Abstract
Human development has been studied for over a century, but the molecular mechanisms underlying human embryogenesis remain largely unknown due to technical difficulties and ethical issues. Accordingly, mice have been used as a model for mammalian development and studied extensively to infer human biology based on the conservation of fundamental processes between the two species. As research has progressed, however, species-specific differences in characteristics between rodents and primates have become apparent. Non-human primates (NHPs) have also been used for biomedical research, and are now attracting attention as a model for human development. Here, we summarize primate species from the evolutionary and genomic points of view. Then we review the current issues and progress in gene modification technology for NHPs. Finally, we discuss recent studies on the early embryogenesis of primates and future perspectives.
Keywords: non-human primates, development, gene modification with NHPs, peri-implantation, embryogenesis
In this article, Nakamura, Tsukiyama, and colleagues summarize primate species from an evolutionary and genomic point of view, and review the current issues and progress in gene modification technology for non-human primates. They also discuss recent studies of the early embryogenesis of primates and future perspectives.
Main text
Rodents have been the predominant model organisms for mammalian biology so far. Mice in particular have numerous advantages that make them an excellent model animal, such as the ease of breeding, short generation time, and relatively large number of offspring. In addition, genome engineering technologies and pluripotent stem cell (PSC) technologies, which are essential for elucidating molecular mechanisms, have long been available in mice. As a result, many remarkable findings have been reported, some of which have contributed to our understanding of human biology as well as to the development of medicines. However, recent studies in rodents and humans have revealed that the gaps between the two species are larger than previously understood. Therefore, an animal model that is closer to humans is desired to infer human biology. In this sense, non-human primates (NHPs) are expected to be the best alternative.
Evolution of rodents and primates
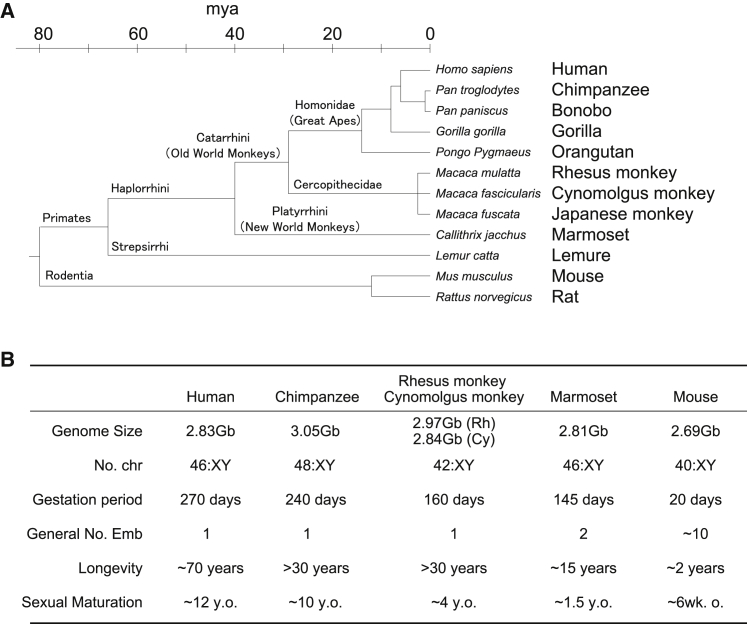
Both primates and rodents belong to the same subclade Euarchontoglires in clade Boreoeutheria, subclass Theria, class Mammalia. They are divided into the orders Primates and Rodents, which are thought to have diverged around 80 million years ago (mya) in the late Cretaceous period (Figure 1A). Primates now consist of more than 300 species, classified roughly into three major categories: New World monkeys/Platyrrhini, Old World monkeys/Catarrhini, and others. Human beings belong to the family Hominidae (also called the Great apes) in one clade of Old World monkeys/Catarrhini (Perelman et al., 2011).
Figure 1.
General description of primates
(A) Simplified phylogenetic tree of primates including rodents. Primates are separated roughly into three groups: New World monkeys, Old World monkeys, and Great apes. Of these, chimpanzees, rhesus and cynomolgus monkeys, and marmosets have been used for biomedical research, while the Great apes are no longer permissible in invasive experiments. mya, million years ago.
(B) Summary table of general information. Generally, primates have evolved to increase their body size and life span, and reduce their litter numbers. However, marmosets and most New World monkeys have small body size and relatively short generation periods. Marmosets also have an interesting feature: the generally produce dizygotic twins sharing a single placental system. y.o., years old; wk.o., weeks old.
Historically, four NHP species have been used for biomedical research with good success: chimpanzees, cynomolgus monkeys, rhesus monkeys, and marmosets (Johnsen et al., 2012). Of these, the evolutionarily closest species to human beings (Homo sapiens) are chimpanzees (Pan troglodytes) in the Great apes, which also includes bonobos (Pan paniscus), gorillas (Gorilla gorilla), and orangutans (Pongo pygmaeus). Chimpanzees and bonobos belong to the same genus, Pan, and it is believed that they and humans diverged around 5–7 mya (Israfil et al., 2011; Prufer et al., 2012) (Figure 1A), whereas gorillas and orangutans are thought to have divided from humans around 6–9 and 12–16 mya, respectively (Israfil et al., 2011; Locke et al., 2011; Scally et al., 2012) (Figure 1A). Currently, chimpanzees and the other Great apes are banned from use in invasive biomedical research in many countries (Johnsen et al., 2012). And since chimpanzees are no longer permissible research models, the primates that are the next-most-closely related to humans are macaques.
Accordingly, cynomolgus (Macaca fascicularis) and rhesus (Macaca mulatta) monkeys, belonging to macaques (genus Macaca) of the Old World monkeys/Catarrhini, have been the most extensively used NHPs for biomedical research. Currently, 23 macaque species are recognized as distinct animals, but mating between, for example, rhesus monkeys and Japanese monkeys, has been observed in Japan (Kawamoto et al., 2004) and this may imply that the differences among macaque species are so small as to be more like the differences among subspecies. NHPs belonging to the Old World monkeys/Catarrhini exist from Africa to the southern part of Eurasia and the Southeast Asian Islands, and macaques and the Great apes are thought to have branched 25–33 mya (Israfil et al., 2011; Locke et al., 2011; Rhesus Macaque Genome et al., 2007) (Figure 1A).
Marmosets belong to the New World monkeys/Platyrrhini, which are mainly native to South America and are thought to have diverged from the Old World monkeys about 40 mya (Marmoset Genome and Analysis, 2014) (Figure 1A). Interestingly, while the other primates have evolved to increase their body size and lifespan, reduce the number of litters (basically becoming singletons), and prolong gestation, marmosets and their close relatives among New World monkeys have undergone reductions in body size from larger primate ancestors and evolved unique reproductive systems to include relatively short gestation and sexual maturation periods and to produce dizygotic twins sharing a single placental system (Figure 1B). As a result, the litters exchange hematopoietic stem cells in utero and have lifelong blood chimerism (Marmoset Genome and Analysis, 2014). Due to such unique characteristics, marmosets are also considered attractive research models and have been used for biomedical research as well.
Genomic information
In the field of current biology, genome sequences are the essential pieces of information with which to understand many biological processes at the molecular level. So far, starting from the human genome in 2003 (International Human Genome Sequencing, 2004), the chimpanzee genome was completed in 2005 (The Chimpanzee Sequencing and Analysis Consortium, 2005), the rhesus monkey genome in 2007 (Rhesus Macaque Genome et al., 2007), the orangutan genome in 2011 (Locke et al., 2011), and the marmoset genome in 2015 (Marmoset Genome and Analysis, 2014; Sato et al., 2015). Currently, the genomes of 18 primate species have been sequenced and are available in public databases.
When the chimpanzee genome was announced in 2005, it revealed surprisingly that there was only a 1%–2% difference in alignable sequences between humans and chimpanzees, with more than 99.5% homology in the protein coding region (The Chimpanzee Sequencing and Analysis Consortium, 2005). These results suggested that the evolution of the protein-coding sequences was not significant enough to explain the species differences among primates. On the other hand, nearly half of the primate genome consists of non-coding sequences and repetitive elements (International Human Genome Sequencing, 2004). Of these, many families of endogenous retrotransposon are uniquely evolved in primate genomes. For example, the Alu element, which is a primate-specific family of short interspersed nuclear elements (SINEs), was specifically acquired in the Old World monkeys, and one endogenous retrovirus family, HERV was also acquired specifically only in humans (The Chimpanzee Sequencing and Analysis Consortium, 2005; Rhesus Macaque Genome et al., 2007). These transposable elements make copies of themselves and transpose to other loci. Therefore, of course they are harmful to the host genome, and they are rapidly inactivated during the evolution of the host genome. Interestingly, however, such transposable elements can also drive the host evolution (Jacques et al., 2013; Kunarso et al., 2010). Bourque and colleagues investigated the binding sites of key transcription factors, POU5f1/Pou5f1 and NANOG/Nanog, in human and mouse embryonic stem cells (ESCs), and found that the binding patterns are markedly different, with only 5% of the regions being homologously occupied. Among the unconserved loci, ∼25% of binding sites were found in transposable elements, indicating that the transposable elements have the potential to dramatically change the transcriptional network (Kunarso et al., 2010). They also investigated the marmoset and chimpanzee genomes and found that those potential transcription factor binding sites on transposable elements were highly species specific, suggesting that the transposable elements would contribute to the genome evolution through the formation of new transcriptional networks (Jacques et al., 2013).
It has recently become possible to identify such species-specific elements due to the development of long-read DNA sequencers. Most NHP genomes generated in the initial stage relied on guidance by the reference human genome. Accordingly, the NHP genomes have been somewhat “humanized.” The advance of the long-read DNA sequencer has enabled us to overcome the problems on genome assembly and to identify structural variations among species (He et al., 2019; Kronenberg et al., 2018). There are 17,000 ape-specific structural variants and many of them are located in enhancer regions. These data suggest that the species differences of phonotypes in primates may not be derived from the differences of protein types but rather from the differences in regulatory elements.
Biomedical research with NHPs
Even though humans and rodents share basic biological processes, the species differences between them are not negligible and are becoming clearer along with advances in research. However, NHPs are often difficult to breed and prohibitively expensive (Johnsen et al., 2012). Colony expansion by captive breeding has also been carried out, and this approach has been promoted by advances in reproductive technology such as hormone treatment approach in NHPs. However, captive breeding is still not easy, due not only to the long gestation and maturation periods, but also to the small number of pregnancies. Thus, NHPs have been used to model particularly serious and widespread diseases, including viral infections such as Ebola, HIV, and hepatitis B/C, which cannot be adequately replicated in mice, as well as for vaccine development and drug safety evaluation (Johnsen et al., 2012). In 2020, NHPs were also used for research related to COVID-19 (Lu et al., 2020). They have also been widely used as a model of higher brain dysfunction that cannot be reproduced in mice. Accordingly, studies using NHPs have been primarily conducted in the areas of adult immunology, physiology, and neurophysiology. Thus, research for developmental biology has been very limited. However, these trends will be changed by the rapid progress in genome-editing technologies, such as the CRISPR-Cas9 system, and the development of stem cell biology based on the human PSCs, as will be discussed in greater detail in other sections, as well as single-cell analysis technologies.
Gene modification in NHPs
As mentioned above, the evaluation of mammalian gene function at the whole-body level had been limited to rodents for both technical and ethical reasons. Because there are no germline-transmittable ESCs or induced pluripotent stem cells (iPSCs) in non-rodent animals, including NHPs, the disruption of specific genes (knockout) and the introduction of genes into specific loci (knockin) was very difficult until the development of CRISPR-Cas9. Now that genome-editing technologies have been developing, however, there is increasing interest in the application of genome editing to various animals, including NHPs and humans. In research on human development, we can use surplus embryos after in vitro fertilization procedures with informed consent, but the supply of such embryos is limited and the permissibility of gene modification in human embryos is still under debate. At the moment, therefore, gene modification in NHPs is the best way to advance our understanding of human biology.
Before the development of CRISPR-Cas9 editing, most reports on gene-modified NHPs involved transgenic (Tg) monkeys produced by viral vectors. Typically, in order to generate Tg mice, linearized vectors are injected into the pronuclei of zygotes. However, most mice generated by this method are genetically mosaic. Therefore, researchers need to generate multiple mouse lines and use them after the F1 generation (the generation after F0). When using mice, it is relatively easy to obtain a large number of lines and select them. However, for large animals such as NHPs, obtaining many lines is impractical in terms of time, cost, and labor because of the long sexual maturity and gestation periods. Thus, it is desirable to analyze the F0 generation (i.e., the first generation). Moreover, Tg mice that have insertions of full-length transgenes can only rarely be obtained in the F0 generation. Therefore, to improve the efficiency of the introduction of full-length transgenes, a viral vector system has been used to generate Tg animals in NHPs.
The first successful generation of Tg animals in NHPs was described in 2001, when GFP-expressing vectors were introduced into rhesus monkey zygotes by retrovirus infection (Chan et al., 2001). Following this report, several technical improvements were achieved, such as the confirmation of germline transmission of lentiviral transgenes (Sasaki et al., 2009), analysis of differences in promoter types (Kim et al., 2007; Seita et al., 2019), analysis of viral injection timing (Kubisch et al., 2008; Seita et al., 2016), and the application of different types of virus (Niu et al., 2010).
Most of the published reports involving gene modification in NHPs were related to studies on disease modeling (Table S1). The pathological recapitulation of human disease is limited in mouse models because there are marked physiological differences between humans and mice. Indeed, many reports have demonstrated the superiority of monkey models over mouse models in this regard (Table S1). For example, duplications of MECP2-containing genomic segments cause a syndrome that shares core symptoms with autism spectrum disorders. It has been difficult to identify autism-like behaviors in the mouse model of MECP2 overexpression. In contrast, monkeys with MECP2 overexpression exhibit autism-like behaviors. These Tg monkeys show an increased frequency of repetitive circular locomotion, increased anxiety, reduced social interaction, and relatively weak cognitive phenotypes (Liu et al., 2016).
In addition, the phenotypic discrepancies between mice and humans are observed in several autosomal dominant diseases, such as autosomal dominant polycystic kidney disease (ADPKD). ADPKD, which is the most common hereditary kidney disease, is caused by PKD1 heterozygous mutations. However, heterozygous deletion of Pkd1 in mice rarely results in the formation of cysts until near the end of life. In contrast, PKD1 heterozygote monkeys exhibit cyst formation perinatally, as in humans (Tsukiyama et al., 2019), highlighting the need for NHP models rather than mouse models. In humans, there are many autosomal dominant diseases and a study of such diseases requires selective production of heterozygotes. To produce heterozygotes selectively, a method for allele-specific targeting using polymorphism has been established (Tsukiyama et al., 2019).
In addition to this, to overcome the difficulties specific for NHPs, many other gene-modification methods have been developed, such as techniques for the specification of gene expression by tissue- or stage-specific promoters, the drug-inducible control of gene expression (Tomioka et al., 2017; Tu et al., 2019), reporter knockin into specific genes (Chu et al., 2019; Cui et al., 2018; Yao et al., 2017, 2018), and floxed allele knockin (Tsukiyama et al., 2019). These technologies can be applied to developmental biology research to, for example, clarify the process by which germ cells and other cell lineages differentiate.
Among the technical advances, the reduction of mosaicism is crucial for gene modification in NHPs. When analysis is performed with F0 animals, if any of the genetically modified individuals have mosaicism, expression of the phenotype may be hindered and phenotype analysis may become difficult. Several research groups have succeeded in reducing mosaicism in knockout monkey production (Midic et al., 2017; Tsukiyama et al., 2019; Tu et al., 2017; Zuo et al., 2017). In general, however, the issue of mosaicism in the generation of Tg or knockin animals remains unresolved. Therefore, the avoidance of mosaicism remains a very important topic in genetic modification in NHPs.
Developmental biology of primates
Along with the progress in gene modification technology, the recent rapid development of molecular biology, especially in methods for single-cell analysis, has provided opportunities to achieve comprehensive analysis even using small amounts of materials. In addition, stem cell technologies using human PSCs have also developed; however, currently it is still difficult to evaluate how much an in vitro model really recapitulates the in vivo process, due to the lack of in vivo information of human development. As a result, the importance of in vivo primate development and research using NHPs has significantly increased.
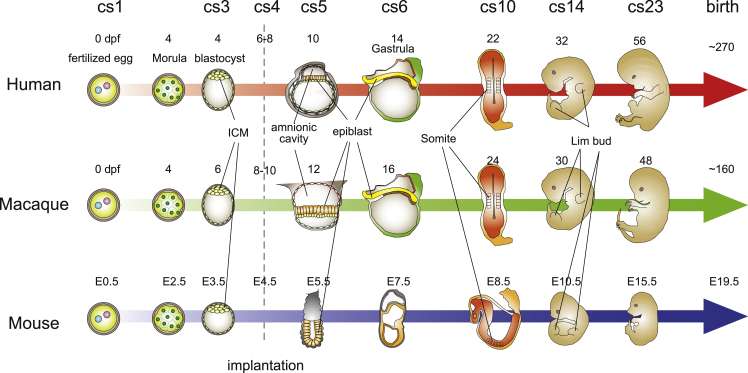
Human embryogenesis can be roughly split into two periods: the embryonic period (from fertilization until around 8 weeks after fertilization) and the fetal period (from the end of the embryonic period until birth). In human embryos, the embryonic period is divided into 23 distinct morphological stages known as Carnegie stages (O'Rahilly and Müller, 2010; 1987). Carnegie stage 1 (cs1) begins with fertilization, while cs2 continues through the cleavage stage. In cs3, at approximately 4 days post-fertilization (dpf), the human embryo forms a blastocyst with three distinct cell populations: the epiblast, hypoblast, and trophectoderm (Rossant and Tam, 2017). At cs4, around 6 dpf, implantation occurs and the embryo starts its dynamic morphological transformation. The trophectoderm begins to invade the uterine endometrium, whereas the epiblast begins lumenogenesis and quickly forms an amnionic cavity surrounded by thin squamous amnionic cells and a thick layer of pseudostratified epithelial epiblast cells. Then, in the late cs5 to early cs6 stage, at around 12 dpf, the posterior side of pluripotent epiblast cells begins dynamic morphogenesis and gastrulation followed by the generation of three germ layer cells: the ectoderm, mesoderm, and endoderm (Rossant and Tam, 2017). While the differentiation and migration of the cells continues, somitogenesis starts at the late cs8 stage around 20 dpf, and then at cs10 the heart begins to beat and the neural tube closes. By the start of cs13, at about 28 dpf, many of the tissue progenitors as well as both the upper and lower limb buds form. The pharyngeal arches also start assembling around this stage and form elements of the face—such as the eyes, nasal pit, mouth, and ears—by cs18. Then each tissue continuously and coordinately develops. By the end of cs23, at around 56 dpf, all of the primordial body parts and tissues are established.
Although these key developmental processes are fundamentally conserved among humans, monkeys, and mice, the timing of when these events occur is very different, as would be expected considering the divergence of their body sizes and gestation periods. After fertilization, the mouse embryo develops to the morula stage at around 2 dpf and implantation takes place at 4 dpf. In humans, the embryo reaches the morula stage at around 4 dpf and implants on the endometrium at around 7–8 dpf. The embryos of macaques, such as cynomolgus and rhesus monkeys, develop to the morula stage at around 4 dpf like humans, but, the morula stage in macaques is 2 days longer than that in humans, and their implantation takes place at around days 9–10 (Heuser and Streeter, 1941; Nakamura et al., 2016; Niakan and Eggan, 2013; Wong et al., 2010). Although this gap between humans and macaques is maintained until around 30 dpf (Sasaki et al., 2016), the correspondence of key developmental events between them is reversed (Figure 2). As explained above, human cs23 begins at 56 dpf, but it takes 48 dpf for macaque embryos to reach cs23, while mouse embryos reach this milestone at around 16 dpf (Figure 2). The human gestation period is about 270 days, whereas those of cynomolgus monkeys and rhesus monkeys are both around 160 days, and that of mice is 20 days (Figure 2). These facts indicate that the fetal period varies widely among species, even among primates, and the timing of key developmental processes does not always diverge proportionally.
Figure 2.
Development of humans, macaques, and mice during the embryonic period
The key developmental processes are fundamentally conserved among humans, monkeys, but mice, and the timing when these events occur is divergent. cs, Carnegie stage; dpf, days post-fertilization; E, embryonic days; ICM, inner cell mass.
The longer fetal period of primates relative to that of mice brings interesting features in the former. For example, in germ cell development, the germ cells of both primates and rodents are first specified as primordial germ cells (PGCs) soon after implantation, and then migrate to the genital ridges through the hindgut endoderm and dorsal mesentery (Saitou and Miyauchi, 2016; Witschi, 1948). In the genital ridges, PGCs proliferate quickly, and while those in the embryonic testis undergo mitotic arrest and differentiation into pro-spermatogonia (Culty, 2009; Saitou and Miyauchi, 2016), those in the embryonic ovaries begin the entry into meiosis and differentiation into oocytes (Kurilo, 1981; Saitou and Miyauchi, 2016). In mice, the onset of such female processes takes place homogeneously with respect to time. For example, mouse PGCs arrive at the embryonic ovaries around 10.5 dpf, and enter into meiosis at around 14.5 dpf. However, in humans, cytological and single-cell transcriptomical analysis showed that these processes proceed in a highly heterogeneous/asynchronous manner. Human PGCs arrived at gonads at around 35 dpf with rapid proliferation, and begin meiosis to differentiate into oocytes from around 100 dpf. However, the cells in the mitotic stage also appear until at least around week 26, 180 dpf (Kurilo, 1981; Li et al., 2017). Therefore, mitotically active germ cells and oocytes in the first prophase of meiosis co-exist for a relatively long time in human embryonic ovaries. Notably, folliculogenesis progresses during the embryonic period, and mature follicles are occasionally formed before birth (Kurilo, 1981). This may indicate that such heterogeneity would be seen in other organs, and there should be species-specific mechanisms for organogenesis that remains to be uncovered.
Recently, two papers revealing the mechanisms underlying the species differences in developmental timing have been published (Matsuda et al., 2020a; Rayon et al., 2020). They investigated and compared different developmental systems between humans and mice; one studied the mechanisms underlying the differentiation kinetics of motor neurons from ESCs (Rayon et al., 2020), and the other focused on the segmentation clock during somitogenesis recapitulated in vitro (Matsuda et al., 2020a). In both cases, the authors confirmed that the developmental pace of mouse cells was faster than that of humans in vitro, just as observed for the two species in vivo. They also investigated the mechanisms that drive the species-specific pace of development. Interestingly, both papers reached the same conclusion: the difference in protein stability was correlated with developmental speed, indicating that species differences are partly created by cell-autonomous mechanisms.
Morphological differences in early development between primates and rodents
Here, we focus on early developments that are crucial in stem cell biology. Following implantation, the mouse epiblast and polar trophectoderm proliferate rapidly and begin lumenogenesis, then eventually form a pro-amnionic cavity by the fusion of the epiblast and trophectoderm lumen (Bedzhov and Zernicka-Goetz, 2014). The amnionic membrane then forms at the most proximal side of the epiblast and separates the amnionic cavity from the yolk sac cavity (Pereira et al., 2011). During this process, the trophectoderm pushes the epiblast, and the epiblast itself also extends to the distal side, causing the embryo to form an elongated shape called a cup shape or egg cylinder. In contrast, although cells of the epiblast and trophectoderm of the primate embryo also proliferate, the primate trophectoderm progressively invades the endometrium and the epiblasts expand and form a flat sheet of cells, resulting in an embryo with a flat morphology known as an embryonic disk (Heuser and Streeter, 1941; Nakamura et al., 2016; O’Rahilly and Müller, 1987; Rossant and Tam, 2017). The amnionic cavity also forms in the primate embryo, but the timing is a little earlier. The mouse amnionic cavity separates after the onset of gastrulation, but in humans the cavity forms before gastrulation and consists only of epiblast-derived cells (Saitou and Miyauchi, 2016). Thus, the developmental processes just after implantation differ greatly between mice and primates, and it is not straightforward to infer the details of human embryogenesis during this period from mouse development.
Differences between primates and rodents at the molecular level
According to studies in humans, cynomolgus monkeys, and marmoset pre-implantation embryos (Boroviak et al., 2015; Nakamura et al., 2016; Petropoulos et al., 2016; Stirparo et al., 2018; Yan et al., 2013), the expression of major transcription factors such as POU5F1/Pou5f1, NANOG/Nanog, and SOX2/Sox2 in the epiblast, and GATA4/Gata4, GATA6/Gata6, and SOX17/Sox17 in the primitive endoderm/hypoblast is preserved, just as it is post-implantation. In this way, the overall expression patterns of key genes are highly conserved among these species. Interestingly, however, the expression patterns of some genes are not conserved even though they play crucial roles in mouse development. For example, in mice, Gata6 is expressed only in the primitive endoderm in pre-implantation embryos, and its deletion mutants exhibit a complete absence of primitive endoderm (Schrode et al., 2014). However, GATA6 is expressed not only in the hypoblast but also in the trophectoderm in the early blastocyst of humans, cynomolgus monkeys and marmosets (Boroviak et al., 2015; Kuijk et al., 2012; Nakamura et al., 2016). Another significant example, Klf2, which is the basic transcription factor expressed in the pre-implantation epiblast of mice, is not expressed at all in monkeys and humans, but another family gene, KLF17, is expressed instead (Blakeley et al., 2015; Nakamura et al., 2016). While the lineage specification until blastocyst development is conserved among primates and rodents, the specification mechanisms may also differ between them. In mice, the primitive endoderm is specified by the Fgf signaling pathway, which is activated by Fgf4 expressed from the epiblast (Nichols et al., 2009; Yamanaka et al., 2010). However, whereas the human epiblast also expresses FGF4, aberrations of this signaling pathway do not disturb lineage specification (Roode et al., 2012).
The expression patterns of key genes also diverge between rodents and primates in the post-implantation stages, as is expected from the morphological differences. In mice, the expression of some so-called naive pluripotency-related genes, such as Zfp42, Dnmt3l, and Prdm14, are tightly controlled and quickly downregulated soon after implantation. However, in primates, they continue to be expressed even after the beginning of gastrulation. Also, while the Fgf, Bmp, and Nodal/activin families play important roles in implantation, Fgf5 is upregulated with implantation in mice, whereas FGF2 is upregulated in monkeys (Nakamura et al., 2016). Interestingly, a comparison of gene expression profiles of the epiblast along with its development, between mice and cynomolgus monkeys revealed that, while the mouse epiblast transforms its pluripotency dramatically day by day, the cynomolgus monkey epiblast keeps its pluripotency for more than 1 week after implantation. The comparison further revealed that the genes related to metabolism, the cytoskeleton, apoptosis, and the cell cycle are highly variable, indicating that in addition to development-related genes, homeostasis-related genes may also be a significant cause of the species differences (Nakamura et al., 2016).
In mammals, ESCs are first established in mice from the pre-implantation embryo (Evans and Kaufman, 1981), and they preserve properties resembling those of the naive pluripotent epiblast. These properties are maintained by activating the LIF and inhibiting the Fgf pathways (Nichols and Smith, 2011). The first primate ESCs (rhesus macaque) were established about 15 years later (Thomson et al., 1995). Soon after that marmoset and human ESCs were established (Thomson et al., 1996, 1998). All these primate ESCs depend on the activation of the FGF pathway but are independent of LIF signaling. Primate ESCs/iPSCs keep the expression of core pluripotency genes, such as POU5F1, NANOG, and SOX2, and also express some of the naive pluripotency markers, i.e., ZFP42, PRDM14, and DNMT3L, as in the post-implantation epiblast. Moreover, even though primate ESCs are also derived from the pre-implantation epiblast, the transcriptome signatures are closest to those of the post-implantation epiblast (Nakamura et al., 2016).
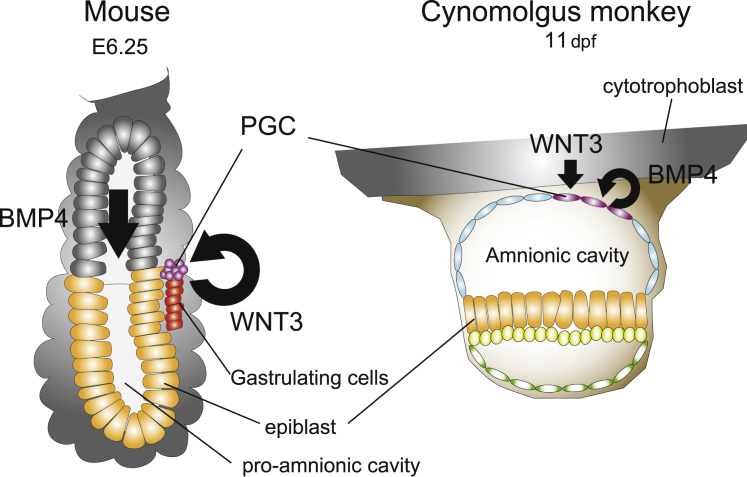
The other striking difference that has been discovered so far between rodents and primates after implantation concerns the manner of PGC specification. Mouse PGCs are specified as a cluster of cells positive for Tfap2c and Prdm1 (also known as Blimp1) at the most posterior edge of the epiblast along with the onset of gastrulation by Bmp and Wnt signaling, which derive from the extraembryonic ectoderm and posterior epiblast (Ohinata et al., 2009; Saitou and Miyauchi, 2016). However, a recent study using cynomolgus monkeys revealed that the cynomolgus PGCs were specified at the most proximal portion of the amnionic membrane (Sasaki et al., 2016). Interestingly, the cynomolgus PGCs appear prior to the onset of gastrulation (Sasaki et al., 2016).
Although there are many differences, as explained above, there are also similar processes between primates and rodents. The cynomolgus amnionic cells themselves express not only BMP4 but also the most likely responsive genes, ID2 and MSX2, and the cytotrophoblast next to the PGC specification site expresses WNT3, indicating that the cynomolgus PGCs are also induced by BMP and WNT signaling (Sasaki et al., 2016) (Figure 3). This is supported by the fact that the cynomolgus and human PGCLCs that are double positive for TFAP2C and BLIMP1 were also induced from ESCs/iPSCs by BMP4 in vitro (Irie et al., 2015; Sakai et al., 2019; Sasaki et al., 2015). In this way, although the molecular mechanisms of epiblast-hypoblast differentiation are not conserved between primates and rodents, the PGC specification mechanisms are conserved, even though the morphologies are very different, suggesting that it is important to observe every biological process carefully.
Figure 3.
Similarity and difference of germ cell specification between mice and cynomolgus monkeys
In mice, PGCs are known to be specified at the most proximal-posterior end of the epiblast by stimulations with Wnt3 and Bmp4, after onset of gastrulation. On the other hand, in cynomolgus monkeys, and perhaps in humans, the WNT and BMP pathways play critical roles in inducing the germ cell fate as in mice, but primate PGCs appear at the top of the amnion prior to gastrulation.
Conclusion and perspective
We have reviewed the evolution of rodents and primates, and the history of biomedical research using NHPs. We also summarized recent reports on genome-editing studies and the early development of primates using cynomolgus monkeys. There are many species differences between rodents and primates, even though the fundamental developmental processes are conserved. Therefore, research using NHPs to infer aspects of human biology are expecetd to become increasingly important.
On the other hand, it is important to note that there are differences among macaques, humans, and the other Great apes. For example, in human and chimpanzee embryos, the entire embryo invades the endometrium upon implantation, whereas the embryos of cynomolgus and rhesus monkeys are only half buried in the endometrium (Elder, 1938; Heuser and Streeter, 1941; Nakamura et al., 2016; O’Rahilly and Müller, 1987). Therefore, it is still necessary to take into account the species differences among primates, and more careful observation and comparison of the biological processes will be required.
As is discussed in greater detail in other chapters, since the establishment of human ESCs/iPSCs, various induction methods of differentiation, including organoid formation, have been developed. The progress has been remarkable, with even the gene expression oscillation during somitogenesis being successfully produced, as mentioned above (Matsuda et al., 2020a, 2020b). Due to this great variety of differentiation methods and their ease of application to experiments, the in vitro human PSC differentiation models are expected to serve as alternatives for post-implantation human materials. In addition to the in vitro model, extended culture systems of pre-implantation human embryos beyond implantation, known as ex vivo culture models, have recently been reported (Deglincerti et al., 2016; Lv et al., 2019; Shahbazi et al., 2016; Xiang et al., 2019; Zhou et al., 2019). The ex vivo culture requires human embryos, but it would provide a model closer to an in vivo system than an in vitro one, suggesting that both in vitro and ex vivo experimental systems together would be a powerful tool for speculating about human post-implantation development.
However, the above models may contain experimental artifacts, so evidence that the models reliably recapitulate in vivo development will be needed. In addition, there are several remaining ethical concerns such as the destruction of human embryos, the prohibition of the continuation of the culture beyond 14 dpf (Warnock, 1985), and insufficient discussion regarding gene-editing experiments using human embryos. On the other hand, although there are still species differences and difficulties with gene editing and the collection of materials, it is possible to perform in vivo experiments with NHPs, and ex vivo culture of cynomolgus monkeys (Ma et al., 2019; Niu et al., 2019). Moreover, several iPSCs of Great apes have been established and made available, and an in vitro model using the iPSCs of Great apes may fill the gaps of species differences that exist even among primates. Thus, the use of cross-platform (in vivo, ex vivo, and in vitro) and cross-species (humans, the Great apes, and other NHPs) analysis and comprehensive comparisons will solve the problems of artifact and species differences, making it possible to approach the true nature of human development. In this regard, we consider that NHPs will play crucial roles in human embryology and will continue to be important for the foreseeable future.
Author contributions
T.N. and T.T. conceived and wrote the manuscript. F.K. performed literature search. M.S. was involved in discussing and revising the contents.
Conflicts of interest
M.S. is a founder of Houjou, Inc., and is an inventor on patent applications relating to induction of germ cells from PSCs filed by Kyoto University. T.N., T.T., and F.K. declare no competing interests.
Acknowledgments
We thank the members of our laboratories for their discussion of this study. The authors are supported in part by Grants-in-Aid for Transformative Research Areas B (20B302), and by Grants-in-Aid for Challenging Research (Pioneering) (20K21370) from the Ministry of Education, Culture, Sports, Science and Technology of Japan (MEXT/JSPS) and by the Japan Agency for Medical Research and Development (AMED-PRIME, 20gm6310013h0001 and 21gm6310013h0002), and by a grant from the Takeda Science Foundation.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.stemcr.2021.03.021.
Contributor Information
Tomonori Nakamura, Email: t-nakamu@anat2.med.kyoto-u.ac.jp.
Tomoyuki Tsukiyama, Email: ttsuki@belle.shiga-med.ac.jp.
Supplemental information
References
- Bedzhov I., Zernicka-Goetz M. Self-organizing properties of mouse pluripotent cells initiate morphogenesis upon implantation. Cell. 2014;156:1032–1044. doi: 10.1016/j.cell.2014.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blakeley P., Fogarty N.M., del Valle I., Wamaitha S.E., Hu T.X., Elder K., Snell P., Christie L., Robson P., Niakan K.K. Defining the three cell lineages of the human blastocyst by single-cell RNA-seq. Development. 2015;142:3151–3165. doi: 10.1242/dev.123547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boroviak T., Loos R., Lombard P., Okahara J., Behr R., Sasaki E., Nichols J., Smith A., Bertone P. Lineage-specific profiling delineates the emergence and progression of naive pluripotency in mammalian embryogenesis. Dev. Cell. 2015;35:366–382. doi: 10.1016/j.devcel.2015.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan A.W., Chong K.Y., Martinovich C., Simerly C., Schatten G. Transgenic monkeys produced by retroviral gene transfer into mature oocytes. Science. 2001;291:309–312. doi: 10.1126/science.291.5502.309. [DOI] [PubMed] [Google Scholar]
- Chu C., Yang Z., Yang J., Yan L., Si C., Kang Y., Chen Z., Chen Y., Ji W., Niu Y. Homologous recombination-mediated targeted integration in monkey embryos using TALE nucleases. BMC Biotechnol. 2019;19:7. doi: 10.1186/s12896-018-0494-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y., Niu Y., Zhou J., Chen Y., Cheng Y., Li S., Ai Z., Chu C., Wang H., Zheng B. Generation of a precise Oct4-hrGFP knockin cynomolgus monkey model via CRISPR/Cas9-assisted homologous recombination. Cell Res. 2018;28:383–386. doi: 10.1038/cr.2018.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culty M. Gonocytes, the forgotten cells of the germ cell lineage. Birth Defects Res. C Embryo Today. 2009;87:1–26. doi: 10.1002/bdrc.20142. [DOI] [PubMed] [Google Scholar]
- Deglincerti A., Croft G.F., Pietila L.N., Zernicka-Goetz M., Siggia E.D., Brivanlou A.H. Self-organization of the in vitro attached human embryo. Nature. 2016;533:251–254. doi: 10.1038/nature17948. [DOI] [PubMed] [Google Scholar]
- Elder J.H. A ten and one-half day chimpanzee embryo, "Yerkes A". J. Am. Med. Assoc. 1938;111:1156–1159. [Google Scholar]
- Evans M.J., Kaufman M.H. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- He Y., Luo X., Zhou B., Hu T., Meng X., Audano P.A., Kronenberg Z.N., Eichler E.E., Jin J., Guo Y. Long-read assembly of the Chinese rhesus macaque genome and identification of ape-specific structural variants. Nat. Commun. 2019;10:4233. doi: 10.1038/s41467-019-12174-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuser C.F., Streeter G.I. Development of the macaque embryo. Contrib. Embryol. 1941;29:15–55. [Google Scholar]
- International Human Genome Sequencing C. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- Irie N., Weinberger L., Tang W.W., Kobayashi T., Viukov S., Manor Y.S., Dietmann S., Hanna J.H., Surani M.A. SOX17 is a critical specifier of human primordial germ cell fate. Cell. 2015;160:253–268. doi: 10.1016/j.cell.2014.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israfil H., Zehr S.M., Mootnick A.R., Ruvolo M., Steiper M.E. Unresolved molecular phylogenies of gibbons and siamangs (family: Hylobatidae) based on mitochondrial, Y-linked, and X-linked loci indicate a rapid Miocene radiation or sudden vicariance event. Mol. Phylogenet. Evol. 2011;58:447–455. doi: 10.1016/j.ympev.2010.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacques P.E., Jeyakani J., Bourque G. The majority of primate-specific regulatory sequences are derived from transposable elements. Plos Genet. 2013;9:e1003504. doi: 10.1371/journal.pgen.1003504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnsen D.O., Johnson D.K., Whitney R.A. Nonhuman Primates in Biomedical Research. 2012. History of the use of nonhuman primates in biomedical research; pp. 1–33. [Google Scholar]
- Kawamoto Y., Hagihara K., Aizawa K. Finding of hybrid individuals between native Japanese macaques and introduced rhesus macaques in the Bousou Peninsula, Chiba, Japan. Primate Res. 2004;20:89–95. [Google Scholar]
- Kim S., Kim G.J., Miyoshi H., Moon S.H., Ahn S.E., Lee J.H., Lee H.J., Cha K.Y., Chung H.M. Efficiency of the elongation factor-1alpha promoter in mammalian embryonic stem cells using lentiviral gene delivery systems. Stem Cells Dev. 2007;16:537–545. doi: 10.1089/scd.2006.0088. [DOI] [PubMed] [Google Scholar]
- Kronenberg Z.N., Fiddes I.T., Gordon D., Murali S., Cantsilieris S., Meyerson O.S., Underwood J.G., Nelson B.J., Chaisson M.J.P., Dougherty M.L. High-resolution comparative analysis of great ape genomes. Science. 2018;360:eaar6343. doi: 10.1126/science.aar6343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubisch H.M., Gagliardi C., Romero D.G., Bunnell B.A., Ratterree M.S. Kinetics of pronuclear development and the effects of vector type and timing of injection on the efficiency of gene transfer into rhesus macaque embryos. Mol. Reprod. Dev. 2008;75:1505–1514. doi: 10.1002/mrd.20901. [DOI] [PubMed] [Google Scholar]
- Kuijk E.W., van Tol L.T., Van de Velde H., Wubbolts R., Welling M., Geijsen N., Roelen B.A. The roles of FGF and MAP kinase signaling in the segregation of the epiblast and hypoblast cell lineages in bovine and human embryos. Development. 2012;139:871–882. doi: 10.1242/dev.071688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunarso G., Chia N.Y., Jeyakani J., Hwang C., Lu X., Chan Y.S., Ng H.H., Bourque G. Transposable elements have rewired the core regulatory network of human embryonic stem cells. Nat. Genet. 2010;42:631–634. doi: 10.1038/ng.600. [DOI] [PubMed] [Google Scholar]
- Kurilo L.F. Oogenesis in antenatal development in man. Hum. Genet. 1981;57:86–92. doi: 10.1007/BF00271175. [DOI] [PubMed] [Google Scholar]
- Li L., Dong J., Yan L., Yong J., Liu X., Hu Y., Fan X., Wu X., Guo H., Wang X. Single-cell RNA-seq analysis maps development of human germline cells and gonadal niche interactions. Cell Stem Cell. 2017;20:858–873 e854. doi: 10.1016/j.stem.2017.03.007. [DOI] [PubMed] [Google Scholar]
- Liu Z., Li X., Zhang J.T., Cai Y.J., Cheng T.L., Chen C., Wang Y., Zhang C.C., Nie Y.H., Chen Z.F. Autism-like behaviours and germline transmission in transgenic monkeys overexpressing MeCP2. Nature. 2016;530:98–102. doi: 10.1038/nature16533. [DOI] [PubMed] [Google Scholar]
- Locke D.P., Hillier L.W., Warren W.C., Worley K.C., Nazareth L.V., Muzny D.M., Yang S.P., Wang Z., Chinwalla A.T., Minx P. Comparative and demographic analysis of orang-utan genomes. Nature. 2011;469:529–533. doi: 10.1038/nature09687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S., Zhao Y., Yu W., Yang Y., Gao J., Wang J., Kuang D., Yang M., Yang J., Ma C. Comparison of nonhuman primates identified the suitable model for COVID-19. Signal. Transduct. Target Ther. 2020;5:157. doi: 10.1038/s41392-020-00269-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv B., An Q., Zeng Q., Zhang X., Lu P., Wang Y., Zhu X., Ji Y., Fan G., Xue Z. Single-cell RNA sequencing reveals regulatory mechanism for trophoblast cell-fate divergence in human peri-implantation conceptuses. PLoS Biol. 2019;17:e3000187. doi: 10.1371/journal.pbio.3000187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H., Zhai J., Wan H., Jiang X., Wang X., Wang L., Xiang Y., He X., Zhao Z.A., Zhao B. In vitro culture of cynomolgus monkey embryos beyond early gastrulation. Science. 2019;366:eaax7890. doi: 10.1126/science.aax7890. [DOI] [PubMed] [Google Scholar]
- Marmoset Genome S., Analysis C. The common marmoset genome provides insight into primate biology and evolution. Nat. Genet. 2014;46:850–857. doi: 10.1038/ng.3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda M., Hayashi H., Garcia-Ojalvo J., Yoshioka-Kobayashi K., Kageyama R., Yamanaka Y., Ikeya M., Toguchida J., Alev C., Ebisuya M. Species-specific segmentation clock periods are due to differential biochemical reaction speeds. Science. 2020;369:1450–1455. doi: 10.1126/science.aba7668. [DOI] [PubMed] [Google Scholar]
- Matsuda M., Yamanaka Y., Uemura M., Osawa M., Saito M.K., Nagahashi A., Nishio M., Guo L., Ikegawa S., Sakurai S. Recapitulating the human segmentation clock with pluripotent stem cells. Nature. 2020;580:124–129. doi: 10.1038/s41586-020-2144-9. [DOI] [PubMed] [Google Scholar]
- Midic U., Hung P.H., Vincent K.A., Goheen B., Schupp P.G., Chen D.D., Bauer D.E., VandeVoort C.A., Latham K.E. Quantitative assessment of timing, efficiency, specificity and genetic mosaicism of CRISPR/Cas9-mediated gene editing of hemoglobin beta gene in rhesus monkey embryos. Hum. Mol. Genet. 2017;26:2678–2689. doi: 10.1093/hmg/ddx154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T., Okamoto I., Sasaki K., Yabuta Y., Iwatani C., Tsuchiya H., Seita Y., Nakamura S., Yamamoto T., Saitou M. A developmental coordinate of pluripotency among mice, monkeys and humans. Nature. 2016;537:57–62. doi: 10.1038/nature19096. [DOI] [PubMed] [Google Scholar]
- Niakan K.K., Eggan K. Analysis of human embryos from zygote to blastocyst reveals distinct gene expression patterns relative to the mouse. Dev. Biol. 2013;375:54–64. doi: 10.1016/j.ydbio.2012.12.008. [DOI] [PubMed] [Google Scholar]
- Nichols J., Silva J., Roode M., Smith A. Suppression of Erk signalling promotes ground state pluripotency in the mouse embryo. Development. 2009;136:3215–3222. doi: 10.1242/dev.038893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols J., Smith A. The origin and identity of embryonic stem cells. Development. 2011;138:3–8. doi: 10.1242/dev.050831. [DOI] [PubMed] [Google Scholar]
- Niu Y., Sun N., Li C., Lei Y., Huang Z., Wu J., Si C., Dai X., Liu C., Wei J. Dissecting primate early post-implantation development using long-term in vitro embryo culture. Science. 2019;366:eaaw5754. doi: 10.1126/science.aaw5754. [DOI] [PubMed] [Google Scholar]
- Niu Y., Yu Y., Bernat A., Yang S., He X., Guo X., Chen D., Chen Y., Ji S., Si W. Transgenic rhesus monkeys produced by gene transfer into early-cleavage-stage embryos using a simian immunodeficiency virus-based vector. Proc. Natl. Acad. Sci. U S A. 2010;107:17663–17667. doi: 10.1073/pnas.1006563107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Rahilly R., Müller F. Developmental stages in human embryos: revised and new measurements. Cells Tissues Organs. 2010;192:73–84. doi: 10.1159/000289817. [DOI] [PubMed] [Google Scholar]
- O’Rahilly R., Müller F. Carnegie Institution of Washington; 1987. Developmental stages in human embryos. [Google Scholar]
- Ohinata Y., Ohta H., Shigeta M., Yamanaka K., Wakayama T., Saitou M. A signaling principle for the specification of the germ cell lineage in mice. Cell. 2009;137:571–584. doi: 10.1016/j.cell.2009.03.014. [DOI] [PubMed] [Google Scholar]
- Pereira P.N., Dobreva M.P., Graham L., Huylebroeck D., Lawson K.A., Zwijsen A.N. Amnion formation in the mouse embryo: the single amniochorionic fold model. BMC Dev. Biol. 2011;11:48. doi: 10.1186/1471-213X-11-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perelman P., Johnson W.E., Roos C., Seuanez H.N., Horvath J.E., Moreira M.A., Kessing B., Pontius J., Roelke M., Rumpler Y. A molecular phylogeny of living primates. PLoS Genet. 2011;7:e1001342. doi: 10.1371/journal.pgen.1001342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petropoulos S., Edsgard D., Reinius B., Deng Q., Panula S.P., Codeluppi S., Plaza Reyes A., Linnarsson S., Sandberg R., Lanner F. Single-cell RNA-seq reveals lineage and X chromosome dynamics in human preimplantation embryos. Cell. 2016;165:1012–1026. doi: 10.1016/j.cell.2016.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prufer K., Munch K., Hellmann I., Akagi K., Miller J.R., Walenz B., Koren S., Sutton G., Kodira C., Winer R. The bonobo genome compared with the chimpanzee and human genomes. Nature. 2012;486:527–531. doi: 10.1038/nature11128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayon T., Stamataki D., Perez-Carrasco R., Garcia-Perez L., Barrington C., Melchionda M., Exelby K., Lazaro J., Tybulewicz V.L.J., Fisher E.M.C. Species-specific pace of development is associated with differences in protein stability. Science. 2020;369:eaba7667. doi: 10.1126/science.aba7667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhesus Macaque Genome S., Analysis C., Gibbs R.A., Rogers J., Katze M.G., Bumgarner R., Weinstock G.M., Mardis E.R., Remington K.A., Strausberg R.L. Evolutionary and biomedical insights from the rhesus macaque genome. Science. 2007;316:222–234. doi: 10.1126/science.1139247. [DOI] [PubMed] [Google Scholar]
- Roode M., Blair K., Snell P., Elder K., Marchant S., Smith A., Nichols J. Human hypoblast formation is not dependent on FGF signalling. Dev. Biol. 2012;361:358–363. doi: 10.1016/j.ydbio.2011.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossant J., Tam P.P.L. New insights into early human development: lessons for stem cell derivation and differentiation. Cell Stem Cell. 2017;20:18–28. doi: 10.1016/j.stem.2016.12.004. [DOI] [PubMed] [Google Scholar]
- Saitou M., Miyauchi H. Gametogenesis from pluripotent stem cells. Cell Stem Cell. 2016;18:721–735. doi: 10.1016/j.stem.2016.05.001. [DOI] [PubMed] [Google Scholar]
- Sakai Y., Nakamura T., Okamoto I., Gyobu-Motani S., Ohta H., Yabuta Y., Tsukiyama T., Iwatani C., Tsuchiya H., Ema M. Induction of the germ-cell fate from pluripotent stem cells in cynomolgus monkeys. Biol. Reprod. 2019;102:620–638. doi: 10.1093/biolre/ioz205. [DOI] [PubMed] [Google Scholar]
- Sasaki E., Suemizu H., Shimada A., Hanazawa K., Oiwa R., Kamioka M., Tomioka I., Sotomaru Y., Hirakawa R., Eto T. Generation of transgenic non-human primates with germline transmission. Nature. 2009;459:523–527. doi: 10.1038/nature08090. [DOI] [PubMed] [Google Scholar]
- Sasaki K., Nakamura T., Okamoto I., Yabuta Y., Iwatani C., Tsuchiya H., Seita Y., Nakamura S., Shiraki N., Takakuwa T. The germ cell fate of cynomolgus monkeys is specified in the nascent amnion. Dev. Cell. 2016;39:169–185. doi: 10.1016/j.devcel.2016.09.007. [DOI] [PubMed] [Google Scholar]
- Sasaki K., Yokobayashi S., Nakamura T., Okamoto I., Yabuta Y., Kurimoto K., Ohta H., Moritoki Y., Iwatani C., Tsuchiya H. Robust in vitro induction of human germ cell fate from pluripotent stem cells. Cell Stem Cell. 2015;17:178–194. doi: 10.1016/j.stem.2015.06.014. [DOI] [PubMed] [Google Scholar]
- Sato K., Kuroki Y., Kumita W., Fujiyama A., Toyoda A., Kawai J., Iriki A., Sasaki E., Okano H., Sakakibara Y. Resequencing of the common marmoset genome improves genome assemblies and gene-coding sequence analysis. Sci. Rep. 2015;5:16894. doi: 10.1038/srep16894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scally A., Dutheil J.Y., Hillier L.W., Jordan G.E., Goodhead I., Herrero J., Hobolth A., Lappalainen T., Mailund T., Marques-Bonet T. Insights into hominid evolution from the gorilla genome sequence. Nature. 2012;483:169–175. doi: 10.1038/nature10842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrode N., Saiz N., Di Talia S., Hadjantonakis A.K. GATA6 levels modulate primitive endoderm cell fate choice and timing in the mouse blastocyst. Dev. Cell. 2014;29:454–467. doi: 10.1016/j.devcel.2014.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seita Y., Tsukiyama T., Azami T., Kobayashi K., Iwatani C., Tsuchiya H., Nakaya M., Tanabe H., Hitoshi S., Miyoshi H. Comprehensive evaluation of ubiquitous promoters suitable for the generation of transgenic cynomolgus monkeys. Biol. Reprod. 2019;100:1440–1452. doi: 10.1093/biolre/ioz040. [DOI] [PubMed] [Google Scholar]
- Seita Y., Tsukiyama T., Iwatani C., Tsuchiya H., Matsushita J., Azami T., Okahara J., Nakamura S., Hayashi Y., Hitoshi S. Generation of transgenic cynomolgus monkeys that express green fluorescent protein throughout the whole body. Sci. Rep. 2016;6:24868. doi: 10.1038/srep24868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahbazi M.N., Jedrusik A., Vuoristo S., Recher G., Hupalowska A., Bolton V., Fogarty N.N.M., Campbell A., Devito L., Ilic D. Self-organization of the human embryo in the absence of maternal tissues. Nat. Cell Biol. 2016;18:700–708. doi: 10.1038/ncb3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirparo G.G., Boroviak T., Guo G., Nichols J., Smith A., Bertone P. Integrated analysis of single-cell embryo data yields a unified transcriptome signature for the human pre-implantation epiblast. Development. 2018;145:dev158501. doi: 10.1242/dev.158501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Chimpanzee Sequencing and Analysis Consortium Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- Thomson J.A., Itskovitz-Eldor J., Shapiro S.S., Waknitz M.A., Swiergiel J.J., Marshall V.S., Jones J.M. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- Thomson J.A., Kalishman J., Golos T.G., Durning M., Harris C.P., Becker R.A., Hearn J.P. Isolation of a primate embryonic stem cell line. Proc. Natl. Acad. Sci. U S A. 1995;92:7844–7848. doi: 10.1073/pnas.92.17.7844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson J.A., Kalishman J., Golos T.G., Durning M., Harris C.P., Hearn J.P. Pluripotent cell lines derived from common marmoset (Callithrix jacchus) blastocysts. Biol. Reprod. 1996;55:254–259. doi: 10.1095/biolreprod55.2.254. [DOI] [PubMed] [Google Scholar]
- Tomioka I., Nogami N., Nakatani T., Owari K., Fujita N., Motohashi H., Takayama O., Takae K., Nagai Y., Seki K. Generation of transgenic marmosets using a tetracyclin-inducible transgene expression system as a neurodegenerative disease model. Biol. Reprod. 2017;97:772–780. doi: 10.1093/biolre/iox129. [DOI] [PubMed] [Google Scholar]
- Tsukiyama T., Kobayashi K., Nakaya M., Iwatani C., Seita Y., Tsuchiya H., Matsushita J., Kitajima K., Kawamoto I., Nakagawa T. Monkeys mutant for PKD1 recapitulate human autosomal dominant polycystic kidney disease. Nat. Commun. 2019;10:5517. doi: 10.1038/s41467-019-13398-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu Z., Yang W., Yan S., Yin A., Gao J., Liu X., Zheng Y., Zheng J., Li Z., Yang S. Promoting Cas9 degradation reduces mosaic mutations in non-human primate embryos. Sci. Rep. 2017;7:42081. doi: 10.1038/srep42081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu Z., Zhao H., Li B., Yan S., Wang L., Tang Y., Li Z., Bai D., Li C., Lin Y. CRISPR/Cas9-mediated disruption of SHANK3 in monkey leads to drug-treatable autism-like symptoms. Hum. Mol. Genet. 2019;28:561–571. doi: 10.1093/hmg/ddy367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnock M. Moral thinking and government policy: the Warnock Committee on Human Embryology. Milbank Mem. Fund Q. Health Soc. 1985;63:504–522. [PubMed] [Google Scholar]
- Witschi E. Migration of germ cells of human embryos from the yolk sac to the primitive gonadal folds. Contrib. Embryol. Carnegie Inst. 1948;32:67–80. [Google Scholar]
- Wong C.C., Loewke K.E., Bossert N.L., Behr B., De Jonge C.J., Baer T.M., Reijo Pera R.A. Non-invasive imaging of human embryos before embryonic genome activation predicts development to the blastocyst stage. Nat. Biotechnol. 2010;28:1115–1121. doi: 10.1038/nbt.1686. [DOI] [PubMed] [Google Scholar]
- Xiang L., Yin Y., Zheng Y., Ma Y., Li Y., Zhao Z., Guo J., Ai Z., Niu Y., Duan K. A developmental landscape of 3D-cultured human pre-gastrulation embryos. Nature. 2019;577:537–542. doi: 10.1038/s41586-019-1875-y. [DOI] [PubMed] [Google Scholar]
- Yamanaka Y., Lanner F., Rossant J. FGF signal-dependent segregation of primitive endoderm and epiblast in the mouse blastocyst. Development. 2010;137:715–724. doi: 10.1242/dev.043471. [DOI] [PubMed] [Google Scholar]
- Yan L., Yang M., Guo H., Yang L., Wu J., Li R., Liu P., Lian Y., Zheng X., Yan J. Single-cell RNA-seq profiling of human preimplantation embryos and embryonic stem cells. Nat. Struct. Mol. Biol. 2013;20:1131–1139. doi: 10.1038/nsmb.2660. [DOI] [PubMed] [Google Scholar]
- Yao X., Liu Z., Wang X., Wang Y., Nie Y.H., Lai L., Sun R., Shi L., Sun Q., Yang H. Generation of knock-in cynomolgus monkey via CRISPR/Cas9 editing. Cell Res. 2018;28:379–382. doi: 10.1038/cr.2018.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao X., Wang X., Hu X., Liu Z., Liu J., Zhou H., Shen X., Wei Y., Huang Z., Ying W. Homology-mediated end joining-based targeted integration using CRISPR/Cas9. Cell Res. 2017;27:801–814. doi: 10.1038/cr.2017.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F., Wang R., Yuan P., Ren Y., Mao Y., Li R., Lian Y., Li J., Wen L., Yan L. Reconstituting the transcriptome and DNA methylome landscapes of human implantation. Nature. 2019;572:660–664. doi: 10.1038/s41586-019-1500-0. [DOI] [PubMed] [Google Scholar]
- Zuo E., Cai Y.J., Li K., Wei Y., Wang B.A., Sun Y., Liu Z., Liu J., Hu X., Wei W. One-step generation of complete gene knockout mice and monkeys by CRISPR/Cas9-mediated gene editing with multiple sgRNAs. Cell Res. 2017;27:933–945. doi: 10.1038/cr.2017.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.