Summary
Non-muscle myosin IIA plays an important role in cell adhesion, cell migration, and tissue architecture. We previously showed that low activity of the heavy chain of non-muscle myosin II Myh9 is beneficial to LGR5+ intestinal stem cell maintenance. However, the function of Myh9 in adult mouse intestinal epithelium is largely unclear. In this study, we used the inducible Villin-creERT2 knockout approach to delete Myh9 in adult mouse intestinal epithelium and observed that homozygous deletion of Myh9 causes colitis-like morphologic changes in intestine, leads to a high sensitivity to dextran sulfate sodium and promotes colitis-related adenoma formation in the colon. Myh9 deletion disturbs cell junctions and impairs intestinal lumen barrier integrity, promoting the necroptosis of epithelial cells. Consistently, these changes can be partially rescued by Ripk3 knockout. Our results indicate that Myh9 is required for the maintenance of intestinal epithelium integrity and the prevention of cell necroptosis.
Key words: Myh9, intestinal stem cells, necroptosis, colitis
Graphical abstract
Highlights
-
•
Homozygous deletion of Myh9 causes colitis-like phenotypes in intestine
-
•
Myh9 deficiency leads to reduced Lgr5+ cell population and Paneth cell dislocation
-
•
Myh9 is critical for maintenance of intestinal epithelium integrity
-
•
Myh9 deletion results in high sensitivity to DSS and colitis adenomas formation
Wang et al. report that Myh9 is required for maintenance of intestinal epithelial integrity mainly by preventing cell necroptosis and inflammation. Myh9 deficiency leads to cell junction disruption and colitis-like phenotypes and related adenomas development.
Introduction
The intestinal epithelium consists of a monolayer of cells, which undergoes rapid turnover and moves from the crypt base to the villi tip. Intestinal stem cells (ISCs), known as columned base cells, localize at the base of crypts and are marked by LGR5 (Barker and Clevers, 2010; Barker et al., 2007). The crypts also harbor an intermediate cell type—trans-amplifying (TA) cells that undergo four to five rounds of cell division before differentiating into the mature epithelial cells, including enterocytes, goblet cells, enteroendocrine cells, and Paneth cells (Barker, 2014). In contrast to the other differentiated cells, Paneth cells reside at the crypt base and are intermingled with LGR5+ ISCs, which are an important part of the niche for LGR5+ ISCs, and contribute to ISC maintenance by producing epidermal growth factor, transforming growth factor α, WNT3A, and the Notch ligand DLL4 (Sato et al., 2011).
People with inflammatory bowel disease are at increased risk of developing colorectal cancer and colitis-associated cancer. In recent years, rapidly increasing incidence of inflammatory bowel disease (IBD) leads to the social concern of intestinal inflammation (Terzic et al., 2010; Waljee et al., 2017). Either disruption of the intestinal lumen barrier or pathogen invention causes colitis. The activation of RIPK3-dependent necroptosis can cause intestinal inflammation (Dannappel et al., 2014; Gunther et al., 2011; Wang et al., 2020; Welz et al., 2011), and cell death in the epithelium has been suggested to be a hallmark of intestinal inflammation (Anderton et al., 2020; Kaser et al., 2010; Patankar and Becker, 2020). Treatment of mice with DSS or combined with azoxymethane (AOM) can generate a rapid and effective model of colitis-associated cancer (Parang et al., 2016). Activation of the RIPK3-MLKL axis has been particularly observed in ulcerative colitis. The molecular mechanisms by which inflammation promotes cancer development remain unclear and could differ from other forms of colorectal cancer (Grivennikov, 2013; Terzic et al., 2010).
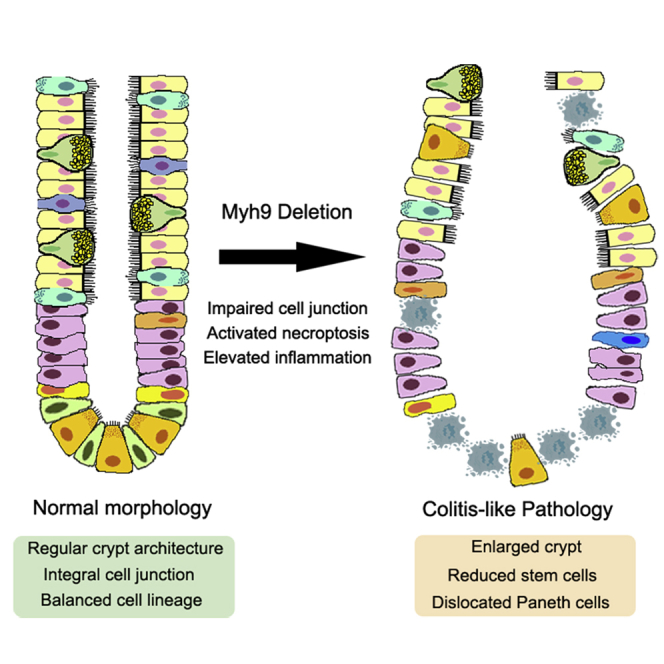
Non-muscle myosin II (NMII) is an actin-binding protein that comprises two heavy chains and two pairs of light chains. In mammals, myosin heavy chain 9 (Myh9) is one of the three genes encoding the NMII heavy chain and plays a critical role in cell adhesion, cell migration, and tissue architecture (Conti and Adelstein, 2008; Vicente-Manzanares et al., 2009). NMII has been indicated to mediate dissociation-induced anoikis of embryonic and induced pluripotent stem cells (Chen et al., 2010; Ohgushi et al., 2010; Walker et al., 2010). Our previous study shows that Myh9 accumulates at epithelial injury sites in mice distal colon after DSS treatment, and Myh9 mono-allelic deletion ameliorates DSS-induced colitis (Zhao et al., 2015). These results suggest that Myh9 plays an important role in the intestinal inflammation process. In this study, to further address the function of Myh9 in the intestinal epithelium of adult mice, by using inducible conditional Myh9 knockout (KO), we found that complete deletion of Myh9 in adult mouse intestinal epithelium causes colitis-like morphologic changes, reduces stem cells, increases necroptosis, leads to a high sensitivity to DSS, and promotes the colitis-related adenoma formation in the colon. Furthermore, inhibition of necroptosis by additional deletion of Ripk3 renders the less pathological characteristics of colitis in mice. Therefore, Myh9 maintains intestinal homeostasis, at least in part, by inhibiting RIPK3 to block necroptosis.
Results
Myh9 deficiency results in impaired villus and abnormal crypts
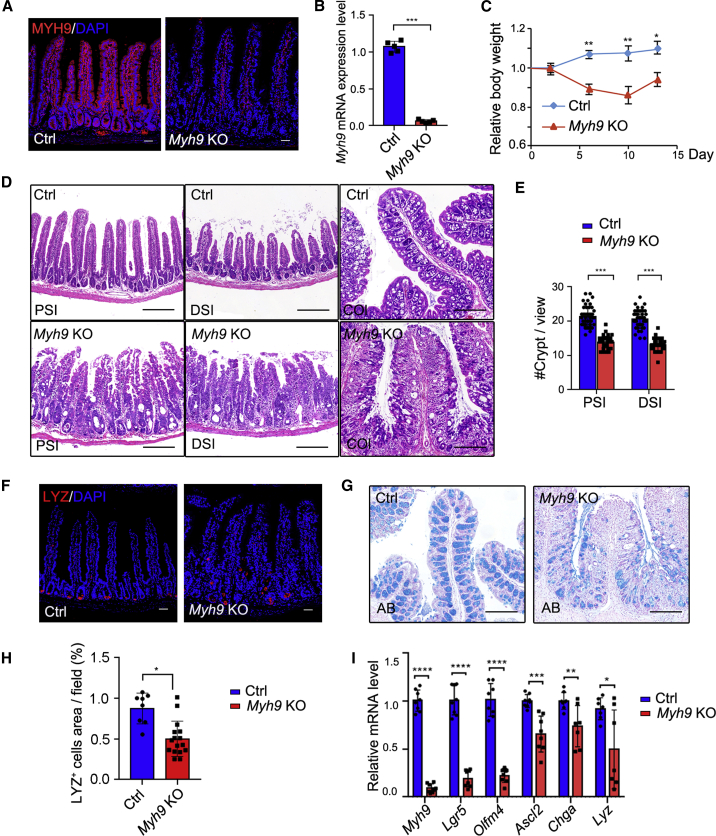
In a previous study, we showed that mono-allelic deletion of Myh9 with Villin-Cre could promote the survival and proliferation of LGR5+ cells (Zhao et al., 2015). As we could not obtain adult mice with Villin-driven homozygous deletion of Myh9 in two alleles, we crossed Myh9fl/fl mice with Vilin-CreERT2 mice to generate inducible conditional knockout mice Myh9fl/fl;Villin-CreERT2 (Myh9-KO). After daily injection of tamoxifen (TAM) for 5 consecutive days, Myh9 was efficiently depleted in knockout mouse intestine as shown by immunofluorescence and qRT-PCR (Figures 1A and 1B). The body weight of Myh9-KO mice started to decrease at day 2 after the TAM injection and remained lower than control mice at day 14 after the TAM injection (Figure 1C). Abnormally expanded crypts and cavities were observed in the small intestine at different time points (Figures 1D and S1A). The crypt number was also decreased in the small intestine (Figure 1E). More seriously for the colon, it is obvious that there are fewer crypts and more infiltrations in the Myh9-KO colon. In the small intestine, Paneth cells were decreased and dislocated in Myh9-KO mice (Figures 1F and 1H). Moreover, the number of goblet cells was dramatically decreased in the colon in Myh9-KO mice (Figure 1G), while they were unchanged in the small intestine (Figures S1B and S1C). qPCR analysis showed that the expression of ISC marker genes, such as Lgr5, Olfm4, and Ascl2, as well as the Paneth cell marker lysozyme (Lyz) and enteroendocrine cell marker chromogranin A (Chga), was reduced in Myh9-KO crypts (Figure 1I). All the morphologic changes, such as abnormal crypts, decreased goblet cells, and body weight loss, in Myh9-KO mice are similar to the phenotypes of DSS-induced colitis mice (Cooper et al., 1993; Wang et al., 2020; Wirtz et al., 2007). However, there was no change in colon length in Myh9-KO mice (Figure S1D).
Figure 1.
Homozygous deletion of Myh9 results in abnormal crypts
(A and B) Immunofluorescence staining (A) and qRT-PCR (B) to detect Myh9 expression in control and Myh9-KO mice at day 5 after the first TAM injection. The data represent mean ± SD derived from three mice for each group.
(C) Relative body weight index of control and Myh9-KO mice after TAM injection. The data represent mean ± SD derived from ten mice for each group.
(D) Hematoxylin and eosin (H&E) staining of small intestine and colon from control and Myh9-KO mice at day 5 after the first TAM injection (PSI/DSI/COL, proximal, distal small intestine, and colon).
(E) Quantification of crypt numbers in the small intestine of control and Myh9-KO mice at day 5 after the first TAM injection. The data represent mean ± SD derived from three mice per group and at least ten views for each mouse.
(F and H) Immunofluorescence staining (F) and quantitative analysis (H) for LYSOZYME in control and Myh9-KO intestinal crypts at day 5 after the first TAM injection. The data represent mean ± SD derived from at least 30 crypts from each mouse (n = 3 mice per group).
(G) Alcian blue staining for goblet cells in control and Myh9-KO colon.
(I) qRT-PCR for intestinal signature genes in control and Myh9-KO mice at day 5 after the first TAM injection.
The data represent mean ± SD derived from seven mice per group. Scale bars, 50 μm in (A and F) and 200 μm in (D and G). ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05, Student's t test (B and F), two-way ANOVA (E and H). See also Figure S1.
Myh9 plays an important role in LGR5+ cell maintenance
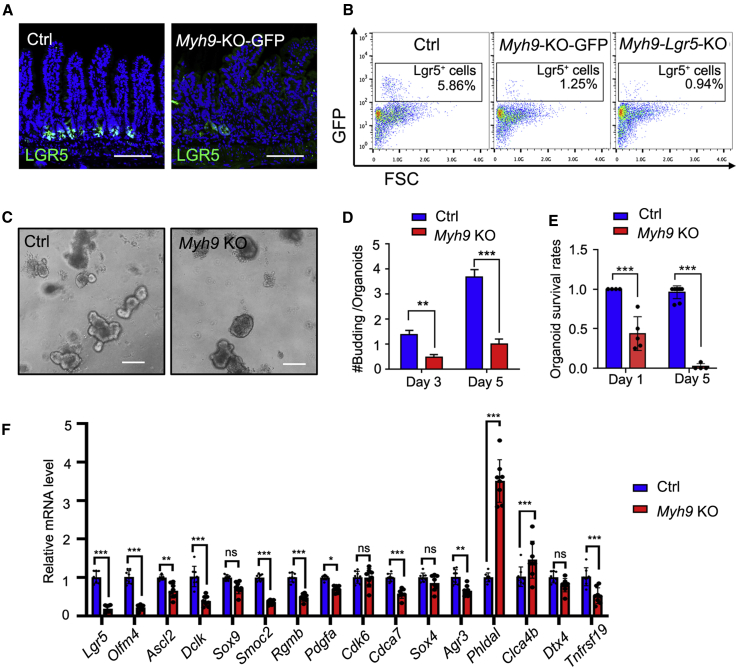
Since LGR5+ ISC marker gene expression was significantly decreased after Myh9 deletion, we then assessed whether Myh9 deficiency affects LGR5+ ISC maintenance. We crossed Myh9fl/fl;Villin-CreERT2 mice with Lgr5-GFP-IRES-CreERT2 mice to generate Myh9-KO-GFP mice. From immunofluorescence and flow cytometry analyses, we detected a significant decrease of LGR5+ cells in Myh9-KO-GFP mice upon TAM-induced Myh9 deletion (Figure 2A). Furthermore, specific Myh9 deletion in LGR5+ ISCs in Myh9fl/fl;Lgr5-GFP-IRES-CreERT2 mice (Myh9-Lgr5-KO) also led to LGR5+ ISC reduction (Figure 2B), indicating that Myh9 is critical for LGR5+ ISC maintenance. To further confirm the role of Myh9 in LGR5+ ISC maintenance, we cultured organoids derived from the crypts of Myh9-KO mice and induced Myh9 deletion by 4-hydroxytamoxifen (4-OH-TAM) on the day after seeding. After 3–5 days of culture, we found fewer buddings in Myh9−/− organoids than in the control (Figures 2C and 2D). Moreover, there were fewer organoids survived after passaging (Figure 2E), which was in contrast to mono-allelic deletion of Myh9, which promoted organoid survival (Zhao et al., 2015). Consistently, the expression of most ISC signature genes was decreased after Myh9 deletion (Figure 2F). These results indicate that an appropriate amount of Myh9 is important for LGR5+ ISC maintenance in vivo and organoid survival in vitro.
Figure 2.
Myh9 deficiency reduces LGR5+ cells
(A) Immunofluorescence staining for LGR5-GFP in small intestine of control and Myh9-KO-GFP mice at day 10 after the first TAM injection.
(B) LGR5+ cell numbers in small intestine of control, Myh9-KO-GFP, and Myh9-Lgr5-KO mice were analyzed by flow cytometry. The data represent mean ± SD derived from at least six mice per group.
(C–E) Organoid cultures of crypts isolated from control or Myh9-KO mice. Representative pictures of organoids (C), quantification of budding numbers on day 3 and 5 after 4-OH-TAM treatment (D), and quantification of survival rates on days 1 and 5 after passaging (E). The data represent mean ± SD derived from at least three wells per group and all experiment repeated with three mice.
(F) qRT-PCR for intestinal stem cell signature genes in control and Myh9-KO mice at day 5 after the first TAM injection.
The data represent mean ± SD derived from seven mice per group. ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05, two-way ANOVA. Scale bars, 200 μm. See also Figure S2.
Although Paneth cells play a critical role in sustaining LGR5+ ISCs (Sato et al., 2011), Myh9 deletion in LGR5+ cells (Myh9-Lgr5-KO) significantly reduced LGR5+ ISCs, to the same extent as in Myh9-KO-GFP mice (Figure 2B), indicating that Myh9 deletion-induced LGR5+ ISC reduction is an LGR5+ cell-autonomous effect. Consistently, Myh9 deletion in LGR5+ cells did not affect Paneth cells (Figure S2A).
Furthermore, we used bromodeoxyuridine (BrdU) to mark proliferation cells and found that Myh9 KO led to more proliferative cells, and that most BrdU-labeled cells in Myh9 KO intestine migrated to the villi zone (Figures S2B and S2C). KI67+ staining also showed a higher proliferation rate in the crypts of small intestine and colon upon Myh9 KO (Figures S2D and S2E). Accumulated works show that, upon epithelium injury, differentiated epithelial cells or progenitors can undergo de-differentiation, re-gain their proliferation ability, and even become ISCs to compensate the lost epithelial cells (Barriga et al., 2017; Li et al., 2016; Liu and Chen, 2020; Liu et al., 2020; Schmitt et al., 2018; Tetteh et al., 2016; Wang et al., 2019; Yan et al., 2017; Yu et al., 2018). The enhanced expression of several regenerative genes, such as Clu, Sca1, and Anxa1 (Figure S2F), suggesting that the expanded proliferating cells could be TA cells or progenitor cells due to the compensation of Myh9 KO-induced damage. These data suggest that Myh9 deficiency reduced LGR5+ cell populations, while resulting in the expansion of proliferating cells.
The Myh9 deficiency-induced decrease of ISCs and Paneth cells is independent of microbiota
The intestinal morphological changes, crypt deformation, reduced LGR5+ ISCs, fewer goblet cells, and dislocated Paneth cells, in Myh9-KO mice resemble pathological characteristics of colitis (Cooper et al., 1993). The transcriptome landscape of epithelium in control and Myh9-KO mice also revealed that the inflammation-related genes were enriched in the small intestinal epithelium of Myh9-KO mice (Figure S3A). Moreover, more neutrophil cells (marked by CD11b) were recruited to the crypt area after Myh9 deletion (Figure S3B). Myh9 KO rendered intestinal epithelium vulnerable to 2.5% DSS-induced colitis as shown by severe body weight loss (Figure S3C), and the Myh9 KO colons were shorter with more erosive areas compared with the control group (Figures S3D and S3E).
Since Myh9 is involved in the connection of actin filament with cell junctions and plays an important role in intestinal epithelium barrier permeability (Naydenov et al., 2016; Vicente-Manzanares et al., 2009), we reasoned that its deficiency might affect cell junction integrity and facilitate bacteria invasion, thus impairing the LGR5+ ISC maintenance and dislocating Paneth cells. Examination with transmission electron microscopy revealed that Myh9 deficiency led to losing cell junctions in crypts (Figure S4A). Moreover, mass spectrometry analysis of anti-MYH9-precipitated proteins identified 380 potential interacting proteins, including cell junction and actin-associated proteins (Figure S4B; Table S1). KEGG pathway analysis showed that the top 5 pathways were tight junction, actin cytoskeleton, adherens junction, leukocyte transendothelial migration, and bacterial invasion (Figure S4C). Thus, inflammation in Myh9-KO mice may be associated with neutrophil infiltration by losing cell junctions.
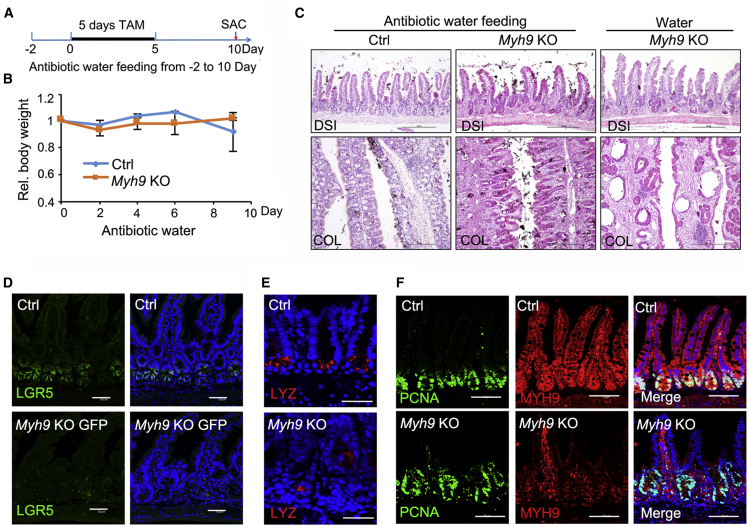
To examine whether the colitis-like phenotypes were caused by bacteria invasion, we fed mice with water containing broad-spectrum antibiotics 2 days before keeping them under germ-free conditions, and then they were injected with TAM before being killed at day 10 (Figure 3A). The antibiotic treatment effectively rescued body weight loss of Myh9-KO mice (Figure 3B). Although the morphologic changes of abnormal crypts were relieved in the intestine (Figure 3C), the reduction of LGR5+ ISCs and Paneth cells and the expansion of proliferation cells were not rescued by antibiotic treatment (Figures 3D–3F). These results suggest that LGR5+ ISC loss, dislocation of Paneth cells, and expansion of proliferating cells in crypts are independent of bacteria invasion.
Figure 3.
Antibiotics treatment has no effect on Myh9 KO-mediated LGR5+ cell reduction
(A and B) Strategy and relative body weight of control and Myh9-KO mice after antibiotics-containing water feeding. The data represent mean ± SD derived from four mice for each group.
(C) H&E staining of distal small intestine and colon from control and Myh9-KO mice with or without antibiotics treatment. Scale bars, 200 μm.
(D) Immunofluorescence staining of LGR5+-GFP cells in control and Myh9-KO-GFP mice under antibiotics treatment. Scale bars, 50 μm.
(E) Immunofluorescence staining of lysozyme in control and Myh9-KO mice under antibiotics treatment. Scale bars, 100 μm.
(F) Immunofluorescence staining of PCNA and MYH9 in control and Myh9-KO mice under antibiotics treatment. Scale bars, 200 μm.
See also Figures S3 and S4 and Table S1.
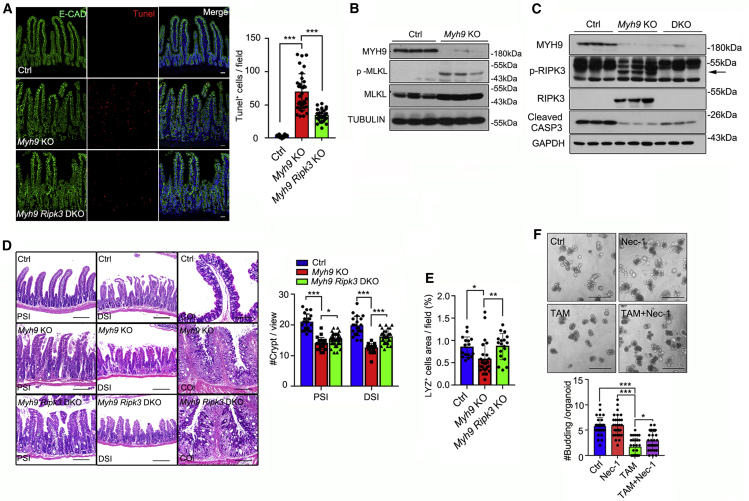
Myh9 deficiency promotes cell necroptosis of intestinal epithelium
To further investigate how Myh9 deficiency leads to abnormal crypt structure and LGR5+ ISC loss, we examined cell death by TUNEL assay and found that there were more labeled cells in the crypts of Myh9-KO mice (Figure 4A). As RIPK3-mediated necroptosis of ISCs is involved in enteritis (Gunther et al., 2011; Welz et al., 2011), we detected the levels of pRIPK3 and pMLKL, and found that both of them were increased in the crypt epithelium of Myh9-KO mice (Figures 4B and 4C) and more pMLKL+ cells were observed in the crypt epithelium of both Myh9-KO mice and Myh9-Lgr5-KO mice (Figures S5A and S5B). Moreover, some PCNA+ and pMLKL+ cells were colocalized in the crypts of Myh9-Lgr5-KO mice, suggesting that necroptosis can also occur in proliferative cells after Myh9 deletion (Figure S5C). In contrast, a significant reduction of cleaved caspase-3 was observed in Myh9 KO crypts (Figure 4C). Previous studies have highlighted that Myh9 mono-allelic deletion activates AKT, which is essential for the survival of LGR5+ stem cells (Zhao et al., 2015). Here, we found that p-AKT remained largely unchanged (Figure S5D). Furthermore, the antibiotics water feeding did not reduce the pRIPK3 and pMLKL level in Myh9-KO mice (Figure S5E).
Figure 4.
Myh9 deficiency enhances necroptosis of cryptic epithelium
(A) TUNEL assay was performed in control, Myh9-KO, and Myh9-Ripk3-DKO small intestine at day 5 after the first TAM injection, with quantification (right) of labeled cells. The data represent mean ± SD derived from at least 30 crypts from 3 mice per group.
(B and C) Intestinal epithelium lysates of control and Myh9-KO mice at day 10 (B) or control, Myh9-KO and Myh9-Ripk3-DKO mice at day 5 (C) after the first TAM injection were subjected to immunoblotting with indicated antibodies.
(D) H&E staining of small intestine and colon and crypt quantitation from control, Myh9-KO, and Myh9-Ripk3-DKO mice at day 5 after the first TAM injection (PSI/DSI/COL: proximal, distal small intestine, and colon). The data represent mean ± SD derived from at least 30 crypts from each mouse (n = 3 mice per group).
(E) Quantitation of LYZ-positive cells in control and Myh9-KO intestinal crypts at day 5 after the first TAM injection. The data represent mean ± SD derived from at least 30 crypts from each mouse (n = 3 mice per group).
(F) Representative pictures of organoids cultured at day 5 after treated with Necrostatin-1, 4-OH-TAM, or both as indicated. Quantification of bud numbers is shown below and the data represent mean ± SD derived from at least three wells per group and all experiments repeated with three mice.
One-way ANOVA (A, E, and F), two-way ANOVA (D). ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05. Scale bars, 200 μm. See also Figure S5.
To confirm that Myh9-deficient epithelial cells undergo the RIPK3-MLKL-dependent necroptosis, we crossed Ripk3−/− mice with Myh9-KO mice to block necroptosis (Myh9-Ripk3-DKO). Compared with Myh9-KO mice, the number of dying cells was greatly reduced in the epithelium of Myh9-Ripk3-DKO mice (Figure 4A). Consequentially, the architecture of crypts and Paneth cells was apparently restored (Figures 4D, 4E, and S5F). However, goblet cell and tumor necrosis factor alpha expression were the same in Myh9-KO and Myh9-Ripk3-DKO mice (Figures S5G and S5H). Furthermore, the inhibitor of RIPK1, Necrostatin-1, could effectively block Myh9 deletion-induced organoid death (Figure 4F). The recruitment of RIPK3 by MYH9 was detectable in the epithelium of control mice; however, genetic ablation of Myh9 caused loss of the interaction with RIPK3 (Table S1). All these data indicate that Myh9 deficiency-induced cell death of the intestinal epithelium is mainly via necroptosis.
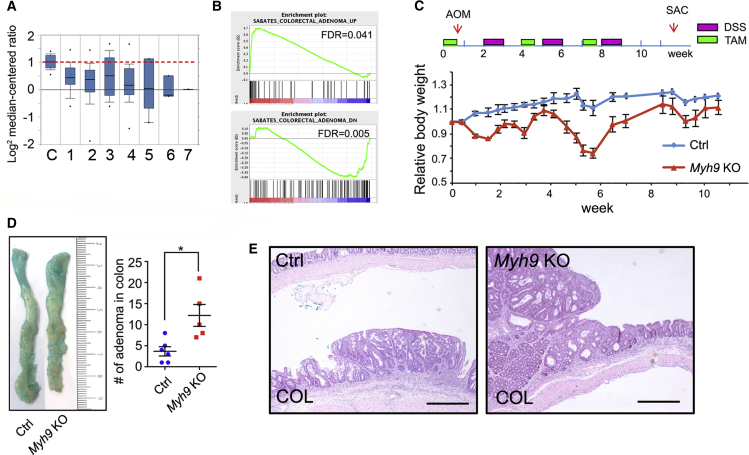
Myh9 deficiency promotes colitis-related adenoma formation in the colon
It has been reported that Myh9 is a tumor suppressor in squamous cell carcinomas (Schramek et al., 2014). Oncomine analysis of TCGA database showed that Myh9 is downregulated in most types of gastrointestinal adenoma/carcinoma (Figure 5A). Moreover, transcriptomic profiling showed that colorectal adenoma-related genes were enriched in Myh9 KO intestinal epithelium, and that these genes showed similar expression patterns in both colorectal adenoma and Myh9 KO epithelium (Figure 5B). As patients with IBD have a high risk of colitis-associated dysplasia and cancer (Grivennikov, 2013; Terzic et al., 2010), we used the AOM/DSS-induced adenoma model to explore the role of Myh9 in gut carcinogenesis. Mice were treated with one-time AOM injection, then five injections with TAM, then with 5 days to recover, 5 days DSS feeding, repeated three times. The AOM-DSS treatment led to loss in body weight in Myh9-KO mice (Figure 5C). The number of adenomas in the colon was significantly increased in Myh9-KO mice (Figure 5D). However, the tumors in Myh9-KO mice were not invasive as shown by H&E staining (Figure 5E). These data suggest that Myh9 plays a critical role in maintaining epithelium integrity and prevents pathological inflammation and then carcinogenesis.
Figure 5.
Myh9 deficiency promotes colitis-related adenoma formation
(A) Oncomine analysis shows that the Myh9 mRNA expression is reduced in multiple types of gastrointestinal tumors in the TCGA database. C, no value (22); 1, cecum adenocarcinoma (22); 2, colon adenocarcinoma (101); 3, colon mucinous adenocarcinoma (22); 4, rectal adenocarcinoma (60); 5, rectal mucinous adenocarcinoma (6); 6, rectosigmoid adenocarcinoma (3); 7, rectosigmoid mucinous adenocarcinoma (1).
(B) GSEA analysis of the transcriptome showing an enrichment of colorectal adenomas in Myh9-KO mice.
(C) Relative body weight of control and Myh9-KO mice upon AOM/DSS treatment. Body weight was normalized to the first day of TAM injection. The data represent mean ± SD derived from six mice for control group and five mice for Myh9 KO group.
(D) Representative colon of control and Myh9-KO mice after AOM/DSS treatment. Alcian blue staining was to improve observable adenomas in the colon. Right: quantification of adenoma numbers. The data represent mean ± SD derived from five mice per group. ∗p < 0.05, Student's t test.
(E) H&E staining shows the adenoma in colon from control and Myh9-KO mice after AOM/DSS treatment. Scale bars, 200 μm.
Discussion
In this study, we demonstrated that inducible homozygous deletion of Myh9 in the intestinal epithelium leads to abnormal crypts, high sensitivity to DSS-induced colitis formation, and colitis-related adenoma development. Myh9 deficiency leads to losing cell junctions in the crypt epithelium and necroptosis of the intestinal epithelium. Blocking necroptosis could partially rescue the phenotype of Myh9-KO mice, including crypt abnormality and reduced Paneth cells.
Our previous study showed that reducing Myh9 expression level or inhibiting its activity could facilitate self-renewal of LGR5+ ISCs by enhancing Akt activation and ameliorate DSS-induced colitis (Zhao et al., 2015). However, this study revealed that homozygous Myh9 deletion in the intestinal epithelium impaired LGR5+ cells. The reason could be that a strong activation of necroptosis signaling overcomes the activated survival signaling. Specific deletion of Myh9 in LGR5+ cells also reduced their cell number, suggesting that Myh9 is critical to protect stem cells from necroptosis. Another possibility for Myh9 deficiency-caused ISC reduction could be insufficient Paneth cell function in the Myh9 KO crypts. Paneth cells constitute the niche to sustain LGR5+ ISCs by providing multiple ligands (Sato et al., 2011; Takeda et al., 2011). We observed that Villin-driven Myh9 deletion reduced the Paneth cell number. As reported previously, either cell junction protein, such as E-cadherin deficiency, or activation of necroptosis, would affect the location and quantity of Paneth cells (Gunther et al., 2011; Schneider et al., 2010; Welz et al., 2011). Interestingly, specific deletion of Myh9 in LGR5+ cells did not affect the number of Paneth cells, suggesting that ISC reduction is mainly caused by Myh9 deficiency inside these cells.
Cell junction integrity is the basis of intestinal lumen barrier stabilization and important for intercellular communication, and protection from pathogen invasion and inflammatory response (Buckley and Turner, 2018; Odenwald and Turner, 2017; Suzuki, 2013). Myh9, a motor protein binding to actin filament, participates in cell adhesion, polarization, and migration (Conti and Adelstein, 2008; Vicente-Manzanares et al., 2009). Consistent with an earlier report (Naydenov et al., 2016), we found that deletion of Myh9 impaired the expression and distribution of cell junction proteins and affected the integrity of the epithelium barrier. Our data are in agreement with the importance of E-cadherin in the proper function of the intestinal epithelial cells by providing mechanical integrity of the adherens junction (Bondow et al., 2012; Mohanan et al., 2018; Schneider et al., 2010).
We also observed increased necroptosis in Myh9 KO crypts. In line with this, Myh9 deletion led to the activation of the necroptosis-related proteins RIPK3 and MLKL. Meanwhile, inhibition of necroptosis by additional deletion of Ripk3 partially rescued the crypt abnormality. Reduced caspase-3 activation suggests that Myh9 deletion could switch cell death from apoptosis to necroptosis, revealing a previously unknown role of Myh9 in involvement of the inflammation and necroptosis. It is noteworthy that further RIPK3 deletion did not fully rescue the cell death phenotype caused by Myh9 deficiency. It would be better to confirm that Myh9-KO-induced cell death is via necroptosis with MLKL deletion. Also, RIPK3 has been shown to have necroptosis-independent cell death (Lawlor et al., 2015). It is unclear whether impaired cell junction integrity is contributed to the enhanced necroptosis in intestinal crypts, and how Myh9 negatively regulates the RIPK3.
Chronic inflammation of the colon is associated with an increased risk of developing colorectal cancer (Grivennikov, 2013; Terzic et al., 2010). Distinct from apoptosis, necroptosis has been considered to be a form of programmed cell death that triggers an inflammatory response (Chan, 2012; Gunther et al., 2013; Wang et al., 2020). Several studies have shown that Myh9 participate in cancer development mainly by regulation of cell junction proteins or stabilizing p53 (Conti et al., 2015; Schramek et al., 2014; Derycke et al., 2011; Ma and Adelstein, 2014). Consistently, we found that Myh9 deletion resulted in deregulation of necroptosis and enhanced inflammation, and promoted colitis-related adenoma formation after AOM/DSS treatment. These data together highlight the role of Myh9 in colitis-associated colorectal cancer.
Experimental procedures
Mice
All animal studies were performed in accordance with the relevant guidelines and regulations and with the approval of the Institutional Animal Care and Use Committee of Tsinghua University. Lgr5-GFP-ires-creERT2 mice and Villin-creERT2 mice were obtained from the Jackson Laboratory. Myh9fl/fl mouse sperms were obtained from MMRRC (MMRRC_036749-JAX). Ripk3−/− mice were a gift from Dr. Xiaodong Wang (National Institute of Biological Sciences, Beijing). For Myh9, Myh9fl/fl mice were crossed with the mice expressing cre-recombinase and treated with five consecutive daily doses of tamoxifen (Sigma, 100 mg kg−1) in corn oil.
Intestinal crypt and LGR5+ cell isolation and culture
Mouse intestine was isolated from 8-week-old mice. For isolation of whole-intestinal epithelial cells, the mouse intestine was cut longitudinally and washed two to three times with ice-cold PBS, then cut into small pieces (3–7 mm long) and incubated for 30 min at 4°C in PBS containing 30 mM EDTA and 1.5 mM dithiothreitol on ice, and collecting suspended cells after gentle vortexing. For isolation of intestinal crypts, mouse small intestine was washed with PBS as above. The villi were scraped using a coverslip. The crypts were released by incubation for 30 min at 4°C in PBS containing 30 mM EDTA and gentle vortexing. The supernatant, which enriched in crypts, was passed through a 70-μm cell strainer (BD) and centrifuged at 800 × g for 5 min. Isolated crypts were counted and pelleted for single-cell purification or organoids culture.
To purify single LGR5+ cells, isolated intestinal crypts were treated with Trypsin (Invitrogen) for 30 min, and then 10% FBS and 50 μL of 10 mg mL−1 DNase was added to the cell solution. The tube was inverted three to four times to thoroughly mix the reagents. After being passed through 70- and 40-μm cell strainers (BD), LGR5+ cells were sorted using FACS (BD FACSAria II). Organoid cultures was performed as described previously (Zhao et al., 2015). The Necrostatin-1 (Selleck) was diluted in DMSO to a final concentration of 10 mM.
DSS-induced colitis
Acute colitis was induced by feeding 8-week-old Myh9-KO mice and control mice with 2.5% DSS (MP Biomedicals) dissolved in drinking water for 5 days and replaced with water.
AOM/DSS-induced tumor
Control mice were co-housed with Myh9-KO mice and injected with 100 mL of 20 mg mL−1 TAM every day for 5 days. At 5 days after the final injection, all mice were given a single intraperitoneal injection of 10 mg kg−1 of AOM (Sigma). One week later, all mice received 2.5% DSS in their drinking water for 7 days and then regular water for 14 days. Two more cycles of TAM and DSS were subsequently administered. Mice were observed daily and weighed every 3 days before being sacrificed approximately 2 weeks after the last DSS cycle. Tumors were counted using a stereomicroscope and colon tissue was subsequently fixed for histology.
Histological analyses
Intestines were washed with PBS, fixed in 10% neutral formalin, paraffin embedded, and sectioned. For immunohistochemistry staining, antigen retrieval was performed by heating slides in 0.01 M citrate buffer (pH 6) in a microwave oven. The sections were incubated with 3% hydrogen peroxide in diH2O to quench endogenous peroxidases. For immunofluorescence staining, paraffin sections were pretreated in 0.01 M citrate buffer (pH 6) in a microwave oven, and incubated in primary antibodies overnight at 4°C, then incubated with FITC- or TRITC-conjugated fluorescent secondary antibodies (1:400, Jackson Laboratory, 715-545-150, 711-025-152) for 30 min at 37°C and counterstained with DAPI. The following antibodies were used: KI67 (1:1,000, Abcam, ab15580), MYH9 (1:400, Proteintech Group, 11128-1-AP), lysozyme (1:50, Santa Cruz, sc-27956), GFP (1:200, Abcam, ab6556), pMLKL (1:200, Abcam, ab196436), CD11b (1:200, Abcam, ab13357), and PCNA (1:200, Santa Cruz, sc56). H&E staining was used according to the manufacturer’s instructions (C0205S, Beyotime). In brief, the sections were rinsed with PBS to remove residual chemicals after being dewaxed by conventional procedures, and then stained with hematoxylin for 2 min and eosin for 20 s. After staining, sections were dehydrated through increasing concentrations of ethanol and xylene and coverslipped with neutral balsam. Alcian blue staining was used to detect goblet cells, and slides were stained with Alcian blue for 15 min and nuclear fast red for 1 min (BA4087B, Baso). A confocal microscope (FV3000, Olympus) was used for imaging the immunofluorescence staining. Other H&E, immunohistochemistry, and Alcian blue staining were imaged with a slide scanning system (KF-PRO-120, Jiang Feng).
TUNEL assay
Formalin-fixed paraffin-embedded slides of the respective genotypes were subjected to cell death assessment using the TUNEL cell death detection kit (Roche).
BrdU assay
For BrdU labeling, mice were injected with 100 mL of 20 mg mL−1 TAM every day for 5 days. Five days after the final TAM injection, all mice were given a single intraperitoneal injection of 100 μL BrdU (BD Pharmingen) 2 or 24 h before killed and intestinal isolation. The BrdU detection was carried out following the directions for the BrdU flow kit from BD Pharmingen.
Immunoblotting
Cells were lysed in TNE buffer with protease inhibitors (Roche). After quantification using a BCA protein assay kit (Pierce), 40 mg of total protein was separated with 7.5% or 10% SDS-PAGE under denaturing conditions and were transferred to nitrocellulose membranes (Bio Trace). The membranes were blocked in 5% milk and then incubated with the primary antibody overnight at 4°C, followed by incubation with a secondary antibody conjugated with horseradish peroxidase (HRP) for 1 h at room temperature. The following antibodies were used for immunoblotting: AKT (1:2,000, Cell Signaling, 9272s), cleaved caspase-3 (1:1,000, Cell Signaling Technology, 9661s), GAPDH (1:2,000, Santa Cruz, SC-137179), MYH9 (1:2,000, Proteintech Group, 11128-1-AP), p-AKT (Ser473) (1:2,000, Cell Signaling Technology, 4060s), TUBULIN (1:1,000, Santa Cruz, sc-8035), RIPK3 (1:1,000, Cell Signaling Technology, 95702s), p-RIPK3 (1:1,000, Abcam, ab195117), MLKL (1:1,000, Abcam, ab172868), p-MLKL (1:1,000, Abcam, ab196436), and HRP-conjugated mouse IgG or rabbit IgG (1:10,000, Cell Signaling Technology, 7074s, 7076s). The experiments were repeated at least three times, and representative data are shown.
qRT-PCR
Total RNA was extracted with RNeasy Mini Kit (QIAGEN) and cDNA was synthesized with Revertra Ace (Toyobo). A Mx3000p qPCR system (Stratagene) was used to perform qRT-PCR. EvaGreen dye (Biotium)-based qRT-PCR was performed to detect gene expressions. A list of primers is given in Table S2.
Mass spectrometry
Whole epithelium lysis isolated from control and Myh9-KO mice at day 5 after the first TAM injection was used for anti-MYH9 immunoprecipitation (Myh9 antibody from Proteintech Group, 11128-1-AP, 1:100 dilution), and the immuneprecipitants were subjected to mass spectrometry analysis.
Transcriptomic profiling
Total RNA was isolated from mouse small intestinal epithelial cells from Ctrl and Myh9-KO mice at day 5 after the administration of TAM using TRIzol Reagent (Life Technologies) according to the manufacturer's instructions. High-throughput sequencing was performed using the Illumina HiSeq 2000. Genes with fold changes of 1.4 were regarded as differentially expressed genes. Gene set enrichment analysis (GSEA) was performed with rank gene based on fold change on the GSEA platform of the Broad Institute (http://www.broadinstitute.org/gsea/index.jsp).
Statistical analysis
All experiments were carried out with at least three biological replicates. Data shown in column graphs represent mean ± SD as indicated in the figure legends. When normality could be assumed, Student's t test analysis was used to compare differences between the two groups, as indicated in the figure legends. Statistical analysis was performed with GraphPad Prism v.6 software. The quantitation of immunoblotting was performed with ImageJ. Each experiment was independently repeated at least three times.
Data and code availability
The accession number for the RNA sequencing is GEO: GSE169225.
Author contributions
S.W., S.L., Y. Liu, Y. Li, and Y.-G.C. conceived the experiments and wrote the manuscript. S.W., S.L., Y. Liu, Y. Li, X.L., and Y.W. performed the experiments. Q.J. and J.-D.H. did the bioinformatic analysis.
Acknowledgments
We thank Drs. Sylvie Robine and Rongwen Xi for Vil-CreERT2 mice, Dr. Xiaodong Wang for Ripk3−/− mice. This work was supported by grants from the National Key Research and Development Program of China (2017YFA0103601) and the National Natural Science Foundation of China (31730056 and 31988101 to Y.-G.C.; 31900550 to Y. Liu.), and the China Postdoctoral Science Foundation to S.W.
Published: April 22, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.stemcr.2021.03.027.
Contributor Information
Yuan Liu, Email: liu-yuan@mail.tsinghua.edu.cn.
Ye-Guang Chen, Email: ygchen@tsinghua.edu.cn.
Supplemental information
References
- Anderton H., Wicks I.P., Silke J. Cell death in chronic inflammation: breaking the cycle to treat rheumatic disease. Nat. Rev. Rheumatol. 2020;16:496–513. doi: 10.1038/s41584-020-0455-8. [DOI] [PubMed] [Google Scholar]
- Barker N. Adult intestinal stem cells: critical drivers of epithelial homeostasis and regeneration. Nat. Rev. Mol. Cell Biol. 2014;15:19–33. doi: 10.1038/nrm3721. [DOI] [PubMed] [Google Scholar]
- Barker N., Clevers H. Leucine-rich repeat-containing G-protein-coupled receptors as markers of adult stem cells. Gastroenterology. 2010;138:1681–1696. doi: 10.1053/j.gastro.2010.03.002. [DOI] [PubMed] [Google Scholar]
- Barker N., van Es J.H., Kuipers J., Kujala P., van den Born M., Cozijnsen M., Haegebarth A., Korving J., Begthel H., Peters P.J. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- Barriga F.M., Montagni E., Mana M., Mendez-Lago M., Hernando-Momblona X., Sevillano M., Guillaumet-Adkins A., Rodriguez-Esteban G., Buczacki S.J.A., Gut M. Mex3a marks a slowly dividing subpopulation of Lgr5+ intestinal stem cells. Cell Stem Cell. 2017;20:801–816 e807. doi: 10.1016/j.stem.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondow B.J., Faber M.L., Wojta K.J., Walker E.M., Battle M.A. E-Cadherin is required for intestinal morphogenesis in the mouse. Dev. Biol. 2012;371:1–12. doi: 10.1016/j.ydbio.2012.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley A., Turner J.R. Cell biology of tight junction barrier regulation and mucosal disease. Cold Spring Harb. Perspect. Biol. 2018;10:a029314. doi: 10.1101/cshperspect.a029314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan F.K. Fueling the flames: mammalian programmed necrosis in inflammatory diseases. Cold Spring Harb. Perspect. Biol. 2012;4:a008805. doi: 10.1101/cshperspect.a008805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G., Hou Z., Gulbranson D.R., Thomson J.A. Actin-myosin contractility is responsible for the reduced viability of dissociated human embryonic stem cells. Cell Stem Cell. 2010;7:240–248. doi: 10.1016/j.stem.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti M.A., Adelstein R.S. Nonmuscle myosin II moves in new directions. J. Cell Sci. 2008;121:11–18. doi: 10.1242/jcs.007112. [DOI] [PubMed] [Google Scholar]
- Conti M.A., Saleh A.D., Brinster L.R., Cheng H., Chen Z., Cornelius S., Liu C., Ma X., Van Waes C., Adelstein R.S. Conditional deletion of nonmuscle myosin II-A in mouse tongue epithelium results in squamous cell carcinoma. Sci. Rep. 2015;5:14068. doi: 10.1038/srep14068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper H.S., Murthy S.N., Shah R.S., Sedergran D.J. Clinicopathologic study of dextran sulfate sodium experimental murine colitis. Lab. Invest. 1993;69:238–249. [PubMed] [Google Scholar]
- Dannappel M., Vlantis K., Kumari S., Polykratis A., Kim C., Wachsmuth L., Eftychi C., Lin J., Corona T., Hermance N. RIPK1 maintains epithelial homeostasis by inhibiting apoptosis and necroptosis. Nature. 2014;513:90–94. doi: 10.1038/nature13608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derycke L., Stove C., Vercoutter-Edouart A.S., De Wever O., Dolle L., Colpaert N., Depypere H., Michalski J.C., Bracke M. The role of non-muscle myosin IIA in aggregation and invasion of human MCF-7 breast cancer cells. Int. J. Dev.Biol. 2011;55:835–840. doi: 10.1387/ijdb.113336ld. [DOI] [PubMed] [Google Scholar]
- Grivennikov S.I. Inflammation and colorectal cancer: colitis-associated neoplasia. Semin. Immunopathol. 2013;35:229–244. doi: 10.1007/s00281-012-0352-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunther C., Martini E., Wittkopf N., Amann K., Weigmann B., Neumann H., Waldner M.J., Hedrick S.M., Tenzer S., Neurath M.F. Caspase-8 regulates TNF-alpha-induced epithelial necroptosis and terminal ileitis. Nature. 2011;477:335–339. doi: 10.1038/nature10400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunther C., Neumann H., Neurath M.F., Becker C. Apoptosis, necrosis and necroptosis: cell death regulation in the intestinal epithelium. Gut. 2013;62:1062–1071. doi: 10.1136/gutjnl-2011-301364. [DOI] [PubMed] [Google Scholar]
- Kaser A., Zeissig S., Blumberg R.S. Inflammatory bowel disease. Annu. Rev. Immunol. 2010;28:573–621. doi: 10.1146/annurev-immunol-030409-101225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawlor K.E., Khan N., Mildenhall A., Gerlic M., Croker B.A., D'Cruz A.A., Hall C., Kaur Spall S., Anderton H., Masters S.L. RIPK3 promotes cell death and NLRP3 inflammasome activation in the absence of MLKL. Nat. Commun. 2015;6:6282. doi: 10.1038/ncomms7282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N., Nakauka-Ddamba A., Tobias J., Jensen S.T., Lengner C.J. Mouse label-retaining cells are molecularly and functionally distinct from reserve intestinal stem cells. Gastroenterology. 2016;151:298–310 e297. doi: 10.1053/j.gastro.2016.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Chen Y.G. Intestinal epithelial plasticity and regeneration via cell dedifferentiation. Cell Regen. 2020;9:14. doi: 10.1186/s13619-020-00053-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Xiong X., Chen Y.G. Dedifferentiation: the return road to repair the intestinal epithelium. Cell Regen. 2020;9:2. doi: 10.1186/s13619-020-00048-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X., Adelstein R.S. The role of vertebrate nonmuscle myosin II in development and human disease. Bioarchitecture. 2014;4:88–102. doi: 10.4161/bioa.29766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanan V., Nakata T., Desch A.N., Levesque C., Boroughs A., Guzman G., Cao Z., Creasey E., Yao J., Boucher G. C1orf106 is a colitis risk gene that regulates stability of epithelial adherens junctions. Science. 2018;359:1161–1166. doi: 10.1126/science.aan0814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naydenov N.G., Feygin A., Wang D., Kuemmerle J.F., Harris G., Conti M.A., Adelstein R.S., Ivanov A.I. Nonmuscle myosin IIA regulates intestinal epithelial barrier in vivo and plays a protective role during experimental colitis. Sci. Rep. 2016;6:24161. doi: 10.1038/srep24161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odenwald M.A., Turner J.R. The intestinal epithelial barrier: a therapeutic target? Nat. Rev. Gastroenterol. Hepatol. 2017;14:9–21. doi: 10.1038/nrgastro.2016.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohgushi M., Matsumura M., Eiraku M., Murakami K., Aramaki T., Nishiyama A., Muguruma K., Nakano T., Suga H., Ueno M. Molecular pathway and cell state responsible for dissociation-induced apoptosis in human pluripotent stem cells. Cell Stem Cell. 2010;7:225–239. doi: 10.1016/j.stem.2010.06.018. [DOI] [PubMed] [Google Scholar]
- Parang B., Barrett C.W., Williams C.S. AOM/DSS model of colitis-associated cancer. Methods Mol. Biol. 2016;1422:297–307. doi: 10.1007/978-1-4939-3603-8_26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patankar J.V., Becker C. Cell death in the gut epithelium and implications for chronic inflammation. Nat. Rev. Gastroenterol. Hepatol. 2020;17:543–556. doi: 10.1038/s41575-020-0326-4. [DOI] [PubMed] [Google Scholar]
- Sato T., van Es J.H., Snippert H.J., Stange D.E., Vries R.G., van den Born M., Barker N., Shroyer N.F., van de Wetering M., Clevers H. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature. 2011;469:415–418. doi: 10.1038/nature09637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt M., Schewe M., Sacchetti A., Feijtel D., van de Geer W.S., Teeuwssen M., Sleddens H.F., Joosten R., van Royen M.E., van de Werken H.J.G. Paneth cells respond to inflammation and contribute to tissue regeneration by acquiring stem-like features through SCF/c-Kit signaling. Cell Rep. 2018;24:2312–2328 e2317. doi: 10.1016/j.celrep.2018.07.085. [DOI] [PubMed] [Google Scholar]
- Schneider M.R., Dahlhoff M., Horst D., Hirschi B., Trulzsch K., Muller-Hocker J., Vogelmann R., Allgauer M., Gerhard M., Steininger S. A key role for E-cadherin in intestinal homeostasis and Paneth cell maturation. PLoS One. 2010;5:e14325. doi: 10.1371/journal.pone.0014325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramek D., Sendoel A., Segal J.P., Beronja S., Heller E., Oristian D., Reva B., Fuchs E. Direct in vivo RNAi Screen unveils myosin IIa as a tumor suppressor of squamous cell carcinomas. Science. 2014;343:309–313. doi: 10.1126/science.1248627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T. Regulation of intestinal epithelial permeability by tight junctions. Cell. Mol. Life Sci. 2013;70:631–659. doi: 10.1007/s00018-012-1070-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda N., Jain R., LeBoeuf M.R., Wang Q.H., Lu M.M., Epstein J.A. Interconversion between intestinal stem cell populations in distinct niches. Science. 2011;334:1420–1424. doi: 10.1126/science.1213214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terzic J., Grivennikov S., Karin E., Karin M. Inflammation and colon cancer. Gastroenterology. 2010;138:2101–2114.e2105. doi: 10.1053/j.gastro.2010.01.058. [DOI] [PubMed] [Google Scholar]
- Tetteh P.W., Basak O., Farin H.F., Wiebrands K., Kretzschmar K., Begthel H., van den Born M., Korving J., de Sauvage F., van Es J.H. Replacement of lost lgr5-positive stem cells through plasticity of their enterocyte-lineage daughters. Cell Stem Cell. 2016;18:203–213. doi: 10.1016/j.stem.2016.01.001. [DOI] [PubMed] [Google Scholar]
- Vicente-Manzanares M., Ma X.F., Adelstein R.S., Horwitz A.R. Non-muscle myosin II takes centre stage in cell adhesion and migration. Nat. Rev. Mol. Cell Biol. 2009;10:778–790. doi: 10.1038/nrm2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waljee A.K., Yang Y., Wu G.D. Aaa IBD 2017: methodologies to collect and analyze clinical data to Support quality improvement initiatives designed to optimize the care of patients with IBD. Gastroenterology. 2017;153:e1–e5. doi: 10.1053/j.gastro.2017.10.020. [DOI] [PubMed] [Google Scholar]
- Walker A., Su H., Conti M.A., Harb N., Adelstein R.S., Sato N. Non-muscle myosin II regulates survival threshold of pluripotent stem cells. Nat. Commun. 2010;1:71. doi: 10.1038/ncomms1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Li H., Wu J., Cai Z.Y., Li B., Ni H., Qiu X., Chen H., Liu W., Yang Z.H. Gut stem cell necroptosis by genome instability triggers bowel inflammation. Nature. 2020;580:386–390. doi: 10.1038/s41586-020-2127-x. [DOI] [PubMed] [Google Scholar]
- Wang Y., Chiang I.L., Ohara T.E., Fujii S., Cheng J., Muegge B.D., Ver Heul A., Han N.D., Lu Q., Xiong S. Long-term culture captures injury-repair cycles of colonic stem cells. Cell. 2019;179:1144–1159 e1115. doi: 10.1016/j.cell.2019.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welz P.S., Wullaert A., Vlantis K., Kondylis V., Fernandez-Majada V., Ermolaeva M., Kirsch P., Sterner-Kock A., van Loo G., Pasparakis M. FADD prevents RIP3-mediated epithelial cell necrosis and chronic intestinal inflammation. Nature. 2011;477:330–334. doi: 10.1038/nature10273. [DOI] [PubMed] [Google Scholar]
- Wirtz S., Neufert C., Weigmann B., Neurath M.F. Chemically induced mouse models of intestinal inflammation. Nat. Protoc. 2007;2:541. doi: 10.1038/nprot.2007.41. [DOI] [PubMed] [Google Scholar]
- Yan K.S., Gevaert O., Zheng G.X.Y., Anchang B., Probert C.S., Larkin K.A., Davies P.S., Cheng Z.F., Kaddis J.S., Han A. Intestinal enteroendocrine lineage cells possess homeostatic and injury-inducible stem cell activity. Cell Stem Cell. 2017;21:78–90 e76. doi: 10.1016/j.stem.2017.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S., Tong K., Zhao Y., Balasubramanian I., Yap G.S., Ferraris R.P., Bonder E.M., Verzi M.P., Gao N. Paneth cell multipotency induced by Notch activation following injury. Cell Stem Cell. 2018;23:46–59 e45. doi: 10.1016/j.stem.2018.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B., Qi Z., Li Y.H., Wang C.K., Fu W., Chen Y.G. The non-muscle-myosin-II heavy chain Myh9 mediates colitis-induced epithelium injury by restricting Lgr5+ stem cells. Nat. Commun. 2015;6:7166. doi: 10.1038/ncomms8166. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number for the RNA sequencing is GEO: GSE169225.