Abstract
Introduction
Kidney renal clear cell carcinoma (KIRC) has a high incidence globally, and its pathogenesis remains unclear. Long non-coding RNA (lncRNA), as a molecular sponge, participates in the regulation of competitive endogenous RNA (ceRNA). We aimed to construct a ceRNA network and screened out possible lncRNAs to predict KIRC prognosis.
Material and methods
All KIRC data were downloaded from the TCGA database and screened to find the possible target lncRNA; a ceRNA network was designed. Next, GO functional enrichment and KEGG pathway of differentially expressed mRNA related to lncRNA were performed. We used Kaplan-Meier curve analysis to predict the survival of these RNAs. We used Cox regression analysis to construct a model to predict KIRC prognosis.
Results
In the KIRC datasets, 1457 lncRNA, 54 miRNA and 2307 mRNA were screened out. The constructed ceRNA network contained 81 lncRNAs, nine miRNAs, and 17 mRNAs differentially expressed in KIRC. Survival analysis of all differentially expressed RNAs showed that 21 lncRNAs, four miRNAs, and two mRNAs were related to the overall survival rate. Cox regression analysis was performed again, and we found that eight lncRNAs were related to prognosis and used to construct predictive models. Three lnRNAs from independent samples were meaningful.
Conclusion
The construction of ceRNA network was involved in the process and transfer of KIRC, and three lncRNAs may be potential targets for predicting KIRC prognosis.
Introduction
Renal cell carcinoma (RCC) is a predominant form of cancer in humans, 3% of all cancers are RCC, with the highest incidence occurring in Western countries. Over the past 20 years, the global and European morbidity rate has increased at a rate of 2% per year. There are an estimated 99,200 new RCC patients in the European region and 39,100 deaths from kidney cancer in 2018 [1]. The incidence of renal clear cell carcinoma (KIRC) is very high, accounting for 80–90% of renal malignancies [2]. Although detection methods have improved with the development of ultrasound and computed tomography, many kidney lumps remain asymptomatic and will not be discovered until late in the disease. Compared with other RCC types, KIRC has a high recurrence and metastasis rate. Moreover, the 5-year survival rate of metastatic patients with KIRC is less than 10% [3]. KIRC also has few biomarkers. Therefore, the detection and identification of new and sensitive biomarkers are essential for improved prediction of progress and prognosis of metastatic KIRC.
Long non-coding RNA (lncRNA) is a subset of transcripts with a length of more than 200 non-protein coding nucleotides [4]. LncRNA occupies a large proportion of ncRNA. They have increasingly been related to many important biological processes, especially lncRNAs. They play important roles in chromatin modification, cell differentiation and proliferation, RNA processes, apoptosis and human diseases [5–9]. MiRNAs are highly conserved and interact with target mRNA to silence genes. In gene expression, other RNA transcripts are regulated by competing for endogenous RNA (ceRNA) through competition for shared miRNAs. As lncRNAs and mRNAs share a common miRNA response element, the relationship network of lncRNA-miRNA-mRNA is essential for regulating RNA expression. Furthermore, lncRNAs function as “molecular sponges” that directly or indirectly competitively binds to miRNA, eventually weakening the effect of miRNAs on mRNA [10–13].
The ceRNA network plays a regulatory role in many cancers of gastric cancers [14, 15], including pancreatic adenocarcinoma [16, 17], lung cancer [17–19] and colorectal cancer [20, 21]. In KIRC, the Cancer Genome Atlas (TCGA) database has been used to prove that several ceRNAs are significantly related to the overall KIRC survival rate. These studies analyzed the KIRC expression data in the TCGA database, screened out differential genes and constructed a ceRNA network, and used their clinical data to perform survival analysis on the key genes screened out. Their analysis results proved that the differential expression of the selected influencing factors impacts the long-term prognosis of patients [22, 23]. Patients with ccRCC with no PTENP1 expression also have a lower survival rate. PTENP1 acts as a competitive endogenous RNA (ceRNA) in ccRCC, thereby inhibiting cancer development [24]. However, they only obtained the prognostic ceRNA and did not study the construction of prediction models in their research. Here, we first screened all differentially expressed lncRNA (DElncRNA), differentially expressed miRNA (DEmiRNA) and differentially expressed mRNA (DEmRNA), and used them to construct a ceRNA network. Based on this, we screened eight possible lncRNAs as prognostic biomarkers for KIRC through survival analysis and COX regression analysis. Additionally, we established and validated a predictive model of KIRC prognosis based on the lncRNAs. The established model may be a potential method to predict the prognosis of KIRC.
Materials and methods
TCGA data collection
RNA sequence data and clinical information were obtained from GDC data portal screening (https://portal.gdc.cancer.gov/) to perform an integrated analysis using the Data Transfer Tool (provided by GDC Apps) according to the published guidelines provided by TCGA (https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga) [25]. TCGA is a landmark cancer genome project with molecular characteristics of more than 20,000 primary cancers and matched normal samples covering 33 cancer types. The data retrieved from these databases help us improve diagnostic levels and cancer treatment. Our current research was based on the requirements of the guidelines proposed in the TCGA database. Since RNA sequencing data was obtained directly from TCGA, the approval of the ethics committee was not required. All original data are in S1 File.
RNA sequence data retrieval and screening of differentially expressed mRNA, miRNA, and lncRNA in KIRC
We used Perl (https://www.perl.org/) and R (https://www.r-project.org/) to analyze and process RNA sequencing data [26, 27]. All the Source codes used are in the S2 File. We merged data from tumor and normal samples through Perl software for subsequent analysis. We collected 611 samples of RNA sequencing data (including 539 KIRC tumor tissues and 72 normal tissues) and 616 samples of miRNA sequencing data (including 545 KIRC tumor tissues and 71 normal tissues; Table 1). To screen for DEmRNA, DEmiRNA, and DElncRNA, we used the “edgeR” package of R software to compare KIRC with normal samples. |log2FC|>2 and P-value < 0.01 was the cut-off standard we determined. We used the “gplots” package to draw the volcano and heat maps of the obtained DEmRNA, DEmiRNA, and DElncRNA.
Table 1. Characteristics and distribution of patients with kidney clear cell carcinoma in the TCGA-KIRC cohort.
| Variables | mRNA-lncRNA (n = 530) | miRNA (n = 516) | |
|---|---|---|---|
| age | <65 | 245 | 326 |
| ≥65 | 285 | 190 | |
| gender | male | 344 | 335 |
| female | 186 | 181 | |
| M | M0 | 420 | 406 |
| M1 | 78 | 78 | |
| MX | 30 | 30 | |
| NA | 2 | 2 | |
| N | N0 | 239 | 228 |
| N1 | 16 | 17 | |
| NX | 275 | 271 | |
| T | T1 | 271 | 259 |
| T2 | 69 | 67 | |
| T3 | 179 | 179 | |
| T4 | 11 | 11 | |
| stage | stage I | 265 | 253 |
| stage II | 57 | 55 | |
| stage III | 123 | 123 | |
| stage IV | 82 | 82 | |
| NA | 3 | 3 | |
NA: Not available.
Correlation analysis of lncRNAs, miRNAs, and mRNAs and construction of the ceRNA network in KIRC
To study the relationship between different RNAs, we used them to assemble a ceRNA network in KIRC. The ceRNA network was based on the theory that ceRNA combined with miRNA through response elements to affect mRNA gene expression. We first used the starBase database to convert the miRNA format. Then we used Perl software to explore DEmiRNA interacting with DElncRNA in the miRcode database (http://www.mircode.org/). Next, miRTarBase (http://mirtarbase.mbc.nctu.edu.tw/php/index.php), miRDB (http://www.mirdb.org/) and TargetScan (http://www.targetscan.org/vert_72/) databases were performed to obtain miRNA targeted mRNAs by Perl software with the criteria for each target gene to appear in three different databases [28–30]. Through the “VennDiagrams” package in R software, the predicted miRNA-target genes and DEmRNA were intersected. Based on the selected miRNA-target genes and the determined lncRNA-miRNA relationship, We was assembled ceRNA network to improve the visual analysis of results. Simultaneously, we used Cytoscape (http://www.cytoscape.org/, verson4.0.2) to visualize the results [31]. All figures were typeset using Microsoft PowerPoint software (https://www.microsoft.com/zh-cn/microsoft-365/powerpoint).
Survival analysis and construction of lncRNA related predictive model
We once again found the patient’s clinical information from the database. The long-term survival of patients with KIRC was analyzed by Kaplan-Meier curve analysis. We used the "survival" software package of R software to analyze the RNA in the ceRNA network. The established P<0.01. We subsequently selected some lncRNAs related to overall survival (P<0.001) according to established criteria as prognostic lncRNA signature candidates for multivariate Cox regression analysis [32]. According to the median risk assessment, patients with KIRC were classified into two cohorts. Receiver operating characteristics (ROC) were used to predict the impact of lncRNA signals (high risk and low risk) on survival [33]. Subsequently, we drew the ROC curve by calculating the area under the curve (AUC) under the binomial exact confidence interval [34].
Functional enrichment analysis of differentially expressed mRNAs in KIRC
To clarify the gene functions, the function enrichment of DEmRNA related to lncRNA in ceRNA network was analyzed by Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) using the Database for Annotation, Visualization, and Integrated Discovery (DAVID) database (https://david.ncifcrf.gov/home.jsp) and the KOBAS 3.0 online tool (http://kobas.cbi.pku.edu.cn/kobas3) [35, 36]. Additionally, the GOCircle, GOChord, and GOCluster drawing functions of the "GOplot" of R software package were used for expression and functional enrichment analyses [37].
Reconstruction of the ceRNA sub-network of prognostic in KIRC
To clarify the regulatory mechanism between the potential prognosis of lncRNA and related miRNA and mRNA, we used Cytoscape to extract RNA with prognosis differential to construct a ceRNA sub-network. We also used the Sankey diagram constructed by the “ggalluival” package for visual analysis. All RNA involved were related to the potential prognosis of lncRNA.
Validation of independent data sets
We selected an independent data set from the Gene Expression Omnibus (GEO) database to verify the results. GSE96574 was used by us for verification. We used Graphpad Prism8 software for statistical analysis and data visualization.
Statistical analysis
All data were expressed as the means± standard deviation (SD). GraphPad Prism 8.0 (GraphPad Software, San Diego, CA, USA) and R software were used for all statistical analyses. Two-tailed Student’s t-test was used to assess the significance of differences between the two groups. The survival analysis results were obtained from the “survival” package. Cox regression analysis was used to screen lncRNA related to prognosis in the ceRNA network and can be used as a target to predict KIRC prognosis. Unless otherwise stated, statistical significance was set at P<0.05.
Results
Screening of differentially expressed RNAs in KIRC
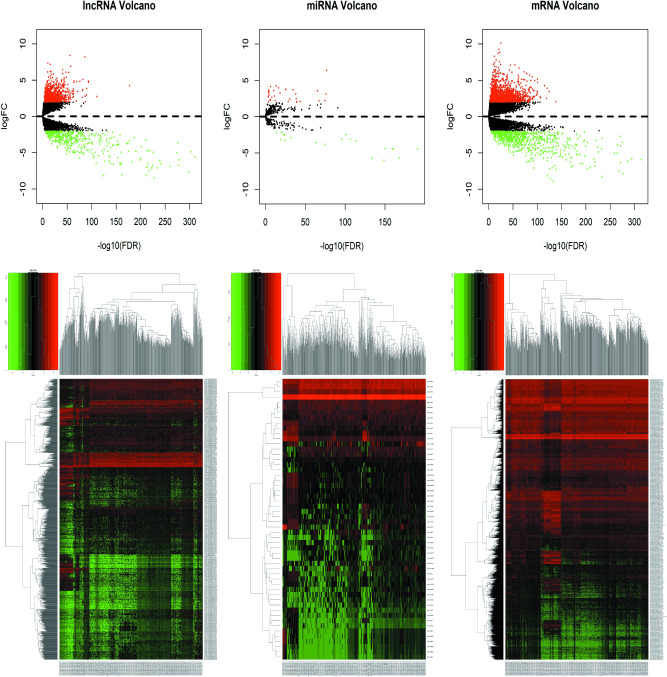
According to the screening conditions (|log2FC| > 2, P-value < 0.01), 2307 DEmRNAs, 1457 DElncRNAs, and 54 DEmiRNAs were screened out. Tables 2–4 showed the top 20 DElncRNAs, DEmiRNAs, and DEmRNAs. A volcano map (Fig 1A–1C) was drawn to show the distribution of DElcnRNA, DEmiRNA, and DEmRNA in the -log10 (FDR) and logFC dimensions. The hierarchical cluster analysis heat map showed all differentially expressed RNA between KIRC tissues and normal tissues (Fig 1D–1F).
Table 2. Top 20 upregulated and downregulated lncRNA in patients with KIRC.
| lncRNA | logFC | PValue | FDR |
|---|---|---|---|
| AP005432.2 | -6.757905074 | 0 | 0 |
| AC019080.1 | -3.886047663 | 0 | 0 |
| LINC01762 | -4.874338157 | 4.24E-307 | 9.35E-304 |
| AC090709.1 | -7.65872183 | 3.27E-302 | 5.77E-299 |
| AP000696.1 | -5.997594408 | 8.83E-283 | 1.30E-279 |
| AC009035.1 | -6.713927435 | 6.55E-281 | 8.26E-278 |
| FAM242C | -3.661469455 | 5.56E-280 | 6.13E-277 |
| AC068631.1 | -5.033045794 | 6.01E-278 | 5.90E-275 |
| AC023421.1 | -6.883709775 | 8.55E-264 | 7.55E-261 |
| LINC01378 | -6.507363007 | 1.51E-245 | 1.21E-242 |
| AC007993.2 | -6.120512627 | 7.66E-243 | 5.63E-240 |
| AC073172.1 | -7.080454189 | 1.02E-237 | 6.90E-235 |
| AC079310.1 | -8.482248422 | 6.71E-232 | 4.23E-229 |
| AC105384.1 | -4.959173524 | 4.96E-229 | 2.92E-226 |
| AL139280.1 | -6.772062579 | 5.11E-228 | 2.82E-225 |
| LINC01571 | -7.099142297 | 4.74E-226 | 2.46E-223 |
| LINC02410 | -4.751856083 | 2.42E-222 | 1.19E-219 |
| AC124017.1 | -7.440808148 | 1.14E-220 | 5.28E-218 |
| AC006441.4 | -4.676557282 | 5.10E-218 | 2.25E-215 |
| LINC01020 | -6.506384384 | 6.96E-218 | 2.93E-215 |
| PVT1 | 4.238882075 | 1.31E-180 | 3.51E-178 |
| SLC16A1-AS1 | 2.729511989 | 1.56E-113 | 1.62E-111 |
| AC010655.2 | 4.842063622 | 3.92E-96 | 3.17E-94 |
| AL391845.2 | 4.704856417 | 6.04E-96 | 4.84E-94 |
| CDKN2B-AS1 | 3.856453462 | 2.86E-92 | 2.12E-90 |
| MIR210HG | 2.956237343 | 6.64E-92 | 4.89E-90 |
| WAKMAR2 | 2.004757779 | 1.31E-89 | 9.12E-88 |
| DARS-AS1 | 2.770230091 | 6.90E-89 | 4.68E-87 |
| TTC21B-AS1 | 8.176350088 | 6.94E-89 | 4.68E-87 |
| LINC02048 | 5.09657003 | 2.15E-84 | 1.33E-82 |
| DGCR9 | 3.577728993 | 5.00E-84 | 3.00E-82 |
| GAS6-AS1 | 3.668197986 | 6.34E-84 | 3.76E-82 |
| SAP30-DT | 2.598462928 | 3.25E-83 | 1.89E-81 |
| AC137834.2 | 4.833231799 | 2.41E-78 | 1.25E-76 |
| AC026369.2 | 3.439940746 | 1.27E-73 | 5.88E-72 |
| LINC00887 | 4.812455488 | 8.95E-72 | 4.01E-70 |
| AC138207.5 | 2.110114795 | 9.00E-71 | 3.87E-69 |
| AC079760.2 | 4.868343482 | 3.36E-66 | 1.32E-64 |
| SFTA1P | 4.67347273 | 4.40E-66 | 1.71E-64 |
| AC019069.1 | 3.108894249 | 2.01E-65 | 7.72E-64 |
FC: Fold change, FDR: False discovery rate.
Table 4. Top 20 upregulated and downregulated miRNA in patients with KIRC.
| miRNA | logFC | PValue | FDR |
|---|---|---|---|
| hsa-mir-508 | -4.413080209 | 1.66E-194 | 7.97E-192 |
| hsa-mir-506 | -5.661839583 | 6.75E-171 | 1.62E-168 |
| hsa-mir-514a-1 | -4.423272889 | 2.21E-163 | 3.53E-161 |
| hsa-mir-514a-3 | -4.448579466 | 3.71E-162 | 4.44E-160 |
| hsa-mir-514a-2 | -4.392090737 | 1.57E-152 | 1.50E-150 |
| hsa-mir-514b | -6.012465545 | 2.85E-151 | 2.28E-149 |
| hsa-mir-934 | -5.744736952 | 1.84E-136 | 1.26E-134 |
| hsa-mir-509-3 | -3.348871994 | 1.86E-116 | 1.11E-114 |
| hsa-mir-509-2 | -3.116798109 | 3.85E-109 | 2.05E-107 |
| hsa-mir-362 | -2.4997835 | 5.73E-104 | 2.75E-102 |
| hsa-mir-509-1 | -3.099261399 | 5.61E-98 | 2.44E-96 |
| hsa-mir-129-1 | -3.810982115 | 7.60E-77 | 2.27E-75 |
| hsa-mir-129-2 | -3.582430837 | 6.41E-66 | 1.33E-64 |
| hsa-mir-200c | -2.886127557 | 1.44E-33 | 1.27E-32 |
| hsa-mir-216b | -3.245682342 | 2.16E-26 | 1.46E-25 |
| hsa-mir-203b | -2.51253395 | 1.07E-25 | 7.04E-25 |
| hsa-mir-138-1 | -2.446678631 | 8.94E-24 | 5.49E-23 |
| hsa-mir-138-2 | -2.188284603 | 4.19E-22 | 2.30E-21 |
| hsa-mir-1251 | -2.16487151 | 1.61E-21 | 8.57E-21 |
| hsa-mir-184 | -3.137223909 | 8.80E-21 | 4.44E-20 |
| hsa-mir-122 | 6.375970805 | 4.90E-79 | 1.80E-77 |
| hsa-mir-210 | 3.106323197 | 2.65E-78 | 9.06E-77 |
| hsa-mir-21 | 2.214057836 | 5.29E-77 | 1.69E-75 |
| hsa-mir-584 | 2.175477131 | 2.38E-69 | 5.70E-68 |
| hsa-mir-155 | 3.563706474 | 2.20E-67 | 4.79E-66 |
| hsa-mir-4772 | 2.073804082 | 1.60E-45 | 2.40E-44 |
| hsa-mir-452 | 2.014972583 | 1.38E-41 | 1.94E-40 |
| hsa-mir-142 | 2.019440538 | 7.28E-41 | 9.97E-40 |
| hsa-mir-224 | 2.453810789 | 1.72E-40 | 2.29E-39 |
| hsa-mir-592 | 3.12806078 | 7.20E-40 | 9.33E-39 |
| hsa-mir-885 | 3.687124949 | 2.37E-36 | 2.42E-35 |
| hsa-mir-3941 | 2.377116049 | 2.76E-32 | 2.32E-31 |
| hsa-mir-6509 | 2.080279385 | 2.92E-30 | 2.33E-29 |
| hsa-mir-4773-1 | 3.717540957 | 3.22E-27 | 2.23E-26 |
| hsa-mir-4773-2 | 3.781228894 | 6.51E-27 | 4.46E-26 |
| hsa-mir-4652 | 3.254741969 | 1.27E-19 | 6.09E-19 |
| hsa-mir-1293 | 3.963710459 | 9.21E-18 | 3.98E-17 |
| hsa-mir-875 | 4.236303576 | 1.30E-14 | 4.78E-14 |
| hsa-mir-3591 | 2.191496424 | 1.88E-13 | 6.67E-13 |
| hsa-mir-374c | 2.832071207 | 1.81E-12 | 5.93E-12 |
FC: Fold change, FDR: False discovery rate.
Fig 1. Volcano and heat maps of all differentially expressed RNA between KIRC and normal tissues.
(a) Volcano map of DElncRNA in KIRC. (b) Volcano map of DEmiRNA in KIRC. (c) Volcano map of DEmRNA in KIRC. (d) Hierarchical clustering heat map of DElncRNA in KIRC. (e) Hierarchical clustering heat map of DEmiRNA in KIRC. (f) Hierarchical clustering heat map of DEmRNA in KIRC. FDR: False discovery rate.
Table 3. Top 20 upregulated and downregulated mRNA in patients with KIRC.
| mRNA | logFC | PValue | FDR |
|---|---|---|---|
| TMEM238L | -7.078308523 | 0 | 0 |
| ACP3 | -5.566157769 | 0 | 0 |
| MFSD4A | -5.237894745 | 0 | 0 |
| SIM2 | -4.49889604 | 0 | 0 |
| GPC5 | -5.82678567 | 5.90E-299 | 1.77E-295 |
| KCNJ10 | -6.175499235 | 6.69E-290 | 1.72E-286 |
| ELF5 | -7.853566134 | 7.87E-281 | 1.77E-277 |
| ADGRF3 | -3.527227811 | 2.91E-279 | 5.83E-276 |
| DDN | -6.340093857 | 1.57E-275 | 2.83E-272 |
| ATP1A1 | -2.749741864 | 3.95E-275 | 6.16E-272 |
| MYLK3 | -4.024276299 | 4.10E-275 | 6.16E-272 |
| CALB1 | -7.504477557 | 4.96E-273 | 6.88E-270 |
| HSPA2 | -3.886122862 | 1.02E-268 | 1.32E-265 |
| IRX2 | -5.021990125 | 1.66E-268 | 1.99E-265 |
| ENPP6 | -5.01847797 | 1.34E-262 | 1.51E-259 |
| SLC12A1 | -8.26530607 | 8.10E-238 | 8.58E-235 |
| MTURN | -3.027205604 | 2.49E-237 | 2.49E-234 |
| SLC4A11 | -4.437954149 | 5.85E-236 | 5.54E-233 |
| TRPV6 | -4.864989485 | 2.91E-230 | 2.50E-227 |
| FGF1 | -4.201610742 | 5.05E-228 | 4.13E-225 |
| DDB2 | 2.049347708 | 3.57E-141 | 6.77E-139 |
| NOL3 | 3.444266195 | 1.49E-127 | 2.01E-125 |
| SPAG4 | 3.90578212 | 4.21E-123 | 5.02E-121 |
| SAP30 | 2.503406987 | 7.80E-122 | 9.07E-120 |
| EGLN3 | 4.276239797 | 9.80E-118 | 1.07E-115 |
| VIM | 2.593518266 | 1.77E-109 | 1.63E-107 |
| ARHGEF39 | 2.742244652 | 2.85E-108 | 2.54E-106 |
| HILPDA | 4.690335522 | 3.29E-103 | 2.63E-101 |
| GABRD | 5.150018388 | 4.66E-103 | 3.68E-101 |
| NETO2 | 3.327264726 | 1.16E-102 | 9.11E-101 |
| VEGFA | 3.503766201 | 2.35E-102 | 1.83E-100 |
| SLC16A3 | 3.061709145 | 4.47E-102 | 3.44E-100 |
| LILRB1 | 2.942896587 | 1.38E-100 | 1.05E-98 |
| CDKN2A | 4.947667844 | 7.98E-97 | 5.55E-95 |
| ARHGAP22 | 2.776844532 | 9.79E-97 | 6.79E-95 |
| AGAP2 | 2.717081495 | 1.43E-96 | 9.84E-95 |
| CXCR4 | 2.800644677 | 7.02E-96 | 4.81E-94 |
| ST8SIA4 | 3.592159173 | 7.67E-96 | 5.23E-94 |
| COL5A3 | 3.566548436 | 3.06E-94 | 2.01E-92 |
| P2RX7 | 2.699430547 | 5.84E-94 | 3.79E-92 |
FC: Fold change, FDR: False discovery rate.
Construction of the ceRNA network in KIRC
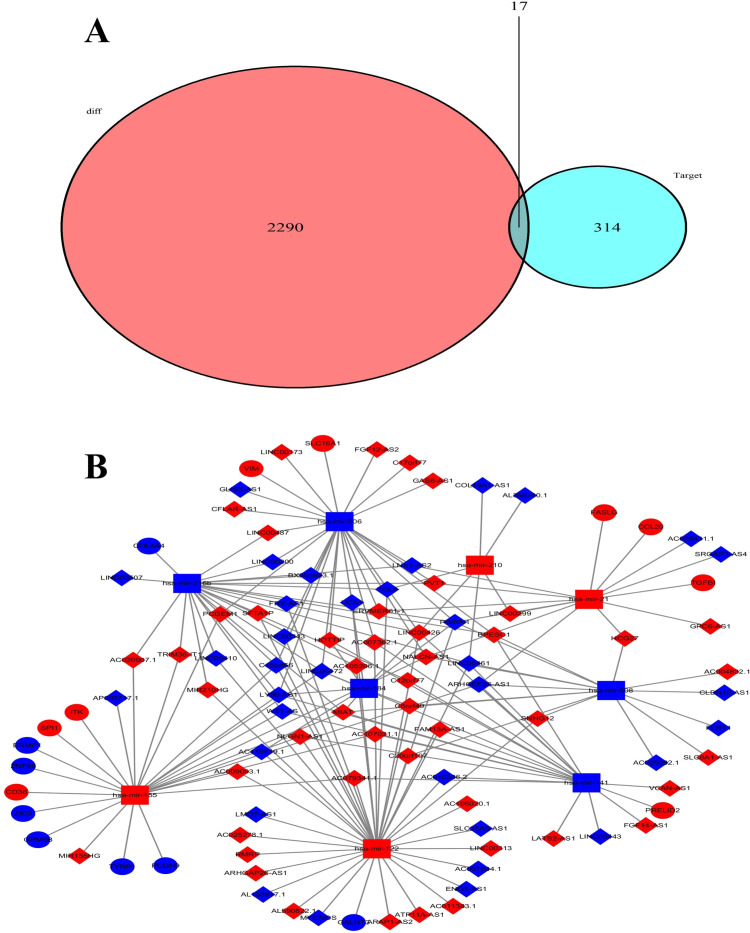
To better explore the interaction between differentially expressed RNA in KIRC, we constructed a lncRNA-mediated ceRNA regulatory network to analyze the interaction between them visually. First, We used the miRCode database and paired 1456 DElncRNAs with Perl software to screen out 161 DElncRNA and miRNA relationship pairs. We screened 9 out of 54 DEmiRNAs to interact with 81 DElncRNAs and used the starBase database to convert the miRNA format. Table 5 showed the relationship between lncRNA and miRNA in patients with KIRC. All nine possible target mRNAs of DEmiRNA were screened using Perl software based on three miRNA databases. According to the specified screening conditions, a total of 337 potential mRNA targets were found. Each target appeared in three different miRNA databases. The representative relationship between miRNA and mRNA in patients with KIRC was shown in Table 6. Then we used the “VennDiagrams” package of R software to hybridize these mRNA targets with 2307 identified DEmRNAs to select 16 cross-over genes in KIRC (Fig 2A). As an open-source software platform, Cytoscape is widely used to integrate any type of attribute data to analyze complex network structures visually. We constructed a KIRC ceRNA regulatory network mediated by lncRNA, including 81 DElncRNAs, nine DEmiRNAs, and 17 DEmRNAs. Furthermore, we used Cytoscape to analyze and process the network visually (Fig 2B). The content of the entire ceRNA network included 107 nodes and 178 edges. The flow chart of the entire construction process is shown in Fig 3.
Table 5. Representative relationships between lncRNAs and miRNAs in patients with kidney renal clear cell carcinoma.
| miRNA | lncRNA |
|---|---|
| hsa-mir-122 | C20orf197,C15orf56,AC105206.1,AC007362.1,LINC00313,TCL6, AC009093.1,SNHG12,RMRP,AL590822.1,AC105020.1,AC025278.1,UCA1, AC010336.2,SLC25A5-AS1,SFTA1P,ARHGAP26-AS1,LINC00343, AC079341.1,NLGN1-AS1,LMO7-AS1,ENO1-AS1,LINC00410,ATP11A-AS1,MYCNOS,FRY-AS1,LINC00426,AC011383.1,AL158817.1,AC007834.1, ARAP1-AS2,LINC00461,MIR210HG,FAM13A-AS1,C8orf49,AC110619.1, PWRN1 |
| hsa-mir-141 | AC007362.1,WT1-AS,AC010336.2,LY86-AS1,MIAT,C12orf77,AC079341.1,NLGN1-AS1,LINC00443,AC107021.1,BPESC1,LINC00472,LATS2-AS1, FGF14-AS1,LINC00426,ARHGEF26-AS1,HOTTIP,LINC00461, FAM13A-AS1,VCAN-AS1 |
| hsa-mir-155 | C15orf56,AC020907.1,WT1-AS,AC009093.1,LY86-AS1,MIAT,AC079341.1,PCGEM1,NLGN1-AS1,AC107021.1,NALCN-AS1,LINC00472,MIR155HG,ARHGEF26-AS1,AP005717.1,TRIM36-IT1 |
| hsa-mir-184 | UCA1,LY86-AS1,MIAT,LINC00472,LINC00426,HOTTIP,C8orf49, AC110619.1,PWRN1 |
| hsa-mir-21 | HCG27,ERVMER61-1,NALCN-AS1,SRGAP3-AS4,GPC6-AS1,LINC00299, ARHGEF26-AS1,AC025431.1,LNX1-AS2,PVT1,PWRN1 |
| hsa-mir-210 | COL18A1-AS1,TCL6,AL356740.1,LINC00299,LINC00426,LINC00461 |
| hsa-mir-216b | C15orf56,AC105206.1,AC020907.1,WT1-AS,TCL6,BX255923.1, LINC00487,LY86-AS1,SFTA1P,MIAT,C12orf77,LINC00200,LINC00410, BPESC1,LINC00472,LINC00426,LINC00461,MIR210HG,LNX1-AS2,PVT1, TRIM36-IT1,LINC00507,PWRN1 |
| hsa-mir-506 | C15orf56,AC007362.1,C17orf77,BX255923.1,LINC00173,LINC00487, UCA1,LY86-AS1,CFLAR-AS1,LINC00343,PCGEM1,LINC00200,FGF12-AS2, ERVMER61-1,BPESC1,NALCN-AS1,LINC00472,GAS6-AS1,GLIS3-AS1, LINC00426,HOTTIP,LNX1-AS2,PVT1,PWRN1 |
| hsa-mir-508 | FLRT1,C20orf197,SNHG12,AC004832.1,HCG27,AC005082.1,CLDN10-AS1,MIAT,SLC6A1-AS1,BPESC1,NALCN-AS1,ARHGEF26-AS1,LINC00461, C8orf49,PWRN1 |
Table 6. Representative relationships between miRNAs and mRNAs in patients with kidney renal clear cell carcinoma.
| miRNA | mRNA |
|---|---|
| hsa-mir-122 | GALNT3 |
| hsa-mir-141 | PRELID2 |
| hsa-mir-155 | ERMP1,PCDH9,TYRP1,SPI1,GPM6B,ITK,CD36,ZNF98,ZIC3 |
| hsa-mir-21 | FASLG,TGFBI,CCL20 |
| hsa-mir-216b | COL4A4 |
| hsa-mir-506 | VIM,SLC16A1 |
Fig 2. Venn diagram of mRNA and ceRNA networks in KIRC.
(a) The intersection of the DEmRNAs and the target mRNAs constituted the Venn diagram. (b) The ceRNA network was composed of DElncRNA, DEmiRNA, and DEmRNA. The diamond was lncRNA, the rectangle was miRNA, and the ellipse was mRNA. Red indicated upregulation, and blue indicated downregulation.
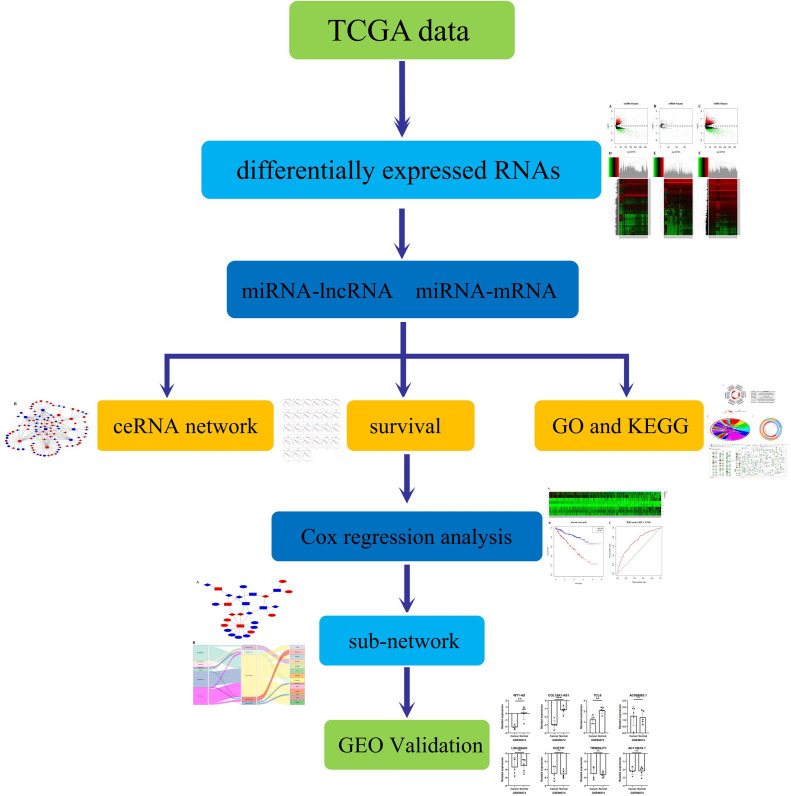
Fig 3. Flow chart of the entire construction process in KIRC.
The screening criteria for DEmRNAs, DEmiRNAs and DElncRNAs were |log2FC| > 2 and P-value < 0.01. The lncRNA-miRNA interaction was predicted through the miRcode database. lncRNAs unrelated to DEmiRNAs were deleted. The mRNAs targeted by miRNA were predicted using miRTarBase miRDB and TargetScan 3 databases. mRNAs unrelated to DEmRNAs were deleted. The ceRNA network was completed.
Screening of survival-related RNA in ceRNA network of KIRC
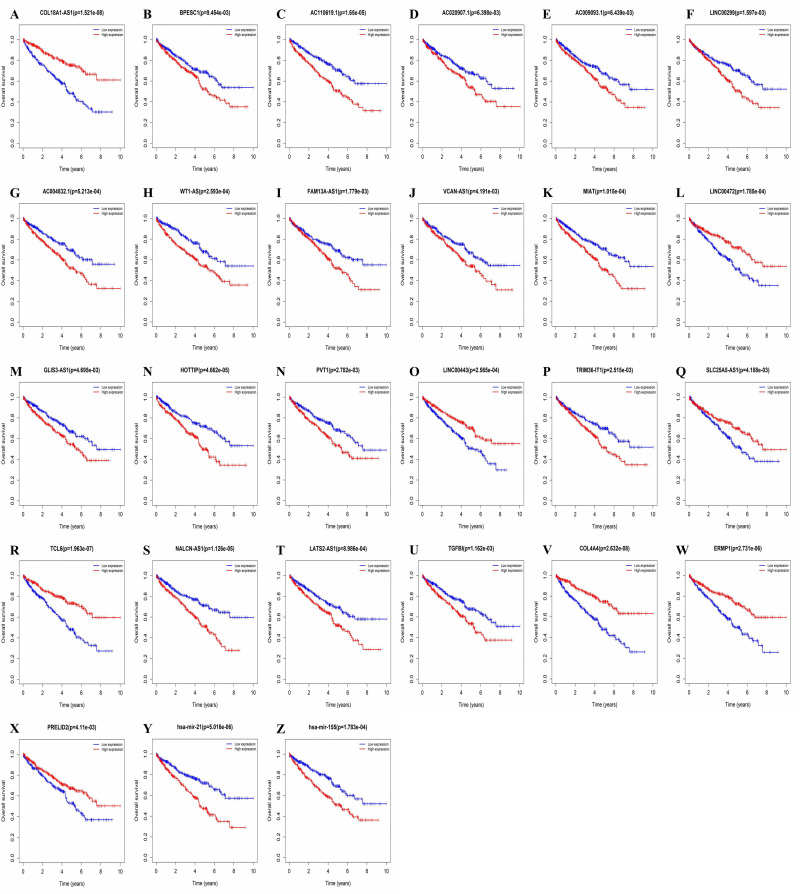
To find lncRNAs, miRNAs, and mRNAs related to prognosis, we used the “Survival” package to evaluate their relationship with the overall survival rate of patients with KIRC. We once again downloaded the clinical data of patients with KIRC from the TCGA database for survival analysis. The results showed that 21 of the 81 DElncRNAs (COL18A1-AS1, BPESC1, AC110619.1, AC020907.1, AC009093.1, LINC00299, AC004832.1, WT1-AS, FAM13A-AS1, VCAN-AS1, MIAT, LINC00472, GLIS3-AS1, HOTTIP, PVT1, LINC00443, TRIM36-IT1, SLC25A5-AS1, TCL6, and NALCN-AS1) were related to overall survival (Fig 4A–4T). Of the 49 DEmRNAs, a total of four (TGFBI, COL4A4, ERMP1, and PRELID2) were confirmed to be related to the overall KIRC survival (Fig 4U–4X). The analysis showed that two DEmiRNAs (hsa-mir-21 and hsa-mir-155) were related to the overall KIRC survival (Fig 4Y and 4Z). The threshold for all survival analyses was set to P-value < 0.01.
Fig 4. Kaplan–Meier curve analysis and overall survival rate of DElncRNA, DEmRNA, DEmiRNA in KIRC.
(a-z) Represented COL18A1-AS1, BPESC1, AC110619.1, AC020907.1, AC009093.1, LINC00299, AC004832.1, WT1-AS, FAM13A-AS1, VCAN-AS1, MIAT, LINC00472, GLIS3-AS1, HOTTIP, PVT1, LINC00443, TRIM36-IT1, SLC25A5-AS1, TCL6, NALCN-AS1, TGFBI, COL4A4, ERMP1, PRELID2, hsa-mir-21, hsa-mir-155.
Functional enrichment and KEGG analysis of differentially expressed mRNA in KIRC
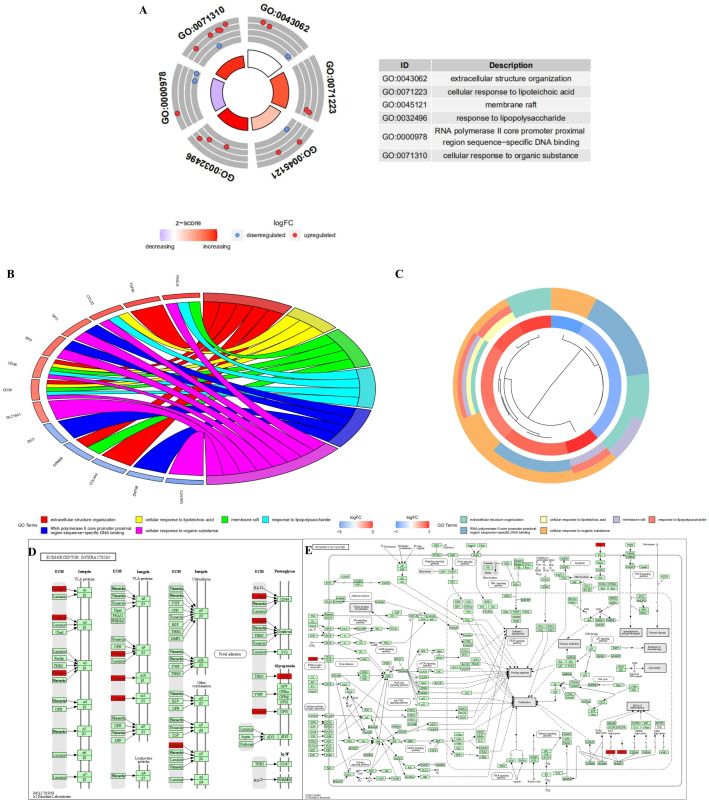
We performed GO and KEGG analyses using the DAVID database and the KOBAS3.0 online tool to clarify the biological function of DEmRNAs related to lncRNAs. The analysis showed that there were six GO projects in total (P<0.05) and seven KEGG pathways (corrected P <0.05; Tables 7 and 8). In detail, six important GO terms were enriched, suggesting that these DEmRNAs played an essential role in tumors (Fig 5A). Fig 5B shows the interaction between statistically different mRNAs and their related GO terms. Fig 5C shows a cluster analysis of statistically significant mRNA and related GO. KEGG analysis suggested that the two most important were the “ECM-receptor interaction” and “Pathways in cancer” pathways, indicating that these DEmRNAs had an essential role in KIRC (Fig 5E and 5F).
Table 7. GO terms of DEmRNA in kidney renal clear cell carcinoma.
| Category | ID | Term | Genes | P |
|---|---|---|---|---|
| BP | GO:0043062 | extracellular structure organization | COL4A4, CD36, TGFBI, GPM6B | 0.003721138 |
| BP | GO:0071223 | cellular response to lipoteichoic acid | CD36, CCL20 | 0.008617542 |
| CC | GO:0045121 | membrane raft | CD36, FASLG, GPM6B | 0.032301155 |
| BP | GO:0032496 | response to lipopolysaccharide | CD36, CCL20, FASLG | 0.033279982 |
| MF | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding | SPI1, ZNF98, ZIC3 | 0.045468359 |
| BP | GO:0071310 | cellular response to organic substance | GALNT3, SLC16A1, CD36, CCL20, SPI1, FASLG | 0.048776595 |
BP: Biological process, CC: Cellular Component, MF: Molecular Function.
Table 8. KEGG pathway of DEmRNA in kidney renal clear cell carcinoma.
| ID | Term | Genes | Corrected P-Value |
|---|---|---|---|
| hsa04512 | ECM-receptor interaction | COL4A4,CD36 | 0.018021432 |
| hsa05200 | Pathways in cancer | SPI1,COL4A4,FASLG | 0.022038886 |
| hsa04062 | Chemokine signaling pathway | CCL20,ITK | 0.032483893 |
| hsa04060 | Cytokine-cytokine receptor interaction | CCL20,FASLG | 0.033020964 |
| hsa05165 | Human papillomavirus infection | COL4A4,FASLG | 0.033605855 |
| hsa04151 | PI3K-Akt signaling pathway | COL4A4,FASLG | 0.03384518 |
| hsa05168 | Herpes simplex virus 1 infection | FASLG,ZNF98 | 0.045381132 |
Fig 5. Functional enrichment and KEGG analysis of DEmRNAs related to lncRNA in KIRC.
(a-c) The GOCircle, GOChord, and GOCluster diagram of DEmRNAs in KIRC. (d and e) The expression pattern of “ECM-receptor interaction” and “pathways in cancer”.
Screening of five important lncRNAs related to prognosis inpatients with KIRC
First, we used Perl software to perform univariate Cox regression analysis, a total of 14 lncRNAs were screened out (p <0.001). Next, the "survival" software package of R software was used to perform the next step of multiple Cox regression analysis on the selected lncRNA. As a result, eight lncRNAs were screened out to build a prediction model. The linear combination of the expression of eight lncRNAs was used to build a predictive model (Table 9).
Table 9. Multivariate cox regression analysis was performed on eight prognostic lncRNAs of patients with KIRC.
| lncRNA | coef | exp(coef) | se(coef) | z | p |
|---|---|---|---|---|---|
| WT1-AS | 0.08275 | 1.08628 | 0.03851 | 2.149 | 0.03164 |
| COL18A1-AS1 | -0.18301 | 0.83276 | 0.05843 | -3.132 | 0.001736 |
| TCL6 | -0.10542 | 0.89995 | 0.03475 | -3.034 | 0.002416 |
| AC009093.1 | 0.10326 | 1.10878 | 0.06151 | 1.679 | 0.093195 |
| LINC00443 | -0.32305 | 0.72394 | 0.08576 | -3.767 | 0.000165 |
| HOTTIP | 0.09631 | 1.1011 | 0.05117 | 1.882 | 0.059799 |
| TRIM36-IT1 | 0.11648 | 1.12353 | 0.07629 | 1.527 | 0.126802 |
| AC110619.1 | 0.10132 | 1.10663 | 0.05841 | 1.735 | 0.082796 |
coef: coefficient, exp: Exponential, se: Standard Error.
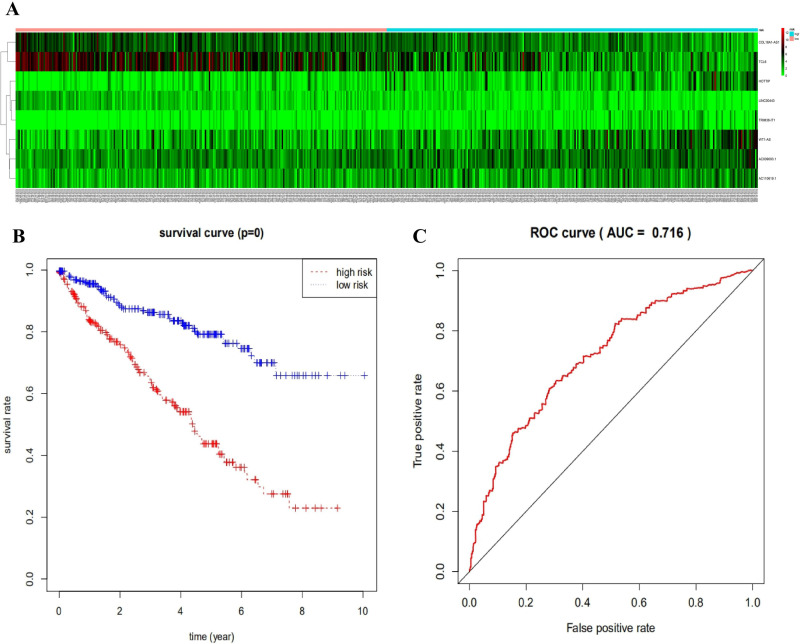
Risk groupings and ROC analysis
The "pheatmap" software package of R software was used to visualize the expression of eight lncRNAs (WT1-AS, COL18A1-AS1, TCL6, AC009093.1, LINC00443, HOTTIP, TRIM36-IT1, AC110619.1; Fig 6A). Based on the median risk, we divided a total of 530 complete survival information samples into two groups, including a high-risk group (n = 265) and a low-risk group (n = 265). Next, we used Kaplan–Meier curves in conjunction with log-rank statistical tests for survival analysis. The “survival” package was used for survival analysis. The results showed that there were differences between the two groups, and the low-risk group was significantly better (Fig 6B). The ROC curve analysis was conducted to test the effect of eight lncRNAs on the overall survival rate of KIRC (Fig 6C).
Fig 6. The prognosis of eight lncRNAs in patients with KIRC.
(a). Risk heat map based on eight lncRNAs from 530 LUSC patients. (b). Kaplan-Meier curve analysis was performed on the overall survival rate of patients with KIRC using eight lncRNAs. (c). ROC curve analysis was performed on the eight prognosis lncRNAs. AUC: Area Under ROC Curve, ROC curve: Receiver operating characteristic curve.
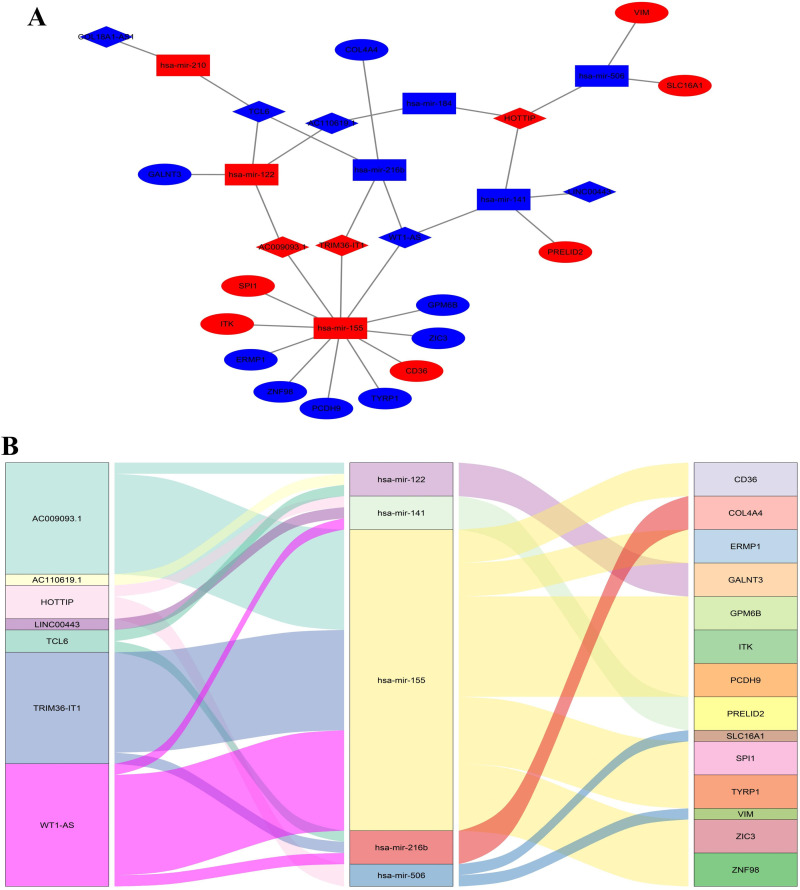
Prognosis-related DElncRNA-mediated ceRNA in sub-network
To better analyze the interaction of RNAs in KIRC, we used Cytoscape to analyze the ceRNA network mediated by prognosis-related DelncRNA. As shown in Fig 7A, only eight DElncRNAs, seven DEmiRNAs, and 14 DEmRNAs were shown in the DElncRNA-mediated ceRNA sub-network. This newly reconstructed sub-network consisted of 29 nodes and 31 edges. We used the Sankey diagram constructed by the “ggalluival” package of R software to visually analyze ceRNA regulatory sub-network mediated by eight lncRNAs related to prognosis (Fig 7B).
Fig 7. The ceRNA sub-network of the DElncRNA-mediated ceRNA network.
(a) The ceRNA sub-network of eight lncRNAs. The diamond was lncRNA, the rectangle was miRNA, and the ellipse was mRNA. Red indicated upregulation, and blue indicated downregulation. (b) The constructed snakey diagram. All involved RNAs were associated with prognosis.
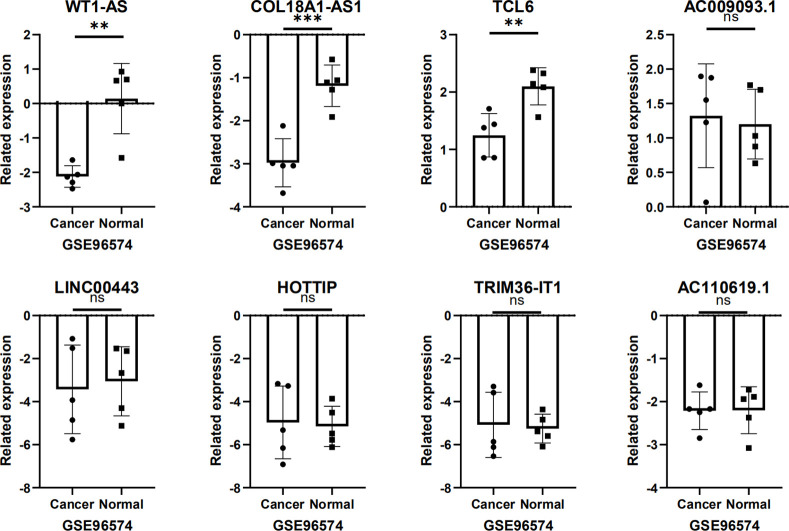
Validation of independent data sets
We validated these eight lncRNAs using the independent dataset GSE96574 in the GEO database. Among them, WT1-AS, COL18A1-AS1, and TCL6 had statistical differences. The results showed that there were three statistical differences (Fig 8).
Fig 8. In the independent dataset GSE96574, the eight lncRNAs selected were verified.
NS: no significance.
Discussion
The recent increase in patients with KIRC highlights the urgent need for more effective prognostic biomarkers to predict the prognosis. lncRNA-related research has attracted extensive attention in the field of cancer. Several studies have shown that lncRNAs related to cancer can be used as a biomarker for the diagnosis or prediction of cancer and plays a vital role in cancer treatment. Although the molecular mechanisms of KIRC protein-coding genes or miRNAs have been studied, little is known about lncRNAs. Few have predicted the prognosis of lncRNA in KIRC and the construction of the ceRNA network. There were no reliable specific lncRNA KIRC biomarkers. Moreover, there remained a lack of RNA sequencing data and analysis related to lncRNA profiles in KIRC. Qin Li et al. studied three hub genes of VHL, SETD2, and TRIP11 from KIRC as possible therapeutic targets and established and verified related ceRNA networks and verified it [38]. Few studies explored lncRNA biomarkers and constructed KIRC-releated ceRNA networks [39].
CeRNAs reveal a new mechanism of RNA interaction and reflects the interactions of coding RNA and non-coding RNA. CeRNAs are regarded as balances; when disrupted, they lead to the disturbance of life activities and the occurrence of diseases, especially in tumor diseases [40]. Many ceRNA network imbalances between these non-coding RNAs play a fundamental role in establishing signs of cancer development. The lncRNA OSER1-AS1promotes the deterioration of non-small cell lung cancer by sponging microRNA‑433‑3p to further increase Smad2 expression [41]. The lncRNA LINC00689 causes miR-526b-3p to sponge and leads to the upregulation of ADAM9 to promote the progression of gastric cancer [42]. These findings open a new window for a better understanding of cancer’s hidden aspects and also highlight novel biomarkers and potentially effective therapeutic targets for cancer. Xiaolong Qi et al. proposed that ceRNA may provide new biomarkers and potentially effective therapeutic targets for breast cancer [43]. Juan Xu et al. constructed mRNA-related ceRNAs in 20 major cancer types and further revealed conserved and reconnected network ceRNA hubs in each cancer. These highly competitive interactions that constitute a conservative and cancer-specific module [44]. Chu Y et al. also found that lncRNA perturbed TLR signaling network performance can serve as a new lncRNA prognostic biomarker in colorectal cancer [45].
Increasing evidence shows that lncRNAs are key members in ceRNA and regulate other RNAs in the ceRNA network [46]. An effective way to study the potential of lncRNAs is by establishing a relationship between the lncRNA of interest and miRNA/mRNA. CASC9 promotes the progression of non-small cell lung cancer through the miR-335-3p/S100A14 axis [47]. The lncRNA SNHG3 promotes the progression of bladder cancer via the miR-515-5p/GINS2 axis [48]. The lnc-HSD17B11-1:1 was found to promote colorectal cancer progression through the lnc-HSD17B11-1:1/miR-338-3p/MACC1 axis [49]. The lncRNA FAM83H-AS1 promotes the progression of esophageal squamous cell carcinoma through miR-10a-5p/Girdin axis [50]. The lncRNA PVT1 also promotes gastric cancer migration by acting as a ceRNA for miR-30a and regulating snails [51]. The lncRNA SNHG17 also promotes the progression of glioma cells by regulating the miR-23b-3p/ZHX1 axis [52]. The STAT3-mediated lncRNA HOXD-AS1 is upregulated by ceRNA and subsequently promotes the metastasis of liver cancer by regulating SOX4 [53]. The lncRNA FAL1 also promotes the proliferation and metastasis of hepatocellular carcinoma cells by acting as a ceRNA of miR-1236 [54]. Therefore, looking for lncRNAs related to prognosis and understanding its regulation mechanism and functional role is vital for KIRC diagnosis and treatment.
It is increasingly challenging increasingly difficult to detect potential human lncRNA-disease associations from these huge biological data sets using traditional biological experimental methods. Therefore, it is important to develop new and effective calculation methods to predict potential human lncRNA diseases. Several experimental methods and computational models have been designed and implemented to identify novel ncRNA-disease associations [55, 56]. For example, some researchers used the principles of machine learning to construct a new type of interpretable analysis to analyze potential biomarkers of tumorigenesis in non-small cell lung cancer. This improves our understanding of the nature of the disease and helps discover more effective diagnosis, prognosis and treatment methods for various cancer types and stages [57]. A semi-supervised interactome network-based method was recently used to explore and predict potential interactions between lncRNA and miRNA. The model will help predict the interaction between lncRNA and miRNA, and successfully demonstrated superiority and good generalizability [58]. In another study, researchers used a combination of incremental principal component analysis and random forest algorithm. By integrating multiple similarity matrices, we proposed a new algorithm based on integrated machine learning technology to predict associated lncRNA diseases [59]. A new type of induction matrix completion model for MiRNA-disease association prediction has been proposed [60, 61]. The study found that the method of Laplacian Regularized Least Squares for LncRNA-Disease Association (LRLSLDA) was further developed in the semisupervised learning framework. It will benefit biomarker identification and drug development. It can be expected that LRLSLDA can become an effective and important biological tool for biomedical research [62]. These studies show that the methods and techniques of computational biology are playing an increasingly important role in medicine and biology [63, 64]. In the future, we need to use more computational biology techniques to research KIRC to find more suitable and effective prediction and analysis models. We searched the KIRC transcriptome and clinical data from the TCGA database. Our research used the “edgeR” package to obtain DEmRNA, DEmiRNA, and DElncRNA. We used the miRcode database to screen the target miRNA corresponding to DElncRNA. Next, we searched and determined the target mRNA of DEmiRNA together across three databases. Finally, the ceRNA network related to lncRNA was established. The new ceRNA network was composed of 81 DElncRNAs, nine DEmiRNAs, and 17 DEmRNAs. This network included 107 nodes and 178 edges. Subsequently, we used survival analysis to find RNA associated with all prognoses. We then analyzed DEmRNAs using the GO and KEGG pathways.
We often need ways to enrich related genes to study the relationship between them, with the increasing availability of useful online tools. The two important online tools are DAVID and KOABS; they can perform functional enrichment analysis and pathway analyses online to study the mechanism of downstream pathways and potential targets [65, 66]. The tools and data processing methods constructed by computer biology have played a huge role in promoting the huge data of medicine and biology and helped cure diseases. GO functional enrichment and KEGG pathway analyses are widely used to evaluate the rich biological functions in multiple coding genes. Our GO analysis showed that most of the DEmRNAs were enriched in extracellular structure organization, cellular response to lipoteichoic acid, membrane raft, response to lipopolysaccharide, RNA polymerase II core promoter proximal region sequence-specific DNA binding, and cellular response to an organic substance. KEGG pathway analysis also showed that most of the DEmRNAs were enriched in ECM-receptor interaction, cancer pathways, chemokine signaling pathway, cytokine-cytokine receptor interaction, human papillomavirus infection, PI3K-Akt signaling pathway, and herpes simplex virus 1 infection.
Due to the heterogeneity between patients, traditional prognostic systems are often incapable of making accurate predictions of risk grouping and clinical outcomes. Therefore, several studies have been constructed new prediction models to carry out the survival expectation of KIRC. Furthermore, we analyzed and screened some DElncRNAs, selected eight lncRNAs related to prognosis, and constructed ROC curves and COX regression models. These eight lncRNAs were screened to construct a predictive model for early diagnosis and prognosis of KIRC. Survival analysis based on the linear combination of 8-lncRNA signals showed that the survival rate of high-risk groups was lower than that of low-risk groups. Therefore, these eight lncRNAs (WT1-AS, COL18A1-AS1, TCL6, AC009093.1, LINC00443, HOTTIP, TRIM36-IT1, and AC110619.1) have a very significant connection with the prognosis of KIRC. Finally, we verified these eight lncRNAs with independent datasets and found that expression of WT1-AS, COL18A1-AS1, and TCL6 were statistically different. However, as the sample size of the independent data set in GEO was small, it did not mean that the other lncRNAs were not statistically different. In the future, a larger amount of sequencing data and experiments are needed to verify these targets.
WT1-AS inhibits the proliferation of cervical squamous cell carcinoma cells by regulating p53 [67]. The overexpression of lncRNA WT1-AS affects non-small cell lung cancer cells by regulating TGF-β1 [68]. WT1-AS promotes hepatoma cell apoptosis by downregulating WT1 [69]. TCL6 inhibits the development of lncRNA in cancer, and in hepatocellular carcinoma directly regulated the PI3K / AKT signaling pathway by binding to miR-106a-5p to affect cancer development [70]. The mechanism and role of several other lncRNAs in cancer remain unclear. Future studies of these lncRNAs should focus on KIRC prognosis-related biomarkers. Our research screened out potential targets for KIRC using bioinformatics and computational biology methods construct a Cox regression analysis model. This will help in the diagnosis and treatment of patients with KIRC going forward, and guide the construction of predictive models and identification of potential targets in other diseases. The TCGA database was the key to analyzing prognostic biomarkers; all of our data were obtained from here. The primary method we used involved bioinformatics, seemingly the most promising way to explore gene and protein pathway and network interactions. However, the function and network of lncRNAs were very complicated. More clinical trials are required to verify our findings in the future. Finally, the biological functions of the eight lncRNAs with KIRC prognosis should be verified through large-scale experiments.
Conclusion
We constructed ceRNA networks using transcriptome sequencing data in the TCGA database and screened out DElncRNsA, DEmiRNAs, and DEmRNAs, which may be related to KIRC pathogenesis. The eight lncRNAs screened in the ceRNA network constructed a predictive model and may play a vital role in the diagnosis and prognosis of patients with KIRC. And three of them by independent samples proved to be meaningful.
Supporting information
(ZIP)
(ZIP)
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
This work was supported by the National Science Foundation for Young Scientists of China (No. 8150020951) and the Natural Science Foundation of Hunan Province (No. 2016JJ4099). LX is supported by the Ministry of Science and Technology of China and the Science and Technology Department of Hunan Province. The funder has no role in research design, data collection and analysis, publication decision, or manuscript preparation.
References
- 1.Ferlay J, Colombet M, Soerjomataram I, Dyba T, Randi G, Bettio M, et al. Cancer incidence and mortality patterns in Europe: Estimates for 40 countries and 25 major cancers in 2018. Eur J Cancer. 2018, 103: 356–387. Epub 2018/08/14. doi: 10.1016/j.ejca.2018.07.005 . [DOI] [PubMed] [Google Scholar]
- 2.Shuch B, Amin A, Armstrong AJ, Eble JN, Ficarra V, Lopez-Beltran A, et al. Understanding pathologic variants of renal cell carcinoma: distilling therapeutic opportunities from biologic complexity. Eur Urol. 2015, 67(1): 85–97. Epub 2014/05/27. doi: 10.1016/j.eururo.2014.04.029 . [DOI] [PubMed] [Google Scholar]
- 3.Rini BI, Campbell SC, Escudier B. Renal cell carcinoma. Lancet. 2009, 373(9669): 1119–1132. Epub 2009/03/10. doi: 10.1016/S0140-6736(09)60229-4 . [DOI] [PubMed] [Google Scholar]
- 4.Lin C, Yang L. Long Noncoding RNA in Cancer: Wiring Signaling Circuitry. Trends Cell Biol. 2018, 28(4): 287–301. Epub 2017/12/25. doi: 10.1016/j.tcb.2017.11.008 ; PubMed Central PMCID: PMC5869122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin W, Zhou Q, Wang CQ, Zhu L, Bi C, Zhang S, et al. LncRNAs regulate metabolism in cancer. Int J Biol Sci. 2020, 16(7): 1194–1206. Epub 2020/03/17. doi: 10.7150/ijbs.40769 ; PubMed Central PMCID: PMC7053319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jin KT, Yao JY, Fang XL, Di H, Ma YY. Roles of lncRNAs in cancer: Focusing on angiogenesis. Life Sci. 2020, 252: 117647. Epub 2020/04/11. doi: 10.1016/j.lfs.2020.117647 . [DOI] [PubMed] [Google Scholar]
- 7.Connerty P, Lock RB, de Bock CE. Long Non-coding RNAs: Major Regulators of Cell Stress in Cancer. Front Oncol. 2020, 10: 285. Epub 2020/04/09. doi: 10.3389/fonc.2020.00285 ; PubMed Central PMCID: PMC7099402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen X, Yan CC, Zhang X, You ZH. Long non-coding RNAs and complex diseases: from experimental results to computational models. Brief Bioinform. 2017, 18(4): 558–576. Epub 2016/06/28. doi: 10.1093/bib/bbw060 ; PubMed Central PMCID: PMC5862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen X, Sun YZ, Guan NN, Qu J, Huang ZA, Zhu ZX, et al. Computational models for lncRNA function prediction and functional similarity calculation. Brief Funct Genomics. 2019, 18(1): 58–82. Epub 2018/09/25. doi: 10.1093/bfgp/ely031 . [DOI] [PubMed] [Google Scholar]
- 10.Wang Y, Hou J, He D, Sun M, Zhang P, Yu Y, et al. The Emerging Function and Mechanism of ceRNAs in Cancer. Trends Genet. 2016, 32(4): 211–224. Epub 2016/02/29. doi: 10.1016/j.tig.2016.02.001 ; PubMed Central PMCID: PMC4805481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang J, Liu L, Li J, Le TD. LncmiRSRN: identification and analysis of long non-coding RNA related miRNA sponge regulatory network in human cancer. Bioinformatics. 2018, 34(24): 4232–4240. Epub 2018/06/30. doi: 10.1093/bioinformatics/bty525 . [DOI] [PubMed] [Google Scholar]
- 12.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011, 146(3): 353–358. Epub 2011/08/02. doi: 10.1016/j.cell.2011.07.014 ; PubMed Central PMCID: PMC3235919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014, 505(7483): 344–352. Epub 2014/01/17. doi: 10.1038/nature12986 ; PubMed Central PMCID: PMC4113481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qi M, Yu B, Yu H, Li F. Integrated analysis of a ceRNA network reveals potential prognostic lncRNAs in gastric cancer. Cancer Med. 2020, 9(5): 1798–1817. Epub 2020/01/11. doi: 10.1002/cam4.2760 ; PubMed Central PMCID: PMC7050084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Piao HY, Guo S, Jin H, Wang Y, Zhang J. LINC00184 involved in the regulatory network of ANGPT2 via ceRNA mediated miR-145 inhibition in gastric cancer. J Cancer. 2021, 12(8): 2336–2350. Epub 2021/03/25. doi: 10.7150/jca.49138 ; PubMed Central PMCID: PMC7974899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weng W, Zhang Z, Huang W, Xu X, Wu B, Ye T, et al. Identification of a competing endogenous RNA network associated with prognosis of pancreatic adenocarcinoma. Cancer Cell Int. 2020, 20: 231. Epub 2020/06/17. doi: 10.1186/s12935-020-01243-6 ; PubMed Central PMCID: PMC7288603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yun Z, Meng F, Li S, Zhang P. Long non-coding RNA CERS6-AS1 facilitates the oncogenicity of pancreatic ductal adenocarcinoma by regulating the microRNA-15a-5p/FGFR1 axis. Aging (Albany NY). 2021, 13(4): 6041–6054. Epub 2021/02/14. doi: 10.18632/aging.202540 ; PubMed Central PMCID: PMC7950275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li R, Yang YE, Jin J, Zhang MY, Liu X, Liu XX, et al. Identification of lncRNA biomarkers in lung squamous cell carcinoma using comprehensive analysis of lncRNA mediated ceRNA network. Artif Cells Nanomed Biotechnol. 2019, 47(1): 3246–3258. Epub 2019/08/01. doi: 10.1080/21691401.2019.1647225 . [DOI] [PubMed] [Google Scholar]
- 19.Cao Q, Dong Z, Liu S, An G, Yan B, Lei L. Construction of a metastasis-associated ceRNA network reveals a prognostic signature in lung cancer. Cancer Cell Int. 2020, 20: 208. Epub 2020/06/11. doi: 10.1186/s12935-020-01295-8 ; PubMed Central PMCID: PMC7271455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang X, Zhou J, Xu M, Yan Y, Huang L, Kuang Y, et al. A 15-lncRNA signature predicts survival and functions as a ceRNA in patients with colorectal cancer. Cancer Manag Res. 2018, 10: 5799–5806. Epub 2018/12/05. doi: 10.2147/CMAR.S178732 ; PubMed Central PMCID: PMC6248371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou Z, Tan F, Pei Q, Li C, Zhou Y, Li Y, et al. lncRNA SNHG4 modulates colorectal cancer cell cycle and cell proliferation through regulating miR-590-3p/CDK1 axis. Aging (Albany NY). 2021, 13. Epub 2021/03/22. doi: 10.18632/aging.202737 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu H, Lu J, Zhao H, Chen Z, Cui Q, Lin Z, et al. Functional Long Noncoding RNAs (lncRNAs) in Clear Cell Kidney Carcinoma Revealed by Reconstruction and Comprehensive Analysis of the lncRNA-miRNA-mRNA Regulatory Network. Med Sci Monit. 2018, 24: 8250–8263. Epub 2018/11/18. doi: 10.12659/MSM.910773 ; PubMed Central PMCID: PMC6251074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao K, Zhang Q, Wang Y, Zhang J, Cong R, Song N, et al. The construction and analysis of competitive endogenous RNA (ceRNA) networks in metastatic renal cell carcinoma: a study based on The Cancer Genome Atlas. Transl Androl Urol. 2020, 9(2): 303–311. Epub 2020/05/19. doi: 10.21037/tau.2020.02.17 ; PubMed Central PMCID: PMC7215020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu G, Yao W, Gumireddy K, Li A, Wang J, Xiao W, et al. Pseudogene PTENP1 functions as a competing endogenous RNA to suppress clear-cell renal cell carcinoma progression. Mol Cancer Ther. 2014, 13(12): 3086–3097. Epub 2014/09/25. doi: 10.1158/1535-7163.MCT-14-0245 ; PubMed Central PMCID: PMC4265235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Z, Li H, Jiang S, Li R, Li W, Chen H, et al. A survey and evaluation of Web-based tools/databases for variant analysis of TCGA data. Brief Bioinform. 2019, 20(4): 1524–1541. Epub 2018/04/05. doi: 10.1093/bib/bby023 ; PubMed Central PMCID: PMC6781580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu W, Islamaj Dogan R, Kwon D, Marques H, Rinaldi F, Wilbur WJ, et al. BioC implementations in Go, Perl, Python and Ruby. Database (Oxford). 2014, 2014. Epub 2014/06/26. doi: 10.1093/database/bau059 ; PubMed Central PMCID: PMC4067548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jalal H, Pechlivanoglou P, Krijkamp E, Alarid-Escudero F, Enns E, Hunink MGM. An Overview of R in Health Decision Sciences. Med Decis Making. 2017, 37(7): 735–746. Epub 2017/01/07. doi: 10.1177/0272989X16686559 . [DOI] [PubMed] [Google Scholar]
- 28.Hsu SD, Lin FM, Wu WY, Liang C, Huang WC, Chan WL, et al. miRTarBase: a database curates experimentally validated microRNA-target interactions. Nucleic Acids Res. 2011, 39(Database issue): D163–169. Epub 2010/11/13. doi: 10.1093/nar/gkq1107 ; PubMed Central PMCID: PMC3013699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wong N, Wang X. miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic Acids Res. 2015, 43(Database issue): D146–152. Epub 2014/11/08. doi: 10.1093/nar/gku1104 ; PubMed Central PMCID: PMC4383922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Riffo-Campos AL, Riquelme I, Brebi-Mieville P. Tools for Sequence-Based miRNA Target Prediction: What to Choose? Int J Mol Sci. 2016, 17(12). Epub 2016/12/13. doi: 10.3390/ijms17121987 ; PubMed Central PMCID: PMC5187787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Doncheva NT, Morris JH, Gorodkin J, Jensen LJ. Cytoscape StringApp: Network Analysis and Visualization of Proteomics Data. J Proteome Res. 2019, 18(2): 623–632. Epub 2018/11/20. doi: 10.1021/acs.jproteome.8b00702 ; PubMed Central PMCID: PMC6800166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sorensen A, Blazhenets G, Rucker G, Schiller F, Meyer PT, Frings L, et al. Prognosis of conversion of mild cognitive impairment to Alzheimer’s dementia by voxel-wise Cox regression based on FDG PET data. Neuroimage Clin. 2019, 21: 101637. Epub 2018/12/17. doi: 10.1016/j.nicl.2018.101637 ; PubMed Central PMCID: PMC6411907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kamarudin AN, Cox T, Kolamunnage-Dona R. Time-dependent ROC curve analysis in medical research: current methods and applications. BMC Med Res Methodol. 2017, 17(1): 53. Epub 2017/04/09. doi: 10.1186/s12874-017-0332-6 ; PubMed Central PMCID: PMC5384160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heller G, Seshan VE, Moskowitz CS, Gonen M. Inference for the difference in the area under the ROC curve derived from nested binary regression models. Biostatistics. 2017, 18(2): 260–274. Epub 2016/09/23. doi: 10.1093/biostatistics/kxw045 ; PubMed Central PMCID: PMC5965312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiao X, Sherman BT, Huang da W, Stephens R, Baseler MW, Lane HC, et al. DAVID-WS: a stateful web service to facilitate gene/protein list analysis. Bioinformatics. 2012, 28(13): 1805–1806. Epub 2012/05/01. doi: 10.1093/bioinformatics/bts251 ; PubMed Central PMCID: PMC3381967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xie C, Mao X, Huang J, Ding Y, Wu J, Dong S, et al. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011, 39(Web Server issue): W316–322. Epub 2011/07/26. doi: 10.1093/nar/gkr483 ; PubMed Central PMCID: PMC3125809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Walter W, Sanchez-Cabo F, Ricote M. GOplot: an R package for visually combining expression data with functional analysis. Bioinformatics. 2015, 31(17): 2912–2914. Epub 2015/05/13. doi: 10.1093/bioinformatics/btv300 . [DOI] [PubMed] [Google Scholar]
- 38.Qin L, Liu Y, Li M, Pu X, Guo Y. The landscape of miRNA-related ceRNA networks for marking different renal cell carcinoma subtypes. Brief Bioinform. 2018. Epub 2018/11/20. doi: 10.1093/bib/bby101 . [DOI] [PubMed] [Google Scholar]
- 39.Yin H, Wang X, Zhang X, Wang Y, Zeng Y, Xiong Y, et al. Integrated analysis of long noncoding RNA associated-competing endogenous RNA as prognostic biomarkers in clear cell renal carcinoma. Cancer Sci. 2018, 109(10): 3336–3349. Epub 2018/08/29. doi: 10.1111/cas.13778 ; PubMed Central PMCID: PMC6172067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qi X, Zhang DH, Wu N, Xiao JH, Wang X, Ma W. ceRNA in cancer: possible functions and clinical implications. J Med Genet. 2015, 52(10): 710–718. Epub 2015/09/12. doi: 10.1136/jmedgenet-2015-103334 . [DOI] [PubMed] [Google Scholar]
- 41.Liu X, Huang S, Guan Y, Zhang Q. Long noncoding RNA OSER1AS1 promotes the malignant properties of nonsmall cell lung cancer by sponging microRNA4333p and thereby increasing Smad2 expression. Oncol Rep. 2020, 44(2): 599–610. Epub 2020/07/07. doi: 10.3892/or.2020.7645 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yin G, Tian P, BuHe A, Yan W, Li T, Sun Z. LncRNA LINC00689 Promotes the Progression of Gastric Cancer Through Upregulation of ADAM9 by Sponging miR-526b-3p. Cancer Manag Res. 2020, 12: 4227–4239. Epub 2020/06/26. doi: 10.2147/CMAR.S231042 ; PubMed Central PMCID: PMC7280092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Abdollahzadeh R, Daraei A, Mansoori Y, Sepahvand M, Amoli MM, Tavakkoly-Bazzaz J. Competing endogenous RNA (ceRNA) cross talk and language in ceRNA regulatory networks: A new look at hallmarks of breast cancer. J Cell Physiol. 2019, 234(7): 10080–10100. Epub 2018/12/12. doi: 10.1002/jcp.27941 . [DOI] [PubMed] [Google Scholar]
- 44.Xu J, Li Y, Lu J, Pan T, Ding N, Wang Z, et al. The mRNA related ceRNA-ceRNA landscape and significance across 20 major cancer types. Nucleic Acids Res. 2015, 43(17): 8169–8182. Epub 2015/08/26. doi: 10.1093/nar/gkv853 ; PubMed Central PMCID: PMC4787795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chu Y, Liu Z, Liu J, Yu L, Zhang D, Pei F. Characterization of lncRNA-Perturbed TLR-Signaling Network Identifies Novel lncRNA Prognostic Biomarkers in Colorectal Cancer. Front Cell Dev Biol. 2020, 8: 503. Epub 2020/07/07. doi: 10.3389/fcell.2020.00503 ; PubMed Central PMCID: PMC7314994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ma Y, Zhang J, Wen L, Lin A. Membrane-lipid associated lncRNA: A new regulator in cancer signaling. Cancer Lett. 2018, 419: 27–29. Epub 2018/01/14. doi: 10.1016/j.canlet.2018.01.008 . [DOI] [PubMed] [Google Scholar]
- 47.Zhao W, Chen T, Zhao Y. Upregulated lncRNA CASC9 Contributes to Progression of Non-Small Cell Lung Cancer Through Inhibition of miR-335-3p and Activation S100A14 Expression. Onco Targets Ther. 2020, 13: 6027–6036. Epub 2020/07/02. doi: 10.2147/OTT.S249973 ; PubMed Central PMCID: PMC7321690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dai G, Huang C, Yang J, Jin L, Fu K, Yuan F, et al. LncRNA SNHG3 promotes bladder cancer proliferation and metastasis through miR-515-5p/GINS2 axis. J Cell Mol Med. 2020. Epub 2020/07/01. doi: 10.1111/jcmm.15564 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang W, Wang B, Wang Q, Zhang Z, Shen Z, Ye Y, et al. Lnc-HSD17B11-1:1 Functions as a Competing Endogenous RNA to Promote Colorectal Cancer Progression by Sponging miR-338-3p to Upregulate MACC1. Front Genet. 2020, 11: 628. Epub 2020/07/01. doi: 10.3389/fgene.2020.00628 ; PubMed Central PMCID: PMC7304498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Feng B, Wang G, Liang X, Wu Z, Wang X, Dong Z, et al. LncRNA FAM83H-AS1 promotes oesophageal squamous cell carcinoma progression via miR-10a-5p/Girdin axis. J Cell Mol Med. 2020. Epub 2020/06/26. doi: 10.1111/jcmm.15530 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang L, Xiao B, Yu T, Gong L, Wang Y, Zhang X, et al. lncRNA PVT1 promotes the migration of gastric cancer by functioning as ceRNA of miR-30a and regulating Snail. J Cell Physiol. 2020. Epub 2020/06/20. doi: 10.1002/jcp.29881 . [DOI] [PubMed] [Google Scholar]
- 52.Ge B, Li G. LncRNA SNHG17 promotes proliferation, migration and invasion of glioma cells by regulating the miR-23b-3p/ZHX1 axis. J Gene Med. 2020: e3247. Epub 2020/07/01. doi: 10.1002/jgm.3247 . [DOI] [PubMed] [Google Scholar]
- 53.Wang H, Huo X, Yang XR, He J, Cheng L, Wang N, et al. STAT3-mediated upregulation of lncRNA HOXD-AS1 as a ceRNA facilitates liver cancer metastasis by regulating SOX4. Mol Cancer. 2017, 16(1): 136. Epub 2017/08/16. doi: 10.1186/s12943-017-0680-1 ; PubMed Central PMCID: PMC5558651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li B, Mao R, Liu C, Zhang W, Tang Y, Guo Z. LncRNA FAL1 promotes cell proliferation and migration by acting as a CeRNA of miR-1236 in hepatocellular carcinoma cells. Life Sci. 2018, 197: 122–129. Epub 2018/02/09. doi: 10.1016/j.lfs.2018.02.006 . [DOI] [PubMed] [Google Scholar]
- 55.Hu H, Zhang L, Ai H, Zhang H, Fan Y, Zhao Q, et al. HLPI-Ensemble: Prediction of human lncRNA-protein interactions based on ensemble strategy. RNA Biol. 2018, 15(6): 797–806. Epub 2018/03/28. doi: 10.1080/15476286.2018.1457935 ; PubMed Central PMCID: PMC6152435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu H, Ren G, Chen H, Liu Q, Yang Y, Zhao Q. Predicting lncRNA–miRNA interactions based on logistic matrix factorization with neighborhood regularized. Knowledge-Based Systems. 2020, 191. Epub 2020/03/05. doi: 10.1016/j.knosys.2019.105261 [DOI] [Google Scholar]
- 57.Farouq MW, Boulila W, Hussain Z, Rashid A, Shah M, Hussain S, et al. A Novel Coupled Reaction-Diffusion System for Explainable Gene Expression Profiling. Sensors (Basel). 2021, 21(6). Epub 2021/04/04. doi: 10.3390/s21062190 ; PubMed Central PMCID: PMC8003942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang L, Liu T, Chen H, Zhao Q, Liu H. Predicting lncRNA-miRNA interactions based on interactome network and graphlet interaction. Genomics. 2021, 113(3): 874–880. Epub 2021/02/16. doi: 10.1016/j.ygeno.2021.02.002 . [DOI] [PubMed] [Google Scholar]
- 59.Zhu R, Wang Y, Liu JX, Dai LY. IPCARF: improving lncRNA-disease association prediction using incremental principal component analysis feature selection and a random forest classifier. BMC Bioinformatics. 2021, 22(1): 175. Epub 2021/04/03. doi: 10.1186/s12859-021-04104-9 ; PubMed Central PMCID: PMC8017839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen X, Wang L, Qu J, Guan N-N, Li J-Q. Predicting miRNA-disease association based on inductive matrix completion. Bioinformatics. 2018, 34(24): 4256–4265. Epub 2018/06/26. doi: 10.1093/bioinformatics/bty503 . [DOI] [PubMed] [Google Scholar]
- 61.Chen X, Xie D, Zhao Q, You ZH. MicroRNAs and complex diseases: from experimental results to computational models. Brief Bioinform. 2019, 20(2): 515–539. Epub 2017/10/19. doi: 10.1093/bib/bbx130 . [DOI] [PubMed] [Google Scholar]
- 62.Chen X, Yan GY. Novel human lncRNA-disease association inference based on lncRNA expression profiles. Bioinformatics. 2013, 29(20): 2617–2624. Epub 2013/09/05. doi: 10.1093/bioinformatics/btt426 . [DOI] [PubMed] [Google Scholar]
- 63.Zhao Q, Yu H, Ming Z, Hu H, Ren G, Liu H. The Bipartite Network Projection-Recommended Algorithm for Predicting Long Non-coding RNA-Protein Interactions. Mol Ther Nucleic Acids. 2018, 13: 464–471. Epub 2018/11/06. doi: 10.1016/j.omtn.2018.09.020 ; PubMed Central PMCID: PMC6205413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhao Q, Zhang Y, Hu H, Ren G, Zhang W, Liu H. IRWNRLPI: Integrating Random Walk and Neighborhood Regularized Logistic Matrix Factorization for lncRNA-Protein Interaction Prediction. Front Genet. 2018, 9: 239. Epub 2018/07/20. doi: 10.3389/fgene.2018.00239 ; PubMed Central PMCID: PMC6040094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009, 4(1): 44–57. Epub 2009/01/10. doi: 10.1038/nprot.2008.211 . [DOI] [PubMed] [Google Scholar]
- 66.Shen S, Kong J, Qiu Y, Yang X, Wang W, Yan L. Identification of core genes and outcomes in hepatocellular carcinoma by bioinformatics analysis. J Cell Biochem. 2019, 120(6): 10069–10081. Epub 2018/12/14. doi: 10.1002/jcb.28290 . [DOI] [PubMed] [Google Scholar]
- 67.Zhang Y, Na R, Wang X. LncRNA WT1-AS up-regulates p53 to inhibit the proliferation of cervical squamous carcinoma cells. BMC Cancer. 2019, 19(1): 1052. Epub 2019/11/07. doi: 10.1186/s12885-019-6264-2 ; PubMed Central PMCID: PMC6836469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jiang X, Wang J, Fang L. LncRNA WT1-AS over-expression inhibits non-small cell lung cancer cell stemness by down-regulating TGF-beta1. BMC Pulm Med. 2020, 20(1): 113. Epub 2020/05/01. doi: 10.1186/s12890-020-1146-6 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lv L, Chen G, Zhou J, Li J, Gong J. WT1-AS promotes cell apoptosis in hepatocellular carcinoma through down-regulating of WT1. J Exp Clin Cancer Res. 2015, 34: 119. Epub 2015/10/16. doi: 10.1186/s13046-015-0233-7 ; PubMed Central PMCID: PMC4604772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Luo LH, Jin M, Wang LQ, Xu GJ, Lin ZY, Yu DD, et al. Long noncoding RNA TCL6 binds to miR-106a-5p to regulate hepatocellular carcinoma cells through PI3K/AKT signaling pathway. J Cell Physiol. 2020, 235(9): 6154–6166. Epub 2020/02/06. doi: 10.1002/jcp.29544 . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(ZIP)
(ZIP)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.