Abstract
The prognosis of diffuse large B-cell lymphoma (DLBCL) has been associated with clinical parameters, cell of origin, and various genetic aberrations. Recently, a NanoString gene expression assay (DLBCL90) was developed, which identifies DLBCL cases with an outcome similar to those with double- or triple-hit DLBCL with both MYC and BCL2 rearrangements. This study validates the predictive ability of the DLBCL90 assay in an independent cohort of patients with the germinal center B-cell subtype DLBCL. A customized targeted sequencing panel was used to analyze the mutational profile in these patients. Cases with a double or triple hit by conventional fluorescence in situ hybridization cytogenetic analysis are known to have a poor prognosis, and the DLBCL90 gene expression signature identified these cases, as well as additional cases that would have otherwise been missed by fluorescence in situ hybridization analysis. Our findings validate use of the DLBCL90 assay for identifying high-risk patients for new and innovative therapies.
Diffuse large B-cell lymphoma (DLBCL) is the most common type of lymphoma and is a biologically heterogeneous disease.1,2 These tumors can be further divided by cell of origin into the activated B-cell and germinal center B-cell (GCB) subtypes of DLBCL.3 The current World Health Organization classification also recognizes a high-grade B-cell lymphoma with MYC and BCL2 and/or BCL6 rearrangements,2 so-called double- or triple-hit lymphoma (HGBL-DH/TH), and such cases have a poor response to conventional therapy and a short survival.4,5 A novel gene expression assay (DLBCL90), recently developed by Ennishi et al,6 reportedly identifies a poor prognosis subgroup of GCB DLBCL that would not be expected to have high-risk disease because of negative conventional fluorescence in situ hybridization (FISH) cytogenetic analysis for DH/TH. They also used this assay to distinguish the DH/TH lymphomas containing a BCL2 translocation (HGBL-DH/TH-BCL2) from the rest of GCB DLBCL. In this study, we validated the predictive ability of the DLBCL90 assay to identify high-risk lymphomas in an independent cohort of patients with GCB DLBCL.
Materials and Methods
Two-hundred and forty-seven patients were identified with de novo DLBCL who were treated with rituximab, cyclophosphamide, doxorubicin hydrochloride, vincristine, and prednisone in the years 2000 to 2016 at the City of Hope National Medical Center and from the province of Manitoba/CancerCare Manitoba. For inclusion, the following criteria were required: complete clinical and laboratory data, adequate diagnostic biopsy material for review, and multiparameter analysis. Patients who had a transformed low-grade B-cell lymphoma, were HIV positive, or on immunosuppressive therapy were excluded. This study was approved by the Institutional Review Board at the City of Hope National Medical Center and the University of Manitoba Research Ethics Board in accordance with the Declaration of Helsinki.
IHC and FISH Cytogenetic Analysis on Tissue Microarrays
Cases with formalin-fixed, paraffin-embedded tissue were stained with hematoxylin and eosin and rereviewed to confirm the diagnosis of DLBCL, not otherwise specified, or high-grade B-cell lymphoma with MYC and BCL2 and/or BCL6 translocations. Immunohistochemistry (IHC) was performed on sections (3 to 4 μm thick) of formalin-fixed, paraffin-embedded tissue microarrays using antibodies to CD20, CD3, CD10, BCL6, MUM1, MYC, and BCL2. The slides were stained on the Ventana Discovery XT platform (Ventana, Tucson, AZ) for MYC and on a Leica Bond III instrument (Leica Biosystems, Chicago, IL) for all other stains. Cases were reviewed and scored independently by three expert hematopathologists (J.Y.S., A.P., and M.N.). FISH cytogenetic analysis for MYC, BCL2, and BCL6 gene rearrangements was performed using the LSI dual-color break-apart probes (Abbott Molecular, Des Plaines, IL). At least 100 nuclei were scored, and rearrangement was defined as the presence of break-apart signals in ≥10% of the nuclei. Double/triple-hit lymphoma had concurrent rearrangements of MYC and BCL2 and/or BCL6 genes.
Mutation Analysis
Tissue blocks in which ≥60% of the surface area consisted of tumor were selected for RNA and DNA extraction using the Qiagen Allprep RNA/DNA formalin-fixed, paraffin-embedded tissue kit (Qiagen, Valencia, CA) and the manufacturer's recommended protocol. A custom targeted panel of 334 genes (designed by W.C.C.), which includes the most frequently mutated genes in B-cell lymphoma, and performed DNA sequencing on an Illumina HiSeq 2500 (Illumina, San Diego, CA) was used (Supplemental Table S1).
Gene Expression Analysis
Two-hundred nanogram of RNA was used on the nCounter platform (NanoString Technologies, Seattle, WA) to determine the cell of origin using the Lymphoma/Leukemia Molecular Profiling Project code set (Lymph2Cx).7 Briefly, the RNA was hybridized to custom code sets overnight at 65°C and processed on the nCounter Prep Station, and gene expression data were acquired on the nCounter Digital Analyzer. The data were then uploaded to the Lymphoma/Leukemia Molecular Profiling Project website (https://llmpp.nih.gov, last accessed January 25, 2021), which generated the cell of origin using the nSolver (NanoString Technologies). In addition, the DLBCL90 double-hit gene expression signature (DHITsig)6 was determined on the nSolver. The DLBCL90 assay identifies cases of GCB DLBCL that are DH/TH with a BCL2 translocation but was not designed to identify DH cases with a BCL6 translocation. The DHITsig scores were defined as DHITsig positive (pos; >–6.3), DHITsig negative (neg; <–15.6), or DHITsig indeterminate (–6.3 to –15.6).
CN Analysis
The Oncoscan Copy Number Variation assay (ThermoFisher, Waltham, MA) was performed according to the manufacturer's directions using 80 ng of DNA. OSCHP files were analyzed with the Chromosome Analysis Suite (ThermoFisher). Briefly, the generation of the OSCHP files entails calculating the log2 ratio, allelic difference, and B-allele frequency, and then identifying normal diploid regions. On the basis of the normal diploid regions, the log2 ratio, allelic difference, and B-allele frequency are recomputed if necessary. Segmentation and visualization of the copy number (CN) abnormalities were performed with Nexus Copy Number 10.0 software (Bio Discovery, El Segundo, CA) using the SNP-FASST2 algorithm. The percentage aberrant genome per case was calculated by taking the total size of the aberrant regions divided by the total size of the genome for chromosomes 1 to 22.
Statistical Analysis
Overall survival (OS) and progression-free survival (PFS) were calculated as the time from initial diagnosis to death attributable to any cause (OS) or to the earliest occurrence of progression or relapse, or death attributable to any cause (PFS). Patients alive at the last contact (OS) or patients alive without progression disease (PFS) were censored at the last contact. OS and PFS were estimated using the Kaplan-Meier product-limit method.8 The Fisher exact test was used for data comparison. Multivariate analysis was performed using Cox proportional hazard regression models for DHITsig and clinical factors (International Prognostic Index). All P values are two sided, and the analyses were performed using SAS 9.4. Survival curves were generated using MedCalc Statistical Software version 19.1.5 (MedCalc Software bv, Ostend, Belgium). Receiver operating characteristic curves were determined using the FISH analysis results and the DHITsig scores.
Results
A total of 247 patients (median age, 60 years; male/female ratio, 1.4:1) had clinical, IHC, FISH, cell of origin, and sequencing data. There were no significant differences in OS between the cohorts from the two centers (P = 0.61). There were 172 cases of GCB DLBCL (69.6%), 63 cases of activated B-cell DLBCL (25.5%), and 12 unclassified cases (4.9%). DH/TH lymphoma comprised 11.2% of the cases, and 13.4% had dual-protein (MYC and BCL2) expression (DPE) but lacked these two translocations. Of the 172 cases that were GCB DLBCL, 92 had additional RNA to perform the DLBCL90 assay. Interestingly, five cases were identified by Lymph2Cx as GCB DLBCL but had a signature by DLBCL90 of primary mediastinal large B-cell lymphoma (PMBL). The Lymph2Cx does not have the ability to identify PMBL by GEP. The clinical histories and histology of these five cases were re-reviewed and the diagnosis of PMBL was confirmed, and these cases were excluded from the study.
Cytogenetic/FISH analysis
Of the remaining 87 cases of GCB DLBCL, there was no significant difference in patient characteristics between DHITsig-neg and DHITsig-pos cases (Table 1). Eleven cases (12.6%) were positive for a DH/TH by FISH analysis, including four cases with MYC/BCL2 translocations, two cases with MYC/BCL6 translocations, and five cases with MYC/BCL2/BCL6 translocations. Of the remaining 76 cases, seven had only a MYC translocation (Figure 1). Significant differences were not seen in OS or PFS for patients with DH/TH lymphoma determined by FISH analysis (MYC/BCL2, MYC/BCL6, or MYC/BCL2/BCL6) compared with those lacking a DH/TH (OS, P = 0.32; PFS, P = 0.09). The differences between MYC/BCL2 and MYC/BCL6 DH lymphomas showed that the two MYC/BCL6 cases had a good OS. The OS and PFS of DH/TH lymphoma with MYC/BCL2 by FISH analysis also did not reach statistical significance when compared with the rest of the cases (OS, P = 0.37; and PFS, P = 0.23).
Table 1.
Clinical Characteristics of Patients With DHITsig-Neg Versus DHITsig-Pos Cases of Diffuse Large B-Cell Lymphoma
| Characteristic | DHITsig-neg cases | DHITsig-pos cases | P value |
|---|---|---|---|
| Patients, N | 72 | 15 | |
| Female sex | 26 (36) | 6 (40) | 0.78 |
| Male sex | 46 (64) | 9 (60) | |
| Aged ≥60 years | 38 (53) | 5 (33) | 0.26 |
| Aged <60 years | 34 (47) | 10 (67) | |
| Stage I or II | 31 (43) | 4 (27) | 0.39 |
| Stage III or IV | 41 (57) | 11 (73) | |
| LDH elevated | 30 (42) | 9 (60) | 0.26 |
| LDH normal | 42 (58) | 6 (40) | |
| Extranodal sites 0–1 | 56 (78) | 11 (73) | 0.74 |
| Extranodal sites ≥2 | 16 (22) | 4 (27) | |
| BM involvement | 3 (5) | 3 (20) | 0.07 |
| No BM involvement | 63 (95) | 12 (80) | |
| B symptoms present | 31 (43) | 5 (33) | 0.57 |
| B symptoms not present | 41 (57) | 10 (67) | |
| IPI low (0–1) | 32 (44) | 4 (27) | 0.40 |
| IPI low-intermediate (2) | 14 (19) | 5 (33) | |
| IPI intermediate (3) | 12 (17) | 4 (27) | |
| IPI high (4) | 14 (19) | 2 (13) |
Data are given as number (percentage) of each group, unless otherwise indicated.
BM, bone marrow; DHITsig, double-hit gene expression signature; IPI, International Prognostic Index; LDH, lactate dehydrogenase; Neg, negative; Pos, positive.
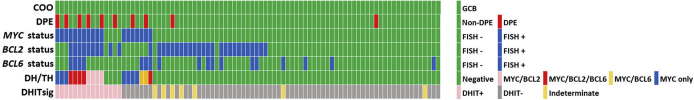
Figure 1.
Cell of origin (COO), fluorescence in situ hybridization (FISH) analysis, and double-hit gene expression signature (DHITsig) status in 87 cases of germinal center B-cell (GCB) diffuse large B-cell lymphoma. DH, double-hit lymphoma; DPE, dual-protein (MYC and BCL2) expression; TH, triple-hit lymphoma.
DPE for MYC and BCL2 by IHC was high in cases that were DHITsig-pos (40%) compared with DHITsig-neg cases (6%). In addition, cases that only had a MYC rearrangement had a high number of DPE (43%) as well as DH/TH lymphomas by FISH analysis (36%). Cases lacking MYC rearrangements or lacking DH/TH did not typically show DPE (4%) (Supplemental Table S2).
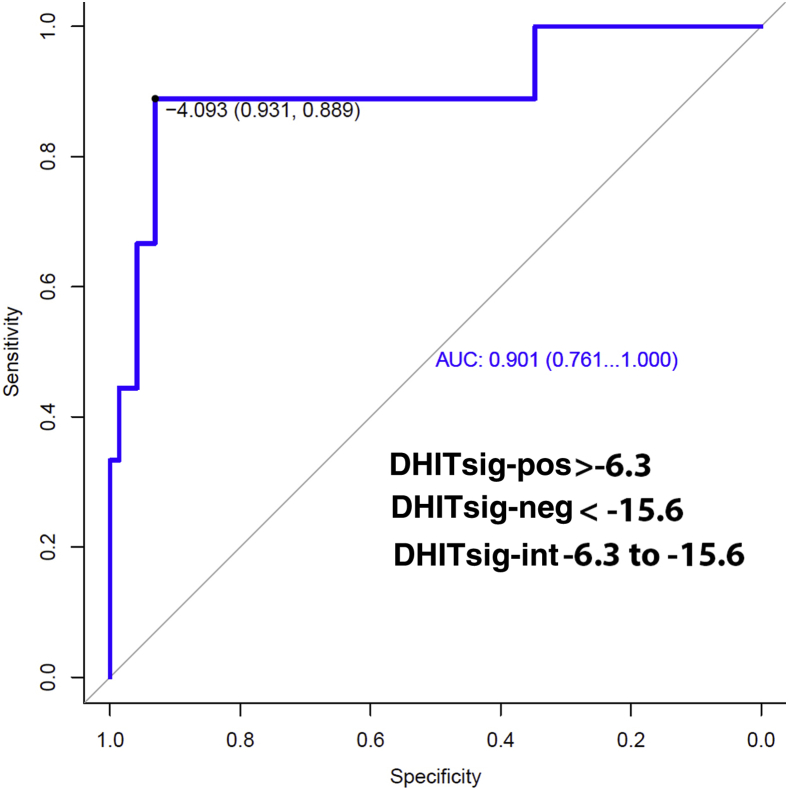
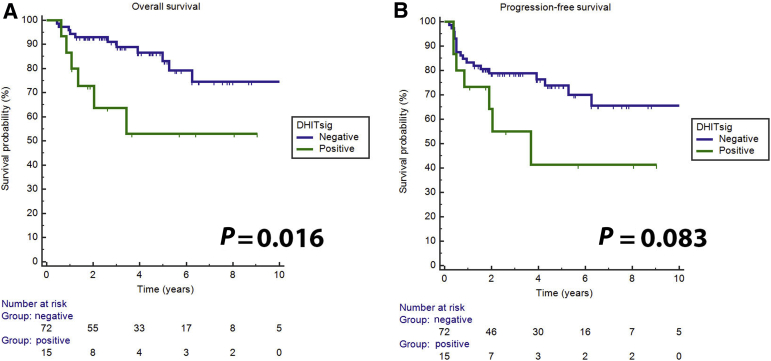
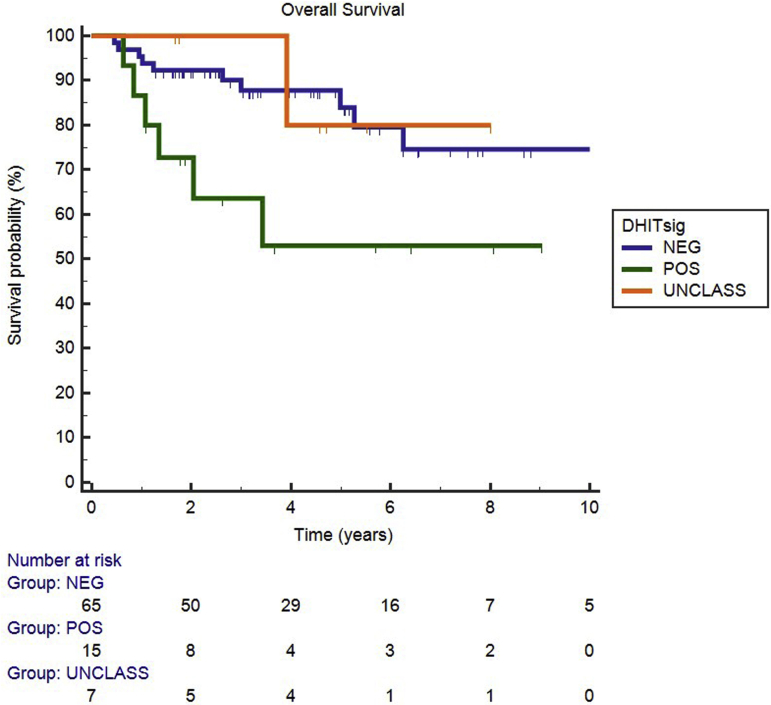
Regarding the DHITsig, 15 cases were DHITsig-pos, 65 were DHITsig-neg, and 7 were DHITsig indeterminate. The OS values for the DHITsig-indeterminate and DHITsig-neg cases were similar (Supplemental Figure S1). Therefore, these cases were grouped together as DHITsig-neg. Of the 15 DHITsig-pos cases, four had TH lymphoma with MYC/BCL2/BCL6, four had DH lymphoma with MYC/BCL2, three had only a MYC translocation, two had only a BCL2 translocation, and two had no translocations detected (Figure 1). There was also one TH lymphoma case that was DHITsig-neg. Review of this patient's clinical history shows that he achieved a remission and was alive at follow-up (4.4 years). The two cases that were MYC/BCL6 DH lymphoma were also DHITsig-neg. The performance of the DHITsig using the FISH analysis results was good, with an area under the receiver operating characteristic curve of 0.901 (95% CI, 0.76–1.00) (Figure 2). For DLBCL, International Prognostic Index has proved to be a powerful prognostic variable, as also noticed in this study (Table 2). DHITsig remained prognostic for OS in multivariate analysis (hazard ratio, 3.0; 95% CI, 1.05–8.6; P = 0.041) but did not reach significance for PFS (hazard ratio, 2.2; 95% CI, 0.87–5.3; P = 0.094) (Table 2). The DHITsig-pos patients had worse OS compared with those who were DHITsig-neg (5-year OS of 53% versus 83%, respectively; P = 0.016). There was also a trend for shorter PFS for the DHITsig-pos cases compared with those who were DHITsig-neg (P = 0.083) (Figure 3).
Figure 2.
Receiver operating characteristic curve using fluorescence in situ hybridization analysis for double- or triple-hit lymphoma and the double-hit gene expression signature (DHITsig) scores. AUC, area under the curve; int, indeterminate; neg, negative; pos, positive.
Table 2.
Cox Proportional Hazard Model Including DHITsig and IPI
| Variable | Overall survival |
Progression-free survival |
||
|---|---|---|---|---|
| P value | Hazard ratio (95% CI) | P value | Hazard ratio (95% CI) | |
| IPI | 0.015 | 0.015 | ||
| Low-intermediate vs low | 0.13 | 3.7 (0.67–20.3) | 0.3554 | 1.8 (0.52–6.3) |
| Intermediate vs low | 0.002 | 11.5 (2.40–54.8) | 0.0095 | 4.4 (1.4–13.7) |
| High vs low | 0.079 | 5.1 (0.83–30.9) | 0.0049 | 5.0 (1.6–1.4) |
| DHITsig (pos vs neg) | 0.041 | 3.0 (1.05–8.6) | 0.0939 | 2.2 (0.87–5.3) |
DHITsig, double-hit gene expression signature; IPI, International Prognostic Index; neg, negative; pos, positive.
Figure 3.
Overall (A) and progression-free (B) survival of double-hit gene expression signature (DHITsig)–positive versus DHITsig-negative cases.
Genomic Analysis of DHITsig-Pos DLBCL
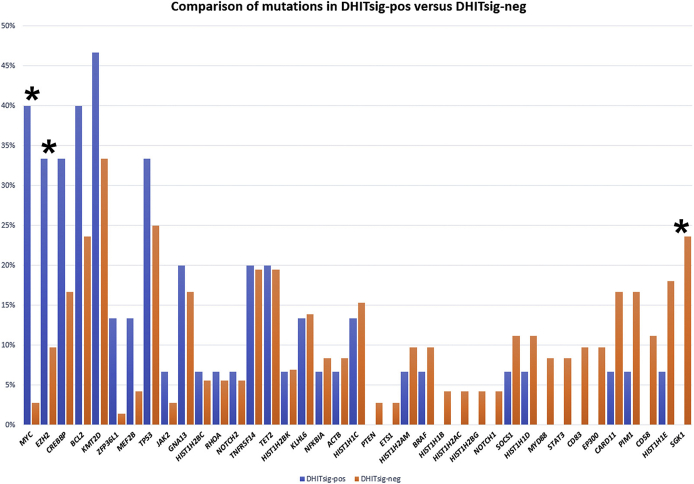
Mutations of MYC (P < 0.001), EZH2 (P = 0.03), CREBBP (not significant), BCL2 (not significant), and KMT2D (not significant) were seen more frequently in the DHITsig-pos cases. Mutations in histone modification genes (HIST1H1E, HIST1H1B, HIST1H2BG, and HIST1H1D) were not common in the DHITsig-pos cases, nor were mutations of SGK1 (P = 0.04) (Figure 4).
Figure 4.
Mutation frequencies of the double-hit gene expression signature–positive (DHITsig-pos) and double-hit gene expression signature–negative (DHITsig-neg) cases. ∗P < 0.05 (MYC, EZH2, and SGK1).
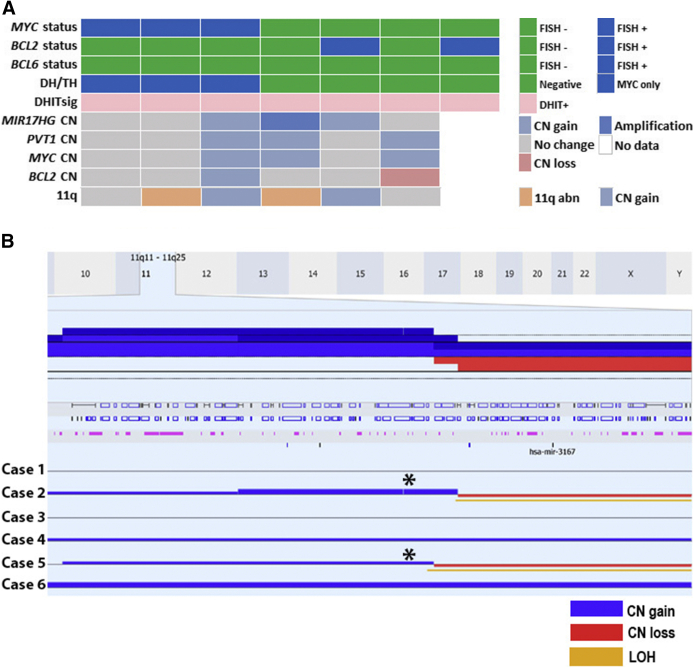
There were seven cases that were DHITsig-pos but lacked a DH/TH by FISH analysis. Of the six cases that were analyzed further with CN analysis, two cases showed CN gains and one case had amplification of MIR17HG. Three cases showed CN gain of MYC, and one case had a CN gain of BCL2. No deletions of PVT1 were seen in these cases (Figure 5A). There were also two cases with the 11q aberration, as seen in Burkitt-like lymphoma with 11q aberration (gain of 11q23.2-23.3 and loss of 11q24.1-ter)2,9 (Figure 5B).
Figure 5.
A: Seven cases are double-hit gene expression signature (DHITsig) positive but lack a double hit (DH) or triple hit (TH) by fluorescence in situ hybridization (FISH) analysis. B: Copy number (CN) analysis of the 11q region in two cases with 11q abnormalities, as seen in Burkitt-like lymphoma with 11q aberrations (cases 2 and 5; asterisks).9 11q abn, 11q abnormalities with gain of 11q23.2-23.3 and loss of 11q24.1-ter9; LOH, loss of heterozygosity.
Discussion
In this study, we validated the NanoString DLBCL90 assay6 and confirmed that the DHITsig has clinical significance in an independent cohort of GCB DLBCL treated with rituximab, cyclophosphamide, doxorubicin hydrochloride, vincristine, and prednisone. The DHITsig-pos patients had a worse OS and PFS compared with those who were DHITsig-neg. This signature is important because it is able to identify a subset of patients lacking the MYC/BCL2 translocations by conventional cytogenetics. Similar to Hilton et al,10 cryptic abnormalities in some of our cases may have contributed to the pathogenesis of these DHITsig-pos lymphomas.
Only half of the DHITsig-pos cases (53%) had either MYC/BCL2 or MYC/BCL2/BCL6 translocations by FISH analysis. Our receiver operating characteristic curve had a similar area under the curve (0.901) to that of the original study (0.907), supporting the ability of the assay to detect HGBL-DH/TH-BCL2.6 Six cases either had a BCL2 translocation without a MYC translocation or a MYC translocation without a BCL2 translocation, and may have had cryptic rearrangements that were missed by conventional FISH analysis.10 Hilton et al10 performed CN analysis and whole genome sequencing on DHITsig-pos cases lacking MYC and/or BCL2 translocations by FISH analysis and found that some of their cases had cryptic rearrangements or CN gains in MYC or MIR17HG, both of which may contribute to the dysregulation of MYC.10 These authors concluded that there were genetic events downstream of MYC and BCL2 that were not detected by FISH analysis, supporting the use of GEP for detecting the double-hit signature. For the six cases analyzed by CN analysis, three cases with CN gain/amplification of MIR17HG, three cases with CN gain of MYC, and one case with CN gain of BCL2 were found. Interestingly, none of these cases showed CN loss of PVT1. There were also two cases that had the genetic abnormalities seen in Burkitt-like lymphoma with 11q aberration,9 one of which also had a concurrent MYC translocation (Figure 5). However, whole genome sequencing was not performed and, therefore, it could not be detremined if there were any cryptic insertions or rearrangements.
Of the DH/TH lymphoma cases, 36% were DPE, which is lower compared with the study by Ennishi et al6 (63%). A recent study by Collinge et al11 found cases with a MYC N11S polymorphism were associated with false-negative MYC IHC staining in MYC rearranged lymphomas, which may be related to disruption of the binding of the antibody epitope. No cases with the N11S polymorphism in MYC were observed in this study, which may be related to technical factors (eg, different IHC platforms, scoring, and age of specimen) or there may be another mechanism in our cases with low IHC expression with MYC. However, this further underlines the importance of using the DHITsig rather than IHC or FISH analysis alone to identify a poor prognostic group.
Before running the DLBCL90 assay, the Lymph2Cx assay was used to identify cases of GCB DLBCL. Interestingly, five cases were identified as GCB DLBCL with Lymph2Cx, but these were classified as PMBL with the DLBCL90 assay. Rereview of these cases confirmed the diagnosis of PMBL rather than GCB DLBCL. This demonstrates that the DLBCL90 assay can identify cases of PMBL in addition to detecting the double-hit signature.
In conclusion, we validated the DHITsig in an independent cohort of GCB DLBCL patients treated with rituximab, cyclophosphamide, doxorubicin hydrochloride, vincristine, and prednisone. This assay identifies conventional double- and triple-hit lymphomas with BCL2 translocation, as well as a poor prognostic group with the DHITsig that would have been missed by conventional FISH analysis. These findings support the use of the DLBCL90 assay to identify high-risk patients with GCB DLBCL for new and innovative therapies.
Footnotes
Supported by the City of Hope National Medical Center Department of Pathology and the Toni Stephenson Lymphoma Center. Research reported in this publication includes work performed in the Pathology Core and Integrated Genomics Core, supported by the National Cancer Institute of the NIH under grant P30CA033572.
Disclosures: D.W.S. is a named inventor on a patent describing the double-hit signature and the DLBCL90 assay.
Supplemental material for this article can be found at http://doi.org/10.1016/j.jmoldx.2021.02.005
Supplemental Data
Supplemental Figure S1.
Overall survival of the germinal center B-cell diffuse large B-cell lymphoma, based on double-hit gene expression signature (DHITsig) (P = 0.055). The unclassified (UNCLASS) are cases that were determined as DHITsig indeterminate. NEG, negative; POS, positive.
References
- 1.Zindy F., Eischen C.M., Randle D.H., Kamijo T., Cleveland J.L., Sherr C.J., Roussel M.F. Myc signaling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes Dev. 1998;12:2424–2433. doi: 10.1101/gad.12.15.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Swerdlow S., Campo E., Harris N.L., Jaffe E.S., Pileri S.A., Stein H., Thiele J., Arber D.A., Hasserjian R.P., Le Beau M.M., Orazi A., Siebert R. International Agency for Research on Cancer; Lyon, France: 2017. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. [Google Scholar]
- 3.Alizadeh A.A., Eisen M.B., Davis R.E., Ma C., Lossos I.S., Rosenwald A., Boldrick J.C., Sabet H., Tran T., Yu X., Powell J.I., Yang L., Marti G.E., Moore T., Hudson J., Jr., Lu L., Lewis D.B., Tibshirani R., Sherlock G., Chan W.C., Greiner T.C., Weisenburger D.D., Armitage J.O., Warnke R., Wilson W., Grever M.R., Byrd J.C., Botstein D., Brown P.O., Staudt L.M. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 4.Horn H., Ziepert M., Becher C., Barth T.F., Bernd H.W., Feller A.C., Klapper W., Hummel M., Stein H., Hansmann M.L., Schmelter C., Moller P., Cogliatti S., Pfreundschuh M., Schmitz N., Trumper L., Siebert R., Loeffler M., Rosenwald A., Ott G. MYC status in concert with BCL2 and BCL6 expression predicts outcome in diffuse large B-cell lymphoma. Blood. 2013;121:2253–2263. doi: 10.1182/blood-2012-06-435842. [DOI] [PubMed] [Google Scholar]
- 5.Johnson N.A., Slack G.W., Savage K.J., Connors J.M., Ben-Neriah S., Rogic S., Scott D.W., Tan K.L., Steidl C., Sehn L.H., Chan W.C., Iqbal J., Meyer P.N., Lenz G., Wright G., Rimsza L.M., Valentino C., Brunhoeber P., Grogan T.M., Braziel R.M., Cook J.R., Tubbs R.R., Weisenburger D.D., Campo E., Rosenwald A., Ott G., Delabie J., Holcroft C., Jaffe E.S., Staudt L.M., Gascoyne R.D. Concurrent expression of MYC and BCL2 in diffuse large B-cell lymphoma treated with rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisone. J Clin Oncol. 2012;30:3452–3459. doi: 10.1200/JCO.2011.41.0985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ennishi D., Jiang A., Boyle M., Collinge B., Grande B.M., Ben-Neriah S., Rushton C., Tang J., Thomas N., Slack G.W., Farinha P., Takata K., Miyata-Takata T., Craig J., Mottok A., Meissner B., Saberi S., Bashashati A., Villa D., Savage K.J., Sehn L.H., Kridel R., Mungall A.J., Marra M.A., Shah S.P., Steidl C., Connors J.M., Gascoyne R.D., Morin R.D., Scott D.W. Double-hit gene expression signature defines a distinct subgroup of germinal center B-cell-like diffuse large B-cell lymphoma. J Clin Oncol. 2019;37:190–201. doi: 10.1200/JCO.18.01583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scott D.W., Wright G.W., Williams P.M., Lih C.J., Walsh W., Jaffe E.S., Rosenwald A., Campo E., Chan W.C., Connors J.M., Smeland E.B., Mottok A., Braziel R.M., Ott G., Delabie J., Tubbs R.R., Cook J.R., Weisenburger D.D., Greiner T.C., Glinsmann-Gibson B.J., Fu K., Staudt L.M., Gascoyne R.D., Rimsza L.M. Determining cell-of-origin subtypes of diffuse large B-cell lymphoma using gene expression in formalin-fixed paraffin-embedded tissue. Blood. 2014;123:1214–1217. doi: 10.1182/blood-2013-11-536433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaplan E.L., Meier P. Nonparametric estimation from incomplete observations. J Am Stat Assoc. 1958;53:457–481. [Google Scholar]
- 9.Salaverria I., Martin-Guerrero I., Wagener R., Kreuz M., Kohler C.W., Richter J., Pienkowska-Grela B., Adam P., Burkhardt B., Claviez A., Damm-Welk C., Drexler H.G., Hummel M., Jaffe E.S., Kuppers R., Lefebvre C., Lisfeld J., Loffler M., Macleod R.A., Nagel I., Oschlies I., Rosolowski M., Russell R.B., Rymkiewicz G., Schindler D., Schlesner M., Scholtysik R., Schwaenen C., Spang R., Szczepanowski M., Trumper L., Vater I., Wessendorf S., Klapper W., Siebert R., Molecular Mechanisms in Malignant Lymphoma Network Project, Berlin-Frankfurt-Munster Non-Hodgkin Lymphoma Group A recurrent 11q aberration pattern characterizes a subset of MYC-negative high-grade B-cell lymphomas resembling Burkitt lymphoma. Blood. 2014;123:1187–1198. doi: 10.1182/blood-2013-06-507996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hilton L.K., Tang J., Ben-Neriah S., Alcaide M., Jiang A., Grande B.M., Rushton C.K., Boyle M., Meissner B., Scott D.W., Morin R.D. The double-hit signature identifies double-hit diffuse large B-cell lymphoma with genetic events cryptic to FISH. Blood. 2019;134:1528–1532. doi: 10.1182/blood.2019002600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collinge B.J., Ben-Neriah S., Chong L.C., Boyle M., Jiang A., Miyata-Takata T., Farinha P., Craig J.W., Slack G.W., Ennishi D., Mottok A., Meissner B., Chavez E.A., Gerrie A.S., Villa D., Freeman C.L., Savage K.J., Sehn L.H., Morin R.D., Mungall A.J., Gascoyne R.D., Marra M.A., Connors J.M., Steidl C., Scott D.W. Impact of MYC and BCL2 structural variants in tumors of DLBCL morphology and mechanisms of false-negative MYC IHC. Blood. 2020 doi: 10.1182/blood.2020007193. 10.1182/blood.2020007193 [Epub ahead of print] doi: [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.