Abstract
Mycovirus infection has been widely shown to attenuate the virulence of phytopathogenic fungi. Valsa mali is an agriculturally important fungus that causes Valsa canker disease in apple trees. In this study, two unrelated mycoviruses [Cryphonectria hypovirus 1 (CHV1, genus Hypovirus, and single-stranded RNA) and Mycoreovirus 1 (MyRV1, genus Mycoreovirus, double-stranded RNA)] that originated from Cryphonectria parasitica (chestnut blight fungus) were singly or doubly introduced into V. mali via protoplast fusion. CHV1 and MyRV1 stably infected V. mali and caused a reduction in fungal vegetative growth and virulence. Co-infection of both viruses further reduced the virulence of V. mali but compromised the stability of CHV1 infection and horizontal transmission through hyphal anastomosis. Infections of MyRV1 and, to a lesser extent, CHV1 up-regulated the transcript expression of RNA silencing-related genes in V. mali. The accumulation of CHV1 (but not MyRV1) was elevated by the knockdown of dcl2, a key gene of the RNA silencing pathway. Similarly, the accumulation of CHV1 and the efficiency of the horizontal transmission of CHV1 during co-infection was restored by the knockdown of dcl2. Thus, CHV1 and MyRV1 are potential biological control agents for apple Valsa canker disease, but co-infection of both viruses has a negative effect on CHV1 infection in V. mali due to the activation of antiviral RNA silencing by MyRV1 infection.
Keywords: mycovirus, hypovirus, mycoreovirus, Valsa mali, hypovirulence, RNA silencing
Introduction
Apple Valsa canker disease is a destructive plant disease that affects apple trees particularly in East Asian countries (Lee et al., 2006; Abe et al., 2007). The elongated cankers occur on the branches and the trunk, leading to the death of the tree and failure of the entire orchard (Chen et al., 1987). This plant disease is widespread in many apple-producing regions in China and results in significant yield losses (Wang et al., 2011; Li et al., 2013). Apple Valsa canker is caused by the plant pathogenic ascomycete fungus, interchangeably referred to as Valsa mali or Valsa ceratosperma due to synonymization of V. mali that was initially identified as a new species for the causative pathogen of apple Valsa canker, to V. ceratosperma. Although V. ceratosperma was later found to be a heterogeneous species complex, further analyses revealed that apple strains of V. mali and V. ceratosperma from East Asia represent the same fungal species (Wang et al., 2011, 2014, 2020). Apple Valsa canker is difficult to control using chemical treatments even though many types of fungicide have been tested (Wang et al., 2009). Currently, strategies for controlling the disease include strict cultivation management and scraping away of lesions before the application of fungicides. However, highly effective prevention and control measures for this disease are not available (Keqiang et al., 2009).
Research in the field of plant pathology has been aiming to the development of sustainable control methods for fungal diseases that have minimal negative effects on the environment and human health (Raymaekers et al., 2020). In this regard, the biological control method which utilizes mycoviruses (fungal viruses) as control agents (virocontrol) is a promising method for the protection of plants against phytopathogenic fungi (Xie and Jiang, 2014; García-Pedrajas et al., 2019). Mycoviruses are widespread among fungal groups (Ghabrial and Suzuki, 2009). Several species of mycoviruses can reduce the virulence of their host, however, most do not affect the host (Hillman et al., 2018). The mycovirus Cryphonectria hypovirus 1 (CHV1) has been successfully used in Europe as a biological control agent to manage chestnut blight caused by the plant pathogenic fungus Cryphonectria parasitica (Rigling and Prospero, 2018). CHV1 infection is associated with attenuated virulence, reduced pigmentation, suppressed asexual sporulation, and altered expression of certain genes of its host (Dawe and Nuss, 2001). CHV1 is a non-segmented, positive-sense, single-stranded RNA virus belonging to the genus Hypovirus of the family Hypoviridae. Its genome (12.7 kilobases) contains two open reading frames (ORFs), ORF A and ORF B. These ORFs encode multifunctional polyproteins such as the papain-like proteases, p29 and p48, which are responsible for proteolytic cleavage in the generation of functional viral proteins (Shapira et al., 1991b). The p29 encoded in CHV1 ORF A is a multifunctional protein which plays a role in the suppression of host pigmentation, sporulation, and RNA silencing (Craven et al., 1993; Sun et al., 2009; Chiba and Suzuki, 2015).
Besides CHV1, Mycoreovirus 1 (MyRV1), isolated from C. parasitica in the United States, is a potential biological control agent. MyRV1 is a member of the genus Mycoreovirus within the family Reoviridae. Its genome contains 11 segments of double-stranded RNA (dsRNA; S1 to S11) ranging from 4127 to 732 base pairs (bp) in length (Hillman et al., 2004). MyRV1 markedly reduces the virulence of its host, but it shows minimal effects on pigmentation and asexual sporulation (Hillman et al., 2004). Mixed infection of CHV1 and MyRV1 in C. parasitica presents a one-way synergism in which CHV1 enhances the replication and vertical transmission of MyRV1 through asexual spores (Sun et al., 2006).
A previous study showed that introduction of CHV1 into V. ceratosperma via biolistic delivery of infectious cDNA clone results in colony morphology changes and reduced virulence (Sasaki et al., 2002). Another study demonstrated that infection of Rosellinia necatrix Mycoreovirus 3, a mycoreovirus originated from the white root rot fungus Rosellinia necatrix in V. ceratosperma causes reduction in fungal virulence (Kanematsu et al., 2010). A more recent study identified a new hypovirus naturally infecting V. ceratosperma, but the infection of this virus is not associated with reduction of fungal virulence (Yaegashi et al., 2012).
RNA silencing is a sequence-specific gene downregulation that is also important as an antiviral defense mechanism in eukaryotes including fungi (Aliyari and Ding, 2009; Chang et al., 2012). In antiviral RNA silencing, viral-derived dsRNAs are cleaved by a Dicer-like protein (DCL) to generate small interfering RNAs that guide an Argonaute (AGO)-containing RNA silencing-induced complex for sequence-specific degradation of a viral RNA target (Ding, 2010). Ascomycete fungi are known to encode two DCLs and a varying number of AGOs, according to the species of fungus (Nakayashiki et al., 2006). The V. mali genome contains two dcl genes (dcl1 and dcl2) and three Argonaute-like (agl) genes (agl1, agl2, and agl3; Feng et al., 2017a,b) but their roles in antiviral defense responses have not been demonstrated.
In this study, we examined the effects of single and double infection of CHV1 and MyRV1 on vegetative growth and the virulence of V. mali. Our results suggest that CHV1 and MyRV1 are potential biological control agents for V. mali but co-infection of both viruses compromised the stability of CHV1 infection. Furthermore, we examined the interaction of CHV1 and MyRV1 with antiviral RNA silencing in V. mali in the context of single and double infection.
Materials and Methods
Fungal Strains and Viruses
The C. parasitica strains EP713 (Nuss et al., 2005) and 9B21 (Hillman et al., 2004), naturally infected with CHV1 and MyRV1, respectively, were provided by Dr. Nobuhiro Suzuki from Okayama University in Japan. The wild-type and dcl2 knockout mutants of V. mali strains were described previously (Feng et al., 2017a).
All fungal strains were grown for 3 to 5 days on potato dextrose agar (PDA, Difco) under benchtop conditions at 24∼26°C for morphological observation or in potato dextrose broth (PDB, Difco) when mycelia were used for the preparation of protoplasts. For maintenance, fungal strains were cultured on regeneration plates (Churchill et al., 1990) and stored at 4°C until further use. To observe the formation of asexual fruiting bodies, the fungal strains were continually cultured on PDA for 4 to 5 additional weeks.
Protoplast Preparation
The protoplasts of all fungal strains were prepared individually using a method described previously (Eusebio-Cope et al., 2009) with a slight modification. The fungal strains grown on PDA medium were cut into small pieces, cultured in 20 ml PDB (50 ml flask) and incubated in the dark at 25°C for 3 days without shaking. The young mycelia were harvested, washed with 0.6 M MgSO4 and suspended in an enzymatic solution containing 3∼6 mg lysine enzyme, 10∼15 mg β-glucuronidase and 6∼12 mg bovine serum albumin in 10 ml of 1.2 M MgSO4. This was followed by incubation at 27°C for 3∼4 h with gentle shaking. The protoplast suspensions were slowly poured into a new 50 ml plastic tube. A 1.25 volume of trapping buffer (0.4 M sorbitol in 100 mM Tris–HCl, pH 7.0) was overlaid onto the protoplast suspensions and centrifuged at 3,500 rpm for 20 min at 4°C to concentrate the protoplasts at the interface. The protoplasts were diluted in 2 volumes of 1 M sorbitol, centrifuged at 3,500 rpm for 20 min at 4°C and washed again with 10 ml of STC (1 M sorbitol, 100 mM CaCl2, 100 mM Tris–HCl, and pH 8.0). Finally, the protoplasts were suspended in a small volume of STC to obtain approximately 1 × 107 protoplasts per ml. This suspension was immediately used or stored at –80°C until further use.
Protoplast Fusion
To introduce CHV1 and MyRV1 into V. mali, the protoplasts prepared from C. parasitica strains EP713 (infected with CHV1) or 9B21 (infected with MyRV1) were fused with V. mali protoplasts using polyethylene glycol (PEG). To obtain doubly infected strain, V. mali protoplasts were simultaneously fused with the protoplasts prepared from both C. parasitica strains EP713 and 9B21. V. mali protoplasts (100 μl 1 × 107 of protoplasts/ml) and C. parasitica protoplasts (100∼200 μl of 1 × 106 protoplasts/ml) were mixed gently and placed on ice for 30 min. After adding 500 μl of PTC solution (40% PEG 4000, 100 mM Tris–HCl pH 8.0, and 100 mM CaCl2) to each protoplast suspension, the mixtures were combined gently and incubated at room temperature for 20 min. The protoplast mixtures were then centrifuged at 3,500 rpm for 5 min at 4°C, resuspended with 100 ml of 1 M sorbitol, divided into 10 aliquot parts and each placed in the center of a petri dish, and then 20∼25 ml of YCDA (0.1% yeast extract, 0.1% casein hydrolysate, 0.5% glucose, and 1.5% agar) was added. The Petri dishes were left on the benchtop for 5 to 7 days and a small piece of hypha from the edges of the colonies was subsequently transferred to new PDA plates and examined for the V. mali phenotype or prepared for dsRNA extraction. To ensure the homogeneity of V. mali isolates obtained from protoplast fusion, the fungal isolates were cultured on PDA plates from single hypha of fungal colonies that grew after regeneration of protoplasts. Basically, V. mali can be isolated due to the faster growth of V. mali on YCDA or PDA medium than that of C. parasitica. Furthermore, the purity of V. mali isolates obtained from the protoplast fusion is confirmed by observation of fungal colony morphology and sequencing of ribosomal-DNA intergenic spacer regions.
Horizontal Transmission of Viruses
The horizontal transmission of viruses through hyphal anastomosis was performed as described previously (Chiba et al., 2013). The virus-infected donor strain was cultured with a virus-free recipient strain at a distance of 1 cm on a PDA plate and incubated at 24∼26°C. After 5 or 7 days of hyphal contact, mycelial plugs were taken from three different positions (near, middle, and far) on the recipient side and cultured on PDA plates layered with cellophane for 4 days before RNA extraction for viral dsRNA analysis or RT-PCR detection.
Phenotypic and Virulence Assays
The growth of fungal colonies on a PDA plate (maintained on a benchtop at 24∼26°C for 4 days) was measured based on the colony area of three replicates. The morphology of the fungi was analyzed and photographed. Virulence has been measured in apples according to the area of the lesions induced by fungal growth (Hillman et al., 1990). Apples (cv. Gala) were purchased from a supermarket and 2-year-old apple twigs were obtained from apple trees planted at an experimental station at Northwest A&F University, China. The fruits and branches were washed with tap water, the surface was wiped with 75% ethanol and they were then rinsed several times with sterile distilled water. Small holes of 6 mm in diameter were created on the surface of the fruits and branches, after which an agar plug from the growing margin of a 4-day-old colony was placed directly on each hole and wrapped with parafilm. The apples were placed in a container with wet tissue paper, and the ends of each twig were covered with wet tissue paper. All samples were maintained at 25°C for 5 or 10 days. The parafilm wraps were removed 2 days after inoculation. After 5 or 10 days of inoculation, lesion sizes were measured as indicators of disease development. Each sample contained five inoculation points, and five apples, and five twigs were used for each strain. Virulence assays were repeated three times independently.
dsRNA Isolation
A rapid, small-scale extraction of dsRNA was performed. Fungal mycelia grown for 4 days were collected from PDA plates layered with cellophane or PDB cultures, then ground into a powder with liquid nitrogen and homogenized with 1 ml of EBA buffer (50 mM Tris–HCl pH 8.5, 50 mM EDTA, 3% SDS, 1% PVPP, and 1% DTT). The suspensions were centrifuged at 4°C for 15 min at 12,000 rpm. The supernatant (600 μl) was mixed with 600 μl of STE (10 mM Tris–HCl pH 8.0, 1 mM EDTA, and 150 mM NaCl), ethanol (up to 16% volume) and a small amount of CC41 cellulose in preparation for column chromatography analysis of dsRNAs. After 40∼60 min of continuous agitation at room temperature, each tube was washed three times with STE-EtOH (16% EtOH; v/v), with vortexing and centrifugation between washes. The dsRNAs were eluted from the dried CC41 by adding 700 μl of STE buffer and centrifuging at 12,000 rpm for 2 min. The eluate was collected and mixed with an equal volume of isopropanol, incubated for 10 min at room temperature and centrifuged at 4°C for 30 min at 12,000 rpm. The dsRNA pellet was washed with 70% ethanol, air-dried at room temperature and dissolved in 30∼50 μl of RNase-free water. Each sample was subjected to 1.4% agarose gel electrophoresis in 1X TBE buffer.
Total RNA Extraction and Viral dsRNA Quantification
Total RNA was prepared from V. mali mycelia cultured in PDA with cellophane for 3 days, as described previously (Sun et al., 2006). The RNA concentration was adjusted to 2 μg/μl using a Nano Photometer (N50 Touch) for use in agarose gel electrophoresis. Viral genomic dsRNA was quantified using densitometry, as described previously (Suzuki et al., 2003). Total RNA samples were analyzed by electrophoresis using a 1.4% agarose gel in a 1X TAE (40 mM Tris/acetate pH 7.8, 1 mM EDTA) buffer system and stained with ethidium bromide. The RNA bands were visualized using a UV lamp under a transilluminator and were photographed digitally at various exposures. The RNA bands were analyzed and quantified using Image J Macro software. Relative amounts of MyRV1 genomic RNA were quantified by measuring the amount of S1, S2, and S3 RNA segments normalized to the amount of host fungal 18S rRNA. A similar method was used to quantify CHV1 genomic RNA accumulation.
RT-PCR and Quantitative PCR Analyses
The single stranded RNAs (ssRNAs) were extracted from fungal mycelia (3 days old) following the procedure described previously (Sun et al., 2006). First-strand cDNA was synthesized with ReverTra Ace reverse transcriptase (Toyobo) using 0.2 μg of ssRNAs as a template (according to the manufacturer’s instructions) and then used for PCR amplification using 2-mixture DNA polymerase (Kangwei) and primers specific for CHV1 p29. Quantitative PCR was performed using a CRX96TM Real-Time PCR Detection System (Bio-Rad) via a Kapa SYBR Fast ABI Prism qPCR mix kit (Kapa Biosystems). The 18S RNA of V. mali was used as an internal control. Three biological replicates were used for each sample and the experiments were repeated three times independently. The primers used in this study are listed in Supplementary Table 1.
Statistic Analysis
One-way ANOVA analysis was performed in Microsoft Excel with post hoc t-tests.
Results and Discussion
CHV1 and MyRV1 Can Infect V. mali
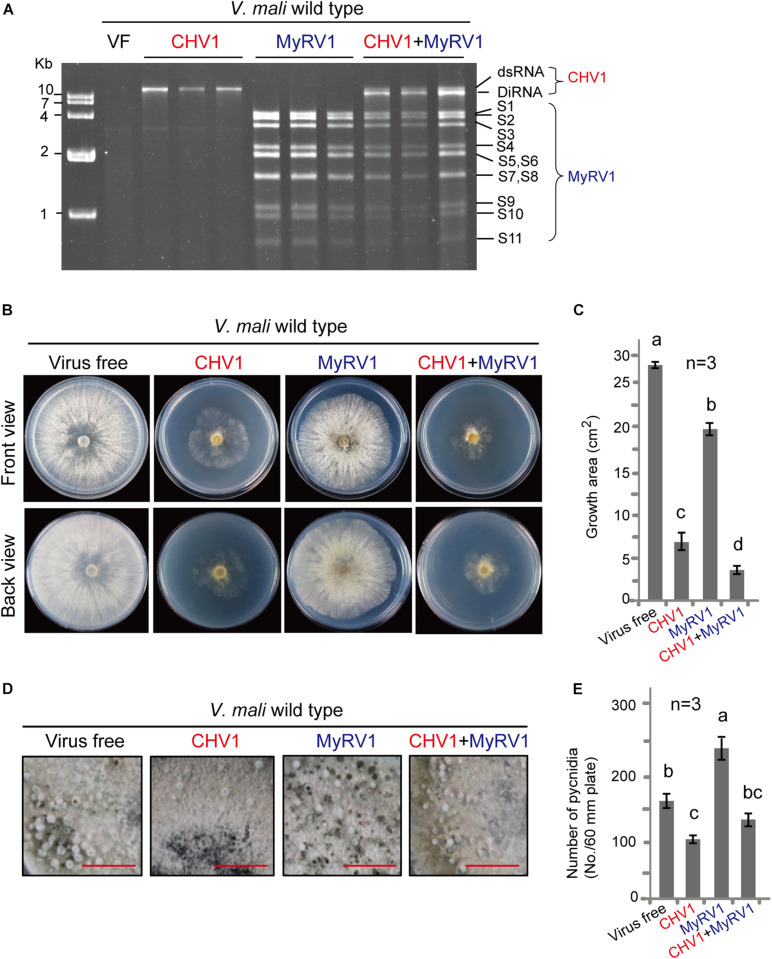
To investigate whether our V. mali strain can host mycoviruses and become hypovirulent, CHV1 and MyRV1 were introduced into V. mali via protoplast fusion using two protoplast fractions prepared from CHV1- or MyRV1-infected C. parasitica and virus-free V. mali. CHV1 and MyRV1 were also simultaneously introduced to V. mali through protoplast fusion using the mixture of protoplasts prepared from C. parasitica that were infected with CHV1 and MyRV1. After protoplast fusion, regeneration of V. mali protoplasts and transfer to new PDA plates, viral dsRNA accumulation in individual V. mali isolates was analyzed at 4 days after transference to a PDA plate. The results showed accumulations of 12.7-kilobase dsRNA of CHV1 or 11 dsRNA genome segments (4,127 to 732 bases) of MyRV1 in V. mali isolates regenerated after protoplast fusion, using corresponding CHV1- or MyRV1-infected C. parasitica. Moreover, simultaneous accumulation of CHV1 and MyRV1 dsRNAs was also detected in V. mali after protoplast fusion using the mixture of protoplasts prepared from C. parasitica that were infected with CHV1 and MyRV1 (Figure 1A). These results indicate that this V. mali strain is a compatible host for CHV1 and MyRV1. Notably, defective interfering RNAs (DI-RNAs) of the CHV1 genome were frequently observed in doubly infected fungi but not in singly infected fungi (Figure 1A). Infection of CHV1 in C. parasitica usually gives rise to DI-RNAs at a very high frequency (Shapira et al., 1991a) and DI-RNAs were also formed in C. parasitica doubly infected with CHV1 and MyRV1 (Supplementary Figure 1). Because dcl2 gene is essential for production of DI-RNAs (Zhang and Nuss, 2008), it is thus possible that generation of CHV1 DI-RNAs during co-infection with MyRV1 in V. mali is due to the upregulation of dcl2 gene expression (further supporting data and discussion are presented below).
FIGURE 1.
Infection of CHV1 and MyRV1 in V. mali. (A) Agarose gel electrophoresis of dsRNAs extracted from V. mali strains singly or doubly infected with CHV1 and MyRV1. The dsRNA samples were analyzed via electrophoresis in an agarose gel stained with ethidium bromide. The CHV1 dsRNA bands that migrated slightly faster than the wild-type CHV1 are defective interference RNA (DI-RNA). (B) Phenotypic growth on PDA medium of representative V. mali strains singly or doubly infected with CHV1 and MyRV1. The colonies were grown on PDA for 4 days and then photographed. (C) The growth areas of fungal strains described in (B). The data are the means ± SD (n = 3). The different letters indicate a significant difference at p < 0.01 (one-way ANOVA). (D) Representative images showing the formation of asexual fruiting bodies (pycnidia) of the fungal strains described in (B). The fungi were cultured on PDA medium for 4–5 weeks until the pycnidia were produced. Bars equal 1 cm. (E) The number of pycnidia counted from the fungal strains described in (D). The data are the means ± SD (n = 3). The different letters indicate a significant difference at p < 0.01 (one-way ANOVA).
CHV1 and MyRV1 Infection Reduced the Growth and Virulence of V. mali
To examine the effect of CHV1 and MyRV1 infection on V. mali, singly and doubly infected fungi, as well as virus-free fungi, were cultured under the same conditions, and the colony growth and morphology were analyzed. V. mali strains infected with either virus showed a reduction in colony size, although the fungus colony infected with CHV1 was much smaller than that infected with MyRV1 and showed an irregular margin and less dense mycelia (Figures 1B,C). Moreover, the MyRV1-infected fungal colony showed a slight increase in brown-colored pigments compared with the CHV1-infected and virus-free V. mali strains. Similarly, in previous studies, MyRV1 infection in C. parasitica resulted in reduced growth of aerial hyphae and enhanced the production of brown pigments (Sun et al., 2006; Tanaka et al., 2011). V. mali co-infected with both viruses exhibited a smaller colony size than when infected with CHV1 alone (Figures 1B,C). Next, the formation of asexual fruiting bodies (pycnidia) of V. mali cultured on a PDA plate was investigated at a later growth stage of the fungus (Figure 1D). CHV1 infection reduced pycnidia production, whereas MyRV1 infection increased pycnidia production (Figure 1E). The V. mali strain infected with both viruses produced a similar number of pycnidia as the virus-free strain (Figure 1E), suggesting that MyRV1 infection compromised the negative effects of CHV1 infection on pycnidia production in V. mali.
For the fungal virulence assay, virus-infected and virus-free V. mali strains (agar plugs) were inoculated in apples and the virulence levels were evaluated by measuring the area of lesions at 5 days after inoculation. The virus-free V. mali strain induced the largest lesions on apples, while the V. mali strains infected with either CHV1 or MyRV1 exhibited significantly smaller canker lesions although CHV1 infection induced a stronger effect than MyRV1 (Figures 2A,C). The doubly infected V. mali strain did not produce obvious lesions in apples (Figures 2A,C), suggesting that double infection strongly reduces the virulence of V. mali. A virulence assay was performed in parallel using apple twigs. In general, the effects of single and double infection on the virulence of V. mali using apple twigs observed at 5 and 10 days after inoculation were similar to those observed in apples (Figures 2B,D and Supplementary Figure 2). Our observations indicate that CHV1 and MyRV1 confer hypovirulence in V. mali and co-infection of these mycoviruses further reduces the virulence of the fungus. The differing levels of hypovirulence observed in this study between CHV1 and MyRV1 in V. mali were opposite to that observed in C. parasitica where MyRV1 showed a stronger effect than CHV1 in attenuating fungal virulence (Sun et al., 2006). Moreover, CHV1 and MyRV1 co-infection in C. parasitica did not markedly reduce the virulence of the fungal host relative to that induced by single infection (Sun et al., 2006). Thus, the molecular mechanisms underlying the hypovirulence caused by CHV1 and MyRV1 may differ between V. mali and C. parasitica.
FIGURE 2.
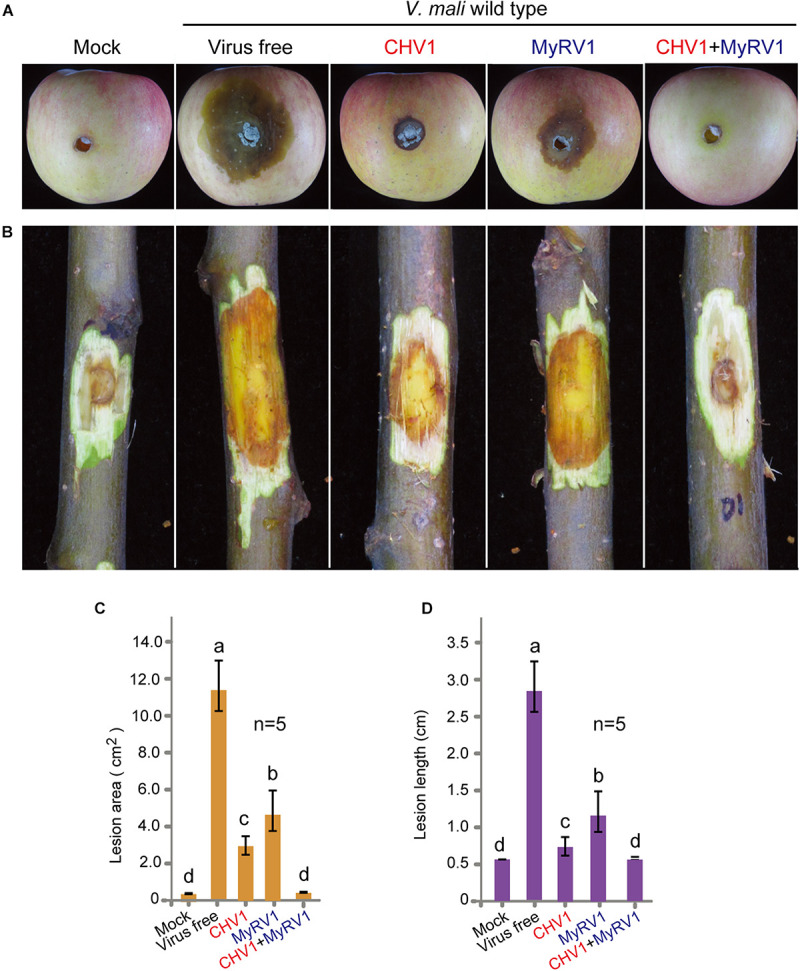
The virulence levels of V. mali strains singly or doubly infected with CHV1 and MyRV1. (A) Representative images showing lesions on apples induced by V. mali strains. The cankers were photographed at 5 days post-inoculation. (B) Representative images showing lesions on apple twigs induced by V. mali strains. The lesions on twigs were photographed at 5 days post-inoculation. (C) Lesion areas on apples measured in the experiment described in (A). The data are the means ± SD (n = 5). The different letters indicate a significant difference at p < 0.01 (one-way ANOVA). (D) Lesion lengths on apple twigs measured in the experiment described in (B). The data are the means ± SD (n = 5). The different letters indicate a significant difference at p < 0.01 (one-way ANOVA).
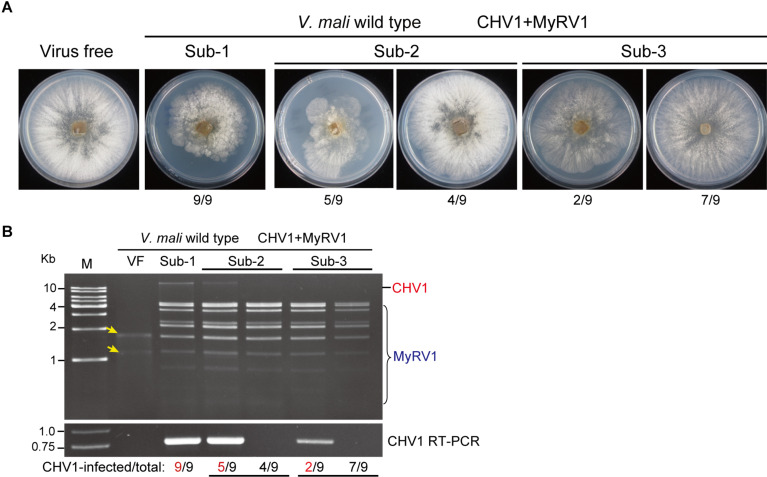
Co-infection With MyRV1 Causes Unstable Infection of CHV1 in V. mali
Subsequent subculturing on PDA plates of V. mali strains that were co-infected with CHV1 and MyRV1 revealed that most fungal strains regained their vegetative growth rate, which contrasts with the small colony size initially exhibited by these strains (Figure 3A). dsRNA and RT-PCR analyses showed that CHV1 was undetectable or its accumulation decreased in the fungal strain that recovered to a higher growth rate, whereas MyRV1 stably accumulated in all strains after fungal subculture (Figure 3B). In the V. mali strains singly infected with CHV1 or MyRV1, stable virus accumulation was observed in subsequent fungal cultures (Supplementary Figure 3). These observations suggest that MyRV1 infection has an antagonistic effect on CHV1 accumulation in V. mali. The interactions between co-infecting viruses can be either synergistic/facilitative or antagonistic while in plant and fungi, RNA silencing mechanism has been shown to be the most common underlying mechanism of both types of virus-virus interactions (Sun et al., 2006; Syller, 2012; Chiba and Suzuki, 2015; Mascia and Gallitelli, 2016; Aulia et al., 2019; Bian et al., 2020).
FIGURE 3.
The accumulation of CHV1 and MyRV1 in co-infected V. mali after subsequent subculturing. (A) The phenotypic growth on PDA medium of representative V. mali strains co-infected with CHV1 and MyRV1 in the first to third fungal subcultures (Sub-1 to Sub-3). The colonies were grown on PDA for 4 days and then photographed. The numbers below the images indicate the number of fungal colonies showing a similar phenotype as presented in the photo per total number of colonies shown. (B) Viral dsRNA and RT-PCR analyses of doubly infected V. mali strains described in (A). The numbers below the lanes indicate the number of samples in which CHV1 was detected or undetected per total number of samples. Yellow-colored arrows mark the traces of ribosomal RNAs in virus-free (VF) sample incorporated during dsRNA isolation.
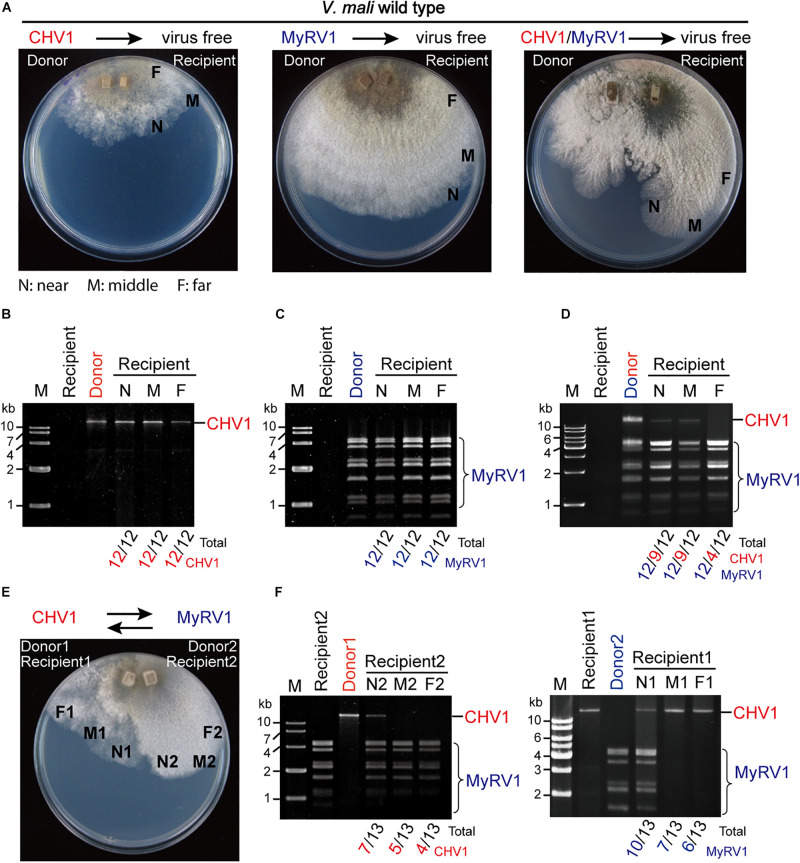
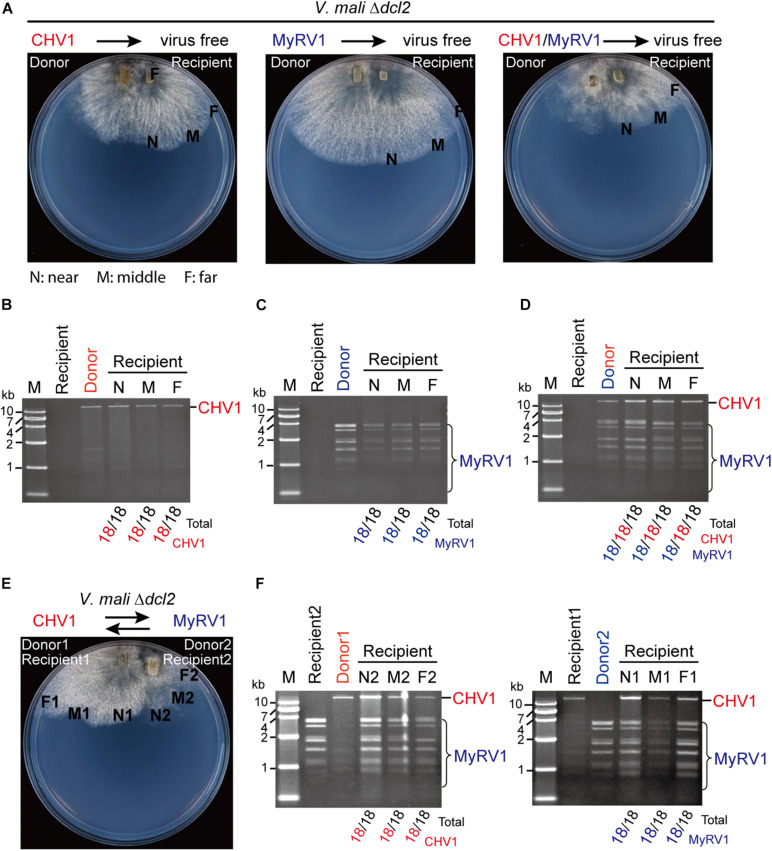
Horizontal Transmission of CHV1 and MyRV1 in V. mali
To examine whether CHV1 and MyRV1 were horizontally transmitted in V. mali via hyphal anastomosis, a virus-infected strain (donor), and a virus-free strain (recipient) were co-cultured on a PDA plate and after 5 days of hyphal fusion, agar plugs taken from the recipient colony side were grown in PDB culture and subjected to dsRNA extraction. As no specific marker gene or antibiotic resistance is available in the recipient strains for selection, the mycelial plugs, which were taken from recipient side may include the donor cells. To discern such possibility, agar plugs were taken from three positions that are near, middle, and far distance from the hyphal fusion areas in the recipient colony. Following hyphal fusion on the PDA plate, the virus-free V. mali strain co-cultured with the CHV1-infected strain showed a CHV1-infected phenotype, while the virus-free strain co-cultured with the MyRV1-infected strain showed no obvious phenotypic change, due to a slightly altered phenotype of MyRV1-infected strain on the PDA plate (Figure 4A). dsRNA analysis showed the accumulation of CHV1 or MyRV1 dsRNAs in all fungal strains taken from the three different positions in the recipient colonies (Figures 4B,C). Intriguingly, in hyphal fusion with the CHV1 and MyRV1 co-infected strain, the virus-free strain did not showed altered phenotype as observed for hyphal fusion using the CHV1-infected strain (Figure 4A). Accordingly, the fungal strains taken from the recipient colonies co-cultured with the CHV1 and MyRV1 co-infected strain as a donor, all accumulated MyRV1 dsRNAs, however, several strains accumulated no or low levels of CHV1 dsRNA, in particular to a higher number when mycelial plugs were taken far from the hyphal fusion areas (Figure 4D). Although in this assay we cannot rule out the possibility of the presence of donor strains in some of fungal strains taken from the recipient colonies, the results of our co-culture assays using up to 12 PDA plates for each single and double infection suggest that co-infection with MyRV1 interferes with the efficient horizontal transmission of CHV1 in V. mali. To further examine whether the co-infection condition affects the horizontal transmission of CHV1 and MyRV1, CHV1- and MyRV1-infected strains were co-cultured on 13 PDA plates to allow for hyphal anastomosis (Figure 4E). dsRNA analysis showed that neither CHV1 nor MyRV1 were efficiently transmitted to the recipient side of the fungus (Figure 4F), suggesting that CHV1 and MyRV1 have an antagonistic effect on each other concerning horizontal transmission.
FIGURE 4.
The efficiency of CHV1 and MyRV1 horizontal transmission via hyphal anastomosis in V. mali. (A) Co-culture on PDA plates of the virus-free V. mali strain and the virus-infected V. mali strain as recipient and donor viruses, respectively. After 1 week of hyphal contact, mycelial plugs were removed from three locations (a near, middle, and far distance from the hyphal fusion areas) in the recipient side, transferred onto new PDA plates and cultured in PDB for dsRNA extraction. (B–D) Detection of viral dsRNAs in recipient strains co-cultured with CHV1-infected (B), MyRV1-infected (C), and CHV1 + MyRV1-infected strains (D). The numbers below the lanes indicate the number of samples in which viral dsRNAs were detected per total number of samples. (E) Co-culture of CHV1-infected and MyRV1-infected V. mali strains on a PDA plate. After 1 week of hyphal contact, mycelial plugs were removed from three locations on both sides, transferred onto new PDA plates and cultured in PDB for dsRNA extraction. (F) Detection of viral dsRNAs in fungal strains obtained in the co-culture experiment described in (E).
Mycoviruses are transmitted vertically from the mycelium to spores or horizontally via hyphal anastomosis (Hillman et al., 2018). Therefore, transmission efficiency is an important factor when evaluating a mycovirus for its potential as a biological control agent. Under natural conditions, the spreading of mycoviruses is limited by vegetative incompatibility among fungal species and strains in a fungal population (Choi et al., 2012), however, some studies on filamentous phytopathogenic fungi have demonstrated the occurrence of virus transmission between vegetatively incompatible fungal hosts (Brusini and Robin, 2013; Hamid et al., 2018). Sclerotinia sclerotiorum Mycoreovirus 4 (genus Mycoreovirus of the family Reoviridae), which originated from the phytopathogenic fungus Sclerotinia sclerotiorum, can suppress host non-self recognition and thus enable the horizontal transmission of heterologous viruses (Wu et al., 2017). Our research group recently proposed a model in which mycoviruses could spread across vegetatively incompatible fungal strains or to different fungal species through plant–fungal-mediated routes facilitated by plant viruses (Bian et al., 2020). Conversely, the present study showed that the efficiency of CHV1 and MyRV1 horizontal transmission is reduced under co-infection conditions. Further investigations should be conducted to assess whether such inhibitory effects on horizontal transmission between co-infected viruses commonly occurs among mycoviruses.
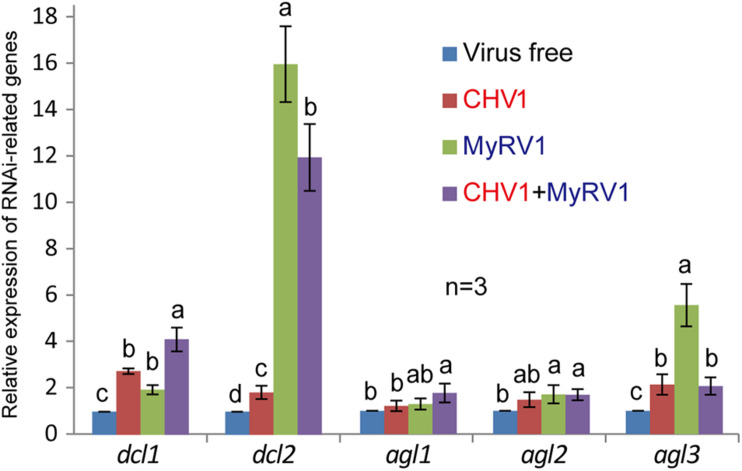
CHV1 and MyRV1 Infection Up-regulates the Transcript Expression of RNA Silencing-Related Genes in V. mali
CHV1 and MyRV1 infection in C. parasitica was shown to increase the transcript expression of dcl2 and agl2 genes, two key components of the antiviral RNA silencing pathway in C. parasitica (Sun et al., 2009; Chiba and Suzuki, 2015; Andika et al., 2017). To examine whether such effects of CHV1 and MyRV1 infection also occur in V. mali, transcript expression of V. mali dcl1, dcl2, agl1, agl2, and agl3 genes were analyzed by quantitative RT-PCR following single and double virus infection. The analyses revealed that single infection and co-infection generally enhanced the transcript expression of all the genes, however, the highest increase in expression was observed for dcl2 and agl3, particularly following MyRV1 infection (Figure 5). This suggests that RNA silencing is activated by CHV1 or MyRV1 infection in V. mali, however, CHV1 may suppress the activation of the RNA silencing response through the activity of the CHV1-encoded p29 silencing suppressor, as previously demonstrated in C. parasitica (Sun et al., 2009; Chiba and Suzuki, 2015). Moreover, much more increase of dcl2 transcript levels following co-infection of CHV1 and MyRV1 than CHV1 single infection supports the notion that generation of CHV1 DI-RNAs during co-infection with MyRV1 (Figure 1A) is related to the high expression of dcl2 gene.
FIGURE 5.
The relative transcript expression of RNA silencing-related genes in V. mali strains singly or doubly infected with CHV1 and MyRV1. The transcript expression of V. mali dcl1, dcl2, agl1, agl2, and agl3 genes following single and double virus infection were analyzed by quantitative RT-PCR. The data are the means ± SD (n = 3). The different letters indicate a significant difference at p < 0.01 (one-way ANOVA).
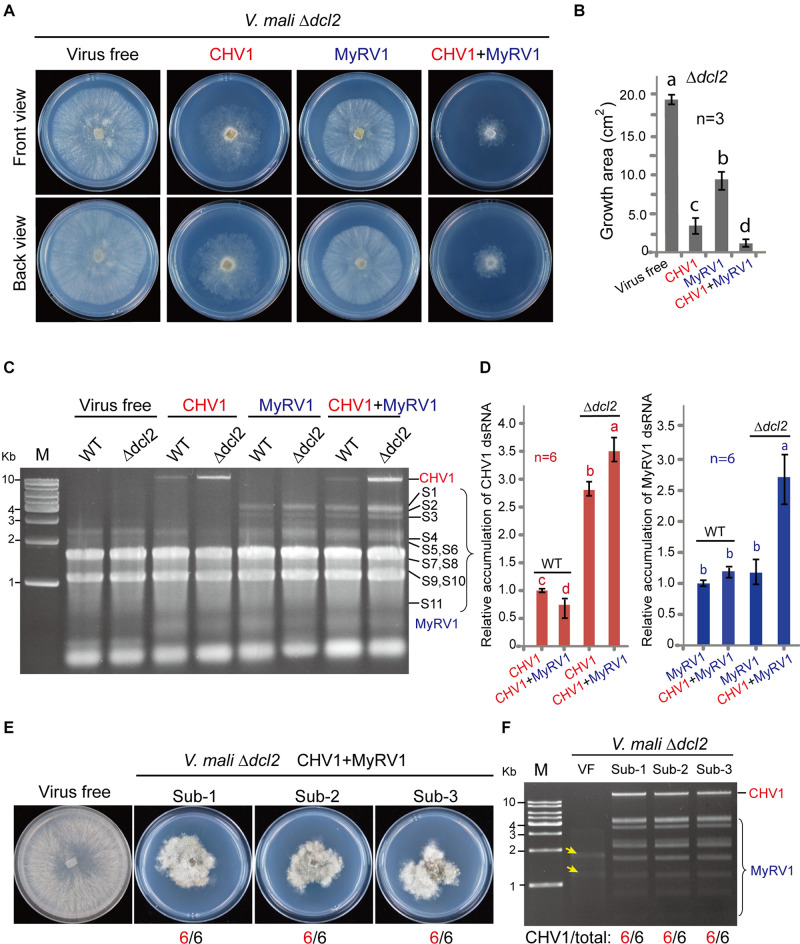
The Inactivation of dcl2 in V. mali Enhances the dsRNA Accumulation and Stability of CHV1
To investigate whether RNA silencing contributes to the inhibition of mycovirus multiplication in V. mali, CHV1 and MyRV1 were introduced to a dcl2 knockout mutant (Δdcl2) of V. mali by hyphal anastomosis. Single infection or co-infection of CHV1 and MyRV1 reduced Δdcl2 growth and produced a similar pattern as that observed in the V. mali wild-type (Figures 6A,B). Next, the relative viral dsRNA accumulation levels in the wild-type and Δdcl2 mutant were compared by extracting the total RNA from the infected fungi. The accumulation of the viral dsRNAs was visualized through agarose gel electrophoresis. CHV1 dsRNA accumulation was enhanced in the Δdcl2 mutant in the case of single infection or co-infection with MyRV1, while MyRV1 dsRNA accumulation levels were similar in the wild-type and Δdcl2 mutant during single infection but were enhanced in the Δdcl2 mutant during co-infection with CHV1 (Figures 6C,D). This suggests that MyRV1 is less affected by antiviral RNA silencing responses in V. mali than CHV1. This is similar to an observation in C. parasitica where the inactivation of dcl2 increased CHV1 RNA accumulation but had no effect on MyRV1 RNA accumulation (Chiba and Suzuki, 2015). It was also observed that co-infection reduced CHV1 dsRNA accumulation relative to that of single infection but had no effect on MyRV1 dsRNA accumulation (Figures 6C,D). The Δdcl2 mutant strains co-infected with CHV1 and MyRV1 maintained restrained vegetative growth in successive fungal subcultures (Figure 6E). Accordingly, CHV1 stably accumulated after successive fungal subculturing in the Δdcl2 mutant co-infected with these two viruses (Figure 6F), similar as the stability of CHV1 and MyRV1 single infection in the Δdcl2 mutant (Supplementary Figure 3). These results indicate that DCL2 is responsible for the suppression of CHV1 accumulation during co-infection with MyRV1. Notably, formation of CHV1 DI-RNAs was not observed in co-infected Δdcl2 mutant (Figure 6F), further supporting the view that a high expression of dcl2 transcripts during co-infection with MyRV1 is responsible for generation of CHV1 DI-RNAs.
FIGURE 6.
The accumulation of CHV1 and MyRV1 in the dcl2 knockout mutant of V. mali. (A) The phenotypic growth on PDA medium of the representative V. mali mutant strains singly or doubly infected with CHV1 and MyRV1. The colonies were grown on PDA for 4 days and then photographed. (B) The growth areas of the fungal strains described in (A). The data are the means ± SD (n = 3). The different letters indicate a significant difference at p < 0.01 (one-way ANOVA). (C) Agarose gel electrophoresis of the total RNA samples extracted from the wild-type and dcl2 knockout mutant of V. mali strains singly or doubly infected with CHV1 and MyRV1. The total RNA samples were analyzed via electrophoresis in agarose gel stained with ethidium bromide. (D) The relative dsRNA accumulation levels of CHV1 and MyRV1 in the wild-type and Δdcl2 knockout mutant. The dsRNA bands detected in the experiment described in (C) were quantified and analyzed using Image J Macro software. The data are the means ± SD (n = 6). The different letters indicate a significant difference at p < 0.01 (one-way ANOVA). (E) The phenotypic growth on PDA medium of the representative V. mali mutant strains co-infected with CHV1 and MyRV1 in the first to third fungal subcultures (Sub-1 to Sub-3). The colonies were grown on PDA for 6 days and then photographed. The numbers below the images indicate the number of fungal colonies showing a similar phenotype as that presented in the photo per total number of colonies. (F) Viral dsRNA analysis of the doubly infected V. mali mutant strains described in (E). The numbers below the lanes indicate the number of samples in which CHV1 dsRNAs were detected per total number of samples. Yellow-colored arrows mark the traces of ribosomal RNAs in virus-free (VF) sample incorporated during dsRNA isolation.
Previous studies have shown that DCL2 (but not DCL1) is critical for antiviral RNA silencing in C. parasitica and Colletotrichum higginsianum (Segers et al., 2007; Campo et al., 2016), while DCL2 and DCL1 are functionally redundant in antiviral RNA silencing in Fusarium graminearum, S. sclerotiorum, and Neurospora crassa (Yu et al., 2018; Neupane et al., 2019; Honda et al., 2020). To a lesser degree, CHV1 or MyRV1 infection also increased the expression of dcl1 transcripts (Figure 5). Further research is required using single and double dcl mutants to assess whether DCL1 also contributes to antiviral RNA silencing in V. mali.
The Inactivation of dcl2 Restores the Horizontal Transmission Efficiency of CHV1 and MyRV1 During Co-infection
The effect of dcl2 inactivation on the horizontal transmission of CHV1 and MyRV1 through hyphal fusion was also examined by co-culturing on 18 PDA plates for each donor and recipient combination. CHV1 and MyRV1 were efficiently transmitted to the virus-free Δdcl2 mutant under single or double infection conditions (Figures 7A–D). Moreover, when CHV1- and MyRV1-infected Δdcl2 mutants were co-cultured on a PDA plate, allowing for hyphal fusion (Figure 7E), both viruses were efficiently transmitted to the recipient side of the fungus (Figure 7F). These observations suggest that the efficiency of transmission of CHV1 is restored by the fact that its accumulation is increased in the Δdcl2 mutant and indirectly, the higher concentration of CHV1 could also enhance the transference of MyRV1 transmission.
FIGURE 7.
The efficiency of CHV1 and MyRV1 horizontal transmission via hyphal anastomosis in the dcl2 knockout mutant of V. mali. (A) Co-culture on PDA plates of the virus-free V. mali mutant strain and the virus-infected V. mali mutant strain as recipient and donor viruses, respectively. After 1 week of hyphal contact, mycelial plugs were taken from three locations (a near, middle, and far distance from the hyphal fusion areas) in the recipient side, transferred onto new PDA plates and cultured in PDB for dsRNA extraction. (B–D) Detection of viral dsRNAs in recipient strains co-cultured with CHV1-infected (B), MyRV1-infected (C), and CHV1 + MyRV1-infected strains (D). The numbers below the lanes indicate the number of samples in which viral dsRNAs were detected per total number of samples. (E) Co-culture on a PDA plate of CHV1-infected and MyRV1-infected V. mali mutant strains. After 1 week of hyphal contact, mycelial plugs were removed from three locations on both sides, transferred onto new PDA plates and cultured in PDB for dsRNA extraction. (F) Detection of viral dsRNAs in fungal strains obtained in the co-culture experiment described in (E).
Multiple infections of mycoviruses in a single host are common in nature and, therefore, interplays between or among mycoviruses may occur (Thapa and Roossinck, 2019). Several studies on virus–virus interaction in C. parasitica demonstrated that an RNA silencing mechanism is implicated in either synergistic or antagonistic effects between viruses. In the case of a one-way interaction between CHV1 and MyRV1, CHV1 enhances MyRV1 replication and induces frequent generation of MyRV1 rearrangements (Sun et al., 2006; Sun and Suzuki, 2008). The p29 RNA silencing suppressor encoded by CHV1 plays an important role in synergism and MyRV1 rearrangement (Sun and Suzuki, 2008; Tanaka et al., 2011). A study on the interactions between two other members of genus Hypovirus and Mycoreovirus showed that Cryphonectria hypovirus 4 facilitates the stable infection and enhanced vertical transmission of Mycoreovirus 2, through suppressing the induction of dcl2 transcripts (Aulia et al., 2019). Chiba and Suzuki (2015) demonstrated that the induction of dcl2 transcripts by co-infection with MyRV1, or a mutant CHV1 lacking the p29 RNA silencing suppressor, leads to suppression of Rosellinia necatrix victorivirus 1 (genus Totivirus of family Totiviridae) multiplication, indicating an RNA silencing-mediated one-way interference between unrelated viruses. Similarly, the results of the present study demonstrated that co-infection with MyRV1 compromised the stability of infection and horizontal transmission of CHV1 in V. mali (Figures 3, 4) and this was associated with highly up-regulated dcl2 transcripts (Figure 5). Moreover, the inactivation of dcl2 restored the stability and horizontal transmission efficiency of CHV1 in the presence of MyRV1 (Figure 6). This suggests that MyRV1 infection has inhibitory effects on CHV1 multiplication and horizontal transmission in V. mali through the activation of antiviral RNA silencing.
Conclusion
Valsa mali causes Valsa canker which penetrates deeply into the phloem and xylem of apple tree; therefore, the disease cannot be controlled effectively through chemical treatments. Research interest in mycoviruses has increased due to their potential use as a biological control agent for crop fungal diseases. Thus, expansion of the host range of hypovirulent mycoviruses and studies on virus-fungal host interactions could facilitate the development of biological control measures for fungal diseases. Hence, we introduced two hypovirulent mycoviruses (CHV1 and MyRV1) which originated from C. parasitica to V. mali using protoplast fusion. Overall, the results of this study revealed that infection with CHV1 and, to a lesser extent, MyRV1 markedly reduced the vegetative growth and virulence of V. mali. Therefore, these mycoviruses are potential biological control agents for Valsa canker disease. Moreover, the results demonstrated that co-infection of CHV1 and MyRV1 conferred stronger hypovirulent effects on V. mali. However, co-infection also suppressed the infection stability and horizontal transmission efficiency of CHV1, which is disadvantageous for its application as a biological control method. The results of this study provide a scientific base for future research on the development of the practical application of mycoviruses as a biological control agent for apple Valsa canker disease in the field.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
LSu designed the experiments. SY, RD, LSa, LH, and JL performed the experimental work. IA and LSu analyzed the data and wrote the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are most grateful to N. Suzuki for kindly providing research materials.
Footnotes
Funding. This work was supported in part by the National Natural Science Foundation of China (31970159), IA at the National Key Research and Development Program of China (2017YFD0201100), and LSu at the National Natural Science Foundation of China (30970163).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.659210/full#supplementary-material
Agarose gel electrophoresis of dsRNAs extracted from C. parasitica strains doubly infected with CHV1 and MyRV1.
(A) Representative images showing lesions on apple twigs induced by V. mali strains. The lesions on twigs were photographed at 10 days post-inoculation. (B) Lesion areas on apples measured in the experiment described in (A). The data are the means ± SD (n = 5). The different letters indicate a significant difference at p < 0.01 (one-way ANOVA).
Agarose gel electrophoresis of dsRNAs extracted from wild-type and Δdcl2 mutant of V. mali strains singly infected with CHV1 or MyRV1. The numbers below the lanes indicate the number of samples in which viral dsRNAs were detected per total number of samples.
List of primers used in this study.
References
- Abe K., Kotoda N., Kato H., Soejima J. (2007). Resistance sources to valsa canker (Valsa ceratosperma) in a germplasm collection of diverse malus species. Plant Breed. 126 449–453. 10.1111/j.1439-0523.2007.01379.x [DOI] [Google Scholar]
- Aliyari R., Ding S. W. (2009). RNA-based viral immunity initiated by the dicer family of host immune receptors. Immunol. Rev. 227 176–188. 10.1111/j.1600-065x.2008.00722.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andika I. B., Jamal A., Kondo H., Suzuki N. (2017). SAGA complex mediates the transcriptional up-regulation of antiviral RNA silencing. Proc. Natl. Acad. Sci. U.S.A. 114 E3499–E3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aulia A., Andika I. B., Kondo H., Hillman B. I., Suzuki N. (2019). A symptomless hypovirus, CHV4, facilitates stable infection of the chestnut blight fungus by a coinfecting reovirus likely through suppression of antiviral RNA silencing. Virology 533 99–107. 10.1016/j.virol.2019.05.004 [DOI] [PubMed] [Google Scholar]
- Bian R., Andika I. B., Pang T., Lian Z., Wei S., Niu E., et al. (2020). Facilitative and synergistic interactions between fungal and plant viruses. Proc. Natl. Acad. Sci. U.S.A. 117 3779–3788. 10.1073/pnas.1915996117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brusini J., Robin C. (2013). Mycovirus transmission revisited by in situ pairings of vegetatively incompatible isolates of Cryphonectria parasitica. J. Virol. Methods 187 435–442. 10.1016/j.jviromet.2012.11.025 [DOI] [PubMed] [Google Scholar]
- Campo S., Gilbert K. B., Carrington J. C. (2016). Small RNA-based antiviral defense in the phytopathogenic fungus Colletotrichum higginsianum. PLoS Pathog. 12:e1005640. 10.1371/journal.ppat.1005640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S.-S., Zhang Z., Liu Y. (2012). RNA interference pathways in fungi: mechanisms and functions. Ann. Rev. Microbiol. 66 305–323. 10.1146/annurev-micro-092611-150138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Li M., Shi X., Wang J. (1987). Studies on the infection period of Valsa mali miyabe et yamada, the causal agent of apple tree canker. Acta Phytopathol. Sin. 17 65–68. [Google Scholar]
- Chiba S., Lin Y.-H., Kondo H., Kanematsu S., Suzuki N. (2013). Effects of defective interfering RNA on symptom induction by, and replication of, a novel partitivirus from a phytopathogenic fungus, Rosellinia necatrix. J. Virol. 87 2330–2341. 10.1128/jvi.02835-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba S., Suzuki N. (2015). Highly activated RNA silencing via strong induction of dicer by one virus can interfere with the replication of an unrelated virus. Proc. Natl. Acad. Sci. U.S.A. 112 E4911–E4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi G. H., Dawe A. L., Churbanov A., Smith M. L., Milgroom M. G., Nuss D. L. (2012). Molecular characterization of vegetative incompatibility genes that restrict hypovirus transmission in the chestnut blight fungus Cryphonectria parasitica. Genetics 190 113–127. 10.1534/genetics.111.133983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill A., Ciuffetti L., Hansen D., Van Etten H., Van Alfen N. (1990). Transformation of the fungal pathogen cryphonectria parasitica with a variety of heterologous plasmids. Curr. Genet. 17 25–31. 10.1007/bf00313245 [DOI] [Google Scholar]
- Craven M., Pawlyk D., Choi G., Nuss D. (1993). Papain-like protease p29 as a symptom determinant encoded by a hypovirulence-associated virus of the chestnut blight fungus. J. Virol. 67 6513–6521. 10.1128/jvi.67.11.6513-6521.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawe A. L., Nuss D. L. (2001). Hypoviruses and chestnut blight: exploiting viruses to understand and modulate fungal pathogenesis. Ann. Rev. Genet. 35 1–29. 10.1146/annurev.genet.35.102401.085929 [DOI] [PubMed] [Google Scholar]
- Ding S.-W. (2010). RNA-based antiviral immunity. Nat. Rev. Immunol. 10 632–644. 10.1038/nri2824 [DOI] [PubMed] [Google Scholar]
- Eusebio-Cope A., Suzuki N., Sadeghi-Garmaroodi H., Taga M. (2009). Cytological and electrophoretic karyotyping of the chestnut blight fungus Cryphonectria parasitica. Fungal Genet. Biol. 46 342–351. 10.1016/j.fgb.2009.01.005 [DOI] [PubMed] [Google Scholar]
- Feng H., Xu M., Liu Y., Dong R., Gao X., Huang L. (2017a). Dicer-Like genes are required for H2O2 and KCl stress responses, pathogenicity and small RNA generation in Valsa mali. Front. Microbiol. 8:1166. 10.3389/fmicb.2017.01166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng H., Xu M., Liu Y., Gao X., Yin Z., Voegele R., et al. (2017b). The distinct roles of argonaute protein 2 in the growth, stress responses and pathogenicity of the apple tree canker pathogen. Forest Pathol. 47:e12354. 10.1111/efp.12354 [DOI] [Google Scholar]
- García-Pedrajas M. D., Cañizares-Nolasco C., Sarmiento-Villamil J. L., Jacquat A. G., Dambolena J. S. (2019). Mycoviruses in biological control: from basic research to field implementation. Phytopathology 109 1828–1839. 10.1094/phyto-05-19-0166-rvw [DOI] [PubMed] [Google Scholar]
- Ghabrial S. A., Suzuki N. (2009). Viruses of plant pathogenic fungi. Ann. Rev. Phytopathol. 47 353–384. 10.1146/annurev-phyto-080508-081932 [DOI] [PubMed] [Google Scholar]
- Hamid M., Xie J., Wu S., Maria S., Zheng D., Assane Hamidou A., et al. (2018). A novel deltaflexivirus that infects the plant fungal pathogen, Sclerotinia sclerotiorum, can be transmitted among host vegetative incompatible strains. Viruses 10:295. 10.3390/v10060295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillman B. I., Annisa A., Suzuki N. (2018). Viruses of plant-interacting fungi. Adv. Virus Res. 100 99–116. 10.1016/bs.aivir.2017.10.003 [DOI] [PubMed] [Google Scholar]
- Hillman B. I., Shapira R., Nuss D. L. (1990). Hypovirulence-associated suppression of host functions in Cryphonectria parasitica can be partially relieved by high light intensity. Phytopathol. 80 950–956. 10.1094/phyto-80-950 [DOI] [Google Scholar]
- Hillman B. I., Supyani S., Kondo H., Suzuki N. (2004). A reovirus of the fungus Cryphonectria parasitica that is infectious as particles and related to the coltivirus genus of animal pathogens. J. Virol. 78 892–898. 10.1128/jvi.78.2.892-898.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda S., Eusebio-Cope A., Miyashita S., Yokoyama A., Aulia A., Shahi S., et al. (2020). Establishment of Neurospora crassa as a model organism for fungal virology. Nat. Comm. 11 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanematsu S., Sasaki A., Onoue M., Oikawa Y., Ito T. (2010). Extending the fungal host range of a partitivirus and a mycoreovirus from Rosellinia necatrix by inoculation of protoplasts with virus particles. Phytopathology 100 922–930. 10.1094/phyto-100-9-0922 [DOI] [PubMed] [Google Scholar]
- Keqiang C., Liyun G., Baohua L. (2009). Investigations on the occurrence and control of apple canker in china. Plant Protec. 35 114–117. [Google Scholar]
- Lee D. H., Lee S. W., Choi K. H., Kim D. A., Uhm J. Y. (2006). Survey on the occurrence of apple diseases in korea from 1992 to 2000. Plant Pathol. J. 22 375–380. 10.5423/ppj.2006.22.4.375 [DOI] [Google Scholar]
- Li Z., Gao X., Du Z., Hu Y., Kang Z., Huang L. (2013). Survey of apple valsa canker in weibei area of shaanxi province. Acta Agri. Boreali-Occidentalis Sin. 1:29. [Google Scholar]
- Mascia T., Gallitelli D. (2016). Synergies and antagonisms in virus interactions. Plant Sci. 252 176–192. 10.1016/j.plantsci.2016.07.015 [DOI] [PubMed] [Google Scholar]
- Nakayashiki H., Kadotani N., Mayama S. (2006). Evolution and diversification of RNA silencing proteins in fungi. J. Mol. Evol. 63 127–135. 10.1007/s00239-005-0257-2 [DOI] [PubMed] [Google Scholar]
- Neupane A., Feng C., Mochama P. K., Saleem H., Marzano S.-Y. L. (2019). Roles of argonautes and dicers on Sclerotinia sclerotiorum antiviral RNA silencing. Front. Plant Sci. 10:976. 10.3389/fpls.2019.00976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuss D., Hillman B., Rigling D., Suzuki N. (2005). Family Hypoviridae. Virus Taxonomy: Eighth Report of the ICTV. 597–601. ∗∗pub Loc&Name. [Google Scholar]
- Raymaekers K., Ponet L., Holtappels D., Berckmans B., Cammue B. P. (2020). Screening for novel biocontrol agents applicable in plant disease management–a review. Biol. Control 144:104240. 10.1016/j.biocontrol.2020.104240 [DOI] [Google Scholar]
- Rigling D., Prospero S. (2018). Cryphonectria parasitica, the causal agent of chestnut blight: invasion history, population biology and disease control. Mol. Plant Pathol. 19 7–20. 10.1111/mpp.12542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki A., Onoue M., Kanematsu S., Suzaki K., Miyanishi M., Suzuki N., et al. (2002). Extending chestnut blight hypovirus host range within diaporthales by biolistic delivery of viral cDNA. Mol. Plant-Microbe Interact. 15 780–789. 10.1094/mpmi.2002.15.8.780 [DOI] [PubMed] [Google Scholar]
- Segers G. C., Zhang X., Deng F., Sun Q., Nuss D. L. (2007). Evidence that RNA silencing functions as an antiviral defense mechanism in fungi. Proc. Natl. Acad. Sci. U.S.A. 104 12902–12906. 10.1073/pnas.0702500104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapira R., Choi G., Hillman B., Nuss D. (1991a). The contribution of defective RNAs to the complexity of viral-encoded double-stranded RNA populations present in hypovirulent strains of the chestnut blight fungus Cryphonectria parasitica. EMBO J. 10 741–746. 10.1002/j.1460-2075.1991.tb08005.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapira R., Choi G. H., Nuss D. L. (1991b). Virus-like genetic organization and expression strategy for a double-stranded RNA genetic element associated with biological control of chestnut blight. EMBO J. 10 731–739. 10.1002/j.1460-2075.1991.tb08004.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L., Nuss D. L., Suzuki N. (2006). Synergism between a mycoreovirus and a hypovirus mediated by the papain-like protease p29 of the prototypic hypovirus CHV1-EP713. J. Gen. Virol. 87 3703–3714. 10.1099/vir.0.82213-0 [DOI] [PubMed] [Google Scholar]
- Sun L., Suzuki N. (2008). Intragenic rearrangements of a mycoreovirus induced by the multifunctional protein p29 encoded by the prototypic hypovirus CHV1-EP713. RNA 14 2557–2571. 10.1261/rna.1125408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q., Choi G. H., Nuss D. L. (2009). A single argonaute gene is required for induction of RNA silencing antiviral defense and promotes viral RNA recombination. Proc. Natl. Acad. Sci. U.S.A. 106 17927–17932. 10.1073/pnas.0907552106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki N., Maruyama K., Moriyama M., Nuss D. L. (2003). Hypovirus papain-like protease p29 functions in trans to enhance viral double-stranded RNA accumulation and vertical transmission. J. Virol. 77 11697–11707. 10.1128/jvi.77.21.11697-11707.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syller J. (2012). Facilitative and antagonistic interactions between plant viruses in mixed infections. Mol. Plant Pathol. 13 204–216. 10.1111/j.1364-3703.2011.00734.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T., Sun L., Tsutani K., Suzuki N. (2011). Rearrangements of mycoreovirus 1 S1, S2 and S3 induced by the multifunctional protein p29 encoded by the prototypic hypovirus cryphonectria hypovirus 1 strain EP713. J. Gen. Virol. 92 1949–1959. 10.1099/vir.0.031138-0 [DOI] [PubMed] [Google Scholar]
- Thapa V., Roossinck M. J. (2019). Determinants of coinfection in the mycoviruses. Front. Cell. Infect. Microbiol. 9:169. 10.3389/fcimb.2019.00169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Gao Z., Huang L., Wei J., Zang R., Kang Z. (2009). Screening fungicide for pathogen inhibition and disease control of apple tree valsa canker. Acta Phytopathol. Sin. 39 549–554. [Google Scholar]
- Wang X., Shi C.-M., Gleason M. L., Huang L. (2020). Fungal species associated with apple valsa canker in east asia. Phytopathol. Res. 2 1–14. [Google Scholar]
- Wang X., Wei J., Huang L., Kang Z. (2011). Re-evaluation of pathogens causing valsa canker on apple in china. Mycologia 103 317–324. 10.3852/09-165 [DOI] [PubMed] [Google Scholar]
- Wang X., Zang R., Yin Z., Kang Z., Huang L. (2014). Delimiting cryptic pathogen species causing apple valsa canker with multilocus data. Ecol. Evol. 4 1369–1380. 10.1002/ece3.1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S., Cheng J., Fu Y., Chen T., Jiang D., Ghabrial S. A., et al. (2017). Virus-mediated suppression of host non-self recognition facilitates horizontal transmission of heterologous viruses. PLoS Pathog. 13:e1006234. 10.1371/journal.ppat.1006234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J., Jiang D. (2014). New insights into mycoviruses and exploration for the biological control of crop fungal diseases. Ann. Rev. Phytopathol. 52 45–68. 10.1146/annurev-phyto-102313-050222 [DOI] [PubMed] [Google Scholar]
- Yaegashi H., Kanematsu S., Ito T. (2012). Molecular characterization of a new hypovirus infecting a phytopathogenic fungus, Valsa ceratosperma. Virus Res. 165 143–150. 10.1016/j.virusres.2012.02.008 [DOI] [PubMed] [Google Scholar]
- Yu J., Lee K.-M., Cho W. K., Park J. Y., Kim K.-H. (2018). Differential contribution of RNA interference components in response to distinct Fusarium graminearum virus infections. J. Virol. 92 e1717–e1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Nuss D. L. (2008). A host dicer is required for defective viral RNA production and recombinant virus vector RNA instability for a positive sense RNA virus. Proc. Natl. Acad. Sci. U.S.A. 105 16749–16754. 10.1073/pnas.0807225105 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Agarose gel electrophoresis of dsRNAs extracted from C. parasitica strains doubly infected with CHV1 and MyRV1.
(A) Representative images showing lesions on apple twigs induced by V. mali strains. The lesions on twigs were photographed at 10 days post-inoculation. (B) Lesion areas on apples measured in the experiment described in (A). The data are the means ± SD (n = 5). The different letters indicate a significant difference at p < 0.01 (one-way ANOVA).
Agarose gel electrophoresis of dsRNAs extracted from wild-type and Δdcl2 mutant of V. mali strains singly infected with CHV1 or MyRV1. The numbers below the lanes indicate the number of samples in which viral dsRNAs were detected per total number of samples.
List of primers used in this study.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.