Abstract
Glioma is the most common type of primary brain cancer in adults. Accumulating studies have reported that long non-coding RNAs (lncRNAs) serve a significant role in the initiation and development of glioma. lncRNA small nucleolar RNA host gene 7 (SNHG7) has been previously demonstrated to serve a role in numerous glioma biological processes, including cell proliferation, invasion and migration. The present study aimed to investigate the role of SNHG7 in glioma through reverse transcription-quantitative PCR, western blotting and cell function assays. The results revealed that SNHG7 expression was upregulated in glioma tissues and cell lines, while microRNA (miR)-138-5p expression was downregulated. Moreover, the knockdown of SNHG7 expression decreased the proliferation of glioma cells. Mechanistic studies demonstrated that SNHG7 downregulated miR-138-5p expression, which subsequently affected the expression levels of its target gene, enhancer of zeste 2 polycomb repressive complex 2 subunit (EZH2). In conclusion, the results of the present study suggested that SNHG7 may act as a competing endogenous RNA to sponge miR-138-5p and modulate EZH2 expression. Thus, SNHG7 may enhance glioma proliferation via modulating the miR-138-5p/EZH2 signaling axis.
Keywords: long non-coding RNA, small nucleolar RNA host gene 7, microRNA-138-5p, enhancer of zeste 2 polycomb repressive complex 2, glioma
Introduction
Glioma, which is the most common type of primary brain and central nervous system (CNS) cancer, originates from supporting or glial cells in the CNS (1). Glioma is classified into two subtypes: Diffuse glioma and non-diffuse glioma, which exhibits slower growth (2). Treatment for glioma mainly comprises surgical resection, chemotherapy and radiation. In addition, immunotherapies and targeted therapies have been developed in recent years in an effort to increase the low survival rate of patients (3). However, further studies on the mechanisms of glioma progression are required to identify and develop novel treatment strategies.
Long non-coding RNAs (lncRNAs) are transcripts of >200 nucleotides in length that lack protein-coding ability (4). lncRNAs have been reported to serve roles in a wide variety of physiological and pathophysiological functions by regulating gene and chromatin dynamics, which subsequently promotes tumorigenesis and metastasis (5). Therefore, lncRNAs have been considered as novel biomarkers and therapeutic targets for various types of cancer (6). Small nucleolar RNA host gene 7 (SNHG7) was first identified in lymphoblastoid cell lines by Chaudhry (7) in 2013. SNHG7 comprises 2,157 nucleotides and is located on chromosome 9q34.3 (8). The expression levels of SNHG7 were reported to be upregulated in numerous types of cancer such as bladder, breast, colorectal, esophageal, gastric and prostate cancer, and were associated with tumor progression, metastasis and the suppression of apoptosis (9). For example, Ren et al (10) discovered that SNHG7 promoted the growth and progression of glioblastoma. However, to the best of our knowledge, the underlying mechanism of the effect of SNHG7 on the proliferation of glioma cells remains poorly understood.
MicroRNAs (miRNAs/miRs) are small non-coding RNAs of 22 nucleotides in length, which also lack protein-coding ability (11). miRNAs downregulate the expression levels of target protein-coding genes by binding to the 3′-untranslated regions (UTRs) of mRNAs, post-transcriptionally (12). An increasing number of studies have demonstrated that miRNAs serve significant roles in the progression of various types of cancer, including glioma. For instance, Wang et al (13) reported that miR-138-5p inhibited the progression of human colorectal cancer by targeting telomerase reverse transcriptase expression. Ou et al (14) reported that miR-138-5p inhibited the proliferation of cervical cancer cells. The lncRNASNP2 database was used in the present study to identify putative binding sites complementary to SNHG7 in miR-138-5p.
Accumulating evidence has demonstrated that lncRNAs can act as competing endogenous RNAs (ceRNAs) by sponging or decoying target miRNAs involved in tumor proliferation (6,15). The aim of the present study was to determine the clinical significance and biological functions of SNHG7 in glioma.
Materials and methods
Patient studies
A total of 20 glioma tissues resected from patients with glioma (11 male and 9 female; mean age ± SD, 55.3±12.5 years; age range, 23–85 years) and 14 normal tissues from brains of traffic accident victims (8 male and 6 female; mean age ± SD, 51.2±15.4 years; age range, 18–82 years) were acquired. The tissues were obtained from the Department of Neurosurgery of Chenzhou First People's Hospital (Chenzhou, China) between January 2016 and December 2018. Samples were frozen in liquid nitrogen immediately after collection. Based on the World Health Organization (WHO) classification system (16), two clinical pathologists pathologically diagnosed and divided the glioma specimens into two groups: Low-grade and high-grade. The low-grade group contained 9 stage I–II tumors, while the high-grade group comprised 11 stage III–IV tumors. Patients had not received chemotherapy or radiotherapy prior to surgical resection. Experiments were performed according to the principles of the Declaration of Helsinki. Experimental protocols were approved by Chenzhou First People's Hospital and written informed consent was obtained from all patients or their relatives prior to participation in the study.
Cell lines and culture
Normal human astrocytes (NHA), glioma LN229, A172 and U251 cell lines, and a glioblastoma cell line of unknown origin, U87 (cat. no. HTB14), were purchased from the American Type Culture Collection. Cells were cultured in DMEM (HyClone; Cytiva) supplemented with 10% FBS (HyClone; Cytiva), and maintained at 37°C in a humidified atmosphere containing 5% CO2.
Cell transfection
Small interfering RNA (siRNA/si) sequences targeting SNHG7 (si-SNHG7-1, 5′-GCCGCUUGUGUU CUUGAUUTT-3′; and si-SNHG7-2, 5′-CCTCTGGTGCCUCGUUCUGGAAACGAUTT-3′) and scrambled si-negative control (NC; 5′-CCCAGUAAUAGGACACCAATT-3′) were purchased from Guangzhou RiboBio Co., Ltd. The miR-138-5p inhibitor (5′-CACACUCGGGUCGAAUGAAGGA-3′), miR-138-5p mimic (5′-AGCUGGUGUUGUGAAUCAGGCCG-3′), scrambled mimic NC (5′-UUCUCCGAACGUGUCACGU-3′) and scrambled inhibitor NC (5′-CAGUACUUUUGUGUAGUA CAA-3′) were obtained from Shanghai GenePharma Co., Ltd. Cell transfection was performed using Lipofectamine® 3000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol.
Reverse transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from cells and tissues using TRIzol® and TRIzol™ LS reagent (both Invitrogen; Thermo Fisher Scientific, Inc.), respectively, according to the manufacturers' protocols. Total RNA was reverse transcribed into cDNA to analyze lncRNA and mRNA expression using the PrimeScript™ RT reagent kit (Takara Bio, Inc.) according to the manufacturer's protocol. qPCR was subsequently performed using a TB Green™ Premix Ex Taq™ II kit (Takara Bio, Inc.), with GAPDH as the internal control. The PCR was performed using the following conditions: Initial denaturation at 95 °C for 30 sec, followed by 40 cycles of 95°C for 5 sec and 60°C for 30 sec, with final annealing and extension at 60°C for 60 sec and 95°C for 15 sec. Total RNA was reverse transcribed into cDNA to determine miRNA expression using a miRcute Plus miRNA First-Strand cDNA kit (Tiangen Biotech Co., Ltd.) according to the manufacturer's protocol. qPCR was subsequently performed using a miRcute Plus miRNA qPCR kit (SYBR Green) (Tiangen Biotech Co., Ltd.), with U6 as the internal control. qPCR was performed on an ABI 7500 Real-Time PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.). The following primer pairs (Sangon Biotech Co., Ltd.) were used for qPCR: SNHG7 forward, 5′-GTTGGGGTGTTGGCATTCTTGTT-3′ and reverse, 5′-TGGTCAGCCTGGTCACTCTGG-3′; EZH2 forward, 5′-AATCAGAGTACATGCGACTGAGA-3′ and reverse, 5′-GCTGTATCCTTCGCTGTTTCC-3′; GAPDH forward, 5′-GTCTCCTCTGACTTCAACAGCG-3′ and reverse, 5′-ACCACCCTGTTGCTGTAGCCAA-3′; miR-138-5p forward, 5′-AGCTGGTGTTGTGAATCAGGCCG-3′ and reverse, 5′-AACGCTTCACGAATTTGCGT-3′; and U6 forward, 5′-CTC GCTTCGGCAGCACA-3′ and reverse, 5′-AACGCTTCACGA ATTTGCGT-3′. The miR-138-5p reverse primer was obtained using the miRcute Plus miRNA qPCR kit (SYBR Green). Expression levels were quantified using the 2−ΔΔCq method (17).
Nucleic and cytoplasmic separation
Nucleic and cytoplasmic fragments were extracted from cells using a PARIS™ kit (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. Briefly, collected cells were lysed in cell fractionation buffer, then the nucleic and cytoplasmic fractions were isolated by centrifugation. The supernatant containing the cytoplasmic fraction was transferred into a clean tube in the absence of RNase. Disruption buffer was used to lyse the pellet containing the nucleic fraction, which was subsequently mixed with the cytoplasmic fraction and lysis/binding solution. Subsequently, 100% ethanol was added the mixture, and the sample was filtered and washed with washing solution. The elution solution was used to obtain cytoplasmic and nucleic mRNAs. U6 and GAPDH were used as the positive controls for the nucleic and cytoplasmic extracts, respectively.
Bioinformatics analysis
Prediction of putative binding sites complementary to SNHG7 in miR-138-5p was performed using the lncRNASNP2 database (http://bioinfo.life.hust.edu.cn/lncRNASNP). Putative targeting sequences between miR-138-5p and EZH2 were predicted using TargetScan 7.2 (http://www.targetscan.org).
Cell Counting Kit-8 (CCK-8) assay
Glioma cells were cultured in 96-well plates and transfected as aforementioned. Following incubation for 24, 48 or 72 h, 20 µl CCK-8 (Dojindo Molecular Technologies, Inc.) was added to each well for 4 h. The absorbance was measured at a wavelength of 490 nm using a Elx800 microplate reader (BioTek Instruments, Inc.; Agilent Technologies, Inc.). According to the absorbance values, cell proliferation was calculated as a percentage of the control.
Flow cytometry
Following transfection for 48 h, U87 and A172 cells were collected for cell cycle analysis. Briefly, cells were fixed in 75% ethanol overnight at −20°C and then stained with PI (BD Biosciences) for 30 min in the dark at 37°C. The cell cycle distribution was analyzed using a flow cytometer (Attune NxT; Thermo Fisher Scientific, Inc.). All data were analyzed using FlowJo v10 software (FlowJo LLC). All experiments were independently performed in triplicate.
Western blotting
Total protein was extracted from glioma cells using a Protein Extraction kit (Nanjing KeyGen Biotech Co., Ltd.) and quantified using a BCA protein assay kit (Sigma-Aldrich; Merck KGaA). Proteins (40 µg/lane) were separated by 10% SDS-PAGE and transferred onto PVDF membranes (Bio-Rad Laboratories, Inc.). The membranes were blocked with 5% skimmed milk for 1.5 h at room temperature. Membranes were subsequently incubated with the following primary antibodies at 4°C overnight: Rabbit polyclonal anti-EZH2 (1:2,000; cat. no. 21800-1-AP; ProteinTech Group, Inc.) and rabbit polyclonal anti-GAPDH (1:1,000; cat. no. 25778; Santa Cruz Biotechnology, Inc.). Following the primary antibody incubation, the membranes were incubated with an HRP-conjugated goat anti-rabbit secondary antibody (1:5,000; cat. no. ab6721; Abcam) at room temperature for 2 h. Protein bands were visualized using the ECL reagent (EMD Millipore).
Dual luciferase reporter assay
Following the prediction of the putative binding sites of miR-138-5p in SNHG7/EZH2, pMIR-REPORT vectors (Guangzhou RiboBio Co., Ltd.) containing wild-type (WT) or mutant (MT) 3′-UTR sequences of SNHG7 or EZH2 were constructed. Cells were co-transfected with miR-138-5p mimic or mimic NC and SNHG7/EZH2-WT or SNHG7/EZH2-MT for 48 h using Lipofectamine® 3000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.). Relative luciferase activity was measured using a Luc-Pair™ Duo-Luciferase assay kit (Shanghai Yeasen Biotechnology Co., Ltd.).
RNA immunoprecipitation (RIP) assay
The RIP assay was performed using a Magna RNA-Binding Protein Immunoprecipitation kit (EMD Millipore) according to the manufacturer's protocol. Whole cell lysates were incubated with anti-IgG negative control antibody (derived from normal mice; EMD Millipore) or anti-Argonaute 2 (Ago2) antibody (from humans; 1:50; EMD Millipore) and RIP buffer containing magnetic beads. Each sample was subsequently incubated with proteinase K buffer. The immunoprecipitated RNAs were isolated and SNHG7 and miR-138-5p expression was detected using RT-qPCR, as aforementioned.
Colony formation assay
Transfected cells were cultured in 6-well plates overnight in medium supplemented with 10% FBS at 37°C. Subsequently, cells were incubated for 14 days at 37°C. Following the incubation, cells were fixed with methanol for 10 min at room temperature and stained with 0.1% crystal violet for 20 min at room temperature. Colonies were counted under a light microscope.
Statistical analysis
Statistical analysis was performed using SPSS 21.0 software (IBM Corp.) and data are presented as the mean ± SD of three independent experiments. Statistical differences between two groups were determined using an unpaired two-tailed Student's t-test, while statistical differences between multiple groups were determined using a one-way ANOVA followed by Tukey's post hoc test. Spearman's rank correlation was used for correlation analysis. P<0.05 was considered to indicate a statistically significant difference.
Results
lncRNA SNHG7 expression levels are upregulated in glioma and promote the proliferation of glioma cells
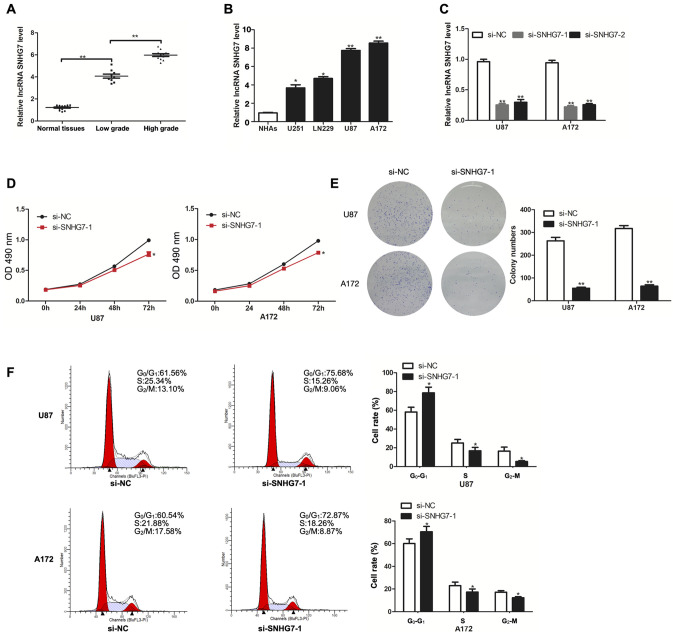
To determine the role of SNHG7 in glioma, the expression levels of SNHG7 in tissues from normal brains, low-grade glioma (stage I–II) and high-grade glioma (stage III–IV) were analyzed. The results revealed that SNHG7 expression levels were significantly upregulated in glioma tissues compared with in normal tissues; in addition, the expression levels of SNHG7 were significantly increased in high grade glioma (stage III–IV) tissues compared with in low-grade glioma (stage I–II) tissues (Fig. 1A). Thus, SNHG7 expression may be positively associated with tumor grade. The expression levels of SNHG7 were also determined in cell lines, including NHAs, U251, LN229, U87 and A172 cells. Compared with in NHAs, SNHG7 expression was significantly upregulated in glioma cell lines, including in U251 (P<0.05) and LN229 (P<0.05), and especially in U87 (P<0.01) and A172 (P<0.01) (Fig. 1B). Therefore, the latter two cell lines (U87 and A172) were chosen for subsequent experiments. RT-qPCR analysis revealed that SNHG7 expression was successfully knocked down in U87 and A172 cell lines following transfection with both si-SNHG7-1 and si-SNHG7-2 (Fig. 1C). The results of the CCK-8 assay demonstrated that SNHG7-knockdown significantly decreased cell proliferation in U87 and A172 cell lines compared with in cells transfected with si-NC (Fig. 1D). In addition, the results of the colony formation assay further indicated that SNHG7-knockdown significantly decreased the number of colonies formed from glioma cells compared with those formed from glioma cells transfected with si-NC (Fig. 1E). Flow cytometry analysis of the cell cycle distribution revealed that SNHG7-knockdown arrested U87 and A172 cells in the G1 phase (Fig. 1F).
Figure 1.
lncRNA SNHG7 expression is upregulated in glioma tissues and cell lines, and promotes proliferation. (A) RT-qPCR analysis of SNHG7 expression in 14 normal tissues, 9 low-grade (stage I–II) glioma tissues and 11 high-grade (stage III–IV) glioma tissues. (B) RT-qPCR analysis of SNHG7 expression in NHAs, U251, LN229, U87 and A172 cell lines. (C) RT-qPCR analysis of SNHG7 expression for transfection efficiency in U87 and A172 cells. (D) Cell proliferation was determined by Cell Counting Kit-8 assay after transfection with si-SNHG7. (E) Colony formation assay was performed to determine colony-forming ability. (F) Flow cytometry was used to evaluate the effects of si-SNHG7 on cell cycle distribution. *P<0.05 and **P<0.01 vs. NHAs or si-NC. OD, optical density; NHAs, normal human astrocytes; RT-qPCR, reverse transcription-quantitative PCR; si, small interfering RNA; NC, negative control; lncRNA; long non-coding RNA; SNHG7, small nucleolar RNA host gene 7.
SNHG7 directly targets miR-138-5p
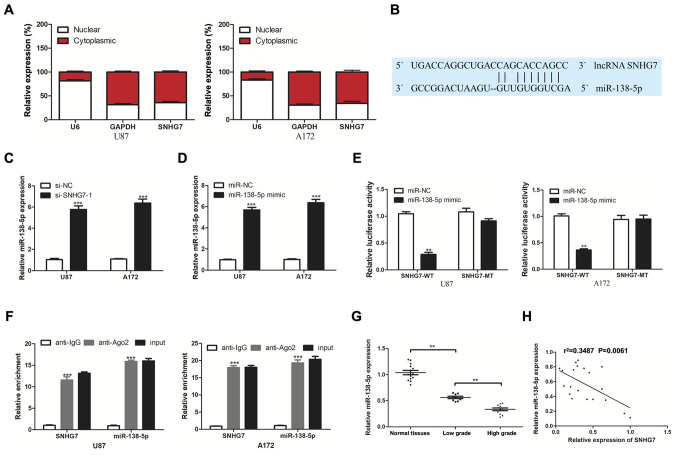
To determine the underlying mechanism of SNHG7 in glioma, RT-qPCR was used to analyze SNHG7 expression in the nucleus and cytoplasm. The results demonstrated that SNHG7 was mainly localized in the cytoplasm of U87 and A172 cell lines (Fig. 2A). Based on these results, it was hypothesized that SNHG7 may function as a ceRNA. Bioinformatics analysis using the lncRNASNP2 database predicted that miR-138-5p contained potential binding sites for SNHG7 (Fig. 2B). miR-138-5p expression levels were upregulated in glioma cells following the transfection of U87 and A172 cells with si-SNHG7-1 compared with si-NC (Fig. 2C). miR-138-5p expression was significantly increased following transfection with the miR-138-5p mimic compared with the miR-NC (Fig. 2D). The results of the dual luciferase reporter assay demonstrated that the relative luciferase activity was significantly decreased following the co-transfection of cells with the miR-138-5p mimic and SNHG7-WT, but not SNHG7-MT vectors (Fig. 2E). These findings indicated the potential association between SNHG7 and miR-138-5p. It was subsequently investigated whether miR-138-5p and SNHG7 existed in the same RNA-induced silencing complex using a RIP assay. The results indicated significant enrichment of SNHG7 in the anti-Ago2 group compared with the anti-IgG group; similar results were obtained for miR-138-5p (Fig. 2F). Additionally, miR-138-5p expression was analyzed in 14 normal brain tissues, nine low-grade glioma (stage I–II) tissues and 11 high-grade glioma (stage III–IV) tissues. miR-138-5p expression levels were significantly downregulated in glioma tissues compared with in normal tissues, and significantly lower expression levels of miR-138-5p were observed in high-grade tissues compared with in low-grade tissues (Fig. 2G). Furthermore, Pearson's correlation analysis identified a negative correlation between miR-138-5p and SNHG7 expression in glioma tissues (Fig. 2H). These findings suggested that SNHG7 may directly target miR-138-5p.
Figure 2.
lncRNA SNHG7 targets miR-138-5p. (A) Nuclear and cytoplasmic SNHG7 percentage was analyzed by RT-qPCR. (B) Bioinformatics prediction of miR-138-5p targeting sites in SNHG7 3′-untranlsated region using lncRNASNP2. (C) Relative miR-138-5p expression was detected via RT-qPCR after transfection with si-NC or si-SNHG7-1. (D) Relative miR-138-5p expression was detected via RT-qPCR after transfection with miR-NC or miR-138-5p mimic. (E) Luciferase activity was analyzed by luciferase reporter assay for the targeting of SNHG7 to miR-138-5p. (F) Enrichment analysis of SNHG7 and miR-138-5p in immunoprecipitates of Ago2 and control IgG by RNA immunoprecipitation. (G) miR-138-5p expression was determined by RT-qPCR in 9 low-grade (stage I–II) glioma tissues, 11 high-grade (stage III–IV) glioma tissues and 14 normal tissues. (H) Negative correlation between SNHG7 and miR-138-5p in glioma tissues. **P<0.01 and ***P<0.001 vs. si-NC, miR-NC or anti-IgG. RT-qPCR, reverse transcription-quantitative PCR; si, small interfering RNA; NC, negative control; lncRNA; long non-coding RNA; SNHG7, small nucleolar RNA host gene 7; miR, microRNA; WT, wild-type; MT, mutant; Ago2, Argonaute 2.
Transfection with the miR-138-5p inhibitor reverses the effect of SNHG7-knockdown in glioma cells
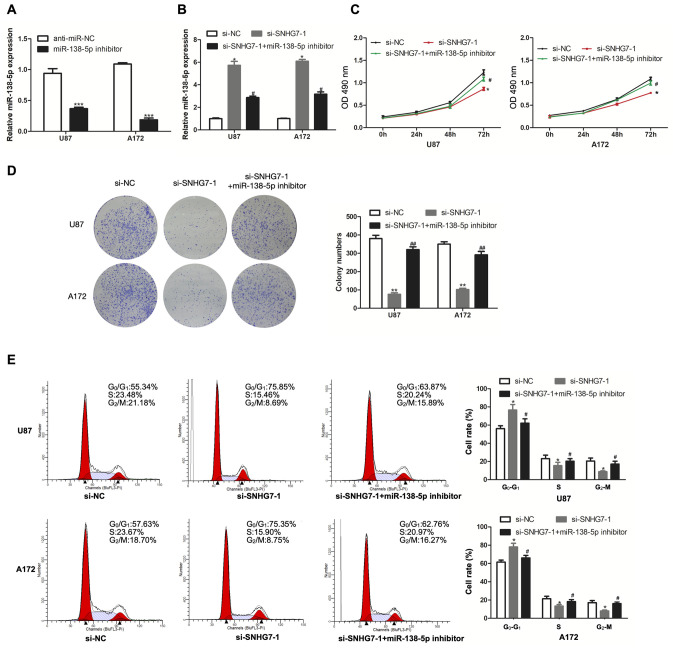
To further investigate whether SNHG7 exerted biological effects on glioma cells by targeting miR-138-5p, miR-138-5p expression was knocked down in U87 and A172 cells by transfection with a miR-138-5p inhibitor, and the transfection efficiency was analyzed using RT-qPCR (Fig. 3A). Compared with the transfection with si-SNHG7 alone, miR-138-5p expression was downregulated to a further extent in U87 and A172 cells co-transfected with si-SNHG7 and miR-138-5p inhibitor (Fig. 3B). The results of the CCK-8 assay demonstrated that transfection with the miR-138-5p inhibitor reversed the SNHG7-knockdown-induced inhibition of proliferation in U87 and A172 cells (Fig. 3C). Notably, the same trend was also observed in the colony formation assay (Fig. 3D). Compared with the si-SNHG7 group, the number of cells in the G1 phase of the cell cycle was decreased in the si-SNHG7 and miR-138-5p inhibitor group (Fig. 3E). Overall, these results suggested that transfection with the miR-138-5p inhibitor may reverse the effect of SNHG7-knockdown in glioma cells.
Figure 3.
miR-138-5p inhibitor reverses the effect of SNHG7-knockdown in glioma cells. (A) Transfection efficiency with miR-138-5p inhibitor was evaluated via RT-qPCR assay. (B) Relative miR-138-5p expression was analyzed via RT-qPCR assay in glioma cells transfected with si-SNHG7 or co-transfected with si-SNHG7 and miR-138-5p inhibitor. (C) Cell proliferation was analyzed by Cell Counting Kit-8 assay in glioma cells after transfection with si-SNHG7 or a combination of si-SNHG7 and miR-138-5p inhibitor. (D) Colony formation assay was performed to evaluate cell proliferation in cells transfected with si-SNHG7 or a combination of si-SNHG7 and miR-138-5p inhibitor. (E) Flow cytometry showed the rate of cell cycle. *P<0.05, **P<0.01 and ***P<0.001 vs. NC groups; #P<0.05 and ##P<0.01 vs. si-SNHG7-1 group. OD, optical density; RT-qPCR, reverse transcription-quantitative PCR; si, small interfering RNA; NC, negative control; SNHG7, small nucleolar RNA host gene 7; miR, microRNA.
SNHG7 regulates EZH2 expression by acting as a ceRNA and sponging miR-138-5p
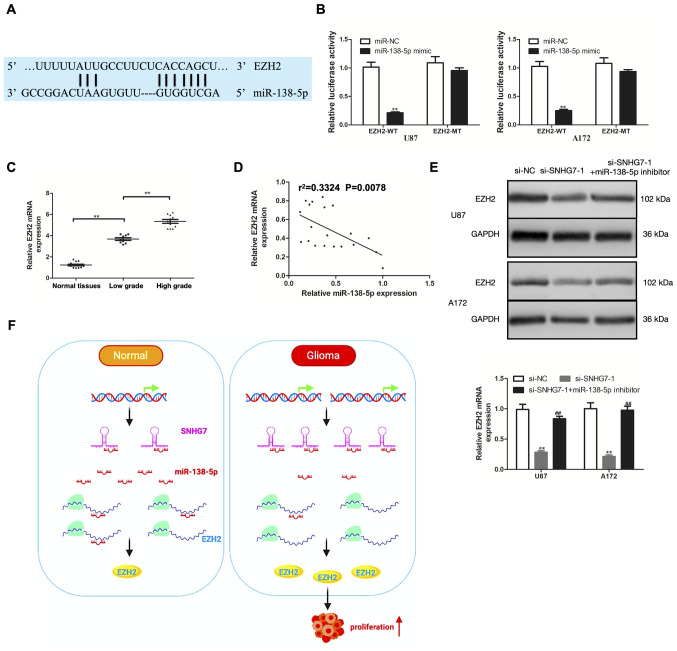
miRNAs are known to act at the post-transcriptional level by directly regulating target gene expression and modulating subsequent protein expression (18). To determine whether SNHG7 regulated EZH2 expression by functioning as a ceRNA and sponging miR-138-5p, bioinformatics analysis was first performed using TargetScan, which predicted the presence of potential miR-138-5p targeting sites in EZH2 (Fig. 4A). According to the results of the dual luciferase reporter assay, the relative luciferase activity was significantly decreased by co-transfection of the miR-138-5p mimic and EZH2-WT vector in U87 and A172 cells, while the relative luciferase activity was unaffected in U87 and A172 cells co-transfected with miR-138-5p mimic and EZH2-MT (Fig. 4B). EZH2 mRNA expression in glioma and normal tissues was subsequently determined using RT-qPCR. The results revealed that EZH2 mRNA expression was significantly upregulated in glioma tissues compared with normal tissues; moreover, compared with in low-grade glioma (stage I–II) tissues, EZH2 mRNA expression was significantly upregulated in high-grade glioma (stage III–IV) tissues (Fig. 4C). In addition, EZH2 mRNA expression was negatively correlated with miR-138-5p expression (Fig. 4D). The role of miR-138-5p in modulating the effects of SNHG7 on EZH2 were further investigated. SNHG7-knockdown markedly downregulated the expression levels of EZH2; however, the expression levels were reversed following the transfection with the miR-138-5p inhibitor (Fig. 4E). These findings suggested that SNHG7 may serve as a ceRNA and may sponge miR-138-5p to modulate EZH2 expression and promote glioma cell proliferation (Fig. 4F).
Figure 4.
SNHG7 regulates EZH2 expression as a competing endogenous RNA by sponging miR-138-5p. (A) Putative binding sequences of EZH2 and miR-138-5p were obtained from TargetScan. (B) Luciferase activity was analyzed by luciferase reporter assay for the targeting of miR-138-5p to EZH2. (C) EZH2 expression was detected by RT-qPCR assay in 14 normal tissues, 9 low-grade (stage I–II) glioma tissues and 11 high-grade (stage III–IV) glioma tissues. (D) Correlation between EZH2 and miR-138-5p was analyzed by Pearson's correlation analysis. (E) EZH2 expression was analyzed by RT-qPCR and western blotting in glioma cells transfected with si-SNHG7 or co-transfected with si-SNHG7 and miR-138-5p inhibitor. (F) Schematic representation of potential mechanism of action. In glioma, SNHG7 may promote glioma cell proliferation by competing with EZH2 to bind miR-138-5p in vitro. **P<0.01 vs. NC groups; ##P<0.01 vs. si-SNHG7-1 group. EZH2, enhancer of zeste homolog 2; RT-qPCR, reverse transcription-quantitative PCR; si, small interfering RNA; NC, negative control; SNHG7, small nucleolar RNA host gene 7; miR, microRNA; WT wild-type; MT, mutant.
Discussion
Glioma is classified into four grades (grade I–IV) according to the level of tumor malignancy by the WHO (19). Standard treatment for glioma includes surgical resection, radiotherapy and chemotherapy. Further potential treatments targeting specific signaling pathways and antigenic profiles of glioma are currently under investigation, which may improve the survival of patients (20). The expression levels of lncRNAs are dysregulated in glioma and are involved in its molecular pathology (10,21).
lncRNAs have been found to influence the prognosis and treatment of numerous types of cancer (22,23). Previous studies have revealed that SNHG7 is involved in the progression of several types of cancer. For example, Zeng et al (24) reported that SNHG7 acted as an oncogene to promote cervical cancer cell proliferation and invasion. In addition, Cheng et al (25) identified an oncogenic role for SNHG7 in promoting the proliferation, migration and invasion of pancreatic cancer cells. Yao et al (26) demonstrated that SNHG7 exerted a tumorigenic role in hepatic cancer cell proliferation and metastasis. The results of the present study revealed that the expression levels of SNHG7 were upregulated in glioma tissues and cells, and were positively associated with tumor grade. Moreover, SNHG7-knockdown decreased the proliferation of glioma cells. These results are consistent with the findings of the aforementioned studies, thereby suggesting that SNHG7 may act as an oncogenic lncRNA to promote cell proliferation in glioma.
It is well established that lncRNAs act as ceRNAs, which sponge and suppress miRNA expression, thereby alleviating their inhibitory effects on mRNA target genes. For example, Wang et al (27) indicated that lncRNA 01121 acted as a ceRNA to promote cell proliferation, migration and invasion in breast cancer. Fan et al (28) discovered that lncRNA FYVE, RhoGEF and PH domain containing 5-antisense RNA 1 upregulated fibroblast growth factor receptor like 1 expression and increased the proliferation of non-small cell lung cancer cells via sponging hsa-miR-107. The present study identified complementary binding sites for SNHG7 in miR-138-5p using the lncRNASNP2 database. In addition, the relative luciferase activity of the SNHG7-WT vector was decreased compared with the SNHG7-MT vector following the co-transfection with the miR-138-5p mimic. The results of the RIP assay revealed a marked enrichment of SNHG7 in the anti-Ago2 group compared with the anti-IgG group, and similar findings were observed for miR-138-5p. In addition, miR-138-5p expression was significantly upregulated in glioma cells transfected with si-SNHG7 compared with si-NC. miR-138-5p expression was significantly downregulated in glioma tissues and was negatively associated with SNHG7 expression. Furthermore, the transfection with the miR-138-5p inhibitor reversed the effects induced by SNHG7-knockdown in glioma cells. The present findings revealed the importance of the association between SNHG7 and miR-138-5p in glioma tumorigenesis. Thus, the current study suggested that SNHG7 may play a carcinogenic role in glioma by promoting cell proliferation, which may be partially rescued by miR-150-5p.
EZH2, which encodes a histone methyltransferase, is an evolutionary conserved gene. EZH2 has been found to have analogous structural motifs and domains (29). As part of the polycomb repressive complex 2, EZH2 catalyzes the tri-methylation of lysine 27 on the Histone H3 protein, which silences downstream target genes (29). Through this mechanism, EZH2 can regulate the cell cycle, cell proliferation, cell differentiation and cancer progression (30). An increasing number of studies have reported that EZH2 plays an oncogenic role in tumor progression (31–33). For instance, Zhang et al (34) revealed that the knockdown of EZH2 inhibited cell cycle progression in glioma cells. Chen et al (35) demonstrated that EZH2, by binding to lncRNA nuclear paraspeckle assembly transcript 1, could promote glioma cell proliferation and invasion. Wu et al (36) indicated that EZH2 was recruited and downregulated by HOXA distal transcript-antisense RNA 2, which facilitated glioma progression. In the present study, the binding sequences between miR-138-5p and EZH2 were identified using TargetScan. The relative luciferase activity of the EZH2-WT vector was decreased compared with the EZH2-MT vector following the co-transfection with the miR-138-5p mimic. The current results indicated that miR-138-5p may target EZH2. In addition, EZH2 mRNA expression was upregulated in glioma tissues and positively associated with tumor grade. Moreover, EZH2 expression was inversely correlated with miR-138-5p expression in glioma tissues. The knockdown of miR-138-5p reversed SNHG7-knockdown-induced downregulation of EZH2 expression and inhibition of cell proliferation in glioma cells.
In conclusion, the findings of the present study suggested that lncRNA SNHG7 may promote the proliferation of glioma by acting as a ceRNA and sponging miR-138-5p to regulate EZH2 expression. The current results indicated that the SNHG7/miR-138-5p/EZH2 signaling axis may represent potential targets for the treatment of glioma. However, the present study had several limitations. For example, the underlying mechanisms of SNHG7 were not investigated in vivo. Therefore, animal experiments should be performed to further explore the mechanisms of SNHG7 in glioma and its potential role as a ceRNA.
Acknowledgements
Not applicable.
Funding Statement
The present study was supported by the National Natural Science Foundation of China (grant no. 82000614), the Natural Science Foundation of Hunan Province (grant no. 2020JJ5876), the Science and Technology Plan Project of Changsha (grant no. kq2004146), the Hunan Provincial Science and Technology Plan Project (grant no. 2019JJ80066) and the Scientific Research Project of Health and Family Planning Commission of Hunan Province of China (grant no. B20-17202).
Funding
The present study was supported by the National Natural Science Foundation of China (grant no. 82000614), the Natural Science Foundation of Hunan Province (grant no. 2020JJ5876), the Science and Technology Plan Project of Changsha (grant no. kq2004146), the Hunan Provincial Science and Technology Plan Project (grant no. 2019JJ80066) and the Scientific Research Project of Health and Family Planning Commission of Hunan Province of China (grant no. B20-17202).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
YD, XM and HZ conceived and designed the experiments. LC and XM performed most of the experiments. ZL collected and analyzed the patient data. YD and HZ statistically analyzed the data. LC wrote the manuscript. YD, XM and HZ confirm the authenticity of all the raw data. All authors read and approved the final version of the manuscript.
Ethics approval and consent to participate
Experimental protocols were approved by Chenzhou First People's Hospital (approval no. 2016-S250) and written informed consent was obtained from all patients or their relatives prior to participation in the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Davis ME. Epidemiology and Overview of Gliomas. Semin Oncol Nurs. 2018;34:420–429. doi: 10.1016/j.soncn.2018.10.001. [DOI] [PubMed] [Google Scholar]
- 2.Wesseling P, Capper D. WHO 2016 Classification of gliomas. Neuropathol Appl Neurobiol. 2018;44:139–150. doi: 10.1111/nan.12432. [DOI] [PubMed] [Google Scholar]
- 3.Bush NA, Chang SM, Berger MS. Current and future strategies for treatment of glioma. Neurosurg Rev. 2017;40:1–14. doi: 10.1007/s10143-016-0709-8. [DOI] [PubMed] [Google Scholar]
- 4.Ulitsky I, Bartel DP. lincRNAs: Genomics, evolution, and mechanisms. Cell. 2013;154:26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sanchez Calle A, Kawamura Y, Yamamoto Y, Takeshita F, Ochiya T. Emerging roles of long non-coding RNA in cancer. Cancer Sci. 2018;109:2093–2100. doi: 10.1111/cas.13642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhan A, Soleimani M, Mandal SS. Long Noncoding RNA and Cancer: A New Paradigm. Cancer Res. 2017;77:3965–3981. doi: 10.1158/0008-5472.CAN-16-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chaudhry MA. Expression pattern of small nucleolar RNA host genes and long non-coding RNA in X-rays-treated lymphoblastoid cells. Int J Mol Sci. 2013;14:9099–9110. doi: 10.3390/ijms14059099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ota T, Suzuki Y, Nishikawa T, Otsuki T, Sugiyama T, Irie R, Wakamatsu A, Hayashi K, Sato H, Nagai K, et al. Complete sequencing and characterization of 21,243 full-length human cDNAs. Nat Genet. 2004;36:40–45. doi: 10.1038/ng1285. [DOI] [PubMed] [Google Scholar]
- 9.Zhou Y, Tian B, Tang J, Wu J, Wang H, Wu Z, Li X, Yang D, Zhang B, Xiao Y, et al. SNHG7: A novel vital oncogenic lncRNA in human cancers. Biomed Pharmacother. 2020;124:109921. doi: 10.1016/j.biopha.2020.109921. [DOI] [PubMed] [Google Scholar]
- 10.Ren J, Yang Y, Xue J, Xi Z, Hu L, Pan SJ, Sun Q. Long noncoding RNA SNHG7 promotes the progression and growth of glioblastoma via inhibition of miR-5095. Biochem Biophys Res Commun. 2018;496:712–718. doi: 10.1016/j.bbrc.2018.01.109. [DOI] [PubMed] [Google Scholar]
- 11.Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 12.Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang X, Zhao Y, Cao W, Wang C, Sun B, Chen J, Li S, Chen J, Cui M, Zhang B, et al. miR-138-5p acts as a tumor suppressor by targeting hTERT in human colorectal cancer. Int J Clin Exp Pathol. 2017;10:11516–11525. [PMC free article] [PubMed] [Google Scholar]
- 14.Ou L, Wang D, Zhang H, Yu Q, Hua F. Decreased Expression of miR-138-5p by lncRNA H19 in Cervical Cancer Promotes Tumor Proliferation. Oncol Res. 2018;26:401–410. doi: 10.3727/096504017X15017209042610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: The Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD, Kleihues P, Ellison DW. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A summary. Acta Neuropathol. 2016;131:803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 17.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Δ Δ C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 18.Beermann J, Piccoli MT, Viereck J, Thum T. Non-coding RNAs in Development and Disease: Background, Mechanisms, and Therapeutic Approaches. Physiol Rev. 2016;96:1297–1325. doi: 10.1152/physrev.00041.2015. [DOI] [PubMed] [Google Scholar]
- 19.Komori T. The 2016 WHO Classification of Tumours of the Central Nervous System: The Major Points of Revision. Neurol Med Chir (Tokyo) 2017;57:301–311. doi: 10.2176/nmc.ra.2017-0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weller M, Wick W, Aldape K, Brada M, Berger M, Pfister SM, Nishikawa R, Rosenthal M, Wen PY, Stupp R, et al. Glioma. Nat Rev Dis Primers. 2015;1:15017. doi: 10.1038/nrdp.2015.17. [DOI] [PubMed] [Google Scholar]
- 21.Pop S, Enciu AM, Necula LG, Tanase C. Long non-coding RNAs in brain tumours: Focus on recent epigenetic findings in glioma. J Cell Mol Med. 2018;22:4597–4610. doi: 10.1111/jcmm.13781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xi J, Sun Q, Ma L, Kang J. Long non-coding RNAs in glioma progression. Cancer Lett. 2018;419:203–209. doi: 10.1016/j.canlet.2018.01.041. [DOI] [PubMed] [Google Scholar]
- 23.Vecera M, Sana J, Lipina R, Smrcka M, Slaby O. Long Non-Coding RNAs in Gliomas: From Molecular Pathology to Diagnostic Biomarkers and Therapeutic Targets. Int J Mol Sci. 2018;19:E2754. doi: 10.3390/ijms19092754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zeng J, Ma YX, Liu ZH, Zeng YL. lncRNA SNHG7 contributes to cell proliferation, invasion and prognosis of cervical cancer. Eur Rev Med Pharmacol Sci. 2019;23:9277–9285. doi: 10.26355/eurrev_201911_19420. [DOI] [PubMed] [Google Scholar]
- 25.Cheng D, Fan J, Ma Y, Zhou Y, Qin K, Shi M, Yang J. lncRNA SNHG7 promotes pancreatic cancer proliferation through ID4 by sponging miR-342-3p. Cell Biosci. 2019;9:28. doi: 10.1186/s13578-019-0290-2. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26.Yao X, Liu C, Liu C, Xi W, Sun S, Gao Z. lncRNA SNHG7 sponges miR-425 to promote proliferation, migration, and invasion of hepatic carcinoma cells via Wnt/β-catenin/EMT signalling pathway. Cell Biochem Funct. 2019;37:525–533. doi: 10.1002/cbf.3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Z, Wang P, Cao L, Li F, Duan S, Yuan G, Xiao L, Guo L, Yin H, Xie D, et al. Long Intergenic Non-Coding RNA 01121 Promotes Breast Cancer Cell Proliferation, Migration, and Invasion via the miR-150-5p/HMGA2 Axis. Cancer Manag Res. 2019;11:10859–10870. doi: 10.2147/CMAR.S230367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fan Y, Li H, Yu Z, Dong W, Cui X, Ma J, Li S. Long non-coding RNA FGD5-AS1 promotes non-small cell lung cancer cell proliferation through sponging hsa-miR-107 to up-regulate FGFRL1. Biosci Rep. 2020;40:BSR20193309. doi: 10.1042/BSR20193309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gan L, Yang Y, Li Q, Feng Y, Liu T, Guo W. Epigenetic regulation of cancer progression by EZH2: From biological insights to therapeutic potential. Biomark Res. 2018;6:10. doi: 10.1186/s40364-018-0122-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gall Trošelj K, Novak Kujundzic R, Ugarkovic D. Polycomb repressive complex's evolutionary conserved function: The role of EZH2 status and cellular background. Clin Epigenetics. 2016;8:55. doi: 10.1186/s13148-016-0226-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu Y, Sun J, Yu J, Ge W, Xiao X, Dai S, Xiang Q. lncRNA CACS15 accelerates the malignant progression of ovarian cancer through stimulating EZH2-induced inhibition of APC. Am J Transl Res. 2019;11:6561–6568. [PMC free article] [PubMed] [Google Scholar]
- 32.Xu M, Chen X, Lin K, Zeng K, Liu X, Xu X, Pan B, Xu T, Sun L, He B, et al. lncRNA SNHG6 regulates EZH2 expression by sponging miR-26a/b and miR-214 in colorectal cancer. J Hematol Oncol. 2019;12:3. doi: 10.1186/s13045-018-0690-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao L, Sun H, Kong H, Chen Z, Chen B, Zhou M. The lncrna-TUG1/EZH2 Axis Promotes Pancreatic Cancer Cell Proliferation, Migration and EMT Phenotype Formation Through Sponging Mir-382. Cell Physiol Biochem. 2017;42:2145–2158. doi: 10.1159/000479990. [DOI] [PubMed] [Google Scholar]
- 34.Zhang K, Sun X, Zhou X, Han L, Chen L, Shi Z, Zhang A, Ye M, Wang Q, Liu C, et al. Long non-coding RNA HOTAIR promotes glioblastoma cell cycle progression in an EZH2 dependent manner. Oncotarget. 2015;6:537–546. doi: 10.18632/oncotarget.2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen Q, Cai J, Wang Q, Wang Y, Liu M, Yang J, Zhou J, Kang C, Li M, Jiang C. Long Noncoding RNA NEAT1, Regulated by the EGFR Pathway, Contributes to Glioblastoma Progression Through the WNT/β-Catenin Pathway by Scaffolding EZH2. Clin Cancer Res. 2018;24:684–695. doi: 10.1158/1078-0432.CCR-17-0605. [DOI] [PubMed] [Google Scholar]
- 36.Wu L, Zhu X, Song Z, Chen D, Guo M, Liang J, Ding D, Wang W, Yan D. Long Non-Coding RNA HOXA-AS2 Enhances The Malignant Biological Behaviors In Glioma By Epigenetically Regulating RND3 Expression. OncoTargets Ther. 2019;12:9407–9419. doi: 10.2147/OTT.S225678. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.