To the editor
Mutations in the annexin A11 gene (ANXA11) have been shown to cause amyotrophic lateral sclerosis (ALS) [6]. Annexin A11 is a Ca2+-dependent phospholipid-binding protein that possesses an N-terminal low-complexity domain and C-terminal repeated annexin domains, being involved in Ca2+ signaling, cell division, apoptosis, and vesicle trafficking [3, 5, 8]. Recent studies have indicated that ALS-related ANXA11 mutations enhance aggregation propensity, leading to dysregulation of intracellular Ca2+ homeostasis and RNA granule transport [3, 5, 6].
The clinical phenotypes of ANXA11-mutated ALS vary, and differ even in patients harboring the same mutation [6, 7, 9]. Autopsy findings of two patients harboring N-terminal ANXA11 mutation have been reported. One patient with p.D40G ANXA11 mutation showed the features of classical ALS [6], while the other with p.G38R ANXA11 mutation showed those of ALS-TDP with frontotemporal lobar degeneration (FTLD)-TDP type A [4, 7]. Both patients showed aggregation of 43 kDa TAR-DNA-binding protein (TDP-43) and annexin A11 in neurons (Table 2). However, the variety of neuropathologic features, including the distribution and morphology of TDP-43- or annexin A11-immunoreactive (ir) inclusions, in patients with ANXA11 mutations remain to be further clarified. Here, we investigated the clinicopathologic features of a Japanese patient with sporadic ALS harboring a novel splicing mutation in the annexin domain of ANXA11, and the functional significance of the mutation.
Table 2.
Clinical and neuropathologic features of patients harboring ANXA11 mutation
| Present patient | Patient with p.D40G [6] | Patient with p.G38R [7] | |
|---|---|---|---|
| Clinical features | |||
| Age at onset (years) | 57 | 72 | 60’sa |
| Disease duration (months) | 19 | 36 | naa |
| Initial symptom | Limb | Bulbar | Limb |
| Cognitive impairment or FTD | − | − | + |
| Neuropathologic phenotype | ALS-TDP | ALS-TDP | ALS-TDP with FTLD-TDP type A |
| Neuronal loss | |||
| Motor cortex | + | na | + |
| Lower motor neurons | + | + | na |
| Frontotemporal cortex | − | na | + |
| Degeneration of CST | + | + | + |
| Bunina body | + | na | + |
| pTDP-43-ir NCIs/GCIs | +/+ | +/na | +/+ |
| Annexin A11-ir NCIs/GCIs | +/few | +/na | +/na |
| Morphology of annexin A11-ir NCIs | Skein-like, filamentous, small round | Skein-like, filamentous, large-caliber and tube-shaped, large basket-like | Tube-shaped, large conglomerate, large round |
| Co-localization of pTDP-43 and annexin A11 | − or + | − | − or + |
+, present; −, absent; a, among 3 patients harboring p.G38R ANXA11 mutation, the autopsied patient was not identified in the reference [7]; FTD, frontotemporal dementia; ALS, amyotrophic lateral sclerosis; FTLD, frontotemporal lobar degeneration; na, not available; CST; corticospinal tract; pTDP-43, phosphorylated 43 kDa TAR DNA-binding protein; ir, immunoreactive
A 57-year-old man, who had no family history of neurologic disorders, presented with progressive limb weakness and stiffness, followed by muscle fasciculations, pyramidal signs, dysphagia and dysarthria. No dementia was noted. He died suddenly 19 months after disease onset. His detailed clinical features are described in the Additional File.
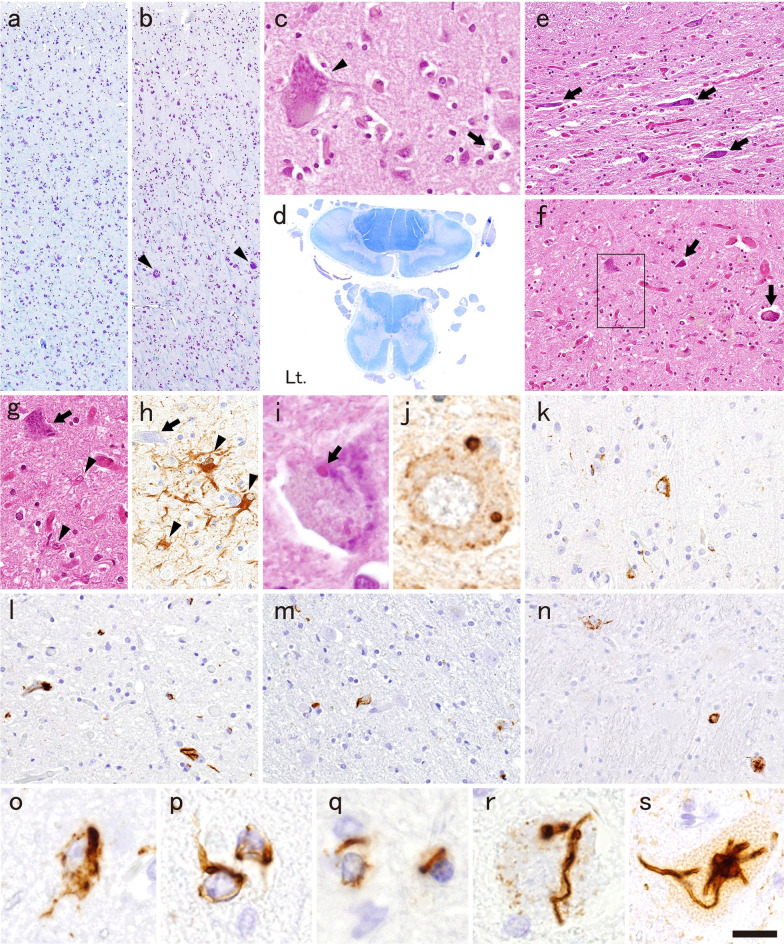
The neuropathologic features were consistent with ALS. Neuronal loss and gliosis were restricted to the motor cortex, brainstem motor nuclei, and spinal anterior horns (Fig. 1a–c, e–h; Table 1). The corticospinal tract in the spinal cord showed severe degeneration (Fig. 1d). Bunina bodies were occasionally found in the lower motor neurons (Fig. 1i, j). Phosphorylated TDP-43 (pTDP-43)-ir neuronal and oligodendroglial cytoplasmic inclusions (NCIs and GCIs, respectively) were evident in the above-mentioned areas (Fig. 1k, o, p, motor cortex; q, white matter adjacent to the motor cortex; l, r, hypoglossal nucleus; s, anterior horn of the lumbar cord) and several others (Fig. 1m, subthalamic nucleus; n, pontine nucleus) but not in the hippocampus or temporal cortex (Table 1). Morphologically, the pTDP-43-ir NCIs were granular and/or filamentous (Fig. 1o, p) in all areas where NCIs were observed, and characteristically those in the lower motor neurons were thick skein-like, or tube-shaped (Fig. 1r, s).
Fig. 1.
Neuropathologic findings. a, b The motor cortex shows neuronal loss and gliosis more prominently in a the lateral side than b the medial side. Some Betz cells are preserved only in the medial side of the motor cortex (arrowheads in b, c). c Macrophage accumulation in a Betz cell-sized hole (arrow in c), indicating ongoing neuronal degeneration, is evident in the medial side. d Myelin pallor in the bilateral lateral columns of the cervical and lumbar cord, and the left anterior column of the cervical cord, which indicate degeneration of the pyramidal tracts. Atrophy of the anterior horns is also evident in the cervical cord. e–h Severe neuronal loss and gliosis in e the cervical anterior horn, and f–h facial nucleus (arrows, atrophic neurons; arrowheads, reactive astrocytes; g and h, high magnification images of the square in f; h, GFAP-immunohistochemistry). i, j Bunina bodies in the remaining motor neurons of the facial nucleus (j, cystatin C-immunohistochemistry). k–s Phosphorylated TDP-43 (pTDP-43)-immunohistochemistry. Several pTDP-43-immunoreactive (ir) neuronal cytoplasmic inclusions (NCIs) and glial cytoplasmic inclusions (GCIs) in k the motor cortex, l hypoglossal nucleus and m subthalamic nucleus. n NCIs in the pontine nucleus. o Granulofilamentous and (left in p) filamentous NCIs and (right in p) GCI in the motor cortex. q GCIs in the white matter adjacent to the motor cortex. r Thick skein-like NCI in the hypoglossal nucleus. (s) Tube-shaped NCI in the anterior horn of the lumbar cord. Bar = 190 µm for a, b; 35 µm for c, g, h; 3 mm for d; 75 µm for e, f; 10 µm for i, j, o–r; 50 µm for k–n; 15 µm for s
Table 1.
Distribution of neuronal loss and pTDP-43- and annexin A11-immunoreactive NCIs and GCIs
| Neuronal loss | pTDP-43 | Annexin A11 | |||
|---|---|---|---|---|---|
| NCIs | GCIs | NCIs | GCIs | ||
| Cerebrum | |||||
| Frontal cortex | − | − | − | − | − |
| Motor cortex | ++ | ++ | ++ | + | − |
| White matter* | +** | NA | ++ | NA | − |
| Temporal cortex | − | − | − | − | − |
| Parietal cortex | − | + | − | − | − |
| Occipital cortex | − | − | − | − | − |
| Subcortical area | |||||
| Ammon/dentate gyrus | − | − | − | − | − |
| Amygdaloid nucleus | − | − | − | − | − |
| Caudate nucleus/putamen | −/− | +/+ | −/+ | −/+ | −/+ |
| Globus pallidus | − | − | − | − | − |
| Thalamus | − | − | − | − | − |
| Subthalamic nucleus | − | + | + | − | − |
| Brainstem | |||||
| Oculomotor nucleus | − | − | + | − | − |
| Substantia nigra | − | − | − | − | − |
| Locus ceruleus | − | + | − | − | − |
| Trigeminal motor nucleus | + | + | − | + | − |
| Facial nucleus | ++ | ++ | + | ++ | − |
| Pontine nucleus | − | + | + | + | − |
| Ambiguous nucleus | + | + | − | − | − |
| Hypoglossal nucleus | ++ | ++ | ++ | ++ | − |
| Inferior olivary nucleus | − | + | + | + | − |
| Reticular formation | − | + | + | + | − |
| Cerebellum | |||||
| Cortex/white matter | −/−** | −/NA | −/+ | −/NA | −/− |
| Dentate nucleus | − | − | − | − | − |
| Spinal cord | |||||
| Anterior horn (C/Th/L) | ++/++/+ | +/+/++ | ++/++/++ | +/+/++ | −/−/+ |
| Lateral funiculus (C/Th/L) | ++/++/+** | NA | −/+/+ | NA | −/−/− |
| Anterior funiculus (C/Th/L) | ++/++/−** | NA | +/+/+ | NA | −/−/− |
*, White matter adjacent to the motor cortex; Neuronal loss (**, degeneration with macrophage infiltration): −, none; +, mild to moderate; ++, severe; NCIs and GCIs: −, none; +, rare (1–4 /10 high-power fields); ++, occasional (≧ 5/10 high-power fields); pTDP-43, phosphorylated 43 kDa TAR DNA-binding protein; NCIs, neuronal cytoplasmic inclusions; GCIs, glial cytoplasmic inclusions; NA, not applicable; C, cervical cord; Th, thoracic cord; L, lumbar cord
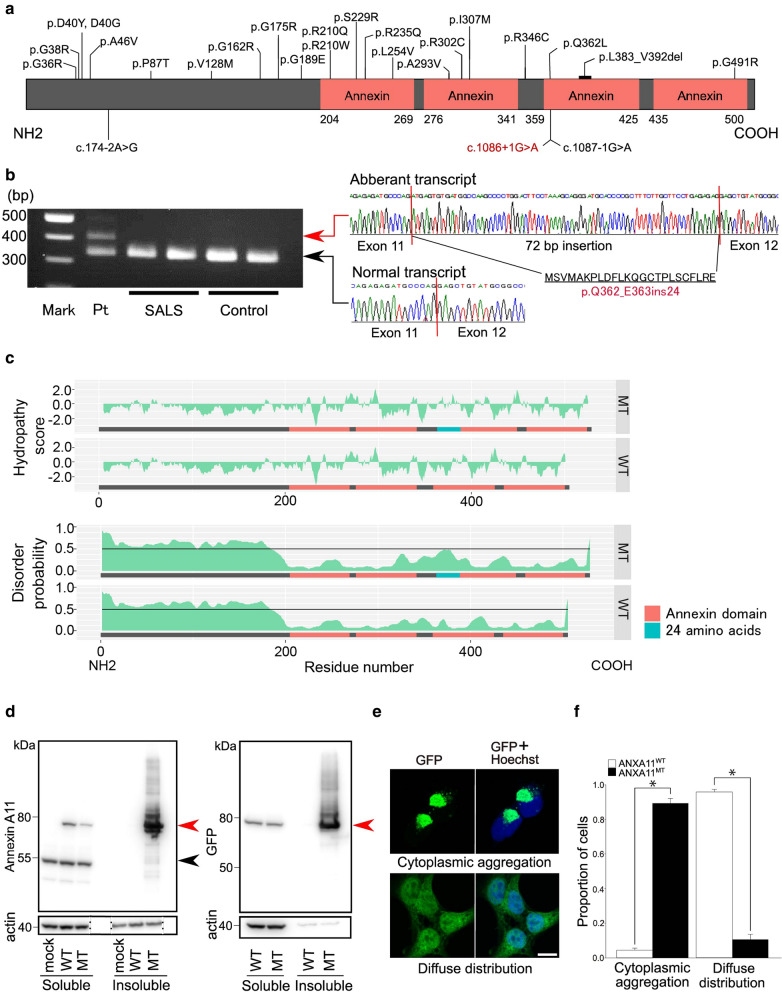
Genetic screening of ALS-related mutations through whole-exome sequencing revealed a heterogeneous splice site mutation, c. 1086+1G>A, in the ANXA11 gene (Fig. 2a), which has an allele frequency of 0.08% in the Human Genetic Variation Database (HGVD), and has not been documented in the Exome Aggregation Consortium (ExAC). On the other hand, no variants which are less than 0.1% frequency were found in any of the other ALS-related genes (See Supplementary methods). A splice site prediction program (NetGene2, http://www.cbs.dtu.dk/services/NetGene2/) correctly recognized the splice donor site of intron 11 in the wild type, but not in the mutant sequence, indicating that the mutation disrupts the splice donor site. Sequencing of the reverse transcription PCR products of ANXA11 mRNA obtained from the autopsied brain showed an aberrant transcript containing a 72-bp insertion between exons 11 and 12 that resulted in the insertion of 24 amino acids (Fig. 2b). Using the structure predictive tool, PSIPRED 4.0 (http://bioinf.cs.ucl.ac.uk/psipred/), the inserted amino acid sequence was predicted to create new alpha helixes. However, no new motifs were identified in the aberrant protein using a motif finder (Pfam 34.0, http://pfam.xfam.org/ and HMMER v3.3.2, http://hmmer.org/). Hydrophobicity prediction [2] using ExPASy (https://web.expasy.org/protscale/) and protein disorder analysis using PrDOS (http://prdos.hgc.jp/cgi-bin/top.cgi) [1] revealed that the aberrant annexin A11 had increased hydrophobicity (Fig. 2c, upper panel) and disorder probability (Fig. 2c, lower panel), respectively, around the inserted 24-amino-acid site compared with wild type. We then undertook cellular experiments using HEK293T cells transfected with the 72-bp inserted mutant, and wild type GFP-tagged ANXA11 (GFP-ANXA11MT and GFP-ANXA11WT, respectively) constructs. Solubility fractionation of the cells transfected with GFP-ANXA11 constructs, followed by Western blotting using antibodies for annexin A11 (Fig. 2d, left) and GFP (Fig. 2d, right) revealed that GFP-annexin A11MT formed detergent-resistant insoluble species. Furthermore, cells expressing GFP-annexin A11WT showed mainly a diffuse nuclear and cytoplasmic distribution of GFP, whereas cells expressing GFP-annexin A11MT showed cytoplasmic aggregation significantly frequently (Fig. 2e, f).
Fig. 2.
Position of the identified ANXA11 mutation and biological characteristics of the mutant annexin A11. a Annexin A11 protein domains and the position of mutations identified in patients with amyotrophic lateral sclerosis (ALS). Mutations found in the previous studies are in black and that of the present patient is in red. b An aberrant ANXA11 mRNA splicing as a consequence of the c.1086 + 1G > A mutation. Gel electrophoresis shows the reverse transcription PCR products from the autopsied brain of the present patient (Pt), patients with sporadic ALS harboring no ANXA11 mutations (SALS), and controls. The lane of Pt contains two different bands. Sequencing results show that the band located at 339 bp implies a normal transcript, and that located at 411 bp implies an aberrant transcript caused by 72-bp insertion between exons 11 and 12, numbered on the basis of ENSEMBL ANXA11-203. The inserted sequence results in a 24-amino-acid insertion. c Hydrophobicity and protein disorder predictions. Upper panel: Hydropathy plot [2] of the aberrant annexin A11 (MT) obtained using ExPASy shows increased hydrophobicity around the inserted 24-amino-acid site. Lower panel: Disorder profile plot of the aberrant annexin A11 (MT) obtained with PrDOS [1] shows increased disorder probability around the inserted 24-amino-acid site compared with that of the wild type (WT), indicating that the aberrant splicing could affect the stability and function of annexin A11. d Solubility fractionation of the cells transfected with GFP-tagged ANXA11 (GFP-ANXA11) constructs, followed by Western blotting using antibodies for (left) annexin A11 and (right) GFP. The wild type annexin A11 protein is predominantly soluble, whereas the mutant construct shows a prominent increase in the insoluble fraction. Red arrowhead: wild type (WT) and mutant (MT) GFP-annexin A11. Black arrowhead: endogenous annexin A11. β-Actin: loading control. e GFP-annexin A11 localization in HEK293T cells. Cells are classified according to GFP-annexin A11 localization patterns, i.e. cytoplasmic aggregation, and diffuse distribution. Bar = 10 µm for all images in e. f Proportion of cells showing each GFP-annexin A11 localization pattern. The proportion of cells showing cytoplasmic aggregation is significantly higher, and that of cells showing diffuse distribution is significantly lower in cells expressing GFP-annexin A11MT than in those expressing GFP-annexin A11WT (*, p < 0.002; Mann–Whitney U test)
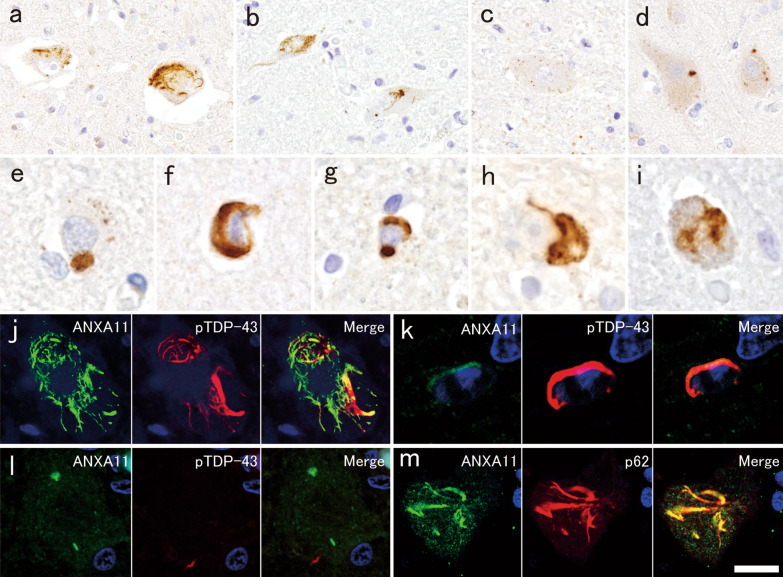
Indeed, the present patient had many annexin A11-ir skein-like NCIs in the lower motor neurons (Fig. 3a, b). In contrast, sparse dot-like cytoplasmic staining (Fig. 3c) and rare filamentous inclusions were observed in the lower motor neurons of ALS patients with no ANXA11 mutation. Such sparse dot-like cytoplasmic staining was also observed in controls, and not only in motor neurons (Additional file 1: Fig. 1). These findings indicated that annexin A11-ir NCIs are not completely specific, but characteristic to ANXA11-mutated patients. In the motor cortex, only a few, irregularly shaped or round, small NCIs were observed (Fig. 3d, e). Filamentous NCIs were also evident in several regions where pTDP-43-ir NCIs were also present (Fig. 3f, h, i, Table 1). Annexin A11 aggregated preferentially in neurons, although a few GCIs were observed (Fig. 3g). Double-labeling immunofluorescence for pTDP-43 and annexin A11 revealed that annexin A11 and pTDP-43 were partially co-localized in skein-like NCIs, and a few GCIs in the anterior horns (Fig. 3j, k), but not in small annexin A11-ir NCIs in the motor cortex (Fig. 3l). Annexin A11-ir skein-like NCIs in the anterior horns were frequently well labeled for p62 (Fig. 3m). Details of methods are in Supplementary methods, and the primary antibodies used are listed in Supplementary table 1 in Additional file 1.
Fig. 3.
Expression of annexin A11 in autopsied CNS tissue. a–i Annexin A11-immunohistochemistry. a Annexin A11-immunoreactive (ir) skein-like neuronal cytoplasmic inclusions (NCIs) in the anterior horn and b hypoglossal nucleus. c Small dot-like staining pattern in an anterior horn cell from a patient with amyotrophic lateral sclerosis without ANXA11 mutation. d Small irregular-shaped and e round NCIs in the motor cortex. f Filamentous NCI and g glial cytoplasmic inclusion (GCI) in the putamen. h Filamentous NCI in the pontine nucleus and i inferior olivary nucleus. j–l Double-label immunofluorescence with annexin A11 and pTDP-43. j Annexin A11 and pTDP-43 are partially co-localized in the skein-like NCI. k Weak immunoreactivity of annexin A11 in pTDP-43-ir GCI. j, k, lumbar anterior horn. l Annexin A11-ir and pTDP-43-negative small, irregularly shaped and short linear NCIs, and pTDP-43-ir and annexin A11-negative dystrophic neurite in the motor cortex. m Annexin A11-ir and p62-ir NCI in the lumbar anterior horn. j–l green, annexin A11; red, pTDP-43, m green, annexin A11; red, p62. Bar = 60 µm for a, b; 30 µm for c, d; 10 µm for e–i, l; 15 µm for j, m; 6 µm for k
The neuropathologic features of this patient were indistinguishable from those of ALS without known mutations, except for the presence of annexin A11-ir NCIs, being similar to the previously reported patient with ANXA11 mutation (Table 2) [6].
On the other hand, genetic screening revealed the novel splicing mutation in the C-terminal of ANXA11, and the in silico analysis and cellular experimental findings indicated that the aberrantly spliced transcript induced cytoplasmic accumulation and enhanced the aggregation propensity of annexin A11, suggesting that the mutation had pathogenicity. Functional studies of ALS-related ANXA11 mutations have shown that both N-terminal and C-terminal mutations induced abnormal phase separation to form aggregates, leading to the functional defects of annexin A11 [3, 5]. Similar toxic-gain-of-function mechanisms might have contributed to ALS pathogenesis in this patient.
Morphologically, the annexin A11-ir NCIs in this patient differed slightly from those in the previous patients with N-terminal ANXA11 mutation [6, 7]. We observed skein-like, filamentous, and small round inclusions but not the conglomerated, round, or basket-like large inclusions that had been present in the previous patients. Such differences might depend on the specific location of each mutation, since the disorder probability is lower in the annexin domain than in the N-terminal low complexity domain (Fig. 2c), which is responsible for protein aggregation [5]. Furthermore, the co-localization of annexin A11 and p62 in NCIs was clear in this patient, but not in the previous patient with N-terminal ANXA11 mutation [7], implying that clear and frequent superposition of annexin A11 and p62 might be specific for the patients with C-terminal ANXA11 mutation.
Interestingly, annexin A11 was aggregated predominantly in neurons and only very sparsely in glial cells, and topographically, in the brainstem motor nuclei and spinal anterior horns rather than the motor cortex, whereas TDP-43 was aggregated in both neurons and glial cells, and frequently in both the upper and lower motor systems. Thus, TDP-43 aggregated even in annexin A11-negative cells, especially glial cells. Similarly, TDP-43 aggregation in annexin A11-negative cells had also been noted in the two previously reported patients with N-terminal ANXA11 mutation [6, 7]. Together with these previous studies, our present findings indicate that annexin A11 aggregation propensity is probably dependent on cell type, and that annexin A11 aggregation is not indispensable for triggering TDP-43 aggregation and neurodegeneration.
Overall, we have confirmed the pathogenicity of this novel mutation in the C-terminal region of ANXA11. Neuropathologic findings suggested a cell type-dependent annexin A11 aggregation propensity. Further studies are needed to confirm this possibility.
Supplementary Information
Additional file 1. Details of methods and case presentation, a list of the primary antibodies used (Supplementary Table 1), and the representative images of annexin A11 immunohistochemistry in CNS tissue of controls (Supplementary Figure 1).
Acknowledgements
Not applicable.
Abbreviations
- ALS
Amyotrophic lateral sclerosis
- pTDP-43
Phosphorylated 43 kDa TAR-DNA-binding protein
Authors' contributions
MT, TI, OO and AK designed research project. MS, MT, HM, YT and AK performed pathological analysis. YH, TI, SA, TK, AY and OO designed the molecular experiments and performed those. JT, GI, TO and OO collected clinical data. KA managed statistical analyses. MS, YH, MT, TI, OO and AK discussed the results and drafted the manuscript for intellectual content.
Funding
Supported in part by JSPS grants-in-aid for Scientific Research to MT (19K07972), AK (19H01061, 19H05559), and TI (17K09750), Grants-in Aid from the Research Committee of CNS Degenerative Diseases, Research on Policy Planning and Evaluation for Rare and Intractable Diseases, Health, Labour and Welfare Sciences Research Grants, the Ministry of Health, Labour and Welfare, Japan to AK and OO.
Availability of data and materials
The datasets used and analyzed during the current study available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of Niigata University (G2018-0033 and G2015-0781). Written informed consent for autopsy including the use of tissues for research purposes was obtained from the patients’ family.
Consent for publication
Family members have consented to publication.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Mari Tada and Tomohiko Ishihara contributed equally to this work.
Contributor Information
Makoto Sainouchi, Email: m.sainouchi@gmail.com.
Yuya Hatano, Email: hatanoyuya@gmail.com.
Mari Tada, Email: tadamari@bri.niigata-u.ac.jp.
Tomohiko Ishihara, Email: ishihara@bri.niigata-u.ac.jp.
Shoichiro Ando, Email: shoando3@gmail.com.
Taisuke Kato, Email: taisuke8077@bri.niigata-u.ac.jp.
Jun Tokunaga, Email: tokunagajun@gmail.com.
Gaku Ito, Email: gakugakuito@gmail.com.
Hiroaki Miyahara, Email: miyahara.hiroaki.926@mail.aichi-med-u.ac.jp.
Yasuko Toyoshima, Email: toyoshima@agano.or.jp.
Akio Yokoseki, Email: yokoseki@bri.niigata-u.ac.jp.
Tetsutaro Ozawa, Email: ozawa@bri.niigata-u.ac.jp.
Kohei Akazawa, Email: akazawa@med.niigata-u.ac.jp.
Osamu Onodera, Email: onodera@bri.niigata-u.ac.jp.
Akiyoshi Kakita, Email: kakita@bri.niigata-u.ac.jp.
References
- 1.Ishida T, Kinoshita K. PrDOS: prediction of disordered protein regions from amino acid sequence. Nucl Acids Res. 2007;35:W460–464. doi: 10.1093/nar/gkm363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 3.Liao Y-C, Fernandopulle MS, Wang G, Choi H, Hao L, Drerup CM, et al. RNA granules hitchhike on lysosomes for long-distance transport, using Annexin A11 as a molecular tether. Cell. 2019;179:147–164.e20. doi: 10.1016/j.cell.2019.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mackenzie IR, Neumann M, Baborie A, Sampathu DM, Du Plessis D, Jaros E, et al. A harmonized classification system for FTLD-TDP pathology. Acta Neuropathol. 2011;122:111–113. doi: 10.1007/s00401-011-0845-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nahm M, Lim SM, Kim YE, Park J, Noh MY, Lee S, et al. ANXA11 mutations in ALS cause dysregulation of calcium homeostasis and stress granule dynamics. Sci Transl Med. 2020;12:eaax3993. doi: 10.1126/scitranslmed.aax3993. [DOI] [PubMed] [Google Scholar]
- 6.Smith BN, Topp SD, Fallini C, Shibata H, Chen H-J, Troakes C, et al. Mutations in the vesicular trafficking protein annexin A11 are associated with amyotrophic lateral sclerosis. Sci Transl Med. 2017;9:eaad9157. doi: 10.1126/scitranslmed.aad9157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Teyssou E, Muratet F, Amador MD, Ferrien M, Lautrette G, Machat S, et al. Genetic screening of ANXA11 revealed novel mutations linked to amyotrophic lateral sclerosis. Neurobiol Aging. 2021;99:102.e11–102.e20. doi: 10.1016/j.neurobiolaging.2020.10.015. [DOI] [PubMed] [Google Scholar]
- 8.Wang J, Guo C, Liu S, Qi H, Yin Y, Liang R, et al. Annexin A11 in disease. Clin Chim Acta. 2014;431:164–168. doi: 10.1016/j.cca.2014.01.031. [DOI] [PubMed] [Google Scholar]
- 9.Zhang K, Liu Q, Liu K, Shen D, Tai H, Shu S, et al. ANXA11 mutations prevail in Chinese ALS patients with and without cognitive dementia. Neurol Genet. 2018;4:e237. doi: 10.1212/NXG.0000000000000237. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Details of methods and case presentation, a list of the primary antibodies used (Supplementary Table 1), and the representative images of annexin A11 immunohistochemistry in CNS tissue of controls (Supplementary Figure 1).
Data Availability Statement
The datasets used and analyzed during the current study available from the corresponding author on reasonable request.