Summary
Human embryonic stem cells cultured in 2D micropatterns with BMP4 differentiate into a radial arrangement of germ layers and extraembryonic cells. Single-cell transcriptomes demonstrate generation of cell types transcriptionally similar to their in vivo counterparts in Carnegie stage 7 human gastrula. Time-course analyses indicate sequential differentiation, where the epiblast arises by 12 h between the prospective ectoderm in the center and the cells initiating differentiation toward extraembryonic fates at the edge. Extraembryonic and mesendoderm precursors arise from the epiblast by 24 h, while nascent mesoderm, endoderm, and primordial germ cell-like cells form by 44 h. Dynamic changes in transcripts encoding signaling components support a BMP, WNT, and Nodal hierarchy underlying germ-layer specification conserved across mammals, and FGF and HIPPO pathways being active throughout differentiation. This work also provides a resource for mining genes and pathways expressed in a stereotyped 2D gastruloid model, common with other species or unique to human gastrulation.
Keywords: stem cell differentiation, gastruloid, gastrulation, epiblast, mesoderm, endoderm, ectoderm, primordial germ cell, WNT, NODAL
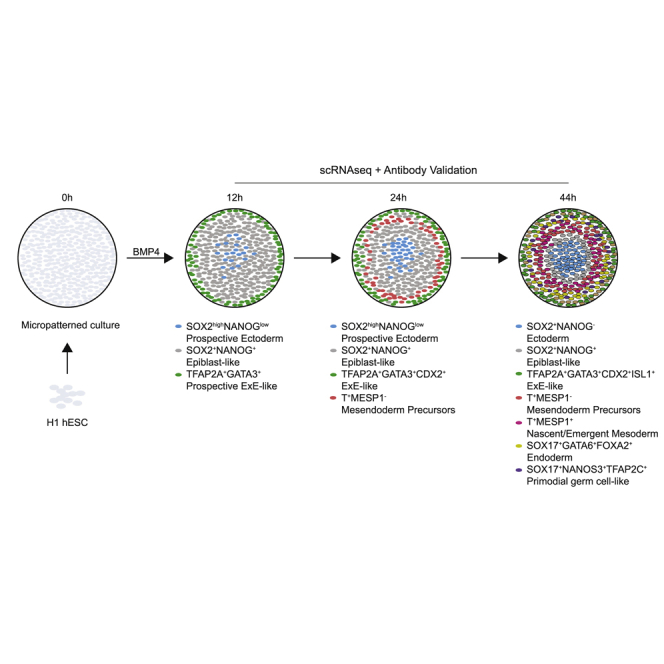
Graphical abstract
Highlights
-
•
hESCs 2D gastruloids form three germ layers, extraembryonic and PGC-like cells
-
•
scRNA-seq shows that 2D gastruloids and CS7 human gastrula have similar cell types
-
•
Sequential emergence of epiblast, ectoderm, ExE, mesoderm, endoderm, and PGC cells
-
•
Conserved BMP, WNT, and Nodal hierarchy underlying germ layer specification
In this article, Solnica-Krezel and colleagues analyze cellular transcriptomes of BMP4-induced hESC 2D micropatterned gastruloids to show they have cellular composition similar to that of Carnegie stage 7 human gastrula. They demonstrate successive emergence of epiblast and ectoderm, followed by extraembryonic and mesendoderm precursors, and finally mesoderm, endoderm, and primordial germ cells. They provide a resource for mining genes underlying human gastrulation.
Introduction
Early mammalian embryogenesis entails sequential specification of extraembryonic (ExE) and embryonic tissues, from which the three germ layers, ectoderm, mesoderm, and endoderm, arise and are shaped into the body plan during gastrulation (Shahbazi and Zernicka-Goetz, 2018). The underlying cellular and molecular mechanisms have been extensively investigated in the mouse and, more recently, in nonhuman primates, in vitro cultured human embryos, and 2D or 3D human embryonic stem cell (hESC) models (Ghimire et al., 2021).
Prior to implantation, the totipotent zygote gives rise to a blastocyst consisting of an inner cell mass (ICM), trophectoderm (TE), and primitive endoderm (PE or hypoblast). While preimplantation lineage segregation processes are generally conserved between mouse and human, differences in timing and molecular mechanisms emerge (Molè et al., 2020). In early human and monkey embryos, blastocyst cells co-express lineage-specific genes, with concurrent establishment of TE, mature hypoblast, and ICM not observed until implantation (Nakamura et al., 2016; Petropoulos et al., 2016; Xiang et al., 2019). After implantation in humans, the ICM forms an epiblast (EPI) and amniotic epithelium surrounding the amniotic cavity. The latter develops later in the mouse and via different mechanisms (Molè et al., 2020). Moreover, post-implantation EPI in humans is shaped like a flat disk, but like a cup in mouse.
Gastrulation starts with the formation of the primitive streak (PS) in the posterior side of the EPI disk. In mouse, ExE cells surrounding the EPI secrete Bmp4, which in turn induces the Wnt3 and Nodal signaling cascade in the EPI (Ben-Haim et al., 2006). WNT and NODAL activities are restricted to the posterior EPI by Lefty1, Cer1, and Dkk1 inhibitors secreted from the anterior visceral endoderm, establishing the anterior-posterior axis (Morgani and Hadjantonakis, 2020). At the posterior EPI, high Nodal and Wnt3 signaling induces the expression of T (TBXT or Brachyury), marking mesoderm precursors, which undergo the epithelial-to-mesenchymal transition (EMT), resulting in the PS formation. As the EMT wave extends the PS anteriorly, mesendoderm precursors enter via the PS and migrate to form mesoderm and endoderm, while the remaining EPI becomes ectoderm (Williams et al., 2012). Fgf8 is necessary for cell movement away from the PS in the mouse (Sun et al., 1999), but low FGF8 expression in Carnegie stage (CS) 7 human gastrula implies that alternative FGF ligands act during human gastrulation (Tyser et al., 2020). Murine Bmp4 mutants fail to express T or form mesoderm and manifest truncated posterior structures (Mishina et al., 1995). Thus, the interplay of the BMP, WNT, NODAL, and FGF pathways underlies germ layer specification and morphogenesis during mouse gastrulation (Morgani and Hadjantonakis, 2020).
Due to the ease of genetic manipulation and the ability to finely control experimental factors, in vitro stem cell systems have emerged as an invaluable tool for understanding the signaling cascades and transcriptional regulatory network underlying cell fate specification and morphogenesis during early human development (Ghimire et al., 2021). These models include 2D micropatterned and 3D gastruloid (Martyn et al, 2018, 2019a; Moris et al., 2020; Warmflash et al., 2014), and 3D early blastocyst (Rivron et al., 2018) and pre-gastrulation EPI (Simunovic et al., 2019) models. The 2D and 3D gastruloid gene expression patterns are similar to those of CS7 (Minn et al., 2020) and CS9 human embryos (Moris et al., 2020), respectively. While both 2D and 3D gastruloids differentiate into germ layers, those of the former are highly reproducible and thus suitable for investigating dynamic gene expression changes underlying cell fate emergence during early human gastrulation (Minn et al., 2020; Warmflash et al., 2014).
As in mouse gastrulation, the BMP, WNT, and NODAL pathways interact in hESC 2D micropatterned gastruloids, which form germ layers and ExE cell types upon BMP4 treatment (Chhabra et al., 2019; Etoc et al., 2016; Martyn et al., 2018; Minn et al., 2020; Tewary et al., 2017; Warmflash et al., 2014). In 2D micropatterned gastruloids, despite global BMP4 treatment, BMP signaling is restricted to the edge and activates WNT, which in turn activates NODAL signaling. WNT and NODAL activities then travel from the edge toward the center as indicated by phosphorylation and/or nuclear localization of the downstream effectors SMAD1, SMAD2/3, and CTNNB1 (β-catenin) (Chhabra et al., 2019; Etoc et al., 2016). However, the spatiotemporal transcriptional dynamics of these pathways' components and the transcription factors underlying cell fate emergence in micropatterned gastruloids are incompletely understood.
Using single-cell RNA sequencing (scRNA-seq), we previously reported that BMP4 treatment of hESCs in 2D micropatterns generates ectoderm, EPI-like cells, PS-like cells, nascent mesoderm, endoderm, human primordial germ cell (PGC)-like cells (hPGCLCs), and ExE-like cells with amnion and TE signatures (Minn et al., 2020). Here, we performed a time course of immunofluorescence (IF) and scRNA-seq on micropatterned cultures at 0, 12, 24, and 44 h of BMP4 application. We show that germ layer formation in 2D gastruloids follows the temporal order of in vivo gastrulation, with EPI and ectoderm precursors forming by 12 h and mesendoderm precursors emerging by 24 h to give rise to nascent mesoderm and endoderm by 44 h, when hPGCLCs are also present. A comparison with CS7 human gastrula (Tyser et al., 2020) showed similarities in transcriptomes and differentiation trajectories of gastruloid cells to those of their in vivo counterparts. Dynamic changes in transcripts encoding signaling components support a BMP, WNT, and NODAL hierarchy underlying germ layer specification conserved across mammals, and FGF and HIPPO signaling being active over the course of differentiation.
Results
H1 hESCs cultured on 500 μm diameter extracellular matrix microdiscs and treated for 44 h with BMP4 in mTeSR, which includes TGF-β and FGF2 ligands, reproducibly recapitulate aspects of gastrulation, namely, the formation of germ layers, and PGC-like and ExE-like cells (Minn et al., 2020; Warmflash et al., 2014). Here, we performed a time-course study of scRNA-seq and IF analyses on these 2D micropatterned cultures at 0 h (just before BMP4 addition) and 12, 24, and 44 h after BMP4 addition (Minn et al., 2020), to investigate cell differentiation trajectories and underlying dynamic gene expression, and deduce cell signaling interactions leading to the formation of these early embryonic cell types. ScRNA-seq datasets for two replicates at each time point were generated from cells pooled from 36 individual colonies per replicate (Figure S1A). In total, 9,167 cells expressing 24,190 genes were analyzed.
Sequential emergence of the seven embryonic and extraembryonic cell types
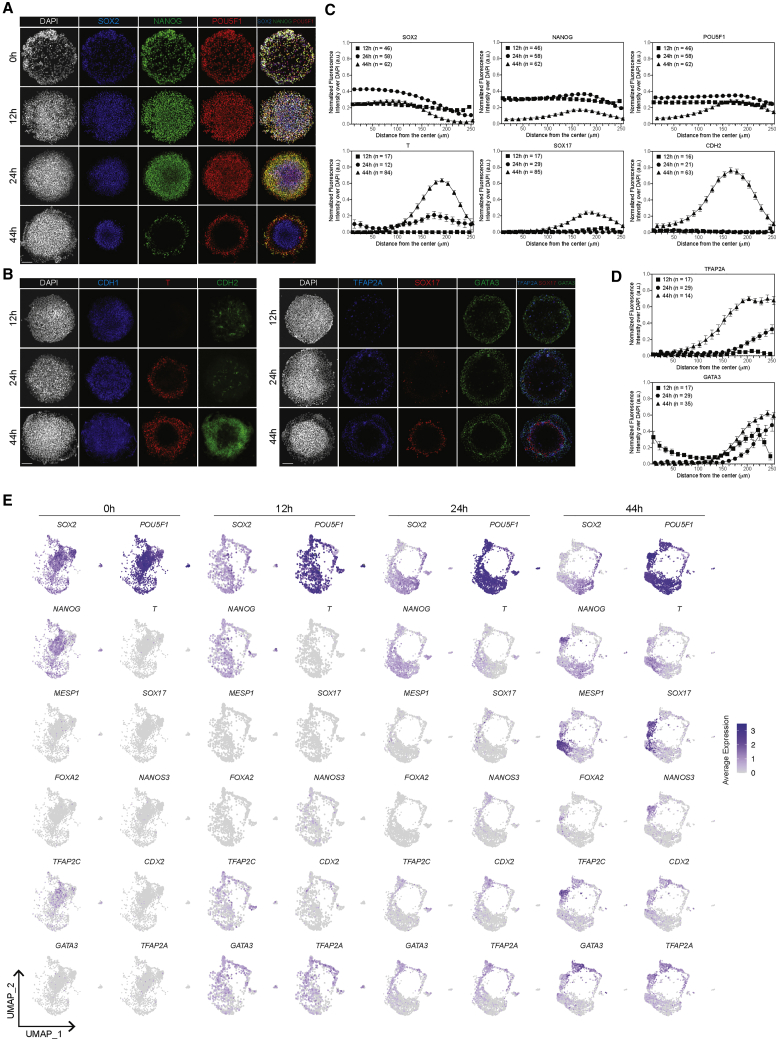
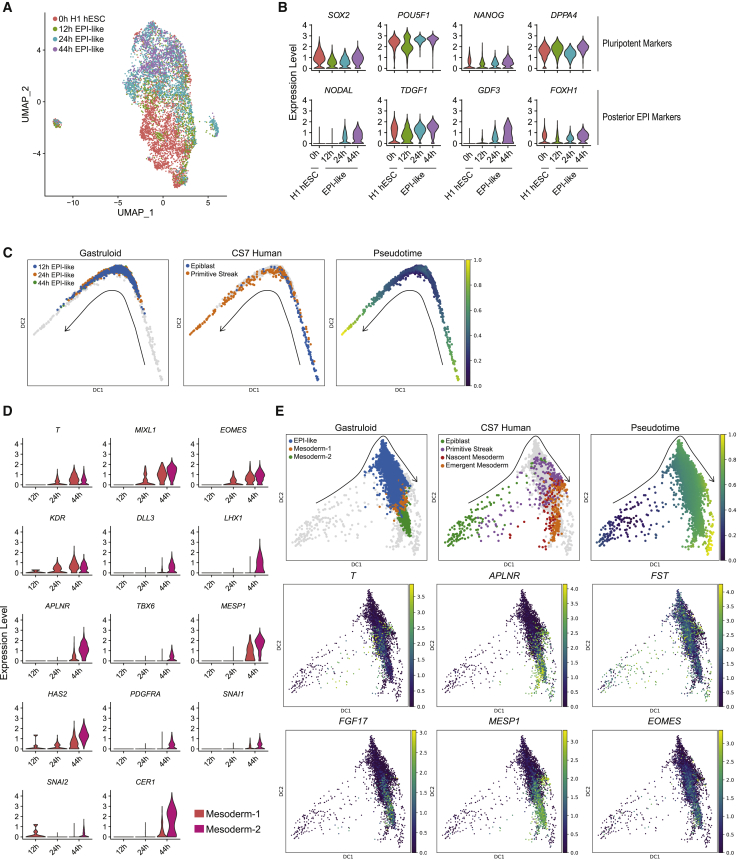
Two-dimensional micropatterned colonies before BMP4 treatment (0 h) expressed POU5F1, SOX2, and NANOG uniformly (Figures 1A and 1E). At 12 h of BMP4 treatment, high pSMAD1 staining was restricted to the colony edge (Figure S1B), consistent with previous reports (Etoc et al., 2016; Warmflash et al., 2014). TE marker GATA3 expression was generally confined to the outer edge, suggesting the initiation of differentiation (Gunne-Braden et al., 2020). However, the amnion marker TFAP2A was expressed in fewer cells and not as confined to the edge as GATA3 (Figure 1B). Neither the PS and mesoderm marker T nor the endoderm marker SOX17 was detected at this stage (Figures 1B and 1E). A few cells in the center were SOX2+ with slightly lower levels of POU5F1 and NANOG, suggestive of a prospective ectoderm identity (Figure 1A). Cells between the GATA3+ edge and the SOX2+POU5F1low center were SOX2+POU5F1highNANOGhigh, indicative of pluripotent or EPI-like characteristics.
Figure 1.
Temporal dynamics of protein and gene expression along the time course of 2D micropatterned gastruloid BMP4 treatment
(A and B) Immunofluorescence images of (A) SOX2, NANOG, and POU5F1 and (B) CDH1, T, CDH2, TFAP2A, SOX17, and GATA3 at the indicated time points. Scale bar, 100 μm.
(C and D) Quantification of normalized fluorescence intensity of (C) SOX2, NANOG, POU5F1, T, SOX17, and CDH2 and (D) TFAP2A and GATA3 after 12, 24, and 44 h of BMP4 treatment. Number of experiments, n = 1 for T, SOX17, CDH2, GATA3, and TFAP2A at 12 and 24 h; n = 1 for TFAP2A at 44 h; n = 2 for GATA3 at 44 h; n = 3 for SOX2, NANOG, and POU5F1 at 12, 24, and 44 h; n = 5 for T and CDH2 at 44 h; n = 6 for SOX17 at 44 h. Data are presented as the mean ± SEM.
(E) Uniform manifold approximation and projection (UMAP) display of indicated marker expression at indicated time points.
At 24 h, SOX2+POU5F1lowNANOG low prospective ectoderm cells were more restricted to the center (Figures 1A and 1C). IF studies showed a ring of T expression but only sporadic SOX17+ cells (Figure 1B), as corroborated by scRNA-seq (Figure 1E). In mouse gastrulae, cells in the forming PS undergo EMT, characterized by a CDH1-to-CDH2 switch. At 24 h, EMT marker CDH2 expression was sporadic and not restricted to T+ cells, while CDH1 expression was observed throughout the gastruloids. The nascent mesoderm marker MESP1 was also absent (Figure 1E). We interpret these results to mean that T+ mesendoderm precursors have emerged, but the EMT has not been initiated by 24 h. The expression of CDX2, GATA3, and TFAP2A was confined to a distinct cell cluster (Figure 1E), and IF staining indicated these cells were confined to the edge (Figures 1B and 1D).
By 44 h, the full complement of ExE, germ layers, and hPGCLCs had formed. CDH2 was restricted to T+ cells in IF, consistent with mesoderm precursors acquiring EMT characteristics (Figure 1B) (Minn et al., 2020). ScRNA-seq distinguished T+ cells into ThighMESP1low and TlowMESP1high, suggestive of PS-like and nascent mesoderm identity, respectively (Figure 1E). A distinct ring of SOX17 expression suggested endoderm formation. ScRNA-seq distinguished SOX17+ cells into SOX17+FOXA2+ endoderm and SOX17+NANOS3+TFAP2C+ hPGCLCs. Co-expression of SOX17 and TFAP2C in IF corroborated the hPGCLC appearance (Figure S1C). Taken together, these studies suggest that EPI-like, prospective ectoderm, and GATA3+ cells that initiate differentiation toward the ExE-like form as early as 12 h. Mesendoderm precursors emerge by 24 h, suggesting the initiation of gastrulation-like events, followed by the formation of mesoderm, endoderm, and hPGCLCs by 44 h.
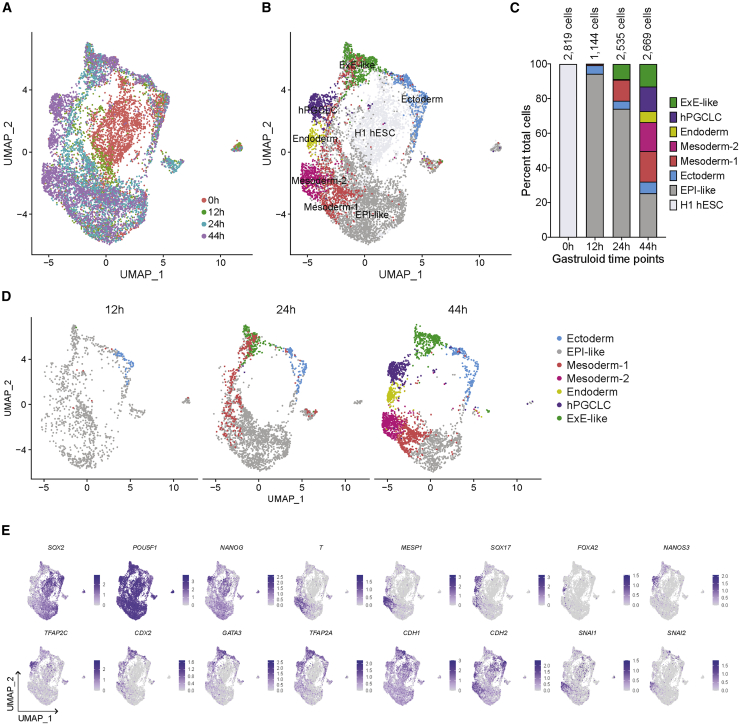
Using canonical markers and monkey and mouse gastrula datasets (Ma et al., 2019; Pijuan-Sala et al., 2019) as references, we previously annotated seven cell types in 44 h gastruloids: EPI-like, ectoderm, PS-like mesoderm-1, nascent mesoderm-like mesoderm-2, endoderm, hPGCLCs, and ExE-like with amnion and TE signatures (Minn et al., 2020). Here, we combined 0, 12, 24, and 44 h datasets using canonical correlation analysis (CCA) in Seurat (Figure 2A) (Stuart et al., 2019). Most 0 h cells formed a distinct cluster, corroborating that differentiation was initiated by 12 h of BMP4 treatment. We used Seurat to compute predicted cell-type scores for cells at 12 and 24 h using the annotated 44 h dataset as reference. Each 12 and 24 h cell with the highest prediction score for a 44 h cell type was assigned the same cell-type annotation (Figures 2B–2D, S2A, and S2B). The annotation of 12 and 24 h micropatterned cultures largely corroborated marker expression analyses described above. Most 12 h cells were predicted to be EPI-like, but the fraction decreased over time (Figure 2C; 94%, 71%, and 25% at 12, 24, and 44 h, respectively), consistent with the emergence of additional differentiated cell types by 44 h. In line with SOX2+POU5F1lowNANOGlow marker expression, 5% of 12 h cells were identified as prospective ectoderm and showed upregulated ectoderm markers VIM, NES, and ID3 (Figure S2C and Table S1). ExE-like cells were observed at 24 h (9%) and increased in proportion by 44 h (13%) (Figure 2C). At 24 and 44 h, predicted mesoderm-1 cells expressed the PS markers T and MIXL1 (Figures 2E and S2C). By 44 h, the proportion of mesoderm-1 cells was reduced, coincident with the emergence of nascent mesoderm-like mesoderm-2. Likewise, endoderm and hPGCLCs did not appear until 44 h.
Figure 2.
Sequential emergence of the seven embryonic and extraembryonic cell types in micropatterned gastruloids
(A and B) UMAP displays of cells at 0, 12, 24, and 44 h of BMP4 treatment clustered by (A) time and (B) cell type.
(C) Bar plot depicting percentage of cell types at the indicated time points.
(D) UMAPs of the seven cell clusters identified at the indicated time points.
(E) Expression of the indicated markers at 0, 12, 24, and 44 h of BMP4 treatment on the UMAP shown in (B).
Together, the IF and scRNA-seq time-course analyses revealed the differentiation dynamics of germ layers and ExE cell types over 44 h of BMP4 treatment in 2D micropatterned gastruloids: (1) at 0 h, untreated cells expressed pluripotent markers and formed a distinct cluster; (2) EPI-like cells predominated at 12 and 24 h but decreased in proportion by 44 h; (3) precursors of ectoderm and GATA3+ cells that initiated differentiation toward the ExE-like fate emerged by 12 h; (4) mesendoderm precursors or mesoderm-1 and ExE-like cells formed by 24 h; and (5) more differentiated cell types, mesoderm-2, endoderm, and hPGCLCs, were primarily observed at 44 h. All annotated clusters expressed corresponding canonical markers (Figures 2E and S2C). The replicates at each time point overlap well, supporting reproducibility of the gene-expression dynamics (Figure S2D).
Comparison of the micropatterned gastruloid cell types with the human Carnegie stage 7 gastrula
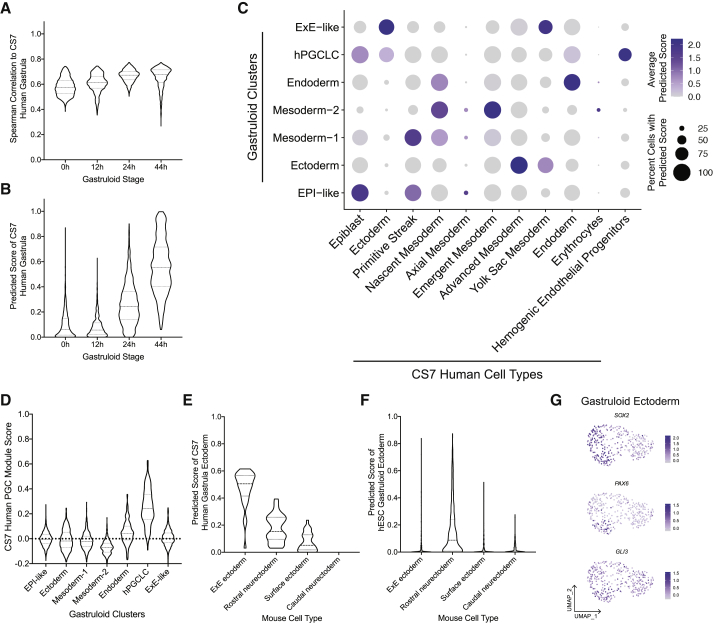
We reported that 2D hESC gastruloids treated with BMP4 for 44 h are transcriptionally similar to E7.0–7.5 mouse and 14–16 dpf (days post-fertilization) cynomolgus monkey gastrula (Minn et al., 2020), likely corresponding to CS7 human gastrulae (O’Rahilly and Müller, 1987). Comparison with the recently published scRNA-seq dataset from a single CS7 (∼16–19 dpf) human gastrula (Tyser et al., 2020) showed the closest correlation in average gene expression and the highest prediction scores of CS7 human gastrula to 44, 24, 12, and 0 h gastruloids, in that order (Figures 3A and 3B). CS7 human gastrula also transcriptionally corresponds to E7.0–7.5 mouse (Figure S3A) and 14–16 dpf cynomolgus monkey (Figure S3B), similar to 44 h hESC gastruloids (Minn et al., 2020). Thus, 2D hESC gastruloids at 44 h may represent the early to mid-gastrulation stage relevant to CS7 human gastrula.
Figure 3.
Comparison of the 2D micropatterned gastruloid cell types with the human Carnegie stage 7 gastrula
(A and B) Comparison of CS7 gastrula with BMP4 micropatterned cultures at the indicated time points using (A) average gene expression correlation and (B) anchor-based prediction.
(C) Prediction score of CS7 cell types in the indicated gastruloid cell types.
(D) Violin plot of the PGC module score in gastruloid cell types.
(E and F) Prediction scores of mouse ectodermal derivatives in ectoderm of (E) CS7 and (F) gastruloid.
(G) Expression of indicated ectoderm makers in the gastruloid ectoderm cluster.
Transcriptional profiling of the CS7 human gastrula defined 11 major cell types (Figure S3C) (Tyser et al., 2020). Using highly variable genes, we applied Seurat to calculate prediction scores of CS7 gastrula cell types in 2D gastruloids (Figure 3C). Gastruloid EPI-like, mesoderm-1, mesoderm-2, and endoderm scored highly for corresponding CS7 gastrula cell types, namely EPI, PS, nascent/emergent mesoderm, and endoderm, respectively. Within the CS7 PS, seven NANOS3+SOX17+Tlow PGCs were identified (Tyser et al., 2020). Similarly, gastruloid hPGCLCs were NANOS3+SOX17+Tlow (Figure S2C). The PGC module score using CS7 PGCs' top 20 differentially expressed genes (DEGs) (Tyser et al., 2020) revealed the highest score in the hPGCLC cluster (Figure 3D), indicating that gastruloid hPGCLCs are similar to CS7 PGCs.
In CS7 gastrula, 29 cells (2.4%) were annotated as ectoderm (Tyser et al., 2020), although its embryonic versus ExE identity was unclear, with cells co-expressing the putative primate amnion markers TFAP2A and ISL1 (Yang et al., 2020; Zheng et al., 2019) but lacking the neuroectoderm regulator SOX2 mRNA (Figure S3D). Thus, CS7 ectoderm cells received high prediction scores for monkey early and late amnion (Figure S3E) and mouse ExE ectoderm (Figure 3E). Correspondingly, we found a high prediction score of gastruloid ExE-like to CS7 ectoderm, monkey amnion and TE derivatives, and mouse ExE ectoderm (Figures S3F). In contrast, gastruloid ectoderm cells had the highest prediction score to mouse rostral neuroectoderm (Figure 3F). SOX2, expressed in neuroectoderm derivatives (Pijuan-Sala et al., 2019), was found in gastruloid ectoderm, but not in CS7 ectoderm (Figure S3D). Early neural markers PAX6 and GLI3 were also detected in gastruloid ectoderm (Figure 3G).
The comparative transcriptomic analysis with CS7 human gastrula suggests that 2D hESC gastruloids recapitulate the formation of EPI, PS, nascent/emergent mesoderm, endoderm, and PGC. However, due to the lack of distinct annotation for ectoderm derivatives and ExE TE or amnion in the CS7 dataset, we cannot conclusively determine a similarity of gastruloid ectoderm and ExE-like to their CS7 counterparts. Nonetheless, additional comparisons suggest a transcriptional similarity of gastruloid ectoderm to mouse rostral neuroectoderm (Figure 3F), and gastruloid ExE-like to monkey TE and amnion (Figure S3F).
Dynamic changes in the expression of signaling pathways' components and their transcriptional targets in the course of 2D gastruloid differentiation
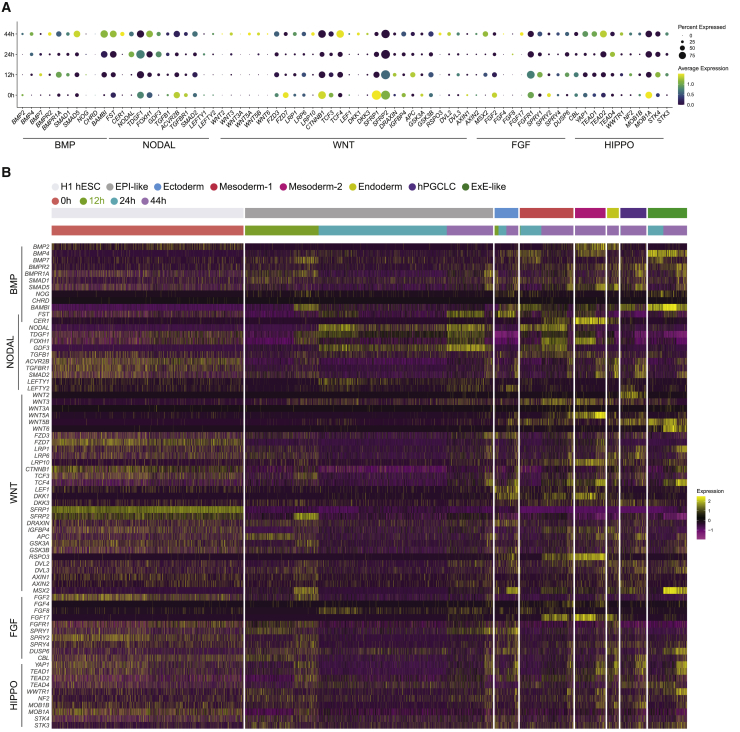
Developmental genetic studies defined conserved signaling cascades underlying the induction and patterning of germ layers in all metazoans (Solnica-Krezel, 2020). In post-implantation mouse embryos, Bmp4 ligands from ExE tissues induce in the adjacent EPI expression of Wnt3, which in turn induces expression of Nodal (Morgani and Hadjantonakis, 2020). As previously reported, despite uniform application of BMP4 to 2D micropatterned hESC cultures, its downstream effector pSMAD1 is largely restricted to the edge of each colony after 12 h, and this pattern persists at 24 and 44 h (Figure S1B) (Etoc et al., 2016). Around 24 h, the BMP4 signaling at the edge initiates WNT signaling, which in turn triggers NODAL signaling, and both WNT and NODAL signaling waves travel inward toward the colony center (Chhabra et al., 2019). Consistent with WNT and NODAL activation after 24 h, we found expression of NODAL, WNT3, and WNT5A in the rings of EPI-like and mesoderm-1/2 clusters at 44 h (Figure S4A). We further interrogated our scRNA-seq datasets to understand the expression of components and downstream targets of these and other key signaling pathways and deduce their activity and potential roles in the emergence of the seven gastruloid cell types. Overall, we found increasing expression of BMP, WNT, NODAL, and HIPPO signaling components from 12 to 44 h, while the reverse dynamics were observed for FGF signaling components (Figure 4A).
Figure 4.
Dynamic changes in the expression of genes encoding signaling pathway components in the course of 2D micropatterned gastruloid differentiation
(A) Temporal gene expression dynamics of the indicated signaling pathway components over the course of BMP4 treatment.
(B) Heatmap showing the expression of components of the indicated pathways in cells for the indicated time points and cell types.
Within the BMP pathway, the BMP2 and BMP7 genes were expressed in the 0 h hESC culture, whereas from 12 to 44 h, we found an increasing expression of genes encoding BMP2 and BMP4 ligands, BMPR2 and BMPR1A receptors, and the SMAD5 downstream effector (Figure 4A). At 44 h, the upregulation of BMP2 was specific to the mesoderm-2 and endoderm clusters (Figures 4B, S4B, and S4C). Consistent with ExE cells being a source of BMP ligands, the ExE-like cells expressed BMP4 at levels higher than other cell types (Figure S4A). Over time, the BMP target gene and negative feedback inhibitor, BAMBI (Onichtchouk et al., 1999), was also increasingly expressed, particularly in the ExE-like. Among genes encoding BMP antagonists, we saw expression of NOG and FST, but low CHRD across all time points in the ectoderm, and CER1 in mesoderm-2. Taken together, at 44 h, the strong expression of FST in the ectoderm, CER1 and BMP2 in mesoderm-2 and endoderm, and BMP4 in ExE-like is consistent with a radial gradient of BMP signaling activity, with low activity in the center, detected as early as 12 h by IF studies of the downstream effector pSMAD1 (Figure S1B).
WNT signaling is induced by BMP in early mouse embryos and 2D hESC gastruloids (Chhabra et al., 2019). We observed few cells expressing genes encoding WNT ligands during differentiation until 44 h, although receptors FZD3 and LRP6, antagonist SFRP1/2, and intercellular components CTNNB1, APC, and GS3KB were broadly expressed (Figures 4A and 4B). At 44 h, we observed expression of WNTs in specific cell populations: WNT2 in hPGCLC, WNT3 in mesoderm-1 and -2, WNT5A in mesoderm-2, WNT5B in hPGCLC and ExE-like, and WNT6 in ExE-like (Figures 4B and S4D). This suggests that different WNT ligands may be involved in signaling between discrete cell types. Among the WNT antagonists, SFRP1 was differentially expressed in ectoderm and EPI-like, and DKK1 in ectoderm and mesoderm-2, whereas SFRP2 was uniformly expressed in all clusters. Expression of WNT ligands and its target AXIN2 is indicative of signaling activity. Thus, expression of WNT ligands in all cell types except in the center ectoderm and the surrounding EPI-like ring, particularly at 44 h, suggests low WNT activity in the gastruloid center.
Within the NODAL pathway, starting at 24 h, we saw upregulation in EPI-like and mesoderm-1/2 of NODAL and GDF3 transcripts encoding the ligand heterodimer proposed to induce mesendoderm (Montague and Schier, 2017). Similar expression patterns were observed for TDGF1 and FOXH1, encoding the NODAL co-receptor and transcriptional effector, respectively (Figure 4B). Antagonist FST was expressed throughout the time course, and highest in the ectoderm (Figures 4B and S4D). NODAL feedback inhibitors LEFTY1 and LEFTY2 were expressed in EPI-like and mesoderm-1 at 24 h, while LEFTY2 also exhibited enrichment in ectoderm (Figure 4B). These RNA expression data at 24 and 44 h suggest high NODAL signaling activity in the middle ring comprising EPI-like, mesoderm, and endoderm, but low activity in the center ectoderm and ExE-like edge.
Within the FGF pathway, at 0 and 12 h, we noted high RNA expression of ligand FGF2, receptor FGFR1, and antagonists SPRY1, DUSP6, and CBL (Ornitz and Itoh, 2015), which all decreased toward 44 h (Figure 4A). Conversely, we found FGF17 transcript levels increasing toward 44 h, mainly in mesoderm-1/2 and endoderm (Figures 4B and S4F). DUSP6, encoding a negative regulator and target gene, was highly expressed in endoderm (Figure S4F). We also noticed selective upregulation in ectoderm of another negative regulator, SPRY1, suggesting a reduced FGF activity in the center. Cells that produce FGF ligands show the strongest signaling response (Morgani and Hadjantonakis, 2020). Hence, the expression at 44 h of FGF ligands (FGF2/17) (Figure 4B), peaking in mesoderm, suggests a circular ring of signaling activity that may play a role in mesoderm and endoderm formation and/or morphogenesis.
Genes encoding HIPPO pathway components were expressed increasingly throughout differentiation, except for the negative regulators STK4 (MST1) and STK3 (MST2), which showed reverse expression dynamics (Figure 4A). Interestingly, we observed high expression in the gastruloid endoderm of YAP1 and TEAD1/2 (Figures 4B and S4G), which were also expressed in CS7 definitive endoderm (DE) (Tyser et al., 2020), implying a potential role for the HIPPO pathway in endoderm formation.
EPI-like cluster represents precursors for gastrulating cells
The gastruloid EPI-like cluster scored highly for EPI in human, monkey, and mouse gastrulae (Figures 3C and S3F). CCA and uniform manifold approximation and projection (UMAP) showed the separation of EPI-like cells from 0 h BMP4-untreated cells (Figure 5A), suggesting an identity distinct from undifferentiated hESCs, but similar to human EPI at gastrulation stages. Toward 44 h, concomitant with the emergence of differentiated cell types, fewer EPI-like cells were identified (Figure 2C). Analyses of untreated 0 h cells and EPI-like cells at 12, 24, and 44 h BMP4 treatment showed expression of the pluripotent markers SOX2, POU5F1, NANOG, and DPPA4 (Figure 5B). We observed expression of the NODAL pathway's components TDGF1 and FOXH1 at 0 h, with increasing expression levels toward 44 h. GDF3 and NODAL expression was appreciably detected only at 24 h and increased by 44 h, suggesting that growing numbers of EPI-like cells at 24 and 44 h are differentiating toward PS-like or mesoderm identity. In support of this, the diffusion map showed an overlap of gastruloid EPI-like along the CS7 EPI-to-PS trajectory (Figure 5C).
Figure 5.
Characterization of gene expression and differentiation trajectory in gastruloid EPI-like and mesoderm clusters
(A) UMAP of untreated cells at 0 h and gastruloid EPI-like cells at 12, 24, and 44 h.
(B) Expression of the indicated markers in the indicated cell types over the gastruloid time course.
(C) Diffusion maps and pseudotime plots of gastruloid EPI-like and CS7 EPI and PS.
(D) Expression of the indicated PS and mesoderm markers in gastruloid mesoderm-1 and -2 clusters over the time course.
(E) Diffusion maps showing the arrangements of gastruloid EPI-like and mesoderm-1/2, and CS7 EPI, PS, and nascent/emergent mesoderm. (Top left) Gastruloid cells shown with CS7 cells in the background, (top center) CS7 cells shown with gastruloid cells in the background, and (top right) a pseudotime plot of both gastruloid and CS7 cells combined. (Bottom) Expression of the indicated posterior EPI and mesoderm markers in gastruloid and CS7 cells along the EPI-to-mesoderm transition.
Gastruloid mesoderm shows transition of primitive streak to nascent mesoderm identity
Over the course of BMP4 treatment, the majority (>99%) of 12 h (only seven cells) and 24 h mesodermal cells were mesoderm-1, whereas at 44 h, 51% were mesoderm-1 and 49% were mesoderm-2 (Figure S5A), suggesting the progression of mesoderm-1 to mesoderm-2. In CS7, T was expressed in PS and nascent mesoderm, but differentiated markers DLL3, MESP1, PDGFRA, and APLNR, as well as EMT markers SNAI1/2, were expressed only in nascent and emergent mesoderm (Figure S5B). At 24 h, only T was expressed in gastruloid mesoderm-1/2, suggestive of PS-like identity. At 44 h, mesoderm-1 expressed relatively higher levels of T, but lower levels of differentiated and EMT markers (Figure 5D). These results suggest a shift in gene expression pattern from PS-like to nascent mesoderm by 44 h. Consistent with this, CCA and UMAP of gastruloid mesoderm-1/2 and CS7 PS and mesoderm cell types showed close clustering of mesoderm-1 to PS and mesoderm-2 to nascent/emergent mesoderm (Figure S5C). A diffusion map of CS7 EPI, PS, and nascent/emergent mesoderm showed sequential ordering along the pseudotime and increasing expression of mesoderm markers (Figure 5E). Similar trajectory, pseudotime arrangement, and marker expression dynamics were observed for gastruloid EPI-like, and mesoderm-1 and -2 cells (Figure 5E), suggesting that mesoderm-1 represents a transitional cell type between EPI-like and mesoderm-2, similar to the transitional nature of PS between EPI and nascent mesoderm in vivo. Hence, we posit that 2D gastruloid formation recapitulates the temporal aspects of in vivo gastrulation, where PS emerges from EPI, and then generates mesoderm and endoderm.
In mouse, Fgf8 is required for the migration of mesodermal cells away from the PS (Sun et al., 1999), yet in primates and hESC gastruloids at 44 h, FGF8 expression is low or absent in mesoderm (Minn et al., 2020). Instead, FGF17 is upregulated in gastruloid mesoderm-2 and CS7 nascent mesoderm, implicating different FGF ligands in primate gastrulation (Figures S5D–S5F). FGF2 was absent in mouse but had high to low expression from EPI to nascent mesoderm transition in human. In gastruloids, we found constant low expression of FGF2 across EPI-like, mesoderm-1, and mesoderm-2 (Figure S5E). Other notable differences between CS7 human and mouse include upregulation of SNAI2 but downregulation of TDGF1 in CS7 EPI to nascent mesoderm transition (Tyser et al., 2020). In mouse, Snai2 exhibits low expression, whereas Tdgf1 is upregulated from EPI to nascent mesoderm transition. Similar to CS7 human, we found upregulation of SNAI2 and slight downregulation of TDGF1 in gastruloid EPI-like to mesoderm-2 transition (Figures S5D–S5F). To probe further similarities and differences in gene expression changes from EPI to nascent mesoderm (EPI-like to mesoderm-2 in gastruloids), we compared upregulated and downregulated DEGs in mouse, human, and gastruloids. We found more shared DEGs between CS7 and hESC gastruloids compared with mouse (Figure S5G; 158 versus 57 upregulated genes and 95 versus 74 downregulated genes). We also found 22 upregulated and 18 downregulated genes common in all datasets (Figure S5H), including known mesodermal markers T, MESP1, SNAI1, and MIXL1. We further identified 136 upregulated and 77 downregulated genes that are shared in CS7 and hESC gastruloids but not in mouse (Figure S5I), arguing that 2D hESC gastruloids can replicate human-specific transcriptional signature of gastrulation.
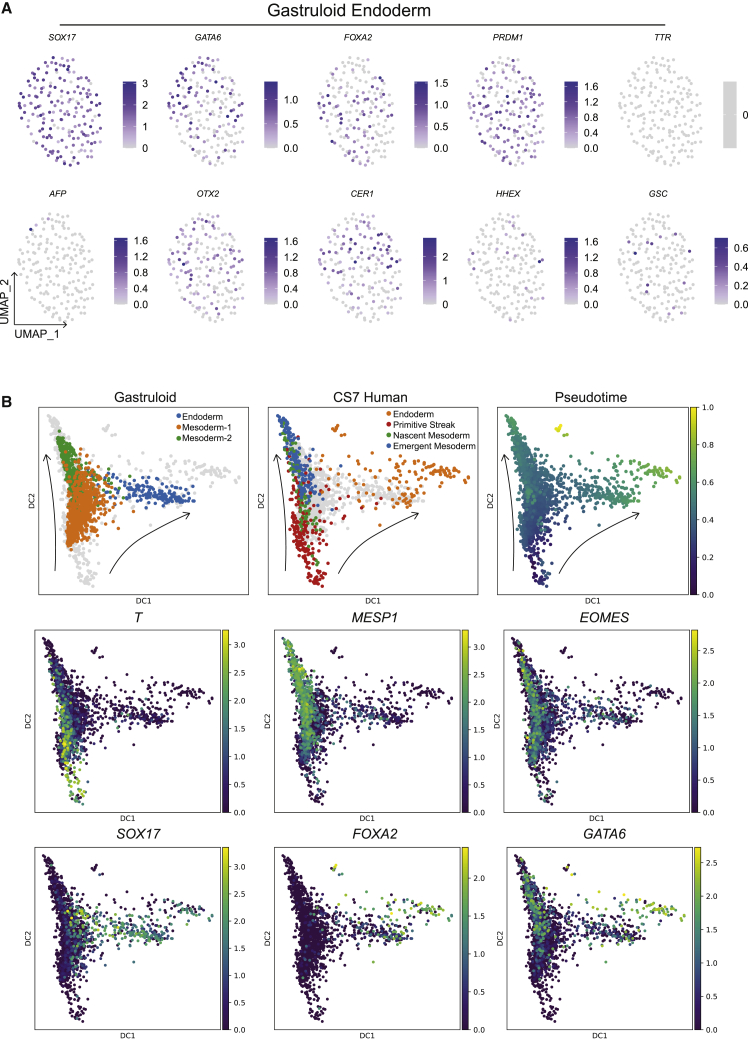
Gastruloid endoderm exhibits similar transcriptional signature to definitive endoderm
IF and scRNA-seq data indicated that the endoderm population emerged by 44 h (Figures 1B and 2D). CS7 endoderm can be discerned into DE and PE, based on TTR and AFP expression in PE (Figures S6A and S6B). Similarly, mouse ExE endoderm expressed TTR and AFP at higher levels than DE (Figure S6C). In contrast, other endoderm markers, SOX17, GATA6, and FOXA2, were expressed in all CS7 and mouse endoderm. Comparison of CS7 endoderm with E7–7.5 mouse endoderm showed the strongest gene expression correlation between respective definitive and ExE endoderm of human and mouse (Figure S6D), suggesting similar transcriptional signatures between human and mouse endoderm derivatives.
Gastruloid endoderm lacked TTR and AFP expression, but expressed SOX17, GATA6, and FOXA2, supporting a DE-like identity. Anterior PS markers OTX2, CER1, HHEX, and GSC, expressed exclusively in CS7 DE (Tyser et al., 2020), were also found in gastruloid endoderm (Figure 6A). Consistently, we found the highest gene expression correlation of gastruloid endoderm to DE of both mouse and human (Figure S6E). A diffusion map of CS7 PS, nascent/emergent mesoderm, and endoderm showed a bifurcating trajectory from PS to mesoderm or endoderm (Figure 6B). Similar trajectory, pseudotime arrangement, and marker expression were observed for gastruloid mesoderm-1 and -2 and endoderm (Figure 6B). Hence, these results support the notion that gastruloid endoderm models PS-derived DE. Given that gastruloid mesoderm-2 and endoderm appear to arise from mesoderm-1, we posit that mesoderm-1 corresponds to mesendodermal precursors.
Figure 6.
Characterization of gene expression and differentiation trajectory in gastruloid endoderm
(A) Expression of endoderm and anterior PS markers.
(B) Diffusion maps showing arrangements of gastruloid mesoderm-1/2 and endoderm, and CS7 PS, nascent/emergent mesoderm, and endoderm. (Top left) Gastruloid cells shown with CS7 cells in the background, (top center) CS7 cells shown with gastruloid cells in the background, and (top right) a pseudotime plot of both gastruloid and CS7 cells combined. (Bottom) Expression of the indicated mesoderm and endoderm markers in gastruloid and CS7 cells along PS-to-mesoderm and PS-to-endoderm trajectories.
Characterization of gastruloid ExE-like cells
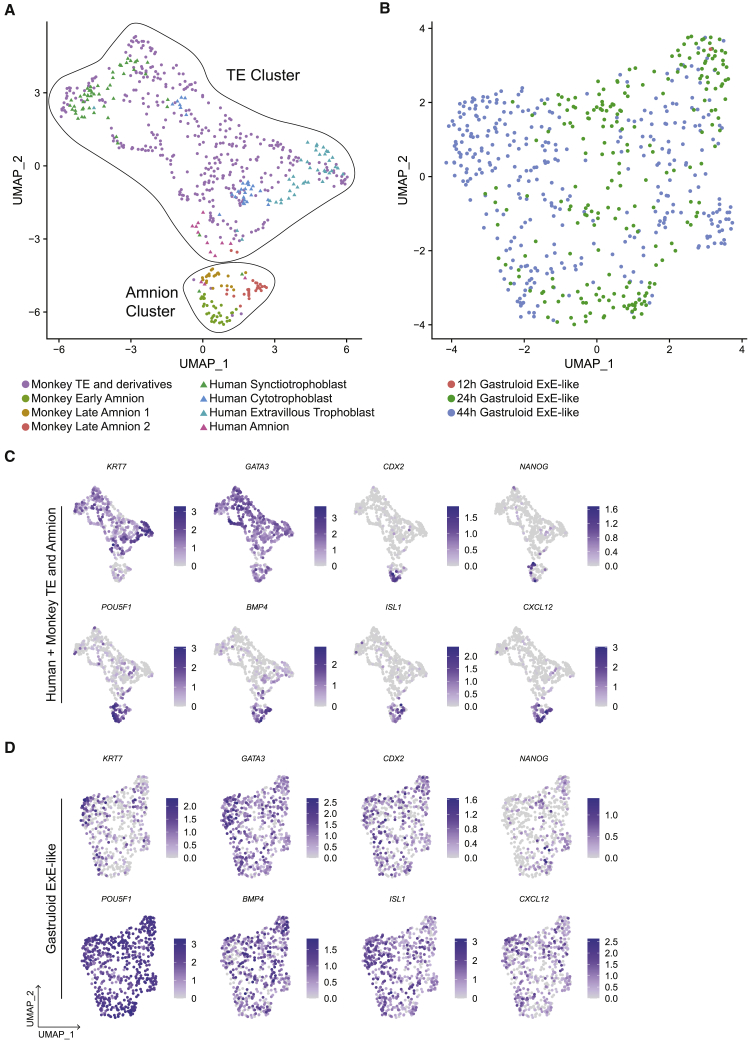
ExE markers GATA3 and TFAP2A were detected by IF and scRNA-seq as early as 12 h (Figures 1A and 1E). However, ExE-like cells were identified only at 24 and 44 h (Figures 2C and 2D). Thus, BMP4 treatment induced GATA3 expression by 12 h in the outermost cells, which were also positive for pSMAD1 (Figure S1B) and later differentiated into ExE-like cells, consistent with GATA3 mediating BMP4-induced differentiation in hESC (Gunne-Braden et al., 2020). We combined TE and amnion datasets from in vitro monkey (Ma et al., 2019) and pre-gastrulation human embryos (Xiang et al., 2019) to determine specific markers for both cell types (Figure 7A). The combined amnion cluster expressed CDX2, NANOG, POU5F1, BMP4, ISL1, and CXCL12, but TE expressed KRT7 and GATA3 (Figure 7C). However, both TE and amnion markers were expressed across all gastruloid ExE-like cells (Figures 7B and 7D). Among DEGs of the combined amnion and TE cluster identified in an unbiased manner, we found 41 TE markers and 54 amnion markers upregulated in the gastruloid ExE-like (Figure S7A). Some markers were expressed at 24 h, and increased toward 44 h, suggesting a progression of ExE-like formation along the time course. Likewise, the prediction score for monkey late amnion-2 and TE increased from 12 to 44 h (Figure S7B). Overall, we interpret these results to mean that gastruloids form an ExE cell type with both amnion and TE signatures, consistent with our analyses at 44 h (Minn et al., 2020), and that both signatures increase over time.
Figure 7.
Transcriptional characterization of gastruloid ExE-like cells
(A and B) UMAPs of (A) in vitro D6–14 human and 11–17 dpf monkey TE and amnion and (B) gastruloid ExE-like cells.
(C and D) Expression of the indicated markers in (C) human and monkey TE and amnion and (D) gastruloid ExE-like cells.
Gastruloid hPGCLCs have transcriptional signatures of gastrulating and amnion cells
In gastruloids, hPGCLCs arise by 44 h of BMP4 treatment, evidenced by SOX17, NANOS3, and TFAP2C co-expression (Figure 2B). These cells exhibited transcriptional signatures similar to those of CS7 and monkey PGCs (Figures S7C–S7E). Over time, in gastruloid hPGCLCs, expression of amnion markers TFAP2A and CDX2, and gastrulating markers T and MIXL1 increased, but that of TE markers GATA3 and KRT7 decreased (Figure S7E). Thus, gastruloid hPGCLCs exhibit gastrulating and amnion transcriptional signatures, as reported for hPGCLCs derived from hESCs in 3D aggregates (Chen et al., 2019). Likewise, we found expression of the gastrulation marker T and ExE markers TFAP2A, GATA3, and CDX2 in monkey PGCs (Figure S7C). However, in CS7 PGCs, only the gastrulation and no amnion signature was observed (Figure S7D). In monkey, PGCs arise in the amnion, with high BMP and WNT signaling, suggesting their underlying role in PGC specification. In 2D micropatterned gastruloids, BMP4 was highly expressed in the ExE-like population, whereas its receptors BMPR1A and BMPR2, as well as target genes SMAD1 and SMAD5, were expressed in hPGCLCs, supporting a potential role of BMP4 signaling from ExE-like populations to hPGCLCs (Figure S4B). Furthermore, complementary expression of WNT ligands (WNT6 and WNT5B) in the ExE-like and receptors (FZD3 and LRP1/6) along with expression of WNT2 and WNT5B in hPGCLCs points to WNT signaling having a role in gastruloid hPGCLC specification (Figure 4C).
Discussion
In vitro hESC models have been recently used to study aspects of human gastrulation, including specification of germ layers, organizer, and morphogenetic processes such as the EMT and tissue elongation (Martyn et al, 2018, 2019b; Moris et al., 2020; Simunovic et al., 2019; Warmflash et al., 2014). Using scRNA-seq, we previously demonstrated that 44 h BMP4 treatment of hESCs cultured on a 2D micropattern induces formation of prospective ectoderm, two types of mesoderm, endoderm, EPI-like cells, ExE-like cells, and hPGCLCs (Minn et al., 2020).
The recently available scRNA-seq dataset of one CS7 human gastrula (Tyser et al., 2020) enabled interrogating the 2D hESC gastruloid model. Cross-comparison with CS7 human, mouse (Pijuan-Sala et al., 2019), and monkey (Ma et al., 2019) gastrulae showed that the global transcriptome of hESC gastruloids at 44 h BMP4 treatment resembles that of CS7, E6.0–6.5 mouse, and 14–16 dpf monkey (Figures 3 and S3) (Minn et al., 2020), consistent with the 2D hESC gastruloid modeling aspects of early to mid-gastrulation. Further comparison with CS7 cell types indicated that 2D hESC gastruloids form cell types that are transcriptionally similar to EPI, PS, nascent/emergent mesoderm, DE, and PGC (Figure 3). However, gastruloid prospective ectoderm is not transcriptionally similar to CS7 ectoderm, possibly due to the latter comprising surface and ExE ectoderm (Tyser et al., 2020). Rather, gastruloid ectoderm is transcriptionally similar to mouse rostral neuroectoderm, and expresses markers of nascent neural tissues such as SOX2, GLI3, and PAX6 (Pijuan-Sala et al., 2019).
The outermost gastruloid cells have ExE identity as indicated by transcriptional similarity with in vitro-cultured D12 and D14 human (Xiang et al., 2019) and 11–17 dpf monkey amnion and TE (Ma et al., 2019). However, distinct amnion or TE identities could not be discerned in gastruloids based on marker gene expression at 44 (Minn et al., 2020) and 24 h time points (this study). Accordingly, a separate study proposed that BMP-treated hESCs are transcriptionally more similar to amnion than TE in the cynomolgus monkey (Chhabra and Warmflash, 2021). These observations are consistent with naive hESCs having a higher potential than primed hESCs to differentiate into ExE lineages and directly give rise to PE (Linneberg-Agerholm et al., 2019) or trophoblast stem cells (Dong et al., 2020).
The emergence of cellular complexity in BMP4-induced 2D gastruloid resembles CS7 human gastrula
Here, we characterized spatiotemporal changes in gene expression over the course of 0,12, 24, and 44 h of BMP4 treatment in 2D micropatterned gastruloids. Our analyses indicate a successive emergence of cell fates, with EPI-like cells, prospective ectoderm, and GATA3+ cells that initiated differentiation by 12 h after BMP4 application, followed by mesendoderm precursors mesoderm-1 and ExE-like by 24 h, and finally, mesoderm-2, endoderm, and hPGCLCs by 44 h. The early emergence of prospective ectoderm is in line with the earlier chromatin accessibility of ectoderm enhancers compared with that of mesendoderm in mouse gastrulae (Argelaguet et al., 2019). The sequential emergence of EPI-like at 12 h, followed by T+MESP1− PS-like mesoderm-1 by 24 h, and T+MESP1+ mesoderm-2 and SOX17+FOXA2+ endoderm by 44 h is consistent with studies in mouse, in which posterior EPI gives rise to PS, where cells undergo EMT, migrate, and differentiate into mesoderm and endoderm (Tam and Behringer, 1997). Moreover, the transition from gastruloid EPI-like to mesoderm-1 to mesoderm-2 follows a similar trajectory from EPI to PS to nascent/emergent mesoderm in CS7 human gastrula (Tyser et al., 2020). Similar bifurcating trajectories from CS7 PS to endoderm and mesoderm were found in gastruloid mesoderm-1 to endoderm and mesoderm-2 (Figures 5C, 5E, and 6B), suggesting that mesoderm-1 corresponds to mesendoderm precursors. Based on these analyses we posit that 2D gastruloids model not only the formation but also the sequential emergence and differentiation trajectories of germ layers and ExE cell types during human gastrulation.
Signaling processes underlying cell fate specification in 2D micropatterned gastruloid
Extensive genetic and embryologic studies in fish, frog, chick, and mouse have identified BMP, FGF, Nodal, and Wnt as key signaling pathways underlying vertebrate gastrulation (Solnica-Krezel, 2020). In post-implantation mouse embryos, Bmp4 from ExE induces Wnt3 in posterior EPI, which in turn activates Nodal and induces expression of T, required for the initiation of gastrulation (Ben-Haim et al., 2006). Bmp, Wnt, and Nodal activities are restricted to the posterior by the key inhibitors Cer1, Lefty1, and Dkk1, secreted by anterior visceral endoderm and EPI. By contrast, BMP4 is uniformly applied in 2D micropatterned gastruloids, yet a similar signaling hierarchy with BMP inducing WNT, and WNT inducing NODAL, has been described (Chhabra et al., 2019; Martyn et al., 2018). Likewise, our scRNA-seq indicates high expression of BMP4 and the negative regulator target gene BAMBI in the edge ExE-like cells, suggestive of signaling activity and a source of BMP ligands, similar to mouse (Lawson et al., 1999) and monkey amnion (Yang et al., 2020). WNT3 was expressed in mesoderm-1/2 and endoderm, and NODAL in mesoderm-1 and EPI-like (Figure 4), suggestive of their signaling activities in the domain between the center and the edge of gastruloids, as previously reported (Chhabra et al., 2019; Martyn et al., 2018). We further showed comprehensive expression patterns of BMP, WNT, NODAL, FGF, and HIPPO pathway genes over the time course of hESC gastruloid differentiation, paving the way for elucidating additional signaling interactions underlying hESC gastruloid formation.
Previous reports on micropatterned cultures using RUES2 hESCs attributed subcellular receptor localization and antagonist NOG expression in the center to a radial BMP4 signaling gradient (Etoc et al., 2016). Consistent with this, we found increasing expression of NOG in the center ectoderm over time (Figure 4B). FST, encoding both BMP and NODAL antagonist, was also high in the center cells (Figure 4). Similar to 2D gastruloids, in the CS7 gastrula, a few cells express NOG, but FST expression is prevalent in EPI, PS, and nascent mesoderm (Tyser et al., 2020).
In monkey, ISL1+ amnion expresses BMP4, and loss of amnion results in impaired BMP4 signaling and PS formation (Yang et al., 2020). Similarly, in 2D gastruloids, ISL1+ ExE-like cells strongly expressed BMP4, while the adjacent mesoderm-1 cells expressed BMPR2 and BMPR1A receptor. Strong expression of CER1, which can inhibit BMP, WNT, and NODAL (Piccolo et al., 1999), was observed in CS7 nascent mesoderm and gastruloid mesoderm-2. However, LEFTY2, expressed in CS7 nascent/emergent mesoderm and mouse nascent mesoderm to restrict NODAL activity to anterior PS and the organizer region, was expressed in few gastruloid cells. Hence, in gastruloids, some aspects of NODAL regulation may not be fully recapitulated, consistent with the absence of the organizer cells in BMP4-induced 2D gastruloid (Martyn et al., 2018; Minn et al., 2020).
The induction of Wnt3 by Bmp4 in posterior EPI is necessary for the induction of T and gastrulation in mouse (Ben-Haim et al., 2006). In addition, Wnt3a, which has an overlapping expression with Wnt3, is necessary for T expression in mouse (Yamaguchi et al., 1999). Although we saw expression of WNT3 and its target gene T in an overlapping pattern, only a few WNT3A-expressing cells were found in gastruloids and CS7 gastrula, suggesting that WNT3 encodes a predominant ligand inducing T expression during human gastrulation. Consistent with WNT activity in gastruloid mesoderm cells, RSPO3, encoding an amplifier for WNT/β-catenin signaling (Kazanskaya et al., 2008), was distinctly expressed in gastruloid mesoderm-2. RSPO3 is also expressed in CS7 nascent mesoderm and mouse mesoderm (Pijuan-Sala et al., 2019; Tyser et al., 2020). CS7 nascent mesoderm and gastruloid mesoderm-2 also share expression of DKK1, encoding a WNT antagonist (Glinka et al., 1998), although its expression is not observed in mouse mesoderm. Overall, our results suggest conservation of WNT3 in inducing T in human and mouse, but also a potential role of DKK1 in human mesoderm differentiation.
FGF signaling, particularly Fgf8-Fgfr1 interaction, is necessary for mesoderm cell migration in mouse gastrulae (Sun et al., 1999). Whereas FGFR1 is broadly expressed in both hESC gastruloid and human CS7, very few cells express FGF8 (Tyser et al., 2020). Rather, nascent mesoderm in CS7 expresses FGF3/17, while in gastruloids, FGF17 was primarily expressed in mesoderm-2, implying different FGF ligands are involved in human gastrulation.
Perspective
Our work extends previous studies on 2D micropatterned gastruloid differentiation by elucidating aspects of in vivo gastrulation that gastruloids model, including temporal emergence of cell types and comprehensive gene expression changes over the course of 44 h BMP4 treatment. Although the CS7 human dataset is an invaluable resource for querying conservation of cell types and gene expression in the hESC gastruloid systems, the dataset is based on one embryo (Tyser et al., 2020), highlighting the importance of comparisons with mouse and monkey. Using studies from all three mammals as reference, we present data suggesting that gastruloids recapitulate temporal emergence and differentiation trajectories of germ layers of in vivo gastrulation. Our data also support conservation of the BMP4, WNT3, and NODAL signaling hierarchy as underlying germ-layer formation, as well as revealing potential features specific to humans, such as DKK1, CER1, and FGF17 in mesoderm development. The datasets generated by our work will inform future functional studies to further elucidate signaling and transcriptional networks underlying hESC gastruloid germ-layer formation and, hence, provide insights into in vivo human gastrulation.
Experimental procedures
Microcontact printing
Polydimethylsiloxane stamps with 66 microcylinders of 500 μm diameter each were fabricated using standard photolithography methods as previously described (Minn et al., 2020).
BMP4 differentiation in 2D micropatterns
H1 hESCs of karyotype 46, XY (WiCell, Madison, WI), routinely cultured in Matrigel-coated plates with mTeSR medium (STEMCELL Technologies, Vancouver, BC, Canada), were differentiated with BMP4 in the micropatterned substrates as previously described (Minn et al., 2020).
Immunofluorescence staining and microscopy
After 12, 24, and 44 h, micropatterned cells were washed with PBS, fixed in 4% paraformaldehyde for 30 min, and rinsed twice with PBS at room temperature. Fixed cells were incubated in the blocking solution (0.1% Triton X and 3% normal donkey serum) for 30 min at room temperature. Primary antibodies diluted in the blocking solution were incubated at 4°C overnight. Cells were then washed with the washing solution (0.1% Tween 20 in PBS) three times, 15 min each, at room temperature. Afterward, secondary antibodies and DAPI at 4 μg/mL (Invitrogen, Carlsbad, CA) in the blocking solution were added for 1 h at room temperature, after which the cells were washed with the washing solution three times, 15 min each, at room temperature.
Images were acquired on an Olympus IX81 inverted spinning disk confocal microscope with 10× or 20× lenses. z-stack images ∼150 μm thick were acquired for samples treated with BMP4 for 44 h, but single 2D images were acquired for 12 and 24 h samples. The z-stack images of 44 h samples were projected into single 2D images before analysis.
IF images were processed and analyzed in Fiji (Schindelin et al., 2012). For fluorescence intensity quantification, each image was converted into binary masks. The Concentric Circles plugin was used to overlay 20 equally spaced concentric circles on each masked image, and average fluorescence intensity was measured along each concentric circle. The fluorescence intensity of each marker at each concentric circle was normalized against that of DAPI from corresponding concentric circles of corresponding images. Last, normalized fluorescence intensity was averaged across multiple colonies per marker and presented.
scRNA-seq and data analysis
Single cell collection and scRNA-seq and data analyses were performed as described previously (Minn et al., 2020) and in more detail in the supplemental information.
Data and code availability
The scRNA-seq datasets described in this article were deposited in the GEO database repository under accession GEO nos. GSE144897 and GSE169074.
Author contributions
Conceptualization, K.T.M., S.A.M., and L.S.K.; methodology, K.T.M., S.D., S.E.W., S.A.M., and L.S.K.; validation, K.T.M.; formal analysis, K.T.M., L.S.K., and S.D.; investigation, K.T.M. and S.A.M.; data curation, S.A.M. and S.E.W.; writing – original draft, K.T.M. and L.S.K.; writing – review & editing, K.T.M., S.D., S.A.M., S.E.W., and L.S.K.
Acknowledgments
We thank Sandra Lam (Washington University School of Medicine in St. Louis) and Steven C. George (University of California in Davis) for their guidance and assistance in fabricating polydimethylsiloxane stamps. We thank Yuheng Fu and Chuner Guo (Washington University School of Medicine in St. Louis) for constructing parts of the data curation libraries. We thank Drs. Blerta Stringa and Florence Marlow for comments on the manuscript. This study was funded by Washington University School of Medicine in St. Louis, and a grant from the Children's Discovery Institute, St. Louis to L.S.K., and a Vallee Scholar Award, an Allen Distinguished Investigator Award (through the Paul G. Allen Frontiers Group), a Sloan Research Fellowship, and a New York Stem Cell Foundation Robertson Investigator award to S.A.M.
Published: April 22, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.stemcr.2021.03.031.
Supplemental information
References
- Argelaguet R., Clark S.J., Mohammed H., Stapel L.C., Krueger C., Kapourani C.-A., Imaz-Rosshandler I., Lohoff T., Xiang Y., Hanna C.W. Multi-omics profiling of mouse gastrulation at single-cell resolution. Nature. 2019;576:487–491. doi: 10.1038/s41586-019-1825-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Haim N., Lu C., Guzman-Ayala M., Pescatore L., Mesnard D., Bischofberger M., Naef F., Robertson E.J.J., Constam D.B. The nodal precursor acting via activin receptors induces mesoderm by maintaining a source of its convertases and BMP4. Dev. Cell. 2006;11:313–323. doi: 10.1016/j.devcel.2006.07.005. [DOI] [PubMed] [Google Scholar]
- Chen D., Sun N., Hou L., Kim R., Faith J., Aslanyan M., Tao Y., Zheng Y., Fu J., Liu W. Human primordial germ cells are specified from lineage-primed progenitors. Cell Rep. 2019;29:4568–4582. doi: 10.1016/j.celrep.2019.11.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chhabra S., Liu L., Goh R., Kong X., Warmflash A. Dissecting the dynamics of signaling events in the BMP, WNT, and NODAL cascade during self-organized fate patterning in human gastruloids. PLoS Biol. 2019;17:e3000498. doi: 10.1371/journal.pbio.3000498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chhabra S., Warmflash A. BMP-treated human embryonic stem cells transcriptionally resemble amnion cells in the monkey embryo. bioRxiv. 2021 doi: 10.1101/2021.01.21.427650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C., Beltcheva M., Gontarz P., Zhang B., Popli P., Fischer L.A., Khan S.A., Park K.M., Yoon E.J., Xing X. Derivation of trophoblast stem cells from naïve human pluripotent stem cells. Elife. 2020;9 doi: 10.7554/eLife.52504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etoc F., Metzger J., Ruzo A., Kirst C., Yoney A., Ozair M.Z., Brivanlou A.H., Siggia E.D. A balance between secreted inhibitors and edge sensing controls gastruloid self-organization. Dev. Cell. 2016;39:302–315. doi: 10.1016/j.devcel.2016.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghimire S., Mantziou V., Moris N., Martinez Arias A. Human gastrulation: the embryo and its models. Dev. Biol. 2021 doi: 10.1016/j.ydbio.2021.01.006. [DOI] [PubMed] [Google Scholar]
- Glinka A., Wu W., Delius H., Monaghan A.P., Blumenstock C., Niehrs C. Dickkopf-1 is a member of a new family of secreted proteins and functions in head induction. Nature. 1998;391:357–362. doi: 10.1038/34848. [DOI] [PubMed] [Google Scholar]
- Gunne-Braden A., Sullivan A., Gharibi B., Sheriff R.S.M., Maity A., Wang Y.F., Edwards A., Jiang M., Howell M., Goldstone R. GATA3 mediates a Fast, Irreversible commitment to BMP4-driven differentiation in human embryonic stem cells. Cell Stem Cell. 2020;26:693–706.e9. doi: 10.1016/j.stem.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazanskaya O., Ohkawara B., Heroult M., Wu W., Maltry N., Augustin H.G., Niehrs C. The Wnt signaling regulator R-spondin 3 promotes angioblast and vascular development. Development. 2008;135:3655–3664. doi: 10.1242/dev.027284. [DOI] [PubMed] [Google Scholar]
- Lawson K.A., Dunn N.R., Roelen B.A.J., Zeinstra L.M., Davis A.M., Wright C.V.E., Korving J.P.W.F.M., Hogan B.L.M. Bmp4 is required for the generation of primordial germ cells in the mouse embryo. Genes Dev. 1999;13:424–436. doi: 10.1101/gad.13.4.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linneberg-Agerholm M., Wong Y.F., Herrera J.A.R., Monteiro R.S., Anderson K.G.V., Brickman J.M. Naïve human pluripotent stem cells respond to Wnt, Nodal and LIF signalling to produce expandable naïve extra-embryonic endoderm. Dev. 2019;146 doi: 10.1242/dev.180620. [DOI] [PubMed] [Google Scholar]
- Ma H., Zhai J., Wan H., Jiang X., Wang X., Wang L., Xiang Y., He X., Zhao Z.-A., Zhao B. In vitro culture of cynomolgus monkey embryos beyond early gastrulation. Science. 2019;366:eaax7890. doi: 10.1126/science.aax7890. [DOI] [PubMed] [Google Scholar]
- Martyn I., Brivanlou A.H., Siggia E.D. A wave of WNT signalling balanced by secreted inhibitors controls primitive streak formation in micropattern colonies of human embryonic stem cells. Development. 2019;146:dev.172791. doi: 10.1242/dev.172791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martyn I., Kanno T.Y., Ruzo A., Siggia E.D., Brivanlou A.H. Self-organization of a human organizer by combined Wnt and Nodal signalling. Nature. 2018;558:132–135. doi: 10.1038/s41586-018-0150-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martyn I., Siggia E.D., Brivanlou A.H. Mapping cell migrations and fates in a gastruloid model to the human primitive streak. Development. 2019;146:dev179564. doi: 10.1242/dev.179564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minn K.T., Fu Y.C., He S., Dietmann S., George S.C., Anastasio M.A., Morris S.A., Solnica-Krezel L. High-resolution transcriptional and morphogenetic profiling of cells from micropatterned human esc gastruloid cultures. Elife. 2020;9:1–34. doi: 10.7554/eLife.59445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishina Y., Suzuki A., Ueno N., Behringer R.R. Bmpr encodes a type I bone morphogenetic protein receptor that is essential for gastrulation during mouse embryogenesis. Genes Dev. 1995;9:3027–3037. doi: 10.1101/gad.9.24.3027. [DOI] [PubMed] [Google Scholar]
- Molè M.A., Weberling A., Zernicka-Goetz M. Academic Press Inc; 2020. Comparative Analysis of Human and Mouse Development: From Zygote to Pre-gastrulationCurrent Topics in Developmental Biology; pp. 113–138. [DOI] [PubMed] [Google Scholar]
- Montague T.G., Schier A.F. Vg1-nodal heterodimers are the endogenous inducers of mesendoderm. Elife. 2017;6 doi: 10.7554/eLife.28183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgani S.M., Hadjantonakis A.K. Signaling regulation during gastrulation: insights from mouse embryos and in vitro systems. Curr. Top. Dev. Biol. 2020:391–431. doi: 10.1016/bs.ctdb.2019.11.011. [DOI] [PubMed] [Google Scholar]
- Moris N., Anlas K., van den Brink S.C., Alemany A., Schröder J., Ghimire S., Balayo T., van Oudenaarden A., Martinez Arias A. An in vitro model of early anteroposterior organization during human development. Nature. 2020;582:410–415. doi: 10.1038/s41586-020-2383-9. [DOI] [PubMed] [Google Scholar]
- Nakamura T., Okamoto I., Sasaki K., Yabuta Y., Iwatani C., Tsuchiya H., Seita Y., Nakamura S., Yamamoto T., Saitou M. A developmental coordinate of pluripotency among mice, monkeys and humans. Nature. 2016;537:57–62. doi: 10.1038/nature19096. [DOI] [PubMed] [Google Scholar]
- O’Rahilly R., Müller F. Carnegie Institution of Washington; 1987. Developmental Stages in Human Embryos : Including a Revision of Streeter’s “Horizons” and a Survey of the Carnegie Collection. [Google Scholar]
- Onichtchouk D., Chen Y.G., Dosch R., Gawantka V., Delius H., Massagué J., Niehrs C. Silencing of TGF-β signalling by the pseudoreceptor BAMBI. Nature. 1999;401:480–485. doi: 10.1038/46794. [DOI] [PubMed] [Google Scholar]
- Ornitz D.M., Itoh N. The fibroblast growth factor signaling pathway. Wiley Interdiscip. Rev. Dev. Biol. 2015;4:215–266. doi: 10.1002/wdev.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petropoulos S., Edsga D., Reinius B., Linnarsson S. Single-cell RNA-seq reveals lineage and X chromosome dynamics in human preimplantation embryos. Cell. 2016;165:1–15. doi: 10.1016/j.cell.2016.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccolo S., Agius E., Leyns L., Bhattacharyya S., Grunz H., Bouwmeester T., De Robertis E.M. The head inducer cerberus is a multifunctional antagonist of Nodal, BMP and Wnt signals. Nature. 1999;397:707–710. doi: 10.1038/17820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pijuan-Sala B., Griffiths J.A., Guibentif C., Hiscock T.W., Jawaid W., Calero-Nieto F.J., Mulas C., Ibarra-Soria X., Tyser R.C.V., Ho D.L.L. A single-cell molecular map of mouse gastrulation and early organogenesis. Nature. 2019;566:490–495. doi: 10.1038/s41586-019-0933-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivron N.C., Frias-Aldeguer J., Vrij E.J., Boisset J.-C.C., Korving J., Vivié J., Truckenmüller R.K., van Oudenaarden A., van Blitterswijk C.A., Geijsen N. Blastocyst-like structures generated solely from stem cells. Nature. 2018;557:106–111. doi: 10.1038/s41586-018-0051-0. [DOI] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahbazi M.N., Zernicka-Goetz M. Deconstructing and reconstructing the mouse and human early embryo. Nat. Cell Biol. 2018;20:878–887. doi: 10.1038/s41556-018-0144-x. [DOI] [PubMed] [Google Scholar]
- Simunovic M., Metzger J.J., Etoc F., Yoney A., Ruzo A., Martyn I., Croft G., You D.S., Brivanlou A.H., Siggia E.D. A 3D model of a human epiblast reveals BMP4-driven symmetry breaking. Nat. Cell Biol. 2019;21:900–910. doi: 10.1038/s41556-019-0349-7. [DOI] [PubMed] [Google Scholar]
- Solnica-Krezel L., editor. Vol. 136. Academic Press; 2020. (Gastrulation: from embryonic pattern to form). Curr. Top. Dev. Biol. [Google Scholar]
- Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., Hao Y., Stoeckius M., Smibert P., Satija R. Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902.e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Meyers E.N., Lewandoski M., Martin G.R. Targeted disruption of Fgf8 causes failure of cell migration in the gastrulating mouse embryo. Genes Dev. 1999;13:1834–1846. doi: 10.1101/gad.13.14.1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam P.P.L., Behringer R.R. Mouse gastrulation: the formation of a mammalian body plan. Mech. Dev. 1997;68:3–25. doi: 10.1016/S0925-4773(97)00123-8. [DOI] [PubMed] [Google Scholar]
- Tewary M., Ostblom J.E., Prochazka L., Zulueta-Coarasa T., Shakiba N., Fernandez-Gonzalez R., Zandstra P.W. A stepwise model of reaction-diffusion and positional information governs self-organized human peri-gastrulation-like patterning. Development. 2017;144:4298–4312. doi: 10.1242/dev.149658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyser R.C.V., Mahammadov E., Nakanoh S., Vallier L., Scialdone A., Srinivas S. A spatially resolved single cell atlas of human gastrulation. bioRxiv. 2020 doi: 10.1101/2020.07.21.213512. [DOI] [Google Scholar]
- Warmflash A., Sorre B., Etoc F., Siggia E.D., Brivanlou A.H. A method to recapitulate early embryonic spatial patterning in human embryonic stem cells. Nat. Methods. 2014;11:847–854. doi: 10.1038/nmeth.3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams M., Burdsal C., Periasamy A., Lewandoski M., Sutherland A. Mouse primitive streak forms in situ by initiation of epithelial to mesenchymal transition without migration of a cell population. Dev. Dyn. 2012;241:270–283. doi: 10.1002/dvdy.23711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang L., Yin Y., Zheng Y., Ma Y., Li Y., Zhao Z., Guo J., Ai Z., Niu Y., Duan K. A developmental landscape of 3D-cultured human pre-gastrulation embryos. Nature. 2019;577:537–542. doi: 10.1038/s41586-019-1875-y. [DOI] [PubMed] [Google Scholar]
- Yamaguchi T.P., Takada S., Yoshikawa Y., Wu N., McMahon A.P. T (Brachyury) is a direct target of Wnt3a during paraxial mesoderm specification. Genes Dev. 1999;13:3185–3190. doi: 10.1101/gad.13.24.3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang R., Goedel A., Kang Y., Si C., Chu C., Zheng Y., Chen Z., Gruber P.J., Xiao Y., Zhou C. Essential amnion signals for primate primitive streak formation resolved by scRNA map. bioRxiv. 2020 doi: 10.1101/2020.05.28.118703. [DOI] [Google Scholar]
- Zheng Y., Xue X., Shao Y., Wang S., Esfahani S.N., Li Z., Muncie J.M., Lakins J.N., Weaver V.M., Gumucio D.L., Fu J. Controlled modelling of human epiblast and amnion development using stem cells. Nature. 2019;573:421–425. doi: 10.1038/s41586-019-1535-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The scRNA-seq datasets described in this article were deposited in the GEO database repository under accession GEO nos. GSE144897 and GSE169074.