Abstract
Objective:
Hepatocyte nuclear factor 4α (HNF4A) has been demonstrated to be an oncogene in gastric cancer (GC). However, the roles of different HNF4A isoforms derived from the 2 different promoters (P1 and P2) and the underlying mechanisms remain obscure.
Methods:
The expression and prognostic values of P1- and P2-HNF4A were evaluated in The Cancer Genome Atlas (TCGA) databases and GC tissues. Then, functional assays of P1- and P2-HNF4A were conducted both in vivo and in vitro. High-throughput RNA-seq was employed to profile downstream pathways in P1- and P2-HNF4A-overexpressing GC cells. The expression and gene regulation network of the candidate target genes identified by RNA-seq were characterized based on data mining and functional assays.
Results:
HNF4A amplification was a key characteristic of GC in TCGA databases, especially for the intestinal type and early stage. Moreover, P1-HNF4A expression was significantly higher in tumor tissues than in adjacent non-tumor tissues (P < 0.05), but no significant differences were found in P2-HNF4A expression (P > 0.05). High P1-HNF4A expression indicated poor prognoses in GC patients (P < 0.01). Furthermore, P1-HNF4A overexpression significantly promoted SGC7901 and BGC823 cell proliferation, invasion and migration in vitro (P < 0.01). Murine xenograft experiments showed that P1-HNF4A overexpression promoted tumor growth (P < 0.05). Mechanistically, RNA-seq showed that the cytokine-cytokine receptor interactions pathway was mostly enriched in P1-HNF4A-overexpressing GC cells. Finally, chemokine (C-C motif) ligand 15 was identified as a direct target of P1-HNF4A in GC tissues.
Conclusions:
P1-HNF4A was the main oncogene during GC progression. The cytokine-cytokine receptor interaction pathway played a pivotal role and may be a promising therapeutic target.
Keywords: Gastric cancer, carcinogenesis, HNF4A, CCL15, transcriptomics
Introduction
Gastric cancer (GC) remains one of the most common malignancies, and one of the most frequent causes of cancer-related deaths1. Due to a lack of specific clinical manifestations in the early stage, most GC patients are generally diagnosed at an advanced stage with an overall 5-year survival of less than 20%, especially in China2. Therefore, increasing an understanding of GC pathogenesis may identify novel strategies for more effective treatments.
Hepatocyte nuclear factor 4α (HNF4A) has been previously shown to be an oncogene in GC3–5. Mechanistically, HNF4A is a target of NF-κB6, and interacts with the Wnt pathway3 and controls tumor metabolism required for GC progression through (isocitrate dehydrogenase 1) IDH15. However, the specific target genes and downstream signaling pathways of HNF4A remain unknown. In addition, a total 9 HNF4A isoforms (HNF4A1-9), transcribed from P1 or P2 promoters, have been shown to display distinct tissue expressions and functions7,8 (Figure 1A). Although both P1- and P2-HNF4A are present in GC tissues9, it remains unclear which splice variant is the most relevant.
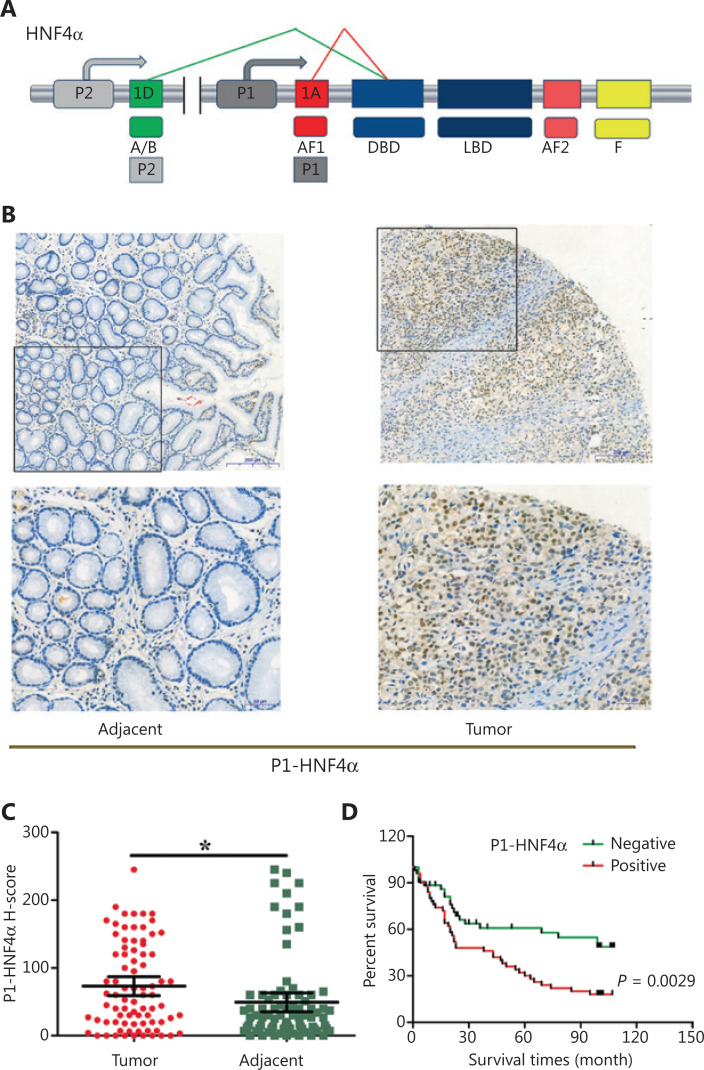
Figure 1.
Expression and prognostic value of P1-HNF4A in gastric cancer (GC). (A) Schematic of the HNF4A gene and its isoforms (P1 and P2). The antigens recognized by isoform-specific (P1 and P2) antibodies are labeled. (B) Representative expression of P1-HNF4A in tumor and adjacent non-tumor tissues of GC patients. Scale bars = 200 μm and 50 μm. (C) H-score of P1-HNF4A in tumor and adjacent non-tumor tissues using tissue microarray and immunohistochemical analyses (*P < 0.05). (D) The prognostic value of P1-HNF4A in GC patients according to negative and positive expression (P < 0.01).
In this study, we initially determined the expressions of both P1- and P2-HNF4A using TCGA databases and tissue microarrays. Then, we performed transcriptome-wide mapping of both P1- and P2-HNF4A targets in GC cell lines. Unbiased analysis revealed that the cytokine-cytokine receptor interaction and PPAR signaling pathway genes were most enriched in P1- and P2-dependent manners. We also identified chemokine (c-c motif) ligand 15 (CCL15), a member of the chemokine family, as a direct target gene of P1-HNF4A, which was required for GC progression.
Materials and methods
Cell culture
The human GC cell lines, AGS, MKN45, BGC823, AZ521, N87, and SGC7901, and the normal gastric epithelial cell line, GES-1 (American Type Culture Collection, Manassas, VA, USA) were cultured in RPMI 1640 medium (Thermo Fisher Scientific, Waltham, MA, USA) at 37 °C with a humidified atmosphere of 5% CO2 supplemented with 10% fetal bovine serum (Biological Industries, Kibbutz Beit Haemek, Israel) and 1% penicillin-streptomycin solution (Thermo Fisher Scientific). All cell line identities were verified through STR DNA profiling and were negative for mycoplasma contamination.
Cell transfection
HNF4A2 (NM_000457) and HNF4A8 (NM_175914) overexpression lentiviruses and a negative control were constructed by GeneCopoeia (Rockville, MD, USA). Human CCL15-shRNA lentivirus and negative controls were constructed by GenePharma Technologies (Shanghai, China) (Table 1). Logarithmic phase cells (1 × 105) were seeded in 24-well culture plates. After adherence, lentiviral vectors were added at a final concentration of ˜50 multiplicity of infection (MOI) with 0.5 μg/mL polybrene for 10 h at 37 °C. Fresh medium was replaced 10 h after transfection. The cells were transferred into 25 cm2 flasks after reaching 70%–80% confluence. Then, the cell culture medium was replaced every 2–3 days with 2 μg/mL puromycin-containing RPMI 1640 medium.
Table 1.
The sequences of shRNAs used in this study
| Gene symbols | Sequences | 5′-3′ |
|---|---|---|
| Human CCL15-121 | Sense | CCUUACACAUGAAGAGGCATT |
| Antisense | UGCCUCUUCAUGUGUAAGGTT | |
| Human CCL15-222 | Sense | CAGCCAGAAAGCACUACAATT |
| Antisense | UUGUAGUGCUUUCUGGCUGTT | |
| Human CCL15-291 | Sense | AGCCCTTGAGTCCGGTGTCTT |
| Antisense | AAGACACCGGACTCAAGGGCT | |
| Negative control | Sense | UUCUCCGAACGUGUCACGUTT |
| Antisense | ACGUGACACGUUCGGAGAATT |
shRNAs, short hairpin RNA; HNF4A, hepatocyte nuclear factor 4α; CCL15, chemokine (C-C motif) ligand 15.
Cell proliferation assay
Cell proliferation was examined using CCK-8 kits (Thermo Fisher Scientific) according to the manufacturer’s instructions, to determine cell growth. A total of 103 target cells were seeded in 96-well plates for the assays in 100 μL of complete medium. Then, 10 μL of CCK-8 reagent was added to the wells and incubated for 2 h. The cultures were assayed each day for 4 continuous days, and the absorbance was read at 450 nm with a reference wavelength at 650 nm using a Varioskan Flash Multimode Reader (Thermo Fisher Scientific). Each experiment was performed in triplicate and repeated 3 times.
Cell migration and invasion assays
Cell migration experiments were performed in a transwell chamber covered with polycarbonate membranes (Corning, NY, NY, USA). Cell invasion assays were performed using chambers coated with Matrigel matrix (BD Science, Sparks, MD, USA). Resuspended cells (1 × 105 cells/well) were added to the upper chambers and cultured for 24 h. Migrated or invaded cells were stained with 0.1% Crystal Violet for 20 min at room temperature. Quantification of migrated or invaded cells was performed using ImageJ software (National Institutes of Health, Bethesda, MD, USA).
Cell cycle assay
Target cells were seeded in 6 cm plates and harvested after reaching approximately 70%–80% confluence. Then, resuspended cells were fixed in 75% ethanol, stained with propidium iodide according to the manufacturer’s protocol (Sigma-Aldrich, St. Louis, MO, USA) and sorted using fluorescence-activated cell sorting (BD Sciences, San Jose, CA, USA). Data were analyzed using ModFit software (BD Sciences).
Murine xenograft model
Animal experiments were approved by the Institutional Animal Care and Use Committee of the Air Force Medical University of PLA (Approval No. 2018-kq-010). Briefly, 6- to 8-week-old BALB/c nude female mice (Shanghai SLAC Laboratory Animal Co., Shanghai, China) were subcutaneously implanted with 1 × 107 SGC7901 cells transfected with a stable control, P1- or P2-HNF4A (n = 5). Then, the animals were euthanized and the tumor volumes were calculated using the formula (V = length × width2 × 0.5) after 2 weeks. The tumor tissues were then harvested and processed for further hematoxylin and eosin staining and immunohistochemistry (IHC) analysis.
Protein lysate preparation and Western blot
Cells were digested with RIPA cell lysis buffer (Beyotime Biotechnology, Shanghai, China) with protease and phosphatase inhibitor cocktails (MCE, Shanghai, China) and were collected using a cell scraper. Total protein was extracted and quantified using the bicinchoninic acid method (Thermo Fisher Scientific). Proteins (20–30 μg) were resolved using 10% PAGE (Bio-Rad, Hercules, CA, USA), transferred to a nitrocellulose membrane (Pall Corporation, Port Washington, NY, USA) at 25 V for 30 min, and blocked for 1 h in 10% nonfat milk in 1× TBS/0.1% (v/v) Tween 20 at room temperature. Primary antibodies (GAPDH, 1:1,000, #5174; Cell Signaling Technology; β-actin, 1:1,000, #4970; Cell Signaling Technology; P1-HNF4A, 1:1,000, ab41898; Abcam; P2-HNF4A, 1:1,000, PP-H6939-00; R & D Systems; and CCL15, 1:1,000, ab219388; Abcam) were added and incubated overnight at 4 °C. Secondary antibodies (goat anti-rabbit IgG, HRP #7074 or goat anti-mouse IgG, HRP, 1:2,000, #7076; Cell Signaling Technology) were incubated at room temperature for 1 h. Signals were detected using Western Lumax Light Sirius HRP substrate reagent (ZETA-Life, San Francisco, CA, USA). All data were normalized to human β-actin or GAPDH. The bands were scanned using a ChemiDocXRS + Imaging System (Bio-Rad).
RNA-seq
Total RNA was extracted using the mirVana miRNA Isolation Kit (Ambion, Austin, TX, USA). RNA integrity was evaluated using the Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). The samples with RNA integrity number (RIN) ≥ 7 were subjected to subsequent analysis. The libraries were constructed using the TruSeq Stranded mRNA LT Sample Prep Kit (Illumina, San Diego, CA, USA). Then, these libraries were sequenced using the Illumina HiSeq X Ten platform, and 125 bp/150 bp paired-end reads were generated.
Transcriptome sequencing and analysis were conducted by OE Biotech (Shanghai, China). Raw data were processed using Trimmomatic (Illumina). Differentially-expressed genes (DEGs) were identified using the DESeq (2012) R package (Bioconductor; http://www.bioconductor.org/). A P value < 0.05 and fold changes > 2 or fold changes < 0.5 was set as the thresholds for significant differential expression. Hierarchical cluster analysis of DEGs was performed to characterize gene expression patterns. Gene Ontology (GO) enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses of DEGs were performed using R based on the hypergeometric distribution.
Quantitative real-time PCR
Total RNA was extracted using the RNeasy Mini Kit (QIAGEN, Duesseldorf, Germany), according to the manufacturer’s instructions. In total, 0.5 μg RNA was synthesized into cDNA using the PrimeScript RT Reagent Kit (TaKaRa, Shiga, Japan) and Mir-X mRNA First-Strand Synthesis Kit (TaKaRa) in a 10 μL volume. Real-time PCR was performed on a CFX96 system using TB Green Premix Ex Taq II (TaKaRa) with 2 μL cDNA and 0.8 μL primers in a final volume of 20 μL. The final PCR conditions were as follows: predenaturation at 95 °C for 10 min, followed by 44 cycles at 95 °C denaturation for 10 s, 60 °C annealing for 20 s, and 72 °C extension for 10 s. The target mRNA gene was normalized to human GAPDH using the 2-ΔΔCT method. Primer sequences are shown in Table 2.
Table 2.
The sequences of primers used in this study
| Gene symbols | Primers | 5′-3′ |
|---|---|---|
| HNF4A | Forward | GTTCAAGGACGTGCTGCTCCTA |
| Reverse | AGGCATACTCATTGTCATCGATCTG | |
| CCL15 | Forward | AGGCCCAGTTCACAAATGA |
| Reverse | CAGTCAGCAGCAAAGTGAAAG | |
| GAPDH | Forward | GCACCGTCAAGGCTGAGAAC |
| Reverse | TGGTGAAGACGCCAGTGGA | |
| CCL15 Promoter 1 | Forward | TAACCTGTGAGCCTTCGATG |
| Reverse | GTGACCTATGATCACACCACTG | |
| CCL15 Promoter 2 | Forward | GGAAGGTCTTCTAGCCTCTAATC |
| Reverse | CACTGCAAACTCCACCTTCTG | |
| CCL15 Promoter 3 | Forward | TAGGCTCAAGTATGTGCACTA |
| Reverse | GGTTTGACCTTTGGATTGGG | |
| CCL15 Promoter 4 | Forward | GAGTAGTCTTGGAAAAGGCAAC |
| Reverse | GTAATACCAGTACTTTGGGAG |
HNF4A, hepatocyte nuclear factor 4α; CCL15, chemokine (C-C motif) ligand 15.
Tissue microarrays and immunohistochemistry
GC tissue microarrays, including 98 cases of gastric tumor and adjunct non-tumor tissues, were obtained from Outdo Biotech (HStmA180Su15; Shanghai, China). Patient characteristics are shown in Table 3. This study was approved by the Institutional Ethics Committee of Xijing Hospital of the Air Force Medical University of PLA (Approval No. KY20183093-1).
Table 3.
Clinical characteristics of the patients analyzed using tissue microarrays
| Characteristics | n | P1-HNF4A Negative |
Positive |
P |
CCL15 Low |
High |
P |
|---|---|---|---|---|---|---|---|
| Age (years) | > 0.05 | > 0.05 | |||||
| < 60 | 32 | 20 | 12 | 16 | 16 | ||
| ≥ 60 | 66 | 27 | 39 | 27 | 39 | ||
| Gender | > 0.05 | > 0.05 | |||||
| Male | 62 | 30 | 32 | 29 | 33 | ||
| Female | 36 | 16 | 20 | 12 | 24 | ||
| TNM stage | > 0.05 | > 0.05 | |||||
| I+II | 39 | 17 | 22 | 20 | 19 | ||
| III+IV | 59 | 29 | 30 | 22 | 37 | ||
| T stage | > 0.05 | > 0.05 | |||||
| T1+T2 | 14 | 5 | 9 | 8 | 6 | ||
| T3+T4 | 84 | 42 | 42 | 34 | 50 | ||
| LN metastasis | > 0.05 | < 0.05 | |||||
| No | 26 | 10 | 16 | 16 | 10 | ||
| Yes | 72 | 36 | 36 | 25 | 47 | ||
| Distant metastasis | > 0.05 | > 0.05 | |||||
| No | 89 | 40 | 49 | 38 | 51 | ||
| Yes | 9 | 6 | 3 | 3 | 6 | ||
| WHO type | > 0.05 | > 0.05 | |||||
| Adenocarcinoma | 47 | 20 | 27 | 15 | 32 | ||
| TA | 17 | 5 | 12 | 4 | 13 | ||
| MA | 10 | 8 | 2 | 5 | 5 | ||
| SRC | 14 | 8 | 6 | 6 | 8 | ||
| UC | 10 | 4 | 6 | 1 | 9 |
TA, tubular adenocarcinoma; MA, mucus adenocarcinoma; SRC, signet ring cell carcinoma; UC, undifferentiated carcinoma.
Sections were dried overnight at 60 °C. Antigen retrieval was performed by heating in 1× citrate for 2 min and then cooling to room temperature. Endogenous peroxidase activity was blocked using 3% H2O2 for 10 min. Slides were incubated with mouse anti-human antibodies against Ki67 (1:1,000, ab92742), P1-HNF4A (1:100, ab41898; Abcam), P2-HNF4A (1:100, PP-H6939-00; R & D Systems), and CCL15 (1:200, ab219388; Abcam) at 4 °C overnight. The sections were then counterstained with hematoxylin staining and dehydrated.
The slides were scanned and viewed using a Pannoramic Viewer (3DHISTECH, Budapest, Hungary). The staining intensity of P1- and P2-HNF4A was semiquantitatively determined using the H-score method10. Only unequivocally stained nuclei were considered positive. H-scores < 50 and ≥ 50 were considered as negative and positive, respectively. H-scores < 150 and ≥ 150 were considered low and high expressions, respectively. CCL15 staining was semiquantitatively performed according to the staining intensity and percentage of positive cells11. IHC scores < 6 and ≥ 6 were considered low and high expressions, respectively.
Dual-luciferase reporter assays
Briefly, 2,000 bp fragments of the human CCL15 promoter were obtained from Ensembl and predicted in the JASPAR database. The wild-type (WT) and corresponding mutational CCL15 promoter fragments covering predicted DR1 sites were PCR-amplified and cloned into the firefly luciferase reporter plasmid, pGL3-basic vector (Promega, Madison, WI, USA). Luciferase activity was then detected using a Dual-Luciferase Reporter Assay Kit (Promega) at 48 h after reporter transfection using Lipofectamine 2000 Transfection Reagent (Invitrogen, Carlsbad, CA, USA) in SGC7901 cells. Firefly luciferase activity was normalized to Renilla luciferase activity, and the final data are presented as the fold induction of luciferase activity compared to that of the negative control.
Chromatin immunoprecipitation (ChIP)
ChIP assays were performed using the EZ ChIP™ Kit (Millipore, Billerica, MA, USA). The cells were cross-linked with 1% formaldehyde for 10 min at 37 °C and quenched with 2.5 M glycine for 5 min at room temperature. DNA was immunoprecipitated from the sonicated cell lysates using HNF4A antibody (Abcam, Cambridge, MA, USA) and subjected to PCR to amplify the HNF4 binding site (Table 2). The amplified fragments were analyzed on an agarose gel. A nonspecific antibody against IgG served as a negative control.
Data mining using public databases
The mRNA expression and DNA copy number of HNF4A in GC were analyzed within the Oncomine (http://oncomine.org), GEPIA (http://gepia.cancer-pku.cn), and Ualcan (http://ualcan.path.uab.edu) databases. The GEPIA and Ualcan databases were used to analyze candidate gene alterations that were identified as being enriched in RNA-seq. We further used the LinkedOmics (http://www.linkedomics.org), cBioPortal (http://cbioportal.org), and GEPIA databases to characterize the relationships between HNF4A and candidate target genes.
Statistical analysis
All cell culture experiments were performed in triplicate to reduce error and ensure reproducibility. All quantitative data are expressed as the mean ± SEM. Differences between two groups were examined using an unpaired Student’s t-test. Differences between multiple groups were compared by one-way analysis of variance with Dunnett’s post hoc tests. All categorical data are expressed as rates. Differences between two groups were examined using the χ2 test. Statistical analysis was performed using SPSS 13.0 statistical software for Windows (SPSS, Chicago, IL, USA) or GraphPad Prism 5.0 (GraphPad Software, San Diego, CA, USA). P < 0.05 was considered statistically significant.
Results
The expression levels of HNF4A in TCGA databases
Initially, the mRNA expression and DNA copy number of HNF4A in GC were analyzed within the Oncomine, GEPIA, and Ualcan databases. Data in the Oncomine database revealed that mRNA expression and DNA copy number variation of HNF4A were significantly higher in GC tissues than in normal tissues in TCGA Gastric, Deng Gastric, Cho Gastric, and DErrico Gastric (P < 0.01), especially for intestinal type adenocarcinomas (P < 0.01) (Supplementary Figure S1). Data in the Ualcan database also showed significantly higher expression levels of HNF4A in tumor tissues than in non-tumor tissues (Supplementary Figure S2A). Early stages showed significantly higher HNF4A expression levels than those in late stages (Supplementary Figure S2B and S2C). Finally, data in the GEPIA database also revealed significantly higher HNF4A expression levels in tumor tissues than in non-tumor tissues (Supplementary Figure S3A). Furthermore, isoform analysis indicated that HNF4A1 and HNF4A2 expressions were significantly higher than HNF4A7 and HNF4A8 expressions (Supplementary Figure S3B). Together, these results showed that HNF4A amplification was common in GC, especially in the intestinal type and early stages. In addition, P1-HNF4A upregulation was more remarkable than P2-HNF4A upregulation.
The expression levels of P1- and P2-HNF4A in gastric tumors and adjacent tissues
Although high HNF4A expression has been shown in GC tissues3–5, previous studies did not distinguish the functions of different HNF4A isoforms. We therefore detected the expression levels of both P1- and P2-HNF4A in gastric tumors and adjacent non-tumor tissues using tumor microarrays. The results indicated that P1-HNF4A expression was significantly increased in tumor tissues compared with non-tumor tissues (P < 0.05) (Figure 1B and 1C). However, no significant difference was noted in P2-HNF4A expression (P > 0.05) (Supplementary Figure S4A and S4B), although the H-score of P2-HNF4A was higher in non-tumor tissues than in tumor tissues (P > 0.05) (Supplementary Figure S4B). Next, we examined P1-HNF4A expression according to cancer clinical features. As shown in Table 2, P1-HNF4A expression levels showed an upregulation in the I+II, T1+T2, M0, and N0 stages, but no significant difference was noted (P > 0.05). Finally, patients with positive P1-HNF4A staining showed a significantly shorter survival compared with negative cases (P < 0.05) (Figure 1D). However, no significant difference was noted between low and high P2-HNF4A expression patients (Supplementary Figure S4C). Together, these results indicated that P1- and P2-HNF4A might play different roles during GC progression. P1-HNF4A expression was the main isoform in gastric carcinogenesis and predicted poor prognoses. However, no significant difference was observed in P2-HNF4A expression levels.
P1- and P2-HNF4A showed distinct functions both in vitro and in vivo
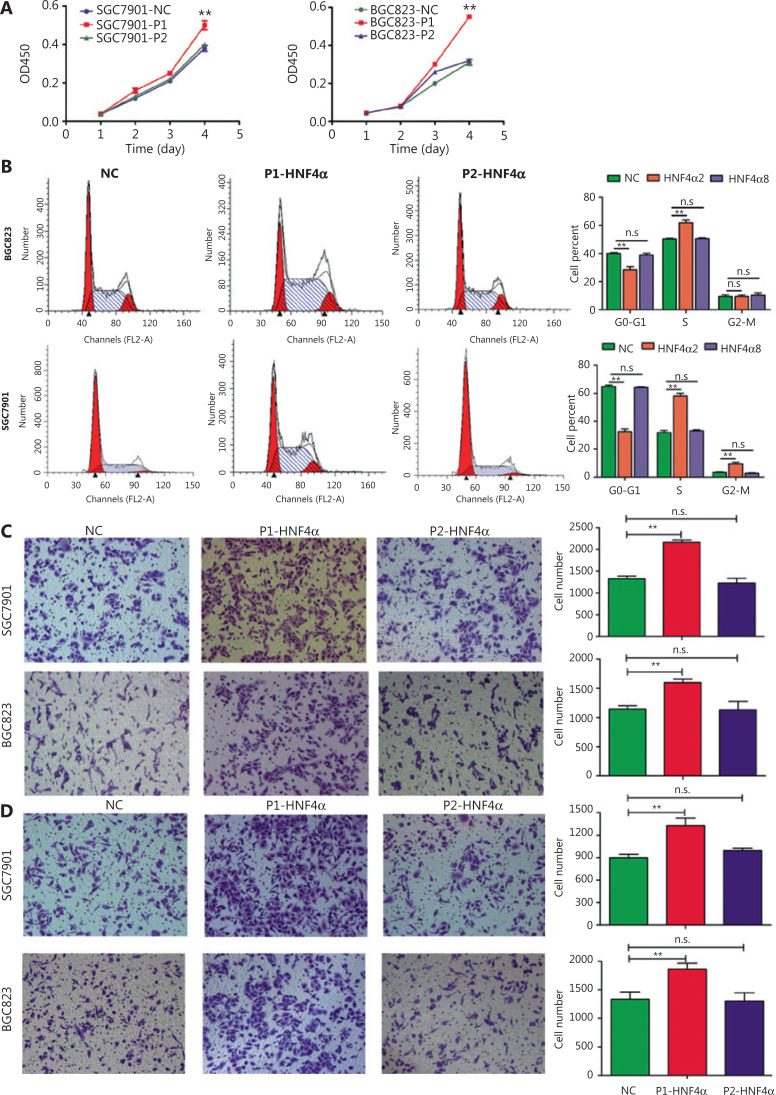
We further examined the roles of both P1- and P2-HNF4A in GC cells in vitro and in vivo. First, we detected the expression levels of P1- and P2-HNF4A in GC cell lines. Western blot results indicated that P1- and P2-HNF4A expression levels showed dramatic differences (Supplementary Figure S5A). Then, we directly introduced P1-HNF4A (HNF4A2) and P2-HNF4A (HNF4A8) into SGC7901 and BGC823 cells, respectively, 2 cell lines with relatively low P1- and P2-HNF4A expressions (Supplementary Figure S5B). The CCK-8 assay indicated that P1-HNF4A overexpression promoted GC cell proliferation (P < 0.01) (Figure 2A). However, no significant difference was noted after P2-HNF4A overexpression (P > 0.05) (Figure 2A). Furthermore, flow cytometry analysis showed that P1-HNF4A overexpression promoted G1/S phase arrest (P < 0.01), and P2-HNF4A overexpression showed no significant effect on the cell cycle (P > 0.05) (Figure 2B). Furthermore, P1- and P2-HNF4A promoted ERK1/2 pathway activation, with a more significant tendency in P1-HNF4A-overexpressing cells (Supplementary Figure S5B).
Figure 2.
The effects of both P1- and P2-HNF4A on malignant phenotypes of gastric cancer cells in vitro. (A) P1-HNF4A or P2-HNF4A overexpression lentivirus were transfected into SGC7901 and BGC823 cells. Cell proliferation was examined using the CCK8 assay. OD450 absorbance was detected every day for a continuous 4 days (**P < 0.01, n.s., nonsignificant, n = 3). (B) The effects of P1- and P2-HNF4A overexpression on SGC7901 and BGC823 cell cycles using a flow cytometry assay (**P < 0.01; n.s., nonsignificant, n = 3). (C) The effects of both P1- and P2-HNF4A overexpression on SGC7901 and BGC823 cell migrations using transwell experiments. Migrated cells were counted using ImageJ (**P < 0.01; n.s., nonsignificant, n = 3) (Crystal Violet staining, 20×). (D) The effects of both P1- and P2-HNF4A overexpression on SGC7901 and BGC823 cell invasion using transwell experiments. Invaded cells were counted by ImageJ (**P < 0.01; n.s., nonsignificant, n = 3).
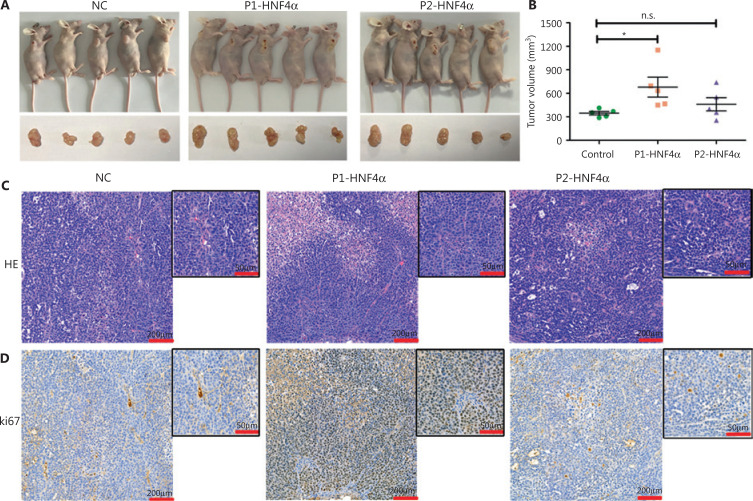
Next, transwell invasion and migration assays also showed that P1-HNF4A overexpression facilitated GC cell invasion and migration (P < 0.01), but P2-HNF4A overexpression showed no effect (P > 0.05) (Figure 2C and 2D). Moreover, we performed an in vivo xenograft mouse experiment using stably transfected SGC7901 cells. The results showed that the average tumor volume in the P1-HNF4A group was significantly larger than that in the control group (P < 0.05) (Figure 3A and 3B). However, no significant difference was found in the average tumor volume in the P2-HNF4A group compared with the control (P > 0.05) (Figure 3A and 3B). In addition, tumors from P1-HNF4A overexpression mice showed more significant Ki67 expression than those from P2-HNF4A overexpression mice (Figure 3C and 3D).
Figure 3.
The roles of both P1- and P2-HNF4A on gastric cancer (GC) progression in vivo using murine xenograft experiments. (A) BALB/c nude mice underwent orthotopic implantation with SGC7901 cells stably transfected by a negative control, P1-HNF4A, and P2-HNF4A overexpression lentiviruses. (B) The tumors were extracted and measured. Tumor volume was calculated using the formula (V = length × width2 × 0.5). The results are expressed as the mean ± SEM (*P < 0.05, n.s., nonsignificant). (C) H&E staining of sections from orthotopic specimens. Scar bars = 200 μm and 50 μm). (D) Ki67 staining of section from orthotopic specimens by immunohistochemistry. Scale bars = 200 μm and 50 μm).
Taken together, the above in vitro and in vivo results indicated that P1-HNF4A could significantly promote GC progression, and that P2-HNF4A showed no significant effect.
RNA-seq showed distinct pathway and gene enrichment in P1- and P2-HNF4A-overexpressing cells
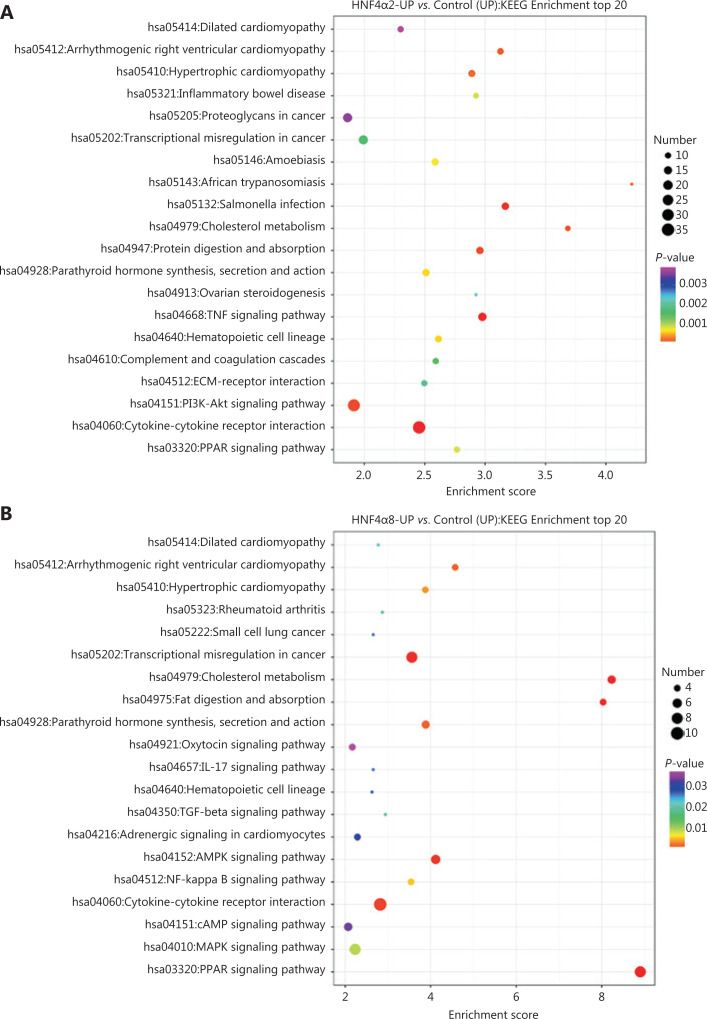
To investigate the underlying mechanisms mediating P1- and P2-HNF4A functions, we performed RNA-seq using P1- and P2-HNF4A-overexpressing SGC7901 cells. For this purpose, we transfected SGC7901 cells with P1-HNF4A, P2-HNF4A and negative control lentiviruses. Increased expression of P1- and P2-HNF4A upon transfection was confirmed by Western blot (Supplementary Figure S5B). Then, we performed RNA-seq to examine the effects of P1- and P2-HNF4A on the SGC7901 transcriptome. We found that both P1- and P2-HNF4A overexpression resulted in considerable changes in gene expression at the mRNA level (Supplementary Figure S6A). Using 2-fold as a cutoff to designate DEGs, we found 310 and 105 genes that were downregulated and 763 and 190 genes that were upregulated in P1- and P2-HNF4A-overexpressing cells, respectively (Supplementary Figure S6B). There was an overall greater effect in P1-HNF4A than in P2-HNF4A in terms of the number of dysregulated genes with large fold changes, which could be because P1-HNF4A typically has a more potent transactivation function than P2-HNF4A. KEGG pathway analysis of the differentially regulated genes showed that there was a significant upregulation of genes involved in the cytokine-cytokine receptor interaction, and a decrease in genes involved in the calcium signaling pathway in P1-HNF4A cells (Figure 4A). In P2-HNF4A cells, there was a significant upregulation of genes involved in the PPAR signaling pathway and a decrease in genes involved in valine, leucine, and isoleucine biosynthesis (Figure 4B).
Figure 4.
RNA-seq screen of target genes and signaling pathways using downstream P1- and P2-HNF4A overexpression in SGC7901 cells. (A) Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis showing enrichment of the top 20 signaling pathways in P1-HNF4A overexpression compared to the negative control in SGC7901 cells. (B) KEGG pathway analysis showing enrichment of the top 20 signaling pathways in P2-HNF4A overexpression compared to the negative control in SGC7901 cells.
CCL15 is a direct target of P1-HNF4A and is functionally required in GC
To test the deregulated genes involved in the cytokine-cytokine receptor interaction pathway in GC development, we examined the expression of these genes within TCGA databases. Data in GEPIA revealed that the expressions of CCL5, CCL15, CCL20, CSF2RA, CXCL1, CXCL8, CXCL16, CXCR4, GDF15, IL1RN, IL2RB, IL2RG, IL22RA1, INHBA, TNFRSF1B, and TNFRSF14 were significantly higher in GC tissues than in normal tissues (Supplementary Figure S7). Data from Ualcan revealed that the expressions of BMP2, CCL5, CCL15, CCL20, IL8, CD70, CSF2RA, CXCL1, CXCL16, CXCR4, TGFB2, GDF15, IL11, IL1R1, IL1RN, IL13RA2, IL32, IL2RB, IL2RG, IL22RA1, IL18R1, IL18RAP, INHBA, INHBB, INHBE, TNFRSF9, TNFRSF1B, and TNFRSF14 were significantly higher in GC tissues than in normal tissues (Supplementary Figure S8).
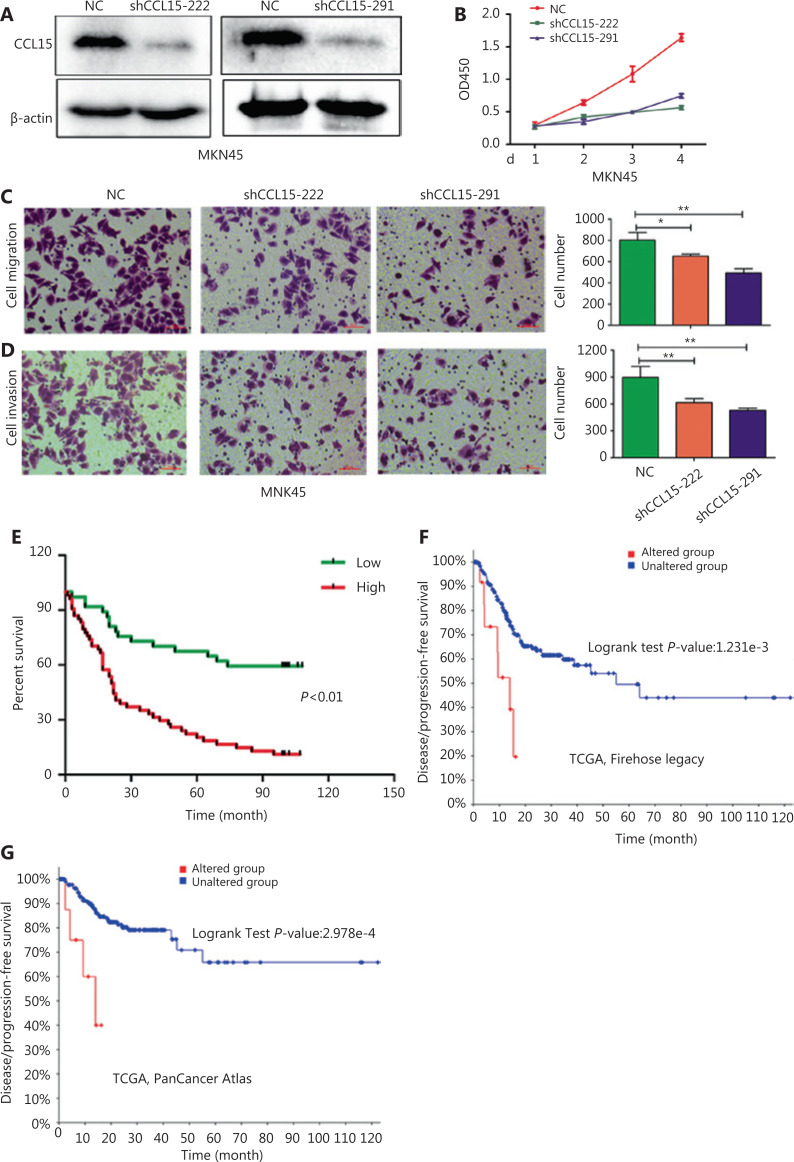
Among the aforementioned gene deregulations in GC, we focused on chemokines, including CCL15, the role of which remained unclear in GC progression. Then, we examined the in vitro role of CCL15 on GC cell proliferation and invasion. Initially, we detected the mRNA expression of CCL15 in the normal gastric epithelial cell line, GES-1, and the GC cell lines, BGC823, MKN45, AZ521, SGC7901, and AGS. Notably, the mRNA level of CCL15 in GC cells far surpassed that in GES-1 cells (Supplementary Figure S9). Then, shRNA-mediated CCL15 knockdown cells were generated in MKN45, a cell line with relatively high CCL15 expression (Figure 5A). Significant reductions in the proliferation rate were observed in shCCL15 cells compared with control cells using the CCK-8 assay (P < 0.01) (Figure 5B). Significantly reduced cell migration and invasion were also observed in shCCL15 cells compared with control cells in the MKN45 cell line (P < 0.01) (Figure 5C and 5D). To determine whether CCL15 was also upregulated in primary GCs, we analyzed CCL15 expression using a GC tissue microarray. The results indicated that CCL15 expression was significantly higher in tumors than in paracancerous tissues (P < 0.01) (Supplementary Figure S10A and S10B). Subgroup analysis indicated that patients with high CCL15 expression showed more lymph node metastasis than those with low CCL15 expression (P < 0.05) (Table 3). Survival analysis showed that patients with high CCL15 expression had a poorer prognosis than patients with low CCL15 expression (P < 0.01) (Figure 5E). We further explored the prognostic value of CCL15 in the cBioportal database. High CCL15 expression showed poorer prognosis in both the Firehose legacy and PanCancer Atlas groups (Figure 5F and 5G). In summary, these results supported CCL15 as an oncogene in GC progression.
Figure 5.
The effects of CCL15 on malignant phenotypes of gastric cancer (GC) cells in vitro. (A) Negative control and sh-CCL15 lentivirus were transfected in MKN45 cells and CCL15 protein expression was detected by Western blot. β-actin was used as internal control. (B) MKN45 cell viability infected with lentivirus expressing shRNAs targeting CCL15 or control shRNA was examined by CCK8, and OD450 was detected at the indicated time (**P < 0.01, n = 3). (C, D) Cell migration and invasion was performed using transwell assays. Migrated (C) or invaded (D) cells were counted by ImageJ (*P < 0.05, **P < 0.01, n = 3) (Crystal Violet staining, 20×). (E) The prognostic value of CCL15 in GC patients using tissue microarrays. (F) The prognostic value of CCL15 in GC patients of the Firehose legacy cohort using the cBioPortal database (Z-Score = 2). (G) The prognostic value of CCL15 in GC patients of the PancancerAtlas cohort using the cBioPortal database (Z-Score = 2).
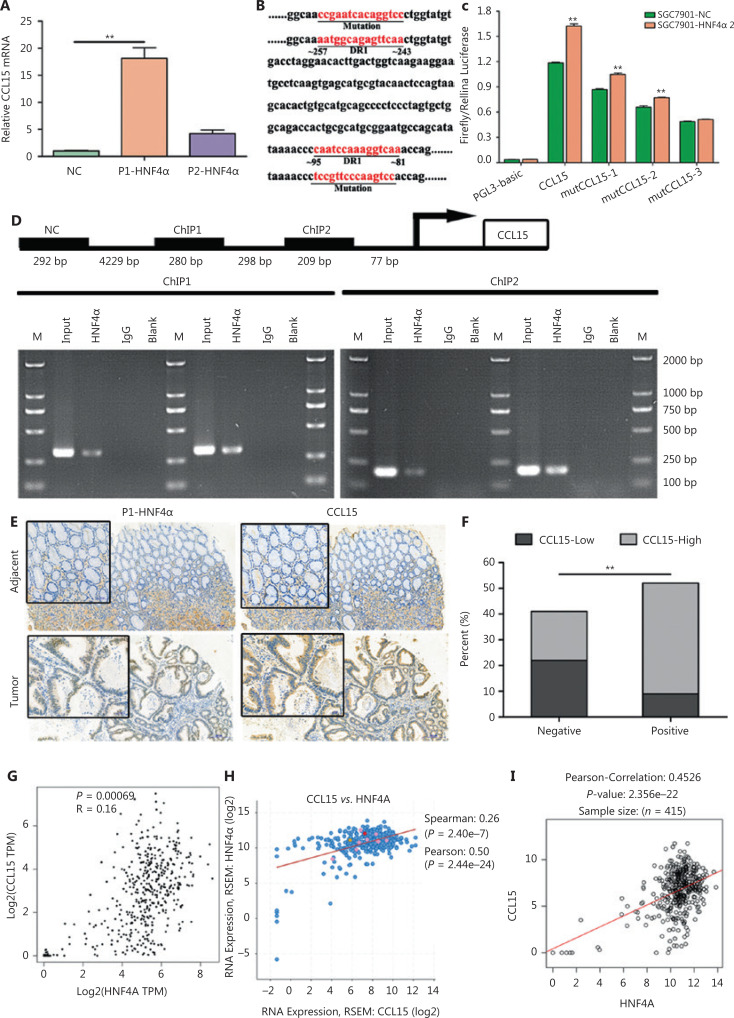
To check if CCL15 acted as a downstream target, we performed qRT-PCR. The results indicated that both P1- and P2-HNF4A could significantly upregulate CCL15 expression, with P1-HNF4A being more prominent (P < 0.01) (Figure 6A). Then, we examined the promoters of CCL15 using JASPAR. The results showed that the promoter of CCL15 contained two predicted DR1 motifs (AGGTCAnAGGTCA) (Figure 6B). Next, we constructed luciferase reporter gene fragments covering the two DR1 sequences (wild-type and mutant) (Figure 6B). The results revealed that HNF4A2 positively regulated the CCL15 promoter containing the two DR1 sites between ˜81˜95 and ˜243˜257 (Figure 6B and 6C). Chromatin immunoprecipitation (ChIP) assays further confirmed that HNF4A2 bound to the speculative sites (˜81˜95 and ˜243˜257) on the CCL15 promoter in SGC7901 cells (Figure 6D). Furthermore, to determine the clinical relevance, we divided the GCs into P1-HNF4A-negative and P1-HNF4A-positive subgroups according to the H-score. CCL15 high expression cases in the P1-HNF4A-positive group were found to be significantly higher than those in the P1-HNF4A-negative group (P < 0.01) (Figure 6E and 6F). We further determined the correlation between HNF4A and CCL15 in TCGA databases. Both data in the GEPIA, cBioportal, and LinkedOmic databases revealed that HNF4A was positively correlated with CCL15 (Figure 6G, 6H, and 6I).
Figure 6.
CCL15 is a direct target of P1-HNF4A in gastric cancer. (A) Relative CCL15 mRNA was examined by qRT-PCR in MKN45 cells transfected with the control, P1-HNF4A, and P2-HNF4A overexpression lentiviruses. GAPDH was used as internal control (**P < 0.01, n = 3). (B) CCL15 promoter fragment (˜2,000 bp) from Ensemble was predicted using JASPAR. Reporter genes containing the classical DR1 motifs or mutational sequences were constructed. (C) Negative control (NC) and the HNF4A2 overexpression plasmid were transiently transfected with CCL15 promoter reporter constructs with the DR1 motif or mutation for 24 h, and luciferase activity was assayed thereafter. The promoter activity was expressed as fold-induction (means ± SEM) compared to that of the NC, n = 3. (**P < 0.01). (D) A chromatin immunoprecipitation assay was performed to show the direct binding of HNF4A2 to the CCL15 promoter. M, Marker. (E) Paraffin sections of human gastric cancer tissues and adjacent non-tumor tissues were immunostained with anti-P1-HNF4A and anti-CCL15 antibodies. H&E staining. Scale bars = 200 μm and 50 μm. (F) The percent of CCL15-low and CCL15-high expression cases in P1-HNF4A negative and positive tissues, respectively according staining score (**P < 0.01). (G) The correlation of CCL15 and HNF4A in the GEPIA database. (H) The correlation of CCL15 and HNF4A in the cBioPortal database. (I) The correlation of CCL15 and HNF4A in the LinkedOmics database.
Discussion
Accumulating evidence has revealed the vital role of HNF4A in GC initiation and progression. However, the functions of different HNF4A isoforms and underlying molecular mechanisms in GC cell growth and invasion have not been completely clarified. In this study, we demonstrated that P1-HNF4A was the main isoform upregulated in GC progression. Furthermore, we revealed that the cytokine-receptor pathway was one of the key downstream signaling pathways. We also demonstrated that CCL15 was the most abundant chemokine downstream of P1-HNF4A in human GC, with significant prognostic value. These findings revealed complex tumor-promoting effects shaped by the HNF4A-CCL15 axis in human GC.
HNF4A is a highly conserved member of the nuclear receptor superfamily, and regulates genes involved in hepatocyte differentiation12,13. It also plays key roles in kidney14, pancreas15, and gut16 development, and regulates the expression of genes involved in metabolism17,18, homeostasis19–21, differentiation22, and immune response23,24. The roles and functions of HNF4A in HCC and CRC have been clearly defined25–27. In contrast, the role of HNF4A in stomach development and GC has only preliminarily been investigated. Recent studies showed that HNF4A is functionally required for GC survival by interacting with the Wnt and NF-κB pathways and regulating tricarboxylic acid (TCA) cycle metabolism3,5,6. In contrast, another recent study showed that the tumor suppressor ITLN1 promoted HNF4A expression by repressing NF-κB in GC28, and high HNF4A expression showed improved survival outcomes28. Similar inconsistent results have also been observed in CRC27,29. These paradoxical conclusions indicated that the functions of HNF4A were characterized with time and spatial distribution characteristics. The expression of HNF4A might be dynamically regulated during GC initiation and progression. Our preliminary results found that HNF4A was upregulated in GC, especially in the early stage, which supported the oncogenic roles of HNF4A.
A total of nine HNF4A splice variants have been generated via two alternative promoters (P1 and P2 promoters) with tissue- and cell-specific expression patterns30,31. P1-driven HNF4A includes HNF4A1-6 isoforms and P2-driven HNF4A includes HNF4A7-9 isoforms respectively30. P1-driven HNF4A is expressed in the adult liver32 and kidney7, whereas P2-driven HNF4A is expressed in the fetal liver, the adult stomach7 and pancreas30. Both P1- and P2-HNF4A are expressed in the adult colon and intestine, but with distinct location distributions30. The unique expression patterns of P1- and P2-driven HNF4A suggested distinct functional roles of different HNF4A isoforms under physiological conditions. Furthermore, previous research has demonstrated that HNF4A isoforms showed significant metabolic differences and varied susceptibilities to colitis-associated colon cancer in exon-swap mice33,34. In gastric carcinogenesis, both P1- and P2-HNF4A were positive in gastric precancerous lesions and tumor tissues9,35, which suggests that the HNF4A gene may exhibit oncogenic activity in GC. However, the relative contributions of the different HNF4A isoforms (P1- and P2-HNF4A) have not been determined.
P1-HNF4A acts as a tumor suppressor both in HCC and CRC, inhibiting cell proliferation and the inflammation pathway25,34. In contrast, P2-HNF4A promotes colitis and colitis-associated colon cancer development34. Previous studies showed that stomach tissues were positive for only P2-HNF4A7. In contrast, Moore et al.19 indicated that HNF4A, especially P1-HNF4A, regulated gastric epithelium homeostasis and was necessary for the maintenance of ZC secretory architecture. In this study, we demonstrated that P1- and P2-HNF4A exhibited significantly different functions during GC progression. However, unlike in HCC36 and CRC37, P1-HNF4A was the main oncogene, while P2-HNF4A expression was irrelevant to GC. Previous studies have demonstrated that P1-HNF4A plays key roles in both intestinal epithelium and liver cell development and differentiation16,23, so it is possible that tissue specific transcription factors might suppress tumorigenesis. In contrast, ectopic expression of lineage-survival oncogenes may promote cancer by reactivating early developmental programs. These results were consistent with the high expression of P1-HNF4A in intestinal-type GC4, so we proposed that the function of HNF4A was tissue-specific, which might account for the distinct roles of P1- and P2-HNF4A in different cancer development. Furthermore, integration of RNA-seq data suggests that this functional difference is due to differential expression of certain target genes, with HNF4A2 upregulating genes involved in cell proliferation and inflammation, and HNF4A8 upregulating genes involved in metabolism. In addition, P1-HNF4A typically has a more potent transactivation function than P2-HNF4A, which displays a similar pattern as in CRC.
Recent evidence suggests the involvement of HNF4A in the pathophysiology of inflammatory diseases, such as inflammatory bowel diseases24,38,39. HNF4A has also been shown to interact with the IL-6/STAT3 and NF-κB pathways6,25. Additionally, HNF4A was involved in IL-1β signal transduction through the regulation of IL-1R6. These results indicated that inflammation regulation might be a potential molecular mechanism involving the functional roles of HNF4A. In this study, we identified CCL15 as a direct P1-HNF4A target gene that promoted GC cell proliferation and invasion. CCL15 has been shown to play a critical role in tumor progression in various cancer types, including liver and colorectal cancers40–42. However, the role of CCL15 in GC remains unclear. Accumulating evidence has suggested that CCL15 may have a crucial role in the progression of tumor cells via the CC chemokine receptor42,43. Previous studies have preliminarily indicated that CCL15 expression is higher in gastric tumor tissues than in normal tissues44,45. In human CRC cells, loss of SMAD4 leads to the upregulation of CCL15 expression40,42,43. Additionally, in human liver cancer cells, both inflammatory factors and epigenetics regulate CCL15 expression41. Mechanistically, CCL15 could promote tumor progression through the only receptor, CCR1, which is expressed by both immune cells such as monocytes41, lymphocytes46, neutrophils40, eosinophils47 and tumor cells41,48. Our results revealed that CCL15 expression was higher in GC tissues than in normal tissues, and predicted a poor prognosis. CCL15 knockdown significantly repressed GC cell proliferation, which indicated that CCL15 might promote GC cell proliferation in an autocrine manner (Supplementary Figure S11). Whether CCL15 promotes GC progression through recruitment of CCR1 immune cells in a paracrine manner requires further exploration. More importantly, our results indicated the positive correlation of P1-HNF4A with CCL15 in a subset of GC patients. Thus, aberrant P1-HNF4A expression in certain GC patients might provide a possible clue for the development of an effective therapy by CCL15 signaling pathway blockade.
Conclusions
In summary, our study preliminarily showed the biological and clinical significance of different HNF4A isoforms in GC. Compared to P2-HNF4A, P1-HNF4A might be the main driver oncogene in GC progression. Mechanistically, P1-HNF4A directly regulated the cytokine-receptor pathway, thereby promoting tumor growth and progression. CCL15 could be regarded as a biomarker to guide therapy in GC patients with a high expression of P1-HNF4A, and pharmaceutical intervention of the CCL15 signaling pathway may provide a promising strategy to improve the prognoses of GC patients.
Supporting Information
Footnotes
Conflict of interest statement No potential conflicts of interest are disclosed.
Grant support
This work was supported by the National Natural Science Foundation of China (Grant No. 81873554) and Shaanxi Foundation for Innovation Team of Science and Technology (Grant No. 2018TD-003).
Availability of data and materials
The RNA-seq datasets generated and analyzed during the current study are available in the NCBI SRA repository (PRJNA612930).
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Strong VE, Wu AW, Selby LV, Gonen M, Hsu M, Song KY, et al. Differences in gastric cancer survival between the U.S. and China. J Surg Oncol. 2015;112:31–7. doi: 10.1002/jso.23940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang HR, Nam S, Kook MC, Kim KT, Liu X, Yao H, et al. HNF4alpha is a therapeutic target that links AMPK to WNT signalling in early-stage gastric cancer. Gut. 2016;65:19–32. doi: 10.1136/gutjnl-2014-307918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chia NY, Deng N, Das K, Huang D, Hu L, Zhu Y, et al. Regulatory crosstalk between lineage-survival oncogenes KLF5, GATA4 and GATA6 cooperatively promotes gastric cancer development. Gut. 2015;64:707–19. doi: 10.1136/gutjnl-2013-306596. [DOI] [PubMed] [Google Scholar]
- 5.Xu C, Ooi WF, Qamra A, Tan J, Chua BY, Ho S, et al. HNF4alpha pathway mapping identifies wild-type IDH1 as a targetable metabolic node in gastric cancer. Gut. 2020;69:231–42. doi: 10.1136/gutjnl-2018-318025. [DOI] [PubMed] [Google Scholar]
- 6.Ma L, Zeng J, Guo Q, Liang X, Shen L, Li S, et al. Mutual amplification of HNF4alpha and IL-1R1 composes an inflammatory circuit in Helicobacter pylori associated gastric carcinogenesis. Oncotarget. 2016;7:11349–63. doi: 10.18632/oncotarget.7239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dean S, Tang JI, Seckl JR, Nyirenda MJ. Developmental and tissue-specific regulation of hepatocyte nuclear factor 4-alpha (HNF4-alpha) isoforms in rodents. Gene Expr. 2010;14:337–44. doi: 10.3727/105221610x12717040569901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tanaka T, Jiang S, Hotta H, Takano K, Iwanari H, Sumi K, et al. Dysregulated expression of P1 and P2 promoter-driven hepatocyte nuclear factor-4alpha in the pathogenesis of human cancer. J Pathol. 2006;208:662–72. doi: 10.1002/path.1928. [DOI] [PubMed] [Google Scholar]
- 9.Takano K, Hasegawa G, Jiang S, Kurosaki I, Hatakeyama K, Iwanari H, et al. Immunohistochemical staining for P1 and P2 promoter-driven hepatocyte nuclear factor-4alpha may complement mucin phenotype of differentiated-type early gastric carcinoma. Pathol Int. 2009;59:462–70. doi: 10.1111/j.1440-1827.2009.02394.x. [DOI] [PubMed] [Google Scholar]
- 10.Zhang Q, Agoston AT, Pham TH, Zhang W, Zhang X, Huo X, et al. Acidic bile salts induce epithelial to mesenchymal transition via VEGF signaling in non-neoplastic Barrett’s cells. Gastroenterology. 2019;156:130–44. doi: 10.1053/j.gastro.2018.09.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu X, Chen B, You W, Xue S, Qin H, Jiang H. The membrane bile acid receptor TGR5 drives cell growth and migration via activation of the JAK2/STAT3 signaling pathway in non-small cell lung cancer. Cancer Lett. 2018;412:194–207. doi: 10.1016/j.canlet.2017.10.017. [DOI] [PubMed] [Google Scholar]
- 12.Colletti M, Cicchini C, Conigliaro A, Santangelo L, Alonzi T, Pasquini E, et al. Convergence of Wnt signaling on the HNF4alpha-driven transcription in controlling liver zonation. Gastroenterology. 2009;137:660–72. doi: 10.1053/j.gastro.2009.05.038. [DOI] [PubMed] [Google Scholar]
- 13.Bolotin E, Liao H, Ta TC, Yang C, Hwang-Verslues W, Evans JR, et al. Integrated approach for the identification of human hepatocyte nuclear factor 4alpha target genes using protein binding microarrays. Hepatology. 2010;51:642–53. doi: 10.1002/hep.23357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marable SS, Chung E, Adam M, Potter SS, Park JS. Hnf4a deletion in the mouse kidney phenocopies Fanconi renotubular syndrome. JCI Insight. 2018;3:e97497. doi: 10.1172/jci.insight.97497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jennings RE, Berry AA, Gerrard DT, Wearne SJ, Strutt J, Withey S, et al. Laser capture and deep sequencing reveals the transcriptomic programmes regulating the onset of pancreas and liver differentiation in human embryos. Stem Cell Rep. 2017;9:1387–94. doi: 10.1016/j.stemcr.2017.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen L, Toke NH, Luo S, Vasoya RP, Fullem RL, Parthasarathy A, et al. A reinforcing HNF4-SMAD4 feed-forward module stabilizes enterocyte identity. Nat Genet. 2019;51:777–85. doi: 10.1038/s41588-019-0384-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Armour SM, Remsberg JR, Damle M, Sidoli S, Ho WY, Li Z, et al. An HDAC3-PROX1 corepressor module acts on HNF4alpha to control hepatic triglycerides. Nat Commun. 2017;8:549. doi: 10.1038/s41467-017-00772-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu Y, Zalzala M, Xu J, Li Y, Yin L, Zhang Y. A metabolic stress-inducible miR-34a-HNF4alpha pathway regulates lipid and lipoprotein metabolism. Nat Commun. 2015;6:7466. doi: 10.1038/ncomms8466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moore BD, Khurana SS, Huh WJ, Mills JC. Hepatocyte nuclear factor 4alpha is required for cell differentiation and homeostasis in the adult mouse gastric epithelium. Am J Physiol Gastrointest Liver Physiol. 2016;311:267–75. doi: 10.1152/ajpgi.00195.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.San RA, Aronson BE, Krasinski SD, Shivdasani RA, Verzi MP. Transcription factors GATA4 and HNF4A control distinct aspects of intestinal homeostasis in conjunction with transcription factor CDX2. J Biol Chem. 2015;290:1850–60. doi: 10.1074/jbc.M114.620211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cattin AL, Le Beyec J, Barreau F, Saint-Just S, Houllier A, Gonzalez FJ, et al. Hepatocyte nuclear factor 4alpha, a key factor for homeostasis, cell architecture, and barrier function of the adult intestinal epithelium. Mol Cell Biol. 2009;29:6294–308. doi: 10.1128/MCB.00939-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Farkas AE, Hilgarth RS, Capaldo CT, Gerner-Smidt C, Powell DR, Vertino PM, et al. HNF4alpha regulates claudin-7 protein expression during intestinal epithelial differentiation. Am J Pathol. 2015;185:2206–18. doi: 10.1016/j.ajpath.2015.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lau HH, Ng N, Loo L, Jasmen JB, Teo A. The molecular functions of hepatocyte nuclear factors – In and beyond the liver. J Hepatol. 2018;68:1033–48. doi: 10.1016/j.jhep.2017.11.026. [DOI] [PubMed] [Google Scholar]
- 24.Babeu JP, Boudreau F. Hepatocyte nuclear factor 4-alpha involvement in liver and intestinal inflammatory networks. World J Gastroenterol. 2014;20:22–30. doi: 10.3748/wjg.v20.i1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hatziapostolou M, Polytarchou C, Aggelidou E, Drakaki A, Poultsides GA, Jaeger SA, et al. An HNF4alpha-miRNA inflammatory feedback circuit regulates hepatocellular oncogenesis. Cell. 2011;147:1233–47. doi: 10.1016/j.cell.2011.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yin C, Wang PQ, Xu WP, Yang Y, Zhang Q, Ning BF, et al. Hepatocyte nuclear factor-4alpha reverses malignancy of hepatocellular carcinoma through regulating miR-134 in the DLK1-DIO3 region. Hepatology. 2013;58:1964–76. doi: 10.1002/hep.26573. [DOI] [PubMed] [Google Scholar]
- 27.Vuong LM, Chellappa K, Dhahbi JM, Deans JR, Fang B, Bolotin E, et al. Differential effects of hepatocyte nuclear factor 4alpha isoforms on tumor growth and T-Cell Factor 4/AP-1 interactions in human colorectal cancer cells. Mol Cell Biol. 2015;35:3471–90. doi: 10.1128/MCB.00030-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li D, Zhao X, Xiao Y, Mei H, Pu J, Xiang X, et al. Intelectin 1 suppresses tumor progression and is associated with improved survival in gastric cancer. Oncotarget. 2015;6:16168–82. doi: 10.18632/oncotarget.3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Darsigny M, Babeu JP, Seidman EG, Gendron FP, Levy E, Carrier J, et al. Hepatocyte nuclear factor-4alpha promotes gut neoplasia in mice and protects against the production of reactive oxygen species. Cancer Res. 2010;70:9423–33. doi: 10.1158/0008-5472.CAN-10-1697. [DOI] [PubMed] [Google Scholar]
- 30.Ko HL, Zhuo Z, Ren EC. HNF4alpha combinatorial isoform heterodimers activate distinct gene targets that differ from their corresponding homodimers. Cell Rep. 2019;26:2549–57. doi: 10.1016/j.celrep.2019.02.033. [DOI] [PubMed] [Google Scholar]
- 31.Guo S, Lu H. Novel mechanisms of regulation of the expression and transcriptional activity of hepatocyte nuclear factor 4alpha. J Cell Biochem. 2019;120:519–32. doi: 10.1002/jcb.27407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li K, Zhang H, Wang Y, Wang Y, Feng M. Differential expression of HNF4alpha isoforms in liver stem cells and hepatocytes. J Cell Biochem. 2006;99:558–64. doi: 10.1002/jcb.20939. [DOI] [PubMed] [Google Scholar]
- 33.Briancon N, Weiss MC. In vivo role of the HNF4alpha AF-1 activation domain revealed by exon swapping. EMBO J. 2006;25:1253–62. doi: 10.1038/sj.emboj.7601021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chellappa K, Deol P, Evans JR, Vuong LM, Chen G, Briancon N, et al. Opposing roles of nuclear receptor HNF4alpha isoforms in colitis and colitis-associated colon cancer. eLife. 2016;5:e10903. doi: 10.7554/eLife.10903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kojima K, Kishimoto T, Nagai Y, Tanizawa T, Nakatani Y, Miyazaki M, et al. The expression of hepatocyte nuclear factor-4alpha, a developmental regulator of visceral endoderm, correlates with the intestinal phenotype of gastric adenocarcinomas. Pathology. 2006;38:548–54. doi: 10.1080/00313020601024011. [DOI] [PubMed] [Google Scholar]
- 36.Fekry B, Ribas-Latre A, Baumgartner C, Deans JR, Kwok C, Patel P, et al. Incompatibility of the circadian protein BMAL1 and HNF4alpha in hepatocellular carcinoma. Nat Commun. 2018;9:4349. doi: 10.1038/s41467-018-06648-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chellappa K, Jankova L, Schnabl JM, Pan S, Brelivet Y, Fung CL, et al. Src tyrosine kinase phosphorylation of nuclear receptor HNF4alpha correlates with isoform-specific loss of HNF4alpha in human colon cancer. Proc Natl Acad Sci U S A. 2012;109:2302–7. doi: 10.1073/pnas.1106799109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chahar S, Gandhi V, Yu S, Desai K, Cowper-Sal-lari R, Kim Y, et al. Chromatin profiling reveals regulatory network shifts and a protective role for hepatocyte nuclear factor 4alpha during colitis. Mol Cell Biol. 2014;34:3291–304. doi: 10.1128/MCB.00349-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meddens CA, Harakalova M, van den Dungen NA, Foroughi AH, Hijma HJ, Cuppen EP, et al. Systematic analysis of chromatin interactions at disease associated loci links novel candidate genes to inflammatory bowel disease. Genome Biol. 2016;17:247. doi: 10.1186/s13059-016-1100-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamamoto T, Kawada K, Itatani Y, Inamoto S, Okamura R, Iwamoto M, et al. Loss of SMAD4 promotes lung metastasis of colorectal cancer by accumulation of CCR1+ tumor-associated neutrophils through CCL15-CCR1 axis. Clin Cancer Res. 2017;23:833–44. doi: 10.1158/1078-0432.CCR-16-0520. [DOI] [PubMed] [Google Scholar]
- 41.Liu LZ, Zhang Z, Zheng BH, Shi Y, Duan M, Ma LJ, et al. CCL15 recruits suppressive monocytes to facilitate immune escape and disease progression in hepatocellular carcinoma. Hepatology. 2019;69:143–59. doi: 10.1002/hep.30134. [DOI] [PubMed] [Google Scholar]
- 42.Liang Q, Tang C, Tang M, Zhang Q, Gao Y, Ge Z. TRIM47 is up-regulated in colorectal cancer, promoting ubiquitination and degradation of SMAD4. J Exp Clin Cancer Res. 2019;38:159. doi: 10.1186/s13046-019-1143-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Itatani Y, Kawada K, Fujishita T, Kakizaki F, Hirai H, Matsumoto T, et al. Loss of SMAD4 from colorectal cancer cells promotes CCL15 expression to recruit CCR1+ myeloid cells and facilitate liver metastasis. Gastroenterology. 2013;145:1064–75. doi: 10.1053/j.gastro.2013.07.033. [DOI] [PubMed] [Google Scholar]
- 44.Camargo MC, Song M, Shimazu T, Charvat H, Yamaji T, Sawada N, et al. Circulating inflammation markers and risk of gastric and esophageal cancers: a case-cohort study within the Japan public health center-based prospective study. Cancer Epidemiol Biomarkers Prev. 2019;28:829–32. doi: 10.1158/1055-9965.EPI-18-1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Raja UM, Gopal G, Shirley S, Ramakrishnan AS, Rajkumar T. Immunohistochemical expression and localization of cytokines/chemokines/growth factors in gastric cancer. Cytokine. 2017;89:82–90. doi: 10.1016/j.cyto.2016.08.032. [DOI] [PubMed] [Google Scholar]
- 46.Dyer DP, Medina-Ruiz L, Bartolini R, Schuette F, Hughes CE, Pallas K, et al. Chemokine receptor redundancy and specificity are context dependent. Immunity. 2019;50:378–89. doi: 10.1016/j.immuni.2019.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fulkerson PC, Schollaert KL, Bouffi C, Rothenberg ME. IL-5 triggers a cooperative cytokine network that promotes eosinophil precursor maturation. J Immunol. 2014;193:4043–52. doi: 10.4049/jimmunol.1400732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li BH, Garstka MA, Li ZF. Chemokines and their receptors promoting the recruitment of myeloid-derived suppressor cells into the tumor. Mol Immunol. 2020;117:201–15. doi: 10.1016/j.molimm.2019.11.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq datasets generated and analyzed during the current study are available in the NCBI SRA repository (PRJNA612930).