Abstract
Radiomics is an emerging area within clinical radiology research. It seeks to take full advantage of all the information contained in multiple medical imaging modalities. With a radiomics approach, medical images are not limited to providing only a qualitative assessment but can also provide quantitative data by parameterizing image features. These parameters can be used to identify regions and volumes of interest and discriminate normal healthy tissue from abnormal or diseased tissue. Radiomics is an interlinked sequence of processes of vital importance that begins with the acquisition and selection of medical images that involve standardization of acquisition protocols and inter-equipment normalization. This is followed by the identification and segmentation of regions or volumes of interest by expert radiologists through the use of computational tools that offer speed while reducing variability and bias. The segmentation process is the most critical stage in radiomics. This sometimes requires the incorporation of a pre-processing stage consisting of advanced techniques (reconstruction processes, filtering, etc.). Thereafter, representative characteristics of the region or volume of interest are extracted by approaches based on statistics, morphological features, and transform-based variables. Next, a statistical selection of the parameters that provide a high association and correlation with the clinical condition of interest is performed. Finally, processes such as data integration, standardization, classification, and mining processes can be applied as needed for particular applications. Ongoing research in radiomics aims to reduce the time and costs involved in interpreting medical images while simultaneously increasing the quality of diagnoses and monitoring of as well as the selection of treatment strategies. The results of many studies combining radiomics with standard medical techniques are highly encouraging, and these new approaches are increasingly used. This review article details the components of radiomics and discusses its applications, challenges, and future directions for this exciting new field of study.
Keywords: Medical Imaging, Radiomics, Segmentation, Tumors, Big Data
Introduction
Etymologically, radiomics comes from the union of the terms radio- (radiation or radiology) and -omics, a common suffix used to form nouns relating to the study of the totality of a field (e.g. genomics, proteomics, metabolomics, transcriptomics). Thus, radiomics is the field that studies a large number of features computationally extracted from radiological images in order to discover hidden features of certain region(s). Radiomics is one of the newest areas in the field of clinical research in radiology; however, its use in other areas such as agriculture, mining, industrial inspection, security, and image retrieval, dates many years prior to its adoption in the field of medicine [1].
There has been an exponential increase in the use of radiomics concepts within publications in clinical research in recent years, mainly in areas of radiology where a large number of medical images distributed across different modalities are generated. The exponential increase in the number of publications on this topic per year, as listed in Science Direct (www.sciencedirect.com), PubMed (www.ncbi.nlm.nih.gov/pubmed/), Science (www.science.gov ), and Nature (www.nature.com), is shown in Figure 1.
Figure 1:
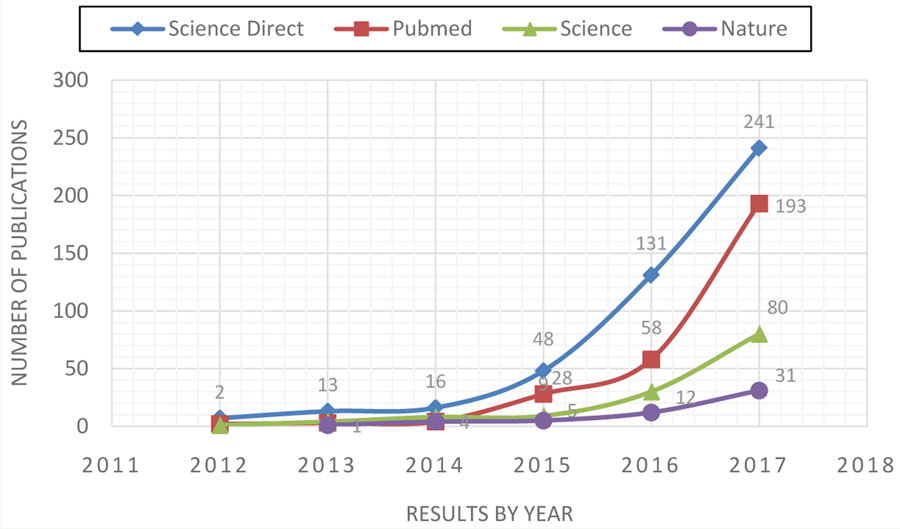
Number of publications, by year, containing the keyword radiomics in some of the most popular and important databases on life science and biomedical topics currently used in medical academic practice. Data were retrieved in February 2018.
One of the first studies in radiomics was reported in 2007 by Segal and coworkers, who performed a retrospective study investigating gene expression in liver cancer by analyzing images [2]. This pioneering publication had several limitations, among them the small number of subjects enrolled and the nascent techniques used to perform certain processes (e.g. its use of manual rather than automated segmentation).
Following that trailblazing publication, many other studies have been carried out based on the principles of radiomics as they apply to different clinical conditions, various patient populations, and using divergent non-invasive medical imaging modalities [3]. These applications were based on multiple medical modalities that include Magnetic Resonance Imaging (MRI) [4], Computed Tomography (CT) [5], Ultrasound (US) [6], Nuclear Medicine Imaging such as Single-Photon Emission Computed Tomography (SPECT) [7], and Positron Emission Tomography (PET) [8].
The use of hybrid imaging (e.g. PET/CT) is quite common in modern medical practice, as practitioners seek complementary functional and anatomical information to facilitate more accurate diagnosis and treatment for each patient [9,10]. These modalities each create images with differing tissue contrast based on the organ being assessed and that organ’s properties and structure. In addition, specific particularities such as spatial resolution, signal to noise ratio, contrast-to-noise ratio, and reconstruction method, among others, are determinants relating to the quality of medical images acquired [11,12].
Images acquired by medical imaging provide precious details about the clinical condition of the patient. However, relevant information is often either not completely visualized or fully understood and appreciated by medical practitioners. Radiomics serves to bridge the gap between image acquisition and interpretation [3], and its wide applicability has gained numerous followers. At this time, workers in the field are primarily researchers who are using radiomics approaches to uncover relevant information hidden in medical images [13], in essence to data mine the images.
Numerous studies support the usefulness of radiomics in identifying and quantifying regions and volumes of interest associated with a variety of organs and pathologies [e.g.14-16]. This review article aims to address the characteristics, applications, and challenges of the emerging field of radiomics. In addition, its relationship with other technical approaches focused on its application to different organs, as well as future directions for the field, will be addressed.
Components and Methods in Radiomics
The workflow of radiomics is a series of consecutive but interconnected steps, involving (a) the acquisition and selection of medical images; (b) identification and segmentation of a Region Of Interest (ROI) and/or Volume Of Interest (VOI); (c) extraction of descriptive texture features from the ROIs or VOIs; (d) statistical selection of the parameter(s) that provide the highest association with the clinical condition assessed; and (e) data integration, standardization, classification, and mining processes (Figure 2). While this is the general methodology, the different forms of implementation and approach to each step pose unique opportunities and challenges.
Figure 2:
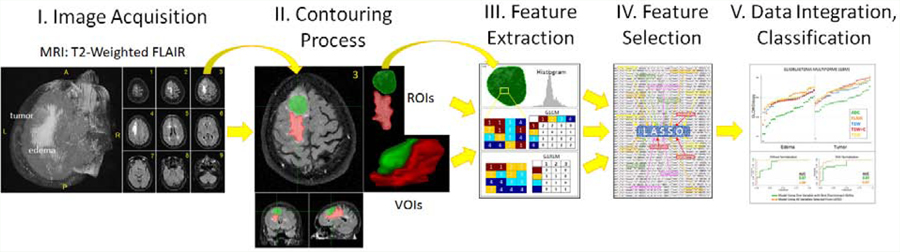
Schematic of the general radiomics workflow: I. acquisition and selection of medical images; II. Contouring of region of interest (ROI) and/or volume of interest (VOI); III. Feature extraction; IV. Reduction and selection process for parameters with the best discriminant ability; and V. data integration, standardization, classification, and mining.
Image acquisition
Contemporary medicine relies on a set of information acquired from multiple sources, including written medical records as well as images and video acquired from multiple devices. Conventional medical imaging methods are used daily in health centers, and the huge amounts of data they provide are one of the most important sources of clinical information. They offer comprehensive support during diagnosis, monitoring, and response-assessment in the routine management of many diseases. However, all medical imaging modalities have inherent advantages and disadvantages, are differentiated in their capacity to offer anatomical or physiological information, and have diverse acquisition protocols and reconstruction parameters, matrix size, noise and artifacts, etc.
Image acquisition is the first step in the radiomics process. Depending on the study scope and the properties of the organ or disease under investigation, a specific modality, multiparametric sequences within a modality, or combination of modalities, may be most appropriate [9,17,18].
For example, CT is the Standard-Of-Care (SoC) for radiation therapy planning. Through this modality, a precise visualization of the geometric positions of tumor and normal tissue in a patient is feasible [19]; however, CT has well-known limitations. CT offers poor soft-tissue contrast, which poses a real challenge when identifying a target-irradiated volume. Functional and multiparametric MRI, however, offers excellent soft-tissue contrast, a major advantage when locating a target volume; thus, MRI and CT are frequently used together for radiation therapy planning [20,21].
If necessary, complementary information from another type of medical modality can also be incorporated.
However, the data integration from different modalities, obtained with divergent medical devices and various acquisition protocols, is not simple. The lack of acquisition protocol standardization and inter-equipment normalization should be taken into consideration when choosing appropriate methods to analyze the data [22,23].
Contouring and segmentation
Before the application of the contouring and segmentation process, some clinical studies incorporate an additional element of pre-processing. Pre-processing consists of a set of state-of-the-art techniques that have the function of improving the data quality of images obtained in the acquisition stage, from which characteristic parameters will be extracted in later stages. Advanced reconstruction techniques [24,25] that are executed on raw data and specialized filters [26–28] are part of the pre-processing stage. It should be noted that the nature of the filters must be consistent with the type of characteristic noise present in each medical imaging modality.
The identification and contouring of one or multiple ROIs or VOIs, depending on the study, is a crucial stage that precedes the extraction of their representative characteristics [29]. Segmentation processes allow obtaining a bidimensional ROI if contouring is performed in a simple slice, or a VOI if whole slices are used [30,31]. In radiomics, this process has commonly been performed manually, relying on the expertise and abilities of a specialist; however, manual contouring is not optimal, because it has the potential to introduce variability and bias [32]. It remains a valid option when a study has a small number of patients and if the professional responsible for the process has significant experience, but when the number of patients is sizable, manual contouring is no longer feasible or cost effective, even when a highly qualified professional performs the process. Semi-automatic or automatic segmentation methods are more appropriate for large cohorts and have the advantage of reducing variability and bias [33].
Semi-automatic segmentation is an agile and reliable process that combines the advantages of automated and manual segmentation. In this form of computer-assisted segmentation, automatic contouring using an algorithm such as Thresholding [34], Snakes [35,36], Level Set [37], Fuzzy Connectedness [38–40], Clustering [41], or Region Growing [42], is followed by minor manual corrections applied by an experienced professional.
Each of these methods has certain advantages and disadvantages, and the choice between them is mainly motivated by the application. Several of these techniques have proven to be robust and efficient in the segmentation of multiple medical images (distinct and/or combined modalities), especially when the images have non-uniform intensities [40,43]. Table 1 offers brief comparisons of the most important advantages and disadvantages of some popular and useful semi-automatic segmentation algorithms.
Table 1:
Advantages and disadvantages of semi-automatic segmentation methods commonly used in medical applications.
| Method | Ref. | Advantages | Disadvantages |
|---|---|---|---|
| Thresholding | 34 | Simplest and faster segmentation approach. Useful to discriminate foreground from the background. | Accurate results are not obtained when there is no significant gray scale difference within the image. Sensitive to noise. |
| Snakes | 35, 36 | Simple to understand and easy to implement. Works well for images with good contrast between regions. | Less robust to noise than other methods. Not suitable for images whose limits are very smooth. |
| Level Set | 37 | Intrinsic, versatile and parameter-free. Works well on images with topological changes and curvature dependence. | Not works properly with complex topology images (poor topological adaptation). Requires heuristic splitting mechanisms and control point regridding mechanisms. |
| Fuzzy Connectedness | 38-40 | Easy to implement based on mathematical concepts. Fast, robust and works well in 3D segmentation. Requires only a seed to work. Nonlinear functions of arbitrary complexity can also be modeled. | Determination of fuzzy membership is not easy. Automatic calculation of the membership (dynamic weights) could cause excessive computational cost, time and/or memory. |
| Clustering | 41 | Eliminates noisy spots. Reduces false blobs. Allows definition of more homogeneous regions. | Computationally expensive. Senstive to normalization or standardization processes. Very sensitive to outliers. |
| Region Growing | 42 | Simple concept. Requires only a few seed points to work. Able to identify the connected regions with the same characteristics. Provides good limit information of the image as well. | Computational cost is considerable. Over-division and voids are caused when the image shows gray scale irregularity and excessive noise. |
Feature extraction
Automated extraction of a large number of quantitative features is possible through the application of different data characterization algorithms to medical images [44]. In general, this approach allows the quantification of morphological properties (shapes), locations, fractal dimensions and texture properties from specific ROIs or VOIs, which are then used for a variety of classification tasks, such as distinguishing normal from abnormal tissue (i.e. tumors).
The parameters used in radiomics applications are grouped into categories based on their statistical [45,46], morphological [47], and transform-based [48] features. Statistical features that describe the properties of an image within the spatial distribution of its gray levels through a variety of statistical parameters (Table 2). Normalization and standardization methods can be optionally used before the extraction of statistical features from images [49,50]. Several studies have described approaches to extract distinct first-order, second-order, and higher-order statistics from images; some methods are outlined in Figure 3, and the parameters involved in each can be seen in Table 2.
Table 2:
Statistical features: Set of first-order statistical descriptors using information directly from the histogram; set of second-order statistical descriptors using Gray Level Co-Occurrence Matrix (GLCM); set of higher-order statistical descriptors using Gray Level Run Length Matrix (GLRLM), Gray Level Size Zone Matrix (GLSZM), Gray-Level Distance Zone Matrix (GLDZM), Neighborhood Gray Tone Difference Matrix (NGTDM), and Neighboring Gray-Level Dependence Matrix (NGLDM).
| Statistic approach | Ref. | Matrix Name | Parameters |
|---|---|---|---|
| First-order | 51, 52 | None | mean, variance, standard deviation, skewness, kurtosis, energy, entropy, uniformity, coarseness, directionality, contrast, percentiles (1-%, 10-%, 50-%, 90-%, and 99-%), absolute gradient |
| Second-order | 53-55 | Gray-level co-occurrence (GLCM) | autocorrelation, contrast or inertia, correlation, cluster prominence, cluster shade, cluster tendency, dissimilarity, angular second moment (energy or uniformity), entropy, inverse difference moment or homogeneity, inverse variance, maximum probability, sum of squares or variance, sum average, sum variance, sum entropy, difference variance, difference entropy, information measures of correlation, inverse difference-normalized, inverse difference moment-normalized |
| Higher-order | 56-58 | Gray-level run length (GLRLM) | short run emphasis, long run emphasis, gray-level non-uniformity, run length non-uniformity, run percentage, low gray-level run emphasis, high gray-level run emphasis, short run low gray-level emphasis, short run high gray-level emphasis, long run low gray level emphasis, long run high gray level emphasis, gray-level variance, run-length variance |
| 59 | Gray-level size zone (GLSZM) | nonuniformity, intensity nonuniformity normalized, size zone nonuniformity normalized, zone percentage, low intensity emphasis, high intensity emphasis, low intensity small area emphasis, high intensity small area emphasis, low intensity large area emphasis, high intensity large area emphasis, intensity variance, size zone variance, zone entropy | |
| 60 | Gray-level distance zone (GLDZM) | distance zone nonuniformity, intensity nonuniformity normalized, distance zone nonuniformity normalized, zone percentage, low intensity emphasis, high intensity emphasis, low intensity small distance emphasis, high intensity small distance emphasis, low intensity large distance emphasis, high intensity large distance emphasis, intensity variance, distance zone variance, distance zone entropy | |
| 51 | Neighborhood gray tone distance (NGTDM) | coarseness, contrast, busyness, complexity, strength | |
| 61 | Neighboring gray level dependence (NGLDM) | nonuniformity, dependence nonuniformity, gray-level nonuniformity normalized, dependence nonuniformity normalized, low gray-level emphasis, high gray-level emphasis, low gray-level small dependence emphasis, high gray-level small dependence emphasis, low gray-level large dependence emphasis, high gray-level large dependence emphasis, gray-level variance, dependence variance, dependence entropy, second moment |
Figure 3:
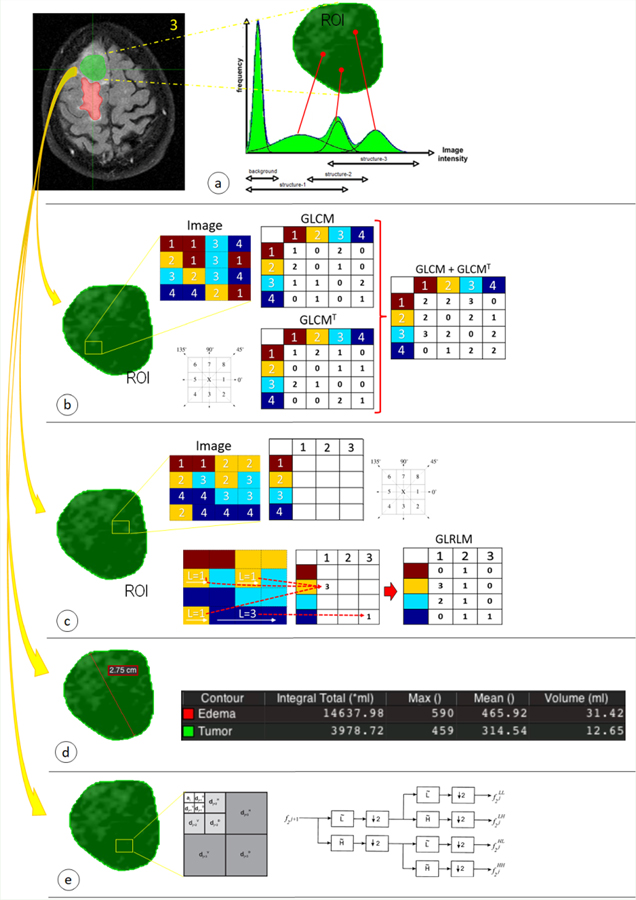
Radiomics features are distributed in different techniques focused primarily on statistical approaches: (a) first-order, (b) second-order and, (c) higher-order statistics. In addition, other methods focused on (d) morphological and (e) transform-based features, are used for extracting information from medical images.
First-order statistics are the simplest way of extracting statistical characteristics directly from the histogram of the medical image [51,52]. The main advantage of this approach is simplicity; it relies on the use of standard descriptors to characterize the data. However, first-order statistics limits robustness in discriminating unique textures in certain applications, as this method does not consider the spatial relationship, interaction, and correlation of neighboring pixel values.
In addition to the features analyzed by first-order statistics, second-order statistics calculates the particular relationship (probability) between two pixels that have similar gray levels [53–55]. The results of these operations are incorporated into a correspondence matrix, the Gray-Level Co-Occurrence Matrix (GLCM).This matrix is calculated for different distances and orientations.
Finally, higher-order statistics is based on a matrix such as the Gray-Level Run Length Matrix (GLRLM) that contains information about the number of runs with pixels of defined grey levels and run lengths in an image [56–58]. This matrix is calculated for different run orientations. An analogous approach is the Gray Level Size Zone Matrix (GLSZM) which quantifies regions of contiguous pixels in the image in a manner similar to GLRLM, except these regions are quantified along the abscissa as size zones rather than run lengths [59]. A size zone is generally defined as a collection of 9-connected pixels (2D) or 26-connected voxels (3D) of the same gray level. There are alternative methods to generate complementary matrices with their own parameters that belong to higher-order statistics: the Gray-Level Distance Zone Matrix (GLDZM) [60], Neighborhood Gray-Tone Difference Matrix (NGTDM) [51], and Neighboring Gray-Level Dependence Matrix (NGLDM) [61].
Morphological features are independent of the gray-level intensity distribution in an image [47,62,63]. Thus, morphological features are calculated directly from the segmented medical image. The main parameters of this group are the size and shape of the ROIs or VOIs. The morphological parameters are volume, surface area and surface area to volume ratio, sphericity, compactness, spherical disproportion, maximum 3D diameter, maximum 2D diameter (slice), maximum 2D diameter (column), maximum 2D diameter (row), major axis, minor axis, least axis, elongation, and flatness.
Transform-based features are calculated using approaches such as Fourier, Gabor and Wavelet transforms [48]. Since an image is constituted by intensity values in the spatial domain, in order to emphasize certain characteristics of the original image a transformation can be made to the frequency domain without losing any information. Fourier [64] and Gabor [65] transforms allow the simultaneous treatment of the spatial and frequency information of the image. Wavelet transforms allow a more detailed analysis of the image components, because it performs a filtering process of the noise contained in the images allowing a better appreciation of certain details [66].
Model-based features, such as fractal models, can be calculated using mathematical approaches [13]. Fractal analysis allows the quantification of self-symmetry through the assessment of repetitive patterns at different scales [48].
Feature reduction and selection
The reduction and selection of the representative parameters are the result of a statistical estimation process based on calculated radiomics parameters [67]. Binomial families, logit links, logistic and linear regression models are often used to reduce and select the radiomics parameters that provide the highest association for distinguishing selected ROIs or VOIs while shrinking irrelevant parameters [68]. Fisher scoring [69], Principal Components Analysis (PCA) [70], and Linear Discriminant Analysis (LDA) [71] are the standard operating procedures for variable selection from groups of variables in linear regression models. Another selection operator used for this purpose is the Least Absolute Shrinkage and Selection Operator (LASSO) with binomial families and logit links, which allows construction of a model specifically tailored to classify diverse ROIs or VOIs [72]. The LASSO procedure is typically performed in R using the glmnet package, while all other statistical procedures are executed using Stata software [73].
Each of these procedures for the reduction and selection of discriminant parameters has attendant advantages and disadvantages (Table 3). Some of these statistical tools are appropriate for certain situations where other methods may be inadequate, obtaining practical decisions, efficient control strategies and easy interpretation of the results. Many disadvantages of the procedures for reduction and selection of parameters can be overcome by using hybrid models (e.g. PCA+LASSO), making the reduction and selection processes more efficient and yielding more accurate results [74].
Table 3:
Advantages and disadvantages of procedures for the reduction and selection of representative features in medical studies with radiomics.
| Procedure | Ref. | Advantages | Disadvantages |
|---|---|---|---|
| Fisher scoring | 69 | Rapid and guaranteed convergence. Easy to interpret. Generates standard errors of all parameters estimates. | Computations become intensive in complex models. Difficult to determine the expected value of the Hessian matrix associated with the difficulty of identifying the appropriate sampling distribution. |
| Principal Component Analysis (PCA) | 70 | Unsupervised and simple technique. Nonparametric. Not computation-intensive. Does not require large amounts of data. | It is necessary to normalize the data before applying PCA to mitigate scale effects. Difficult to evaluate the covariance matrix accurately. |
| Linear Discriminant Analysis (LDA) | 71 | Easy and intuitive to use and understand. Maximizes the separation between classes while minimizing dispersion within the class. | Only models relationships between linear dependent and independent variables. Very sensitive to the anomalies in the data. |
| Least Absolute Shrinkage and Selection Operator (LASSO) | 72 | Reduces and selects variables simultaneously for better prediction and model interpretation. | Tends to select more covariates than expected, promoting a conflict between the correct selection and the optimal prediction. |
Data Integration, Classification, and the Data Mining Process
The data integration, classification, and mining process make use of the parameters that have been determined to best represent the object of study (e.g. tumor). These valuable data can be analyzed in a variety of ways and for different purposes. Figure 4 shows an example of this process.
Figure 4:
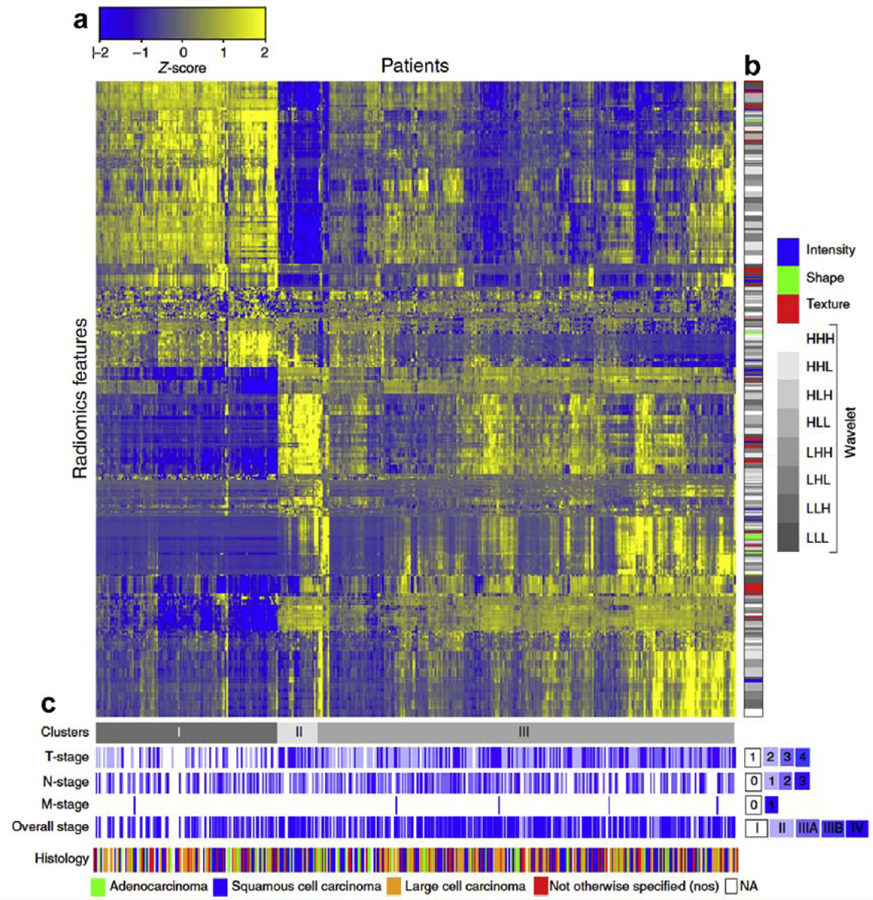
Heat map that includes the process of data integration and clustering generated in a study conducted by Alerts H. and coworkers, entitled “Decoding tumor phenotype by noninvasive imaging using a quantitative radiomics approach” [94]. It is possible to identify some elements in the image such as: (a) direct association between the patients enrolled and the radiomics feature expressions (indicating similarity through a scale of scores), (b) clinical parameters of the patients associated with the different radiomics feature expressions and, (c) classification of patients according to different stages of the assessed disease.
Recently, a group of computational techniques intended to perform decision-making and related tasks in a manner similar to human intelligence, have become important in many areas of research and development [75]. Medical imaging research has not been an exception, and advanced algorithms have been applied to automatically classify imaged tissue as healthy or abnormal [76].
Well-known techniques such as clustering [77], decision tree [78], deep learning [79], machine learning [80], and data mining [81], among others, are ideal for radiomics assessment. There is no one perfect combination of techniques, since the use of each of these approaches is related to the details of the application [82]. All these techniques are in constant and committed expansion, contributing with their advantages, strengths, and robustness not only in processes of detection and discrimination of particular tissues [78,80] but also in procedures of automatic segmentation [79], as well as other procedures that affect medical applications. The growth of these high-level automated approaches has allowed precision medicine to also be highly personalized medicine.
Discussion and Summary
New advances in scanners, better reconstruction methods, and filtering algorithms, among other advances, have improved the quality and resolution of medical images. Thus, functional and anatomical information from medical imaging modalities offer specialists clear advantages in making the correct clinical diagnosis in patients with complicated pathologies.
Although the human visual system is quite efficient, it has clear limitations. When a person executes an image assessment, the human eye can discriminate only a dozen different intensities [83]. Thus, there are certain limits and a level of information loss for even the most qualified professional. These limitations play a role against when a qualitative diagnosis is performed by directly observing the medical image, as well as when a manual contouring process is executed. It has been established that the contouring process is the most critical and relevant component for the success of a radiomics approach; contouring and segmentation form the basis for all analysis of radiomics parameters [29]. Some studies show that 3D radiomics features have higher performance than 2D [84,85]; this is due to the higher information density. The process of extracting radiomics features is not limited to the information contained in the ROI (a single slice) but includes information contained in the entire VOI (a set of slices) [86].
Radiomics features, computer-extracted textures, offer a novel approach to medical image analysis. They have played an important role in several medical applications, offering important support in the organization, diagnosis, and characterization of lesions and a variety of tumor types [13,82]. A number of studies focused on different diseases associated with important organs have been published in prestigious radiology and oncology journals. Several studies using radiomics approach have examined organs including the liver [2,87,88], brain [89–91], breast [92–94], heart [48], lung [95–98], kidney [99,100], rectum [101,102], and prostate [103–106].
Radiomics uses several well-known concepts and approaches including analysis of statistical, morphological, and transform-based features. Several hundred parameters can be extracted from each ROI or VOI in a few seconds using these methods. The application of advanced algorithms of identification, selection, and reduction of the representative radiomics features that provide the highest association for distinguishing selected ROIs or VOIs while shrinking irrelevant parameters is essential. Individual approaches can work well, and the use of combined or hybrid models is increasingly common.
The diversity of image types, acquisition procedures, and methods of reconstruction, among other protocols, are just a few examples of the substantial number of details with which each research group in radiomics must contend. This suggests standardization should be sought in order that the investigations may be oriented towards a common horizon. A proposed solution to this problem is the generation of common databases with free access, with different modalities of medical images and organs of interest [44]. Data validation and standardization will allow research laboratories to compare their proposals and results, enabling radiomics to realize its true potential as a support system for clinical diagnosis and treatment.
Conclusion and Future Outlook
Radiomics is a relatively new discipline, currently in the midst of simultaneous expansion and standardization. This process will require a series of new studies to give radiomics technical and clinical validation.
The strength, versatility, and applicability of radiomics as an approach to the extraction and representation of medical ROI or VOI through selected parameters has been amply demonstrated in the clinical and academic fields. Radiomics is a non-invasive diagnostic tool used to complement existing methods of evaluating the characteristics and behavior of the disease. In addition, this valuable tool can help improve the diagnosis, surveillance and/or prognosis of multiple diseases.
In the future, achieving further consensus from the research community is expected to plan and develop protocols to achieve standardization and characterization for all medical imaging modalities, scanner manufacture, image acquisition procedures, reconstruction, pre-processing, contouring, robust extraction of informative features, feature analysis and classification, and data mining.
Acknowledgments
The preparation of this review article was supported by the Department of Radiology at The University of Mississippi Medical Center.
Funding
The present studies were funded by a UMMC Intramural Grant (to CMH and PPC) and partially funded by NIH Grant CA138510–01 (to PPC). Candace M Howard is partially supported by the National Institute of General Medical Sciences of the National Institutes of Health under Award Number 5U54GM115428. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health All authors have read the journal’s policy on disclosure of potential conflict of interest and all the authors declare that they have no any financial or personal relationship with organizations that could potentially be perceived as influencing the described research.
References
- 1.Pietikäinen M, Ojala T. Texture Analysis in Industrial Applications. Image Technology 1996; 337–359. [Google Scholar]
- 2.Segal E, Sirlin CB, Ooi C, Adler AS, Gollub J, Chen X, et al. Decoding global gene expression programs in liver cancer by noninvasive imaging. Nat Biotechnol 2007; 25: 675–680. [DOI] [PubMed] [Google Scholar]
- 3.Lambin P, Leijenaar RTH, Deist TM, Peerlings J, de Jong EEC, van Timmeren J, et al. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol 2017; 14: 749–762. [DOI] [PubMed] [Google Scholar]
- 4.Wibmer A, Hricak H, Gondo T, Matsumoto K, Veeraraghavan H, Fehr D, et al. Haralick texture analysis of prostate MRI: utility for differentiating non-cancerous prostate from prostate cancer and differentiating prostate cancers with different Gleason scores. Eur Radiol 2015; 25: 2840–2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huynh E, Coroller TP, Narayan V, Agrawal V, Hou Y, Romano J, et al. CT based radiomic analysis of stereotactic body radiation therapy patients with lung cancer. Radiother Oncol 2016; 120: 258–266. [DOI] [PubMed] [Google Scholar]
- 6.Jeon JH, Choi JY, Lee S, Ro YM. Multiple ROI selection based focal liver lesion classification in ultrasound images. Expert Syst Appl 2013; 40:450–457. [Google Scholar]
- 7.Rajkumar V, Goh V, Siddique M, Robson M, Boxer G, et al. Texture analysis of (125)I-A5B7 anti-CEA antibody SPECT differentiates metastatic colorectal cancer model phenotypes and anti-vascular therapy response. Br J Cancer 2015; 112: 1882–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tixier F, Le Rest CC, Hatt M, Albarghach N, Pradier O, Metges JP, et al. Intratumor heterogeneity characterized by textural features on baseline 18F-FDG PET images predicts response to concomitant radiochemotherapy in esophageal cancer. J Nucl Med 2011; 52: 369–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hricak H, Choi BI, Scott AM, Sugimura K, Muellner A, von Schulthess GK, et al. Global Trends in Hybrid Imaging. Radiology 2010; 257: 498–506. [DOI] [PubMed] [Google Scholar]
- 10.Vaidya M, Creach KM, Frye J, Dehdashti F, Bradley JD, El Naqa I. Combined PET/CT image characteristics for radiotherapy tumor response in lung cancer. Radiother Oncol 2012; 102: 239–345. [DOI] [PubMed] [Google Scholar]
- 11.Watson SR, Dormer JD, Fei B. Imaging technologies for cardiac fiber and heart failure: a review. Heart Fail Rev 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wagner HN, Conti PS. Advances in medical imaging for cancer diagnosis and treatment. Cancer 1991; 67: 1121–1128. [DOI] [PubMed] [Google Scholar]
- 13.Lee G, Lee HY, Ko ES, Jeong WK. Radiomics and imaging genomics in precision medicine. Precis Future Med 2017; 1:10–31. [Google Scholar]
- 14.Vallières M, Kay-Rivest E, Perrin LJ, Liem X, Furstoss C, Aerts HJWL, et al. Radiomics strategies for risk assessment of tumour failure in head-and-neck cancer. Sci Rep 2017; 7:10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tanadini-Lang S, Bogowicz M, Veit-Haibach P, Huellner M, Pauli C, Shukla V, et al. Exploratory Radiomics in Computed Tomography Perfusion of Prostate Cancer. Anticancer Res 2018; 38:685–690. [DOI] [PubMed] [Google Scholar]
- 16.Wu W, Parmar C, Grossmann P, Quackenbush J, Lambin P, Bussink J, et al. Exploratory Study to Identify Radiomics Classifiers for Lung Cancer Histology. Front Oncol 2016; 6:71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bowen SR, Yuh WTC, Hippe DS, Wu W, Partridge SC, Elias S, et al. Tumor radiomic heterogeneity: Multiparametric functional imaging to characterize variability and predict response following cervical cancer radiation therapy. J. Magn. Reson. Imaging 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hsu CY, Doubrovin M, Hua CH, Mohammed O, Shulkin B, Kaste S, et al. Radiomics Features Differentiate Between Normal and Tumoral High-Fdg Uptake. Sci Rep 2018; 8: 3913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pereira GC, Traughber M, Muzic RF Jr. The Role of Imaging in Radiation Therapy Planning: Past, Present, and Future. Biomed Res Int 2014; 2014: 231090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Farjam R Tyagi Neelam, Veeraraghavan H, Apte A, Zakian K, Hunt MA, Deasy JO . Multiatlas approach with local registration goodness weighting for MRI-based electron density mapping of head and neck anatomy. Med. Phys 2017; 44: 3706–3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shiradkar R, Podder TK, Algohary A, Viswanath S, Ellis RJ, Madabhushi A. Radiomics based targeted radiotherapy planning (Rad-TRaP): a computational framework for prostate cancer treatment planning with MRI. Radiat Oncol 2016; 11: 148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Silva LAB, Ribeiro LS, Santos M, Neves N, Francisco D, Costa C, et al. Normalizing Heterogeneous Medical Imaging Data to Measure the Impact of Radiation Dose. J Digit Imaging 2015; 28: 671–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dorfman GS. Standardization of Clinical Trial Image Acquisition is Essential for Establishing Clinical Utility. Quantitative Imaging Biomarkers Alliance - Newsletter 2012; 4:1. [Google Scholar]
- 24.Matej S, Daube-Witherspoon ME, Karp JS. Analytic TOF PET reconstruction algorithm within DIRECT data partitioning framework. Phys Med Biol 2016; 61:3365–3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xi Y, Zhao J, Bennett JR, Stacy MR, Sinusas AJ, Wang G. Simultaneous CT-MRI Reconstruction for Constrained Imaging Geometries Using Structural Coupling and Compressive Sensing. IEEE Trans Biomed Eng 2016; 63: 1301–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mascarenhas NDA, Santos CAN, Cruvinel PE. Transmission Tomography under Poisson noise using the Anscombe Transform and a Wiener Filter of the projections. Nucl Instr Meth Phys Res 1999; 423: 265–271. [Google Scholar]
- 27.Coupe P, Hellier P, Kervrann C, Barillot C. Nonlocal means-based speckle filtering for ultrasound images. IEEE Trans Image Process 2009; 18: 2221–2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Attivissimo F, Cavone G, Lanzolla AML, Spadavecchia M. A Technique to Improve the Image Quality in Computer Tomography. IEEE Trans Instrum Meas 2010; 59: 1251–1257. [Google Scholar]
- 29.Gillies RJ, Kinahan PE, Hricak H. Radiomics: images are more than pictures, they are data. Radiology 2016; 278: 563–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ng F, Kozarski R, Ganeshan B, Goh V. Assessment of tumor heterogeneity by CT texture analysis: can the largest cross-sectional area be used as an alternative to whole tumor analysis? Eur J Radiol 2013; 82: 342–348. [DOI] [PubMed] [Google Scholar]
- 31.Fave X, Cook M, Frederick A, Zhang L, Yang J, Fried D, et al. Preliminary investigation into sources of uncertainty in quantitative imaging features. Comput Med Imaging Graph 2015; 44: 54–61. [DOI] [PubMed] [Google Scholar]
- 32.Rios Velazquez E, Aerts HJ, Gu Y, Goldgof DB, De Ruysscher D, Dekker A, et al. A semiautomatic CT-based ensemble segmentation of lung tumors: comparison with oncologists’ delineations and with the surgical specimen. Radiother Oncol 2012; 105: 167–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parmar C, Rios Velazquez E, Leijenaar R, Jermoumi M, Carvalho S, Mak RH, et al. Robust Radiomics Feature Quantification Using Semiautomatic Volumetric Segmentation. PLoS ONE 2014; 9: e102107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Otsu N Threshold Selection Method from Gray-Level Histograms. IEEE Trans Syst Man Cybern 1979; 9: 62–66. [Google Scholar]
- 35.Xu C, Prince JL. Generalized Gradient Vector Flow External Forces for Active Contours. Signal Processing 1998; 71: 131–139. [Google Scholar]
- 36.Kass M, Witkin A, Terzopoulos D. Snakes: Active Contour Models. Int J Comput Vis 1988; 1: 321–331. [Google Scholar]
- 37.Osher S, Fedkiw R. Level Set Methods and Dynamic Implicit Surfaces 2003; Springer-Verlag, New York: 273. [Google Scholar]
- 38.Nyúl LG, Falcão AX, Udupa JK. Fuzzy-connected 3D image segmentation at interactive speeds. Graph Models 2002; 64: 259–281. [Google Scholar]
- 39.Udupa JK, Saha PK. Fuzzy Connectedness and Image Segmentation. Proc IEEE 2003; 91: 1649–1669. [Google Scholar]
- 40.Pednekar AS; Kakadiaris IA. Image Segmentation Based on Fuzzy Connectedness using Dynamic Weights. IEEE Trans Image Process 2006; 15: 1555–1562. [DOI] [PubMed] [Google Scholar]
- 41.Jobin Christ MC, Parvathi RMS. Segmentation of Medical Image using Clustering and Watershed algorithms. Am J Appl Sci 2011; 8: 1349–1352. [Google Scholar]
- 42.Justice RK, Stokely EM, Strobel JS, Ideker RE, Smith WM. Medical image segmentation using 3D seeded region growing. Proc. SPIE 3034. Medical Imaging 1997. [Google Scholar]
- 43.Mustafa ID, Hassan MA. A Comparison between Different Segmentation Techniques used in Medical Imaging. American Journal of Biomedical Engineering 2016; 6: 59–69. [Google Scholar]
- 44.Lambin P, Rios-Velazquez E, Leijenaar R, Carvalho S, van Stiphout RG, Granton P, et al. Radiomics: Extracting more information from medical images using advanced feature analysis. Eur J Cancer 2012; 48: 441–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Paget R Texture Modelling and Synthesis. In Handbook of Texture Analysis, Mirmehdi M, Xie X, Suri J (eds). Imperial College Press, London. 2008; 424. [Google Scholar]
- 46.Xie X, Mirmehdi M. A Galaxy of Texture Features. In Handbook of Texture Analysis 2008. [Google Scholar]
- 47.Ahonen I, Härmä V, Schukov HP, Nees M, Nevalainen J. Morphological Clustering of Cell Cultures Based on Size, Shape and Texture Features. Stat Biopharm Res 2016; 8: 217–228. [Google Scholar]
- 48.Kolossvary M, Kellermayer M, Merkely B, Maurovich-Horvat P. Cardiac Computed Tomography Radiomics: A Comprehensive Review on Radiomic Techniques. J Thorac Imaging 2018; 33: 26–34. [DOI] [PubMed] [Google Scholar]
- 49.Materka A Texture analysis methodologies for magnetic resonance imaging. Dialogues Clin Neurosci 2004; 6: 243–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nyúl LG, Udupa JK. On standardizing the MR image intensity scale. Magn Reson Med 1999; 42: 1072–1081. [DOI] [PubMed] [Google Scholar]
- 51.Amadasun M, King R. Textural Features Corresponding to Textural Properties. IEEE Trans Syst Man Cybern 1989; 19: 1264–1274. [Google Scholar]
- 52.Tuceryan M, Jain AK Chapter 2.1: Texture Analysis. In The Handbook of Pattern Recognition and Computer Vision, Second Edition. Chen CH, Pau LF, Wang PSP, eds. World Scientific Publishing Co, River Edge, NJ. 1998; 207–248. [Google Scholar]
- 53.Haralick RM, Shanmugam K, Dinstein I. Textural Features of Image Classification, IEEE Trans Syst Man Cybern 1973; SMC-3: 610–621. [Google Scholar]
- 54.Soh L- K, Tsatsoulis C. Texture Analysis of SAR Sea Ice Imagery Using Gray Level Co-Occurrence Matrices, IEEE Trans Geosci Remote Sens 1999; 37: 780–785. [Google Scholar]
- 55.Clausi DA. An analysis of co-occurrence texture statistics as a function of grey level quantization. Can J Remote Sensing 2002; 28: 45–62. [Google Scholar]
- 56.Chu A, Sehgal CM, Greenleaf JF. Use of gray value distribution of run lengths for texture analysis. Pattern Recognit Lett 1990; 11: 415–419. [Google Scholar]
- 57.Dasarathy BV, Holder EB. Image characterization based on joint gray level-run length distribution. Pattern Recognit Lett 1991; 12: 497–502. [Google Scholar]
- 58.Galloway MM. Texture Analysis using Gray Level Run Lengths. Comput Vis Graph Image Process 1975; 4: 172–179. [Google Scholar]
- 59.Thibault G, Fertil B, Navarro C, Pereira S, Cau P, Levy N, et al. Texture indexes and gray level size zone matrix application to cell nuclei classification. Int J Pattern Recogn 2009; 27: 1357002–1357002. [Google Scholar]
- 60.Thibault G, Angulo J, Meyer F. Advanced statistical matrices for texture characterization: application to cell classification. IEEE Trans Biomed Eng 2014; 61:630–637. [DOI] [PubMed] [Google Scholar]
- 61.Sun C, Wee WG. Neighboring gray level dependence matrix for texture classification. Comput Vis Graph Image Process 1983; 23: 341–352. [Google Scholar]
- 62.Cameron A, Modhafar A, Khalvati F, Lui D, Shafiee MJ, Wong A, et al. Multiparametric MRI prostate cancer analysis via a hybrid morphological-textural model. Conf Proc IEEE Eng Med Biol Soc 2014:3357–3360. [DOI] [PubMed] [Google Scholar]
- 63.Burrell HC, Pinder SE, Wilson AR, Evans AJ, Yeoman LJ, Elston CW, et al. The positive predictive value of mammographic signs: a review of 425 non-palpable breast lesions. Clin Radiol 1996; 51:277–281. [DOI] [PubMed] [Google Scholar]
- 64.Cuthbert L, Huynh VM. Statistical analysis of optical Fourier transform patterns for surface texture assessment. Meas Sci Technol 1992; 3: 740–745. [Google Scholar]
- 65.Li L, Shan T, Xue L, Wang J, Chen X. Study on Woven Fabric Texture Based on Fourier Transform and Gabor Transform. Key Eng Mater 2016; 671: 369–377. [Google Scholar]
- 66.Chang T, Jay Kuo CC. Texture analysis and classification with tree-structured wavelet transform. IEEE Trans Image Process 1993; 2: 429–440. [DOI] [PubMed] [Google Scholar]
- 67.Materka A, Benoit-Cattin H, Bezy-Wendling J, Collewet G, et al. Statistical methods. In Texture Analysis for Magnetic Resonance Imaging eds. Med4publishing, Prague. 2006; 81–105. [Google Scholar]
- 68.Lindsey JK, Jones B. Choosing among generalized linear models applied to medical data. Stat Med 1998; 17: 59–68. [DOI] [PubMed] [Google Scholar]
- 69.Kagan A, Rao CR. Some properties and applications of the efficient Fisher score. J Stat Plan Inference 2003; 116: 343–352. [Google Scholar]
- 70.Kim KI, Jung K, Park SH, Kim HJ. Texture classification with kernel principal component analysis. Electron Lett 2000; 36: 1021–1022. [Google Scholar]
- 71.Witjes H, Rijpkema M, van der Graaf M, Melssen W, Heerschap A, Buydens L. Multispectral Magnetic Resonance Image Analysis Using Principal Component and Linear Discriminant Analysis. J Magn Reson Imaging 2003; 17: 261–269. [DOI] [PubMed] [Google Scholar]
- 72.Tibshirani R Regression shrinkage and selection via the lasso. J R Stat Soc B 1996; 58: 267–288. [Google Scholar]
- 73.Rabe-Hesketh S, Everitt BS. A Handbook of Statistical Analyses Using Stata Fourth edition. 2006; Chapman & Hall/CRC Press, Boca Raton: 354. [Google Scholar]
- 74.Zhang Y, Zhang H, Chen X, Lee SW, Shen D. Hybrid High-order Functional Connectivity Networks Using Resting-state Functional MRI for Mild Cognitive Impairment Diagnosis. Sci Rep 2017; 7: 6530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pascual DG. Artificial Intelligence Tools: Decision Support Systems in Condition Monitoring and Diagnosis. Chapter 4. Input and Output Data & Chapter 7. Cluster-Based Techniques 2015; CRC Press, Boca Raton: 549. [Google Scholar]
- 76.Kumar R Srivastava R, Srivastava S. Detection and Classification of Cancer from Microscopic Biopsy Images Using Clinically Significant and Biologically Interpretable Features. J Med Eng 2015; 2015: 457906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang Q, Xiao Y, Suo J, Shi J, Yu J, Guo Y, et al. Sonoelastomics for Breast Tumor Classification: a Radiomics Approach with Clustering-Based Feature Selection on Sonoelastography. Ultrasound Med Biol 2017; 3: 1058–1069. [DOI] [PubMed] [Google Scholar]
- 78.Shofty B, Artzi M, Ben Bashat D, Liberman G, Haim O, Kashanian A, et al. MRI radiomics analysis of molecular alterations in low-grade gliomas. Int J Comput Assist Radiol Surg 2017; 1–9. [DOI] [PubMed] [Google Scholar]
- 79.Kontos D, Summers RM, Giger ML. Radiomics and Deep Learning. J Med Imag 2017; 4: 041301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Giger ML, Yongyi Y, Jovan BG, Grigori Y, Stephen SC. Machine Learning in Medical Imaging. IEEE Signal Process Mag 2010. Jul; 27: 25–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wei K, Su H, Zhou G, Zhang R, Cai P, Fan Y, et al. Potential Application of Radiomics for Differentiating Solitary Pulmonary Nodules. OMICS J Radiol 2016; 5: 218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Parekh V, Jacobs MA. Radiomics: a new application from established techniques. Expert Rev Precis Med Drug Dev 2016; 1: 207–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tabrett DR, Latham K. Factors influencing self-reported vision-related activity limitation in the visually impaired. Invest Ophthalmol Vis Sci 2011; 52: 5293–5302. [DOI] [PubMed] [Google Scholar]
- 84.Shen C, Liu Z, Guan M, Song J, Lian Y, Wang S, et al. 2D and 3D CT Radiomics Features Prognostic Performance Comparison in Non-Small Cell Lung Cancer. Transl Oncol 2017; 10: 886–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yip SSF, Aerts HJWL. Applications and limitations of radiomics. Phys Med Biol 2016; 61: 150–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hatt M, Majdoub M, Vallières M, Tixier F, Le Rest CC, Groheux D, et al. 18F-FDG PET uptake characterization through texture analysis: investigating the complementary nature of heterogeneity and functional tumor volume in a multi–cancer site patient cohort. J. Nucl Med 2015; 56: 38–44. [DOI] [PubMed] [Google Scholar]
- 87.Ahn SJ, Kim JH, Park SJ, Han JK. Prediction of the therapeutic response after FOLFOX and FOLFIRI treatment for patients with liver metastasis from colorectal cancer using computerized CT texture analysis. Eur J Radiol 2016; 85: 1867–1874. [DOI] [PubMed] [Google Scholar]
- 88.Echegaray S, Gevaert O, Shah R, Kamaya A, Louie J, Kothary N, et al. Core samples for radiomics features that are insensitive to tumor segmentation: method and pilot study using CT images of hepatocellular carcinoma. J Med Imaging (Bellingham) 2015; 2:041011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yu J, Shi Z, Lian Y, Li Z, Liu T, Gao Y, et al. Noninvasive IDH1 mutation estimation based on a quantitative radiomics approach for grade II glioma. Eur Radiol 2017; 27: 3509–3522. [DOI] [PubMed] [Google Scholar]
- 90.Lopez CJ, Nagornaya N, Parra NA, Kwon D, Ishkanian F, Markoe AM, et al. Association of radiomics and metabolic tumor volumes in radiation treatment of glioblastoma multiforme. Int J Radiat Oncol Biol Phys 2017; 97: 586–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Diehn M, Nardini C, Wang DS, McGovern S, Jayaraman M, Liang Y, et al. Identification of noninvasive imaging surrogates for brain tumor gene-expression modules. Proc Natl Acad Sci USA 2008; 105: 5213–5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gibbs P, Turnbull LW. Textural analysis of contrast-enhanced MR images of the breast. Magn Reson Med 2003; 50: 92–98. [DOI] [PubMed] [Google Scholar]
- 93.Holli K, Laaperi AL, Harrison L, Luukkaala T, Toivonen T, Ryymin P, et al. Characterization of breast cancer types by texture analysis of magnetic resonance images. Acad Radiol 2010; 17: 135–141. [DOI] [PubMed] [Google Scholar]
- 94.Teruel JRHM, Heldahl MG, Goa PE, Pickles M, Lundgren S, Bathen TF, et al. Dynamic contrast-enhanced MRI texture analysis for pretreatment prediction of clinical and pathological response to neoadjuvant chemotherapy in patients with locally advanced breast cancer. NMR Biomed 2014; 27: 887–896. [DOI] [PubMed] [Google Scholar]
- 95.Balagurunathan Y, Kumar V, Gu Y, Kim J, Wang H, Liu Y, et al. Test-retest reproducibility analysis of lung CT image features. J Digit Imaging 2014; 27: 805–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gao X, Chu C, Li Y, Lu P, Wang W, Liu W, et al. The method and efficacy of support vector machine classifiers based on texture features and multiresolution histogram from 18F FDG PET CT images for the evaluation of mediastinal lymph nodes in patients with lung cancer. Eur J Radiol 2015; 84: 312–317. [DOI] [PubMed] [Google Scholar]
- 97.Al-Kadi OS, Watson D. Texture analysis of aggressive and nonaggressive lung tumor CE CT images. IEEE Trans Biomed Eng 2008; 55: 1822–1830. [DOI] [PubMed] [Google Scholar]
- 98.Grossmann P, Stringfield O, El-Hachem N, Bui MM, Rios Velazquez E, Parmar C, et al. Defining the biological basis of radiomic phenotypes in lung cancer. Elife 2017; 6: e23421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Farber NJ, Wu Y, Zou L, Belani P, Singer EA. Challenges in RCC Imaging: Renal Insufficiency, Post-Operative Surveillance, and the Role of Radiomics. Kidney Cancer J 2015; 13: 84–90. [PMC free article] [PubMed] [Google Scholar]
- 100.Farber NJ, Kim CJ, Modi PK, Hon JD, Sadimin ET, Singer EA. Renal cell carcinoma: the search for a reliable biomarker. Transl Cancer Res 2017; 6: 620–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Nie K, Shi L, Chen Q, Hu X, Jabbour SK, Yue N, et al. Rectal cancer: assessment of neoadjuvant chemoradiation outcome based on radiomics of multiparametric MRI. Clin Cancer Res 2016; 22: 5256–5264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hu P, Wang J, Zhong H, Zhou Z, Shen L, Hu W, et al. Reproducibility with repeat CT in radiomics study for rectal cancer. Oncotarget 2016; 7: 71440–71446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wibmer A, Hricak H, Gondo T, Matsumoto K, Veeraraghavan H, Fehr D, et al. Haralick texture analysis of prostate MRI: utility for differentiating non-cancerous prostate from prostate cancer and differentiating prostate cancers with different Gleason scores. Eur Radiol 2015; 25: 2840–2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ginsburg SB, Algohary A, Pahwa S, Gulani V, Ponsky L, Aronen HJ, et al. Radiomic features for prostate cancer detection on MRI differ between the transition and peripheral zones: preliminary findings from a multi-institutional study. J Magn Reson Imaging 2017; 184–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Shiradkar R, Podder TK, Algohary A, Viswanath S, Ellis RJ, Madabhushi A. Radiomics based targeted radiotherapy planning (Rad-TRaP): a computational framework for prostate cancer treatment planning with MRI. Radiat Oncol 2016; 11: 148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Cameron A, Khalvati F, Haider MA, Wong A. MAPS: a quantitative radiomics approach for prostate cancer detection. IEEE Trans Biomed Eng 2016; 63: 1145–1156. [DOI] [PubMed] [Google Scholar]


