Abstract
Tissue engineering has markedly matured since its early beginnings in the 1980s. In addition to the original goal to regenerate damaged organs, the field has started to explore modeling of human physiology “in a dish”. Induced pluripotent stem cell (iPSC) technologies now enable studies of organ regeneration and disease modeling in a patient-specific context. We discuss the potential of “organ-on-a-chip” systems to study regenerative therapies with focus on three distinct organ systems: cardiac, respiratory, and hematopoietic. We propose that the combinatorial studies of human tissues at these two scales would help realize the translational potential of tissue engineering.
Keywords: organ-on-a-chip, tissue engineering, regenerative medicine, pluripotent stem cells, precision medicine, bioengineering
In this Review, Vunjak-Novakovic and colleagues discuss the potential of “organ-on-a-chip” systems to study regenerative therapies, focusing on three distinct organ systems: cardiac, respiratory, and hematopoietic. They propose that combinatorial studies of human tissues at these two scales would help realize the translational potential of tissue engineering with stem cells.
In the early years of the field, tissue engineering was focused on the development and eventual application of lab-grown tissues for regenerative medicine, using a variety of cell sources, materials, and culture systems. More recently, human “Organs-on-a-chip” (OOCs), also known as microphysiological systems (MPS), have emerged as an entirely new approach to recapitulate organ physiology during development and disease (Fig. 1). These models utilize human iPSC-derived or primary cells, and span a range of technologies, from self-assembling organoids, to bioengineered tissues and microfluidic organs-on-a-chip (Bhatia and Ingber, 2014; Hofer and Lutolf, 2021; Ronaldson-Bouchard and Vunjak-Novakovic, 2018). In this review, we refer to these technologies as OOCs unless specified differently.
Figure 1. In vitro models.
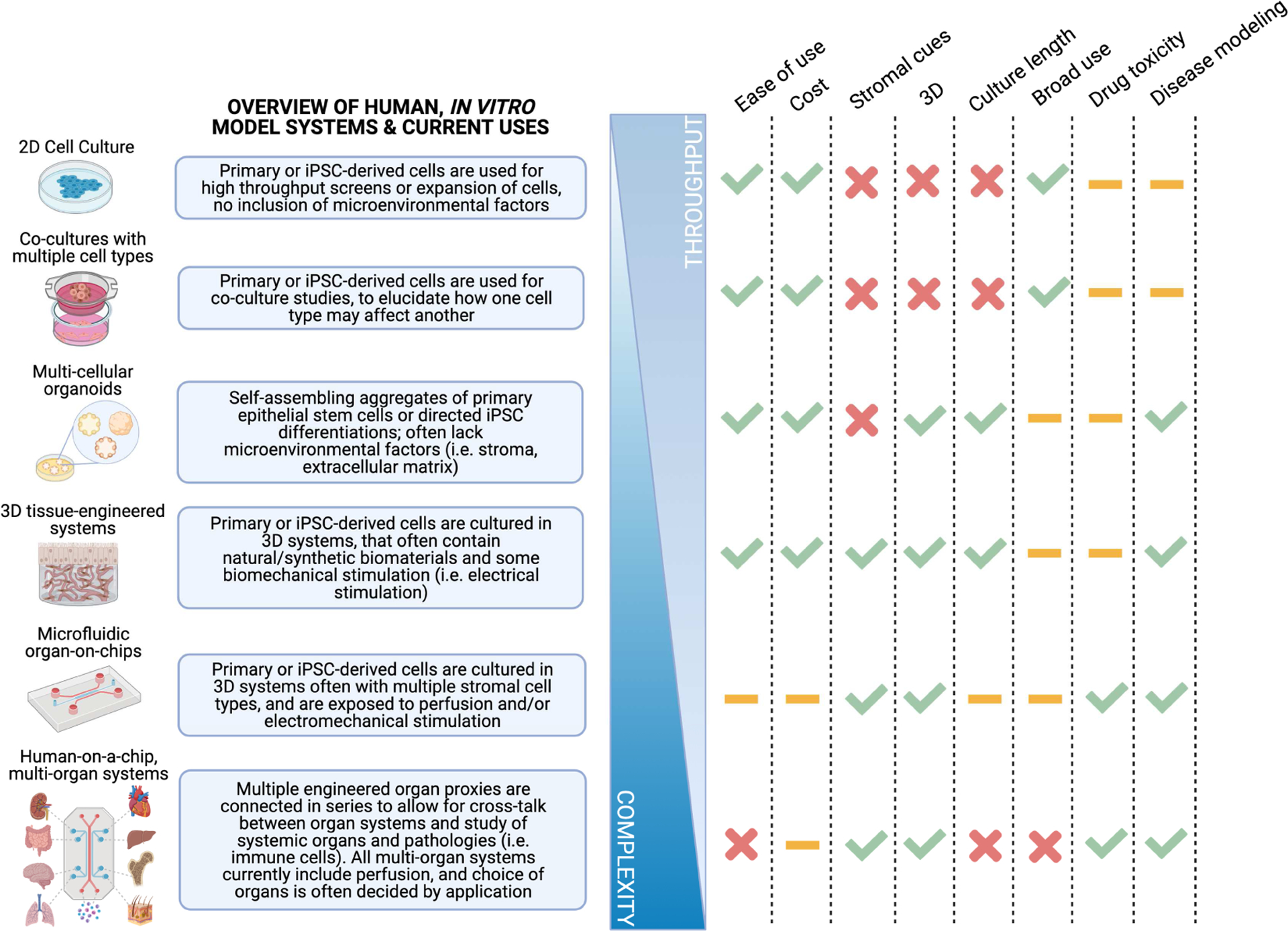
A range of models, from 2D cell culture to organoids and human OOC systems, enable a number of applications for modeling drug toxicity, disease phenotypes, and organ function while balancing the inverse relationship between complexity and throughput. Key: high (checkmark), medium (dash), low (X) frequencies.
An increasing number of groups are now developing OOC models of the heart, lung, liver, marrow, kidney, gut, and brain, among other organ systems, in the form of minimally functional tissue units capable of modeling in vitro some aspects of human physiology in health and disease, as well as the effects and mechanisms of drug actions (Low et al., 2020; Sharma et al., 2020).
From early works that have used decellularized bone grafts to recent stem cell therapies, regenerative engineering has also faced numerous challenges to reach the clinic. Biomaterials, either with or without cellular sources, have been employed to promote regeneration of some but not all organs, with skin, bone, and vessels being largely successful (Amini et al., 2012; Chen et al., 2020; MacNeil, 2007; Pashneh-Tala et al., 2015). There have been major advances in using human cells, primary and iPSC-derived, to help guide regenerative function in various organ systems. Major challenges in this area, including large-scale cell production, effective cell engraftment, and tissue vascularization, are currently being addressed through new bioengineering approaches (Fleischer et al., 2020).
We discuss progress in two complementary areas of tissue engineering: OOC models and regenerative medicine, that show immense potential for convergence and synergy (Fig. 2). Micro-scaled OOCs can be valuable for studying the underlying biology of injury and regeneration, with the aid of real-time monitoring using biosensors and imaging techniques that allow for understanding dynamic changes in cell and tissue functions (Fig. 1). OOCs enable researchers to de-couple the effects of cell type, cell-cell, cell-microenvironment, and tissue-tissue interactions, as well as to advance our fundamental understanding of native organ function and the effects of treatments. For example, dosing strategies can be easily manipulated in vitro to mitigate or enhance inflammation, or to optimize cell dosage in cell therapy applications. We focus here on the potential use of bioengineered models in vitro for studying new approaches in regenerative medicine in tandem with animal models, using examples from three organ systems: cardiac, respiratory, and hematopoietic (Fig. 2).
Figure 2.
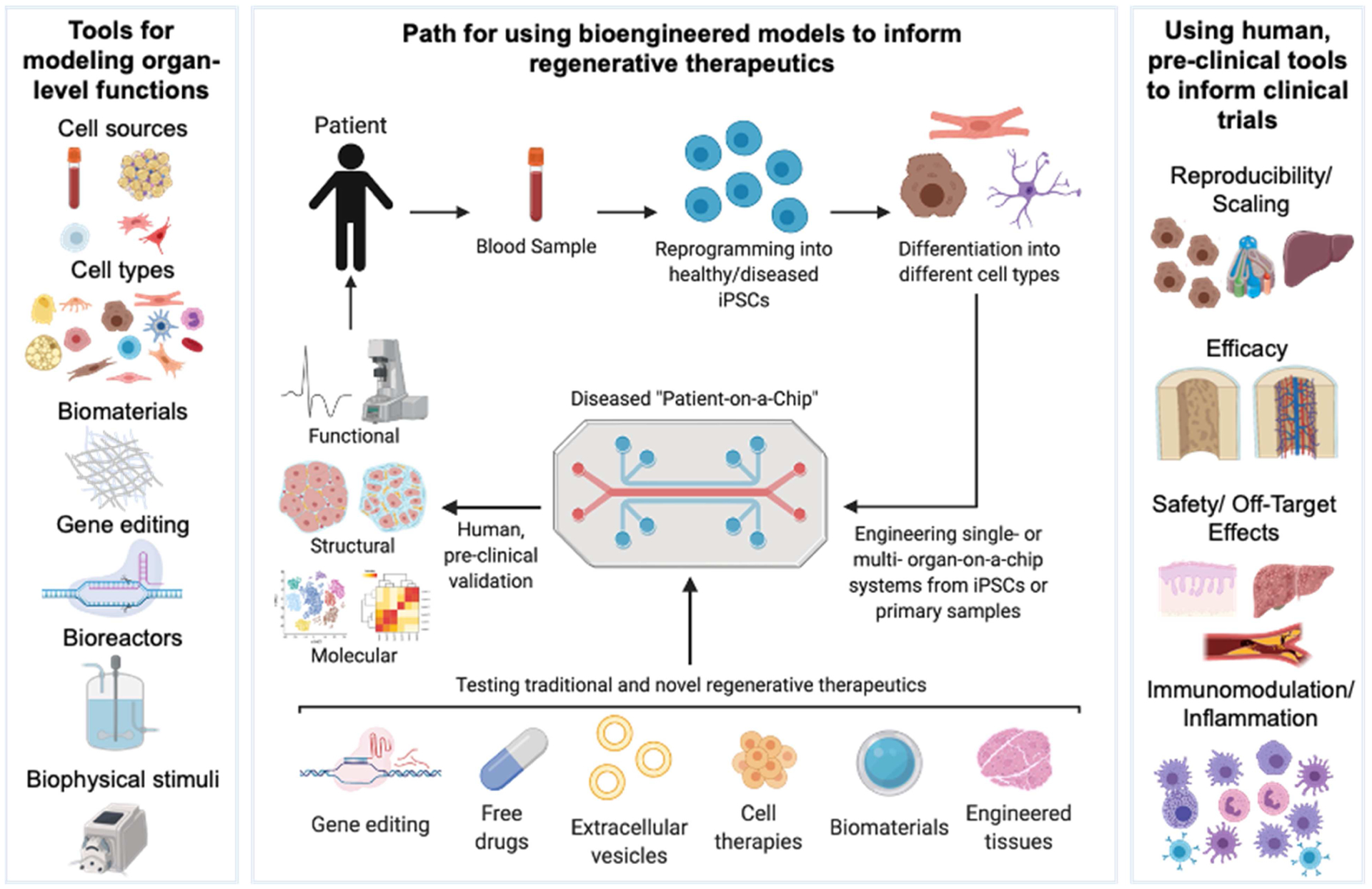
A framework for using engineered human tissues for testing regenerative therapies.
Bioengineering human physiological functions on the micro and macro scale
Modeling the biological complexity of the body in vitro has been widely pursued, but still remains a difficult challenge. Recapitulating cell microenvironment, cell-cell interactions, and spatial organizations seen in native human tissues brings major challenges. While acknowledging that it is simply impossible for every single biological characteristic to be faithfully represented in vitro, the field is trying to answer the long-standing question of “how simple is complex enough?” In the bioengineering community, it is often understood that with higher biological fidelity, we often see more realistic functional outputs, but generally at the expense of longer protocols for tissue assembly and maturation. In some applications, however, establishing just the basic functional outputs can be sufficient, as for in vitro studies of toxicity and disease modeling. In contrast to tissue implants, which require stable multi-scale vasculature perfused with blood, OOCs are often able to recapitulate functional validity of an organ system with minimal or no vascularization. Using patient-derived cells and materials, OOC model systems can help understand the mechanistic differences between the patient groups, to study sex differences, identify groups more susceptible to a specific disease, or classify patients into subpopulations to predict prognosis. Other in vitro models, like organoids, have emerged over the past decade to study self-organizing tissues from iPSC or primary cell sources, including applications in gut epithelia, the brain, and vasculature (Lancaster et al., 2013; O’Neill et al., 2021; Sato et al., 2009; Wimmer et al., 2019). These models are high throughput and are starting to become commonplace in many research institutions, though often lack the environmental milieu, controlled spatial organization, and maturity necessary for mimicking adult human physiology (Fig. 1). An important goal for the field is to design the bioengineering tools in the context of biological questions of interest.
The field of tissue engineering and regenerative medicine has steadily developed, as shown by the number of papers published in each field each year (Fig. 3A). Since the mid-2000s, the advent of iPSCs and encompassing technologies has led to an exponential growth of OOC models (Fig. 3B). Going forward, OOCs can provide testing opportunities for understanding the efficacy and safety of regenerative therapeutics in human tissue models, and assessing the mechanistic basis of the therapy prior to its testing in human patients (Fig. 3B–H).
Figure 3. OOCs and related technologies for testing regenerative therapies in vitro.
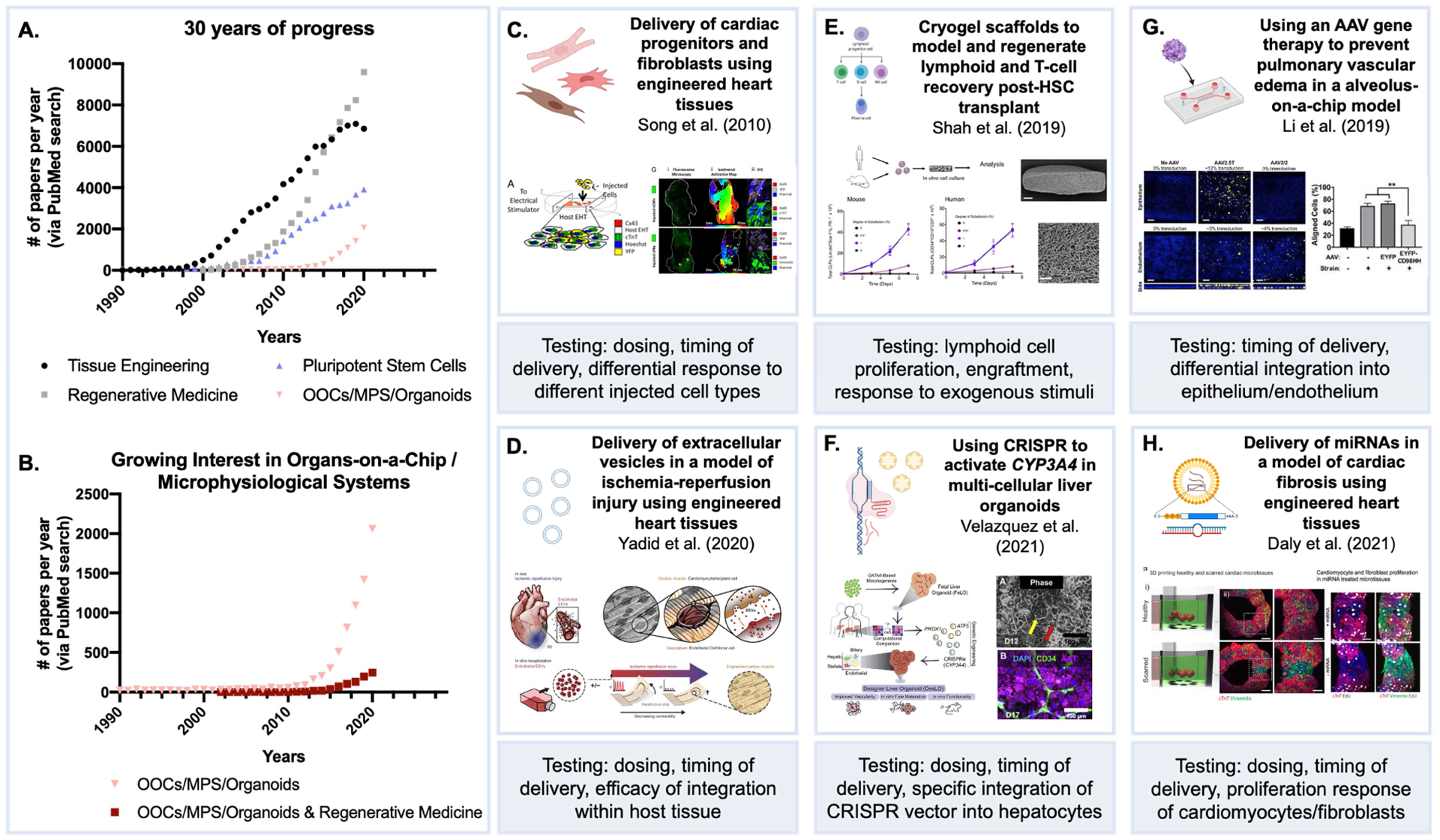
(A, B) Growth of the field (NIH PubMed database). (C) Engineered cardiac models for studying engraftment of injected cardiomyocyte progenitors, testing timing of delivery and efficacy of different cell types on cardiac muscle function (Song et al., 2010); Copyright (2010) National Academy of Sciences. (D) Extracellular vesicles (EVs) for hypoxia-induced myocardial infarction repair (Yadid et al., 2020); Reproduced with permission, AAAS. (E) Lymphoid progenitor expansion in vitro can mimic testing cryogel scaffolds for hematopoietic reconstitution in vivo (Shah et al., 2019). (F) Testing efficacy of CRISPR/Cas9 vector systems in liver organoids (Velazquez et al., 2021); Reproduced with permission. (G) Validating cell-specific integration of adeno-associated viruses with OOC models (Li et al., 2019). (H) Elucidating the effects of specific miRNAs on cardiac muscle repair (Daly et al., 2021); Reproduced under the terms of the Creative Commons License.
Cardiovascular System: Modeling Cardiac Muscle to Advance Cardiac Regeneration
Cardiovascular diseases are the leading cause of death in the Western world. Over the past few decades, major strides have been made in cardiac tissue engineering to address this clinical need. From early work in understanding the behavior of animal-derived cardiomyocytes to recent developments in vascularizing complex human muscle tissue and deriving ventricular and atrial cardiac cells and tissues (Fleischer et al., 2017; Goldfracht et al., 2020; Ogle et al., 2016). Cardiac muscle is a dense tissue rich in collagen and elastin fibers, containing a variety of cell sub-populations such as cardiomyocytes, cardiac fibroblasts, endothelial cells, smooth muscle cells and macrophages. While cardiomyocytes are not the largest cell population in the heart, they are the most studied, and are most critical for cardiac regeneration as the non-proliferative, terminally differentiated cells driving the contractile behavior of the muscle. In the states of injury (e.g., following myocardial infarction) or disease (e.g., cardiomyopathies), these cells are unable to restore the functional capacity of healthy muscle. Following injury, the non-parenchymal cells, mainly fibroblasts, tend to proliferate and activate into myofibroblasts, to further exacerbate fibrotic disease (Kapoun Ann et al., 2004). Despite major advances in the field, there is still a critical need to better understand the underlying molecular mechanisms of disease and develop effective strategies to promote regeneration and restore heart function.
Cardiac Tissue Engineering
Successful engineering of cardiac muscle began from studying embryonic chick cardiomyocytes seeded within collagen hydrogel and their organization into spontaneously beating tissues in the late 1990s (Akins et al., 1999; Carrier et al., 1999; Eschenhagen et al., 1997). Many groups continued to develop methods for engineering cardiac tissues, using different cell sources (in particular from neonatal rat or chick embryo cardiomyocytes), varying the cell density, and developing advanced biomaterials and bioreactor designs to guide functional tissue assembly. It has been shown that tissues engineered from animal-derived cells can be used both for electrophysiological studies (Bursac et al., 1999) and for transplantation to improve cardiac function in animal models of myocardial infarction (Leor et al., 2000).
External stimuli were recognized as very important for synchronized contractions and maturation of engineered tissues. In some of the first studies using engineered cardiac tissues, Fink et al. demonstrated that mechanical stimulation, including unidirectional cyclic stretch, induced cardiomyocyte alignment, hypertrophy and led to enhanced functional properties of cardiac tissues grown from neonatal rat cardiomyocytes in vitro (Fink et al., 2000). Auxotonic mechanical stimulation can be applied by culturing engineered cardiac tissue between flexible pillars or posts, enhancing tissue contractile properties (Boudou et al., 2012; Zimmermann et al., 2006). In separate studies, electrical stimulation designed to induce macroscopic contractions, increased cell alignment and coupling, in addition to improved ultrastructure, gene expression and functional tissue properties (Radisic et al., 2004).
In the mid-2000s, a new age of human stem cell derived cardiomyocytes started with directed ESC differentiation into mesodermal lineages in vitro (Laflamme et al., 2007; Yang et al., 2008). Lessons learned from early work using animal-derived cardiomyocytes paved the way towards successful studies in engineering cardiac tissues from ESC-derived sources (Caspi et al., 2007). Recently after, the Kamp and Gepstein groups independently published derivation of functional fetal-like cardiomyocytes from human iPSC sources (Zhang et al., 2009; Zwi et al., 2009). Following differentiation, these cells were seeded into 3D scaffolds, co-cultured with other cell types such as MSCs, ECs, or epicardial cells (Bargehr et al., 2019), and subjected to external stimulation to promote maturation (Tulloch et al., 2011).
Models of cardiac muscle
Heart physiology is inherently different between animals and humans, limiting the capability of animal models – especially rodents – to faithfully recapitulate clinical outcome and serve as tools for mechanistic studies. Monolayer cultures of iPSC-CMs provide a simple way to study some aspects of cardiac biology. Co-culture of iPSC-CMs with other cell types such as MSCs, fibroblasts, and epicardial cells were shown to improve the iPSC-CM phenotype. However, the cell maturity remained far below the adult human cardiomyocytes (Yoshida et al., 2018; Zhang et al., 2019). While 2D cultures enable parallel studies of various cell types of the heart at in vitro high throughput (Sharma et al., 2017), 3D models are required for advanced maturation and physiological relevance. Aggregation of iPSCs into 3D cardiac microspheres have been reported to predict drug toxicity at high throughput and model cardiac disease (Giacomelli et al., 2020; Hoang et al., 2018; Mills et al., 2019; Mills et al., 2017).
Using lessons learned from regenerative medicine, 3D models of the heart are engineered to generate miniature functional units recapitulating different aspects of the cardiac muscle in a simplified manner, often not requiring complex vascular networks that limit large-scale regenerative efforts. Emphasis is placed on generating simple and robust models, guiding tissue anisotropy and electrical coupling to promote tissue maturation, and developing platforms that will enable real-time measurements and capture the interactions of the heart with other organ models (Fig. 4).
Figure 4. Engineered cardiac models for testing cardiac regeneration therapies.
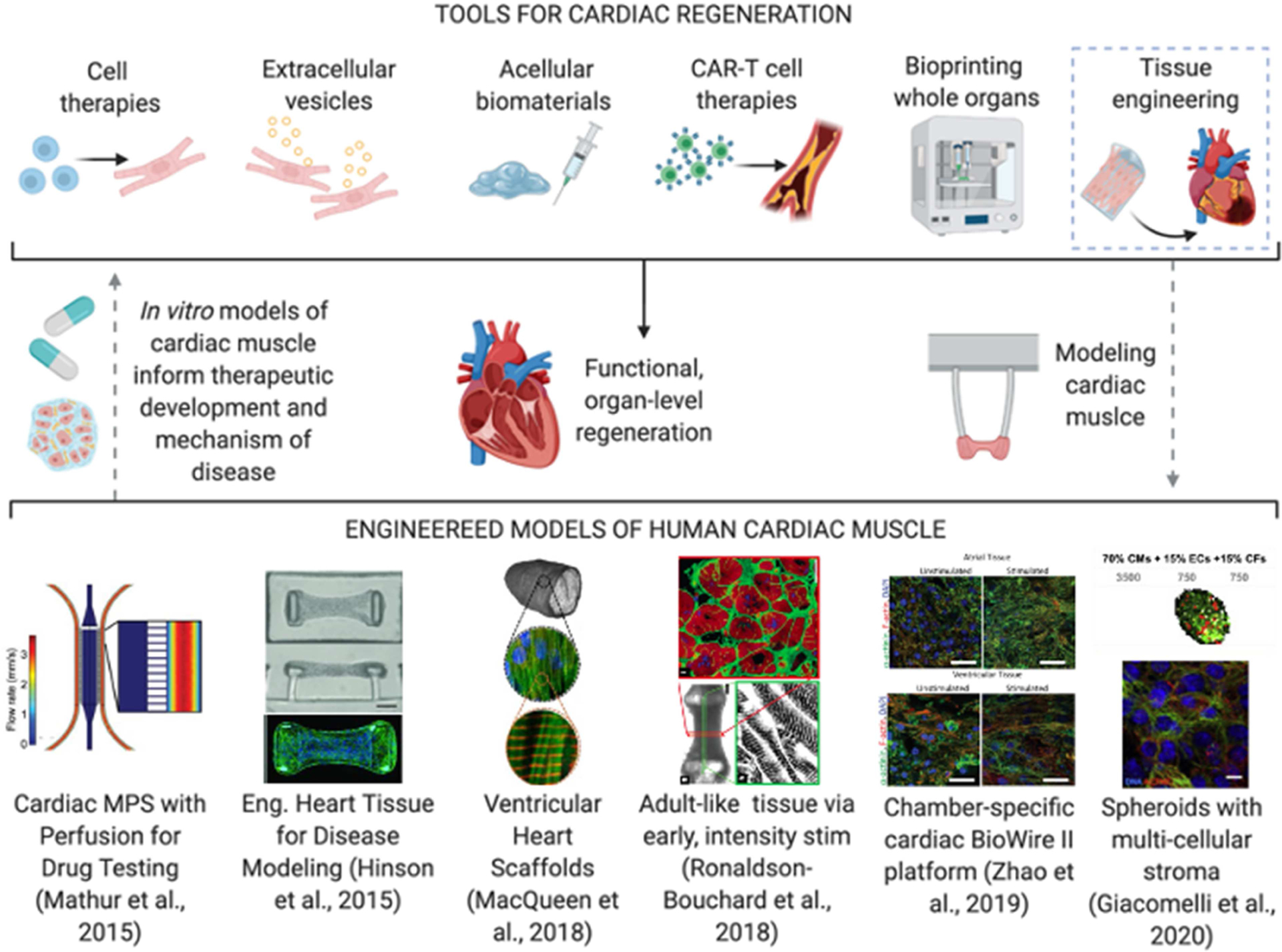
Top: dotted lines demonstrate the envisioned pathway of using bioengineered tools to make mimetic models of cardiac muscle and use such models to inform new therapeutic developments. Bottom: representative examples of recent cardiac muscle models: (Mathur et al., 2015), Reproduced under the terms of the Creative Commons License; (Hinson et al., 2015), Copyright 2015, American Association for the Advancement of Science (AAAS). (MacQueen et al., 2018); (Ronaldson-Bouchard et al., 2018), Reproduced with permission; (Zhao et al., 2019a), Reproduced with permission; (Giacomelli et al., 2020), Reproduced under the terms of the Creative Commons License.
In one of the early studies in the field, Kit Parker’s group demonstrated the use of thin film-based cantilevers to support generation of anisotropic cardiac sheets in a “heart-on-a-chip” platform (Grosberg et al., 2011). However, while this platform provides an improved model of heart physiology compared to 2D cell cultures, it is not scalable to 3D. To promote the assembly of functional 3D tissue models, several groups have utilized anchoring pillars, posts, and wires to stretch cardiac tissues in order to provide mechanical loading (Boudou et al., 2012; Ronaldson-Bouchard et al., 2018; Schaaf et al., 2011; Tiburcy et al., 2017). Mathur and colleagues fabricated a microfluidic device consisting of anisotropic cardiac tissues and controlled perfusion to mimic diffusion transport to the cardiac muscle (Mathur et al., 2015). Nunes et al. demonstrated the use of a “Biowire” model to electrically stimulate and pace cardiomyocytes cultured on a wire, showing increased ultrastructural organization, calcium handling, and conduction velocity (Nunes et al., 2013). Zhang and Khademhosseini endothelialized engineered cardiac muscle by bioprinting, and harnessed this model in studies of vascular changes in myocardium (Zhang et al., 2016b). Several teams have subsequently coupled cardiac tissues with biosensors and advanced imaging techniques to dynamically monitor functional responses (Bhise et al., 2014; Feiner et al., 2016; Sidorov et al., 2017; Zhang et al., 2017; Zhao et al., 2019b).
Cardiac toxicity, which is a major cause of drug withdrawal, motivated the need for reliable human tissue models for drug screening. Long-time cultures (>4 weeks) were conducted by several groups to study disease progression and drug responses using gene-edited iPSC lines (Ma et al., 2018; Mathur et al., 2015).
By using iPSCs, a multitude of genetic cardiac disorders, including the dilated cardiomyopathy, PRKAG2 cardiomyopathy, Barth syndrome, and Timothy syndrome, were recapitulated in vitro for mechanistic, developmental, and therapeutic studies (Hinson et al., 2016; Sun et al., 2012; Yazawa et al., 2011). For example, in a study investigating the significance of titin-based mutations in iPSC-derived cardiomyocytes, their active role in relaying sarcomere deterioration was identified as a cause for dilated cardiomyopathy (Hinson et al., 2015). A different study in a pillar-based model demonstrated the importance of full length-titin and MHC-β in sarcomerogenesis (Chopra et al., 2018). Cardiac fibrosis models have been developed by several groups in recent years, highlighting the opportunities to study mechanisms of cardiac disease progression and remodeling and evaluate the utility of antifibrotic drugs (Mastikhina et al., 2020; Nunes et al., 2013; van Spreeuwel et al., 2017). In studies integrating the cardiac and liver tissues, several groups have demonstrated drug metabolism along with known cardiotoxic side effects (McAleer et al., 2019; Pires de Mello et al., 2020).
To generate a cardiac model with adult-like characteristics, numerous groups have promoted tissue and cell maturation by long-term culture, addition of supporting cells, new media formulations, and mechanical, electrical and hydrodynamic stimulation (Eng et al., 2016; Feyen et al., 2020; Jackman et al., 2016; Nunes et al., 2013; Shadrin et al., 2017; Tiburcy et al., 2017; Zhao et al., 2019a). To build upon some of this work, our lab has investigated maturation of iPSC-derived cardiomyocytes in vitro (Ronaldson-Bouchard et al., 2018), by culturing early stage iPSC-cardiomyocytes (immediately after the initiation of spontaneous contractions) in fibrin hydrogels anchored between two pillars to support auxotonic contractions. The resulting cell-hydrogel constructs were exposed to an intensity training regimen at the gradually increasing electrical stimulation frequency starting from 2 Hz to 6Hz over three weeks of culture, and then stabilized at 2 Hz for an additional week in culture. This regime of stimulation markedly improved the molecular, structural, and functional tissue properties towards more mature phenotype. Other groups also reported additional methodologies for maturing iPSC-derived cardiomyocytes more recently (Feyen et al., 2020; Giacomelli et al., 2020), derivation of ventricular-like and atrial-like cardiac tissues (MacQueen et al., 2018; Patel et al., 2016; Zhao et al., 2019a), and producing sinoatrial node pacemaker cells from iPSCs (Goldfracht et al., 2020; Protze et al., 2017). The directed cell differentiation is especially important for the cardiac muscle, as some drugs and diseases are known to affect different subpopulations of cardiac cells or regions of the cardiac muscle. For example, specific drugs target the atrial chambers and lead to atrial fibrillation, while some diseases, such as Systemic Lupus Erythematosus, are known to affect mainly the left ventricle (Wijetunga and Rockson, 2002).
The choice of a specific cardiac model system should be based on the biological question at hand. 2D and microsphere co-cultures of human cells are highly suitable for high throughput studies. While microspheres are more physiologically relevant than 2D cultures (Ravenscroft et al., 2016), both systems do not capture the 3D complexity of cardiac tissues and do not incorporate architectural elements (i.e. stroma, ECM). Microfluidic channels in heart-on-a-chip models enable modeling of drug transport (Mathur et al., 2015); however, these systems are primarily fabricated out of polydimethylsiloxane (PDMS), which non-selectively absorbs hydrophobic molecules including oxygen and many drugs (Bhatia and Ingber, 2014). Engineered cardiac tissues in OOC platforms can capture more closely the complexity of the heart, and their enhanced maturity is especially important to predict physiological responses, such as calcium handling and force generation (Ronaldson-Bouchard et al., 2018; Zhao et al., 2019a).
Cardiac repair therapies
Cell delivery studies began in the early 1990s with the goal to restore cardiac function post-myocardial infarction and treat other cardiovascular disorders such as cardiomyopathy and congenital heart disease (Reinlib and Field, 2000). Many cell types have been used, including skeletal muscle cells, hematopoietic cells, endothelial cells, MSCs, epicardial cells and the ESC- and iPSC-derived cardiomyocytes, with the anticipation that each type of cell delivery may benefit the damaged myocardium in a certain way (Ishida et al., 2019; Orlic et al., 2001; Yanamandala et al., 2017; Ye et al., 2014). However, the mechanistic basis for cell therapy still remains unclear. Successful delivery of ESC-derived cardiomyocytes has been achieved in non-human primates by the Murry group, with indications of success that included remuscularization of the heart and improved left ventricle function (Chong et al., 2014; Liu et al., 2018b). However, arrhythmias and low retention of cells have appeared in late preclinical trials. Emerging technologies, such as gene therapies to overexpress cell cycle activator CCND2 in iPSC-derived cardiomyocytes, has proven to increase remuscularization and angiogenesis of injured cardiac muscle once implanted in vivo (Zhu et al., 2018).
Engineered cardiac tissues can overcome the challenge of poor cell engraftment. One of the first studies that designed engineered cardiac tissue for transplantation in animal models was reported by Leor and Cohen in 2000, leading the way in assessing tissue engraftment and efficacy of repair of the damaged cardiac muscle (Leor et al., 2000). Similar to the in vitro models of the heart, many groups have used the recent success in iPSC-derived cardiomyocytes to engineer cardiac patches for regeneration (Bejleri et al., 2018; Jackman et al., 2018; Tiburcy et al., 2017; Zhang et al., 2018). Menasché and colleagues reported the first clinical study of ESC-derived cardiac progenitor cells in a 68-year-old patient, followed by a larger clinical trial in 6 patients with severe ischemic left ventricular dysfunction, suggesting the safety and efficacy of using pluripotent stem cells (Menasché et al., 2015; Menasché et al., 2018). While these clinical studies represent a major breakthrough for the field, critical challenges, such as patch vascularization and immune rejection, that still remain and should be addressed to ensure clinical translation (Edri et al., 2019; Fleischer et al., 2020).
Over the past five years, many groups have been attempting to incorporate vasculature into engineered cardiac tissues to ensure the immediate supply of oxygen and nutrients to the implanted cells (Gao et al., 2017; Ye et al., 2014). Zhang and Radisic demonstrated the development of an “AngioChip” system for engineered vasculature in highly-dense, millimeter-thick tissues that enables direct anastomosis to host for immediate perfusion (Zhang et al., 2016a). Future developments in this space will rely on vascularizing tissues and on their integration with host vasculature, using different bioengineering approaches, including 3D bioprinting (Lee et al., 2019; Noor et al., 2019; Skylar-Scott et al., 2019).
Other groups have studied the role of cell secreted micro-RNAs (miRNAs) in cardiac repair (Eulalio et al., 2012; Shao et al., 2017). Namely, the delivery of the secretome and extracellular cargo of regenerative cells has been indicated as a potential mechanism for alleviating cardiac damage following myocardial infarction (Liu et al., 2021). EVs containing miRNAs that are secreted by human ESCs, iPSCs, ECs, MSCs and iPSC-derived CMs have all been implicated in regeneration of the heart after exogenous delivery (Adamiak et al., 2018; Akbar et al., 2017; An et al., 2017; Bian et al., 2014; Khan et al., 2015). Recently, our group reported that extracellular vesicles collected from human iPSC-derived cardiomyocytes were superior to those collected from iPSCs in promoting a pro-regenerative phenotype in infarcted hearts, suggesting that the cardiogenic phenotype of cells used to produce EVs is important for treating infarcted hearts (Liu et al., 2018a). Another group has shown beneficial effects of EVs secreted by vasculogenic cells in treating infarcted heart models (Yadid et al., 2020). However, the engraftment of EVs, their off-target effects, and mechanisms of action (prevention or treatment of cardiac fibrosis) still remain as unknowns in translational cardiac repair.
Harnessing emerging bioengineering tools for cardiac regeneration
A new wave of bioengineering approaches to cardiac regeneration have appeared following advances in other fields. Over the past decade, gene editing technologies have enabled therapies based on manipulating diseased cells in vivo. Although much more work needs to be done, some groups have attempted functionalizing viral carriers into engineered scaffolds for controlled viral vector release (Gu et al., 2017). Qian and Srivastava reported in 2012 the selective reprogramming of cardiac fibroblasts into cardiomyocyte-like cells in situ (Qian et al., 2012), using transcriptional expression of Gata4, Mef2c, and Tbx5, though challenges remain in efficacy and functional maintenance long-term (Tani et al., 2018). For example, by understanding the role of ECM-derived proteins in development, Bassat and Tzahor uncovered the role of agrin in promoting cardiomyocyte proliferation and muscle regeneration post-MI, demonstrating the importance of the microenvironmental cues in promoting in vivo regeneration (Bassat et al., 2017). Recently, Aghajanian and Epstein demonstrated engineering of CD8+ T cells to target cardiac fibroblasts after infarction-induced fibrosis in mice (Aghajanian et al., 2019). This study, although at a proof-of-concept level, showed that targeting fibroblast activation protein via chimeric antigen receptors (CAR) on T cells could advance immunotherapy past their recent applications in treating cancer.
Outlook
Stem cell and cardiac tissue engineering therapies are currently not routinely used in the clinic (Menasché et al., 2018). Although these therapies have demonstrated capability to prevent or reduce heart injury, the precise mechanisms remain largely unknown (Curfman, 2019; Vagnozzi et al., 2020). Gaining a better mechanistic understanding of these processes can greatly advance the field towards clinical translation.
Recent developments in modeling the healthy and injured myocardium using OOCs (Giacomelli et al., 2020; Mastikhina et al., 2020; Ronaldson-Bouchard et al., 2018; Wang et al., 2019; Zhao et al., 2019a) suggest that these in vitro systems could emulate some aspects of injury and study regeneration therapies, in conjunction with animal models. The 2010 collaborative effort from the Zandstra and Radisic groups shows one of the first forays into the concept of using an engineered model to study regenerative cell therapy (Fig. 3C) (Song et al., 2010). In MI, engineered models of ischemia-reperfusion injury can help inform the administration of new drugs and biologics, by mimicking what is seen in the clinic (hours to days after heart attack) (Chen and Vunjak-Novakovic, 2018). Recent applications using exosome-based therapies in engineered models of MI have enabled the optimization of exosome delivery regimens necessary to decrease acute fibrosis (Wagner et al., 2020; Yadid et al., 2020). Models of disease are constantly improving, with cardiac organoids being used to model important hallmarks of MI - including metabolic shifts, fibrosis, and changes to functional calcium handling (Richards et al., 2020). In recent study of iPSC-derived cardiac microtissues by the Mummery group, cardiac stromal cells, endothelial cells and fibroblasts, were all found to be contributing to cardiomyocyte maturation, and reducing progression of cardiac injury (Giacomelli et al., 2020). Of particular note, the specificity of cell types from cardiovascular lineages, like in cardiac fibroblasts as compared to dermal fibroblasts, was important for better sarcomere formation in vitro. Understanding how stromal to parenchymal cell ratios in engineered models may parallel in vivo organ makeup is an ongoing effort in the field. In studying early gestational development, Drakhlis et al. recently demonstrated that human heart-forming organoids could model the structural complexity of early heart and foregut development, with potential to model genetic mutations causing cardiac malformation, another area where OOCs can serve as a template for regenerative interventions in congenital disease (Drakhlis et al., 2021).
OOC systems could also help identify novel mechanisms of action for regenerative therapies, while decoupling the effects of multiple factors, an important aspect that is difficult to achieve in animal models (Fig 4). Some of the outstanding questions of interest include:
How are regenerative treatments affecting tissue function over time (e.g., by attenuating myofibroblast activation and fibrosis, promoting cardiomyocyte survival, inducing vascularization)?
What are the underlying mechanisms of repair in different cell types, and what is the route of action (paracrine factors, direct contact)?
What is the timeframe for therapy? Are certain therapies more advantageous during the inflammatory phase, while others are beneficial during inflammatory resolution?
What role do immune cells play in the progression of MI and regeneration?
What are the changes that occur in the cardiac ECM following regenerative therapies (e.g., stiffening, degradation, deposition of specific proteins)?
Overall, modeling of cardiac injury and regeneration using OOCs would help advance future work and clinical translation.
Respiratory System: Modeling alveoli to advance lung regeneration
The lung airway is made of a dense network of branching units, from the proximal bifurcation of the mainstem bronchi to the most distal oxygen-exchanging alveoli, that are intertwined with the equally complex vascular network (Basil et al., 2020), resulting in gas exchange surface of nearly 200 m2. With a highly complex structure, about 23 airway generations, over 40 cell types, and 150 different native ECM components, recapitulating lung physiology and functionality for disease modeling or regeneration remains a major challenge in the field. Some of the first reports in the OOC field were from the Ingber group, where a model of the airway epithelium, with functional oxygen exchange and barrier protection metrics, was developed in 2010 (Huh et al., 2010). Many groups since then have recapitulated one or more of the functional metrics for bioengineered airway, and used these model systems to study systemic diseases. To date, only few groups have attempted large-scale, clinical-grade techniques for restoring lung function. As the lung is an extremely complex organ, several groups have relied on advances in bioengineering technologies that restore functional lungs starting from damaged or diseased donor lungs (Basil et al., 2020).
Models of lung physiology
Huh and Ingber demonstrated in 2010 the ability to create a perfusable alveolar epithelial-endothelial barrier in vitro (Fig. 5). This simple in vitro model was shown to recapitulate the organ-level functions of the lung, such as responses to bacterial infection, inflammation, and mechanical behavior simulating breathing (Huh et al., 2010). Since this seminal work, Ingber’s group has demonstrated the potential to use the “lung-on-a-chip” to study drug-induced pulmonary edema, intravascular thrombosis, and non-small-cell lung carcinoma (Hassell et al., 2017; Huh et al., 2012; Jain et al., 2018; Jain et al., 2016). These devices were benchmarked against clinically observed drug responses. One of the first and highly successful commercial ventures in the OOC space, a company Emulate, Inc., has emerged from this early work. Prior to this, many groups had studied primary human lung epithelium using 3D air-liquid-interface (ALI) models, that were able to recapitulate mucus secretion, cilia formation, and ECM remodeling (Pezzulo et al., 2011; Yamaya et al., 1992). Raredon and Niklason demonstrated that the use of a rolling bioreactor to establish a constantly dynamic ALI culture was important for increasing the number of ciliated cells, mucus secretion, and barrier function in differentiating epithelial cells (Raredon et al., 2015).
Figure 5. Engineered alveolar models for testing novel therapies for macro-scale lung regeneration.
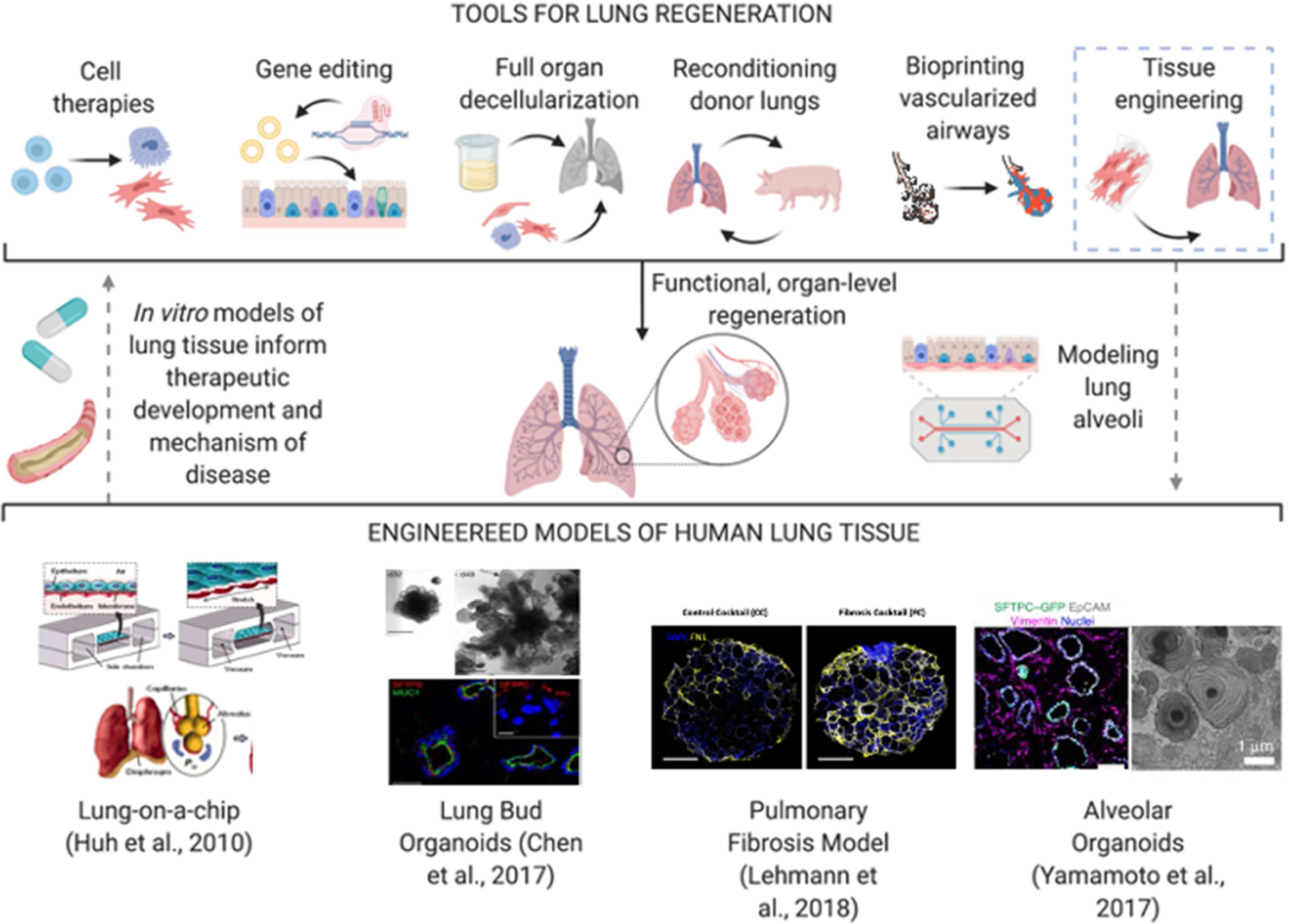
Top: Dotted lines demonstrate the envisioned pathway of using bioengineered tools to make mimetic models of the lung and use such models to inform new and refined therapeutic development. Bottom: Representative examples of lung tissue models: (Huh et al., 2010), Copyright 2010, AAAS; (Chen et al., 2017); (Lehmann et al., 2018), Reproduced under the terms of the Creative Commons License; (Yamamoto et al., 2017).
Starting from the donor lungs, many groups have established engineered systems to study the lung ex vivo for drug testing and disease progression in organotypic cultures. Königshoff and colleagues demonstrated the effective culture of primary human lung slices for up to 21 days, using samples from both healthy donors and patients with idiopathic pulmonary fibrosis (IPF), allowing enough time to assess efficacy of potential therapeutics (Alsafadi et al., 2017; Bailey et al., 2020; Lehmann et al., 2018). This approach takes advantage of using primary tissue samples to preserve alveolar architecture, as complex lung structures are often neglected in in vitro lung models.
To establish a source of human cells for lung regeneration, Snoeck and colleagues developed a protocol for highly efficient differentiation of lung progenitors and airway epithelial cells from pluripotent stem cell sources. In 2014, their group was able to generate basal, goblet, Clara, ciliated, type I and type II alveolar epithelial cells from ESC and iPSC sources, with effective functional metrics and progenitor proliferation in vitro (Huang et al., 2015; Huang et al., 2014). These studies were instrumental for providing a milieu for human, patient-specific drug testing and disease modeling in vitro (Snoeck, 2015). The group developed a 3D lung bud organoid model, capable of effectively producing mesodermal and pulmonary endodermal populations (Chen et al., 2017). These organoids had mobile cilia, secreted airway mucus, were maintained in vitro for over 170 days, engrafted with host vasculature and formed tubules following transplantation in immunodeficient mice. In addition, it was demonstrated that lung organoids were effective for disease modeling of viral infection, fibrosis, and genetic diseases of the lung (Porotto et al., 2019; Strikoudis et al., 2019; Yamamoto et al., 2017).
Organoids can be used for high throughput studies of disease modeling and drug screening. However, these models often lack essential cellular components, such as immune and vascular progenitors, which play major roles in homeostasis and response to injury. Like for most iPSC-derived models, maturation is a major concern, reaching only fetal-like status in even long-term (weeks to months) culture. Establishing gas exchange in lung organoids remains a challenge, as lack of integrated vasculature prevents this essential lung function (Tian et al., 2020). Further, lack of tissue-specific ECM overlooks the scarring that often progresses in diseases like IPF. 3D tissue ALI cultures, including those within microfluidic chips, incorporate perfusion flow to mimic the endothelial-epithelial barrier, but only at medium-to-low throughput, which limits their widespread use. Validation metrics include barrier function/permeability, multipotent epithelial cell maintenance and differentiation, and ability to respond to acute injury. As with any organ system, the balance between throughput and complexity is essential, with the question of interest often informing the choice of human model system.
Lung regeneration
Approaches to regenerating whole lungs have ranged from repopulating decellularized scaffolds with lung epithelial cells to clinical devices for normothermic ex vivo lung perfusion (EVLP), which aim to maintain lungs outside the body prior to transplant. The range of techniques have crossed the boundaries of repairing damaged donor lungs from the “top down” (in the case of EVLP) to creating new organs or tissues from the “bottom up” (in the case of tissue engineering) (Pinezich and Vunjak-Novakovic, 2019). In newer approaches, cell and gene delivery methods are aiming to target specific replacement of damaged epithelium without invasive transplantation (Dorrello and Vunjak-Novakovic, 2020).
In the early 2000s, bioengineers developed methodologies to remove the epithelial and endothelial cells from the lung matrix while maintaining the native lung architecture and ECM components (Gilbert et al., 2006). Ott et al. developed a protocol for bioreactor-based perfusion-decellularization of the heart, that maintained perfusable vascular channels and then recellularized the heart matrix with cardiomyocytes and endothelial cells over four weeks of bioreactor culture (Ott et al., 2008). In Ott and Vacanti’s follow up work in the lung, and in parallel studies by Peterson and Niklason, it was demonstrated that entire lungs can be decellularized, with preservation of intact hierarchical branching, and then recellularized by infusing epithelial and endothelial cells (Ott et al., 2010; Petersen et al., 2010). These engineered lungs were perfused through the vascular channels and ventilated through the airway at physiological conditions. Following implantation, the lungs survived for only a short period of time (several hours), largely due to the lack of functional vasculature. These studies demonstrated both the potential of this approach and the need to improve and extend the lung functionality. These seminal papers enabled the field to use decellularization of whole organs (heart, lungs, etc.), to obtain fully biological scaffolds for organ engineering. To maintain blood supply to the lung, Dorrello et al. demonstrated selective de-epithelization of the lung with the maintenance of intact vasculature and branching hierarchy (Dorrello et al., 2017). After re-seeding de-epithelialized lungs, human pulmonary epithelial cells were shown to engraft and expand, suggesting potential application in epithelial disease treatment and transplantation of bioengineered human lungs. Other targeted delivery methods have been explored for controlled delivery of cells, drugs, and reagents to restricted regions of the lungs, for spatial reorganization of damaged lung regions in vivo (Kim et al., 2017; Kim et al., 2015).
In the context of donor organ recovery, EVLP bioreactors have been widely explored and notably, translated into clinical use. Keshavjee and colleagues demonstrated the extension of normothermic EVLP to up to 12 hours (Cypel et al., 2008). In tandem, the group tested anti-inflammatory cytokine IL-10 on ex vivo perfused lungs, to avoid graft dysfunction and retain gas exchange functionality of alveoli prior to transplantation (Cypel et al., 2009). Our group has extended the duration of extracorporeal support of donor lungs on ventilation and cross-circulation with a recipient from 6–12 hours to as long as 100 hours (Hozain et al., 2020b; Huang et al., 2014). Using a host swine model as a bioreactor, donor lungs rejected for transplant were recovered and maintained ex vivo (Hozain et al., 2020b). These techniques enable a number of future bioengineering therapies in donor lungs, including cell and gene delivery, and can be extended to severely damaged lungs after gastric aspiration injury (Guenthart et al., 2019; Kim et al., 2015). The efficacy of cell therapy of the lung is yet to be proven in humans, though the technology holds immense potential for treatment of diseases that affect the epithelium (Ghadiri et al., 2016). In a more recent study by our group, we have extended the use of cross-circulation to regeneration of human donor lungs to the levels needed for transplantation, in the first study of its kind (Hozain et al., 2020a). Notably, donor lung vasculature remained intact for the duration of the study, with stable blood-gas barriers and no signs of angiopathy. In many cases of whole organ lung repair, maintaining stable and functional vasculature remains a major challenge, especially in regards to anastomosis to host tissue once implanted. Throughout the study, Hozain et al. assessed the metabolic and immunogenic reactions between the swine host and the human lungs. Using in vitro OOC models could be useful in these cases to test the xenogenic contributions of the host on the human tissue, and to assess any adverse reactions prior to transplantation.
Harnessing bioengineering tools for lung regeneration
Gene therapies targeting epithelial cells would allow for targeted correction of single-gene defects in inherited lung disorders. In vivo correction of the CFTR gene has been explored for treating Cystic Fibrosis (Griesenbach et al., 2016). Firth et al. demonstrated the ability to use CRISPR to correct iPSCs derived from cystic fibrosis patients with a deletion of F508 in the CFTR gene. After correcting this common mutation, they were able to generate airway epithelial cells from the corrected iPSCs, suggesting the potential for generating healthy, patient-matched lung epithelia for genetic lung diseases (Firth et al., 2015). Osman et al. recently developed polyethylene-glycol (PEG)-coated nanoparticles for increasing the efficacy of transfection in mucus-coated membranes, like the lung epithelia, enabling better in situ options for gene therapies (Osman et al., 2018). With major developments in high-resolution 3D biomanufacturing techniques, many groups, including the Feinberg and Lewis teams, have independently pursued the integration of perfusable vasculature into bioprinting of human organs, in the heart and kidney, as examples (Homan et al., 2019; Lee et al., 2019; Skylar-Scott et al., 2019). Of note, the Stevens and Miller groups recently reported the formation of highly complex, vascularized alveolar hydrogel structures using stereolithography (Grigoryan et al., 2019). Capable of ventilation and oxygenation in vitro, red blood cells flowed in and out of the alveolar structures, showing potential functional benefit of these engineered constructs in recapitulating critical gas-exchange properties of the lungs. Advances in biomanufacturing are helping target one of the most difficult challenges in lung regeneration, whereas scalable, multi-lobular lungs continue to remain a long-range goal for the bioprinting field.
Outlook
Advances in lung regeneration in situ and ex vivo show promise for restoring lung function in pre-clinical models. With advances in gene editing and bioengineering cell therapies, there is a large space for collaborative efforts to target genetic lung diseases, such as in cystic fibrosis, and restore functional lung epithelium. Many groups are developing strategies for studying genetic and acute lung diseases in vitro, using primary tissue samples for high biological fidelity and patient-specific models. In such diseased microenvironments, the roles of specific cellular components (epithelial cells, macrophages, and fibroblasts) and extracellular remodeling processes (collagen deposition, and MMP secretion) can be studied using OOC models, to facilitate therapeutic development (Fig. 5) (Castellani et al., 2018).
Models of the lung, including those that recapitulate alveolar gas exchange, can serve as tools for studying the effects of new therapeutics targeting epithelia. For example, these models could help (i) investigate how the microbiomes of healthy and diseased lungs impact their ability to respond to injury and therapeutics, (ii) how the innate and adaptive immune components remodel the alveolar microenvironments during injury and repair, (iii) what is the role of therapeutic MSCs in the epithelial-to-mesenchymal transition (Basil et al., 2020). With an organ as complex as the lung, it is essential to understand and decouple the roles of specific cell types, extracellular matrix, and environmental contaminants in the context of injury or disease. Lung progenitor cell populations are crucial for the natural regenerative processes, and replacing damaged epithelium comes with questions about maintaining the structure and function of this mechanically-dependent organ. For example, in cases of targeted replacement of epithelial cells, an engineered lung model can help determine whether damaged epithelial cells can be replaced to drive lung remodeling while the innate vasculature remains intact. Another interesting question is whether CRISPR-corrected iPSC-derived epithelial cells from CF patients are able to engraft and replace damaged mucosal epithelium (Firth et al., 2015). In most cases, precise alveolar architecture may be required for reliable in vitro testing, similar to the model described by the Stevens and Miller groups (Grigoryan et al., 2019).
Parallel experiments in large animal models that are close to human lung size and physiological function and in patient-specific OOC models, would help drive the development of new treatment modalities, speed up the route to translation, and provide personalized approaches to lung regeneration. OOC models could help mitigate lung injuries due to ischemia and gastric aspiration by serving as a test-bed for studying candidate therapies (Guenthart et al., 2019). With the emergence of cross-circulation system to regenerate damaged lungs, there is a need to better understand xenogeneic relationships between the porcine host and human lung, with both a presence and absence of immune components, which may exacerbate damage (Guenthart et al., 2019; Hozain et al., 2020a; Hozain et al., 2020b). Although there is potential to synergize OOC models with regenerative lung therapeutics (Fig. 5) there are many outstanding questions:
With over 40 types of cells with many different functions in the lung, how can we elucidate the effects of different treatment modalities on specific groups of cells?
What are the effects of regenerative therapies on the stromal and endothelial cell populations of the lung?
How do selective cell replacement therapies alter the alveolar microenvironment?
What is the time window for therapy during acute and chronic lung injury?
What are the immunological effects of autologous versus allogenic cell therapies? And what role do lung-resident immune cell populations play in it?
In the age of the SARS-CoV-2 (COVID-19) pandemic, engineered model systems are being repurposed for mechanistic studies of viral infection and therapeutic discovery and validation in human tissue platforms (Lamers et al., 2020; Monteil et al., 2020). For the lung, where the majority of fatalities result from acute respiratory distress syndrome (ARDS), epithelial organoids or lung OOC systems are proving to be excellent tools for disease modeling and therapeutic discovery (Abo et al., 2020; Mulay et al., 2020; Si et al., 2021). With the field of lung regeneration at its early stages, there is immense potential for using OOC models for mechanistic and proof-of-concept studies of bioengineered human lung tissues.
Blood and Immune System: Modeling the Bone Marrow for Regenerative Hematopoiesis
In hematopoiesis, it is widely accepted that a single blood stem cell has potential to regenerate the entire blood and immune system. Commonly found in the bone marrow, hematopoietic stem and progenitor cells (HSPCs) are regulated by a number of microenvironmental cues that regulate their self-renewal and differentiation, with ability to respond to stress signals throughout the body during times of injury. The bone marrow niche is highly complex, often requiring a variety of stromal cellular components to allow for proliferation of HSCs and differentiation of progenitors into downstream blood and myeloid/lymphoid lineages (Morrison and Scadden, 2014).
In recent years, different cellular components, including endothelial cells, adipocytes, and osteoblasts, have all been shown to regulate hematopoiesis in both health and disease (Coutu et al., 2017; Kobayashi et al., 2010; Naveiras et al., 2009; Zhou et al., 2017). In treating hematological disorders and cancers of the blood, full-body irradiation and subsequent hematopoietic stem cell transplant had been the traditional options for reducing or curing symptoms of disease (Copelan, 2006), with more recent developments in small molecule and CAR-T therapies reaching the clinic in recent years (June et al., 2018). Early success with HSC transplantation (HSCT) in the 1950s has allowed for the first successful stem cell transplants for regenerative therapies still ongoing to this date; however, approximately 25% of these transplants fail to engraft, causing downstream complications for patients. In addition, immunological rejection, like graft-versus-host disease (GVHD), can emerge as complications to immunosuppressants. With the large number of blood disorders and the growing number of blood and immune cell therapies in pursuit, approaches to studying these treatments in human, in vitro models may lead to more mimetic efficacy and safety research, as well as studying the mechanism of cell delivery, engraftment, and survival in the host tissue context.
Models of the hematopoietic niche
The murine hematopoietic niche has long been studied for preventing the onset of blood cancers and age-related exhaustion of hematopoietic cells (Doulatov et al., 2012). In fact, much of our understanding of the human hematological disorders comes from mouse models that are still a gold standard in the field. Also, many engineered models to study in vitro diseases of the blood and marrow rely on using murine hematopoietic cells (Fig. 6). The necessity of stroma in these models has been demonstrated already in the initial co-culture experiments leading the way into the in vitro platforms to study hematopoietic support (Bianco et al., 2008; Friedenstein et al., 1974; Majumdar et al., 2000; Walenda et al., 2010). Butler and Rafii were the first to show that cord blood-derived CD34+ HSPCs expanded significantly better in co-culture with endothelial cells than in monocultures (Butler et al., 2012).
Figure 6. Engineered models of the bone marrow for testing hematopoietic regeneration.
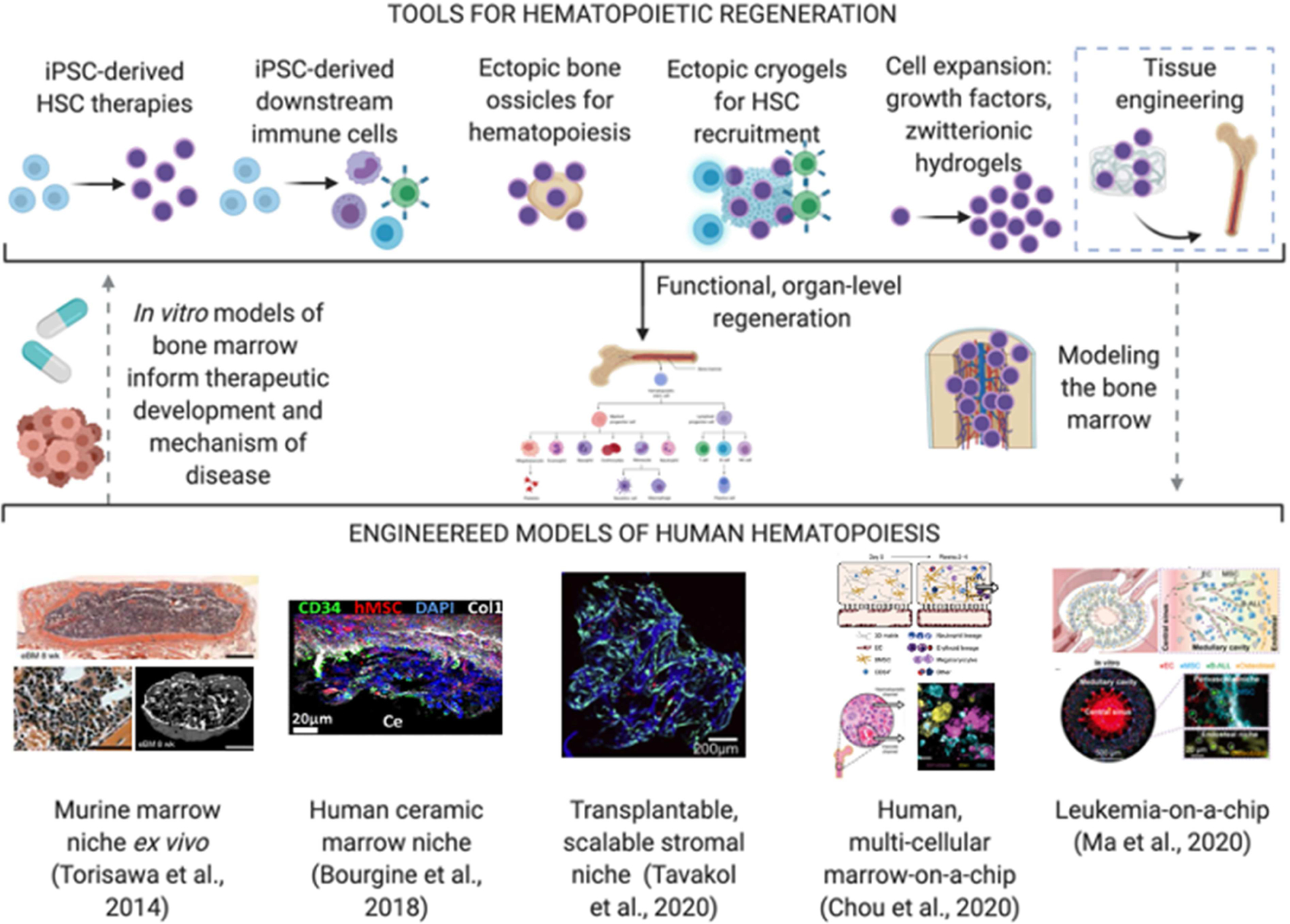
Top: Dotted lines demonstrate the envisioned pathway of using bioengineered tools to make mimetic models of the bone marrow and use such models to inform new and refined therapeutic development. Bottom: representative examples of hematopoietic models: (Torisawa et al., 2014); (Bourgine et al., 2018), Reproduced with permission, Copyright 2018, National Academy of Sciences; (Tavakol et al., 2019), Reproduced with Permission, Copyright 2019, Elsevier; (Chou et al., 2020); (Ma et al., 2020), Reproduced with permission, Copyright 2020, AAAS.
Other early approaches, such as the use of mesenchymal stromal cell spheroids to expand HSCs in vitro were developed to aid in expansion and engraftment in vivo (Isern et al., 2013). In one of the first studies using ectopic extramedullary hematopoietic niches, or sites of hematopoiesis other than the host bone marrow, Tavassoli and Crosby showed in 1968 that hematopoiesis was established in subcutaneous rat tissues after transplantation of stromal cells. The use of ectopic sites has paved a way for a number of hematopoietic models (Tavassoli and Crosby, 1968). Studies of hematopoiesis were markedly enhanced by the development of human ectopic hematopoietic niches through scaffold-free transplantation of MSC-derived cartilage tissues and selective differentiation into osteogenic/hematopoietic lineages via developmentally inspired approaches, like endochondral ossification, subcutaneously in animals (Scotti et al., 2013). Xenotransplantation of humanized stromal “ossicles” in mice have been used in studies of leukemia, affording a useful model system to study healthy and leukemic cell engraftment in a system capable for easy biopsy accessibility from subcutaneous tissue (Reinisch et al., 2016).
In the first “bone marrow-on-a-chip” model, Torisawa and Ingber established in 2014 murine hematopoiesis by implanting a PDMS mold with bone-inducing materials subcutaneously, explanting the engineered tissue with recruited stromal and hematopoietic cells, and testing the metrics for radio- and cyto-toxicity of the marrow (Torisawa et al., 2014). This work used murine cells with limited culture times and is yet to be extended to wide-scale application of human models. In 2018, the first fully human OOC model of hematopoiesis was developed by the Martin group, using MSC-derived osteoblasts and cord-blood HSPCs in a ceramic scaffold, to demonstrate hematopoietic support and response to injury in vitro (Bourgine et al., 2018). In a follow up study, a multi-cellular, human model was developed using endothelial cells and MSCs within fibrin hydrogels to support cord-blood derived HSPCs (Chou et al., 2020). This study demonstrated the clinical utility of the marrow model to study Shwachman-Diamond syndrome, as well as various toxicities of hematopoietic cells in response to radiation or chemotherapeutic exposure. The human models, though at their early stages, have opened the door to studies of diseases of blood stem cells and their progenitors, and off-target measures of marrow toxicity.
In modeling immune cell function in vitro, bioengineering approaches have been explored to study single cell types in response to various stimuli or disease states. Spiller and colleagues have shown the utility of macrophages in driving vascularization of tissue engineered materials, elucidating the role of macrophage subtype differentiation in early wound healing (Graney et al., 2020; Spiller et al., 2014). Other groups have explored the in vitro formation of neutrophil extracellular traps in response to material stiffness, modeling the in vivo response to biomaterial implantation (Abaricia et al., 2021). For studying more complex adaptive immune responses in vitro, several groups have developed lymphoid organs for modeling B and T cell responses in vitro. In recent years, generation of a lymph node-on-a-chip device enabled studies of dendritic and T cell interactions, including priming T cell activation in response to varying shear stresses (Moura Rosa et al., 2016). Similarly, the feasibility of deriving B cell organoids for modeling the germinal center response in helping propagate and differentiate naïve B cells was reported (Purwada et al., 2015). Possible inclusion of secondary immune organs has been discussed in several excellent reviews (Irimia and Wang, 2018; Kim et al., 2019).
Like all OOCs, benchmarking hematopoietic and immune models to human patho(physiology) remains a challenge, as murine responses have been studied more extensively than human models (Doulatov et al., 2012). Validation metrics includes the ability to maintain hematopoiesis over time with multipotent colony forming potential, transcriptional similarity to adult BM-HSPCs, and cytokine-directed differentiation into multiple blood lineages. Capturing the complexities of the hematopoietic niche remains a futile challenge, as it is still not possible to study the entire hematopoietic landscape in one model system. Simple models of the BM can be incredibly important in studying basic immune function and myelopoiesis, often providing a native, self-repopulating niche of immune progenitors to respond to injury. However, the engineered models of the BM generally lack maturation of progenitors to distinct differentiated cells, especially to adaptive immune cells. Integrating models of the BM with secondary lymphoid organs may prove effective at mimicking immune responses, after these models are benchmarked and validated against whole organism responses.
Regenerative hematopoiesis
HSC transplants have resulted in the survival and recovery of millions of patients over the past 50 years (Singh and McGuirk, 2016). However, there is still a need for therapeutic cells that can be expanded in vitro while retaining capacity to engraft in vivo. Although hematopoietic cells are phenotypically HSCs via antigen identification, not all of these cells are functionally supporting blood regeneration. In vitro, colony-forming-unit (CFU) assays are often used to identify the multipotent, colony-forming cells. In vivo, the gold-standard for HSC functionality is engraftment in a host marrow niche after primary and secondary transplantation. Therefore, it is opportune to use these assays to validate the efficacy of HSCs derived from different sources and expand these cells in vitro. Many efforts over the past two decades have focused on deriving HSCs from ESC and iPSCs, using transcription factor overexpression, isolation from teratomas, and growth factor delivery (Irion et al., 2010; Lengerke et al., 2009; Wang et al., 2005).
Many groups have identified the need to induce a vascular phenotype, starting with a hemogenic endothelial population as found during development, prior to hematopoietic commitment (Kennedy et al., 2012; Sandler et al., 2014; Sturgeon et al., 2014). Sugimura and Daley demonstrated in 2017 effective generation of engraftable, functional HSCs from human iPS and ES cells by differentiation into a hemogenic endothelium fate and subsequent expression of seven transcription factors (ERG, HOXA5, HOXA9, HOXA10, LCOR, RUNX1 and SPI1) (Sugimura et al., 2017). These cells were able to produce myeloid, B, and T cells and engraft in primary and secondary mouse transplantation, with the efficacy shown in half of the experiments. This translational improvement further enables modeling of patient-specific blood disorders in vitro towards developing therapeutic applications for autologous HSCT (Doulatov et al., 2017; Georgomanoli and Papapetrou, 2019). Of note, mutations in iPSCs derived from acute myeloid leukemia patients are retained after reprograming, with gene expression signatures and methylation changes reacquired once differentiated to hematopoietic cells (Chao et al., 2017; Kotini et al., 2017). More recently, Wang and Papapetrou demonstrated the gradual clonal evolution of leukemia from pre-malignant states (clonal hematopoiesis and myelodysplastic syndrome) to malignant acute myeloid leukemia using gene editing of iPSCs (Wang et al., 2021). As in native hematopoiesis, the stromal microenvironment from engineered models is still required to study chemotherapeutics, as leukemic blasts often alter their environment to propagate resistance and relapse (Rashidi and DiPersio, 2016). In response to these needs, in vitro models of leukemia are beginning to be explored (de la Puente et al., 2015; Ma et al., 2020).
Recently, Lis et al. demonstrated conversion of adult murine endothelial populations into HSCs through temporary overexpression of transcription factor-encoding genes Fosb, Gfi1, Runx1, and Spi1, potentially opening up a new source of cells that is independent of pluripotent cells (Barcia Durán et al., 2018; Lis et al., 2017). Though the promise that more directed differentiation from adult cell populations would alleviate the lengthy iPSC timeline and risk of inefficient pluripotent differentiation capacity, the potential for future malignant transformation of EC-derived HSCs has not been explored past 20 weeks in vivo. Importantly, the loss of epigenetic endothelial properties over longer periods of time was not confirmed and future studies are needed to address this important question. In addition to cell sourcing, maintaining and expanding HSCs in vitro has been a challenge. Different approaches have used different components to expand HSCs in vitro (high cytokine/growth factor culture media, hydrogels, co-cultures) in a way that maintains their functional capacity (Bello et al., 2018; Blank et al., 2008; Conneally et al., 1997; Soffer-Tsur et al., 2017; Zonari et al., 2017), though this still remains a translational limitation in the field.
With early successes in differentiating iPSCs into hematopoietic progenitors, further advances in generating downstream immune cell types have reinvigorated the field for clinically-applicable, iPSC-based cell therapies. In 2012, T lymphocyte progenitors were generated from iPSC-derived definitive hematopoietic cells, independent of activin/nodal signaling and with potential to drive myeloid, erythroid, and lymphoid differentiations in vitro (Kennedy et al., 2012). Shortly after, the generation of iPSC-derived T cells was demonstrated, this time with the addition of engineered CARs targeted to CD19, with anti-tumor activity both in vitro and in vivo, demonstrating feasibility in using patient-specific T cells for cancer therapies (Themeli et al., 2013). A similar approach was harnessed to generate natural killer (NK) cell-specific-CAR-expressing iPSC-NK cells, with increased anti-tumor efficacy in comparison to primary NK cells and T-antigen-specific CAR-iPSC-NK-cells (Li et al., 2018).
In addition, iPSC-derived NK-CAR-specific NK cells are important for maintaining the populations of cells that may be manipulated for gene editing more easily than their primary cell counterparts, as well as for unlimited generation of NK cells in vitro prior to in vivo transfusion. By knocking down CISH, a negative regulator for IL-15 signaling, in iPSC-NK cells, Zhu et al. demonstrated more efficient metabolic activity and anti-tumor persistence in a xenograft model of acute myeloid leukemia (Zhu et al., 2020). CAR-iPSC-NK cells have numerous alternative benefits, as they are less prone to GVHD than CAR-iPSC-T cells, though both technologies are still being evaluated for safety, efficacy, and scalability for future “off-the-shelf” cell immunotherapies (Depil et al., 2020; Iriguchi et al., 2021). While clinical trials in primary-based NK and CAR-T cell therapies are underway and have already shown success in some cancers, including leukemia (Davila et al., 2014; Park et al., 2018), iPSC-derived alternatives are just beginning to reach phase I/II clinical trials. Fate Therapeutics has been gaining momentum with the development of iPSC-derived NK cells and gene edited iPSC-derived NK cells for treatment of hematological malignancies, among other cancers. These major advancements in cell therapies are expected to reach clinical translation faster than HSC therapies, which can have immense implications in treating not only cancers, but also autoimmune diseases and heart disease, among other applications.
Harnessing new bioengineering tools for hematopoietic regeneration
In the past year, a few breakthrough studies have generated great potential for the expansion of HSCs for transplantation. Wilkinson and Yamazaki identified that high concentrations of thrombopoietin, in addition to using polyvinyl alcohol as albumin replacement, resulted in a ~200–900- fold expansion of functional HSCs (Wilkinson et al., 2019). This work demonstrates that in vitro culture, although heterogenous in its clonal repopulation capacity, may be useful for expanding HSCs prior to transplantation, to increase engraftment capability for competitive HSCT. In parallel, Bai et al. showed that using a 3D zwitterionic hydrogel markedly increased the efficacy of HSC expansion in vitro, by decreasing the production of reactive oxygen species (Bai et al., 2019).
Shah et al. took advantage of ectopic, extramedullary hematopoietic compartments, using engineered bone marrow cryogels to enhance T cell generation in hosts undergoing allogenic HSCT (Shah et al., 2019). At the time of HSCT, cryogels were implanted subcutaneously, serving as a depot for specifying lymphoid progenitors into mature donor CD4+ T cells. These collaborative efforts from the Mooney and Scadden groups demonstrated the use of cryogel scaffolds to enhance donor T-Cell proliferation and T-cell receptor diversification post HSCT, resulting in reduced GVHD and the need for excessive immunosuppressants for HSCT recipients (Shah et al., 2019).
In mechanistic studies of hematopoietic engraftment and differentiation in vivo, Dong et al. showed recently the ability to track the differentiation timeline of single HSCs via single-cell transcriptomic analysis, revealing the fate decisions and timelines of transplanted hematopoietic cells, previously inaccessible to HSCT patients (Dong et al., 2020). Single-cell analysis has enabled the discovery of novel transcription factors essential for self-renewal and quiescence in HSCs (Rodriguez-Fraticelli et al., 2020). By using a combination of CRISPR-Cas9 gene editing and transcriptomic analysis, bioengineered systems to study blood and immune cell function can be further optimized to study hematopoietic regeneration in a patient-specific context. These technologies are likely to enable new efforts to expand cord blood- or pluripotent-derived HSCs prior to transplant.
In expanding downstream blood and immune populations for transplantation, maintaining the delicate phenotype of functional iPSC-derived blood and immune cells ex vivo remains a challenge. Recently, Montel-Hagen et al. showed the maintenance and maturation of early T cells derived from iPSCs in artificial thymic organoids, producing naïve CD3+, CD8+ and CD3+CD4+ T cells that aligned functionally and molecularly to primary single-positive T cells (Montel-Hagen et al., 2019). In generation of large numbers of iPSC-derived myeloid cells, stirred bioreactors are also being used to generate downstream cells, including macrophages, for eventual use in cell therapies targeting acute infection (Ackermann et al., 2018). Similarly, turbulence-controlled bioreactors were shown to help promote thrombopoiesis and scalable culture of iPSC-derived megakaryocytes and platelets (Ito et al., 2018). Efforts to develop “off-the-shelf” cells at scalable quantities and engineer multi-cellular immune niches to drive maturation in vitro are further summarized elsewhere (Smerchansky and Kinney, 2020).
Outlook
Innovative use of biomaterials for regenerative hematopoiesis is starting to allow for ex vivo expansion of hematopoietic cells, potentially serving as a test bed for studying disease mechanisms and effects of drugs in vitro (Fig. 6). For example, the teams of Naveiras and Braschler recently co-cultured MSCs and HSPCs on 3D collagen-coated cryogel particles for efficient cytokine-free hematopoietic culture in vitro and subsequent transplantation in vivo (Tavakol et al., 2019). With facile aggregation of co-cultured particles and subcutaneous injection without the need for surgery, these engineered niches retained hematopoietic and supported stromal populations for up to 12 weeks in mice. These methodologies suggest new opportunities to study hematopoiesis; however, further validation in human subjects should be conducted to validate the clinical feasibility of these approaches.
OOCs could provide a test-bed for in vitro studies of hematopoietic proliferation and engraftment to inform preclinical and clinical trials. Studies of competitive hematopoiesis and whether healthy, regenerative cells can out-perform diseased, malignant cells could be conducted using the OOC models of the marrow (Fig. 6). In addition, OOC models provide a powerful tool to understanding the underlying signaling pathways and cellular interactions involved in these processes and shed new light on the mechanisms of repair.
OOC models could help determine the effective timelines for immunotherapy, the need for alignment with the traditional radio/chemotherapy, and the numbers and phenotypical state of antigen-specific immune cells necessary for complete remission of hematological cancers. Though in vivo work will continue to be essential, iPSC-derived NK and T cells in tumor spheroid models can provide a toolset for optimizing the route of cell delivery and efficacy, and for studies of the underlying mechanisms of action (Cichocki et al., 2020). In microfluidic OOCs, recent work by Khademhosseini and colleagues demonstrated the efficacious use of an anti-PD-1 immune-checkpoint inhibitor in studying immune-breast tumor spheroid interactions (Jiang et al.). iPSC-derived cells, if used in such integrated immune models, still do not benchmark closely to adult immune responses. Future work must build on these types of studies for design of integrated, multi-organ systems with multiple immune components, like a living bone marrow, for studies of how progenitor and downstream immune-interactions may influence disease models or regenerative strategies in a human context (Graney et al., 2021). Open questions remaining include:
By what mechanism, whether by direct cell-to-cell contact or paracrine signaling, do stromal populations interact with hematopoietic cells during health and disease?
What role do extracellular matrix components play in hematopoietic malignancies?
What is the timeline of hematopoietic regeneration and how do niche components, including endothelial cells and adipocytes, contribute to this regeneration?
How can we mimic both adaptive and innate immune arms in the in vitro models of the blood and immune system?
Can we design testing platforms to study immunotherapies using human biopsies?
It is advantageous to use engineered hematopoietic models, in tandem with animal models and clinical data, to study the progression of disease, responses to environmental stimuli, and the regenerative capabilities in HSCT preparations in a human-relevant context.
Future Perspectives
With major advancements in stem cell biology, bioengineers can now use patient-specific cells to generate human tissues in vitro. In combination with natural or synthetic biomaterials, stem cells are being used to engineer both micro- or macro-scale tissues for a variety of purposes. In the future, standardization and scaling up of cell production will be essential for translational advances of larger scale tissues or increased throughput of tissue models. As cells derived from iPSC sources are immature and difficult to maintain in vitro, better methods are needed to modulate cell phenotypes into committed, matured cells and precise benchmarks should be defined (Sharma et al., 2020). Another critical challenge in the use of iPSCs is understanding the effects of their epigenetic and genomic changes during reprogramming, and how these changes might propagate over long periods of time in vivo or in OOC models (Pera, 2011).
Engineered systems that can begin to emulate native tissue functions, and in particular OOCs, can be useful in many ways to recapitulate organ phenotypes for drug testing and disease modeling, and to study organ- and tissue-level functional characteristics in vitro (Fig. 1). OOCs with multiple organ systems are of interest for studying organ-organ interactions that are important for studies of systemic conditions. For example, a drug in the body is being metabolized in liver prior to reaching other tissue compartments, a process that could be recapitulated using OOCs. In recent years, there has been an increasing focus on multi-organ interactions, including a number of groups focused on examining the on- and off-target effect of anti-cancer therapeutics in devices with as few as two and as many as ten coupled organs (Chramiec et al., 2020; Edington et al., 2018; Herland et al., 2020; McAleer et al., 2019). The Griffith and Ingber groups have both demonstrated the feasibility of coupling multiple tissue systems by transferring culture medium between tissue compartments, enabling modeling pharmacokinetic-pharmacodynamic relationships. Recently, it was demonstrated that an OOC model of the reproductive tracts was able to produce the hormonal profile of the female menstrual cycle (Xiao et al., 2017). With the increasing interest in sex-specific studies and the recent NIH requirement to include sex as a biological variable, it would be of interest to include hormone secreting reproductive organs to better emulate female and male physiology.
There is also a need to optimize OOC models to study vascular-tissue interactions, mimic perfusion of blood through a tissue, drug distribution dynamics, and eventually model drug and cell infiltration into the injury sites. In a model of the Blood-Brain-Barrier, Park et al. demonstrated the role of the brain microvascular endothelium, coupled with pericyte and astrocyte supporting cells, in regulating transport of drugs, antibodies, and peptides (Park et al., 2019). George and Kamm have done this effectively for studying tumor metastasis (Jeon et al., 2015; Moya et al., 2013). Since there are significant differences in endothelium phenotype between different tissues, future research would benefit from developing tissue-specific vasculature and mimicking in vivo functionality within each tissue compartment (Fleischer et al., 2020).
Using different types of cells and tissues in the same OOC poses a number of challenges, in particular with respect to the use of the same culture medium for all cell/tissue types (common medium). Future work will likely need to find ways to appropriately prepare and selectively maintain essential media compositions to support diverse tissue types.
Although OOC systems offer advantages including humanized and personalized design, animal models will still be essential to understanding the effects of different therapeutic measures on an entire organism and for cognitive and behavioral studies. Parallel studies will be essential in understanding both mechanisms of therapeutic action, by decoupling different variables (OOCs), and complex organism-wide components (animal models); the United States Food & Drug Administration (FDA) is already working towards integrating OOC models within their therapeutic approval pipeline (Ingber, 2020).
As the field grows, interest is developing for integrating immunity into OOCs to increase the biological fidelity, especially in recapitulating disease phenotypes. To date, many microfluidic OOC systems that study immune interactions with a circulatory component have included only one or few cell types, including examples of circulating monocytes (McAleer et al., 2019), neutrophils (Benam et al., 2016), and regulatory/helper T cells (Trapecar et al., 2021). As many studies of physiological regeneration have demonstrated the important roles of the innate and adaptive immune system in regeneration (Chung et al., 2017; Dick et al., 2019; Tang et al., 2017), engineered OOC systems need to recapitulate aspects of immunity in order to effectively test potential adverse reactions to regenerative strategies (Sadtler et al., 2016). Sadtler and Elisseeff have reported on the varying immune responses elicited from both biological and synthetic scaffolds in mice, with responses ranging from a few days to a few months post-implantation showing increasing neutrophil infiltration in synthetic scaffolds, especially those with higher material stiffnesses (Sadtler et al., 2019). Dvir and colleagues have shown the feasibility of generating omentum-based hydrogels with iPSC-derived cells to immunogenically match implants to the donor (Edri et al., 2019). These native biomaterials were assessed for their immunogenicity after incubation with separate fractions of myeloid/lymphoid lineages from the donor. Mimicking these regenerative biomaterial strategies using complex OOC systems with functional immune components would be an effective next step in assessing human responses to regenerative tools.
Despite their utility, OOCs cannot capture complex organism-wide behaviors, unexpected organ-organ interactions, and effects of environmental cues. However, the level of control that can be attained is unparalleled by other methodologies, and thus enables researchers to decouple numerous factors and gain mechanistic insights into tissue homeostasis, disease, and regeneration.
Ongoing considerations:
Establishing benchmarks by the scientific community is critical for each OOC system and can help standardize model systems between institutions
“How simple is complex enough?” – choice of model system is dependent on a specific case-by-case application basis and the biological question
Level of maturity for iPSC-derived cells and tissues need to defined and reported in models
Choice of iPSC cell sources and population groups can enable personalized medicine; autologous “all iPSC-derived” models may not always be necessary
Advances in stem cell differentiations and biofabrication techniques are required for the fabrication of mature tissues at higher throughput
Development and integration of biosensors and “real-time” readouts is required for serial data collection to obtain a better understanding of regeneration or disease progression
Scaling organ systems in “Human-on-a-Chip” models will better recapitulate physiological organ size and functional load in the body
Refining which organs or organ systems are necessary to include in “Human-on-a-Chip” models for studying a specific disease phenotype or drug response – and whether omission of one engineered model may affect the efficacy of a potential therapeutic
Collaborative networks, including funding agencies, can help promote synergy between those developing regenerative therapeutics and those developing models of disease
Conclusion
Novel and repurposed regenerative therapies are increasingly relying on the information obtained in OOC systems, as these human models offer important validation data prior to early clinical trials. In addition, regenerative therapy can be mechanistically understood using OOCs, as many factors can be decoupled, allowing for systematic understanding of how a treatment is affecting each individual component and what components are the key players in this process. In parallel, OOCs can also be used to test drugs for safety, which in turn would speed up, de-risk, and lower the cost of clinical trials. Patient-specific factors, such as sex and genetic background, can be accounted for in OOC models. These testing platforms can help identify target populations for up-and-coming technologies, as personalized medicine has been emerging in a number of treatment plans. With significant progress in recent years, the use of micro-scale tissue models to study macro-scale organ regeneration is of increasingly high interest for translation of regenerative technologies.
ACKNOWLEDGMENTS
The authors gratefully acknowledge funding support of NIH (grants EB025765, EB027062, HL076485, HL120046, CA249799), NSF (grant ERC 16478; Graduate Research Fellowship DGE1644869), NYSTEM (grant C32606GG), Columbia SEAS-HICCC Pilot Grant, and Blavatnik Foundation STAR grant. All figures made in part from Biorender.com.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
G.V.-N is a co-founder of Tara Biosystems, a Columbia University startup company commercializing organs-on-a-chip with human heart muscle, as well as EpiBone, a Columbia University startup company commercializing human bone grafts for regeneration.
REFERENCES
- Abaricia JO, Shah AH, and Olivares-Navarrete R (2021). Substrate Stiffness Induces Neutrophil Extracellular Traps (NETs) Formation through Focal Adhesion Kinase Activation. Biomaterials, 120715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abo KM, Ma L, Matte T, Huang J, Alysandratos KD, Werder RB, Mithal A, Beermann ML, Lindstrom-Vautrin J, Mostoslavsky G, et al. (2020). Human iPSC-derived alveolar and airway epithelial cells can be cultured at air-liquid interface and express SARS-CoV-2 host factors. bioRxiv. [Google Scholar]
- Ackermann M, Kempf H, Hetzel M, Hesse C, Hashtchin AR, Brinkert K, Schott JW, Haake K, Kühnel MP, Glage S, et al. (2018). Bioreactor-based mass production of human iPSC-derived macrophages enables immunotherapies against bacterial airway infections. Nature Communications 9, 5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamiak M, Cheng G, Bobis-Wozowicz S, Zhao L, Kedracka-Krok S, Samanta A, Karnas E, Xuan YT, Skupien-Rabian B, Chen X, et al. (2018). Induced Pluripotent Stem Cell (iPSC)-Derived Extracellular Vesicles Are Safer and More Effective for Cardiac Repair Than iPSCs. Circ Res 122, 296–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aghajanian H, Kimura T, Rurik JG, Hancock AS, Leibowitz MS, Li L, Scholler J, Monslow J, Lo A, Han W, et al. (2019). Targeting cardiac fibrosis with engineered T cells. Nature 573, 430–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akbar N, Digby JE, Cahill TJ, Tavare AN, Corbin AL, Saluja S, Dawkins S, Edgar L, Rawlings N, Ziberna K, et al. (2017). Endothelium-derived extracellular vesicles promote splenic monocyte mobilization in myocardial infarction. JCI Insight 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akins RE, Boyce RA, Madonna ML, Schroedl NA, Gonda SR, McLaughlin TA, and Hartzell CR (1999). Cardiac organogenesis in vitro: reestablishment of three-dimensional tissue architecture by dissociated neonatal rat ventricular cells. Tissue Eng 5, 103–118. [DOI] [PubMed] [Google Scholar]
- Alsafadi HN, Staab-Weijnitz CA, Lehmann M, Lindner M, Peschel B, Königshoff M, and Wagner DE (2017). An ex vivo model to induce early fibrosis-like changes in human precision-cut lung slices. American Journal of Physiology-Lung Cellular and Molecular Physiology 312, L896–L902. [DOI] [PubMed] [Google Scholar]
- Amini AR, Laurencin CT, and Nukavarapu SP (2012). Bone Tissue Engineering: Recent Advances and Challenges. 40, 363–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An M, Kwon K, Park J, Ryu DR, Shin JA, Lee Kang J, Choi JH, Park EM, Lee KE, Woo M, et al. (2017). Extracellular matrix-derived extracellular vesicles promote cardiomyocyte growth and electrical activity in engineered cardiac atria. Biomaterials 146, 49–59. [DOI] [PubMed] [Google Scholar]
- Bai T, Li J, Sinclair A, Imren S, Merriam F, Sun F, O’Kelly MB, Nourigat C, Jain P, Delrow JJ, et al. (2019). Expansion of primitive human hematopoietic stem cells by culture in a zwitterionic hydrogel. Nat Med 25, 1566–1575. [DOI] [PubMed] [Google Scholar]
- Bailey KE, Pino C, Lennon ML, Lyons A, Jacot JG, Lammers SR, Königshoff M, and Magin CM (2020). Embedding of Precision-Cut Lung Slices in Engineered Hydrogel Biomaterials Supports Extended Ex Vivo Culture. Am J Respir Cell Mol Biol 62, 14–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barcia Durán JG, Lis R, Lu TM, and Rafii S (2018). In vitro conversion of adult murine endothelial cells to hematopoietic stem cells. Nat Protoc 13, 2758–2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bargehr J, Ong LP, Colzani M, Davaapil H, Hofsteen P, Bhandari S, Gambardella L, Le Novère N, Iyer D, Sampaziotis F, et al. (2019). Epicardial cells derived from human embryonic stem cells augment cardiomyocyte-driven heart regeneration. Nature biotechnology 37, 895–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basil MC, Katzen J, Engler AE, Guo M, Herriges MJ, Kathiriya JJ, Windmueller R, Ysasi AB, Zacharias WJ, Chapman HA, et al. (2020). The Cellular and Physiological Basis for Lung Repair and Regeneration: Past, Present, and Future. Cell Stem Cell 26, 482–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassat E, Mutlak YE, Genzelinakh A, Shadrin IY, Baruch Umansky K, Yifa O, Kain D, Rajchman D, Leach J, Riabov Bassat D, et al. (2017). The extracellular matrix protein agrin promotes heart regeneration in mice. Nature 547, 179–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejleri D, Streeter BW, Nachlas ALY, Brown ME, Gaetani R, Christman KL, and Davis ME (2018). A Bioprinted Cardiac Patch Composed of Cardiac-Specific Extracellular Matrix and Progenitor Cells for Heart Repair. Adv Healthc Mater 7, e1800672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bello AB, Park H, and Lee SH (2018). Current approaches in biomaterial-based hematopoietic stem cell niches. Acta Biomater 72, 1–15. [DOI] [PubMed] [Google Scholar]
- Benam KH, Villenave R, Lucchesi C, Varone A, Hubeau C, Lee HH, Alves SE, Salmon M, Ferrante TC, Weaver JC, et al. (2016). Small airway-on-a-chip enables analysis of human lung inflammation and drug responses in vitro. Nat Methods 13, 151–157. [DOI] [PubMed] [Google Scholar]
- Bhatia SN, and Ingber DE (2014). Microfluidic organs-on-chips. Nature Biotechnology 32, 760–772. [DOI] [PubMed] [Google Scholar]
- Bhise NS, Ribas J, Manoharan V, Zhang YS, Polini A, Massa S, Dokmeci MR, and Khademhosseini A (2014). Organ-on-a-chip platforms for studying drug delivery systems. Journal of Controlled Release 190, 82–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian S, Zhang L, Duan L, Wang X, Min Y, and Yu H (2014). Extracellular vesicles derived from human bone marrow mesenchymal stem cells promote angiogenesis in a rat myocardial infarction model. J Mol Med (Berl) 92, 387–397. [DOI] [PubMed] [Google Scholar]
- Bianco P, Robey PG, and Simmons PJ (2008). Mesenchymal stem cells: revisiting history, concepts, and assays. Cell Stem Cell 2, 313–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blank U, Karlsson G, and Karlsson S (2008). Signaling pathways governing stem-cell fate. Blood 111, 492–503. [DOI] [PubMed] [Google Scholar]
- Boudou T, Legant WR, Mu A, Borochin MA, Thavandiran N, Radisic M, Zandstra PW, Epstein JA, Margulies KB, and Chen CS (2012). A microfabricated platform to measure and manipulate the mechanics of engineered cardiac microtissues. Tissue Eng Part A 18, 910–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgine PE, Klein T, Paczulla AM, Shimizu T, Kunz L, Kokkaliaris KD, Coutu DL, Lengerke C, Skoda R, Schroeder T, et al. (2018). In vitro biomimetic engineering of a human hematopoietic niche with functional properties. Proc Natl Acad Sci U S A 115, E5688–e5695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bursac N, Papadaki M, Cohen RJ, Schoen FJ, Eisenberg SR, Carrier R, Vunjak-Novakovic G, and Freed LE (1999). Cardiac muscle tissue engineering: toward an in vitro model for electrophysiological studies. Am J Physiol 277, H433–444. [DOI] [PubMed] [Google Scholar]
- Butler JM, Gars EJ, James DJ, Nolan DJ, Scandura JM, and Rafii S (2012). Development of a vascular niche platform for expansion of repopulating human cord blood stem and progenitor cells. Blood 120, 1344–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrier RL, Papadaki M, Rupnick M, Schoen FJ, Bursac N, Langer R, Freed LE, and Vunjak-Novakovic G (1999). Cardiac tissue engineering: cell seeding, cultivation parameters, and tissue construct characterization. Biotechnol Bioeng 64, 580–589. [DOI] [PubMed] [Google Scholar]
- Caspi O, Lesman A, Basevitch Y, Gepstein A, Arbel G, Habib IH, Gepstein L, and Levenberg S (2007). Tissue engineering of vascularized cardiac muscle from human embryonic stem cells. Circ Res 100, 263–272. [DOI] [PubMed] [Google Scholar]
- Castellani S, Di Gioia S, di Toma L, and Conese M (2018). Human Cellular Models for the Investigation of Lung Inflammation and Mucus Production in Cystic Fibrosis. Anal Cell Pathol (Amst) 2018, 3839803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao MP, Gentles AJ, Chatterjee S, Lan F, Reinisch A, Corces MR, Xavy S, Shen J, Haag D, Chanda S, et al. (2017). Human AML-iPSCs Reacquire Leukemic Properties after Differentiation and Model Clonal Variation of Disease. Cell stem cell 20, 329–344.e327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Wu JY, Kennedy KM, Yeager K, Bernhard JC, Ng JJ, Zimmerman BK, Robinson S, Durney KM, Shaeffer C, et al. (2020). Tissue engineered autologous cartilage-bone grafts for temporomandibular joint regeneration. Science Translational Medicine 12, eabb6683. [DOI] [PubMed] [Google Scholar]
- Chen T, and Vunjak-Novakovic G (2018). Human Tissue-Engineered Model of Myocardial Ischemia–Reperfusion Injury. Tissue Engineering Part A 25, 711–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y-W, Huang SX, de Carvalho ALRT, Ho S-H, Islam MN, Volpi S, Notarangelo LD, Ciancanelli M, Casanova J-L, Bhattacharya J, et al. (2017). A three-dimensional model of human lung development and disease from pluripotent stem cells. Nature Cell Biology 19, 542–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong JJH, Yang X, Don CW, Minami E, Liu Y-W, Weyers JJ, Mahoney WM, Van Biber B, Cook SM, Palpant NJ, et al. (2014). Human embryonic-stem-cell-derived cardiomyocytes regenerate non-human primate hearts. Nature 510, 273–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopra A, Kutys ML, Zhang K, Polacheck WJ, Sheng CC, Luu RJ, Eyckmans J, Hinson JT, Seidman JG, Seidman CE, et al. (2018). Force Generation via β-Cardiac Myosin, Titin, and α-Actinin Drives Cardiac Sarcomere Assembly from Cell-Matrix Adhesions. Dev Cell 44, 87–96.e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou DB, Frismantas V, Milton Y, David R, Pop-Damkov P, Ferguson D, MacDonald A, Vargel Bölükbaşı Ö, Joyce CE, Moreira Teixeira LS, et al. (2020). On-chip recapitulation of clinical bone marrow toxicities and patient-specific pathophysiology. Nat Biomed Eng 4, 394–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chramiec A, Teles D, Yeager K, Marturano-Kruik A, Pak J, Chen T, Hao L, Wang M, Lock R, Tavakol DN, et al. (2020). Integrated human organ-on-a-chip model for predictive studies of anti-tumor drug efficacy and cardiac safety. Lab on a Chip 20, 4357–4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung L, Maestas DR, Housseau F, and Elisseeff JH (2017). Key players in the immune response to biomaterial scaffolds for regenerative medicine. Advanced Drug Delivery Reviews 114, 184–192. [DOI] [PubMed] [Google Scholar]
- Cichocki F, Bjordahl R, Gaidarova S, Mahmood S, Abujarour R, Wang H, Tuininga K, Felices M, Davis ZB, Bendzick L, et al. (2020). iPSC-derived NK cells maintain high cytotoxicity and enhance in vivo tumor control in concert with T cells and anti–PD-1 therapy. Science Translational Medicine 12, eaaz5618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conneally E, Cashman J, Petzer A, and Eaves C (1997). Expansion in vitro of transplantable human cord blood stem cells demonstrated using a quantitative assay of their lympho-myeloid repopulating activity in nonobese diabetic–scid/scid mice. Proceedings of the National Academy of Sciences 94, 9836–9841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copelan EA (2006). Hematopoietic Stem-Cell Transplantation. New England Journal of Medicine 354, 1813–1826. [DOI] [PubMed] [Google Scholar]
- Coutu DL, Kokkaliaris KD, Kunz L, and Schroeder T (2017). Three-dimensional map of nonhematopoietic bone and bone-marrow cells and molecules. Nat Biotechnol 35, 1202–1210. [DOI] [PubMed] [Google Scholar]
- Curfman G (2019). Stem Cell Therapy for Heart Failure: An Unfulfilled Promise? JAMA 321, 1186–1187. [DOI] [PubMed] [Google Scholar]
- Cypel M, Liu M, Rubacha M, Yeung JC, Hirayama S, Anraku M, Sato M, Medin J, Davidson BL, de Perrot M, et al. (2009). Functional Repair of Human Donor Lungs by IL-10 Gene Therapy. Science Translational Medicine 1, 4ra9–4ra9. [DOI] [PubMed] [Google Scholar]
- Cypel M, Yeung JC, Hirayama S, Rubacha M, Fischer S, Anraku M, Sato M, Harwood S, Pierre A, Waddell TK, et al. (2008). Technique for Prolonged Normothermic Ex Vivo Lung Perfusion. The Journal of Heart and Lung Transplantation 27, 1319–1325. [DOI] [PubMed] [Google Scholar]
- Davila ML, Riviere I, Wang X, Bartido S, Park J, Curran K, Chung SS, Stefanski J, Borquez-Ojeda O, Olszewska M, et al. (2014). Efficacy and toxicity management of 19–28z CAR T cell therapy in B cell acute lymphoblastic leukemia. Sci Transl Med 6, 224ra225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Puente P, Muz B, Gilson RC, Azab F, Luderer M, King J, Achilefu S, Vij R, and Azab AK (2015). 3D tissue-engineered bone marrow as a novel model to study pathophysiology and drug resistance in multiple myeloma. Biomaterials 73, 70–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depil S, Duchateau P, Grupp SA, Mufti G, and Poirot L (2020). ‘Off-the-shelf’ allogeneic CAR T cells: development and challenges. Nature Reviews Drug Discovery 19, 185–199. [DOI] [PubMed] [Google Scholar]
- Dick SA, Macklin JA, Nejat S, Momen A, Clemente-Casares X, Althagafi MG, Chen J, Kantores C, Hosseinzadeh S, Aronoff L, et al. (2019). Self-renewing resident cardiac macrophages limit adverse remodeling following myocardial infarction. Nature Immunology 20, 29–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong F, Hao S, Zhang S, Zhu C, Cheng H, Yang Z, Hamey FK, Wang X, Gao A, Wang F, et al. (2020). Differentiation of transplanted haematopoietic stem cells tracked by single-cell transcriptomic analysis. Nature Cell Biology 22, 630–639. [DOI] [PubMed] [Google Scholar]
- Dorrello NV, Guenthart BA, O’Neill JD, Kim J, Cunningham K, Chen Y-W, Biscotti M, Swayne T, Wobma HM, Huang SXL, et al. (2017). Functional vascularized lung grafts for lung bioengineering. Science Advances 3, e1700521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorrello NV, and Vunjak-Novakovic G (2020). Bioengineering of Pulmonary Epithelium With Preservation of the Vascular Niche. Front Bioeng Biotechnol 8, 269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doulatov S, Notta F, Laurenti E, and Dick John E. (2012). Hematopoiesis: A Human Perspective. Cell Stem Cell 10, 120–136. [DOI] [PubMed] [Google Scholar]
- Doulatov S, Vo LT, Macari ER, Wahlster L, Kinney MA, Taylor AM, Barragan J, Gupta M, McGrath K, Lee HY, et al. (2017). Drug discovery for Diamond-Blackfan anemia using reprogrammed hematopoietic progenitors. Sci Transl Med 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drakhlis L, Biswanath S, Farr C-M, Lupanow V, Teske J, Ritzenhoff K, Franke A, Manstein F, Bolesani E, Kempf H, et al. (2021). Human heart-forming organoids recapitulate early heart and foregut development. Nature Biotechnology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edington CD, Chen WLK, Geishecker E, Kassis T, Soenksen LR, Bhushan BM, Freake D, Kirschner J, Maass C, Tsamandouras N, et al. (2018). Interconnected Microphysiological Systems for Quantitative Biology and Pharmacology Studies. Scientific Reports 8, 4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edri R, Gal I, Noor N, Harel T, Fleischer S, Adadi N, Green O, Shabat D, Heller L, Shapira A, et al. (2019). Personalized Hydrogels for Engineering Diverse Fully Autologous Tissue Implants. Advanced Materials 31, 1803895. [DOI] [PubMed] [Google Scholar]
- Eng G, Lee BW, Protas L, Gagliardi M, Brown K, Kass RS, Keller G, Robinson RB, and Vunjak-Novakovic G (2016). Autonomous beating rate adaptation in human stem cell-derived cardiomyocytes. Nature Communications 7, 10312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eschenhagen T, Fink C, Remmers U, Scholz H, Wattchow J, Weil J, Zimmermann W, Dohmen HH, Schäfer H, Bishopric N, et al. (1997). Three-dimensional reconstitution of embryonic cardiomyocytes in a collagen matrix: a new heart muscle model system. Faseb j 11, 683–694. [DOI] [PubMed] [Google Scholar]
- Eulalio A, Mano M, Dal Ferro M, Zentilin L, Sinagra G, Zacchigna S, and Giacca M (2012). Functional screening identifies miRNAs inducing cardiac regeneration. Nature 492, 376–381. [DOI] [PubMed] [Google Scholar]
- Feiner R, Engel L, Fleischer S, Malki M, Gal I, Shapira A, Shacham-Diamand Y, and Dvir T (2016). Engineered hybrid cardiac patches with multifunctional electronics for online monitoring and regulation of tissue function. Nat Mater 15, 679–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feyen DAM, McKeithan WL, Bruyneel AAN, Spiering S, Hörmann L, Ulmer B, Zhang H, Briganti F, Schweizer M, Hegyi B, et al. (2020). Metabolic Maturation Media Improve Physiological Function of Human iPSC-Derived Cardiomyocytes. Cell Reports 32, 107925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink C, Ergün S, Kralisch D, Remmers U, Weil J, and Eschenhagen T (2000). Chronic stretch of engineered heart tissue induces hypertrophy and functional improvement. The FASEB Journal 14, 669–679. [DOI] [PubMed] [Google Scholar]
- Firth AL, Menon T, Parker GS, Qualls SJ, Lewis BM, Ke E, Dargitz CT, Wright R, Khanna A, Gage FH, et al. (2015). Functional Gene Correction for Cystic Fibrosis in Lung Epithelial Cells Generated from Patient iPSCs. Cell Rep 12, 1385–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleischer S, Feiner R, and Dvir T (2017). Cutting-edge platforms in cardiac tissue engineering. Current Opinion in Biotechnology 47, 23–29. [DOI] [PubMed] [Google Scholar]
- Fleischer S, Tavakol DN, and Vunjak-Novakovic G (2020). From Arteries to Capillaries: Approaches to Engineering Human Vasculature. Advanced Functional Materials n/a, 1910811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedenstein AJ, Chailakhyan RK, Latsinik NV, Panasyuk AF, and Keiliss-Borok IV (1974). STROMAL CELLS RESPONSIBLE FOR TRANSFERRING THE MICROENVIRONMENT OF THE HEMOPOIETIC TISSUES: Cloning In Vitro and Retransplantation In Vivo. Transplantation 17, 331–340. [DOI] [PubMed] [Google Scholar]
- Gao L, Kupfer ME, Jung JP, Yang L, Zhang P, Da Sie Y, Tran Q, Ajeti V, Freeman BT, Fast VG, et al. (2017). Myocardial Tissue Engineering With Cells Derived From Human-Induced Pluripotent Stem Cells and a Native-Like, High-Resolution, 3-Dimensionally Printed Scaffold. Circ Res 120, 1318–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgomanoli M, and Papapetrou EP (2019). Modeling blood diseases with human induced pluripotent stem cells. Dis Model Mech 12, dmm039321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghadiri M, Young PM, and Traini D (2016). Cell-based therapies for the treatment of idiopathic pulmonary fibrosis (IPF) disease. Expert Opin Biol Ther 16, 375–387. [DOI] [PubMed] [Google Scholar]
- Giacomelli E, Meraviglia V, Campostrini G, Cochrane A, Cao X, van Helden RWJ, Krotenberg Garcia A, Mircea M, Kostidis S, Davis RP, et al. (2020). Human-iPSC-Derived Cardiac Stromal Cells Enhance Maturation in 3D Cardiac Microtissues and Reveal Non-cardiomyocyte Contributions to Heart Disease. Cell stem cell 26, 862–879.e811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert TW, Sellaro TL, and Badylak SF (2006). Decellularization of tissues and organs. Biomaterials 27, 3675–3683. [DOI] [PubMed] [Google Scholar]
- Goldfracht I, Protze S, Shiti A, Setter N, Gruber A, Shaheen N, Nartiss Y, Keller G, and Gepstein L (2020). Generating ring-shaped engineered heart tissues from ventricular and atrial human pluripotent stem cell-derived cardiomyocytes. Nature Communications 11, 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graney PL, Ben-Shaul S, Landau S, Bajpai A, Singh B, Eager J, Cohen A, Levenberg S, and Spiller KL (2020). Macrophages of diverse phenotypes drive vascularization of engineered tissues. Science Advances 6, eaay6391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graney PL, Tavakol DN, Chramiec A, Ronaldson-Bouchard K, and Vunjak-Novakovic G (2021). Engineered Models of Tumor Metastasis with Immune Cell Contributions. iScience, 102179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griesenbach U, Davies JC, and Alton E (2016). Cystic fibrosis gene therapy: a mutation-independent treatment. Curr Opin Pulm Med 22, 602–609. [DOI] [PubMed] [Google Scholar]
- Grigoryan B, Paulsen SJ, Corbett DC, Sazer DW, Fortin CL, Zaita AJ, Greenfield PT, Calafat NJ, Gounley JP, Ta AH, et al. (2019). Multivascular networks and functional intravascular topologies within biocompatible hydrogels. Science 364, 458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosberg A, Alford PW, McCain ML, and Parker KK (2011). Ensembles of engineered cardiac tissues for physiological and pharmacological study: heart on a chip. Lab Chip 11, 4165–4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X, Matsumura Y, Tang Y, Roy S, Hoff R, Wang B, and Wagner WR (2017). Sustained viral gene delivery from a micro-fibrous, elastomeric cardiac patch to the ischemic rat heart. Biomaterials 133, 132–143. [DOI] [PubMed] [Google Scholar]
- Guenthart BA, O’Neill JD, Kim J, Queen D, Chicotka S, Fung K, Simpson M, Donocoff R, Salna M, Marboe CC, et al. (2019). Regeneration of severely damaged lungs using an interventional cross-circulation platform. Nature Communications 10, 1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassell BA, Goyal G, Lee E, Sontheimer-Phelps A, Levy O, Chen CS, and Ingber DE (2017). Human Organ Chip Models Recapitulate Orthotopic Lung Cancer Growth, Therapeutic Responses, and Tumor Dormancy In Vitro. Cell Rep 21, 508–516. [DOI] [PubMed] [Google Scholar]
- Herland A, Maoz BM, Das D, Somayaji MR, Prantil-Baun R, Novak R, Cronce M, Huffstater T, Jeanty SSF, Ingram M, et al. (2020). Quantitative prediction of human pharmacokinetic responses to drugs via fluidically coupled vascularized organ chips. Nature Biomedical Engineering. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinson JT, Chopra A, Lowe A, Sheng CC, Gupta RM, Kuppusamy R, O’Sullivan J, Rowe G, Wakimoto H, Gorham J, et al. (2016). Integrative Analysis of PRKAG2 Cardiomyopathy iPS and Microtissue Models Identifies AMPK as a Regulator of Metabolism, Survival, and Fibrosis. Cell Rep 17, 3292–3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinson JT, Chopra A, Nafissi N, Polacheck WJ, Benson CC, Swist S, Gorham J, Yang L, Schafer S, Sheng CC, et al. (2015). HEART DISEASE. Titin mutations in iPS cells define sarcomere insufficiency as a cause of dilated cardiomyopathy. Science (New York, NY) 349, 982–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang P, Wang J, Conklin BR, Healy KE, and Ma Z (2018). Generation of spatial-patterned early-developing cardiac organoids using human pluripotent stem cells. Nat Protoc 13, 723–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofer M, and Lutolf MP (2021). Engineering organoids. Nature Reviews Materials. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homan KA, Gupta N, Kroll KT, Kolesky DB, Skylar-Scott M, Miyoshi T, Mau D, Valerius MT, Ferrante T, Bonventre JV, et al. (2019). Flow-enhanced vascularization and maturation of kidney organoids in vitro. Nature Methods 16, 255–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hozain AE, O’Neill JD, Pinezich MR, Tipograf Y, Donocoff R, Cunningham KM, Tumen A, Fung K, Ukita R, Simpson MT, et al. (2020a). Xenogeneic cross-circulation for extracorporeal recovery of injured human lungs. Nat Med 26, 1102–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hozain AE, Tipograf Y, Pinezich MR, Cunningham KM, Donocoff R, Queen D, Fung K, Marboe CC, Guenthart BA, O’Neill JD, et al. (2020b). Multiday maintenance of extracorporeal lungs using cross-circulation with conscious swine. The Journal of Thoracic and Cardiovascular Surgery 159, 1640–1653.e1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang SX, Green MD, de Carvalho AT, Mumau M, Chen YW, D’Souza SL, and Snoeck HW (2015). The in vitro generation of lung and airway progenitor cells from human pluripotent stem cells. Nat Protoc 10, 413–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang SX, Islam MN, O’Neill J, Hu Z, Yang YG, Chen YW, Mumau M, Green MD, Vunjak-Novakovic G, Bhattacharya J, et al. (2014). Efficient generation of lung and airway epithelial cells from human pluripotent stem cells. Nat Biotechnol 32, 84–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh D, Leslie DC, Matthews BD, Fraser JP, Jurek S, Hamilton GA, Thorneloe KS, McAlexander MA, and Ingber DE (2012). A human disease model of drug toxicity-induced pulmonary edema in a lung-on-a-chip microdevice. Sci Transl Med 4, 159ra147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh D, Matthews BD, Mammoto A, Montoya-Zavala M, Hsin HY, and Ingber DE (2010). Reconstituting organ-level lung functions on a chip. Science 328, 1662–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingber DE (2020). Is it Time for Reviewer 3 to Request Human Organ Chip Experiments Instead of Animal Validation Studies? Advanced Science 7, 2002030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iriguchi S, Yasui Y, Kawai Y, Arima S, Kunitomo M, Sato T, Ueda T, Minagawa A, Mishima Y, Yanagawa N, et al. (2021). A clinically applicable and scalable method to regenerate T-cells from iPSCs for off-the-shelf T-cell immunotherapy. Nature Communications 12, 430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irimia D, and Wang X (2018). Inflammation-on-a-Chip: Probing the Immune System Ex Vivo. Trends Biotechnol 36, 923–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irion S, Clarke RL, Luche H, Kim I, Morrison SJ, Fehling H-J, and Keller GM (2010). Temporal specification of blood progenitors from mouse embryonic stem cells and induced pluripotent stem cells. Development 137, 2829–2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isern J, Martin-Antonio B, Ghazanfari R, Martin AM, Lopez JA, del Toro R, Sanchez-Aguilera A, Arranz L, Martin-Perez D, Suarez-Lledo M, et al. (2013). Self-renewing human bone marrow mesenspheres promote hematopoietic stem cell expansion. Cell Rep 3, 1714–1724. [DOI] [PubMed] [Google Scholar]
- Ishida M, Miyagawa S, Saito A, Fukushima S, Harada A, Ito E, Ohashi F, Watabe T, Hatazawa J, Matsuura K, et al. (2019). Transplantation of Human-induced Pluripotent Stem Cell-derived Cardiomyocytes Is Superior to Somatic Stem Cell Therapy for Restoring Cardiac Function and Oxygen Consumption in a Porcine Model of Myocardial Infarction. Transplantation 103, 291–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y, Nakamura S, Sugimoto N, Shigemori T, Kato Y, Ohno M, Sakuma S, Ito K, Kumon H, Hirose H, et al. (2018). Turbulence Activates Platelet Biogenesis to Enable Clinical Scale Ex Vivo Production. Cell 174, 636–648.e618. [DOI] [PubMed] [Google Scholar]
- Jackman CP, Carlson AL, and Bursac N (2016). Dynamic culture yields engineered myocardium with near-adult functional output. Biomaterials 111, 66–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackman CP, Ganapathi AM, Asfour H, Qian Y, Allen BW, Li Y, and Bursac N (2018). Engineered cardiac tissue patch maintains structural and electrical properties after epicardial implantation. Biomaterials 159, 48–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain A, Barrile R, van der Meer AD, Mammoto A, Mammoto T, De Ceunynck K, Aisiku O, Otieno MA, Louden CS, Hamilton GA, et al. (2018). Primary Human Lung Alveolus-on-a-chip Model of Intravascular Thrombosis for Assessment of Therapeutics. Clin Pharmacol Ther 103, 332–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain A, van der Meer AD, Papa A-L, Barrile R, Lai A, Schlechter BL, Otieno MA, Louden CS, Hamilton GA, Michelson AD, et al. (2016). Assessment of whole blood thrombosis in a microfluidic device lined by fixed human endothelium. Biomed Microdevices 18, 73–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon JS, Bersini S, Gilardi M, Dubini G, Charest JL, Moretti M, and Kamm RD (2015). Human 3D vascularized organotypic microfluidic assays to study breast cancer cell extravasation. Proceedings of the National Academy of Sciences 112, 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Ren L, Tebon P, Wang C, Zhou X, Qu M, Zhu J, Ling H, Zhang S, Xue Y, et al. Cancer-on-a-Chip for Modeling Immune Checkpoint Inhibitor and Tumor Interactions. Small n/a, 2004282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- June CH, O’Connor RS, Kawalekar OU, Ghassemi S, and Milone MC (2018). CAR T cell immunotherapy for human cancer. Science 359, 1361–1365. [DOI] [PubMed] [Google Scholar]
- Kapoun Ann M, Liang F, O’Young G, Damm Deborah L, Quon D, White RT, Munson K, Lam A, Schreiner George F, and Protter Andrew A (2004). B-Type Natriuretic Peptide Exerts Broad Functional Opposition to Transforming Growth Factor-β in Primary Human Cardiac Fibroblasts. Circulation Research 94, 453–461. [DOI] [PubMed] [Google Scholar]
- Kennedy M, Awong G, Sturgeon Christopher M., Ditadi A, LaMotte-Mohs R, Zúñiga-Pflücker Juan C., and Keller G (2012). T Lymphocyte Potential Marks the Emergence of Definitive Hematopoietic Progenitors in Human Pluripotent Stem Cell Differentiation Cultures. Cell Reports 2, 1722–1735. [DOI] [PubMed] [Google Scholar]
- Khan M, Nickoloff E, Abramova T, Johnson J, Verma SK, Krishnamurthy P, Mackie AR, Vaughan E, Garikipati VN, Benedict C, et al. (2015). Embryonic stem cell-derived exosomes promote endogenous repair mechanisms and enhance cardiac function following myocardial infarction. Circ Res 117, 52–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Guenthart B, O’Neill JD, Dorrello NV, Bacchetta M, and Vunjak-Novakovic G (2017). Controlled delivery and minimally invasive imaging of stem cells in the lung. Scientific Reports 7, 13082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, O’Neill JD, Dorrello NV, Bacchetta M, and Vunjak-Novakovic G (2015). Targeted delivery of liquid microvolumes into the lung. Proc Natl Acad Sci U S A 112, 11530–11535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Shah SB, Graney PL, and Singh A (2019). Multiscale engineering of immune cells and lymphoid organs. Nat Rev Mater 4, 355–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi H, Butler JM, O’Donnell R, Kobayashi M, Ding BS, Bonner B, Chiu VK, Nolan DJ, Shido K, Benjamin L, et al. (2010). Angiocrine factors from Akt-activated endothelial cells balance self-renewal and differentiation of haematopoietic stem cells. Nat Cell Biol 12, 1046–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotini AG, Chang C-J, Chow A, Yuan H, Ho T-C, Wang T, Vora S, Solovyov A, Husser C, Olszewska M, et al. (2017). Stage-Specific Human Induced Pluripotent Stem Cells Map the Progression of Myeloid Transformation to Transplantable Leukemia. Cell stem cell 20, 315–328.e317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laflamme MA, Chen KY, Naumova AV, Muskheli V, Fugate JA, Dupras SK, Reinecke H, Xu C, Hassanipour M, Police S, et al. (2007). Cardiomyocytes derived from human embryonic stem cells in pro-survival factors enhance function of infarcted rat hearts. Nat Biotechnol 25, 1015–1024. [DOI] [PubMed] [Google Scholar]
- Lamers MM, Beumer J, van der Vaart J, Knoops K, Puschhof J, Breugem TI, Ravelli RBG, Paul van Schayck J, Mykytyn AZ, Duimel HQ, et al. (2020). SARS-CoV-2 productively infects human gut enterocytes. Science 369, 50–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster MA, Renner M, Martin C-A, Wenzel D, Bicknell LS, Hurles ME, Homfray T, Penninger JM, Jackson AP, and Knoblich JA (2013). Cerebral organoids model human brain development and microcephaly. Nature 501, 373–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A, Hudson AR, Shiwarski DJ, Tashman JW, Hinton TJ, Yerneni S, Bliley JM, Campbell PG, and Feinberg AW (2019). 3D bioprinting of collagen to rebuild components of the human heart. Science 365, 482. [DOI] [PubMed] [Google Scholar]
- Lehmann M, Buhl L, Alsafadi HN, Klee S, Hermann S, Mutze K, Ota C, Lindner M, Behr J, Hilgendorff A, et al. (2018). Differential effects of Nintedanib and Pirfenidone on lung alveolar epithelial cell function in ex vivo murine and human lung tissue cultures of pulmonary fibrosis. Respir Res 19, 175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengerke C, Grauer M, Niebuhr NI, Riedt T, Kanz L, Park IH, and Daley GQ (2009). Hematopoietic development from human induced pluripotent stem cells. Ann N Y Acad Sci 1176, 219–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leor J, Aboulafia-Etzion S, Dar A, Shapiro L, Barbash IM, Battler A, Granot Y, and Cohen S (2000). Bioengineered cardiac grafts: A new approach to repair the infarcted myocardium? Circulation 102, Iii56–61. [DOI] [PubMed] [Google Scholar]
- Li Y, Hermanson DL, Moriarity BS, and Kaufman DS (2018). Human iPSC-Derived Natural Killer Cells Engineered with Chimeric Antigen Receptors Enhance Anti-tumor Activity. Cell stem cell 23, 181–192.e185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lis R, Karrasch CC, Poulos MG, Kunar B, Redmond D, Duran JGB, Badwe CR, Schachterle W, Ginsberg M, Xiang J, et al. (2017). Conversion of adult endothelium to immunocompetent haematopoietic stem cells. Nature 545, 439–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Lee BW, Nakanishi K, Villasante A, Williamson R, Metz J, Kim J, Kanai M, Bi L, Brown K, et al. (2018a). Cardiac recovery via extended cell-free delivery of extracellular vesicles secreted by cardiomyocytes derived from induced pluripotent stem cells. Nature biomedical engineering 2, 293–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Wang B, Zhang X, Lock R, Nash T, and Vunjak-Novakovic G (2021). Cell type–specific microRNA therapies for myocardial infarction. Science Translational Medicine 13, eabd0914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YW, Chen B, Yang X, Fugate JA, Kalucki FA, Futakuchi-Tsuchida A, Couture L, Vogel KW, Astley CA, Baldessari A, et al. (2018b). Human embryonic stem cell-derived cardiomyocytes restore function in infarcted hearts of non-human primates. Nat Biotechnol 36, 597–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low LA, Mummery C, Berridge BR, Austin CP, and Tagle DA (2020). Organs-on-chips: into the next decade. Nature Reviews Drug Discovery. [DOI] [PubMed] [Google Scholar]
- Ma C, Witkowski MT, Harris J, Dolgalev I, Sreeram S, Qian W, Tong J, Chen X, Aifantis I, and Chen W (2020). Leukemia-on-a-chip: Dissecting the chemoresistance mechanisms in B cell acute lymphoblastic leukemia bone marrow niche. Science Advances 6, eaba5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z, Huebsch N, Koo S, Mandegar MA, Siemons B, Boggess S, Conklin BR, Grigoropoulos CP, and Healy KE (2018). Contractile deficits in engineered cardiac microtissues as a result of MYBPC3 deficiency and mechanical overload. Nature Biomedical Engineering 2, 955–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacNeil S (2007). Progress and opportunities for tissue-engineered skin. Nature 445, 874–880. [DOI] [PubMed] [Google Scholar]
- MacQueen LA, Sheehy SP, Chantre CO, Zimmerman JF, Pasqualini FS, Liu X, Goss JA, Campbell PH, Gonzalez GM, Park S-J, et al. (2018). A tissue-engineered scale model of the heart ventricle. Nature Biomedical Engineering 2, 930–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumdar MK, Thiede MA, Haynesworth SE, Bruder SP, and Gerson SL (2000). Human Marrow-Derived Mesenchymal Stem Cells (MSCs) Express Hematopoietic Cytokines and Support Long-Term Hematopoiesis When Differentiated Toward Stromal and Osteogenic Lineages. Journal of Hematotherapy & Stem Cell Research 9, 841–848. [DOI] [PubMed] [Google Scholar]
- Mastikhina O, Moon BU, Williams K, Hatkar R, Gustafson D, Mourad O, Sun X, Koo M, Lam AYL, Sun Y, et al. (2020). Human cardiac fibrosis-on-a-chip model recapitulates disease hallmarks and can serve as a platform for drug testing. Biomaterials 233, 119741. [DOI] [PubMed] [Google Scholar]
- Mathur A, Loskill P, Shao K, Huebsch N, Hong S, Marcus SG, Marks N, Mandegar M, Conklin BR, Lee LP, et al. (2015). Human iPSC-based Cardiac Microphysiological System For Drug Screening Applications. Scientific Reports 5, 8883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAleer CW, Long CJ, Elbrecht D, Sasserath T, Bridges LR, Rumsey JW, Martin C, Schnepper M, Wang Y, Schuler F, et al. (2019). Multi-organ system for the evaluation of efficacy and off-target toxicity of anticancer therapeutics. Science Translational Medicine 11, eaav1386. [DOI] [PubMed] [Google Scholar]
- Menasché P, Vanneaux V, Hagège A, Bel A, Cholley B, Cacciapuoti I, Parouchev A, Benhamouda N, Tachdjian G, Tosca L, et al. (2015). Human embryonic stem cell-derived cardiac progenitors for severe heart failure treatment: first clinical case report. Eur Heart J 36, 2011–2017. [DOI] [PubMed] [Google Scholar]
- Menasché P, Vanneaux V, Hagège A, Bel A, Cholley B, Parouchev A, Cacciapuoti I, Al-Daccak R, Benhamouda N, Blons H, et al. (2018). Transplantation of Human Embryonic Stem Cell-Derived Cardiovascular Progenitors for Severe Ischemic Left Ventricular Dysfunction. J Am Coll Cardiol 71, 429–438. [DOI] [PubMed] [Google Scholar]
- Mills RJ, Parker BL, Quaife-Ryan GA, Voges HK, Needham EJ, Bornot A, Ding M, Andersson H, Polla M, Elliott DA, et al. (2019). Drug Screening in Human PSC-Cardiac Organoids Identifies Pro-proliferative Compounds Acting via the Mevalonate Pathway. Cell Stem Cell 24, 895–907.e896. [DOI] [PubMed] [Google Scholar]
- Mills RJ, Titmarsh DM, Koenig X, Parker BL, Ryall JG, Quaife-Ryan GA, Voges HK, Hodson MP, Ferguson C, Drowley L, et al. (2017). Functional screening in human cardiac organoids reveals a metabolic mechanism for cardiomyocyte cell cycle arrest. Proceedings of the National Academy of Sciences 114, E8372–E8381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteil V, Kwon H, Prado P, Hagelkrüys A, Wimmer RA, Stahl M, Leopoldi A, Garreta E, Hurtado Del Pozo C, Prosper F, et al. (2020). Inhibition of SARS-CoV-2 Infections in Engineered Human Tissues Using Clinical-Grade Soluble Human ACE2. Cell 181, 905–913.e907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montel-Hagen A, Seet CS, Li S, Chick B, Zhu Y, Chang P, Tsai S, Sun V, Lopez S, Chen HC, et al. (2019). Organoid-Induced Differentiation of Conventional T Cells from Human Pluripotent Stem Cells. Cell Stem Cell 24, 376–389.e378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison SJ, and Scadden DT (2014). The bone marrow niche for haematopoietic stem cells. Nature 505, 327–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moura Rosa P, Gopalakrishnan N, Ibrahim H, Haug M, and Halaas Ø (2016). The intercell dynamics of T cells and dendritic cells in a lymph node-on-a-chip flow device. Lab Chip 16, 3728–3740. [DOI] [PubMed] [Google Scholar]
- Moya ML, Hsu Y-H, Lee AP, Hughes CCW, and George SC (2013). In vitro perfused human capillary networks. Tissue Eng Part C Methods 19, 730–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulay A, Konda B, Garcia G, Yao C, Beil S, Sen C, Purkayastha A, Kolls JK, Pociask DA, Pessina P, et al. (2020). SARS-CoV-2 infection of primary human lung epithelium for COVID-19 modeling and drug discovery. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naveiras O, Nardi V, Wenzel PL, Hauschka PV, Fahey F, and Daley GQ (2009). Bone-marrow adipocytes as negative regulators of the haematopoietic microenvironment. Nature 460, 259–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noor N, Shapira A, Edri R, Gal I, Wertheim L, and Dvir T (2019). 3D Printing of Personalized Thick and Perfusable Cardiac Patches and Hearts. Advanced Science 6, 1900344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes SS, Miklas JW, Liu J, Aschar-Sobbi R, Xiao Y, Zhang B, Jiang J, Massé S, Gagliardi M, Hsieh A, et al. (2013). Biowire: a platform for maturation of human pluripotent stem cell-derived cardiomyocytes. Nat Methods 10, 781–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill JD, Pinezich MR, Guenthart BA, and Vunjak-Novakovic G (2021). Gut bioengineering strategies for regenerative medicine. American Journal of Physiology-Gastrointestinal and Liver Physiology 320, G1–G11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogle BM, Bursac N, Domian I, Huang NF, Menasché P, Murry CE, Pruitt B, Radisic M, Wu JC, Wu SM, et al. (2016). Distilling complexity to advance cardiac tissue engineering. Sci Transl Med 8, 342ps313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlic D, Kajstura J, Chimenti S, Jakoniuk I, Anderson SM, Li B, Pickel J, McKay R, Nadal-Ginard B, Bodine DM, et al. (2001). Bone marrow cells regenerate infarcted myocardium. Nature 410, 701–705. [DOI] [PubMed] [Google Scholar]
- Osman G, Rodriguez J, Chan SY, Chisholm J, Duncan G, Kim N, Tatler AL, Shakesheff KM, Hanes J, Suk JS, et al. (2018). PEGylated enhanced cell penetrating peptide nanoparticles for lung gene therapy. J Control Release 285, 35–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott HC, Clippinger B, Conrad C, Schuetz C, Pomerantseva I, Ikonomou L, Kotton D, and Vacanti JP (2010). Regeneration and orthotopic transplantation of a bioartificial lung. Nat Med 16, 927–933. [DOI] [PubMed] [Google Scholar]
- Ott HC, Matthiesen TS, Goh SK, Black LD, Kren SM, Netoff TI, and Taylor DA (2008). Perfusion-decellularized matrix: using nature’s platform to engineer a bioartificial heart. Nat Med 14, 213–221. [DOI] [PubMed] [Google Scholar]
- Park JH, Rivière I, Gonen M, Wang X, Sénéchal B, Curran KJ, Sauter C, Wang Y, Santomasso B, Mead E, et al. (2018). Long-Term Follow-up of CD19 CAR Therapy in Acute Lymphoblastic Leukemia. New England Journal of Medicine 378, 449–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park T-E, Mustafaoglu N, Herland A, Hasselkus R, Mannix R, FitzGerald EA, Prantil-Baun R, Watters A, Henry O, Benz M, et al. (2019). Hypoxia-enhanced Blood-Brain Barrier Chip recapitulates human barrier function and shuttling of drugs and antibodies. Nature Communications 10, 2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pashneh-Tala S, MacNeil S, and Claeyssens F (2015). The Tissue-Engineered Vascular Graft—Past, Present, and Future. Tissue Engineering Part B: Reviews 22, 68–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel NM, Yazdi IK, Tasciotti E, and Birla RK (2016). Optimizing cell seeding and retention in a three-dimensional bioengineered cardiac ventricle: The two-stage cellularization model. Biotechnology and Bioengineering 113, 2275–2285. [DOI] [PubMed] [Google Scholar]
- Pera MF (2011). The dark side of induced pluripotency. Nature 471, 46–47. [DOI] [PubMed] [Google Scholar]
- Petersen TH, Calle EA, Zhao L, Lee EJ, Gui L, Raredon MB, Gavrilov K, Yi T, Zhuang ZW, Breuer C, et al. (2010). Tissue-engineered lungs for in vivo implantation. Science (New York, NY) 329, 538–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pezzulo AA, Starner TD, Scheetz TE, Traver GL, Tilley AE, Harvey B-G, Crystal RG, Paul B McCray J, and Zabner J (2011). The air-liquid interface and use of primary cell cultures are important to recapitulate the transcriptional profile of in vivo airway epithelia. American Journal of Physiology-Lung Cellular and Molecular Physiology 300, L25–L31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinezich M, and Vunjak-Novakovic G (2019). Bioengineering approaches to organ preservation ex vivo. Exp Biol Med (Maywood) 244, 630–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires de Mello CP, Carmona-Moran C, McAleer CW, Perez J, Coln EA, Long CJ, Oleaga C, Riu A, Note R, Teissier S, et al. (2020). Microphysiological heart–liver body-on-a-chip system with a skin mimic for evaluating topical drug delivery. Lab on a Chip 20, 749–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porotto M, Ferren M, Chen YW, Siu Y, Makhsous N, Rima B, Briese T, Greninger AL, Snoeck HW, and Moscona A (2019). Authentic Modeling of Human Respiratory Virus Infection in Human Pluripotent Stem Cell-Derived Lung Organoids. mBio 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Protze SI, Liu J, Nussinovitch U, Ohana L, Backx PH, Gepstein L, and Keller GM (2017). Sinoatrial node cardiomyocytes derived from human pluripotent cells function as a biological pacemaker. Nature Biotechnology 35, 56–68. [DOI] [PubMed] [Google Scholar]
- Purwada A, Jaiswal MK, Ahn H, Nojima T, Kitamura D, Gaharwar AK, Cerchietti L, and Singh A (2015). Ex vivo engineered immune organoids for controlled germinal center reactions. Biomaterials 63, 24–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian L, Huang Y, Spencer CI, Foley A, Vedantham V, Liu L, Conway SJ, Fu JD, and Srivastava D (2012). In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature 485, 593–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radisic M, Park H, Shing H, Consi T, Schoen FJ, Langer R, Freed LE, and Vunjak-Novakovic G (2004). Functional assembly of engineered myocardium by electrical stimulation of cardiac myocytes cultured on scaffolds. Proceedings of the National Academy of Sciences 101, 18129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raredon MS, Ghaedi M, Calle EA, and Niklason LE (2015). A Rotating Bioreactor for Scalable Culture and Differentiation of Respiratory Epithelium. Cell Med 7, 109–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashidi A, and DiPersio JF (2016). Targeting the leukemia-stroma interaction in acute myeloid leukemia: rationale and latest evidence. Ther Adv Hematol 7, 40–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravenscroft SM, Pointon A, Williams AW, Cross MJ, and Sidaway JE (2016). Cardiac Non-myocyte Cells Show Enhanced Pharmacological Function Suggestive of Contractile Maturity in Stem Cell Derived Cardiomyocyte Microtissues. Toxicological Sciences 152, 99–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinisch A, Thomas D, Corces MR, Zhang X, Gratzinger D, Hong WJ, Schallmoser K, Strunk D, and Majeti R (2016). A humanized bone marrow ossicle xenotransplantation model enables improved engraftment of healthy and leukemic human hematopoietic cells. Nat Med 22, 812–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinlib L, and Field L (2000). Cell Transplantation as Future Therapy for Cardiovascular Disease? Circulation 101, e182–e187. [DOI] [PubMed] [Google Scholar]
- Richards DJ, Li Y, Kerr CM, Yao J, Beeson GC, Coyle RC, Chen X, Jia J, Damon B, Wilson R, et al. (2020). Human cardiac organoids for the modelling of myocardial infarction and drug cardiotoxicity. Nature Biomedical Engineering 4, 446–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Fraticelli AE, Weinreb C, Wang S-W, Migueles RP, Jankovic M, Usart M, Klein AM, Lowell S, and Camargo FD (2020). Single-cell lineage tracing unveils a role for TCF15 in haematopoiesis. Nature. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronaldson-Bouchard K, Ma SP, Yeager K, Chen T, Song L, Sirabella D, Morikawa K, Teles D, Yazawa M, and Vunjak-Novakovic G (2018). Advanced maturation of human cardiac tissue grown from pluripotent stem cells. Nature 556, 239–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronaldson-Bouchard K, and Vunjak-Novakovic G (2018). Organs-on-a-Chip: A Fast Track for Engineered Human Tissues in Drug Development. Cell Stem Cell 22, 310–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadtler K, Singh A, Wolf MT, Wang X, Pardoll DM, and Elisseeff JH (2016). Design, clinical translation and immunological response of biomaterials in regenerative medicine. Nature Reviews Materials 1, 16040. [Google Scholar]
- Sadtler K, Wolf MT, Ganguly S, Moad CA, Chung L, Majumdar S, Housseau F, Pardoll DM, and Elisseeff JH (2019). Divergent immune responses to synthetic and biological scaffolds. Biomaterials 192, 405–415. [DOI] [PubMed] [Google Scholar]
- Sandler VM, Lis R, Liu Y, Kedem A, James D, Elemento O, Butler JM, Scandura JM, and Rafii S (2014). Reprogramming human endothelial cells to haematopoietic cells requires vascular induction. Nature 511, 312–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ, et al. (2009). Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459, 262–265. [DOI] [PubMed] [Google Scholar]
- Schaaf S, Shibamiya A, Mewe M, Eder A, Stöhr A, Hirt MN, Rau T, Zimmermann W-H, Conradi L, Eschenhagen T, et al. (2011). Human Engineered Heart Tissue as a Versatile Tool in Basic Research and Preclinical Toxicology. PLoS One 6, e26397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scotti C, Piccinini E, Takizawa H, Todorov A, Bourgine P, Papadimitropoulos A, Barbero A, Manz MG, and Martin I (2013). Engineering of a functional bone organ through endochondral ossification. Proc Natl Acad Sci U S A 110, 3997–4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shadrin IY, Allen BW, Qian Y, Jackman CP, Carlson AL, Juhas ME, and Bursac N (2017). Cardiopatch platform enables maturation and scale-up of human pluripotent stem cell-derived engineered heart tissues. Nat Commun 8, 1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah NJ, Mao AS, Shih TY, Kerr MD, Sharda A, Raimondo TM, Weaver JC, Vrbanac VD, Deruaz M, Tager AM, et al. (2019). An injectable bone marrow-like scaffold enhances T cell immunity after hematopoietic stem cell transplantation. Nat Biotechnol 37, 293–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao L, Zhang Y, Lan B, Wang J, Zhang Z, Zhang L, Xiao P, Meng Q, Geng YJ, Yu XY, et al. (2017). MiRNA-Sequence Indicates That Mesenchymal Stem Cells and Exosomes Have Similar Mechanism to Enhance Cardiac Repair. Biomed Res Int 2017, 4150705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A, Burridge PW, McKeithan WL, Serrano R, Shukla P, Sayed N, Churko JM, Kitani T, Wu H, Holmström A, et al. (2017). High-throughput screening of tyrosine kinase inhibitor cardiotoxicity with human induced pluripotent stem cells. Science Translational Medicine 9, eaaf2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A, Sances S, Workman MJ, and Svendsen CN (2020). Multi-lineage Human iPSC-Derived Platforms for Disease Modeling and Drug Discovery. Cell Stem Cell 26, 309–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si L, Bai H, Rodas M, Cao W, Oh CY, Jiang A, Moller R, Hoagland D, Oishi K, Horiuchi S, et al. (2021). A human-airway-on-a-chip for the rapid identification of candidate antiviral therapeutics and prophylactics. Nature Biomedical Engineering. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidorov VY, Samson PC, Sidorova TN, Davidson JM, Lim CC, and Wikswo JP (2017). I-Wire Heart-on-a-Chip I: Three-dimensional cardiac tissue constructs for physiology and pharmacology. Acta Biomaterialia 48, 68–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh AK, and McGuirk JP (2016). Allogeneic Stem Cell Transplantation: A Historical and Scientific Overview. Cancer Research 76, 6445–6451. [DOI] [PubMed] [Google Scholar]
- Skylar-Scott MA, Uzel SGM, Nam LL, Ahrens JH, Truby RL, Damaraju S, and Lewis JA (2019). Biomanufacturing of organ-specific tissues with high cellular density and embedded vascular channels. Science Advances 5, eaaw2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smerchansky ME, and Kinney MA (2020). Engineered multicellular niches for pluripotent stem cell–derived immunotherapy. Current Opinion in Biomedical Engineering 16, 19–26. [Google Scholar]
- Snoeck HW (2015). Modeling human lung development and disease using pluripotent stem cells. Development 142, 13–16. [DOI] [PubMed] [Google Scholar]
- Soffer-Tsur N, Peer D, and Dvir T (2017). ECM-based macroporous sponges release essential factors to support the growth of hematopoietic cells. Journal of Controlled Release 257, 84–90. [DOI] [PubMed] [Google Scholar]
- Song H, Yoon C, Kattman SJ, Dengler J, Massé S, Thavaratnam T, Gewarges M, Nanthakumar K, Rubart M, Keller GM, et al. (2010). Interrogating functional integration between injected pluripotent stem cell-derived cells and surrogate cardiac tissue. Proceedings of the National Academy of Sciences 107, 3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiller KL, Anfang RR, Spiller KJ, Ng J, Nakazawa KR, Daulton JW, and Vunjak-Novakovic G (2014). The role of macrophage phenotype in vascularization of tissue engineering scaffolds. Biomaterials 35, 4477–4488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strikoudis A, Cieślak A, Loffredo L, Chen YW, Patel N, Saqi A, Lederer DJ, and Snoeck HW (2019). Modeling of Fibrotic Lung Disease Using 3D Organoids Derived from Human Pluripotent Stem Cells. Cell Rep 27, 3709–3723.e3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturgeon CM, Ditadi A, Awong G, Kennedy M, and Keller G (2014). Wnt signaling controls the specification of definitive and primitive hematopoiesis from human pluripotent stem cells. Nature Biotechnology 32, 554–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimura R, Jha DK, Han A, Soria-Valles C, da Rocha EL, Lu YF, Goettel JA, Serrao E, Rowe RG, Malleshaiah M, et al. (2017). Haematopoietic stem and progenitor cells from human pluripotent stem cells. Nature 545, 432–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun N, Yazawa M, Liu J, Han L, Sanchez-Freire V, Abilez OJ, Navarrete EG, Hu S, Wang L, Lee A, et al. (2012). Patient-specific induced pluripotent stem cells as a model for familial dilated cardiomyopathy. Sci Transl Med 4, 130ra147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J, Cui X, Caranasos TG, Hensley MT, Vandergriff AC, Hartanto Y, Shen D, Zhang H, Zhang J, and Cheng K (2017). Heart Repair Using Nanogel-Encapsulated Human Cardiac Stem Cells in Mice and Pigs with Myocardial Infarction. ACS nano 11, 9738–9749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tani H, Sadahiro T, and Ieda M (2018). Direct Cardiac Reprogramming: A Novel Approach for Heart Regeneration. Int J Mol Sci 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavakol DN, Tratwal J, Bonini F, Genta M, Campos V, Burch P, Hoehnel S, Béduer A, Alessandrini M, and Naveiras O (2019). Injectable, scalable 3D tissue-engineered model of marrow hematopoiesis. Biomaterials, 119665. [DOI] [PubMed] [Google Scholar]
- Tavassoli M, and Crosby WH (1968). Transplantation of Marrow to Extramedullary Sites. Science 161, 54–56. [DOI] [PubMed] [Google Scholar]
- Themeli M, Kloss CC, Ciriello G, Fedorov VD, Perna F, Gonen M, and Sadelain M (2013). Generation of tumor-targeted human T lymphocytes from induced pluripotent stem cells for cancer therapy. Nature Biotechnology 31, 928–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L, Gao J, Garcia IM, Chen HJ, Castaldi A, and Chen Y-W (2020). Human pluripotent stem cell-derived lung organoids: Potential applications in development and disease modeling. WIREs Developmental Biology n/a, e399. [DOI] [PubMed] [Google Scholar]
- Tiburcy M, Hudson JE, Balfanz P, Schlick S, Meyer T, Chang Liao ML, Levent E, Raad F, Zeidler S, Wingender E, et al. (2017). Defined Engineered Human Myocardium With Advanced Maturation for Applications in Heart Failure Modeling and Repair. Circulation 135, 1832–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torisawa Y. s., Spina CS, Mammoto T, Mammoto A, Weaver JC, Tat T, Collins JJ, and Ingber DE (2014). Bone marrow–on–a–chip replicates hematopoietic niche physiology in vitro. Nature Methods 11, 663–669. [DOI] [PubMed] [Google Scholar]
- Trapecar M, Wogram E, Svoboda D, Communal C, Omer A, Lungjangwa T, Sphabmixay P, Velazquez J, Schneider K, Wright CW, et al. (2021). Human physiomimetic model integrating microphysiological systems of the gut, liver, and brain for studies of neurodegenerative diseases. Sci Adv 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulloch NL, Muskheli V, Razumova MV, Korte FS, Regnier M, Hauch KD, Pabon L, Reinecke H, and Murry CE (2011). Growth of engineered human myocardium with mechanical loading and vascular coculture. Circ Res 109, 47–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagnozzi RJ, Maillet M, Sargent MA, Khalil H, Johansen AKZ, Schwanekamp JA, York AJ, Huang V, Nahrendorf M, Sadayappan S, et al. (2020). An acute immune response underlies the benefit of cardiac stem cell therapy. Nature 577, 405–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Spreeuwel ACC, Bax NAM, van Nierop BJ, Aartsma-Rus A, Goumans M-JTH, and Bouten CVC (2017). Mimicking Cardiac Fibrosis in a Dish: Fibroblast Density Rather than Collagen Density Weakens Cardiomyocyte Function. J Cardiovasc Transl Res 10, 116–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velazquez JJ, LeGraw R, Moghadam F, Tan Y, Kilbourne J, Maggiore JC, Hislop J, Liu S, Cats D, Chuva de Sousa Lopes SM, et al. (2021). Gene Regulatory Network Analysis and Engineering Directs Development and Vascularization of Multilineage Human Liver Organoids. Cell Syst 12, 41–55.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner KT, Nash TR, Liu B, Vunjak-Novakovic G, and Radisic M (2020). Extracellular Vesicles in Cardiac Regeneration: Potential Applications for Tissues-on-a-Chip. Trends in Biotechnology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walenda T, Bork S, Horn P, Wein F, Saffrich R, Diehlmann A, Eckstein V, Ho AD, and Wagner W (2010). Co-culture with mesenchymal stromal cells increases proliferation and maintenance of haematopoietic progenitor cells. Journal of Cellular and Molecular Medicine 14, 337–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang EY, Rafatian N, Zhao Y, Lee A, Lai BFL, Lu RX, Jekic D, Davenport Huyer L, Knee-Walden EJ, Bhattacharya S, et al. (2019). Biowire Model of Interstitial and Focal Cardiac Fibrosis. ACS Central Science 5, 1146–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Pine AR, Kotini AG, Yuan H, Zamparo L, Starczynowski DT, Leslie C, and Papapetrou EP (2021). Sequential CRISPR gene editing in human iPSCs charts the clonal evolution of myeloid leukemia and identifies early disease targets. Cell Stem Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Yates F, Naveiras O, Ernst P, and Daley GQ (2005). Embryonic stem cell-derived hematopoietic stem cells. Proceedings of the National Academy of Sciences of the United States of America 102, 19081–19086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijetunga M, and Rockson S (2002). Myocarditis in systemic lupus erythematosus. The American Journal of Medicine 113, 419–423. [DOI] [PubMed] [Google Scholar]
- Wilkinson AC, Ishida R, Kikuchi M, Sudo K, Morita M, Crisostomo RV, Yamamoto R, Loh KM, Nakamura Y, Watanabe M, et al. (2019). Long-term ex vivo haematopoietic-stem-cell expansion allows nonconditioned transplantation. Nature 571, 117–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimmer RA, Leopoldi A, Aichinger M, Wick N, Hantusch B, Novatchkova M, Taubenschmid J, Hämmerle M, Esk C, Bagley JA, et al. (2019). Human blood vessel organoids as a model of diabetic vasculopathy. Nature 565, 505–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao S, Coppeta JR, Rogers HB, Isenberg BC, Zhu J, Olalekan SA, McKinnon KE, Dokic D, Rashedi AS, Haisenleder DJ, et al. (2017). A microfluidic culture model of the human reproductive tract and 28-day menstrual cycle. Nat Commun 8, 14584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadid M, Lind JU, Ardoña HAM, Sheehy SP, Dickinson LE, Eweje F, Bastings MMC, Pope B, O’Connor BB, Straubhaar JR, et al. (2020). Endothelial extracellular vesicles contain protective proteins and rescue ischemia-reperfusion injury in a human heart-on-chip. Science Translational Medicine 12, eaax8005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y, Gotoh S, Korogi Y, Seki M, Konishi S, Ikeo S, Sone N, Nagasaki T, Matsumoto H, Muro S, et al. (2017). Long-term expansion of alveolar stem cells derived from human iPS cells in organoids. Nat Methods 14, 1097–1106. [DOI] [PubMed] [Google Scholar]
- Yamaya M, Finkbeiner WE, Chun SY, and Widdicombe JH (1992). Differentiated structure and function of cultures from human tracheal epithelium. American Journal of Physiology-Lung Cellular and Molecular Physiology 262, L713–L724. [DOI] [PubMed] [Google Scholar]
- Yanamandala M, Zhu W, Garry DJ, Kamp TJ, Hare JM, Jun HW, Yoon YS, Bursac N, Prabhu SD, Dorn GW 2nd, et al. (2017). Overcoming the Roadblocks to Cardiac Cell Therapy Using Tissue Engineering. J Am Coll Cardiol 70, 766–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Soonpaa MH, Adler ED, Roepke TK, Kattman SJ, Kennedy M, Henckaerts E, Bonham K, Abbott GW, Linden RM, et al. (2008). Human cardiovascular progenitor cells develop from a KDR+ embryonic-stem-cell-derived population. Nature 453, 524–528. [DOI] [PubMed] [Google Scholar]
- Yazawa M, Hsueh B, Jia X, Pasca AM, Bernstein JA, Hallmayer J, and Dolmetsch RE (2011). Using induced pluripotent stem cells to investigate cardiac phenotypes in Timothy syndrome. Nature 471, 230–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye L, Chang YH, Xiong Q, Zhang P, Zhang L, Somasundaram P, Lepley M, Swingen C, Su L, Wendel JS, et al. (2014). Cardiac repair in a porcine model of acute myocardial infarction with human induced pluripotent stem cell-derived cardiovascular cells. Cell Stem Cell 15, 750–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S, Miyagawa S, Fukushima S, Kawamura T, Kashiyama N, Ohashi F, Toyofuku T, Toda K, and Sawa Y (2018). Maturation of Human Induced Pluripotent Stem Cell-Derived Cardiomyocytes by Soluble Factors from Human Mesenchymal Stem Cells. Molecular Therapy 26, 2681–2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Montgomery M, Chamberlain MD, Ogawa S, Korolj A, Pahnke A, Wells LA, Massé S, Kim J, Reis L, et al. (2016a). Biodegradable scaffold with built-in vasculature for organ-on-a-chip engineering and direct surgical anastomosis. Nat Mater 15, 669–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Tao R, Campbell KF, Carvalho JL, Ruiz EC, Kim GC, Schmuck EG, Raval AN, da Rocha AM, Herron TJ, et al. (2019). Functional cardiac fibroblasts derived from human pluripotent stem cells via second heart field progenitors. Nature Communications 10, 2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Wilson GF, Soerens AG, Koonce CH, Yu J, Palecek SP, Thomson JA, and Kamp TJ (2009). Functional cardiomyocytes derived from human induced pluripotent stem cells. Circ Res 104, e30–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Zhu W, Radisic M, and Vunjak-Novakovic G (2018). Can We Engineer a Human Cardiac Patch for Therapy? Circ Res 123, 244–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YS, Aleman J, Shin SR, Kilic T, Kim D, Mousavi Shaegh SA, Massa S, Riahi R, Chae S, Hu N, et al. (2017). Multisensor-integrated organs-on-chips platform for automated and continual in situ monitoring of organoid behaviors. Proceedings of the National Academy of Sciences 114, E2293–E2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YS, Arneri A, Bersini S, Shin S-R, Zhu K, Goli-Malekabadi Z, Aleman J, Colosi C, Busignani F, Dell’Erba V, et al. (2016b). Bioprinting 3D microfibrous scaffolds for engineering endothelialized myocardium and heart-on-a-chip. Biomaterials 110, 45–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Rafatian N, Feric NT, Cox BJ, Aschar-Sobbi R, Wang EY, Aggarwal P, Zhang B, Conant G, Ronaldson-Bouchard K, et al. (2019a). A Platform for Generation of Chamber-Specific Cardiac Tissues and Disease Modeling. Cell 176, 913–927.e918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Wang EY, Davenport LH, Liao Y, Yeager K, Vunjak-Novakovic G, Radisic M, and Zhang B (2019b). A Multimaterial Microphysiological Platform Enabled by Rapid Casting of Elastic Microwires. Advanced Healthcare Materials 8, 1801187. [DOI] [PubMed] [Google Scholar]
- Zhou BO, Yu H, Yue R, Zhao Z, Rios JJ, Naveiras O, and Morrison SJ (2017). Bone marrow adipocytes promote the regeneration of stem cells and haematopoiesis by secreting SCF. Nat Cell Biol 19, 891–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Blum RH, Bernareggi D, Ask EH, Wu Z, Hoel HJ, Meng Z, Wu C, Guan KL, Malmberg KJ, et al. (2020). Metabolic Reprograming via Deletion of CISH in Human iPSC-Derived NK Cells Promotes In Vivo Persistence and Enhances Anti-tumor Activity. Cell Stem Cell 27, 224–237.e226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W, Zhao M, Mattapally S, Chen S, and Zhang J (2018). CCND2 Overexpression Enhances the Regenerative Potency of Human Induced Pluripotent Stem Cell–Derived Cardiomyocytes. Circulation Research 122, 88–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann WH, Melnychenko I, Wasmeier G, Didié M, Naito H, Nixdorff U, Hess A, Budinsky L, Brune K, Michaelis B, et al. (2006). Engineered heart tissue grafts improve systolic and diastolic function in infarcted rat hearts. Nat Med 12, 452–458. [DOI] [PubMed] [Google Scholar]
- Zonari E, Desantis G, Petrillo C, Boccalatte FE, Lidonnici MR, Kajaste-Rudnitski A, Aiuti A, Ferrari G, Naldini L, and Gentner B (2017). Efficient Ex Vivo Engineering and Expansion of Highly Purified Human Hematopoietic Stem and Progenitor Cell Populations for Gene Therapy. Stem Cell Reports 8, 977–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwi L, Caspi O, Arbel G, Huber I, Gepstein A, Park IH, and Gepstein L (2009). Cardiomyocyte differentiation of human induced pluripotent stem cells. Circulation 120, 1513–1523. [DOI] [PubMed] [Google Scholar]


