Summary
Despite heterogeneity across the six layers of the mammalian cortex, all excitatory neurons are generated from a single founder population of neuroepithelial stem cells. However, how these progenitors alter their layer competence over time remains unknown. Here, we used human embryonic stem cell-derived cortical progenitors to examine the role of fibroblast growth factor (FGF) and Notch signaling in influencing cell fate, assessing their impact on progenitor phenotype, cell-cycle kinetics, and layer specificity. Forced early cell-cycle exit, via Notch inhibition, caused rapid, near-exclusive generation of deep-layer VI neurons. In contrast, prolonged FGF2 promoted proliferation and maintained progenitor identity, delaying laminar progression via MAPK-dependent mechanisms. Inhibiting MAPK extended cell-cycle length and led to generation of layer-V CTIP2+ neurons by repressing alternative laminar fates. Taken together, FGF/MAPK regulates the proliferative/neurogenic balance in deep-layer corticogenesis and provides a resource for generating layer-specific neurons for studying development and disease.
Keywords: stem cells, neurogenesis, cortex, lamination, Notch, fibroblast growth factor 2, MAPK signaling, human neural development
Highlights
-
•
FGF/MAPK regulates the proliferative/neurogenic balance in deep-layer corticogenesis
-
•
FGF/MAPK signaling maintains the progenitor pool and generates layer-VI neurons
-
•
MAPK inhibition prolongs cell cycle to yield layer-V neurons, repressing other fates
-
•
Protocols to generate layer-specific cortical neurons to study development and disease
Despite heterogeneity across human cortical layers, all excitatory neurons evolve from a single founder stem cell population. How these progenitors alter layer competence over time is unknown. Using human pluripotent stem cell-derived cortical progenitors, we demonstrate that FGF/MAPK signaling regulates the proliferative/neurogenic balance in deep-layer corticogenesis. Modulating FGF/MAPK enables generation of layer-specific neurons to study development and disease.
Introduction
Neuroepithelial stem cells (NESCs) of the developing telencephalon give rise to the diverse populations of neurons and glia within the cortex. NESCs divide symmetrically to generate daughter radial glia (RG) and asymmetrically to generate neurons directly (Betizeau et al., 2013; Lui et al., 2011; Nowakowski et al., 2017). As development proceeds, RG within the ventricular zone (VZ) preferentially divide asymmetrically to generate a neuron and an intermediate progenitor cell (IPC). IPCs migrate basally to populate the superficial germinal zones of the inner and outer subventricular zones (ISVZ/OSVZ), where they undergo further rounds of division before terminal neurogenesis (Lui et al., 2011). The expansion of progenitor diversity underlies the complexity and enlargement of the primate cortex, particularly in the upper layers (Betizeau et al., 2013). During development, apical RG (aRG) undergo transcriptional changes that alter their developmental potential from deep- to upper-layer fates (Nowakowski et al., 2017; Telley et al., 2019). These spatiotemporal events during corticogenesis are tightly orchestrated, resulting in early-born neurons becoming deep-layer (DL) infragranular neurons (layers V/VI), while later-born precursors become upper-layer (UL) supragranular neurons (layers I–III) (Lui et al., 2011). Our understanding of human cortical development has been bolstered by differentiation of human pluripotent stem cells (hPSCs) into cortical progenitors (Espuny-Camacho et al., 2013; Shi et al., 2012). However, despite significant advances over the past two decades (Lui et al., 2011), how progenitor diversity and neuronal identity are linked remains elusive and is complicated by the lack of synchronicity across progenitor populations and the ability of human cortical progenitors to fluidly transition between NESC and IPC states.
Fibroblast growth factor (FGF) signaling has pleiotropic roles in cortical development (Iwata and Hevner, 2009). In rodents, Fgf2 signaling shortens G1 length and increases proliferative divisions (Lukaszewicz et al., 2002), while Fgf receptor (Fgfr) loss can lead to premature exhaustion of the proliferating progenitor pool and acceleration of neurogenesis, partly via loss of Notch (Rash et al., 2011). Strikingly, this loss of Fgf signaling alters the timing but not the competency of progenitors to form both DL and UL neurons (Rash et al., 2011). In addition, FGF-MAPK (mitogen-activated protein kinase) signaling is enriched in human cortical development compared with rodents and may play a role in formation of the OSVZ (Heng et al., 2017). Notch signaling also regulates self-renewal, via Notch ligand Delta-like 1 (DLL1)-induced upregulation of cyclin D1 and suppression of pro-neural genes. Notch pathway activation is mutually active and oscillatory among aRG, later shifting to a lateral inhibition mode in the neurogenic state. Taken together, FGF and Notch signaling act in concert to maintain RG fate and regulate the progression from proliferative to neurogenic divisions, modulating the size of the cortex via the cell-cycle program. However, if and how these extrinsic cues influence the shift in layer competence during human development remains unknown.
Here, we assessed the impact of Notch and FGF signaling on laminar fate using a PAX6 hPSC reporter line to specifically identify changes within the committed dorsal forebrain progenitor population. Our results show that Notch inhibition in early PAX6+ progenitors led to the rapid generation of functional TBR1+ DL neurons. Continued FGF2 exposure maintained early progenitor identity, giving rise exclusively to TBR1+ neurons when cells exit the cell cycle. In contrast, inhibition of FGF signaling (via MEK inhibition) biased CTIP2+, layer-V neurons. Together, these results shed light on the pathways involved in the sequential generation of cortical neuron subtypes and provide a platform for the in vitro derivation of homogeneous human cortical layer populations.
Results
PAX6+ cortical progenitors generate deep-layer and upper-layer neurons
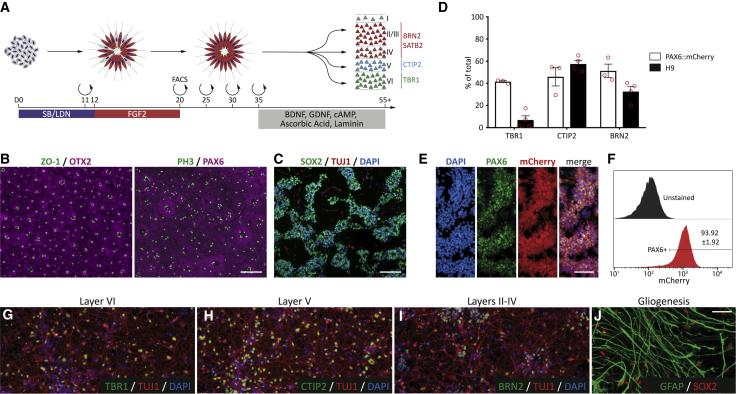
We first validated that both DL and UL neurons could be generated using traditional two-dimensional (2D) monolayer culture conditions (Figure 1A). Dual-SMAD differentiation of H9 embryonic stem cells (ESCs) robustly induced neuroectodermal specification, shown by the expression of forebrain (OTX2) and dorsal (PAX6) markers, and the appearance of rosettes with basal mitoses (PH3+) following FGF2 addition (Figure 1B). By day 25 (D25), heterogeneous cultures of SOX2+ progenitors and TUJ1+ neurons were observed, indicating that a subset of cortical progenitors was capable of terminal neurogenesis at this time (Figure 1C). Replating of these cultures at D35 for terminal differentiation yielded both DL and UL neuronal identities as shown by TBR1+, CTIP2+, and BRN2+ expression at D55. H9 human ESCs (hESCs) consistently generated mixed populations, albeit with low expression of TBR1 (Figure 1D). We hypothesized that the low proportion of TBR1+ neurons within H9 cultures was due to the mixed nature of the progenitor pool, which may be too mature at D20 to consistently generate these earliest born layer-VI neurons. Therefore, to select a specific progenitor population, we utilized a HES3:PAX6mCherry reporter line (Figures 1E and 1F). PAX6 is limited to the VZ, ISVZ, and OSVZ during primate neurogenesis, allowing for specific analysis and isolation of dorsal forebrain progenitors from heterogeneous cortical cultures (Manuel et al., 2015). mCherry expression was readily observed in rosettes, and reporter specificity was validated by staining with a PAX6 antibody (Figure 1E). At D20, PAX6+ progenitors were isolated for further differentiation and analysis (Figure 1F). As expected, PAX6+ progenitors gave rise to both DL and UL neurons, including layer-VI TBR1+, layer-V CTIP2+, and layers-II–IV BRN2+ by D55, as well as GFAP+ astrocytes following extended (beyond D80) culture (Figures 1G–1J), similar to previous studies (Espuny-Camacho et al., 2013; Shi et al., 2012). Common to these differentiations was a residual population of SOX2+ progenitors (Figure 1J), highlighting the asynchronicity of cortical differentiation. By D55, mCherry+ progenitors gave rise to roughly equal numbers of TBR1+, CTIP2+, and BRN2+, in contrast to H9 (Figure 1D), validating PAX6+ isolation to probe the role of signaling pathways in human DL corticogenesis.
Figure 1.
PAX6+ cortical progenitors form both deep- and upper-layer cortical neurons
(A) Schematic showing dual-SMAD neural induction and FGF2 expansion to generate dorsal telencephalic progenitors from hPSCs. HES3:PAX6mCherry progenitors were FACS isolated at day 20 (D20) and matured until D55 for analysis.
(B) H9-derived cortical progenitors express forebrain (OTX2+) and dorsal (PAX6+) markers and self-organize into rosettes with ZO-1+ lumens and basal PH3+ mitoses.
(C) Asynchronous maturation generates cultures of SOX2+ progenitors and TUJ1+ neurons.
(D) Quantification of TBR1+ (layer VI), CTIP2+ (layer V), and BRN2+ (layers II–IV) neurons for two hPSC lines (n = 3–4 independent experiments, data are mean ± SEM).
(E) The HES3:PAX6mCherry reporter mirrored PAX6 protein expression in vitro.
(F) FACS isolation and quantification of PAX6+ cortical progenitors at D20 (n = 5).
(G–I) Long-term culture (D55) yielded cortical neurons expressing markers of layers VI (G), V (H), and II–IV (I).
(J) Glial differentiation (GFAP/SOX2) was observed after extended culture (>D80).
Scale bars, 100 μm.
Notch inhibition rapidly initiates cell-cycle exit and differentiation
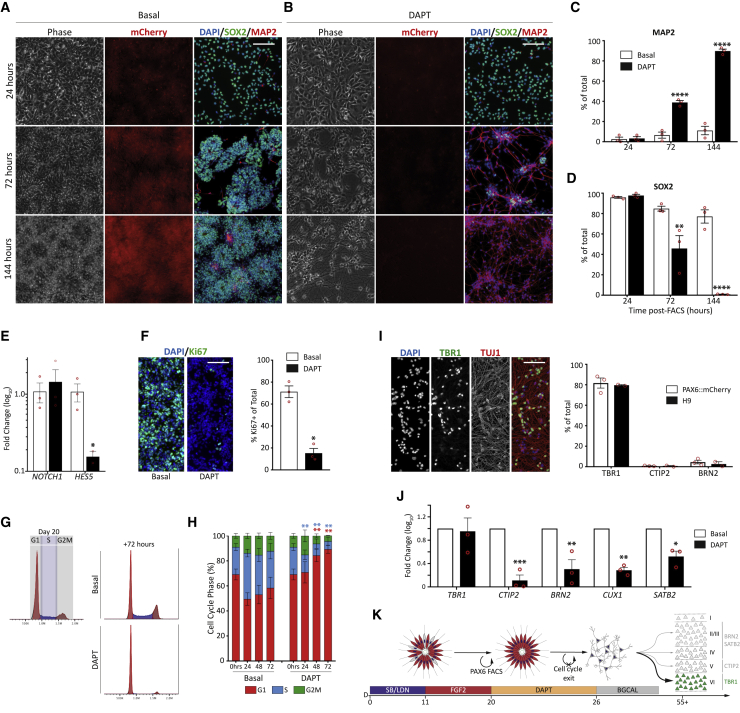
Notch signaling regulates proliferative or neurogenic fate decisions in cortical progenitors and is routinely used to promote cell-cycle exit in vitro (Borghese et al., 2010; Fiddes et al., 2018; Mizutani et al., 2007; Niclis et al., 2017; Rash et al., 2011; Suzuki et al., 2018). To identify the window of DL neurogenesis in our system, we treated homogeneous PAX6+ cultures immediately following fluorescence-activated cell sorting (FACS) with the γ-secretase inhibitor N-[N-(3,5-difluorophenacetyl)-L-alanyl]-S-phenylglycine t-butyl ester (DAPT). DAPT prevents the final cleavage of transmembrane Notch, decreasing the level of the active Notch intracellular domain. Under Basal conditions, mCherry+/SOX2+ rosettes appeared rapidly after sorting (Figure 2A) and the proportion of neurogenic divisions remained low after 6 days (144 h), shown by the low proportion of MAP2+ neurons (11.1% ± 4.2% MAP2+) and presence of SOX2+ progenitors (Figures 2A–2D). DAPT-treated cultures did not reform rosettes, lost mCherry and SOX2 expression, and generated MAP2+ neurons (89.8% ± 1.9%) (Figures 2B–2D). Repression of the Notch target gene HES5 48 h after DAPT addition confirmed inhibition of signaling downstream of NOTCH1 (Figure 2E). To validate the rapid cell-cycle exit of cortical progenitors, we assessed progenitor cell-cycle dynamics. The proportion of Ki67+ cycling progenitors was reduced 48 h after DAPT addition (DAPT: 15.3% ± 4.3%; Basal: 71.2% ± 5.3% Ki67+/DAPI) (Figure 2F). Analysis of DNA content using FACS (Figure 2G) at defined intervals after DAPT treatment (0, 24, 48 and 72 h) revealed inhibition of G1/S-phase transitions as early as 24 h after treatment; an effect that peaked at 72 h (Figures 2G and 2H, blue bars; DAPT: 6.4% ± 3.3%; Basal: 29.4% ± 6.2%). An increase in the proportion of cells in G1 was also observed (Figure 2H, red bars; DAPT: 89.3% ± 3.2%; Basal: 58.4% ± 8.6%). Analysis of the identity of DAPT-induced neurons after maturation (D55) revealed almost exclusive generation of TBR1+ layer-VI neurons (Figure 2I), an effect that was consistent for both HES3:PAX6mCherry and H9 lines (Figure 2I). The loss of other neuronal phenotypes at D55 was confirmed using qPCR (Figure 2J). Taken together, Notch inhibition efficiently crystallized cortical progenitor fate, demonstrating that rapid cell-cycle exit of young D20 PAX6+ cortical progenitors prevents progression to the layer-V, CTIP2+ identity (Figure 2K).
Figure 2.
Notch inhibition of PAX6+ progenitors generates TBR1+, early-born cortical neurons
(A) Under basal conditions, FACS-isolated progenitors generated PAX6-mCherry+/SOX2+ rosettes, which persisted for >7 days and showed few MAP2+ neurons.
(B) DAPT treatment downregulated mCherry, rapidly generated neurons, and depleted SOX2+ progenitors.
(C and D) Quantification for MAP2+ (C) neurons or SOX2+ (D) progenitors ± DAPT (n = 3 independent experiments).
(E) DAPT had no effect on NOTCH1 but decreased downstream HES5 expression after 48 h, confirming inhibitor activity (n = 3 independent experiments).
(F) DAPT rapidly reduced Ki67+ cells after 48 h (n = 3 independent experiments).
(G) Example FACS-based cell-cycle analysis in Basal and DAPT cultures (n = 3 independent experiments).
(H) DAPT reduced the number of progenitors entering S phase within 24 h.
(I) Neurons generated after DAPT treatment were almost exclusively TBR1+ at D55 (n = 3 independent experiments).
(J) qRT-PCR confirmed that DAPT treatment reduced CTIP2, BRN2, CUX1, and SATB2 expression (n = 3 independent experiments).
(K) Schematic of DAPT-induced cell-cycle exit showing that D20 progenitors are competent to generate early-born, layer-VI TBR1+ neurons.
Data are mean ± SEM; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. Scale bars, 100 μm.
Loss of FGF signaling induces layer-V-like CTIP2+ neurogenesis
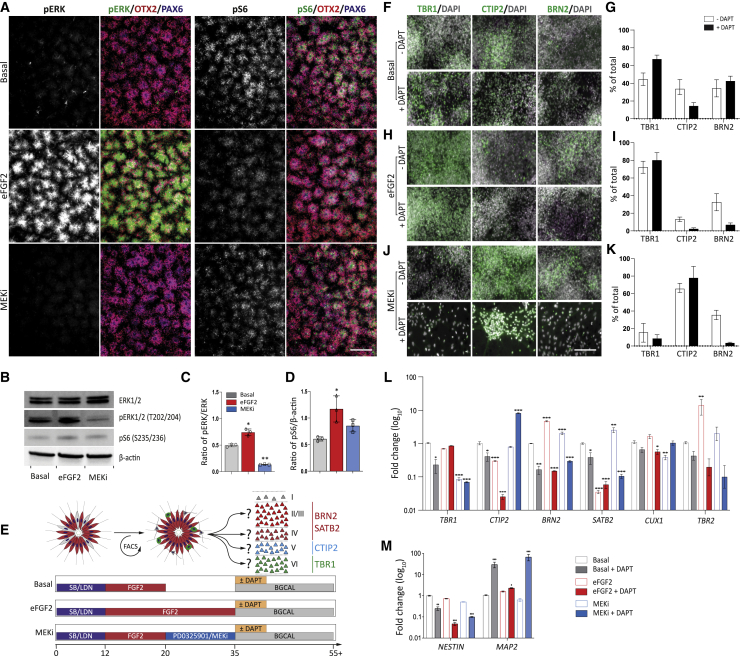
We next sought to investigate the impact of cell-cycle promotion by probing the FGF-MAPK pathway. In contrast to Notch, FGF2 is routinely used in vitro to promote proliferation; however, its effect on temporal specification remains unknown (Espuny-Camacho et al., 2013; Shi et al., 2012). We first analyzed the acute effect of FGF2 on phosphorylated ERK (pERK) and phosphorylated ribosomal protein S6 (pS6) proteins, downstream elements of the FGF2-associated MAPK and phosphatidylinositol 3-kinase (PI3K) signaling cascades, respectively (Figure 3A). Under Basal conditions, rare OTX2+/PAX6+/pERK+ progenitors could be observed in cortical rosettes while the majority of progenitors expressed pS6. Extended FGF2 treatment (from D12 to D20 to D12 to D35, henceforth eFGF2) increased pERK but not pS6 expression (Figure 3A). MEK inhibition blocked endogenous pERK expression. Immunoblotting confirmed FGF2-induced ERK and S6 phosphorylation, while MEKi blocked ERK but not S6 phosphorylation (Figures 3B–3D).
Figure 3.
eFGF2 or MEK inhibition alters cortical layer phenotype
(A) Under Basal conditions few phosphorylated ERK+ (pERK+) progenitors are observed, while the majority of cells are phosphorylated S6+ (pS6+). FGF2 treatment increased pERK expression, while MEK inhibition (MEKi) blocked pERK expression.
(B–D) Western blot showing upregulated pERK and pS6 after 24 h of FGF2 treatment (B). MEKi abolished pERK expression but had no effect on pS6. Western blot quantification of pERK (C) and pS6 (D) (n = 3 independent experiments).
(E) Differentiation conditions of cortical progenitors showing Basal, eFGF2, and MEKi conditions (±DAPT).
(F–K) Sorted PAX6+ progenitors under Basal conditions (±DAPT) (F and G), or treated with eFGF2 (H and I) or PD0325901 (J and K) were assessed for laminar fate at D55. (G) Quantification showed that progenitors under Basal conditions generated DL and UL neurons, with DAPT at D35 biasing TBR1+ neurons. (I) eFGF2 treatment generated TBR1+ neurons almost exclusively ± DAPT. (K) MEKi-treated progenitors predominantly generated CTIP2+ neurons ± DAPT, while in the absence of DAPT UL BRN2+ cells were observed (n = 3 independent experiments).
(L) qRT-PCR analysis of cortical genes confirmed cell quantifications (n = 3 independent experiments). Relative to Basal, eFGF2 (−DAPT) increased BRN2 and decreased SATB2 expression, suggestive of an IPC population rather than UL fate. MEKi (+DAPT) decreased the alternative deep (TBR1) and superficial (BRN2) laminar fates. In contrast, MEKi (−DAPT) increased BRN2 and SATB2 expression, reflective of UL fates.
(M) Increased NESTIN and decreased MAP2 gene expression confirmed cell-cycle exit and maturation in DAPT cultures (n = 3 independent experiments).
Data are mean ± SEM, one-way ANOVA with Dunnett's correction, n = 3; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. Scale bars, 100 μm (A) and 50 μm (F–J).
Having established that MAPK signaling could be modulated within these cortical progenitor cultures, we next examined the consequences on layer specification (Figure 3E). Neurons born after eFGF2 treatment almost exclusively became early-born layer-VI TBR1+ cortical neurons by D55 (Figures 3F–3I, open bars), phenocopying DAPT-treated D20 progenitors (Figure 2I). Upon forced cell-cycle exit at D35, both Basal cultures and eFGF-treated cultures retained their laminar fates (Figures 3F–3I, black bars). Strikingly, MEKi, targeted at blocking endogenous MAPK signaling, resulted in a later-born, layer-V CTIP2+ phenotype, at the expense of TBR1+ (Figures 3J and 3K), irrespective of DAPT treatment. Importantly, the shift from TBR1+ to CTIP2+ neurogenesis under eFGF2 or MEKi conditions, respectively, was reproducible in H9-derived cultures (Figure S1). Transcriptional analysis at D55 confirmed the laminar fate biases—notably conserved TBR1 expression and downregulation of CTIP2 and BRN2 after eFGF2, and CTIP2 expression at the expense of TBR1 and BRN2 after MEKi (Figure 3L). SATB2 gene expression was decreased after eFGF2 (±DAPT), suggesting that the observed BRN2 cells (Figure 3H) were IPCs (supported by increased TBR2 levels) and not UL fated neurons (Figure 3L). In contrast, under MEKi conditions SATB2 was elevated and SATB2+ neurons were increased (Figure S2), suggestive of enhanced UL neuron specification. CUX1 expression remained low for all culture conditions, suggesting that longer culture periods (beyond D55) are required to attain this uppermost fate (Espuny-Camacho et al., 2013; Shi et al., 2012). Confirming DAPT maturation, NESTIN was downregulated in eFGF2- and MEKi-treated cultures (after subsequent DAPT addition) while MAP2 (a cytoskeletal protein marking post-mitotic neurons) was upregulated (Figure 3M).
To confirm that eFGF2- and MEKi-derived neurons were functional, we carried out electrophysiological analysis of maturing cultures (D80 and later) (Figure S3). DAPT-treated eFGF2 or MEKi cultures showed lower resting membrane potentials (Figure S3A). Neurons from all groups were capable of action potential (AP) generation in response to depolarizing current injections. However, neurons derived from each group exhibited varying input frequency relationships indicating a mix of both of mature (AP trains) and immature (AP collapse) development (Figures S3B and S3C). Spontaneous excitatory postsynaptic currents (EPSCs) were increased after MEKi (Figure S3D), reflective of greater neural connectivity.
Antagonism of FGF-MAPK signaling promotes neurogenesis
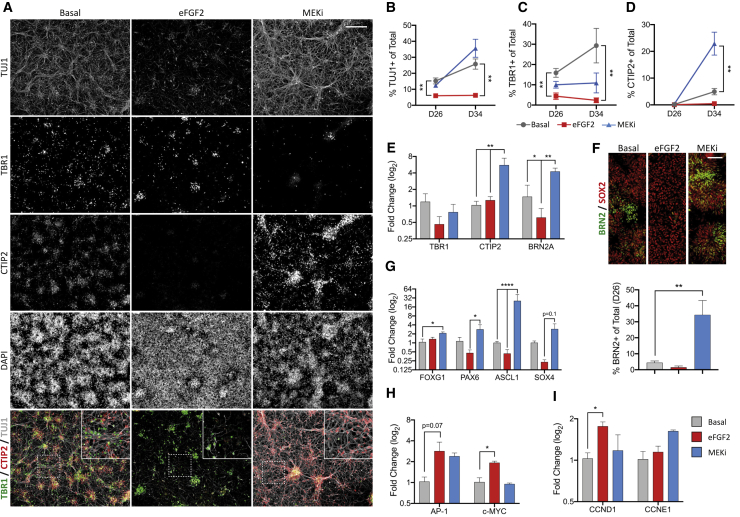
Laminar identity is directed cell autonomously during cortical progenitor expansion within the VZ/SVZ (Bayraktar and Doe, 2013; Dominguez et al., 2013; Lui et al., 2011), as well as non-cell autonomously during post-mitotic maturation (Leone et al., 2015; McKenna et al., 2015; Ozair et al., 2018; Toma et al., 2014). To understand how eFGF2 or MEKi generate distinct neuronal phenotypes, we characterized cultures during ongoing maturation between D26 and D34 (5 and 13 days post treatment, respectively). Under Basal and MEKi conditions, cultures increasingly generated TUJ1+ neurons (Basal: 15.2% ± 1.9% at D26, 25.8% ± 3.2% at D34; MEKi: 12.5% ± 1.1% at D26, 35.6% ± 5.7% at D34) (Figures 4A and 4B). This effect was blocked after eFGF2 treatment (eFGF2: 6.0% ± 1.3% at D26, 6.2% ± 1.0% at D34) (Figures 4A and 4B). At D26 the majority of generated neurons were early-born TBR1+, regardless of culture condition (Figure 4C), reflective of appropriate DL neuronal identity in the absence of DAPT addition. At this stage CTIP2+ neurons were absent from all groups (<1%), indicating that progenitors were developmentally immature (Figure 4D). At D34 the proportion of both TBR1+ and CTIP2+ neurons increased under Basal conditions, albeit to a limited extent for CTIP2. In contrast, CTIP2+ cells remained absent following eFGF2 treatment but were highly enriched after MEKi (Basal: 4.9% ± 1.2% and MEKi: 22.9% ± 4.3%) (Figure 4D). qPCR profiling at D26 revealed an upregulation of CTIP2 in response to MEKi (Figure 4E), preceding the appearance of these neurons at D35 (Figures 4A and 4D). Interestingly, while BRN2+/TUJ1+ neurons were rare in all conditions at both D26 and D34 (data not shown), BRN2 was upregulated (Figure 4E) and BRN2+/SOX2+ cycling progenitors were highly enriched after MEKi ( at D26, Basal: 4.5% ± 1.0%; eFGF2 1.5% ± 09%; MEKi: 34.4% ± 8.9%) (Figure 4F), indicative of a progenitor pool likely targeted for UL neurogenesis.
Figure 4.
MEK inhibition accelerates the development of CTIP2-competent progenitors and the pro-neural gene network
(A) Under Basal conditions, TBR1+ and CTIP2+ cells are present at D34, showing that progenitors can generate both layers. In contrast, eFGF2 treatment limits neurogenesis and only few TBR1+ neurons are born. After MEKi, CTIP2+ neurons predominate at the expense of TBR1. Inset shows magnified section.
(B–D) Quantification at D26 and D35 for TUJ1+ (B), TBR1+ (C), and CTIP2+ (D) neurons (n = 3–5 independent experiments, one-way ANOVA with Dunnett's correction).
(E) qRT-PCR at D26 showed increased CTIP2 and BRN2A mRNA after acute MEKi.
(F) BRN2+/SOX2+ progenitors were rare in eFGF2 conditions and increased following MEKi.
(G) The pro-neural genes FOXG1, PAX6, and ASCL1 were increased at D26 after MEKi, concomitant with increased CTIP2+ neurogenesis.
(H) eFGF2 induced AP-1 and c-MYC, downstream of FGFR activation.
(I) eFGF2 also increased levels of CCND1, required for G1/S-phase transition (n = 3 independent experiments).
Data are mean ± SEM; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. Scale bars, 200 μm (A) and 50 μm (F).
Reflective of chronic regulation of FGF signaling in these cultures, significant increases in the pro-neural genes FOXG1, PAX6, and ASCL1 were observed after 5 days of MEKi treatment (D26, Figure 4G), while eFGF2 upregulated cell-cycle gene expression (c-MYC and CCND1, Figures 4H and 4I). Changes in mCherry expression further confirmed progenitor behavior in response to eFGF2 or MEKi conditions (Figure S2). Unlike eFGF2 treatment, changes in neurogenic competence in response to MEKi were not accompanied by alterations in CCND1 and CCNE1, two genes responsible for cell-cycle entry and S-phase transition, respectively (Figure 4I). Together, these data show that in early progenitors, MEKi-mediated upregulation of pro-neural genes does not force cell-cycle exit.
eFGF2 maintains cortical progenitor identity
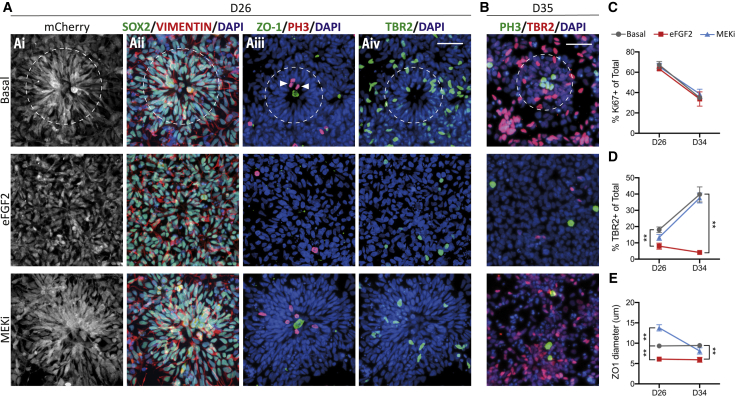
2D rosettes recapitulate key features of human corticogenesis (Espuny-Camacho et al., 2013; Shi et al., 2012). Notably, apicobasal polarity of PAX6-mCherry+/SOX2+/Vimentin+ RG cells (Figures 5Ai and Aii), radial orientation around a lumen (Figure 5Ai), apical mitoses (Figure 5Aiii), and emergence of basally dividing TBR2+ IPCs (Figure 5Aiv). Under Basal conditions at D26, 88.2% ± 1.7% of cells were SOX2+ progenitors (Figure 5Aii) and 67.2% ± 3.3% were proliferating Ki67+ progenitors, a proportion which dropped to 35.1% ± 1.9% by D35 (Figure 5C). Neither eFGF2 nor MEKi altered Ki67+ numbers (Figure 5C). In contrast, eFGF2 showed a diminished proportion of TBR2+ IPCs at D26 (Basal: 18.1% ± 1.7%; eFGF2: 7.9% ± 1.9%; MEKi: 13.3% ± 1.8%), an effect that was more pronounced by D35 (Basal: 39.8% ± 4.6%; eFGF2: 4.1% ± 1.0%; MEKi: 37.6% ± 3.1%) (Figures 5A, 5B, and 5D). Morphologically, eFGF2 treatment consistently decreased ZO1+ lumen diameter (Figures 5Aiii and 5E). Importantly, caspase-3 analysis confirmed that eFGF2 or MEKi did not alter progenitor dynamics via increased apoptosis (Figure S3).
Figure 5.
Progenitor dynamics highlight immaturity of eFGF2-treated progenitors
(A) Cortical rosettes under Basal conditions recapitulated in vivo cortical development, showing apicobasal polarity (Ai and Aii), ZO1+ lumens (Aiii), PH3+ apical mitoses (white arrows, Aiii), and TBR2+ IPCs located basal to the lumen (Aiv). Rosette organization and TBR2+ IPCs were reduced after eFGF2 treatment and unaffected by MEKi. White circles highlight rosettes.
(B) At D35, IPCs were increased in Basal and MEKi conditions but were rare after eFGF2.
(C) No difference in Ki67+ cycling progenitors was observed.
(D) TBR2 quantification at D26 and D35 shows that eFGF2-treated cultures did not generate IPCs.
(E) MEKi generated larger rosettes, shown by increased ZO-1 lumen diameter (n = 3–5 independent experiments, one-way ANOVA with Dunnett's correction).
Data are mean ± SEM; ∗p < 0.05, ∗∗p < 0.01. Scale bar, 50 μm.
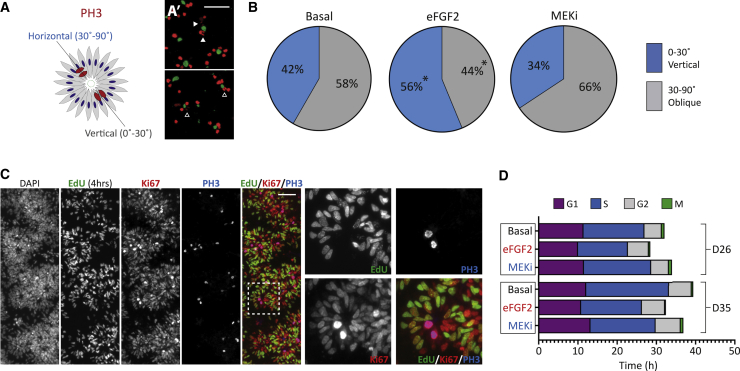
The observation that eFGF2 cultures failed to generate TBR2+ IPCs led us to speculate that eFGF2 was maintaining an early cortical progenitor identity. Key changes in cell-cycle kinetics accompany maturation of cortical progenitors, including increases in asymmetric divisions and cell-cycle length (Arai et al., 2011; Betizeau et al., 2013; Noctor et al., 2004; Subramanian et al., 2017; Turrero Garcia et al., 2016). We next assessed the cleavage plane of recently divided cells at the ventricular surface of D26 cultures (Figure 6A). Both vertical and oblique divisions were observed under Basal conditions, while eFGF2 increased the proportion of proliferative/vertical divisions (Basal: 42%; eFGF2: 56%; MEKi: 34%), at the expense of neurogenic/oblique divisions (Basal: 58%; eFGF2: 44%; MEKi: 66%) (Figure 6B). We further probed cell-cycle kinetics using a cumulative 5-ethynyl-2′-deoxyuridine (EdU) labeling protocol (Nowakowski et al., 1989) that enabled estimates of cell-cycle phase lengths (Figures 6C and S6). At D26 and D35 we observed consistent EdU labeling, across all three conditions, which was restricted to the cycling Ki67+ progenitor pool (Figure 6C). As previously shown in rodents and primates (Arai et al., 2011; Betizeau et al., 2013; Subramanian et al., 2017; Turrero Garcia et al., 2016), under Basal conditions the total cell-cycle length increased over cortical development from 32.1 h at D26 to 39.4 h at D35, via extension of the S phase (+5.6 h) (Figures 6D and S6A), concurrent with increasing neurogenic divisions. We found that eFGF2 cultures consistently had shorter cell-cycle lengths (Figures 6D and S6A). Taken together, eFGF2 inhibits development of cortical progenitors by promoting proliferative divisions and limiting development of multiple neurogenic and intermediate characteristics.
Figure 6.
Cell-cycle analysis highlights progenitor characteristics of eFGF2-treated cultures
(A) Quantification method for determining the cleavage angle of dividing PH3+ progenitors.
(A′) Images showing horizontal (filled arrows) or vertical (hollow arrows) cleavage angles (n = 103 cells).
(B) Quantification of cleavage angle under Basal, eFGF, and MEKi conditions. eFGF2-treated progenitors preferentially underwent proliferative divisions, shown by the increase in vertical cleavage planes.
(C) EdU assessment of cell-cycle kinetics with EdU labeling cells in S phase and Ki67 total cycling pool. Inset shows magnified region. Note the two PH3+ mitotic progenitors are EdU− at this time point (4 h), showing that they had exited S phase prior to EdU addition.
(D) Representation of G1, S, G2, and M phase and total cell-cycle length under Basal, eFGF2, or MEKi conditions, highlighting that eFGF2 shortened the cell cycle (representative of n = 3 independent experiments).
Scale bar, 50 μm.
eFGF2 acts via MAPK/ERK signaling
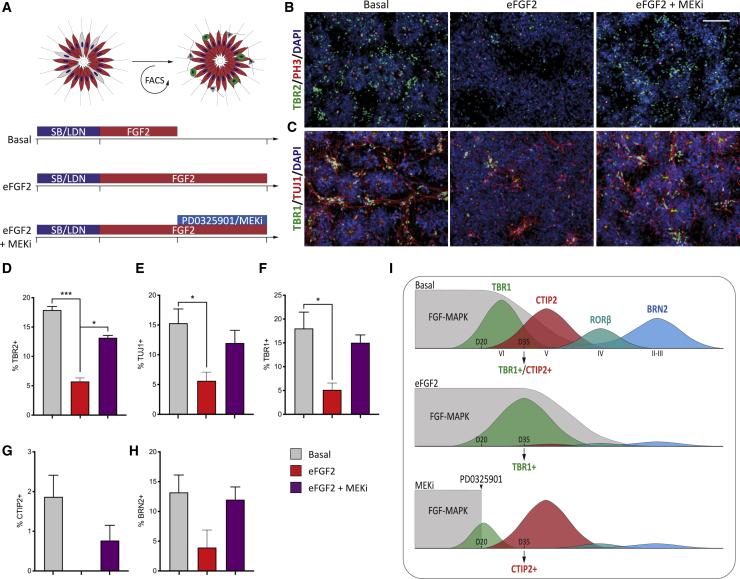
To confirm that eFGF2-induced maintenance of progenitor identity was dependent on MAPK signaling, we analyzed progenitors in the combined presence of eFGF2 and MEKi (Figure 7A). Addition of MEKi to FGF2-treated cultures was sufficient to block FGF2-induced upregulation of pERK (Figures S7A–S7C), while pS6 levels remained upregulated, highlighting inhibitor specificity. eFGF2 conditions blocked TBR2+ IPC and TUJ1+ neuron generation at D26, an effect that could be reversed by MEKi, resulting in cultures resembling Basal conditions (TBR2+ IPCs: Basal 17.9% ± 0.6%; eFGF2 5.7% ± 0.6%; eFGF2 + MEKi 13.2% ± 0.4%; and TUJ1+ neurogenesis: Basal 15.3% ± 2.4%; eFGF2 5.6% ± 1.5%; eFGF2 + MEKi 12.0% ± 2.1%) (Figures 7B–7E). In addition, eFGF2 + MEKi-treated progenitors generated comparable TBR1+, CTIP2+, and BRN2+ neurons to Basal conditions (Figures 7F–7H). We validated these changes using qPCR against cortical layer, pro-neural, and cell-cycle specific gene sets (Figures S7D–S7F). MEKi abolished FGF2 induction of downstream target genes AP-1 and c-MYC. However, eFGF2 + MEKi-treated progenitors did not completely phenocopy Basal progenitors, indicating that FGF2 may be acting through multiple downstream pathways, including via pS6-induced programs, which are unchanged after MEKi addition (Figures S7D–S7F). To verify that MEK inhibition was acting downstream of FGF2 signaling, we treated cultures with the FGF receptor (FGFR) inhibitor SU5402. Results showed comparable upregulation of pro-neural, layer-V, and UL (CTIP2 and BRN2) gene expression for both SU5402- and MEKi-treated cultures (Figures S7G and S7H). We conclude that FGF2 solidifies early cortical progenitor identity by blocking temporal maturation and that this effect is mediated by the MAPK pathway (Figure 7I).
Figure 7.
eFGF2 acts via MEK to maintain progenitor identity
(A) Experimental diagram of mechanism experiments. Cortical progenitors were exposed to eFGF2 (±MEKi) to analyze the role of MEK.
(B) eFGF2-treated cultures did not generate TBR2+ IPCs, an effect that was reversed by MEKi.
(C) Likewise, eFGF2 cultures generated limited TBR1+ neurons at D26, in contrast to Basal conditions. This effect was also reversed by MEKi.
(D–H) Quantification at D26 for TBR2 (D), TUJ1 (E), TBR1 (F), CTIP2 (G), and BRN2 (H) under Basal, eFGF2 and eFGF2+ MEKi conditions (n = 3 independent experiments).
(I) Putative role of FGF2 signaling during cortical maturation and laminar specification.
Scale bar, 50 μm.
Discussion
Utilizing a PAX6 reporter line to isolate dorsal telencephalic progenitors, we confirmed the sequential acquisition of cortical neurons in vitro. Manipulation of FGF-MAPK signaling, by extending FGF2 treatment or MEK inhibition, slowed or accelerated the development of progressive layer competence, respectively. eFGF2 signaling resulted in the maintenance of the progenitor phenotype, reflected in gene expression, cell-cycle kinetics, cell-fate decisions, and laminar competence. In contrast, MEKi accelerated the acquisition of layer-V (CTIP2+) competence at the expense of TBR1+ layer-VI neurons (Figure 7I). We demonstrate that this effect is associated with the upregulation of a pro-neural gene network including ASCL1 and PAX6.
FGFR1/3 signaling is critical for controlling proliferation and cortical size in rodents, with loss leading to premature depletion of the progenitor pool and decreased cortical volume (Maric et al., 2007; Raballo et al., 2000; Rash et al., 2011). In contrast, FGFR2 signaling controls the maturation of RG from NESCs (Sahara and O'Leary, 2009; Yoon et al., 2004). FGF2, which binds FGFR1 and FGFR3, exerts a mitogenic effect on cortical progenitors by altering cell-cycle and fate decisions, consistent with its role in vivo (Lukaszewicz et al., 2002; Maric et al., 2007). Moreover, FGF2 is routinely used to promote proliferation and neural progenitor cell expansion in culture. The findings reported here suggest that FGF2 exerts a similar effect on human cortical progenitors as that previously shown in the rodent. In particular, sustained FGF2 (eFGF2) blocked the developmental progression of RG, such that older cultures (D35) remained competent only to generate TBR1+ neurons (Figure 7I). eFGF2 cultures failed to undergo neurogenic divisions to generate TBR2+ IPCs or TUJ1+ neurons and instead cells preferentially underwent proliferative, symmetrical divisions, similar to those in the mouse (Kang et al., 2009). Consequently, we propose that FGF-MAPK is crucial for mediating the expansion and subsequent differentiation of human cortical progenitors but not laminar competence. Of particular interest, this result suggests that laminar competence is not linked to the number of divisions, as has been shown in the mouse (Ohtsuka and Kageyama, 2019; Okamoto et al., 2016). Instead, eFGF2 shortens cell-cycle length and blocks laminar progression, supporting cell-cycle length as a critical mediator in facilitating maturation (Calegari et al., 2005; Dehay and Kennedy, 2007; Ohtsuka and Kageyama, 2019; Pilaz et al., 2009; Watanabe et al., 2015).
While the repression-derepression model within neocortical neurons is widely accepted, the factors that govern temporal progression in RG and, thus, layer competence, have remained relatively unknown. In mouse, self-renewal is inhibited by Bcl6, which is a convergent repressor of Wnt, Notch, Fgf, and Shh signaling and drives neurogenesis (Bonnefont et al., 2019). Ascl1 and Neurog2 are required for progression from default Tbr1 to Ctip2 neurogenesis in rodents by modulating Foxg1 (Dennis et al., 2017), which itself determines the sequence of layer generation by repressing Tbr1 (Toma et al., 2014). In addition, RAS/ERK signaling has been linked to pro-neural lineage decisions during development and disease (Dee et al., 2016; Li et al., 2014) and generation of basal RG (bRG), and is increased in human VZ progenitors (Heng et al., 2017). Consistent with a role of FGF2 in delaying progenitor maturation, we observed a downregulation of the pro-neural genes ASCL1, PAX6, and SOX4 after eFGF2 treatment. In the mouse, Ascl1 is sufficient to specify Ctip2+ neurogenesis in DL progenitors, and its loss results in decreased Ctip2+ neurogenesis and expansion of Tbr1+ neurons into layer V (Dennis et al., 2017). Here, MEKi rapidly led to decreases in PAX6 and ASCL1, exhaustion of TBR1+ neurogenesis, and acceleration of CTIP2+ neurogenesis. Whether human cortical progenitor signaling converges through Bcl6 as in the mouse is unknown (Bonnefont et al., 2019). Our results suggest that FGF-MAPK may provide a context-dependent switch during laminar specification, whereby loss may act to break TBR1 repression of CTIP2 and thus drive temporal maturation. Indeed, the appearance of BRN2+ progenitors (and elevated BRN2 and SATB2 mRNA) after MEKi treatment suggests an acceleration rather than a specific increase in layer-V neurogenesis. Further studies will be necessary to confirm the role of FGF-MAPK regulation in human corticogenesis in vivo.
The primate OSVZ contributes disproportionately to layers II–IV and is accompanied by an expanded repertoire of progenitors, including multiple RG and IPC subtypes (Betizeau et al., 2013; Dehay et al., 2015; Lui et al., 2011; Reillo et al., 2011). In contrast, 2D hPSC models generate disproportionately low numbers of UL neurons. Here we have focused on the role of FGF-MAPK signaling in DL neurogenesis, as these layers are generated in a robust temporal sequence in vitro. In the mouse, FGF2 expands the NESC progenitor pool (Vaccarino et al., 1999), blocks RG/IPC transitions (Kang et al., 2009), and is downregulated by the mid-stage of corticogenesis (Raballo et al., 2000), allowing appropriate time for DL neurogenesis, while FGF-MAPK signaling is crucial for aRG-to-bRG transitions during OSVZ expansion (Heng et al., 2017). Thus, MEKi may simultaneously accelerate temporal maturation while blocking bRG generation and UL neurogenesis. FGF-MAPK signaling may have a biphasic role in cortical development, first promoting NESC/RG expansion and subsequently being required for OSVZ expansion and UL neurogenesis in accordance with observed biphasic changes in cell-cycle length during primate corticogenesis (Betizeau et al., 2013). Studies utilizing organoid culture systems, which more fully recapitulate OSVZ and laminar organization, may illuminate the role of FGF-MAPK signaling in human UL neurogenesis.
Finally, while traditional hPSC-derived cortical differentiation systems produce asynchronous, heterogeneous populations of pyramidal neurons and glia, we demonstrate homogeneous differentiation of specific DL projection neurons (layer IV or V) that may prove valuable for the study, modeling and therapeutic development for laminar-specific disorders (Espuny-Camacho et al., 2013; Shi et al., 2012).
Experimental procedures
hPSC maintenance and differentiation
hESC lines H9 (WA09, WiCell) and HES3 (ES03) reporter line HES3:PAX6mCherry (Bellmaine et al., 2017) were maintained as previously described (de Luzy et al., 2019). Cortical induction was achieved using an established protocol with adaptations (Shi et al., 2012). hPSCs were seeded into Laminin-521-coated (5 μg/mL) plates at 0.375 × 106/cm2 in mTeSR1 medium with10 μM Rock inhibitor Y-27632 (Tocris Bioscience). Twenty-four hours later, cells were switched to dual-SMAD inhibition for 11 days in “cortex medium” supplemented with 100 nM LDN193189 (Stemgent) and 10 μM SB431542 (R&D Systems). Cortex medium consisted of 1:1 DMEM/F12 and Neurobasal with 0.5× B27, 0.5× N2, 0.5× ITSA, 1× GlutaMAX, 0.5× penicillin/streptomycin, and 50 μM 2-mercaptoethanol (Life Technologies). On D11, progenitors were passaged and reseeded 1:2.5. The following day, cultures were transferred to cortex medium containing 20 ng/mL fibroblast growth factor 2 (FGF2; R&D Systems) for 8 days before cell sorting. Following FACS purification, cells were plated at 0.45 × 106/cm2 in cortex medium for further culture, or for cell-cycle exit at 0.2 × 106/cm2 in cortex medium supplemented with the γ-secretase inhibitor DAPT (10 μM; Sigma-Aldrich) on day 20 or day 35, henceforth known as “exit medium.” Exit medium was changed every day for 4 days and every second day thereafter. For ongoing culture, 1 day after FACS, PAX6+ progenitors were maintained in either cortex medium, 20 ng/mL FGF2-supplemented cortex medium, 1 μM MEK inhibitor-supplemented cortex medium (PD0325901; Sigma-Aldrich), FGF2 + MEKi- or SU5402-supplemented medium subsequently referred to as Basal, eFGF2, MEKi, eFGF2 + MEKi, or SU5402, respectively. One week following final plating, neurons were switched to maturation medium containing 1:1 DMEM/F12 and Neurobasal, 1× B27, 1× N2, 1× ITSA, 1× NEAA, 1× GlutaMAX, and 0.5× penicillin/streptomycin, with 40 ng/mL brain-derived neurotrophic factor (R&D Systems), 40 ng/mL glial cell line-derived neurotrophic factor, 0.05 mM N6,2′-O-dibutyryladenosine 3′,5′-cyclic monophosphate (Tocris Bioscience), 200 nM ascorbic acid (Sigma-Aldrich), and 1 μg/mL laminin (Sigma-Aldrich).
Flow cytometry
FACS was carried out using previously described methods (de Luzy et al., 2019). Dissociated D20 HES3:PAX6mCherry progenitor cultures, labeled with 0.5 μg/mL 4′,6-diamidino-2-phenylindole (DAPI; Sigma-Aldrich), were purified using an FACS Aria III (BD Biosciences) or MoFlo XDP (Beckman Coulter) using a 100-μm nozzle at 20 psi. mCherry positivity was gated using time-matched H9-derived cortical progenitors. DNA content analysis was carried out using an FxCycle kit (Invitrogen) as per manufacturer's instructions. Cells were prefixed by dropwise addition of ice-cold ethanol to a final concentration of 70%. For univariate analysis, a cell-cycle model was fitted using FlowJo software.
Cumulative EdU cell-cycle analysis
D20 FACS-purified PAX6+ progenitors were seeded at 0.45 × 106/cm2 in cortex medium. Twenty-four hours later, cultures were switched to cortex medium containing 20 ng/mL FGF2 or 1 μM MEKi. EdU (5 μM) was added to wells 20, 16, 8, 6, 4, 3, 2, 1, and 0.5 h prior to fixation in 4% (w/v) paraformaldehyde. Cultures were permeabilized in 0.3% (v/v) Triton X-100 for 20 min before visualization via a click reaction (Click-iT EdU kit; Life Technologies). Azide-bound Alexa-488 or -555 were used to visualize EdU incorporation. The growth fraction, defined as the average maximal percentage of EdU+/Ki67+ of the total Ki67+ population (16-h and/or 20-h pulse lengths dependent on total cell-cycle length), varied between 85% and 100% of Ki67+ progenitors. For analysis of TC − TS and TS length, linear regression models were plotted during the linear phase of EdU accumulation (0.5- to 8-h pulse lengths). TG2 was calculated as the half-maximal time of EdU accumulation within mitotic (PH3+) progenitors, which represents the average transit duration from S phase to mitosis (for details of calculation see Arai et al., 2011; Nowakowski et al., 1989; Turrero Garcia et al., 2016).
Gene expression analysis
For qPCR, cortical cells were pelleted, resuspended in 40 μL of RNAlater (Thermo Fisher), and stored at −80°C for later RNA isolation using an ISOLATE-II Mini Kit (Bioline). After extraction, RNA were converted to 500 ng of cDNA using the SuperScript VILO cDNA synthesis kit (Thermo Fisher). Upon completion,10 ng of cDNA was amplified using in-house designed primers and PowerUp SYBR Master Mix (Thermo Fisher) under the following conditions: 50°C for 2 min, 95°C for 2 min, and 40 cycles of 95°C for 15 s and 60°C for 1 min. All gene expression data were determined using 2−ΔΔCt from at least three independent biological replicates. Table S1 presents a list of all primers.
Immunocytochemistry, microscopy, and quantification
Immunocytochemistry was performed as previously described (de Luzy et al., 2019) (see Table S2 for a list of primary antibodies). Nuclei were counterstained with DAPI. Images were captured on a Zeiss Axio ObserverZ.1 upright epifluorescence microscope or a Zeiss LSM780 confocal microscope. For quantification of laminar specification, tiled images across wells were generated with at least five fields of view from multiple wells quantified for TBR1/BRN2/CTIP2 cell number. For cleavage plane analysis, a tangential line was drawn at the rosette center. A second intersecting line was drawn through clearly dividing PH3+ cells and the angle between the two lines calculated and binned as vertical (0°–30°) or horizontal/oblique (30°–90°).
Electrophysiology
Whole-cell recordings were performed on HES3:PAX6mCherry-derived neurons at D80. Cells were patched in an external solution containing 125 mM NaCl, 3 mM KCl, 1.2 mM KH2PO4, 1.2 mM MgSO4, 25 mM NaHCO3, 10 mM dextrose, and 2 mM CaCl2 (300 mOsmol, bubbled with 95% O2, 5% CO2, at 32°C), and using recording pipettes (3–6 MΩ) filled with an internal solution containing 6 mM NaCl, 4 mM NaOH, 130 mM K (OH and gluconate), 11 mM EGTA, 1 mM CaCl2, 10 mM HEPES, 1 mM MgCl2, and 0.1% biocytin (pH 7.3) (290 mOsmol, ECl −68.8 mV). In voltage-clamp mode (VH = −60 mV), EPSCs were recorded for 1 min. In current-clamp mode (I = 0 pA), resting membrane potential was recorded over 1 min. Thereafter, membrane potential was adjusted to −60 mV in each cell for step protocols (20 × 10 pA cumulative steps from −20 pA).
Author contributions
Conceptualization, C.W.G., J.C.N., C.P.J.H., and C.L.P.; Investigation, C.W.G., C.P.J.H., V.P. and S.J.M; Analysis, C.W.G. and C.P.J.H.; Provision of resources, L.H.T. and C.L.P.; Writing – original draft, C.W.G.; Writing – review and editing, C.W.G., C.P.J.H., and C.L.P.; Funding, C.L.P.
Acknowledgments
Experiments were performed at the Florey Institute of Neuroscience and Mental Health. The authors thank Andrew G. Elefanty and Edouard G. Stanley for the HES3:PAX6mCherry reporter line and acknowledge the support of the flow-cytometry facilities at the Melbourne Brain Centre and Murdoch Children's Research Institute. C.W.G. was supported by an Australian Postgraduate Award. V.P. was supported by a University of Melbourne International Scholarship. C.P. was supported by a National Health and Medical Research Council Australia (NHMRC) Senior Research Fellowship. This work was supported by project grants awarded by the NHMRC, Australia and the Australian Research Council. The Florey Institute of Neuroscience and Mental Health acknowledges the strong support from the Victorian Government and in particular the funding from the Operational Infrastructure Support Grant.
Published: April 8, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.stemcr.2021.03.014.
Supplemental information
References
- Arai Y., Pulvers J.N., Haffner C., Schilling B., Nusslein I., Calegari F., Huttner W.B. Neural stem and progenitor cells shorten S-phase on commitment to neuron production. Nat. Commun. 2011;2:154. doi: 10.1038/ncomms1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayraktar O.A., Doe C.Q. Combinatorial temporal patterning in progenitors expands neural diversity. Nature. 2013;498:449–455. doi: 10.1038/nature12266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellmaine S.F., Ovchinnikov D.A., Manallack D.T., Cuddy C.E., Elefanty A.G., Stanley E.G., Wolvetang E.J., Williams S.J., Pera M. Inhibition of DYRK1A disrupts neural lineage specification in human pluripotent stem cells. eLife. 2017;6:e24502. doi: 10.7554/eLife.24502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betizeau M., Cortay V., Patti D., Pfister S., Gautier E., Bellemin-Menard A., Afanassieff M., Huissoud C., Douglas R.J., Kennedy H. Precursor diversity and complexity of lineage relationships in the outer subventricular zone of the primate. Neuron. 2013;80:442–457. doi: 10.1016/j.neuron.2013.09.032. [DOI] [PubMed] [Google Scholar]
- Bonnefont J., Tiberi L., van den Ameele J., Potier D., Gaber Z.B., Lin X., Bilheu A., Herpoel A., Velez Bravo F.D., Guillemot F. Cortical neurogenesis requires Bcl6-mediated transcriptional repression of multiple self-renewal-promoting extrinsic pathways. Neuron. 2019;103:1096–1108 e1094. doi: 10.1016/j.neuron.2019.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borghese L., Dolezalova D., Opitz T., Haupt S., Leinhaas A., Steinfarz B., Koch P., Edenhofer F., Hampl A., Brustle O. Inhibition of notch signaling in human embryonic stem cell-derived neural stem cells delays G1/S phase transition and accelerates neuronal differentiation in vitro and in vivo. Stem Cells. 2010;28:955–964. doi: 10.1002/stem.408. [DOI] [PubMed] [Google Scholar]
- Calegari F., Haubensak W., Haffner C., Huttner W.B. Selective lengthening of the cell cycle in the neurogenic subpopulation of neural progenitor cells during mouse brain development. J. Neurosci. 2005;25:6533–6538. doi: 10.1523/JNEUROSCI.0778-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Luzy I.R., Niclis J.C., Gantner C.W., Kauhausen J.A., Hunt C.P.J., Ermine C., Pouton C.W., Thompson L.H., Parish C.L. Isolation of LMX1a ventral midbrain progenitors improves the safety and predictability of human pluripotent stem cell-derived neural transplants in Parkinsonian disease. J. Neurosci. 2019;39:9521–9531. doi: 10.1523/JNEUROSCI.1160-19.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dee A., Li K., Heng X., Guo Q., Li J.Y. Regulation of self-renewing neural progenitors by FGF/ERK signaling controls formation of the inferior colliculus. Development. 2016;143:3661–3673. doi: 10.1242/dev.138537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehay C., Kennedy H. Cell-cycle control and cortical development. Nat. Rev. Neurosci. 2007;8:438–450. doi: 10.1038/nrn2097. [DOI] [PubMed] [Google Scholar]
- Dehay C., Kennedy H., Kosik K.S. The outer subventricular zone and primate-specific cortical complexification. Neuron. 2015;85:683–694. doi: 10.1016/j.neuron.2014.12.060. [DOI] [PubMed] [Google Scholar]
- Dennis D.J., Wilkinson G., Li S., Dixit R., Adnani L., Balakrishnan A., Han S., Kovach C., Gruenig N., Kurrasch D.M. Neurog2 and Ascl1 together regulate a postmitotic derepression circuit to govern laminar fate specification in the murine neocortex. Proc. Natl. Acad. Sci. U S A. 2017;114:E4934–E4943. doi: 10.1073/pnas.1701495114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez M.H., Ayoub A.E., Rakic P. POU-III transcription factors (Brn1, Brn2, and Oct6) influence neurogenesis, molecular identity, and migratory destination of upper-layer cells of the cerebral cortex. Cereb Cortex. 2013;23:2632–2643. doi: 10.1093/cercor/bhs252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espuny-Camacho I., Michelsen K.A., Gall D., Linaro D., Hasche A., Bonnefont J., Bali C., Orduz D., Bilheu A., Herpoel A. Pyramidal neurons derived from human pluripotent stem cells integrate efficiently into mouse brain circuits in vivo. Neuron. 2013;77:440–456. doi: 10.1016/j.neuron.2012.12.011. [DOI] [PubMed] [Google Scholar]
- Fiddes I.T., Lodewijk G.A., Mooring M., Bosworth C.M., Ewing A.D., Mantalas G.L., Novak A.M., van den Bout A., Bishara A., Rosenkrantz J.L. Human-specific NOTCH2NL genes affect notch signaling and cortical neurogenesis. Cell. 2018;173:1356–1369 e1322. doi: 10.1016/j.cell.2018.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heng X., Guo Q., Leung A.W., Li J.Y. Analogous mechanism regulating formation of neocortical basal radial glia and cerebellar Bergmann glia. eLife. 2017;6:e23253. doi: 10.7554/eLife.23253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata T., Hevner R.F. Fibroblast growth factor signaling in development of the cerebral cortex. Dev. Growth Differ. 2009;51:299–323. doi: 10.1111/j.1440-169X.2009.01104.x. [DOI] [PubMed] [Google Scholar]
- Kang W., Wong L.C., Shi S.H., Hebert J.M. The transition from radial glial to intermediate progenitor cell is inhibited by FGF signaling during corticogenesis. J. Neurosci. 2009;29:14571–14580. doi: 10.1523/JNEUROSCI.3844-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leone D.P., Heavner W.E., Ferenczi E.A., Dobreva G., Huguenard J.R., Grosschedl R., McConnell S.K. Satb2 regulates the differentiation of both Callosal and subcerebral projection neurons in the developing cerebral cortex. Cereb. Cortex. 2015;25:3406–3419. doi: 10.1093/cercor/bhu156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Mattar P., Dixit R., Lawn S.O., Wilkinson G., Kinch C., Eisenstat D., Kurrasch D.M., Chan J.A., Schuurmans C. RAS/ERK signaling controls proneural genetic programs in cortical development and gliomagenesis. J. Neurosci. 2014;34:2169–2190. doi: 10.1523/JNEUROSCI.4077-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui J.H., Hansen D.V., Kriegstein A.R. Development and evolution of the human neocortex. Cell. 2011;146:18–36. doi: 10.1016/j.cell.2011.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukaszewicz A., Savatier P., Cortay V., Kennedy H., Dehay C. Contrasting effects of basic fibroblast growth factor and neurotrophin 3 on cell cycle kinetics of mouse cortical stem cells. J. Neurosci. 2002;22:6610–6622. doi: 10.1523/JNEUROSCI.22-15-06610.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manuel M.N., Mi D., Mason J.O., Price D.J. Regulation of cerebral cortical neurogenesis by the Pax6 transcription factor. Front. Cell Neurosci. 2015;9:70. doi: 10.3389/fncel.2015.00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maric D., Fiorio Pla A., Chang Y.H., Barker J.L. Self-renewing and differentiating properties of cortical neural stem cells are selectively regulated by basic fibroblast growth factor (FGF) signaling via specific FGF receptors. J. Neurosci. 2007;27:1836–1852. doi: 10.1523/JNEUROSCI.5141-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna W.L., Ortiz-Londono C.F., Mathew T.K., Hoang K., Katzman S., Chen B. Mutual regulation between Satb2 and Fezf2 promotes subcerebral projection neuron identity in the developing cerebral cortex. Proc. Natl. Acad. Sci. U S A. 2015;112:11702–11707. doi: 10.1073/pnas.1504144112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizutani K., Yoon K., Dang L., Tokunaga A., Gaiano N. Differential Notch signalling distinguishes neural stem cells from intermediate progenitors. Nature. 2007;449:351–355. doi: 10.1038/nature06090. [DOI] [PubMed] [Google Scholar]
- Niclis J.C., Gantner C.W., Alsanie W.F., McDougall S.J., Bye C.R., Elefanty A.G., Stanley E.G., Haynes J.M., Pouton C.W., Thompson L.H. Efficiently specified ventral midbrain dopamine neurons from human pluripotent stem cells under xeno-free conditions restore motor deficits in Parkinsonian rodents. Stem Cell Transl. Med. 2017;6:937–948. doi: 10.5966/sctm.2016-0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor S.C., Martinez-Cerdeno V., Ivic L., Kriegstein A.R. Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat. Neurosci. 2004;7:136–144. doi: 10.1038/nn1172. [DOI] [PubMed] [Google Scholar]
- Nowakowski R.S., Lewin S.B., Miller M.W. Bromodeoxyuridine immunohistochemical determination of the lengths of the cell cycle and the DNA-synthetic phase for an anatomically defined population. J. Neurocytol. 1989;18:311–318. doi: 10.1007/BF01190834. [DOI] [PubMed] [Google Scholar]
- Nowakowski T.J., Bhaduri A., Pollen A.A., Alvarado B., Mostajo-Radji M.A., Di Lullo E., Haeussler M., Sandoval-Espinosa C., Liu S.J., Velmeshev D. Spatiotemporal gene expression trajectories reveal developmental hierarchies of the human cortex. Science. 2017;358:1318–1323. doi: 10.1126/science.aap8809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsuka T., Kageyama R. Regulation of temporal properties of neural stem cells and transition timing of neurogenesis and gliogenesis during mammalian neocortical development. Semin. Cell Dev. Biol. 2019;95:4–11. doi: 10.1016/j.semcdb.2019.01.007. [DOI] [PubMed] [Google Scholar]
- Okamoto M., Miyata T., Konno D., Ueda H.R., Kasukawa T., Hashimoto M., Matsuzaki F., Kawaguchi A. Cell-cycle-independent transitions in temporal identity of mammalian neural progenitor cells. Nat. Commun. 2016;7:11349. doi: 10.1038/ncomms11349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozair M.Z., Kirst C., van den Berg B.L., Ruzo A., Rito T., Brivanlou A.H. hPSC modeling reveals that fate selection of cortical deep projection neurons occurs in the subplate. Cell Stem Cell. 2018;23:60–73 e66. doi: 10.1016/j.stem.2018.05.024. [DOI] [PubMed] [Google Scholar]
- Pilaz L.J., Patti D., Marcy G., Ollier E., Pfister S., Douglas R.J., Betizeau M., Gautier E., Cortay V., Doerflinger N. Forced G1-phase reduction alters mode of division, neuron number, and laminar phenotype in the cerebral cortex. Proc. Natl. Acad. Sci. U S A. 2009;106:21924–21929. doi: 10.1073/pnas.0909894106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raballo R., Rhee J., Lyn-Cook R., Leckman J.F., Schwartz M.L., Vaccarino F.M. Basic fibroblast growth factor (Fgf2) is necessary for cell proliferation and neurogenesis in the developing cerebral cortex. J. Neurosci. 2000;20:5012–5023. doi: 10.1523/JNEUROSCI.20-13-05012.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rash B.G., Lim H.D., Breunig J.J., Vaccarino F.M. FGF signaling expands embryonic cortical surface area by regulating Notch-dependent neurogenesis. J. Neurosci. 2011;31:15604–15617. doi: 10.1523/JNEUROSCI.4439-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reillo I., de Juan Romero C., Garcia-Cabezas M.A., Borrell V. A role for intermediate radial glia in the tangential expansion of the mammalian cerebral cortex. Cereb. Cortex. 2011;21:1674–1694. doi: 10.1093/cercor/bhq238. [DOI] [PubMed] [Google Scholar]
- Sahara S., O'Leary D.D. Fgf10 regulates transition period of cortical stem cell differentiation to radial glia controlling generation of neurons and basal progenitors. Neuron. 2009;63:48–62. doi: 10.1016/j.neuron.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Kirwan P., Smith J., Robinson H.P., Livesey F.J. Human cerebral cortex development from pluripotent stem cells to functional excitatory synapses. Nat. Neurosci. 2012;15:477–486. doi: 10.1038/nn.3041. S471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian L., Bershteyn M., Paredes M.F., Kriegstein A.R. Dynamic behaviour of human neuroepithelial cells in the developing forebrain. Nat. Commun. 2017;8:14167. doi: 10.1038/ncomms14167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki I.K., Gacquer D., Van Heurck R., Kumar D., Wojno M., Bilheu A., Herpoel A., Lambert N., Cheron J., Polleux F. Human-specific NOTCH2NL genes expand cortical neurogenesis through delta/notch regulation. Cell. 2018;173:1370–1384.e16. doi: 10.1016/j.cell.2018.03.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telley L., Agirman G., Prados J., Amberg N., Fievre S., Oberst P., Bartolini G., Vitali I., Cadilhac C., Hippenmeyer S. Temporal patterning of apical progenitors and their daughter neurons in the developing neocortex. Science. 2019;364:eaav2522. doi: 10.1126/science.aav2522. [DOI] [PubMed] [Google Scholar]
- Toma K., Kumamoto T., Hanashima C. The timing of upper-layer neurogenesis is conferred by sequential derepression and negative feedback from deep-layer neurons. J. Neurosci. 2014;34:13259–13276. doi: 10.1523/JNEUROSCI.2334-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turrero Garcia M., Chang Y., Arai Y., Huttner W.B. S-phase duration is the main target of cell cycle regulation in neural progenitors of developing ferret neocortex. J. Comp. Neurol. 2016;524:456–470. doi: 10.1002/cne.23801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaccarino F.M., Schwartz M.L., Raballo R., Nilsen J., Rhee J., Zhou M., Doetschman T., Coffin J.D., Wyland J.J., Hung Y.T. Changes in cerebral cortex size are governed by fibroblast growth factor during embryogenesis. Nat. Neurosci. 1999;2:246–253. doi: 10.1038/6350. [DOI] [PubMed] [Google Scholar]
- Watanabe N., Kageyama R., Ohtsuka T. Hbp1 regulates the timing of neuronal differentiation during cortical development by controlling cell cycle progression. Development. 2015;142:2278–2290. doi: 10.1242/dev.120477. [DOI] [PubMed] [Google Scholar]
- Yoon K., Nery S., Rutlin M.L., Radtke F., Fishell G., Gaiano N. Fibroblast growth factor receptor signaling promotes radial glial identity and interacts with Notch1 signaling in telencephalic progenitors. J. Neurosci. 2004;24:9497–9506. doi: 10.1523/JNEUROSCI.0993-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.