward injured neurons. This study has established a protocol for the rapid conversion of hiPSCs into functional iMGs, which should facilitate functional studies of human microglia using different disease models and also help with drug discovery.
Keywords: induced pluripotent stem cells, induced microglia, SPI1/PU.1, CEBPA, reprogramming
Graphical abstract
Highlights
-
•
Efficient generation of human iMGs from iPSCs by forced expression of SPI1 and CEBPA
-
•
The transcriptome profile of iMGs resembles that of human primary microglia
-
•
The iMG cells possess appropriate physiological functioning
-
•
An iN-iMG co-culture model is established for studying neuron-microglia interactions
Chen et al. report a novel approach that produces human microglia (iMG) from induced pluripotent stem cells within 10 days by forced expression of SPI1 and CEBPA together. These iMGs exhibit appropriate physiological functions and can be co-cultured with hiPSC-derived neurons, allowing the study of in vitro neuron-microglia interactions under both normal and disease conditions.
Introduction
Microglia are the innate immune cells of the central nervous system (CNS), and account for 10%–15% of all glial cells (von Bartheld et al., 2016); they play important roles in brain homeostasis and functioning. During development, microglia regulate neural progenitor cell death and survival, and also contribute to the maintenance of normal neurogenesis, oligodendrogenesis, and myelinogenesis (Shigemoto-Mogami et al., 2014; Ueno et al., 2013). They also control axon tract fasciculation and modulate synaptogenesis and neural circuit formation via the release of diffusible factors (Miyamoto et al., 2016; Schafer et al., 2012). During brain development, their regulation of cell genesis and synapse pruning is accompanied by the ability to eliminate redundant neuronal precursor cells and promote developmental neuronal apoptosis via the removal of cell corpses by phagocytosis (Cunningham et al., 2013; Paolicelli et al., 2011). In the adult brain, microglia behave as the brain's primary immune cells and carry out various major innate immune functions, including release of inflammatory cytokines, phagocytosis of pathogens, and removal of cell debris. Microglia, via highly dynamic processes, constantly survey the local environment at a speed estimated to cover the entire parenchyma within a few hours (Davalos et al., 2005; Nimmerjahn et al., 2005). Moreover, microglia play a critical role in a number of neurological disorders, including schizophrenia, depression, Alzheimer's disease, Parkinson's disease, and amyotrophic lateral sclerosis (Perry et al., 2010; Sellgren et al., 2019; Yirmiya et al., 2015). This highlights a need to improve our understanding of their functioning in healthy and diseased brains. Nevertheless, studying human microglia is challenging, because primary cells are available only in limited amounts from human fetal/adult CNS tissue. Therefore, there is a need for a renewable source of human microglia.
Microglia are derived from myeloid progenitor cells that originate from the embryonic yolk sac (Takahashi et al., 1989). Myeloid precursor cells exit the yolk sac blood islands at the onset of blood circulation and colonize the neuroepithelium, starting from embryonic day (E)9.5 in the mouse embryo, to give rise to microglia (McGrath et al., 2003). The blood-brain barrier starts to form from E13.5, and this isolates the developing brain from any contribution from cells produced by fetal liver hematopoiesis (Daneman et al., 2010). During development, various transcription factors (TFs), including RUNX1 (Runt-related transcription factor 1), SPI1 (SFFV pro-viral integration 1), IRF8 (interferon regulatory factor 8), TAL1 (stem cell leukemia/T cell acute lymphoblastic leukemia 1), and SALL1 (Sal-like protein 1), orchestrate the commitment of yolk sac myeloid precursors into brain microglia (Buttgereit et al., 2016; Kierdorf et al., 2013; Rapino et al., 2013; Smith et al., 2013; Wehrspaun et al., 2015). These TFs act in a combinatorial manner to promote an appropriate cell fate and to maintain microglial identity. In addition, factors such as IL-34 (interleukin 34) and TGF-β (transforming growth factor β) are released in discrete regions of the brain to promote microglial function and terminal differentiation (Butovsky et al., 2014; Wang et al., 2012). Embryonic microglia undergo slow proliferation during the embryonic stage, but proliferation increases dramatically during the early postnatal stage. Throughout adulthood the microglial population is homeostatic and is maintained via local self-renewal (Ginhoux et al., 2010).
Since the isolation of human embryonic stem cells (hESCs) and the development of human induced pluripotent stem cell (hiPSC) technology, various protocols have been developed to generate microglia from hiPSCs via recapitulation in vitro of appropriate molecular signals and re-creation of the events occurring during in vivo microglial development (Abud et al., 2017; Claes et al., 2019; Douvaras et al., 2017; Haenseler et al., 2017; Muffat et al., 2016; Pandya et al., 2017). However, these protocols remain inefficient, are quite variable in terms of their microglia yield, and, most importantly, require >40 days for the cells to differentiate into functional microglia. It has been reported that PSCs can be converted into specific cell types in a short time by turning on a “master regulator,” such as a TF at the top of the gene regulation hierarchy (Davis and Rebay, 2017). Using distinct sets of TFs, fibroblasts or PSCs can be reprogrammed into a number of cell types found in the brain, including glutamatergic neurons, dopaminergic neurons, GABAergic neurons, serotonergic neurons, motoneurons (Caiazzo et al., 2011; Son et al., 2011; Xu et al., 2016; Yang et al., 2017; Zhang et al., 2013), and astrocytes and oligodendrocytes (Tcw et al., 2017; Yang et al., 2013). Therefore, alternative and better approaches to inducing microglia differentiation from hiPSCs were explored, and we used a Tet-On system to examine whether induced expression of “pro-microglial” genes in hiPSCs can initiate microglial differentiation.
By screening seven of the TFs involved in defining microglial cell fate during embryogenesis, we found that overexpression of two genes, SPI1 and CEBPA, in hiPSCs led to the generation of IBA1-positive microglia-like cells within 10 days. The transcriptome profile of these hiPSC-derived microglia-like cells (induced microglia, or iMGs) resembles human primary microglia, and they show similar physiological functioning, including lipopolysaccharide/interferon-γ (LPS/IFN-γ)-induced inflammatory responses, phagocytic ability, and ADP/ATP-evoked signaling/migration. In addition, we also developed a rapid protocol for co-culturing hiPSC-derived neurons (iNs) with iMGs using our reprogramming. The interaction between iMGs and iNs was assessed using time-lapse imaging and laser ablation. Taken together, the results of this study establish a protocol to rapidly convert hiPSCs into functional iMGs, creating a useful tool for research into human microglia, both in the healthy brain and in the disease brain.
Results
Identification of the minimal set of transcription factors that allows hiPSC-to-MG conversion
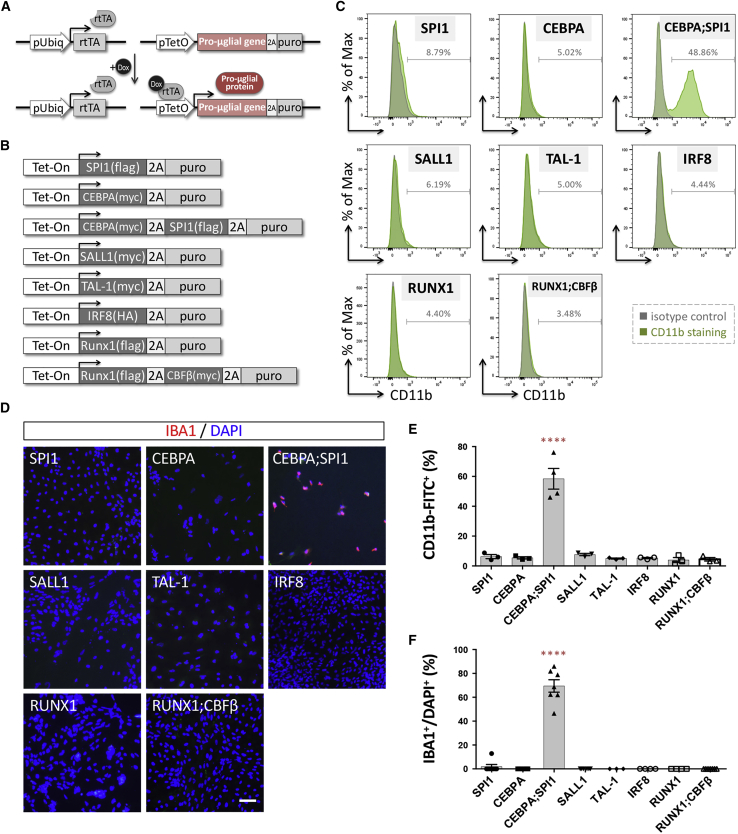
hESCs and hiPSCs can be converted into functional neurons in less than 2 weeks by forced expression of neurogenin 2 (NGN2), a pro-neural gene encoding a TF of the basic helix-loop-helix class (Zhang et al., 2013). Inspired by this, we adopted a similar procedure and established a protocol to examine whether forced expression of a "pro-microglial" gene in hiPSCs might initiate microglial differentiation (Figure 1A). We selected seven candidate TFs known to play pivotal roles in the development/maintenance of microglial identity (Buttgereit et al., 2016; Kierdorf et al., 2013; Rapino et al., 2013; Smith et al., 2013; Wehrspaun et al., 2015) (Figure 1B). We used a lentiviral delivery system to constitutively express reverse tetracycline-controlled transactivator (rtTA) and the inducible expression of the candidate TFs driven by a TetO promoter. The tetracycline-inducible expression of protein candidates in hiPSCs was verified by western blotting (Figure S1B). Interestingly, we found that overexpressing either of the candidate factors was able to rapidly induce morphological changes in hiPSCs under phase-contrast microscopy (Figure S1C). After 1 week of differentiation, cells overexpressing SPI1 and its cofactor, CCAAT/enhancer-binding protein α (CEBPA), showed a microglia-like morphology that resembled mouse primary cultured microglia and the BV2 microglial cell line (Figures 2B and S1D). To further confirm the microglial identity of these hiPSC-derived cells, expression of the microglial surface marker CD11b was assessed by flow cytometry analysis. Only a combination of SPI1 and CEBPA, among all seven candidate TF combinations, induced a significant number of CD11b+ cells (Figures 1C and 1E, and Table S1). Moreover, using the same immunolabeling procedure, expression of a typical microglial marker, IBA1, was exclusively found in cells overexpressing both SPI1 and CEBPA (Figures 1D and 1F). Because this conversion was based on forced expression of these two lineage-specific TFs and might have skipped the step-to-step transitions that occur during early embryonic development, we refer to the resulting microglia-like cells as iMGs.
Figure 1.
Ectopic expression of pro-microglial factors in hiPSCs induces expression of various microglial markers
(A) The lentiviral vectors used for pro-microglial (pro-μglial) gene-mediated conversion of hiPSCs to microglia. hiPSCs were sequentially transduced with lentivirus expressing rtTA and then a Tet-On promoter-driven pro-microglial gene linked to puromycin resistance by T2A.
(B) Eight candidate transcription factor constructs involved in defining microglial cell fate during embryogenesis from the literature.
(C) Flow cytometry analysis shows CD11b expression in N2-iPS cells (iN2) ectopically expressing the candidate genes. Green, CD11b antibody conjugated to fluorescein isothiocyanate (FITC); gray, isotype control. The figure is representative of three independent experiments.
(D) Analysis of IBA1 immunoreactivity (red) of iN2 cells ectopically expressing the candidate genes. Cell nuclei are stained with DAPI. Scale bar, 50 μm.
(E) Quantification of CD11b+ cells present among iN2 cells ectopically expressing the candidate genes. Data are presented as means ± standard error of the mean (SEM; n = 3–4 batches of independent differentiation). ∗∗∗∗p < 0.0001 by one-way ANOVA with Tukey's post hoc test.
(F) Quantification of the IBA1+ cells present among hiPSCs ectopically expressing the candidate genes. Data are presented as means ± SEM (n = 3–7 batches of independent differentiation). ∗∗∗∗p < 0.0001 by one-way ANOVA with Tukey's post hoc test.
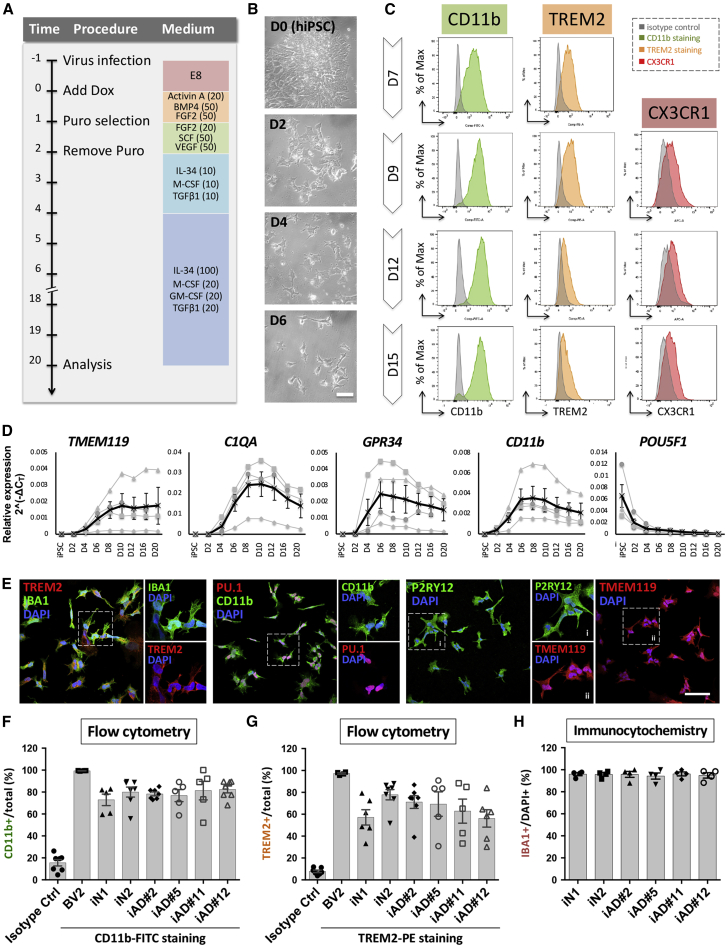
Figure 2.
Differentiation of hiPSC-derived microglia-like cells
(A) Flow diagram of generation of induced microglia. Numbers in parentheses indicate the concentration of human recombinant proteins in ng/mL.
(B) Representative images of hiPSC-derived cells during differentiation. iN2s differentiated into microglia-like cells within 1 week. Scale bar, 50 μm.
(C) Flow cytometry analysis shows the kinetics of expression of CD11b, TREM2, and CX3CR1 by the induced microglia (N2-iMG) at 7 to 15 days of differentiation. Green, CD11b antibody conjugated to FITC; orange, TREM2 antibody conjugated to phycoerythrin; red, CX3CR1 antibody conjugated to Alexa Fluor 647; gray, isotype control. The figure is representative of three independent experiments.
(D) Time course of mRNA expression levels of key microglial markers (TMEM119, C1QA, GPR34, CD11b) and a stem cell marker (POU5F1) by RT-qPCR (normalized against a reference gene, RPL13A). Data points in black are the means of four batches of N2-iMG induction together with the SEM.
(E) Representative images of N2-iMG cells immunostained for microglial markers IBA1, TREM2, CD11b, TMEM119, P2RY12, PU.1/SPI1, and DAPI. Scale bar, 50 μm.
(F and G) Quantification of the CD11b+ (F) or TREM2+ (G) cells present among iMG cells by flow cytometry. Data are the means ± SEM (n = 5–7 batches of independent differentiation).
(H) The differentiation efficiency is shown as IBA1+ over total DAPI+ cells. Data are presented as means ± SEM (n = 4 batches of independent differentiation and 30–70 cells in each batch).
Optimizing the culture medium composition for hiPSC-to-iMG conversion
During pilot experiments, forced expression of both SPI1 and CEBPA converted hiPSCs into iMGs at a yield of nearly 70% after less than 2 weeks (Figure 1F). To further optimize the medium and growth factors needed for efficient differentiation of hiPSCs into iMGs, we tested a range of culture medium ingredients that might aid differentiation. Previous evidence had indicated that BMP4, activin A, VEGF, and SCF are critical for initiating the microglial ontogeny of primitive hematopoietic progenitors, and that the ligands of the CSF1 receptor, IL-34 and M-CSF, GM-CSF, as well as TGF-β, are able to induce microglial morphology and maintain their cell survival (Schilling et al., 2001; Wang et al., 2012; Wei et al., 2010) (Figures S2A–S2C). Because recapitulation of microglial ontogeny through hematopoiesis under hypoxic circumstances is critical to microglia development in vitro (Abud et al., 2017), we also examined two more conditions during the first 2 days of induction, namely hypoxic cell culture (5% O2 and 5% CO2) and the addition of β-mercaptoethanol (β-ME), a potent reducing agent used to reduce the levels of oxygen radicals (Figure S2D). By flow cytometry analysis of microglial surface marker expression of CD11b and TREM2, it was found that the added components under the Standard A condition, with the addition of 50 μM β-ME during the first 2 days of differentiation, induced the highest number of iMGs (red frames in Figures S2B and S2E). This medium and an appropriate differentiation protocol were used for all subsequent experiments (Figure 2A).
Characterization of iMGs
To further support the microglial identity of our iMGs, we assessed the dynamics of microglial marker expression by flow cytometry between 7 and 15 days after iMG induction. The percentage of cells expressing the microglial surface markers, CD11b, TREM2, and CX3CR1, increased over this time, reaching a maximum after 9 days (Figure 2C). The temporal expression of various other classical microglial markers, namely TMEM119, C1QA, GPR34, and CD11b, was also examined by qRT-PCR. We found that there were clear increases in expression of these microglial markers after 6 days of induction and that they reached their highest levels on either day 8 or day 10. Furthermore, and importantly, markers for neurons (MAP2 and RBFOX3), astrocytes (ALDH1L1 and GFAP), and oligodendrocytes (MAG) were barely detectable in iMGs (Figure S3A). The stem cell marker POU5F1 was almost absent from day 2 onward (Figure 2D). These results reveal that forced expression of both SPI1 and CEBPA in hiPSCs, plus appropriate growth conditions, is capable of converting hiPSCs into iMGs within 10 days, much faster than any other currently available method for generating microglia-like cells from hiPSCs or hESCs.
Furthermore, immunostaining at 9 days after induction showed that various typical microglial markers, IBA1, TREM2, CD11b, PU.1/SPI1, P2RY12, and TMEM119, were all expressed by these iMGs (Figure 2E). Quantification of the IBA1+ cells present showed that our protocol yielded iMGs of high purity (>93%, n = 5) (Figure 2H). To access the reproducibility of our protocol, we examined a panel of six hiPSC lines derived from healthy and diseased individuals (Wu et al., 2019) and successfully generated iMGs from all lines. The resulting iMGs expressed typical microglial markers by qRT-PCR, flow cytometry, and immunostaining analysis (Figures 2F–2H and S3). On average, the yields ranged from 4.1% to 25.5% of the number of input hiPSCs, and the final populations' cellular homogeneity was almost pure monocultures of iMGs at day 9 based on immunostaining of IBA1+ (Figures 2H and S3C). The yield of iMG cells varied across the lines without correlation to sex, age of biopsy, or donor disease status, although the genetic background of the hiPSC donor did seem to affect the kinetics of differentiation and the iMG cell yield. It can be concluded that our iMG differentiation protocol is both efficient and reproducible across several hiPSC lines.
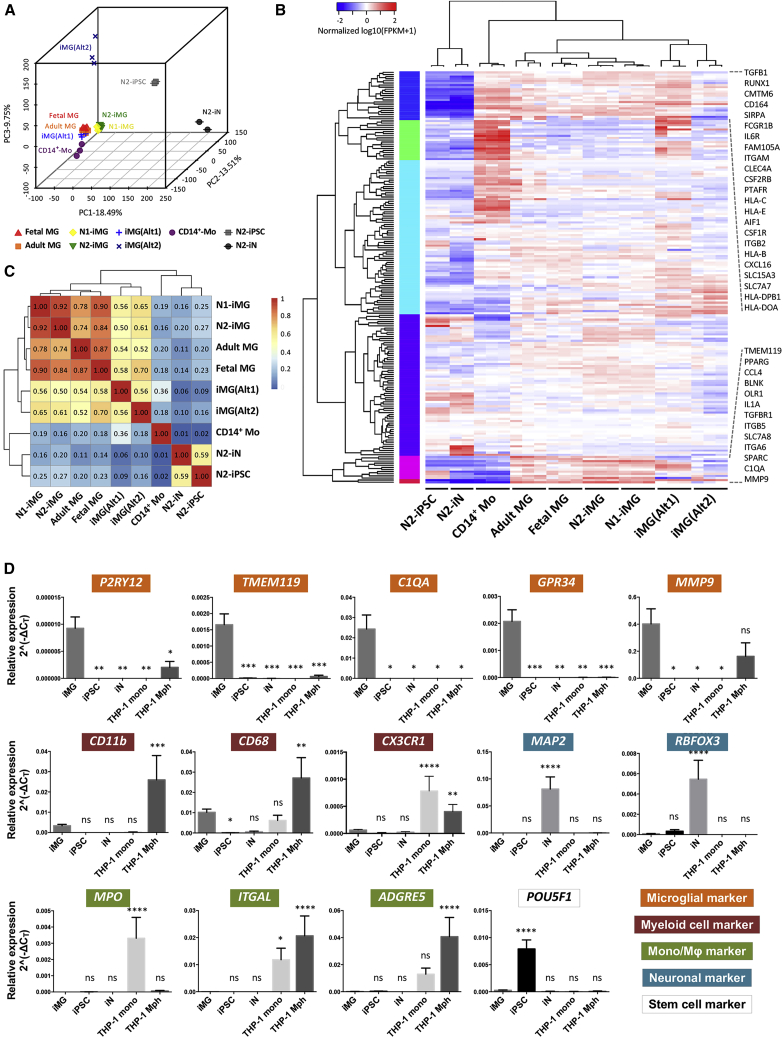
Transcriptome profiling confirms iMG identity
To further validate the microglial phenotype of our iMGs, we compared the whole-transcriptome profile of the iMGs derived from two independent hiPSC lines (N1-iMG and N2-iMG), on day 11 or day 12, with hiPSCs (N2-iPSC), iNs (N2-iN), and those previously reported for human primary fetal microglia (fetal MG), adult microglia (adult MG), adult blood-derived CD14+/CD16− monocytes (CD14+-Mo), and two hiPSC-derived microglia profiles created by alternative methods, iMG(Alt1) (Abud et al., 2017) and iMG(Alt2) (Brownjohn et al., 2018). Principal component analysis was used to assess the similarity of the above cell types. For the first two principal components (PC1 and PC2), human fetal/adult MGs, iMG(Alt1), and CD14+-Mo were located in close proximity to one another, possibly because of the same culture methods, the same ethnicity and similar genetic background, and/or the same sequencing method; in addition, these four samples were from the same databank/laboratory (Figures 3A and S4B) (Abud et al., 2017). Based on PC2 and PC3, the analyzed samples separated into six distinct groups; these consisted of (1) human fetal/adult MGs and iMG(Alt1) from the same databank, (2) N1-iMG and N2-iMG, (3) CD14+ monocytes, (4) iMG(Alt2), (5) N2-iPSC, and (6) N2-iN. Our N1/N2-iMGs showed a clear separation from the hiPSCs and iNs, and were closer to the fetal and adult microglia populations. Moreover, a clustered heatmap and Spearman correlation matrix of 195 selected microglia/monocyte/macrophage-related genes showed that our iMG cells had an expression pattern similar to that of human fetal/adult MGs and are distinct from CD14+ monocytes (Figures 3B and 3C, and Table S3). Importantly, differential analyses between N1-iMG, N2-iMG, fetal/adult MGs, iMG(Alt1), iMG(Alt2), and CD14+ monocytes revealed that our iMG cells express many major microglial genes, including P2RY12, TMEM119, GPR34, C1QA, MMP9, SALL1, CD11b, CX3CR1, MERTK, and PROS1, at expression levels similar to those of human fetal and adult MGs (Figure S4C). The transcriptome profiling findings thus confirm that our iMGs closely resemble human fetal and adult microglia and are quite distinct from monocytes.
Figure 3.
iMG cells express consensus microglial markers
(A) Three-dimensional principal component analysis of N1- and N2-iMG cells (yellow and green, respectively) by whole-transcriptome sequencing of protein coding genes. The profiles of hiPSC (N2-iPSC), iN (N2-iN), and iMG cells derived from two hiPSC lines with different genetic backgrounds, N1-iMG and N2-iMG, were merged with the dataset from Abud et al. (2017) and Brownjohn et al. (2018), including cultured human primary microglia from adult and fetal microglia, iMG cells from two different methods (iMG(Alt1) and iMG(Alt2)), and CD14+ peripheral blood monocytes. Each spot represents one independently differentiated cell batch and each cell type is coded (different colors and shapes).
(B) Heatmap of 195 microglial, myeloid, and other immune-related genes. A pseudo-color is used to present the log10-transformed FPKM values (FPKM+1).
(C) Spearman correlation matrix for correlations between different cell DeSeq2 rlog-transformed raw counts of genes used in (B). Median rlog gene counts of the biological replicates were used as input. The color shows the strength and direction of the correlation.
(D) Expression of key microglial markers (P2RY12, TMEM119, C1QA, GPR34, MMP9), myeloid cell markers (CD11b, CD68, CX3CR1), monocyte and macrophage markers (MPO, ITGAL, ADGRE5), neuronal markers (MAP2, RBFOX3), and a stem cell marker (POU5F1) in N2-iMG cells after 9–12 days of induction (obtained by qRT-PCR). Fold change was calculated using the ΔCT method with RPL13A as an endogenous control. Data are means ± SEM (n = 3–11 independent experiments). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001; ns, not significant. All compared with the iMG sample by one-way ANOVA with Fisher's least-significant difference (LSD) multiple comparisons.
We next used qRT-PCR analysis to identify the molecules that are specific to microglia and are not present in CNS neurons or other peripheral myeloid cells. By comparing our iMGs with iNs, THP1 (a human monocytic cell line), and THP1-derived macrophages, we found that iMG cells expressed canonical microglial genes, such as TMEM119, P2RY12, GPR34, C1QA, and MMP9, but had lower or no expression of three monocyte/macrophage markers, MPO, ITGAL, and ADGRE5; two neuronal markers, MAP2 and RBFOX3; and the stem cell marker POU5F1 (Figure 3D). Thus, the forced expression of both SPI1 and CEBPA, together with the addition of appropriate growth factors and cytokines to the culture, is able and sufficient to activate in vitro a microglial gene program in hiPSCs.
Functional characterization of the iMGs
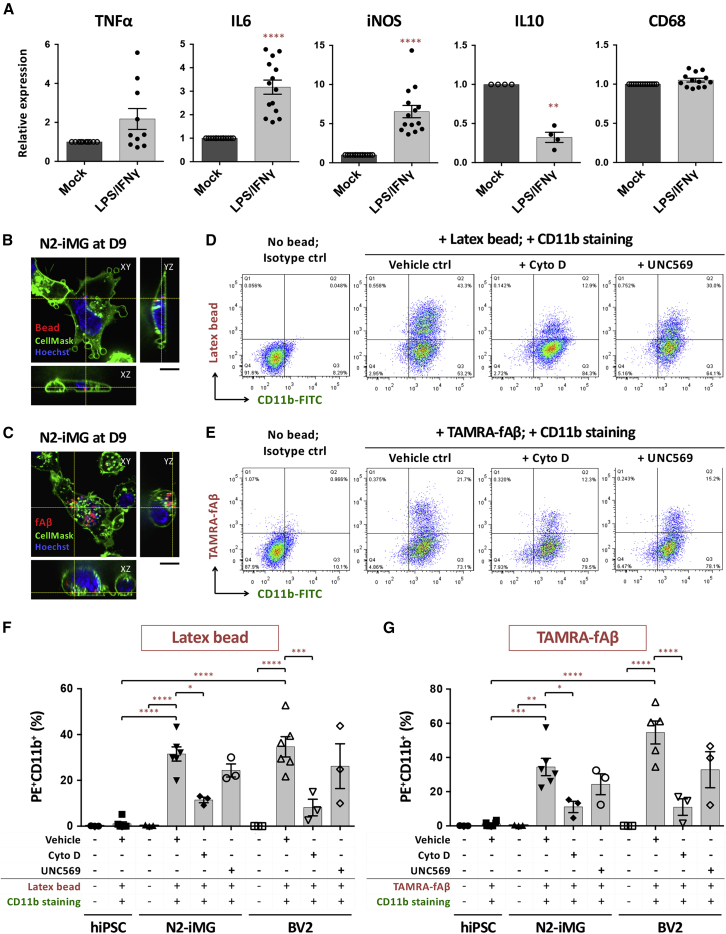
Microglia are considered to be the CNS's first-line defense. When activated, microglia undergo morphological changes, becoming ameboid in shape; they then migrate toward an injured area, releasing inflammatory cytokines, and eventually remove foreign substances and/or cell debris by phagocytosis. One of the most characteristic features of microglia is their rapid reaction to neuroinflammation (Nimmerjahn et al., 2005). Therefore we examined whether our iMG cells exhibit a similar response in vitro. Using qRT-PCR analysis, we found that a 6-h treatment of iMGs with LPS and IFN-γ brought about a marked elevation in the expression of various genes involved in the inflammatory response, including pro-inflammatory cytokine (IL-6) and the inducible nitric oxide synthase (iNOS) (Figure 4A). The changes in protein abundance confirmed those at the transcript level (Figure S5). On the other hand, expression of the anti-inflammatory cytokine IL-10 was greatly decreased. Notably, expression of the phagocytic cell marker CD68 was not affected by this treatment. These results indicate that our iMGs, just like microglia in vivo, exhibit appropriate physiological responses to inflammation in vitro.
Figure 4.
iMG cells exhibit appropriate physiological responses to LPS and IFN-γ challenge and are able to engulf microspheres or fibrillar Aβ
(A) qRT-PCR analysis of inflammation-related gene expression in N2-iMG cells after LPS/IFN-γ stimulation for 6 h. Data are means ± SEM (TNF-α, n = 10; IL-6, n = 14; iNOS, n = 14; IL-10, n = 4; CD68, n = 13). ∗∗p < 0.01, ∗∗∗∗p < 0.0001 compared with the mock-treatment group (ratio paired t test).
(B and C) Representative spinning-disc confocal microscopy images of phagocytosis by N2-iMG cells (day 9) of latex beads (B, red) or TAMRA-labeled fibrillar Aβ (fAβ) (C, red). Cells were incubated with substrates for 1 h. The plasma membrane and nuclei of the live cells were stained with CellMask deep red (green) and Hoechst 33,342 (blue), respectively. The z-axis images at the vertical and horizontal yellow lines were extracted from 3D images, and indicate right and bottom positions, respectively. Scale bar, 10 μm.
(D and E) Flow cytometry analysis of fluorescent microsphere bead (D) or TAMRA-fAβ (E) uptake by N2-iMGs at day 9.
(F and G) Quantitative results for percentage of CD11b+ cells with fluorescent beads (F) or fAβs (G). Data are means ± SEM (n = 3–6 independent experiments). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001 by one-way ANOVA using Tukey's multiple comparisons test. Cyto D, cytochalasin D.
In addition to the above inflammatory responses, microglial phagocytosis plays an essential role in the clearance from the CNS of pathogens, extracellular protein aggregates, and apoptotic cell debris (Fu et al., 2014). To examine whether our iMG cells have phagocytic ability, we incubated these cells with fluorescent latex microspheres or fibrillar amyloid β-peptide 1–42 (fAβ) for 1 h, labeled them with the microglial surface marker CD11b, and then analyzed these cells by flow cytometry. Compared with control hiPSCs, our iMGs could internalize both the microbeads and fAβ to similar extents compared with the BV2 cell line, which is known to be phagocytotic (Rangaraju et al., 2018) (Figures 4B–4G and Videos S1 and S2). The phagocytosis was markedly reduced by pre-incubating the iMGs with 10 μM cytochalasin D, which disrupts actin polymerization during phagocytosis (Figures 4D–4G). Moreover, as human and mouse microglia highly express the tyrosine kinase receptor MerTK, which is an essential regulator of the phagocytic clearance of myelin and apoptotic cells (Healy et al., 2016; Zizzo et al., 2012), we explored whether our iMG phagocytosis uses this pathway. We found that the number of iMG cells engulfing microbeads or fAβ was slightly decreased by MerTK inhibitor UNC569 pretreatment, indicating that MerTK does not play a crucial role in iMG phagocytosis when ligands are not present on the substrate. Thus SPI1/CEBPA-induced iMGs are able to actively phagocytose extracellular substances in a manner similar to that of bona fide brain microglia.
The plasma membrane and nuclei of the live cells were stained with CellMask deep red (green) and Hoechst 33342 (blue), respectively. Scale bar, 10 μm.
The plasma membrane and nuclei of the live cells were stained with CellMask deep red (green) and Hoechst 33342 (blue), respectively. Scale bar, 10 μm.
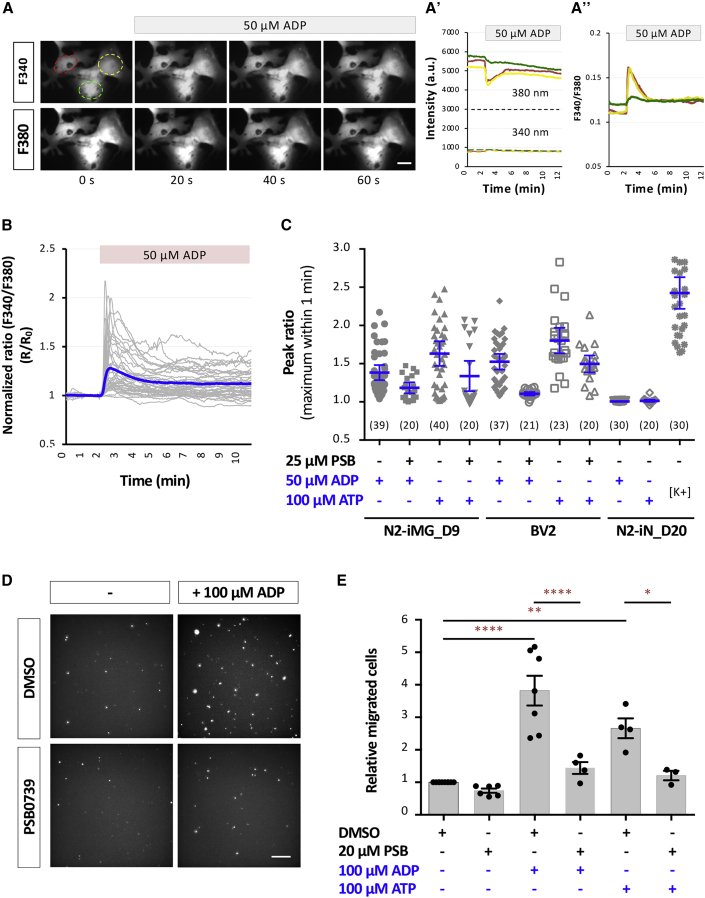
Nucleotide-evoked calcium signaling and migration by iMGs
Endogenous nucleotides are key messengers during the microglial activation process (Davalos et al., 2005; Koizumi et al., 2007). An accumulation of extracellular nucleotides as a result of injury-induced cell death activates microglial P2X and P2Y receptors and this initiates intracellular signaling cascades that regulate microglial functions. Adenine nucleotides, such as ATP and ADP, induce elevation of intracellular calcium ([Ca2+]i) transients and this serves as a signal to differentiate microglia from other cell types. We found that either ADP or ATP is able to rapidly induce Ca2+ elevation, with a plateau being reached within 20 s in N1- and N2-iMG cells (Figures 5A–5C). This increase was largely abolished by pre-incubating iMGs with a potent selective antagonist (PSB0739, 25 μM) of the P2RY12 receptor, which is expressed by microglia and senses extracellular nucleotides (Haynes et al., 2006). However, none of the iNs responded to ADP or ATP, but rather reliably underwent K+-induced depolarization. Thus our iMGs exhibit a nucleotide-triggered response similar to that of microglia in vivo.
Figure 5.
iMG cells are physiologically functional and exhibit ADP-evoked and ATP-evoked Ca2+ transients and migration
(A) Representative images showing an example of [Ca2+]i transients following addition of ADP to N2-iMG cells loaded with the Ca2+ indicator Fura-2/AM. Time-course changes in the fluorescence intensity at 340 and 380 nm (A′) and the ratio of F340/F380 (A″) were measured from three N2-iMG cells shown on the left. Scale bar, 5 μm.
(B) Time-lapse changes in fluorescence intensity produced by adding ADP to N2-iMG cells. Gray traces indicate the changes in fluorescence intensity ratio (F340/F380) of each individual cell. Blue trace is the mean ratio change of each time point.
(C) Quantitative results of the amplitudes of the [Ca2+]i transients. Maximum amplitude of the [Ca2+]i transient of each responsive cell is presented as a dot in the corresponding category. Data are pooled from three independent experiments and are means ±95% CI. The number in parentheses indicates the number of cells in each group.
(D) Representative fluorescence images showing ADP-induced N2-iMG cell migration into a Transwell chamber (8 μm). The cells migrating from the top compartment to the bottom of the Transwell membrane were fixed, DAPI stained, and observed using an inverted microscope. Scale bar, 50 μm.
(E) Quantitative results of (D). Data are means ± SEM (n = 4–5 fields for each replicate and involve 3–7 independent experiments for each condition). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗∗p < 0.0001 by one-way ANOVA with Tukey's multiple comparisons.
The nucleotides and their metabolites function as chemoattractants for microglia, allowing them to find damaged CNS cells (Honda et al., 2001). Using a Transwell assay, we found that the number of iMG cells migrating across the membrane was significantly higher in the presence of 100 μM ADP/ADP in the bottom (attractant) chamber, compared with the control (Figures 5D and 5E). Furthermore, the increased migration was mostly inhibited by pre-incubating the iMGs with PSB0739 (50 μM). These findings demonstrate that our iMGs respond appropriately to the presence of chemoattractive stimuli via their microglial P2RY12 receptors.
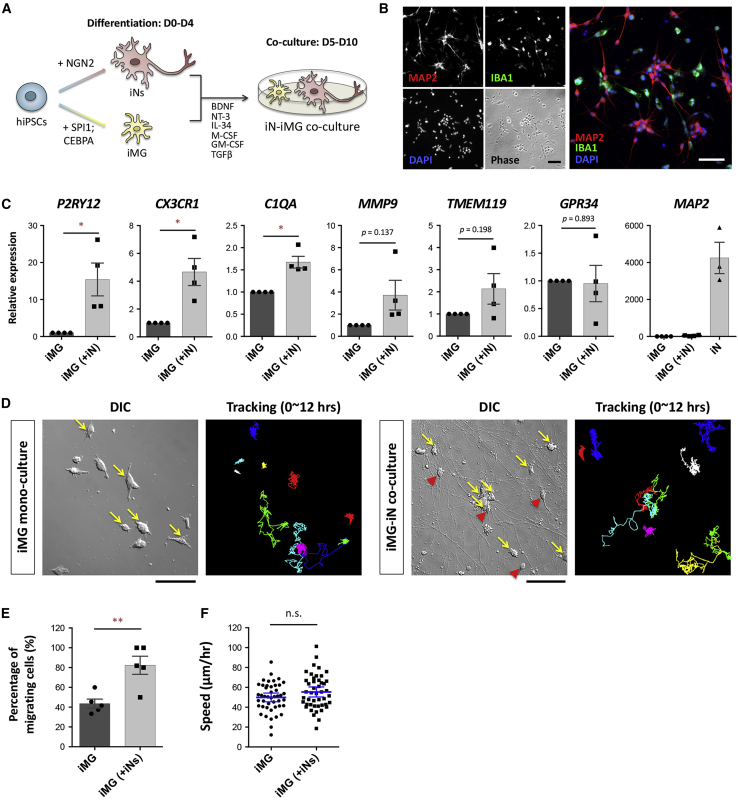
Establishing a co-culture model containing iN and iMG cells
Microglia are increasingly being recognized as key players in physiology/pathology. To demonstrate the potential applications of iMGs in brain research, we developed an efficient neuron-microglia co-culture system by combining our protocol for microglial induction and a well-established protocol for neuronal induction via the expression of NGN2 (Zhang et al., 2013). Briefly, iNs and iMGs were first prepared separately, and then the iMGs were harvested and seeded on day 4 into a mono-culture of iNs in a compatible co-culture medium (Figure 6A). On day 5, we found that the iNs were exhibiting a spindle-shaped or round-shaped soma with extensive neurites, and that the iMGs were irregularly shaped without a long process (Figures 6B and 6D). The identities of the cells were confirmed by immunostaining with the neuronal marker MAP2 and the microglial marker IBA1 (Figure 6B). Double immunostaining showed no overlap between the MAP2+ cells and the IBA1+ cells. Thus an iN-iMG co-culture model from hiPSCs could be successfully established in only 10 days.
Figure 6.
Co-culture of iMG cells with hiPSC-derived neurons promotes iMG maturation
(A) Schematic overview of the co-culture protocol for iN and iMG cells generated from the same hiPSC line. The hiPSCs carrying the NGN2 or SPI1/CEBPA transgene were induced to form neurons (iNs) or microglia (iMG) separately. Next, the iN and iMG cells were co-cultured under compatible conditions from day 5 to day 10.
(B) Representative images of the N2-iN and N2-iMG co-culture immunolabeled for MAP2 (red) and IBA1 (green) at day 10. Cell nuclei are stained with DAPI (blue). Scale bar, 50 μm.
(C) Expression levels by qRT-PCR of key microglial markers (P2RY12, CX3CR1, C1QA, MMP9, TMEM119, GPR34) and the somatodendritic marker MAP2 in mono-cultured or co-cultured N2-iMG cells at 9–12 days of induction. Fold changes in target genes were calculated using the 2ˆ(-ΔΔCT) method with RPL13A as an endogenous control and relative to the expression levels found in mono-cultured iMGs. Data are means ± SEM (n = 4 independent experiments). ∗p < 0.05 by paired t test.
(D) An example of time-lapse differential interference contrast (DIC) imaging of N2-iMG cultures with or without N2-iNs (recorded for 12 h). Yellow arrows in the DIC images mark the migrating iMG cells, defined as cells with a displacement length of over two cell bodies between two continuous frames over 12 h. Red arrowheads indicate iNs. The right-side images show the cell trace results and are presented as the color-coded trajectories of each cell over 12 h. Scale bar, 100 μm.
(E) Percentage of migrating N2-iMG cells in mono-culture or in co-culture (12-h recording period). Data are means ± SEM (n = 5 independent fields in each group). ∗∗p < 0.01 by unpaired t test.
(F) The speed of migration of the N2-iMG cells in the mono-culture or co-culture (12-h recording period). Data are means ± SEM (n = 45 and 46 cells from 5 fields in the N2-iMG and N2-iMG(+N2-iN) groups, respectively). n.s., not significant by unpaired t test.
Consistent with the idea that neurons are able to influence the gene expression and functioning of microglia (Dubbelaar et al., 2018), we found that expression of important microglial genes was altered in iMGs after co-culture with iNs. The results from a qRT-PCR analysis showed that iMGs isolated from iN-iMG co-culture via CD11b-coated magnetic beads expressed three key microglia-specific genes, P2RY12, CX3CR1, and C1QA, at much higher levels than iMGs cultured alone (Figure 6C). Furthermore, three other microglial genes, MMP9, TMEM119, and GPR34, did not show a consistent increase. These results show that iMGs, when co-cultured with iNs, express the expected microglial genes to a similar or an even higher extent compared with iMGs cultured alone, suggesting that iMG maturation and homeostasis are modulated by the environment created by iN co-culture. These findings also demonstrate that iN-iMG co-culture will be a useful model when studying in vitro neuron-microglia interactions under normal or diseased conditions.
The co-culture environment modulates microglial motility
To better understand how iN and iMG cells interact with one another over time, we performed time-lapse imaging for 12–14 h on day 9 (see Figure 6D, and Videos S3 and S4). The iMGs in co-culture were highly motile and continually underwent dynamic changes in shape, whereas the iNs in co-culture had a relatively stable shape over the entire imaging period. When iMGs were cultured alone, although they moved constantly, most seemed to have their own territory. Next, we quantified the percentage of migrating iMG cells with a migration distance between two successive frames (5 min) of more than 2× their soma length over 12 h. The results showed that more co-cultured iMGs (82.36% ± 9.15%; n = 5 fields) moved this long distance than mono-cultured iMGs (43.59% ± 4.58%; n = 5 fields) (Figure 6E). However, the average speed of migration was not significantly different between the two migrating iMGs (Figure 6F). This told us that co-cultured iMGs have a morphology and dynamic behavior that is what would be expected of microglia in the CNS, in that they are continually sensing and responding to their neuronal environment, changing as necessary their shape and location. This contrasts with the quiescent neurons, which transduce signals via their polarized axons/dendrites and synapses.
Scale bar, 50 μm
Scale bar, 50 μm.
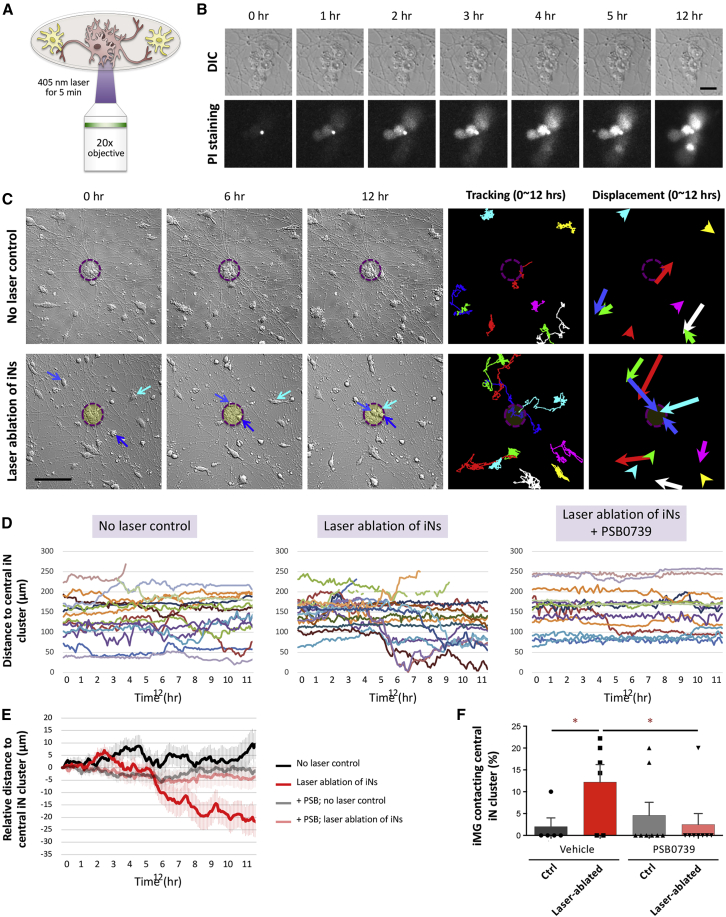
iMGs respond and migrate toward an injured neuron cluster
Using our practicable iN-iMG co-culture model, we examined whether our iMGs are functionally competent when interacting with iNs. Microglia are known to act as scavengers and clean up CNS debris and dead cells in order to maintain homeostasis (Szepesi et al., 2018). On neuronal cell death, a series of “find-me” and “eat-me” signals are released that trigger microglia to approach the dead cells and execute phagocytosis. To evaluate the scavenging response of iMGs, we induced cell death by exposing iN clusters (10–20 cells) to 405-nm laser light for 5 min; this was followed by time-lapse imaging at 5-min intervals for 12 h (Figure 7A). After laser ablation, we observed obvious changes in the morphology of the central iN clumps, namely flattening and spreading out, which was accompanied within 4 h by an apparent increase in propidium iodide-positive signals. This confirms that the laser light did lead to the death of targeted iNs without damaging the surrounding iNs and iMGs (Figures 7B and 7C, laser-targeted region shown as dashed magenta circle; Video S5). Using the resulting time-lapse images, we found that the surrounding iMGs moved from their initial positions during the first 2 h after laser ablation. Notably, several of these iMGs migrated toward the dead iN clusters at 4 h after laser application (Figures 7C and 7D; Videos S6 and S7). Moreover, when the migrating distance of the iMGs toward the central iN clumps in the laser ablated and control groups was compared using 12-h tracking data, in the non-laser control group, the average distance from the iMGs to their central iN clump did not change. On the other hand, the average distance of the iMGs in relation to the center of a clump of dead iNs in the laser-ablated groups decreased (Figures 7D and 7E). This decrease in the distance between the iMGs and the dead iNs occurred at 4–6 h after laser ablation, indicating that the iMG cells had begun to detect soluble chemoattractants and were migrating toward the dead neurons. During the 12-h recording period, 12.20% ± 4.01% (n = 6 fields) of the migrating iMG cells approached the dead neurons in the laser-ablation group, while almost no iMGs moved toward the central iN clumps in the control group (Figure 7F). The migration of iMGs induced by the dying neurons was almost completely blocked by adding PSB0739, a P2RY12 antagonist that only slightly affects cell motility (Figure S6). This suggests that the chemoattractants released by the wounded iNs are the adenine nucleotides ADP and ATP. As a whole, our findings show that iMG cells can detect their surroundings and respond and migrate toward dead neurons in our iN-iMG co-culture system. Thus the SPI1/CEBPA-induced iMGs function physiologically and display the dynamic characteristics of migration, both of which are similar to microglia in vivo. Finally, our findings clearly support the idea that our co-culture scheme is a relevant model for studying neuron-microglia interactions associated with normal and diseased brains.
Figure 7.
iMG cells respond to and migrate toward an injured neuron cluster
(A) Experimental design of the laser-induced neuronal injury.
(B) Cell death of N2-iNs was monitored using propidium iodide (PI) staining. Each selected iN cluster was exposed to 405-nm laser light for 5 min and immediately examined by time-lapse imaging (sampling rate of 1/300 Hz) for PI signal. Scale bar, 20 μm.
(C) An example of the time-lapse DIC imaging of co-cultures with or without laser-induced neuronal injury (recorded for 12 h). Blue arrows in the DIC images mark an iMG cell migrating toward the central iN cluster. The two-panel images on the right show the results of cell traces, which are presented as a color-coded trajectory for each cell over 12 h. The displacement of each cell is shown on the right. Scale bar, 100 μm.
(D) Measurements of the distance between each iMG cell and the central iN cluster in the control and laser ablation groups for each time point. Different cells are coded by color and are from a representative experiment.
(E) The graph depicts the results of relative distance changes over time for the four conditions. Data are means ± SEM (n = 46 N2-iMG cells in 5 fields [no laser control]; n = 58 N2-iMG cells in 6 fields [laser ablation]; n = 41 N2-iMG cells in 8 fields [no laser control and PSB application]; n = 33 N2-iMG cells in 8 fields [laser ablation and PSB application]).
(F) The percentage of iMG cells that are able to contact the central iN cluster over the 12-h recording period. Data are means ± SEM (n = 5–8 fields). ∗p < 0.05 by one-way ANOVA with Fisher's LSD multiple comparisons.
Scale bar, 20 μm
Scale bar, 50 μm
Scale bar, 50 μm.
Discussion
Microglia are important to brain physiology and pathology. Here we have developed an efficient method to generate human microglia-like cells from hiPSCs by forced expression of two reprogramming factors, SPI1 and CEBPA. The reprogrammed iMG cells show robust expression of microglia-specific markers (IBA1, P2RY12, TMEM119, GPR34, C1QA, and MMP9). Whole-transcriptome analysis showed that the iMG cells resemble primary human microglia and do not resemble monocytes. A functional assessment demonstrated that these iMG cells possess appropriate physiological functioning, including LPS/IFN-γ-induced inflammatory responses, and phagocytic ability, as well as ADP/ATP-evoked signaling/migration. When co-cultured with iNs, the iMG cells had an active “surveying” phenotype involving dynamic cross talk between microglia and neurons. Thus, this study has established a protocol for rapidly converting hiPSCs into functional iMG cells; this will facilitate studies of human microglia, aid modeling of human diseases, and help with drug discovery.
Several groups have reported carrying out stepwise differentiation of microglia from the PSC stage using procedures recapitulating the microglial ontogeny via either embryonic body (EB) formation or the progenitor stage. In 2016 Muffat and colleagues described how they differentiated human ESCs or iPSCs into neuralized EBs and cystic EBs (Muffat et al., 2016). When these cells were kept in culture for 30 days, they matured into microglia-like cells; the total culture time was around 75 days. Recently, other groups have published protocols whereby ESCs and iPSCs were first differentiated into myeloid progenitor cells, rather than using EB formation, by serial exposure to defined media (Abud et al., 2017; Douvaras et al., 2017). The myeloid progenitor cells obtained were purified, replated, and further differentiated into microglia-like cells in a medium containing IL-34, M-CSF, and TGF-β1. The entire time needed to generate mature microglia from the PSC stage in these cases varied from 40 to 60 days. Using other protocols, ESC/iPSC-derived microglia were obtained that expressed microglial markers and resembled human fetal primary microglia based on their gene expression profile, as well as exhibiting inflammatory responses on LPS challenge. Here, we established an efficient protocol for generating human microglia-like cells from hiPSCs using two reprogramming factors, SPI1 and CEBPA, in combination. Differentiation of iPSCs via reprogramming seems to allow the cells to skip the progenitor stage of primitive development and thus shorten the time needed for differentiation. This resulted in our iMGs showing robust expression of the microglial markers after only 9 days of induction, which is very much faster than any other currently available method. Our iMGs also exhibited bona fide physiological functioning, including inflammatory properties, phagocytic ability, and ADP/ATP-evoked signaling/migration; these are comparable to the functioning of EB-derived and progenitor-derived microglia. Apparently, the combination of two TFs, SPI1 and CEBPA, acts as a master regulator during the differentiation of microglia and induces not only microglial specificity, but also an accelerated maturation process.
To address specific issues regarding MG lineage commitment in our culture, we tested a range of defined differentiation conditions. First, to generate hiPSC-derived microglia by reprogramming, we initially performed literature data mining to select a first pool of seven candidate TFs that have important roles in microglial development; these were our pro-microglial TF candidates. After 10 days of induction of an infected cell culture, the microglial surface marker CD11b was assessed by flow cytometry. No single TF could elicit any significant CD11b expression (Figures 1 and S2). We then combined two or three factors to pinpoint the best-performing combination giving the highest CD11b+ and IBA1+ cell conversion efficiency (Table S1). Among all tested groups, only cells overexpressing both SPI1 and its cofactor CEBPA induced a significant number of CD11b+ cells. No further increase in CD11b+ cells was seen with any other TF combination. Thus, the expression of SPI1 and CEBPA is critical for iMG differentiation from the PSC stage, as no CD11b+ cells were detected in the absence of doxycycline (Dox) induction (medium condition B in Figures S2D and S2E). It has been reported that C/EBPα (encoded by the CEBPA gene), in collaboration with PU.1/SPI1, reverses lymphoid cells into myeloid progenitors to allow eventual differentiation in vitro into monocytes/granulocytes (Di Tullio et al., 2011). Our findings also suggest that the expression of both PU.1/SPI1 and C/EBPα is essential for microglia development during cell fate decision-making.
In addition, to optimize the medium and growth factors needed for the efficient conversion of hiPSCs into iMGs, we tested a variety of recombinant signaling proteins known to control MG differentiation during embryonic development. Unexpectedly, the proportion of CD11b+ cells in the differentiation medium with Dox, but without any growth factors, was as high as that in the medium including complete factors, while the total number of live cells was much fewer in the “no factors” group (Figures S3 S2A–S2C). We found that GM-CSF slightly promotes iMG cell survival under our conditions but does not further increase the fraction of CD11b+ cells. Collectively, these findings demonstrate that the combined expression of SPI1 and CEBPA plays a crucial role in iMG conversion from hiPSCs, and that the growth factors help sustain cell viability at each differentiation step. It should be noted that Dox was needed in the culture medium to induce transgene expression during the entire differentiation process. This was because the population of CD11b+ iMGs was slightly reduced when defined medium without Dox was used after day 4 (condition C in Figure S2D). Hypothetically, after having passed the initiation gateway of reprogramming, the cells should enter a balanced state under their endogenous transcriptional regulatory network, and at this point the overexpressed reprogramming factors are no longer necessary. Nevertheless, it should be noted that the critical time point of MG determination remains unclear, and it may range from the beginning to the end of our incubation period. Based on the time-course marker changes, as measured by qRT-PCR and flow cytometry (Figures 2C and 2D), iMGs began to show increased expression of microglia-specific markers at 6 days after induction. This suggests that Dox may be removable from the culture from that time point onward.
There is a fast-growing interest in microglia, as they are increasingly implicated in various neurodevelopmental disorders and in a number of neurodegenerative diseases. Here we report an efficient method to differentiate hiPSCs into microglia, thus making available a renewable source of human microglia. This will allow the key genes involved in microglia functioning and microglia dysfunction to be investigated, as well as healthy and diseased brains to be explored. We have also, in this study, demonstrated potential applications of iMGs when studying neuron-microglia interactions in vitro, which will significantly further our knowledge of the molecular and functional mechanisms underlying microglia activation over a broad range of CNS development, during homeostatic functioning of the brain, and when studying various neurological disease models.
Experimental procedures
hiPSC lines and maintenance
The two control hiPSC lines, NTUH-iPSC-01-05 and NTUH-iPSC-02-02 (iN1 and iN2, respectively), were purchased from BCRC/FIRDI, Taiwan. The four AD-iPSC lines were generated by the Brain Research Center of National Yang-Ming University, Taiwan (Wu et al., 2019). The uses of the hiPSC lines followed the Policy Instructions of the Ethics of Human Embryo and Embryonic Stem Cell Research guidelines in Taiwan. In addition, approval from the institutional review boards of National Yang-Ming University was obtained. Human iPSCs were routinely maintained in Essential 8 medium (Gibco) on vitronectin (VTN-N, Gibco)-coated dishes following the manufacturer's instructions.
Differentiation of microglia-like cells from hiPSCs
hiPSCs that had been infected with both FUW-rtTA and pTetO-CEBPA-T2A-SPI1-T2A-Puro were seeded on day −1. On day 0, the cells had reached ≥70% confluency; at this point the culture medium was replaced with DMEM/F12 (Gibco) supplemented with N2 (Gibco) and non-essential amino acids (NEAA, Gibco), BMP4 (50 ng/mL), bFGF (50 ng/mL), and activin-A (20 ng/mL). Dox (2 μg/mL) was added to promote TetO downstream gene expression and remained until the end of the experiment. On day 1, the medium was replaced with DMEM/F12/N2/NEAA containing human VEGF (50 ng/mL), SCF (50 ng/mL), and FGF2 (20 ng/mL). Puromycin (1 μg/mL) was also added for 24 h to select for successfully infected cells. On day 2, the medium was replaced with DMEM/F12/N2/NEAA containing human IL-34 (10 ng/mL), M-CSF (10 ng/mL), and TGF-β1 (10 ng/mL). After day 4, half of the medium was replaced with fresh DMEM/F12/N2/NEAA containing human IL-34 (100 ng/mL), M-CSF (20 ng/mL), GM-CSF (20 ng/mL), and TGF-β1 (20 ng/mL).
Data and code availability
RNA-sequencing data have been deposited in the NCBI database under accession no. GSE163984.
All detailed experimental procedures are available in the supplemental information.
Author contributions
Conceptualization, S.-W.C., M.-J.F., and Y.-H.W.; methodology, S.-W.C., Y.-S.H., S.-C.C., Y.-S.C., and Y.-H.W.; formal analysis, S.-W.C., Y.-S.H., S.-C.C., and Y.-H.W.; investigation, S.-W.C. and Y.-H.W.; writing – original draft, S.-W.C. and Y.-H.W.; writing – review & editing, S.-W.C., J.-L.F., N.-J.C., Y.-S.C., M.-J.F., and Y.-H.W.; resources, J.-L.F., N.-J.C., M.-J.F., and Y.-H.W.; supervision, M.-J.F. and Y.-H.W.; project administration, Y.-H.W.; funding acquisition, J.-L.F., M.-J.F., and Y.-H.W.
Acknowledgments
We thank the Human Disease iPSC Service Consortium (funded by the Ministry of Science and Technology (MOST) of Taiwan, MOST 108-2319-B-001-004) for iPSC generation and technical support. We also thank the Clinical and Industrial Genomic Application Development Service Center of the National Core Facility for Biopharmaceuticals, Taiwan (MOST 108-2319-B-010-001) for the Sanger sequencing, and Dr. Mu-ming Poo and Dr. Jenn-Yah Yu for their discussions and critical reading of the manuscript. This work was financially supported by the Brain Research Center of National Yang-Ming University from the Featured Areas Research Center Program within the framework of the Higher Education Sprout Project by the Ministry of Education (MOE) in Taiwan, and by MOST grants (MOST 106-2321-B-075-001, MOST 107-2221-E-075-006, and MOST 108-2321-B-075-001) to J.-L.F., as well as MOST 108-2320-B-010-042 to Y.-H.W.
Published: April 8, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.stemcr.2021.03.010.
Supplemental information
References
- Abud E.M., Ramirez R.N., Martinez E.S., Healy L.M., Nguyen C.H.H., Newman S.A., Yeromin A.V., Scarfone V.M., Marsh S.E., Fimbres C. iPSC-derived human microglia-like cells to study neurological diseases. Neuron. 2017;94:278–293.e9. doi: 10.1016/j.neuron.2017.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownjohn P.W., Smith J., Solanki R., Lohmann E., Houlden H., Hardy J., Dietmann S., Livesey F.J. Functional studies of missense TREM2 mutations in human stem cell-derived microglia. Stem Cell Reports. 2018;10:1294–1307. doi: 10.1016/j.stemcr.2018.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butovsky O., Jedrychowski M.P., Moore C.S., Cialic R., Lanser A.J., Gabriely G., Koeglsperger T., Dake B., Wu P.M., Doykan C.E. Identification of a unique TGF-beta-dependent molecular and functional signature in microglia. Nat. Neurosci. 2014;17:131–143. doi: 10.1038/nn.3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttgereit A., Lelios I., Yu X., Vrohlings M., Krakoski N.R., Gautier E.L., Nishinakamura R., Becher B., Greter M. Sall1 is a transcriptional regulator defining microglia identity and function. Nat. Immunol. 2016;17:1397–1406. doi: 10.1038/ni.3585. [DOI] [PubMed] [Google Scholar]
- Caiazzo M., Dell'Anno M.T., Dvoretskova E., Lazarevic D., Taverna S., Leo D., Sotnikova T.D., Menegon A., Roncaglia P., Colciago G. Direct generation of functional dopaminergic neurons from mouse and human fibroblasts. Nature. 2011;476:224–227. doi: 10.1038/nature10284. [DOI] [PubMed] [Google Scholar]
- Claes C., Van Den Daele J., Boon R., Schouteden S., Colombo A., Monasor L.S., Fiers M., Ordovas L., Nami F., Bohrmann B. Human stem cell-derived monocytes and microglia-like cells reveal impaired amyloid plaque clearance upon heterozygous or homozygous loss of TREM2. Alzheimers Dement. 2019;15:453–464. doi: 10.1016/j.jalz.2018.09.006. [DOI] [PubMed] [Google Scholar]
- Cunningham C.L., Martinez-Cerdeno V., Noctor S.C. Microglia regulate the number of neural precursor cells in the developing cerebral cortex. J. Neurosci. 2013;33:4216–4233. doi: 10.1523/JNEUROSCI.3441-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daneman R., Zhou L., Kebede A.A., Barres B.A. Pericytes are required for blood-brain barrier integrity during embryogenesis. Nature. 2010;468:562–566. doi: 10.1038/nature09513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davalos D., Grutzendler J., Yang G., Kim J.V., Zuo Y., Jung S., Littman D.R., Dustin M.L., Gan W.B. ATP mediates rapid microglial response to local brain injury in vivo. Nat. Neurosci. 2005;8:752–758. doi: 10.1038/nn1472. [DOI] [PubMed] [Google Scholar]
- Davis T.L., Rebay I. Master regulators in development: Views from the Drosophila retinal determination and mammalian pluripotency gene networks. Dev. Biol. 2017;421:93–107. doi: 10.1016/j.ydbio.2016.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Tullio A., Vu Manh T.P., Schubert A., Castellano G., Mansson R., Graf T. CCAAT/enhancer binding protein alpha (C/EBP(alpha))-induced transdifferentiation of pre-B cells into macrophages involves no overt retrodifferentiation. Proc. Natl. Acad. Sci. U S A. 2011;108:17016–17021. doi: 10.1073/pnas.1112169108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douvaras P., Sun B., Wang M., Kruglikov I., Lallos G., Zimmer M., Terrenoire C., Zhang B., Gandy S., Schadt E. Directed differentiation of human pluripotent stem cells to microglia. Stem Cell Reports. 2017;8:1516–1524. doi: 10.1016/j.stemcr.2017.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubbelaar M.L., Kracht L., Eggen B.J.L., Boddeke E. The Kaleidoscope of microglial phenotypes. Front. Immunol. 2018;9:1753. doi: 10.3389/fimmu.2018.01753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu R., Shen Q., Xu P., Luo J.J., Tang Y. Phagocytosis of microglia in the central nervous system diseases. Mol. Neurobiol. 2014;49:1422–1434. doi: 10.1007/s12035-013-8620-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginhoux F., Greter M., Leboeuf M., Nandi S., See P., Gokhan S., Mehler M.F., Conway S.J., Ng L.G., Stanley E.R. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science. 2010;330:841–845. doi: 10.1126/science.1194637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haenseler W., Sansom S.N., Buchrieser J., Newey S.E., Moore C.S., Nicholls F.J., Chintawar S., Schnell C., Antel J.P., Allen N.D. A highly efficient human pluripotent stem cell microglia model displays a neuronal-Co-culture-Specific expression profile and inflammatory response. Stem Cell Reports. 2017;8:1727–1742. doi: 10.1016/j.stemcr.2017.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes S.E., Hollopeter G., Yang G., Kurpius D., Dailey M.E., Gan W.B., Julius D. The P2Y12 receptor regulates microglial activation by extracellular nucleotides. Nat. Neurosci. 2006;9:1512–1519. doi: 10.1038/nn1805. [DOI] [PubMed] [Google Scholar]
- Healy L.M., Perron G., Won S.Y., Michell-Robinson M.A., Rezk A., Ludwin S.K., Moore C.S., Hall J.A., Bar-Or A., Antel J.P. MerTK is a functional regulator of myelin phagocytosis by human myeloid cells. J. Immunol. 2016;196:3375–3384. doi: 10.4049/jimmunol.1502562. [DOI] [PubMed] [Google Scholar]
- Honda S., Sasaki Y., Ohsawa K., Imai Y., Nakamura Y., Inoue K., Kohsaka S. Extracellular ATP or ADP induce chemotaxis of cultured microglia through Gi/o-coupled P2Y receptors. J. Neurosci. 2001;21:1975–1982. doi: 10.1523/JNEUROSCI.21-06-01975.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kierdorf K., Erny D., Goldmann T., Sander V., Schulz C., Perdiguero E.G., Wieghofer P., Heinrich A., Riemke P., Holscher C. Microglia emerge from erythromyeloid precursors via Pu.1- and Irf8-dependent pathways. Nat. Neurosci. 2013;16:273–280. doi: 10.1038/nn.3318. [DOI] [PubMed] [Google Scholar]
- Koizumi S., Shigemoto-Mogami Y., Nasu-Tada K., Shinozaki Y., Ohsawa K., Tsuda M., Joshi B.V., Jacobson K.A., Kohsaka S., Inoue K. UDP acting at P2Y6 receptors is a mediator of microglial phagocytosis. Nature. 2007;446:1091–1095. doi: 10.1038/nature05704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath K.E., Koniski A.D., Malik J., Palis J. Circulation is established in a stepwise pattern in the mammalian embryo. Blood. 2003;101:1669–1676. doi: 10.1182/blood-2002-08-2531. [DOI] [PubMed] [Google Scholar]
- Miyamoto A., Wake H., Ishikawa A.W., Eto K., Shibata K., Murakoshi H., Koizumi S., Moorhouse A.J., Yoshimura Y., Nabekura J. Microglia contact induces synapse formation in developing somatosensory cortex. Nat. Commun. 2016;7:12540. doi: 10.1038/ncomms12540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muffat J., Li Y., Yuan B., Mitalipova M., Omer A., Corcoran S., Bakiasi G., Tsai L.H., Aubourg P., Ransohoff R.M. Efficient derivation of microglia-like cells from human pluripotent stem cells. Nat. Med. 2016;22:1358–1367. doi: 10.1038/nm.4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimmerjahn A., Kirchhoff F., Helmchen F. Resting microglial cells are highly dynamic surveillants of brain parenchyma in vivo. Science. 2005;308:1314–1318. doi: 10.1126/science.1110647. [DOI] [PubMed] [Google Scholar]
- Pandya H., Shen M.J., Ichikawa D.M., Sedlock A.B., Choi Y., Johnson K.R., Kim G., Brown M.A., Elkahloun A.G., Maric D. Differentiation of human and murine induced pluripotent stem cells to microglia-like cells. Nat. Neurosci. 2017;20:753–759. doi: 10.1038/nn.4534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paolicelli R.C., Bolasco G., Pagani F., Maggi L., Scianni M., Panzanelli P., Giustetto M., Ferreira T.A., Guiducci E., Dumas L. Synaptic pruning by microglia is necessary for normal brain development. Science. 2011;333:1456–1458. doi: 10.1126/science.1202529. [DOI] [PubMed] [Google Scholar]
- Perry V.H., Nicoll J.A., Holmes C. Microglia in neurodegenerative disease. Nat. Rev. Neurol. 2010;6:193–201. doi: 10.1038/nrneurol.2010.17. [DOI] [PubMed] [Google Scholar]
- Rangaraju S., Raza S.A., Li N.X., Betarbet R., Dammer E.B., Duong D., Lah J.J., Seyfried N.T., Levey A.I. Differential phagocytic properties of CD45(low) microglia and CD45(high) brain mononuclear phagocytes-activation and age-related effects. Front. Immunol. 2018;9:405. doi: 10.3389/fimmu.2018.00405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapino F., Robles E.F., Richter-Larrea J.A., Kallin E.M., Martinez-Climent J.A., Graf T. C/EBPalpha induces highly efficient macrophage transdifferentiation of B lymphoma and leukemia cell lines and impairs their tumorigenicity. Cell Rep. 2013;3:1153–1163. doi: 10.1016/j.celrep.2013.03.003. [DOI] [PubMed] [Google Scholar]
- Schafer D.P., Lehrman E.K., Kautzman A.G., Koyama R., Mardinly A.R., Yamasaki R., Ransohoff R.M., Greenberg M.E., Barres B.A., Stevens B. Microglia sculpt postnatal neural circuits in an activity and complement-dependent manner. Neuron. 2012;74:691–705. doi: 10.1016/j.neuron.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilling T., Nitsch R., Heinemann U., Haas D., Eder C. Astrocyte-released cytokines induce ramification and outward K+ channel expression in microglia via distinct signalling pathways. Eur. J. Neurosci. 2001;14:463–473. doi: 10.1046/j.0953-816x.2001.01661.x. [DOI] [PubMed] [Google Scholar]
- Sellgren C.M., Gracias J., Watmuff B., Biag J.D., Thanos J.M., Whittredge P.B., Fu T., Worringer K., Brown H.E., Wang J. Increased synapse elimination by microglia in schizophrenia patient-derived models of synaptic pruning. Nat. Neurosci. 2019;22:374–385. doi: 10.1038/s41593-018-0334-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigemoto-Mogami Y., Hoshikawa K., Goldman J.E., Sekino Y., Sato K. Microglia enhance neurogenesis and oligodendrogenesis in the early postnatal subventricular zone. J. Neurosci. 2014;34:2231–2243. doi: 10.1523/JNEUROSCI.1619-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A.M., Gibbons H.M., Oldfield R.L., Bergin P.M., Mee E.W., Faull R.L., Dragunow M. The transcription factor PU.1 is critical for viability and function of human brain microglia. Glia. 2013;61:929–942. doi: 10.1002/glia.22486. [DOI] [PubMed] [Google Scholar]
- Son E.Y., Ichida J.K., Wainger B.J., Toma J.S., Rafuse V.F., Woolf C.J., Eggan K. Conversion of mouse and human fibroblasts into functional spinal motor neurons. Cell Stem Cell. 2011;9:205–218. doi: 10.1016/j.stem.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szepesi Z., Manouchehrian O., Bachiller S., Deierborg T. Bidirectional microglia-neuron communication in health and disease. Front. Cell Neurosci. 2018;12:323. doi: 10.3389/fncel.2018.00323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Yamamura F., Naito M. Differentiation, maturation, and proliferation of macrophages in the mouse yolk sac: a light-microscopic, enzyme-cytochemical, immunohistochemical, and ultrastructural study. J. Leukoc. Biol. 1989;45:87–96. doi: 10.1002/jlb.45.2.87. [DOI] [PubMed] [Google Scholar]
- Tcw J., Wang M., Pimenova A.A., Bowles K.R., Hartley B.J., Lacin E., Machlovi S.I., Abdelaal R., Karch C.M., Phatnani H. An efficient platform for astrocyte differentiation from human induced pluripotent stem cells. Stem Cell Reports. 2017;9:600–614. doi: 10.1016/j.stemcr.2017.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno M., Fujita Y., Tanaka T., Nakamura Y., Kikuta J., Ishii M., Yamashita T. Layer V cortical neurons require microglial support for survival during postnatal development. Nat. Neurosci. 2013;16:543–551. doi: 10.1038/nn.3358. [DOI] [PubMed] [Google Scholar]
- von Bartheld C.S., Bahney J., Herculano-Houzel S. The search for true numbers of neurons and glial cells in the human brain: a review of 150 years of cell counting. J. Comp. Neurol. 2016;524:3865–3895. doi: 10.1002/cne.24040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Szretter K.J., Vermi W., Gilfillan S., Rossini C., Cella M., Barrow A.D., Diamond M.S., Colonna M. IL-34 is a tissue-restricted ligand of CSF1R required for the development of Langerhans cells and microglia. Nat. Immunol. 2012;13:753–760. doi: 10.1038/ni.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehrspaun C.C., Haerty W., Ponting C.P. Microglia recapitulate a hematopoietic master regulator network in the aging human frontal cortex. Neurobiol. Aging. 2015;36:2443.e9–2443.e20. doi: 10.1016/j.neurobiolaging.2015.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei S., Nandi S., Chitu V., Yeung Y.G., Yu W., Huang M., Williams L.T., Lin H., Stanley E.R. Functional overlap but differential expression of CSF-1 and IL-34 in their CSF-1 receptor-mediated regulation of myeloid cells. J. Leukoc. Biol. 2010;88:495–505. doi: 10.1189/jlb.1209822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu P.C., Fann M.J., Tran T.T., Chen S.C., Devina T., Cheng I.H., Lien C.C., Kao L.S., Wang S.J., Fuh J.L. Assessing the therapeutic potential of Graptopetalum paraguayense on Alzheimer's disease using patient iPSC-derived neurons. Sci. Rep. 2019;9:19301. doi: 10.1038/s41598-019-55614-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Jiang H., Zhong P., Yan Z., Chen S., Feng J. Direct conversion of human fibroblasts to induced serotonergic neurons. Mol. Psychiatry. 2016;21:62–70. doi: 10.1038/mp.2015.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang N., Chanda S., Marro S., Ng Y.H., Janas J.A., Haag D., Ang C.E., Tang Y., Flores Q., Mall M. Generation of pure GABAergic neurons by transcription factor programming. Nat. Methods. 2017;14:621–628. doi: 10.1038/nmeth.4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang N., Zuchero J.B., Ahlenius H., Marro S., Ng Y.H., Vierbuchen T., Hawkins J.S., Geissler R., Barres B.A., Wernig M. Generation of oligodendroglial cells by direct lineage conversion. Nat. Biotechnol. 2013;31:434–439. doi: 10.1038/nbt.2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yirmiya R., Rimmerman N., Reshef R. Depression as a microglial disease. Trends Neurosci. 2015;38:637–658. doi: 10.1016/j.tins.2015.08.001. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Pak C., Han Y., Ahlenius H., Zhang Z., Chanda S., Marro S., Patzke C., Acuna C., Covy J. Rapid single-step induction of functional neurons from human pluripotent stem cells. Neuron. 2013;78:785–798. doi: 10.1016/j.neuron.2013.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zizzo G., Hilliard B.A., Monestier M., Cohen P.L. Efficient clearance of early apoptotic cells by human macrophages requires M2c polarization and MerTK induction. J. Immunol. 2012;189:3508–3520. doi: 10.4049/jimmunol.1200662. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The plasma membrane and nuclei of the live cells were stained with CellMask deep red (green) and Hoechst 33342 (blue), respectively. Scale bar, 10 μm.
The plasma membrane and nuclei of the live cells were stained with CellMask deep red (green) and Hoechst 33342 (blue), respectively. Scale bar, 10 μm.
Scale bar, 50 μm
Scale bar, 50 μm.
Scale bar, 20 μm
Scale bar, 50 μm
Scale bar, 50 μm.
Data Availability Statement
RNA-sequencing data have been deposited in the NCBI database under accession no. GSE163984.
All detailed experimental procedures are available in the supplemental information.