Abstract
The development of engineered T cells to treat acute myeloid leukemia (AML) is challenging due to difficulty in target selection and the need for robust T-cell expansion and persistence. We designed a T cell stimulated to kill AML cells based on recognition of the AML-associated surface marker CLEC12A, via secretion of a CLEC12AxCD3 bispedfic "engager" molecule (CLEC12A-ENG). CLEC12A-ENG T cells are spedfically activated by CLEC12A, are not toxic to hematopoietic progenitor cells, and exhibit antigen-dependent AML killing. Next we coupled stimulation of T-cell survival to triggering of a chimeric IL7 receptor with an ectodomain that binds a second AML-associated surface antigen, CD123. The resulting T cells, identified as CLEC12A-ENG.CD123IL7Rα T cells, demonstrate improved activation upon dual target recognition, kill AML, and exhibit antitumor activity in xenograft models. Enhanced T-cell activation conferred by CD123.IL7Ra was dependent both on recognition of the CD123 target and on IL7Ra-mediated downstream signaling. Expression of a chimeric IL7R targeted to a second tumor-assodated antigen (TAA) should improve T-cell activity not only against hematologic malignancies, but perhaps against all cancers.
Introduction
Acute myeloid leukemia (AML) is a malignancy with poor prognosis in children and adults (1, 2). Intensive chemotherapy and myeloablative conditioning followed by stem cell transplant are therapeutic strategies that can improve survival (2). Increasing cumulative doses of cytotoxic agents, which may improve antileukemia efficacy, also result in long-term toxicity to normal tissues. In addition, elderly adults, who represent the highest number of new AML diagnoses annually, may be unfit for intensive chemotherapy protocols and the associated side effects (3,4). The toxicities of targeted T-cell immunotherapy agents do not generally overlap with those of cytotoxic chemotherapy. Therefore, T-cell immunotherapy may be ideal to add to AML treatment regimens to enhance antitumor efficacy. One challenge to the development of targeted AML immunotherapy includes the lack of a single tumor-discriminating target. A second challenge shared by all T-cell-based cell therapies is the difficulty in maintaining T-cell persistence in vivo. Long-term survival of transferred T cells is necessary to ensure leukemia-free remissions and effective antitumor immunosurveillance. We have sought to solve both problems simultaneously by programming T-cell cytotoxicity to be dependent on recognition of an AML-associated antigen and by ensuring T-cell survival by binding of a second target.
The capability of targeted T cells to safely and specifically bind AML is restricted by the limited number of tumor-associated surface antigens acceptable for chimeric antigen receptor (CAR) design. The ideal antigen for anti-AML targeting has not yet been discovered and may not exist. Malignant and nonmalignant myeloid cells share expression of surface antigens (5). On-target, off-cancer toxicity is expected when targeting any of these. Even so, CD123 and CLEC12A have both been used in clinical trials as AML target antigens (6) due to expression on a large percentage of myeloid leukemias (7–9). We have designed T cells that receive separate activation signals from CD123 or CLEC12A. Our CLEC12A-ENG.CD123IL7Rα T cells are designed to acquire signal 1 [T-cell receptor (TCR) activation] from CLEC12Aand signal 3 (cytokine stimulation) from CD 123. Triggering of TCR activation is achieved via the use of a secreted bispecific "engager" molecule that binds to the CD3ε subunit of the TCR and the CLEC12A surface Drotein on AML cells.
Although in our design, a second conditional activation signal could be costimulation, or signal 2, we chose instead to couple signal 1 with signal 3. One hurdle preventing durable disease control with T-cell therapy is its association with transferred T-cell persistence. Without sufficient tumor-specific cells, the targeted malignancy is likely to recur (10,11). Human peripheral T cells are maintained by mechanisms that preserve a diverseT-cell pool. The cytokine IL7 is integral to T-cell homeostasis (12,13). IL7 is also a nonredundant cytokine necessary for maintenance of memory T cells and for support of their intermittent proliferative bursts (12). For these reasons, we chose to use a chimeric IL7R to convey signal 3 to our engineered cells.
In T cells modified to secrete a bispecific CLEC12AxCD3 binding molecule (CLEC12A-ENG), we demonstrate antigen-specific T-cell activation and AML cytotoxicity. To enhance CLEC12A-ENGT-cell survival, we introduced an additional modification: expression of an anti-CD123.IL7Rα chimeric receptor. T cells secreting CLEC12A-ENG and expressing CD123.IL7Rα display enhanced activation in response to target AML cells, increased proliferation in vivo, and improved antitumor responses.
Materials and Methods
Cells and culture conditions
The 293T, MV-4–11, OCI-AML-3, and K562 cell lines were purchased from the ATCC and cultured in DMEM (Thermo Fisher Scientific; 293T, MV-4–11), RPMI (Thermo Fisher Scientific; OCIAML-3), or Iscove’s Modified Dulbecco’s Medium (IMDM, Thermo Fisher Scientific; K562) supplemented with 10% FBS (Thermo Fisher Scientific). All cells were maintained in a humidified atmosphere containing 5% C02 at 37°C. K562 cells expressing CD 123 (K562.CD123), CLEC12A (K562.CLEC12A), or CD123 and CLEC12A (K562.CD123.CLEC12A) were generated by first subdoning the frill length human CD123 (NCBI Accession: NP_002174.1) or CLEC12A coding sequence (NCBI Accession: NP_612210.4) into a pCDH lentiviral backbone (System Biosdences, Inc.). VSVG-pseudotyped lentiviral partides were produced according to the manufacturer’s instructions. K562 cells were then transduced with the vims and antigen expression verified with flow cytometric analyses. Cells were isolated via FACS and expression verified prior to use. All cells used for cytotoxidty analysis were generated by transdudng the cell line with a retroviral vector encoding an enhanced GFP firefly ludferase fusion gene (GFP.ffLuc; ref. 14). GFP-positive cells were sorted and maintained in the appropriate culture medium. Ludferase expression was confirmed using D-ludferin and quantification of bioluminescence. The identity of all progeny of cell lines generated and used in this study were authenticated using ATCC (Manassas, VA) human STR profiling cell authentication service. MV-4–11 and MV-4-ll.ffLuc were authenticated in 2016. OCIAML-3 was not authenticated, though the progeny OCI-AMh3. ffLuc were, in 2016. K562 cells were authenticated in 2016 and K562.ffLucin 2017. Following authentication, cells were stored in a cell bank and freshly thawed for each set of experiments. Cdls were not kept in culture for longer than one month. Mycoplasma testing was routinely performed and was not found positive in any cell line used for generation of data in this manuscript at any time.
Construction of viral vectors
The CLEC12A-ENG construct was produced using GeneArt (Thermo Fisher Scientific) gene synthesis of a CLEC12A-specific scFv (21.16; ref. 15), which was then subcloned into a pSFG retroviral vector encoding a CD3ε-binding element and mOrange downstream of an IRES sequence (ref. 16,Fig. 1A). ACD19-ENG construct was shared by Dr. Stephen Gottschalk (St. Jude Children’s Research Hospital, Memphis, TN), also in a pSFG backbone (17). CD123.IL7Rα CAR constructs were generated using GeneArt (Thermo Fisher Scientific) gene synthesis of a CD123specific scFv (26292; ref. 18) linked to an IgGl hinge (GenBank: AAA58693.1) and the transmembrane and intracellular domain of IL7Rα (NCBI: NP_002176.2, amino adds 240–459). IL7Rα mutants were generated using the Q5 Site-directed Mutagenesis Kit (New England Biolabs). Primers for mutagenesis were obtained from Integrated DNA Technologies (Supplementary Fig. S1). All newly generated plasmids were verified by Sanger sequencing performed by the University of Michigan DNA Sequencing Core. RD114-pseudotyped retroviral particles were generated as described previously (16).
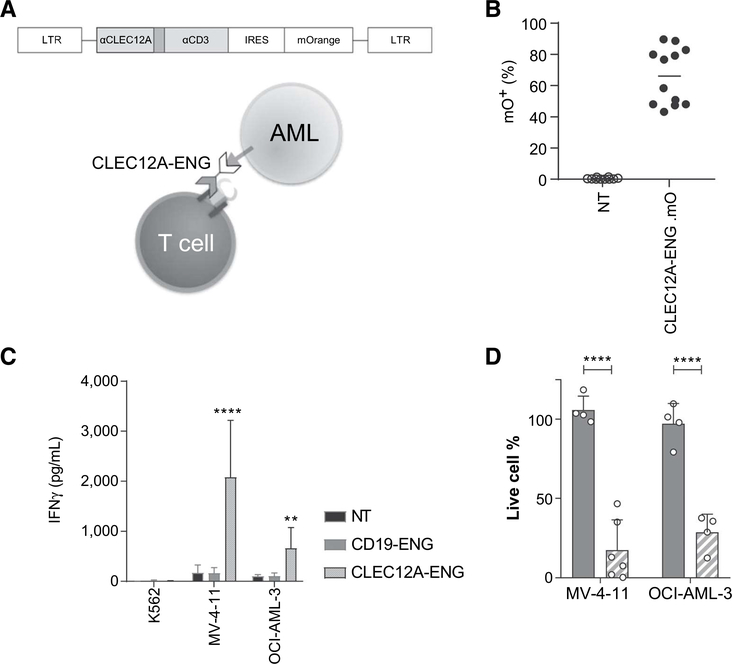
Figure 1.
CLEC12A-targeting with CLEC12A-ENG T cells has specific antileukemic activity in vitro. A, Schematic of construct used for targeting and method of T-cell activation. B, Transduction efficiency of CLEC12A-ENG T cells, (n = 10–12 independent T-cell donors). C, IFNγ secreted by CLEC12A-ENG T cells after overnight culture with CLEC12A−(K562) or CLEC12A+ (MV-4–11, OCI-AML-3) AML cell lines. Unmodified (NT) and CD19-specific T cells (CD19-ENG) included as controls. (n = 4–6 independent donors, difference noted between CLEC12A-ENG and NTT cells.**, P< 0.01; ****,P<0.0001). D, CLEC12A+ (MV-4–11, OCI-AML-3) target cells modified to stably express ffLuc were cultured for 3 days with NT, CD19-ENG (solid bar), or CLEC12A-ENG (striped bar) T cells. D-luciferin was added and live cell percentage determined by BLI. Cells normalized to those in NT T-cell condition. T-cell:target cell ratio was 1:1. Dots representative of individual T-cell donors, (n = 4–6; ****, P< 0.0001).
Generation of engineered T cells
Blood cells were purchased in the form of huffy coats (Carter BloodCare). Peripheral blood mononuclear cells (PBMC) were isolated using a Ficoll density gradient and were then used according to a University of Michigan Institutional Review Board (IRB)-approved protocol. Cells were activated by stimulation on OKT3 (Miltenyi Biotec) and anti-CD28 (for costimulation, Becton Dickinson) coated nontissue culture-treated 24-well plates. Recombinant human IL7 (10 ng/mL, predinical biorepository, NCI) andIL15 (5 ng/mL, National Cancer Institute) were added to cultures on day 2, and the following day, cells were transduced with retroviral particles immobilized on RetroNectin (Clontech Laboratories). In experiments using ludferase-expressing T cells, cells were transduced on subsequent days, first with the T-celltargeting moiety and second with the retrovirus carrying an enhanced GFP.firefly Ludferase fusion gene (14). T cells were maintained and expanded in the presence of IL7 and IL15. Cdls were analyzed for expression of mOrange or artifidal receptor by using flow cytometric analysis 5 to 7 days posttransduction.
Flow cytometry
Fluorochrome-conjugated isotype controls, anti-CD 123, antiCLEC12A, anti-His, and anti-CD3 were purchased from BD Biosciences. Recombinant human CD123-His was produced by Sino Biological. Analysis was performed on at least 20,000 cells per sample using an Attune NxT instrument (Thermo Fisher Scientific) and analyzed with Flojo software. Antigen quantification was performed using QuantiBrite beads (BD Biosciences) per manufacturer protocol. Each quantification was performed in triplicate.
Cytokine secretion assay
T cells were plated with target cells at a 1:1 ratio. Following 24 hours of culture, the supernatant was harvested and analyzed for the presence of IFNγ or IL2 using ELISA kits (R&D Systems) according to the manufacturer’s instruction. Percent relative change was calculated with formula (Y-X)/X*100.
Cytotoxicity assay
Target cells containing stable expression of firefly luciferase were incubated with T cells at 1:1, 1:2, 1:5, and 1:10 effector-totarget (E:T) ratios. Targets in media alone were used for assessment of spontaneous death. After 18 hours of culture, bioluminescence was measured. Mean percentage of specific lysis of triplicate samples was calculated as 100* (spontaneous deathexperimental death)/(spontaneous death-background). Cytotoxicity assays were incubated for 3 days: on day 0, cells were plated at a 1:1 E:T ratio. On day 3, cells were treated with ludferin and bioluminescence was measured and compared with an identical condition containing unmodified T cells.
Xenograft mouse model
All animal experiments were conducted in accordance with and with the approval of the University of Michigan Institutional Animal Care and Use Committee (LACUC). Briefly, NSG (NOD.Cg-Prkdcscid/Il2igtmlWjl/SzJ, Jackson Laboratory) mice were engrafted with AML cell lines (MV-4-11, MV-4-11 ffLuc, OCI-AML-3.ffLuc) via intravenous tail vein injection. One week following leukemia injection, mice were given 106 unmodified or modified T cells, as described. Mice were followed weekly with bioluminescent imaging (BLI). Animals were imaged using the IVIS system (IVIS, Xenogen Corp.) 10 minutes alter intraperitoneal injection of D-Luciferin (Thermo Fisher Scientific). Images were analyzed using Living Image 4.5.1 software (PerkinElmer). Peripheral blood was drawn via the facial vein and analyzed via FACS. Mice were euthanized when they either lost >20% of body weight, exhibited hind leg paralysis, or were otherwise deemed of clinical concern in collaboration with the veterinary team of the University of Michigan Unit for Laboratory Animal Medicine.
Colony-forming unit assays
Healthy donor bone marrow was obtained under a University of Michigan IRB-approved protocol. Bone marrow mononuclear cells (BMMC) were isolated by standard density gradient centrifugation and cyropreserved. BMMCs were incubated with media alone, unmodified, or modified T cells as described at a 5:1 E:T ratio for 6 hours, then plated in MethoCult (Classic) media (Stemcell Technologies) per manufacturer’s instruction. After 10 to 14 days, colonies were manually counted.
Western blotting
T cells were plated overnight in RPMI + 10% FBS and glutamax without additional cytokine. T cells were lysed in RIPA lysis buffer with protease (cOmplete) and phosphatase (PhosSTOP) inhibitor cocktails (MilliporeSigma) on ice. Electrophoresis was conducted using Novex WedgeWell 10% Bis-Tris Mini Gels (Thermo Fisher Scientific) and protein transferred to polyvinylidene difluoride (PVDF) membrane. Western blot analysis was performed with the following antibodies: rabbit anti-human p-STAT5 (done D47E7, Cell Signaling Technology), STAT5 (done D2605, Cell Signaling Technology), and GAPDH (done 6C5, Invitrogen).
Statistical analysis
GraphPad Prism 7 software (GraphPad Software) was used for statistical analysis. Measurement data were presented using descriptive statistics as noted in text, including, mean, median, range, ± SE. For comparison between two groups, two-tailed t test was used with correction for multiple comparisons using the Holm-Sidak method. For comparisons including more than two groups, two-way ANOVA corrected for comparison using the method of Sidek was incorporated. Survival of mice was estimated by the Kaplan-Meier method and differences in survival between groups were calculated by the log-rank (Mantel-Cox) and GehanBreslow-Wilcoxon test.
Results
CLEC12A-ENG T cells recognize and kill CLEC12A+ AML targets
We designed and synthesized a γ-retroviral construct containing the coding sequence of a CLEC12AxCD3 bispedfic engager molecule (CLEC12A-ENG) and that of the fluorescent protein mOrange (Fig. 1A). Next, we used retroviral transduction of primary activated T cells to induce constitutive production and secretion of CLEC12A-ENG. To verify T-cell modification, cells were analyzed via FACS for expression of mOrange. Reprodudble transduction effidendes were achieved (Fig. 1B, median: 67.5%, range 43.2%-89.6%, n = 12 independent T-cell donors). CLEC12A expression on a panel of AML cell lines (MV-4-11, OCI-AML-3, Molm-13) was measured with FACS analysis. CLEC12A expression was variable between the tested lines, and overall was found to be less than 5,000 molecules/cell (Supplementary Fig. S2). Despite this relatively low surface expression, CLEC12A-ENG T cells were specifically activated by CLEC12A+ target cells (MV-4-11, OCI-AML-3) as measured by IFNγ secretion. T cells engineered to secrete a bispedfic engager molecule targeted to the antigen CD19 (CD19-ENG) were not activated by target cells. Similarly, CLEC12A-ENG T cells were not activated by the CLEC12A− K562 cell line (Fig. 1C). CLEC12A-ENG T cells were also found to be spedfically cytotoxic to CLEC12A+ AML cells (Fig. 1D).
CLEC12A-ENG T cells demonstrate specific in vivo antitumor activity
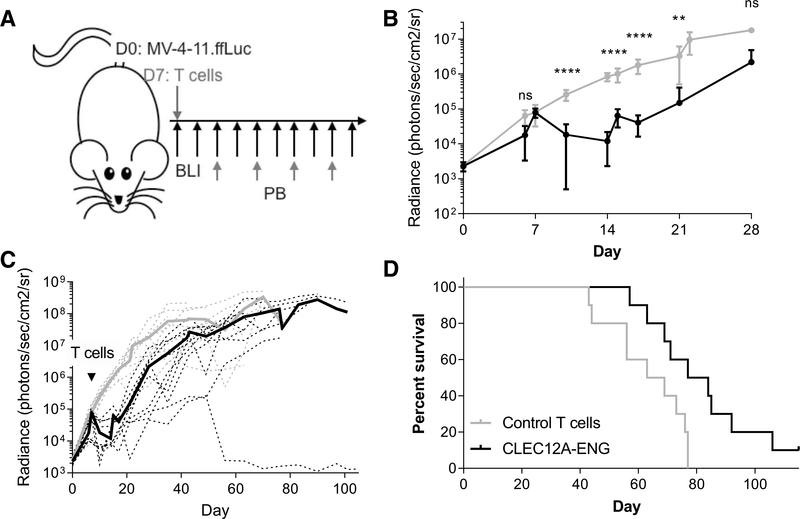
To evaluate in vivo antitumor activity of CLEC12A-ENG T cells, we established a human xenograft mouse model in NOD.CgPrkdcscidIL2rgtm1wil/SzJ (NSG) mice. The human CLEC12A+AML cell line, MV-4–11, was first modified to stably express firefly ludferase (ffLuc). These cells were then used to engraft NSG mice via tail vein injection. One week following leukemia injection, mice received control T cells or CLEC12A-ENG T cells (Fig. 2A). Leukemic progression was monitored by bioluminescence imaging following intraperitoneal injection of D-Luciferin. CLEC12A-ENG T cells demonstrated antitumor efficacy that led to improved median survival in the treated mice (Fig. 2B–D). Murine peripheral blood was monitored every 2 weeks starting on day 21 or 35 post-AML injection. Analysis of our initial cohorts of mice treated with CLEC12A-ENG T cells suggests correlation of antitumor responsiveness to T-cell expansion in peripheral blood. The single mouse that remained disease-free forthe course of the experiment (mouse #2) demonstrated enhanced T-cell expansion and persistence, primarily between days 49 and 63 (Supplementary Fig. S3).
Figure 2.
CLEC12A-ENG T cells exhibit antitumor efficacy in vivo. A, Schematic of MV-4–11 Xenograft model. NSG mice were engrafted with 1e6 MV-4–11.ffLuc i.v. on day 0. On day 7,10e6 T cells (control or CLEC12A-ENG) were administered. Leukemia was followed with BLI and T cells tracked in peripheral blood every 2 weeks beginning on day 21. B and C, Quantitation of sequential BLI of mice (n = 5 mice per group per experiment, two independent experiments, with a different T-cell donor for each experiment). B, Leukemia burden over first 28 days between control (gray line) and CLEG2A-ENG-treated (black line) mice. ****, P< 0.001; **, P< 0.01. C, Extended BLI data from B. Dashed lines represent each individual mouse, solid lines are average BLI of group. Gray lines, control mice; black lines, treated mice.D, Kaplan-Meier survival analysis, median survival: control T cells, 66 days; CLEC12A-ENG T cells, 80.5 days (P = 0.01).
Cytotoxicity unaffected by coexpression of CD123.IL7Rα and CLEC12A-ENG
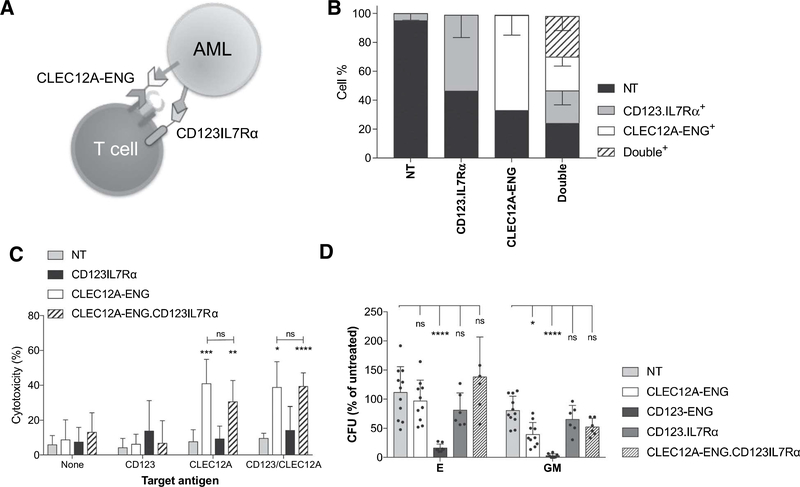
Although CLEC12A-ENG T cells had anti-AML activity, leukemia eventually progressed. We, therefore, asked whether the provision of signal 3 in the form of a chimeric IL7Rα could improve the anti-AML activity of CLEC12A-ENG T cells. We designed a chimeric receptor composed of an extracellular domain consisting of a scFv specific for the AML-associated antigen CD123 (16, 18) and the transmembrane and intracellular domains of IL7Ra (CD123.IL7Rtx; Fig. 3A). Cells were either single transduced with retroviral vectors encoding CLEC12A-ENG or CD123.IL7Rα, or double transduced with both vectors simultaneously. Single transduction resulted in a median of 60% ENG+ and 54.5% CD123. IL7Ra+ cells (range 56%-81% and 36.1%-66.6%, respectively). Double transduction resulted in a median of 22.7% ENG+ (range: 17.5%-29.9%), 24.9% CD123.IL7Rα+ (range: 11.9%-30.6%), and 25.9% double-positive cells (range: 19.8%-38.7%; Fig. 3B). We further modified the CLEC12A-ENG construct (Fig. 1A), replacing the mOrange sequence with our CD123.IL7Ra to allowforthe expression of both synthetic proteins after a single transduction. To assess the inhibition of antigen-dependent cytotoxicity in CLEC12A-ENG.CD123IL7Rα T cells, we cocultured effector cells with targets expressing either CD123 or CLEC12A alone, or both antigens together (Supplementary Fig. S4). Coexpression of CD123.IL7Rα with the CLEC12A-ENG did not diminish the CLEC12A-spedfic cytotoxicity. In a short-term cytotoxicity assay, both CLEC12A-ENG and CLEC12A-ENG.CD123IL7Rα T cells were equally effective at killing target cells expressing CLEC12A (Fig. 3C). We did not observe additional cytotoxicity secondary to CD123 binding by CD123.IL7Rα. Expression of CD123.IL7Rα did not cause toxicity to hematopoietic progenitor cells at a high (5:1) E:T ratio (Fig. 3D). T cells engineered to secrete a bispecific CD123xCD3 molecule (CD123-ENG) were used as a positive control for progenitor cell toxicity due to CD 123 binding.
Figure 3.
Coupling of CD123 recognition to IL7R signaling does not cause hematopoietic toxicity. A, Schematic of CLEC12A-ENG.CD123IL7Rα T-cell targeting of CLEC12A+CD123+AML B, Transduction efficiency of NT, singly modified CD123.IL7Rα, CLEC12A-ENG, and dual modified CLEC12A-ENG.CD123IL7Rα T cells. (n = 4 independent T-cell donors). C, Cytotoxicity of T cells versus CLEC12A+, CD123+, or CLECI2A+CD123+ target cells. E:T ratio, 1:5. Significant differences measured in comparison to NTT cells or as indicated, (n = 6 T-cell donors, *, P< 0.05; **, P< 0.01; ***, P< 0.001; ****, P< 0.0001). D, Colony-forming unit (CFU) assays performed with 5:1 ratio of T cells to BMMCs. CD123-ENG T cells used as positive control, (n = 5 BMMC donors, 4 T-cell donors. Data normalized to "no T cell" conditions, differences relative to NT T-cell control (*, P < 0.05; ****, P< 0.0001).
IL7Rα signaling and CD123 binding enhance CD123.IL7Rα T-cell activation
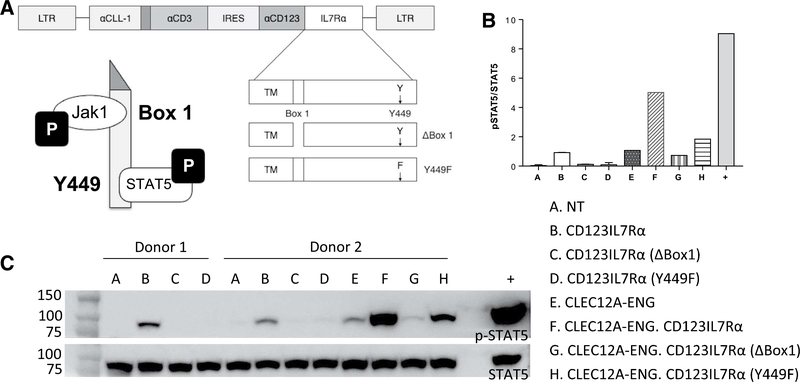
Having established that CD123.IL7Rα expression is feasible, we next evaluated for improved effector function. To test for intact IL7Rα signaling, we created two additional chimeric receptors. In one, Box 1 is deleted to prevent Jakl binding (CD123.IL7Rα.ABox1). In a second molecule, tyrosine 449 is mutated to phenylalanine (Y449F), which prevents Y449 phosphorylation and subsequent STAT5 binding (CD123.IL7Ra. Y449F; Fig. 4A; refs. 19–22). We evaluated STAT5 phosphorylation in T cells expressing the three CD123.IL7Rα with Western blotting. Independent of binding to CD 123, we found constitutive STAT5 phosphorylation in T cells expressing unmutated chimeric IL7Rα (Fig. 4B and C). Deletion of Box1 or mutation of Y449 to phenylalanine (Y449F) rendered p-STAT5 undetectable. In cells expressing CLEC12A-ENG, CD 123. IL7Rα expression increased p-STAT5 levels. IL7Rα mutation reduced STAT5 phosphorylation (Fig. 4B and C). Therefore, CD123.IL7Rα expression induces downstream STAT5 phosphorylation dependent on known motifs in the corresponding endogenous IL7R
Figure 4.
Expression of CD123.IL7Ra stimulates enhanced T-cell activation. A, Schematic of mutant IL7Rα receptors. B, STAT5 activation demonstrated downstream of IL7Rα receptor in engineered T cells. Cells were starved of cytokine, lysed, and Western blot analysis performed. Densitometry analysis calculated as p-STAT5/ STATS, (n = 1–2 donors of each T-cell type). C, Representative Western blot analysis for phospho-STAT5 and STAT5. Positive control (+): NT T cells treated with 10 ng/mL rhlL7 for 30 minutes prior to lysis.
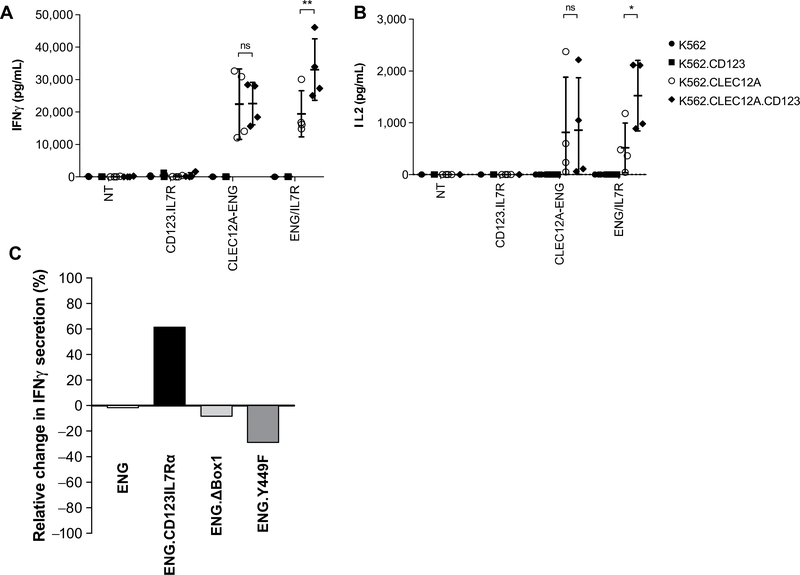
To assess functional activation, we cultured effector cells witn targets expressing either CD123 or CLEC12A alone, or both antigens together (Supplementary Fig. S4). CLEC12A-ENG.CD123IL7Rα T cells were stimulated to secrete more cytokine when challenged with targets expressing both CLEC12A and CD123 (Fig. 5A and B). In contrast, the presence of the CD123 target antigen did not stimulate additional IFNγ or secretion by CLEC12A-ENG cells. Use of our CD123.IL7Rα mutants diminished this augmented T-cell activation state (Fig. 5C). In sum, T cells expressing CLEC12A-ENG and CD123.IL7Rα secrete more inflammatory cytokine when target cells express both CLEC12A and CD123. CLEC12A-ENG T secretes the same amount of cytokine regardless of target cell CD 123 expression. Mutation of certain CD123.IL7Rα elements prevented optimal T-cell activation following dual-target recognition.
Figure 5.
Enhanced CLEC12A-ENG.CD123IL7Rα T-cell activation is secondary to CD123+ target recognition and intact IL7Rα elements. IFNγ (A) and IL2 (B) secreted into supernatant following the culture of indicated effector and target cells, (n = 4 T-cell donors, *, P< 0.05; **, P< 0.01). C, Relative change in IFNγ secretion in T cells cultured with K562.CLEC12A versus K562.CLEC12A.CD123 (n = 1 T-cell donor).
CLEC12A-ENG.CD123IL7Rα T cells have improved in vivo expansion and efficacy
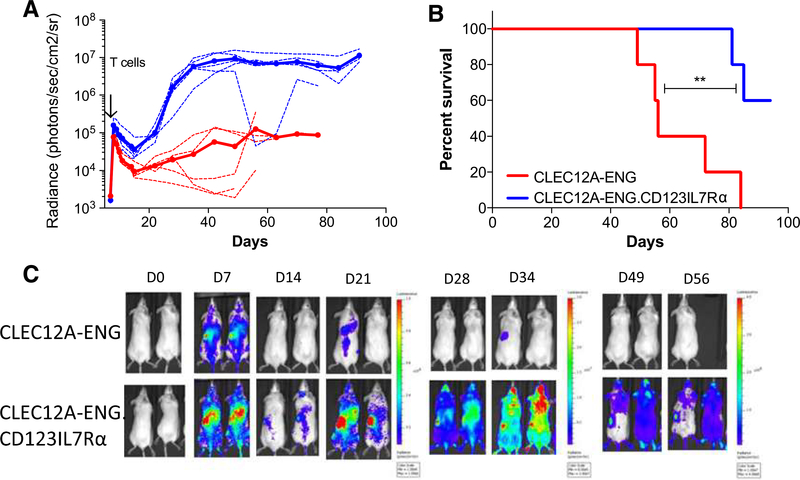
To evaluate the capacity for the CD123.IL7Rα to improve CLEC12A-ENG T-cell expansion and antitumor activity in vivo, we used our CLEC12A+/CD123+ MV-4–11 xenograft model. To facilitate T-cell tracking, we engineered ffLuc expression in CLEC12A-ENG and CLEC12A-ENG.CD123IL7Rα T cells (Supplementary Fig. S5). Both T-cell expansion and overall survival were enhanced in mice treated with CLEC12A-ENG.CD123IL7Rα T cells as compared with animals treated with CLEC12A-ENG T cells (median overall survival: not reached vs. 56 days, P < 0.01; Fig. 6A–C). Improved survival in CLEC12A-ENG.CD123IL7Rαtreated mice was also seen in a second AML xenograft using OCI-AML-3.ffLuc leukemia cells. Our MV-4–11.ffLuc AML model suggests a trend, but did not meet significance when evaluating the improved survival of the treatment group (Supplementary Fig. S6). Given the different overall survival seen in mice engrafted with MV-4–11.ffLuc as compared with those engrafted with unmodified MV-4–11, we tested the lines for phenotypic differences. We noted decreased expression of CLEC12A in our MV-411 cell line (Supplementary Fig. S7).
Figure 6.
CD123.IL7Ra T cells have improved expansion and in vivo antileukemia control. A, T cells were transduced with ffLuc to enable tracking of expansion and persistence versus unlabeled MV-4–11. (n = 5 mice each group, one T-cell donor used for cohort; solid line, mean; dotted lines, individual mice). B, Kaplan-Meier survival analysis, median survival: CLEC12A-ENG, 56 days; CLEC12A-ENG.CD123IL7Ra, undefined. (**, P<0.01). C, Representative images of mice.
Discussion
We have described the generation and antigen-specific activation of engineered cells designed to respond to the AML-associated target antigens, CLEC12A and CD123. In our CLEC12A-ENG.CD123IL7Ra T cells, binding of CLEC12A stimulated the T-cell activation and specific cytotoxicity. Expression of a CD 123. IL7Ra coupled IL7Ra downstream activation and improved antitumor activity in a CD 123-dependent fashion. CLEC12A-ENG. CD123IL7RaT cells demonstrated improved expansion potential and antitumor activity in a xenoeraft model of human AML.
Engineered T-cell therapy for AML has not been as successful as that for lymphoblastic leukemia. This is partly due to difficulty in taiget selection because of shared antigen expression on leukemic blasts and normal myeloid cells (5). We chose CD123 and CLEC12A as our AMDassociated antigen targets based on the validated expression on a majority of AML (7, 8) and the likely enrichment on leukemia-initiating cells (9, 23). CLEC12A is expressed on peripheral blood and bone marrow myeloid cells and a minority of committed progenitor cells (8, 24). CAR-T cells targeting CLEC12A have demonstrated anti-AML activity in preclinical studies (25–27). CD123 is the a chain of the IL3 receptor and is expressed on hematopoietic stem and progenitor cells as well as endothelial cells (7, 23, 28, 29). CD123 has been or is being tested in early phase CAR-T cell trials (NCT02623582, NCT02159495, NCT03203369, and NCT03190278) and has thus far demonstrated an acceptable safety profile. Even so, the targeting of CD 123 with engineered T cells is toxic to nonmalignant hematopoietic progenitor cells (16, 30, 31). To overcome this hurdle, we coupled the T-cell "signal 3” to CD 123 binding. Cytotoxicity was not trigged by CD 123, but is instead secondary to CLEC12A recognition. Thus, our approach mitigates "on-target, off-cancer" toxicity to CD123+ normal cells.
A second limitation to achieving durable antitumor responses following T-cell adoptive transfer is the correlation of efficacy with the persistence of transferred cells (10, 11). By coupling CD123 recognition to downstream IL7Ra activation, we wanted to promote T-cell survival. In unmodified T cells, IL7 signaling is critical to homeostatic preservation of the peripheral T-cell pool (12, 13). Bispedfic T-eell engager molecules trigger TCRs. TCR activation can strengthen IL7 downstream activation, whereas IL7 activation can strengthen TCR responiveness (32). The mechanism of this cooperative effect is not fully described, but may involve TCR-mediated formation of lipid rafts and dustering of IL7 receptors in the membrane (33). Our experimental results support synergistic activation. We have shown low-density CLEC12A expression on AML cell lines. CAR-T activation state correlates with target antigen density (34–37). In addition to CLEC12A, other tumor-assodated antigens (TAA) also demonstrate low-density expression. We believe our dual ENG/IL7Rα strategy has potential to target other low-density TAA In addition, CLEC12A-ENG.CD123IL7Rα T cells allow for biological investigation of TCR/IL7Rα synergistic activation.
Our choice of the IL7 receptor to provide signal 3 follows the work of others who have used chimeric IL7 receptors to boost T-cell adoptive transfer (38,39). Our work differs in that we have coupled the intentional TAA recognition to IL7Rα activation. We hypothesized that in CD123.IL7Rα T cells, downstream IL7Rα activation would follow CD 123 binding. We were surprised to find tonic, taiget-independent, constitutive STAT5 phosphorylation in modified cells. Mutation of known IL7R elements prevented STAT5 activation, confirming that this was secondary to the activation state of our engineered receptor. Adoptive transfer of cells with a constitutively active survival signal is not safe. Further analysis of the longevity of these cells without cytokine or antigen support is needed. Addition of a safety switch may be necessary to guard against cellular immortality prior to clinical translation. Although we describe a constitutively present taigetindependent activation state, we also discovered the amplification of inflammatory cytokine secretion reliant on CD 123 binding.
As an alternate to cytokine stimulation, or signal 3, we could have chosen to engineer costimulation, or signal 2, downstream of CD 123 binding. Expression of CARs in combination with costimulatory receptors has been reported to stimulate optimal engineered T-cell activity (40–42). We have sought to eliminate the potential for CAR costimulatory receptor inhibitory interactions by pairing single CAR expression with bispedfic molecule secretion. Previous study has shown combinatorial TCR activation via bispedfic antibody secretion paired with engineered costimulation to be superior to TCR activation alone (43). As an alternate approach, we chose to explore whether a stimulatory signal transduced by the IL7R would improve persistence and antitumor activity of AML-specific CLEC12A-ENG T cells. We found enhanced activation, in vivo expansion, and leukemia control when using CLEC12A-ENG.CD 123IL7Rα T cells in our functional studies. Whether these would be superior to CLEC12A-ENG T cells with engineered costimulation is an outstanding question.
Our in vivo T-cell tracking supports our hypothesis that CD123IL7Rα expression improves T-cell antitumor activity. In this study, using MV-4–11, we found CLEC12A-ENG. CD123IL7Rα T cells to be more effective than CLEC12A-ENG T cells in AML control. In contrast we found no survival advantage in CLEC12A-ENG.CD123IL7Rα-treated animals engrafted with MV-4–11.ffLuc, though there was a trend toward improved survival. Treatment of mice in a second xenograft model of human AML confirmed the superiority of CLEC12A-ENG.CD123IL7Rα in antitumor responsiveness. We would like to highlight the difference in overall survival in untreated mice engrafted with MV-4–11 and MV-4–11.ffLuc. Despite the confirmed genetic origin of these cell lines, we found variation in CLEC12A expression. This does not explain the lost potency of our MV-4–11.ffLuc, but does indicate potential selection of a sub-done in our modification and FACS-based sorting of the MV-4–11.ffLuc. Further comparative evaluation of our MV-4–11 and MV-4–11.ffLuc is necessary to understand differential responsiveness in these models. Nonetheless, we suggest that provision of an IL7Rα survival signal in engineered cells stimulates enhanced activation and improves antitumor activity of transduced cells.
A limitation or our analysis or in vivo T-cell expansion is the possibility of xenogeneic GVHD stimulating an antigen-independent T-cell response. As we performed no selection prior to the injection of our cells, mice received a mixed cell population consisting of unmodified, GFP.ffLuc+ only, CLEC12A-ENG+ (or CLEC12A−ENG.CD123IL7Rα+) only, and double-positive cells. GFP.ffLuc+-only cells have the potential for xenogeneic activation, independent of CLEC12A-ENG or CD123.IL7Rα expression. The observed delayed T-cell proliferation may signify xenogeneic TCR stimulation supported by antigen-independent CD 123. IL7Rα downstream signaling. Alternatively, the expansion may also correlate to the circulation time of the T cells prior to extravasation and proliferation to a detectable concentration at sites of bulky disease. Tracking of engineered T-cell behavior is ongoing in our laboratory. We hope to further understand the kinetics of T-cell/tumor cell localization and the specificity of T-cell expansion. We are additionally focused on enhancing the specificity and duration of antitumor responses.
The work reported herein represents a potential solution to two challenges that stand in the way of further development of engineered T-cell therapy for AML: target selection and limited T-cell survival. We have shown that coupling of TCR and IL7Ra activation improves anti-AML activity in vitro and in vivo. The principles of our combinatorial dual-antigen recognition design can be applied to other tumor types with low antigen density. One can envisage the local stimulation of T-cell survival and the potentiation of tumor-specific attack, driven by antigen in the tumor microenvironment. These findings contribute to the understanding of T-cell immunobiology and highlight the potential of T-cell immunotherapies for clinical use.
Supplementary Material
Acknowledgments
We would like to thank Stephen Gottschalk, Shannon Carty, and Pavan Reddy for helpful discussions, review, and advice. This work was supported by the Damon Runyon Cancer Research Foundation, The Gorman Family Foundation, the Jake Wetchler Foundation, and the Ravitz Family Foundation (to C.L. Bonifant). K. Huang was supported as part of the Alex’s Lemonade Stand Foundation Pediatric Oncology Student Training Program. Research reported in this publication was supported by the NCI of the NIH under Award Number P30CA046592 by the use of the Rogel Cancer Center Preclinical Imaging & Computational Analysis Shared Resource.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Disclosure of Potential Conflicts of Interest
C.L. Bonifant has ownership interests in pending patents describing engineered T-cell technology as anticancer treatment. No potential conflicts of interest were disclosed by the other authors.
Note: Supplementary data for this article are available at Cancer Immunology Research Online (http://cancerimmunolres.aacrjournals.org/).
Disclaimer
Publisher's Disclaimer: The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
References
- 1.Zwaan CM, Kolb EA, Reinhardt D, Abrahamsson J, Adachi S, Aplenc R, et al. Collaborative efforts driving progress in pediatric acute myeloid leukemia. J Clin Oncol 2015;33:2949–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O’Donnell MR, Tallman MS, Abboud CN, Altman JK, Appelbaum FR, Aiber DA, et al. Acute myeloid leukemia, version 3.2017, NCCN dinical practice guidelines in oncology. J Natl Compr Cane Netw 2017;15:926–57. [DOI] [PubMed] [Google Scholar]
- 3.Appelbaum FR, Gundacker H, Head DR, Slovak ML, Willman CL, Godwin JE, et al. Age and acute myeloid leukemia. Blood 2006;107:3481–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.NCI. Cancer stat facts: acute myeloid leukemia (AML). Available form: https://seer.cancer.gov/statfacts/html/amyl.html2011-2015.
- 5.Pema F, Berman SH, Soni RK, Mansilla-Soto J, Eyquem J, Hamieh M, et al. Integrating proteomics and transcriptomics for systematic combinatorial chimeric antigen receptor therapy of AML. Cancer Cell 2017;32:506–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonifant CL, Velasquez MP, Gottschalk S. Advances in immunotherapy for pediatric acute myeloid leukemia. Expert Opin Biol Ther 2018;18:51–63. [DOI] [PubMed] [Google Scholar]
- 7.Munoz L, Nomdedeu JF, Lopez O, Camicer MJ, Bellido M, Aventin A, et al. Interleukin-3 receptor alpha chain (CD 123) is widely expressed in hematologic malignancies. Haematologica 2001;86:1261–9. [PubMed] [Google Scholar]
- 8.Bakker AB, van den Oudenrijn S, Bakker AQ, Feller N, van Meijer M, Bia JA, et al. C-type lectin-like molecule-1: a novel myeloid cell surface marker associated with acute myeloid leukemia. Cancer Res 2004;64:8443–50. [DOI] [PubMed] [Google Scholar]
- 9.van Rhenen A, van Dongen GA, Kelder A, Rombouts EJ, Feller N, Moshaver B, et al. The novel AML stem cell associated antigen CLD1 aids in discrimination between normal and leukemic stem cells. Blood 2007; 110:2659–66. [DOI] [PubMed] [Google Scholar]
- 10.Maude SL, Frey N, Shaw PA, Aplenc R, Barrett DM, Bunin NJ, et al. Chimeric antigen receptor T cells for sustained remissions in leukemia. N Engl J Med 2014;371:1507–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Porter DL, Hwang WT, Frey NV, Lacey SF, Shaw PA, Loren AW, et al. Chimeric antigen receptor T cells persist and induce sustained remissions in relapsed refractory chronic lymphocytic leukemia. Sd Transl Med 2015;7: 303ra139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Surh CD, Sprent J. Homeostasis of naive and memory T cells. Immunity 2008;29:848–62. [DOI] [PubMed] [Google Scholar]
- 13.Takada K, Jameson SC. Naive T cell homeostasis: from awareness of space to a sense of place. Nat Rev Immunol 2009;9:823–32. [DOI] [PubMed] [Google Scholar]
- 14.Vera J, Savoldo B, Vigouroux S, Biagi E, Pule M, Rossig C, et al. T lymphocytes redirected against the kappa light chain of human immunoglobulin efficiently kill mature B lymphocyte-derived malignant cells. Blood 2006;108:3890–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao X, Singh S, Pardoux C, Zhao J, Hsi ED, Abo A, et al. Targeting C-type lectin-like molecule-1 for antibody-mediated immunotherapy in acute myeloid leukemia. Haematologica 2010;95:71–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bonifant CL, Szoor A, Torres D, Joseph N, Velasquez MP, Iwahori K, et al. CD 123-engager T cells as a novel immunotherapeutic for acute myeloid leukemia. Mol Ther 2016;24:1615–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Velasquez MP, Torres D, Iwahori K, Kakarla S, Arber C, Rodriguez-Cruz T, et al. T cells expressing CD19-spedfic engager molecules for the immunotherapy of CD19-positive malignancies. Sd Rep 2016;6:27130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Du X, Ho M, Pastan I. New immunotoxins targeting CD 123, a stem cell antigen on acute myeloid leukemia cells. J Immunother 2007;30:607–13. [DOI] [PubMed] [Google Scholar]
- 19.Venkitaraman AR, Cowling RJ. Interleukin-7 induces the assodation of phosphatidylinositol 3-kinase with the alpha chain of the interleukin-7 receptor. Eur J Immunol 1994;24:2168–74. [DOI] [PubMed] [Google Scholar]
- 20.Lin JX, Migone TS, Tsang M, Friedmann M, Weatherbee JA, Zhou L, et al. The role of shared receptor motifs and common Stat proteins in the generation of cytokine pleiotropy and redundancy by IL-2, IL-4, IL-7, IL-13, and IL-15. Immunity 1995;2:331–9. [DOI] [PubMed] [Google Scholar]
- 21.Jiang Q, Ii WQ, Hofmeister RR, Young HA, Hodge DR, Keller JR, et al. Distinct regions of the interleukin-7 receptor regulate different Bcl2 family members. Mol Cell Biol 2004;24:6501–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rosenthal LA, Winestock KD, Finbloom DS. IL-2 and IL-7 induce heterodimerization of STAT5 isoforms in human peripheral blood T lymphoblasts. Cell Immunol 1997;181:172–81. [DOI] [PubMed] [Google Scholar]
- 23.Taussig DC, Pearce DJ, Simpson C, Rohatiner AZ, Lister TA, Kelly G, et al. Hematopoietic stem cells express multiple myeloid markers: implications for the origin and targeted therapy of acute myeloid leukemia. Blood 2005; 106:4086–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bill M, Bvan Kooten Niekerk P, SWoll P, Laine Herborg L, Stidsholt Roug A, Hokland P, et al. Mapping the CLEC12A expression on myeloid progenitors in normal bone marrow; implications for understanding CLEC12Arelated cancer stem cell biology. J Cell Mol Med 2018;22:2311–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tashiro H, Sauer T, Shum T, Parikh K, Mamonkin M, Omer B, et al. Treatment of acute myeloid leukemia with T cells expressing chimeric antigen receptors directed to C-type lectin-like molecule 1. Mol Ther 2017; 25:2202–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laborda E, Mazagova M, Shao S, Wang X, Quirino H, Woods AK, et al. Development of A chimeric antigen receptor targeting C-type lecdn-like molecule-1 for human acute myeloid leukemia. Int J Mol Sd 2017;18:pii: E2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang J, Chen S, Xiao W, Li W, Wang L, Yang S, et al. CAR-T cells targeting CLL-1 as an approach to treat acute myeloid leukemia. J Hematol Oncol 2018;11:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Korpelainen EI, Gamble JR, Vadas MA, Lopez AF. IL-3 receptor expression, regulation and function in cells of the vasculature. Immunol Cell Biol 1996;74:1–7. [DOI] [PubMed] [Google Scholar]
- 29.Korpelainen EI, Gamble JR, Smith WB, Dottore M, Vadas MA, Lopez AF. Interferon-gamma upregulates interleukin-3 (ID3) receptor expression in human endothelial cells and synergizes with lh-3 in stimulating major histocompatibility complex dass II expression and cytokine production. Blood 1995;86:176–82. [PubMed] [Google Scholar]
- 30.Pizzitola I, Anjos-Afonso F, Rouault-Pierre K, Lassailly F, Tettamanti S, Spinelli O, et al. Chimeric antigen receptors against CD33/CD123 antigens effidently target primary acute myeloid leukemia cells in vivo. Leukemia 2014;28:1596–605. [DOI] [PubMed] [Google Scholar]
- 31.Gill S, Tasian SK, Ruella M, Shestova O, Li Y, Porter DL, et al. Predinical targeting of human acute myeloid leukemia and mydoablation using chimeric antigen receptor-modified T cells. Blood 2014;123:2343–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deshpande P, Cavanagh MM, Le Saux S, Singh K, Weyand CM, Goronzy JJ. IL-7- and ID 15-mediated TCR sensitization enables T cell responses to selfantigens. J Immunol 2013;190:1416–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cho JH, Kim HO, Surh CD, Sprent J. T cell receptor-dependent regulation of lipid rafts controls naive CD8+ T cell homeostasis. Immunity 2010;32: 214–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alvarez-Vallina L, Russdl SJ. Effident discrimination between different densities of target antigen by tetracycline-regulatable T bodies. Hum Gene Ther 1999;10:559–63. [DOI] [PubMed] [Google Scholar]
- 35.Chmielewski M, Hombach A, Heuser C, Adams GP, Abken H. T cell activation by antibody-like immunoreceptors: increase in affinity of the single-chain fragment domain above threshold does not increase T cell activation against antigen-positive target cells but decreases selectivity. J Immunol 2004;173:7647–53. [DOI] [PubMed] [Google Scholar]
- 36.Turatti F, Figini M, Balladore E, Alberti P, Casalini P, Marks JD, et al. Redirected activity of human antitumor chimeric immune receptors is governed by antigen and receptor expression levels and affinity of interaction. J Immunother 2007;30:684–93. [DOI] [PubMed] [Google Scholar]
- 37.Fry TJ, Shah NN, Orentas RJ, Stetler-Stevenson M, Yuan CM, Ramakrishna S, et al. CD22-targeted CAR-T cells induce remission in B-ALLthat is naive or resistant to CD 19-targeted CAR immunotherapy. Nat Med 2018;24:20–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shum T, Omer B, Tashiro H, Kruse RL, Wagner DL, Parikh K, et al. Constitutive signaling from an engineered IL7 receptor promotes durable tumor elimination by tumor-redirected T cells. Cancer Discov 2017;7: 1238–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mohammed S, Sukumaran S, Bajgain P, Watanabe N, Heslop HE, Rooney CM, et al. Improving chimeric antigen receptor-modified T cell function by reversing the immunosuppressive tumor microenvironment of pancreatic cancer. Mol Ther 2017;25:249–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao Z, Condomines M, van der Stegen SJC, Pema F, Kloss CC, Gunset G, et al. Structural design of engineered costimulation determines tumor rejection kinetics and persistence of CAR T cells. Cancer Cell 2015;28: 415–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stephan MT, Ponomarev V, Brentj ens RJ, Chang Al I, Dobrenkov KV, Heller G, et al. T cell-encoded CD80 and 4–1BBL induce auto- and transcostimulation, resulting in potent tumor rejection. Nat Med 2007;13:1440–9. [DOI] [PubMed] [Google Scholar]
- 42.Kloss CC, Condomines M, Cartellieri M, Bachmann M, Sadelain M. Combinatorial antigen recognition with balanced signaling promotes selective tumor eradication by engineered T cells. Nat Biotechnol 2013; 31:71–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Velasquez MP, Szoor A, Vaidya A, Thakkar A, Nguyen P, Wu MF, et al. CD28 and 41BB costimulation enhances the effector function of CD 19-specific engager T cells. Cancer Immunol Res 2017;5:860–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.