Summary
Emerging technologies in stem cell engineering have produced sophisticated organoid platforms by controlling stem cell fate via biomaterial instructive cues. By micropatterning and differentiating human induced pluripotent stem cells (hiPSCs), we have engineered spatially organized cardiac organoids with contracting cardiomyocytes in the center surrounded by stromal cells distributed along the pattern perimeter. We investigated how geometric confinement directed the structural morphology and contractile functions of the cardiac organoids and tailored the pattern geometry to optimize organoid production. Using modern data-mining techniques, we found that pattern sizes significantly affected contraction functions, particularly in the parameters related to contraction duration and diastolic functions. We applied cardiac organoids generated from 600 μm diameter circles as a developmental toxicity screening assay and quantified the embryotoxic potential of nine pharmaceutical compounds. These cardiac organoids have potential use as an in vitro platform for studying organoid structure-function relationships, developmental processes, and drug-induced cardiac developmental toxicity.
Keywords: cardiac organoids, cell micropatterning, embryotoxicity, in vitro embryo model, data mining, human induced pluripotent stem cells
Graphical abstract
Highlights
-
•
Micropattern-based geometric confinement directs cardiac organoid development
-
•
Cardiac organoid structure-function relationships are guided by organoid size
-
•
Cardiac organoids can be used as an in vitro embryotoxicity assessment tool
Advances in organoid engineering have produced sophisticated organoid platforms by controlling stem cell fate via biomaterial instructive cues. Ma and colleagues show that micropatterning is a versatile tool to engineer cardiac organoids with different architectures and cardiac functions. Furthermore, the cardiac organoid development was sensitive to numerous drug compounds and demonstrated potential as an embryotoxicity screening platform.
Introduction
Recent progress in stem cell-based organoid technology offers unique opportunities to in vitro recapitulate biological processes of organogenesis into spatially organized tissue structures that resemble the architecture and functions of specific tissues (Fatehullah et al., 2016). Integration of organoid technology and microfabrication has provided promising ways to guide self-organization and spatial pattern formation of developing biological tissues (Brassard and Lutolf, 2019). For example, Microwell technology has been widely applied to control the aggregate sizes of human pluripotent stem cells (hPSCs) for generating brain (Lancaster et al., 2017), kidney (Czerniecki et al., 2018), and blastocyst (Rivron et al., 2018) organoids of a desired architecture. Microfluidic systems could precisely control the localization of morphogen source and gradient to guide spatial hPSC differentiation and organization into in vitro synthetic embryonic tissues, such as in germ layer patterning (Manfrin et al., 2019), as well as amniotic and epiblast layer separation (Zheng et al., 2019). Surface micropatterning techniques were also able to provide geometric confinement for modeling gastrulation, where patterned PSCs differentiated to form concentric rings indicative of specific germ layers (Morgani et al., 2018; Warmflash et al., 2014). These examples illustrate the critical need for spatiotemporal engineering of cell microenvironments to guide the structure and function of stem cell organoids to specific biological tissues.
Despite extensive efforts in controlling stem cell lineage specification via biophysical inputs, there are few studies focusing on how 2D patterned cell colonies could give rise to organoids with defined structure-function relationships. Understanding these relationships requires comprehensive analysis of multiple variables simultaneously, which increases the data dimensionality for analysis and visualization. As methods to detect tissue functions become more large scale and multidimensional, analysis requires more advanced analytics to effectively comprehend the functional outcomes. Data dimensionality reduction techniques, which are the basis of bioinformatics, are still underexplored for analyzing tissue-level structural and functional properties. The combination of tissue engineering, organoid technology, and advanced data-mining techniques would potentially provide the capability to discover trends and relationships to guide new engineering designs with a spectrum of biological structures and functions.
hPSC-derived organoids exhibit characteristics of specific tissue lineages at their early developmental stages, thus providing great potential as in vitro assays of developmental drug toxicity. In vivo animal models and in vitro mouse embryonic stem cell tests (mEST) are widely implemented by pharmaceutical companies as biological assays for embryotoxicity screening (Seiler and Spielmann, 2011). To overcome species barriers that impose limitations in traditional drug screening, hPSC technology has been proposed to replace the mEST for better predictions of human-specific developmental toxicity (Kumar et al., 2012). However, most 2D stem cell-based assays lack the capability of morphological scoring of 3D tissue morphogenesis. This undermines the predictability of drug-induced teratogenicity, which potentially manifests as structural malformations in prenatal development. Moreover, traditional organoid technology exhibits relatively random positioning of tissue regions of specific cell types, and these regions are not reliably spatially positioned relative to one another. Heterogeneity in organoid formation also makes it difficult for embryotoxicity drug testing purposes with high consistency and reproducibility. Advances in microtechnologies have produced organoids with consistent 3D morphology and spatial structure, enabling robust morphological scoring of in vitro tissue formation, which is crucial for screening drug-induced developmental toxicity.
In this work, we optimized cardiac organoid production from 2D micropatterned human induced PSCs (hiPSCs) by generating organoid arrays with different geometries (Hoang et al., 2018). Organoids generated from genome-engineered GCaMP6f hiPSCs were used for integrated functional analysis of calcium transient and contraction motion. Using data-mining tools, we established relationships among a multitude of organoid metrics of tissue structure and contractile functions. We also explored the inter-dependency among these parameters associated with the geometric sizes, which highlighted that biophysical microenvironment could modulate tissue structure and cardiac function. Next, we implemented this model for evaluating cardiac developmental toxicity by quantifying the drug effects based on cardiac differentiation, contractile behaviors and 3D tissue morphology. We also compared drug-induced cardiac developmental toxicity on hiPSC-based cardiac organoids to effects on in vivo cardiac development using whole embryo culture (WEC) of living Danio rerio (zebrafish) embryos. We envision the human cardiac organoid model as a versatile platform to assess cardiac organogenesis and developmental toxicity, which can be adopted for pharmaceutical development and fetal safety assessments.
Results
Cardiac differentiation of patterned hiPSCs produces spatially organized cardiac organoids
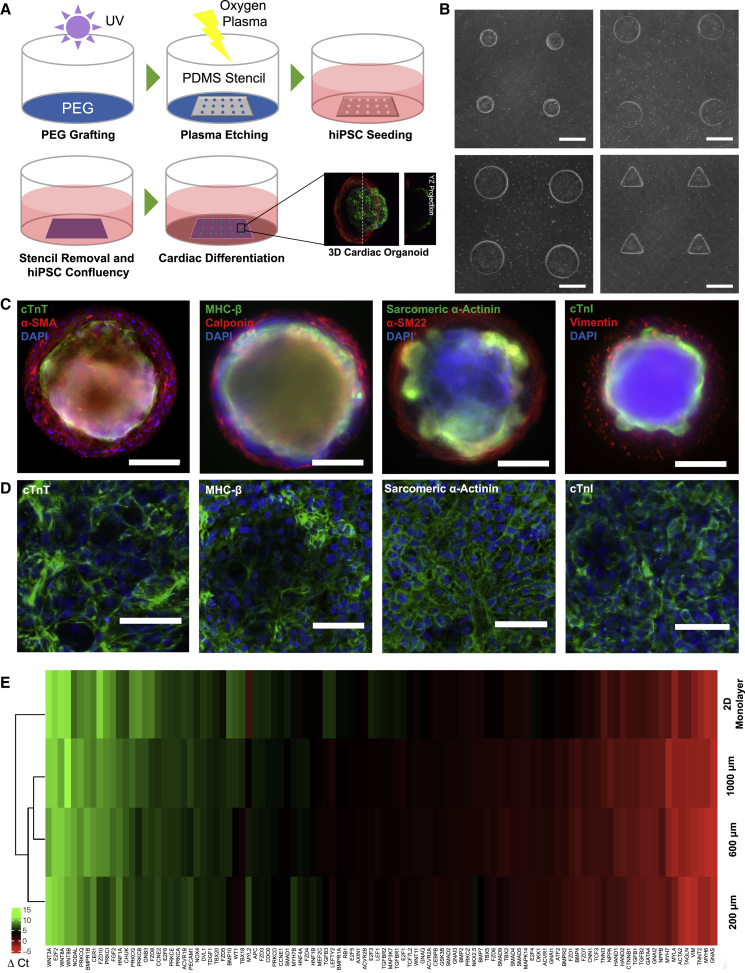
Controlling biophysical microenvironments through cell-patterning techniques has been used for regulating stem cell differentiation and modeling mammalian embryonic development (Hwang et al., 2008; Ma et al., 2015; Shao et al., 2015; Warmflash et al., 2014). Geometric confinement to hiPSCs was provided by a poly(ethylene glycol) (PEG)-based micropatterned substrate created by oxygen plasma etching (Figure 1A). hiPSCs seeded on the micropatterned substrates (Figure 1B). Upon confluency, hiPSCs were differentiated via small-molecule modulation of the Wnt/β-catenin pathway (Figures S1A and S1B) (Lian et al., 2012). Patterned hiPSCs were able to proliferate, conform to the pattern geometry and retain pluripotency, as indicated by positive immunofluorescence of OCT4, NANOG, E-cadherin, SOX2, and SSEA4 (Figure S1C). This approach generated robust contracting cardiac organoids arrays of 50–100 organoids within 20 days.
Figure 1.
Human iPSC micropatterning and differentiation into cardiac organoids
(A) The procedure schematic for micropatterning hiPSCs on standard tissue culture plate using a selective PEG-based etching method.
(B) Bright-field microscopy images of arrays of micropatterned hiPSCs with different sizes and shapes. Scale bar, 400 μm.
(C) Spatial-organized cardiac organoids showed cardiac muscle on the top center with cardiomyocyte-specific proteins (cardiac troponin T [cTnT], β-myosin heavy chain [MHC-β], sarcomeric α-actinin, and cardiac troponin I [cTnI]) and smooth muscle-like cells on the perimeter of the organoids with stromal cell markers (smooth muscle actin [α-SMA], calponin, transgelin [α-SM22]), and vimentin from 600 μm organoids. Scale bar, 200 μm.
(D) Maximum intensity confocal projections of cardiomyocytes on the cardiac organoids. Scale bar, 50 μm.
(E) Gene expression profile of cardiogenesis was compared between organoids of varying sizes with 2D monolayer differentiation. ΔCt values used in this gene map were calculated relative to the average Ct of GAPDH. HPRT1 and GUSB housekeeping controls.
At early differentiation stages, we verified positive expression of mesoderm marker BRA (day 1) (Figure S1D), and cardiac progenitor markers ISL1 (day 8), and NKX2.5 and GATA4 (day 10) (Figures S1E–S1G). Expression of early cardiac progenitor makers was distributed across the entire pattern before the cardiac organoids began contracting around day 12. As the contraction became robust over time, the cardiac tissue compacted toward the center, revealing the underneath stromal cells. At day 20 of differentiation, cardiomyocytes were primarily differentiated on the center top of the organoids, demonstrated by multiple cardiac-specific markers (Figures 1C and 1D): cardiac troponin T, myosin heavy chain β, sarcomeric α-actinin, and cardiac troponin I, while smooth muscle-like stromal cells along the pattern perimeter, indicated by positive expression of α-smooth muscle actin, calponin, α-SM22, and vimentin.
Next, we compared the gene expression profile between cardiac organoids from three different pattern sizes (200, 600, and 1,000 μm in diameter) and traditional 2D monolayer differentiation. In general, gene expression showed upregulation of cardiac-specific genes and downregulation of WNT signaling at day 20 differentiation, although the gene expression profile had closer similarity among different organoids than 2D differentiation (Figure 1E). The organoids also showed significantly higher gene expression related to transforming growth factor β (TGF-β) signaling (TGFB1, TGFB2, TGFB3, TGFBR1, and TGFBR1). This might relate to a high content of stromal cells in the organoids under geometric confinement on the patterned substrate (Figure S1H). To compare the organoids generated from different pattern sizes, we found that organoids of 600 and 1,000 μm showed upregulation of cardiac-specific genes (MYL4, MYH7, and NKX2.5), while smaller 200 μm organoids had higher expression of stromal cell genes (ACTA2, TAGLN, and VIM). These results illustrated that high geometric confinement from smaller patterns promoted the differentiation into supportive cell types.
Pattern geometry dictates structural morphology and contractile physiology of cardiac organoids
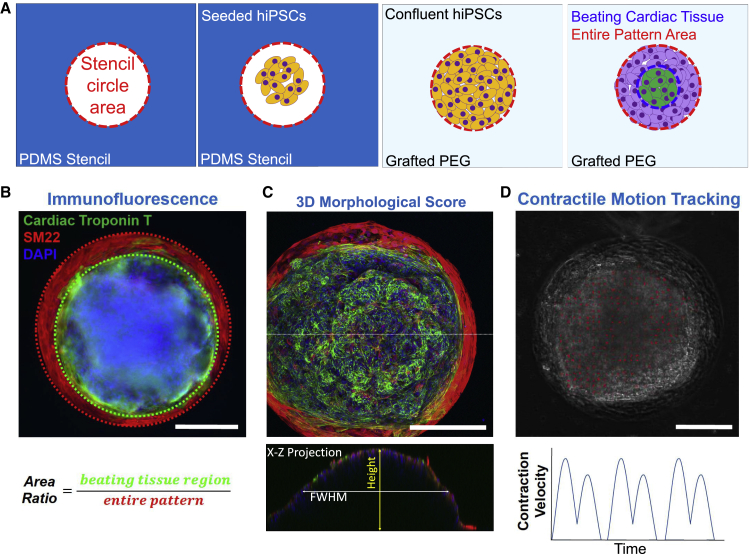
Using immunofluorescence staining, we measured the cardiac tissue distribution by calculating the area ratio between the areas of cardiomyocyte differentiation and entire pattern area (Figures 2A and 2B). Using confocal microscopy and 3D image reconstruction (Videos S1 and S2), we assessed the three-dimensionality of cardiac organoids by measuring the height and full-width at half-maximum (FWHM) (Figure 2C). To study the contractile functions of cardiac organoids, we generated the cardiac organoids from genome-edited hiPSC line with GCaMP6f reporter (Chen et al., 2013; Huebsch et al., 2016; Mandegar et al., 2016), which provided us the capability to assess the cardiac contractions using bright-field motion tracking analysis (Huebsch et al., 2015) (Figure 2D; Video S3). Motion data were integrated with fluorescent calcium flux (Figures S2A and S2B; Video S4) for a comprehensive characterization of contraction functions. t-SNE plots, together with a heatmap, were generated using quantified contractile function parameters from individual organoids with different pattern sizes: area ratio between GCaMP6 fluorescence and pattern area, beat rate, maximum calcium flux, pulse duration of calcium flux cycle (τ0, τ50, and τ75), peak-to-peak interval, maximum contraction velocities, and relaxation velocities (Figure S2C).
Figure 2.
Cardiac organoid characterization metrics
(A) Schematic illustration of metrics used to calculate the area ratio. Red dotted outline illustrates the organoid area, which is equivalent to the stencil pattern area. Blue dotted outline illustrates the area of contracting cardiac tissue.
(B) Immunofluorescence staining used to compute the area ratio, which is the area of cardiac tissue (cTnT) coverage relative to the area of the entire pattern.
(C) Confocal imaging was used to capture z-stacks of the tissues, which were reconstructed for 3D morphological scoring of the height and full-width at half-maximum (FWHM).
(D) Motion tracking analysis based on vector computation of pixel movement generates contraction velocity waveforms representing beat cycles of the cardiac organoids. Graph shown represents a schematic illustration of contraction motion waveform and no data are associated. Scale bars, 200 μm.
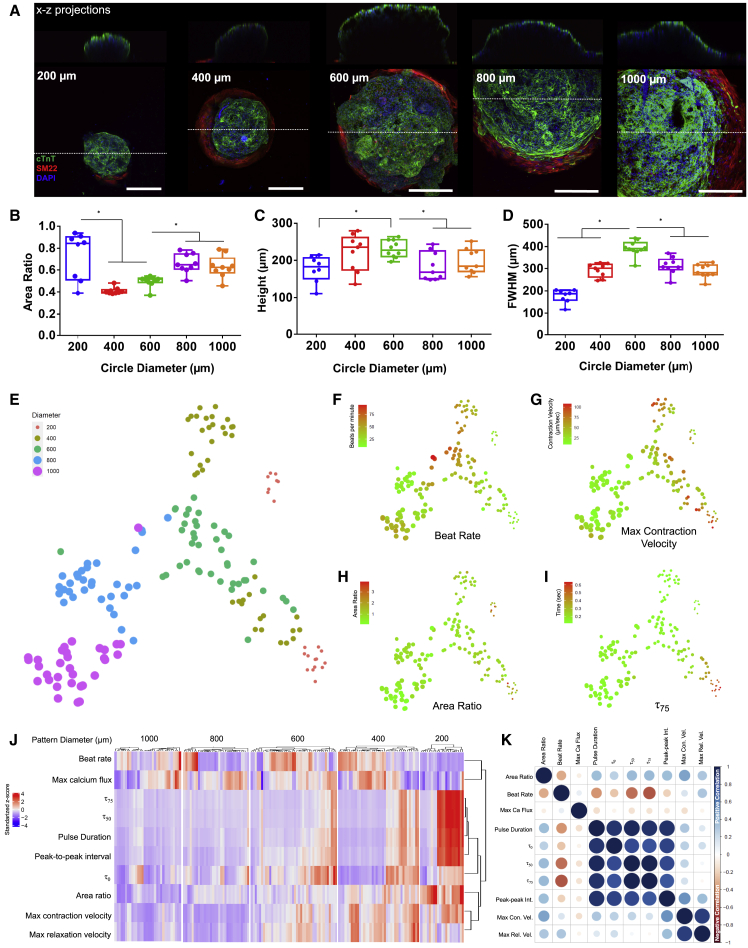
To examine the effects of pattern size on cardiac organoid development, we generated organoids from circular patterns ranging from 200 to 1,000 μm in diameter (Figures 3 and 4A). The 200 μm patterns did not reliably produce the organoids with a relatively low organoid production efficiency (∼20%), in comparison with all other sizes (∼80%). Albeit with the largest variability, 200 μm circle patterns produced the cardiac organoids with the largest area ratio of cardiac muscles among all the sizes (Figure 4B). Regarding large sizes (800 and 1,000 μm in diameter), these patterns produced the organoids with a greater area ratio, but lower organoid height than the ones from 400 to 600 μm circles (Figure 4C). In addition, the FWHM steadily increased with increasing pattern size, peaked at 600 μm patterns, and decreased at larger sizes (Figure 4D). This indicated that 600 μm patterns produced the cardiac organoids with high consistency and large 3D morphology.
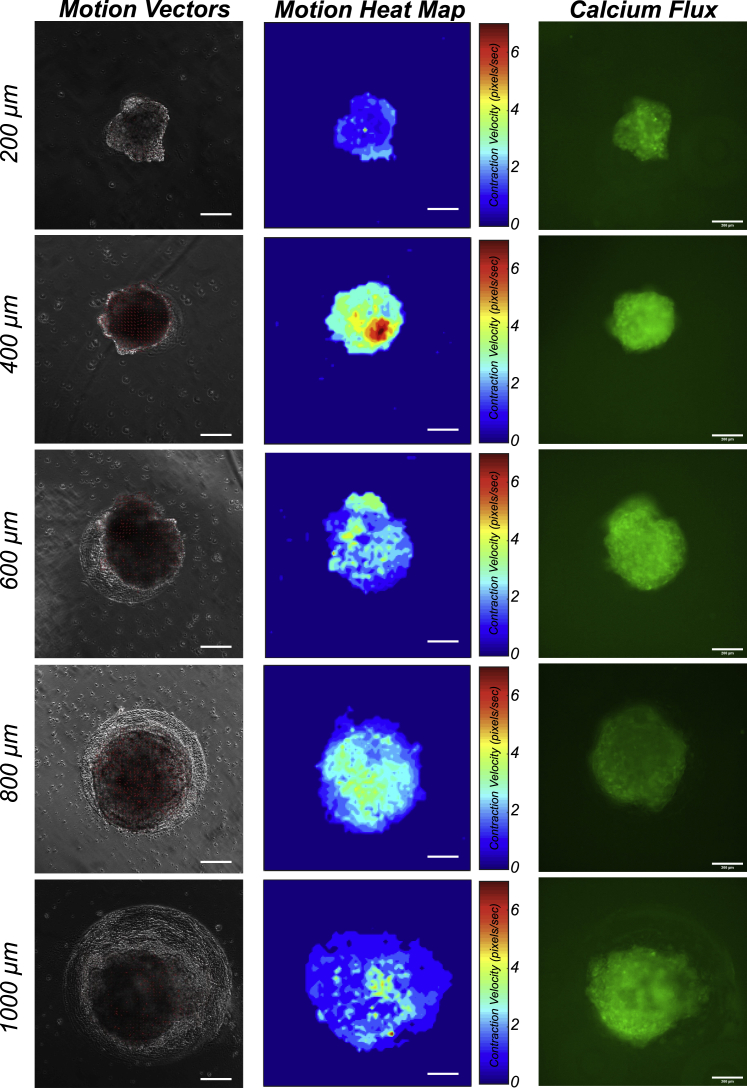
Figure 3.
Contraction functions of cardiac organoids ranging in size from 200 μm (top) to 1,000 μm (bottom) in diameter
Contraction functions were assessed using contraction motion tracking (left), which computes contraction magnitude as a function of the contraction velocity (middle). Fluorescent calcium flux (right) was measured to further characterize contraction behavior by classifying the calcium handling properties. Scale bars, 200 μm.
Figure 4.
Pattern size effects on cardiac organoid characteristics
(A) Cardiac organoids generated from the circular patterns with different sizes (200–1,000 μm in diameter) showed spatial-organization of cardiomyocytes and stromal cells and 3D dome shape in the X-Z plane. Scale bar, 200 μm.
(B–D) (B) The area ratio between cardiomyocyte staining and entire pattern size showed larger coverage but higher variation in cardiac muscle differentiation on the cardiac organoids of 200, 800, and 1,000 μm patterns (n = 9, ∗p ≤ 0.0001 between 200 and 400/600 μm; ∗p ≤ 0.0001 between 400/600 and 800/1,000 μm). The cardiac organoids of 600 μm patterns exhibited better 3D morphology with largest values in (C) height (n = 8, ∗p = 0.0126 between 200 and 600 μm; ∗p = 0.0126 between 600 and 800/1,000 μm) and (D) FWHM (n = 8, ∗p ≤ 0.0001 between 600 and 200/400 μm; ∗p ≤ 0.0001 between 600 and 800/1,000 μm).
(E–I) (E) Contractile function parameters were used to cluster individual organoids with different pattern sizes and generate a t-SNE plot to evaluate organoid-to-organoid correlations. t-SNE gradients were plotted for (F) beat rate, (G) maximum contraction velocity, (H) τ75, and (I) area ratio.
(J) Heatmap illustrating all contractile functions parameters relative to pattern size showed strong correlation between size and prolonged contraction duration, especially for the organoids of small patterns.
(K) Correlation plot illustrated proportional relationships between contraction duration parameters, but inverse correlations between contraction duration and contraction rate. All boxplots show the minimum, maximum, median, and 25th and 75th percentiles, and statistical analysis was performed based on analysis of variance (ANOVA) with Tukey's multiple comparison test. Data were pooled from three independent experiments.
From the t-SNE plot (Figure 4E) generated from data mining of contraction function parameters (Figure S2C), organoids of larger sizes (600, 800, and 1,000 μm) were better clustered with high consistency. Organoids from 200 to 400 μm patterns showed significant organoid divergence with separated clusters, indicating less consistency in organoid properties. We also used t-SNE plots to illustrate the gradients of each parameter across the organoid sample distribution (Figures 4F–4I and S3A–S3F). Organoids of larger patterns exhibited higher beat rate (Figure 4F), while smaller patterns exhibited higher contraction and relaxation velocities (Figures 4G and S3A). Consistent with the confocal structural analysis, the 200 μm organoids produced the largest area ratio of cardiac tissue relative to pattern size (Figure 4H). In these t-SNE plots, the divergence of small organoids from 200 to 400 μm pattern primarily resulted from the significant differences on parameters of contraction duration (peak-to-peak intervals, pulse duration, τ50, τ75) (Figures 4I and S3B–S3D). The metrics of τ0 and maximum calcium flux exhibited no apparent trends among different organoid sizes (Figures S3E and S3F).
These parameters for the cardiac organoids of different sizes were then compared in a heatmap (Figure 4J). Organoids larger than 200 μm exhibited comparable trends of beat rate and maximum calcium flux, indicating efficient calcium handling and consistent beat frequency. Meanwhile, the contraction duration parameters of peak-to-peak intervals, pulse duration, τ50, and τ75 showed high levels of size dependency. The prolongation of contraction duration, especially regarding calcium decay in the cardiac organoids from small patterns, is prone to arrhythmia related to the abnormal diastolic functions (Periasamy and Janssen, 2008; Weber et al., 2006). From the correlation matrix (Figure 4K), strongest correlations were observed between parameters of calcium analysis (τ0, τ50, τ75, pulse duration), which had negative correlation with beat rate (Figures S3G and S3H). Furthermore, there was a strong positive correlation between calcium decay from calcium flux analysis and peak-to-peak interval from contractile motion analysis. More importantly, area ratio from morphological analysis is positively correlated with calcium decay and contractile motion velocities from different functional analysis, indicating a structure-function relationship between the relative tissue size and cardiac contraction cycles.
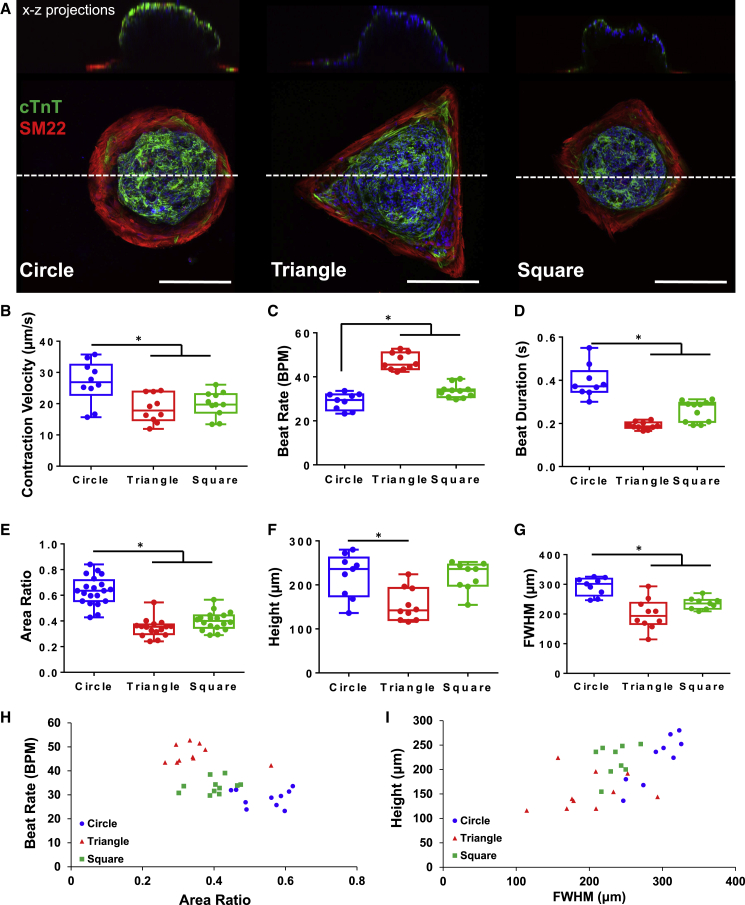
In addition, we compared circle, square, and triangle organoids with the same area (Figure 5A). Cardiac organoids from circular patterns exhibited significantly higher contraction velocity, lower beat rate, and longer beat duration (Figures 5B–5D). Furthermore, circle patterns produced the organoids with larger area ratio, height, and FWHM than squares and triangles (Figures 5E–5G). Overall, the circular pattern geometry promoted more robust formation of cardiac organoids with large size and high contractile functions. These observations illustrated that the development of cardiac organoids was affected by pattern shape, in which the angular geometries presented high physical constraint to the cells and promoted a higher degree of differentiation into the stromal cell types, rather than cardiomyocytes. By plotting the structure-function correlations and structure-structure correlations, we observed a decline in the beat rate that corresponded to the increasing area ratio (Figures 5H and 5I) as a negative correlation between the cardiac tissue size and the beat rate.
Figure 5.
Polygonal geometry effects on cardiac organoid development
(A–D) (A) Z-Projection of confocal images of cardiac organoids generated in circular, triangular, and square shapes. Scale bars, 200 μm. Triangular and square organoids exhibited (B) significantly decreased contraction velocity (n ≥ 10, ∗p = 0.0024), but (C) significantly faster beat rate (n ≥ 9, ∗p ≤ 0.0001), and (D) longer beat duration (n ≥ 9, ∗p ≤ 0.0001) relative to circular organoids.
(E) Morphological scoring of cardiac organoids showed that circle shapes produced organoids that were significantly larger in area ratio (n ≥ 16, ∗p ≤ 0.0001) than triangle and square organoids.
(F and G) (F) Only triangle organoids produced significantly decreased tissue height (n ≥ 9, ∗p = 0.0013), (G) while both triangular and square organoids had significantly smaller FWHM (n ≥ 9, ∗p ≤ 0.0001).
(H) Structure-function correlation plot illustrate that area ratio is negatively correlated to beat rate for these three geometric shapes.
(I) Structure-structure correlation plot illustrate that organoid structure is also strongly correlated with geometry where circular organoids had the largest 3D structures. In all panels, boxplots show the minimum, maximum, median, and 25th and 75th percentiles, and statistical ANOVA with Tukey's multiple comparison test. Data were pooled from three independent experiments.
Cardiac organoids as an in vitro assay for cardiac developmental toxicity
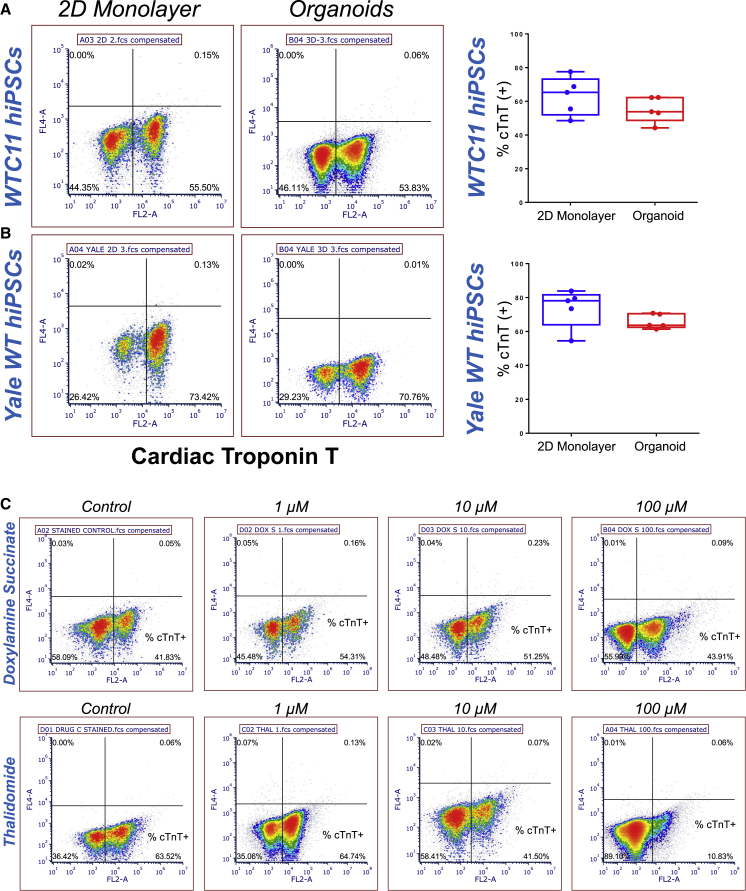
The heart is the first functional organ to form, thus cardiac differentiation is often used as a key evaluation for developmental toxicity (Tandon and Jyoti, 2012). Since 600 μm diameter circular patterns gave robust organoid production, large 3D morphology, consistent contractile functions, and high level of cardiac-specific differentiation, arrays of cardiac organoids from this geometry were used to test the capabilities of this platform to be implemented as a cardiac developmental toxicity assay. Using flow cytometry, we quantified the consistency of cardiac differentiation efficiency of organoids and 2D monolayer differentiation from five different batches (n = 5) across ten passages of hiPSCs from two different lines. Both organoids of 600 μm and 2D differentiation produced over 50% cells that were positive for cardiac troponin T (Figures 6A and 6B). These results illustrated that cardiac organoids can be generated consistently across multiple cell batches and multiple hiPSC lines. Furthermore, the result of ∼50% cTnT+ cells from flow cytometry was consistent with the area ratio calculation, where 600 μm organoids exhibited an average area ratio of approximately 0.5 (Figure 4B). Since the percentage of cTnT+ cells that composed organoids was comparable with the area ratio metric, we inferred that the area ratio is an appropriate metric to approximate cardiac differentiation efficiency for the drug-screening purposes.
Figure 6.
Flow cytometry quantification of cardiac differentiation efficiency
(A) 2D monolayer differentiation and organoid differentiation using WTC11 hiPSCs show that over 50% cells are cTnT+ cardiomyocytes.
(B) Reproducibility was demonstrated using a second hiPSC line (Yale WT), which also resulted in over 50% of cells expressing cTnT. Sample size n = 5.
(C) Flow cytometry density plots quantifying cardiac differentiation efficiency of organoids treated with doxylamine succinate and thalidomide. Doxylamine succinate treatment concentrations did not affect the percentage of cells expressing cTnT. Thalidomide treatment caused a considerable reduction in cTnT+ cells with increasing concentration. Data were pooled from at least three independent experiments.
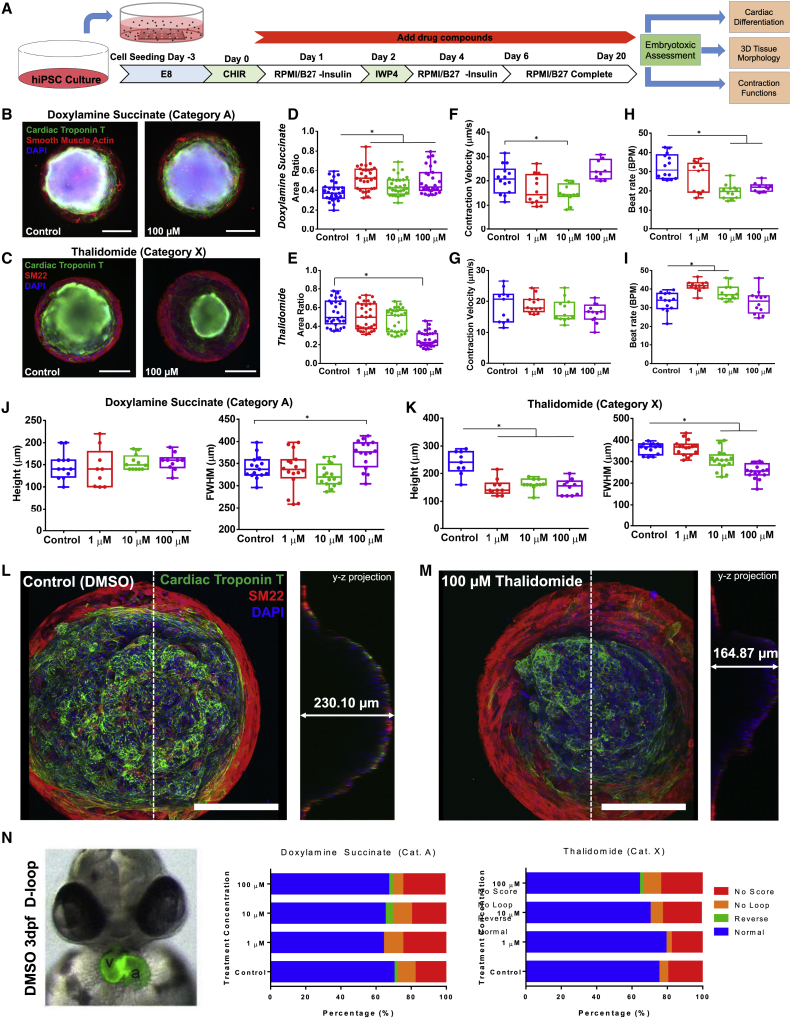
Nine drugs (Table S1) used in this study covered the entire spectrum of the past FDA pregnancy category system that ranked drugs from A (safe) to X (toxic) based on predicted teratogenic risk. First, we compared two drugs between category A (doxylamine succinate) and category X (thalidomide). The cardiac differentiation efficiency remained consistent for doxylamine succinate treatment, whereas thalidomide resulted in a clear decrease in cardiac differentiation with each increasing dose (Figure 6C). Next we performed a more comprehensive embryotoxicity assessment characterizing differentiation (area ratio), 3D tissue morphology, and contraction functions (Figure 7A). Doxylamine succinate (category A, Figure 7B) treatment had no negative effects on cardiac area ratio, whereas thalidomide (category X, Figure 7C) had produced the abnormal cardiac organoids with less cardiac tissue relative to the untreated controls (Figures 7D and 7E). Doxylamine succinate had prominent effects on the contractile functions with lower contraction velocity and slower beat rate (Figures 7F and 7H) due to its anticholinergic effects as an H1 receptor antagonist. Generally used as a sleeping aid, doxylamine succinate is also prescribed as an analgesic to reduce muscle tension. It is possible that the reduction of muscle contraction due to doxylamine treatment led to less tissue compaction, which resulted in an increase of area ratio. In contrast, there was no significant effect from thalidomide on contraction velocity (Figure 7G), although high dosage led to high variability in the beat rate (Figure 7I). More importantly, high concentration of thalidomide (100 μM) impaired the 3D morphology of the cardiac organoids with significantly lower height and FWHM relative to organoids treated with doxylamine succinate (Figure 7J) and to the controls (Figures 7K–7M). The impairment of cardiac tissue is potentially caused by fibroblast growth factor antagonism of this drug, resulting in decreased muscle development. These results indicated that exposure of a well-known teratogen resulted in severe impairment to hiPSC differentiation and organization into 3D cardiac organoids, which confirmed that this organoid model was sensitive to morphological defects as a result of drug exposure.
Figure 7.
Cardiac organoids as a developmental toxicity screening assay
(A–H) (A) The timeline of cardiac organoid generation and continuous drug exposure. Epi-fluorescent microscopy images of the cardiac organoids for comparison between untreated controls with (B) doxylamine succinate (category A drug) and (C) thalidomide (category X drug). The area ratio of cardiac muscle coverage showed (D) an increase with doxylamine succinate (n = 29, ∗p ≤ 0.0001), but a decrease with thalidomide at high concentration (n ≥ 28, ∗p ≤ 0.0001). The contraction velocity of cardiac contractile motion showed no drug effect from (F) either doxylamine succinate (G) or thalidomide. The beat rate showed (H) a decrease with doxylamine succinate (n ≥ 9, ∗p ≤ 0.0001), and an increase with thalidomide at 1 and 10 μM concentrations (n ≥ 12, ∗p ≤ 0.0001).
(J) The cardiac organoids under doxylamine succinate exposure showed no drug effects on the 3D organoid morphology at all the tested concentrations.
(K) Thalidomide exposure induced significant structural impairment on 3D organoid formation, showed as lower height (n ≥ 9, ∗p ≤ 0.0001) and FWHM (n ≥ 14, ∗p ≤ 0.0001) at high concentrations. Representative confocal projections of cardiac organoids at (K) untreated control and (M) 100 μM thalidomide exposure showed severe abnormal organoid formation for quantitative morphological scoring.
(N) Cardiac looping scored as a cardiac developmental toxicity evaluation in the zebrafish whole-embryo culture assay (zWEC) showed no significant drug effect from doxylamine succinate exposure, and a very mild toxicity level from high dosage exposure of thalidomide. A sample size of n > 40 embryos were analyzed for these treatment groups. In all panels, boxplots show the minimum, maximum, median, and 25th and 75th percentiles, and statistical ANOVA with Dunnett's multiple comparison test against controls. All data, including organoids and embryos, were pooled from three independent experiments.
Three antibiotic drugs tested on the cardiac organoids showed increased developmental toxicity with the increase of their risk classification in the pregnancy category. Amoxicillin (category B antibiotic) in mammalian cells has been shown to induce DNA lesions as a result of amoxicillin-induced oxidative stress (Li et al., 2007). Amoxicillin showed no clear toxic effect on either structure or functions of the cardiac organoids at all three tested concentrations (Figure S4A). Rifampicin (category C antibiotic) targeting bacterial RNA and DNA for its effectiveness, has been shown to inhibit protein synthesis in mammalian cells (Buss et al., 1978). Rifampicin showed severe developmental toxicity at a high concentration (100 μM) with no organoid formation (Figure S4B). Doxycycline (category D antibiotics) inhibits the synthesis of bacterial proteins by binding to the 30S ribosomal subunit, but also showed adverse effects on mitochondrial ribosomes within mammalian cells (Ahler et al., 2013). Doxycycline treatment resulted in severe impairment on cardiac differentiation and organoid formation even at a moderate concentration (10 μM) (Figure S5A).
We then tested other category D dugs with various therapeutic applications. Lithium carbonate (category D antidepressant), which inhibits the PKC signaling for its psychiatric medication purpose, appeared to inhibit phosphatidylinositol cycle and Wnt pathway activation, based on mEST assays (Shaikh Qureshi et al., 2014). Exposure of lithium did not affect the contractile functions of the cardiac organoids but exhibited mild toxicity to the organoid formation measured by area ratio and FWHM (Figure S5B). Phenytoin (category D anticonvulsant) protects against seizures via voltage-dependent antagonism of voltage-gated sodium channels. Phenytoin exposure produced smaller cardiac organoids than controls, but still maintained structural integrity (Figure S5C). Due to its inhibition on sodium channels, the cardiac organoids stopped beating at a high concentration (100 μM), until the drug was removed to allow the organoids to recover the contractile functions. Tretinoin (all-trans-retinoic acid, category D retinoid) treatment completely abolished the cardiac differentiation at a concentration as low as 10 μM, but still produced the organoids with comparable height and overall size as controls (Figure S6A).
Overexposure to retinoids was shown to result in birth defects from retinoic acid deficiency with decreased levels of retinoic acid-producing enzymes (Lee et al., 2012). Hence, we also tested isotretinoin (13-cis-retinoic acid, category X retinoid) (Figures S6B and S6C) on the cardiac organoids. Similar to tretinoin, isotretinoin produced large organoids at all tested concentrations, but totally abolished the cardiac differentiation at even lower concentration at 1 μM. This implied that retinoids caused severe impairment to the cardiac differentiation, but not tissue growth. Overall, these results verified that this cardiac organoid model was sensitive to the drug-induced cardiac developmental toxicity and offered the capability of morphological scoring based on the 3D tissue formation, which is often not available from other stem cell-based in vitro assays.
Last, we compared the developmental toxicity of these drugs between the cardiac organoid model and zebrafish whole embryo culture (zWEC), a well-established toxicity assay with promising potential to screen for teratogenicity (Lantz-McPeak et al., 2015). We used transgenic Tg(myl7:GFP) (Huang et al., 2003) with GFP only in cardiomyocytes, which allowed us to readily score myocardial development and heart tube looping at 48 h post fertilization (Sarmah and Marrs, 2016) (Figures 7N and S7A). Live zebrafish embryos were collected and exposed to the identical drugs and concentrations used in the cardiac organoid assay. Consistent with the organoid model, exposure with doxylamine succinate (category A) had negligible effect on the zebrafish embryonic heart development, as there was a considerable proportion of embryos exhibiting normal D-looped heart across all concentrations. However, the effects of thalidomide on zebrafish embryonic heart development were not as significant as we observed in the organoid model. We found that the proportion of normal heart looping did not notably decrease at higher concentrations of thalidomide (Figure 7N). Rifampicin (Figure S7C) and phenytoin (Figure S7B) showed mild embryotoxic effects to the zebrafish embryos at the highest concentration, while amoxicillin (Figure S7D), lithium carbonate (Figure S7E), and doxycycline (Figure S7F) showed moderate toxicity. Retinoids, tretinoin (Figure S7G), and isotretinoin (Figure S7H), at low concentration (0.1 μM), resulted in embryos with smaller heart size and abnormal morphology. At higher concentrations, the hearts failed to develop, as indicated by 0% of embryos expressing the GFP transgene. Upon comparing the organoid model with the zWEC model, we saw developmental toxicity that was comparable for between these two systems for most of drugs (doxylamine succinate, amoxicillin, lithium carbonate, phenytoin, tretinoin, and isotretinoin). However, rifampicin, doxycycline, and thalidomide showed distinct mismatch between these two model systems. We infer that this might be due to a number of factors, including species differences, method of drug exposure, and differences in the range of effective treatment concentrations.
Discussion
The majority of in vitro cardiac tissue models focus on accurate recapitulation of physiologically relevant tissue structures of adult human heart, which are generally achieved by populating pre-fabricated 3D biomaterial scaffolds with pre-differentiated hiPSC-derived cardiomyocytes (hiPSC-CMs) (Ogle et al., 2016; Rodrigues et al., 2018; Turnbull et al., 2018). These adult-mimicking model systems are designed to enhance the maturity of hiPSC-CMs for the purpose of drug screening and disease modeling, but not designed for studying dynamic cellular self-organization occurring along with the cardiac differentiation process. In contrast, stem cell-derived organoids are designed to resemble the early developing organs through self-organization of differentiating cells into spatial-distinct tissue-specific structures. However, cardiac organoids are still largely generated by aggregating pre-differentiated hiPSC-CMs with other stromal cells (Richards et al., 2017), instead of originating from directed stem cell differentiation. Our technique to generate cardiac organoids started with 2D micropatterned hiPSC colonies, allowing for cell self-organization into 3D tissue structures during the differentiation process under geometric confinement. Our new approach opens the possibility to create cardiac organoids that could resemble the similarity to a certain extent of biological process of tissue self-assembly and morphogenesis during early heart formation.
Commonly reported birth defects are heart related, and the potential for generating cardiac defects is a primary concern in determining drug developmental toxicity (Pamies et al., 2011). The cardiac organoid model allowed us to evaluate human-specific drug-induced developmental toxicity based on its disruption of forming correct 3D organoid structures and developing normal cardiac contractile functions. By exposing the cardiac organoids to a range of drugs with different risk, we found an overall increase of teratogenic severity on cardiac organoid formation, corresponding to an increase of test concentrations, and an increase of risk category from A to X. Especially, category D drugs (phenytoin, lithium, doxycycline, and tretinoin) showed diverse effects on developmental toxicity. Surprisingly, exposure of lithium only showed mild developmental toxicity of slight reduction in cardiac differentiation and organoid formation, although there has been a long debate of this drugs' developmental toxicity, especially resulting in congenital heart defects (Hoberman et al., 1990; Shaikh Qureshi et al., 2014).
Our drug response results of thalidomide were generally consistent with published works of embryotoxicity using both whole embryo and in vitro stem cell models. Previous work showed reduced cardiac differentiation efficiency in a hiPSC embryotoxicity test due to thalidomide treatment (Aikawa et al., 2014). A recent study demonstrated that thalidomide inhibited early mesoderm differentiation in chick embryos (Belair et al., 2020), which is the transitional stage during cardiac differentiation. From an embryoid body (EB)-based cardiac differentiation assay, thalidomide treatment showed decreased EB volume, similar to the decreased organoid height and FWHM from our cardiac organoids. Retinoids actually act as morphogens in embryogenesis, guiding gastrulation and body axis formation (Piersma et al., 2017). Overdose of retinoids has been linked to cardiovascular and skeletal developmental malformations (Collins and Mao, 1999). From iPSC models, retinoids impair metabolic responses involved in cell proliferation and differentiation (Palmer et al., 2017). In our study, retinoids impaired the cardiac differentiation but promoted the formation of giant tissues. It is possible that progenitor cells in our retinoid-treated organoids retained a high proliferative capacity to give rise large tissue growth, but inhibited the terminal differentiation of cardiomyocytes (Drowley et al., 2020). Another possibility is that the cells were directed to endoderm lineages, as exposure to these compounds was shown to severely disrupt mesoderm formation (Liu et al., 2018).
Embryotoxicity assays based on WEC have been invaluable in drug toxicology for decades, because they can study drug effects on whole systematic biological processes (Augustine-rauch et al., 2010). Generally, WEC assays focus on drug toxicity on structural and morphological features, such as limb and appendage malformations, but suffer from species differences that can lead to inaccurate predictions in humans (He et al., 2014). In contrast, stem cell-based assays, including mEST and newly developed in vitro platforms using human pluripotent stem cells, offer a cheaper and less-invasive methods to measure drug toxicity on mammalian and human cell differentiation (Seiler and Spielmann, 2011). However, they cannot characterize tissue morphogenesis and organ formation. In a comparison of triazole exposure to rat WEC, zebrafish WEC, and mEST, the zebrafish tests showed the best correlation, followed by mEST tests, regarding their toxicity levels relative to in vivo studies conducted in industry (de Jong et al., 2011). Rat WEC had the lowest correlation scores, which was likely caused by differences in drug exposure times calculated for each system, illustrating the challenges in embryotoxicity model comparisons. Other studies on embryotoxicity indicated a comparable result between WEC and mEST models, but poor correlation with in vivo reports (Dimopoulou et al., 2018; Inoue et al., 2016; Strikwold et al., 2012). Furthermore, these works suggest that a combination of different testing systems can provide better predictivity of embryotoxic potential. One study integrated mEST and zebrafish WEC to understand biological mechanisms of triclosan on early development (Chen et al., 2015), and found that triclosan causes developmental defects via disruption of pluripotent markers. In relation to the apparent heterogeneity of the cardiac organoids, we envision that the cardiac organoids can serve as complementary tests to current well-established assays to assess teratogenicity in both cell differentiation and tissue morphogenesis, which can provide a comprehensive risk-assessment toolkit to better predict drug toxicity on fetal health.
Recently, fully 3D heart organoids have been reported that form from Matrigel-embedded hPSC aggregates and develop into early heart and foregut endoderm tissues (Drakhlis et al., 2021). Although our method to micropattern hiPSCs and generate organoids is versatile and reproducible for various applications, one primary limitation is that the organoids are artificially attached to the substrate, instead of embodying a fully 3D culture system. Stimuli-responsive biomaterials can be explored in future to facilitate the on-demand detachment of organoids from the surface with proper triggering (e.g., temperature changes). Furthermore, the maturity of cardiac organoids has not been assessed, although we think the cardiomyocytes within our organoids are less mature compared with other engineered tissue model systems that applied different external stimulations to promote the maturation (Kolanowski et al., 2020; Nunes et al., 2013; Ronaldson-Bouchard et al., 2018). In addition, the presence of stromal cells in cardiac microtissues has been shown to promote maturity as well (Hookway et al., 2019). Currently, our model is purposed to studying early developmental events and drug effects on embryonic cardiogenesis, instead of mimicking adult-like physiology and drug responses. Future work to expand this study, such as lineage tracking, fate mapping, and single-cell genomic sequencing of organoids at different developmental stages, will reveal parallelism between human cardiac development and cardiac organoid formation through comprehensive molecular evidence. In the perspective of embryotoxicity assay development, our cardiac-based model system presents a major limitation in that it only focuses on cardiac differentiation as key assessment variables but cannot evaluate the developmental toxicity to a wide range of organ types (Seiler and Spielmann, 2011). Developmental toxicology can encompass a wide range of organ targets, depending on the molecular target of the specific drug, but may not particularly result in cardiac defects (Xu et al., 2020). Finally, many drug compounds are safe in their native chemical makeups, but undergo metabolism to produce toxic metabolites, which could lead to misclassifications of drug safety unless the screening can be achieved in a multi-organ system.
Experimental procedures
Cardiac organoid generation
Surface micropatterning on tissue culture polystyrene was carried out using the selective etching approach described previously (Hoang et al., 2018) (see supplemental information for detailed procedures). hiPSCs were cultured using standard PSC culture methods and seeded onto microfabricated wells at a density of approximately 0.63 × 105 cells/cm2. Cardiac differentiation was initiated approximately 3 days after seeding (day 0) when the micropatterns reached confluency, and performed via small-molecule modulation of the Wnt/β-catenin pathway (Lian et al., 2012) with GSK3 inhibitor CHIR99021 (day 0) and WNT pathway inhibitor IWP4 (day 2).
Drug treatment
Drug stocks were prepared in water or DMSO, depending on solubility, and diluted to working concentrations in culture medium. Once initiated, the drugs were supplied continuously throughout the differentiation into cardiac organoids to mimic the continuous drug exposure during fetal development. Samples were terminated on day 20 for motion tracking analysis and for fluorescence/confocal imaging.
See supplemental information for detailed procedures, additional methods, and vendor information.
Data and code availability
Data requests and correspondence should be addressed to Z.M.
Author contributions
Z.M. and P.H. conceived the study and designed the experiments. P.H., S.S., and A.M.A. performed surface patterning and organoid generation-related experiments. S.S. acquired the data of GCaMP6f organoids and performed the calcium flux analysis. A.K. performed data mining on contractile function data from cardiac organoids. T.S.W. performed flow cytometry analysis. A.G.E.-C., A.R.G., W.L., and M.I.K. generated and provided the Yale WT hiPSC line for cell line comparison. P.H. performed all organoid-based drug toxicity studies. P.H. and J.D.A. performed zebrafish whole-embryo experiments. P.H. and S.M.L. collected and analyzed bulk motion tracking and image analysis datasets. P.H. and Z.M. wrote the manuscript with discussion and improvements from all authors. Z.M. supervised the project development and funded the study.
Acknowledgments
We would like to thank the members of the Amack lab for their guidance with whole embryo experiments. This work was supported by the NIH NICHD (R01HD101130), NSF (CBET-1804875 and CBET-1943798), and SU Collaboration for Unprecedented Success and Excellence (CUSE) Grant. Z.M. acknowledges the support from Lush Prize Young Researchers at Americas. P.H. acknowledges support from the National Science Foundation Integrative Graduate Education and Research Traineeship (NSF IGERT) DMR-DGE-1068780 and the American Heart Association Predoctoral Fellowship (AHA 19PRE34380591).
Ethics statement
All experiments using live zebrafish embryos were approved and performed in accordance with IACUC guidelines and regulations as directed by SUNY Upstate Medical University.
Published: May 11, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.stemcr.2021.03.013.
Supplemental information
References
- Ahler E., Sullivan W.J., Cass A., Braas D., York A.G., Bensinger S.J., Graeber T.G., Christofk H.R. Doxycycline alters metabolism and proliferation of human cell lines. PLoS One. 2013;8:e64561. doi: 10.1371/journal.pone.0064561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aikawa N., Kunisato A., Nagao K., Kusaka H., Takaba K., Ohgami K. Detection of thalidomide embryotoxicity by in vitro embryotoxicity testing based on human iPS cells. J. Pharmacol. Sci. 2014;124:201–207. doi: 10.1254/jphs.13162fp. [DOI] [PubMed] [Google Scholar]
- Augustine-rauch K., Zhang C.X., Panzica-kelly J.M. In vitro developmental toxicology assays: a review of the state of the science of rodent and zebrafish whole embryo culture and embryonic stem cell assays. Birth Defects Res. C Embryo Today. 2010;98:87–98. doi: 10.1002/bdrc.20175. [DOI] [PubMed] [Google Scholar]
- Belair D.G., Lu G., Waller L.E., Gustin J.A., Collins N.D., Kolaja K.L. Thalidomide inhibits human iPSC mesendoderm differentiation by modulating CRBN-dependent degradation of SALL4. Sci. Rep. 2020;10:2864. doi: 10.1038/s41598-020-59542-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brassard J.A., Lutolf M.P. Engineering stem cell self-organization to build better organoids. Cell Stem Cell. 2019;24:860–876. doi: 10.1016/j.stem.2019.05.005. [DOI] [PubMed] [Google Scholar]
- Buss W.C., Morgan R., Guttmann J., Barela T., Stalter K. Rifampicin inhibition of protein synthesis in mammalian cells. Science. 1978;200:432 LP–434. doi: 10.1126/science.644307. [DOI] [PubMed] [Google Scholar]
- Chen T.-W., Wardill T.J., Sun Y., Pulver S.R., Renninger S.L., Baohan A., Schreiter E.R., Kerr R.A., Orger M.B., Jayaraman V. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature. 2013;499:295–300. doi: 10.1038/nature12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Xu B., Han X., Mao Z., Chen M., Du G., Talbot P., Wang X., Xia Y. The effects of triclosan on pluripotency factors and development of mouse embryonic stem cells and zebrafish. Arch. Toxicol. 2015;89:635–646. doi: 10.1007/s00204-014-1270-2. [DOI] [PubMed] [Google Scholar]
- Collins M.D., Mao G.E. Teratology of retinoids. Annu. Rev. Pharmacol. Toxicol. 1999;39:399–430. doi: 10.1146/annurev.pharmtox.39.1.399. [DOI] [PubMed] [Google Scholar]
- Czerniecki S.M., Cruz N.M., Harder J.L., Menon R., Annis J., Otto E.A., Gulieva R.E., Islas L.V., Kim Y.K., Tran L.M. High-throughput screening enhances kidney organoid differentiation from human pluripotent stem cells and enables automated multidimensional phenotyping. Cell Stem Cell. 2018;22:929–940.e4. doi: 10.1016/j.stem.2018.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimopoulou M., Verhoef A., Gomes C.A., van Dongen C.W., Rietjens I.M.C.M., Piersma A.H., van Ravenzwaay B. A comparison of the embryonic stem cell test and whole embryo culture assay combined with the BeWo placental passage model for predicting the embryotoxicity of azoles. Toxicol. Lett. 2018;286:10–21. doi: 10.1016/j.toxlet.2018.01.009. [DOI] [PubMed] [Google Scholar]
- Drakhlis L., Biswanath S., Farr C.-M., Lupanow V., Teske J., Ritzenhoff K., Franke A., Manstein F., Bolesani E., Kempf H. Human heart-forming organoids recapitulate early heart and foregut development. Nat. Biotechnol. 2021 doi: 10.1038/s41587-021-00815-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drowley L., McPheat J., Nordqvist A., Peel S., Karlsson U., Martinsson S., Müllers E., Dellsén A., Knight S., Barrett I. Discovery of retinoic acid receptor agonists as proliferators of cardiac progenitor cells through a phenotypic screening approach. Stem Cells Transl. Med. 2020;9:47–60. doi: 10.1002/sctm.19-0069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatehullah A., Tan S.H., Barker N. Organoids as an in vitro model of human development and disease. Nat. Cell Biol. 2016;18:246–254. doi: 10.1038/ncb3312. [DOI] [PubMed] [Google Scholar]
- He J.H., Gao J.M., Huang C.J., Li C.Q. Zebrafish models for assessing developmental and reproductive toxicity. Neurotoxicol. Teratol. 2014;42:35–42. doi: 10.1016/j.ntt.2014.01.006. [DOI] [PubMed] [Google Scholar]
- Hoang P., Wang J., Conklin B.R., Healy K.E., Ma Z. Generation of spatial-patterned early-developing cardiac organoids using human pluripotent stem cells. Nat. Protoc. 2018;13:723–737. doi: 10.1038/nprot.2018.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoberman A.M., Deprospo J.R., Lochry E.A., Christian M.S. Developmental toxicity study of orally administered lithium hypochlorite in rats. Int. J. Toxicol. 1990;9:367–379. [Google Scholar]
- Hookway T.A., Matthys O.B., Mendoza-Camacho F.N., Rains S., Sepulveda J.E., Joy D.A., Mcdevitt T.C. Phenotypic variation between stromal cells differentially impacts engineered cardiac tissue function. Tissue Eng. - Part A. 2019;25:773–785. doi: 10.1089/ten.tea.2018.0362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C.J., Tu C.T., Hsiao C. Der, Hsieh F.J., Tsai H.J. Germ-line transmission of a myocardium-specific GFP transgene reveals critical regulatory elements in the cardiac myosin light chain 2 promoter of zebrafish. Dev. Dyn. 2003;228:30–40. doi: 10.1002/dvdy.10356. [DOI] [PubMed] [Google Scholar]
- Huebsch N., Loskill P., Mandegar M.A., Marks N.C., Sheehan A.S., Ma Z., Mathur A., Nguyen T.N., Yoo J.C., Judge L.M. Automated video-based analysis of contractility and calcium flux in human-induced pluripotent stem cell-derived cardiomyocytes cultured over different spatial scales. Tissue Eng. Part C Methods. 2015;21:467–479. doi: 10.1089/ten.tec.2014.0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huebsch N., Loskill P., Deveshwar N., Spencer C.I., Judge L.M., Mandegar M.A., Fox C.B., Mohamed T.M.A., Ma Z., Mathur A. Miniaturized iPS-cell-derived cardiac muscles for physiologically relevant drug response analyses. Sci. Rep. 2016;6:24726. doi: 10.1038/srep24726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang N.S., Varghese S., Elisseeff J. Controlled differentiation of stem cells. Adv. Drug Deliv. Rev. 2008;60:199–214. doi: 10.1016/j.addr.2007.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue A., Nishimura Y., Matsumoto N., Umemoto N., Shimada Y., Maruyama T., Kayasuga K., Morihara M., Katagi J., Shiroya T. Comparative study of the zebrafish embryonic toxicity test and mouse embryonic stem cell test to screen developmental toxicity of human pharmaceutical drugs. Fundam. Toxicol. Sci. 2016;3:79–87. [Google Scholar]
- de Jong E., Barenys M., Hermsen S.A.B., Verhoef A., Ossendorp B.C., Bessems J.G.M., Piersma A.H. Comparison of the mouse embryonic stem cell test, the rat whole embryo culture and the zebrafish embryotoxicity test as alternative methods for developmental toxicity testing of six 1,2,4-triazoles. Toxicol. Appl. Pharmacol. 2011;253:103–111. doi: 10.1016/j.taap.2011.03.014. [DOI] [PubMed] [Google Scholar]
- Kolanowski T.J., Busek M., Schubert M., Dmitrieva A., Binnewerg B., Pöche J., Fisher K., Schmieder F., Grünzner S., Hansen S. Enhanced structural maturation of human induced pluripotent stem cell-derived cardiomyocytes under a controlled microenvironment in a microfluidic system. Acta Biomater. 2020;102:273–286. doi: 10.1016/j.actbio.2019.11.044. [DOI] [PubMed] [Google Scholar]
- Kumar K.K., Aboud A.A., Bowman A.B. The potential of induced pluripotent stem cells as a translational model for neurotoxicological risk. Neurotoxicology. 2012;33:518–529. doi: 10.1016/j.neuro.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster M.A., Corsini N.S., Wolfinger S., Gustafson E.H., Phillips A.W., Burkard T.R., Otani T., Livesey F.J., Knoblich J.A. Guided self-organization and cortical plate formation in human brain organoids. Nat. Biotechnol. 2017;35:659–666. doi: 10.1038/nbt.3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lantz-McPeak S., Guo X., Cuevas E., Dumas M., Newport G.D., Ali S.F., Paule M.G., Kanungo J. Developmental toxicity assay using high content screening of zebrafish embryos. J. Appl. Toxicol. 2015;35:261–272. doi: 10.1002/jat.3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee L.M.Y., Leung C.-Y., Tang W.W.C., Choi H.-L., Leung Y.-C., McCaffery P.J., Wang C.-C., Woolf A.S., Shum A.S.W. A paradoxical teratogenic mechanism for retinoic acid. Proc. Natl. Acad. Sci. 2012;109:13668 LP–13673. doi: 10.1073/pnas.1200872109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P.-Y., Chang Y.-C., Tzang B.-S., Chen C.-C., Liu Y.-C. Antibiotic amoxicillin induces DNA lesions in mammalian cells possibly via the reactive oxygen species. Mutat. Res. 2007;629:133–139. doi: 10.1016/j.mrgentox.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Lian X., Hsiao C., Wilson G., Zhu K., Hazeltine L.B., Azarin S.M., Raval K.K., Zhang J., Kamp T.J., Palecek S.P. Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling. Proc. Natl. Acad. Sci. U S A. 2012;109:E1848–E1857. doi: 10.1073/pnas.1200250109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Van Bortle K., Zhang Y., Zhao M.T., Zhang J.Z., Geller B.S., Gruber J.J., Jiang C., Wu J.C., Snyder M.P. Disruption of mesoderm formation during cardiac differentiation due to developmental exposure to 13-cis-retinoic acid. Sci. Rep. 2018;8:1–11. doi: 10.1038/s41598-018-31192-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z., Wang J., Loskill P., Huebsch N., Koo S., Svedlund F.L., Marks N.C., Hua E.W., Grigoropoulos C.P., Conklin B.R. Self-organizing human cardiac microchambers mediated by geometric confinement. Nat. Commun. 2015;6:7413. doi: 10.1038/ncomms8413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandegar M.A., Huebsch N., Frolov E.B., Shin E., Truong A., Olvera M.P., Chan A.H., Miyaoka Y., Holmes K., Spencer C.I. CRISPR interference efficiently induces specific and reversible gene silencing in human iPSCs. Cell Stem Cell. 2016;18:541–553. doi: 10.1016/j.stem.2016.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manfrin A., Tabata Y., Paquet E.R., Vuaridel A.R., Rivest F.R., Naef F., Lutolf M.P. Engineered signaling centers for the spatially controlled patterning of human pluripotent stem cells. Nat. Methods. 2019;16:640–648. doi: 10.1038/s41592-019-0455-2. [DOI] [PubMed] [Google Scholar]
- Morgani S.M., Metzger J.J., Nichols J., Siggia E.D., Hadjantonakis A.K. Micropattern differentiation of mouse pluripotent stem cells recapitulates embryo regionalized cell fate patterning. eLife. 2018;7:1–35. doi: 10.7554/eLife.32839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes S.S., Miklas J.W., Liu J., Aschar-Sobbi R., Xiao Y., Zhang B., Jiang J., Massé S., Gagliardi M., Hsieh A. Biowire: a platform for maturation of human pluripotent stem cell-derived cardiomyocytes. Nat. Methods. 2013;10:781–787. doi: 10.1038/nmeth.2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogle B.M., Bursac N., Domian I., Huang N.F., Menasché P., Murry C.E., Pruitt B., Radisic M., Wu J.C., Wu S.M. Distilling complexity to advance cardiac tissue engineering. Sci. Transl. Med. 2016;8:342ps13. doi: 10.1126/scitranslmed.aad2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer J.A., Smith A.M., Egnash L.A., Colwell M.R., Donley E.L.R., Kirchner F.R., Burrier R.E. A human induced pluripotent stem cell-based in vitro assay predicts developmental toxicity through a retinoic acid receptor-mediated pathway for a series of related retinoid analogues. Reprod. Toxicol. 2017;73:350–361. doi: 10.1016/j.reprotox.2017.07.011. [DOI] [PubMed] [Google Scholar]
- Pamies D., Martínez C.E., Sogorb M.A., Vilanova E. Elsevier Inc.; 2011. Mechanism-Based Models in Reproductive and Developmental Toxicology. [Google Scholar]
- Periasamy M., Janssen P.M.L. Molecular basis of diastolic dysfunction. Heart Fail. Clin. 2008;4:13–21. doi: 10.1016/j.hfc.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piersma A.H., Hessel E.V., Staal Y.C. Retinoic acid in developmental toxicology: teratogen, morphogen and biomarker. Reprod. Toxicol. 2017;72:53–61. doi: 10.1016/j.reprotox.2017.05.014. [DOI] [PubMed] [Google Scholar]
- Richards D.J., Coyle R.C., Tan Y., Jia J., Wong K., Toomer K., Menick D.R., Mei Y. Inspiration from heart development: biomimetic development of functional human cardiac organoids. Biomaterials. 2017;142:112–123. doi: 10.1016/j.biomaterials.2017.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivron N.C., Frias-Aldeguer J., Vrij E.J., Boisset J.C., Korving J., Vivié J., Truckenmüller R.K., Van Oudenaarden A., Van Blitterswijk C.A., Geijsen N. Blastocyst-like structures generated solely from stem cells. Nature. 2018;557:106–111. doi: 10.1038/s41586-018-0051-0. [DOI] [PubMed] [Google Scholar]
- Rodrigues I.C.P., Kaasi A., Maciel Filho R., Jardini A.L., Gabriel L.P. Cardiac tissue engineering: current state-of-the-art materials, cells and tissue formation. Einstein (Sao Paulo). 2018;16:eRB4538. doi: 10.1590/S1679-45082018RB4538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronaldson-Bouchard K., Ma S.P., Yeager K., Chen T., Song L.J., Sirabella D., Morikawa K., Teles D., Yazawa M., Vunjak-Novakovic G. Advanced maturation of human cardiac tissue grown from pluripotent stem cells. Nature. 2018;556:239–243. doi: 10.1038/s41586-018-0016-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarmah S., Marrs J.A. Zebrafish as a vertebrate model system to evaluate effects of environmental toxicants on cardiac development and function. Int. J. Mol. Sci. 2016;17:2123. doi: 10.3390/ijms17122123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiler A.E.M., Spielmann H. The validated embryonic stem cell test to predict embryotoxicity in vitro. Nat. Protoc. 2011;6:961–978. doi: 10.1038/nprot.2011.348. [DOI] [PubMed] [Google Scholar]
- Shaikh Qureshi W.M., Latif M.L., Parker T.L., Pratten M.K. Lithium carbonate teratogenic effects in chick cardiomyocyte micromass system and mouse embryonic stem cell derived cardiomyocyte—possible protective role of myo-inositol. Reprod. Toxicol. 2014;46:106–114. doi: 10.1016/j.reprotox.2014.03.009. [DOI] [PubMed] [Google Scholar]
- Shao Y., Sang J., Fu J. On human pluripotent stem cell control: the rise of 3D bioengineering and mechanobiology. Biomaterials. 2015;52:26–43. doi: 10.1016/j.biomaterials.2015.01.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strikwold M., Woutersen R.A., Spenkelink B., Punt A., Rietjens I.M.C.M. Relative embryotoxic potency of p-substituted phenols in the embryonic stem cell test (EST) and comparison to their toxic potency in vivo and in the whole embryo culture (WEC) assay. Toxicol. Lett. 2012;213:235–242. doi: 10.1016/j.toxlet.2012.07.005. [DOI] [PubMed] [Google Scholar]
- Tandon S., Jyoti S. Embryonic stem cells: an alternative approach to developmental toxicity testing. J. Pharm. Bioallied Sci. 2012;4:96–100. doi: 10.4103/0975-7406.94808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbull I.C., Mayourian J., Murphy J.F., Stillitano F., Ceholski D.K., Costa K.D. Cardiac tissue engineering models of inherited and acquired cardiomyopathies. Methods Mol. Biol. 2018;1816:145–159. doi: 10.1007/978-1-4939-8597-5_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warmflash A., Sorre B., Etoc F., Siggia E.D., Brivanlou A.H. A method to recapitulate early embryonic spatial patterning in human embryonic stem cells. Nat. Methods. 2014;11:847–854. doi: 10.1038/nmeth.3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber T., Auer J., O’Rourke M.F., Punzengruber C., Kvas E., Eber B. Prolonged mechanical systole and increased arterial wave reflections in diastolic dysfunction. Heart. 2006;92:1616–1622. doi: 10.1136/hrt.2005.084145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T., Wu L., Xia M., Simeonov A., Huang R. Systematic identification of molecular targets and pathways related to human organ level toxicity. Chem. Res. Toxicol. 2020 doi: 10.1021/acs.chemrestox.0c00305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Xue X., Shao Y., Wang S., Esfahani S.N., Li Z., Muncie J.M., Lakins J.N., Weaver V.M., Gumucio D.L. Controlled modelling of human epiblast and amnion development using stem cells. Nature. 2019;573:421–425. doi: 10.1038/s41586-019-1535-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data requests and correspondence should be addressed to Z.M.