Key Points
Question
Are there associations between candidate noncardiovascular diseases and statin treatment using genetic variants in the 3-hydroxy-3-methylglutaryl coenzyme A reductase (HMGCR) gene that decrease low-density lipoprotein cholesterol levels as an instrumental variable?
Findings
In a mendelian randomization approach cohort study including 53 385 individuals, using variants in the HMGCR gene affecting low-density lipoprotein cholesterol levels replicated the association between statin use and increased type 2 diabetes risk. However, no strong associations between 3-hydroxy-3-methylglutaryl coenzyme A reductase inhibition by statins and other diseases were noted.
Meaning
In this cohort study, statin treatment was associated with an increased risk of type 2 diabetes, but there appeared to be no strong indication of other pleiotropic outcomes of the low-density lipoprotein cholesterol level–decreasing outcomes with this drug class on the clinical phenotypes studied.
Abstract
Importance
Observational studies suggest that statins, which inhibit 3-hydroxy-3-methylglutaryl coenzyme A (HMG-CoA) reductase, may be associated with beneficial effects in many noncardiovascular diseases.
Objective
To construct a weighted HMG-CoA reductase (HMGCR) gene genetic risk score (GRS) using variants in the HMGCR gene affecting low-density lipoprotein cholesterol as an instrumental variable for mendelian randomization analyses to test associations with candidate noncardiovascular phenotypes previously associated with statin use in observational studies.
Design, Setting, and Participants
This cohort study included 53 385 unrelated adults of European ancestry with genome-wide genotypes available from BioVU (a practice-based biobank, used for discovery) and 30 444 unrelated adults with European ancestry available in the Electronic Medical Records and Genomics (eMERGE; a research consortium that conducts genetic research using electronic medical records, used for replication). The study was conducted from February 6, 2015, through April 31, 2019; data analysis was performed from August 26, 2019, through December 22, 2020.
Interventions
An HMGCR GRS was calculated.
Main Outcomes and Measures
The association between the HMGCR GRS and the presence or absence of 22 noncardiovascular phenotypes previously associated with statin use in clinical studies.
Results
Of the 53 385 individuals in BioVU, 29 958 (56.1%) were women; mean (SD) age was 59.9 (15.6) years. The finding between the HMGCR GRS and the noncardiovascular phenotypes of interest in this cohort was significant only for type 2 diabetes. An HMGCR GRS equivalent to a 10-mg/dL decrease in the low-density lipoprotein cholesterol level was associated with an increased risk of type 2 diabetes (odds ratio [OR], 1.09; 95% CI, 1.04-1.15; P = 5.58 × 10−4). The HMGCR GRS was not associated with other phenotypes; the closest were increased risk of Parkinson disease (OR, 1.30; 95% CI, 1.07-1.58; P = .007) and kidney failure (OR, 1.18; 95% CI, 1.05-1.34; P = .008). Of the 30 444 individuals in eMERGE, 16 736 (55.0%) were women; mean (SD) age was 68.7 (15.4) years. The association between the HMGCR GRS and type 2 diabetes was replicated in this cohort (OR, 1.09; 95% CI, 1.01-1.17; P = .02); however, the HMGCR GRS was not associated with Parkinson disease (OR, 0.93; 95% CI, 0.75-1.16; P = .53) and kidney failure (OR, 1.18; 95% CI, 0.98-1.41; P = .08) in the eMERGE cohort.
Conclusions and Relevance
A mendelian randomization approach using variants in the HMGCR gene replicated the association between statin use and increased type 2 diabetes risk but provided no strong evidence for pleiotropic effects of statin-induced decrease of the low-density lipoprotein cholesterol level on other diseases.
This cohort study uses a mendelian randomization approach with variants in the HMGCR gene to evaluate associations between statin therapy and noncardiovascular diseases.
Introduction
Statins (HMG coenzyme A [HMG-CoA] reductase inhibitors) are first-line medications for lowering circulating low-density lipoprotein cholesterol (LDL-C) levels, and they reduce the risk of cardiovascular disease (CVD) substantially.1 However, statins have also been reported to have many beneficial and deleterious associations with the risk of non-CVD conditions, suggesting that they have multiple effects of therapeutic importance. More than a quarter of US adults aged 40 years or older use statin therapy to prevent CVD.2 Given this widespread use, it is important to understand the spectrum of both deleterious and beneficial effects of long-term statin treatment in diseases and conditions beyond the cardiovascular system.
In addition to their effects on CVD, statins may have associations with other diseases (ie, pleiotropy) with both increases (eg, type 2 diabetes, myopathy, and rhabdomyolysis) and decreases (eg, cancers and infection) reported in the risk of many diseases.3 However, most studies reporting statin associations with diseases other than CVD are inconclusive. A comprehensive review of meta-analyses of 112 observational studies and 144 randomized clinical trials evaluated the strength of evidence for 278 noncardiovascular outcomes reported to be associated with statins.3 The authors found none of these statin-disease associations to be conclusive; however, 23 associations were suggestive or highly suggestive, requiring additional study.
Studies to evaluate statins and the risk of non-CVD are challenging. Placebo-controlled, randomized clinical trials of statins in CVD are generally of short duration and thus often unable to address the effects of long-term statin therapy on other diseases. Observational studies, on the other hand, are faced with a methodologic challenge—patients who receive statins are substantially different from those who do not—thus, any differences in outcomes observed between patients using and not using statin therapy could be due to residual confounding. Mendelian randomization (MR) is an approach that might provide less-biased insights.
An MR approach to study statin therapy makes use of the fact that there are naturally occurring variants in the gene HMGCR (OMIM 142910) that encodes the enzyme, HMG-CoA reductase, that statins target. Because those genetic variants are inherited randomly and remain unchanged, an MR approach may be less-vulnerable to reverse causation and residual confounding and might provide evidence consistent with a causal effect between statin treatment and candidate outcomes. This approach has shown an association between statin treatment and an increased risk of type 2 diabetes.4
The availability of deidentified genetic and clinical information in large biobanks provides the opportunity to use an MR approach to define the outcomes of modulating HMG-CoA activity associated with the risk of many diseases. Herein, we apply an MR approach using genetic variants in HMGCR in 2 large, practice-based biobanks: the biobank at Vanderbilt University (BioVU) and the Electronic Medical Records and Genomics (eMERGE) network. We constructed a weighted genetic risk score (GRS) for HMGCR using variants that affect LDL-C levels and tested the association of the GRS with 22 candidate outcomes previously reported to be associated with statins.
Methods
The study was conducted from February 6, 2015, through April 31, 2019; data analysis was performed from August 26, 2019, through December 22, 2020. The study was approved by Vanderbilt University Medical Center institutional review board, which waived the need for informed consent owing to the use of deidentified data. This study followed the Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) reporting guideline.
Data were obtained from BioVU, a deidentified DNA biobank, linked to the deidentified electronic health record (EHR) for patients at Vanderbilt University Medical Center. BioVU has been described in detail previously.5,6,7 Information available in BioVU includes diagnostic and procedure codes, demographic characteristics, clinical notes, problem lists, laboratory test values, and medications.8,9,10 We included adults aged 18 years and older of European ancestry with genome-wide genotypes available.
The eMERGE network is a consortium that uses DNA biorepositories with EHR systems for large-scale, high-throughput genetic research and has been described previously.11,12,13 We replicated BioVU findings in eMERGE data on adults of European ancestry aged 18 years and older after removing individuals common to both cohorts.
Genetic Analysis
We selected candidate disease phenotypes that have previously been associated with statins in randomized clinical trials on the drugs or observational studies.3 In both BioVU and eMERGE cohorts, we established the presence or absence of 22 noncardiovascular phenotypes. We selected only phenotypes with more than 100 cases in BioVU to maintain statistical power and those that could be defined using PheCodes. For example, disease-associated mortality was not studied because defining such phenotypes in the EHR would require development and validation of a case-by-case protocol. We identified phenotypes using PheCodes, a phenotyping system based on clinical diagnosis codes (International Classification of Diseases, Ninth Revision, Clinical Modification [ICD-9-CM] and International Statistical Classification of Diseases, 10th Revision, Clinical Modification [ICD-10-CM]).14,15 A PheCode amalgamates related ICD codes mapping to a distinct disease or trait.14,15 A case was defined as an individual with 2 or more occurrences of the PheCode of interest in the EHR. Controls were individuals without that code. Individuals with 1 mention of the code or with related codes were excluded from the analysis to limit misclassification.
To evaluate possible associations between statin treatment and noncardiovascular effects (ie, pleiotropy), we used a previously reported genetic instrument for HMGCR.16 Specifically, we constructed a weighted GRS of HMGCR using 6 genetic variants within the HMGCR region that have been independently associated with LDL-C levels (eTable 1 in the Supplement). This HMGCR GRS is an instrumental variable not for statin use (ie, likelihood of receiving a statin) but rather for the probability of a statin effect (ie, lower LDL-C level). In other words, individuals with an HMGCR GRS that estimated the probability of lower enzyme activity would estimate the probability of the effects of statin therapy in a population. We also constructed an HMGCR GRS in the eMERGE cohort, using 5 of 6 single-nucleotide variants (rs2006760 was not successfully imputed in the eMERGE cohort and was therefore excluded from analysis). We calculated the HMGCR GRS by summing LDL-C increasing alleles, weighted by the effect size, but to reflect the same directional effects as statin therapy, we adjusted the HMGCR GRS to reflect a standard decrement of 10 mg/dL (to convert to millimoles per liter, multiply by 0.0259) so that a high GRS would be associated with lower LDL-C levels, as occurs with statin therapy.
Statistical Analysis
We conducted logistic regression in assessing the association between the HMGCR GRS and candidate phenotypes using R, version 3.6.0 (R Foundation for Statistical Computing). We included age at the most recent medical encounter, sex, and length of EHR (defined as the time between each patient’s first and most recent medical encounter) as covariates for all candidate phenotype analyses, except for cancer of the prostate (PheCode 185), analyzed only in men, and miscarriage (PheCode 634), analyzed only in women. In addition, 10 principal components were used for adjustment in BioVU and 5 principal components for eMERGE, as available. For each candidate phenotype, we estimated odds ratios (ORs) and 95% CIs per decrease of 10 mg/dL in LDL-C levels between the HMGCR GRS and the phenotype of interest. For the 22 prespecified phenotypes, P < .002 (05/22 phenotypes) was considered significant in the BioVU cohort. In the eMERGE replication cohort, a Bonferroni-adjusted value of P < .05 was considered significant. All statistical tests were 2-sided. We also conducted Pearson correlation coefficient testing using R, version 3.6.0 (R Foundation for Statistical Computing).
In sensitivity analyses, we adjusted for statin use, with individuals receiving statin therapy defined as patients with 1 or more exposure to a statin documented in the EHR. In addition, we stratified the HMGCR GRS into quartiles and compared individuals in lower LDL-C level quartiles (estimated by HMGCR GRS) with those in the highest estimated LDL-C level quartile. Genotype quality control and imputation are described in the eMethods in the Supplement.
Results
The BioVU Discovery Cohort
We calculated an HMGCR GRS for 53 385 unrelated adults of European ancestry in BioVU who had a mean (SD) age of 59.93 (15.61) years; a total of 29 958 women (56.11%) and 23 427 men (43.89%) were included. The mean (SD) length of available EHRs was 10.24 (7.01) years (Table 1).
Table 1. Cohort Demographic Characteristics.
| Characteristic | Patients, mean (SD) | |
|---|---|---|
| BioVU (n = 53 385) | eMERGE (n = 30 444) | |
| Sex, No. (%) | ||
| Women | 29 958 (56.11) | 16 736 (55.0) |
| Men | 23 427 (43.89) | 13 708 (45.0) |
| Age, y | 59.93 (15.61) | 68.65 (15.43) |
| EHR length, y | 10.24 (7.01) | 17.11 (9.89) |
| Statin therapy, No. (%) of patients | 25 625 (48.0) | 16 060 (52.8) |
Abbreviations: BioVU, biobank at Vanderbilt University; EHR, electronic health record; eMERGE, Electronic Medical Records and Genomics network.
As expected, the HMGCR GRS was associated with a diagnosis of hypercholesterolemia. For a 10-mg/dL HMGCR GRS–estimated decrease of the LDL-C level, the OR of having the PheCode of hypercholesterolemia was 0.91 (95% CI, 0.85-0.98; P = .009) (eTable 2 and the eFigure in the Supplement). In a subset of patients (n = 29 652) with both HMGCR GRS and measured LDL-C levels, the HMGCR GRS correlated with measured LDL-C levels (correlation coefficient, −0.03; P = 5.60 × 10−8).
The finding between the HMGCR GRS and the prespecified noncardiovascular phenotypes of interest was significant only for type 2 diabetes (Table 2) (OR, 1.10; 95% CI, 1.04-1.15; P = 5.58 × 10−4). The finding was also significant after further adjusting for statin use (OR, 1.13; 95% CI, 1.07-1.19; P = 8.50 × 10−6) (eTable 3 in the Supplement).
Table 2. Findings for HMGCR Genetic Risk Score and Candidate Noncardiovascular Phenotypes in the Biobank at Vanderbilt Universitya.
| PheCode | Description | Odds ratio (95% CI) | P value | No. (%) | ||
|---|---|---|---|---|---|---|
| Total | Cases | Controls | ||||
| 250.2 | Type 2 diabetes | 1.10 (1.04-1.15) | 5.58 × 10−4 | 47 757 | 9210 (19.3) | 38 547 (80.7) |
| 332 | Parkinson disease | 1.30 (1.07-1.58) | .007 | 41 193 | 553 (1.3) | 40 640 (98.7) |
| 585.32 | Kidney disease | 1.18 (1.05-1.34) | .008 | 39 811 | 1308 (3.3) | 38 503 (96.7) |
| 772.4 | Rhabdomyolysis | 1.38 (0.96-1.97) | .08 | 46 140 | 159 (0.3) | 45 981 (99.7) |
| 202.2 | Non-Hodgkin lymphoma | 1.21 (0.96-1.52) | .11 | 51 062 | 367 (0.7) | 50 695 (99.3) |
| 151 | Cancer of stomach | 0.86 (0.71-1.05) | .13 | 45 818 | 509 (1.1) | 45 309 (98.9) |
| 800.1 | Fracture of neck or femur | 1.11 (0.93-1.31) | .24 | 45 613 | 679 (1.5) | 44 934 (98.5) |
| 770 | Myalgia and myositis, unspecified | 1.05 (0.96-1.13) | .28 | 50 460 | 3202 (6.3) | 47 258 (93.7) |
| 185 | Cancer of prostate | 0.94 (0.84-1.06) | .31 | 18 262 | 1786 (9.8) | 16 476 (90.2) |
| 577.1 | Acute pancreatitis | 1.09 (0.91-1.31) | .33 | 51 731 | 596 (1.2) | 51 135 (98.8) |
| 38 | Septicemia | 1.04 (0.96-1.13) | .38 | 46 724 | 3049 (6.5) | 43 675 (93.5) |
| 634 | Miscarriage; stillbirth | 0.88 (0.65-1.18) | .39 | 27 815 | 234 (0.8) | 27 581 (99.2) |
| 8.52 | Intestinal infection due to Clostridioides difficile | 0.94 (0.79-1.10) | .43 | 51 807 | 672 (1.3) | 51 135 (98.7) |
| 585.1 | Acute kidney failure | 1.02 (0.96-1.09) | .46 | 44 949 | 6446 (14.3) | 38 503 (85.7) |
| 70.3 | Viral hepatitis C | 0.96 (0.85-1.10) | .58 | 44 776 | 1168 (2.6) | 43 608 (97.4) |
| 366.2 | Senile cataract | 1.02 (0.95-1.09) | .67 | 50 734 | 4469 (8.8) | 46 265 (91.2) |
| 290.1 | Dementias | 1.03 (0.89-1.20) | .68 | 42 515 | 890 (2.1) | 41 625 (97.9) |
| 153.2 | Colon cancer | 0.98 (0.88-1.10) | .77 | 43 091 | 1510 (3.5) | 41 581 (96.5) |
| 38.3 | Bacteremia | 0.99 (0.89-1.10) | .83 | 45 543 | 1868 (4.1) | 43 675 (95.9) |
| 155.1 | Malignant neoplasm of liver, primary | 1.02 (0.83-1.25) | .84 | 45 770 | 461 (1.0) | 45 309 (99.0) |
| 359.2 | Myopathy | 0.99 (0.86-1.13) | .86 | 47 572 | 1028 (2.2) | 46 544 (97.8) |
| 743.11 | Osteoporosis | 1.00 (0.92-1.08) | .97 | 45 863 | 3410 (7.4) | 42 453 (92.6) |
The analyses were adjusted for sex, age at most recent visit, electronic health record length, and 10 principal components for ancestry. Cancer of the prostate was analyzed only in men; miscarriage or stillbirth was analyzed only in women. The HMGCR GRS was standardized for a decrement of 10 mg/dL in the low-density lipoprotein cholesterol level (to convert to millimoles per liter, multiply by 0.0259).
The findings between the HMGCR GRS and increased risk of Parkinson disease (OR, 1.30; 95% CI, 1.07-1.58; P = .007) and kidney failure (OR, 1.18; 95% CI, 1.05-1.34; P = .008) (Table 2) did not meet the Bonferroni-adjusted level of statistical significance (P < .002). In the prespecified sensitivity analysis, after adjusting for statin use (eTable 3 in the Supplement), the findings for Parkinson disease (OR, 1.30; 95% CI, 1.07-1.58; P = .007) and kidney failure (OR, 1.21; 95% CI, 1.07-1.37; P = .003) did not change markedly. Because type 2 diabetes is a common and important risk factor for developing kidney disease, we conducted a post hoc analysis and further adjusted the finding with kidney failure for the presence or absence of type 2 diabetes; was not statistically significant (OR, 1.10; 95% CI, 0.96-1.27; P = .16) (eTable 4 in the Supplement).
No other prespecified phenotypes were associated with the HMGCR GRS (Table 2). The HMGCR GRS was not associated with an increased risk of rhabdomyolysis, which is a well-documented statin adverse effect (OR, 1.38; 95% CI, 0.96-1.97; P = .08) (Table 2). The finding was not significant after adjustment for statin use (OR, 1.39; 95% CI, 0.97-1.99; P = .07) (eTable 3 in the Supplement).
We also compared individuals in different HMGCR GRS quartiles (Figure). Compared with those in the HMGCR GRS-estimated highest LDL-C level quartile, individuals in the lowest estimated LDL-C level quartile were more likely to have type 2 diabetes (OR, 1.11; 95% CI, 1.04-1.19; P = .001) (Figure, A) and the findings for kidney failure (OR, 1.26; 95% CI, 1.08-1.47; P = .004) (Figure, B) and Parkinson disease (OR, 1.44; 95% CI, 1.13-1.84; P = .003) (Figure, C) were not significant for the P value threshold of .002.
Figure. Association Between HMGCR Genetic Risk Score (GRS) Quartiles and Candidate Phenotypes.
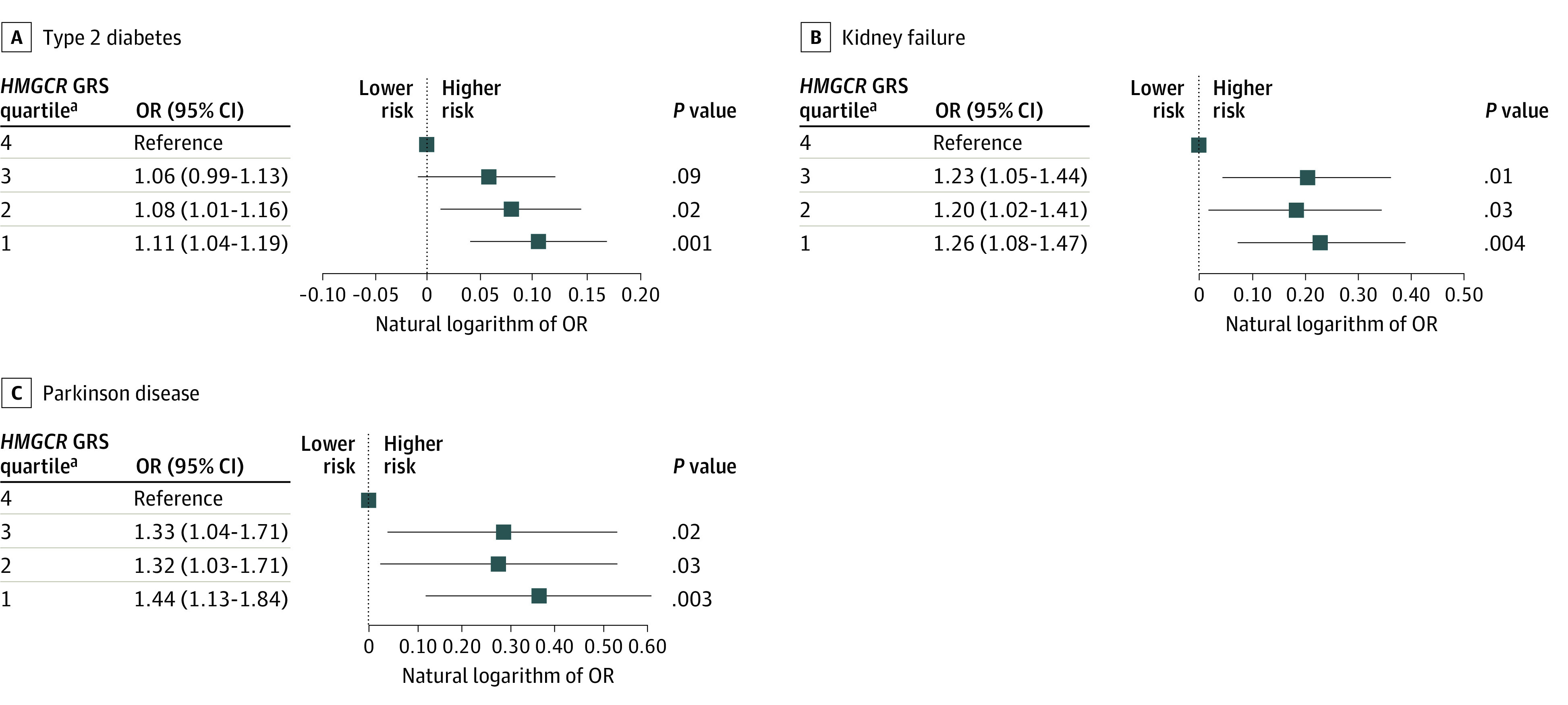
aIndividuals in the lowest estimated LDL-C level quartile were more likely to have diseases.
Replication in eMERGE Cohort
We calculated an HMGCR GRS for 30 444 unrelated adults of European ancestry in the eMERGE cohort (Table 1) and found that, for a 10-mg/dL HMGCR GRS–estimated decrease in the LDL-C level, the OR of having a diagnosis of hypercholesterolemia was 0.85 (95% CI, 0.77-0.92; P = 2.25 × 10−4) (eTable 2 in the Supplement). We tested HMGCR GRS and type 2 diabetes, kidney failure, and Parkinson disease in eMERGE. The finding with type 2 diabetes was statistically significant (OR, 1.09; 95% CI, 1.01-1.17; P = .02), and the findings for kidney failure (OR, 1.18; 95% CI, 0.98-1.41; P = .08) and Parkinson disease (OR, 0.93; 95% CI, 0.75-1.16; P = .53) (Table 3) were not significant. The ORs estimated for both type 2 diabetes and kidney failure in eMERGE (Table 3) were similar to those observed in BioVU (Table 2). In the prespecified sensitivity analysis, after adjusting for statin use in eMERGE (eTable 5 in the Supplement), the finding for type 2 diabetes was statistically significant (OR, 1.12; 95% CI, 1.04-1.21; P = .004), and the finding with kidney failure was significant (OR, 1.24; 95% CI, 1.03-1.49; P = .02). However, in the post hoc analysis, the finding between the HMGCR GRS and kidney failure was not statistically significant after adjusting for type 2 diabetes status (OR, 1.08; 95% CI, 0.88-1.32; P = .48) (eTable 4 in the Supplement).
Table 3. Findings Between HMGCR Genetic Risk Score and Candidate Phenotypes in Electronic Medical Records and Genomics Networka.
| PheCode | Description | P value | Odds ratio (95% CI) | No. | |
|---|---|---|---|---|---|
| Cases | Controls | ||||
| 250.2 | Type 2 diabetes | .02 | 1.09 (1.01-1.17) | 6877 | 17 920 |
| 585.32 | Kidney failure | .08 | 1.18 (0.98-1.41) | 858 | 18 355 |
| 332 | Parkinson disease | .53 | 0.93 (0.75-1.16) | 538 | 19 330 |
The analyses were adjusted for sex, age at most recent visit, electronic health record length, and 5 principal components for ancestry. The HMGCR GRS was standardized for a decrement of 10 mg/dL in the low-density lipoprotein cholesterol level (to convert to millimoles per liter, multiply by 0.0259).
Discussion
We used an HMGCR GRS as an instrumental variable for statin-induced lowering of the LDL-C level due to inhibition of HMG-CoA to evaluate statin treatment and 22 noncardiovascular diseases for which statins have been reported to have beneficial or detrimental outcomes in previous clinical trials or observational studies.3 The major finding of our study was that HMGCR GRS was not associated with the risk for most diseases; the reported association between statin treatment and the increased risk of type 2 diabetes was confirmed,4,16 and the next 2 strongest findings in BioVU—Parkinson disease and kidney failure—were not significant in eMERGE or were explained by type 2 diabetes status.
Although statins have been prescribed since 1987 and studied extensively, a modest increase in the risk of new-onset type 2 diabetes was only recognized in 2008.17,18 and has since been replicated in large-scale meta-analyses of randomized clinical trials.18,19,20,21 Reports from genetic analyses4,16 and well-controlled observational studies22 suggest that the increased type 2 diabetes risk is an on-target effect of statins: a result of a statin-induced decrease in the LDL-C level. By applying an MR approach in the EHR, we replicated the association between HMGCR and risk of type 2 diabetes and found that every 10-mg/dL estimated decrease of the LDL-C level was associated with an approximately 9% increase in the risk of type 2 diabetes—an estimate comparable with a previous MR report.16 However, the cardiovascular benefits of statins in clinical practice outweigh the small increased risk of type 2 diabetes. Nevertheless, particularly for patients with high type 2 diabetes risk (eg, patients with overweight or obesity), health care professionals could encourage a healthy diet and lifestyle and consider clinical monitoring for the development of type 2 diabetes.
The mechanisms underlying the increased risk of type 2 diabetes with statins are unclear; however, several hypotheses have been suggested. For example, in pancreatic cells, inhibition of cholesterol biosynthesis leads to impaired insulin secretion, and this mechanism can be reversed by cholesterol repletion23; also, statins can decrease insulin signaling through several mechanisms24 and decrease glucose uptake through reduced expression of glucose transporter 4.24
Because cholesterol is an essential constituent of the myelin encircling neurons in the brain, long-term low levels of LDL-C were thought to influence neuronal health and increase the risk of neurologic diseases, such as dementia and Parkinson disease.25 However, randomized clinical trials and observational studies have yielded inconsistent findings,26,27,28,29 and statins have been associated with both increased risk of cognitive dysfunction and decreased risk of dementia. Such disparate findings are likely due to residual confounding or indication bias. In the present study, we applied an MR approach to limit confounding and evaluated Parkinson disease and dementia. In BioVU, the finding between HMGCR GRS indicated probable lowering of the LDL-C levels and increased risk of Parkinson disease was not statistically significant. However, the Parkinson disease signal was also not significant in the eMERGE cohort. In a previous report, Benn et al30 used an MR approach with only 1 single-nucleotide variant from HMGCR and found no support for the proposed association between statin treatment and Parkinson disease.
Statins have generally been found to be renoprotective or neutral in studies of both acute kidney injury or progression of chronic kidney disease; a meta-analysis of 57 studies with 143 888 participants found no benefit of statin treatment on risk of kidney failure, but statins had small beneficial effects on the rate of decline in glomerular filtration rate and proteinuria.31 In contrast, a retrospective observational study using a propensity score–matched cohort of 6342 individuals receiving or not receiving statin therapy found an increased risk of both acute and chronic kidney disease in those receiving statin therapy.32 In the present study, the association between HMGCR GRS and kidney failure was close to statistical significance; however, this association was likely affected by confounding because it was not significant after adjusting for type 2 diabetes—a potent risk factor for kidney disease.
We found no significant association between the HMGCR GRS and increase risk of rhabdomyolysis. There was also no signal for myopathy, which is another muscle adverse effect commonly ascribed to statin. It is difficult to study muscle problems in EHRs because mild symptoms may not be well documented by diagnosis codes. Rhabdomyolysis is a well-documented but rare statin adverse effect; however, most cases of rhabdomyolysis, even in patients who receive statin therapy, are due to surgery, trauma, or other factors rather than statins.33 It remains controversial whether rhabdomyolysis is an off-target or on-target statin adverse effect. Both genetic and observational studies of rhabdomyolysis are difficult to conduct because the condition is rare. A study in a very large cohort would be needed to elucidate the association between statin use and rhabdomyolysis.
Strengths and Limitations
This study has several strengths. First, we conducted a comprehensive evaluation of 22 candidate phenotypes in 2 large cohorts: BioVU and eMERGE. Second, we applied an MR approach using genetic variants in the HMGCR gene, which is a method that is less susceptible to confounding and reverse causation than observational studies. Although the HMGCR GRS explains only a relatively small amount of variation in LDL-C levels, it is a genetic instrument whose utility has been demonstrated.16
This study also has several limitations. First, in addition to associations with diseases, statins have been reported to affect mortality related to diseases such as cancer. Disease-specific mortality is difficult to study in the EHR; thus, we did not include such outcomes. Second, we constructed an HMGCR GRS based on the associations of genetic variants with LDL-C levels as an instrumental variable for statins; however, we may not detect off-target statin effects (ie, statin effects that are not mediated by decreasing LDL-C levels). Until the genetic determinants of statin effects unrelated to their decreases in LDL-C levels are characterized it will not be possible to use an MR approach to address the potential off-target risks or benefits of statin therapy. Third, we did not test the differential outcomes of statins in patients who received them at different ages because MR uses a GRS that represents lifelong risk. Fourth, for 12 phenotypes in BioVU, we had more than 1000 cases, but for uncommon phenotypes, such as rhabdomyolysis (n = 159), our ability to detect a genetic signal was limited. Fifth, we focused on individuals of European ancestry and do not know whether the observations apply to other racial/ethnic groups because the prevalence of noncardiovascular outcomes (eg, type 2 diabetes) varies between different ancestries and the association between HMGCR variant alleles and LDL-C may differ in populations of varying ancestry.34
Conclusions
We applied an MR approach in this cohort study using genetic variants in the HMGCR gene that affect LDL-C levels and assessed GRS and candidate phenotypes in 2 large cohorts to evaluate disease and modulation of HMG-CoA activity. We replicated the finding between statin use and increased risk of type 2 diabetes, but there was no strong indication of other pleiotropic outcomes of statins related to their LDL-C level–decreasing effects.
eMethods. Genotype Quality Control and Imputation
eReferences
eTable 1. List of Genetic Variants in the HMGCR GRS
eTable 2. Associations Between HMGCR GRS and Hypercholesterolemia
eTable 3. Sensitivity Analysis of Associations Between HMGCR GRS and Candidate Phenotypes in BioVU Adjusted for Statin Use
eTable 4. Sensitivity Analysis for Association Between HMGCR GRS and End Stage Renal Disease, Further Adjusting Type 2 Diabetes Status
eTable 5. Sensitivity Analysis of Associations Between HMGCR GRS and Candidate Phenotypes in eMERGE Adjusting for Statin Use
eFigure. Associations Between HMGCR GRS Quartiles and Hypercholesterolemia
References
- 1.Delahoy PJ, Magliano DJ, Webb K, Grobler M, Liew D. The relationship between reduction in low-density lipoprotein cholesterol by statins and reduction in risk of cardiovascular outcomes: an updated meta-analysis. Clin Ther. 2009;31(2):236-244. doi: 10.1016/j.clinthera.2009.02.017 [DOI] [PubMed] [Google Scholar]
- 2.US Centers for Disease Control and Prevention. Prescription cholesterol-lowering medication use in adults aged 40 and over: United States, 2003–2012. Published June 7, 2019. Accessed October 2, 2020. https://www.cdc.gov/nchs/products/databriefs/db177.htm
- 3.He Y, Li X, Gasevic D, et al. Statins and multiple noncardiovascular outcomes: umbrella review of meta-analyses of observational studies and randomized controlled trials. Ann Intern Med. 2018;169(8):543-553. doi: 10.7326/M18-0808 [DOI] [PubMed] [Google Scholar]
- 4.Swerdlow DI, Preiss D, Kuchenbaecker KB, et al. ; DIAGRAM Consortium; MAGIC Consortium; InterAct Consortium . HMG-coenzyme A reductase inhibition, type 2 diabetes, and bodyweight: evidence from genetic analysis and randomised trials. Lancet. 2015;385(9965):351-361. doi: 10.1016/S0140-6736(14)61183-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feng Q, Wei W-Q, Chaugai S, et al. Association between low-density lipoprotein cholesterol levels and risk for sepsis among patients admitted to the hospital with infection. JAMA Netw Open. 2019;2(1):e187223. doi: 10.1001/jamanetworkopen.2018.7223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feng Q, Wei W-Q, Chaugai S, et al. A genetic approach to the association between PCSK9 and sepsis. JAMA Netw Open. 2019;2(9):e1911130-e1911130. doi: 10.1001/jamanetworkopen.2019.11130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ritchie MD, Denny JC, Crawford DC, et al. Robust replication of genotype-phenotype associations across multiple diseases in an electronic medical record. Am J Hum Genet. 2010;86(4):560-572. doi: 10.1016/j.ajhg.2010.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wei W-Q, Denny JC. Extracting research-quality phenotypes from electronic health records to support precision medicine. Genome Med. 2015;7(1):41. doi: 10.1186/s13073-015-0166-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu H, Stenner SP, Doan S, Johnson KB, Waitman LR, Denny JC. MedEx: a medication information extraction system for clinical narratives. J Am Med Inform Assoc. 2010;17(1):19-24. doi: 10.1197/jamia.M3378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Denny JC, Bastarache L, Ritchie MD, et al. Systematic comparison of phenome-wide association study of electronic medical record data and genome-wide association study data. Nat Biotechnol. 2013;31(12):1102-1110. doi: 10.1038/nbt.2749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McCarty CA, Chisholm RL, Chute CG, et al. ; eMERGE Team . The eMERGE Network: a consortium of biorepositories linked to electronic medical records data for conducting genomic studies. BMC Med Genomics. 2011;4:13. doi: 10.1186/1755-8794-4-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gottesman O, Kuivaniemi H, Tromp G, et al. ; eMERGE Network . The Electronic Medical Records and Genomics (eMERGE) network: past, present, and future. Genet Med. 2013;15(10):761-771. doi: 10.1038/gim.2013.72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mosley JD, Benson MD, Smith JG, et al. Probing the virtual proteome to identify novel disease biomarkers. Circulation. 2018;138(22):2469-2481. doi: 10.1161/CIRCULATIONAHA.118.036063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wei W-Q, Bastarache LA, Carroll RJ, et al. Evaluating phecodes, clinical classification software, and ICD-9-CM codes for phenome-wide association studies in the electronic health record. PLoS One. 2017;12(7):e0175508. doi: 10.1371/journal.pone.0175508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu P, Gifford A, Meng X, et al. Mapping ICD-10 and ICD-10-CM codes to phecodes: workflow development and initial evaluation. JMIR Med Inform. 2019;7(4):e14325. doi: 10.2196/14325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ference BA, Robinson JG, Brook RD, et al. Variation in PCSK9 and HMGCR and risk of cardiovascular disease and diabetes. N Engl J Med. 2016;375(22):2144-2153. doi: 10.1056/NEJMoa1604304 [DOI] [PubMed] [Google Scholar]
- 17.Ridker PM, Pradhan A, MacFadyen JG, Libby P, Glynn RJ. Cardiovascular benefits and diabetes risks of statin therapy in primary prevention: an analysis from the JUPITER trial. Lancet. 2012;380(9841):565-571. doi: 10.1016/S0140-6736(12)61190-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ridker PM, Danielson E, Fonseca FAH, et al. ; JUPITER Study Group . Rosuvastatin to prevent vascular events in men and women with elevated C-reactive protein. N Engl J Med. 2008;359(21):2195-2207. doi: 10.1056/NEJMoa0807646 [DOI] [PubMed] [Google Scholar]
- 19.Sattar N, Preiss D, Murray HM, et al. Statins and risk of incident diabetes: a collaborative meta-analysis of randomised statin trials. Lancet. 2010;375(9716):735-742. doi: 10.1016/S0140-6736(09)61965-6 [DOI] [PubMed] [Google Scholar]
- 20.Preiss D, Seshasai SRK, Welsh P, et al. Risk of incident diabetes with intensive-dose compared with moderate-dose statin therapy: a meta-analysis. JAMA. 2011;305(24):2556-2564. doi: 10.1001/jama.2011.860 [DOI] [PubMed] [Google Scholar]
- 21.Navarese EP, Buffon A, Andreotti F, et al. Meta-analysis of impact of different types and doses of statins on new-onset diabetes mellitus. Am J Cardiol. 2013;111(8):1123-1130. doi: 10.1016/j.amjcard.2012.12.037 [DOI] [PubMed] [Google Scholar]
- 22.Feng Q, Wei W-Q, Chung CP, et al. Relationship between very low low-density lipoprotein cholesterol concentrations not due to statin therapy and risk of type 2 diabetes: a US-based cross-sectional observational study using electronic health records. PLoS Med. 2018;15(8):e1002642. doi: 10.1371/journal.pmed.1002642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xia F, Xie L, Mihic A, et al. Inhibition of cholesterol biosynthesis impairs insulin secretion and voltage-gated calcium channel function in pancreatic beta-cells. Endocrinology. 2008;149(10):5136-5145. doi: 10.1210/en.2008-0161 [DOI] [PubMed] [Google Scholar]
- 24.Ward NC, Watts GF, Eckel RH. Statin toxicity. Circ Res. 2019;124(2):328-350. doi: 10.1161/CIRCRESAHA.118.312782 [DOI] [PubMed] [Google Scholar]
- 25.Strom BL, Schinnar R, Karlawish J, Hennessy S, Teal V, Bilker WB. Statin therapy and risk of acute memory impairment. JAMA Intern Med. 2015;175(8):1399-1405. doi: 10.1001/jamainternmed.2015.2092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bernick C, Katz R, Smith NL, et al. ; Cardiovascular Health Study Collaborative Research Group . Statins and cognitive function in the elderly: the Cardiovascular Health Study. Neurology. 2005;65(9):1388-1394. doi: 10.1212/01.wnl.0000182897.18229.ec [DOI] [PubMed] [Google Scholar]
- 27.Huang X, Alonso A, Guo X, et al. Statins, plasma cholesterol, and risk of Parkinson’s disease: a prospective study. Mov Disord. 2015;30(4):552-559. doi: 10.1002/mds.26152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trompet S, van Vliet P, de Craen AJM, et al. Pravastatin and cognitive function in the elderly: results of the PROSPER study. J Neurol. 2010;257(1):85-90. doi: 10.1007/s00415-009-5271-7 [DOI] [PubMed] [Google Scholar]
- 29.Bai S, Song Y, Huang X, et al. Statin use and the risk of parkinson’s disease: an updated meta-analysis. PLoS One. 2016;11(3):e0152564. doi: 10.1371/journal.pone.0152564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Benn M, Nordestgaard BG, Frikke-Schmidt R, Tybjærg-Hansen A. Low LDL cholesterol, PCSK9 and HMGCR genetic variation, and risk of Alzheimer’s disease and Parkinson’s disease: mendelian randomisation study. BMJ. 2017;357:j1648. doi: 10.1136/bmj.j1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Su X, Zhang L, Lv J, et al. Effect of statins on kidney disease outcomes: a systematic review and meta-analysis. Am J Kidney Dis. 2016;67(6):881-892. doi: 10.1053/j.ajkd.2016.01.016 [DOI] [PubMed] [Google Scholar]
- 32.Acharya T, Huang J, Tringali S, Frei CR, Mortensen EM, Mansi IA. Statin use and the risk of kidney disease with long-term follow-up (8.4-year study). Am J Cardiol. 2016;117(4):647-655. doi: 10.1016/j.amjcard.2015.11.031 [DOI] [PubMed] [Google Scholar]
- 33.Floyd JS, Heckbert SR, Weiss NS, Carrell DS, Psaty BM. Use of administrative data to estimate the incidence of statin-related rhabdomyolysis. JAMA. 2012;307(15):1580-1582. doi: 10.1001/jama.2012.489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feng Q, Wei W-Q, Levinson RT, Mosley JD, Stein CM. Replication and fine-mapping of genetic predictors of lipid traits in African-Americans. J Hum Genet. 2017;62(10):895-901. doi: 10.1038/jhg.2017.55 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eMethods. Genotype Quality Control and Imputation
eReferences
eTable 1. List of Genetic Variants in the HMGCR GRS
eTable 2. Associations Between HMGCR GRS and Hypercholesterolemia
eTable 3. Sensitivity Analysis of Associations Between HMGCR GRS and Candidate Phenotypes in BioVU Adjusted for Statin Use
eTable 4. Sensitivity Analysis for Association Between HMGCR GRS and End Stage Renal Disease, Further Adjusting Type 2 Diabetes Status
eTable 5. Sensitivity Analysis of Associations Between HMGCR GRS and Candidate Phenotypes in eMERGE Adjusting for Statin Use
eFigure. Associations Between HMGCR GRS Quartiles and Hypercholesterolemia


