Abstract
Type 2 diabetes increases the risk various types of cancer and is associated with a poor prognosis therein. There is also evidence that the disease is associated with cancer metastasis. Centrosome amplification can initiate tumorigenesis with metastasis in vivo and increase the invasiveness of cancer cells in vitro. Our previous study reported that type 2 diabetes promotes centrosome amplification via the upregulation and centrosomal translocation of Rho-associated protein kinase 1 (ROCK1), which suggests that centrosome amplification is a candidate biological link between type 2 diabetes and cancer development. In the present study, functional proteomics analysis was used to further investigate the molecular pathways underlying centrosome amplification by targeting ROCK1 binding partners. High glucose, insulin and palmitic acid were used to induce centrosome amplification, and immunofluorescent staining was employed to visualize centrosomal alterations. Combined with immunoprecipitation, mass spectrometry-based proteomics analysis was used to identify ROCK1 binding proteins, and protein complex disruption was achieved by siRNA-knockdown. In total, 1,148 ROCK1 binding proteins were identified, among which 106 proteins were exclusively associated with the treated samples, 193 were only associated with the control samples, and 849 were found in both the control and treated samples. Of the proteins with evidence of centrosomal localization, Dynactin subunit 2 (DCTN2) was confirmed to be localized to the centrosomes. Treating the cells with high glucose, insulin and palmitic acid increased the protein levels of ROCK1 and DCTN2, promoted their binding with each other, and triggered centrosome amplification. Disruption of the protein complex by knocking down ROCK1 or DCTN2 expression partially attenuated centrosome amplification, while simultaneous knockdown of both proteins completely inhibited centrosome amplification. These results suggested ROCK1-DCTN2 binding as a signal for the regulation of centrosome homeostasis, which is key for diabetes-associated centrosome amplification, and enriches our knowledge of centrosome biology. Therefore, the ROCK1-DCTN2 complex may serve as a target for inhibiting centrosome amplification both in research or future therapeutic development.
Keywords: centrosome amplification, type 2 diabetes, Rho-associated protein kinase 1, dynactin subunit 2, colon cancer cells, functional proteomics
Introduction
Diabetes is a chronic disease with increasing prevalence worldwide (1), and exerts significant socioeconomic burden on society (2). Diabetes is classified into type 1 and 2, and type 2 diabetes accounts for the majority of clinical cases (1). According to the latest dataset from the World Health Organization, the number of diabetes cases increased from 108 million in 1980 to 422 million in 2014 (3). In 2016, ~1.6 million mortalities were reportedly directly caused by diabetes (4). In the United States alone, the economic burden of diabetes, undiagnosed diabetes and prediabetic complications, was estimated to be 404 billion dollars in 2017 (5). Patients with type 2 diabetes have increased levels of blood glucose and free fatty acids such as palmitic acid. Moreover, hyperinsulinemia occurs, at least at certain stages in disease development, due to insulin resistance (6). In addition to classical vascular complications such as nephropathy and retinopathy, accumulating evidence suggests that cancer is a complication of type 2 diabetes, which led to a joint meeting of oncologists and diabetologists in 2010. These experts reviewed the evidence and released a consensus report stating that type 2 diabetes increases the risk of cancer at various sites, including the colon (7). Moreover, type 2 diabetes has been associated with a poor cancer prognosis (8), as well as metastasis (9). At present, cancer accounts for a large proportion of the mortality of patients with type 2 diabetes (10). However, why and how type 2 diabetes promotes cancer development remains unknown.
The centrosome is the smallest organelle in the eukaryotic cell, which serves as a microtubule-organization center, and is a key regulator of cell division. Centrosome amplification is defined as >2 centrosomes in a single cell. It has been reported to occur in various cancer types, and is recognized as a typical feature of cancer cells (11). Experimental data have indicated that centrosome amplification is associated with tumorigenesis and tumor metastasis. For example, genetically modified cells with centrosome amplification can form tumors in recipient animals, with the potential for metastasis (12,13), while genetically modified mice with centrosome amplification can spontaneously develop tumors (14,15). Furthermore, in Barrett's esophagus tumorigenesis, a multistep progression model of human cancer, centrosome amplification occurs in a stepwise manner before neoplasia (16). It has also been shown that genetically modified cells with centrosome amplification exhibit increased invasive potential (17). However, the obvious limitation is that genetically modified models do not accurately reflect reality, as human cancers are not considered to commonly be caused by transgenesis.
Since type 2 diabetes is associated with DNA damage (18), which can cause centrosome amplification (19), it was hypothesized that type 2 diabetes may favor centrosome amplification. Indeed, when compared between non-diabetic subjects and diabetic patients, a 2.6-fold increase in centrosome amplification was observed in the peripheral blood mononuclear cells of diabetic patients (6). In vitro experiments have identified that the pathophysiological factors in type 2 diabetes, including high glucose, insulin and free fatty acids (such as palmitic acid), are potent inducers, and that the centrosomal translocation of Rho-associated protein kinase 1 (ROCK1) is a signal for diabetes-associated centrosome amplification (6). Moreover, inhibition of ROCK1 partially attenuated centrosome amplification (6,20). These results suggest that centrosome amplification may be a candidate mechanism for the association between type 2 diabetes and cancer development, and that type 2 diabetes may present a natural model to investigate the roles of centrosome amplification in cancer. However, to confirm this conclusion with substantial evidence, the following observations should be considered: i) Non-cancerous cells with centrosome amplification induced by high glucose, insulin and palmitic acid, can form tumors in immunodeficient mice; ii) treated non-cancerous cells are unable to form tumors in vivo when centrosome amplification is inhibited, such as by interfering with the associated signaling pathways; iii) treatment with high glucose, insulin and palmitic acid promotes cancer cell metastasis; and iv) the metastatic potential of treated cancer cells is suppressed when centrosome amplification is inhibited. Therefore, the identification of key signaling pathway(s) that effectively inhibit centrosome amplification is required.
Protein kinase ROCK1 is a key regulator of the actin cytoskeleton and cellular polarity, which serves various functional roles via the phosphorylation of its substrates, including death-associated protein kinase 3, glial fibrillary acidic protein and LIM domain kinase 1 (21). ROCK1 is the target of the small GTPases, the Ras homolog family member A (RhoA), RhoB and RhoC. More specifically, ROCK1 promotes cellular proliferation (22), regulates apoptosis (23) and enhances migration and invasiveness (24). In humans, the ROCK1 genotype was found to be associated with colon cancer in men (25), and is a candidate prognostic marker for vulvar cancer (26). Our previous studies revealed that ROCK1 signaling promotes diabetes-associated centrosome amplification (6,20). However, inhibiting ROCK1 only partially suppresses centrosome amplification, indicating that other factors are involved. Therefore, the aim of the present study was to investigate ROCK1 binding proteins in order to identify key signaling molecules involved in centrosome amplification.
Materials and methods
Reagents and cell lines
All chemical reagents were purchased from Sigma-Aldrich (Merck KGaA). Cell culture media and supplements were obtained from Gibco (Thermo Fisher Scientific, Inc.). HCT116 human colon cancer cells were kindly gifted by Dr B. Vogelstein of the Johns Hopkins University School of Medicine (Baltimore, USA). IEC-6, which is a non-cancerous immortalized rat colon epithelial cell line, was purchased from the Chinese Academy of Sciences Beijing Cell Bank. The cells were maintained in the lab in low-glucose medium.
Cell culture and treatments
Cells were cultured in DMEM (low glucose, 5 mM) supplemented with 50 U/ml penicillin, 50 µg/ml streptomycin and 10% (v/v) fetal calf serum, and maintained at 37°C (5% CO2) in a humidified incubator. Cells in the experimental group were treated with high glucose (15 mM), insulin (5 nM) and palmitic acid (150 µM). Cells treated for 48 h were used for the assessment of the centrosome amplification level; cells treated for 30 h were used for immunoprecipitation and western blot analyses. Palmitic acid, the most common saturated free fatty acid, is frequently used to investigate the biological activities of free fatty acids, particularly their adverse effects (27). Palmitic acid was conjugated to BSA; both reagents were mixed (molar ratio, 3:1) and incubated for 1 h before use.
Immunofluorescent staining and confocal microscopy
Cells were seeded on a cover slip into a 6-well plate, at a density of 5×104 cells per well. Following treatment with high glucose, insulin and palmitic acid, the cells were fixed in equal volumes of 100% cold methanol and 100% acetone (6 min; −20°C), washed three times with PBS (10 min each time), incubated with 0.1% Triton X-100 (15 min) and then with 3% BSA (60 min) at room temperature. The cells were subsequently incubated with antibodies against the following: γ-tubulin (cat. no. ab27074; mouse; Abcam; 1:500), ROCK1 (cat. no. 4035; rabbit; Cell Signaling Technology, Inc.; 1:500) and dynactin subunit 2 (DCTN2; cat. no. D122213; rabbit; Sangon Biotech Co., Ltd.; 1:1,000). Cells were washed twice with PBS, and incubated with anti-mouse fluorescent secondary antibody (cat. no. ab150116; Abcam; 1:500) and anti-rabbit fluorescent secondary antibody (cat. no. 4412; Goat; Cell Signaling Technology, Inc.; 1:1,000) in 3% BSA for 60 min at room temperature. Finally, the cells were mounted with mounting medium containing DAPI. Confocal microscopy was performed using a Zeiss LSM880 microscope (Carl Zeiss AG), and image processing was performed using Zen software (3.2; Carl Zeiss AG). The number of centrosome signals in each cell was determined by manually counting 300 cells in each specimen using a fluorescence microscope (magnification, ×1,000). Centrosome amplification was defined as cases with >5% of cells having ≥3 centrosomes per cell.
Co-immunoprecipitation (CoIP)
CoIP was performed under cold conditions (4°C or on ice) and ice-cold solutions were used, except in the sample heating step. A total of 5×106 cells per sample were harvested and lysed in 500 µl precooled CoIP buffer (50 mM Tris/HCl pH 8.0, 150 mM NaCl, 5 mM EDTA, 0.5% NP-40, 1 mM phenylmethylsulphonyl fluoride, 2 µg/ml leupeptin and 2 µg/ml pepstatin A) under non-denaturing conditions. After 24 h, cells seeded into 10-cm dishes were washed three times with ice cold PBS (5 min each time) and then lysed in 0.5 ml lysis buffer (Sangon Biotech Co., Ltd.) and collected in a 1.5-ml centrifuge tube, which was followed by a 30-min incubation on ice and 14,000 × g centrifugation step for 10 min. The supernatant was harvested and a volume equivalent to 200 µg total cellular protein was incubated with 20 µl Protein G Plus/Protein A agarose suspension (cat. no. IP05; EMD Millipore) for 2 h with gentle shaking. The agarose beads were removed by centrifugation, and the supernatant was collected and incubated overnight with primary antibodies against DCTN2 (cat. no. D122213; Sangon Biotech Co., Ltd.) or ROCK1 (cat. no. 4035; Cell Signaling Technology, Inc.), also with gentle shaking. Subsequently, 30 µl agarose beads were added to each sample, and incubated for 4 h with gentle shaking. Finally, the beads were collected by centrifugation, re-suspended in 2X loading buffer, heated to 100°C and then centrifuged once more (14,000 × g, 1 min). The heated sample was used for western blot analysis for the detection of the target proteins. Up to 40 µl sample was subjected to western blot analysis, as aforementioned. Whole cell lysate (40 µg) was used as the positive control for detection of the target proteins, and IgG was used as the negative control.
Proteomics analysis
Samples from three different CoIP experiments were used for the proteomic identification of binding partners. Experiments were performed on a Q Exactive mass spectrometer linked to an Easy n-liquid chromatography (LC) system (Thermo Fisher Scientific, Inc.). In total, 6 µl per fraction was injected for nano-LC-mass spectrometry (MS)/MS analysis. The peptide mixture (5 µg) was loaded onto the C18-reversed phase column (Thermo Scientific Easy Column, 10 cm long, 75 µm inner diameter, 3 µm resin; Thermo Fisher Scientific, Inc.) in buffer A (0.1% formic acid), and was separated with a linear gradient of buffer B (80% acetonitrile and 0.1% formic acid) at a flow rate of 250 nl/min, which was controlled by an IntelliFlow device over 140 min. MS data were acquired using a data-dependent top 10 method, dynamically selecting the most abundant precursor ions from the survey scan (300–1800 m/z) for high-energy collision-induced dissociation fragmentation. Determination of the target value was based on predictive automatic gain control. The dynamic exclusion duration was 60 sec. Survey scans were acquired at a resolution of 70,000 at 200 m/z, and the resolution for the higher-energy collisional dissociation spectra was set to 17,500 at 200 m/z. The normalized collision energy was 30 eV and the underfill ratio, which specifies the minimum percentage of the target value likely to be reached at maximum fill time, was defined as 0.1%. The instrument was run with the peptide recognition mode enabled.
Bioinformatics annotation of proteomics analysis results
To determine the biological and functional properties of the identified proteins, Gene Ontology (GO) analysis was conducted by searching the GO database (http://www.geneontology.org). Functional category analysis was performed with protein2go and go2protein for annotation. Visualization and Integrated Discovery v6.7 (28) was used for functional enrichment analysis of GO terms. A false discovery rate <0.01 was selected as the cut-off criterion.
Western blotting
Cells were lysed in RIPA buffer (Sigma-Aldrich; Merck KGaA). The protein concentration in the samples was determined using a Bradford protein assay (Bio-Rad Laboratories, Inc.). Equal amounts of cell lysate (30 µg/lane) were separated via 10% polyacrylamide gel electrophoresis, and transferred onto PVDF membranes, which were blocked using 5% w/v non-fat milk containing TBS-Tween 20 (TBST; 0.05% v/v) for 1 h at room temperature. The membranes were incubated with antibodies against DCTN2 (cat. no. D122213; Sangon Biotech Co., Ltd., Shanghai, China) and ROCK1 (cat. no. 4035; Cell Signaling Technology, Inc.) overnight at 4°C, washed with TBST, and then incubated with anti-rabbit secondary antibody (cat. no. 7074; Cell Signaling Technology, Inc.) at room temperature for 1 h. β-actin (cat. no. D191047; Sangon Biotech Co., Ltd.; 1:1,000) was detected as a reference protein for equal sample loading. ECL reagents (cat. no. 32106; Thermo Fisher Scientific, Inc.) were used to visualize the protein bands, which were captured on X-ray films. Image Lab 5.0 software (Bio-Rad Laboratories, Inc.) was used for quantitative analysis.
Knockdown of protein expression
Small interfering (si)RNA technology was used to knockdown protein expression. The pre-designed siRNA oligonucleotides were obtained from Sangon Biotech Co., Ltd.: ROCK1 forward, 5′-UGAUCUUGUAGCUCCCGCAUGUGUC-3′ and reverse, 5′-GACACAUGCGGGAGCUACAAGAUCA-3′; DCTN2 forward, 5′-GCACAAGUGUGGAACACAU-3′ and reverse, 5′-AUGUGUUCCACACUUGUGC-3′ and non-coding siRNA control, forward, 5′-UUCUCCGAACGUGUCACGUTT-3′ and reverse 5′-ACGUGACACGUUCGGAGAATT-3′. Cells (5×104 per well) were seeded into a 6-well plate and cultured at 37°C for 24 h, and then were transfected with 200 nM siRNA oligonucleotides using Lipofectamine® 2000 transfection reagent (Thermo Fisher Scientific, Inc.) at 37°C for 6 h, according to the manufacturer's instructions. Transfection efficiency was evaluated by western blot analysis 24 h after transfection.
Bioinformatics analysis for RNA expression of DCTN2 and ROCK1 in colon cancer
RNA expression data of DCTN2 and ROCK1 in normal and colon cancer tissues were acquired in CPTAC from UALCAN (http://ualcan.path.uab.edu). Prognosis correlation data was acquired from the HUMAN PROTEIN ATLAS (https://www.proteinatlas.org). Best value was selected for prognosis analysis. Based on the FPKM value of each gene, patients were classified into two groups and association between prognosis (survival) and gene expression (FPKM) was examined. The best expression cut-off refers the FPKM value that yields maximal difference with regard to survival between the two groups at the lowest log-rank P-value.
Statistical analysis
Statistical analyses were performed using SPSS version 21 (IBM Corp.), and the data are expressed as the mean ± SD. Differences between two groups were assessed by Student's t-test, and multiple group comparisons were performed using ANOVA followed by Tukey's post hoc test. P<0.05 was considered to indicate a statistically significant difference.
Results
High glucose, insulin and palmitic acid trigger centrosome amplification
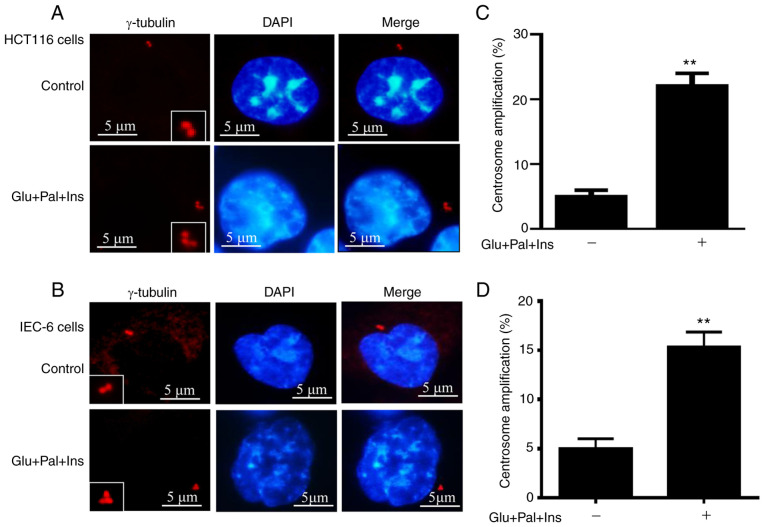
HCT116 and IEC-6 cells were treated with high glucose, insulin and palmitic acid, which was found to increase centrosome amplification. Representative images of centrosomes and centrosome amplification in HCT116 and IEC-cells are displayed in Fig. 1A and B, respectively. Compared with the control samples, an increased percentage of centrosome amplification (~4.6- and 3.2-fold) was observed in treated HCT116 (5 vs. ~23%; P<0.01; Fig. 1C) and IEC-6 cells (5 vs. ~16%; P<0.01; Fig. 1D), respectively. The results confirmed that the pathophysiological factors of type 2 diabetes cause centrosome amplification.
Figure 1.
High Glu, Ins and Pal trigger centrosome amplification. HCT116 and IEC-6 cells were treated for 48 h, and centrosomes were visualized via immunofluorescent staining using an antibody against γ-tubulin. Representative images of centrosomes are shown in (A) HCT116 and (B) IEC-6 cells. Percentages of cells with centrosome amplification was increased in (C) HCT116 and (D) IEC-6 cells following treatment. In total, 200 cells were evaluated for centrosome quantification. Percentage of centrosome amplification=(the number of cells with centrosome amplification/200) ×100. **P<0.01. Glu, glucose, 15 mM; Ins, insulin, 5 nM; Pal, palmitic acid, 150 µM.
Identification of ROCK1 binding proteins using CoIP and high MS-based proteomics analysis
Our previous study revealed that treatment with high glucose, insulin and palmitic acid increased the expression level and centrosomal translocation of ROCK1, which promotes centrosome amplification (15). To characterize ROCK1 binding partners, CoIP was performed using a ROCK1 antibody, and binding proteins were identified using MS. In total, 106 proteins were identified exclusively from the treated samples, 193 proteins exclusively from the control samples and 849 proteins from both the control and treated samples (Table SI).
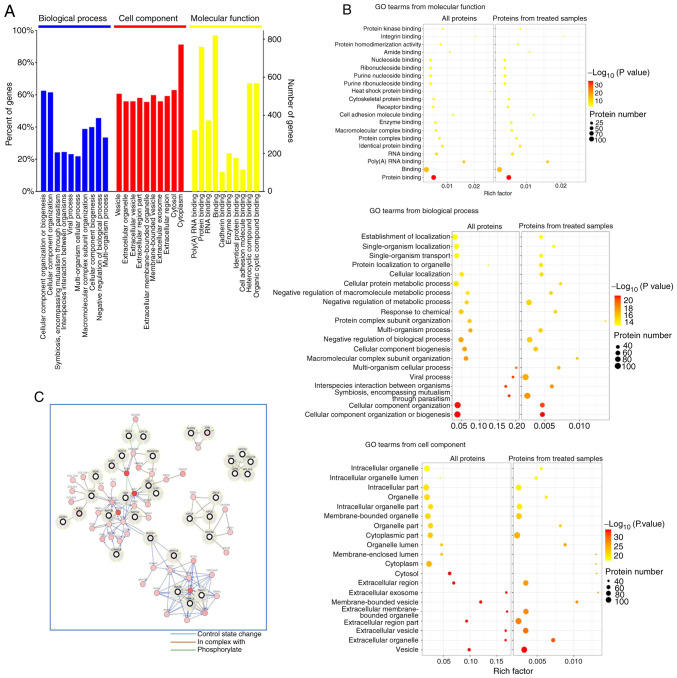
GO clustering analysis identified the enrichment of these proteins in biological processes, molecular functions and cellular components. Within the biological process category, the majority of the proteins were involved in ‘cellular component organization’ or ‘biogenesis and cellular component organization’. For molecular function, the data indicated that most of the proteins were associated with ‘protein binding’, ‘heterocyclic compound binding’ and ‘organic cyclic compound binding’ (Fig. 2A). Moreover, most proteins were located in the ‘cytoplasm’ and ‘cytosol’ (Fig. 2A). The GO term analysis was also used to reveal specific meaningful terms, especially in the treatment condition. Notably, it was found that ‘protein binding’ in molecular function was significantly enriched in all of the identified proteins, and in those found only in the treated samples (Fig. 2B). The differentially expressed proteins that had not been shown to interact with ROCK1 were clustered into three complexes (Fig. 2C). Considering the centrosomal localization of ROCK1, proteins associated with centrosomal localization were investigated (Table I), from which DCTN2 was selected for further investigation. This protein was selected for analysis as it can interact with microtubules, which are associated with the centrosome. These analyses identified various candidate binding partners of ROCK1, which provided the background information for the study that confirmed the binding between ROCK1 and specific partners, signaling for the centrosome amplification.
Figure 2.
Identification of ROCK1 binding proteins using coimmunoprecipitation in combination with mass spectrometry-based proteomic analysis. (A) GO annotation of the identified proteins, which were enriched in different terms. (B) Deepened analysis of GO annotation revealed significant enrichment of the term ‘protein binding’. (C) Differentially expressed proteins were primarily clustered in three complexes. GO, Gene Ontology; ROCK1, Rho-associated protein kinase 1.
Table I.
Centrosomal proteins identified by nano-LC-MS&MS.
| Protein name | Protein description | Protein ID | Sample present in |
|---|---|---|---|
| BCCIP | BRCA2 and CDKN1A-interacting protein | Q9P287 | T |
| DCTN2 | Dynactin subunit 2 | Q13561 | T |
| KIF2A | Kinesin-like protein KIF2A | O00139 | T |
| KIF2C | Kinesin-like protein KIF2C | Q99661 | |
| PLEKHG6 | Pleckstrin homology domain-containing family G member 6 | Q3KR16 | T |
| TPR | Nucleoprotein TPR | P12270 | T |
| ZW10 | Centromere&kinetochore protein zw10 homolog | O43264 | T |
| ACTR1A | Alpha-centractin | P61163 | C |
| AURKB | Aurora kinase B | Q96GD4 | C |
| MEAF6 | Chromatin modification-related protein MEAF6 | Q9HAF1 | C |
| NUDT21 | Cleavage and polyadenylation specificity factor subunit 5 | O43809 | C |
| NUMA1 | Nuclear mitotic apparatus protein 1 | Q14980 | C |
| RANBP1 | Ran-specific GTPase-activating protein | P43487 | C |
| TOP2A | DNA topoisomerase 2-alpha | P11388 | C |
| CALM3 | Calmodulin-3 | P0DP25 | C&T |
| HSPA1B | Heat shock 70 kDa protein 1B | P0DMV9 | C&T |
| CCT8 | T-complex protein 1 subunit theta | P50990 | C&T |
| NPM1 | Nucleophosmin | P06748 | C&T |
| HSPA6 | Heat shock 70 kDa protein 6 | P17066 | C&T |
| CCT4 | T-complex protein 1 subunit delta | P50991 | C&T |
| TCP1 | T-complex protein 1 subunit alpha | P17987 | C&T |
| CCT5 | T-complex protein 1 subunit epsilon | P48643 | C&T |
| MDH1 | Malate dehydrogenase, cytoplasmic | P40925 | C&T |
| DHX9 | ATP-dependent RNA helicase A | Q08211 | C&T |
| RUVBL2 | RuvB-like 2 | Q9Y230 | C&T |
| C14orf166 | UPF0568 protein C14orf166 | Q9Y224 | C&T |
| CROCC | Rootletin | Q5TZA2 | C&T |
| MAPRE1 | Microtubule-associated protein RP&EB family member 1 | Q15691 | C&T |
| PAFAH1B1 | Platelet-activating factor acetyl hydrolase IB subunit alpha | P43034 | C&T |
C, control sample; T, treated sample; C&T, both control and treated sample; LC, liquid chromatography; MS, mass spectrometry.
DCTN2 is localized to the centrosome
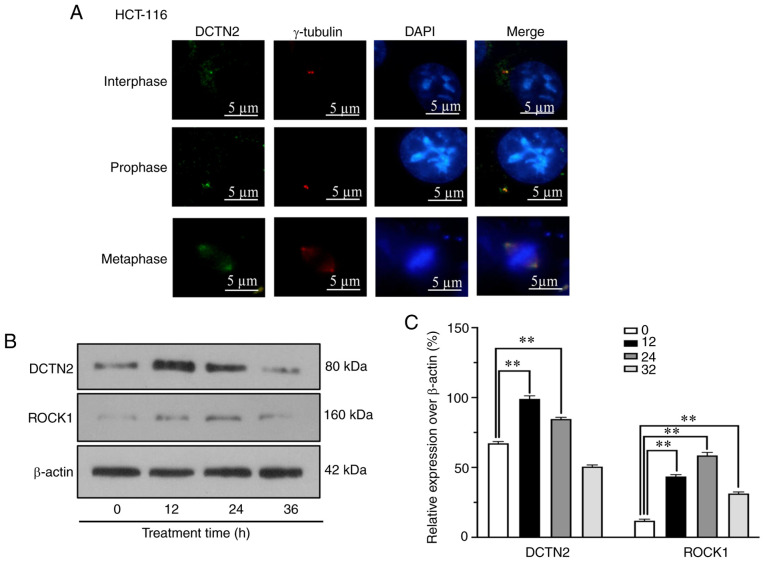
Immunofluorescent staining was conducted to evaluate the centrosomal localization of DCTN2, as ROCK1 was already shown to be transported to the centrosome following treatment with high glucose, insulin and palmitic acid (15). As presented in Fig. 3A, DCTN2 was localized to only one centrosome in HCT116 cells at the early stage of cell division, and was then distributed between both centrosomes thereafter. DCTN2 also appeared in the spindles, particularly the spindle area closest to the centrosomes. When multipolar division occurred due to centrosome amplification, DCTN2 was present in multipolar spindles. It was also demonstrated that upon treatment, the protein levels of ROCK1 and DCTN2 were upregulated, which peaked at 12 h, and then declined at 24 h (Fig. 3B and C). The results confirm that DCTN2 is indeed a centrosomal protein.
Figure 3.
Centrosomal localization and the treatment-induced upregulation of the of ROCK1 and DCTN2 expression in HCT116 cells. (A) DCTN2 was localized to the centrosome in ~100% of the 300 cells counted. Treatment with high glucose, insulin and palmitic acid increased the expression levels of (B) ROCK1 and (C) DCTN2. **P<0.01. ROCK1, Rho-associated protein kinase 1; DCTN2, dynactin subunit 2.
High glucose, insulin and palmitic acid increase the binding between DCTN2 and ROCK1
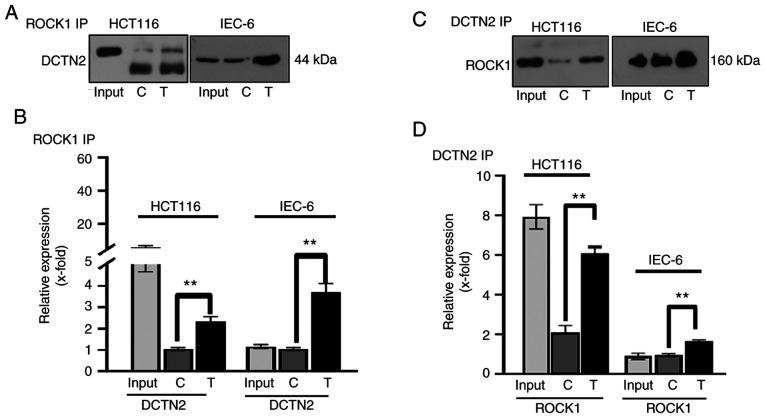
CoIP was performed in combination with western blot analysis to evaluate binding between ROCK1 and DCTN2 in HCT116 and IEC-6 cells, and to investigate whether the experimental treatment enhanced this protein binding. Similar protein binding profiles were obtained from both the HCT116 and IEC-6 cell lines. An anti-ROCK1 antibody was able to pull down DCTN2, and the protein expression level of DCTN2 pulled down by the ROCK1 antibody was increased upon treatment with high glucose, insulin and palmitic acid (Fig. 4A and B). Similarly, the antibody against DCTN2 was able to pull down ROCK1, which was enhanced by the experimental treatment (Fig. 4C and D). The results indicated that high glucose, insulin and palmitic acid enhanced the binding between ROCK1 and DCTN2 in non-cancerous (IEC-6) and cancerous (HCT116) colon cells.
Figure 4.
ROCK1 and DCTN2 form a protein complex in HCT116 and IEC-6 cells. (A and B) ROCK1 antibody pulled down DCTN2, which was enhanced by the experimental treatment. (C and D) DCTN2 antibody pulled down ROCK1, which was enhanced by the experimental treatment. Lower band below DCTN2 is likely to be the IgG used for IP, which was visualized by the secondary antibody used during western blot analysis. **P<0.01 vs. control. C, control sample; T, treated sample; IP, immunoprecipitation; ROCK1, Rho-associated protein kinase 1; DCTN2, dynactin subunit 2.
Disruption of the ROCK1-DCTN2 protein complex inhibits centrosome amplification
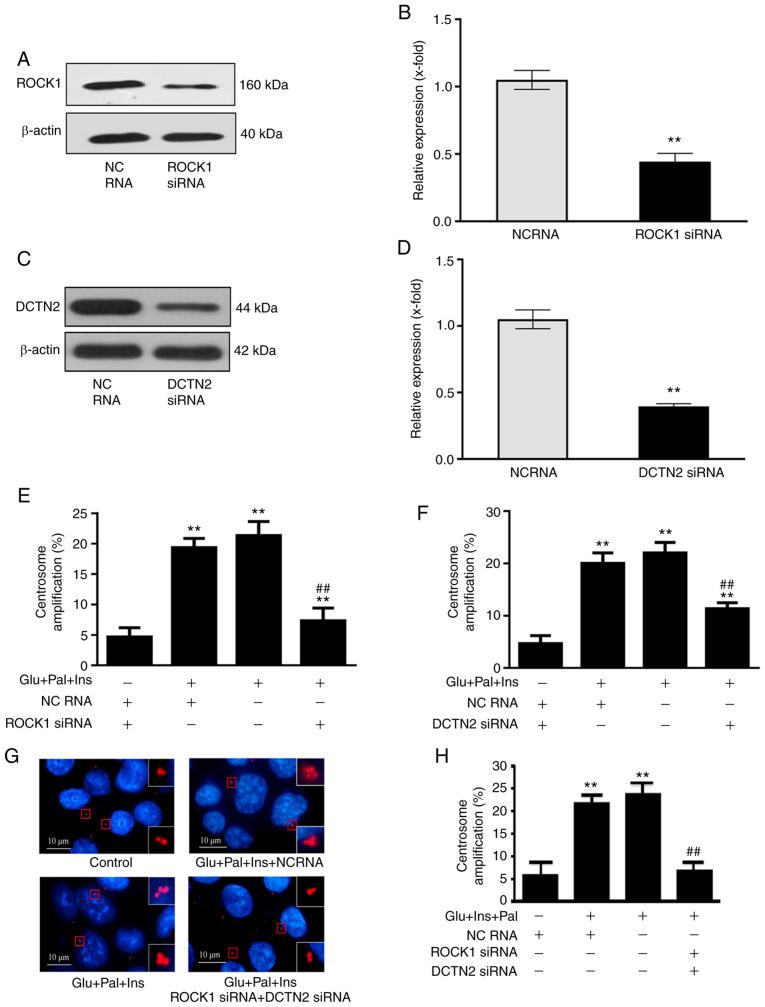
Next, the ROCK1-DCTN2 protein complex was disrupted by knocking down the expression of ROCK1 and DCTN2, and the effect on centrosome amplification in HCT116 cells was investigated. Knockdown of ROCK1 (Fig. 5A and B) or DCTN2 (Fig. 5C and D) partially inhibited the centrosome amplification triggered by the experimental treatment (Fig. 5E and F). Notably, simultaneous knockdown of these two proteins inhibited of centrosome amplification to a large degree (Fig. 5G and H). The results suggested that the binding of these proteins mediated diabetes-associated centrosome amplification.
Figure 5.
Disruption of the ROCK1-DCTN2 protein complex inhibits centrosome amplification in HCT116 cells. siRNA-knockdown decreased the expression levels of (A and B) ROCK1 and (C and D) DCTN2. (E and F) Individually, knockdown of ROCK1 and DCTN2 partially attenuated centrosome amplification. (G and H) Co-transfection with ROCK1 and DCTN2 siRNAs inhibited centrosome amplification to a considerable degree. In total, 200 cells used for centrosome quantification. Percentage of centrosome amplification=(the number of cells with centrosome amplification/200) ×100. **P<0.01 vs. control; ##P<0.01 vs. samples treated high glucose, insulin and palmitic acid. Glu, high glucose; Pal, palmitic acid; Ins, insulin; NC, non-coding RNA; ROCK1, Rho-associated protein kinase 1; DCTN2, dynactin subunit 2; siRNA, small interfering RNA.
Expression levels of ROCK1 and DCTN2 in colon cancer
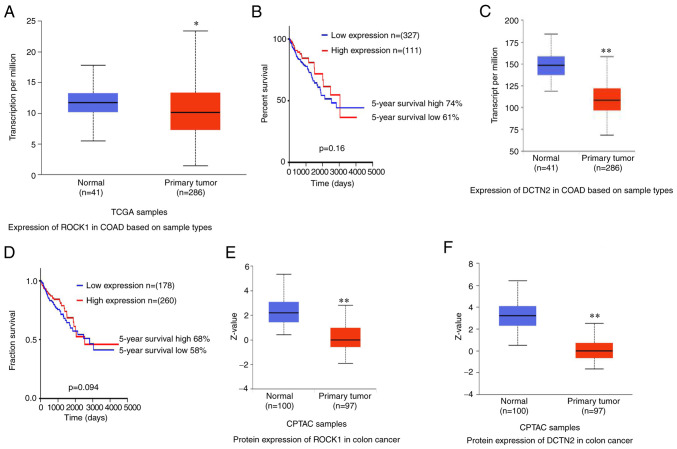
The expression levels of ROCK1 and DCTN2 in colon cancer and their correlation with prognosis were analyzed using UALCAN and the HUMAN PROTEIN ATLAS. ROCK1 mRNA expression was higher in normal colon tissues compared with cancer tissues (P<0.05). However, ROCK1 expression was not associated with prognosis, and it did not predict the 5-year survival rate (P>0.05; Fig. 6A and B). Furthermore, the expression level of DCTN2 was lower in cancer tissues compared with normal colon tissues (P<0.01), and was not associated with prognosis (P>0.05; Fig. 6C and D). The expression levels of ROCK1 and DCTN2 were also analyzed using Clinical Proteomic Tumor Analysis Consortium, and were also decreased in cancer tissues in comparison with normal tissues (both P<0.001; Fig. 6E and F). It would be interesting to investigate whether binding between ROCK1 and DCTN2, rather than the expression levels, is associated colon cancer prognosis.
Figure 6.
Analysis of mRNA expression levels of ROCK1 and DCTN2 and their association with prognosis in colon cancer. (A) mRNA expression level of ROCK1 in human normal colon tissues and colon cancer samples. (B) Prognosis analysis of ROCK1 expression in human colon adenocarcinoma. (C) mRNA expression level of DCTN2 in normal colon and colon adenocarcinoma tissues. (D) Prognosis analysis of DCTN2 expression in human colon adenocarcinoma. mRNA expression levels of (E) ROCK1 and (F) DCTN2 in normal colon and colon adenocarcinoma tissues in CPTAC samples. ROCK1, Rho-associated protein kinase 1; DCTN2, dynactin subunit 2; CPTAC, Clinical Proteomic Tumor Analysis Consortium. *P<0.05 vs. normal, **P<0.01 vs. normal.
Discussion
The results of the present study demonstrated that treating HCT116 human colon cancer cells and non-cancerous IEC-6 cells with high glucose, insulin and palmitic acid, increased the expression levels and binding interaction between ROCK1 and DCTN2, which resulted in centrosome amplification. These results confirmed those of our previous study, indicating that ROCK1 is an initiating signal for diabetes-associated centrosome amplification (6); notably, knockdown of both ROCK1 and DCTN2 blocked centrosome amplification, suggesting that the formation of the ROCK1-DCTN2 protein complex is a key signaling event. It was indicated that the binding of the two proteins was more important than their expression levels, as these levels were not associated with prognosis, although their individual roles in cancer development should be further investigated. It should be noted that binding between the two proteins should be confirmed using other methodologies, such as far-western blot analysis, yeast two-hybrids and fluorescence resonance energy transfer analyses. Whether the increased binding in response to the treatment occurred in the centrosomes should also be tested. Moreover, immunofluorescent staining in combination with confocal imaging is not accurate enough to differentiate the level of protein complex in the centrosome before and after treatment. The present study was unable to predict the binding model between ROCK1 and DCTN2 using existing databases, since the three-dimensional structure of DCTN2 is not available. In addition, data from experiments using other technologies are required to further support the current view that ROCK1 and DCTN2 form a complex under the present experimental conditions.
DCTN2 is a 50 kDa subunit of dynactin, which is a multiprotein complex associated with dynein. Cytoplasmic dynein is a molecular motor responsible for minus-end-directed movement along microtubules (29). DCTN2 is also required for efficient autophagy under hypertonic stress, where it is likely to act within dynein (30), supporting the notion that dynein serves a regulatory role in autophagy (31). In a Norwegian family, the DCTN2 gene was found to be associated with Charcot-Marie-Tooth disease (32), suggesting that it contributes to the pathogenesis of this inherited neuropathy. The DCTN2 is located on chromosome 12q13-15, which is a region prone to stable amplification in various cancer types. For example, in osteosarcoma, DCTN2 is upregulated in SJSA-1 cells compared with SJRH30 cells, which is associated with diminished centrosomes (33), suggesting that it serves a role in centrosome homeostasis. In glioma, amplification of 12q13-21 is observed even in low grade tumors, leading to the upregulation of genes including DCTN2 (34), thereby suggesting a role for this region in glioma development. Furthermore, low DCTN2 mRNA expression is associated with favorable prognosis in cutaneous melanoma (35), and its upregulation is associated with colon cancer (36). These results suggest that enhanced expression of DCTN2 is not a desirable event. The present study demonstrated a new functional role of ROCK1 and DCTN2, with particular relevance in cancer development.
How the ROCK1-DCTN2 complex promotes centrosome amplification remains unknown. Our previous study identified nucleophosmin 1 (NPM) as a signal for centrosome amplification (37). NPM phosphorylation and dissociation from the centrosome is known to cause centrosome amplification (29). Therefore, it would be interesting to determine whether the protein complex targets NPM, directly or indirectly. It also remains unknown whether the ROCK1-DCTN2 complex interacts with other recognized protein mediators of centrosome amplification, such as polo kinase 4. Both ROCK1 and DCTN2 are localized to one centrosome, at least at some stage in centrosome dynamics (6). This raises the issue of whether a centrosome harboring these proteins is classified as a mother or daughter centrosome.
As a key signaling complex, The ROCK1-DCTN2 provides a target for the effective inhibition of centrosome amplification, which allows for the production of cells with defective centrosome amplification upon stable inhibition of cellular signal(s). This may facilitate investigation into the roles of diabetes-associated centrosome amplification in tumorigenesis and cancer cell metastasis, where it is required to show that inhibiting centrosome amplification prevents diabetes-associated tumor development. These results may promote the assessment of centrosome amplification in tumorigenesis and metastasis in cancer in general. The ROCK1-DCTN2 complex serves as a promising target for the potential future development of intervention protocols targeting centrosome amplification. In addition, centrosome amplification has other pathological consequences (38), such as disturbed cilia signaling (39), and the dysregulation of cilia function is associated with kidney, retinal and neural defects. In particular, ciliopathy is implicated in the development of a retinal-renal ciliopathy (18,40). Moreover, amplification of the centrosome disrupts renal development and causes cystogenesis (16). Thus, it would be beneficial to determine whether the ROCK1-DCTN2 protein complex triggers centrosome amplification to promote other diseases, such as diabetic retinopathy, nephropathy and neuropathy.
In conclusion, the results of the present suggested that the binding between ROCK1 and DCTN2 regulates centrosome homeostasis, which is a key signal for the centrosome amplification triggered by pathophysiological factors in type 2 diabetes, such as high glucose, insulin and palmitic acid. The ROCK1-DCTN2 protein complex may therefore be a favorable target for inhibiting centrosome amplification in future research and therapeutic development.
Supplementary Material
Acknowledgements
Not applicable.
Funding Statement
The present study was supported by grants from the Shanxi Health Commission Research Project (grant no. 09509) and Jiangsu Normal University (grant no. 9212418102), as well as the Department of Science and Technology of the Jiangsu Province (grant no. BX2019029).
Funding
The present study was supported by grants from the Shanxi Health Commission Research Project (grant no. 09509) and Jiangsu Normal University (grant no. 9212418102), as well as the Department of Science and Technology of the Jiangsu Province (grant no. BX2019029).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
YFL and SCL contributed to the grant application, study design and manuscript preparation. PW, LJS, JWW and GYS performed the experiments. PW and YFL were responsible for the statistical analysis. YFL and SCL confirm the authenticity of all the raw data. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Zheng Y, Ley SH, Hu FB. Global aetiology and epidemiology of type 2 diabetes mellitus and its complications. Nat Rev Endocrinol. 2018;14:88–98. doi: 10.1038/nrendo.2017.151. [DOI] [PubMed] [Google Scholar]
- 2.Bommer C, Heesemann E, Sagalova V, Manne-Goehler J, Atun R, Barnighausen T, Vollmer S. The global economic burden of diabetes in adults aged 20–79 years: A cost-of-illness study. Lancet Diabetes Endocrinol. 2017;5:423–430. doi: 10.1016/S2213-8587(17)30097-9. [DOI] [PubMed] [Google Scholar]
- 3.Nazir MA, AlGhamdi L, AlKadi M, AlBeajan N, AlRashoudi L, AlHussan M. The burden of diabetes, its oral complications and their prevention and management. Open Access Maced J Med Sci. 2018;6:1545–1553. doi: 10.3889/oamjms.2018.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.World Health Organization. Fact sheets. http://www.who.int/news-room/fact-sheets/detail/diabetes. [Jun 29;2019 ]; [Google Scholar]
- 5.Dall TM, Yang W, Gillespie K, Mocarski M, Byrne E, Cintina I, Beronja K, Semilla AP, Iacobucci W, Hogan PF. The economic burden of elevated blood glucose levels in 2017: Diagnosed and undiagnosed diabetes, gestational diabetes mellitus, and prediabetes. Diabetes Care. 2019;42:1661–1668. doi: 10.2337/dc18-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang P, Lu YC, Wang J, Wang L, Yu H, Li YF, Kong A, Chan J, Lee S. Type 2 diabetes promotes cell centrosome amplification via AKT-ROS-dependent signalling of ROCK1 and 14-3-3σ. Cell Physiol Biochem. 2018;47:356–367. doi: 10.1159/000489812. [DOI] [PubMed] [Google Scholar]
- 7.Giovannucci E, Harlan DM, Archer MC, Bergenstal RM, Gapstur SM, Habel LA, Pollak M, Regensteiner JG, Yee D. Diabetes and cancer: A consensus report. Diabetes Care. 2010;33:1674–1685. doi: 10.2337/dc10-0666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu B, Wu X, Wu B, Pei D, Zhang L, Wei L. The relationship between diabetes and colorectal cancer prognosis: A meta-analysis based on the cohort studies. PLoS One. 2017;12:e0176068. doi: 10.1371/journal.pone.0176068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Y, You X, Liu H, Xu M, Dang Q, Yang L, Huang J, Shi W. High KIF2A expression predicts unfavorable prognosis in diffuse large B cell lymphoma. Ann Hematol. 2017;96:1485–1491. doi: 10.1007/s00277-017-3047-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Landman GW, Ubink-Veltmaat LJ, Kleefstra N, Kollen BJ, Bilo HJ. Increased cancer mortality in type 2 diabetes (ZODIAC-3) Anticancer Res. 2008;28:1373–1375. [PubMed] [Google Scholar]
- 11.D'Assoro AB, Lingle WL, Salisbury JL. Centrosome amplification and the development of cancer. Oncogene. 2002;21:6146–6153. doi: 10.1038/sj.onc.1205772. [DOI] [PubMed] [Google Scholar]
- 12.Basto R, Brunk K, Vinadogrova T, Peel N, Franz A, Khodjakov A, Raff JW. Centrosome amplification can initiate tumorigenesis in flies. Cell. 2008;133:1032–1042. doi: 10.1016/j.cell.2008.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li J, Xuan JW, Khatamianfar V, Valiyeva F, Moussa M, Sadek A, Yang BB, Dong BJ, Huang YR, Gao WQ. SKA1 over-expression promotes centriole over-duplication, centrosome amplification and prostate tumourigenesis. J Pathol. 2014;234:178–189. doi: 10.1002/path.4374. [DOI] [PubMed] [Google Scholar]
- 14.Fan G, Sun L, Shan P, Zhang X, Huan J, Zhang X, Li D, Wang T, Wei T, Zhang X, et al. Loss of KLF14 triggers centrosome amplification and tumorigenesis. Nat Commun. 2015;6:8450. doi: 10.1038/ncomms9450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Levine MS, Bakker B, Boeckx B, Moyett J, Lu J, Vitre B, Spierings DC, Lansdorp PM, Cleveland DW, Lambrechts D, et al. Centrosome amplification is sufficient to promote spontaneous tumorigenesis in mammals. Dev Cell. 2017;40:313–322.e5. doi: 10.1016/j.devcel.2016.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dionne LK, Shim K, Hoshi M, Cheng T, Wang J, Marthiens V, Knoten A, Basto R, Jain S, Mahjoub MR. Centrosome amplification disrupts renal development and causes cystogenesis. J Cell Biol. 2018;217:2485–2501. doi: 10.1083/jcb.201710019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Godinho SA, Picone R, Burute M, Dagher R, Su Y, Leung CT, Polyak K, Brugge JS, Théry M, Pellman D. Oncogene-like induction of cellular invasion from centrosome amplification. Nature. 2014;510:167–171. doi: 10.1038/nature13277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee SC, Chan JC. Evidence for DNA damage as a biological link between diabetes and cancer. Chin Med J (Engl) 2015;128:1543–1548. doi: 10.4103/0366-6999.157693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dodson H, Bourke E, Jeffers LJ, Vagnarelli P, Sonoda E, Takeda S, Earnshaw WC, Merdes A, Morrison C. Centrosome amplification induced by DNA damage occurs during a prolonged G2 phase and involves ATM. EMBO J. 2004;23:3864–3873. doi: 10.1038/sj.emboj.7600393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He QJ, Wang P, Liu Q, Wu Q, Li YF, Wang J, Lee SC. Secreted Wnt6 mediates diabetes-associated centrosome amplification via its receptor FZD4. Am J Physiol Cell Physiol. 2020;318:C48–C62. doi: 10.1152/ajpcell.00091.2019. [DOI] [PubMed] [Google Scholar]
- 21.Hartmann S, Ridley AJ, Lutz S. The function of Rho-associated kinases ROCK1 and ROCK2 in the pathogenesis of cardiovascular disease. Front Pharmacol. 2015;6:276. doi: 10.3389/fphar.2015.00276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liang H, Zhang C, Guan H, Liu J, Cui Y. LncRNA DANCR promotes cervical cancer progression by upregulating ROCK1 via sponging miR-335-5p. J Cell Physiol. 2019;234:7266–7278. doi: 10.1002/jcp.27484. [DOI] [PubMed] [Google Scholar]
- 23.Tsai NP, Wei LN. RhoA/ROCK1 signaling regulates stress granule formation and apoptosis. Cell Signal. 2010;22:668–675. doi: 10.1016/j.cellsig.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen S, Liu Z, Lu S, Hu B. EPEL promotes the migration and invasion of osteosarcoma cells by upregulating ROCK1. Oncol Lett. 2019;17:3133–3140. doi: 10.3892/ol.2019.9975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zucchini C, Martinelli M, De Sanctis P, Rodia MT, Mattei G, Ugolini G, Montroni I, Ghignone F, Solmi R. Possible gender-related modulation by the ROCK1 gene in colorectal cancer susceptibility. Pathobiology. 2015;82:252–258. doi: 10.1159/000439405. [DOI] [PubMed] [Google Scholar]
- 26.Akagi EM, Lavorato-Rocha AM, Maia Bde M, Rodrigues IS, Carvalho KC, Stiepcich MM, Baiocchi G, Sato-Kuwabara Y, Rogatto SR, Soares FA, Rocha RM. ROCK1 as a novel prognostic marker in vulvar cancer. BMC Cancer. 2014;14:822. doi: 10.1186/1471-2407-14-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beeharry N, Lowe JE, Hernandez AR, Chambers JA, Fucassi F, Cragg PJ, Green MH, Green IC. Linoleic acid and antioxidants protect against DNA damage and apoptosis induced by palmitic acid. Mutat Res. 2003;530:27–33. doi: 10.1016/S0027-5107(03)00134-9. [DOI] [PubMed] [Google Scholar]
- 28.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 29.Chan N, Lim TM. Cytoplasmic nucleophosmin has elevated T199 phosphorylation upon which G2/M phase progression is dependent. Sci Rep. 2015;5:11777. doi: 10.1038/srep11777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nunes P, Ernandez T, Roth I, Qiao X, Strebel D, Bouley R, Charollais A, Ramadori P, Foti M, Meda P, et al. Hypertonic stress promotes autophagy and microtubule-dependent autophagosomal clusters. Autophagy. 2013;9:550–567. doi: 10.4161/auto.23662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu M, Li XX, Chen Y, Pitzer AL, Zhang Y, Li PL. Enhancement of dynein-mediated autophagosome trafficking and autophagy maturation by ROS in mouse coronary arterial myocytes. J Cell Mol Med. 2014;18:2165–2175. doi: 10.1111/jcmm.12326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Braathen GJ, Høyer H, Busk ØL, Tveten K, Skjelbred CF, Russell MB. Variants in the genes DCTN2, DNAH10, LRIG3, and MYO1A are associated with intermediate charcot-marie-tooth disease in a Norwegian family. Acta Neurol Scand. 2016;134:67–75. doi: 10.1111/ane.12515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bransfield KL, Askham JM, Leek JP, Robinson PA, Mighell AJ. Phenotypic changes associated with DYNACTIN-2 (DCTN2) over expression characterise SJSA-1 osteosarcoma cells. Mol Carcinog. 2006;45:157–163. doi: 10.1002/mc.20151. [DOI] [PubMed] [Google Scholar]
- 34.Fischer U, Keller A, Leidinger P, Deutscher S, Heisel S, Urbschat S, Lenhof HP, Meese E. A different view on DNA amplifications indicates frequent, highly complex, and stable amplicons on 12q13-21 in glioma. Mol Cancer Res. 2008;6:576–584. doi: 10.1158/1541-7786.MCR-07-0283. [DOI] [PubMed] [Google Scholar]
- 35.Wang Q, Wang X, Liang Q, Wang S, Liao X, Li D, Pan F. Prognostic value of dynactin mRNA expression in cutaneous melanoma. Med Sci Monit. 2018;24:3752–3763. doi: 10.12659/MSM.910566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang S, Wang Q, Zhang X, Liao X, Wang G, Yu L, Zhang W, Zhou Q, Hu S, Yuan W. Distinct prognostic value of dynactin subunit 4 (DCTN4) and diagnostic value of DCTN1, DCTN2, and DCTN4 in colon adenocarcinoma. Cancer Manag Res. 2018;10:5807–5824. doi: 10.2147/CMAR.S183062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Han X, Hou S, Yang A. Correlation between IGFs-related proteins expression and incidence of colorectal cancer in diabetic patients and related mechanisms. Med Sci Monit. 2016;22:848–854. doi: 10.12659/MSM.895837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marthiens V, Rujano MA, Pennetier C, Tessier S, Paul-Gilloteaux P, Basto R. Centrosome amplification causes microcephaly. Nat Cell Biol. 2013;15:731–740. doi: 10.1038/ncb2746. [DOI] [PubMed] [Google Scholar]
- 39.Mahjoub MR, Stearns T. Supernumerary centrosomes nucleate extra cilia and compromise primary cilium signaling. Curr Biol. 2012;22:1628–1634. doi: 10.1016/j.cub.2012.06.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Otto EA, Hurd TW, Airik R, Chaki M, Zhou W, Stoetzel C, Patil SB, Levy S, Ghosh AK, Murga-Zamalloa CA, et al. Candidate exome capture identifies mutation of SDCCAG8 as the cause of a retinal-renal ciliopathy. Nat Genet. 2010;42:840–850. doi: 10.1038/ng.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.