Abstract
Coronavirus disease 2019 (COVID-19) is an infectious disease with first symptoms similar to the flu. COVID-19 appeared first in China and very quickly spreads to the rest of the world, causing then the 2019-20 coronavirus pandemic. In many cases, this disease causes pneumonia. Since pulmonary infections can be observed through radiography images, this paper investigates deep learning methods for automatically analyzing query chest X-ray images with the hope to bring precision tools to health professionals towards screening the COVID-19 and diagnosing confirmed patients. In this context, training datasets, deep learning architectures and analysis strategies have been experimented from publicly open sets of chest X-ray images. Tailored deep learning models are proposed to detect pneumonia infection cases, notably viral cases. It is assumed that viral pneumonia cases detected during an epidemic COVID-19 context have a high probability to presume COVID-19 infections. Moreover, easy-to-apply health indicators are proposed for estimating infection status and predicting patient status from the detected pneumonia cases. Experimental results show possibilities of training deep learning models over publicly open sets of chest X-ray images towards screening viral pneumonia. Chest X-ray test images of COVID-19 infected patients are successfully diagnosed through detection models retained for their performances. The efficiency of proposed health indicators is highlighted through simulated scenarios of patients presenting infections and health problems by combining real and synthetic health data.
Keywords: Chest X-ray images (CXR), Pneumonia detection, Health scoring system, Coronavirus disease, COVID-19, Radiology
Introduction and motivation
COVID-19, initially named 2019-nCoV, appeared first in China and very quickly spreads to the rest of the world causing then the 2019-20 coronavirus pandemic. At the date of the preprint of this work (April 19th, 2020) [1], there have been 83,805 controversial confirmed cases in China, more than 1,000,000 confirmed cases in Europe and 2,355,676 confirmed cases all around the world. To date (April 29th, 2021), there have been 149,744,454 global cases all around the world.1
In many cases, this disease causes pneumonia. Characteristics of such an infection can be observed by radiologists. Also, deep learning methods can be helpful for operating deep analysis on query radiography images. Thanks to artificial intelligence, early stage and precision diagnosis can be done. Moreover, augmented intelligence can also play a key role for detecting and preventing the spread of novel coronavirus [2].
In this pandemic, the effective screening of COVID-19 is an arduous task in practice. Standard screening test kits called Reverse Transcription-Polymerase Chain Reaction (RT-PCR) are often unavailable. Moreover, the RT-PCR test is highly sensitive. It was found that deep-based Computed Tomography (CT) images analysis could be more reliable than RT-PCR test in early-stage diagnostic [3–5]. Notably, where RT-PCR test can turn out negative, deep CT image analysis can already predict true positives in certain cases. False negatives to RT-PCR test can lead to non-negligible propagation of this disease.
At this time, American College of Radiology recommendations for the use of chest radiography and CT for Suspected COVID-19 infection [6] point that generally, the findings on chest imaging in COVID-19 are not specific, and overlap with other infections, including influenza, H1N1, SARS and MERS. Notably, being in the midst of the current flu season with a much higher prevalence of influenza in the U.S. than COVID-19, further limits the specificity of CT. Besides, the use of radiography equipment requires high disinfection needs after each use which can make massive tests laborious and time-consuming. In practice, for hygienic reasons, chest X-rays (CXR) are often frontally taken with patients on a stretcher or bed, lying down or at best sitting. Such constraints often conduct to chest X-rays with poor quality and real issues in term of analysis.
Nevertheless, CT images of lung and chest X-ray images offer additional data for screening COVID-19. Notably, AI technology is already deployed in China for radiography examination and radiomics-like analysis from CT images [7]. AI technology can also facilitate remote operations and help to face the lack of expert radiologists. At this date, many AI tools and radiography image datasets are private resources. The access to publicly open COVID-19-related sets of lung CT images towards conducting deep learning experiments is relatively limited. Some open access X-ray image sets of chest are publicly available.
Research works are highly needed in areas of image detection, evaluation and making-decision techniques, and radiography examinations in order to highlight proof of concepts and scientific truths about this misunderstood COVID-19 disease. The goal of this paper is twofold: i) to present deep learning models tailored with chest X-ray images for detecting pneumonia infection cases such as viral cases towards screening COVID-19, ii) to propose easy-to-apply health indicators for evaluating detected pneumonia infection cases with an estimator of infection and predictions of patient status. This study is presented with the aim of supporting radiologists and other clinicians. In no case this preliminary study could be substituted to a medical advice.
The next section presents some investigated deep learning based image detection architectures and analysis strategies. Section “Experimental study” shows a set of experiments to evaluate the performance of the considered architectures and “Conclusion” concludes the paper.
Proposed approach for pneumonia analysis
CNN-based detection and evaluation of infected patients
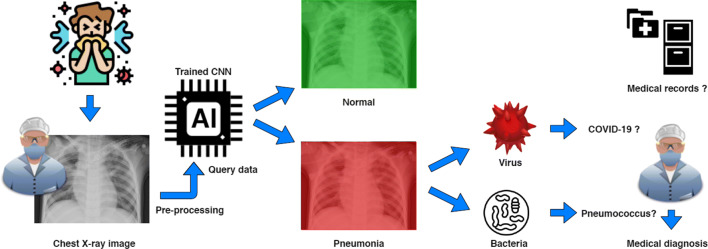
In [8, 9], authors emphasized that the COVID-19 is a viral disease and not a bacterial one. Respectively, an efficient classifier is designed to automatically detect if a query chest X-ray image is Normal, Bacterial or Viral by assuming that a COVID-19 infected patient, tested during an epidemic period, has a high probability to be a true positive when the classification output is Virus (see Fig. 1). Nevertheless, it is worth mentioning that a severe viral respiratory infection can lead to a secondary pneumonia of bacterial nature [10]. For this reason, our classifier aims to be useful at early stage of COVID-19 pulmonary symptoms.
Fig. 1.
Global workflow using deep learning for automatic detection of infection towards supporting COVID-19 screening from chest X-ray images. In a COVID-19 epidemic context, a detected viral pneumonia can particularly presume a COVID-19 infection
The pulmonary infections can be more directly visible in CT images than in chest X-Ray images [11]. Nevertheless, detection of COVID-19 from chest X-ray images is investigated since they represent widespread resources that are often analyzed upstream of CT scans.
At this time, chest images of COVID-19 cases are relatively limited in open access. Several CNN-based classification architectures such as [12, 13] are trained by using small chest image sets of COVID-19 cases. In such cases, it is hard to statistically build meaningful architecture. However, this could permit to observe first behaviors and trends of directly tailored CNN-based classification models. In our case, we adopt another strategy which consists of training CNN-based classification architectures over larger chest image sets of pneumonia and normal cases, and particularly evaluating them by using COVID-19 images.
Tailored CNN models
A set of tailored models based on CNNs have been designed to take three set of image categories (e.g.; normal case, viral pneumonia case and bacterial case) as input and to output the predicted probability for each of the categories. The trained models exploit the CNN backbones ResNet34, ResNet50 and DenseNet169 through the fastai library and a fully connected head, with a single hidden layer, as a classifier.
Besides, a trained model exploits the CNN reference backbone VGG-19. In addition, a dual use model (Inception ResNetV2 - RNN) is prepared for i) characterizing categories of input split images by getting a hidden layer output of a fin-tuned Inception ResNetV2 architecture, ii) predicting final categories of split images (image blocks) using a bidirectional Long Short-Term Memory (RNN-LSTM) architecture. For these last ones, a Keras and TensorFlow workflow is used.
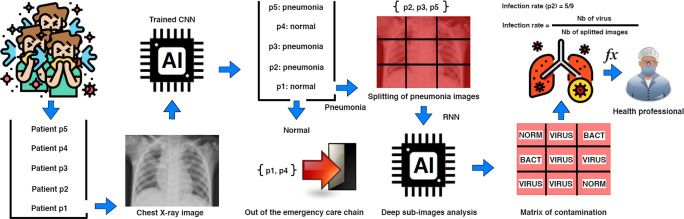
Specifically, the prediction stage of the dual-use model operates at a second level analysis of the data. A sequence of sub-images is first generated while entirely covering the images by directly positioning a regular grid onto the original query chest X-ray images. Precisely, the image is split into a set of image blocks that correspond to grid cells (see Fig. 2). This operation enhances the size of the training set while limiting the loss of image details. This loss often occurs when the original images are resized for fitting inputs of standard deep learning architectures. Then, each image block is given as input to the RNN for providing a set of local predictions (radiographic pattern) towards estimating health indicators such as a CNN-based infection ratio (a use is described in Estimation of CNN-based health indicators). The grid discretization (matrix of contamination) should be tuned according to the obtained predictive performance of the considered architecture.
Fig. 2.
Global workflow using deep learning based for automatic estimation of a CNN-based infection rate indicator from chest X-ray images
Data preparation and model inputs
For our experiments, we exploited chest X-ray images from the Chest X-Ray Images (Pneumonia) dataset.2 This dataset is related to the paper [14] on the identification of medical diagnoses and treatable diseases by image-based deep learning. This dataset contains 5,863 children X-ray images divided in two categories, namely Normal and Pneumonia. The Pneumonia category is composed of pneumonia images that are labeled either bacterial or viral (see illustrations in Fig. 6 of [14]).
The Chest X-Ray Images (Pneumonia) dataset is reorganized into three classes; namely normal, bacterial pneumonia and viral pneumonia (see samples in Fig. 3a, b and c, respectively). Each training set contains 1345 images and each test set contains 148 images. Since this dataset was composed of pulmonary images having heterogeneous and large sizes; and to deal with reasonable computational times during the CNN training experiments, all the images were resized to a unique dimension and rescaled into smaller images (e.g.; size 310 × 310) to fit with standard inputs of tailored architectures. For the last tailored model using RNN, a preliminary split of the original image precedes the resizing step.
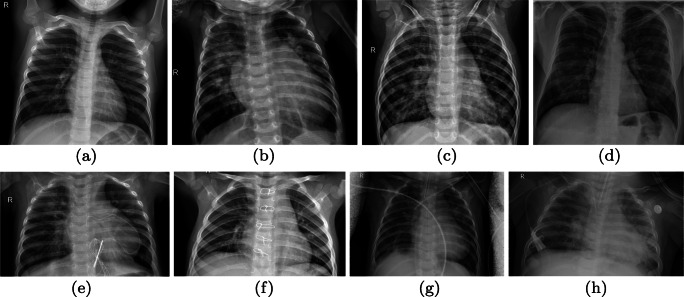
Fig. 3.
Chest X-ray samples from the test datasets. Row 1 shows image categories. Row 2 shows various artifacts captured with chest X-rays such as writings (e.g.; letter “R”) and medical devices (e.g., tubes, sensors)
Regarding the tests, we added a test set (blind test) that is composed of a single class containing 145 chest X-ray images of COVID-19 infected patients (see sample in Fig. 3d). This test set has been constituted by filtering the heterogeneous COVID-19 Image Data Collection dataset [15]; folder containing a mix of CT and X-ray images with a variety of infection types.
We emphasize that the exploited images often contain artifacts such as described in [16] that can make difficult the training stage of machine learning models. Such artifacts are shown in Fig. 3e, f, g and h.
At this time, we consider that the quantity of available COVID-19 is still too limited for building a reliable detector that can discern between Non-COVID-19 viral pneumonia and COVID-19 viral pneumonia. In our case, the 145 chest X-ray images are specifically used as a test set towards ideally detecting them as viral pneumonia. As previously mentioned, we assume that a COVID-19 infected patient, tested during an epidemic period, has a high probability to be a true positive when the classification output is viral pneumonia (Fig. 1).
Estimation of CNN-based health indicators
Based on statistical tools (logistic regression and statistical tests) and realistic data, studies on COVID-19-related death risk factor have been proposed in [17, 18]. In this paper, we sketch a simple measure to provide to health professionals an estimator for evaluating the chance of a patient to survive COVID-19 considering risk factors; namely age, comorbidity and the infection rate indicator (Fig. 2). For each risk factor, we associate a score which represents a penalty (a large value decreases the chance of a patient to escape fatality). The proposed measure F is the addition of scores divided by a critical threshold . Beyond , there is no chance to be recovered. Formally, F is expressed as follows:
where measures the risk due to the patient’s age, measures the risk related to the CNN-derived infection rate measured from the X-ray chest image of a patient, measures the risk associated with comorbidities of a patient that can lead to the development of complications.
More precisely, let us give an example to concretely compute the measure F. First, we point out that the scoring system used hereafter must be adjusted by health professionals to match with reality.
In this example, we use the values of fatality risk-ratio during COVID-19 epidemic in Hubei, China [19, 20]. Proportionally to these values, we define penalty scores (), see Table 1.
Table 1.
Scores related to age
| Age | Fatality risk ratio (Hubei, China) | Associated score () |
|---|---|---|
| ≥ 80 | 18% | 100 |
| 70–79 | 9.8% | 54.4 |
| 60–69 | 4.6% | 25.5 |
| 50–59 | 1.3% | 7.2 |
| 40–49 | 0.4% | 2.2 |
| 30–39 | 0.18% | 1 |
| 20–29 | 0.09% | 0.5 |
| 10–19 | 0.02% | 0.1 |
| 0–9 | 0.01 | 0.05 |
Then, we define a Virality score () related to the infection rate of X-ray image (CXR score). can be the probability of the concerned class that is directly provided by the used CNN. A more refined formulation of is proposed to scrutinize the X-ray image and to consider radiographic infection patterns. Each image is divided in n sub-images where n = 9 (value for which our RNN effectively performs). After this multi-regional analysis, each sub-image will be in status virus, bacteria or normal (see Fig. 2). A score equal to 100 is assigned to the X-ray image when the n sub-images are infected by a virus. Proportionally, the infection rate of the image is: , where N is the number of the virus infected sub-images.
The third risk factor () is related to diseases of a patient in addition to COVID-19. We use the values of case fatality rate during COVID-19 epidemic in China [21, 22]. Proportionally to these values, we define penalty scores (), see Table 2.
Table 2.
Scores related to disease
| Disease | Case fatality rate | Associated |
|---|---|---|
| (China) | score () | |
| Cardiovascular disease | 10.5% | 100 |
| Diabetes | 7.3% | 69.5 |
| Chronic respiratory | 6.3% | 60 |
| Hypertension | 6 % | 57,1 |
| Cancer | 5.6 % | 53,3 |
| No health condition | 0.9% | 8.6 |
In this example, we set the critical threshold to 200, then beyond this limit, one cannot escape fatality. The measure F can be used as follows. If F ≥ 1 then the hope to escape fatality is null. Varying F from 0 to 1, the patient gradually moves away from the hope of recovering.
The value 200 assigned to is obtained by taking reference a patient having a cardiovascular disease and aged over 80 years. We assume that such a patient cannot fight against COVID-19. Therefore, a person having COVID-19 which reaches the score 200 cumulates too many factors to overcome the illness.
Finally, we point out that the term of our measure F can be tuned:
- can be modified to take into account a time factor. Indeed, a serie of X-ray images is observed for each COVID-19 inpatient pointed in [11, 23, 24]. Accordingly, can be measured at two timely spaced X-rays t1 and t2 to take into account the time kinetics of symptom onset and disease progression for the infected patient. For this latter case, the infection rate can be redefined as follows:
where f(t) = (100/n) × N(t) is the infection rate of the chest X-ray image captured at a moment t, N(t) is the number of detected viral sub-images at a moment t, t1 is inferior to t2, δ is threshold fixed to 20, bonus and malus are a gain and a penalty fixed to (− 20 × f(t2))/100 and (20 × f(t2))/100, respectively. can be replaced by another multi-regional CXR score, named Brixia score, that has been proposed in [25] for grading the infection according to observed lung abnormalities.
Experimental study
Performance of tailored CNN models
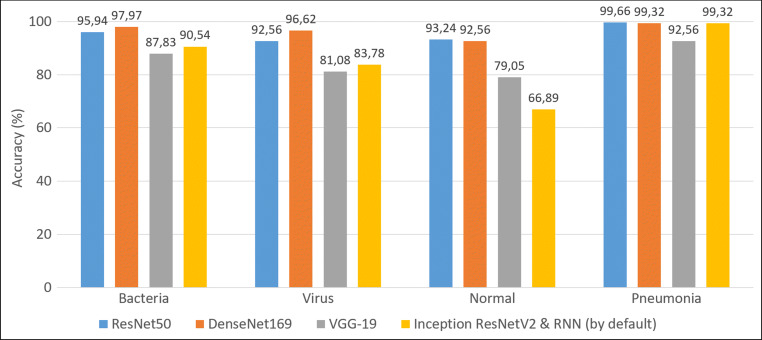
Table 3 presents performance for classification of normal and infection cases by using tailored CNN-based architectures. The DenseNet169 architecture has reached best performance with an average classification accuracy of 95.72% from the Chest X-Ray Images (Pneumonia) dataset.[2] It is worth mentioning that similarly to the work from [13], the DenseNet architecture outperforms the other architectures. The classification accuracies are 97.97%, 96.62% and 92.57% for the class bacterial, virus and normal, respectively (see Table 4). The associated confusion matrix is shown in Table 5. The performance of our DenseNet model is competitive with performances obtained by [14] in average classification accuracy of bacterial and viral cases (90.7%). The histogram depicted in Fig. 4 provides a detailed overview about accuracy results obtained with architectures of different natures for the classification of chest X-ray images with 4 categories.
Table 3.
Comparison of average accuracies obtained on classification using some tailored CNN-based architectures
| Data/Network | Average accuracy (%) | ||||
|---|---|---|---|---|---|
| ResNet34 | ResNet50 | DenseNet169 | VGG-19 | Inception | |
| ResNetV2 & RNN | |||||
| Image size | (310 × 310), (310 × 273) | (310 × 310), (310 × 273) | (310 × 310), (310 × 273) | (224 × 224) | (300 × 300) |
| Raw resizing | 93.69 | 93.47 | 91.89 | 82.66 | – |
| Original ratio with padding | 90.54 | 93.92 | 95.72 | – | – |
| Split in 9 & raw resizing | – | – | – | 80.40 | |
Bold entries correspond to values of interest with respect to contextual information such as best performances
Table 4.
Classification performance obtained by testing our best trained architectures with two query image sets
| Accuracy (%) | ||||
|---|---|---|---|---|
| Query test set/Class ouput | Bacteria | Virus | Normal | Pneumonia |
| Dataset Chest X-Ray Images (Pneumonia)a by [14] (2-stage binary training outputs) | 90.7 (Bact. vs Vir.) | 92.8 (Norm. vs Pneu.) | ||
| 3-class dataset Chest X-Ray Images (Pneumonia)a by DenseNet169 | 97.97 | 96.62 | 92.57 | – |
| 3-class dataset Chest X-Ray Images (Pneumonia)a by VGG-19 | 87.84 | 81.08 | 79.05 | – |
| 3-class dataset Chest X-Ray Images (Pneumonia)a by Inception ResNetV2 & RNN (majority voting) | 86.49 | 84.46 | 63.52 | – |
| 3-class dataset Chest X-Ray Images (Pneumonia)a by Inception ResNetV2 & RNN (by default) | 90.54 | 83.78 | 66.89 | – |
| Sensitivity (%) | ||||
| Query test set/Class ouput | Virus | Pneumonia | ||
| 1-class blind set of varied COVID-19 images by DenseNet169 | 45.51 | 88.27 | ||
| 1-class blind set of varied COVID-19 images by Inception ResNetV2 & RNN (majority voting) | 60.64 | 95.12 | ||
| 1-class blind set of varied COVID-19 images by Inception ResNetV2 & RNN (by default) | 51.72 | 99.3 | ||
a Chest X-Ray Images (Pneumonia) dataset: https://www.kaggle.com/paultimothymooney/chest-xray-pneumonia
The first row reflects classification results of [14]
Bold entries correspond to values of interest with respect to contextual information such as best performances
Table 5.
Confusion matrix with DenseNet169
| Actual/Predicted | Bacteria | Normal | Virus |
|---|---|---|---|
| Bacteria | 145 | 1 | 2 |
| Normal | 0 | 137 | 11 |
| Virus | 4 | 1 | 143 |
Bold entries correspond to values of interest with respect to contextual information such as best performances
Fig. 4.
Histogram of classification accuracy obtained for each class by using varied architecture types with the Chest X-Ray Images (Pneumonia) dataset
As can be seen in Table 4, our RNN-based architecture is particularly sensitive to pneumonia cases with the blind COVID-19 test set since it detects pneumonia at 99.3% for at least 24 COVID-19 infected patients using default setting. Also, it promisingly detects viral infection for 60.64% considering majority voting in sequences. We stress the fact that the 145 COVID-19 images which have been extracted from [15] are highly heterogeneous. Notably, these extracted images come from at least 24 different hospitals over the world. The RNN output results show a particularly robust pneumonia detection of COVID-infected patients and satisfying viral detection in view of the diversity of exploited radiography sources.
In [15], a histogram shows that significant image quantity has been acquired during the first week of the start of symptoms or hospitalization. Since the quasi-totality of pneumonia are detected, our models should be able to operate at an early detection stage.
Also, a histogram shows a significant image distribution in term of age in between 20 and 80 years old. Since the Chest X-Ray Images (Pneumonia) dataset[2] is principally collected from children (5,232 chest X-ray images) and since the quasi-totality of pneumonia are detected, models trained on children chest X-ray image database may be relevant for detecting pneumonia from adult chest X-ray images.
Projection with the CNN-based health indicators
Table 6 shows the RNN-derived infection rates estimated from real pairs of successive X-ray images for COVID-19 infected patients. Table 7 details examples of F calculated for 9 patients from synthetic data. Table 8 gives the distribution of the F values for a patient aged between 60 and 69 considering the possible diseases (rows) and rates of infection (columns from 0 to 9). The corresponding distribution is illustrated in Fig. 5.
Table 6.
CNN-derived infection rates estimated from real pairs of successive X-ray images for 5 COVID-19 infected patients
| Image pairs | f(t1) | Elapsed days | f(t2) | Observations | |
|---|---|---|---|---|---|
| (t2 − t1) | |||||
| (1,3), Fig. 5 of [11] | 9/9 | 7 | 9/9 | elderly man | f(t2) |
| “improvements” | |||||
| (1,2), Fig. 1 of [23] | 1/9 | 5 | 2/9 | 67-y-old woman | f(t2) |
| “wires, attenuation” | |||||
| (1,3), Fig. 2 of [23] | 5/9 | 8 | 5/9 | 36-y-old man | f(t2) |
| “death” | |||||
| (1,2), case 1, Fig. 5 of [24] | 6/9 | 1 | 9/9 | “worse status” | f(t2)+malus |
| (1,2), case 2, Fig. 5 of [24] | 7/9 | 4 | 8/9 | “worse status” | f(t2) |
Table 7.
Examples of F values for 9 patients from synthetic data
| Patients | Aggravation factors: values | S1 | S2 | S3 | F |
|---|---|---|---|---|---|
| Patient 1 | Age: 65 | 25.5 | |||
| nb. of infected sub-image: 3 | 33.33 | 0.79415 (79.41%) | |||
| Cardiovascular disease | 100 | ||||
| Patient 2 | Age: 55 | 7.2 | |||
| nb. of infected sub-image: 4 | 44.44 | 0.7582 (75.82 %) | |||
| Cardiovascular disease | 100 | ||||
| Patient 3 | Age: 75 | 54.4 | |||
| nb. of infected sub-image: 1 | 11.11 | 0.59405 (59.40%) | |||
| Cancer | 53.3 | ||||
| Patient 4 | Age: 82 | 100 | |||
| nb. of infected sub-image: 1 | 11.11 | 1.05 (> 100%) | |||
| Cardiovascular disease | 100 | ||||
| Patient 5 | Age: 34 | 1 | |||
| nb. of infected sub-image: 5 | 55.55 | 0.63025 (63.02%) | |||
| Diabetes | 69.5 | ||||
| Patient 6 | Age: 18 | 0.1 | |||
| nb. of infected sub-image: 7 | 77.77 | 0.68935 (68.93%) | |||
| Chronic respiratory | 60 | ||||
| Patient 7 | Age: 13 | 0.1 | |||
| nb. of infected sub-image: 4 | 44.44 | 0.2657 (26.57%) | |||
| No health condition | 8.6 | ||||
| Patient 8 | Age: 56 | 7.2 | |||
| nb. of infected sub-image: 2 | 22.22 | 0.4326 (43.26%) | |||
| Hypertension | 57,1 | ||||
| Patient 9 | Age: 47 | 2.2 | |||
| nb. of infected sub-image: 3 | 33.33 | 0.2206 (22.06%) | |||
| No health condition | 8.6 |
Table 8.
Distribution of F values for a patient aged between 60 and 69 considering the possible diseases (rows) and rates of infection (columns from 0 to 9)
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | |
|---|---|---|---|---|---|---|---|---|---|---|
| No health condition | 17,05 | 22,60 | 28,16 | 33,71 | 39,27 | 44,82 | 50,38 | 55,93 | 61,49 | 67,04 |
| Cancer | 39,4 | 44,95 | 50,51 | 56,06 | 61,62 | 67,17 | 72,73 | 78,28 | 83,84 | 89,39 |
| Hypertension | 41,3 | 46,85 | 52,41 | 57,96 | 63,52 | 69,07 | 74,63 | 80,18 | 85,74 | 91,29 |
| Chronic respiratory | 42,75 | 48,30 | 53,86 | 59,41 | 64,97 | 70,52 | 76,08 | 81,63 | 87,19 | 92,74 |
| Diabetes | 47,5 | 53,05 | 58,61 | 64,16 | 69,72 | 75,27 | 80,83 | 86,38 | 91,94 | 97,49 |
| Cardiovascular | 62,75 | 68,30 | 73,86 | 79,41 | 84,97 | 90,52 | 96,08 | 101,63 | 107,19 | 112,74 |
Bold entries correspond to values of interest with respect to contextual information such as best performances
Fig. 5.
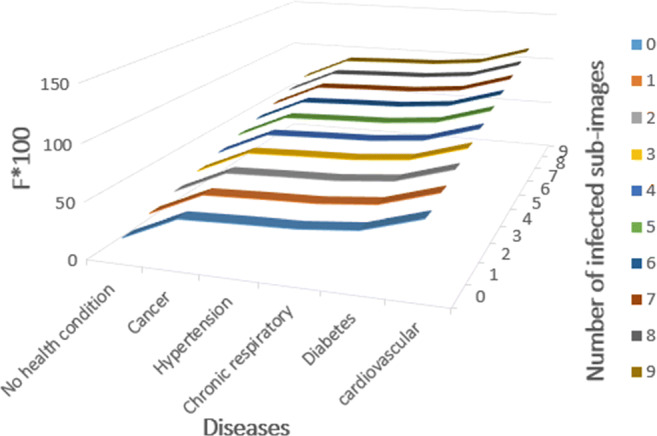
Graph obtained by representing the distribution of F values for a patient aged between 60 and 69 considering the possible diseases and rates of infection given in Table 8
The simulation shows that a patient having 65 years old, a cardiovascular disease and rate of infection greater than 6 has no hope to recover the health. The Patient 1 of Table 7 that has 65 years old, a cardiovascular disease and rate of infection of 3 should recover the health (F = 79.41%).
Conclusion
A comparative study is provided for performance evaluation of tailored deep learning models for the detection of pneumonia cases. Tailored models have shown promising performances since they all exceeded 84% of average accuracy on pneumonia detection cases for the pneumonia reorganized dataset.[2] Hence, a patient that has a pneumonia during the epidemic context has a high probability to be detected by these models. In particular, the InceptionResNetV2 model has detected the minimum of false negatives to the pneumonia on the COVID-19 blind test set (0.7%). Moreover, we have shown in our experiments that the transfer of knowledge from pediatric chest X-ray training towards infection screening of adults can be efficient. Additionally, an attempt based on realistic scenarios is done to provide easy-to-apply health indicators for evaluating infection rate and aggravation risk to the COVID-19 pneumonia. Future works may exploit our models to discern between COVID-19 viral and non-COVID-19 viral pneumonia once chest X-ray images of COVID-19 will be accessible in sufficient quantity. This should permit to specifically identify COVID-19 infected patients even in a non-epidemic context. Furthermore, reliability of proposed models must be cross-checked by RT-PCR tests and clinical tests before deployment.
Declarations
Ethics approval
This article does not contain any studies with human participants performed by any of the authors.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Online map of COVID-19 Global Cases by the CSSE center at Johns Hopkins University: https://coronavirus.jhu.edu/map.html
Chest X-Ray Images (Pneumonia) dataset: https://www.kaggle.com/paultimothymooney/chest-xray-pneumoniahttps://www.kaggle.com/paultimothymooney/chest-xray-pneumonia
This article is part of the Topical Collection on Image & Signal Processing
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Karim Hammoudi, Email: karim.hammoudi@uha.fr.
Halim Benhabiles, Email: halim.benhabiles@junia.com.
Mahmoud Melkemi, Email: mahmoud.melkemi@uha.fr.
Fadi Dornaika, Email: fadi.dornaika@ehu.eus.
Ignacio Arganda-Carreras, Email: ignacio.arganda@ehu.eus.
Dominique Collard, Email: dominique.collard@junia.com.
Arnaud Scherpereel, Email: arnaud.scherpereel@chru-lille.fr.
References
- 1.Hammoudi K., Benhabiles H., Melkemi M., Dornaika F., Arganda-Carreras I., Collard D., Scherpereel A. (2020) Deep learning on chest x-ray images to detect and evaluate pneumonia cases at the era of covid-19. arXiv:https://arxiv.org/abs/2004.03399 [DOI] [PMC free article] [PubMed]
- 2.Long J.B., Ehrenfeld J.M. The role of Augmented Intelligence (AI) in detecting and preventing the spread of novel coronavirus. J. Med. Syst. 44(3):59. 10.1007/s10916-020-1536-6. https://app.dimensions.ai/details/publication/pub.1124592049 and https://link.springer.com/content/pdf/10.1007/s10916-020-1536-6.pdf [DOI] [PMC free article] [PubMed]
- 3.Xie X., Zhong Z., Zhao W., Zheng C., Wang F., Liu J. Chest CT for typical 2019-nCoV pneumonia: relationship to negative RT-PCR testing. Radiology 200343. 10.1148/radiol.2020200343. PMID: 32049601 [DOI] [PMC free article] [PubMed]
- 4.Xu X., Jiang X., Ma C., Du P., Li X., Lv S., Yu L., Chen Y., Su J., Lang G., Li Y., Zhao H., Xu K., Ruan L., Wu W. (2020) Deep learning system to screen coronavirus disease 2019 pneumonia. arXiv:https://arxiv.org/abs/2002.09334 [DOI] [PMC free article] [PubMed]
- 5.How does covid-19 appear in the lungs? (2020). https://www.itnonline.com/content/how-does-covid-19-appear-lungs
- 6.Acr recommendations for the use of chest radiography and computed tomography (ct) for suspected covid-19 infection (2020). https://www.acr.org/Advocacy-and-Economics/ACR-Position-Statements/Recommendations-for-Chest-Radiography-and-CT-for-Suspected-COVID19-Infection
- 7.Artificial intelligence assisted radiology technologies aid covid-19 fight in China (2020). https://www.itnonline.com/article/artificial-intelligence-assisted-radiology-technologies-aid-covid-19-fight-china
- 8.Treatment for coronavirus disease (covid-19) (2020). https://www.healthline.com/health/coronavirus-treatment#available-treatment
- 9.Covid-19: what we know so far about the 2019 novel coronavirus (2020). https://www.uchicagomedicine.org/forefront/prevention-and-screening-articles/wuhan-coronavirus
- 10.Hunter, P. EMBO reports n/a(n/a), e50334. 10.15252/embr.202050334
- 11.Ng MY, Lee EY, Yang J, Yang F, Li X, Wang H, Lui MMS, Lo CSY, Leung B, Khong PL, Hui CKM, Yuen KY, Kuo MD. Imaging profile of the COVID-19 infection: radiologic findings and literature review. Radiol.: Cardiothoracic Imaging. 2020;2(1):e200034. doi: 10.1148/ryct.2020200034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang L., Wong A. (2020) Covid-net: a tailored deep convolutional neural network design for detection of covid-19 cases from chest radiography images. arXiv:2003.09871v1 [DOI] [PMC free article] [PubMed]
- 13.Karim M. R., Dohmen T., Rebholz-Schuhmann D., Decker S., Cochez M., Beyan O. (2020) Deepcovidexplainer: explainable covid-19 predictions based on chest x-ray images. arXiv:2004.04582
- 14.Kermany D, Goldbaum M, Cai W, Valentim C, Liang HY, Baxter S, McKeown A, Yang G, Wu X, Yan F, Dong J, Prasadha M, Pei J, Ting M, Zhu J, Li C, Hewett S, Dong J, Ziyar I, Zhang K. Identifying medical diagnoses and treatable diseases by image-based deep learning. Cell. 2018;172:1122. doi: 10.1016/j.cell.2018.02.010. [DOI] [PubMed] [Google Scholar]
- 15.Cohen J. P., Morrison P., Dao L. (2020) Covid-19 image data collection. arXiv:2003.11597
- 16.Hunter TB, Taljanovic MS, Tsau PH, Berger WG, Standen JR. Medical devices of the chest. RadioGraphics. 2004;24(6):1725. doi: 10.1148/rg.246045031. [DOI] [PubMed] [Google Scholar]
- 17.Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z, Xiang J, Wang Y, Song B, Gu X, Guan L, Wei Y, Li H, Wu X, Xu J, Tu S, Zhang Y, Chen H, Cao B. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Caramelo B. O. F., Ferreira N (2020) MedRxiv. 10.1101/2020.02.24.20027268
- 19.Belluz J. (2020) Did the coronavirus get more deadly? The death rate, explained. https://www.vox.com/2020/3/5/21165973/coronavirus-death-rate-explained
- 20.Riou J., Hauser A., Counotte M.J., Althaus C.L. (2020) medRxiv. 10.1101/2020.03.04.20031104. https://www.medrxiv.org/content/early/2020/03/06/2020.03.04.20031104
- 21.Roser M., Ritchie H., Ortiz-Ospina E. (2020) Coronavirus disease COVID-19—statistics and research. https://ourworldindata.org/coronavirus
- 22.T.N.C.P.E.R.E. Team (2020) The epidemiological characteristics of an outbreak of 2019 novel coronavirus diseases COVID-19 - China, 2020. https://cdn.onb.it/2020/03/COVID-19.pdf.pdf [PMC free article] [PubMed]
- 23.Zhang J. J., Dong X., Cao Y. Y., Yuan Y. D., Yang Y. B., Yan Y. Q., Akdis C. A., Gao Y. D. Allergy n/a(n/a). 10.1111/all.14238
- 24.Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y., Xia J., Yu T., Zhang X., Zhang L. (2020) Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet 395. 10.1016/S0140-6736(20)30211-7 [DOI] [PMC free article] [PubMed]
- 25.Borghesi A., Maroldi R. (2020) COVID-19 outbreak in Italy: experimental chest X-ray scoring system for quantifying and monitoring disease progression. Radiol. Med. 125. 10.1007/s11547-020-01200-3 [DOI] [PMC free article] [PubMed]