Summary
Sequestosome-1 (SQSTM1/p62) is involved in cellular processes such as autophagy and metabolic reprogramming. Mutations resulting in the loss of function of SQSTM1 lead to neurodegenerative diseases including frontotemporal dementia. The pathogenic mechanism that contributes to SQSTM1-related neurodegeneration has been linked to its role as an autophagy adaptor, but this is poorly understood, and its precise role in mitochondrial function and clearance remains to be clarified. Here, we assessed the importance of SQSTM1 in human induced pluripotent stem cell (iPSC)-derived cortical neurons through the knockout of SQSTM1. We show that SQSTM1 depletion causes altered mitochondrial gene expression and functionality, as well as autophagy flux, in iPSC-derived neurons. However, SQSTM1 is not essential for mitophagy despite having a significant impact on early PINK1-dependent mitophagy processes including PINK1 recruitment and phosphorylation of ubiquitin on depolarized mitochondria. These findings suggest that SQSTM1 is important for mitochondrial function rather than clearance.
Keywords: iPSC disease modeling, FTD, SQSTM1, mitochondria
Highlights
-
•
SQSTM1 is dispensable for cortical neuron differentiation, modeled with human iPSCs
-
•
Expression of bioenergetic genes is altered in human cortical neurons lacking SQSTM1
-
•
Loss of SQSTM1 causes aberration in mitochondrial functionality
-
•
SQSTM1 affects mitophagic processes but is not required for mitochondrial clearance
Sequestosome-1 (SQSTM1/p62) deficiency leads to several neurodegenerative diseases wherein mitochondrial dysfunction is observed in neurons. Whether SQSTM1 contributes to pathogenesis by playing a role in mitochondrial function and clearance remains controversial. Poon and colleagues generated cortical neurons depleted of SQSTM1 from human induced pluripotent stem cells and demonstrated that SQSTM1 is a regulator of mitochondrial respiratory capacity but is dispensable for neuronal mitophagy.
Introduction
SQSTM1 is a multifunctional protein and an important regulator of cellular homeostasis whereby it is involved in a wide range of cellular processes including ubiquitin-mediated autophagy, oxidative stress response, and metabolic reprogramming (Sanchez-Martin and Komatsu, 2018). Loss of SQSTM1 can lead to childhood- or adolescence-onset neurodegenerative disorder (Haack et al., 2016; Muto et al., 2018), frontotemporal dementia (FTD) (Le Ber et al., 2013), and amyotrophic lateral sclerosis (ALS) (Fecto et al., 2011; Hirano et al., 2013; Rubino et al., 2012). The pathogenic mechanism that contributes to SQSTM1-related neurodegeneration remains poorly understood, and patients with mutations in SQSTM1 are rare (∼1% of all ALS and up to 3% of all FTD cases) (Pytte et al., 2020; Rubino et al., 2012; van der Zee et al., 2014). Previous reports have implied mitochondrial dysfunction in the pathophysiological events associated with SQSTM1 deficiency. However, data supporting the mitochondrial role of SQSTM1 have been inconsistent and appear to be cell-type specific (Bartolome et al., 2017; Calvo-Garrido et al., 2019). With respect to mitochondrial function, fibroblasts from FTD patients with loss-of-function SQSTM1 mutations (A381V and K238del) display reduced mitochondrial respiration, increased cytosolic reactive oxygen species production, reduced complex 1 activity of the electron transport chain (ETC), and lowered mitochondrial membrane potential (Bartolome et al., 2017). SQSTM1 knockout (KO) mouse models also showed impairment in mitochondrial oxidative phosphorylation (OXPHOS) and reduced ATP production (Seibenhener et al., 2013), whereas overexpression of SQSTM1 rescued ATP production (Seibenhener et al., 2013). However, a study of patients with childhood-onset neurodegenerative disease and loss-of-function SQSTM1 nonsense mutation (p.Arg96∗) reported a mild reduction in ETC enzyme activities and normal mitochondrial ATP production rates in patient-derived neuroepithelial stem cell models (Calvo-Garrido et al., 2019).
There is some evidence that SQSTM1 has a role in mitophagy. In mammalian cells, the mitochondrial kinase PINK1 accumulates selectively at the surface of damaged mitochondria where it phosphorylates ubiquitin and recruits, phosphorylates, and activates the E3 ubiquitin ligase Parkin (PRKN). Shortly after its recruitment, PRKN ubiquitinates a wide range of outer mitochondrial membrane proteins and induces the aggregation and perinuclear localization of mitochondria (Narendra et al., 2010; Vives-Bauza et al., 2010). This leads to the further recruitment of autophagy receptors such as SQSTM1, triggering the engulfment of damaged mitochondria in autophagosomes that ultimately leads to fusion with lysosomes where the targeted mitochondria become degraded (McWilliams and Muqit, 2017). Knockdown of SQSTM1 in HeLa cells overexpressing PRKN was shown to have no effect on PRKN translocation to damaged mitochondria, but resulted in a significant loss of mitochondrial clearance, suggesting that SQSTM1 is an essential adaptor protein for PINK1-PRKN-dependent mitophagy (Geisler et al., 2010). These data were challenged by a later study showing that SQSTM1 is not necessary for selective mitophagy despite SQSTM1 recruitment to depolarized mitochondria (Narendra et al., 2010). Calvo-Garrido et al. (2019) also reported that SQSTM1 is dispensable for mitophagy in fibroblasts from a patient with childhood-onset neurodegenerative disease caused by a nonsense SQSTM1 mutation. They also assessed mitophagy in neurons derived from the patient's fibroblasts, but the role of SQSTM1 in mitochondrial clearance process remained unclear from their neurons as their mitophagy induction protocol did not lead to significant loss of mitochondrial markers following mitochondrial depolarization (Calvo-Garrido et al., 2019).
To determine the role of SQSTM1 in mitochondrial health in neurons, we used CRISPR/Cas9 to generate induced pluripotent stem cell (iPSC) lines with mutated SQSTM1 producing premature termination of translation. We then assessed mitochondrial function in the iPSC-derived cortical neurons with or without SQSTM1 using high-content screening systems. We also established treatment regimens to induce mitochondrial depolarization and biochemical methods to detect mitophagy. Our data suggest that SQSTM1 is not required for cortical neuron differentiation and mitochondria distribution in iPSC-derived neurons. SQSTM1 is, however, a regulator of mitochondrial functionality and modulates expression of genes underlying mitochondrial respiration. In addition, SQSTM1 affects early processes of PINK1-dependent mitophagy but is dispensable for mitochondria clearance.
Results
Generation and validation of human iPSC-derived cortical neurons depleted of SQSTM1
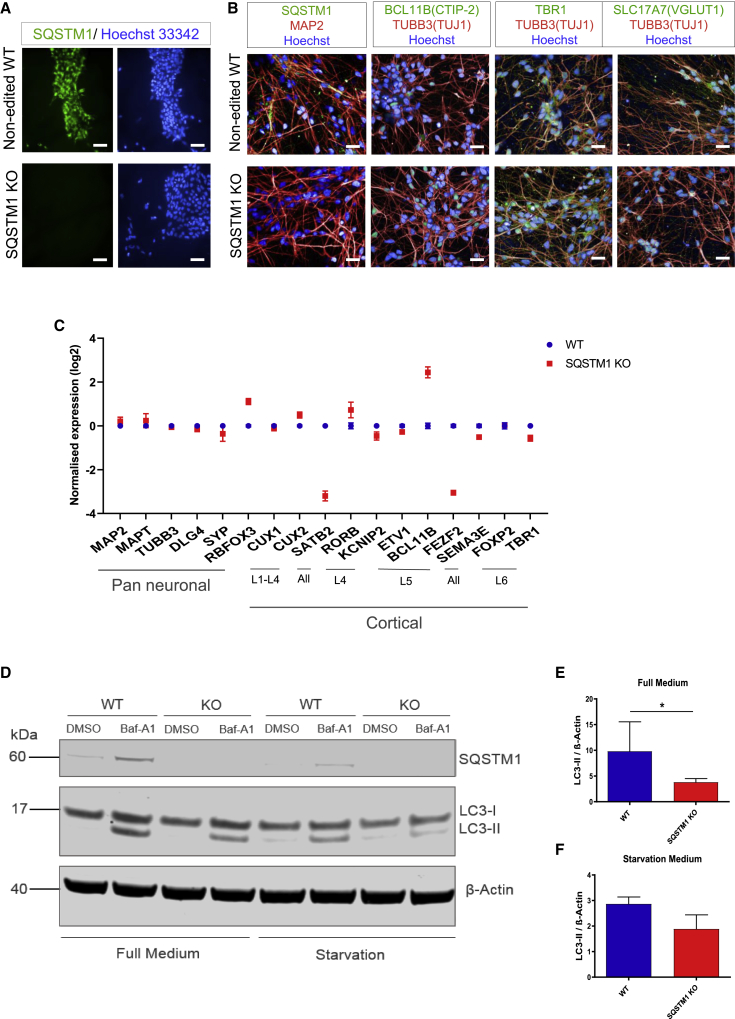
To investigate the effect of SQSTM1 loss of function in human neurons derived from iPSCs, we used CRISPR/Cas9 to generate homozygous SQSTM1 KO iPSC lines on two different genetic backgrounds. The iPSCs were transfected by nucleofection of the CRISPR-Cas9 ribonucleoprotein complexes targeting exon 1 of SQSTM1 followed by single-cell seeding of transfected iPSCs and clonal selection. Homozygous KO state was confirmed with insertion of frameshift mutations and premature termination codons (Figures S1A, S1B, and S1E), leading to nonsense-mediated decay and loss of SQSTM1 protein as shown by immunofluorescence staining (Figure 1A). Genomic integrity of edited iPSC clones was confirmed by karyotype G-banding analysis and array comparative genome hybridization (Figures S1C and S1D).
Figure 1.
Generation and validation of human iPSCs and iPSC-derived cortical neurons depleted of SQSTM1
(A) Representative fluorescence images of SQSTM1 KO and unedited WT iPSCs (RBi001-A) immunostained against SQSTM1 (green) and Hoechst (blue). Scale bars, 50 μm.
(B) Representative fluorescence images of cortical neurons, after 80 days of differentiation, immunostained against SQSTM1 (green), MAP2 (red), CTIP2 (green), TUBB3/TUJ1 (red), TBR1 (green), SLC17A7/VGlut1 (green), and Hoechst (blue). Scale bars, 25 μm.
(C) Relative mRNA expression of markers for cortical brain regions and pan-neuronal genes. Dots indicate expression mean ± SEM of neurons generated from two independent neural differentiation.
(D) Representative western blot analysis of LC3-II under normal or starvation conditions, treated with or without Baf-A1 overnight. β-Actin was used as a loading control.
(E and F) Bar graphs showing the band intensity ratio of LC3-II to β-actin detected in iPSC-derived neurons cultured in full medium (E) or starvation medium (F). Data are expressed as mean ± SEM of three independent experiments. Statistical differences were tested by unpaired two-tailed t test with Welch's correction. ∗p < 0.05.
SQSTM1 KO and isogenic control iPSCs (i.e., unedited wild type [WT]) were subsequently differentiated into cortical neurons (Shi et al., 2012). Cortical neurons generated from the SQSTM1 KO iPSCs were morphologically identical to those derived from the unedited iPSCs, as judged by immunohistochemistry using the neuronal markers TUBB3 (β-III tubulin, also known as TUJ1), MAP2, BCL11B(CTIP-2), TBR1, and SLC17A7 (Figure 1B). The neuronal differentiation capacity between the SQSTM1 KO and WT was also very similar, as reflected at the gene expression level where SQSTM1 KO iPSC-derived neurons expressed the appropriate pan-neuronal genes and the repertoire of markers of the six cortical layers (Klingler, 2017) (Figure 1C). Taken together, these results demonstrate that the differentiation capacity of iPSCs into cortical neurons appears to be unaffected by the loss of SQSTM1.
We further validated our SQSTM1 KO neuronal model by examining whether the depletion of SQSTM1 reduces autophagy flux using LC3-II, as previously reported in immortalized non-neuronal cell lines (van der Zee et al., 2014; Viiri et al., 2010). Neurons were treated in the absence or presence of bafilomycin A1 (Baf-A1), and with or without nutrients including amino acids and glucose (starvation medium). The absence of SQSTM1 resulted in reduction in LC3-II level following Baf-A1 treatment compared with WT cortical neurons (Figure 1D), and this was observed in both normal and starvation medium (Figures 1E and 1F), suggestive of impaired autophagic flux. We confirmed our findings in another set of KO and isogenic control lines derived from a different background (Figures S2A–S2C).
Depletion of SQSTM1 does not affect mitochondrial distribution but alters OXPHOS-related gene expression
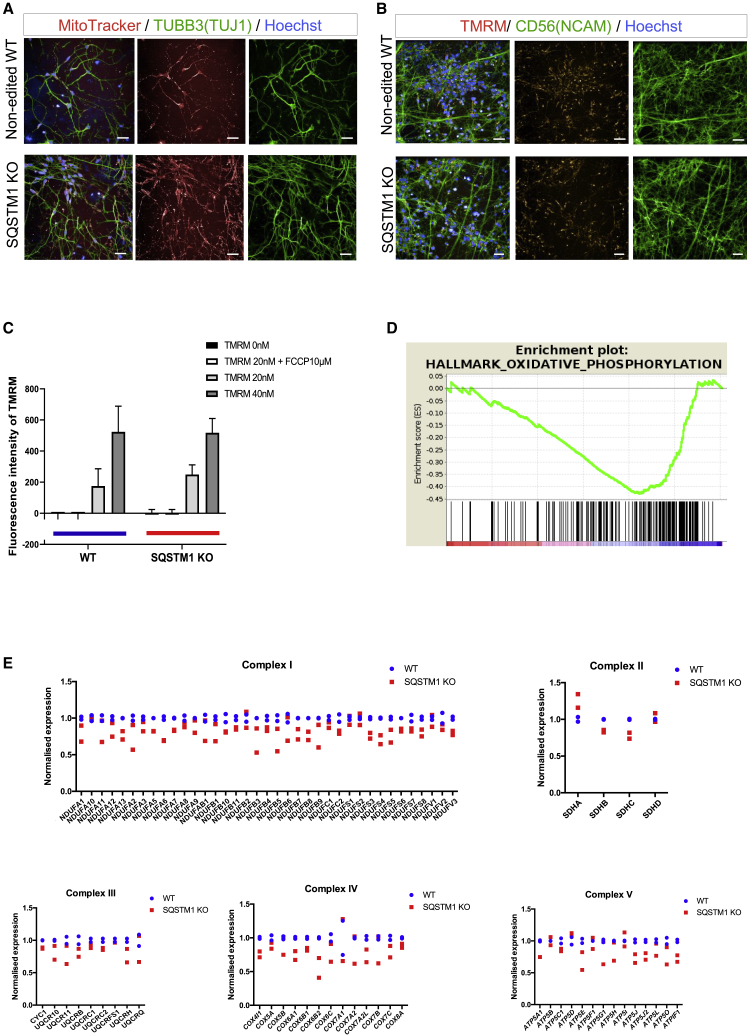
There is growing evidence supporting the correlation between SQSTM1 expression and mitochondrial function (Bartolome et al., 2017; Seibenhener et al., 2013). To examine the effects of SQSTM1 depletion on mitochondrial functionality, we first visualized mitochondria localization in SQSTM1 KO and WT cortical neurons using a MitoTracker Deep Red FM, a far red-fluorescent dye that stains mitochondria in live cells. Mitochondria from neurons depleted of SQSTM1 appeared to be distributed throughout the neurons at levels similar to WT (Figure 2A). Normal distribution of mitochondria in KO neurons was confirmed with immunocytochemistry and the anti-mitochondrial protein TOMM 20 (Figure S2D). We further assessed mitochondrial health, which is reflected in the mitochondrial membrane potential, using tetramethylrhodamine methyl ester (TMRM), an orange fluorescent dye that is sequestered by active mitochondria with intact membrane potentials (Figure 2B). Neurons were treated with two different concentrations of TMRM dye and exhibited similar proportional increase in TMRM fluorescence irrespective of genotype, indicating that basal mitochondrial membrane potential was not altered in the absence of SQSTM1 (Figure 2C). Addition of 10 μM mitochondrial uncoupler carbonyl cyanide-p-trifluoromethoxyphenylhydrazone (FCCP) resulted in complete mitochondrial depolarization, as shown by the lack of TMRM fluorescence signals from both the KO and WT neurons (Figure 2C).
Figure 2.
Depletion of SQSTM1 does not alter mitochondrial distribution and membrane potential but downregulates the expression of mitochondrial respiration-associated genes
(A) Representative fluorescence images of MitoTracker Deep Red FM (red) retention in mitochondria of TUBB3/TUJ1 (green) iPSC-derived neurons in the absence or presence of SQSTM1. Scale bars, 25 μm.
(B) Representative images of tetramethylrhodamine methyl ester (TMRM; orange) and CD56/NCAM (green) live staining in iPSC-derived neurons with or without SQSTM1. Scale bars, 25 μm.
(C) TMRM fluorescence signal intensity was quantified after treating WT and SQSTM1 KO neurons with 0, 20, or 40 nM TMRM and with or without 10 μM uncoupling agent FCCP. Bars represent mean ± SEM of three technical replicates. Statistical difference between SQSTM1 KO and WT was calculated using Welch's t test.
(D) GSEA enrichment plot of the oxidative phosphorylation pathway. The x axis is genes (vertical black lines) and y axis represents enrichment score (ES), which represents the enrichment of the oxidative phosphorylation pathway at the bottom of the ranked gene list. The colored bar at the bottom represents the degree of correlation of genes with the SQSTM1 KO phenotype (red for positive and blue for negative correlation).
(E) Relative mRNA expression of genes that composed the mitochondrial electron transport chain (complexes I–V) in neurons derived with and without SQSTM1. Dots indicate mean datapoints from each of two independent neural differentiations for each line (SQSTM1 KO and WT).
We next examined the impact of SQSTM1 deletion on mitochondrial gene expression and performed RNA sequencing (RNA-seq) on cortical neurons with or without SQSTM1. Based on RNA-seq data, we identified 1,370 and 1,509 genes significantly (corrected p < 0.01) downregulated and upregulated, respectively, in SQSTM1 KO neurons (Figure S2E). Gene set enrichment analysis (GSEA) predicted “oxidative phosphorylation” as significantly enriched in downregulated genes (normalized enrichment score = −1.71 and false discovery rate q value = 0.007) (Figure 2D). Interestingly, the leading-edge genes contributing to the core enrichment for this pathway were enriched in mitochondrial ETC complex genes. Furthermore, 80% of the genes composing the mitochondrial ETC complexes were downregulated in SQSTM KO neurons compared with their WT counterparts. Expression of selective ETC genes was validated in KO neurons derived from iPSCs of a different genetic background using quantitative real-time RT-PCR (Figure S2F). Together, our data indicates that SQSTM1 depletion alters mitochondrial OXPHOS-related gene expression but not mitochondrial distribution and membrane potential.
Loss of SQSTM1 impairs mitochondrial spare respiratory capacity
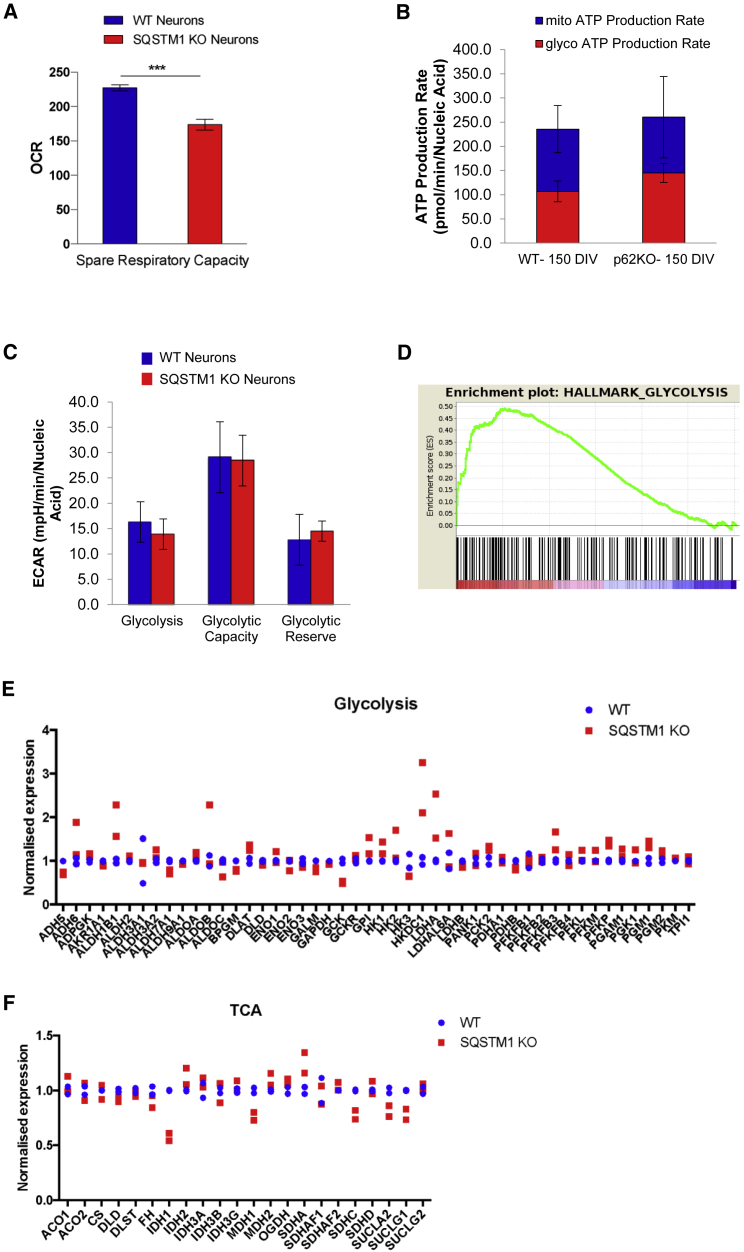
Decreased expression of OXPHOS genes in the absence of SQSTM1 pointed to possible alterations in energy metabolism. We evaluated real-time energy metabolism in SQSTM1 KO and WT cortical neurons using a live cell Mitochondrial Bioenergetics assay (Agilent Seahorse). A small but significant deficit in spare respiratory capacity was detected repeatedly in SQSTM1 KO neurons (Figure 3A; repeated three times over different days). We confirmed the perturbed SQSTM1 KO bioenergetic profile and decreased spare respiratory capacity in a KO line on a different background (Figure S3; repeated three times over different days). We also examined other parameters, such as oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) as indicators of mitochondrial respiration and glycolysis, respectively. However, the observed signal for these readouts was quite variable, making interpretation difficult (Figure S3A). This was less of an issue with our secondary line (UOXFi005-A), for which we did not observe any differences between KO and WT lines for these parameters (Figure S3B).
Figure 3.
Deletion of SQSTM1 has small but significant effect on mitochondrial spare respiratory capacity
(A) Results of spare respiratory capacity from Seahorse XF Mito Stress Test performed on cortical neurons derived from SQSTM1 KO and WT iPSCs (RBi001-A). Data presented as mean ± SEM of three independent experiments and statistical differences were tested by Welch's t test. ∗∗∗p < 0.001.
(B) Seahorse XF Real-Time ATP Rate Assay in iPSC-derived neurons with or without SQSTM1. Basal ATP production rates from mitochondrial (blue) and glycolytic (red) pathways were quantified and are summarized in the stacked bar chart. Data are presented as the mean ± SEM for eight technical replicates, and statistical differences were tested by Welch's t test.
(C) Results of Seahorse XF Glycolysis Stress Test from cortical neurons derived with or without SQSTM1 KO. Individual parameters of glycolytic function including basal glycolysis, glycolytic capacity, and glycolytic reserve, were obtained from the extracellular acidification rates (ECAR). Data are presented as mean ± SEM for six technical replicates, and statistical differences were tested by Welch's t test.
(D) GSEA enrichment plot of the glycolysis pathway. The x axis is genes (vertical black lines) and y axis represents enrichment score (ES), which represents the enrichment of the glycolysis pathway at the top of the ranked gene list. The colored bar at the bottom represents the degree of correlation of genes with the SQSTM1 KO phenotype (red for positive and blue for negative correlation).
(E) Relative mRNA expression of glycolytic genes in neurons derived from SQSTM1 KO and WT iPSCs (RBi001-A). Dots indicate mean datapoints from each of two independent neural differentiations for each line (SQSTM1 KO and WT).
(F) Relative mRNA expression of TCA genes in neurons derived from SQSTM1 KO and WT iPSCs (RBi001-A). Dots indicate mean datapoints from each of two independent neural differentiations for each line (SQSTM1 KO and WT).
Spare respiratory capacity, an indicator of cell fitness and cell's ability to response to increased energy demand, was also assessed. To further support the observation that basal levels of ATP were unaffected in the absence of SQSTM1, we performed an ATP rate assay for real-time kinetic quantification of ATP production from mitochondria and glycolysis simultaneously. No significant differences in mitochondrial and glycolysis-mediated ATP production rates were observed between the SQSTM1 KO and WT cortical neurons (Figure 3B). We also assessed key parameters of glycolytic flux including basal glycolysis, glycolytic capacity, and glycolytic reserve in our SQSTM1 KO neurons and detected no difference in these parameters when compared with their WT counterparts (Figure 3C). At the transcriptional level, we found enrichment of glycolysis in upregulated genes from GSEA (Figure 3D). One of the differentially expressed genes was lactate dehydrogenase A (LDHA), which catalyzes the conversion of pyruvate into lactate, and overexpression of this gene was reported to promote glycolysis and inhibit mitochondrial respiration (Figure 3E) (Yetkin-Arik et al., 2019). Upregulation of LDHA was detected in SQSTM1 KO neurons, and this was also observed by Calvo-Garrido et al. (2019) in SQSTM1-deficient patients' lines. Also, selective genes that are related to the tricarboxylic acid (TCA) cycle were downregulated in KO neurons compared with WT (Figure 3F). One of TCA genes, isocitrate dehydrogenase 1 (IDH1), has been previously shown to regulate mitochondrial spare respiratory capacity (Izquierdo-Garcia et al., 2015). Taken together, these data indicate that the loss of SQSTM1 does not inhibit glycolysis and mitochondrial respiration in iPSC-derived cortical neurons. Depletion of SQSTM1, however, diminishes spare respiratory capacity, which is regarded as an important aspect of mitochondrial function. This suggests that cortical neurons depend on SQSTM1 to ramp up mitochondrial respiration in order to meet the higher energy demand under stress.
SQSTM1 regulates the PINK1 mitophagy process but is not essential for the clearance of damaged mitochondria
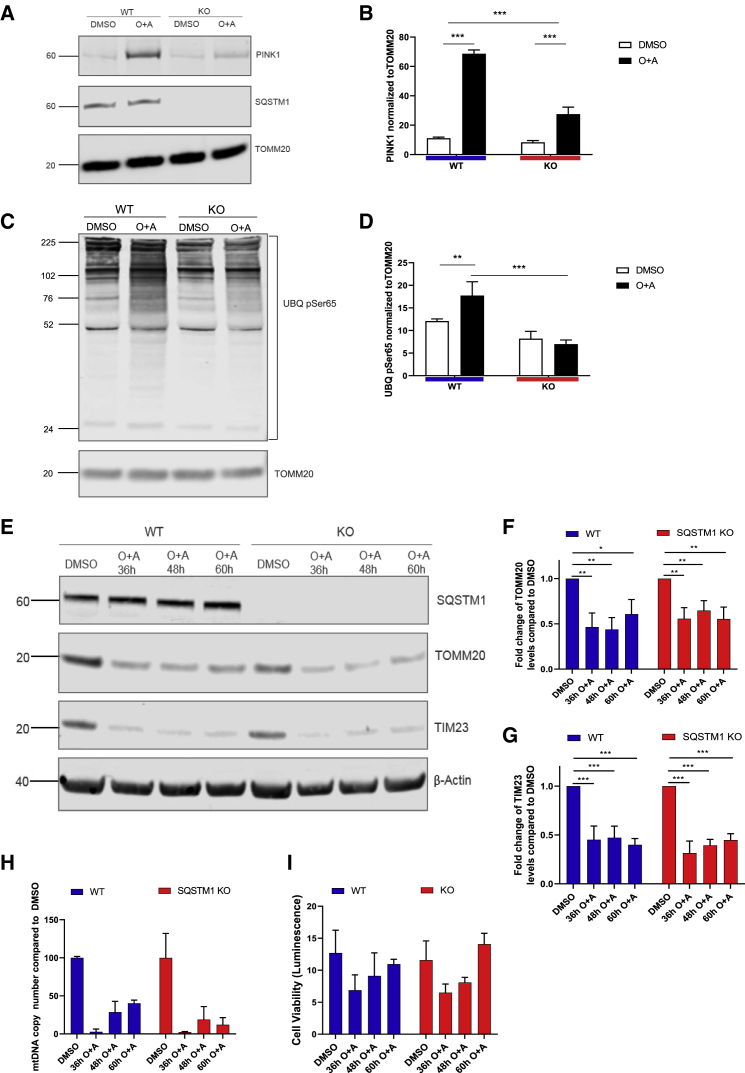
As autophagy flux and mitochondrial spare respiratory capacity were affected in SQSTM1 KO neurons, we next sought to determine whether SQSTM1 regulates mitophagy. Oligomycin and antimycin A (O + A, both at 1 μM) were added to iPSC-derived neuronal cultures to induce mitochondrial depolarization and mitophagy. After 16 h of O + A treatment, a reduced amount of PINK1 was detected in mitochondria-enriched fractions of SQSTM1 KO neurons compared with their WT counterparts (Figures 4A and 4B). PINK1 has been previously reported to phosphorylate ubiquitin specifically at Ser65 (UBQ pSer65) on depolarized mitochondria and activate PRKN, which then triggers the mitophagy pathway to clear damaged mitochondria (Kazlauskaite et al., 2014; Okatsu et al., 2015; Shiba-Fukushima et al., 2014; Soutar et al., 2018). Using western blotting, we showed that diminished PINK1 accumulation at mitochondria resulted in a reduced level of ubiquitinated mitochondrial-enriched proteins containing UBQ pSer65 compared with WT (Figures 4C and 4D). To further determine whether SQSTM1 regulates mitophagy, we assessed mitochondrial clearance 36–60 h after O + A treatment. Mitochondrial clearance was demonstrated by the reduced levels of mitochondrial markers, TOMM20 and TIM23, which are the respective translocases of the outer and inner mitochondrial membranes, when comparing O + A treatment groups with dimethyl sulfoxide (DMSO) controls (Figure 4E). Western blotting showed similar reduction in the levels of mitochondrial markers in SQSTM1 KO and WT cortical neurons following O + A treatments compared with DMSO controls (Figures 4F and 4G). These data were corroborated in KO and WT neurons derived from iPSCs of a different genetic background (Figure S4).
Figure 4.
SQSTM1 is not required for mitophagy in iPSC-derived cortical neurons
(A) Immunoblots probed for mitochondrial components PINK1, SQSTM1, and TOMM20 after cells were treated with 1 μM oligomycin and antimycin A.
(B) Bar graph displays quantified images showing PINK1 normalized to TOMM20 (loading control). Data are expressed as mean ± SEM of three independent experiments. Statistical differences were tested by two-tailed t test. ∗∗∗p < 0.001.
(C) Immunoblot for Ser65 (UBQ pSer65) and TOMM20 (using the same samples as examined in A).
(D) Bar graph displays quantified images showing UBQ pSer65 normalized to TOMM20 (loading control). Data are expressed as mean ± SEM of three independent experiments. Statistical differences were tested by two-tailed t test. ∗∗∗p < 0.001.
(E) Immunoblot for TOMM20, TIM23, SQSTM1, and β-actin from neurons treated for 36–60 h with 1 μM oligomycin and 1 μM antimycin A (or DMSO as control).
(F and G) Bar graphs display quantified images showing fold change versus DMSO control of TOMM20 and TIM23 versus β-actin loading control. Data are expressed as mean ± SEM of three independent experiments. Statistical differences were tested by two-tailed t test. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
(H) Quantitative mitochondrial DNA copy number in neurons, treated for 36–60 h with 1 μM oligomycin and 1 μM antimycin A (or DMSO as control), as determined by real-time PCR. Data are expressed as the mean ratio of mitochondrial-to-nuclear genes ± SEM of three technical replicates.
(I) Cellular viability in neurons, treated for 36–60 h with 1 μM oligomycin and 1 μM antimycin A (or DMSO as control), analyzed luminometrically using the RealTime-Glo MT Cell Viability Assay. Data are expressed as mean ± SEM of three technical replicates.
We further confirmed these observations by measuring the level of mitochondrial DNA relative to the nuclear DNA in neurons following O + A treatments and observed a similar pattern of decrease in mitochondrial DNA levels in SQSTM1 KO and WT neurons (Figure 4H). In addition, no difference in cell viability was observed between the SQSTM1 KO and WT neurons following different durations of O + A treatments as assessed by the RealTime-Glo MT Cell Viability Assay (Figure 4I). Altogether, these data demonstrate that although SQSTM1 regulates PINK1 accumulation at depolarized mitochondria, it may not be essential for mitochondrial protein degradation following mitochondrial depolarization. This suggests an additional mechanism(s) underlying damaged mitochondrial clearance in cortical neurons, independent of SQSTM1 and PINK1.
Discussion
Loss-of-function mutations in SQSTM1 are associated with several neurodegenerative disorders, and mitochondrial dysfunction has been implicated in the pathogenesis of these disorders (Haack et al., 2016; Le Ber et al., 2013; Muto et al., 2018; Pytte et al., 2020). Previous research has suggested that SQSTM1 acts as a regulator of mitochondrial function and clearance (Bartolome et al., 2017; Geisler et al., 2010; Seibenhener et al., 2013). However, there have been some inconsistent reports from different cell lines regarding the exact role of SQSTM1 (Calvo-Garrido et al., 2019; Narendra et al., 2010). In this study, we focused on the effects of SQSTM1 on mitochondrial homeostasis in human iPSC-based neuronal systems, as these may be a closer approximation to the biological process in patients.
Our data indicate that SQSTM1 is a modulator of neuronal bioenergetics, whereby the loss of SQSTM1 alters the expression of several metabolic genes and reduced spare respiratory capacity. Spare respiratory capacity, also known as reserve respiratory capacity, is the cell's ability to generate extra ATP via oxidative phosphorylation in case of a sudden increase in energy demand and to overcome stress such as oxidative stress (Desler et al., 2012; Hill et al., 2009). Decline in spare respiratory capacity has been associated with aging and neurodegenerative diseases (Desler et al., 2012). Here we demonstrated that SQSTM1 modulates spare respiratory capacity in cortical neurons, which are the cells most affected in FTD. Depletion of spare respiratory capacity upon SQSTM1 loss can sensitize the neurons to surges in ATP demand and decrease the ability to cope with oxidative stress, and may increase the risk of cell death if this ATP demand is not met (Desler et al., 2012). Although the mechanism by which SQSTM1 exerts its effect on neuronal bioenergetics is not fully known, our RNA-seq analysis and GSEA revealed that metabolic pathways such as OXPHOS and glycolysis are perturbed by loss of SQSTM1 in iPSC-derived cortical neurons. Furthermore, our bioinformatics analysis indicates that SQSTM1 affects the transcription of the majority of the ETC genes involved in mitochondrial respiration and spare respiratory capacity (Desler et al., 2012). Other metabolic genes associated with mitochondrial spare respiratory capacity such IDH1 and LDHA were also differentially expressed in the SQSTM1 KO compared with WT neurons. IDH is an enzyme involved in the citric acid cycle in intermediary metabolism. A reduced level of IDH1, as detected in the SQSTM1 KO neurons, has been previously shown to lower spare respiratory capacity by decreasing pyruvate dehydrogenase activity and reprogramming pyruvate metabolism (Izquierdo-Garcia et al., 2015). The expression of the pro-glycolytic gene LDHA was found to be elevated in the SQSTM1 KO neurons in our study and in a similar KO system established by Calvo-Garrido et al. (2019). The overexpression of LDHA is expected to downregulate mitochondrial spare respiratory capacity. based on previous findings by Yetkin-Arik et al. (2019). The expression of LDHA and several other metabolic genes are regulated by the mammalian target of rapamycin (mTOR) pathway and the nuclear factor κB (NF-κB) signaling pathway (Albensi, 2019; Manerba et al., 2018). SQSTM1 may exert its transcriptional control via direct interaction with key regulators of mTOR and NF-κB signaling pathways including Raptor and RIP1/TRAF6, respectively (Puissant et al., 2012). In the future, it will be interesting to find out whether the SQSTM1 iPSCs differentiated to different lineages display similar vulnerabilities, or are specific to cells of a specific faith.
In addition to the transcriptional regulatory mechanisms associated with SQSTM1, we also examined the role of SQSTM1 in mitophagy. We focused on PINK1-dependent mitophagy, where the mechanism involving PINK1 and PRKN is the best characterized. While the removal of SQSTM1 did influence autophagy, the rate of mitochondrial clearance upon induction of mitochondrial damage was unaffected between SQSTM1 KO and WT neurons. This finding is consistent with observations from Calvo-Garrido et al. (2019) in their neuronal models depleted of SQSTM1. However, the earlier stages of the mitophagy process seemed to be have been affected by the loss of SQSTM1. These include reduced accumulation of PINK1 and the subsequent PINK1-mediated phosphorylation of ubiquitin on depolarized mitochondria in SQSTM1 KO compared with WT neurons. SQSTM1 is not the only autophagy receptor associated with PINK1-mediated mitophagy. To date, four other autophagy receptors have been identified: optineurin (OPTN), neighbor of BRCA1 gene 1 (NBR1), calcium-binding and coiled-coil domain-containing protein 2 (CALCOCO2, NDP52), and Tax1-binding protein 1 (TAX1BP1). Like SQSTM1, these receptors have a ubiquitin binding domain that binds to polyubiquitinated proteins on the mitochondrial surface, and an LC3-interacting region (LIR) motif that binds LC3-II present on the autophagosome structures. CALCOCO2 and OPTN appear to play a more central role in mitophagy than SQSTM1 (Lazarou et al., 2015; Yamano et al., 2020). In addition to autophagy adaptors, mitophagy can be mediated by proteins in the outer mitochondrial membrane such as B cell lymphoma 2 nineteen kilodalton interacting protein 3 (BNIP3), its analog Nix (BNIP3L), and Bcl-2-like protein 13 (Bcl2-L-13) (Murakawa et al., 2015; Schweers et al., 2007; Zhang and Ney, 2009). These proteins contain the LIR motif, which allows them to serve as mitochondrial membrane receptors bringing the mitochondria to the autophagosome structures for degradation (Murakawa et al., 2015; Rogov et al., 2017). It is possible that these receptors play a compensatory role in mitophagy in the absence of SQSTM1. Future work is also needed to elucidate the consequence of reduction in PINK1 accumulation upon SQSTM1 depletion. There is growing evidence for the role of PINK1 in regulating mitochondrial dynamic and function, independent of mitophagy. For instance, PINK1 regulates ETC complex I activity by phosphorylating complex I subunit NdufA10S250 (Morais et al., 2014) and may regulate mitochondrial fission by phosphorylating Drp1S616 (Han et al., 2020). PINK1 and PRKN have also been implicated in the partial degradation of damaged mitochondrial proteins through the formation of mitochondrial-derived vesicles that bud off mitochondria and fuse with lysosomes for degradation (McLelland et al., 2014). Identifying both upstream and downstream signaling mechanisms that trigger PINK1-regulated cellular processes will help explain the nature of the insults affecting mitochondrial function in neurodegenerative diseases with SQSTM1 deficiency.
Taken together, our data highlight the importance of SQSTM1 in mitochondrial function but not mitophagy in the pathophysiological events associated with its deficiency. Our transcriptomics analysis suggests that impairment in multiple components of energy metabolism may be a key mechanism contributing to the risk and/or pathophysiology of neurodegenerative diseases with SQSTM1 loss-of-function mutations. In addition to insights gained on mitochondrial homeostasis, we also showed that SQSTM1 is not required for neural differentiation, in contrast to what was previously reported by Calvo-Garrido et al. (2019). Discrepancies between our two studies, which may explain some of this, were the use of human iPSCs in our study versus control neuroepithelial-like stem cells in the Calvo-Garrido et al. (2019) study, for the CRISPR/Cas9-mediated knockout of SQSTM1, and the use of the Shi et al. (2012) protocol for differentiation to cerebral cortex neurons in our study (mostly glutamatergic) versus the use of a neuroepithelial-like stem cell protocol in the Calvo-Garrido et al. (2019) study (which produces neurons of predominantly GABAergic phenotype with some glia [Falk et al., 2012]). Thus, it is possible that depending on cellular backgrounds, differentiation method, and time points examined, SQSTM1 KO may display impaired neuronal differentiation. This is potentially an interesting area for future exploration. Nonetheless, in our hands the cortical neurons generated from SQSTM1 KO iPSCs resembled those derived from their WT parents at the morphological and immunohistochemical level at our point of examination, 80 days after differentiation. This result was repeated on a cell line from a different background. Our findings are further supported by in vivo and clinical data. SQSTM1 KO mice are viable with normal neuronal development (Durán et al., 2004, Kwon et al., 2012). In accordance with this, patients with loss-of-function SQSTM1 mutations were reported to have normal brain development, and their disease manifested later in life (Haack et al., 2016; Muto et al., 2018). One common observation in both patients and SQSTM1 KO mice was the accumulation of hyperphosphorylated MAPT (microtubule-associated protein tau) (Haack et al., 2016; Ramesh Babu et al., 2008). SQSTM1 has been previously reported to work in concert with the membrane-damage sensor galectin-8 and CALCOCO2 in promoting the autophagic degradation of seeded MAPT, thereby preventing its propagation throughout the brain (Falcon et al., 2018). Future work should examine other arms of the autophagy pathway and cargo, including MAPT removal mediated by SQSTM1 in human iPSC-derived neurons that have metabolic constraints and vulnerabilities similar to those detected in patients.
Experimental procedures
Cell culture
The control human iPSC lines RBi001-A and UOXFi005-A were obtained from the European Bank for Induced Pluripotent Stem Cells. Human iPSCs were cultured in complete Essential 8 Flex Medium (Thermo Fisher Scientific) on human embryonic stem cell-qualified Matrigel-coated plates (Corning) and passaged at a split ratio of 1:3 as cell clusters using Versene solution (Thermo Fisher Scientific). Expanded iPSCs were banked in complete Essential 8 Flex Medium with 10% DMSO at −150°C.
CRISPR/Cas9-mediated knockout of SQSTM1
The CRISPR-Cas9 crRNA (5′-ACC GTG AAG GCC TAC CTT CT-3′) was designed to target exon 1 of SQSTM1 and synthesized by Integrated DNA Technologies (IDT). The crRNA was mixed with the CRISPR-Cas9 tracrRNA (IDT) in equimolar concentrations at 100 μM. The crRNA/tracrRNA duplex was mixed with the Alt-R S.p. Cas9 Nuclease V3 (IDT) at 120 pmol:104 pmol, respectively, to form the CRISPR-Cas9 ribonucleoprotein complexes. Human iPSCs grown to 80% confluence in complete StemFlex medium were enzymatically dissociated into single cells using StemPro Accutase cell dissociation reagent (Thermo Fisher Scientific). The CRISPR-Cas9 ribonucleoprotein complexes were delivered into the iPSCs using a 4D-Nucleofector System (Lonza). After nucleofection, cells were seeded at 1 million cells per 10-cm dish containing complete StemFlex medium and Revita (1:100, Thermo Fisher Scientific). Following expansion, single clones were manually picked under the EVOS XL Core cell imaging system (Thermo Fisher Scientific) and transferred into individual 96 wells for expansion. Clonal lines were split and cells in one of the 96 wells was treated with 50 μM chloroquine (Cell Signaling Technology) overnight (which caused SQSTM1 to accumulate by blocking the autophagic process). The iPSC clones were then screened for the absence of SQSTM1 by immunohistochemistry using anti-SQSTM1 antibody (Cell Signaling Technology, 88588) and imaged on the Opera Phenix high-content screening system (PerkinElmer). Clones lacking SQSTM1 expression were subsequently confirmed by western blot analysis using a second SQSTM1 antibody (BD Biosciences, 610832) and Sanger sequencing for the presence of insertion of frameshift mutations and premature termination codons. Cytogenetic analyses including G-banding karyotyping and array comparative genome hybridization were performed on the edited and unedited iPSC lines to assess cell quality and the presence of abnormalities (Cell Guidance Systems).
Human iPSC cortical differentiation
Differentiation of iPSCs into forebrain cortical neurons was based on the (Shi et al. (2012) protocol. In brief, iPSCs were grown to 100% confluence and Essential 8 Flex Medium was replaced with neural induction medium, consisting of neural maintenance medium supplemented with 10 μM SB431542 (Tocris) and 1 μM dorsomorphin (Tocris). The neural maintenance medium consisted of neurobasal medium (Thermo Fisher Scientific), DMEM/F-12, GlutaMAX supplement (Thermo Fisher Scientific), N-2 supplement (Thermo Fisher Scientific), 50× B-27 supplement (Thermo Fisher Scientific), 10 mg/mL insulin (Sigma), 50 mM 2-mercaptoethanol (Thermo Fisher Scientific), non-essential amino acids (Thermo Fisher Scientific), 100 nM sodium pyruvate (Sigma), and 200 mM GlutaMAX Supplement (Thermo Fisher Scientific).The neural induction medium was changed daily during which the iPSCs converted into a neural precursors-formed neuroepithelial layer. At days 14 and 21, precursors were passaged using Dispase (STEMCELL Technologies) and plated in Matrigel-coated wells in neural maintenance medium. Neural precursors were dissociated into single cells using Accutase and expanded in neural maintenance medium containing 20 ng/mL human recombinant basic fibroblast growth medium (STEMCELL). For further differentiation into cortical neurons, precursors were seeded at 100,000 cells/cm2 in plates coated with 0.01% poly-L-ornithine (Sigma-Aldrich) and 20 μg/mL laminin (Sigma-Aldrich) and in neural maintenance medium. The final passage was performed at day 35 using Accutase, and neurons were plated at a final density of 150,000 cells/cm2 and maintained in new poly-L-ornithine- and laminin-coated plates with neural maintenance medium until day 80.
Western blotting
Cell lysates were prepared in M-PER mammalian protein extraction reagent (Thermo Fisher Scientific) with cOmplete EDTA-free protease inhibitor cocktail (Roche) and PhosSTOP phosphatase inhibitors (Roche). Pierce BCA protein assay (Thermo Fisher Scientific) was used to quantitate total protein in cell lysates, which were then subjected to western blot according to the NuPAGE technical guide (Thermo Fisher Scientific, manual part no. IM-1001). The primary antibodies used were anti-SQSTM1/p62 (BD Biosciences, 610832), LC3B (Novus Biologicals, NB100-2220), PINK1 (Cell Signaling Technology, 6946), TOMM20 (Thermo Fisher Scientific, PA5-52843), TIM23 (BD Biosciences, 611222), PMPCB (Proteintech, 16064-1-AP), phospho-ubiquitin (Ser65) (Cell Signaling Technology, 37642), β-actin (Abcam, ab8224), and GAPDH (Abcam, ab9485). Membranes were incubated with primary antibodies overnight at 4▫C, washed, and incubated with secondary antibodies, IR dye 800 CW donkey anti-rabbit and IR dye 680 RD donkey anti-mouse (LI-COR), for 1 h at room temperature. Membranes were imaged on LI-COR Odyssey CLx, and Image Studio Lite Ver 5.2 was used to measure band intensities.
Immunocytochemistry
Cells were fixed in Image-iT Fixative Solution (4% formaldehyde, methanol-free) (Thermo Fisher Scientific) for 15 min. After three washes with phosphate-buffered saline (PBS) and one wash with 0.1% Triton X-100 in PBS (PBST), cells were blocked in 1% bovine serum albumin and 5% normal goat serum in PBST. Cells were incubated in primary antibodies in blocking solution overnight at 4°C. The primary antibodies used were anti-SQSTM1antibody (Cell Signaling Technology, 88588), MAP2 (Abcam, ab183830), TBR1 (Abcam, ab31940), TUJ1/ neuron-specific class III β-tubulin (Abcam, ab78078 and ab229590), CTIP2 (Abcam, ab18465), VGLUT1 (Thermo Fisher Scientific, 48-2400), and TOMM20 (Abcam, ab56783). After three washes in PBST, cells were incubated with secondary antibodies, goat anti-mouse/rabbit Alexa Fluor 488/647 (Thermo Fisher Scientific), in blocking solution for 1 h in darkness. Hoechst 33342 was added to the cells as a nuclear counterstain at 0.1 μg/mL and the cells were washed three times in PBS before being imaged on the Opera Phenix high-content screening system (PerkinElmer) at 20× and 40× magnification. To visualize mitochondrial localization, human iPSC-derived cortical neurons with or without SQSTM1 were treated with 50 nM MitoTracker Deep Red FM (Thermo Fisher Scientific) in neurobasal medium minus phenol red (Thermo Fisher Scientific) for 15 min at 37°C, fixed, co-stained with TUJ1, and imaged as detailed above.
Measurement of mitochondrial membrane potential
Human iPSC-derived cortical neurons with or without SQSTM1 were cultured in neurobasal medium (minus phenol red) that contained Alexa Fluor 488 Mouse anti-Human CD56/neural cell adhesion molecule (1:50, BD Biosciences, 557699), neuro background suppressor (1:10, Thermo Fisher Scientific), and 10 μM carbonyl cyanide m-chlorophenyl hydrazone or DMSO for 1 h at 37°C. Neurons were then loaded with 0, 20, or 40 nM tetramethylrhodamine methyl ester perchlorate (TMRM) (Thermo Fisher Scientific) for 30 min at 37°C and counterstained with Hoechst 33342. Cells were washed with pre-warmed neurobasal medium (minus phenol red) with neuro background suppressor twice and imaged with the Opera Phenix high-content screening system (PerkinElmer). Fluorescence signal from TMRM was detected with a tetramethylrhodamine-isothiocyanate filter set (absorbance peak at 548 nm and emission peak at 574 nm). TMRM fluorescence signal intensity was quantified per pixel after thresholding to remove background signal.
Mitochondrial function analysis
Human iPSC-derived cortical precursors with or without SQSTM1 were seeded in XFe96 microplates (Agilent) at a density of 3 × 104 cells/well and matured in culture for 6 weeks. On the day of the assay, the culture medium was replaced with unbuffered XF DMEM medium (pH 7.4) supplemented with 10 mM glucose, 2 mM glutamine, and 1 mM pyruvate. Neurons containing microplates were placed in a non-CO2 incubator at 37°C for 45 min prior to the assay. Mitochondrial function was assessed using the Seahorse XF Cell Mito Stress Test (Agilent). In brief, test of mitochondrial function was initiated by three baseline OCR measurements. Additional OCR measurements were acquired after sequential injections of the 2.5 μM oligomycin, 1 μM carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone/FCCP, and 0.5 μM rotenone and antimycin A. OCR readouts were normalized to the amount of cellular nucleic acid content. After the last OCR measurement, cells in the XFe96 microplate were frozen at −80°C overnight, treated with lysis butter and nucleic acid detection reagents containing green fluorescent dyes from the CyQUANT Cell Proliferation Assay Kit (Thermo Fisher Scientific), and imaged on a fluorescence microplate reader.
ATP analysis
Label-free, real-time kinetic quantification of cellular ATP production rates was obtained using the Seahorse XFe96 extracellular flux analyzer (Agilent). Data were obtained under basal conditions and after serial addition of mitochondrial inhibitors, 1.5 μM oligomycin, and 0.5 μM rotenone + antimycin A. Mitochondrial and glycolytic ATP production rates were determined using the Seahorse XF Real-Time ATP Rate Assay Report Generator (Agilent). Details on the series of calculations used to transform the OCR and ECAR data to ATP production rates are provided in the Seahorse XF Real-Time ATP Rate Assay user guide.
Glycolysis
Key parameters of glycolytic function were assessed using the ECAR readouts from the Seahorse XFe96 extracellular flux analyzer (Agilent). Neurons were incubated in the Seahorse XF Base Medium (with 2 mM glutamine and without glucose or pyruvate). Glucose (10 mM) was injected and the rate of glycolysis under basal conditions measured. The second injection was 1 μM oligomycin (for cellular maximum glycolytic capacity measurement). The final injection was 50 mM 2-deoxyglucose (for glycolytic reserve measurement). Glycolytic reserve was determined by subtracting the ECAR of basal glycolysis from the ECAR of maximum glycolytic capacity.
Assessment of mitophagy
Human iPSC-derived neurons with or without SQSTM1 were treated with 1 μM oligomycin and 1 μM antimycin A (or DMSO as control) in modified neural maintenance medium wherein the original B-27 was replaced with B27 minus antioxidants (Thermo Fisher Scientific). Every 12 h, 50% of the medium was removed and replaced with fresh medium with compounds. Stages of the PINK1 mitophagy process were assessed biochemically following different 1 μM oligomycin and antimycin A treatment durations from 0, 16, 36, 48, and 60 h, whereby cell lysates were collected for downstream experiments including western blotting, measurement of mitochondrial DNA copy number, and cell viability assessment.
Mitochondria were isolated by centrifugation and assessed for PINK1 accumulation and Ser65 phosphorylation of ubiquitinated mitochondrial proteins after 16 h of 1 μM oligomycin and antimycin A treatment. Post-treated cells were washed with PBS before addition of mitochondrial homogenization buffer (250 mM sucrose, 1 mM edetate disodium salt dehydrate/EDTA, 10 mM Tris [pH 7.4]) supplemented with protease and phosphatase inhibitors. Cell plates were then frozen at −80°C overnight and thawed on ice the next day. Cell lysates were then scraped into 1.5-mL centrifuge tubes, triturated 20 times, and frozen again at −80°C for 1h. Thawed cell lysates were centrifuged at 1,500 g for 10 min, and the supernatants (enriched for mitochondria minus cell debris) were transferred into clean centrifuge tubes. Subsequent centrifugation at 12,000 × g for 10 min pelleted the mitochondria, and the pellets were washed twice with mitochondrial homogenization buffer to remove any cytoplasmic contamination. Isolated mitochondria pellets were suspended in 1× NuPAGE LDS sample buffer supplemented with NuPAGE Reducing Agent prior to SDS-PAGE and western blotting.
Measurement of mitochondrial DNA copy number
The relative number of copies of human mitochondrial DNA in iPSC-derived cortical neurons was determined by real-time PCR using the human mitochondrial DNA (mtDNA) Monitoring Primer Set (Takara). Pre-validated primer pairs were supplied for the amplification of four regions: two primer pairs for detecting mtDNA (ND1, ND5) and two primer pairs for detecting nuclear DNA content (SLC O 2B1, SERPINA1). Real-time PCR assays were set up according to the manufacturer's protocol. The Ct values were uploaded to the mtDNA Copy Number Calculation Tool provided by the manufacturer, and the ratio of mtDNA-to-nuclear DNA for each sample was determined.
Cell viability assay
The RealTime-Glo MT Cell Viability Assay (Promega) was used to determine cell viability and was performed according to the manufacturer's protocol.
Whole-transcriptome mRNA sequencing and analysis
RNA from the RBi00-1A iPSC-derived cortical neurons with or without SQSTM1 (generated from two independent replicates) was extracted using an RNeasy Plus Mini Kit (Qiagen). Library preparation, RNA with PolyA selection, and sequencing with Illumina HiSeq (2 × 150 bp) were performed by GENEWIZ. Sequence reads were trimmed to remove possible adapter sequences and nucleotides with poor quality using Trimmomatic v.0.36 (Bolger et al., 2014). The trimmed reads were mapped to the human GRCh38 reference genome using the STAR aligner v.2.5.2b (Dobin et al., 2013). Raw read counts for each sample were calculated from mapped reads at the gene level using the HTSeq-count tool from the Python package HTSeq (Anders et al., 2015), with an unstranded library and position-sorted settings. DESeq2 R package (Love et al., 2014) was then used to perform differential expression and statistical analysis. Genes with adjusted p values of <0.01 were considered as significantly differentially expressed genes between iPSC-derived cortical neurons with and without SQSTM1. GSEA was used to identify biological pathways enriched in up- and downregulated genes using HALLMARK gene sets obtained from the Molecular Signature Database (MSigDB).
Quantitative real-time RT-PCR
Total RNA was extracted using an RNeasy Mini Kit (Qiagen), and 1 μg of RNA was reverse-transcribed using a High-Capacity RNA-to-cDNA Kit (Thermo Fisher Scientific). Quantitative real-time RT-PCR was performed using the ViiA 7 instrument, TaqMan gene expression assays, and TaqMan gene expression master mix (Thermo Fisher Scientific).
Statistical analysis
Statistical tests of quantitative data were calculated using GraphPad Prism 8 software. Statistical significance for comparison of means between SQSTM1 KO and WT was performed by unpaired two-tailed t test with Welch's correction. Significance evaluation is marked on figures as ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001.
Data and code availability
RNA-seq data are deposited in ArrayExpress under the accession number E-MTAB-9383.
Author contributions
Conceptualization, A.P., G.A.O., H.P.-F., S.W., L.A.D., and J.M.M.; Cell culture, A.P.; Methodology, A.P., G.A.O., H.P.-F., S.W., and H.S.; Investigation, A.P.; Analysis, A.P., S.S., H.S., and J.M.M.; Writing – original draft, A.P., H.S., and J.M.; Writing – review & editing, all co-authors; Supervision, H.P.-F., S.W., and J.M.M.
Conflicts of interests
This work was funded by Astex Therapeutics (436 Cambridge Science Park Milton Road, Cambridge CB4 0QA, UK) and at the time of work, A.P., H.S., S.S., G.A.O., L.A.D., and J.M.M. were all employees.
Acknowledgments
This work was performed as part of Astex's Sustaining Innovation Post-doctoral Program. The authors would like to thank Carla Bento, Marc Soutar, and Daniela Melandri for their technical support. We would also like to express our gratitude to Cerevance (418 Cambridge Science Park, Milton Road, Cambridge CB4 0PZ, UK) for access to their Seahorse XFe96 instrument.
Published: April 22, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.stemcr.2021.03.030.
Supplemental information
References
- Albensi B.C. What is nuclear factor kappa B (NF-κB) doing in and to the mitochondrion? Front. Cell Dev. Biol. 2019;7:154. doi: 10.3389/fcell.2019.00154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S., Pyl P.T., Huber W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolome F., Esteras N., Martin-Requero A., Boutoleau-Bretonniere C., Vercelletto M., Gabelle A., Le Ber I., Honda T., Dinkova-Kostova A.T., Hardy J. Pathogenic p62/SQSTM1 mutations impair energy metabolism through limitation of mitochondrial substrates. Sci. Rep. 2017;7:1666. doi: 10.1038/s41598-017-01678-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo-Garrido J., Maffezzini C., Schober F.A., Clemente P., Uhlin E., Kele M., Stranneheim H., Lesko N., Bruhn H., Svenningsson P. SQSTM1/p62-Directed metabolic reprogramming is essential for normal neurodifferentiation. Stem Cell Rep. 2019;12:696–711. doi: 10.1016/j.stemcr.2019.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desler C., Hansen T.L., Frederiksen J.B., Marcker M.L., Singh K.K., Juel Rasmussen L. Is there a link between mitochondrial reserve respiratory capacity and aging? J. Aging Res. 2012;2012:192503. doi: 10.1155/2012/192503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durán A., Serrano M., Leitges M., Flores J.M., Picard S., Brown J.P., Moscat J., Diaz-Meco M.T. The atypical PKC-interacting protein p62 is an important mediator of RANK-activated osteoclastogenesis. Dev Cell. 2004;6:303–309. doi: 10.1016/s1534-5807(03)00403-9. [DOI] [PubMed] [Google Scholar]
- Falcon B., Noad J., McMahon H., Randow F., Goedert M. Galectin-8-mediated selective autophagy protects against seeded tau aggregation. J Biol Chem. 2018;293:2438–2451. doi: 10.1074/jbc.M117.809293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk A., Koch P., Kesavan J., Takashima Y., Ladewig J., Alexander M., Pollard S. Capture of neuroepithelial-like stem cells from pluripotent stem cells provides a versatile system for in vitro production of human neurons. PLoS one. 2012;7:e29597. doi: 10.1371/journal.pone.0029597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fecto F., Yan J., Vemula S.P., Liu E., Yang Y., Chen W., Zheng J.G., Shi Y., Siddique N., Arrat H. SQSTM1 mutations in familial and sporadic amyotrophic lateral sclerosis. Arch. Neurol. 2011;68:1440–1446. doi: 10.1001/archneurol.2011.250. [DOI] [PubMed] [Google Scholar]
- Geisler S., Holmstrom K.M., Skujat D., Fiesel F.C., Rothfuss O.C., Kahle P.J., Springer W. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat. Cell Biol. 2010;12:119–131. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- Haack T.B., Ignatius E., Calvo-Garrido J., Iuso A., Isohanni P., Maffezzini C., Lönnqvist T., Suomalainen A., Gorza M., Kremer L.S. Absence of the autophagy adaptor SQSTM1/p62 causes childhood-onset neurodegeneration with ataxia, dystonia, and gaze palsy. Am. J. Hum. Genet. 2016;99:735–743. doi: 10.1016/j.ajhg.2016.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han H., Tan J., Wang R., Wan H., He Y., Yan X., Guo J., Gao Q., Li J., Shang S. PINK1 phosphorylates Drp1(S616) to regulate mitophagy-independent mitochondrial dynamics. EMBO Rep. 2020:e48686. doi: 10.15252/embr.201948686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill B.G., Dranka B.P., Zou L., Chatham J.C., Darley-Usmar V.M. Importance of the bioenergetic reserve capacity in response to cardiomyocyte stress induced by 4-hydroxynonenal. Biochem. J. 2009;424:99–107. doi: 10.1042/bj20090934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano M., Nakamura Y., Saigoh K., Sakamoto H., Ueno S., Isono C., Miyamoto K., Akamatsu M., Mitsui Y., Kusunoki S. Mutations in the gene encoding p62 in Japanese patients with amyotrophic lateral sclerosis. Neurology. 2013;80:458–463. doi: 10.1212/WNL.0b013e31827f0fe5. [DOI] [PubMed] [Google Scholar]
- Izquierdo-Garcia J.L., Viswanath P., Eriksson P., Cai L., Radoul M., Chaumeil M.M., Blough M., Luchman H.A., Weiss S., Cairncross J.G. IDH1 mutation induces reprogramming of pyruvate metabolism. Cancer Res. 2015;75:2999–3009. doi: 10.1158/0008-5472.Can-15-0840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazlauskaite A., Kondapalli C., Gourlay R., Campbell D.G., Ritorto M.S., Hofmann K., Alessi D.R., Knebel A., Trost M., Muqit M.M. Parkin is activated by PINK1-dependent phosphorylation of ubiquitin at Ser65. Biochem. J. 2014;460:127–139. doi: 10.1042/bj20140334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klingler E. Development and organization of the evolutionarily conserved three-layered olfactory cortex. eNeuro. 2017;4 doi: 10.1523/eneuro.0193-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon J., Han E., Bui C.B., Shin W., Lee J., Lee S., Choi Y.B., Lee A.H., Lee K.H., Park C. Assurance of mitochondrial integrity and mammalian longevity by the p62-Keap1-Nrf2-Nqo1 cascade. EMBO Rep. 2012;13:150–156. doi: 10.1038/embor.2011.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarou M., Sliter D.A., Kane L.A., Sarraf S.A., Wang C., Burman J.L., Sideris D.P., Fogel A.I., Youle R.J. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature. 2015;524:309–314. doi: 10.1038/nature14893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Ber I., Camuzat A., Guerreiro R., Bouya-Ahmed K., Bras J., Nicolas G., Gabelle A., Didic M., De Septenville A., Millecamps S. SQSTM1 mutations in French patients with frontotemporal dementia or frontotemporal dementia with amyotrophic lateral sclerosis. JAMA Neurol. 2013;70:1403–1410. doi: 10.1001/jamaneurol.2013.3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manerba M., Di Ianni L., Govoni M., Comparone A., Di Stefano G. The activation of lactate dehydrogenase induced by mTOR drives neoplastic change in breast epithelial cells. PLoS One. 2018;13:e0202588. doi: 10.1371/journal.pone.0202588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLelland G.L., Soubannier V., Chen C.X., McBride H.M., Fon E.A. Parkin and PINK1 function in a vesicular trafficking pathway regulating mitochondrial quality control. EMBO J. 2014;33:282–295. doi: 10.1002/embj.201385902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McWilliams T.G., Muqit M.M. PINK1 and Parkin: emerging themes in mitochondrial homeostasis. Curr. Opin. Cell Biol. 2017;45:83–91. doi: 10.1016/j.ceb.2017.03.013. [DOI] [PubMed] [Google Scholar]
- Morais V.A., Haddad D., Craessaerts K., De Bock P.J., Swerts J., Vilain S., Aerts L., Overbergh L., Grünewald A., Seibler P. PINK1 loss-of-function mutations affect mitochondrial complex I activity via NdufA10 ubiquinone uncoupling. Science. 2014;344:203–207. doi: 10.1126/science.1249161. [DOI] [PubMed] [Google Scholar]
- Murakawa T., Yamaguchi O., Hashimoto A., Hikoso S., Takeda T., Oka T., Yasui H., Ueda H., Akazawa Y., Nakayama H. Bcl-2-like protein 13 is a mammalian Atg32 homologue that mediates mitophagy and mitochondrial fragmentation. Nat. Commun. 2015;6:7527. doi: 10.1038/ncomms8527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muto V., Flex E., Kupchinsky Z., Primiano G., Galehdari H., Dehghani M., Cecchetti S., Carpentieri G., Rizza T., Mazaheri N. Biallelic SQSTM1 mutations in early-onset, variably progressive neurodegeneration. Neurology. 2018;91:e319–e330. doi: 10.1212/wnl.0000000000005869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra D., Kane L.A., Hauser D.N., Fearnley I.M., Youle R.J. p62/SQSTM1 is required for Parkin-induced mitochondrial clustering but not mitophagy; VDAC1 is dispensable for both. Autophagy. 2010;6:1090–1106. doi: 10.4161/auto.6.8.13426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okatsu K., Koyano F., Kimura M., Kosako H., Saeki Y., Tanaka K., Matsuda N. Phosphorylated ubiquitin chain is the genuine Parkin receptor. J. Cell Biol. 2015;209:111–128. doi: 10.1083/jcb.201410050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramesh Babu J., Lamar Seibenhener M., Peng J., Strom A.L., Kemppainen R., Cox N., Zhu H., Wooten M.C., Diaz-Meco M.T., Moscat J., Wooten M.W. Genetic inactivation of p62 leads to accumulation of hyperphosphorylated tau and neurodegeneration. J Neurochem. 2008;106:107–120. doi: 10.1111/j.1471-4159.2008.05340.x. [DOI] [PubMed] [Google Scholar]
- Puissant A., Fenouille N., Auberger P. When autophagy meets cancer through p62/SQSTM1. Am. J. Cancer Res. 2012;2:397–413. [PMC free article] [PubMed] [Google Scholar]
- Pytte J., Anderton R.S., Flynn L.L., Theunissen F., Jiang L., Pitout I., James I., Mastaglia F.L., Saunders A.M., Bedlack R. Association of a structural variant within the SQSTM1 gene with amyotrophic lateral sclerosis. Neurol. Genet. 2020;6:e406. doi: 10.1212/nxg.0000000000000406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogov V.V., Suzuki H., Marinković M., Lang V., Kato R., Kawasaki M., Buljubašić M., Šprung M., Rogova N., Wakatsuki S. Phosphorylation of the mitochondrial autophagy receptor Nix enhances its interaction with LC3 proteins. Sci. Rep. 2017;7:1131. doi: 10.1038/s41598-017-01258-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubino E., Rainero I., Chio A., Rogaeva E., Galimberti D., Fenoglio P., Grinberg Y., Isaia G., Calvo A., Gentile S. SQSTM1 mutations in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Neurology. 2012;79:1556–1562. doi: 10.1212/WNL.0b013e31826e25df. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Martin P., Komatsu M. p62/SQSTM1—steering the cell through health and disease. J. Cell Sci. 2018;131 doi: 10.1242/jcs.222836. [DOI] [PubMed] [Google Scholar]
- Schweers R.L., Zhang J., Randall M.S., Loyd M.R., Li W., Dorsey F.C., Kundu M., Opferman J.T., Cleveland J.L., Miller J.L. NIX is required for programmed mitochondrial clearance during reticulocyte maturation. Proc. Natl. Acad. Sci. U S A. 2007;104:19500–19505. doi: 10.1073/pnas.0708818104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seibenhener M.L., Zhao T., Du Y., Calderilla-Barbosa L., Yan J., Jiang J., Wooten M.W., Wooten M.C. Behavioral effects of SQSTM1/p62 overexpression in mice: support for a mitochondrial role in depression and anxiety. Behav. Brain Res. 2013;248:94–103. doi: 10.1016/j.bbr.2013.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Kirwan P., Livesey F.J. Directed differentiation of human pluripotent stem cells to cerebral cortex neurons and neural networks. Nat. Protoc. 2012;7:1836–1846. doi: 10.1038/nprot.2012.116. [DOI] [PubMed] [Google Scholar]
- Shiba-Fukushima K., Arano T., Matsumoto G., Inoshita T., Yoshida S., Ishihama Y., Ryu K.Y., Nukina N., Hattori N., Imai Y. Phosphorylation of mitochondrial polyubiquitin by PINK1 promotes Parkin mitochondrial tethering. PLoS Genet. 2014;10:e1004861. doi: 10.1371/journal.pgen.1004861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soutar M.P.M., Kempthorne L., Miyakawa S., Annuario E., Melandri D., Harley J., O'Sullivan G.A., Wray S., Hancock D.C., Cookson M.R. AKT signalling selectively regulates PINK1 mitophagy in SHSY5Y cells and human iPSC-derived neurons. Sci. Rep. 2018;8:8855. doi: 10.1038/s41598-018-26949-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Zee J., Van Langenhove T., Kovacs G.G., Dillen L., Deschamps W., Engelborghs S., Matěj R., Vandenbulcke M., Sieben A., Dermaut B. Rare mutations in SQSTM1 modify susceptibility to frontotemporal lobar degeneration. Acta Neuropathol. 2014;128:397–410. doi: 10.1007/s00401-014-1298-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viiri J., Hyttinen J.M., Ryhanen T., Rilla K., Paimela T., Kuusisto E., Siitonen A., Urtti A., Salminen A., Kaarniranta K. p62/sequestosome 1 as a regulator of proteasome inhibitor-induced autophagy in human retinal pigment epithelial cells. Mol. Vis. 2010;16:1399–1414. [PMC free article] [PubMed] [Google Scholar]
- Vives-Bauza C., Zhou C., Huang Y., Cui M., de Vries R.L., Kim J., May J., Tocilescu M.A., Liu W., Ko H.S. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc. Natl. Acad. Sci. U S A. 2010;107:378–383. doi: 10.1073/pnas.0911187107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamano K., Kikuchi R., Kojima W., Hayashida R., Koyano F., Kawawaki J., Shoda T., Demizu Y., Naito M., Tanaka K. Critical role of mitochondrial ubiquitination and the OPTN-ATG9A axis in mitophagy. J. Cell Biol. 2020;219 doi: 10.1083/jcb.201912144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yetkin-Arik B., Vogels I.M.C., Nowak-Sliwinska P., Weiss A., Houtkooper R.H., Van Noorden C.J.F., Klaassen I., Schlingemann R.O. The role of glycolysis and mitochondrial respiration in the formation and functioning of endothelial tip cells during angiogenesis. Sci. Rep. 2019;9:12608. doi: 10.1038/s41598-019-48676-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Ney P.A. Role of BNIP3 and NIX in cell death, autophagy, and mitophagy. Cell Death Differ. 2009;16:939–946. doi: 10.1038/cdd.2009.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data are deposited in ArrayExpress under the accession number E-MTAB-9383.