Abstract
Background
Malignant melanoma of the female genital tract is relatively uncommon and accounts for 3–7% of all melanoma localizations. This study aimed to identify driver genes in melanoma of the female genital tract with the purpose of enhancing understanding of disease pathogenesis and identifying potential new therapeutic targets to develop effective therapies.
Methods
KIT (CD117) and BRAF expression were detected immunohistochemically. Polymerase Chain Reaction (PCR) and Sanger sequencing techniques were performed to identify the mutational status of BRAF, NRAS, KRAS, NF1, KIT, PDGFRA and SF3B1 on 19 melanomas of the female genital tract, paired with 25 cutaneous melanomas, 18 acral melanomas and 11 melanomas of nasal cavity.
Results
Somatic variant analysis identified KRAS (6/19; 32%) as the most commonly mutated gene, followed by KIT (4/19; 21%), SF3B1 (3/19; 16%) and NRAS (1/19; 5%). None of the cases were found to harbor BRAF, NF1 and PDGFRA mutations in melanomas of the female genital tract. However, none of the cases were found to harbor SF3B1 and KIT mutations in cutaneous melanomas, acral melanomas and melanomas of nasal cavity. Recurrent KIT mutations, as well as mutations in the less frequently mutated genes NRAS and SF3B1, were exclusively detected in vulvovaginal melanomas, but not in tumors arising in the cervix. However, recurrent KRAS mutations were detected in similar frequencies in tumors of the vulva, vagina, and cervix. Additionally, recurrent KRAS and KIT mutations occurred predominantly in polygonal and epithelioid cell types of melanoma in the female genital tract. Immunohistochemistry revealed moderate or strong cytoplasmic CD117 expression in 6 of the 19 cases (31.6%).
Conclusions
We observed that gynecologic melanoma harbored distinct mutation rates in the KIT, BRAF, SF3B1, KRAS, and NRAS genes. Our findings support the notion that gynecologic melanoma is a distinct entity from non-gynecologic melanoma, and these findings offer insights into future therapeutic options for these patients.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12885-021-08427-x.
Keywords: Malignant melanoma, Female genital tract, Mutation, Targeted therapy, Driver genes
Introduction
Primary malignant melanomas arise from melanocytes which present in the genital mucosal epithelium of the female genital tract are extremely rare, accounting for 3–7% of all melanoma localizations [1, 2]. The most common site of melanomas of the female genital tract is the vulva (75%), followed by the vagina, and 20% of cases are multifocal [3]. Other sites such as cervix, uterus and ovary are very rare. Patients more frequently present with early metastatic spread and poor prognosis, and the 5-year overall survival rate is approximately 10% [4]. In recent years, many of the advances made in the treatment of cutaneous melanoma with the aim of improving overall survival, but not readily applicable to mucosal melanomas [5, 6].
Mucosal melanomas are not related to sun exposure or other known environmental factors; therefore, mucosal melanomas have been shown to have distinct molecular profiles compared with cutaneous melanoma arising on sun-exposed sites [7, 8]. Some common activating driver mutations in genes identified in cutaneous melanoma, such as mutated BRAF V600E [9], have less frequently identified in mucosal melanoma [10–19]. To better understand the mutational profile and identify potential new therapeutic targets of mucosal melanoma, several investigators have performed next-generation sequencing on a small number of mucosal melanomas from different anatomical sites and compared the results with cutaneous melanoma [7, 20–23]. Hayward and colleagues [21] found that acral and mucosal melanomas were dominated by structural changes and mutation signatures of unknown etiology that were not previously identified in melanoma, such as in SF3B1. Newell and colleageues [22] also found that mucosal melanomas had a low point mutation burden and high numbers of structural variants; the significantly mutated genes in mucosal melanomas included NRAS, BRAF, NF1, KIT, SF3B1, TP53, SPRED1, ATRX, HLA-A and CHD8 genes. These studies have provided the foundation for understanding the molecular profiles of mucosal melanoma and expanded our knowledge of this rare disease. The distinct mutation profiles of cutaneous and mucosal melanoma are striking and support a model of different developmental pathways.
To better understand the mutational profile and provide important new clues into the molecular changes that occur in melanomas of the female genital tract, our study analyzed the molecular drivers of 19 melanomas of the female genital tract (vulva, vagina and cervix), paired with 25 cutaneous melanomas, 18 acral melanomas and 11 melanomas of nasal cavity. Our findings expand this knowledge by contributing the large cohort of mucosal melanoma of the female genital tract known to date with validated mutations and may lead to a better understanding of the molecular drivers, and hence improved therapeutics for these rare forms of melanoma.
Material and methods
Case collection and histological assessment
Nineteen melanomas of the female genital tract, 25 cutaneous melanomas, 18 acral melanomas and 11 melanomas of nasal cavity were obtained from the case files of the Department of Pathology of Fujian Cancer Hospital, China, from October 2010 to September 2019. The biopsy material was fixed in 10% formalin and embedded in paraffin after routine histological tissue processing. Formalin-fixed paraffin-embedded tissue (FFPE) sections (3–4 μm thick) were stained with hematoxylin-eosin for microscopic examination. All cases were carefully reviewed independently by two melanoma pathologists to confirm the histological diagnosis according to the criteria of the 2014 WHO Classification of Tumors Female Reproductive Organ, and extensive review of medical records was performed. This study was carried out in accordance with the Declaration of Helsinki and written informed consent was obtained from the patients or their legal guardians. The study protocol was approved by the institutional review boards of Fujian Cancer Hospital.
Immunohistochemistry
Immunohistochemistry assays were performed on diagnosed patient tissues available in the form of FFPE tissue blocks using Bond-III Autostainer (Leica Biosystems, Melbourne, Australia) in the Department of Pathology’s clinical immunohistochemistry laboratory. Immunostaining on paraffin sections was done for CK, EMA, S-100, HMB45, Melan-A, SOX-10, Ki67, KIT (CD117) and BRAF (using antibody clones from Maixin Biotech Co., Ltd., Fuzhou China). Conditions for individual immunohistochemistry assays including antigen retrieval and antibody dilutions varied according to the antibody used and were determined by standard optimization and validation procedures. Positive and negative staining controls were included as appropriate.
DNA isolation
Five pieces (5-μm-thick sections) were cut from FFPE tumor tissues. The sections were deparaffinized and manually microdissected according to standard procedures. Genomic DNA was isolated using the QIAamp FFPE DNA Tissue Kit (Qiagen, Germantown, MD, USA) according to the manufacturer’s instructions. Samples were quantified using the Qubit DNA high sensitivity assay (Life Technologies, Carlsbad, USA).
Sanger sequencing
Nested PCR was performed to amplify BRAF exon 15; NRAS exons 2, 3 and 4; KRAS exons 2, 3 and 4; KIT exons 9, 11, 13, 17 and 18; PDGFRA exons 12 and 18; NF1 exon 22; and SF3B1 exon 14 on 19 melanomas of the female genital tract, as previously described [24–26]. PCR was carried out using AmpliTaq Gold polymerase (Applied Biosystems, Weiterstadt, Germany) according to the manufacturer’s instructions under the following conditions: 95 °C for 5 min followed by 38 cycles of denaturation for 30 s at 94 °C, annealing for 30 s at 58 °C and extension for 60 s at 72 °C. The primer pairs for BRAF, NRAS, KRAS, PDGFRA, KIT, NF1 and SF3B1 were designed using Primer Premier 5. The PCR products were routinely purified and sequenced in both directions using the BigDye Terminator version 3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA). At least two independent PCR and sequencing experiments were conducted to confirm mutations.
Next-generation sequencing assay
The panel of targeted genes was designed on the basis of large-scale mutation-profiling studies on melanomas covering BRAF, NRAS, KRAS, NF1, KIT, PDGFRA and SF3B1 [21, 22]. The genomes of 25 cutaneous melanomas, 18 acral melanomas and 11 melanomas of nasal cavity with eligible formalin-fixed and paraffin-embedded samples were sequenced using Nextseq. 500 (Illumina, Inc., USA) instrument. The raw data were aligned and analyzed for the detection of insertions/deletions and single-nucleotide variants. The detail process of NGS library preparation, capture-based targeted DNA sequencing and data analysis were described in supplementary material.
Results
Clinicopathologic findings of patients with melanomas of the female genital tract
This study included 19 patients with primary melanomas of the female genital tract, with a median patient age of 53.0 years and an average age of 55.4 years at diagnosis (range from 34 to 84 years). The primary melanomas of the female genital tract included five vulvar (26.3%), eight vaginal (42.1%) and six cervical melanomas (31.6%). Six (31.6%) patients had T4 disease, two (10.5%) had T3 disease, three (15.8%) patients had T1 disease and the remaining eight (42.1%) patients had T2 disease. The clinicopathological features of the patients are summarized in Table 1.
Table 1.
Cumulative clinical data and mutational status for 19 patients with mucosal melanoma of female genital tract
| Clinical information | Mutational status | CD117 expression |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patients | Tumor location | Age at diagnosis | Clinical stage | histological subtype | SF3B1 | KIT | PDGFRA | BRAF | NRAS | KRAS | NF1 | |
| P01 | vagina | 56 | I | spindle | – | – | – | – | – | – | – | |
| P02 | vagina | 40 | II | mixed type | – | – | – | – | – | – | – | |
| P03 | vulva | 50 | III | mixed type | – | – | – | – | p.Q61 * | – | – | |
| P04 | vagina | 53 | II | epithelioid | – | p.S476N p.G498V | – | – | – | – | – | + |
| P05 | vulva | 53 | I | epithelioid | – | p.L640P | – | – | – | – | – | + |
| P06 | vagina | 77 | II | epithelioid | – | – | – | – | – | – | – | |
| P07 | cervix | 48 | I | epithelioid | – | – | – | – | – | – | – | |
| P08 | vagina | 84 | II | epithelioid | – | – | – | – | – | – | – | |
| P09 | vagina | 52 | II | epithelioid | – | p.D810H | – | – | – | p.G13D | – | + |
| P10 | cervix | 42 | IV | spindle | – | – | – | – | – | p.G13D | – | |
| P11 | vulva | 59 | II | epithelioid | p.R625H | – | – | – | – | p.G13D | – | |
| P12 | cervix | 55 | IV | epithelioid | – | – | – | – | – | p.G12D | – | + |
| P13 | vulva | 74 | IV | mixed type | – | – | – | – | – | – | – | |
| P14 | vulva | 72 | IV | spindle | p.R625H | – | – | – | – | – | – | |
| P15 | vagina | 56 | IV | epithelioid | – | – | – | – | – | p.G12D | – | |
| P16 | cervix | 42 | II | spindle | – | – | – | – | – | – | – | |
| P17 | cervix | 34 | III | mixed type | – | – | – | – | – | – | – | |
| P18 | vagina | 52 | II | epithelioid | p.R625H | p.V852A | – | – | – | – | – | + |
| P 19 | cervix | 54 | IV | epithelioid | – | – | – | – | – | p.G13D | – | + |
the variant type: SNV; mutation type: missense_variant; mixed type: both epithelioid and spindle tumor cells
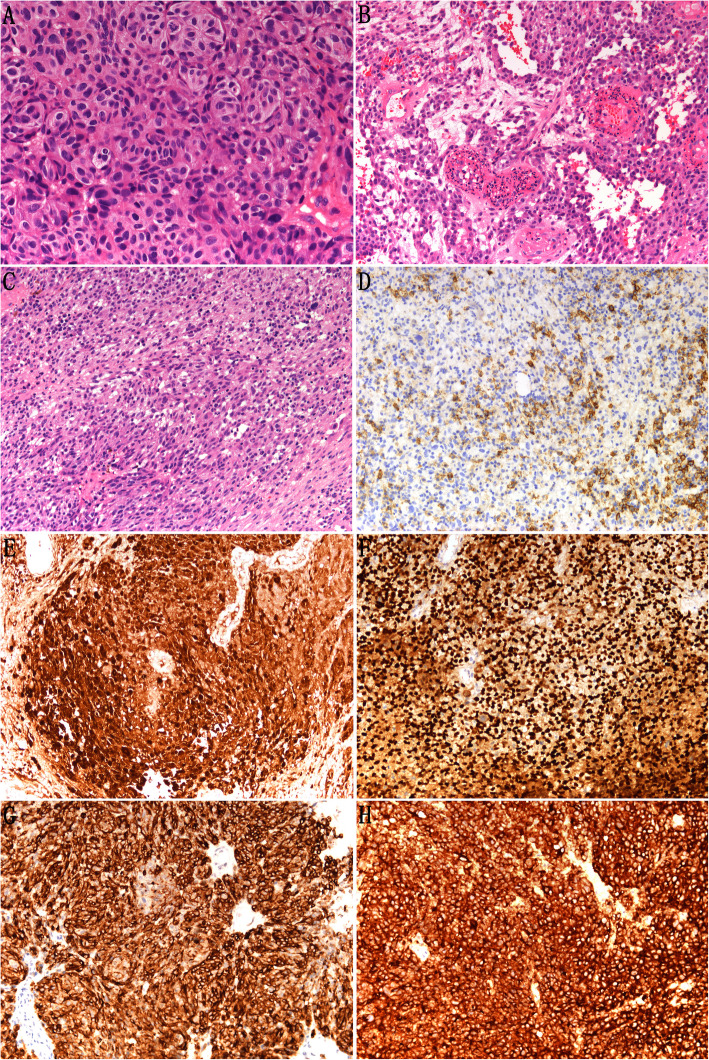
Microscopic examination revealed that the tumor cells were distributed in solid islets, nests or band-like formations. Tumor cells were polygonal, epithelioid or spindle in shape and rarely of small cell type, with oval, pleomorphic, hyperchromic nuclei (Fig. 1A–C). In most cases, diffuse or focal tumor cells contained dark-brown intracellular melanin pigment. No melanin pigment was found microscopically in a few cases. Mitoses were easy to see. The average number of mitotic figures in the tumor cells per square millimeter was 3–5. Immunohistochemical analysis revealed that CK was negative in most cases and weakly focally positive in three cases (3/19, 15.8%) (Fig. 1D and Table 1). The tumor cells were strongly positive for S-100 and SOX-10 protein (Fig. 1 E–F). Positive Melan-A and HMB-45 protein expressions were also detected, with somewhat lower intensity of staining (Fig.1 G–H). Tumor cells were negative for EMA (data not shown). These findings supported the diagnosis of melanoma.
Fig. 1.
Representative hematoxylin and eosin staining and immunostaining of mucosal melanoma of the female genital tract. Microscopic examination revealed that the tumor cells were distributed in solid islets, nests, or band-like formations. Tumor cells were polygonal (A), epithelioid (B), or spindle in shape (C) (× 200 magnification). Immunohistochemical analysis revealed that CK expression was negative in most cases, but weakly focally positive in three of the 19 cases (D) (× 200 magnification). Tumor cells were strongly positive for Melan-A (E), HMB45 (F), S100 (G) and SOX-10 (H) protein in vulvar malignant melanomas (× 200 magnification)
CD117 and BRAF expression in patients with melanomas of the female genital tract
Immunohistochemistry revealed moderate or strong cytoplasmic CD117 expression in 6 of the 19 cases (31.6%) (as shown in Fig. 2A). The remaining cases (68.4%) were negative for CD117 (as shown in Fig. 2B). Interestingly, none of the cases were positive for BRAF (as shown in Fig. 2C).
Fig. 2.
Representative immunostaining of CD117 and BRAF in mucosal melanomas of the female genital tract. (A) Representative staining showing tumor cells strongly positive for CD117. (B) Representative staining showing tumor cells negative for CD117. (C) Representative staining showing tumor cells negative for BRAF
BRAF, NRAS, KRAS, NF1, KIT, PDGFRA and SF3B1 gene mutation analysis
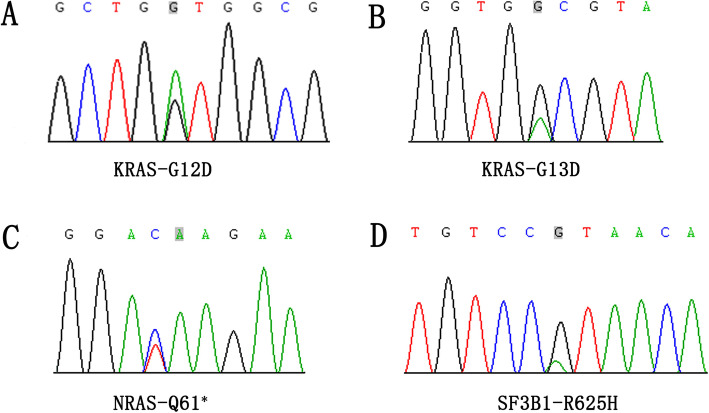
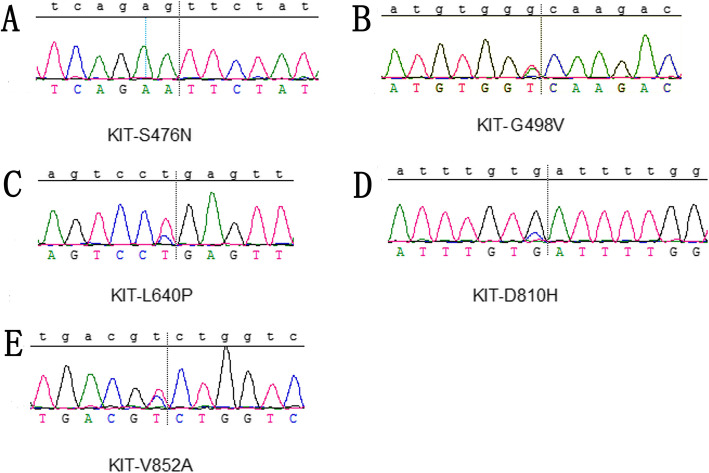
We examined gene mutations in the 19 melanomas of the female genital tract (listed in Table 1). We identified six cases with KRAS mutations (four p.G13D, two p.G12D) and one cases with NRAS mutation (p.Q61*) (Fig. 3 A–C). In total, 37% of tumors showed either a KRAS or NRAS mutation (32% KRAS mutation, 5% NRAS mutation). The SF3B1 p.R625H hotspot mutation was detected in 16% (3/19) of the mucosal melanomas of the female genital tract (Fig. 3D). Notably, recurrent KIT mutations (p.S476N, p.G498V, p.L640P, p.D810H, p.V852A) were found in 21% (4/19) of the melanomas of the female genital tract; one sample was found to harbor both p.S476N and p.G498V mutations (Fig. 4). In addition, one case had both KRAS and C-KIT mutations. All four tumors with recurrent KIT mutations showed strong CD117 immunostaining. In contrast, oncogenic driver mutations in BRAF, which are commonly identified in cutaneous melanoma, were not detected in any sample (data not shown). None of the cases were found to harbor NF1 and PDGFRA mutations (data not shown).
Fig. 3.
Representative examples of KRAS (A, G12D; B, G13D), NRAS (C, Q61*) and SF3B1 (D, R625H) mutations identified in mucosal melanoma of the female genital tract
Fig. 4.
Representative examples of KIT (A, S476N; B, G498V; C, L640P; D, D810H; E. V852A) mutations identified in mucosal melanoma of the female genital tract
Notably, recurrent KIT mutations, as well as less frequent gene mutations in NRAS and SF3B1, were exclusively detected in vulvovaginal melanomas, but not in tumors arising in the cervix (Table 1). However, recurrent KRAS mutations were detected at a similar frequency in tumors of the vulva, vagina and cervix. Additionally, high numbers of KRAS and KIT mutations were identified with frequencies varying according to histological subtype. Interestingly, the recurrent KRAS and KIT mutations occurred predominantly in polygonal and epithelioid cell subtypes, but rarely in spindle cells, in melanoma of the female genital tract.
We next examined gene mutations in 25 cutaneous melanomas, 18 acral melanomas and 11 melanomas of nasal cavity (listed in Supplementary Table 1). We identified BRAF V600E (11/25; 44%) as the most commonly mutated gene, followed by NRAS (2/25; 8%), and KRAS (1/25; 4%) in cutaneous melanomas. While, BRAF V600E mutation was less frequent in acral melanomas (1/18;6%). In total, 28% (3/18) of acral melanomas showed either a KRAS or NRAS mutation (11% KRAS mutation, 17% NRAS mutation). Interestingly, the BRAF V600E hotspot mutation was not detected in the melanomas of nasal cavity. We identified five cases with NRAS mutations (5/11;45%) and two cases with KRAS mutations (2/11;18%) in the melanomas of nasal cavity. Notably, none of the cases were found to harbor SF3B1, NF1, KIT and PDGFRA mutations in cutaneous melanomas, acral melanomas and melanomas of nasal cavity. In a word, compared with gynecologic melanoma, non-gynecologic melanoma harbored distinct mutation rates in KIT, BRAF, SF3B1, KRAS and NRAS genes.
Discussion
Primary malignant melanomas of the female genital tract are extremely rare. The clinical behavior and molecular characteristics of these melanomas have not been well explored. Although melanoma of the female genital tract is an aggressive disease with histological resemblance to melanomas of other sites, recent studies found the heterogeneity of molecular biology of melanoma of different sites. Up to date, relatively little information is known about the molecular alterations that drive melanoma of the female genital tract [10–18]. To better understand the mutational profile and offer insights into future therapeutic options for patients with melanomas of the female genital tract, our study analyzed the histological and genetic characteristics of 19 melanomas of the female genital tract (vulva, vagina and cervix), paired with 25 cutaneous melanomas, 18 acral melanomas and 11 melanomas of nasal cavity.
Activating V600E or V600K mutations in BRAF kinase have been observed in up to 62% of melanomas arising in sun-exposed skin. However, in melanomas arising on mucosal surfaces or non-sun-exposed skin, BRAF mutations are infrequently reported [11]. Previous studies showed that BRAF was mutated in 0 to 33% of patients with vulvar and vaginal melanomas with sample sizes ranging from 1 to 51 cases [27–29]. In our study, oncogenic driver mutations in BRAF V600E, which were commonly identified in 44% cutaneous melanoma, were not detected in the melanomas of female genital tract. Our finding is similar to most published data on vulvovaginal melanomas [10–18]. The differences between our findings and some published studies reporting on BRAF mutations in urogenital melanomas or vulvovaginal melanomas are unclear. We doubt the small number of samples in our series (19 patients) could account for this discrepancy. One explanation may lie in the use of different mutation screening methods, which vary in sensitivity. In our study, Sanger sequencing (covering exon 15) was used to detect BRAF mutation. In contrast, Hou and colleagues [27] used a combination of next-generation sequencing (covering exons 1–18) and Sanger sequencing (covering exons 11 and 15). In addition, many of their samples were metastatic and may have harbored mutations that differed from the molecular makeup of the primary tumor. Notably, only some of the BRAF-mutant vulvar and vaginal melanomas in the literature harbored BRAF V600E mutations [27–29]. A literature search of the remaining BRAF variant-mutant including BRAF G469A, D594E, D594N, D594H in vulvovaginal melanomas revealed possible inactivating mutations that were less likely to respond to vemurafenib, which is the FDA-approved selective inhibitor of the V600E mutant BRAF kinase used to treat patients who have metastatic or unresectable melanoma with BRAF mutations [27]. Our results indicate that none of the patients with melanomas of the female genital tract can be treated with vemurafenib.
According to the literatures, KRAS mutations are common in pancreas, colon and lung cancers [30], whereas NRAS mutations are common in myeloid leukemias and cutaneous melanomas [30–32]. However, we identified six KRAS mutations and one NRAS mutation in 19 melanomas of the female genital tract. In total, 37% of tumors showed either a KRAS or NRAS mutation (32% KRAS, 5% NRAS). As reported previously, the mutations were found to be mutually exclusive. In our study, the prevalence of KRAS mutation in melanomas of the female genital tract was notably higher than melanomas of other sites, whereas the prevalence of NRAS mutation in melanomas of the female genital tract was notably lower compared with the prevalence in melanomas arising in nasal cavity, where mutation rates of up to 45%. Our finding is similar to the published data on esophageal melanomas, which harbored NRAS mutations in 30% of cases [33]. Recurrent KRAS or NRAS mutation contribute to poor prognosis [30–32]. However, in recent years, MEK inhibition was shown to demonstrate therapeutic activity in NRAS-mutated melanoma in clinical trials, opening a novel therapeutic era for these tumors [34].
KIT mutations have been observed in varying frequencies in melanomas arising at different primary sites [18, 20]. KIT protein expression or overexpression as detected by immunohistochemistry has been reported to show some correlation with KIT gene mutations but has been insufficient to predict response to KIT-targeted therapy with imatinib [16, 18]. In our study, moderate or strong cytoplasmic KIT expression was detected in 6 of the 19 cases (31.6%), and KIT mutations were observed in 21% (4/19) of the mucosal melanomas of female genital tract. All four tumors with KIT mutations showed strong KIT immunostaining. This finding shows that KIT protein expression correlated with KIT mutation. The frequency of KIT mutation in our series was much higher than rates reported in studies on non-gynecologic melanoma [27]. Interestingly, KIT mutations were associated with histological subtype and tumor site. Notably, recurrent KIT mutations were exclusively detected in vulvovaginal melanomas, but not in tumors arising in the cervix, and KIT mutation varied immensely between vulvar and vaginal sites, with 20% (1/5) of vulvar samples harboring the mutation compared with only 37.5% (3/8) of vaginal samples. This further highlights our conclusion that mucosal melanomas of the female genital tract have a genetic profile that is distinct from that of mucosal melanomas from different anatomical sites. In addition, we found that KIT mutations occurred predominantly in polygonal and epithelioid cell subtypes, but rarely in spindle cells. However, our findings are different from those of Hou and colleagues [27] that showed that vulvar melanoma may be associated with a much higher KIT mutation rate than vaginal melanoma. The differences between our findings and published studies on KIT mutations in vulvar and vaginal melanomas could be due to the small numbers of samples in our series, different methodology or ethnic difference. It is also possible that vulvar tumors were regarded as melanomas of non-sun-exposed areas. In addition, co-mutations of KIT and NF1 have been reported in mucosal melanoma, although they are rare [35]. However, in our study, none of the cases were found to harbor NF1 and PDGFRA mutations in melanomas of the female genital tract, as well as in cutaneous melanomas, acral melanomas and melanomas of nasal cavity. I think the small number of samples in our series could account for this discrepancy.
SF3B1 mutations have been identified in subsets of solid tumors, as well as in myelodysplastic syndrome and chronic lymphocytic leukemia [36–38]. Recently, SF3B1 was identified as a significantly mutated gene in mucosal melanoma, especially in uveal, female genital and anorectal melanomas [20–23]. Our study also found that the SF3B1 R625 hotspot mutation occurred in 16% of the mucosal melanomas of the female genital tract and not detected in cutaneous melanomas, acral melanomas and melanomas of nasal cavity. SF3B1 mutations have different prognostic associations in different types of cancers. In uveal melanoma, SF3B1 mutations are associated with a better prognosis, whereas in other mucosal melanomas [36], SF3B1 mutations are correlated with a worse prognosis [25]. However, the study of these rare tumors was underpowered to detect statistically significant differences, and larger studies are required to address this issue. This finding suggests that SF3B1 might be exploited as a novel prognostic and/or therapeutic target in melanomas of the female genital tract.
Conclusion
We observed that gynecologic melanoma harbored distinct mutation rates in c-KIT, BRAF, SF3B1, KRAS and NRAS genes compared with non-gynecologic melanoma. Our findings support the notion that gynecologic melanoma is a distinct entity from non-gynecologic melanoma, especially cutaneous melanomas. Although our results are preliminary, they highlight the unique molecular landscape of gynecologic melanoma within the spectrum of melanoma malignancies, and these findings offer insights into future therapeutic options for these patients.
Supplementary Information
Acknowledgments
We thank Gabrielle White Wolf, PhD, from Liwen Bianji, Edanz Editing China (www.liwenbianji.cn/ac), for editing the English text of a draft of this manuscript.
Authors’ contributions
Y-j C contributed to study design, experimental studies and data acquisition. L-f K contributed to literature research and data interpretation. W-w Z contributed to manuscript preparation and data analysis. J-p Lu contributed to manuscript revision and review. Y-p C contributed to guarantor of integrity of entire study and manuscript editing. The author(s) read and approved the final manuscript.
Funding
This study was funded by the Natural Science Foundation of Fujian Province (grant number 2018 J01277), Joint Funds for the Innovation of Science and Technology, Fujian province (grant number 2017Y9081) and Science and Technology Program of Fujian Province, China (grant number 2018Y2003).
Availability of data and materials
The data used to support the findings of this study are available from the corresponding author by request.
Declarations
Ethics approval and consent to participate
This study was carried out in accordance with the Declaration of Helsinki and written informed consent was obtained from the patients or their legal guardians. The study protocol was approved by the institutional review boards of Fujian Cancer Hospital.
Consent for publication
Not applicable.
Competing interests
All authors declare that they have no conflict of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yuan-jun Cai and Long-feng Ke contributed equally to this work as the co-first authors.
References
- 1.Nobbenhuis MA, Lalondrelle S, Larkin J, et al. Management of melanomas of the gynaecological tract. Curr Opin Oncol. 2014;26(5):508–513. doi: 10.1097/CCO.0000000000000104. [DOI] [PubMed] [Google Scholar]
- 2.Sanchez A, Rodriguez D, Allard CB, et al. Primary genitourinary melanoma: epidemiology and disease-specific survival in a large population-based cohort. Urol Oncol. 2016;34:166. e 7–1166.e14. doi: 10.1016/j.urolonc.2015.11.009. [DOI] [PubMed] [Google Scholar]
- 3.Mihajlovic M, Vlajkovic S, Jovanovic P, Stefanovic V. Primary mucosal melanomas: a comprehensive review. Int J Clin Exp Pathol. 2012;5(8):739–753. [PMC free article] [PubMed] [Google Scholar]
- 4.Gadducci A, Carinelli S, Guerrieri ME, et al. Melanoma of the lower genital tract: Prognostic factors and treatment modalities. Gynecol Oncol. 2018;150(1):180–9. [DOI] [PubMed]
- 5.Del Vecchio M, Di Guardo L, Ascierto PA, et al. Efficacy and safety of ipilimumab 3 mg/kg in patients with pretreated, metastatic, mucosal melanoma. Eur J Cancer. 2014;50(1):121–127. doi: 10.1016/j.ejca.2013.09.007. [DOI] [PubMed] [Google Scholar]
- 6.Spencer KR, Mehnert JM. Mucosal melanoma: epidemiology, biology and treatment. Cancer Treat Res. 2016;167:295–320. doi: 10.1007/978-3-319-22539-5_13. [DOI] [PubMed] [Google Scholar]
- 7.Furney SJ, Turajlic S, Stamp G, Nohadani M, Carlisle A, Thomas JM, Hayes A, Strauss D, Gore M, van den Oord J, Larkin J, Marais R. Genome sequencing of mucosal melanomas reveals that they are driven by distinct mechanisms from cutaneous melanoma. J Pathol. 2013;230(3):261–269. doi: 10.1002/path.4204. [DOI] [PubMed] [Google Scholar]
- 8.Curtin JA, Fridlyand J, Kageshita T, Patel HN, Busam KJ, Kutzner H, Cho KH, Aiba S, Bröcker EB, LeBoit PE, Pinkel D, Bastian BC. Distinct sets of genetic alterations in melanoma. N Engl J Med. 2005;353(20):2135–2147. doi: 10.1056/NEJMoa050092. [DOI] [PubMed] [Google Scholar]
- 9.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W, Davis N, Dicks E, Ewing R, Floyd Y, Gray K, Hall S, Hawes R, Hughes J, Kosmidou V, Menzies A, Mould C, Parker A, Stevens C, Watt S, Hooper S, Wilson R, Jayatilake H, Gusterson BA, Cooper C, Shipley J, Hargrave D, Pritchard-Jones K, Maitland N, Chenevix-Trench G, Riggins GJ, Bigner DD, Palmieri G, Cossu A, Flanagan A, Nicholson A, Ho JWC, Leung SY, Yuen ST, Weber BL, Seigler HF, Darrow TL, Paterson H, Marais R, Marshall CJ, Wooster R, Stratton MR, Futreal PA. Mutations of the BRAF gene in human cancer. Nature. 2002;417(6892):949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 10.Aulmann S, Sinn HP, Penzel R, Gilks CB, Schott S, Hassel JC, Schmidt D, Kommoss F, Schirmacher P, Kommoss S. Comparison of molecular abnormalities in vulvar and vaginal melanomas. Mod Pathol. 2014;27(10):1386–1393. doi: 10.1038/modpathol.2013.211. [DOI] [PubMed] [Google Scholar]
- 11.Cohen Y, Rosenbaum E, Begum S, Goldenberg D, Esche C, Lavie O, Sidransky D, Westra WH. Exon 15 BRAF mutations are uncommon in melanomas arising in nonsun-exposed sites. Clin Cancer Res. 2004;10(10):3444–3447. doi: 10.1158/1078-0432.CCR-03-0562. [DOI] [PubMed] [Google Scholar]
- 12.van Engen-van Grunsven AC, Kusters-Vandevelde HV, De Hullu J, et al. NRAS mutations are more prevalent than KIT mutations in melanoma of the female urogenital tract–a study of 24 cases from the Netherlands. Gynecol Oncol. 2014;134(1):10–14. doi: 10.1016/j.ygyno.2014.04.056. [DOI] [PubMed] [Google Scholar]
- 13.Yun J, Lee J, Jang J, et al. KIT amplification and gene mutations in acral/mucosal melanoma in Korea. APMIS. 2011;119(6):330–335. doi: 10.1111/j.1600-0463.2011.02737.x. [DOI] [PubMed] [Google Scholar]
- 14.Satzger I, Schaefer T, Kuettler U, Broecker V, Voelker B, Ostertag H, Kapp A, Gutzmer R. Analysis of c-KIT expression and KIT gene mutation in human mucosal melanomas. Brit J Cancer. 2008;99(12):2065–2069. doi: 10.1038/sj.bjc.6604791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tseng D, Kim J, Warrick A, Nelson D, Pukay M, Beadling C, Heinrich M, Selim MA, Corless CL, Nelson K. Oncogenic mutations in melanomas and benign melanocytic nevi of the female genital tract. J Am Acad Dermato. 2014;71(2):229–236. doi: 10.1016/j.jaad.2014.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carvajal RD, Antonescu CR, Wolchok JD, Chapman PB, Roman RA, Teitcher J, Panageas KS, Busam KJ, Chmielowski B, Lutzky J, Pavlick AC, Fusco A, Cane L, Takebe N, Vemula S, Bouvier N, Bastian BC, Schwartz GK. KIT as a therapeutic target in metastatic melanoma. Jama. 2011;305(22):2327–2334. doi: 10.1001/jama.2011.746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Edwards RH, Ward MR, Wu H, Medina CA, Brose MS, Volpe P, Nussen-Lee S, Haupt HM, Martin AM, Herlyn M, Lessin SR, Weber BL. Absence of BRAF mutations in UV-protected mucosal melanomas. J Med Genet. 2004;41(4):270–272. doi: 10.1136/jmg.2003.016667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beadling C, Jacobson-Dunlop E, Hodi FS, le C, Warrick A, Patterson J, Town A, Harlow A, Cruz F, III, Azar S, Rubin BP, Muller S, West R, Heinrich MC, Corless CL. KIT gene mutations and copy number in melanoma subtypes. Clin Cancer Res. 2008;14(21):6821–6828. doi: 10.1158/1078-0432.CCR-08-0575. [DOI] [PubMed] [Google Scholar]
- 19.Postow MA, Hamid O, Carvajal RD. Mucosal melanoma: pathogenesis, clinical behavior, and management. Curr Oncol Rep. 2012;14(5):441–448. doi: 10.1007/s11912-012-0244-x. [DOI] [PubMed] [Google Scholar]
- 20.Hintzsche JD, Gorden NT, Amato CM, Kim J, Wuensch KE, Robinson SE, Applegate AJ, Couts KL, Medina TM, Wells KR, Wisell JA, McCarter MD, Box NF, Shellman YG, Gonzalez RC, Lewis KD, Tentler JJ, Tan AC, Robinson WA. Whole-exome sequencing identifies recurrent SF3B1 R625 mutation and comutation of NF1 and KIT in mucosal melanoma. Melanoma Res. 2017;27(3):189–199. doi: 10.1097/CMR.0000000000000345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hayward NK, Wilmott JS, Waddell N, Johansson PA, Field MA, Nones K, Patch AM, Kakavand H, Alexandrov LB, Burke H, Jakrot V, Kazakoff S, Holmes O, Leonard C, Sabarinathan R, Mularoni L, Wood S, Xu Q, Waddell N, Tembe V, Pupo GM, de Paoli-Iseppi R, Vilain RE, Shang P, Lau LMS, Dagg RA, Schramm SJ, Pritchard A, Dutton-Regester K, Newell F, Fitzgerald A, Shang CA, Grimmond SM, Pickett HA, Yang JY, Stretch JR, Behren A, Kefford RF, Hersey P, Long GV, Cebon J, Shackleton M, Spillane AJ, Saw RPM, López-Bigas N, Pearson JV, Thompson JF, Scolyer RA, Mann GJ. Whole-genome landscapes of major melanoma subtypes. Nature. 2017;545(7653):175–180. doi: 10.1038/nature22071. [DOI] [PubMed] [Google Scholar]
- 22.Newell F, Kong Y, Wilmott JS, Johansson PA, Ferguson PM, Cui C, Li Z, Kazakoff SH, Burke H, Dodds TJ, Patch AM, Nones K, Tembe V, Shang P, van der Weyden L, Wong K, Holmes O, Lo S, Leonard C, Wood S, Xu Q, Rawson RV, Mukhopadhyay P, Dummer R, Levesque MP, Jönsson G, Wang X, Yeh I, Wu H, Joseph N, Bastian BC, Long GV, Spillane AJ, Shannon KF, Thompson JF, Saw RPM, Adams DJ, Si L, Pearson JV, Hayward NK, Waddell N, Mann GJ, Guo J, Scolyer RA. Whole-genome landscape of mucosal melanoma reveals diverse drivers and therapeutic targets. Nat Commun. 2019;10(1):3163–3178. doi: 10.1038/s41467-019-11107-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou R, Shi C, Tao W, Li J, Wu J, Han Y, Yang G, Gu Z, Xu S, Wang Y, Wang L, Wang Y, Zhou G, Zhang C, Zhang Z, Sun S. Analysis of mucosal melanoma whole-genome landscapes reveals clinically relevant genomic aberrations. Clin Cancer Res. 2019;25(12):3548–3560. doi: 10.1158/1078-0432.CCR-18-3442. [DOI] [PubMed] [Google Scholar]
- 24.Schilling B, Bielefeld N, Sucker A, et al. Lack of SF3B1 R625 mutations in cutaneous melanoma. Diagn Pathol. 2013;8(1):87. doi: 10.1186/1746-1596-8-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quek C, Rawson RV, Ferguson PM, Shang P, Silva I, Saw RPM, Shannon K, Thompson JF, Hayward NK, Long GV, Mann GJ, Scolyer RA, Wilmott JS. Recurrent hotspot SF3B1 mutations at codon 625 in vulvovaginal mucosal melanoma identified in a study of 27 Australian mucosal melanomas. Oncotarget. 2019;10(9):930–941. doi: 10.18632/oncotarget.26584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Omholt K, Grafstrom E, Kanter-Lewensohn L, et al. KIT pathway alterations in mucosal melanomas of the vulva and other sites. Clin Cancer Res. 2011;17(12):3933–3942. doi: 10.1158/1078-0432.CCR-10-2917. [DOI] [PubMed] [Google Scholar]
- 27.Hou JY, Baptiste C, Hombalegowda RB, Tergas AI, Feldman R, Jones NL, Chatterjee-Paer S, Bus-Kwolfski A, Wright JD, Burke WM. Vulvar and vaginal melanoma: a unique subclass of mucosal melanoma based on a comprehensive molecular analysis of 51 cases compared with 2253 cases of nongynecologic melanoma. Cancer. 2016;123(8):1333–1344. doi: 10.1002/cncr.30473. [DOI] [PubMed] [Google Scholar]
- 28.Rouzbahman M, Kamel-Reid S, Al Habeeb A, et al. Malignant melanoma of vulva and vagina: a Histomorphological review and mutation analysis-a single-center study. J Low Genit Tract Dis. 2015;19(4):350–353. doi: 10.1097/LGT.0000000000000142. [DOI] [PubMed] [Google Scholar]
- 29.Wong CW, Fan YS, Chan TL, Chan AS, Ho LC, Ma TK, Yuen ST, Leung SY, Cancer Genome Project BRAF and NRAS mutations are uncommon in melanomas arising in diverse internal organs. J Clin Pathol. 2005;58(6):640–644. doi: 10.1136/jcp.2004.022509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bos JL. Ras oncogenes in human cancer: a review. Cancer Res. 1989;49:4682–4689. [PubMed] [Google Scholar]
- 31.Omholt K, Platz A, Kanter L, Ringborg U, Hansson J. NRAS and BRAF mutations arise early during melanoma pathogenesis and are preserved throughout tumor progression. Clin Cancer Res. 2003;9(17):6483–6488. [PubMed] [Google Scholar]
- 32.Van Elsas A, Zerp SF, van der Flier S, et al. Relevance of ultraviolet-induced N-ras oncogene point mutations in development of primary human cutaneous melanoma. Am J Pathol. 1996;149(3):883–893. [PMC free article] [PubMed] [Google Scholar]
- 33.Sekine S, Nakanishi Y, Ogawa R, Kouda S, Kanai Y. Esophageal melanomas harbor frequent NRAS mutations unlike melanomas of other mucosal sites. Virchows Arch. 2009;454(5):513–517. doi: 10.1007/s00428-009-0762-6. [DOI] [PubMed] [Google Scholar]
- 34.Ascierto PA, Schadendorf D, Berking C, Agarwala SS, van Herpen CML, Queirolo P, Blank CU, Hauschild A, Beck JT, St-Pierre A, Niazi F, Wandel S, Peters M, Zubel A, Dummer R. MEK162 for patients with advanced melanoma harbouring NRAS or Val600 BRAF mutations: a non-randomised, open-label phase 2 study. Lancet Oncol. 2013;14(3):249–256. doi: 10.1016/S1470-2045(13)70024-X. [DOI] [PubMed] [Google Scholar]
- 35.Maertens O, Johnson B, Hollstein P, Frederick DT, Cooper ZA, Messiaen L, Bronson RT, McMahon M, Granter S, Flaherty K, Wargo JA, Marais R, Cichowski K. Elucidating distinct roles for NF1 in melanomagenesis. Cancer Discov. 2013;3(3):338–349. doi: 10.1158/2159-8290.CD-12-0313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Furney SJ, Pedersen M, Gentien D, Dumont AG, Rapinat A, Desjardins L, Turajlic S, Piperno-Neumann S, de la Grange P, Roman-Roman S, Stern MH, Marais R. SF3B1 mutations are associated with alternative splicing in uveal melanoma. Cancer Discov. 2013;3(10):1122–1129. doi: 10.1158/2159-8290.CD-13-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Papaemmanuil E, Cazzola M, Boultwood J, Malcovati L, Vyas P, Bowen D, Pellagatti A, Wainscoat JS, Hellstrom-Lindberg E, Gambacorti-Passerini C, Godfrey AL, Rapado I, Cvejic A, Rance R, McGee C, Ellis P, Mudie LJ, Stephens PJ, McLaren S, Massie CE, Tarpey PS, Varela I, Nik-Zainal S, Davies HR, Shlien A, Jones D, Raine K, Hinton J, Butler AP, Teague JW, Baxter EJ, Score J, Galli A, Della Porta MG, Travaglino E, Groves M, Tauro S, Munshi NC, Anderson KC, el-Naggar A, Fischer A, Mustonen V, Warren AJ, Cross NC, Green AR, Futreal PA, Stratton MR, Campbell PJ, Chronic Myeloid Disorders Working Group of the International Cancer Genome Consortium Somatic SF3B1 mutation in myelodysplasia with ring sideroblasts. N Engl J Med. 2011;365(15):1384–1395. doi: 10.1056/NEJMoa1103283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ellis MJ, Ding L, Shen D, Luo J, Suman VJ, Wallis JW, van Tine BA, Hoog J, Goiffon RJ, Goldstein TC, Ng S, Lin L, Crowder R, Snider J, Ballman K, Weber J, Chen K, Koboldt DC, Kandoth C, Schierding WS, McMichael JF, Miller CA, Lu C, Harris CC, McLellan MD, Wendl MC, DeSchryver K, Allred DC, Esserman L, Unzeitig G, Margenthaler J, Babiera GV, Marcom PK, Guenther JM, Leitch M, Hunt K, Olson J, Tao Y, Maher CA, Fulton LL, Fulton RS, Harrison M, Oberkfell B, du F, Demeter R, Vickery TL, Elhammali A, Piwnica-Worms H, McDonald S, Watson M, Dooling DJ, Ota D, Chang LW, Bose R, Ley TJ, Piwnica-Worms D, Stuart JM, Wilson RK, Mardis ER. Whole-genome analysis informs breast cancer response to aromatase inhibition. Nature. 2012;486(7403):353–360. doi: 10.1038/nature11143. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data used to support the findings of this study are available from the corresponding author by request.