Abstract
Dysregulation of long non-coding RNA (IncRNA) antisense non-coding RNA in the INK4 locus (ANRIL) is associated with the risk of myocardial infarction (MI). Therefore, the present study aimed to determine the mechanisms underlying this association, which is currently poorly understood, to the best of our knowledge. The current study used an in vitro myocardial ischemia and reperfusion (MI/R) model, in which H9c2 cardiomyocytes were exposed to hypoxia/reoxygenation (H/R), which demonstrated that ANRIL expression was downregulated and that ANRIL positively regulated sirtuin 1 (SIRT1) expression following H/R injury. Subsequently, it was demonstrated that ANRIL upregulated SIRT1 expression by sponging microRNA-181a (miR-181a). In addition, ANRIL overexpression reduced lactate dehydrogenase release and apoptosis of H9c2 cardiomyocytes exposed to H/R, indicating that ANRIL prevented H/R-induced cardiomyocyte injury. Moreover, both miR-181a overexpression and SIRT1 knockdown significantly decreased the protective effects of ANRIL on H/R-induced cardiomyocyte injury, thus demonstrating that SIRT1 upregulation via sponging miR-181a is a critical mechanism that mediates the function of ANRIL. These results provided a novel mechanistic insight into the role of ANRIL in H/R-injured cardiomyocytes and suggested that the ANRIL/miR-181a/SIRT1 axis may be a therapeutic target for reducing MI/R injury.
Keywords: long non-coding RNA antisense non-coding RNA in the INK4 locus, hypoxia/reoxygenation injury, cardiomyocyte, microRNA-181a, sirtuin 1
Introduction
Myocardial infarction (MI) remains one of the leading causes of mortality and disability worldwide (1). At present, the most effective approach for reducing MI injury and size is timely myocardial reperfusion (2). However, unsatisfactorily, reperfusion causes further myocardial injury and cardiomyocyte death, causing a phenomenon denominated myocardial ischemia and reperfusion (MI/R) injury (3). MI/R injury contributes to disease progression in patients with MI. However, some mechanical and pharmacological therapeutic strategies for preventing MI/R injury have been shown to be effective in improving clinical outcomes (2).
Recently, MI pathology has been increasingly associated with long non-coding RNAs (lncRNAs) (4,5), a novel class of endogenous cellular RNAs that regulate gene expression through various means, including acting as microRNA (miRNA) sponges (6). The antisense non-coding RNA in the INK4 locus (ANRIL) can regulate cell proliferation, senescence and apoptosis involved in some cardiovascular diseases and human cancers (7). In particular, ANRIL was newly identified as a dysregulated lncRNA in patients with MI compared with healthy controls (8). In addition, an ANRIL polymorphism (rs1333049:C>G) is associated with MI (9). Consistently, another previous study also demonstrated that ANRIL gene variants contribute to MI risk (10). These reports suggest that ANRIL may play a functional role in the pathology of MI. However, to the best of our knowledge, at present, no evidence has been provided to mechanistically link ANRIL with MI/R injury.
In the present study, the role of ANRIL in the regulation of MI/R injury was investigated using a hypoxia/reoxygenation (H/R) in vitro experimental model. It was found that the expression of ANRIL was downregulated after H/R injury and that ANRIL positively regulated sirtuin 1 (SIRT1) expression by sponging miR-181a. This regulatory mechanism may be responsible for the protective role of ANRIL against H/R-induced cardiomyocyte injury. These data could offer a novel mechanistic insight into understanding the association between ANRIL and MI pathology.
Materials and methods
Cell culture and H/R treatment
H9c2 cardiomyocytes (cat. no. CRL-1446) were purchased from the American Type Culture Collection and then cultured in DMEM (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% (v/v) FBS (Gibco; Thermo Fisher Scientific, Inc.), 100 U/ml penicillin and 100 mg/ml streptomycin (Gibco; Thermo Fisher Scientific, Inc.) in an incubator at 37°C with 5% CO2. In vitro H/R treatment was used to mimic MI/R injury, as described previously (11). Briefly, H9c2 cardiomyocytes were cultured in glucose-free DMEM with an oxygen-free atmosphere (95% N2 and 5% CO2) at 37°C for 4 h, followed by incubation with normal medium (4.5 mg/ml glucose) and atmosphere (95% air and 5% CO2) at 37°C for 2 to 24 h, depending on the experimental purposes.
Transfection
ANRIL-overexpressing plasmids were constructed by inserting the ANRIL coding sequence into the pcDNA3.1 vector (Invitrogen; Thermo Fisher Scientific, Inc.). Oligonucleotides, including miR-181a mimic (5′-AACAUUCAACGCUGUCGGUGAGU-3′), negative control (NC) mimic (5′-UUCUCCGAACGUGUCACGUTT−3′), miR-181a antagomir (5′-ACUCACCGACAGCGUUGAAUGUU-3′) and NC antagomir (5′-CAGUACUUUUGUGUAGUACAA−3′), were purchased from Guangzhou RiboBio Co., Ltd. Small interfering (si)RNA targeting SIRT1 (siSIRT1, 5′-CCCUGUAAAGCUUUCAGAATT−3′) and NC siRNA (siNC; 5′-UUCUCCGAACGUGUCACGUTT−3′) were obtained from Shanghai GenePharma Co., Ltd. When the cell density reached 50–60% confluence, cell transfection was performed with Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocols. Equal amounts of empty pcDNA3.1 vector (4 µg/well), NC mimic, NC antagomir (50 or 150 nM) or siNC (10 or 20 nM) diluted in the same volume of transfection reagents were transfected as controls, depending on the experimental purposes. The efficiency was examined 2 days after transfection. For H/R treatment, cells were cultured under normoxia or exposed to H/R (hypoxia for 4 h followed by reoxygenation for 8 h) at 2 days after transfection. The transfection efficiency for all transfections was effective under normoxic or H/R treatment conditions.
Reverse transcription-quantitative PCR (RT-qPCR) analysis
Total RNA from H9c2 cells was extracted using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. ANRIL and SIRT1 expression levels were determined via RT-qPCR analysis using the One Step PrimeScript™ RT-RNA kit (Takara Biotechnology Co., Ltd.) according to the manufacturer's protocol. miR-181a expression was determined using the TaqMan™ MicroRNA Reverse Transcription kit and TaqMan Universal Master Mix II (Applied Biosystems; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocols. The thermocycling conditions were as follows: Initial denaturation at 95°C for 5 min, followed by 40 cycles of 95°C for 15 sec and 60°C for 1 min. β-actin served as an internal control for ANRIL and SIRT1. U6 served as an internal control for miR-181a. Data were calculated using the 2−ΔΔCq method (12). The following primer sequences were used for the qPCR: ANRIL forward, 5′-CAAGCCACGTTGGAAGATGC-3′ and reverse, 5′-AGAGTGTGTAGCAGCTGACG-3′; miR-181a forward, 5′-CACTCCAGCTGGGAACATTCAACGCTGTCGG-3′ and reverse, 5′-TGGTGTCGTGGAGTCG-3′; U6 forward, 5′-TGGGGTTATACATTGTGAGAGGA-3′ and reverse, 5′-GTGTGCTACGGAGTTCAGAGGTT-3′; SIRT1 forward, 5′-CCAGATCCTCAAGCCATG−3′ and reverse, 5′-TTGGATTCCTGCAACCTG−3′; and β-actin forward, 5′-CATGCCATCCTGCGTCTGGA-3′ and reverse, 5′-CCACATCTGCTGGAAGGTGG-3′.
Immunoblotting
Total cellular proteins were extracted using RIPA lysis buffer (Beyotime Institute of Biotechnology) supplemented with the protease inhibitor cocktail (Roche Diagnostics GmbH). The protein concentration was determined using a BCA protein assay (Pierce; Thermo Fisher Scientific, Inc.). Total proteins (30 µg per well) were separated via 10% SDS-PAGE electrophoresis, and then separated proteins were transferred to polyvinylidene difluoride membranes, which were then blocked for 1 h with 5% non-fat milk at room temperature and incubated overnight at 4°C with primary antibodies against SIRT1 (1:1,000; cat. no. ab7343; Abcam), β-actin (1:5,000; cat. no. sc-47778; Santa Cruz Biotechnology, Inc.), cleaved caspase-3 (1:500; cat. no. ab49822; Abcam), Bcl-2 (1:500; cat. no. sc-7382; Santa Cruz Biotechnology, Inc.) and Bax (1:2,000; cat. no. NBP1-28566; Novus Biologicals, LLC). After washing, the membranes were further incubated at room temperature for 1 h with a horseradish peroxidase-conjugated secondary antibody (1:10,000; cat. no. ab6721; Abcam). Protein bands were visualized with the Immobilon Western Chemiluminescent HRP Substrate (EMD Millipore). Protein expression was semi-quantified using Image Lab software (version 5.1; Bio-Rad Laboratories, Inc.).
H/R injury detection
The terminal deoxynucleotidyl-transferase mediated dUTP nick-end labeling (TUNEL) method was carried out to evaluate cell apoptosis using the TUNEL assay kit (OriGene Technologies, Inc.) according to the manufacturer's protocols. Briefly, cells were fixed with 4% paraformaldehyde solution for 1 h at room temperature and then stained with TUNEL reagent for 1 h at 37°C. The nuclei were stained with 1 µg/ml DAPI for 15 min at room temperature. Cells were mounted with mounting solution (Beyotime Institute of Biotechnology), and images were captured with the LSM510META fluorescence microscope (Carl Zeiss AG; magnification, ×100). TUNEL-positive cells were considered as apoptotic cells and counted in 12 random fields from each treatment group. Cell apoptosis was expressed as the number of apoptotic cells divided by the total number of cells. Lactate dehydrogenase (LDH) release was determined using an LDH cytotoxicity detection kit (cat. no. A020; Nanjing Jiancheng Bioengineering Institute) following the manufacturer's instructions. The absorbance at 450 nm was measured using the Synergy HT microplate reader (BioTek Instruments, Inc.). The results were calculated as the LDH activity in the medium divided by the LDH activity in total cell lysates.
RNA immunoprecipitation (RIP) assay
RIP experiments were performed using a Magna RIP RNA Binding Protein Immunoprecipitation kit (cat. no. 17-700; EMD Millipore) according to the manufacturer's protocol. The Argonaute 2 (AGO2)-RIP assay was conducted using lysates of H9c2 cells. Briefly, 1×107 cells were pelleted at 3,000 × g for 10 min at 4°C and resuspended with an equal volume of RIP lysis buffer (100 µl) containing protease and RNase inhibitors. Cell lysates (100 µl) were incubated with 5 µg control IgG antibody (cat. no. ab182931; Abcam) or anti-AGO2 antibody (cat. no. ab32381; Abcam)-coated protein A/G magnetic beads at 4°C overnight, with constant rotation. After treating with proteinase K at 55°C for 30 min, the immunoprecipitated RNAs were extracted using a RNeasy MinElute Cleanup kit (Qiagen, Inc.) and reverse transcribed as described above for the RT-qPCR experiments. The expression levels of miR-181a and ANRIL were analyzed using RT-qPCR as described above.
Luciferase reporter assay
The putative targets of miRNAs were predicted using in silico computational tools, including starBase (v2.0; http://starbase.sysu.edu.cn/starbase2) and TargetScan (http://www.targetscan.org/vert_72). The wild-type (wt) 3′untranslated region (UTR) of ANRIL and SIRT1 containing the putative miR-181a binding sites was amplified by PCR and inserted into the pmirGLO Dual-luciferase miRNA Target Expression Vector (Promega Corporation). The clone primers are as follows: ANRIL forward, 5′-ATCGATAGCGATAAGATCTCATTGCTCTATCCGCCAA-3′ and reverse, 5′-CGCAAGCGCAAAGAGTAGTTCAAAACTGACATTCAGC-3′; and SIRT1 forward, 5′-CAAACAAATCATAGTGTAATAA-3′ and reverse, 5′-CGCAAGCGCAAAGAGACAATCTATTTTACCAACCTAT−3′. DNA was synthesized from 293T cells via TaqFast DNA polymerase (cat. no. T828421; MACKLIN) and isolated using a DNA purification kit (cat. no. DP304; Tiagen Biotech Co., Ltd.) according to the manufacturer's protocol. Those with putative mutated binding sites (ANRIL-mut and SIRT1-mut) were generated using the Site-Directed Mutagenesis System (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. The luciferase assay was performed by co-transfecting reporter vectors along with NC mimic (NC-mimic) or miR-181a mimic into 293T cells using Lipofectamine 2000 reagent according to the manufacturer's protocols. Each treatment was performed five times. Then, 2 days after transfection, firefly luciferase activity was measured using the Dual-Luciferase Reporter Assay System (Promega Corporation), according to the manufacturer's instructions. The Renilla luciferase activity served as a normalization control for each well.
Statistical analysis
All results from at least three replicates are expressed as the mean ± SD. P-values were calculated using one-way analysis of variance followed by Tukey's post hoc test using SPSS 19.0 software (IBM Corp.). P<0.05 was considered to indicate a statistically significant difference.
Results
ANRIL and SIRT1 transcription levels are decreased following H/R injury
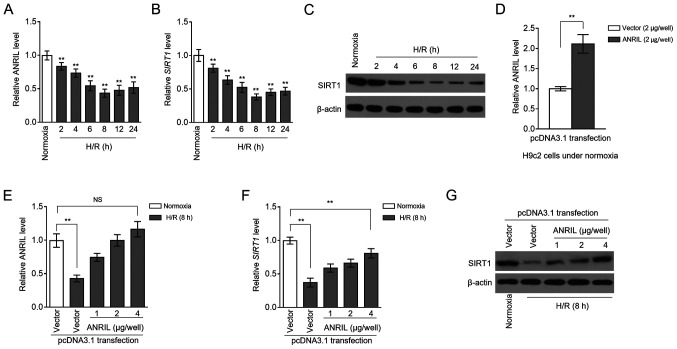
MI/R injury is an insurmountable problem that arises during reperfusion therapy for patients with MI (2). Despite the discovery of the association between ANRIL polymorphisms and the risk of MI (9,10), the expression pattern of ANRIL after MI/R injury is still unknown. To explore this issue, MI/R injury was mimicked using a H/R model with H9c2 cardiomyocytes cultured in vitro (13). RT-qPCR analysis showed that ANRIL transcription was decreased in H9c2 cells after H/R treatment, compared with the normoxia control (Fig. 1A). This result suggested that ANRIL may be functionally involved in H/R-induced cardiomyocyte pathology. Similarly, the transcription of SIRT1 was also downregulated in H/R-treated H9c2 cells (Fig. 1B). This was consistent with the immunoblotting analysis, which revealed that SIRT1 protein expression also decreased following H/R treatment (Fig. 1C). In order to determine whether ANRIL was associated with SIRT1 downregulation in H/R-treated H9c2 cells, ANRIL was overexpressed in H9c2 cells by transfecting pcDNA3.1-ANRIL plasmids. In fact, the decreased ANRIL level in H/R-treated H9c2 cells was completely recovered after transfection compared with the control (Fig. 1D and E). Moreover, consistent with restoring ANRIL expression, the downregulation of SIRT1 expression was significantly reversed at both the mRNA (Fig. 1F) and protein (Fig. 1G) expression levels following ANRIL overexpression. Together, these results suggested that a reduction in the expression of ANRIL contributed to the downregulation of SIRT1 expression in H9c2 cardiomyocytes following H/R injury.
Figure 1.
ANRIL upregulates SIRT1 expression following H/R injury. (A-C) H9c2 cardiomyocytes were cultured under a normoxic condition (normoxia) or exposed to hypoxia for 4 h followed by reoxygenation (H/R) for 2 to 24 h, as indicated. (A) ANRIL and (B) SIRT1 mRNA expression levels were determined by reverse transcription-quantitative PCR analysis. β-actin served as an internal control. Results are expressed as relative to the normoxia group. (C) SIRT1 protein expression was detected by an immunoblotting assay. β-actin served as a loading control. Results are representative of three independent experiments. (D-G) H9c2 cardiomyocytes were transiently transfected with pcDNA3.1-vector or pcDNA3.1-ANRIL plasmids (1, 2 or 4 µg per well). At 2 days after transfection, cells were cultured under normoxia or exposed to H/R (4 h hypoxia followed by 8 h reoxygenation) as indicated. The expression of (D) ANRIL following transfection was detection. (E) ANRIL and (F) SIRT1 mRNA expression levels, and (G) SIRT1 protein expression were determined. Data are presented as the mean ± SD. n=3. **P<0.01 vs. control group. NS, not significant; ANRIL, antisense non-coding RNA in the INK4 locus; SIRT1, sirtuin 1; H/R, hypoxia/reoxygenation.
ANRIL upregulates SIRT1 expression via sponging miR-181a
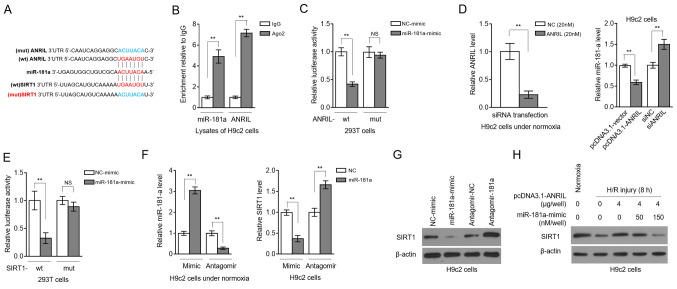
LncRNAs can serve as competitive endogenous RNAs to sponge miRNAs and, thus, regulate the expression of downstream targets (14). To understand how ANRIL regulates SIRT1 expression, the computational tools starBase (v2.0) (15) and TargetScan (http://www.targetscan.org/vert_72/) were used to predict candidate miRNAs. miR-181a was found to share common complementary binding sites with the 3′-UTR of both ANRIL and SIRT1 (Fig. 2A). Besides, RIP showed that ANRIL and miR-181a were simultaneously enriched in beads conjugated to Ago2 antibodies in contrast to IgG control antibodies (Fig. 2B). Luciferase reporter assays revealed that, compared with the NC mimics, the miR-181a mimics transfection significantly reduced the luciferase activity of the wt, but not from the mut ANRIL construct (Fig. 2C). The efficiency of siRNA targeting ANRIL was confirmed in H9c2 cells under normoxia (Fig. 2D, left panel). In addition, ANRIL overexpression in H9c2 cells resulted in decreased miR-181a expression, and inversely, ANRIL knockdown via siRNA transfection elevated miR-181a expression (Fig. 2D, right panel). Therefore, these results revealed that ANRIL is capable of sponging miR-181a.
Figure 2.
ANRIL upregulates SIRT1 expression by sponging miR-181a. (A) Putative binding sites (red) between miR-181a and the 3′-UTR of ANRIL and SIRT1. Mutant sequences (blue) of ANRIL were depicted at the top. (B) RNA-binding protein immunoprecipitation assay was conducted to detect the enrichment of miR-181a and ANRIL by using Ago2 antibody to precipitate the lysates of H9c2 cells. Isotype IgG antibody was used as a control. Results are expressed as relative to IgG control (n=3). (C) 293T cells were co-transfected with NC-mimic or miR-181a mimic along with wt or mut ANRIL luciferase reporter plasmids. Luciferase activity was measured at 2 days after transfection. Results are expressed as relative to NC transfection (n=5). (D) H9c2 cells were transfected with pcDNA3.1-vector or pcDNA3.1-ANRIL plasmids, or si-NC or siANRIL as indicated. ANRIL (left) and miR-181a (right) expression levels were determined at 2 days after transfection. (E) 293T cells were co-transfected with NC-mimic or miR-181a mimic along with wt or mut SIRT1 luciferase reporter plasmids. Luciferase activity was measured at 2 days after transfection. Results are expressed as relative to NC transfection (n=5). (F and G) H9c2 cells were transfected with NC-mimic or miR-181a mimic, or antagomir-NC or antagomir-181a. At 2 days after transfection, the (F) mRNA expression levels of miR-181a and SIRT1, and the (G) protein expression levels of SIRT1 were determined (n=3). (H) H9c2 cells were transfected with pcDNA3.1-vector or pcDNA3.1-ANRIL plasmids (4 µg per well) along with or without 50 or 150 nM miR-181a mimic. At 2 days after transfection, cells were cultured under normoxia or exposed to H/R (4 h hypoxia followed by 8 h reoxygenation). SIRT1 protein expression was determined by an immunoblotting assay (n=3). Data are presented as the mean ± SD. **P<0.01. NS, not significant; ANRIL, antisense non-coding RNA in the INK4 locus; SIRT1, sirtuin 1; miR, microRNA; UTR, untranslated region; NC, negative control; wt, wild-type; mut, mutant; si/siRNA, small interfering RNA; H/R, hypoxia/reoxygenation.
Luciferase reporter assays also showed that miR-181a targeted the 3′-UTR of SIRT1 (Fig. 2E), indicating that SIRT1 is a target for miR-181 in H9c2 cells. Then, miR-181a was overexpressed or inhibited in H9c2 cells by transfecting mimics or antagomir, respectively, and transfection efficiency was confirmed by RT-qPCR analysis (Fig. 2F, left). The transcription level of SIRT1 was decreased by miR-181a overexpression, and, conversely, SIRT1 transcription was elevated upon miR-181a inhibition (Fig. 2F, right). Similar results were obtained at the SIRT1 protein level (Fig. 2G). These results reinforced the evidence that miR-181a targets and suppresses SIRT1 expression, at least in H9c2 cells. Remarkably, the ANRIL-attenuated decline of SIRT1 expression in H/R-treated H9c2 cells was completely reversed in the presence of miR-181a overexpression (Fig. 2H). Thus, these data showed that miR-181a was an intermediary that controlled ANRIL positive regulation over SIRT1 after H/R injury, as observed in Fig. 1D-F.
ANRIL exhibits protective effects against H/R injury
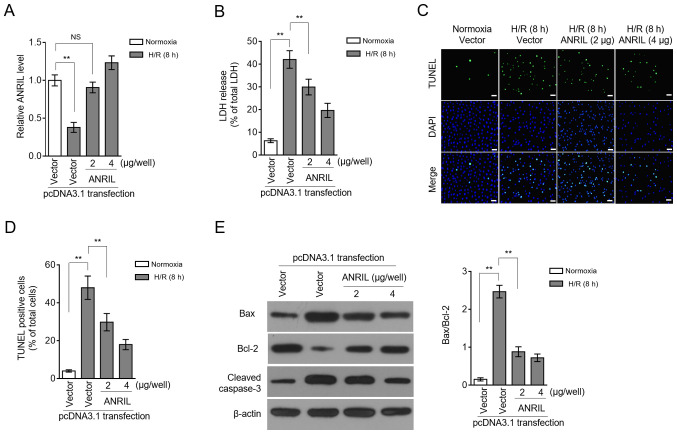
LDH release and apoptosis are two commonly used indicators for H/R injury (16,17). Restoration of the ANRIL transcription level by enforced overexpression (Fig. 3A) caused a large decrease in the LDH release induced by H/R injury (Fig. 3B), suggesting that ANRIL may have the ability to reduce H/R injury. This was reinforced by the observation that ANRIL overexpression decreased the apoptotic cell number, which had been increased in H9c2 cells upon H/R injury (Fig. 3C and D). The ANRIL-inhibited apoptosis of H9c2 cells was evidenced by the reduced expression of Bax and cleaved caspase-3, and simultaneously, by the elevated expression of Bcl-2 (Fig. 3E). These data showed that ANRIL reduced H/R-induced LDH release and apoptosis in H9c2 cells and, therefore, that ANRIL had protective effects against H/R injury.
Figure 3.
ANRIL protects against H/R injury. (A-E) H9c2 cells were transiently transfected with pcDNA3.1-vector or pcDNA3.1-ANRIL plasmids (2 or 4 µg per well). At 2 days after transfection, cells were cultured under normoxia or exposed to H/R (4 h hypoxia followed by 8 h reoxygenation). (A) ANRIL expression was determined via reverse transcription-quantitative PCR analysis. β-actin served as an internal control. Results are expressed as relative to normoxia group. (B) The release of LDH from cells was measured and expressed as a percentage of the total LDH activity (n=5). (C and D) Apoptosis was detected using a TUNEL assay and defined as a percentage of TUNEL positive cells (green) of the total number of cells (DAPI, blue) (n=12). Scale bar, 100 µm. (E) The expression levels of Bax, Bcl-2 and cleaved caspase-3 were determined using an immunoblotting assay (n=3). Data are presented as the mean ± SD. **P<0.01. NS, not significant; LDH, lactate dehydrogenase; ANRIL, antisense non-coding RNA in the INK4 locus; H/R, hypoxia/reoxygenation.
ANRIL protects against H/R injury by regulating the miR-181a/SIRT1 axis
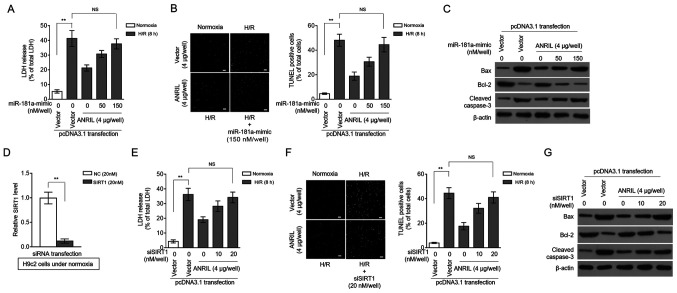
SIRT1 was previously reported to exhibit protective activities against MI/R injury in various pathological scenarios (18–21). Based on the SIRT1 regulation via miR-181a and ANRIL revealed by the present study, it was investigated whether miR-181a and SIRT1 could contribute to the protective effects of ANRIL against H/R injury. Indeed, in H9c2 cells induced with H/R injury, miR-181a overexpression significantly reversed the ANRIL-induced decrease in LDH release (Fig. 4A), as well as the ANRIL inhibitory effects on the apoptotic cell number (Fig. 4B) and on the expression of pro-apoptotic markers (Fig. 4C). Moreover, SIRT1 knockdown via siRNA (Fig. 4D) also reversed the ANRIL alleviating effects on H/R-induced LDH release (Fig. 4E) and cell apoptosis (Fig. 4F and G) in H9c2 cells. These results indicated that miR-181a could antagonize the protective effects of ANRIL against H/R injury, which was associated with decreased SIRT1 expression. Along with the aforementioned observation that ANRIL upregulated SIRT1 expression via sponging miR-181a, these data established that ANRIL exerted its protective effects against H/R injury by regulating the miR-181a/SIRT1 axis.
Figure 4.
ANRIL protects against H/R injury via the miR-181a/SIRT1 axis. (A-C) H9c2 cells were transiently transfected with pcDNA3.1-vector or pcDNA3.1-ANRIL plasmids (4 µg per well) along with or without 50 or 150 nM miR-181a mimic. At 2 days after transfection, cells were cultured under normoxia or exposed to H/R (4 h hypoxia followed by 8 h reoxygenation). (A) The release of LDH from cells was measured and expressed as a percentage of the total LDH activity (n=5). (B) Apoptosis was detected using a TUNEL assay and defined as a percentage of TUNEL positive cells (green) of the total number of cells (DAPI, blue) (n=12). Scale bar, 100 µm. (C) The expression levels of Bax, Bcl-2 and cleaved caspase-3 were analyzed. (D) The efficiency of siRNA targeting SIRT1 was confirmed in H9c2 cells under normoxia. (E-G) H9c2 cells were transiently transfected with pcDNA3.1-vector or pcDNA3.1-ANRIL plasmids (4 µg per well) along with or without 10 or 20 nM siSIRT1. At 2 days after transfection, cells were cultured under normoxia or exposed to H/R (4 h hypoxia followed by 8 h reoxygenation). (E) LDH release, (F) cell apoptosis (scale bar, 100 µm) and (G) expression of Bax, Bcl-2 and cleaved caspase-3 were analyzed as described in A-C. Data are presented as the mean ± SD. **P<0.01. NS, not significant; ANRIL, antisense non-coding RNA in the INK4 locus; H/R, hypoxia/reoxygenation; SIRT1, sirtuin 1; miR, microRNA; LDH, lactate dehydrogenase; si/siRNA, small interfering RNA; NC, negative control.
Discussion
In recent years, the association between lncRNAs and MI/R injury has been increasingly recognized, which offers novel perspectives in the discovery of potential therapeutic targets to reduce MI/R injury (22). In the present study, it was found that the expression levels of the lncRNA ANRIL and SIRT1 were synchronously decreased in H9c2 cardiomyocytes after H/R-induced injury in vitro. Besides, it was demonstrated that ANRIL upregulated SIRT1 expression by sponging miR-181a. These data, from a mechanistic point of view, may explain how ANRIL regulates SIRT1 expression in this in vitro experimental system used to mimic MI/R injury. Furthermore, the present study linked the protective effects of ANRIL to miR-181a/SIRT1 regulation, providing a novel mechanistic insight into the protective role of ANRIL observed following H/R injury. As a whole, this study may have identified ANRIL as being a novel regulator in MI/R injury pathogenesis and also highlighted its potential to be explored as a therapeutic target for reducing MI/R injury.
The present study observed that ANRIL was downregulated in H9c2 cells after H/R-induced injury, which was in line with a previous clinical study that reported lower ANRIL expression in 414 patients with MI compared with healthy volunteers (8). However, the exact underlying mechanism of how ANRIL expression is regulated after H/R injury is still unclear and needs to be elucidated by further investigation. Of note, in the present study a decrease in the expression of SIRT was observed to be in sync with the downregulation of ANRIL. Thus, it was concluded that the decrease in SIRT1 expression following H/R injury could be attributed, at least partially, to ANRIL, since the recovery of ANRIL normal expression level restored its expression. Consistently, the repressed SIRT1 expression was also found in the mouse myocardium following I/R injury and cardiomyocytes exposed to H/R in vitro (23). In addition, it was previously found that SUV39H1 binding to the SIRT1 promoter suppresses SIRT1 transcription (24). Therefore, the present findings uncovered a novel regulatory mechanism by which SIRT1 was modulated in response to H/R injury.
ANRIL can regulate gene expression through a direct epigenetic mechanism or by recruiting the polycomb repression complex 2 (25,26). Besides, ANRIL can also act as a miRNA sponge to execute its biological activities, such as miR-186 (27) and let-7 (28). It was demonstrated in the present study that ANRIL upregulated SIRT1 by sponging miR-181a, thus identifying another ANRIL target. Remarkably, two recent investigations reinforce the present data by also documenting that ANRIL inhibits cell senescence of vascular smooth muscle cells (29) and promotes lymphangiogenesis (30) by regulating miR-181a. In conjunction with these previous studies, the present results suggested that the negative regulation of miR-181a by ANRIL may be a critical mechanism by which ANRIL exerts its versatile activities.
ANRIL knockdown has been demonstrated to aggravate H2O2-induced cell injury (31). On the other hand, ANRIL upregulation protects lens epithelial cells against H2O2-induced cell injury (32). Moreover, in PC-12 cells, ANRIL reduces oxygen and glucose deprivation-induced injury (33), indicating a protective role of ANRIL implicated in the pathogenesis of cellular oxidative and ischemic damage. In line with these studies, the present study found that ANRIL protected against H/R injury in H9c2 cells. This function of ANRIL may be associated with its modulation of the miR-181a/SIRT1 axis since both miR-181a and SIRT1 knockdown diminished the protective effects of ANRIL. These data highlighted the important role of SIRT1 in mediating ANRIL protection against H/R injury. To date, it is known that SIRT1 exhibits cardioprotective effects on H/R injury by comprehensive means, including inhibition of oxidative and endoplasmic reticulum stress (34,35), and orchestration of autophagy induction (36). Examining the specific contribution of these underlying mechanisms to the effects of ANRIL is one of the future research topics that deserves further investigation.
Finally, a major limitation of the present study is that all the observations were obtained only from H9c2 cells. The differences in physiological aspects between mice and humans combined with the complexity of cell involvement during MI/R injury, require future studies to explore ANRIL activities involved in MI/R injury using other physiologically relevant approaches and materials in order to evaluate effects within a more complex system than just in vitro cultured cardiomyocytes, such as other appropriate types of cardiac cells and tissues and animal models.
In conclusion, the present study established a key role of the downstream regulation of the miR-181a/SIRT1 signaling axis in mediating ANRIL protection against H/R injury in cardiomyocytes, suggesting that targeting this axis may have a potential therapeutic benefit for patients with MI/R injury.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
BS conceived and designed the study. BS and DW prepared the manuscript, ensured its legitimacy and revised the final draft of the manuscript. BS, DW, GY, XS, SW, SJ, JZ, LL and XW performed the experiments and analyzed the data. BS and DW confirm the authenticity of all the raw data. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Anderson JL, Morrow DA. Acute myocardial infarction. N Engl J Med. 2017;376:2053–2064. doi: 10.1056/NEJMra1606915. [DOI] [PubMed] [Google Scholar]
- 2.Hausenloy DJ, Yellon DM. Myocardial ischemia-reperfusion injury: A neglected therapeutic target. J Clin Invest. 2013;123:92–100. doi: 10.1172/JCI62874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yellon DM, Hausenloy DJ. Myocardial reperfusion injury. N Engl J Med. 2007;357:1121–1135. doi: 10.1056/NEJMra071667. [DOI] [PubMed] [Google Scholar]
- 4.Guo Y, Luo F, Liu Q, Xu D. Regulatory non-coding RNAs in acute myocardial infarction. J Cell Mol Med. 2017;21:1013–1023. doi: 10.1111/jcmm.13032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ong SB, Katwadi K, Kwek XY, Ismail NI, Chinda K, Ong SG, Hausenloy DJ. Non-coding RNAs as therapeutic targets for preventing myocardial ischemia-reperfusion injury. Expert Opin Ther Targets. 2018;22:247–261. doi: 10.1080/14728222.2018.1439015. [DOI] [PubMed] [Google Scholar]
- 6.Long Y, Wang X, Youmans DT, Cech TR. How do lncRNAs regulate transcription? Sci Adv. 2017;3:eaao2110. doi: 10.1126/sciadv.aao2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Congrains A, Kamide K, Ohishi M, Rakugi H. ANRIL: Molecular mechanisms and implications in human health. Int J Mol Sci. 2013;14:1278–1292. doi: 10.3390/ijms14011278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vausort M, Wagner DR, Devaux Y. Long noncoding RNAs in patients with acute myocardial infarction. Circ Res. 2014;115:668–677. doi: 10.1161/CIRCRESAHA.115.303836. [DOI] [PubMed] [Google Scholar]
- 9.Ahmed W, Ali IS, Riaz M, Younas A, Sadeque A, Niazi AK, Niazi SH, Ali SH, Azam M, Qamar R. Association of ANRIL polymorphism (rs1333049:C>G) with myocardial infarction and its pharmacogenomic role in hypercholesterolemia. Gene. 2013;515:416–420. doi: 10.1016/j.gene.2012.12.044. [DOI] [PubMed] [Google Scholar]
- 10.Cheng J, Cai MY, Chen YN, Li ZC, Tang SS, Yang XL, Chen C, Liu X, Xiong XD. Variants in ANRIL gene correlated with its expression contribute to myocardial infarction risk. Oncotarget. 2017;8:12607–12619. doi: 10.18632/oncotarget.14721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li X, Dai Y, Yan S, Shi Y, Han B, Li J, Cha L, Mu J. Down-regulation of lncRNA KCNQ1OT1 protects against myocardial ischemia/reperfusion injury following acute myocardial infarction. Biochem Biophys Res Commun. 2017;491:1026–1033. doi: 10.1016/j.bbrc.2017.08.005. [DOI] [PubMed] [Google Scholar]
- 12.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 13.Zhang YQ, Tang Y, Wu AL, Zhu HB. Salvianolic acid A displays cardioprotective effects in in vitro models of heart hypoxia/reoxygenation injury. J Asian Nat Prod Res. 2010;12:899–915. doi: 10.1080/10286020.2010.510115. [DOI] [PubMed] [Google Scholar]
- 14.Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505:344–352. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li JH, Liu S, Zhou H, Qu LH, Yang JH. StarBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42((Database Issue)):D92–D97. doi: 10.1093/nar/gkt1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He Y, Li C, Ma Q, Chen S. Esculetin inhibits oxidative stress and apoptosis in H9c2 cardiomyocytes following hypoxia/reoxygenation injury. Biochem Biophys Res Commun. 2018;501:139–144. doi: 10.1016/j.bbrc.2018.04.195. [DOI] [PubMed] [Google Scholar]
- 17.Wang X, Ha T, Hu Y, Lu C, Liu L, Zhang X, Kao R, Kalbfleisch J, Williams D, Li C. MicroRNA-214 protects against hypoxia/reoxygenation induced cell damage and myocardial ischemia/reperfusion injury via suppression of PTEN and Bim1 expression. Oncotarget. 2016;7:86926–86936. doi: 10.18632/oncotarget.13494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Becatti M, Taddei N, Cecchi C, Nassi N, Nassi PA, Fiorillo C. SIRT1 modulates MAPK pathways in ischemic-reperfused cardiomyocytes. Cell Mol Life Sci. 2012;69:2245–2260. doi: 10.1007/s00018-012-0925-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang Y, Duan W, Lin Y, Yi W, Liang Z, Yan J, Wang N, Deng C, Zhang S, Li Y, et al. SIRT1 activation by curcumin pretreatment attenuates mitochondrial oxidative damage induced by myocardial ischemia reperfusion injury. Free Radic Biol Med. 2013;65:667–679. doi: 10.1016/j.freeradbiomed.2013.07.007. [DOI] [PubMed] [Google Scholar]
- 20.Shalwala M, Zhu SG, Das A, Salloum FN, Xi L, Kukreja RC. Sirtuin 1 (SIRT1) activation mediates sildenafil induced delayed cardioprotection against ischemia-reperfusion injury in mice. PLoS One. 2014;9:e86977. doi: 10.1371/journal.pone.0086977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ding M, Lei J, Han H, Li W, Qu Y, Fu E, Fu F, Wang X. SIRT1 protects against myocardial ischemia-reperfusion injury via activating eNOS in diabetic rats. Cardiovasc Diabetol. 2015;14:143. doi: 10.1186/s12933-015-0299-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu SY, Tang L, Zhou SH. Long noncoding RNAs: New players in ischaemia-reperfusion injury. Heart Lung Circ. 2018;27:322–332. doi: 10.1016/j.hlc.2017.09.011. [DOI] [PubMed] [Google Scholar]
- 23.Guo Y, Zhang L, Li F, Hu CP, Zhang Z. Restoration of sirt1 function by pterostilbene attenuates hypoxia-reoxygenation injury in cardiomyocytes. Eur J Pharmacol. 2016;776:26–33. doi: 10.1016/j.ejphar.2016.02.052. [DOI] [PubMed] [Google Scholar]
- 24.Yang G, Zhang X, Weng X, Liang P, Dai X, Zeng S, Xu H, Huan H, Fang M, Li Y, et al. SUV39H1 mediated SIRT1 trans-repression contributes to cardiac ischemia-reperfusion injury. Basic Res Cardiol. 2017;112:22. doi: 10.1007/s00395-017-0608-3. [DOI] [PubMed] [Google Scholar]
- 25.Yap KL, Li S, Muñoz-Cabello AM, Raguz S, Zeng L, Mujtaba S, Gil J, Walsh MJ, Zhou MM. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol Cell. 2010;38:662–674. doi: 10.1016/j.molcel.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kotake Y, Nakagawa T, Kitagawa K, Suzuki S, Liu N, Kitagawa M, Xiong Y. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene. Oncogene. 2011;30:1956–1962. doi: 10.1038/onc.2010.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang JJ, Wang DD, Du CX, Wang Y. Long noncoding RNA ANRIL promotes cervical cancer development by acting as a sponge of miR-186. Oncol Res. 2018;26:345–352. doi: 10.3727/096504017X14953948675449. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 28.Wang Y, Cheng N, Luo J. Downregulation of lncRNA ANRIL represses tumorigenicity and enhances cisplatin-induced cytotoxicity via regulating microRNA let-7a in nasopharyngeal carcinoma. J Biochem Mol Toxicol. 2017:31. doi: 10.1002/jbt.21904. doi: 10.1002/jbt.21904. [DOI] [PubMed] [Google Scholar]
- 29.Tan P, Guo YH, Zhan JK, Long LM, Xu ML, Ye L, Ma XY, Cui XJ, Wang HQ. LncRNA-ANRIL inhibits cell senescence of vascular smooth muscle cells by regulating miR-181a/Sirt1. Biochem Cell Biol. 2019;97:571–580. doi: 10.1139/bcb-2018-0126. [DOI] [PubMed] [Google Scholar]
- 30.He ZY, Wei TH, Zhang PH, Zhou J, Huang XY. Long noncoding RNA-antisense noncoding RNA in the INK4 locus accelerates wound healing in diabetes by promoting lymphangiogenesis via regulating miR-181a/Prox1 axis. J Cell Physiol. 2019;234:4627–4640. doi: 10.1002/jcp.27260. [DOI] [PubMed] [Google Scholar]
- 31.Li R, Yin F, Guo YY, Zhao KC, Ruan Q, Qi YM. Knockdown of ANRIL aggravates H2O2-induced injury in PC-12 cells by targeting microRNA-125a. Biomed Pharmacother. 2017;92:952–961. doi: 10.1016/j.biopha.2017.05.122. [DOI] [PubMed] [Google Scholar]
- 32.Qi D, Wang M, Zhang D, Li H. Tanshinone IIA protects lens epithelial cells from H2O2-induced injury by upregulation of lncRNA ANRIL. J Cell Physiol. 2019 Jan 30; doi: 10.1002/jcp.28189. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 33.Liu B, Cao W, Xue J. LncRNA ANRIL protects against oxygen and glucose deprivation (OGD)-induced injury in PC-12 cells: Potential role in ischaemic stroke. Artif Cells Nanomed Biotechnol. 2019;47:1384–1395. doi: 10.1080/21691401.2019.1596944. [DOI] [PubMed] [Google Scholar]
- 34.Li YP, Wang SL, Liu B, Tang L, Kuang RR, Wang XB, Zhao C, Song XD, Cao XM, Wu X, et al. Sulforaphane prevents rat cardiomyocytes from hypoxia/reoxygenation injury in vitro via activating SIRT1 and subsequently inhibiting ER stress. Acta Pharmacol Sin. 2016;37:344–353. doi: 10.1038/aps.2015.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang H, Wang C, Zhang L, Lv J, Ni H. Rutin alleviates hypoxia/reoxygenation-induced injury in myocardial cells by up-regulating SIRT1 expression. Chem Biol Interact. 2019;297:44–49. doi: 10.1016/j.cbi.2018.10.016. [DOI] [PubMed] [Google Scholar]
- 36.Qiu R, Li W, Liu Y. MicroRNA-204 protects H9C2 cells against hypoxia/reoxygenation-induced injury through regulating SIRT1-mediated autophagy. Biomed Pharmacother. 2018;100:15–19. doi: 10.1016/j.biopha.2018.01.165. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.