Abstract
The nucleus is the organelle in the cell that contains the genome and its associate proteins which is collectively called chromatin. New work has shown that chromatin and its compaction level, dictated largely through histone modification state, provides rigidity to protect and stabilize the nucleus. Alterations in chromatin, its mechanics, and downstream loss of nuclear shape and stability are hallmarks of human disease. Weakened nuclear mechanics and abnormal morphology have been shown to cause rupturing of the nucleus which results in nuclear dysfunction including DNA damage. Thus, the rigidity provided by chromatin to maintain nuclear mechanical stability also provides its own protection from DNA damage via compartmentalization maintenance.
Keywords: euchromatin, Nuclear mechanics, Chromatin, nuclear morphology, DNA damage, heterochromatin
The nucleus must house and protect the genome contained within it. One way that the nucleus protects the genome and its organization is to maintain shape stability and compartmentalization through being the strongest organelle in the cell. The physical properties that protect the nucleus are provided by two major mechanical components, chromatin filling the interior and lamins forming a thin meshwork at the periphery (reviewed in (Stephens et al., 2018a)). While focus has been placed on lamins, chromatin is arising as a major mechanical component that dominates an entire force response regime (short extensions < 30% strain) independent of lamin contribution (Stephens et al., 2017). Thus, chromatin, or the DNA backbone and associate proteins, provides mechanical support to the nucleus that directly protects the DNA from damage and protects overall nuclear function.
Recent findings have now clearly shown that weakened nuclear mechanics can cause nuclear ruptures that lead to increased DNA damage among other nuclear dysfunctions. This work has been pioneered in the lamin field (Denais et al., 2016; Irianto et al., 2017; Pfeifer et al., 2018; Raab et al., 2016; Xia et al., 2018). However, studies are showing that either direct chromatin perturbations or secondary effects on chromatin from lamin mutations can result in nuclear ruptures and increased DNA damage. The general picture is that chromatin provides mechanical support for the nucleus to resist forces, namely compressive and push/pulling forces from the cytoskeleton. Upon decreased stiffness the nucleus succumbs to these antagonistic forces causing abnormal deformations, blebbing, and rupture of the nucleus resulting in exchange of nuclear and cytosolic contents and, in many cases, a nuclear bleb that remains prone to rupture. This rupture results in titrating away DNA damage repair factors as well as allowing DNA cutting enzymes from the cytosol to enter the nucleus (Maciejowski et al., 2015; Xia et al., 2018). New studies call attention to the growing field of chromatin-based nuclear mechanics and its role in maintaining genome integrity through maintenance of nuclear compartmentalization.
Chromatin histones and their modification state are a major mechanical component
Chromatin histone modification state compaction dictates chromatin-based nuclear mechanics. The main compaction of the DNA is provided by wrapping the DNA around a histone octamer core of the nucleosome (Luger et al., 1997). Nucleosome spacing and thus the compaction of the chromatin fiber is dictated by modifications to the histone tails and the presence of linker histone 1 (H1) (Ou et al., 2017). These modifications can generally be separated into two types, eu- and hetero-chromatin. Euchromatin is decompact chromatin which is transcriptionally active or accessible and is generally marked by acetylated histone tails. Heterochromatin is generally methylated histone tails that result in compact chromatin that is transcriptionally silent. Thus, the basic compaction of the chromatin fiber filling about 1/3 the volume of the nucleus provides mechanical support.
Many different experiments have verified that altering chromatin histone modification state to increase euchromatin and cause relative decompaction results in decreased nuclear rigidity. Use of histone deacetylase inhibitors (HDACi) causes an increase in acetylated histone tails and thus a general increase in decompact euchromatin. Two of these major drugs are Trichostatin A (TSA) (Yoshida et al., 1990) and valproic acid (VPA) (Marchion et al., 2005), which have been used for decades to decompact chromatin. Atomic force microscopy (AFM) studies revealed that decreased chromatin compaction via TSA treatment resulted in decreased nuclear stiffness by ~35% and greater penetration of the AFM tip pushing on the cell and nucleus (Krause et al., 2013). These results have been recapitulated multiple times showing that increased euchromatin via TSA or VPA treatment resulted in a similar 35–50% decrease in nuclear stiffness via two separate micromanipulation force measurement techniques, isolated nucleus (Stephens et al., 2017) and in cell (Shimamoto et al., 2017), as well as recent AFM-SPIM (Hobson et al., 2020). Thus, chromatin decompaction via increased euchromatin results in a weaker nucleus.
Chromatin-based nuclear mechanics can also be modulated through changing levels of heterochromatin. A broad histone methyltransferase inhibitor 3-deazaneplanocin-A DZNep provides means to decrease levels of methylated histones and thus bulk levels of both constitutive and facultative heterochromatin (Miranda et al., 2009). Decreased levels of heterochromatin causes a 35% decrease in nuclear spring constant measured by micromanipulation, similar to increasing levels of euchromatin (Stephens et al., 2018b). Oppositely, heterochromatin levels can be increased by using a broad histone demethylase inhibitor called methylstat to increase levels of methylated histones and thus facultative and constitutive heterochromatin (Luo et al., 2011). Methylstat treated nuclei with increased levels of heterochromatin provide a ~40% increase in nuclear stiffness (Stephens et al., 2018b). Increased heterochromatin levels in HT29 CSK knockdown cells showed an increase in the nuclear spring constant compared to wild-type, recapitulating these findings through a different means to increase heterochromatin levels (Stephens et al., 2017). Alterations of constitutive heterochromatin via H3K9 methylation levels have also been shown to alter nuclear mechanics to similar levels (Nava et al., 2020). Finally, mechanosensitive ion channels can downstream signal to increase methyltransferase activity, heterochromatin levels, and nuclear mechanics (Stephens et al., 2019b). Currently, many labs are focused on how mechanotransduction/sensation pathways can modulate eu/heterochromatin levels in development and to possibly fight against human disease, which we will discuss later.
An alternative way to alter chromatin compaction is via histone 1 (H1) which stabilizes the nucleosome and higher-order chromatin structure/compaction (Hergeth and Schneider, 2015). Histone 1 is a linker histone located outside of the core nucleosome binding to the entry and exit sites of DNA wrapped around the nucleosome core. Depletion of H1 via over expression of an antagonist HGMN5 results in decompaction and decreased chromatin mechanics measured via atomic force microscopy (Furusawa et al., 2015). Another H1 competitor HMGA1 also alters/decreases nuclear mechanics in breast cancer model cell lines, which the authors state may provide increased invasiveness (Senigagliesi et al., 2019). Both of these perturbations result in a ~30% decrease, similar to HDACi treatments that increase decompact euchromatin. Histone H1 has been shown to compact DNA under force and/or during chromatin assembly (Xiao et al., 2012). Alterations to the chromatin histone modification state as well as H1 dynamics are well documented in human disease and now known to alter nuclear mechanical stiffness and protection.
Alterations to DNA itself can change the mechanical strength that chromatin provides to the nucleus. Addition of divalent ions to induce chromatin condensation is the most extreme case of increasing chromatin’s mechanical contribution. Micropipette aspirations high extension force measurement regime measured an increased nuclear stiffness from divalent ion (Mg2+) chromatin condensation experiments (Pajerowski et al., 2007). Similarly, micromanipulation experiments resulted in a four-fold change upon divalent ion chromatin condensation (Shimamoto et al., 2017; Stephens et al., 2017). However, it should be noted that micropipette aspiration remains unable to resolve the increase in nuclear mechanics caused by DNA intercalating agents such as Hoechst which are known to increase nuclear mechanical strength by 50–100% in atomic force microscopy (AFM) (Saito et al., 2005), single (Neelam et al., 2015) and dual (Stephens et al., 2017) micropipette micromanipulation, and magnetic tweezers on double stranded DNA (Lipfert et al., 2010). This difference in sensitivity to chromatin mechanics stems from the ability to measure short extensions via micromanipulation and compressions via atomic force microscopy whereas micropipette aspiration measures local, fast, and high deformations (50–200%), which are more sensitive to lamin mechanics (Vaziri and Mofrad, 2007). Overall, chromatin’s mechanical contribution to the cell nucleus is based on DNA condensation, H1 dynamics, and histone modification states.
Chromatin dictates the short extension and compression mechanical regime of the nucleus
Chromatin dominates the mechanical resistance of the nucleus to small deformations independently. Micromanipulation force measurement experiments have provided, for the first time, the separation of the two major mechanical components of the nucleus, chromatin and lamins (Stephens et al., 2017). Using micromanipulation’s physiologically relevant slow extension (45 nm/sec) at both short and long extensions provided a novel nuclear force response with two regimes. The initial/short force extension regime is linear until 30% strain or ~3 μm. At this point (>30% strain, denoted as the long extension force response regime) the nucleus strain stiffens to a new stronger linear force response. The long extension force response regime is 1.5–2.0-fold stronger than the initial/short force response regime. These force vs. extension lines provide a force per unit length spring constant in nN/μm, which corresponds to 0.2–1 kPa, which agrees with many other force measurements including atomic force microscopy (AFM) and micropipette aspiration. Altering histone modification state alters short extension force response and the absolute force response of the long extension but not the strain stiffening. Depletion of or cell type specific lower levels of lamin A/C results in no change in the short extension nuclear spring constant but causes strain thinning (a weaker spring constant) at longer extensions. Recent work using AFM and light sheet volumetric imaging revealed that nuclear volume change dominates initial/short indentations and is modulated by chromatin compaction while the surface area change dominates at high indentions and is controlled by lamin A/C (Hobson et al., 2020). Thus, a second complementary force measurement approach is recapitulating this key finding in a completely different manner. The ability to differentiate the contribution of the two major mechanical components of the nucleus will provide unprecedented understanding of human diseases which display changes in both chromatin and lamins, abnormal nuclear mechanics, and abnormal morphology that disrupt key cellular functions.
The two separate regimes are due to geometry and the physical principles of these two different components as determined by molecular dynamics simulations (Banigan et al., 2017). Chromatin which fills the nucleus acts as a 3D polymer gel which responds to initial extension through its interconnectedness and bulk behavior. The lamina, however, located at the periphery as a thin meshwork behaves as a 1D polymer which is easy to bend via its short persistence length (Turgay et al., 2017) but hard to stretch (Panorchan et al., 2004). These differences are clearly visible in experiments digesting away the interior chromatin with MNase which resulted in a floppy/loose lamina that provides no force response to initial extensions but contributes when stretched out at long extensions (Banigan et al., 2017; Belaghzal et al., 2019; Shimamoto et al., 2017; Stephens et al., 2017). This experiment also shows that chromatin contributes ~90%, most if not all, of the nuclear mechanical resistance to short extensions. Furthermore, cell seeding experiments show that the nuclear lamina is wrinkled until the actin compression stretches the nucleus at high deformations (Li et al., 2015). Finite element analysis continuum modeling agrees with these findings and experimental findings as the nuclear interior volume change (chromatin) is activated initially by compression and the nuclear periphery (lamin A) is not stretched until higher deformations (Hobson et al., 2020). These two separate regimes may also serve functional roles, where chromatin is within all nuclei which incur at least small forces while lamin A is only present in substantial amounts in cells in mechanical demanding environments/tissues (Swift et al., 2013). Overall, the two regime force response is now supported by numerous studies including complementary experimental force measurements during extension (micromanipulation) and compression (AFM), as well as complementary simulations of the experiments via molecular dynamics and finite element analysis continuum modeling, respectively.
Chromatin-chromatin and chromatin-lamin linkers as possible mechanical components
Experiments and simulations show that beyond chromatin and lamins, chromatin-chromatin crosslinking proteins and chromatin-lamin linking proteins are also key components in nuclear mechanics. Simulations of nucleus uniaxial extension reveal that chromatin and lamins are insufficient to recapitulate experimentally measured two regime force-extension data (Banigan et al., 2017). Instead, mechanical coupling is required through both chromatin crosslinking and chromatin to lamin linkers to recapitulate experimental data. This idea was not necessarily novel, as mechanics papers have been investigating possible linker proteins as mechanical contributors since the advent of the mechanobiology field (Guilak et al., 2000). In fact, there are many proteins that fill these roles that have been or remain to be studied, providing an exciting time in nuclear mechanics.
Recent studies have shown that the chromatin is highly crosslinked by key nuclear functions and proteins. Here we will define crosslinking as proteins that bind to and/or connect different parts to the chromatin allowing force transmission and physically resisting deformation across the bulk chromatin polymer and nucleus. The existence of these linking proteins exists through biochemical binding assays (such as heterochromatin protein 1, HP1; (Machida et al., 2018)) and chromosome conformation capture/Hi-C (cohesin and CTCF; (Dekker and Mirny, 2016)) which reports distant genomic regions being held in proximity by chromatin proteins to form loops. A novel study has recently revealed that the genome is crosslinked about every 10–25 kb (Belaghzal et al., 2019). A technique labeled “liquid chromatin” used restriction endonucleases to digest the isolated nuclei interior DNA frequently (4 bp cutter DpnII) or less frequently (6 bp cutter HindIII) and assayed for chromosome contacts via Hi-C and force resistance via micromanipulation force measurements. Frequently digested chromatin (cut less than every 6 kb) revealed a significant loss of chromosome contacts, especially in euchromatin, and a 60% decrease in the chromatin-based nuclear spring constant. Alternatively, digestion every 10–25 kb revealed insignificant change in both chromosome interactions and chromatin-based nuclear stiffness suggesting chromatin crosslinks are physically holding the bulk chromatin together. Thus, chromatin within the nucleus forms a crosslinked polymer network that behaves mechanically like a gel.
There are many interesting possible chromatin-chromatin crosslinkers that might function to both organize and provide nuclear stiffness. Initial unpublished studies from the 4D Nucleome consortium show that HP1α, a known physical crosslinker via heterochromatin binding and dimer function, provides chromatin-based nuclear stiffness that is separate from heterochromatin (unpublished). Many recent publications show that RNA acts as a chromatin crosslinker with or without CCCTC-binding factor (CTCF), evidenced by Hi-C and other data (Hansen et al., 2019; Saldana-Meyer et al., 2019). RNA may even dictate heterochromatin organization (Thakur et al., 2019). RNA’s capacity to act as a crosslinker is due to the fact that many chromatin-binding proteins also bind RNA (D et al., 2016). Thus, RNA can physically tether chromatin proteins to hold together distant regions of chromatin. Other major Hi-C or loop forming proteins like zinc finger CTCF, structural maintenance of chromosome (SMC) protein complexes cohesin and condensin, multi-subunit mediator, and more may also provide mechanical support to the nucleus. Furthermore, studies will be needed to reveal what proteins are responsible for the chromatin-chromatin crosslinking mechanics and if their loss or disruption leads to abnormal nuclear morphology and rupture which causes DNA damage, which we will discuss in the following section.
Chromatin peripheral linkers are also another large group of proteins relevant to nuclear biology. The need for mechanical tethering of chromatin to the periphery was shown in yeast, which do not have lamins. Loss of chromatin tethering to the nuclear envelope via deletion of inner nuclear membrane (INM) proteins resulted in decreased nuclear mechanics and increased nuclear envelope fluctuations (Schreiner et al., 2015). These nuclear membrane proteins are orthologues to the mammalian LEM domain proteins, named for the three proteins in this group Lem2, Emerin, and Mam1 (Gonzalez et al., 2012; King et al., 2006). Other proteins that might be important chromatin-lamina linkers are Linker of Nucleoskeleton and Cytoskeleton (LINC) proteins such as Nesprins (Kandert et al., 2007) and Sun1/2 (Chi et al., 2012; Gilbert et al., 2018), which have both been shown to influence nuclear mechanics and shape. There are many other chromatin peripheral linker proteins such as LBR localizing heterochromatin to the periphery (Buxboim et al., 2017; Solovei et al., 2013), LAP2α (Zheng et al., 2000), BAF which binds chromatin and lamins (Loi et al., 2016), Nucleoskeleton proteins spectrin (Wren et al., 2012) and NuMA (Jayaraman et al., 2017), and others that while implicated to provide mechanical strength have yet to be fully tested. Since many of these proteins aid heterochromatin localization to the periphery, it will be important for future work to determine if this peripheral localization is important for heterochromatin’s mechanical contribution. Overall, understanding the relative contributions of chromatin-chromatin and chromatin-lamina linkers vs. chromatin and lamins remains a relatively open field for inquiry, especially as it relates to nuclear morphology and compartmentalization maintenance.
Chromatin mechanics dictates function through nucleus compartmentalization
Abnormal nuclear morphology has been a diagnostic indicator of human disease since the invention of the microscope. The Pap smear was developed in the 1930s to diagnose cervical cancer through screening for large and abnormally shaped nuclei (Papanicolaou and Traut, 1997). Abnormal nuclear morphology is prevalent across the human disease spectrum including heart disease and advanced aging (Goldman et al., 2004), muscular dystrophy (Fidziańska et al., 1998), neurological disorders (Macfarlane et al., 2015), and a large subset of cancers (Gisselsson et al., 2001) (lung (Jia et al., 2019), renal (Delahunt et al., 2011), prostate (Helfand et al., 2012), cervical, breast (Lu et al., 2018; Radhakrishnan et al., 2017), and leukemia (Bessho et al., 1981)). It has taken until the last 20 years to realize that morphological changes occur through altered physical properties of the tissues, cells, and nuclei. It has taken us until the past 10 years to discover that mechanically perturbed nuclei causing abnormal shapes ultimately disrupt basic nuclear functions through the rupturing of the nucleus. The loss of nucleus compartmentalization due to these ruptures results in major functional consequences, including DNA damage (Xia et al., 2018), altered transcription (De Vos et al., 2011), and loss of cell cycle control (Pfeifer et al., 2018). These associated nuclear dysfunctions could be contributing factors to human disease.
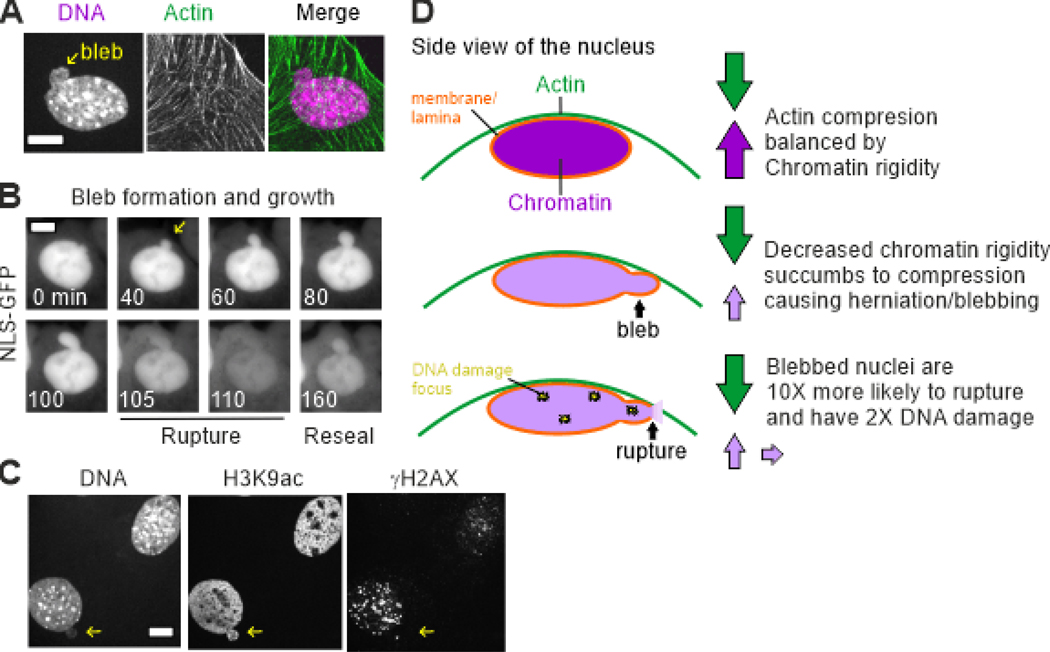
Decreased chromatin-based nuclear mechanics results in abnormal nuclear ruptures and morphology. Loss of chromatin-based rigidity has been shown to result in nuclear ruptures and increased DNA damage, independent of changes to lamins (Stephens et al., 2018b). The current model is that the cytoskeleton compresses the nucleus while chromatin resists this compression to maintain compartmentalization measured commonly by maintenance of fluorescently labeled nuclear localization signal within the nucleus (NLS-GFP; Figure 1, A and D). However, a weakened nucleus succumbs to compressive forces herniating at the major axis of the nucleus to form a bleb and rupturing the nuclear membrane resulting in spilling of nuclear contents (NLS-GFP) into the cytoplasm (Hatch and Hetzer, 2016; Le Berre et al., 2012; Tamiello et al., 2013). The capacity for nuclear ruptures occurs in the previously listed examples of decreased nuclear mechanics caused by perturbations to chromatin and histone modification states. Depletion of H1 resulted in an increased incidence of nuclei rupturing under shear stress (Furusawa et al., 2015). Increased euchromatin and decreased heterochromatin has more directly been shown to result in the formation of nuclear blebs and ruptures via NLS-GFP (Stephens et al., 2018b; Stephens et al., 2019b) (Figure 1B). More recently, it has been directly shown that nuclear blebs that arise due to rupturing of the nucleus are highly associated with a two-fold increase in DNA damage (Stephens et al., 2019b) (Figure 1, C and D). It should also be noted that rescuing nuclear stiffness and morphology by increasing heterochromatin also rescues DNA damage levels to wild-type levels, in either chromatin or lamin perturbations (Stephens et al., 2019b). Thus, the independent nature for which chromatin histone modification state can cause or rescue abnormal nuclear shape suggest chromatin’s mechanical properties protect the nucleus from DNA damage and dysfunction in the short extension force response regime of the nucleus.
Figure 1.
Decreased chromatin-based nuclear rigidity results in nuclear blebbing, rupture, and DNA damage. (A) An example image of a blebbed nucleus due to decreased chromatin-based nuclear rigidity caused by treatment with histone methyltransferase inhibitor DZNep to decrease levels of heterochromatin. An example image of chromatin labeled with Hoechst and actin labeled with phalloidin. The image shows how actin cables run over the top of the nucleus. (B) Example time-lapse of nuclear bleb formation and rupture shown by imaging nuclear localization signal green fluorescence protein (NLS-GFP). Time is in minutes. Rupture is indicated by the spilling of NLS-GFP into the cytoplasm. (C) Example image showing that blebbed nuclei display higher levels of DNA damage marker γH2AX. (D) Side view schematic showing that chromatin rigidity resists actin (shown), and other cytoskeletal forces, to maintain normal nuclear shape and compartmentalization. Loss of chromatin-based nuclear rigidity results in actin compression forces overcoming nuclear rigidity forces resulting in a herniation and rupture of the nucleus leading to spilling of nuclear contents thereby allowing cytosolic contents into the nucleus, DNA damage, transcription changes, and cell cycle disruption.
Nuclear ruptures are transient events that can be associated with nuclear blebs or abnormal morphology depending on the cell type. First, it should be noted that recent evidence suggests that chromatin tethering to the nuclear envelope is important to resealing ruptures via the chromatin binding protein BAF and its recruitment of membrane proteins (Halfmann et al., 2019). Furthermore, blebs that form coincident with nuclear ruptures can be reabsorbed in the nuclear body via chromatin condensation (Robijns et al., 2016). Blebs that are not reabsorbed are ten-fold more likely to result in nuclear rupture and nuclei containing blebs display two-fold more DNA damage (Stephens et al., 2019b). Specifically, nuclear rupture causes DNA damage because of loss of DNA damage repair factors to the cytoplasm (Irianto et al., 2017; Xia et al., 2018). Alternatively, massive nuclear deformations alone have been shown to cause DNA damage via constricted migration (Denais et al., 2016) or AFM force probing (Hobson and Lammerding, personal communication), likely through local displacement of DNA damage repair factors. Though DNA damage in these cases without rupture result in less DNA damage compared cases of documented nuclear rupture via loss of NLS-GFP.
Alterations in histone modification state are not the only chromatin perturbations that result in abnormal nuclear morphology and ruptures. Interestingly, depletion of commonly known cancer and chromatin protein p53 or chromatin protein Rb results in nuclear ruptures but no significant change in nuclear shape (Yang et al., 2017). Depletion of either a SWI/SNF chromatin remodeler Brg1 (Imbalzano et al., 2013) or a heterochromatin-lamin binding protein PRR14 (Poleshko et al., 2013) results in abnormal nuclear morphology but studies have yet to address nuclear rupture or altered chromatin-based nuclear mechanics. Beyond p53 other cancer relevant molecules p63 depletion (Rapisarda et al., 2017) and miR-29b blockade (Kriegel et al., 2018) result in abnormal nuclear shapes. Furthermore, depletion of many nucleolar proteins such as NOP53 (Lee et al., 2018), nucleophosmin (Amin et al., 2008), and fibrillarin (Amin et al., 2007) cause abnormal nuclear morphology. However, the cause and consequence remain unclear for these nucleolar proteins as effects may be due to failed mitosis that impact the daughter nuclei or simply decreased chromatin-based nuclear mechanics that cause nuclear ruptures. Similarly, long-term depletion of cohesin causes highly abnormally shaped nuclei that are likely to be due to mitotic errors (Cremer et al., 2019). Disruption of chromocenters via D1/HMGA1 perturbations can also result in abnormal nuclear morphology, ruptures, and increased DNA damage (Jagannathan et al., 2018). Finally, newly published gene screen reveals that knockdown of many chromatin proteins results in abnormal nuclear morphology (Tamashunas et al., 2020). Thus, there are many possible chromatin proteins that could provide mechanical support to the nucleus, maintaining shape and compartmentalization, and protect the genome from DNA damage. The field of chromatin-based nuclear mechanics currently offers many unanswered questions that are ready for new scientists to tackle.
Alterations in nuclear shape that result in nuclear ruptures from lamin mutations might be due to decreased levels of heterochromatin (reviewed in (Stephens et al., 2019a)). Lamin mutations have been shown to alter chromatin-lamin interactions (Scaffidi and Misteli, 2006). Furthermore, well-known lamin mutant progerin, that causes the advanced aging disease Progeria, results in loss of constitutive heterochromatin (McCord et al., 2013; Shumaker et al., 2006; Stephens et al., 2018b). Rheology measurements which provide indirect force measurements show that chromatin motion increases suggesting decreased chromatin-based nuclear mechanics (Booth et al., 2015). Lamin B depletion, which occurs in senescence (Shimi et al., 2011), displays nuclear rupture (Vargas et al., 2012) and increased DNA damage (Stephens et al., 2019b) also presents decreased levels of facultative heterochromatin (Camps et al., 2014; Stephens et al., 2018b). Importantly, these alterations to decrease levels of heterochromatin provide a means to cause these effects. Alternatively, changes in these lamins provide no clear mechanism resulting in nuclear blebs and ruptures as progerin (Schape et al., 2009) and lamin B depletion (Lammerding et al., 2006) result in increased or no change lamin mechanics, respectively. Additional experiments are required to continue to separate the relative roles of chromatin and lamins in nuclear mechanics, morphology, rupture, and resulting DNA damage.
Using chromatin mechanics as a possible therapeutic approach
Cellular mechanosensitive pathways have recently been shown to rescue nuclear mechanics, morphology, rupture, and DNA damage. Mechano-transduction/-sensitive pathways are pathways in which extra- or intracellular forces elicit a change in cell behavior. Mechanosensitive pathways are activated in mesenchymal stem cell differentiation (Heo et al., 2016; Le et al., 2016). It is important to note that stem cells have relatively decompact chromatin and no lamin A (Meshorer and Misteli, 2006), meaning they must adapt efficiently to extracellular force to protect themselves, possibly through auxetic behavior (Pagliara et al., 2014). Mechanotransduction partially relies on mechanosensitive ion channels in the plasma membrane that open under force to signal among other things an increase in chromatin compaction and heterochromatin levels (Heo et al., 2016; Heo et al., 2015; Stephens et al., 2019b). This pathway exists across multiple cell types that were able to activate mechanosensitive ion changes through changes in ion levels to cause cell membrane stretching and mechanosensitive ion channel opening (Stephens et al., 2019b). Activation of mechanosensitive ion channels over long time-intervals (day/s) resulted in increased heterochromatin due to histone methyltransferases and ultimately increased chromatin mechanical rigidity resulting in stiffer nuclei. This increase in nuclear stiffness resulted in maintained shape, less nuclear ruptures, and presented less DNA damage in cells with either chromatin or lamin perturbations (Stephens et al., 2019b). Even activation of this pathway which relies on F-actin force transmission, inactivated by drug Y-27632 a ROCK inhibitor, rescues UV induced DNA damage through a yet fully uncovered mechanism (Nagayama and Fukuei, 2019). This data suggests that increased chromatin compaction and mechanics may be sufficient to decrease DNA damage from possible nuclear rupture.
Altered levels of both eu-/heterochromatin can also be induced via other mechanosensitive pathways in the cell. Cells grown in 3D matrix result in weaker nuclei due to activation of a specific histone methyltransferase in the cell that increases the H3K4me3 euchromatin marker (Wang et al., 2018). Another example is that cellular compression causing reversible chromatin condensation via unbinding from actin and nuclear import of histone deacetylase 3 (Damodaran et al., 2018). However, it remains unclear how a single histone modification enzyme could have such strong effects when there are many different types of eu/heterochromatin markers.
The ability to modulate chromatin compaction and histone modification state in a cell native manner provides a unique opportunity for possible therapeutic approaches. Use of broad histone posttranslational enzyme inhibitors could cause massive off target effects if used as therapeutics. Alternatively, modulating mechanosensitive pathways allows for changing histone modification state in a way that the cell is accustom to. This pathway provides a way to increase chromatin-based nuclear mechanics, maintain nuclear shape, and compartmentalization, as has been proof of principal tested (Stephens et al., 2019b). Alternatively, this pathway could dial chromatin mechanics down in specific cells with the aim to kill the cells by severely destabilizing the nucleus. These same ideas are being considered for lamins as well (please refer to the review by Xie, Walker, and Irianto in this special issue). Thus, mechanosensitive cellular pathways dictating the mechanics of either or both major nuclear mechanical components, chromatin and lamins, could provide therapeutic approaches to treat the many human disease presenting abnormal nuclear morphology.
Acknowledgments
Thank you to Pierre-Alexandre Vidi and Andrew Seeber for helpful discussions. This work is supported by Pathway to Independence Award NIHGMS K99GM123195 awarded to A.D. Stephens.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amin MA, Matsunaga S, Ma N, Takata H, Yokoyama M, Uchiyama S, and Fukui K. 2007. Fibrillarin, a nucleolar protein, is required for normal nuclear morphology and cellular growth in HeLa cells. Biochem Biophys Res Commun. 360:320–326. [DOI] [PubMed] [Google Scholar]
- Amin MA, Matsunaga S, Uchiyama S, and Fukui K. 2008. Depletion of nucleophosmin leads to distortion of nucleolar and nuclear structures in HeLa cells. Biochem J. 415:345–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banigan EJ, Stephens AD, and Marko JF. 2017. Mechanics and Buckling of Biopolymeric Shells and Cell Nuclei. Biophysical journal. 113:1654–1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belaghzal H, Borrman T, Stephens AD, Lafontaine DL, Venev SV, Weng Z, Marko JF, and Dekker J. 2019. Compartment-dependent chromatin interaction dynamics revealed by liquid chromatin Hi-C. bioRxiv:704957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessho F, Fujiu M, and Kinumaki H. 1981. Acute nonlymphocytic leukemia showing abnormal nuclear lobulation. Am J Clin Pathol. 75:684–692. [DOI] [PubMed] [Google Scholar]
- Booth EA, Spagnol ST, Alcoser TA, and Dahl KN. 2015. Nuclear stiffening and chromatin softening with progerin expression leads to an attenuated nuclear response to force. Soft Matter. 11:6412–6418. [DOI] [PubMed] [Google Scholar]
- Buxboim A, Irianto J, Swift J, Athirasala A, Shin JW, Rehfeldt F, and Discher DE. 2017. Coordinated increase of nuclear tension and lamin-A with matrix stiffness outcompetes lamin-B receptor that favors soft tissue phenotypes. Molecular biology of the cell. 28:3333–3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camps J, Wangsa D, Falke M, Brown M, Case CM, Erdos MR, and Ried T. 2014. Loss of lamin B1 results in prolongation of S phase and decondensation of chromosome territories. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 28:3423–3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi YH, Chen CY, and Jeang KT. 2012. Reversal of laminopathies: the curious case of SUN1. Nucleus. 3:418–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremer M, Brandstetter K, Maiser A, Rao SSP, Schmid V, Mitra N, Mamberti S, Klein K-N, Gilbert DM, Leonhardt H, Cardoso MC, Aiden EL, Harz H, and Cremer T. 2019. Cohesin depleted cells pass through mitosis and reconstitute a functional nuclear architecture. bioRxiv:816611. [Google Scholar]
- D GH, Kelley DR, Tenen D, Bernstein B, and Rinn JL. 2016. Widespread RNA binding by chromatin-associated proteins. Genome Biol. 17:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damodaran K, Venkatachalapathy S, Alisafaei F, Radhakrishnan AV, Sharma Jokhun D, Shenoy VB, and Shivashankar GV. 2018. Compressive force induces reversible chromatin condensation and cell geometry dependent transcriptional response. Molecular biology of the cell:mbcE18040256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vos WH, Houben F, Kamps M, Malhas A, Verheyen F, Cox J, Manders EM, Verstraeten VL,van Steensel MA, Marcelis CL, van den Wijngaard A, Vaux DJ, Ramaekers FC, and Broers JL. 2011. Repetitive disruptions of the nuclear envelope invoke temporary loss of cellular compartmentalization in laminopathies. Human molecular genetics. 20:4175–4186. [DOI] [PubMed] [Google Scholar]
- Dekker J, and Mirny L. 2016. The 3D Genome as Moderator of Chromosomal Communication. Cell. 164:1110–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delahunt B, Sika-Paotonu D, Bethwaite PB, William Jordan T, Magi-Galluzzi C, Zhou M, Samaratunga H, and Srigley JR. 2011. Grading of clear cell renal cell carcinoma should be based on nucleolar prominence. Am J Surg Pathol. 35:1134–1139. [DOI] [PubMed] [Google Scholar]
- Denais CM, Gilbert RM, Isermann P, McGregor AL, te Lindert M, Weigelin B, Davidson PM, Friedl P, Wolf K, and Lammerding J. 2016. Nuclear envelope rupture and repair during cancer cell migration. Science. 352:353–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fidziańska A, Toniolo D, and Hausmanowa-Petrusewicz I. 1998. Ultrastructural abnormality of sarcolemmal nuclei in Emery-Dreifuss muscular dystrophy (EDMD). J Neurol Sci. 159:88–93. [DOI] [PubMed] [Google Scholar]
- Furusawa T, Rochman M, Taher L, Dimitriadis EK, Nagashima K, Anderson S, and Bustin M. 2015. Chromatin decompaction by the nucleosomal binding protein HMGN5 impairs nuclear sturdiness. Nat Commun. 6:6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert HT, Mallikarjun V, Dobre O, Jackson MR, Pedley R, Gilmore AP, Richardson SM, and Swift J. 2018. Nuclear decoupling is part of a rapid protein-level cellular response to high-intensity mechanical loading. bioRxiv:317404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gisselsson D, Björk J, Höglund M, Mertens F, Dal Cin P, Akerman M, and Mandahl N. 2001. Abnormal nuclear shape in solid tumors reflects mitotic instability. Am J Pathol. 158:199–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman RD, Shumaker DK, Erdos MR, Eriksson M, Goldman AE, Gordon LB, Gruenbaum Y, Khuon S, Mendez M, Varga R, and Collins FS. 2004. Accumulation of mutant lamin A causes progressive changes in nuclear architecture in Hutchinson-Gilford progeria syndrome. Proceedings of the National Academy of Sciences of the United States of America. 101:8963–8968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez Y, Saito A, and Sazer S. 2012. Fission yeast Lem2 and Man1 perform fundamental functions of the animal cell nuclear lamina. Nucleus. 3:60–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilak F, Tedrow JR, and Burgkart R. 2000. Viscoelastic properties of the cell nucleus. Biochem Biophys Res Commun. 269:781–786. [DOI] [PubMed] [Google Scholar]
- Halfmann CT, Sears RM, Katiyar A, Busselman BW, Aman LK, Zhang Q, O’Bryan CS, Angelini TE, Lele TP, and Roux KJ. 2019. Repair of nuclear ruptures requires barrier-to-autointegration factor. The Journal of Cell Biology. 218:2136–2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen AS, Hsieh TS, Cattoglio C, Pustova I, Saldana-Meyer R, Reinberg D, Darzacq X, and Tjian R. 2019. Distinct Classes of Chromatin Loops Revealed by Deletion of an RNA-Binding Region in CTCF. Mol Cell. 76:395–411 e313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch EM, and Hetzer MW. 2016. Nuclear envelope rupture is induced by actin-based nucleus confinement. J Cell Biol. 215:27–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfand BT, Wang Y, Pfleghaar K, Shimi T, Taimen P, and Shumaker DK. 2012. Chromosomal regions associated with prostate cancer risk localize to lamin B-deficient microdomains and exhibit reduced gene transcription. J Pathol. 226:735–745. [DOI] [PubMed] [Google Scholar]
- Heo SJ, Driscoll TP, Thorpe SD, Nerurkar NL, Baker BM, Yang MT, Chen CS, Lee DA, and Mauck RL. 2016. Differentiation alters stem cell nuclear architecture, mechanics, and mechano-sensitivity. Elife. 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo SJ, Thorpe SD, Driscoll TP, Duncan RL, Lee DA, and Mauck RL. 2015. Biophysical Regulation of Chromatin Architecture Instills a Mechanical Memory in Mesenchymal Stem Cells. Sci Rep. 5:16895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hergeth SP, and Schneider R. 2015. The H1 linker histones: multifunctional proteins beyond the nucleosomal core particle. EMBO Rep. 16:1439–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobson CM, Kern M, O’Brien ET, Stephens AD, Falvo MR, and Superfine R. 2020. Correlating nuclear morphology and external force with combined atomic force microscopy and light sheet imaging separates roles of chromatin and lamin A/C in nuclear mechanics. Molecular biology of the cell:mbcE20010073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imbalzano KM, Cohet N, Wu Q, Underwood JM, Imbalzano AN, and Nickerson JA. 2013. Nuclear shape changes are induced by knockdown of the SWI/SNF ATPase BRG1 and are independent of cytoskeletal connections. PloS one. 8:e55628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irianto J, Xia Y, Pfeifer CR, Athirasala A, Ji J, Alvey C, Tewari M, Bennett RR, Harding SM, Liu AJ, Greenberg RA, and Discher DE. 2017. DNA Damage Follows Repair Factor Depletion and Portends Genome Variation in Cancer Cells after Pore Migration. Curr Biol. 27:210–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagannathan M, Cummings R, and Yamashita YM. 2018. A conserved function for pericentromeric satellite DNA. Elife. 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaraman S, Chittiboyina S, Bai Y, Abad PC, Vidi PA, Stauffacher CV, and Lelievre SA. 2017. The nuclear mitotic apparatus protein NuMA controls rDNA transcription and mediates the nucleolar stress response in a p53-independent manner. Nucleic acids research. 45:11725–11742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Y, Vong JS-L, Asafova A, Garvalov BK, Caputo L, Cordero J, Singh A, Boettger T, Günther S, Fink L, Acker T, Barreto G, Seeger W, Braun T, Savai R, and Dobreva G. 2019. Lamin B1 loss promotes lung cancer development and metastasis by epigenetic derepression of RET. Journal of Experimental Medicine. 216:1377–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandert S, Luke Y, Kleinhenz T, Neumann S, Lu W, Jaeger VM, Munck M, Wehnert M, Muller CR, Zhou Z, Noegel AA, Dabauvalle MC, and Karakesisoglou I. 2007. Nesprin-2 giant safeguards nuclear envelope architecture in LMNA S143F progeria cells. Human molecular genetics. 16:2944–2959. [DOI] [PubMed] [Google Scholar]
- King MC, Lusk CP, and Blobel G. 2006. Karyopherin-mediated import of integral inner nuclear membrane proteins. Nature. 442:1003–1007. [DOI] [PubMed] [Google Scholar]
- Krause M, Te Riet J, and Wolf K. 2013. Probing the compressibility of tumor cell nuclei by combined atomic force-confocal microscopy. Physical biology. 10:065002. [DOI] [PubMed] [Google Scholar]
- Kriegel AJ, Terhune SS, Greene AS, Noon KR, Pereckas MS, and Liang M. 2018. Isomer-specific effect of microRNA miR-29b on nuclear morphology. The Journal of biological chemistry. 293:14080–14088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammerding J, Fong LG, Ji JY, Reue K, Stewart CL, Young SG, and Lee RT. 2006. Lamins A and C but not lamin B1 regulate nuclear mechanics. The Journal of biological chemistry. 281:25768–25780. [DOI] [PubMed] [Google Scholar]
- Le Berre M, Aubertin J, and Piel M. 2012. Fine control of nuclear confinement identifies a threshold deformation leading to lamina rupture and induction of specific genes. Integr Biol (Camb). 4:1406–1414. [DOI] [PubMed] [Google Scholar]
- Le HQ, Ghatak S, Yeung CY, Tellkamp F, Gunschmann C, Dieterich C, Yeroslaviz A, Habermann B, Pombo A, Niessen CM, and Wickstrom SA. 2016. Mechanical regulation of transcription controls Polycomb-mediated gene silencing during lineage commitment. Nat Cell Biol. 18:864–875. [DOI] [PubMed] [Google Scholar]
- Lee S, Ahn YM, Kim JY, Cho YE, and Park JH. 2018. Downregulation of NOP53 Ribosome Biogenesis Factor Leads to Abnormal Nuclear Division and Chromosomal Instability in Human Cervical Cancer Cells. Pathol Oncol Res. [DOI] [PubMed] [Google Scholar]
- Li Y, Lovett D, Zhang Q, Neelam S, Kuchibhotla RA, Zhu R, Gundersen GG, Lele TP, and Dickinson RB. 2015. Moving Cell Boundaries Drive Nuclear Shaping during Cell Spreading. Biophysical journal. 109:670–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipfert J, Klijnhout S, and Dekker NH. 2010. Torsional sensing of small-molecule binding using magnetic tweezers. Nucleic acids research. 38:7122–7132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loi M, Cenni V, Duchi S, Squarzoni S, Lopez-Otin C, Foisner R, Lattanzi G, and Capanni C. 2016. Barrier-to-autointegration factor (BAF) involvement in prelamin A-related chromatin organization changes. Oncotarget. 7:15662–15677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Romo-Bucheli D, Wang X, Janowczyk A, Ganesan S, Gilmore H, Rimm D, and Madabhushi A. 2018. Nuclear shape and orientation features from H&E images predict survival in early-stage estrogen receptor-positive breast cancers. Lab Invest. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, and Richmond TJ. 1997. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 389:251–260. [DOI] [PubMed] [Google Scholar]
- Luo X, Liu Y, Kubicek S, Myllyharju J, Tumber A, Ng S, Che KH, Podoll J, Heightman TD, Oppermann U, Schreiber SL, and Wang X. 2011. A selective inhibitor and probe of the cellular functions of Jumonji C domain-containing histone demethylases. J Am Chem Soc. 133:9451–9456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macfarlane MD, Looi JC, Walterfang M, Spulber G, Velakoulis D, Styner M, Crisby M, Orndahl E, Erkinjuntti T, Waldemar G, Garde E, Hennerici MG, Bäzner H, Blahak C, Wallin A, and Wahlund LO. 2015. Shape abnormalities of the caudate nucleus correlate with poorer gait and balance: results from a subset of the LADIS study. Am J Geriatr Psychiatry. 23:59–71.e51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machida S, Takizawa Y, Ishimaru M, Sugita Y, Sekine S, Nakayama JI, Wolf M, and Kurumizaka H. 2018. Structural Basis of Heterochromatin Formation by Human HP1. Mol Cell. 69:385–397 e388. [DOI] [PubMed] [Google Scholar]
- Maciejowski J, Li Y, Bosco N, Campbell PJ, and de Lange T. 2015. Chromothripsis and Kataegis Induced by Telomere Crisis. Cell. 163:1641–1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchion DC, Bicaku E, Daud AI, Sullivan DM, and Munster PN. 2005. Valproic acid alters chromatin structure by regulation of chromatin modulation proteins. Cancer research. 65:3815–3822. [DOI] [PubMed] [Google Scholar]
- McCord RP, Nazario-Toole A, Zhang H, Chines PS, Zhan Y, Erdos MR, Collins FS, Dekker J, and Cao K. 2013. Correlated alterations in genome organization, histone methylation, and DNA-lamin A/C interactions in Hutchinson-Gilford progeria syndrome. Genome research. 23:260–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meshorer E, and Misteli T. 2006. Chromatin in pluripotent embryonic stem cells and differentiation. Nat Rev Mol Cell Biol. 7:540–546. [DOI] [PubMed] [Google Scholar]
- Miranda TB, Cortez CC, Yoo CB, Liang G, Abe M, Kelly TK, Marquez VE, and Jones PA. 2009. DZNep is a global histone methylation inhibitor that reactivates developmental genes not silenced by DNA methylation. Mol Cancer Ther. 8:1579–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagayama K, and Fukuei T. 2019. Cyclic stretch-induced mechanical stress to the cell nucleus inhibits ultraviolet radiation-induced DNA damage. Biomech Model Mechanobiol. [DOI] [PubMed] [Google Scholar]
- Nava MM, Miroshnikova YA, Biggs LC, Whitefield DB, Metge F, Boucas J, Vihinen H, Jokitalo E, Li X, Garcia Arcos JM, Hoffmann B, Merkel R, Niessen CM, Dahl KN, and Wickstrom SA. 2020. Heterochromatin-Driven Nuclear Softening Protects the Genome against Mechanical Stress-Induced Damage. Cell. 181:800–817 e822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neelam S, Chancellor TJ, Li Y, Nickerson JA, Roux KJ, Dickinson RB, and Lele TP. 2015. Direct force probe reveals the mechanics of nuclear homeostasis in the mammalian cell. Proceedings of the National Academy of Sciences of the United States of America. 112:5720–5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou HD, Phan S, Deerinck TJ, Thor A, Ellisman MH, and O’Shea CC. 2017. ChromEMT: Visualizing 3D chromatin structure and compaction in interphase and mitotic cells. Science. 357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagliara S, Franze K, McClain CR, Wylde G, Fisher CL, Franklin RJM, Kabla AJ, Keyser UF, and Chalut KJ. 2014. Auxetic nuclei in embryonic stem cells exiting pluripotency. Nat Mater. 13:638–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajerowski JD, Dahl KN, Zhong FL, Sammak PJ, and Discher DE. 2007. Physical plasticity of the nucleus in stem cell differentiation. Proceedings of the National Academy of Sciences of the United States of America. 104:15619–15624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panorchan P, Schafer BW, Wirtz D, and Tseng Y. 2004. Nuclear envelope breakdown requires overcoming the mechanical integrity of the nuclear lamina. The Journal of biological chemistry. 279:43462–43467. [DOI] [PubMed] [Google Scholar]
- Papanicolaou GN, and Traut HF. 1997. The diagnostic value of vaginal smears in carcinoma of the uterus. 1941. Arch Pathol Lab Med. 121:211–224. [PubMed] [Google Scholar]
- Pfeifer CR, Xia Y, Zhu K, Liu D, Irianto J, Garcia VMM, Millan LMS, Niese B, Harding S, Deviri D, Greenberg RA, and Discher DE. 2018. Constricted migration increases DNA damage and independently represses cell cycle. Molecular biology of the cell. 29:1948–1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poleshko A, Mansfield KM, Burlingame CC, Andrake MD, Shah NR, and Katz RA. 2013. The human protein PRR14 tethers heterochromatin to the nuclear lamina during interphase and mitotic exit. Cell Rep. 5:292–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raab M, Gentili M, de Belly H, Thiam HR, Vargas P, Jimenez AJ, Lautenschlaeger F, Voituriez R, Lennon-Dumenil AM, Manel N, and Piel M. 2016. ESCRT III repairs nuclear envelope ruptures during cell migration to limit DNA damage and cell death. Science. 352:359–362. [DOI] [PubMed] [Google Scholar]
- Radhakrishnan A, Damodaran K, Soylemezoglu AC, Uhler C, and Shivashankar GV. 2017. Machine Learning for Nuclear Mechano-Morphometric Biomarkers in Cancer Diagnosis. Sci Rep. 7:17946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapisarda V, Malashchuk I, Asamaowei IE, Poterlowicz K, Fessing MY, Sharov AA, Karakesisoglou I, Botchkarev VA, and Mardaryev A. 2017. p63 Transcription Factor Regulates Nuclear Shape and Expression of Nuclear Envelope-Associated Genes in Epidermal Keratinocytes. J Invest Dermatol. 137:2157–2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robijns J, Molenberghs F, Sieprath T, Corne TD, Verschuuren M, and De Vos WH. 2016. In silico synchronization reveals regulators of nuclear ruptures in lamin A/C deficient model cells. Sci Rep. 6:30325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito M, Takamura Y, and Tamiya E. 2005. Nanoscale time-lapse AFM imaging in solution for DNA aggregation. NanoBiotechnology. 1:361–368. [Google Scholar]
- Saldana-Meyer R, Rodriguez-Hernaez J, Nishana M, Jacome-Lopez K, Nora EP, Bruneau BG, Furlan-Magaril M, Skok J, and Reinberg D. 2019. RNA interactions with CTCF are essential for its proper function. bioRxiv:530014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaffidi P, and Misteli T. 2006. Lamin A-dependent nuclear defects in human aging. Science. 312:1059–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schape J, Prausse S, Radmacher M, and Stick R. 2009. Influence of lamin A on the mechanical properties of amphibian oocyte nuclei measured by atomic force microscopy. Biophysical journal. 96:4319–4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiner SM, Koo PK, Zhao Y, Mochrie SG, and King MC. 2015. The tethering of chromatin to the nuclear envelope supports nuclear mechanics. Nat Commun. 6:7159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senigagliesi B, Penzo C, Severino LU, Maraspini R, Petrosino S, Morales-Navarrete H, Pobega E, Ambrosetti E, Parisse P, Pegoraro S, Manfioletti G, Casalis L, and Sgarra R. 2019. The High Mobility Group A1 (HMGA1) Chromatin Architectural Factor Modulates Nuclear Stiffness in Breast Cancer Cells. Int J Mol Sci. 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimamoto Y, Tamura S, Masumoto H, and Maeshima K. 2017. Nucleosome-nucleosome interactions via histone tails and linker DNA regulate nuclear rigidity. Molecular biology of the cell. 28:1580–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimi T, Butin-Israeli V, Adam SA, Hamanaka RB, Goldman AE, Lucas CA, Shumaker DK, Kosak ST, Chandel NS, and Goldman RD. 2011. The role of nuclear lamin B1 in cell proliferation and senescence. Genes & development. 25:2579–2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shumaker DK, Dechat T, Kohlmaier A, Adam SA, Bozovsky MR, Erdos MR, Eriksson M, Goldman AE, Khuon S, Collins FS, Jenuwein T, and Goldman RD. 2006. Mutant nuclear lamin A leads to progressive alterations of epigenetic control in premature aging. Proceedings of the National Academy of Sciences of the United States of America. 103:8703–8708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solovei I, Wang AS, Thanisch K, Schmidt CS, Krebs S, Zwerger M, Cohen TV, Devys D, Foisner R, Peichl L, Herrmann H, Blum H, Engelkamp D, Stewart CL, Leonhardt H, and Joffe B. 2013. LBR and lamin A/C sequentially tether peripheral heterochromatin and inversely regulate differentiation. Cell. 152:584–598. [DOI] [PubMed] [Google Scholar]
- Stephens AD, Banigan EJ, Adam SA, Goldman RD, and Marko JF. 2017. Chromatin and lamin A determine two different mechanical response regimes of the cell nucleus. Molecular biology of the cell. 28:1984–1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens AD, Banigan EJ, and Marko JF. 2018a. Separate roles for chromatin and lamins in nuclear mechanics. Nucleus. 9:119–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens AD, Banigan EJ, and Marko JF. 2019a. Chromatin’s physical properties shape the nucleus and its functions. Curr Opin Cell Biol. 58:76–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens AD, Liu PZ, Banigan EJ, Almassalha LM, Backman V, Adam SA, Goldman RD, and Marko JF. 2018b. Chromatin histone modifications and rigidity affect nuclear morphology independent of lamins. Molecular biology of the cell. 29:220–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens AD, Liu PZ, Kandula V, Chen H, Almassalha LM, Herman C, Backman V, O’Halloran T, Adam SA, Goldman RD, Banigan EJ, and Marko JF. 2019b. Physicochemical mechanotransduction alters nuclear shape and mechanics via heterochromatin formation. Molecular biology of the cell. 30:2320–2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swift J, Ivanovska IL, Buxboim A, Harada T, Dingal PC, Pinter J, Pajerowski JD, Spinler KR, Shin JW, Tewari M, Rehfeldt F, Speicher DW, and Discher DE. 2013. Nuclear lamin-A scales with tissue stiffness and enhances matrix-directed differentiation. Science. 341:1240104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamashunas AC, Tocco VJ, Matthews J, Zhang Q, Atanasova KR, Paschall L, Pathak S, Ratnayake R, Stephens AD, Luesch H, Licht JD, and Lele TP. 2020. High-throughput gene screen reveals modulators of nuclear shape. Molecular biology of the cell:mbcE19090520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamiello C, Kamps MA, van den Wijngaard A, Verstraeten VL, Baaijens FP, Broers JL, and Bouten CC. 2013. Soft substrates normalize nuclear morphology and prevent nuclear rupture in fibroblasts from a laminopathy patient with compound heterozygous LMNA mutations. Nucleus. 4:61–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur J, Fang H, Llagas T, Disteche CM, and Henikoff S. 2019. Architectural RNA is required for heterochromatin organization. bioRxiv:784835. [Google Scholar]
- Turgay Y, Eibauer M, Goldman AE, Shimi T, Khayat M, Ben-Harush K, Dubrovsky-Gaupp A, Sapra KT, Goldman RD, and Medalia O. 2017. The molecular architecture of lamins in somatic cells. Nature. 543:261–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas JD, Hatch EM, Anderson DJ, and Hetzer MW. 2012. Transient nuclear envelope rupturing during interphase in human cancer cells. Nucleus. 3:88–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaziri A, and Mofrad MR. 2007. Mechanics and deformation of the nucleus in micropipette aspiration experiment. J Biomech. 40:2053–2062. [DOI] [PubMed] [Google Scholar]
- Wang P, Dreger M, Madrazo E, Williams CJ, Samaniego R, Hodson NW, Monroy F, Baena E, Sanchez-Mateos P, Hurlstone A, and Redondo-Munoz J. 2018. WDR5 modulates cell motility and morphology and controls nuclear changes induced by a 3D environment. Proceedings of the National Academy of Sciences of the United States of America. 115:8581–8586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wren NS, Zhong Z, Schwartz RS, and Dahl KN. 2012. Modeling nuclear blebs in a nucleoskeleton of independent filament networks. Cell Mol Bioeng. 5:73–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Y, Ivanovska IL, Zhu K, Smith L, Irianto J, Pfeifer CR, Alvey CM, Ji J, Liu D, Cho S, Bennett RR, Liu AJ, Greenberg RA, and Discher DE. 2018. Nuclear rupture at sites of high curvature compromises retention of DNA repair factors. J Cell Biol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao B, Freedman BS, Miller KE, Heald R, and Marko JF. 2012. Histone H1 compacts DNA under force and during chromatin assembly. Molecular biology of the cell. 23:4864–4871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Maciejowski J, and de Lange T. 2017. Nuclear Envelope Rupture Is Enhanced by Loss of p53 or Rb. Mol Cancer Res. 15:1579–1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida M, Kijima M, Akita M, and Beppu T. 1990. Potent and specific inhibition of mammalian histone deacetylase both in vivo and in vitro by trichostatin A. The Journal of biological chemistry. 265:17174–17179. [PubMed] [Google Scholar]
- Zheng R, Ghirlando R, Lee MS, Mizuuchi K, Krause M, and Craigie R. 2000. Barrier-to-autointegration factor (BAF) bridges DNA in a discrete, higher-order nucleoprotein complex. Proceedings of the National Academy of Sciences of the United States of America. 97:8997–9002. [DOI] [PMC free article] [PubMed] [Google Scholar]