Abstract
Cigarette smoking is an important source of human exposure to toxicants and carcinogens and contributes significantly to cancer morbidity and mortality worldwide. Acrolein, a widespread environmental pollutant, is present in relatively high amounts in cigarette smoke and can react directly with DNA to form DNA adducts, which serve as important biomarkers for the assessment of exposure to acrolein and its potential role in smoking related cancer. Etheno-DNA adducts are promutagenic DNA lesions that can derive from exogenous chemicals as well as endogenous sources, including lipid peroxidation. In this study, we developed a combined method for the quantitation of (6R/S)-3-(2′-deoxyribos-1′-yl)-5,6,7,8,-tetrahydro-6-hydroxypyrimido[1,2-a]purine-10(3H)-one (α-OH-Acr-dGuo), (8R/S)-3-(2′-deoxyribos-1′-yl)-5,6,7,8,-tetrahydro-8-hydroxypyrimido[1,2-a]purine-10(3H)-one (γ-OH-Acr-dGuo), 1,N6-etheno-dAdo (εdAdo) and 3,N4-etheno-dCyd (εdCyd) adducts in oral rinse and cytobrush DNA from smokers and non-smokers by liquid chromatography-nanoelelctrospray ionization-high resolution tandem mass spectrometry (LC-NSI-HRMS/MS). For oral rinse samples, there was a statistically significant difference between the levels of α-OH-Acr-dGuo, γ-OH-Acr-dGuo, εdAdo and εdCyd in smokers (12.1 ± 17.9, 163 ± 227, 182 ± 568, and 194 ± 400 adducts/109 nucleotides, respectively) and non-smokers (1.85 ± 2.08, 5.95 ± 4.23, 7.69 ± 11.7, and 6.07 ± 10.9 adducts/109 nucleotides, respectively). For cytobrush samples, there was a statistically significant difference between the levels of γ-OH-Acr-dGuo and εdAdo in smokers (259 ± 540, and 82.9 ± 271 adducts/109 nucleotides, respectively) and non-smokers (7.37 ± 5.09, and 16.2 ± 30.2 adducts/109 nucleotides, respectively), but not for α-OH-Acr-dGuo and εdCyd. Our results demonstrate that oral mucosa cells are an excellent source of material for evaluating DNA adducts to be used as biomarkers of tobacco smoke exposure and molecular changes potentially related to cancer.
Graphical Abstract
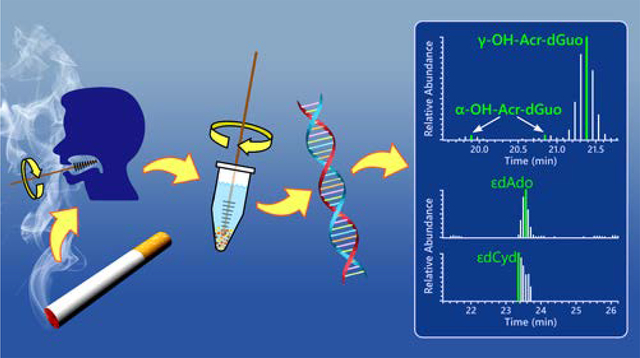
Introduction
There are 1.1 billion tobacco smokers in the world1 and 32 million adult cigarette smokers in the U.S.2 Therefore, smoking continues to be a major source of human exposure to relatively high amounts of toxicants and carcinogens resulting in substantially increased incidences of oral cavity cancer, lung cancer, and at least 14 other types of cancer as well as numerous other diseases.3, 4 A major goal of our research program is to develop biomarkers which can be used to determine individual risk for cancer in people who use tobacco products, as an avenue to early detection and prevention of cancer. DNA adduct levels have the potential to become biomarkers of cancer risk in smokers because they are central to the carcinogenic process.5 While certain exposure biomarkers such as total nicotine equivalents and total NNAL are related to cancer risk, DNA adducts may increase the specificity of the relationship by providing an integration of exposure, absorption, distribution, metabolism and DNA repair.6, 7 However, there are major challenges involved in the quantitative analysis of DNA adducts in accessible human tissues. Oral cells are one readily available source of material for assessing DNA adducts and molecular changes potentially associated with cancer.5
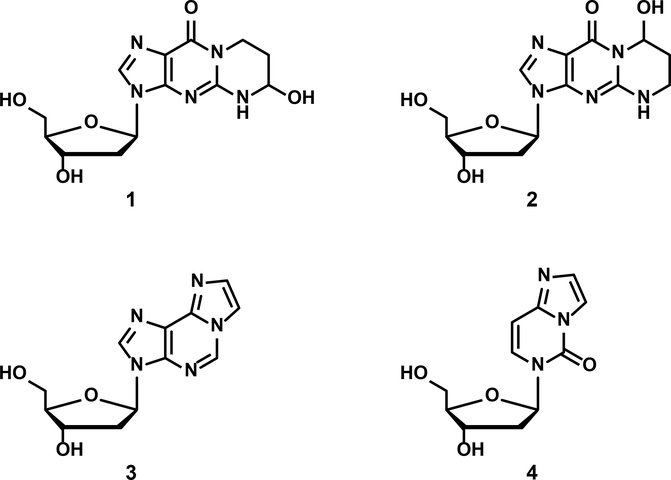
The study reported here focuses on two pairs of exocyclic oral cell DNA adducts. The first pair is formed from acrolein – (6R/S)-3-(2′-deoxyribos-1′-yl)-5,6,7,8,-tetrahydro-6-hydroxypyrimido[1,2-a]purine-10(3H)-one (α-OH-Acr-dGuo, 1) and (8R/S)-3-(2′-deoxyribos-1′-yl)-5,6,7,8,-tetrahydro-8-hydroxypyrimido[1,2-a]purine-10(3H)-one (γ-OH-Acr-dGuo, 2). The second pair is produced in DNA as a result of lipid peroxidation - 1,N6-etheno-dAdo (ƐdAdo, 3) and 3,N4-etheno-dCyd (ƐdCyd, 4)8–16 (Figure 1).
Figure 1.
Structures of α-OH-Acr-dGuo (1), γ-OH-Acr-dGuo (2), εdAdo (3), and εdCyd (4).
Acrolein, with a mean level in mainstream smoke of American brands of 177 μg per cigarette using the Health Canada smoking conditions,17 is one of the most toxic compounds in cigarette smoke, but there is scant evidence for its carcinogenicity.18–20 Acrolein is considered by IARC as “not classifiable as to its carcinogenicity in humans (Group 3)” based on inadequate evidence for its carcinogenicity in laboratory animals.20 Nevertheless, data from humans indicates that acrolein forms DNA adducts at similar mutational hotspots in the TP53 gene as found in smoking-related lung cancers, recalling observations with benzo[a]pyrene diol epoxide.21–23 These studies, along with its ease of reactivity with DNA to form the moderately mutagenic adducts 1 and 2, have rekindled interest in the possible carcinogenic effects of acrolein. The etheno adducts 3 and 4 are promutagenic DNA lesions that can be formed as a result of lipid peroxidation, a consequence of the oxidative damage and inflammation produced by cigarette smoking, alcohol consumption, exposure to PM2.5, and related causes of the inflammatory response.10–14
Several previous studies have quantified these DNA adducts by mass spectrometry and related techniques in oral cells or saliva. Nath et al. analyzed gingival tissue DNA by a 32P-postlabelling-HPLC method and found significantly higher levels of adduct 2 in smokers than in non-smokers (1360 vs 460 adducts/109 unmodified DNA bases).24 Bessette et al. quantified several DNA adducts in buccal cell DNA from smokers and reported levels of 1 and 2 greater than 500 adducts per 109 unmodified DNA bases.25 Chen and Lin analyzed human saliva for adducts 1 plus 2 and reported a mean of 1040 adducts/109 nucleotides; in the same analysis, they quantified 3 and 4 with mean levels of 990 ± 500 and 720 ± 490 adducts/109 nucleotides, respectively.13 Li et al. reported levels of adduct 2 of 1500 adducts/109 nucleotides and 2000 adducts/109 nucleotides in smokers and non-smokers, respectively, showing a non-significant difference.26
Artifact formation can occur easily in analyses of adducts 1 and 2 due to the ubiquitous occurrence of acrolein in H2O, laboratory reagents, and other common sources. In previous studies, we and others have developed methods to avoid artifact formation, for example by including glutathione as a scavenger during analysis.27, 28 In this study, we developed an artifact-free method for simultaneous quantitation by liquid chromatography-nanoelelctrospray ionization-high resolution tandem mass spectrometry (LC-NSI-HRMS/MS) of adducts 1–4 in oral cells of smokers and non-smokers.
Experimental Section
Chemicals and Enzymes
Acr-dGuo and [13C1015N5]Acr-dGuo (comprising both adducts 1 and 2) were prepared as described29 by reaction of acrolein with fully labeled dGuo (Toronto Research Chemicals); εdAdo, [13C5]εdAdo (labeled with 13C in the five carbons of the deoxyribose ring), εdCyd and [13C15N2]εdCyd (labeled with 13C in position 5, and with 15N in positions 4 and 6) were purchased from Toronto Research Chemicals (North York, Ontario, Canada). Reagents for DNA isolation were obtained from Qiagen (Germantown, MD). Calf thymus DNA, micrococcal nuclease and phosphodiesterase II were purchased from Worthington Biochemical Co. (Lakewood, NJ). Alkaline phosphatase and other solvents were obtained from Sigma-Aldrich Chemical Co. (Milwaukee, WI).
Study Design and Oral Cell Collection
Subjects were recruited through the Tobacco Research Programs, University of Minnesota, at the Delaware Clinical Research Unit, and at the Minnesota State Fair through the University of Minnesota research and outreach programs. The study was approved by the University of Minnesota Institutional Review Board. Study participants were adults (more than 18 years old) who were non-smokers or smokers of at least 10 cigarettes/day. Eligible volunteers were asked to brush their teeth with a pre-pasted disposable toothbrush and to wait 15 min between brushing and collecting oral rinse samples. While waiting, participants were asked to take a breathalyzer test to measure their blood alcohol concentration, a carbon monoxide breath test to confirm their smoking status, and to complete a set of self-report questionnaires about their smoking, medication and alcohol use, demographics, and medical history. Oral rinse samples were collected by vigorously swishing 10 mL of sterile normal saline from cheek to cheek for at least 30 sec, then spitting the rinse into a sterile collection tube. After 15 min, cheek cell samples were collected by using a soft-bristled cytobrush to brush the oral mucosa inside the right cheek from top to bottom approximately 30 times. A separate cytobrush was used for collecting cells from the left cheek. The cells were collected by shaking each brush in its own separate 15 mL sterile polypropylene tube containing 5 mL of sterile saline solution. After the collection, oral rinse and cytobrush samples were centrifuged at 2,000g for 15 min to pellet the cells and the pellets were transferred to 1.5 mL centrifuge tubes. For this transfer, we used 200 μL of sterile saline solution for the oral rinse samples and 100 μL each for the cytobrush cell pellets obtained from each subject’s right and left cheek, which were then combined. The samples were stored at −20 °C until DNA isolation and analysis. In this study, we analyzed oral rinse and cheek cell samples from 19 smokers and 20 non-smokers.
DNA Isolation from Oral Cells
DNA isolation was performed using the DNA purification from oral rinse and cytobrush protocols from Qiagen (Qiagen, Valencia, CA) with some modifications. Briefly, samples were thawed and centrifuged at 15,000g for 2 min to pellet the oral cells. After pouring off the supernatant, oral rinse and cytobrush cells were lysed using 1000 μL and 300 μL of cell lysis solution containing 10 mM glutathione (GSH), respectively, and incubated for 20 min at room temperature. GSH was used to inhibit the artifactual formation of Acr-dGuo (1 and 2), εdAdo (3) and εdCyd (4). This was followed by a 15 min treatment at room temperature with 10 μL of proteinase K for oral rinse samples and 2 μL of proteinase K for cytobrush samples, and a 30 min treatment at 37 °C with 6 μL of RNase for oral rinse samples and 2 μL of RNase for cytobrush samples. Then, 340 μL and 100 μL of protein precipitation solution was added to oral rinse and cytobrush cells, respectively, and the samples were centrifuged at 16,000 g for 10 min. DNA was precipitated by pouring the oral rinse supernatant into a 5 mL tube containing 1460 μL of 100% ice-cold 2-propanol and the cytobrush supernatant into a 1.5 mL tube containing 450 μL of 100% ice-cold 2-propanol. The precipitated oral rinse DNA pellet was washed with 1000 μL of 70% ice-cold 2-propanol and then 1000 μL of 100% ice-cold 2-propanol, and the cytobrush DNA with 300 μL of 70% ice-cold 2-propanol and then 300 μL of 100% ice-cold 2-propanol. DNA was dried and stored at −20 °C until analysis.
DNA Hydrolysis and Sample Purification
The isolated DNA (0.5 – 50 μg) was dissolved in 200 μL of 10 mM sodium succinate/5 mM CaCl2 buffer (pH 7.0) containing 5mM GSH to which 10 fmol of [13C1015N5]Acr-dGuo ([13C1015N5]1 and 2), [13C5]εdAdo ([13C5]3), and [13C15N2]εdCyd ([13C15N2]4) were added as internal standards. To monitor the possible artifactual formation of Acr-dGuo (1 and 2), εdAdo (3) and εdCyd (4), some samples were spiked with 5 μg of [15N]DNA isolated from cultures of E. Coli grown in 15N-containing media. Protocols for [15N]DNA generation and isolation from bacteria are presented in the Supporting Information. A buffer blank (200 μL) was prepared each time and used as a negative control to evaluate possible contamination; a sample of calf thymus DNA (20 μg in 200 μL of buffer) was also included in each set of samples as a positive control. The mixture was heated at 100 °C for 30 min and cooled to room temperature. Then, the DNA was enzymatically hydrolyzed by incubation with 7.5 units of micrococcal nuclease and 0.045 units of phosphodiesterase II at 37 °C overnight. Then, 15 units of alkaline phosphatase (from calf intestine) were added, and the mixture was incubated at 37 °C overnight. The resulting mixtures were partially purified by centrifugal filtration (Ultracel 10K, Millipore). A 10 μL aliquot was removed for dGuo quantitation, and the remaining hydrolysate was purified using a solid-phase extraction (SPE) cartridge [Strata-X, 33 μm, 30 mg/1 mL (Phenomenex, Torrance, CA)] activated with 3 mL of MeOH and 3 mL of H2O containing 0.1 mM GSH. After the samples were applied, the cartridges were washed with 6 mL of H2O containing 0.1 mM GSH and 1 mL 5% CH3OH in H2O containing 0.1mM GSH, and the analytes were eluted into high recovery glass vials (Thermo Scientific, San Jose, CA) with 1 mL 35% CH3OH in H2O containing 0.1 mM GSH. Prior to elution, 0.65 μL of 100 mM GSH was added in each vial. The eluants were evaporated to dryness and stored at 4 °C.
Analysis of Acr-dGuo (1 and 2), εdAdo (3), and εdCyd (4) by LC-NSI-HRMS/MS
Samples were dissolved in 20 μL of H2O for LC-NSI-HRMS/MS analysis. The analysis was carried out on an Orbitrap Fusion Lumos instrument (Thermo Scientific, San Jose, CA) interfaced with a UPLC system (Ultimate 3000 RSLCnano UPLC, Thermo Scientific, Waltham, MA) using nanoelectrospray ionization. The UPLC was equipped with a 5 μL loop and the separation was performed using a capillary column (75 μm ID, 20 cm length, 15 μm orifice) prepared by hand packing a commercially available fused-silica emitter (New Objective, Woburn MA) with Luna C18 bonded separation media (Phenomenex, Torrance, CA). The mobile phase consisted of 2 mM NH4OAc and CH3CN. The gradient started at 1% CH3CN for 6 min at a flow rate of 0.9 μL/min, increased to 13% CH3CN in 20 min at a flow rate of 0.3 μL/min and then to 30% CH3OH in 1 min, holding at this composition for 4 min. The gradient was then returned to 1% CH3CN in 1 min and the system was re-equilibrated at this mobile phase composition for 1 min at a flow rate of 0.9 μL/min before the next injection. The source temperature was set at 300 °C and the spray voltage was static at 2200 V. The maximum injection time was 400 ms and the normalized automatic gain control (AGC) was set at 50%. The precursor ions were isolated by the quadrupole with an isolation width of m/z 1.5 and fragmented by higher energy collisional dissociation (HCD) at 20% for Acr-dGuo (1 and 2), [13C1015N5]Acr-dGuo ([13C1015N5]1 and 2), εdAdo (3) and [13C5]εdAdo ([13C5]3) and 30% for εdCyd (4) and [13C15N2]εdCyd ([13C15N2]4) and the fragment ions were detected by the Orbitrap detector at a resolution of 60,000. The ion transitions monitored with accurate mass extracted were: m/z 324.1 → m/z 208.0829 ([M + H]+ → [BH]+) for adducts 1 and 2, m/z 339.1 → m/z 218.0849 for [13C1015N5]1 and 2, m/z 276.1 → m/z 160.0618 for adduct 3, m/z 281.1 → m/z 160.0618 for [13C5]3, m/z 252.1 → m/z 136.0505 for adduct 4 and m/z 255.1 → m/z 139.0480 for [13C15N2]4. To monitor the artifactual formation of these adducts in the [15N]DNA added to the samples before the enzymatic hydrolysis, the following ion transitions were included in the method: m/z 329.1 → m/z 213.0681 for [15N5]Acr-dGuo ([15N5]1 and 2), m/z 281.1 → m/z 165.0469 for [15N5]εdAdo ([15N5]3), and m/z 255.1 → m/z 139.0416 for [15N3]εdCyd ([15N3]4).
Calibration curves were prepared using standard solutions of adducts 1 and 2, [13C1015N5]1 and 2, 3, [13C5]3, 4, and [13C15N2]4). A constant amount each of [13C1015N5]1 and 2, [13C5]3 and [13C15N2]4 (200 amol/μL) was mixed with different amounts of adducts 1–4 (5–500 amol/μL).
Quantitation of dGuo was performed on a HPLC (Agilent Technologies, Palo Alto, CA) system with a UV detector operated at 254 nm. A 0.5 mm × 250 mm capillary Luna 5 μm C18 column (Phenomenex, Torrance, CA) was used. The elution program was a gradient from 5% to 22% CH3OH in H2O in 20 min at a flow rate of 10 μL/min, increased to 80% CH3OH over 5 min, and then returned to 5% CH3OH in 5 min followed by 5 min re-equilibration. Calibration curves (1–100 μg/mL dGuo) were prepared and run after each batch of samples.
Method Validation
The limit of detection (LOD) was established using standard solutions of adducts 1–4. The limit of quantitation (LOQ), accuracy and precision of the method were determined by analyzing calf thymus DNA enriched with different amounts of Acr-dGuo (1 and 2), εdAdo (3) and εdCyd (4) standards (0.1, 0.4, 1, 2, and 4 fmol for adduct 1; 0.1, 0.3, 1, 3, and 6 fmol for adduct 2; 0.1, 0.6, 1, 3, and 6 fmol for adducts 3 and 4). Each sample was analyzed in triplicate. Background levels of adducts 1–4 in calf thymus DNA were determined by analyzing three non-spiked samples; these amounts were subtracted from each amount detected in the spiked samples. The LOQ was defined as the lowest Acr-dGuo (1 and 2), εdAdo (3) and εdCyd (4) amount added to calf thymus DNA that produced a coefficient of variation lower than 20%. Accuracy was determined by comparing added and measured amounts of the adducts at each level. Precision was determined as intra-day and inter-day coefficients of variation (% CV) for the triplicate samples analyzed on three separate days.
In addition to the method validation studies, we investigated intra-day and inter-day variation of DNA adduct levels obtained by oral rinse. Samples were collected from three non-smoking subjects on the same day at three time points (9:30 am, 12:30 pm, 3:30 pm) and on three separate days, and analyzed for adducts 1–4.
Statistical Analysis
Statistical analyses were performed using SigmaPlot 12.5 (Systat Software, San Jose, CA, https://systatsoftware.com/products/sigmaplot/). The non-parametric Mann-Whitney U test was used to compare (1 and 2), εdAdo (3) and εdCyd (4) levels between smokers and non-smokers. Statistical significance was set at p<0.05.
Results
Development of the Analytical Procedure
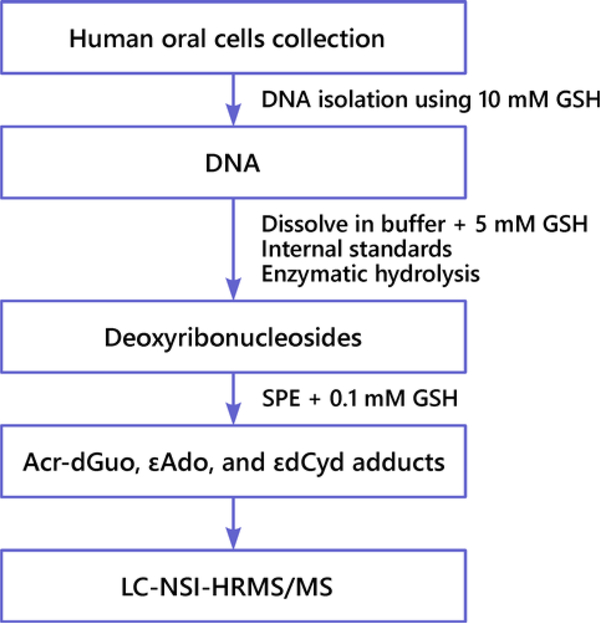
We developed a method for the quantitation of Acr-dGuo (1 and 2), εdAdo (3) and εdCyd (4) adducts in human oral cells. The protocol is outlined in Figure 2.
Figure 2.
Scheme of the LC-NSI-HRMS/MS method for analysis of Acr-dGuo (1 and 2), εdAdo (3) and εdCyd (4) in human oral cells.
Artifactual formation of adducts 1–4 can occur during DNA isolation, hydrolysis and SPE clean-up steps, potentially leading to an overestimation of the adducts in DNA samples. In a previous study, we developed an artifact-free method for the analysis of adducts 1 and 2 in lung tissue DNA and GSH was chosen as scavenger to inhibit the artifact during sample analysis.30 In the study reported here, we modified the method to simultaneously analyze adducts 1–4 in human oral cells. We carefully monitored and controlled the artifactual formation of adducts 1–4 and tested different GSH concentrations during each step of the analysis. Consistent with our previous study,30 there was no difference in the amounts of adducts 1 and 2 measured in oral cell DNA extracted with 5 or 10mM GSH, or no scavenger present, indicating minimal artifactual formation during DNA isolation and purification. However, we observed artifactual formation of adducts 3 and 4 during this step. Adduct 3 levels in DNA extracted with 10mM GSH were ∼ 50% and ∼ 70% lower than levels measured in DNA extracted with 5 mM GSH and no scavenger present, respectively. Adduct 4 levels in DNA extracted with 10mM GSH were ∼ 30% and ∼ 50% lower than levels measured in DNA extracted with 5 mM GSH and no scavenger present, respectively.
When the scavenger was omitted during the hydrolysis and SPE steps, the amounts of adducts 1–4 measured were significantly higher and highly variable, indicating artifactual formation during these two steps. Most of the artifact of adduct 2 was formed during the SpeedVac drying step, while adduct 1 was formed to a much lesser extent during both the hydrolysis and drying steps. The artifactual formation of adducts 3 and 4 was observed mainly during the hydrolysis step. Therefore, we tested different GSH concentrations in calf thymus DNA during DNA hydrolysis (0.5, 5, and 10 mM GSH) and used 0.1 mM GSH during the SPE step, as reported in our previous study.30 There was no significant difference in the levels of adducts 1–4 measured in DNA treated with 0.5, 5 or 10 mM GSH; however, we measured slightly lower levels of adducts 3 and 4 when using 5 mM GSH during the hydrolysis step. Aminoguanidine (AG, 0.5 and 5 mM) was also tested as a scavenger to inhibit artifactual formation of adducts 3 and 4 during both the hydrolysis and SPE clean-up steps, but it was ineffective. Therefore, GSH (5mM) was chosen as scavenger for the DNA hydrolysis step. As previously reported for adducts 1 and 2,30 to further investigate and ensure an efficient inhibition of the artifactual formation of adducts 3 and 4 by using GSH, we analyzed purified dAdo and dCyd following the same protocol. Commercial dAdo and dCyd contained trace amounts of adducts 3 and 4, respectively, and were purified by HPLC before use. We measured the levels of 3 and 4 artificially formed during sample analysis. The results showed that by using 5 mM GSH during DNA hydrolysis and 0.1 mM GSH during the SPE step, the artifactual formation of adducts 3 and 4 was less than 25% of the levels measured in human oral cell samples. It should be noted that this artifact monitoring method provides the maximum levels formed during the sample work-up since real DNA samples contain other matrix components to further scavenge acrolein and other aldehydes and therefore lower artifact formation, as shown below by good intra- and inter-day CV% for positive controls. Further details of the method development and validation are presented in the Supporting Information.
The LC-NSI-HRMS/MS method used here was based on the method previously developed30 for the analysis of adducts 1 and 2 in lung tissue DNA, with some modifications. The analysis was carried out on an Orbitrap Fusion Lumos instrument and the gradient was modified to make the run time shorter (33 min). Fragmentation ions for adducts 3 and 4 and internal standards [13C5]3 and [13C15N2]4, were included. Using HCD fragmentation in the LC-NSI-HRMS/MS system, the product ion spectra of adducts 3 and 4 had several major ions. Because the highest signal intensities in the spectra of both adducts 3 and 4 were m/z 160.0618 and m/z 136.0505, respectively, these ions were selected for quantitative analysis.
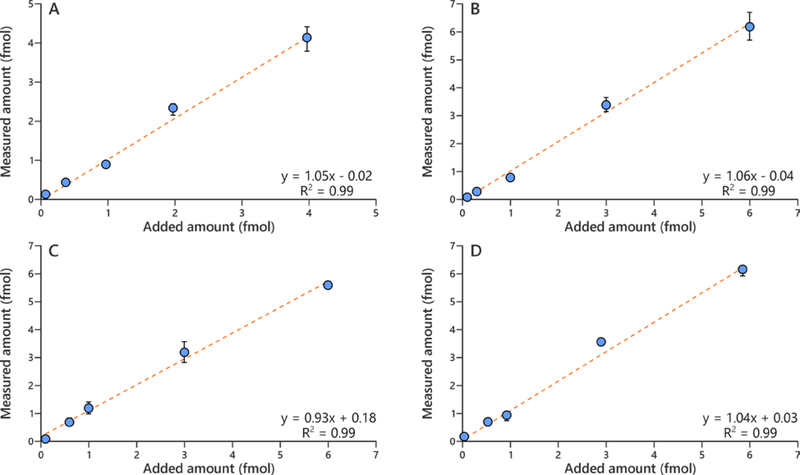
An LOD of 2 amol on-column for adducts 1 and 2, and 1 amol on-column for adducts 3 and 4 was achieved using standard solutions of adducts 1–4. The concentration range for the calibration curves and validation experiments were chosen to cover the range of the levels of adducts 1–4 found in human oral cell samples. The calibration curves for adducts 1–4 showed good linearity within the low concentration range (R2 > 0.99). The assay accuracy was calculated as a percentage of added amount of adducts 1–4 to 20 μg of calf thymus DNA and the average accuracy was 101% for adduct 1, 99% for adduct 2, 108% for adduct 3 and 105% for adduct 4 (n = 5), and good linearity was observed across the tested concentration ranges (Figure 3).
Figure 3.
Relationship between added amount and measured amount of (A) α-OH-Acr-dGuo (1), (B) γ-OH-Acr-dGuo (2)(C), εdAdo (3), and (D) εdCyd (4) in human oral cells.
The assay precision and the LOQ were determined by analyzing 20 μg of calf thymus DNA (n = 12). The intra-day and inter-day CV were both within 20% (Table 1) and an assay LOQ of 1 adduct/109 nucleotides for adducts 1–4 was achieved based on a CV of 3% for adduct 1, 16% for adducts 2 and 4, and 6% for adduct 3.
Table 1.
Assay precision determined by analyzing calf thymus DNA (n = 12).
| Adduct | Calf Thymus DNA adducts/109 nucleotides | CV% |
|
|---|---|---|---|
| Intra-day | Inter-day | ||
| α-OH-Acr-dGuo (1) | 12.1 ± 1.3 | 8 | 12 |
| γ-OH-Acr-dGuo (2) | 12.6 ± 2.3 | 12 | 9 |
| εdAdo (3) | 21.8 ± 5.7 | 11 | 12 |
| εdCyd (4) | 36.9 ± 3.5 | 10 | 9 |
Quantitation of Acr-dGuo (1 and 2), εdAdo (3), and εdCyd (4) in Human Oral Cells
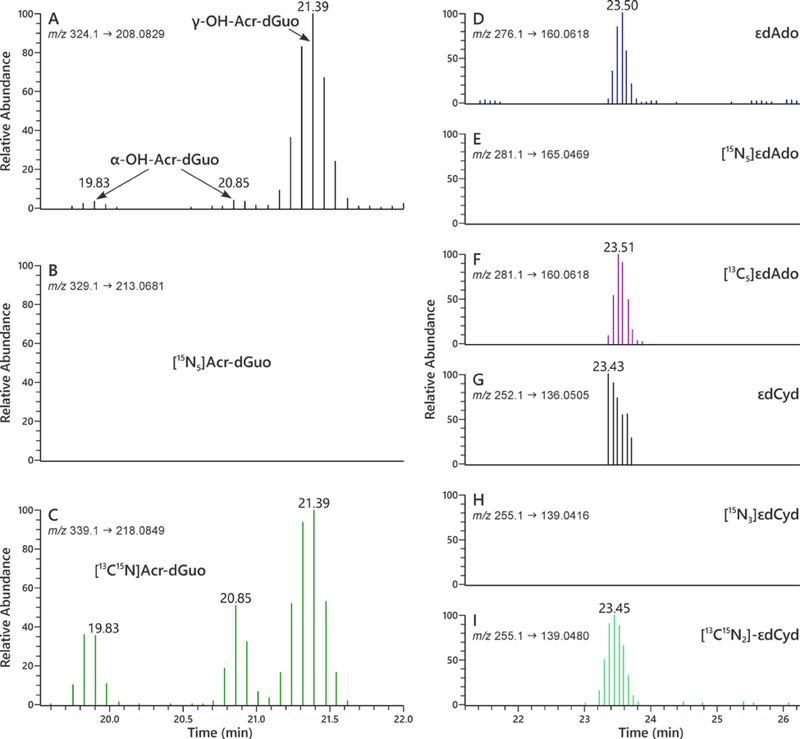
The method was applied to the analysis of Acr-dGuo (1 and 2), εdAdo (3), and εdCyd (4) in oral cells from 19 smokers and 20 non-smokers. Typical extracted ion chromatograms for the quantitation of adducts 1–4 in human oral cells using LC-NSI-HRMS/MS are presented in Figure 4.
Figure 4.
LC-NSI-HRMS/MS chromatograms obtained upon analysis of human oral cell DNA obtained from an oral rinse sample for Acr-dGuo (1 and 2), εdAdo (3), and εdCyd (4). SRM was carried out at (A) m/z 324.1 → m/z 208.0829 for adducts 1 and 2, (B) m/z 329.1 → m/z 213.0681 for spiked 15N bacterial DNA adducts [15N5]1 and 2, and (C) m/z 339.1 → m/z 218.0849 for internal standard adducts [13C1015N5]1 and 2; SRM was carried out at (D) m/z 276.1 → m/z 160.0618 for adduct 3, (E) m/z 281.1 → m/z 165.0469 for spiked 15N bacterial DNA adduct [15N5]3, and (F) m/z 281.1 → m/z 160.0618 for internal standard adduct [13C5]3; SRM was carried out at (G) m/z 252.1 → m/z 136.0505 for adduct 4, (H) m/z 255.1 → m/z 139.0416 for spiked 15N bacterial DNA adduct [15N3]4, and (I) m/z 255.1 → m/z 139.0480 for internal standard adduct [13C15N2]4. In each case, the transition monitored represents loss of the deoxyribose ring with transfer of a proton to the base, e.g. ([M + H]+ → [BH]+).
DNA was isolated from oral rinse and cytobrush samples and analyzed for adducts 1–4. [15N]DNA isolated from cultures of E. Coli grown in 15N-containing media was spiked in some samples to monitor the artifactual formation of these adducts during the DNA hydrolysis and SPE steps. As shown in Figure 4, we did not observe artifactual formation of adducts 1–4 in [15N]DNA, further confirming that there was minimal artifactual formation of adducts 1–4 in oral cell DNA during sample analysis.
The levels of adducts 1–4 in oral cell DNA from smokers and non-smokers are presented in Tables 2 (oral rinse) and 3 (cytobrush) for the same subjects. Adducts 1 and 4 were not detected in the majority of cytobrush samples and in oral rinse samples from non-smokers.
Table 2.
Levels Acr-dGuo (1 and 2), εdAdo (3), and εdCyd (4) in oral rinse DNA from smokers (A) and non-smokers (B).
| A. Smokers Oral rinse sample | DNA (μg) a | α-OH-Acr-dGuo (1) | γ-OH-Acr-dGuo (2) | εdAdo (3) | εdCyd (4) |
|
adducts/109 nucleotides | |||||
| 1 | 13.1 | 4.01 | 91.6 | 5.84 | 3.06 |
| 2 | 17.9 | 5.19 | 170 | 9.76 | 4.77 |
| 3 | 8.23 | 67.5 | 211 | 52.3 | 37.4 |
| 4 | 24.9 | ND b | 254 | 17.4 | 9.89 |
| 5 | 2.51 | ND | ND | 12.2 | 6.91 |
| 6 | 16.9 | 51.9 | 365 | 48.7 | 45.9 |
| 7 | 9.74 | ND | 10.8 | 9.32 | 736 |
| 8 | 6.36 | 5.06 | 1020 | 15.9 | 21.8 |
| 9 | 5.52 | 22.9 | 74.9 | 12.5 | ND |
| 10 | 7.31 | ND | 82.2 | 15.6 | 46.3 |
| 11 | 17.2 | 6.58 | 140 | 2490 | 15.9 |
| 12 | 4.23 | ND | 87.8 | 307 | 1510 |
| 13 | 4.35 | 14.5 | 167 | 337 | 791 |
| 14 | 6.92 | 8.43 | 82.9 | 39.9 | 387 |
| 15 | 22.1 | 4.59 | 51.8 | ND | ND |
| 16 | 30.8 | 18.6 | 170 | 14.8 | 23.6 |
| 17 | 11.2 | 7.83 | 14.9 | 45.9 | 31.6 |
| 18 | 33.5 | ND | 14.9 | 12.5 | 3.06 |
| 19 | 26.3 | ND | 89.1 | 9.69 | 5.35 |
| Mean ± S.D.c | 14.2 ± 9.49 | 12.1 ± 17.9 | 163 ± 227 | 182 ± 568 | 194 ± 400 |
| B. Non-smokers Oral rinse sample | DNA (μg) a | α-OH-Acr-dGuo (1) | γ-OH-Acr-dGuo (2) | εdAdo (3) | εdCyd (4) |
|
adducts/109 nucleotides | |||||
| 1 | 1.54 | ND b | ND | ND | ND |
| 2 | 5.15 | ND | 10.1 | 3.74 | 12.7 |
| 3 | 9.81 | ND | 10.6 | 6.49 | 5.07 |
| 4 | 14.3 | 2.11 | 8.66 | 3.86 | 3.83 |
| 5 | 11.8 | 3.18 | 13.8 | 3.97 | 4.32 |
| 6 | 13.1 | ND | ND | 11.8 | 10.6 |
| 7 | 18.1 | ND | 7.21 | 6.64 | 5.96 |
| 8 | 18.9 | ND | ND | 55.9 | 50.2 |
| 9 | 20.9 | ND | 4.29 | 3.79 | 3.41 |
| 10 | 18.6 | ND | 8.31 | 5.58 | 5.01 |
| 11 | 31.2 | ND | 1.32 | 1.42 | ND |
| 12 | 30.4 | ND | 4.52 | 2.48 | ND |
| 13 | 9.48 | ND | 5.75 | 7.63 | ND |
| 14 | 23.3 | ND | 3.32 | 3.29 | ND |
| 15 | 6.12 | ND | 5.86 | 5.56 | ND |
| 16 | 27.3 | ND | 0.99 | 1.44 | ND |
| 17 | 9.32 | ND | ND | 3.69 | ND |
| 18 | 5.87 | ND | ND | 3.95 | ND |
| 19 | 14.8 | ND | 14.5 | 4.84 | ND |
| 20 | 23.3 | ND | 4.84 | 8.58 | ND |
| Mean ± S.D.c | 15.6 ± 8.61 | 1.85 ± 2.08 | 5.95 ± 4.23 | 7.69 ± 11.7 | 6.07 ± 10.9 |
calculated on the basis of the dGuo quantified by HPLC-UV
ND: not detected
calculated using ½ LOQ of the adduct for ND values
Table 3.
Levels Acr-dGuo (1 and 2), εdAdo (3), and εdCyd (4) in cytobrush DNA from smokers (A) and non-smokers (B). There is a spacing problem in part B
| A. Smokers Cytobrush sample | DNA (μg) a | α-OH-Acr-dGuo (1) | γ-OH-Acr-dGuo (2) | εdAdo (3) | εdCyd (4) |
| adducts/109 nucleotides | |||||
| 1 | 1.59 | ND b | 51.7 | 15.1 | ND |
| 2 | 2.65 | 38.9 | 1560 | 44.9 | ND |
| 3 | 1.88 | ND | 35.7 | 18.7 | ND |
| 4 | 1.64 | ND | 314 | 1200 | nd c |
| 5 | 2.17 | ND | ND | 12.1 | ND |
| 6 | 1.08 | 34.8 | 302 | 58.7 | 108 |
| 7 | 3.99 | ND | 15.6 | 9.24 | ND |
| 8 | 0.59 | 44.9 | 1950 | 58.2 | ND |
| 9 | 2.09 | ND | 146 | 16.3 | 32.9 |
| 10 | 0.66 | ND | 216 | 47.5 | ND |
| 11 | 2.44 | ND | 20.5 | 14.9 | ND |
| 12 | 2.86 | ND | 34.6 | 12.2 | 351 |
| 13 | 2.94 | ND | 83.2 | 9.05 | 14.1 |
| 14 | 2.16 | ND | 12.1 | 14.8 | 30.1 |
| 15 | 2.81 | ND | 20.8 | 11.9 | ND |
| 16 | 2.39 | ND | 23.5 | 11.8 | 49.2 |
| 17 | 3.02 | ND | 76.2 | 7.21 | 16.2 |
| 18 | 3.81 | ND | 29.5 | 10.7 | ND |
| 19 | 4.72 | ND | 19.7 | 6.33 | ND |
| Mean ± S.D. d | 2.27 ± 0.94 | 11.8 ± 13.1 | 259 ± 540 | 82.9 ± 271 | 36.9 ± 79.9 |
| B. Non-smokers Cytobrush sample | DNA (μg) a | α-OH-Acr-dGuo (1) | γ-OH-Acr-dGuo (2) | εdAdo (3) | εdCyd (4) |
| adducts/109 nucleotides | |||||
| 1 | 0.83 | ND b | ND | ND | ND |
| 2 | 2.83 | ND | ND | 9.19 | ND |
| 3 | 3.71 | ND | 16.5 | 8.91 | ND |
| 4 | 0.85 | ND | ND | 19.3 | ND |
| 5 | 1.76 | ND | ND | 11.3 | ND |
| 6 | 2.59 | ND | 10.8 | 14.4 | 33.3 |
| 7 | 1.31 | ND | ND | 11.4 | 1060 |
| 8 | 1.39 | ND | ND | 11.6 | ND |
| 9 | 1.44 | ND | ND | 10.7 | ND |
| 10 | 1.97 | ND | 11.3 | 7.41 | ND |
| 11 | 5.06 | ND | ND | 5.07 | ND |
| 12 | 9.52 | ND | ND | 4.62 | ND |
| 13 | 2.59 | ND | ND | 7.98 | ND |
| 14 | 3.58 | ND | ND | 11.4 | ND |
| 15 | 3.49 | ND | ND | 6.62 | ND |
| 16 | 5.26 | ND | ND | 5.12 | ND |
| 17 | 4.56 | ND | ND | 143 | 246 |
| 18 | 5.83 | ND | ND | 4.06 | ND |
| 19 | 2.92 | ND | ND | 7.69 | ND |
| 20 | 4.33 | ND | ND | 5.94 | ND |
| Mean ± S.D. d | 3.29 ± 2.10 | ND | 7.37 ± 5.09 | 16.2 ± 30.2 | 72.2 ± 238 |
calculated on the basis of the dGuo quantified by HPLC-UV
ND: not detected
nd: not determined
calculated using ½ LOQ of the adduct for ND values
For oral rinse samples, there was a statistically significant difference between smokers and non-smokers for adducts 1–4 (p=0.001 for adduct 1 and p<0.001 for adducts 2–4); for cytobrush samples, there was a statistically significant difference between smokers and non-smokers for adducts 2 (p<0.001) and 3 (p=0.009). We did not observe a consistent difference in any of the adduct levels between male and female smokers or male and female non-smokers and there was at most a weak correlation between adducts 1–4 levels in oral rinse and cytobrush samples from the same subjects.
We investigated intra-day and inter-day variation of adducts 1–4 obtained by oral rinse samples collected from three non-smoking subjects on the same day at 3 time points (9:30 am, 12:30 pm, 3:30 pm) and on 3 separate days. Adduct 1 was not detected in any of the samples analyzed in this experiment. The overall intra-individual variation was 21% for adduct 2, 27% for adduct 3, and 24% for adduct 4. The overall inter-day variation was 61% for adduct 2, 56% for adduct 3, and 63% for adduct 4.
Discussion
We present a validated LC-NSI-HRMS/MS method for the combined analysis of Acr-dGuo (1 and 2), εdAdo (3), and εdCyd (4) in oral cells of smokers and non-smokers. The accuracy, precision, and detection limits of the new method were established thus providing a path for the quantitation of these important DNA modifications. This is apparently the first application of HRMS for quantitative analysis of oral cell DNA adducts. The results of this study clearly demonstrate the effect of cigarette smoking on levels of adducts 2 and 3, which were significantly higher in smokers than non-smokers in samples collected by both oral rinse and cytobrush. Levels of adducts 1 and 4 were also significantly higher in smokers vs. non-smoker DNA samples obtained by oral rinse but not in the cytobrush DNA samples.
The 27-fold difference of adduct 2 between smokers’ and non-smokers’ oral rinse samples and the 36-fold difference of adduct 2 between smokers’ and non-smokers’ cytobrush samples were highly significant (p<0.001). These results contrast with our previous studies of acrolein-DNA adducts using similar mass spectrometry based methods in which we found no significant differences when comparing these adduct levels in lung tissue or leukocytes from smokers vs. non-smokers.28, 30 The data from these studies are summarized in Table 4 and compared with the data reported here.
Table 4.
Comparison of acrolein-DNA adducts with previous studies.
| α-OH-Acr-dGuo (1) adducts/109 nucleotides |
γ-OH-Acr-dGuo (2) adducts/109 nucleotides |
|||
|---|---|---|---|---|
| DNA Sample Type | Smokers | Non-smokers | Smokers | Non-smokers |
| Lung30 | 8.6 ± 2.7 | 10.3 ± 4.7 | 19.9 ± 14.3 | 14.7 ± 6.4 |
| Leukocyte28 | ND | ND | 7.0 ± 2.5 | 9.7 ± 5.5 |
| Oral rinse | 12.1 ± 17.9 | 1.85 ± 2.08 | 163 ± 227 | 5.95 ± 4.23 |
| Cytobrush | 1.8 ± 13.1 | ND | 259 ± 540 | 7.37 ± 5.09 |
We also observed that the levels of adduct 2 in oral cell DNA from smokers were significantly higher than in lung or leukocyte DNA. These results as well as the differences between smokers and non-smokers in levels of adduct 2 undoubtedly reflect the fact that the oral cavity is the first site of exposure to cigarette smoke. Levels of acrolein – averaging 177 μg per cigarette in one study, and a range of 139–213 μg per cigarette in another, both under Health Canada smoking conditions, and 30.8–82.6 μg per cigarette under ISO conditions – are very high.17, 31 Acrolein reacts directly with DNA to form adducts 1 and 2; no metabolism is required.8 Clearly, adduct 2 in oral cell DNA is an exposure biomarker for acrolein. The extent to which it may also be a biomarker of effect requires further study.
Glutathione is important in the detoxification of acrolein. Thus, cigarette smokers have 4–5 times higher levels of urinary 3-hydroxypropyl mercapturic acid, the major end product of glutathione detoxification of acrolein, than non-smokers, and levels of this metabolite rapidly decrease after smoking cessation.32, 33 We have previously speculated that glutathione detoxification of acrolein is efficient in smokers, thus controlling levels of adduct 2 in lung and leukocyte DNA such that they did not differ from non-smokers, but that was not observed here.28, 30 Cellular glutathione concentrations have been reported to be higher in human lung tissue (11.6 ± 3.0 nmol/mg protein) than in human oral mucosa (1 – 4.80 nmol/mg protein).34–36 The higher glutathione concentrations in lung tissue compared to oral tissue may play some role in the lower levels of adduct 2 in lung DNA compared to oral mucosa DNA, but further studies are needed.
Levels of adduct 2 in oral cells collected both by oral rinse and cytobrush were higher than those of adduct 1. These results are consistent with most other studies that have quantified these adducts by mass spectrometry in various tissues, but differ from our results in the analysis of DNA from human lung, in which comparable amounts of adducts 1 and 2 were found.13, 24, 26, 28–30, 37 Amino acids and proteins can facilitate the formation of adduct 2 compared to 1.37 It is unclear whether this mechanism is involved in the differential levels of adducts 1 and 2 in lung versus oral cells or leukocytes, or whether other mechanisms facilitate formation of adduct 1 in human lung.
As noted in the Introduction, several previous studies have investigated levels of acrolein adducts 1 and 2 in oral tissues and saliva. The amounts reported were generally consistent with those found here. The study by Nath et al. using gingival tissue reported significantly higher levels of adduct 2 in smokers than non-smokers, consistent with our results, while that of Li et al. found no difference in levels of adduct 2 in salivary DNA from smokers compared to non-smokers.24, 26 The other studies did not present comparative data.13, 25 A recent study using immunochemical methods without apparent artifact prevention reported levels of adduct 2 considerably higher than in any of these studies.38
Levels of adduct 3 were also significantly higher in smokers than in non-smokers, for samples collected by both oral rinse and cytobrush, while those of adduct 4 were higher only in the oral rinse samples. Our data are somewhat lower compared to the only previous report of these adduct levels in the oral cavity, based on data from human saliva. Thus, Chen and Lin found levels of 99 ± 50 and 72 ± 49 adducts 3 and 4, per 108 normal nucleotides, respectively, in human salivary DNA samples from healthy volunteers; smoking status was not reported.13
Lipid peroxidation and related oxidative processes as well as inflammation are closely associated with cigarette smoking and lead to the formation of etheno-DNA adducts. Thus, cigarette smoking consistently produces elevated levels of 8-iso-PGF2α, an F2 isoprostane and biomarker of oxidative damage and PGE-M, a biomarker of inflammation, among others.39–41 The elevated levels of adduct 3 and 4 observed in the oral cells of smokers in this study are consistent with these previous studies. However, there was quite substantial variation in the levels of adduct 3 and 4 in both smokers and non-smokers, suggesting that other factors influencing the inflammatory response likely contribute, and further studies are needed. As mentioned above, during sample collection, participants were asked to take a breathalyzer test to measure their blood alcohol concentration and to provide some personal information, including alcohol consumption. Alcohol can be one of the factors that influences levels of adducts 3 and 4 and that may have contributed to the high variation observed in this study, but our relatively small sample size prevents further analysis of this variable.
Inter-subject variations of adducts 1–4 were large and far greater than the intra-subject variations which ranged from 27 – 63%, depending on the adduct and the day measured. This indicates that the inter-subject variations were due to inter-individual differences in exposure and endogenous factors affecting the DNA adduct levels.
The classical initiation promotion and co-carcinogenicity models of tobacco carcinogenesis were established by decades of research demonstrating that cigarette smoke contains both types of activities, and that initiating and promoting/co-carcinogenic activities were both required for its carcinogenicity.42 Thus, tumor initiators such as benzo[a]pyrene and other polycyclic aromatic hydrocarbons as well as tumor promoters and co-carcinogens combined to demonstrate the effects of tobacco smoke as a complete carcinogen in both rodent inhalation and skin painting studies. The results presented here potentially recapitulate the initiation/promotion model of tobacco carcinogenesis in the human oral mucosa by demonstrating elevated DNA adduct formation in cigarette smokers by a genotoxic compound (acrolein) and products of inflammation (adducts 3 and 4). The weakness in this argument is that acrolein, while highly toxic, is a relatively weak mutagen and has only limited or no demonstrated carcinogenicity.9, 20 In contrast to acrolein, oral cell DNA adducts of mutagenic oral cavity carcinogens such as N’-nitrosonornicotine and dibenzo[a,l]pyrene could provide the genotoxic stimulus to act together with etheno-DNA adducts and other inflammatory/oxidative damage processes to result in oral cavity cancer as observed in smokers.43–45
There was a poor correlation between adduct levels obtained by cytobrush and mouthwash from the same individuals. This undoubtedly reflects the methods of collection. Mouthwash will capture saliva and perhaps certain cells that are being shed whereas collection with a cytobrush will likely provide a more homogeneous cell population. The contribution of bacterial DNA to the data reported here is also unknown.
In summary, this study establishes a highly reliable LC-NSI-HRMS-MS method for the quantitation of DNA adducts 1–4 in human oral cell DNA. The results clearly demonstrate elevated levels of acrolein-DNA adduct γ-OH-Acr-dGuo (2) and εdAdo (3) in oral cells from smokers vs. non-smokers independent of the method of collection while levels of α-OH-Acr-dGuo (1) and εdCyd (4) were higher in smokers compared to non-smokers only in the samples collected by mouthwash. These results provide potentially important leads for the use of oral cell DNA adducts as biomarkers of tobacco smoke exposure and biological effects.
Supplementary Material
Acknowledgements
The authors thank Yingchun Zhao for his help with mass spectrometry, carried out in the Analytical Biochemistry Shared Resources of the Masonic Cancer Center, University of Minnesota and Bob Carlson for his editorial assistance.
Funding
This study was supported by grant P01-CA-138338 from the National Cancer Institute. Mass spectrometry was carried out in the Analytical Biochemistry Shared Resource of the Masonic Cancer Center, supported in part by Cancer Center Support Grant CA-077598.
Footnotes
Supporting Information
Protocols for [15N]DNA generation and isolation from bacteria. Further details of artifact control studies.
References
- 1.World Health Organization. Tobacco Fact Sheet, https://www.who.int/news-room/fact-sheets/detail/tobacco (accessed May 27, 2020).
- 2.Centers for Disease Control and Prevention. Current Cigarette Smoking Among Adults in the United States, https://www.cdc.gov/tobacco/data_statistics/fact_sheets/adult_data/cig_smoking/index.htm (accessed May 27, 2020).
- 3.U.S. Department of Health and Human Services. (2014) The Health Consequences of Smoking: 50 Years of Progress. A Report of the Surgeon General, U.S. Dept. of Health and Human Services, Centers for Disease Control and Prevention, Coordinating Center for Health Promotion, National Center for Chronic Disease Prevention and Health Promotion, Office on Smoking and Health, Atlanta, GA. [Google Scholar]
- 4.International Agency for Research on Cancer. (2012) Personal Habits and Indoor Combustions, IARC Monographs on the Evaluation of Carcinogenic Risks to Humans, v. 100E. IARC, Lyon, FR. [PMC free article] [PubMed] [Google Scholar]
- 5.Hecht SS (2017) Oral cell DNA adducts as potential biomarkers for lung cancer susceptibility in cigarette smokers. Chem Res Toxicol 30, 367–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Murphy SE, Park SL, Balbo S, Haiman CA, Hatsukami DK, Patel Y, Peterson LA, Stepanov I, Stram DO, Tretyakova N, Hecht SS, and Le Marchand L (2018) Tobacco biomarkers and genetic/epigenetic analysis to investigate ethnic/racial differences in lung cancer risk among smokers. NPJ Precis Oncol 2, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ma B, Stepanov I, and Hecht SS (2019) Recent studies on DNA adducts resulting from human exposure to tobacco smoke. Toxics 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chung FL, Young R, and Hecht SS (1984) Formation of cyclic 1,N2-propanodeoxyguanosine adducts in DNA upon reaction with acrolein or crotonaldehyde. Cancer Res 44, 990–995. [PubMed] [Google Scholar]
- 9.Minko IG, Kozekov ID, Harris TM, Rizzo CJ, Lloyd RS, and Stone MP (2009) Chemistry and biology of DNA containing 1,N2-deoxyguanosine adducts of the α,ß-unsaturated aldehydes acrolein, crotonaldehyde, and 4-hydroxynonenal. Chem. Res. Toxicol 22, 759–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu Y, Cui Y, Niedernhofer LJ, and Wang Y (2016) Occurrence, biological consequences, and human health relevance of oxidative stress-induced DNA damage. Chem Res Toxicol 29, 2008–2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Linhart K, Bartsch H, and Seitz HK (2014) The role of reactive oxygen species (ROS) and cytochrome P-450 2E1 in the generation of carcinogenic etheno-DNA adducts. Redox Biol 3, 56–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Monien BH, Schumacher F, Herrmann K, Glatt H, Turesky RJ, and Chesne C (2015) Simultaneous detection of multiple DNA adducts in human lung samples by isotope-dilution UPLC-MS/MS. Anal Chem 87, 641–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen HJ, and Lin WP (2011) Quantitative analysis of multiple exocyclic DNA adducts in human salivary DNA by stable isotope dilution nanoflow liquid chromatography-nanospray ionization tandem mass spectrometry. Anal Chem 83, 8543–8551. [DOI] [PubMed] [Google Scholar]
- 14.Nair U, Bartsch H, and Nair J (2007) Lipid peroxidation-induced DNA damage in cancer-prone inflammatory diseases: A review of published adduct types and levels in humans. Free Radical Biology & Medicine 43, 1109–1120. [DOI] [PubMed] [Google Scholar]
- 15.Nair J, Barbin A, Velic I, and Bartsch H (1999) Etheno DNA-base adducts from endogenous reactive species. Mutat. Res 424, 59–69. [DOI] [PubMed] [Google Scholar]
- 16.Chung FL, Chen HJC, and Nath RG (1996) Lipid peroxidation as a potential source for the formation of exocyclic DNA adducts. Carcinogenesis 17, 2105–2111. [DOI] [PubMed] [Google Scholar]
- 17.Bodnar JA, Morgan WT, Murphy PA, and Ogden MW (2012) Mainstream smoke chemistry analysis of samples from the 2009 US cigarette market. Regul Toxicol Pharmacol 64, 35–42. [DOI] [PubMed] [Google Scholar]
- 18.Haussmann HJ (2012) Use of hazard indices for a theoretical evaluation of cigarette smoke composition. Chem Res Toxicol 25, 794–810. [DOI] [PubMed] [Google Scholar]
- 19.Yeager RP, Kushman M, Chemerynski S, Weil R, Fu X, White M, Callahan-Lyon P, and Rosenfeldt H (2016) Proposed mode of action for acrolein respiratory toxicity associated with inhaled tobacco smoke. Toxicol Sci 151, 347–364. [DOI] [PubMed] [Google Scholar]
- 20.International Agency for Research on Cancer. (1995) Acrolein, In IARC Monographs on the Evaluation of Carcinogenic Risks to Humans , v. 63 pp 337–372, IARC, Lyon, FR. [PMC free article] [PubMed] [Google Scholar]
- 21.Feng Z, Hu W, Hu Y, and Tang M-S (2006) Acrolein is a major cigarette-related lung cancer agent. Preferential binding at p53 mutational hotspots and inhibition of DNA repair. Proc. Natl. Acad. Sci. USA 103, 15404–15409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Denissenko MF, Pao A, Tang M, and Pfeifer GP (1996) Preferential formation of benzo[a]pyrene adducts at lung cancer mutational hot spots in P53. Science 274, 430–432. [DOI] [PubMed] [Google Scholar]
- 23.Pfeifer GP, Denissenko MF, Olivier M, Tretyakova N, Hecht SS, and Hainaut P (2002) Tobacco smoke carcinogens, DNA damage and p53 mutations in smoking-associated cancers. Oncogene 21, 7435–7451. [DOI] [PubMed] [Google Scholar]
- 24.Nath RG, Ocando JE, Guttenplan JB, and Chung FL (1998) 1,N2-Propanodeoxyguanosine adducts: Potential new biomarkers of smoking-induced DNA damage in human oral tissue. Cancer Res 58, 581–584. [PubMed] [Google Scholar]
- 25.Bessette EE, Goodenough AK, Langouet S, Yasa I, Kozekov ID, Spivack SD, and Turesky RJ (2009) Screening for DNA adducts by data-dependent constant neutral loss-triple stage mass spectrometry with a linear quadrupole ion trap mass spectrometer. Anal. Chem 81, 809–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li X, Liu L, Wang H, Chen J, Zhu B, Chen H, Hou H, and Hu Q (2017) Simultaneous analysis of six aldehyde-DNA adducts in salivary DNA of nonsmokers and smokers using stable isotope dilution liquid chromatography electrospray ionization-tandem mass spectrometry. J Chromatogr B 1060, 451–459. [DOI] [PubMed] [Google Scholar]
- 27.Emami A, Dyba M, Cheema AK, Pan J, Nath RG, and Chung FL (2008) Detection of the acrolein-derived cyclic DNA adduct by a quantitative 32P-postlabeling/solid-phase extraction/HPLC method: blocking its artifact formation with glutathione. Anal. Biochem 374, 163–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang S, Balbo S, Wang M, and Hecht SS (2011) Analysis of acrolein-derived 1,N2-propanodeoxyguanosine adducts in human leukocyte DNA from smokers and nonsmokers. Chem. Res. Toxicol 24, 119–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang S, Villalta PW, Wang M, and Hecht SS (2007) Detection and quantitation of acrolein-derived 1,N2-propanodeoxyguanosine adducts in human lung by liquid chromatography-electrospray ionization-tandem mass spectrometry. Chem. Res. Toxicol 20, 565–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang J, Balbo S, Villalta PW, and Hecht SS (2019) Analysis of acrolein-derived 1,N2-propanodeoxyguanosine adducts in human lung DNA from smokers and nonsmokers. Chem Res Toxicol 32, 318–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cecil TL, Brewer TM, Young M, and Holman MR (2017) Acrolein yields in mainstream smoke from commercial cigarette and little cigar tobacco products. Nicotine Tob Res 19, 865–870. [DOI] [PubMed] [Google Scholar]
- 32.Alwis KU, deCastro BR, Morrow JC, and Blount BC (2015) Acrolein exposure in U.S. tobacco smokers and non-tobacco users: NHANES 2005–2006. Environ Health Perspect 123, 1302–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carmella SG, Chen M, Han S, Briggs A, Jensen J, Hatsukami DK, and Hecht SS (2009) Effects of smoking cessation on eight urinary tobacco carcinogen and toxicant biomarkers. Chem Res Toxicol 22, 734–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wong DY, Hsiao YL, Poon CK, Kwan PC, Chao SY, Chou ST, and Yang CS (1994) Glutathione concentration in oral cancer tissues. Cancer Lett 81, 111–116. [DOI] [PubMed] [Google Scholar]
- 35.Blair SL, Heerdt P, Sachar S, Abolhoda A, Hochwald S, Cheng H, and Burt M (1997) Glutathione metabolism in patients with non-small cell lung cancers. Cancer Res 57, 152–155. [PubMed] [Google Scholar]
- 36.Wang TW, Liu JH, Tsou HH, Liu TY, and Wang HT (2019) Identification of acrolein metabolites in human buccal cells, blood, and urine after consumption of commercial fried food. Food Sci Nutr 7, 1668–1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chung FL, Wu MY, Basudan A, Dyba M, and Nath RG (2012) Regioselective formation of acrolein-derived cyclic 1,N(2)-propanodeoxyguanosine adducts mediated by amino acids, proteins, and cell lysates. Chem Res Toxicol 25, 1921–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weng MW, Lee HW, Park SH, Hu Y, Wang HT, Chen LC, Rom WN, Huang WC, Lepor H, Wu XR, Yang CS, and Tang MS (2018) Aldehydes are the predominant forces inducing DNA damage and inhibiting DNA repair in tobacco smoke carcinogenesis. Proc Natl Acad Sci U S A 115, E6152–E6161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McElroy JP, Carmella SG, Heskin AK, Tang MK, Murphy SE, Reisinger SA, Jensen JA, Hatsukami DK, Hecht SS, and Shields PG (2019) Effects of cessation of cigarette smoking on eicosanoid biomarkers of inflammation and oxidative damage. PLoS One 14, e0218386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Milne GL, Yin H, and Morrow JD (2008) Human biochemistry of the isoprostane pathway. J Biol Chem 283, 15533–15537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mohebati A, Milne GL, Zhou XK, Duffield-Lillico AJ, Boyle JO, Knutson A, Bosworth BP, Kingsley PJ, Marnett LJ, Brown PH, Akpa EG, Szabo E, and Dannenberg AJ (2013) Effect of zileuton and celecoxib on urinary LTE4 and PGE-M levels in smokers. Cancer Prev Res 6, 646–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.U.S. Department of Health and Human Services. (2010) How Tobacco Smoke Causes Disease: The Biology and Behavioral Basis for Smoking-Attributable Disease: A Report of the Surgeon General. Ch. 5, U.S. Department of Health and Human Services, Washington, D.C. [Google Scholar]
- 43.Khariwala SS, Ma B, Ruszczak C, Carmella SG, Lindgren B, Hatsukami DK, Hecht SS, and Stepanov I (2017) High level of tobacco carcinogen-derived DNA damage in oral cells is an independent predictor of oral/head and neck cancer risk in smokers. Cancer Prev Res (Phila) 10, 507–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.El-Bayoumy K, Chen KM, Zhang SM, Sun YW, Amin S, Stoner G, and Guttenplan JB (2017) Carcinogenesis of the oral cavity: Environmental causes and potential prevention by black raspberry. Chem Res Toxicol 30, 126–144. [DOI] [PubMed] [Google Scholar]
- 45.Balbo S, James-Yi S, Johnson CS, O’Sullivan G, Stepanov I, Wang M, Bandyopadhyay D, Kassie F, Carmella S, Upadhyaya P, and Hecht SS (2013) (S)-N’-Nitrosonornicotine, a constituent of smokeless tobacco, is a powerful oral cavity carcinogen in rats. Carcinogenesis 34, 2178–2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.