Abstract
Background
This study sought to explore the expression pattern in prostate cancer (PCa) tissues, as well as the regulatory effects of circ_0061140 on the proliferative potential of PCa cells.
Methods
A quantitative real-time polymerase chain reaction (qRT-PCR) analysis was undertaken to detect circ_0061140 levels in 43 paired PCa tissues and adjacent normal tissues. After the knockdown of circ_0061140, changes in the proliferative potential of PCa cells and tumor growth in nude mice with PCa were detected. Finally, the relationship of circ_0061140 and miR-1193 in the development of PCa was assessed.
Results
The results showed that circ_0061140 was upregulated in PCa tissues. PCa patients with higher Gleason score or larger sized tumors expressed higher levels of circ_0061140. Additionally, the knockdown of circ_0061140 inhibited the proliferative potential of PCa cells. MiR-1193 was the target gene binding circ_0061140, and its level was negatively regulated by circ_0061140. Finally, rescue experiments showed that miR-1193 was regulated by circ_0061140 in the development of PCa.
Conclusions
Circ_0061140 is upregulated in PCa tissues, and is closely linked to Gleason score and tumor size in PCa. Additionally, it causes PCa cells to proliferate by negatively regulating miR-1193.
Keywords: Circ_0061140, miR-1193, prostate cancer (PCa), malignant development
Introduction
Prostate cancer (PCa) is a common malignancy in men. In developed countries, PCa ranks first among all male malignancies for incidence, and its mortality rate is also high (1-3). In China, the incidence of PCa has markedly increased with rising economic levels, lifestyle changes, aging, and improvements in medical standards (4,5). In early PCa, the symptoms are atypical, and they only gradually become obvious at the middle or advanced stages. Thus, exploration of the mechanism of PCa development could be of great significance in elevating the detective and therapeutic efficacy for the disease (6-8).
Circular RNA (circRNA) is a newly discovered non-coding RNA with a special closed structure. It is abundantly distributed in the cytoplasm of eukaryotes, and is mainly produced by the variable shear processing of precursor messenger RNAs (mRNAs) (9,10). In recent years, circRNAs have been discovered to have vital regulatory functions. Serving as competing endogenous RNAs (ceRNA), circRNAs sponge target microRNAs (miRNAs) to suppress their inhibitory effect on target genes (11-13). MiRNAs are non-coding RNAs of 19-25 nucleotides in length. They can regulate the expression levels of target gene mRNA before and after transcription, and have extremely important biological functions. Some evidence suggests that circRNA homeodomain-interacting protein kinase 3 (circHIPK3) downregulates miR-124/miR-29b expression and is associated with the status of gastric cancer (14). Circ_0044516 downregulates the expression of miR-29a-3p in PCa, and plays an important role in the survival and metastasis of PCa cells (15). Thus, through their regulation of miRNAs, circRNAs play an important role in a variety of cancers.
Additionally, circRNAs exert biological functions by binding to RNA-binding proteins or interacting with other RNAs (13,16). Notably, circRNAs are conserved in different species, and exert tissue specificity. Compared to linear RNAs, circRNAs are more stable due to their resistance to ribonuclease (RNase) (17-19). Circ_0061140 has been reported to have multiple functions in the regulation of cell phenotypes (20).
Following a gene chip profiling analysis of differentially expressed circRNAs in PCa, circ_0061140 was selected as the focus of the present study. Its expression levels in collected PCa samples were determined, and its regulatory effects on the development of PCa were examined.
We present the following article in accordance with the ARRIVE Guidelines reporting checklist (available at http://dx.doi.org/10.21037/tau-20-1477).
Methods
Patients and samples
A total of 43 paired PCa tissues and adjacent normal tissues were surgically resected from patients undergoing radical prostatectomy for PCa in the Urology Department of the Sir Run Run Shaw Hospital, Zhejiang University School of Medicine (Zhejiang, China). Patients with recurrent or metastatic PCa were excluded. The clinical data of the patients were recorded.
Cell culture
PCa cell lines (PC-3, DU-145, 22RV1, Lncap and C4-2) and the prostatic matrix immortalized cell line (WPMY-1) were purchased from the American Type Culture Collection (ATCC) (Manassas, VA, USA). The cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) (Gibco, Rockville, MD, USA) containing 10% fetal bovine serum (FBS) (Gibco, Rockville, MD, USA), 100 UI/mL penicillin, and 100 µg/mL streptomycin in a 5% CO2 incubator at 37 °C. Cell passaging was conducted using 1×tyrpsin + EDTA (ethylenediaminetetraacetic acid).
Transfection
After being cultured to 50−70% confluence in 6-well plates, cells were transfected with plasmids constructed by GenePharma (Shanghai, China), using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA), and collected for use 48 hours later.
Cell proliferation assay
Cells were inoculated in a 96-well plate with 2×103 cells per well, and a Cell Counting Kit-8 (CCK-8) assay (Dojindo Laboratories, Kumamoto, Japan) was carried out to measure cell proliferation. At appointed times, the absorbance value of each sample was recorded at 490 nm, and viability curves was plotted.
Colony formation assay
Cells were inoculated in a 6-well plate with 200 cells per well and cultured for 2 weeks. The culture medium was replaced once in the first week and twice in the second week. Upon completion of the culture, the visible colonies were washed in phosphate-buffered saline (PBS), fixed in methanol for 20 minutes, and dyed with 0.1% crystal violet for 20 minutes. Finally, images were captured, and the number of colonies was calculated.
Quantitative real time-polymerase chain reaction
RNA extracted with TRIzol reagent (Invitrogen, Carlsbad, CA, USA) was purified though treatment with DNase I, and reverse-transcribed into complementary DNA (cDNA) using Primescript RT Reagent (TaKaRa, Otsu, Shiga, Japan). The obtained cDNA was subjected to quantitative real-time polymerase chain reaction (qRT-PCR) using SYBR®Premix Ex Taq™ (TaKaRa, Otsu, Shiga, Japan). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and U6 served as the internal references. Each sample was analyzed in triplicate, and relative level was calculated using the 2-ΔΔCt method. circ_0061140: forward: 5'-GGGGACGCAGAGCCCAG-3', reverse: 5'-GTGCAGGTTCCGAGGT-3', GAPDH: forward: 5'-AATGGGCAGCCGTTAGGAAA-3', reverse: 5'-TGAAGGGGTCATTGATGGCA-3', miR-1193: forward: 5'-CGUGCAGUGGCCAGAU-3', reverse: 5'-GGUAUGGGACGAUCCAUGA-3', U6: forward: 5'-CTCGCTTCGGCAGCACA-3', reverse: 5'-AACGCTTCACGAATTTGCGT-3'.
Luciferase assay
HEK293T cells were inoculated in a 24-well plate and then co-transfected with miR-1193 mimic/NC mimic and circ_0061140-WT/circ_0061140-MUT using Lipofectamine 2000. After 48 hours, the cells were lysed to determining the relative level of luciferase activity.
In vivo xenograft model
This study was approved by the Key Laboratory for Analytical Science of Food Safety and Biology, Ministry of Education, Fuzhou University. Ten male nude mice (8 weeks old) were randomly assigned to 1 of 2 groups (n=5 per group). The mice were subcutaneously administered with 22RV1 cells transfected with sh-NC or sh-circ_0061140 via the armpit. Another 10 mice were administered with DU-145 cells transfected with sh-NC or sh-circ_0061140 using the same method. Five weeks later, the mice were sacrificed and the PCa tissues were collected. Tumor volume = (width2 × length)/2.
Statistical analysis
GraphPad Prism 5 V5.01 (La Jolla, CA, USA) was used for the data analyses. Data were expressed as mean ± standard deviation. Differences between 2 groups were analyzed using the t-test, and the chi-square test was employed to analyze the relationships between the level of circ_0061140 and clinical data of PCa. Pearson’s correlation test was undertaken to evaluate the relationship between 2 genes in PCa tissues. A statistically significant difference was indicated by P<0.05.
Ethics
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The experiments involving human were approved by the Medical Ethics Committee of the Sir Run Run Shaw Hospital, Zhejiang University Medical College (No.: 20200803-37) and informed consent was obtained from each participant. The experiments involving animal were approved by the Key Laboratory for Analytical Science of Food Safety and Biology, Ministry of Education, Fuzhou University, and were conducted in accordance with the “Guide to the Care and Use of Experimental Animals” (2016), and national or institutional guidelines on the care and use of animals.
Results
Circ_0061140 was highly expressed in PCa
We first examined the differential expression of circ_0061140 in PCa tissues and normal tissues. Compared to that in adjacent normal tissues, the expression of circ_0061140 in PCa tissues (P<0.001, Figure 1A,B). Similarly, circ_0061140 was also upregulated in PCa cell lines (Figure 1C, 22RV1, P=0.008; Lncap, P=0.032; PC-3, P=0.021; DU-145, P=0.005; C4-2, P=0.015). Thus, circ_0061140 may aggravate the development of PCa as an oncogene.
Figure 1.
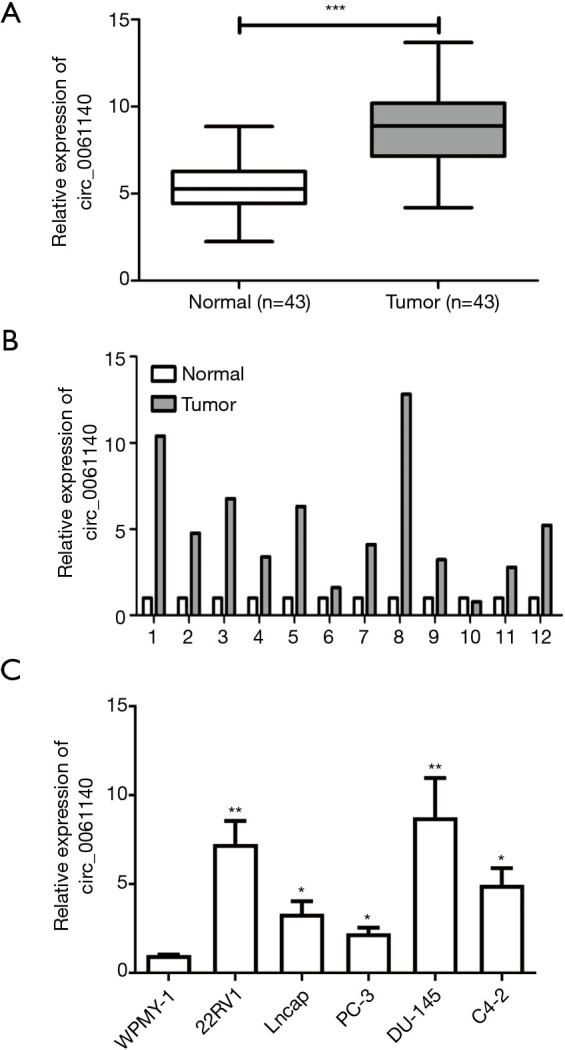
Circ_0061140 is highly expressed in PCa. (A, B) Differential expressions of circ_0061140 in PCa tissues and adjacent normal tissues. (C) Expression levels of circ_0061140 in PCa cell lines. Data are expressed as mean ± SD. *P<0.05, **P<0.01, ***P<0.001.
Circ_0061140 expression was correlated with the Gleason Score and overall survival in PCa
Based on the median level of circ_0061140, 43 patients with PCa were classified into 2 groups. An analysis of the clinical data of the PCa patients, showed that circ_0061140 expression was positively correlated to the Gleason score and tumor size (Table 1).
Table 1. Association of circ_0061140 expression with clinicopathological characteristics of prostate cancer.
| Parameters | Number of cases | circ_0061140 expression | P value | |
|---|---|---|---|---|
| Low (%) | High (%) | |||
| Age (years) | 0.553 | |||
| <60 | 19 | 12 | 7 | |
| ≥60 | 24 | 13 | 11 | |
| Gleason score | 0.019 | |||
| ≤7 | 21 | 16 | 5 | |
| >7 | 22 | 9 | 13 | |
| Tumor size (cm) | 0.001 | |||
| <1 | 22 | 18 | 4 | |
| ≥1 | 21 | 7 | 14 | |
| Lymph node metastasis | 0.234 | |||
| No | 26 | 17 | 9 | |
| Yes | 17 | 8 | 9 | |
Knockdown of Circ_0061140 suppressed the proliferative potential of PCa
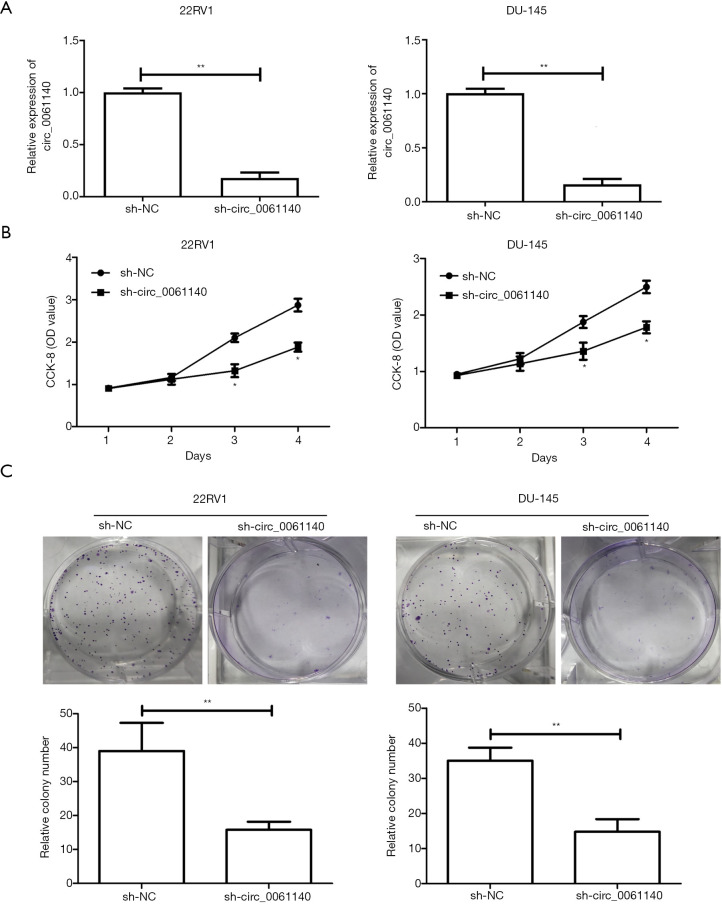
A knockdown model of circ_0061140 was constructed using 22RV1 and DU-145 cells. The transfection efficacy of sh-circ_0061140 was tested (Figure 2A, 22RV1, P=0.006; DU-145, P=0.003). The CCK-8 results revealed that the viability of PCa cells was markedly decreased after knockdown of circ_0061140 (Figure 2B, 22RV1, Day 3, P=0.026; 22RV1, Day 4, P=0.017; DU-145, Day 3, P=0.011; DU-145, Day 4, P=0.022). Furthermore, PCa cells transfected with sh-circ_0061140 formed fewer colonies than the controls (Figure 2C, 22RV1, P=0.007; DU-145, P=0.003). Thus, the proliferative potential of circ_0061140 in PCa was demonstrated.
Figure 2.
Knockdown of circ_0061140 suppresses the proliferative potential of PCa cells. (A) Transfection efficacy of sh-circ_0061140 in 22RV1 and DU-145 cells. (B) CCK-8 assay results show the viability of 22RV1 and DU-145 cells transfected with sh-NC or sh-circ_0061140. (C) Colony formation assay results show the number of 22RV1 and DU-145 cell colonies after transfection with sh-NC or sh-circ_0061140 (magnification: 10×). Data are expressed as mean ± SD. *P<0.05, **P<0.01.
In vivo function of Circ_0061140 in the tumorigenesis of PCa
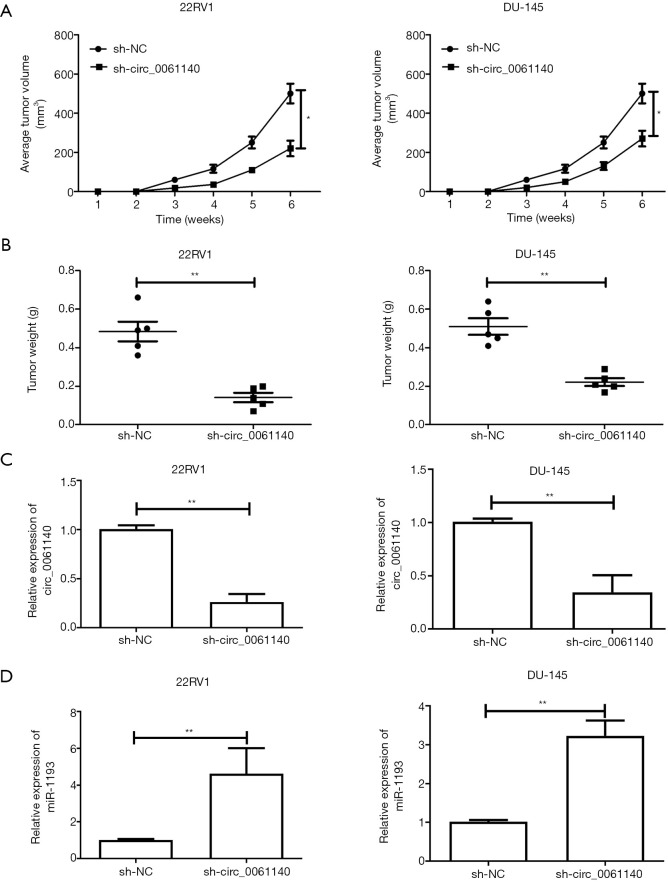
Nude mice were administered with PCa cells transfected with either sh-NC or sh-circ_0061140. The average tumor volume of mice with in vivo knockdown of circ_0061140 was much lower than that of the control group (Figure 3A, 22RV1, P=0.023; DU-145, P=0.031). As expected, the tumor weight was also lower in mice with circ_0061140 knockdown than in the control mice (Figure 3B, 22RV1, P=0.007; DU-145, P=0.001). PCa tissues were collected from the nude mice to prepare homogenate. Downregulation of circ_0061140 (Figure 3C, 22RV1, P=0.003; DU-145, P=0.005) and upregulation of miR-1193 (Figure 3D, 22RV1, P=0.008; DU-145, P=0.005) were observed in mice with circ_0061140 knockdown. Together, these results suggested that circ_0061140 stimulated the tumorigenesis of PCa.
Figure 3.
In vivo function of circ_0061140 in the tumorigenesis of PCa. (A) Volume of PCa tumors extracted from nude mice administered with 22RV1 and DU-145 cells transfected with sh-NC or sh-circ_0061140. (B) Weight of PCa tumors extracted from nude mice administered with 22RV1 and DU-145 cells transfected with sh-NC or sh-circ_0061140. (C) Expression levels of circ_0061140 in PCa tissues extracted from nude mice administered with 22RV1 and DU-145 cells transfected with sh-NC or sh-circ_0061140. (D) Expression levels of miR-1193 in PCa tissues extracted from nude mice administered with 22RV1 and DU-145 cells transfected with sh-NC or sh-circ_0061140. Data are expressed as mean ± SD. *P<0.05, **P<0.01.
Circ_0061140 bound to MiR-1193
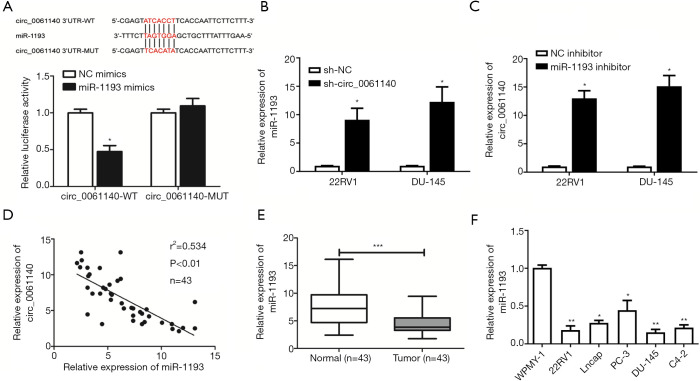
Bioinformatics analysis predicted binding sites in the promoter regions of circ_0061140 and miR-1193. Subsequently, the luciferase assay results revealed that the overexpression of miR-1193 markedly decreased luciferase activity in cells transfected with circ_0061140-WT vector, which confirmed the specific binding of circ_0061140 to miR-1193 (Figure 4A, P=0.016). In 22RV1 and DU-145 cells transfected with sh-circ_0061140, the level of miR-1193 was upregulated compared to that in the control group (Figure 4B, 22RV1, P=0.048; DU-145, P=0.011). Further, knockdown of miR-1193 upregulated circ_0061140 in PCa cells (Figure 4C, 22RV1, P=0.031; DU-145, P=0.026). A negative correlation was discovered between the expression levels of circ_0061140 and miR-1193 in PCa tissues (Figure 4D), while miR-1193 was downregulated in both PCa tissues (Figure 4E, P<0.001) and PCa cell lines (Figure 4F, 22RV1, P=0.005; Lncap, P=0.033; PC-3, P=0.013; DU-145, P=0.003; C4-2, P=0.001).
Figure 4.
Circ_0061140 binds to miR-1193. (A) Luciferase activity in HEK293T cells co-transfected with miR-1193 mimic/NC mimic and circ_0061140-WT/circ_0061140-MUT. (B) Expression levels of miR-1193 in 22RV1 and DU-145 cells transfected with sh-NC or sh-circ_0061140. (C) Expression levels of circ_0061140 in 22RV1 and DU-145 cells transfected with NC inhibitor or miR-1193 inhibitor. (D) A negative correlation between the expression levels of circ_0061140 and miR-1193 in PCa tissues. (E) Differential expression of miR-1193 in PCa tissues and adjacent normal tissues. (F) Expression levels of miR-1193 in PCa cell lines. Data are expressed as mean ± SD. *P<0.05, **P<0.01, ***P<0.001.
MiR-1193 was regulated by circ_0061140 in the development of PCa
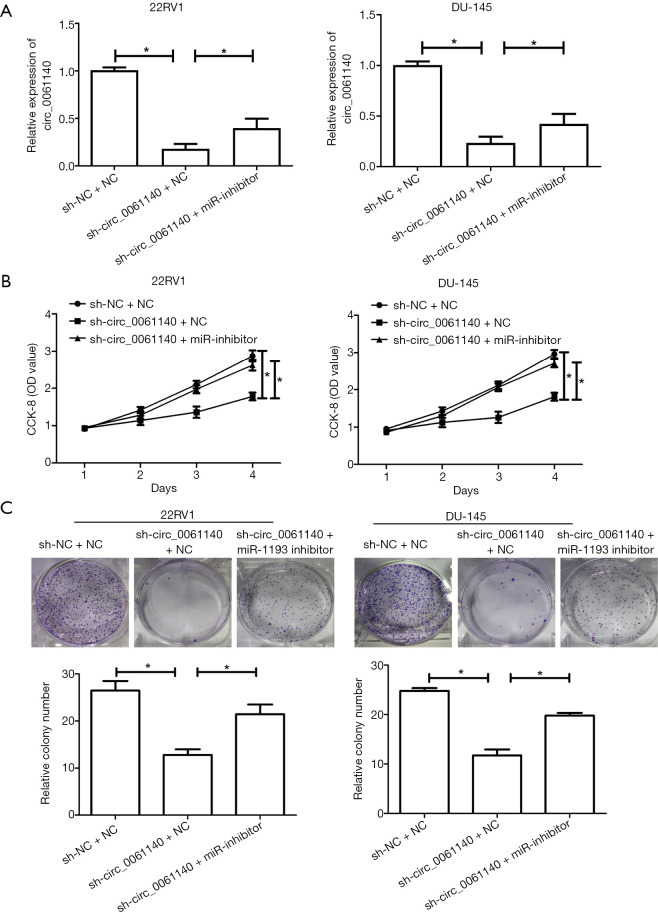
Rescue experiments were conducted to illustrate the involvement of miR-1193 in the development of PCa. Co-transfection with miR-1193 inhibitor and sh-circ_0061140 partially reversed the downregulation circ_0061140 in PCa cells (Figure 5A, 22RV1, sh-NC+NC vs. sh-circ_0061140+NC, P=0.024; 22RV1, sh-circ_0061140+NC vs. sh-circ_0061140+miR-1193 inhibitor, P=0.011; DU-145, sh-NC+NC vs. sh-circ_0061140+NC, P=0.033; DU-145, sh-circ_0061140+NC vs. sh-circ_0061140+miR-1193 inhibitor, P=0.013). Notably, to a certain extent, silencing of miR-1193 reversed the attenuation of viability (Figure 5B, 22RV1, sh-NC+NC vs. sh-circ_0061140+NC, P=0.016; 22RV1, sh-circ_0061140+NC vs. sh-circ_0061140+miR-1193 inhibitor, P=0.045; DU-145, sh-NC+NC vs. sh-circ_0061140+NC, P=0.010; DU-145, sh-circ_0061140+NC vs. sh-circ_0061140+miR-1193 inhibitor, P=0.014) and clonality (Figure 5C, 22RV1, sh-NC+NC vs. sh-circ_0061140+NC, P=0.014; 22RV1, sh-circ_0061140+NC vs. sh-circ_0061140+miR-1193 inhibitor, P=0.022; DU-145, sh-NC+NC vs. sh-circ_0061140+NC, P=0.017; DU-145, sh-circ_0061140+NC vs. sh-circ_0061140+miR-1193 inhibitor, P=0.021) in PCa cells with circ_0061140 knockdown.
Figure 5.
MiR-1193 is regulated by circ_0061140 in the development of PCa. (A) Expression levels of circ_0061140 in 22RV1 and DU-145 cells transfected with sh-NC+NC, sh-circ_0061140+NC or sh-circ_0061140+miR-1193 inhibitor. (B) CCK-8 assay results show the viability of 22RV1 and DU-145 cells transfected with sh-NC+NC, sh-circ_0061140+NC or sh-circ_0061140+miR-1193 inhibitor. (C) Colony formation assay results show the number of 22RV1 and DU-145 cells colonies after transfection with sh-NC+NC, sh-circ_0061140+NC or sh-circ_0061140+miR-1193 inhibitor (magnification: 10×). Data are expressed as mean ± SD. *P<0.05.
Discussion
CircRNAs, which are stably expressed in eukaryotes, display tissue specificity and evolutionary conservation (9-11). CircRNAs were first discovered in the viroids of higher plants by Sanger et al. (21) in 1976, and were initially thought to be produced by erroneously variable shear, however, with the rapid development of sequencing and bioinformatics, the vital functions of circRNAs have since been identified (12,13,16,17).
Previous studies (20,22) have shown circ_0061140 to be involved in tumor progression. The present study showed that circ_0061140 is upregulated in PCa tissues, and that it might be an oncogene in the development of PCa. During PCa development, the surrounding micro-environment stimulates tumor cells, producing changes in cell polarity, which eventually enhances the potential of tumor cells. In the present study, the knockdown of circ_0061140 decreased the viability and colony number of PCa cells, which suggests that it inhibited their proliferative potential. Further, the in vivo results showed that knockdown of circ_0061140 markedly reduced the tumor weight and size of nude mice with PCa. Notably, circ_0061140 slowed down the development of PCa by inducing malignant progression.
Some studies have shown that circRNAs can regulate the selective splicing of miRNAs and regulate the expression of target genes by acting as miRNA sponges (13). Compared to linear RNAs, circRNAs are large in number, characterized by structural stability and tissue specificity, and contain more miRNA binding sites, thus, circRNAs make ideal sponges for miRNA molecules (23). In endometrial cancer, circ_0061140 acts as a molecular sponge for miR-149-5p and promotes STAT3 expression, which in promotes disease progression (24). Circ_0061140 is upregulated in ovarian cancer, and promotes the growth and metastasis of ovarian cancer cells by competitively binding to miR-370 (20). It is also significantly expressed in bladder cancer tissue, and can specifically bind to microRNA-1234 to enhance the proliferation and invasion abilities of bladder cancer cells (25).
However, the regulatory mechanism of circ_0061140 in the proliferation of PCa is still unclear. The present study may provide a basis for novel targeted therapies for PCa based on the circRNA-miRNA interaction. Through bioinformatics prediction and luciferase assay verification, we confirmed the binding relationship between circ_0061140 and miR-1193.
The abnormal expression of miR-1193, a recently discovered miRNA, has been reported in many types of solid malignancies, and it has been considered as a tumor suppressor (26-28). The findings of the present study showed that miR-1193 was expressed at a low level in PCa, and its expression was negatively regulated by circ_0061140. Notably, miR-1193 was confirmed to be involved in the development of PCa, and to be regulated by circ_0061140. Thus, intervention with circ_0061140 and miR-1193could play a potential role in the diagnosis and treatment of PCa.
Conclusions
Circ_0061140 is upregulated in PCa tissues and is closely linked to the Gleason score and tumor size of patients with PCa. Additionally, it stimulates PCa cell proliferation by negatively regulating miR-1193.
Supplementary
The article’s supplementary files as
Acknowledgments
Funding: This study was funded by the Health Science and Technology Planning Project of Hangzhou (No. 20190682).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The experiments involving human were approved by the Medical Ethics Committee of the Sir Run Run Shaw Hospital, Zhejiang University Medical College (No.: 20200803-37) and informed consent was obtained from each participant. The experiments involving animal were approved by the Key Laboratory for Analytical Science of Food Safety and Biology, Ministry of Education, Fuzhou University, and were conducted in accordance with the “Guide to the Care and Use of Experimental Animals” (2016), and national or institutional guidelines on the care and use of animals.
Footnotes
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at http://dx.doi.org/10.21037/tau-20-1477
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tau-20-1477
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tau-20-1477). The authors have no conflicts of interest to declare.
References
- 1.Barry MJ, Simmons LH. Prevention of prostate cancer morbidity and mortality: primary prevention and early detection. Med Clin North Am 2017;101:787-806. 10.1016/j.mcna.2017.03.009 [DOI] [PubMed] [Google Scholar]
- 2.Rebbeck TR. Prostate cancer genetics: Variation by race, ethnicity, and geography. Semin Radiat Oncol 2017;27:3-10. 10.1016/j.semradonc.2016.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang G, Zhao D, Spring DJ, et al. Genetics and biology of prostate cancer. Genes Dev 2018;32:1105-40. 10.1101/gad.315739.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fenner A. Prostate cancer: ERSPC calculator recalibrated for China. Nat Rev Urol 2017;14:66. 10.1038/nrurol.2016.259 [DOI] [PubMed] [Google Scholar]
- 5.Reulen RC, de Vogel S, Zhong W, et al. Physical activity and risk of prostate and bladder cancer in China: the South and East China case-control study on prostate and bladder cancer. PLoS One 2017;12:e0178613. 10.1371/journal.pone.0178613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grozescu T, Popa F. Prostate cancer between prognosis and adequate/proper therapy. J Med Life 2017;10:5-12. [PMC free article] [PubMed] [Google Scholar]
- 7.Smith ZL, Eggener SE, Murphy AB. African-American prostate cancer disparities. Curr Urol Rep 2017;18:81. 10.1007/s11934-017-0724-5 [DOI] [PubMed] [Google Scholar]
- 8.Ramnarine VR, Kobelev M, Gibb EA, et al. The evolution of long noncoding RNA acceptance in prostate cancer initiation, progression, and its clinical utility in disease management. Eur Urol 2019;76:546-59. 10.1016/j.eururo.2019.07.040 [DOI] [PubMed] [Google Scholar]
- 9.Luo J, Liu H, Luan S, Li Z. Guidance of circular RNAs to proteins' behavior as binding partners. Cell Mol Life Sci 2019;76:4233-43. 10.1007/s00018-019-03216-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kristensen LS, Andersen MS, Stagsted L, et al. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet 2019;20:675-91. 10.1038/s41576-019-0158-7 [DOI] [PubMed] [Google Scholar]
- 11.Arnaiz E, Sole C, Manterola L, et al. CircRNAs and cancer: biomarkers and master regulators. Semin Cancer Biol 2019;58:90-9. 10.1016/j.semcancer.2018.12.002 [DOI] [PubMed] [Google Scholar]
- 12.Pandey PR, Munk R, Kundu G, et al. Methods for analysis of circular RNAs. Wiley Interdiscip Rev RNA 2020;11:e1566. 10.1002/wrna.1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rong D, Sun H, Li Z, et al. An emerging function of circRNA-miRNAs-mRNA axis in human diseases. Oncotarget 2017;8:73271-81. 10.18632/oncotarget.19154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheng J, Zhuo H, Xu M, et al. Regulatory network of circRNA-miRNA-mRNA contributes to the histological classification and disease progression in gastric cancer. J Transl Med 2018;16:216. 10.1186/s12967-018-1582-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li T, Sun X, Chen L. Exosome circ_0044516 promotes prostate cancer cell proliferation and metastasis as a potential biomarker. J Cell Biochem 2020;121:2118-26. 10.1002/jcb.28239 [DOI] [PubMed] [Google Scholar]
- 16.Du WW, Zhang C, Yang W, et al. Identifying and characterizing circRNA-protein interaction. Theranostics 2017;7:4183-91. 10.7150/thno.21299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu T, Wang Y, Fan Y, et al. CircRNAs in cancer metabolism: a review. J Hematol Oncol 2019;12:90. 10.1186/s13045-019-0776-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Diallo LH, Tatin F, David F, et al. How are circRNAs translated by non-canonical initiation mechanisms? Biochimie 2019;164:45-52. 10.1016/j.biochi.2019.06.015 [DOI] [PubMed] [Google Scholar]
- 19.Yin Y, Long J, He Q, et al. Emerging roles of circRNA in formation and progression of cancer. J Cancer 2019;10:5015-21. 10.7150/jca.30828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen Q, Zhang J, He Y, et al. Hsa_circ_0061140 knockdown reverses FOXM1-mediated cell growth and metastasis in ovarian cancer through miR-370 sponge activity. Mol Ther Nucleic Acids 2018;13:55-63. 10.1016/j.omtn.2018.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Sanger HL, Klotz G, Riesner D, et al. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci U S A 1976;73:3852-6. 10.1073/pnas.73.11.3852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao X, Cai Y, Xu J. Circular RNAs: biogenesis, mechanism, and function in human cancers. Int J Mol Sci 2019;20:3926. 10.3390/ijms20163926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou R, Wu Y, Wang W, et al. Circular RNAs (circRNAs) in cancer. Cancer Lett 2018;425:134-42. 10.1016/j.canlet.2018.03.035 [DOI] [PubMed] [Google Scholar]
- 24.Liu Y, Chang Y, Cai Y. Hsa_circ_0061140 promotes endometrial carcinoma progression via regulating miR-149-5p/STAT3. Gene 2020;745:144625. 10.1016/j.gene.2020.144625 [DOI] [PubMed] [Google Scholar]
- 25.Feng F, Chen AP, Wang XL, et al. Circ_0061140 promotes metastasis of bladder cancer through adsorbing microRNA-1236. Eur Rev Med Pharmacol Sci 2020;24:5310-9. [DOI] [PubMed] [Google Scholar]
- 26.Zhang J, Jing L, Tan S, et al. Inhibition of miR-1193 leads to synthetic lethality in glioblastoma multiforme cells deficient of DNA-PKcs. Cell Death Dis 2020;11:602. 10.1038/s41419-020-02812-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang J, Li C, Xu L, Yang C, Zhang X. MiR-1193 was sponged by LINC00963 and inhibited cutaneous squamous cell carcinoma progression by targeting SOX4. Pathol Res Pract 2019;215:152600. 10.1016/j.prp.2019.152600 [DOI] [PubMed] [Google Scholar]
- 28.Zhang B, Lin Y, Bao QF, et al. MiR-1193 Inhibits the Malignancy of Cervical Cancer Cells by Targeting Claudin 7 (CLDN7). Onco Targets Ther 2020;13:4349-58. 10.2147/OTT.S247115 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as