Abstract
Recently, developmental systems are investigated with increasing technological power. Still, open questions remain, especially concerning self-organization capacity and its control. Here, we present three areas where synthetic biology tools are used in top-down and bottom-up approaches for studying and constructing developmental systems. First, we discuss how synthetic biology tools can improve stem cell-based organoid models. Second, we discuss recent studies employing user-defined perturbations to study embryonic patterning in model species. Third, we present “toy models” of patterning and morphogenesis using synthetic genetic circuits in non-developmental systems. Finally, we discuss how these tools and approaches can specifically benefit the field of embryo models.
Keywords: synthetic biology, synthetic development, synthetic morphogenesis, synthetic patterning, developmental biology, synthetic signaling, synthetic receptors, embryo
With this Perspective, Morsut and Ho aim to stimulate the stem cell and developmental biology community with the latest applications of synthetic biology for developmental systems: improvements in organoids protocol, rescue of patterning mutants in model organisms, and construction of “toy models” in non-developmental cell lines. Finally, current and potential applications in embryo models are discussed.
Main text
Introduction
Synthetic biology is defined as the application of an engineering paradigm of systems design to biological systems to produce predictable and robust systems. We use “synthetic biology tools” here to specifically indicate the use of artificially designed genetic parts and networks. In this sense, the field began with the generation of transcriptional circuits in bacteria (Elowitz and Leibler, 2000; Gardner et al., 2000). Advanced techniques and tools have since been developed that allow engineering in eukaryotic systems, first in single cells and then in multicellular contexts (Cameron et al., 2014). One of the most recent advances in synthetic biology is the application of synthetic biology approaches to multicellular, developmental systems, a field dubbed synthetic development or synthetic morphogenesis (Ebrahimkhani and Ebisuya, 2019; Johnson et al., 2017; Santorelli et al., 2019; Schlissel and Li, 2020; Toda et al., 2019; Velazquez et al., 2018). One breakthrough in this area was the development of synthetic signaling pathways, often based on synthetic input-output devices that are controllable by the user: from optogenetic tools (Johnson and Toettcher, 2018; Kowalik and Chen, 2017; Toettcher et al., 2011) to synthetic receptors (Morsut et al., 2016). With these, researchers have generated synthetic signaling pathways that can trigger complex cell behaviors like differentiation, migration, and target cell elimination (e.g., chimeric antigen receptor T [CAR-T] cells) (Roybal et al., 2016a, 2016b). One way these synthetic tools have been used is for building simplified in vitro systems; for example, for patterning of bacterial communities (Basu et al., 2005). How these tools have been used in developmental systems is the focus of this perspective.
When we use “developmental systems” in this perspective, we mean metazoan multicellular systems, either reconstructed or natural, in vivo or in vitro, that display some form of patterning, morphogenesis, or differentiation: in short, development. Embryo models are one example of such developmental systems and are the focus of the journal issue of which this perspective is part. The specifics of different embryo models are described in detail in other reviews of the issue and will not be discussed in depth here. These models collectively are on their way to change radically how we learn about developmental biology on one side, and how disease modeling and therapeutic screenings are performed on the other (Schutgens and Clevers, 2020). We focus here on how synthetic, artificial, genetic circuits from synthetic biology have been integrated in developmental systems, the beginning of what has been done for embryo models, and what we imagine could be done in embryo models in the future.
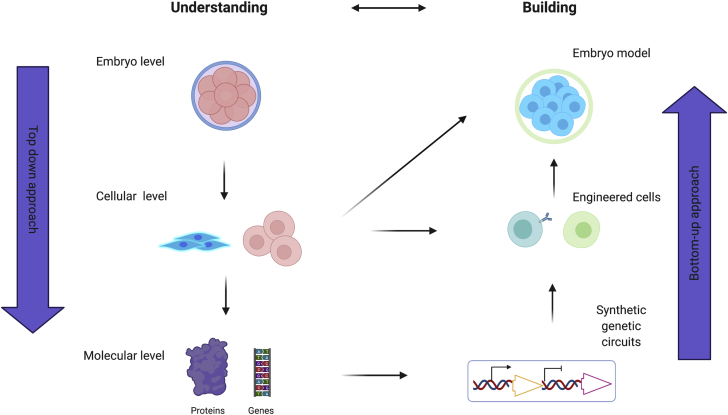
For the sake of discussion, we distinguish two opposite sets of approaches when dealing with developmental systems: top down and bottom up (Figure 1). On one hand, a top-down approach is being used by embryologists to understand existing examples of embryogenesis. Scientists have long been interested in how organs and tissues form during embryogenesis. The field entered the molecular and genetic era with a breakthrough in 1979–1980: a large-scale mutagenesis study performed in the fruit fly, Drosophila melanogaster, allowed identification of a large number of genes involved in embryo patterning (Wieschaus and Nüsslein-Volhard, 2016). Since then, the study of various model organisms elucidated key principles of cellular and molecular phenomena underpinning developmental transitions. In this context, the top-down approach consists of scaling down the object of study: looking from a whole-embryo level to the tissue level and underlying cellular and molecular mechanisms. The top-down approach is analytic, in that you want to get as much information as possible about what the different players are doing during development. Recent breakthroughs in this area include increasingly quantitative approaches as well as advanced real-time imaging techniques (Betzig et al., 2006; Chen et al., 2014; Planchon et al., 2011). To gain cause-effect understanding, the top-down approach is also perturbative, such that an embryo is manipulated/perturbed with a tool, and the system reaction is analyzed in terms of its components. Many tools for perturbation at the different levels of organization have been developed toward finer understanding of embryogenesis and are reviewed elsewhere (Castanon and González-Gaitán, 2011; Johnson and Toettcher, 2018; Kowalik and Chen, 2017; Krueger et al., 2019; Ouyang and Chen, 2010; Shestopalov and Chen, 2008). Among the perturbative tools, synthetic approaches such as optogenetics and synthetic morphogens have also been used to rescue patterning mutants and will are described later in the section “rescue of mutants of model species embryos with synthetic biology molecular tools.” In general, top-down approaches, with their perturbative nature, give information on the necessity of genes or processes for developmental transitions.
Figure 1.
Overview of the complementary approaches to study complexity in developmental biology
On the left, the reductionist top-down approach is shown; whole embryos are scaled down into component cells, and down to component molecules. On the right, the reconstructive bottom-up approach aims to build in vitro models from elementary components at different levels in the hierarchical scale. Reconstitution beginning from the cellular level would include organoids and embryo models, while reconstitution beginning from the molecular level would include the use of synthetic circuits for engineering developmental transitions and patterned assemblies. The two approaches represent a schematic guide and inform each other. For example, a basal understanding from top-down approaches is necessary to attempt reconstructive approaches. Reconstructive approaches can then, in themselves, help further understanding. While useful to think about these approaches as distinct, the two are not fully separable.
In contrast, a bottom-up approach is being used to build developmental systems in a way that is different from what is currently present in nature. A bottom-up approach is constructive, based on synthesis. Here, an attempt is made to reconstruct the complexity seen in real systems in vitro. Depending on which level is engineered/constructed, this can take different forms. In one form, the building blocks are stem cells and growth factors/culture conditions. In organoid research, for example, the autonomous self-organization of stem cells is steered with the external cues. This approach has yielded organoids for various organs of the body (Clevers, 2016; Serra et al., 2019). When the goal is to construct an embryo, the resulting constructs have taken the name of “embryo models” (Li et al., 2019; Rivron et al., 2018). If the level of intervention is the genetic circuits, the bottom-up approach can take the form of introducing synthetic genetic circuits in non-developmental systems (e.g., fibroblast cell lines) to generate new, simple models of self-organization displaying various degrees of self-organization (we call these models “toy models”, see section “building toy models of developmental transitions for understanding basic principles of patterning and morphogenesis”). Regardless of the level of intervention, an advantage of these in vitro simplified systems is enhanced tractability and accessibility, as well as greater control in the make-up of the experimental system (e.g., ratio of component cells, scaffold, delivery of drugs or growth factors, high-content imaging). In general, the constructive approach can provide useful information on minimal requirements for a circuit/cell/growth factor to perform a certain function.
Given the complexity of the process of embryonic development, it is not surprising that the community is using these approaches in complementary and intersecting ways (Castanon and González-Gaitán, 2011). For example, systems built via a bottom-up approach, such as embryo models, are now being analyzed and perturbed with tools from a top-down approach (Pezzulo and Levin, 2016).
We describe here three intersections of synthetic biology and developmental systems:
-
1.
Synthetic biology can be used to build better, more controlled, stem cell-derived model systems (e.g., organoids) (Figure 2);
-
2.
Synthetic biology approaches are providing new tools that can be used to address specific questions about developmental systems in model organisms (Figure 3);
-
3.
Synthetic biology can be used in non-developmental systems (e.g., fibroblast cell lines) to build toy models for constructive study of developmental principles (Figure 4).
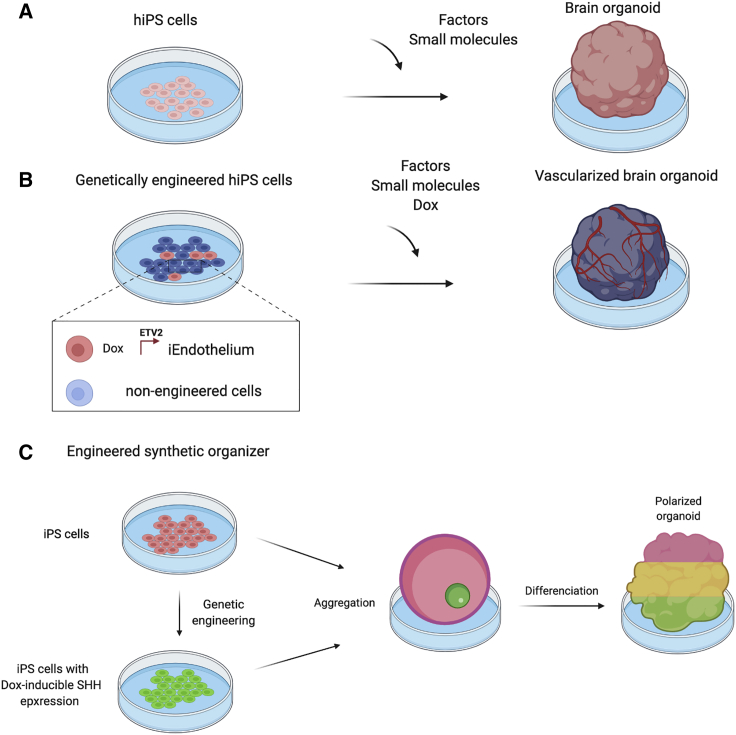
Figure 2.
Building better in vitro organoids using synthetic biology
(A) Cortical organoids can be generated from hiPSCs using a cocktail of small molecules and specific signaling factors.
(B) Combining normal and genetically engineered hiPSCs that turn into endothelial cells upon Dox induction can be used to make vascularized cortical organoids.
(C) Combining normal and genetically engineered hiPSCs that secrete the morphogen Shh upon Dox induction can be used to make dorsoventrally patterned brain organoids.
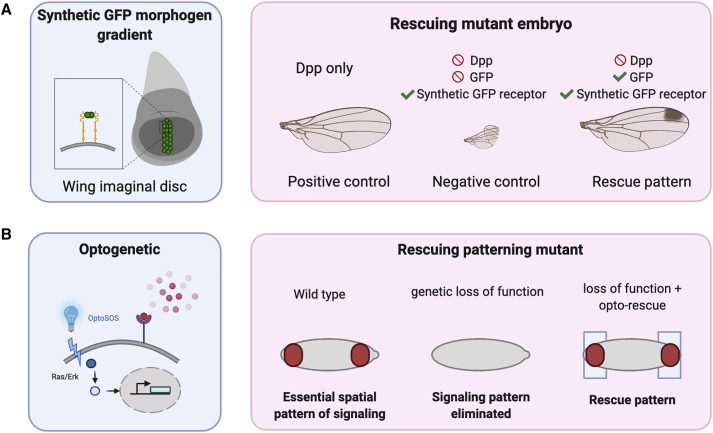
Figure 3.
Rescuing embryonic patterning in model organisms using synthetic approaches
(A) Engineered synthetic morphogen GFP can rescue pattern mutant in Drosophila wing imaginal disc.
(B) Optogenetic technique can be used to control endogenous ERK signaling pathway to rescue loss of function mutants in Drosophila early patterning.
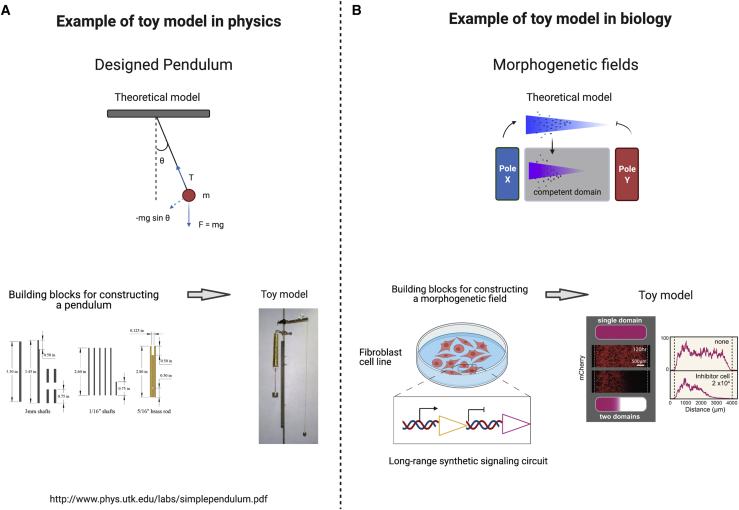
Figure 4.
Building developmental toy models in non-developmental systems with synthetic genetic circuits
(A) Pendulum is an example of a toy model in physics that has been built to understand basic physical forces such gravity and tension. These parameters can be modulated, making the pendulum a good tool to probe basic physical systems. Picture of toy model extracted from http://www.phys.utk.edu/labs/simplependulum.pdf.
(B) Example of a developmental biology toy model. Toda et al. (2020) engineered a synthetic morphogen gradient leading to morphogenetic patterning of mCherry responses by using GFP ligands. Range of the gradient can be changed, and sources of morphogens can be adjusted. Picture of the biological toy model is from Toda et al. (2020).
In the following three sections we describe these approaches in developmental systems that are not embryo models, to show their potential. In the last section, we describe the first results of using synthetic biology in embryo models and speculate on how other studies in embryo models could be carried out with the tools and approaches of synthetic biology (Figure 5).
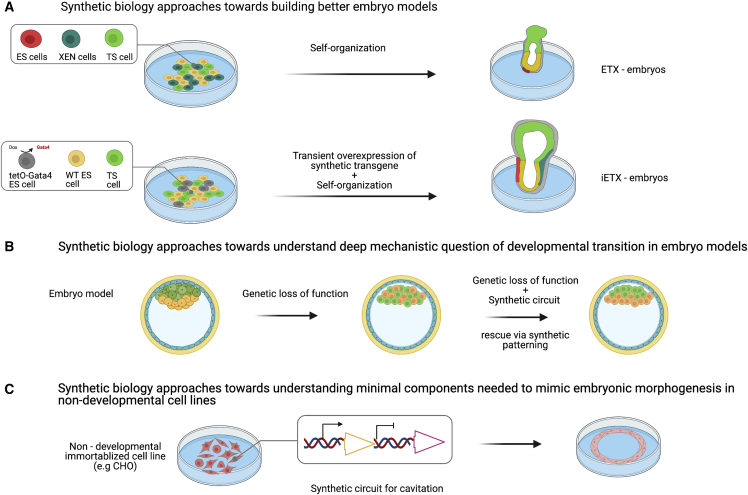
Figure 5.
Application of synthetic biology tools and approaches to embryo models
(A) Increasing developmental potential of embryo models: transient overexpression of Gata4 transcription factor enabled generation of a more developmentally potent mouse embryo model (Amadei et al., 2021).
(B) Potential application of synthetic rescue of a developmental mutation in embryo model.
(C) Potential description of a toy model for cavitation of an assembly of non-developmental cells CHO thanks to synthetic circuits.
Using synthetic biology to enhance organoid construction
Within the bottom-up approach for constructing developmental systems in vitro, one of the most inspiring and successful approaches is the organoid: using stem cells, and deciding the number and kind of different cell types, as well as the mechanical and/or chemical environment, it has been possible to build a number of different organ-like structures in vitro (Brassard and Lutolf, 2019; Schutgens and Clevers, 2020; Takebe and Wells, 2019). The first reports of the self-organization capacity of stem cells in vitro have been astonishing to see (Sasai, 2013). While promising, current challenges for the field of in vitro organogenesis include reproducibility, functionality, scalability, terminal differentiation, control of organ-to-organ interaction, and germ-layer cross-talk, but these are only a few of the areas in which increased controls are needed. Organogenesis is a highly choreographed and complex process that unfolds in a very controlled spatiotemporal fashion. Many properties of organogenesis emerge from the interactions between and within different levels of organization. It is, therefore, not surprising that improved organoids have been developed with the introduction of multiple initial cell types (Takebe and Wells, 2019), as well as by merging organoids of different regions; for example, for the brain (Marton and Pașca, 2020).
As the genetic level is one of the crucial low-level languages that underlies the emergent properties of morphogenesis, methods to control emergent properties with genetic programs seem particularly well suited to provide user-defined control on higher levels of organization. A few simple examples utilizing transcription factor overexpression demonstrate the power and potential of this approach, which we expect to produce more results in the future as more complex perturbations are deployed.
Transcription factors have been identified as master fate controllers, epitomized in the phenomenon of reprogramming (Chronis et al., 2017; Soufi et al., 2015; Takahashi and Yamanaka, 2016). Control of transcription factor overexpression has been heavily used in the field of stem cell differentiation, to produce differentiated cell types for therapies and for in vitro study of diseases (Peterson, 1993). Some of the fastest and most efficient protocols for production of specific cell types employ transcription factor-mediated reprogramming alongside chemical factor media formulations; for example, overexpression of Ngn2 for production of neurons (Busskamp et al., 2014; Zhang et al., 2013).
In a multicellular context, one of the first examples of organoid protocols improved by genetic engineering showed that complex liver organoids could be generated by overexpression of Gata6 transcription factors in a non-uniform fashion in a population of stem cells (reviewed also in Ebrahimkhani and Ebisuya, 2019; Guye et al., 2016; Johnson et al., 2017). In a follow-up study, informed by computational analyses to identify key factors, genetic manipulation of human induced pluripotent stem cells (hiPSCs) improved the maturity and vascularity of fetal liver organoids in vitro (Velazquez et al., 2021). Interestingly, the overexpression of a factor from the same family, Gata4, has been used in an embryo model system to generate a more faithful multi-lineage egg-cylinder model (see more detail in section “challenges and opportunities in applying synthetic developmental biology to embryo models”).
A similar approach is to engineer stem cells to produce master transcription factors under the control of drug-inducible promoters. For example, ETV2 has been used to generate endothelial cells from hiPSCs when overexpressed (Elcheva et al., 2014; Lange et al., 2020; Liu et al., 2015; Veldman et al., 2013). These ETV2 cells can then be mixed with hiPSCs differentiating toward other cell types; for example, neuronal fates in a brain cortical organoid protocol (Figures 2A and 2B). The subpopulation of engineered cells with ETV2 expression go on to form endothelial precursors that can fuse to each other to form microvasculature cells (Cakir et al., 2019). In the context of brain organoids, this approach has been recently used to generate organoids with more complex cellular architecture, by mixing cells with dox-inducible factors for specific neuronal and oligodendrocyte subtypes (Skylar-Scott et al., 2020).
Another key principle of developmental biology that is being used to improve organoid protocols is that of organizers. Organizers are transient tissues that orchestrate development at specific developmental transitions (Meinhardt, 2006; Nakamura et al., 2005). A famous example is the node that orchestrates gastrulation (De Robertis, 2009). During organoid differentiation in vitro, it would be helpful to have user-designed organizer tissues to help direct development. This principle has been successfully used by Studer and coworkers to give positional identity to forebrain organoids. In vivo, the signal to pattern the developing brain comes from the notochord and has been identified as the morphogen Shh (Jessell, 2000; Lupo et al., 2006). In the organoid study, the authors genetically engineered a population of stem cells to overexpress Shh under a dox-inducible promoter and pre-aggregate them to form a coherent organizer (Cederquist et al., 2019). When normal stem cells are co-cultivated around the organizer in a forebrain organoid protocol, and the production of Shh is activated by adding Dox, the resulting organoid displays dorsoventral patterning along the axis dictated by the synthetic organizer (Figure 2C).
As described, these systems are using genetic engineering and, as such, fall into our definition of synthetic biology, but are definitively of the simplest kind. Even so, they show the potential of this approach to improve construction of stem cell-derived in vitro systems. One challenge that is always present in these in vitro protocols is how much the need for control can be balanced with allowing cells to display their own endogenous self-organization (a dilemma common in the control of complex systems; Solé, 2016). It is tempting to speculate that, in the next generation of organoids, we will see convergence of more complex synthetic biology tools to help control differentiation in time and space.
Rescue of mutants of model species embryos with synthetic biology molecular tools
One way to gain understanding of the molecular underpinnings of developmental transitions has been the analysis of the consequences of mutations in certain genes. This approach provided direct evidence that genetic manipulations could alter morphogenesis (Wieschaus and Nüsslein-Volhard, 2016). While initially mutations where constitutive and expressed by all the cells of the embryo, refined tools allowed for increased precision. One crucial example is the recombinase-based systems, e.g., Cre-LoxP (Lewandoski, 2001; Metzger, 1999), that allow genetic perturbations in a cell type in a time-dependent manner. Instead of mutating genes, another approach is the use of small-molecule drugs to perturb function of proteins with temporal and spatial precision (Kowalik and Chen, 2017; Ouyang and Chen, 2010; Shestopalov and Chen, 2008). Perturbations of genes and proteins have elucidated the necessity of specific genes and molecules in certain processes: basically, a process of de-construction. If a gene is removed from an embryo, and the consequence is that it does not undergo gastrulation, the gene is understood to be implicated in the process of gastrulation. Rescue experiments, where the gene or gene product is provided in another form to rescue the mutant phenotype, are strong controls that help to confirm these findings. Recently, the rescue approach has been used with synthetic biology tools. A mutation generated a de-constructed embryo that could not progress past a certain step in development. A rescue experiment was constructed that consisted in delivering synthetic versions of the control mechanism. In this sense, these experiments are a bottom-up/constructivist approach inside a de-constructed real embryo, typically used for top-down approaches.
Genetically encoded synthetic signaling pathways have been recently used to rescue patterning mutants. This approach has been used to address a fundamental question about morphogenetic fields. Signaling molecules, termed morphogens, form a gradient due to diffusion from a localized source, providing complex positional information that guides the growth of functional tissue during development. Cells that can respond to the morphogenetic signal, when exposed to these morphogen gradients, acquire specific cell fates in a dose-dependent manner in time and space. Over the past decades, numerous perturbative studies have shown the capital involvement of morphogen gradients in tissue patterning in different species (Briscoe and Small, 2015), and especially in the fly wing (Lecuit and Cohen, 1998; Nellen et al., 1996; Teleman and Cohen, 2000). Although many details of the system are known, and the role of Dpp gradient as a morphogen is established, one outstanding question in the field was whether the system could work by diffusion only, as opposed to another mechanism for passing the signal between cells (e.g., long-range filopodia, secreted vesicles). To address this question Stapornwongkul et al. (2020) devised a new way to activate the Dpp pathway with genetically encoded synthetic ligands, which allowed them to prove that cells do not read something special about Dpp, but simply its gradient (Figure 3A). Specifically, the authors generated a new synthetic receptor that recognizes GFP as input and activates the Dpp pathway in response (GFP→Dpp receptor). These synthetic receptors were generated by grafting anti-GFP nanobodies into the extracellular domain of natural anti-DPP receptors. Since the anti-DPP receptors are activated by dimerization, when these synthetic receptors bind to GFP-dimers they activate endogenous Dpp target genes. The authors showed that they can generate a GFP gradient in the imaginal disc by engineering fly embryos with transgenes for expression of soluble GFP protein downstream of a promoter that is active only in the vertical midline of the developing imaginal disc. This gives rise to a bell-shaped gradient centered on the vertical midline, currently believed to be the endogenous shape of the Dpp morphogen gradient. With both a capacity to generate a gradient of synthetic signal and the capacity to induce Dpp responses, the authors were able to perform rescue experiments. Using fly embryos without Dpp, which do not form proper wings, they expressed synthetic GFP→Dpp receptors under ubiquitous promoters, thereby giving all cells of the imaginal wing disc the capacity to activate Dpp responses when binding GFP signals. Coupled with the GFP gradient created by midline expression, wing development is largely rescued in these mutants. The fact that wing formation is rescued, even imperfectly, in these embryos is a very impressive result. It proves that morphogenesis can be achieved via diffusion of the morphogen. It also shows that you can drive development with a synthetic pathway: cells do not read something special about Dpp, but simply its gradient. Additionally, the researchers went on to introduce further modifications in the gradient-forming capacity of the system with GPI-anchored anti-GFP nanobodies, to show that a better-looking gradient of Dpp responses induces a better-looking wing. This work shows that synthetic pathways can be used to direct development, thereby providing tools for testing underlying principles in what can be very decisive ways.
Another set of tools that have been used for these synthetic developmental rescue experiments is optogenetics, the use of light to activate genetically engineered light-responsive pathways. Optogenetic tools for activation of action potentials have been used widely in neuroscience; similar molecular technologies allow the perturbation of developmental pathways (De Renzis, 2020; Guglielmi et al., 2015; Hartmann et al., 2020; Izquierdo et al., 2018; Krueger et al., 2019). In a landmark paper, the Toettcher laboratory performed an optogenetic-mediated rescue of a Drosophila patterning mutant (Johnson et al., 2020), which demonstrates the power of this approach (Figure 3B). In this system, it was known that receptor tyrosine kinase ligands are the patterning signals (Goyal et al., 2018; Johnson et al., 2020; Pae et al., 2017). The optogenetic tool enables a new level of investigation into which features of signaling patterns carry essential information: The instantaneous protein concentration? The area under the curve? The total duration of signaling above a threshold? To address these questions, the authors used their previously developed, blue-light-activatable ERK response (Johnson and Toettcher, 2018). It constitutes a membrane-localized component that bears an optogenetically activatable recruiting domain for SOS, a transducer of ERK signals that is activated by membrane localization. A switch from darkness to light induces SOS membrane localization within seconds, followed by Erk activation and expression of Erk-dependent target genes, whereas a switch to darkness triggers a rapid reversal of this process, returning Erk activity and gene expression to baseline within minutes. Thus, the tool is an optogenetically controlled signaling pathway. The authors generated mutant flies containing Opto-SOS but lacking the endogenous ERK receptors that would normally activate SOS. In the dark, these flies mimic the phenotype of the receptor mutants, and fail to gastrulate. When exposed to light, the Opto-SOS mutants recreate the spatial pattern of SOS activation usually driven by endogenous ERK activity and these flies undergo gastrulation and normal development all the way to adult flies. This experimental setup allowed the researchers to further investigate the consequences of varying spatial and temporal aspects of the signal. They discovered that three distinct developmental switches are triggered at successively longer illumination times, and that graded spatial domains of signaling activation are important for certain developmental phenomena but not for others (Johnson et al., 2020).
Together, these studies are a testament to the advancement of synthetic biology tools to the point where they can rescue developmental patterning, providing insightful information on developmental mechanisms.
Building toy models of developmental transitions for understanding basic principles of patterning and morphogenesis
In the bottom-up approach, reconstitution of patterning and morphogenesis in toy models that are less complex than the real embryos have been attempted. It is a classic style of physics, where the simplest example that illustrates a principle is constructed to understand a system more fully. The philosophical discussion of why models are useful in science is very interesting, but beyond the scope of this piece (a starting point for the interested reader is Roman and Stephan, 2020). For developmental systems, models include the French flag model of morphogenetic patterning and the clock-and-wavefront model of somitogenesis (Cooke and Zeeman, 1976). Another class of models are the material models, which include Watson and Crick's metal model of DNA (Schaffner, 1969). Their simplicity and the fact that they are available for real or mental manipulation makes them amenable to more accessible understanding and experimentation. Toy models can bridge the gap between reality and theoretical description. One example is the pendulum, which is often used for exploring motion, gravity, and periodic behaviors in mechanics (Figure 4A). Pendulums have been used in several iterations and modifications to describe different phenomena, from convection to non-linear dynamics (Hu et al., 2019).
In this sense, a new wave of toy models in patterning and morphogenesis have become available in recent years thanks to synthetic biology tools. We call them toy models here to distinguish them from other kinds of models (e.g., embryo models, or organoids). We define them here as mammalian multicellular systems that, driven by synthetic genetic circuits, undergo patterning and/or morphogenesis starting from non-developmental cell lines (e.g., CHO, L929, and human embryonic kidney 293 [HEK293]). We are aware this naming may be controversial, as it may convey a sense of them being less important. On the contrary, our intention here is to highlight them as a worthy of their own category of study; we and others have high regard of toy models in general and in other disciplines in particular (Luczak, 2017; Nguyen, 2020) and think that learning through playing is one of the highest forms of human behavior. We also hope to add to increased discussion in the field on nomenclature and epistemological status of our models and arguments.
For developmental systems, we distinguish three classes of toy models, depending on what the engineered component is: synthetic signaling pathways, synthetic patterning circuits, synthetic morphogenetic circuits.
Synthetic versions of existing developmental signaling pathways have been generated. One class of synthetic pathway is generated by engineering existing pathways to control them better. These can be used to study more precisely the functions of existing pathways that they mimic. Examples include synthetic Shh (Li et al., 2018) and synthetic Dpp (Stapornwongkul et al., 2020) pathways. In the example of Shh, Li et al. (2018) were able to reconstitute both linear and radial morphogen gradients in vitro using a synthetic version of the Hedgehog pathway in the mouse fibroblast NIH/3T3 cell line. Having a toy model of the pathway at hand enabled more systematic investigation of architectural features of the signal transduction pathway underlying rapid formation of a robust signaling gradient (Stapornwongkul et al., 2018). Other developmental pathways could benefit from the same approach by focusing on elementary modules of development by isolating the primary and basic functions of developmental processes.
A second class of synthetic pathways is instead inspired by endogenous pathways but has more user-defined input and outputs, a feature defined as orthogonality in the field. These synthetic pathways can be used for orthogonal control of synthetic patterning and morphogenesis (see below). One example, a synthetic Notch pathway, which has been used for encoding contact-dependent signaling in L929 and Madin-Darby Canine Kidney (MDCK) cell lines (as well as a number of other primary cells including neurons and T cells; Morsut et al., 2016). SynNotch schemes have also been used recently for encoding morphogenetic-like signaling in L929 fibroblasts (Toda et al., 2020). Another avenue for introducing orthogonal signaling pathways in cell lines is to borrow from other organisms, as with the example of synthetic auxin signaling. Auxin is a small molecule produced by plant cells that is used as a key hormone for regulating their morphogenesis (Woodward, 2005). It works as a dimerizing element that targets one of the partners for degradation. It had already been shown that externally provided auxin can be used to regulate gene expression in mammalian cells (Lambrus et al., 2018). In a new study, it was shown that, by providing the genes encoding the enzymes to produce auxin from precursor to mammalian cells, cells can produce auxin and release it in the extracellular space. This allowed the authors to set up a system of source and graded responses of an artificial reporter gene (Ma et al., 2020).
An increasingly large toolkit of synthetic receptors inspired by design principles of natural signaling pathways is being developed for applications in cell therapies (Scheller and Fussenegger, 2019). We expect that these receptor systems will be helpful for developmental systems as well in the coming years.
Synthetic patterning circuits have been implemented in non-developmental systems via synthetic signaling pathways, or by deploying natural pathways in cell lines that do not endogenously engage in that type of signaling. Initial successes in this area came from the latter class, thanks to pioneering work by the Ebisuya laboratory. They used the overexpression of signaling circuits constructed with components of the Notch signaling pathway to program patterning in cells that do not natively engage in Notch signaling. The two classic Notch-mediated patterning circuits, lateral inhibition (Collier et al., 1996) and lateral induction (Hartman et al., 2010), have been implemented in CHO cells and a combination of CHO and MDCK cell lines respectively (Matsuda et al., 2012, 2015). More recently, using natural Nodal pathways elements and the Nodal inhibitor Lefty, the same group was able to implement reaction-diffusion patterning in the HEK293 cell line (Sekine et al., 2018). This kind of patterning was first proposed by Alan Turing as a chemical system of interacting and diffusible molecules that give rise to various stable patterns (Turing, 1990). Instead of using natural pathways in cell lines that do not engage in that signaling, another way to generate patterns in non-developmental systems has been to use completely orthogonal synthetic pathways. For example, synNotch has been used to generate contact-dependent edge detection and multi-layer cascades in MDCK cells (Morsut et al., 2016). More recently, morphogenetic signaling circuits based on synNotch has been developed and used to control morphogenetic-like patterning in L929 fibroblasts (Toda et al., 2020) (Figure 4B). Using one pole as a source of GFP and the other as an inhibitory anti-GFP, the authors showed that GFP can act like a morphogen in a competent domain, leading to gene expression patterns similar to many tissue systems in vivo. Additionally, the authors added another layer of complexity to the design by engineering both positive and negative feedback loops. This provides a new toy model for morphogenetic signaling that could be used to address questions about gradients in more complex systems.
Finally, synthetic morphogenetic systems have been generated, where morphogenesis is driven by user-introduced genetic circuits in non-developmental cell lines. Overexpression and mixing of cells with different members of the cadherin family of adhesion proteins enabled creation of spatial patterns in 2D and 3D using the HEK293 cell line (Cachat et al., 2016, 2017). Combining changes in cadherin expression downstream of synNotch-based signaling circuits has been used to program self-organized multicellular structures such as spheroids composed of two- and three-layer structures in L929 fibroblast cell lines (Toda et al., 2018). In particular, the combination of contact-dependent signaling with downstream induction of cadherin-family adhesion molecules allowed the generation of robust, multi-layered structures. These simplified intercellular programs were sufficient to yield assemblies with hallmarks of natural developmental systems: robust self-organization into multidomain structures, well-choreographed sequential assembly, cell-type divergence, symmetry breaking, and the capacity for regeneration upon injury.
These studies show that tissue patterning and morphogenesis can be directed in vitro by implementing synthetic signaling circuits in non-developmental cell lines.
Overall, a synthetic toolkit can offer tight control of variable parameters such as gradient range and density of anchoring cells to modulate the gradient shape. As opposed to in vivo systems composed of complex interactions with different morphogens and cell types, synthetic model systems provide a simplified context and modulable parameters to generate synthetic developmental trajectories. As with the pendulum in physics, synthetic gradients appear to be an adequate toy model that can be used to study basic principles governing gradient formation. These toy models could help to understand developmental biology concepts and challenge them by testing hypotheses. In the future, these toy models may even be used to interface in more complex way with natural systems, either as perturbative tools, rescue systems, synthetic organizers, or even therapeutic entities.
Challenges and opportunities in applying synthetic developmental biology to embryo models
We have described how synthetic biology tools are being used for developmental systems. In this last section we present how these tools and approaches have been or could be applied to embryo models.
For construction of better embryo models, simple artificial genetic circuits have been used in a recent study by the Zernicka-Goetz group (Amadei et al., 2021) (Figure 5A). The embryo model that was improved with genetic engineering is called ETX, a model that partially recapitulates mouse development by combining embryonic stem cells (ESCs), trophoblast stem cells (TS), and extra-embryonic endoderm (XEN) stem cells (Sozen et al., 2018). It was shown that ETX embryos are transcriptionally similar to the mouse embryo at around gastrulation stage, but they were unable to recapitulate all the complex morphogenetic events that characterize murine gastrulation. To address this, the authors replaced the XEN cells with ESCs transiently expressing the transcription factor Gata4, which drives extra-embryonic endoderm fate. Although the expression of Gata4 is only transient, the resulting embryos, dubbed induced ETX (iETX) embryos, represent an improvement in maturation and recapitulation of natural morphogenesis. The movies of gastrulating iETX embryos are rather remarkable in their display of gastrulating embryonic and extra-embryonic mesoderm, as well in the finer details of the anterior signaling center cell movements. It is possible that this enhanced developmental capacity of iETX derives from increased competency of the genetically engineered ESCs compared with the XEN cells. Having better embryo model systems like these enables study of the establishment of anterior-posterior patterning and gastrulation in an in vitro system. Particularly interesting is the fact that even transient overexpression of transcription factors could provide cells with increased developmental potential and self-organization capacity. In this sense, this study parallels the work on Gata6 overexpression for generation of liver organoids mentioned in section “using synthetic biology to enhance organoid construction” (Guye et al., 2016).
Overall, the approach of transcription factor overexpression in stem cells is ripe with potential and could be improved by novel technologies for synthetic transcription factors. Thus far, most transcription factor overexpression studies rely on the tetracycline-controlled induction of overexpression, which limits the number of transcription factors that can be overexpressed independently in a user-controlled, timed fashion. Synthetic transcription factors have been developed recently, both based on Crispr/dCas9 system, as well as on zinc-finger DNA-binding motifs (Donahue et al., 2020; Israni et al., 2021), some with interesting small-molecule induction regimes that, if proved to work reliably in stem cells, would pave the way to increased user-controllability of multiple transcription factor overexpression. In terms of which transcription factors to overexpress, recent efforts to explore the capacity of the whole library of transcription factors present in mammalian cells have been explored (Ng et al., 2020), and could be used as a starting point for transcription factor selection.
Embryo models could likewise be subjected to the deconstruction/reconstitution studies described in section “rescue of mutants of model species embryos with synthetic biology molecular tools.” We imagine that embryo models will become model systems in their own right, and that the same approaches used for model organisms like Drosophila could be applied to embryo models. For example, an embryo model could be made defective in its morphogenesis via a genetic mutation in a developmental patterning gene (Figure 5B). The developmental potential of the embryo model could be then rescued via synthetic signaling. This approach would allow the study of developmental transitions in increased detail under more controlled conditions.
Finally, synthetic toy models could be built in non-developmental systems that construct pattern and/or shapes similar to those displayed by embryo models. For example, a stepwise model that describes the first events during mammalian embryo formation has been proposed in a recent article (intriguingly titled “Instructions for Assembling the Early Mammalian Embryo”; White et al., 2018). Each step (or chain of steps) could be isolated and its reconstruction attempted in non-developmental systems using existing or novel synthetic patterning and morphogenetic circuits (Figure 5C). As initial steps are dominated by cellular polarization and assembly of cell-cell junctions, recent synthetic efforts that generated planar polarization (Loza et al., 2017) and cell-cell adhesion (Appleton et al., 2019; Glass and Riedel-Kruse, 2018) seem to suggest that it could be soon possible to assemble a structure that resembles the early mammalian embryo starting from a non-developmental cell line, using synthetic biology.
Despite all the potential, some current limitations are looming and could limit progress. Examples include silencing of synthetic constructs. It has been noticed that transgene expression can be negatively affected by epigenetic context and differentiation status (Alhaji et al., 2019), and that the situation is even more complex for larger circuit integration (Zimak et al., 2021). Although small-molecule modifiers of epigenetic silencing have been used successfully in vitro, it is likely that these would affect the developmental potential of stem cells also thereby altering efforts to mimic developmental transitions if applied to embryo models. Another possibility for controlling epigenetic silencing could lie in synthetic factors that could insulate or control the epigenetic status of user-defined genetic loci (Bintu et al., 2016; Park et al., 2016, 2019; Van et al., 2021). In the future, complex synthetic circuits could contain sensors for epigenetic silencing that trigger synthetic reversion of silencing itself.
Finally, we feel the need to introduce the classic synthetic biology of surpassing nature, with a cautionary twist. The narrative basically says that mastering a scientific field suggests using our acquired basic knowledge to go further and overcome the limits of natural systems. In this narrative, synthetic biology is compared with chemistry, which can be seen as a basic science interested in how atoms can assemble into molecules and how molecules can interact with each other to create different and more complex molecules. After deciphering the basic principles of the atom and its laws in molecules, chemists are now able to synthesize new molecules that did not exist previously in nature. These molecules can now be designed and formulated for different ends, such as therapeutic, food, and cosmetic applications. Molecular biology can be said to have followed the same path with the invention of synthetic proteins that can control cellular behaviors. It may only be a question of time until the same approach is applied to embryo models, and synthetic biology could enable the encoding of novel, non-natural developmental trajectories. In this last application, it reaches a stage where ethical implications cannot (and should not) be ignored. It has already been shown that editing of human embryos is technically feasible, although we agree with the community that at this point it should not be a goal of the field. Engaging in discussion with professional bioethicists and with the larger public will be critical to moving the field forward in a fashion that should, in the common interest, be sustainable for all.
Acknowledgments
We express gratitude to all the members of the Morsut laboratory and especially to Marion Johnson, as well as two anonymous reviewers, for their insightful comments on earlier versions of the manuscript. Figures 1, 2, 3, and 5 have been created by C.H. with BioRender.com. This work has been supported by funding to L.M.: R00 EB021030-03 from NIH/NIBIB, R35 GM138256 from NIH/NIGMS, CBET-2034495 RECODE from NSF.
Authors contributions
C.H. and L.M. conceived and wrote the article.
Declaration of interests
L.M. is an inventor of synNotch patent US9670281B2 and receive royalty payments from licensing to Gilead Inc. through UCSF.
References
- Alhaji S.Y., Ngai S.C., Abdullah S. Silencing of transgene expression in mammalian cells by DNA methylation and histone modifications in gene therapy perspective. Biotechnol. Genet. Eng. Rev. 2019;35:1–25. doi: 10.1080/02648725.2018.1551594. [DOI] [PubMed] [Google Scholar]
- Amadei G., Lau K.Y.C., De Jonghe J., Gantner C.W., Sozen B., Chan C., Zhu M., Kyprianou C., Hollfelder F., Zernicka-Goetz M. Inducible stem-cell-derived embryos capture mouse morphogenetic events in vitro. Dev. Cell. 2021;56:366–382.e9. doi: 10.1016/j.devcel.2020.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appleton E., Mehdipour N., Daifuku T., Briers D., Haghighi I., Moret M., Chao G., Wannier T., Chiappino-Pepe A., Huang J. 2019. Genetic Design Automation for Autonomous Formation of Multicellular Shapes from a Single Cell Progenitor (Synthetic Biology)http://biorxiv.org/lookup/doi/10.1101/807107 bioRxiv. [Google Scholar]
- Basu S., Gerchman Y., Collins C.H., Arnold F.H., Weiss R. A synthetic multicellular system for programmed pattern formation. Nature. 2005;434:1130–1134. doi: 10.1038/nature03461. [DOI] [PubMed] [Google Scholar]
- Betzig E., Patterson G.H., Sougrat R., Lindwasser O.W., Olenych S., Bonifacino J.S., Davidson M.W., Lippincott-Schwartz J., Hess H.F. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- Bintu L., Yong J., Antebi Y.E., McCue K., Kazuki Y., Uno N., Oshimura M., Elowitz M.B. Dynamics of epigenetic regulation at the single-cell level. Science. 2016;351:720–724. doi: 10.1126/science.aab2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brassard J.A., Lutolf M.P. Engineering stem cell self-organization to build better organoids. Cell Stem Cell. 2019;24:860–876. doi: 10.1016/j.stem.2019.05.005. [DOI] [PubMed] [Google Scholar]
- Briscoe J., Small S. Morphogen rules: design principles of gradient-mediated embryo patterning. Development. 2015;142:3996–4009. doi: 10.1242/dev.129452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busskamp V., Lewis N.E., Guye P., Ng A.H., Shipman S.L., Byrne S.M., Sanjana N.E., Murn J., Li Y., Li S. Rapid neurogenesis through transcriptional activation in human stem cells. Mol. Syst. Biol. 2014;10:760. doi: 10.15252/msb.20145508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cachat E., Liu W., Martin K.C., Yuan X., Yin H., Hohenstein P., Davies J.A. 2- and 3-dimensional synthetic large-scale de novo patterning by mammalian cells through phase separation. Sci. Rep. 2016;6:20664. doi: 10.1038/srep20664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cachat E., Liu W., Davies J.A. Synthetic self-patterning and morphogenesis in mammalian cells: a proof-of-concept step towards synthetic tissue development. Eng. Biol. 2017;1:71–76. [Google Scholar]
- Cakir B., Xiang Y., Tanaka Y., Kural M.H., Parent M., Kang Y.-J., Chapeton K., Patterson B., Yuan Y., He C.-S. Engineering of human brain organoids with a functional vascular-like system. Nat. Methods. 2019;16:1169–1175. doi: 10.1038/s41592-019-0586-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron D.E., Bashor C.J., Collins J.J. A brief history of synthetic biology. Nat. Rev. Microbiol. 2014;12:381–390. doi: 10.1038/nrmicro3239. [DOI] [PubMed] [Google Scholar]
- Castanon I., González-Gaitán M. Integrating levels of complexity: a trend in developmental biology. Curr. Opin. Cell Biol. 2011;23:647–649. doi: 10.1016/j.ceb.2011.10.003. [DOI] [PubMed] [Google Scholar]
- Cederquist G.Y., Asciolla J.J., Tchieu J., Walsh R.M., Cornacchia D., Resh M.D., Studer L. Specification of positional identity in forebrain organoids. Nat. Biotechnol. 2019;37:436–444. doi: 10.1038/s41587-019-0085-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B.-C., Legant W.R., Wang K., Shao L., Milkie D.E., Davidson M.W., Janetopoulos C., Wu X.S., Hammer J.A., Liu Z. Lattice light-sheet microscopy: imaging molecules to embryos at high spatiotemporal resolution. Science. 2014;346:1257998. doi: 10.1126/science.1257998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chronis C., Fiziev P., Papp B., Butz S., Bonora G., Sabri S., Ernst J., Plath K. Cooperative binding of transcription factors orchestrates reprogramming. Cell. 2017;168:442–459.e20. doi: 10.1016/j.cell.2016.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H. Modeling development and disease with organoids. Cell. 2016;165:1586–1597. doi: 10.1016/j.cell.2016.05.082. [DOI] [PubMed] [Google Scholar]
- Collier J.R., Monk N.A.M., Maini P.K., Lewis J.H. Pattern formation by lateral inhibition with feedback: a mathematical model of delta-notch intercellular signalling. J. Theor. Biol. 1996;183:429–446. doi: 10.1006/jtbi.1996.0233. [DOI] [PubMed] [Google Scholar]
- Cooke J., Zeeman E.C. A clock and wavefront model for control of the number of repeated structures during animal morphogenesis. J. Theor. Biol. 1976;58:455–476. doi: 10.1016/s0022-5193(76)80131-2. [DOI] [PubMed] [Google Scholar]
- De Renzis S. Morphogenesis: guiding embryonic development with light. Curr. Biol. 2020;30:R998–R1001. doi: 10.1016/j.cub.2020.07.048. [DOI] [PubMed] [Google Scholar]
- De Robertis E.M. Spemann’s organizer and the self-regulation of embryonic fields. Mech. Dev. 2009;126:925–941. doi: 10.1016/j.mod.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donahue P.S., Draut J.W., Muldoon J.J., Edelstein H.I., Bagheri N., Leonard J.N. The COMET toolkit for composing customizable genetic programs in mammalian cells. Nat. Commun. 2020;11:779. doi: 10.1038/s41467-019-14147-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebrahimkhani M.R., Ebisuya M. Synthetic developmental biology: build and control multicellular systems. Curr. Opin. Chem. Biol. 2019;52:9–15. doi: 10.1016/j.cbpa.2019.04.006. [DOI] [PubMed] [Google Scholar]
- Elcheva I., Brok-Volchanskaya V., Kumar A., Liu P., Lee J.-H., Tong L., Vodyanik M., Swanson S., Stewart R., Kyba M. Direct induction of haematoendothelial programs in human pluripotent stem cells by transcriptional regulators. Nat. Commun. 2014;5:4372. doi: 10.1038/ncomms5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elowitz M.B., Leibler S. A synthetic oscillatory network of transcriptional regulators. Nature. 2000;403:335–338. doi: 10.1038/35002125. [DOI] [PubMed] [Google Scholar]
- Gardner T.S., Cantor C.R., Collins J.J. Construction of a genetic toggle switch in Escherichia coli. Nature. 2000;403:339–342. doi: 10.1038/35002131. [DOI] [PubMed] [Google Scholar]
- Glass D.S., Riedel-Kruse I.H. A synthetic bacterial cell-cell adhesion toolbox for programming multicellular morphologies and patterns. Cell. 2018;174:649–658.e16. doi: 10.1016/j.cell.2018.06.041. [DOI] [PubMed] [Google Scholar]
- Goyal Y., Schüpbach T., Shvartsman S.Y. A quantitative model of developmental RTK signaling. Dev. Biol. 2018;442:80–86. doi: 10.1016/j.ydbio.2018.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guglielmi G., Barry J.D., Huber W., De Renzis S. An optogenetic method to modulate cell contractility during tissue morphogenesis. Dev. Cell. 2015;35:646–660. doi: 10.1016/j.devcel.2015.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guye P., Ebrahimkhani M.R., Kipniss N., Velazquez J.J., Schoenfeld E., Kiani S., Griffith L.G., Weiss R. Genetically engineering self-organization of human pluripotent stem cells into a liver bud-like tissue using Gata6. Nat. Commun. 2016;7:10243. doi: 10.1038/ncomms10243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman B.H., Reh T.A., Bermingham-McDonogh O. Notch signaling specifies prosensory domains via lateral induction in the developing mammalian inner ear. Proc. Natl. Acad. Sci. U S A. 2010;107:15792–15797. doi: 10.1073/pnas.1002827107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann J., Krueger D., De Renzis S. Using optogenetics to tackle systems-level questions of multicellular morphogenesis. Curr. Opin. Cell Biol. 2020;66:19–27. doi: 10.1016/j.ceb.2020.04.004. [DOI] [PubMed] [Google Scholar]
- Hu Z., Legeza V., Dychka I., Onai M. Non-linear model of the damping process in a system with a two-mass pendulum absorber. Int. J. Intell. Syst. Appl. 2019;11:67–72. doi: 10.5815/ijisa.2019.01.07. [DOI] [Google Scholar]
- Israni D.V., Li H.-S., Gagnon K.A., Sander J.D., Roybal K.T., Joung J.K., Wong W.W., Khalil A.S. 2021. Clinically-driven design of synthetic gene regulatory programs in human cells (Synthetic Biology)http://biorxiv.org/lookup/doi/10.1101/2021.02.22.432371 bioRxiv. [Google Scholar]
- Izquierdo E., Quinkler T., De Renzis S. Guided morphogenesis through optogenetic activation of Rho signalling during early Drosophila embryogenesis. Nat. Commun. 2018;9:2366. doi: 10.1038/s41467-018-04754-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessell T.M. Neuronal specification in the spinal cord: inductive signals and transcriptional codes. Nat. Rev. Genet. 2000;1:20–29. doi: 10.1038/35049541. [DOI] [PubMed] [Google Scholar]
- Johnson H.E., Toettcher J.E. Illuminating developmental biology with cellular optogenetics. Curr. Opin. Biotechnol. 2018;52:42–48. doi: 10.1016/j.copbio.2018.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson H.E., Djabrayan N.J.V., Shvartsman S.Y., Toettcher J.E. Optogenetic rescue of a patterning mutant. Curr. Biol. 2020;30:3414–3424.e3. doi: 10.1016/j.cub.2020.06.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson M.B., March A.R., Morsut L. Engineering multicellular systems: using synthetic biology to control tissue self-organization. Curr. Opin. Biomed. Eng. 2017;4:163–173. doi: 10.1016/j.cobme.2017.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalik L., Chen J.K. Illuminating developmental biology through photochemistry. Nat. Chem. Biol. 2017;13:587–598. doi: 10.1038/nchembio.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger D., Izquierdo E., Viswanathan R., Hartmann J., Pallares Cartes C., De Renzis S. Principles and applications of optogenetics in developmental biology. Development. 2019;146:dev175067. doi: 10.1242/dev.175067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambrus B.G., Moyer T.C., Holland A.J. Applying the auxin-inducible degradation system for rapid protein depletion in mammalian cells. Methods Cell Biol. 2018;144:107–135. doi: 10.1016/bs.mcb.2018.03.004. [DOI] [PubMed] [Google Scholar]
- Lange L., Hoffmann D., Schwarzer A., Ha T.-C., Philipp F., Lenz D., Morgan M., Schambach A. Inducible forward programming of human pluripotent stem cells to hemato-endothelial progenitor cells with hematopoietic progenitor potential. Stem Cell Rep. 2020;14:122–137. doi: 10.1016/j.stemcr.2019.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecuit T., Cohen S.M. Dpp receptor levels contribute to shaping the Dpp morphogen gradient in the Drosophila wing imaginal disc. Dev. Camb. Engl. 1998;125:4901–4907. doi: 10.1242/dev.125.24.4901. [DOI] [PubMed] [Google Scholar]
- Lewandoski M. Conditional control of gene expression in the mouse. Nat. Rev. Genet. 2001;2:743–755. doi: 10.1038/35093537. [DOI] [PubMed] [Google Scholar]
- Li P., Markson J.S., Wang S., Chen S., Vachharajani V., Elowitz M.B. Morphogen gradient reconstitution reveals Hedgehog pathway design principles. Science. 2018;360:543–548. doi: 10.1126/science.aao0645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Zhong C., Yu Y., Liu H., Sakurai M., Yu L., Min Z., Shi L., Wei Y., Takahashi Y. Generation of blastocyst-like structures from mouse embryonic and adult cell cultures. Cell. 2019;179:687–702.e18. doi: 10.1016/j.cell.2019.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F., Li D., Yu Y.Y.L., Kang I., Cha M., Kim J.Y., Park C., Watson D.K., Wang T., Choi K. Induction of hematopoietic and endothelial cell program orchestrated by ETS transcription factor ER 71/ETV 2. EMBO Rep. 2015;16:654–669. doi: 10.15252/embr.201439939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loza O., Heemskerk I., Gordon-Bar N., Amir-Zilberstein L., Jung Y., Sprinzak D. A synthetic planar cell polarity system reveals localized feedback on Fat4-Ds1 complexes. eLife. 2017;6:e24820. doi: 10.7554/eLife.24820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luczak J. Talk about toy models. Stud. Hist. Philos. Sci. B Stud. Hist. Philos. Mod. Phys. 2017;57:1–7. [Google Scholar]
- Lupo G., Harris W.A., Lewis K.E. Mechanisms of ventral patterning in the vertebrate nervous system. Nat. Rev. Neurosci. 2006;7:103–114. doi: 10.1038/nrn1843. [DOI] [PubMed] [Google Scholar]
- Ma Y., Budde M.W., Mayalu M.N., Zhu J., Murray R.M., Elowitz M.B. 2020. Synthetic Mammalian Signaling Circuits for Robust Cell Population Control (Synthetic Biology)http://biorxiv.org/lookup/doi/10.1101/2020.09.02.278564 bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marton R.M., Pașca S.P. Organoid and assembloid technologies for investigating cellular crosstalk in human brain development and disease. Trends Cell Biol. 2020;30:133–143. doi: 10.1016/j.tcb.2019.11.004. [DOI] [PubMed] [Google Scholar]
- Matsuda M., Koga M., Nishida E., Ebisuya M. Synthetic signal propagation through direct cell-cell interaction. Sci. Signal. 2012;5:ra31. doi: 10.1126/scisignal.2002764. [DOI] [PubMed] [Google Scholar]
- Matsuda M., Koga M., Woltjen K., Nishida E., Ebisuya M. Synthetic lateral inhibition governs cell-type bifurcation with robust ratios. Nat. Commun. 2015;6:6195. doi: 10.1038/ncomms7195. [DOI] [PubMed] [Google Scholar]
- Meinhardt H. Primary body axes of vertebrates: generation of a near-Cartesian coordinate system and the role of Spemann-type organizer. Dev. Dyn. 2006;235:2907–2919. doi: 10.1002/dvdy.20952. [DOI] [PubMed] [Google Scholar]
- Metzger D. Engineering the mouse genome by site-specific recombination. Curr. Opin. Biotechnol. 1999;10:470–476. doi: 10.1016/s0958-1669(99)00012-9. [DOI] [PubMed] [Google Scholar]
- Morsut L., Roybal K.T., Xiong X., Gordley R.M., Coyle S.M., Thomson M., Lim W.A. Engineering customized cell sensing and response behaviors using synthetic Notch receptors. Cell. 2016;164:780–791. doi: 10.1016/j.cell.2016.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura H., Katahira T., Matsunaga E., Sato T. Isthmus organizer for midbrain and hindbrain development. Brain Res. Rev. 2005;49:120–126. doi: 10.1016/j.brainresrev.2004.10.005. [DOI] [PubMed] [Google Scholar]
- Nellen D., Burke R., Struhl G., Basler K. Direct and long-range action of a DPP morphogen gradient. Cell. 1996;85:357–368. doi: 10.1016/s0092-8674(00)81114-9. [DOI] [PubMed] [Google Scholar]
- Ng A.H.M., Khoshakhlagh P., Rojo Arias J.E., Pasquini G., Wang K., Swiersy A., Shipman S.L., Appleton E., Kiaee K., Kohman R.E. A comprehensive library of human transcription factors for cell fate engineering. Nat. Biotechnol. 2020;39:510–519. doi: 10.1038/s41587-020-0742-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen J. It’s not a game: accurate representation with toy models. Br. J. Philos. Sci. 2020;71:1013–1041. [Google Scholar]
- Ouyang X., Chen J.K. Synthetic strategies for studying embryonic development. Chem. Biol. 2010;17:590–606. doi: 10.1016/j.chembiol.2010.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pae J., Cinalli R.M., Marzio A., Pagano M., Lehmann R. GCL and CUL3 control the switch between cell lineages by mediating localized degradation of an RTK. Dev. Cell. 2017;42:130–142.e7. doi: 10.1016/j.devcel.2017.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park M., Keung A.J., Khalil A.S. The epigenome: the next substrate for engineering. Genome Biol. 2016;17:183. doi: 10.1186/s13059-016-1046-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park M., Patel N., Keung A.J., Khalil A.S. Engineering epigenetic regulation using synthetic read-write modules. Cell. 2019;176:227–238.e20. doi: 10.1016/j.cell.2018.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson M. Transcription factor based therapeutics: drugs of the future? Trends Biotechnol. 1993;11:11–18. doi: 10.1016/0167-7799(93)90069-L. [DOI] [PubMed] [Google Scholar]
- Pezzulo G., Levin M. Top-down models in biology: explanation and control of complex living systems above the molecular level. J. R. Soc. Interf. 2016;13:20160555. doi: 10.1098/rsif.2016.0555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planchon T.A., Gao L., Milkie D.E., Davidson M.W., Galbraith J.A., Galbraith C.G., Betzig E. Rapid three-dimensional isotropic imaging of living cells using Bessel beam plane illumination. Nat. Methods. 2011;8:417–423. doi: 10.1038/nmeth.1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivron N.C., Frias-Aldeguer J., Vrij E.J., Boisset J.-C., Korving J., Vivié J., Truckenmüller R.K., van Oudenaarden A., van Blitterswijk C.A., Geijsen N. Blastocyst-like structures generated solely from stem cells. Nature. 2018;557:106–111. doi: 10.1038/s41586-018-0051-0. [DOI] [PubMed] [Google Scholar]
- Roman F., Stephan H. Metaphysics Research Lab, Stanford University; 2020. Models in Science. [Google Scholar]
- Roybal K.T., Williams J.Z., Morsut L., Rupp L.J., Kolinko I., Choe J.H., Walker W.J., McNally K.A., Lim W.A. Engineering T cells with customized therapeutic response programs using synthetic Notch receptors. Cell. 2016;167:419–432.e16. doi: 10.1016/j.cell.2016.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roybal K.T., Rupp L.J., Morsut L., Walker W.J., McNally K.A., Park J.S., Lim W.A. Precision tumor recognition by T cells with combinatorial antigen-sensing circuits. Cell. 2016;164:770–779. doi: 10.1016/j.cell.2016.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santorelli M., Lam C., Morsut L. Synthetic development: building mammalian multicellular structures with artificial genetic programs. Curr. Opin. Biotechnol. 2019;59:130–140. doi: 10.1016/j.copbio.2019.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasai Y. Next-generation regenerative medicine: organogenesis from stem cells in 3D culture. Cell Stem Cell. 2013;12:520–530. doi: 10.1016/j.stem.2013.04.009. [DOI] [PubMed] [Google Scholar]
- Schaffner K.F. The Watson-Crick model and reductionism. Br. J. Philos. Sci. 1969;20:325–348. [Google Scholar]
- Scheller L., Fussenegger M. From synthetic biology to human therapy: engineered mammalian cells. Curr. Opin. Biotechnol. 2019;58:108–116. doi: 10.1016/j.copbio.2019.02.023. [DOI] [PubMed] [Google Scholar]
- Schlissel G., Li P. Synthetic developmental biology: understanding through reconstitution. Annu. Rev. Cell Dev. Biol. 2020;36:339–357. doi: 10.1146/annurev-cellbio-020620-090650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schutgens F., Clevers H. Human organoids: tools for understanding biology and treating diseases. Annu. Rev. Pathol. Mech. Dis. 2020;15:211–234. doi: 10.1146/annurev-pathmechdis-012419-032611. [DOI] [PubMed] [Google Scholar]
- Sekine R., Shibata T., Ebisuya M. Synthetic mammalian pattern formation driven by differential diffusivity of Nodal and Lefty. Nat. Commun. 2018;9:5456. doi: 10.1038/s41467-018-07847-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serra D., Mayr U., Boni A., Lukonin I., Rempfler M., Challet Meylan L., Stadler M.B., Strnad P., Papasaikas P., Vischi D. Self-organization and symmetry breaking in intestinal organoid development. Nature. 2019;569:66–72. doi: 10.1038/s41586-019-1146-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shestopalov I.A., Chen J.K. Chemical technologies for probing embryonic development. Chem. Soc. Rev. 2008;37:1294. doi: 10.1039/b703023c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skylar-Scott M.A., Huang J.Y., Lu A., Ng A.H.M., Duenki T., Nam L.L., Damaraju S., Church G.M., Lewis J.A. 2020. An Orthogonal Differentiation Platform for Genomically Programming Stem Cells, Organoids, and Bioprinted Tissues (Bioengineering)http://biorxiv.org/lookup/doi/10.1101/2020.07.11.198671 bioRxiv. [Google Scholar]
- Solé R. Synthetic transitions: towards a new synthesis. Philos. Trans. R. Soc. B Biol. Sci. 2016;371:20150438. doi: 10.1098/rstb.2015.0438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soufi A., Garcia M.F., Jaroszewicz A., Osman N., Pellegrini M., Zaret K.S. Pioneer transcription factors target partial DNA motifs on nucleosomes to initiate reprogramming. Cell. 2015;161:555–568. doi: 10.1016/j.cell.2015.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sozen B., Amadei G., Cox A., Wang R., Na E., Czukiewska S., Chappell L., Voet T., Michel G., Jing N. Self-assembly of embryonic and two extra-embryonic stem cell types into gastrulating embryo-like structures. Nat. Cell Biol. 2018;20:979–989. doi: 10.1038/s41556-018-0147-7. [DOI] [PubMed] [Google Scholar]
- Stapornwongkul K.S., Salbreux G., Vincent J.P. Developmental biology: morphogen in a dish. Curr. Biol. 2018;28:R755–R757. doi: 10.1016/j.cub.2018.05.047. [DOI] [PubMed] [Google Scholar]
- Stapornwongkul K.S., de Gennes M., Cocconi L., Salbreux G., Vincent J.P. Patterning and growth control in vivo by an engineered GFP gradient. Science. 2020;39:510–519. doi: 10.1126/science.abb8205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Yamanaka S. A decade of transcription factor-mediated reprogramming to pluripotency. Nat. Rev. Mol. Cell Biol. 2016;17:183–193. doi: 10.1038/nrm.2016.8. [DOI] [PubMed] [Google Scholar]
- Takebe T., Wells J.M. Organoids by design. Science. 2019;364:956–959. doi: 10.1126/science.aaw7567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teleman A.A., Cohen S.M. Dpp gradient formation in the Drosophila wing imaginal disc. Cell. 2000;103:971–980. doi: 10.1016/s0092-8674(00)00199-9. [DOI] [PubMed] [Google Scholar]
- Toda S., Blauch L.R., Tang S.K.Y., Morsut L., Lim W.A. Programming self-organizing multicellular structures with synthetic cell-cell signaling. Science. 2018;361:156–162. doi: 10.1126/science.aat0271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda S., Brunger J.M., Lim W.A. Synthetic development: learning to program multicellular self-organization. Curr. Opin. Syst. Biol. 2019;14:41–49. [Google Scholar]
- Toda S., McKeithan W.L., Hakkinen T.J., Lopez P., Klein O.D., Lim W.A. Engineering synthetic morphogen systems that can program multicellular patterning. Science. 2020;370:327–331. doi: 10.1126/science.abc0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toettcher J.E., Voigt C.A., Weiner O.D., Lim W.A. The promise of optogenetics in cell biology: interrogating molecular circuits in space and time. Nat. Methods. 2011;8:35–38. doi: 10.1038/nmeth.f.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turing A.M. The chemical basis of morphogenesis. 1953. Bull. Math. Biol. 1990;52:153–197. doi: 10.1007/BF02459572. discussion 119-152. [DOI] [PubMed] [Google Scholar]
- Van M.V., Fujimori T., Bintu L. Nanobody-mediated control of gene expression and epigenetic memory. Nat. Commun. 2021;12:537. doi: 10.1038/s41467-020-20757-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velazquez J.J., LeGraw R., Moghadam F., Tan Y., Kilbourne J., Maggiore J.C., Hislop J., Liu S., Cats D., Chuva de Sousa Lopes S.M. Gene regulatory network analysis and engineering directs development and vascularization of multilineage human liver organoids. Cell Syst. 2021;12:41–55.e11. doi: 10.1016/j.cels.2020.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velazquez J.J., Su E., Cahan P., Ebrahimkhani M.R. Programming morphogenesis through systems and synthetic biology. Trends Biotechnol. 2018;36:415–429. doi: 10.1016/j.tibtech.2017.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldman M.B., Zhao C., Gomez G.A., Lindgren A.G., Huang H., Yang H., Yao S., Martin B.L., Kimelman D., Lin S. Transdifferentiation of fast skeletal muscle into functional endothelium in vivo by transcription factor Etv2. PLoS Biol. 2013;11:e1001590. doi: 10.1371/journal.pbio.1001590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White M.D., Zenker J., Bissiere S., Plachta N. Instructions for assembling the early mammalian embryo. Dev. Cell. 2018;45:667–679. doi: 10.1016/j.devcel.2018.05.013. [DOI] [PubMed] [Google Scholar]
- Wieschaus E., Nüsslein-Volhard C. The Heidelberg screen for pattern mutants of Drosophila : a personal account. Annu. Rev. Cell Dev. Biol. 2016;32:1–46. doi: 10.1146/annurev-cellbio-113015-023138. [DOI] [PubMed] [Google Scholar]
- Woodward A.W. Auxin: regulation, action, and interaction. Ann. Bot. 2005;95:707–735. doi: 10.1093/aob/mci083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Pak C., Han Y., Ahlenius H., Zhang Z., Chanda S., Marro S., Patzke C., Acuna C., Covy J. Rapid single-step induction of functional neurons from human pluripotent stem cells. Neuron. 2013;78:785–798. doi: 10.1016/j.neuron.2013.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimak J., Wagoner Z.W., Nelson N., Waechtler B., Schlosser H., Kopecky M., Wu J., Zhao W. Epigenetic silencing directs expression heterogeneity of stably integrated multi-transcript unit genetic circuits. Sci. Rep. 2021;11:2424. doi: 10.1038/s41598-021-81975-1. [DOI] [PMC free article] [PubMed] [Google Scholar]